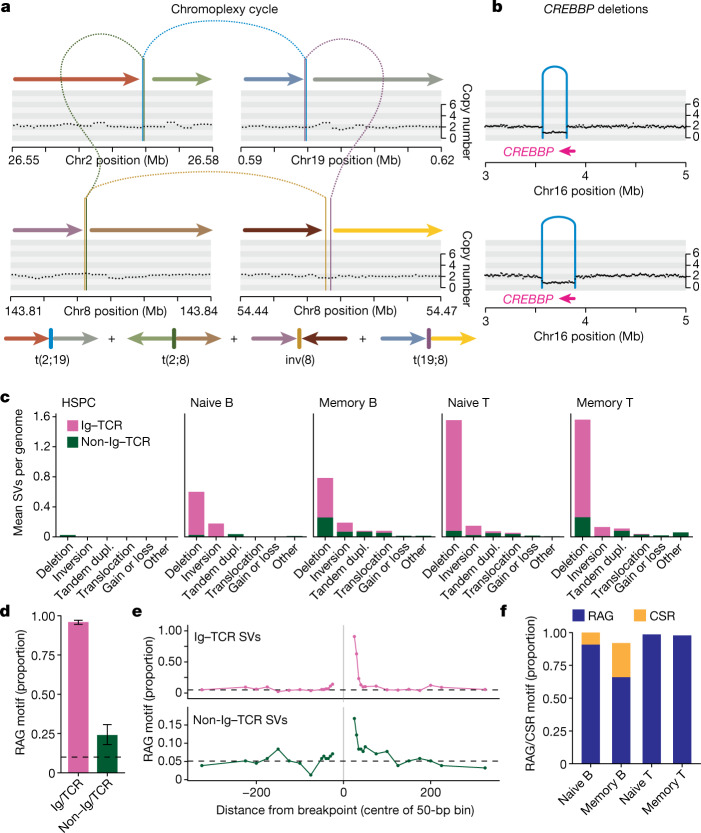
Fig. 4. Structural variation burden and off-target RAG-mediated deletion.
a, Top, chromoplexy cycle (sample PD40667sl, donor KX002). Black points represent the corrected read depth along the chromosome and arcs denote structural variants. Bottom, the final genomic configuration of the four derivative chromosomes is shown as coloured arrows. b, CREBBP deletions (samples PD40521po, donor KX001 and BMH1_PlateB1_E2, donor AX001). c, Burden of structural variants per cell type. Dupl., duplication. d, The proportion of deletions with an RSS (RAG) motif within 50 bp of the breakpoint for Ig–TCR (0.96) and non-Ig–TCR (0.24) regions. The black dashed line represents the genomic background rate of RAG motifs. Error bars represent 95% bootstrap confidence intervals. n = 889 Ig–TCR structural variants and 253 non-Ig–TCR structural variants. e, Proportion of deletions with an RSS (RAG) motif as a function of distance from the breakpoint, with a positive distance representing bases interior to the deletion, and a negative value representing bases exterior to the breakpoint. The black dashed line represents the genomic background rate of RAG motifs. f, The proportion of deletions with an RSS (RAG) or switch (CSR) motif.