Abstract
Era is an essential membrane-associated GTPase that is present in bacteria and mycoplasmas. Era appears to play an important role in the regulation of the bacterial cell cycle. In this study, we expressed the native and glutathione S-transferase (GST) fusion forms of Streptococcus pneumoniae Era in Escherichia coli and purified both proteins to homogeneity. We showed that RNA was copurified with the GST-Era protein of S. pneumoniae during affinity purification and remained associated with the protein after removal of the GST tag by thrombin cleavage. The thrombin-treated and untreated GST-Era proteins could bind and hydrolyze GTP and exhibited similar kinetic properties (dissociation constant [kD], Km, and Vmax). However, the native Era protein purified by using different chromatographic columns had a much lower GTPase activity than did GST-Era, although it had a similar kD. In addition, RNA was not associated with the protein. Purified GST-Era protein was shown to be present as high (600-kDa)- and low (120-kDa)-molecular-mass forms. The high-molecular-mass form of GST-Era was associated with RNA and exhibited a very high GTPase activity. Approximately 40% of purified GST-Era protein was associated with RNA, and removal of the RNA resulted in a significant reduction in GTPase activity. The RNA associated with GST-Era was shown to be predominantly 16S rRNA. The native Era protein isolated directly from S. pneumoniae was also present as a high-molecular-mass species (600 kDa) complexed with RNA. Together, our results suggest that 16S rRNA is associated with Era and might stimulate its GTPase activity.
The era gene encodes an essential membrane-associated GTPase that binds GTP and GDP and hydrolyzes GTP to GDP (1, 5, 6, 8, 13, 16, 24, 36, 39, 40). Homologues of Era have been identified in all bacterial genomes sequenced to date (43). All era genes from bacteria tested have been shown to complement Escherichia coli mutants deficient in expression of Era (32, 43, 44). Era was originally thought to be a member of the Ras GTPase family based on sequence similarity of the protein to the GTP binding domain of Ras (1). Recently, a human homologue of Era which has significant sequence identity to the entire length of the Era protein but not to Ras proteins has been identified (6). Thus, Era is distinct from Ras (6, 8).
The exact role of Era in the cell remains elusive. Genetic studies with E. coli suggest that Era is involved in cell cycle regulation and possibly ribosome assembly (5, 6, 13, 28–30). Limiting the cellular levels of Era or reducing the GTPase activity of Era causes cells to arrest growth just prior to cell division (5, 6, 13). Analysis of a cold-sensitive era mutant of E. coli showed that ribosome assembly was altered in this mutant (28–30). Like that of rRNA, the expression level of Era increases as the growth rate of the cell increases (6). These observations suggest that Era may have a multifunctional role in the regulation of the cell cycle and translation. In addition, a mutation in the Era GTPase domain of E. coli suppressed temperature-sensitive mutations affecting DNA replication and chromosome partitioning but not cell division (5, 6). It is believed that Era may function as a timing element in the regulation of cell division (6).
GTPases are widespread in both eukaryotes and prokaryotes (2, 3, 23). Cellular GTPases are known to be involved in a wide range of regulatory mechanisms, such as translation, cell differentiation, and cell cycle regulation (2, 3, 23). The GTPases are generally considered to be regulatory switches that are converted from an active state to an inactive state by hydrolysis of GTP to GDP, thereby regulating downstream effector enzymes in various cellular processes (2, 3, 23).
Two bacterial GTPases that are known to bind RNA are Ffh and EF-G (19, 25, 37). The Ffh protein is a homologue of the mammalian signal recognition particle which is involved in bacterial protein secretion (10, 19, 25), and EF-G is a translational elongation factor (26, 37). The binding of RNA to the GTPases may have a regulatory role (11, 17). Recently, a large number of enzymes that use nucleotides as substrates have also been shown to bind RNA (14). In some cases, the mechanisms of RNA binding to the enzymes are known, but in many cases, the mechanisms are not understood (14, 34).
In this study, we present evidence that 16S rRNA is associated with the Streptococcus pneumoniae Era protein and might stimulate its GTPase activity. We found that about 40% of the S. pneumoniae glutathione S-transferase (GST)–Era protein produced in E. coli was present as a high-molecular-mass species complexed with RNA, which possessed a high level of GTPase activity. Removal of the RNA resulted in a significant reduction of the GTPase activity of the enzyme. Approximately 70% of the Era protein from the crude extract of S. pneumoniae was also found to exist as a complex with RNA.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The following E. coli strains were used in this study: E. coli BL21(DE3) plysS [F− dcm ompT hsdS (rB− mB−) gal λ(DE3) (plysS Camr)] (Stratagene, La Jolla, Calif.) and LY41 [E. coli BL21(DE3) plysS/pLY41 (GST-era+ Ampr)], LY42 [E. coli BL21(DE3) plysS/pLY42 (era+ Ampr)] (42), and LY160 [E. coli BL21(DE3) plysS/pGEX-2T (GST+ Ampr)] (Pharmacia LKB Biotechnology, Alameda, Calif.). S. pneumoniae R6 (hex), a penicillin-sensitive laboratory strain, was kindly provided by A. Tomasz (Rockefeller University).
All E. coli strains that express the S. pneumoniae era gene were first grown overnight at 37°C with vigorous shaking in Luria-Bertani medium (Bio 101, Inc., La Jolla, Calif.) supplemented with 100 μg of ampicillin per ml. The overnight cultures (4%) were inoculated into 1.25 liters each of fresh Luria-Bertani medium (Bio 101) containing ampicillin and then induced with 0.8 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (Gibco BRL, Gaithersburg, Md.) at an optical density at 600 nm of 0.5 to 0.6 for 3 h at 33°C. Cells were harvested by centrifugation at 4,000 × g for 10 min and washed with 20 mM Tris-HCl (pH 8.0)–5 mM MgCl2. S. pneumoniae was grown in brain heart infusion broth (Difco Laboratories, Detroit, Mich.) at 37°C without shaking. Cells were harvested by centrifugation as described above and washed with 20 mM potassium phosphate, pH 7.2.
Purification of Era and GST proteins of S. pneumoniae.
The GST-Era protein was purified from cells of E. coli LY41 by glutathione-Sepharose affinity column chromatography (Pharmacia LKB Biotechnology) as follows. Cells of LY41 were resuspended in 20 mM Tris-HCl (pH 7.5)–140 mM NaCl–5 mM MgCl2. The resulting suspension was disrupted by being passed twice through a 20 K French pressure cell (Aminco Laboratories, Inc., Rochester, N.Y.) and centrifuged at 180,000 × g for 60 min. The supernatant fraction collected was loaded onto a 10-ml glutathione-Sepharose column (Pharmacia). The column was washed with 100 ml of the Tris-HCl buffer and eluted with 10 mM glutathione in the same buffer. All fractions were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (20), and those containing GST-Era were collected and dialyzed by using a dialysis tubing (molecular mass cutoff, 50 kDa; Sigma Chemical Company, St. Louis, Mo.) in 4 liters of 20 mM Tris-HCl (pH 8.0)–5 mM MgCl2 at 4°C overnight. After dialysis, glycerol was added to the Era preparation at a final concentration of 16% (vol/vol), and the resulting preparation was stored in aliquots at −70°C. The protein concentration was determined by using a Bradford protein assay kit (Bio-Rad, Hercules, Calif.) with bovine serum albumin (BSA) as a standard (4).
The native (nontagged) Era protein of S. pneumoniae was purified from cells of E. coli LY42 by using a three-step column chromatographic method (42). Briefly, the crude extract of E. coli LY42 was loaded onto a source Q column (Pharmacia) equilibrated with 20 mM Tris-HCl (pH 8.0)–5 mM MgCl2 (buffer A) and eluted with a gradient of 0 to 1,000 mM KCl in buffer A. The fractions containing the Era protein were collected, dialyzed in 20 mM potassium phosphate (pH 7.5) and subjected to chromatography on a hydroxyapatite column (Bio-Rad) equilibrated with the phosphate buffer. The Era protein was eluted with a gradient of 20 to 700 mM potassium phosphate, pH 7.5. The fractions containing Era were collected, dialyzed in buffer A, and subjected to chromatography on a heparin column (Bio-Rad) equilibrated with buffer A. The Era protein was eluted with a gradient of 0 to 1,000 mM KCl in buffer A. The fractions that contained Era were collected, dialyzed by using a dialysis tubing (molecular mass cutoff, 25 kDa; Sigma) in 4 liters of buffer A overnight at 4°C, and stored in small aliquots at −70°C as described above.
GST was purified from LY160 by using a glutathione-Sepharose affinity column as described above for the GST-Era protein.
Determination of RNA association with S. pneumoniae Era.
To separate the nucleic acids associated with the S. pneumoniae GST-Era protein from the enzyme, the purified protein was loaded onto a Mono Q column (Pharmacia Biotech, Piscataway, N.J.) equilibrated with buffer A. The column was washed with buffer A and eluted with a linear gradient of 0 to 1,000 mM KCl in buffer A. Fractions (1 ml each) were collected. The peak fractions containing Era or RNA were treated with RNase A (Sigma) or DNase I (Gibco BRL) as indicated in the figure legends. These fractions were also analyzed by agarose gel electrophoresis and spectrophotometric scanning (200 to 600 nm) with a Bio-Spec 1601 spectrophotometer (Shimadzu, Columbia, Md.).
Purified GST protein was also subjected to analyses by column chromatography, agarose gel electrophoresis, and spectrophotometric scanning as described above.
To establish if Era isolated directly from S. pneumoniae was also associated with RNA, a crude extract of S. pneumoniae was prepared in buffer A as described above. Half of this crude extract was treated with RNase A (1 mg/ml), and the other half of the sample was not treated. The RNase A-treated or untreated sample was incubated at room temperature for 2 h and loaded onto a HiLoad 16/60 Superdex 200 column (Pharmacia) equilibrated with buffer A. The column was eluted with buffer A. The molecular mass of each species resolved was determined by calibrating the column with the following protein standards (Sigma): blue dextran (2,000 kDa), alcohol dehydrogenase (150 kDa), BSA (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa). Fractions (15 μl each) were subjected to SDS-PAGE (20), and then the proteins were transferred to polyvinylidene difluoride membranes and subjected to Western blotting analysis with polyclonal antibodies prepared against SDS-denatured native Era protein of S. pneumoniae (42). Intensities of individual Era bands were quantified by using an imaging densitometer (model GS-700; Bio-Rad).
To establish further that RNA was associated with Era, thrombin-treated and untreated GST-Era and native Era proteins of S. pneumoniae were chromatographed under the conditions described above. Each protein peak resolved was tested for its GTP hydrolysis activity as described below and subjected to RNase A and DNase I treatments, agarose gel electrophoresis, and scanning analysis as described above.
To remove the GST fusion portion of the GST-Era protein, GST-Era was completely cleaved with thrombin protease (14 U of thrombin/mg of GST-Era in 1× phosphate-buffered saline buffer) after an overnight incubation at 4°C. Half of the thrombin-treated protein preparation was passed through a glutathione-Sepharose column for the removal of the GST fusion part of the protein as described above. The other half of the preparation was not purified further. The resulting preparations were used for further studies as described above.
Analysis of RNA associated with the S. pneumoniae Era protein.
Approximately 4 mg each of purified GST-Era and native Era proteins of S. pneumoniae or purified GST protein was extracted with an equal volume of phenol-chloroform-isoamyl alcohol, and RNA was precipitated with ethanol (35). The RNA (pellet) collected was air dried and then resuspended in 200 μl of diethyl pyrocarbonate-treated water. The RNA collected (10 μl each) was analyzed by agarose gel electrophoresis (1.5% agarose containing 0.5 μg of ethidium bromide per ml) (35). E. coli rRNAs were isolated from E. coli LY42 by phenol-chloroform extraction (35).
To directly visualize RNA associated with the protein, approximately 10 μg of purified proteins was run on an agarose gel as described above. Purified Era proteins and extracted RNA were treated with 1 mg of RNase A (Boehringer Mannheim, Indianapolis, Ind.) per ml or 0.2 mg of DNase I (Gibco BRL) per ml for 15 min at room temperature prior to electrophoresis.
Determination of Era GTPase activity.
The GTPase activity of the S. pneumoniae Era protein was assayed by using both thin-layer chromatography and high-pressure liquid chromatography (HPLC). For the thin-layer chromatography assay, reaction mixtures (20 μl each) contained 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 1 mM dithiothreitol, 100 mM NaCl, 1 mg of BSA per ml, and 10 μM GST-Era. Reactions were initiated by adding 10 μM [α-32P]GTP (100 Ci/mmol; NEN-Dupont), and the reaction mixtures were incubated at 23°C for 60 min for routine assays and for 0 to 90 min for kinetic analyses. After incubation, 2 μl of each sample was spotted onto a polyethyleneimine cellulose thin-layer plate (Selecto Scientific, Norcross, Ga.) and dried. The polyethyleneimine plate was developed in 0.75 M KH2PO4 (8). The GDP produced was quantified with a PhosphorImage Analyzer (Molecular Dynamics). For the HPLC assay, reaction mixtures (300 μl each) containing 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 1.15 μM GST-Era or native Era were incubated at 23 or 37°C. To initiate the reactions, GTP was added at concentrations of 40 to 2,000 μM. Aliquots (100 μl each) were removed at time zero and after a 30-min incubation, and the reactions were stopped by adding 5 μl of 1 N HCl. Then, 50 μl of each aliquot was injected into a 4.6- by 250-mm ODS-AQ HPLC column (YMC, Inc.) and separated under isocratic conditions (79 mM potassium phosphate [pH 6.0], 4 mM tetrabutyl ammonium hydrogen sulfate, 21% methanol). The GDP produced was quantified by comparing its peak areas with those of GDP standards. Since GDP is bound to the protein when purified, the total amount of GDP produced after a 30-min incubation was calculated by subtracting the GDP present at the start of the reaction.
Assay of Era GTP binding activity.
The dissociation constant (kD) of the S. pneumoniae Era protein for GTP was determined by using a filter binding assay. Reaction mixtures (50 μl each) contained 25 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 0.2 mg of BSA per ml, 1.3 μM GST-Era or native Era, and 1 to 100 μM [3H]GTP (0.6 Ci/mmol). The reaction mixtures were incubated at room temperature for 30 min and then filtered through a MANP NOB Multiscreen filter plate (Millipore, Bedford, Mass.). The plate was washed three times with 200 μl of 50 mM Tris-HCl (pH 7.5)–5 mM MgCl2 per well, and then 30 μl of Optiphase Supermix scintillant (Wallac Oy, Turku, Finland) was added to each well and the radioactivity was counted with a Wallac Microbeta scintillation counter. All kD values were determined by Scatchard plots.
Reconstitution of Era protein and RNA.
To reconstitute the S. pneumoniae native Era and GST-Era proteins whose RNA had been removed, the proteins were mixed with RNA extracted from purified GST-Era protein in 50 mM Tris-HCl (pH 7.5)–5 mM MgCl2, and the mixtures were incubated for 0 to 24 h at 4, 23, or 37°C. After incubation, GTPase activities were tested by the HPLC method. RNA or RNA oligonucleotides tested included E. coli tRNA (Sigma), E. coli rRNA (Sigma), RNA extracted from GST-Era, and poly(A), poly(C), poly(U), poly(G), and poly(AGU) (Sigma). In addition, phenol-chloroform-extracted RNA from GST-Era was heat denatured at 95°C for 5 min, immediately chilled on ice, and then mixed with the proteins as described above. GTPase activities were tested as described above.
To refold Era protein in the presence of RNA, GST-Era was denatured in 4 M urea and mixed with phenol-chloroform-extracted RNA from GST-Era. The refolding preparation was dialyzed against 4 liters of 50 mM Tris-HCl (pH 7.5)–5 mM MgCl2 overnight at 4°C.
Cloning and sequencing of the RNA associated with GST-Era of S. pneumoniae.
The synthesis of the first- and second-strand cDNAs of the RNA associated with GST-Era was performed by using the SuperScript Choice System for cDNA Synthesis (Gibco BRL) according to the manufacturer’s directions. Two different RNA templates, phenol-chloroform-extracted RNA and GST-Era protein that was still associated with RNA, were used for cDNA synthesis. For the first- and second-strand syntheses, 8 μl of extracted RNA or 5 μl of GST-Era was mixed with 100 ng of random hexamer primers (Gibco BRL). After the second-strand synthesis, the DNA fragments were extracted with phenol-chloroform, precipitated with ethanol as described above, and directly used for ligation to pUC18 by using a Ready-To-Go pUC18 SmaI/BAP+ ligase kit (Pharmacia). The ligation mixtures were then transformed into E. coli XL1-Blue MRF (Stratagene). Colonies were randomly picked, and their plasmid DNA was isolated by using a Wizard miniprep kit (Promega). DNA sequences of the plasmids isolated were determined by using PE-ABI Prism Dye Terminator Cycle Sequencing Ready Reaction fluorescence-based chemistry. Sequence data was collected on an ABI377 instrument and analyzed by using PE-ABI Sequence Analysis version 3.0 software. Data was edited by using Squercher version 3.0.
RESULTS
Kinetic comparison of the GST-Era and native Era proteins of S. pneumoniae.
To characterize the kinetic properties of the S. pneumoniae Era protein, we expressed the native Era and GST-Era proteins of S. pneumoniae in E. coli, purified both to homogeneity (see Fig. 1 and 4), and determined their GTP hydrolysis activities. To our surprise, GST-Era protein exhibited a specific activity (150 mmol/min/mol) that was 12-fold higher than the specific activity (12 mmol/min/mol) of native Era protein when assayed at 500 μM GTP. The specific activity determined for the native Era protein of S. pneumoniae was very similar to those reported for the E. coli and Streptococcus mutans Era proteins (8, 41). Thus, the GST-Era protein of S. pneumoniae appears to be significantly more active than the native form of the enzyme and also than the E. coli and S. mutans enzymes.
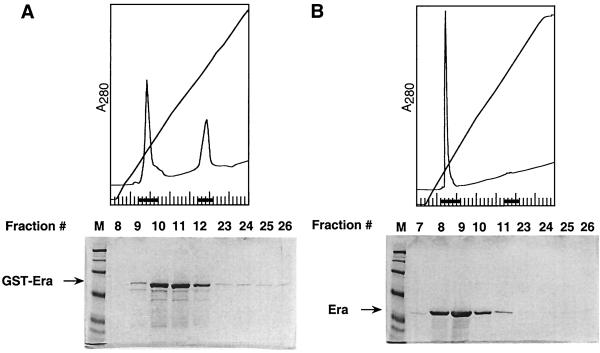
FIG. 1.
Analysis of the native Era and GST-Era proteins of S. pneumoniae by ion-exchange column chromatography. Purified GST-Era (A) and native Era (B) protein preparations were subjected to chromatography on a Mono Q column as described in Materials and Methods. Fractions indicated by black dots were analyzed by SDS–10% PAGE and stained with Coomassie blue. Lanes M, markers.
FIG. 4.
Analysis of the S. pneumoniae native Era and GST-Era proteins by gel filtration column chromatography. Purified GST-Era (A) and native Era (B) proteins were subjected to gel filtration chromatography as described in Materials and Methods. Each fraction was analyzed by SDS-PAGE. Fractions indicated by black dots were analyzed by SDS–10% PAGE and stained with Coomassie blue.
To investigate if the fusion of GST to Era might have altered the kinetic properties of the S. pneumoniae Era protein, we removed the GST fusion part of the protein by thrombin cleavage and determined the kinetic properties (kD, Km, and Vmax) of native Era, GST-Era, and thrombin-treated GST-Era proteins. The kD values of the three Era proteins for GTP were very similar (Table 1) and also were in good agreement with the kD values (3 to 6 μM) reported for E. coli Era (8). The Km and Vmax values of the thrombin-treated and untreated GST-Era proteins were virtually identical (Table 1) but significantly higher than those of native Era protein (Table 1) when GTP was used as a substrate. In addition, when the GST fusion portion of the protein was removed from the thrombin-treated GST-Era protein preparation by use of a glutathione-Sepharose affinity column, this Era protein also exhibited Km and Vmax values identical to those of the starting proteins (data not shown). Thus, these results clearly establish that the fusion of GST to Era did not alter the kinetic properties of the S. pneumoniae Era protein and that GST-Era protein is more active than native Era protein.
TABLE 1.
Kinetic properties of the native Era, GST-Era, and thrombin-treated GST-Era proteinsa
| Protein | kd (μM) | Km (μM) | Vmax (mmol/min/mol) |
|---|---|---|---|
| Native Era | 3.00 ± 0.45 | 50.90 ± 17.00 | 10.64 ± 5.00 |
| GST-Era | 3.73 ± 0.90 | 456.00 ± 27.00 | 295.00 ± 31.00 |
| Thrombin-treated GST-Era | 3.50 | 526.50 ± 5.00 | 229.50 ± 3.00 |
The native Era protein was purified by using a three-step purification scheme as described in Materials and Methods. The GST-Era protein was affinity purified by using a glutathione-Sepharose column as described in Materials and Methods. The thrombin-treated GST-Era protein preparation was not further purified. The GTPase activities of these Era proteins were assayed by measuring their ability to hydrolyze GTP or GDP by using HPCL (see Materials and Methods).
As we will show below, RNA is associated with purified GST-Era protein but not with purified native Era protein. Removal of the RNA associated with GST-Era reduced its GTPase activity to a level similar to that of the purified native Era protein.
RNA is copurified with the Era protein of S. pneumoniae.
During the course of analyzing a purified GST-Era protein preparation by ion-exchange chromatography, we unexpectedly observed two peaks, one eluted at a low salt concentration (150 mM KCl) and one eluted at a high salt concentration (650 mM KCl) (Fig. 1A). SDS-PAGE analysis showed that the fractions of the low-salt peak contained GST-Era (Fig. 1A). Surprisingly, we could barely detect the GST-Era protein in the fractions of the high-salt peak (Fig. 1A). The GTP hydrolysis activity of the GST-Era protein from the low-salt peak fractions was significantly reduced to a level similar to that of native Era protein (data not shown). Similar column profiles and specific activities were obtained for thrombin-treated GST-Era protein with or without affinity removal of the GST fusion portion of the protein (data not shown). We then examined purified native Era protein under the identical conditions. Unlike purified GST-Era protein, purified native Era protein exhibited only a single peak that was eluted at a low salt concentration (Fig. 1B). This suggested that the material that was associated with the native Era protein had been removed during its purification. Together, these results suggested that the high-salt peak contained the material that might be required for high GTPase activity of the GST-Era protein.
To investigate the nature of the material in the high-salt peak fractions, we scanned these fractions spectrophotometrically and found that they exhibited a maximum at 260 nm. This suggested that the material in the fractions was nucleic acid in nature. We also found that the native Era and GST-Era proteins exhibited different maxima, at 280 and 260 nm, respectively (data not shown). These findings indicated that the high-salt peak fractions contained the material, possibly nucleic acid, that was copurified with GST-Era protein.
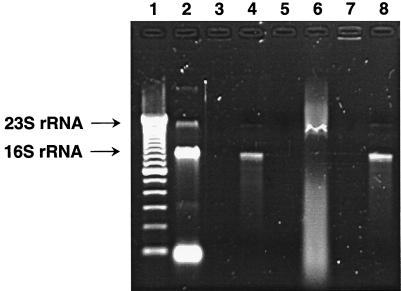
To determine whether nucleic acids were associated with GST-Era, we analyzed the purified GST-Era protein and phenol-chloroform-extracted material from the protein by agarose gel electrophoresis. The phenol-chloroform-extracted material appeared as two distinct bands in the gel after staining with ethidium bromide (Fig. 2). The major band (≈95%) and the minor band (≈5%) exhibited mobilities corresponding to those of E. coli 16S rRNA and 23S rRNA, respectively (Fig. 2), thus suggesting that the S. pneumoniae Era was mainly associated with the E. coli 16S rRNA. The purified GST-Era protein also appeared as a broad band with a mobility slower than that of 16S rRNA (Fig. 2). This mobility shift suggests that the GST-Era protein is associated with nucleic acids, possibly 16S rRNA. To test this further, we performed RNase and DNase treatments. We found that treatment with RNase, but not DNase, of both GST-Era and its phenol-chloroform-extracted material eliminated the bands, thus indicating that RNA, but not DNA, was associated with GST-Era (Fig. 2 and data not shown). In contrast, the purified native Era protein and its phenol-chloroform-extracted material lacked any RNA detectable by ethidium bromide staining (data not shown). These findings suggested that RNA was copurified with GST-Era and that the RNA associated with the native Era protein had been removed during purification.
FIG. 2.
Analysis of RNA association with the Era protein of S. pneumoniae. E. coli rRNAs were isolated as described in Materials and Methods. All samples were electrophoresed on a 1.5% agarose gel containing ethidium bromide. Lane 1, DNA standards (100-bp increments from bottom to top; Gibco BRL); lanes 2 and 3, E. coli rRNA untreated and treated with RNase A, respectively; lanes 4 and 5, phenol-chloroform-extracted material from a purified GST-Era protein preparation untreated and treated with RNase A, respectively; lanes 6 and 7, a purified GST-Era protein preparation untreated and treated with RNase A, respectively; lane 8, phenol-chloroform-extracted material from a purified GST-Era protein preparation treated with DNase.
We also examined purified GST protein preparations for their possible association with RNA and GTPase activity. We found no evidence that either RNA or GTPase activity was associated with purified GST protein. First, GST yielded only one peak eluted at a low salt concentration during ion-exchange chromatography (data not shown). GST also appeared as a single peak with an estimated molecular mass of 50 kDa during gel filtration chromatography (data not shown). Second, spectrophotometric analysis of the protein or its phenol-chloroform-extracted material showed no significant absorbance at 260 nm (data not shown). Third, the purified GST protein did not exhibit a detectable GTPase activity when assayed. Together, these results suggest that neither RNA nor GTPase activity is associated with the GST protein.
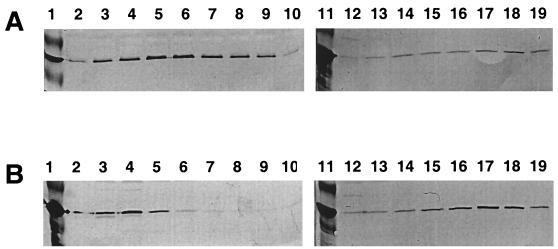
To further establish that RNA is associated with Era and is required for stimulation of its GTPase activity, we subjected a crude extract of S. pneumoniae to gel filtration column chromatography. We reasoned that if RNA is associated with Era and required for its high GTPase activity, then the Era-RNA complex should be present as a higher-molecular-mass species possessing a high level of GTPase activity. As shown in Fig. 3A, the Era protein isolated directly from the S. pneumoniae cells exhibited two peaks, one located in the void volume fractions from the column, with an estimated molecular mass of 600 kDa, and one eluted at a position with a molecular mass of about 32 kDa (monomer). The amount of Era present in the void volume fractions of the peak was shown to be about 3 times more than that in the fractions of the low-molecular-mass peak (Fig. 3A). In addition, when the crude extract of S. pneumoniae was treated with RNase A and chromatographed, the Era protein was also present in two peaks similar to those of the untreated sample (Fig. 3B). However, the amount of Era present in the void volume fractions of the peak was approximately 3 times less than that in the fractions of the low-molecular-mass peak (Fig. 3B). Thus, the RNase treatment converted a significant amount of the Era complex to the monomeric form. Together, these findings establish that the Era protein is complexed with RNA in S. pneumoniae. The GTPase activity of Era in the void volume fractions could not be determined because of contamination from other proteins.
FIG. 3.
Analysis of Era-RNA complex formation in a crude extract of S. pneumoniae by gel filtration column chromatography. A crude extract of S. pneumoniae was prepared, untreated (A) or treated with RNase A (1 mg/ml) (B), and subjected to chromatography on a gel filtration column as described in Materials and Methods. The presence of Era in the fractions collected was detected by Western blotting analysis with polyclonal antibodies prepared against the native Era protein of S. pneumoniae (27). The intensity of each band was quantified by scanning as described in Materials and Methods. Lanes 1 and 11, molecular mass markers and purified Era of S. pneumoniae (30 ng), respectively; lanes 2 to 10, fractions 23 to 31, respectively; lanes 12 to 19, fractions 39 to 46, respectively.
When purified GST-Era protein was chromatographed under the identical conditions, it exhibited two peaks; the major one eluted in the void volume fractions, with an estimated molecular mass of 600 kDa, and the minor one eluted at a position indicating a molecular mass of 120 kDa, a dimer of GST-Era (Fig. 4A). As judged by SDS-PAGE, approximately 40% of the total GST-Era protein was associated with RNA which was all eluted in the void volume fractions (Fig. 4A). RNase treatment significantly reduced the major peak and the specific activity of the GST-Era derived from the fractions of this major peak (data not shown). The specific activity for the GST-Era protein was determined to be 200 mmol/min/mol, but the specific activities for the GST-Era proteins collected from the high- and the low-molecular-mass peaks (Fig. 4A) were 600 and 25 mmol/min/mol, respectively. Clearly, removal of the RNA associated with the GST-Era protein resulted in a significant reduction in its GTPase activity. When the thrombin-treated GST-Era protein was chromatographed, similar column profiles and specific activities were obtained (data not shown). In contrast, a 32-kDa monomeric enzyme but no high-molecular-mass species was detected when the native Era protein purified by using different chromatographic columns was chromatographed under these conditions (Fig. 4B). However, when a crude extract of E. coli containing the expressed native Era of S. pneumoniae was chromatographed, this Era protein also exhibited two peaks similar to those of the purified GST-Era protein (data not shown), indicating that the RNA associated with the native Era protein was removed during purification. Together, these results establish that RNA is associated with Era and might stimulate the GTPase activity of the enzyme.
Effect of RNase treatment on GTPase activity of S. pneumoniae GST-Era.
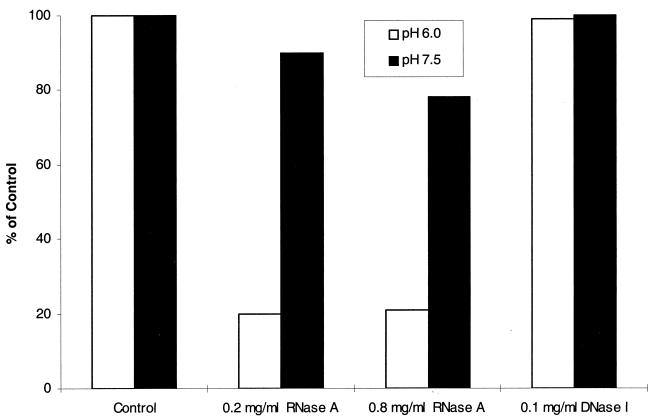
If RNA was copurified with Era and was necessary for stimulation of its GTPase activity, then removal of the RNA associated with Era by RNase treatment should significantly reduce its GTPase activity. As shown in Fig. 5, RNase A, but not DNase I, decreased the GTPase activity of the enzyme at both pH 6.0 and 7.5. Interestingly, a more pronounced reduction of the GTPase activity was observed at pH 6.0 (Fig. 5), although the specific activities of the protein were similar at both pH 6.0 and 7.5 (153 and 197 mmol/min/mol, respectively). This observation suggests that at lower pH, either Era adopted a conformation whereby the RNA was more accessible to RNase A treatment or RNase A was more active. These results support the possibility that RNA might be responsible for stimulation of the GTPase activity of the S. pneumoniae Era protein.
FIG. 5.
Effects of RNase A and DNase I treatments on the GST-Era GTPase activity of S. pneumoniae. Purified GST-Era protein (200 μg/ml) was treated with RNase A (200 or 800 μg/ml) or DNase I (100 μg/ml) at 37°C for 30 min. The RNase A- or DNase I-treated and untreated (control) GST-Era proteins (10 μg each) were then tested for their GTPase activities at 37°C for 30 min by the HPLC method as described in Materials and Methods. After the RNase A and DNase I treatments, parts of the GST-Era preparations were also analyzed by agarose gel electrophoresis (see Materials and Methods), and RNA was not detectable by ethidium bromide staining (data not shown).
The RNA associated with Era is rRNA.
In order to determine the identity of the RNA associated with Era, we extracted the RNA associated with GST-Era by phenol-chloroform treatment. The extracted RNA and purified GST-Era–RNA complex preparation were used as templates for cDNA synthesis with random DNA primers. The DNA fragments generated were cloned into a plasmid and sequenced. The results of this analysis showed that the RNA associated with Era was predominantly E. coli 16S rRNA with a small portion of 23S rRNA. This is consistent with the results obtained from agarose gel electrophoresis of the RNA associated with Era (Fig. 2).
In vitro reconstitution of Era with RNA.
In an effort to restore the high GTPase activity of S. pneumoniae GST-Era or native Era protein whose RNA had been removed by column chromatography, we attempted to reconstitute the proteins with RNA. The addition of the purified native Era protein to E. coli rRNA, poly(U), poly(C), poly(G), or poly(A) (Sigma) failed to restore the high GTPase activity of the protein (data not shown). The attempts to reconstitute the purified native Era and GST-Era proteins with the RNA derived from the purified GST-Era protein preparation (Fig. 4A) under denaturing or nondenaturing conditions also failed (data not shown). These results suggest that either the RNA associated with Era is incorporated into the enzyme during its synthesis or the binding of the RNA to the Era enzyme after its synthesis requires another factor(s) or proper folding in the cell.
The RNA associated with GST-Era of S. pneumoniae does not possess GTPase activity.
To further understand the role of the RNA associated with GST-Era in the stimulation of GTPase activity of the enzyme, we examined the RNA associated with the protein for its possible GTPase activity. To separate the RNA from the protein, a purified GST-Era protein preparation was subjected to Mono Q column chromatography (see Materials and Methods), and the fractions containing RNA were collected. Part of the collected fractions was passed through a glutathione-Sepharose column to remove any possible remaining GST-Era associated with RNA (see Materials and Methods), and the other part of the fractions was not purified further. Examination of both RNA preparations did not show a detectable level of GTPase activity (data not shown). Thus, this result indicated that the elevated GTPase activity of GST-Era probably did not result from the direct contribution of any GTPase activity of the RNA.
We also isolated RNA (mainly rRNAs) directly from E. coli cells by phenol-chloroform extraction (see Materials and Methods) and examined the isolated RNAs for their GTPase activity. Like the RNA associated with GST-Era, the total RNA isolated did not exhibit any GTPase activity under the assay conditions used.
DISCUSSION
In this study, we have shown that Era, a cell-cycle-regulatory GTPase, is associated with 16S rRNA and that the RNA might enhance the GTPase activity of Era. The RNA associated with the S. pneumoniae Era protein was initially detected in purified GST-Era fusion protein that had been expressed in E. coli. Subsequently, we found that approximately 70% of Era was present as a species of at least 600 kDa in crude extracts of S. pneumoniae. The treatment of the crude extract with RNase converted a significant amount of the high-molecular-mass complex of Era to the monomeric form. Since the E. coli 16S rRNA is known to be 1,542 nucleotides in length with an estimated molecular mass of 510 kDa (7), the predicted molecular mass for an Era-16S rRNA complex should be around 600 kDa (see below), consistent with the value (or void volumes) estimated by our gel filtration column analysis. Similarly, the purified GST-Era preparation also appears as a high-molecular-mass species complexed with RNA. Agarose gel electrophoresis established that the RNA was physically associated with the Era protein and was retarded relative to free 16S rRNA. Furthermore, the Era-RNA complex exhibited a maximum absorption at 260 nm. There is also evidence that Era is not directly associated with 30S ribosomes, since the Era-RNA complex was isolated from the supernatant fractions of the crude extracts that had been centrifuged at 180,000 × g for 60 min (see Materials and Methods). Under these conditions, ribosomes had been removed from the supernatant fractions (15, 18, 27, 31). When associated with RNA, the GST-Era protein of S. pneumoniae exhibited a GTPase activity significantly higher than that of the native Era or GST-Era protein that was free of RNA. In addition, the RNA associated with GST-Era alone or the total RNA isolated directly from E. coli cells did not appear to possess a detectable level of GTPase activity. Finally, RNase treatment, but not DNase treatment, reduced the GTPase activity of purified GST-Era protein. Therefore, we conclude that rRNA is associated with the S. pneumoniae Era protein and might be responsible for the stimulation of the GTPase activity.
By contrast, the purified S. pneumoniae native Era protein that was produced in E. coli did not contain any RNA and exhibited a lower GTPase activity. This enzyme was in fact shown to be associated with RNA when produced in E. coli. However, it should be noted that this enzyme was purified by using three different chromatographic columns (including an ion-exchange column) rather than an affinity column as used for the GST-Era protein (see Materials and Methods). The RNA associated with this native Era had been removed during purification, which is consistent with our observation that the RNA was easily removed from the GST-Era by ion-exchange chromatography.
Several lines of evidence appear to suggest that the stimulation of the GTPase activity of the S. pneumoniae GST-Era protein by 16S rRNA is an intrinsic property of the enzyme. First, RNase treatment significantly reduced the GTPase activity of the enzyme. Second, the GST-Era and GST proteins were expressed in the same E. coli strain and purified under identical conditions (see Materials and Methods). Yet, only the purified GST-Era protein preparation was associated with 16S rRNA and exhibited a GTPase activity much higher than that of the Era protein that was free of RNA. The purified GST protein preparation did not exhibit a detectable GTPase activity. In addition, the RNA associated with purified GST-Era protein did not appear to exhibit any GTPase activity. Finally, the GST-Era protein of S. pneumoniae was purified to apparent electrophoretic homogeneity (Fig. 1 and 4). Thus, the observed stimulation of Era GTPase activity by RNA is probably not a result of contamination with other GTPases. However, we could not conclusively rule out the possibilities that other proteins exhibiting GTPase activities were specifically associated with Era or 16S rRNA and that their association with the protein or RNA was disrupted during further column chromatographic purification.
There is also evidence that the Era-RNA complex formation is functionally significant. Our preliminary studies have indicated that the E. coli Era protein is also associated with 16S rRNA which stimulates its GTPase activity (data not shown). The E. coli Era protein (purified as a His-tagged form), when associated with the RNA, exhibited a level of GTPase activity similar to that of the S. pneumoniae GST-Era protein (data not shown). It should be noted that 16S rRNAs from S. pneumoniae and E. coli are 98% identical at the sequence level (reference 7 and data not shown). In addition, we have established that the S. pneumoniae era and GST-era genes can complement an E. coli mutant defective in production of Era, even without overexpression (42). Thus, the observed complex of the S. pneumoniae GST-Era and E. coli 16S rRNA is physiologically functional in E. coli. Furthermore, we were unable to reconstitute purified Era proteins in vitro with different RNA preparations, which suggests that the complex already exists in the bacterial cells before their disruption for purification.
There are two other observations that link Era to rRNA. First, the overexpression of the E. coli ksgA gene, which encodes 16S rRNA methyltransferase (33), was shown to suppress a cold-sensitive mutation in the E. coli era gene (22). This suggests that newly synthesized 16S rRNA is normally undermethylated by KsgA in the cell and that the association of the mutant Era with 16S rRNA is influenced by methylation of rRNA. Second, in E. coli the era gene is part of a transcriptional unit with rnc (40), which encodes the RNase III that is responsible for the initial processing of the 30S precursor rRNA (9). Also, expression of RNase III and Era, like that of ribosomal proteins and RNAs, is positively growth rate regulated (6, 18).
The role of rRNA in the function of Era is not known. Era appears to play a role in cell cycle control (5, 6, 13). However, we do not know whether the RNA-bound or unbound form of Era is responsible, nor do we know the mechanism of this regulation. A reduction in the GTPase activity of Era is inhibitory to cell growth, and overproduction of the mutant Era protein with reduced GTPase activity restored cell growth (5). Thus, it seems possible that the RNA-bound form of Era, which might have the highest GTPase activity, is in an active state that signals cell division. The RNA-free form of the enzyme may be an inactive state that signals a stop of cell division. Thus, the function of Era might be to sense the level of newly synthesized free rRNA in the cell, 16S rRNA in particular. Era has also been shown to partition between the cytoplasm and the cytoplasmic membrane of the bacterial cell (21, 41, 42). The C-terminal domain of Era appears to be required for membrane association (42). The C-terminal domain of Era also appears to resemble the KH domain of the pre-mRNA binding K protein, which is known to bind RNA (38). Thus, it is conceivable that rRNA binds to the C terminus of Era. This binding may block the ability of Era to bind to the membrane, which may serve as a mechanism to sequester the RNA-free form of Era. Finally, since cold-sensitive mutations in the E. coli era gene have been shown to affect ribosome assembly (28–30), the interaction of Era with rRNA may play a role in ribosome synthesis and cell cycle progression.
Finally, the kD value of the S. pneumoniae GST-Era protein is similar to that of E. coli Era (8), but its Km and Vmax values are significantly higher than those reported previously (8, 41). At low GTP concentrations (2.5 to 40 μM), a Km value of 15 μM was obtained, which is in good agreement with the published Km value for E. coli Era (8). However, at high GTP concentrations (40 to 2,000 μM), the Km and Vmax values obtained were 10- to 20-fold higher (Table 1). The unusual kinetic properties of the S. pneumoniae Era protein observed suggest that the GTPase activity of the enzyme may be regulated through the interaction of the enzyme with the bound RNA or through the autophosphorylation of the enzyme (39) at higher GTP concentrations.
ACKNOWLEDGMENTS
We thank D. L. Court and H. Watson for critical reading of the manuscript and P. Treadway for providing S. pneumoniae cells.
REFERENCES
- 1.Ahnn J, March P E, Takiff H E, Inouye M. A GTP-binding protein of Escherichia coli has homology to yeast RAS proteins. Proc Natl Acad Sci USA. 1986;83:8849–8853. doi: 10.1073/pnas.83.23.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 3.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Britton R A, Powell B S, Court D L, Lupski J R. Characterization of mutations affecting the Escherichia coli essential GTPase Era that suppress two temperature-sensitive dnaG alleles. J Bacteriol. 1997;179:4575–4582. doi: 10.1128/jb.179.14.4575-4582.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britton R A, Powell B S, Dasgupta S, Sun Q, Margolin W, Lupski J R, Court D L. Cell cycle arrest in Era GTPase mutants: a potential growth rate-regulated checkpoint in Escherichia coli. Mol Microbiol. 1998;27:739–750. doi: 10.1046/j.1365-2958.1998.00719.x. [DOI] [PubMed] [Google Scholar]
- 7.Brosius J, Palmer M L, Kennedy P J, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen S-M, Takiff H E, Barber A H, Dubois G C, Bardwell J C A, Court D C. Expression and characterization of RNaseIII and Era proteins: products of the rnc operon of Escherichia coli. J Biol Chem. 1990;265:2888–2895. [PubMed] [Google Scholar]
- 9.Court D L. RNA processing and degradation by RNase III. In: Belasco J, Brawerman G, editors. Control of messenger RNA stability. San Diego, Calif: Academic Press; 1993. pp. 71–116. [Google Scholar]
- 10.de Gier J W, Scotti P A, Saaf A, Valent Q A, Kuhn A, Luirink J, Von Heijne G. Differential use of the signal recognition particle translocase targeting pathway for inner membrane protein assembly in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:4646–4651. doi: 10.1073/pnas.95.25.14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farmery M, Macao B, Larsson T, Samuelsson T. Binding of GTP and GDP induces a significant conformational change in the GTPase domain of Ffh, a bacterial homologue of the SRP54 kDa subunit. Biochim Biophys Acta. 1998;1385:61–68. doi: 10.1016/s0167-4838(98)00045-4. [DOI] [PubMed] [Google Scholar]
- 12.Gollop N, March P E. Localization of the membrane binding sites of Era in Escherichia coli. Res Microbiol. 1991;142:301–307. doi: 10.1016/0923-2508(91)90045-c. [DOI] [PubMed] [Google Scholar]
- 13.Gollop N, March P E. A GTP-binding protein (Era) has an essential role in growth rate and cell cycle control in Escherichia coli. J Bacteriol. 1991;173:2265–2270. doi: 10.1128/jb.173.7.2265-2270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hentze M W. Enzymes as RNA-binding proteins: a role for (di)nucleotide binding domains? Trends Biochem Sci. 1994;19:101–103. doi: 10.1016/0968-0004(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 15.Horie K, Wada A, Fukutome H. Conformational studies of Escherichia coli ribosomes with the use of acridine orange as a probe. J Biochem (Tokyo) 1981;90:449–461. doi: 10.1093/oxfordjournals.jbchem.a133492. [DOI] [PubMed] [Google Scholar]
- 16.Inada T, Kawakami K, Chen S, Takiff H E, Court D L, Nakamura Y. Temperature-sensitive lethal mutant of Era, a G protein in Escherichia coli. J Bacteriol. 1989;171:5017–5024. doi: 10.1128/jb.171.9.5017-5024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jagath J R, Rodnina M V, Lentzen G, Wintermeyer W. Interaction of guanine nucleotides with the signal recognition particle from Escherichia coli. Biochemistry. 1998;37:15408–15413. doi: 10.1021/bi981523a. [DOI] [PubMed] [Google Scholar]
- 18.Keener J, Nomura M. Regulation of ribosome synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 1417–1431. [Google Scholar]
- 19.Kurita K, Honda K, Suzuma S, Takamatsu H, Nakamura K, Yamane K. Identification of a region of Bacillus subtilis Ffh, a homologue of mammalian SRP54 protein, that is essential for binding to small cytoplasmic RNA. J Biol Chem. 1996;271:13140–13146. doi: 10.1074/jbc.271.22.13140. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y P, Sharer J D, March P E. GTPase-dependent signaling in bacteria: characterization of a membrane-binding site for Era in Escherichia coli. J Bacteriol. 1994;176:44–49. doi: 10.1128/jb.176.1.44-49.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Q, Inouye M. The gene for 16S rRNA methyltransferase (ksgA) functions as a multicopy suppressor for a cold-sensitive mutant of Era, an essential Ras-like GTP-binding protein in Escherichia coli. J Bacteriol. 1998;180:5243–5249. doi: 10.1128/jb.180.19.5243-5246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.March P E. Membrane associated GTPases in bacteria. Mol Microbiol. 1992;6:1253–1257. doi: 10.1111/j.1365-2958.1992.tb00845.x. [DOI] [PubMed] [Google Scholar]
- 24.March P E, Lerner C G, Ahnn J, Cui X, Inouye M. The Escherichia coli Ras-like protein (Era) has GTPase activity and is essential for cell growth. Oncogene. 1988;2:539–544. [PubMed] [Google Scholar]
- 25.Miller J D, Bernstein H D, Walter P. Interaction of E. coli Ffh/4.5S ribnucleoprotein and FtsY mimics that of mammalian signal recognition particle and its receptor. Nature. 1994;356:657–659. doi: 10.1038/367657a0. [DOI] [PubMed] [Google Scholar]
- 26.Moazed D, Noller H F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 27.Mougel M, Philippe C, Ebel J-M, Ehresmann B, Ehresmann C. The E. coli 16S rRNA binding site of ribosomal protein S15: higher-order structure in the absence and in the presence of the protein. Nucleic Acids Res. 1988;16:2825–2839. doi: 10.1093/nar/16.7.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nashimoto, H. 1993. Non-ribosomal proteins affecting the assembly of ribosomes in Escherichia coli, p. 185–195. In H. K. Nierhaus, F. Franceschi, A. R. Subramanian, V. A. Erdmann, and B. Wittmann-Liebold, B. (ed.), The translational apparatus. Plenum Press, New York, N.Y.
- 29.Nashimoto H, Miura A, Saito H, Uchida H. Suppressors of temperature-sensitive mutations in a ribosomal protein gene, rpsL (S12), of Escherichia coli. Mol Gen Genet. 1985;199:381–387. doi: 10.1007/BF00330746. [DOI] [PubMed] [Google Scholar]
- 30.Nashimoto H, Uchida H. DNA sequencing of the Escherichia coli ribonuclease III gene and its mutations. Mol Gen Genet. 1985;201:25–29. doi: 10.1007/BF00397981. [DOI] [PubMed] [Google Scholar]
- 31.Noll M, Hapke B, Schreier M H, Noll H. Structural dynamics of bacterial ribosomes. I. Characterization of vacant couples and their relation to complexed ribosomes. J Mol Biol. 1973;75:281–294. doi: 10.1016/0022-2836(73)90021-1. [DOI] [PubMed] [Google Scholar]
- 32.Pillutla C R, Sharer J D, Gulati P S, Wu E, Yamashita Y, Lerner C G, Inouye M, March P E. Cross-species complementation of the indispensable Escherichia coli era gene highlights amino acid regions essential for activity. J Bacteriol. 1995;177:2194–2196. doi: 10.1128/jb.177.8.2194-2196.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poldermans B, Roza L, Van Knippenberg P H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3′-end of 16S ribosomal RNA of Escherichia coli. III. Purification and properties of the methylating enzyme and methylase-30S interactions. J Biol Chem. 1979;254:9094–9100. [PubMed] [Google Scholar]
- 34.Preiss T, Sang A E, Chrzanowska-Lightowlers Z M, Lightowlers R N. The mRNA-binding protein COLBP is glutamate dehydrogenase. FEBS Lett. 1995;367:291–296. doi: 10.1016/0014-5793(95)00569-u. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Sato T, Wu J, Kuramitsu H. The sgp gene modulates stress responses of Streptococcus mutans: utilization of an antisense RNA strategy to investigate essential gene functions. FEMS Microbiol Lett. 1998;159:241–245. doi: 10.1111/j.1574-6968.1998.tb12867.x. [DOI] [PubMed] [Google Scholar]
- 37.Shibata T, Fuji Y, Nakamura Y, Nakamura K, Yamane K. Identification of protein synthesis elongation factor G as a 4.5 S RNA-binding protein in Escherichia coli. J Biol Chem. 1996;271:13162–13168. doi: 10.1074/jbc.271.22.13162. [DOI] [PubMed] [Google Scholar]
- 38.Siomi H, Matunis M J, Michael W M, Dreyfuss G. The pre-mRNA binding K-protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sood P, Lerner C G, Shimamoto T, Lu Q, Inouye M. Characterization of the autophosphorylation of Era, an essential GTPase in Escherichia coli. Mol Microbiol. 1994;12:201–208. doi: 10.1111/j.1365-2958.1994.tb01009.x. [DOI] [PubMed] [Google Scholar]
- 40.Takiff H E, Chen S-M, Court D L. Genetic analysis of the rnc operon of Escherichia coli. J Bacteriol. 1989;171:2581–2590. doi: 10.1128/jb.171.5.2581-2590.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J, Cho M-I, Kuramitsu H K. Expression, purification, and characterization of a novel G protein, SGP, from Streptococcus mutans. Infect Immun. 1995;63:2516–2521. doi: 10.1128/iai.63.7.2516-2521.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao G, Meier T I, Peery R B, Skatrud P L. Biochemical and genetic characterization of the carboxyl-terminal domain of Era from Streptococcus pneumoniae. Microbiology. 1999;145:791–800. doi: 10.1099/13500872-145-4-791. [DOI] [PubMed] [Google Scholar]
- 43.Zuber M, Hoover T A, Dertzbaugh M T, Court D L. A Francisella tularensis DNA clone complements Escherichia coli defective for the production of Era, an essential Ras-like GTP-binding protein. Gene. 1997;189:31–34. doi: 10.1016/s0378-1119(96)00813-x. [DOI] [PubMed] [Google Scholar]
- 44.Zuber M, Hoover T A, Powell B S, Court D L. Analysis of the rnc locus of Coxiella burnetii. Mol Microbiol. 1990;14:291–300. doi: 10.1111/j.1365-2958.1994.tb01290.x. [DOI] [PubMed] [Google Scholar]