Abstract
Extracellular vesicles (EVs) represent small, membrane-enclosed particles that are derived from parent cells and are secreted into the extracellular space. Once secreted, EVs can then travel and communicate with nearby or distant cells. Due to their inherent stability and biocompatibility, these particles can effectively transfer RNAs, proteins, and chemicals/metabolites from parent cells to target cells, impacting cellular and pathological processes. EVs have been shown to respond to disease-causing agents and impact target cells. Given that disease-causing agents span environmental contaminants, pathogens, social stressors, drugs, and other agents, the translation of EV methods into public health is now a critical research gap. This paper reviews approaches to translate EVs into exposure science, toxicology, and public health applications, highlighting blood as an example due to its common use within clinical, epidemiological, and toxicological studies. Approaches are reviewed surrounding the isolation and characterization of EVs and molecular markers that can be used to inform EV cell-of-origin. Molecular cargo contained within EVs are then discussed, including an original analysis of blood EV data from Vesiclepedia. Methods to evaluate functional consequences and target tissues of EVs are also reviewed. Lastly, the expanded integration of these approaches into future public health applications is discussed, including the use of EVs as promising biomarkers of exposure, effect, and disease.
1. INTRODUCTION
1.1. Introduction to Extracellular Vesicles (EVs)
1.1.1. What are EVs?
All cells, including healthy and diseased, release small membrane-enclosed particles called extracellular vesicles (EVs) that can travel into the extracellular space and exist in almost all bodily tissues/fluids [1–3]. These particles contain an outer lipid bilayer but are distinguished from cells because they cannot replicate; meaning, they do not contain a functional nucleus that allows for cellular replication. Instead, these particles serve as transient cargo carriers of many types of molecules between cells.
EVs were originally identified as early as the 1940s, though inconsistent terminology coupled with technological limitations made them difficult to characterize and report clearly until recently [3]. To provide a brief history, EVs may have been first described by scientists Chargaff and West, who hypothesized that small, minute breakdown products of blood contributed to blood clotting properties [4]. It was later that Wolf et al. in 1967 found through electron microscopy that small platelet-derived particulate matter acted as coagulant material [5]. This study also found that these particles contained lipids originating from platelets and referred to them as “platelet-dust” [5]. Two notable papers were then published over a decade later, in 1983, that simultaneously identified small vesicles (~50 nm) that were released from maturing blood reticulocytes into surrounding extracellular areas [6,7]. These small vesicles were found to play a role in transmitting blood plasma iron transporters, transferrins, between cells and extracellular environments of rat and sheep reticulocytes cultured in vitro [6,7]. Since these landmark studies, there has been an abundance of published research supporting the wide variety of roles EVs play in general cell health and disease.
When referring to these small particles, the term “EV” was not implemented until recently, where other umbrella terms have been coined over the past few decades. This evolving terminology supports the sudden expansion of EV research, which undoubtably contributed to the development of contradicting terminologies across labs and research consortia. However, this point of confusion is being directly addressed through the International Society for Extracellular Vesicles (ISEV), which have now produced two guidance documents (one in 2014 and one in 2018), to bring consensus to EV terminology and common aspects for reporting EV study findings. The terminology used in this review aligns with the most recent guidelines published by ISEV in 2018, where the term EV is endorsed as “the generic term for particles naturally released from the cell that are delimited by a lipid bilayer and cannot replicate, i.e., do not contain a functional nucleus” [8].
1.1.2. Overview of Biogenesis and Release Mechanisms that Define EV Subtypes
EV subtypes are defined by their mechanism of biogenesis, or in other words, how each particle is formed and released from the parent cell (Figure 1). EV subtypes span three major classes: exosomes, microvesicles, and apoptotic bodies that are produced by processes that are distinct and independent from each other [2,9–11]. Once generated, these EV subtypes are difficult to differentiate, as they overlap in size and composition. Exosomes are formed within endosomal compartments which are organelles that aid in the process of endocytosis (i.e., the uptake of biological matter via the cellular membrane). Exosomes are released via secretion when these intracellular compartments fuse with the plasma membrane. Exosomes range in size of ~30–200 nm. In contrast, microvesicles are formed and released directly from a cell’s plasma membrane. Microvesicles range in size of ~100–2000 nm [2,9,10]. Apoptotic bodies are generated from cellular apoptotic disintegration. Apoptosis refers to programmed cell death that occurs as an essential and controlled part of the cell cycle to maintain tissue homeostasis. As the cell breaks down into small parts, apoptotic bodies are released which typically range in size of ~1–5 um [11]. Of the three major EV subtypes, apoptotic bodies are the only subtype required to generate from dying cells, by definition; although it is important to note that other EV subtypes can be released during other cell death processes including primary necrosis, secondary necrosis, pyroptosis and necroptosis. Dying-cell-derived EVs can also be produced from ruptured membrane fragments that reform extracellularly into EVs [11]. Other terms that are sometimes used to refer to EV subtypes include microparticles, ectosomes (sometimes interchangeable with microvesicles), and oncosomes, among other possible names [8,9].
Figure 1. Mechanisms of EV biogenesis, resulting in three defined classes of EVs (i.e., exosomes, microvesicles, and apoptotic bodies) with overlapping ranges in size, composition, and function.
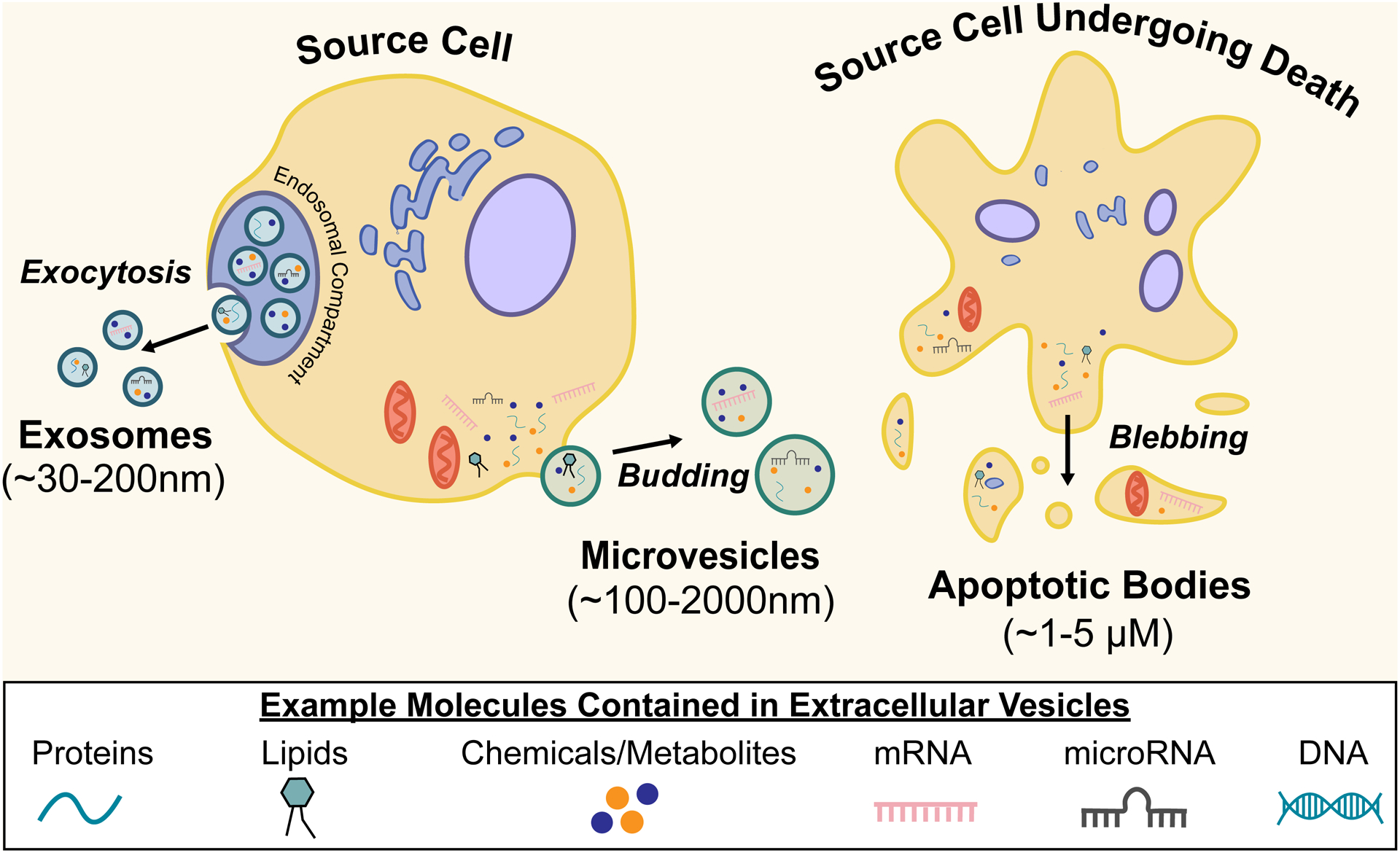
Note that dying-cell-derived EVs can be generated through any of the below mechanisms.
1.1.3. Overview on EV Content
EVs can contain important molecules that are well established regulators of cell function and overall health. Genetic material can be contained within EVs spanning DNA and RNA-based molecules, including mRNAs, microRNAs, various non-coding RNAs, and mitochondrial RNAs [12]. Other types of molecules that can be contained within EVs include lipids, proteins, chemicals, and/or chemical metabolites.
An interesting aspect of EV content is that the loading of specific cargo within EVs was once thought to be completely stochastic, based on random events and the local environment that happened to be in proximity during EV biogenesis [2]. However, there is now a propensity of evidence showing that EV content is not random; rather, the loading of molecular content within EVs is based on highly regulated processes [2,13–16]. EV loading is now recognized to reflect what the cell is experiencing in response to physiological and pathological status and potential changes in response to insults. EV content loading is further recognized as regulated with purposeful consequences towards downstream events and target cells [2,13–16]. This relatively recent shift in understanding has further accelerated interest surrounding EVs as carefully regulated transporters of molecular signals capable of influencing nearby and distal cells.
1.1.4. Potential Consequences of EV Release and Changes to Target Cells
Once EVs are generated and released into the extracellular/pericellular environment, these particles can communicate with nearby cells or potentially travel and communicate to distant recipient cells. EVs can then impart beneficial, neutral, or detrimental effects to target cells [17]. The majority of secreted EVs remain locally and can remain inert/inactive without encountering target cells, or they can actively influence local processes. For example, for tissues undergoing cell death/injury, EVs often remain associated with the pericellular matrix. In these conditions, EVs have been shown to promote clearance of cell debris and tissue repair while also sometimes contributing to inflammation and the propagation of immune signaling that overlaps with several cell death regulatory mechanisms [11]. In some instances, EVs can also travel to distant recipient cells via blood-mediated transport [11,18], which supports the utility of evaluating these particles in human blood samples for public health research applications.
EV-mediated communication can occur by a variety of mechanisms, including cell surface contact-mediated interactions, direct membrane fusion, endocytosis, and phagocytosis [11,18]. Other mechanisms of EV uptake are more dependent upon the lipid content surrounding EVs, where depending on the conditions, EVs can physically fuse together with target cell membranes [19]. After EV materials are transferred to target cells, these molecules can remain functional and influence target cell biology. Much of what we know surrounding EV-induced biological changes has resulted from cancer research. For example, tumor-derived EVs have been shown to contain certain RNA molecules that activate the important receptor, such as toll-like receptor 3 (TLR3), in lung cells, resulting in increased neutrophil infiltration and stimulation of chemokine secretion that promote lung tumor metastasis [20]. In addition to cancer, molecular contents within EVs have also been reported to alter processes involved in other many disease outcomes, including cardiovascular, liver, metabolic, and neurodegenerative disease, among many others [17,21–24]. In summary, EVs can remain inert or potentially influence target cells locally or distally, transfer important molecular cargo contained within them, and alter target cell biology, potentially influencing disease. Further examples of these important intercellular communications are detailed throughout this review in the context of exposure science, toxicology, and public health.
1.2. Current Review Examines Approaches to Integrate EVs into Public Health Research
Strategies for analyzing EVs in biological samples have been reviewed, with publications occurring largely within the past few of years [2,3,8,9,12,25]. These reviews and corresponding original research articles have primarily been generated from molecular biology, genetics, and medical fields. However, there remains a critical gap in knowledge and communication of these approaches towards public health research. The field of public health encompasses clinical, epidemiological, and toxicological research aimed at identifying causes of human diseases and implementing measures to reduce the impact of disease-causing agents. Disease-causing agents can span environmental contaminants, pathogens, social stressors, and drugs, among others. Research in exposure science, toxicology, and public health relies heavily upon human biomonitoring efforts, clinical studies, and other controlled in vivo experimentation. One of the most commonly utilized biological samples in public health research is peripheral blood, as it represents an important tissue that is relatively easy to collect in comparison to other tissues. Molecular information within blood and other biological samples can also be leveraged to inform the overall health of an individual, as is common in clinical research and medical practice. Blood and biological fluid based analyses can also inform potential molecular mediators involved in disease, as is common for epidemiological and toxicological research. The advantage of sampling EVs is that they are present in all biological fluids. Much of the blood-focused methods discussed in this review overlap with those that can be used to analyze other biological fluids applicable to public health research, including less invasive sampling of body fluids such as saliva, sweat, tears, and urine. Because EVs are present in all biological fluids and influence other target tissues, approaches to evaluate EVs in blood and other biological samples have the potential to significantly enhance ongoing public health research efforts. This review focuses on these approaches to help bridge the gap between these advanced molecular biology techniques and approaches to expand research on exposure science, toxicology, and public health.
2. APPROACHES TO ISOLATE BLOOD EVS FOR PUBLIC HEALTH RESEARCH
2.1. Blood Composition
Blood is one of the most common biological samples incorporated into public health research, and is composed of mixed cell types and other circulating matter, including red blood cells, white blood cells, platelets, and plasma. When conducting EV research, it is important to isolate and enrich for EVs amongst this complex biological fluid. This is particularly important given that blood (in particular, plasma/serum) contains non-EV particles that can range in size similar to EVs (e.g., lipoprotein, soluble cytokines, and other lipid-like particles) [26,27].
2.2. Preparation of Plasma and Serum
EVs are abundantly present in bodily fluids including blood components plasma and serum, which are commonly collected in exposure science, toxicology, and public health research. Several methods have been developed to isolate EVs from plasma and begin with collection of whole blood into anticoagulant-treated tubes. Several anticoagulants have been used to collect blood for analysis of EVs, including ethylenediaminetetraacetic acid (EDTA), sodium fluoride/potassium oxalate, and sodium citrate with or without additives such as adenosine and dipyridamole or dextrose [28–30]. Selection of anticoagulant is multifactorial and should depend on the intended downstream analysis. For example, citrate is one of the most commonly used anti-coagulants in EV isolation and is compatible with various downstream analyses such as RNA sequencing. Additionally, use of citrate has previously recommended by the International Society on Thrombosis and Haemostasis for use in isolation of EVs [31]. In contrast, heparin should not be used prior to EV isolation due to its interference with polymerase chain reaction methods commonly used in gene-specific expression analyses [29]. Heparin has also been reported to prevent EV uptake by cells which can lead to the inaccurate finding of increased EV numbers in the blood [8,32]. To isolate EVs from plasma, methods commonly require platelet-poor plasma. Platelet removal is essential because platelets release EVs on activation and fragment during a freeze–thaw cycle [29]. This is achieved via centrifugation to pellet cellular components, and collection of the resulting supernatant, representing platelet-poor plasma. Resulting plasma can be used to proceed to EV isolation.
Serum is collected from whole blood, typically by allowing blood to clot at room temperature and removing the clot by centrifuging. Remaining supernatant (serum) can then be used for EV isolation [29,33]. Methods used to prepare serum have been shown to result in enrichment of platelet derived EVs, making plasma the preferred source of EVs in many studies [30]. However, some argue for the utility of serum-based EVs as containing informative miRNA disease biomarkers [33]. Therefore, careful consideration should be given when selecting EV sample source, where further data will continue to inform the advantages and disadvantages surrounding sample preparation in exposure science, toxicology, and public health research.
2.3. Methods for Isolation of EVs
A variety of methods have been developed to isolate EVs, each with potential advantages and limitations depending on the study purpose. Current methods include ultracentrifugation (e.g., differential centrifugation and isopycnic density centrifugation), size exclusion techniques, immunoaffinity, and precipitation. These methods leverage various properties of EVs such as their density, size, surface proteins and receptors, and solubility. Given that individual methods isolate based primarily on one EV characteristic, each method will vary in yield, potential co-eluting contaminants, and downstream applicability. Additionally, many methods aim to isolate exosomes as the specific subtype of EV of interest; however, because of overlapping particle properties (Figure 1), it is important to recognize potential limitations in each isolation technique.
Two of the most common EV isolation techniques include ultracentrifugation (UC) and size-based isolation. UC currently accounts for over 50% of all published isolation methods due to its relative ease and efficiency [34]. UC isolates EVs by leveraging size and density differences of each component within the original sample. This method is typically performed in two steps: first, a low-speed centrifugation removes cells, cell debris, and other large particles, and then second, a high-speed centrifugation (≥ 100,000 × g) separates EVs. EVs can then be resuspended in medium such as phosphate buffered saline and stored for future analyses [29,35]. Size-based isolation techniques typically refer to methods such as ultrafiltration (UF) and size exclusion chromatography (SEC). UF isolates EVs based on their size using membrane filters with defined size or molecular weight exclusion limits [35]. Similarly, SEC utilizes a porous stationary phase to elute macromolecules and particulate matter out and retain small molecules in the gel to later be eluted by mobile phase [36]. For both UC and size-based isolation methods, similar sized particles can potentially co-elute, acting as potential sources of contamination. Overall, these methods have high yields, are relatively easy to perform, and after the upfront purchase of instrumentation (e.g., ultracentrifuge), these methods can be quite cost-efficient [29,32,36].
Additional methods for isolating EVs include immunoaffinity and co-precipitation [29]. Immunoaffinity is highly specific and leverages proteins and receptors in the membrane of EVs to separate and purify them from heterogenous mixtures [34]. Co-precipitation methods utilize EV solubility properties to isolate them from biological fluids. For these methods, a water-excluding polymer such as polyethylene glycol can be added to the sample to exclude EVs from a solution (via void volume exclusion) resulting in precipitated fractions of EVs can then be collected via centrifugation [37]. Various kits are available for exosome enrichment which have been tested for accuracy based on their performance measures using approaches described below. Several of these kits utilize water excluding polymers such as ExoQuick (System BioScience), Exosome Isolation Reagent (Life Technologies), and Total Exosome Isolation Reagent (ThermoFisher) to isolate intact EVs. Given that no isolation technique works perfectly, it is important to further characterize EVs that are isolated from biological samples.
3. APPROACHES TO CHARACTERIZE EVS
A variety of methods are now available to characterize EV samples isolated/enriched from blood and/or other biological samples of interest for exposure science, toxicology, and public health applications. Physical properties that are commonly measured include EV size, concentration, and morphology captured using high-resolution microscopy, light scattering, electrical charge, and/or flow cytometry. EV presence can also be evaluated through biomarkers, which at this point, largely encompass protein markers that can inform the purity of EVs against similarly sized particles that can co-elute during EV isolation protocols. It is notable that at this point, the majority of EV-related studies depend upon the use of multiple complementary methods to characterize EVs within their samples of interest. These methods have been previously reviewed [8,9,25] and are summarized here to inform the design of ongoing/future research in public health.
3.1. Methods for EV Physical Characterization
3.1.1. Microscopy Methods for EV Physical Characterization
EVs can be structurally characterized using imaging techniques based on microscopy that include scanning electron microscopy (SEM), transmission electron microscopy (TEM), cryo-electron microscopy (CEM), and atomic force microscopy (AFM). SEM is the most established of these methods, commonly used in molecular laboratories, and results in images of EVs within a sample that have been typically stained with a contrasting agent (i.e., a counterstain), fixed, and dehydrated prior to imaging. Images are produced via scanning of the fixed surface with electrons, resulting in 3D images of surface topography and elemental sample composition. TEM is a similar method, though results are typically higher resolution than those produced via scanning electron microscopy. TEM can also notably be integrated with immunogold labeling to result in the potential parallel characterization of specific molecules [25]. CEM is similar to the two aforementioned methods except that instead of fixing/staining samples prior to analysis, samples are frozen and analyzed at low temperatures. This leads to improved morphology analyses, where the 3D structure of EVs has been verified as round [38]. Another type of microscopy method is AFM, where physical attribute data are obtained via mechanical measurements of deflection over the samples’ surface to inform molecular surface topography as well as stiffness and adhesion properties. Altogether, microscopy-based methods can provide important information regarding EV size and morphology, and typically result in high-resolution measures (>5 nm) of EVs on an individual basis [9]. These methods have the notable limitation of not providing information across all EVs within a sample, and thus cannot provide concentration measures or collective size distributions that can be used to trace back concentration/size estimates within originating blood, tissues, or other biological samples.
3.1.2. Light Scattering Methods for EV Physical Characterization
EVs can be characterized for concentration and size distribution using bulk- and individual EV-based light scattering methods. A type of bulk method is dynamic light scattering, where EVs are suspended, and the bulk amount of scattered light is measured as the particles continuously move due to Brownian motion physics. Resulting light intensities and correlative fluctuation rates can be used to derive hydrodynamic particle diameters, yielding particle size and number distribution [9]. A similar light scattering-based method is nanoparticle tracking analysis (NTA) [39]; though for this method, scattering measures are obtained at the individual particle-level, in contrast to bulk particle measures. Through NTA methods, the path of each individual particle is recorded and used to determine particle velocity and diffusivity, which are measures that are used to calculate particle size and number distribution. These values can be used to characterize EV size distributions and quantify the number of EVs in a given sample volume. NTA methods can also notably incorporate protein profiling through immunofluorescent labeling methods [9].
3.1.3. Flow Cytometry Methods for EV Physical Characterization
Flow cytometry methods include flow cytometry for larger EVs (typically ≥ 300 nm) and other more high-resolution flow cytometry methods for smaller particles [2,8,40]. Flow cytometry traditionally operates by suspending a sample in a fluidics system, representing a hydrodynamically focused fluid stream, that then is passed through multiple laser beams. The sample’s particles are thus illuminated and their associated fluorescent light and scattered light are measured through an optical system [40]. High-resolution flow cytometers often differ from more standard flow cytometers by having higher powered lasers with more focused beams, a more stable fluid stream with a smaller diameter, and higher sensitivity optical detectors [40]. Single cell EV analysis methods have recently been incorporated through flow cytometry, specifically through the enabling of chemical-based signal amplification methods [2]. These methods result in EV quantitation and size distribution information. Flow cytometry methods are also advantageous when coupled with fluorescence-based measurements to measure select proteins [40].
3.1.4. Tunable Resistive Pulse Sensing for EV Physical Characterization
Tunable resistive pulse sensing represents an additional method for the evaluation of EV size distribution and concentration, that measures EVs >30 nm on an individual particle basis [41]. This method measures changes in electrical currents as individual particles pass through a polyurethane membrane containing size-adjustable nanopores. The amount in which the electrical current drops during the passing of each particle through the membrane is recorded and used to derive each particle’s associated volume. One limitation of this approach is that it becomes difficult to analyze complex biofluids containing heterogenous EV populations which block pores of certain sizes, although this method is quite efficient when analyzing purified samples [9,25].
3.1.5. Limitations of EV Physical Characterization Methods
Certain methods described have been shown to outperform others when measuring select EV size ranges, notably causing some researchers to advocate for characterization based on multiple platforms [42]. Notably, there are evolving guidelines to inform which physical characteristics should be reported when publishing molecular data from isolated EVs [8]. As research surrounding EVs continues to grow, methods and associated best practices for characterizing EVs will continue to develop and should be considered when designing studies in public health research.
3.2. Methods to Evaluate EV Presence through Protein Markers
Particles that are isolated and physically characterized are often further analyzed for select proteins to further support that they are, indeed, EVs as opposed to other similarly sized particles that may be present within biological samples. Protein markers have been postulated to differentiate between EVs vs other particles and sometimes inform EV subtype. These methods and associated markers have been previously reviewed [8,9] and are summarized here.
3.2.1. Types of Protein Markers to Evaluate EV Presence
Because EVs are secreted/released from cells through budding or endosomal transition mechanisms, they maintain parts of the originating cells’ membrane and cytosol. Transmembrane and cytosolic proteins from the originating cell are therefore transferred in this process and can be leveraged to evaluate EV presence. The presence of EVs is commonly evaluated through multiple platforms, including the detection of at least one transmembrane protein known to be embedded in the plasma membrane. Transmembrane proteins that are commonly measured for these purposes include members of the tetraspanin family of transmembrane proteins, such as CD9 (CD9 molecule), CD63 (CD63 molecule), and CD81 (CD81 molecule) [8,9]. The presence of these molecules suggests the presence of the lipid-bilayer outer membrane structure, indicating that the isolated particles may have originated from cellular multi-vesicular bodies/endosomal organelles, consistent with EV formation. The detection of proteins commonly present in cellular cytosol that are transferred into EVs can also be measured, such as ALIX (ALG-2 interacting protein X) and TSG101 (tumor susceptibility 101). To evaluate the purity of EV isolation, common protein contaminants that may be isolated at the same time as EVs can be measured, such as apolipoproteins, albumin, and uromodulin [8,9].
3.2.2. Methods to Evaluate Protein Markers of EV Presence
Methods used to evaluate protein markers in EV-based analyses include immunostaining approaches, including immunoblotting and immunohistochemical (IHC) analysis, imaging-based light scattering (e.g., NTA) and imaging flow cytometry analyses. Immunoblotting and IHC approaches are standard molecular biology techniques that have previously been described in the context of EV research allowing for semiquantitative measures of protein abundance [9]. A notable aspect of EV IHC analyses is that they can now be performed with further efficiency through assay miniaturization, for instance by leveraging microfluidics (e.g., ExoChip) [43] and spot-printing of antibody panels (e.g., EV arrays) [44,45]. In some instances, these surface-based methods for detecting proteins even allow for the recovery of EVs for further downstream processing and analyses [9].
Immunosorbent analyses can also be performed using immune-magnetic beads to capture and measure EV-associated proteins. These methods have several advantages compared to immunostaining methods, including the potential detection of proteins with increased efficiency, relatively simple labeling and washing protocols, shorter assay run times, and assay flexibility that allows for subsequent analyses using the same samples. Methods can incorporate microfluidics and/or multiplexing to allow for the simultaneous detection of many surface markers [46]. Methods include the integrated microfluidic exosome analysis platform, the ExoScreen assay, and single EV analysis based on multiplexing [9,25]. Some of the most efficient approaches are those that couple protein-based measures with EV physical analyses. For example, NTA can be coupled with antibody and other dye-labeling techniques (e.g., cell membrane labeling) to obtain important phenotyping information alongside particle size distributions of labeled vs non-labeled particles. Similar information can be obtained through flow cytometry methods, where high-resolution flow cytometry can provide information on surface marker distributions at a single EV-level [46]. These technologies are currently being developed and refined and should serve as promising platforms in public health research.
3.2.3. Limitations of Protein Markers to Evaluate EV Presence
The use of protein markers to evaluate EV presence has important limitations, resulting in data that are largely qualitative when informing EV type. To date, proteins have yet to be identified that can be used to characterize all types of EVs, nor have any been identified as present consistently across every EV within a particular EV subtype [9]. Recent evidence, for example, has highlighted proteins once thought of as classical exosome markers (e.g., CD9 or CD81) are also included in other EV subtypes and are not so clearly subtype-specific [47,48]. Overall, these current limitations must be considered when designing EV-related studies and interpreting resulting data, particularly when analyzing heterogenous tissues (e.g., blood) as is common in exposure science, toxicology, and public health studies.
4. EVALUATING EVS BASED ON CELL-OF-ORIGIN
It is well established that information from originating cells (aka “parent” cells or “host” cells) can be packaged into EVs, including lipids, proteins, and nucleic acids [12]. These molecules can then be functional within recipient cells upon arrival. In addition, some markers within the plasma membrane of originating cells can be packaged onto EVs, if they are generated through budding mechanisms [12]. EV content can therefore include information that can be used to inform cell-of-origin. The utility of cell-of-origin markers may be promising, particularly when multiple molecular evidence streams are combined using computational models to potentially improve cell-of-origin predictions. These markers can be incorporated into clinical diagnosis and treatment applications, drug delivery applications, as well as future applications postulated by this review surrounding public health.
4.1. Protein Markers Informing Cell-of-Origin
Several proteins have been identified to inform cell-of-origin for EVs (Table 1). For example, proteins that are present within the cell surface membrane of the host cell, such as transmembrane or GPI-anchored proteins, are often transferred to the surface membrane of the EV during EV biogenesis. When these surface proteins are associated with certain cell types, they can thus be used to inform cell-of-origin. Some examples of cell surface proteins that can inform cell-of-origin include cluster of differentiation (CD) proteins, including CD9, CD14, CD53, and CD56 (also referred to as neural cell adhesion molecule [NCAM]) that are associated with certain immune cells. Other proteins that inform cell-of-origin include those that are located internally within cells (e.g., cytosolic proteins). It is important to note that most of these example proteins are merely associated with the listed tissue/cell type, and are very rarely exclusive to those tissues/cell types. Because of this current limitation, there is an overall lack of definitive EV markers that are completely cell-specific. This limitation, combined with the inherent heterogeneity of EV subtypes, support the need for improved cell-of-origin markers and other coinciding measures that could further strengthen the identification of EV cell-of-origins through multifactorial data analysis and prediction modeling.
Table 1. Example protein markers that have been used to inform cell-of-origin.
References are listed to include those that recognized the use of each protein marker towards informing a certain cell-of-origin. These examples do not represent an exhaustive list of all proteins and corresponding potential cells-of-origin. As discussed in section 4, these markers, in themselves, cannot definitely identify which type of cell emitted a specific EV; rather, this information can be used to inform cell-of-origin in concert with additional data streams and study design context.
| Associated Protein Marker Symbol | Cell-of-Origin | Associated Protein Marker Full Name | Reference(s) |
|---|---|---|---|
| A4/APP | Neurons | Amyloid beta / Amyloid precursor protein | [8,93] |
| AChE-E | Erythrocytes | Acetylcholinesterase-erythrocyte | [8,94] |
| AChE-S | Neurons | Acetylcholinesterase-synaptic | [8,94] |
| CD2 | Natural killer cells | CD2 molecule | [46] |
| CD8 | Natural killer cells | Cluster of differentiation 8 | [46] |
| CD9 | Eosinophils | CD9 molecule | [95] |
| CD11b | Non-resident monocytes/macrophages | Integrin subunit alpha M | [96] |
| CD14 | Monocytes | Monocyte differentiation antigen 14 | [8,97] |
| CD16 | Macrophage subsets | CD16 antigen | [98] |
| CD29 / FNRB | Hepatocytes | CD29 antigen / Fibronectin receptor subunit beta | [27,99] |
| CD29 / FNRB | Mesenchymal stromal cells | CD29 antigen / Fibronectin receptor subunit beta | [100] |
| CD37 | Leukocytes | Leukocyte antigen CD37 | [8,101] |
| CD41b / CD42a | Platelets | Glycoprotein (Gp) IIb/IIIa integrin | [8,46] |
| CD44 | Mesenchymal stromal cells | CD44 molecule | [100] |
| CD45 / PTPRC / LCA | Immune cells | CD45 antigen / Protein tyrosine phosphatase, receptor type, C / Lymphocyte common antigen | [8] |
| CD56 / NCAM | Natural killer cells | CD56 antigen / Neural cell adhesion molecule | [46] |
| CD61 / GPIIIa | Platelets | CD61 antigen / Platelet glycoprotein IIIa | [27,46,102] |
| CD63 | Epithelial cells | CD63 molecule | [95] |
| CD66b / CEACAM8 | Granulocytes/eosinophils | CD66b antigen / Carcinoembryonic antigen-related cell adhesion molecule 8 | [98] |
| CD73 | Mesenchymal stromal cells | 5’-nucleotidase ecto | [100] |
| CD81 | Epithelial cells | CD81 molecule | [95] |
| CD90 | Mesenchymal stromal cells | Cluster of differentiation 90 | [100] |
| CD105 | Mesenchymal stromal cells | Endoglin | [100] |
| CD105 | Endothelial cells | Endoglin | [74] |
| CRABP1 | Neural cells | Cellular retinoic acid binding protein 1 | [27,103] |
| EPCAM | Epithelial cells | Epithelial cell adhesion molecule | [8,27,104] |
| GYPA | Red blood cells | Glycophorin A | [8,105] |
| Ly6G | Neutrophils | Lymphocyte antigen 6 complex, locus G | [106] |
| PECAM1 | Endothelial cells | Platelet and endothelial cell adhesion molecule 1 | [8,107] |
| PLAP | Placental cells | Placental alkaline phosphatase | [89,108] |
| TSPAN8 | Epithelial cells | Tetraspanin-8 | [8,27,109] |
4.2. Lipid Markers Informing Cell-of-Origin
One of the major components of EVs is the surrounding lipid bilayer, which is derived from the parent cell, allowing it to potentially inform cell-of-origin. One side of phospholipid bilayers consists of two hydrophilic phosphate heads and a hydrophobic tail, with specific contents and configurations showing variety across cells and resulting EVs [2]. Studies have described the percentage of different lipid classes in several different cell types such as Oli-neu cells, human B-cells, mast cells, dendritic cells, and reticulocytes [49]. Enrichment of select classes of lipids have been identified in EVs, including cholesterol, glycosphingolipids, phosphatidylserine, and sphingomyelin. The changes in amounts of lipid classes have been quantified across select cell types, with several showing lipid class commonalities indicating that similar enrichment can happen in multiple types of cells. Certain phospholipids can also notably be distributed differentially according to cellular (and subsequence EV) surface sides [50]. Unlike RNA and proteomic sorting mechanisms, studying lipid composition has been difficult due to their inherent variability and limitations surrounding analytical methods [12]. Future research will likely expand upon the potential utility of lipid content in informing EV cell-of-origin.
4.3. Nucleic Acid Markers Informing Cell-of-Origin
EVs notably contain nucleic acids, including small RNAs (e.g., miRNAs) and mRNA molecules, which can also display patterns that are reliant upon cell-of-origin. In addition, there are certain RNA-based molecules that are expressed at relatively higher levels in certain cell types that could be leveraged for such purposes. Even molecules that are known to be highly expressed in certain cell types above others could be analyzed for suitability in informing cell-of-origin. Although the methods are available to link potential RNA molecules to specific parent cells, there have been an overall lack of studies evaluating the utility of using RNA-based molecules for informing or predicting EV cell-of-origin. One example study found that HEK293T cells released EVs that were enriched for specific miRNAs in comparison to other cell types [51]. Another recent example study employed computational methods based on gene expression profiles coupled with deconvolution analyses to estimate relative proportions of EV tissue origins in blood plasma samples [52]. Authors noted a large fraction of EVs attributable to blood cells as well as liver tissue from patients with hepatic disease, though it is notable that these methods are still in their infancy and require further validation. We have demonstrated utility using mRNA and miRNA-based signatures in machine learning and computational models to predict environmental exposures and toxicological outcomes [53,54], further supporting the potential of this field to leverage such signatures in EV research. Given the variable nature of RNA profiles across tissues [55], it is evident that such profile data could be leveraged to continue to develop computational models that predict, or otherwise inform, EV cell-of-origin.
4.4. Limitations and Future Directions Surrounding Cell-of-Origin EV Markers in Public Health Studies
Collectively, there is a current lack of specific EV-based biomarkers that are capable of clearly estimating cells-of-origin with high precision and/or confidence across tissues, including blood [12]. Most of the information surrounding cell-of-origin information centers around protein markers, largely encompassing cell surface markers that are largely specific to certain cell types. As this field continues to grow, additional layers of information will undoubtably contribute to the increased predictive capabilities of biomarkers in informing cell-of-origin, particularly in blood samples. Additional information that will likely bolster these predictive capabilities include other types of molecular profiles (e.g., RNA-based molecules, chemicals/metabolites) combined with enhanced EV property characterizations to allow for predictive modeling-based approaches. The combination of diverse data streams will contribute towards the improved elucidation of where EVs come from within investigated samples of relevance to exposure science, toxicology, and public health research.
5. EVALUATING EV CONTENT, FUNCTIONAL CONSEQUENCES, AND TARGET TISSUES IN PUBLIC HEALTH STUDIES
5.1. Evaluating EV Content
EVs contain important molecular cargo that are loaded based on highly regulated processes, resulting in the intercellular transmission of various molecules. These molecules include nucleic acids (e.g., mRNAs and miRNAs), proteins, lipids, and chemicals/metabolites [8,12]). Methods to evaluate expression/concentration signatures of these important molecules in research related to public health applications have been previously reviewed [56–60]. A challenge in EV research when evaluating these molecular signatures surrounds the limited amount of material within isolated EV samples. Therefore, approaches that can extract measures from small sample inputs are optimal, particularly when using samples of limited quantity which are typical of exposure science, toxicology, and public health research.
Molecular profiling data that have been published on EV content have been aggregated, in part, through the Vesiclepedia online database [61]. This online resource serves as a collection of nucleic acids, proteins, lipids, and chemical metabolites that have been identified in EVs, manually curated from previous studies, comprising mostly online peer-reviewed publications. This database currently contains data from over 1250 EV studies, with over 38,000 nucleic acid records, 1000 protein records, and 600 lipid/metabolite records [61].
To provide some examples relevant to public health research, we extracted and summarized human blood EV content molecular data within Vesiclepedia. Specifically, all molecular-level data were downloaded from Vesiclepedia (v4.1) and filtered for records that reflected publications listed in the ‘Plasma’ browse category [62]. Data were filtered for ‘Human’ species and organized to highlight the most frequently measured molecules. Fibrinogen alpha chain (FGA), platelet endothelial cell adhesion molecule (PECAM1), and haptoglobin-related protein (HPR) were identified as the most commonly measured EV-based plasma proteins across studies in humans recorded within Vesiclepedia. Functional analysis of commonly measured proteins identified immune response, blood coagulation, and hemostasis as the top-ranking functions associated with EV proteins (Figure 2). Notably, some of these proteins represent ubiquitous molecules present in blood that tend to stick to EVs regardless of isolation protocol (e.g., FGA) [63]. Similar to the protein results, the most commonly measured EV-based mRNAs included FGA, HPR, and PECAM1 (Figure 3). The most commonly measured lipid was phosphatidylserine, a component of the cell membrane, present in all EVs. Lastly, the most common miRNAs found in plasma EVs include let-7a, miR-103, and miR-142–3p (Figure 3). These miRNAs are notably involved in DNA damage response (let-7a), glucose homeostasis/insulin stimulus (miR-103), inflammatory processes (miR-142–3p), and cancer (let-7a, miR-103, and miR-142–3p) [64–66]. Data within Vesiclepedia, as well as other EV-related repositories, will continue to expand in the coming years, serving as valuable resources to merge EV data across samples and research applications.
Figure 2. The most common proteins that have been measured in EVs circulating in human blood and their related biological functions.
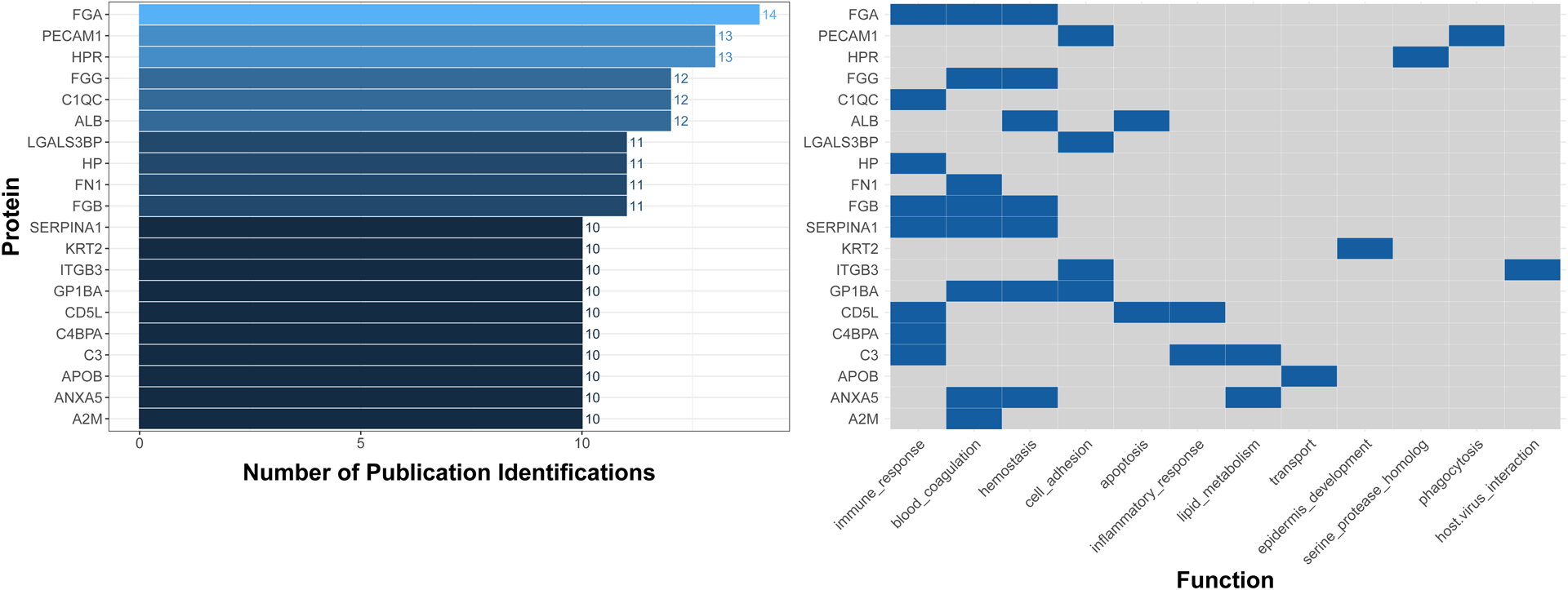
The illustrated proteins include those that have been identified in 10 or more publications aggregated from the Vesiclepedia database. Molecules were further visualized according to functional categories based on Gene Ontology Biological Processes obtained from the Universal Protein Resource Knowledgebase [92]. Abbreviations: A2M, alpha-2-macroglobulin; ALB, albumin; ANXA5, annexin A5; APOB, apolipoprotein B; C1QC, complement C1q subcomponent subunit C; C3, complement C3; C4BPA, c4b-binding protein alpha chain; CD5L, CD5 antigen-like; FGA, fibrinogen alpha chain; FGB, fibrinogen beta chain; FGG, fibrinogen gamma chain; FN1, fibronectin; GP1BA, platelet glycoprotein 1b alpha chain; HP, haptoglobin; HPR, haptoglobin-related protein; ITGB3, integrin beta-3; KRT2, kertain, type II cytoskeletal 2 epidermal; LGALS3BP, Galectin-3-binding protein; PECAM1, platelet endothelial cell adhesion molecule; SERPINA1, alpha-1-antitrypsin.
Figure 3. The most common lipids, mRNAs, and miRNAs and that have been measured in EVs circulating in human blood.
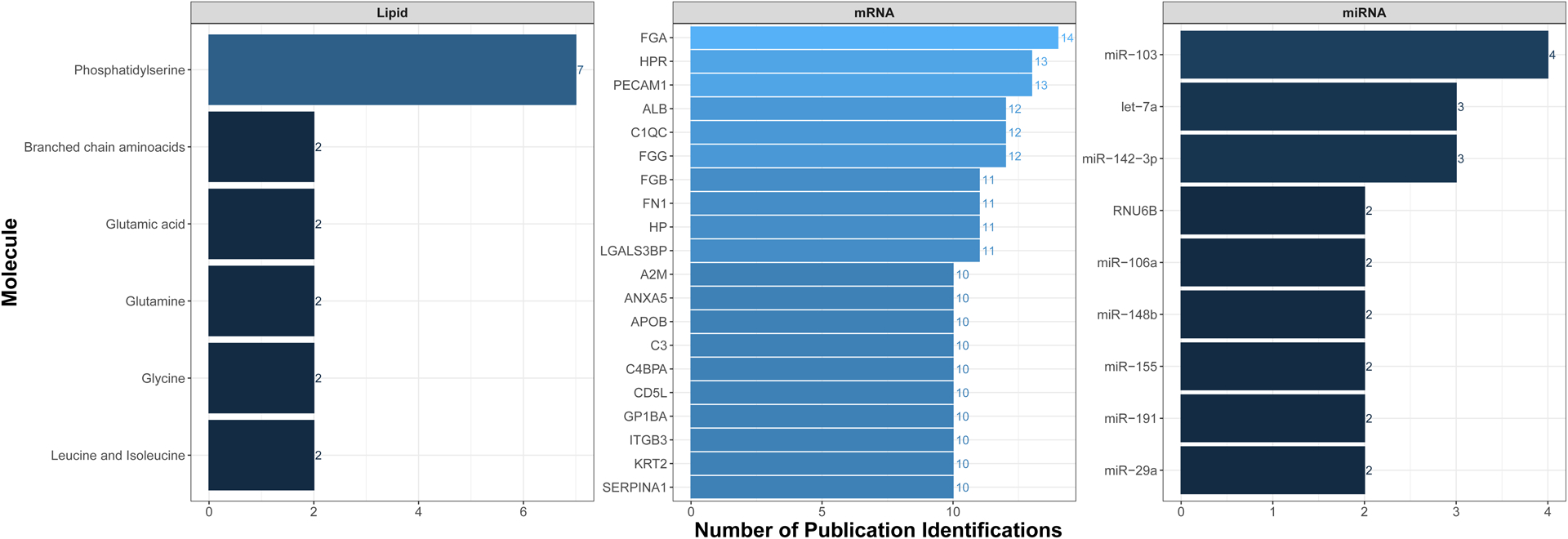
The illustrated molecules include those that have been identified across the highest number of publications aggregated from the Vesiclepedia database.
Few studies have compared molecular profiles between full biological samples, analyzed in bulk, vs EVs that are isolated from the same full biological samples. One notable study compared miRNA content in whole plasma vs EVs isolated from plasma in patients with prostate cancer [67]. Authors noted clear differences in miRNA expression profiles between full plasma vs EVs isolated from plasma [67]. These obvious differences in molecular profiles further support the differential loading of molecular cargo within EVs vs general extracellular compartments. Furthermore, these findings support the importance of isolating EVs from biological samples and analyzing their contents, as opposed to evaluating whole tissue/blood samples collectively, to better characterize purposeful communication signals between cells.
5.2. Evaluating Functional Consequences of EVs
In addition to understanding the molecular content of EVs, it is of critical importance to also understand the biological function of EVs, as well as potential consequences of changes in EV content that may occur in response to exogenous insults and/or changes in disease state that are commonly investigated in exposure science, toxicology, and public health research. One strategy that researchers can employ to evaluate the functional consequences of EVs is through targeted inhibition of EV secretion, using agents that block EV secretion within controlled experimental designs. Comparing biological events that occur in the presence vs absence of these agents imparts useful information towards understanding EV consequences that can be leveraged towards exposure science, toxicology, and public health applications.
Scientists have explored methods to inhibit EV release, both as research tools and potential therapeutics. Several pharmacological inhibitors have been used to block EV release, each targeting various subcategories or biological properties. Some of the most commonly used inhibitors include calpeptin, GW4869, imipramine, manumycin A, pantethine, and Y27632 [68]. These inhibitors can be classified into two main groups: (1) those that affect EV trafficking (e.g., calpeptin, manumycin A, and Y2763); and (2) those that primarily effect lipid metabolism and thus EV biogenesis (e.g., GW4869, imipramine, and pantethine). These inhibitors can be further classified based on the primary EV subtype that is targeted and blocked. In general, studies may select GW4869 or manumycin A to target exosomes; or calpeptin, imipramine, pantethine, or Y27632 to target microvesicles. As an example, one in vitro study used the exosome secretion blocker GW4869 and found that it reduced the levels of pro-angiogenic miRNAs in mesenchymal stem cell-derived conditioned media [69]. These results suggested that exosomes may aid in promoting angiogenesis, and thus, inhibition of exosome secretion in these instances may be a useful therapeutic strategy in the future [69]. In addition to pharmaceuticals, some studies have implemented gene editing knockdown techniques to target specific genes known to influence EV release. For example, several studies have targeted Rab proteins to limit EV release due to their essential role in EV biogenesis and secretion [12,70]. One specific study found that inhibition of Rab27a gene expression by shRNA decreased exosome count by more than 50% and led to decreased tumor growth and metastasis in 4T1 carcinoma cells [70]. Similar methods involving pharmaceutical or gene knock-down EV blockers may be modified in future studies to target exposure-induced exosomes to reduce impacts of environmental toxicants.
The translation of findings produced from controlled, in vitro study designs to in vivo human biology requires further testing, which the medical research community is starting to address. A limited number of pre-clinical in vivo studies indicate that many compounds have the ability to block or limit the release of exosomes and/or microvesicles. For example, sulfisoxazole, an antibiotic that is approved by the Food and Drug Administration (FDA), was reported to inhibit small EVs from breast cancer cells in mice, leading to anti-tumor and anti-metastatic effects [71]. Additionally, the use of the drug, dimethyl amiloride, has led to decreased production of exosomes in three mouse tumor models, as well as increased efficacy of chemotherapeutic treatment [72].
The same study reported similar results in humans using amiloride, a drug typically used for treatment of high blood pressure, as an EV blocker among patients with colorectal metastatic cancer. Other therapeutic agents have demonstrated capabilities towards blocking EV assembly/loading/section, include clopidogrel, glibenclamide, imatinib, and indomethacin [68]. Given their current use in medicine for other treatments, these agents serve as potential candidates for pre-clinical evaluation.
5.3. Evaluating Target Tissues of EVs
There is mounting evidence supporting the specificity of EVs in targeting specific cellular properties associated with individual tissues throughout the body. For example, EVs have been demonstrated to serve as effective drug delivery carriers between tissues within controlled in vitro and in vivo experimental settings [12]. Therapeutic mediators transferred via EVs have included siRNA, mRNA, miRNAs, and classic small molecule therapeutics including curcumin, paclitaxel, rhodamine 123, and doxorubicin. For example, incubation of the murine tumor cell line, EL-4, with curcumin resulted in the successful incorporation of curcumin into EVs released from these cells [73]. These in vitro-derived EVs were interestingly found to capture curcumin with stability and bioavailability in vivo. Specifically, these EVs were found to preferentially target brain tissue in mice, after nasal EV treatment, resulting in the protection against lipopolysaccharide-induced septic shock [73]. Still, the preferential targeting of specific EVs to specific target cell types/tissues requires further studies within medical and toxicological settings.
An approach that researchers can employ to more specifically address target tissue specificity associated with EVs is through mRNA/protein-based reporter tagging. A recent notable example surrounds the development of a mouse model with CRISPR-Cas9-mediated genome editing that enables the specific labeling of circulating CD63+ EVs from any cell type when crossed with lineage-specific Cre recombinase driver mice [74]. This group provided a case study of this application through the incorporation of a common green fluorescent protein, emerald GFP, specifically within developing vascular tissue of tamoxifen-treated pregnant mice. Circulating plasma emerald GFP-labeled EVs were also noted to contain additional EV (CD9 and CD81) and endothelial cell (CD105) protein markers and were noted to contain specific miRNAs [74]. It is notable that these types of models can be evaluated through imaging techniques, spanning magnetic resonance imaging (MRI), computed tomography scanning (CT scan), and other image-based analyses (e.g., flow cytometry-based microscopy). These types of strategies can impart important information surrounding where an EV is going throughout the body and imparting beneficial, neutral, or detrimental effects on recipient cell targets.
6. APPLYING EV RESEARCH TOWARDS ADDITIONAL PUBLIC HEALTH APPLICATIONS: A LOOK TOWARDS THE FUTURE
The field of exposure science, toxicology, and public health will significantly benefit from findings produced through EV research. Here, we summarize some ways in which EVs can be immediately incorporated into public health applications to improve our knowledge of chemical-disease relationships, disease etiology, and therapeutic interventions. EVs can specifically be used as new biomarkers of chemical exposure, biomarkers of disease, molecules to use for therapeutic interventions, and molecules to better understand factors contributing to overall public health (Figure 4). Specific attributes of EVs that can be leveraged for these purposes include EV size, concentration, and content, all representing novel data points that could serve as informative biomarkers of exposure and disease. These measurements can be acquired from blood samples, as this biological sample represents one of the most common type of sample acquired in clinical, epidemiological, and toxicological settings. Applications highlight the utility of blood-based EVs, though these approaches also extend towards additional biological samples, including potential tissue samples or less invasive biological fluid samples.
Figure 4. Overview of example utilities of EV research in environmental science, toxicology, and public health applications.
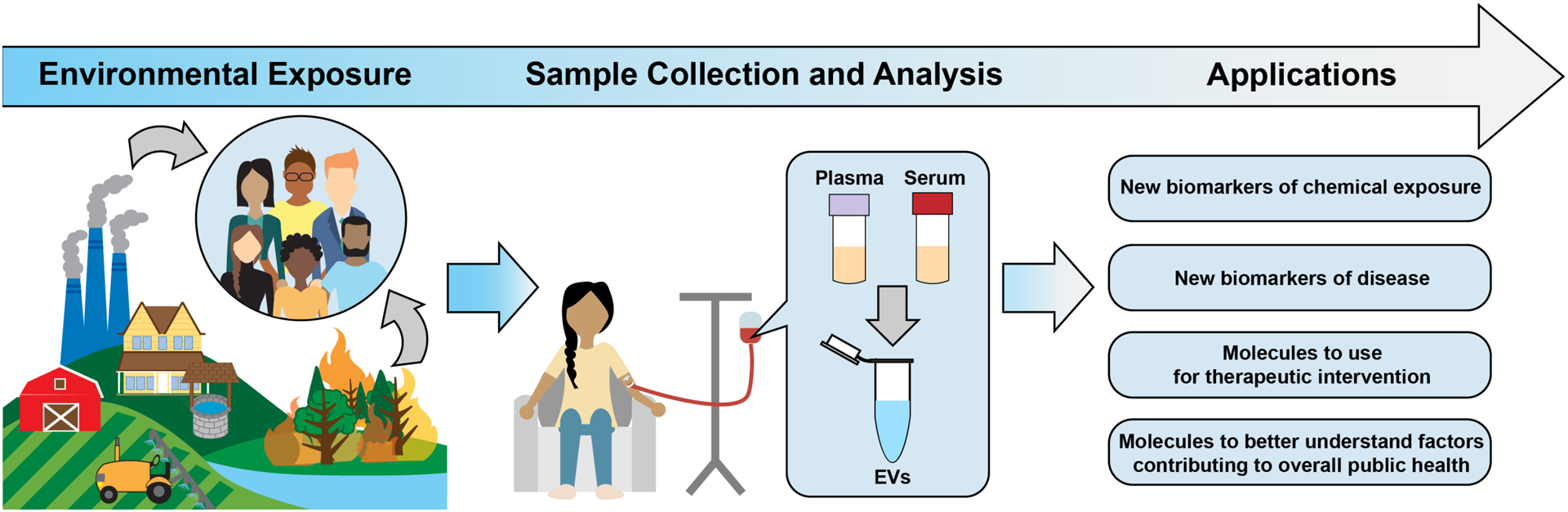
Shown here are example approaches to integrate EVs within human cohort evaluations, resulting in applications to better understand disease etiology, biomarkers of disease, and solutions to better protect public health.
6.1. Using EVs as Exposure Biomarkers
As described throughout this review, the molecular contents within isolated EVs differ between the molecular contents present in whole tissues analyzed in bulk. It is therefore important to specifically characterize molecular profiles within EVs, as opposed to (or in addition to) analyzing bulk samples. In the context of blood samples, this means that isolating and analyzing EVs from whole plasma or serum samples can impart more specific, and biologically meaningful, information surrounding molecular biomarkers of exposure and/or effect.
In relation to environmental exposure assessments, emerging evidence supports the utility of evaluating EVs as exposure biomarkers in humans. These data have been primarily generated in human studies relating air toxicant exposures to alterations in miRNA expression signatures derived from EVs. For example, several potential EV molecular biomarkers have been identified in humans in response to cigarette smoking in chronic lung diseases [75]. These putative biomarkers included certain miRNAs (e.g., miR-191, miR-200b-5p, miR-378a, mi-379, miR-7, let-7e, let-7g) and proteins (leucine-rich alpha-2-glycoprotein 1, μ-calpain, and m-calpain) within EVs isolated from human plasma, serum, bronchoalveolar lavage fluid, cell supernatant, and/or urine samples. These biomarkers were also notably linked to lung cancer or chronic obstructive pulmonary disease [75]. Another study in humans has been presented in the form of a conference abstract, and provides preliminary evidence supporting the use of extracted saliva EVs and their associated miRNA expression in relation to ambient air pollution exposure in residents that bike within New York City [76].
Studies carried out in vivo and in vitro also provide evidence supporting the use of EVs as exposure biomarkers. For example, one study reported that exposure to particulate matter caused EVs to be released in a dose- and time-dependent manner within immortalized and primary human macrophages [77]. In addition, EVs have been evaluated for alterations associated with polycylic aromatic hydrocarbon (PAH) exposure, both in vitro, in human endothelial cells, and in vivo, in urine samples from PAH-exposed rats [78]. PAH exposure was found to increase EV production/release in both models. EVs have also been evaluated in relation to irradiation exposure in mice [79]. Here, mice were exposed to clinically-relevant cranial (head only) irradiation, and circulating plasma samples collected two week post-exposure to detect signs of sustained systemic injury. EVs within these plasma samples were found to display differential metabolomic and lipidomic profiles involved in inflammation in exposed vs. unexposed mice. These data support an EV-mediated mechanism for sustained impacts of exposure conditions occurring systemically, through blood [79]. Collectively, there is still limited data in this research area, particularly in relation to environmental science and public health; though these important emerging studies support the utility of incorporating EVs as novel biomarkers of exposure across models and target tissues.
6.2. Expanding the Use of EVs as Disease Biomarkers and Mechanisms for Therapeutic Interventions to Improve Public Health
As discussed throughout this review, extensive research has been conducted characterizing the role of EVs in human disease. Of particular interest towards public health, EV cargo can be evaluated to inform the health status of individuals by informing/predicting pathological state [2,12]. EV cargo such as RNA, miRNA, proteins, and lipids have been studied in association with a multitude of human disease outcomes. For example, EVs originating from cancer sites are quickly gaining attention due to an abundance of evidence supporting their role in tumor progression, establishment of pre-metastatic niche, and spreading to the secondary site [12,80]. Example molecules within EVs that have been implicated as potential biomarkers of cancer include Epidermal Growth Factor Receptor (EGFR) [81], Caveolin-1 (Cav-1) [82], and MET proto-oncogene, receptor tyrosine kinase (MET) [83], among others. Other EV-based biomarkers have been proposed from studies that isolated EVs from blood for diseases such as breast cancer [84], Alzheimer’s disease [24], and diabetes [21]. While research on EVs as disease biomarkers is abundant in the medical field, it has yet to be adequately incorporated into public health research. Future studies may evaluate EV cargo in human cohort studies as potential indicators of general health, as well as EV cargo changes resulting from human exposures to inform mechanism of action and potential therapeutic interventions.
While EV cargo can provide useful information towards disease state, the actual physical properties of EVs can also serve as biomarkers of disease and thus, be incorporated into public health research. For example, EV size and concentration have been shown to vary at different stages of cancer, suggesting that these parameters are in themselves useful for clinical diagnostics and informing an individual’s health status [85–87]. Studies suggest that cancer aggressiveness is associated with increased EV release and thus may be a useful diagnostic marker [82,88]. Several other diseases have been associated with altered EV release in blood such as diabetes [21], nonalcoholic fatty liver disease [22], and cardiovascular disease [23]. EV size and concentration data can therefore be used alongside EV molecular profile as multi-factorial contributors to exposure and/or disease biomarkers that translate to public health applications.
Many researchers are also experimenting with the use of EVs as tools for therapeutic intervention, which in public health research, could be leveraged for solution-oriented interventions in collaboration with medical communities. Given that many studies have reported disease-derived EVs, potential therapies may include EV blockers to reduce release and uptake. As detailed in section 5.2, EV blockers have the potential for improving disease outcomes influenced by EV communication, particularly in relation to cancer growth and metastasis. EVs also show promise as therapeutic delivery vehicles, as discussed in detail in section 5.3. The ability of EVs to transfer their content to target tissues makes them promising candidates for drug and gene delivery applications, especially given their biocompatibility with target cells. Future studies may translate and expand these methods to target and repair environmental exposure induced cell injury and associated health outcomes in the public health research setting.
6.3. EVs for Understanding General Public Health
Changes in EV profiles occur during general cellular processes that regularly occur in the human body; and thus, EVs can be used to inform normal healthy responses to changes that occur naturally in humans. For example, changes in EVs are observed within maternal blood throughout pregnancy. Fetal placental tissues have been identified as a rich source of EVs within circulating maternal blood, serving as a route of feto-maternal communication [89]. Release of EVs associated with placenta tissue seems to even occur in a time-dependent manner over the course of pregnancy, as characterized through the evaluation of blood EVs that are PARP+. Specifically, PARP+ EVs in maternal plasma have been shown in several studies to increase with increasing gestational age throughout the first and second trimesters, and then maintain a constant concentration during the third trimester, demonstrating a time-dependent relationship alongside placental growth [89]. Though it is important to note that PARP expression occurs in other tissues, as well as the placenta, which could have influenced this trend. Of further relevance to maternal/child health, EVs have been identified and characterized within breast milk, and studies are beginning to link molecular profiles within breast milk EVs to child health outcomes [90]. Evaluation of EV-based indices could therefore inform whether or not healthy changes are occurring during and after pregnancy, potentially informing overall health of mothers and children. These techniques extend beyond maternal/child health research, where EVs are now being characterized in urine samples of humans across various health statuses [91]. Although the majority of EV research focuses on diseased patients, future applications of EV-based findings will inform broader health implications across overall public health.
7. CONCLUSIONS
In conclusion, the recent expansion surrounding EV discoveries will significantly impact many areas of exposure science, toxicology, and human health research. This review aimed to contribute to the translation of the new molecular biological techniques into the public health research arena, including clinical, epidemiological, and toxicological evaluations. These techniques can be leveraged to better understand chemical-disease relationships, disease mechanisms, and therapeutic interventions. EV content was once considered stochastic; though current evidence supports the loading of molecular content within EVs as highly regulated and purposeful in communicating cellular processes and resulting biological functions. Because of this, EVs can be used as biomarkers of exposure, effect, and disease that can contribute increased specificity in addition to existing measures. Future research efforts will include the continued expansion of technological development, allowing for increased sensitivity to allow for improved assessment of EV samples that inherently have low sample yields. Additional data are needed to improve molecular markers and computational models to inform/predict EV subtypes, cell-of-origin, and target tissue, to span both biological and chemical signatures. As these methods and associated findings expand, EV research will continue to impact our knowledge of human disease and public health.
Impact statement:
Extracellular vesicles (EVs) represent small, cell-derived structures consisting of molecules that can serve as biomarkers of exposure, effect, and disease. This review lays a novel foundation for integrating EVs, a rapidly advancing molecular biological tool, into the field of public health research including epidemiological, toxicological, and clinical investigations. This article represents an important advancement in public health and exposure science as it is among the first to translate EVs into this field.
ACKNOWLEDGEMENTS
FUNDING
This study was supported by grants from the National Institutes of Health (NIH) from the National Institute of Environmental Health Sciences (1R21ES031740, P42ES031007). Support was additionally provided through the Institute for Environmental Health Solutions (IEHS) at the University of North Carolina Gillings School of Global Public Health.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Yekula A, Muralidharan K, Kang KM, Wang L, Balaj L, Carter BS. From laboratory to clinic: Translation of extracellular vesicle based cancer biomarkers. Methods. 2020;177:58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang G, Lin G, Zhu Y, Duan W, Jin D. Emerging technologies for profiling extracellular vesicle heterogeneity. Lab Chip. 2020;20(14):2423–37. [DOI] [PubMed] [Google Scholar]
- 3.Bazzan E, Tinè M, Casara A, Biondini D, Semenzato U, Cocconcelli E, et al. Critical Review of the Evolution of Extracellular Vesicles’ Knowledge: From 1946 to Today. Int J Mol Sci. 2021;22(12):6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chargaff E, West R. The biological significance of the thromboplastic protein of blood. J Biol Chem. 1946;166(1):189–97. [PubMed] [Google Scholar]
- 5.Wolf P The nature and significance of platelet products in human plasma. Br J Haematol. 1967;13(3):269–88. [DOI] [PubMed] [Google Scholar]
- 6.Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983;97(2):329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33(3):967–78. [DOI] [PubMed] [Google Scholar]
- 8.Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartjes TA, Mytnyk S, Jenster GW, van Steijn V, van Royen ME. Extracellular Vesicle Quantification and Characterization: Common Methods and Emerging Approaches. Bioengineering (Basel). 2019;6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vesiclepedia. A community compendium for extracellular vesicles 2021. [cited 2021 May 1]. Available from: http://www.microvesicles.org/.
- 11.Baxter AA. Stoking the Fire: How Dying Cells Propagate Inflammatory Signalling through Extracellular Vesicle Trafficking. Int J Mol Sci. 2020;21(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Corbett AL, Taatizadeh E, Tasnim N, Little JP, Garnis C, et al. Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019;3(1):011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, et al. Reassessment of Exosome Composition. Cell. 2019;177(2):428–45 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8(5):1432–46. [DOI] [PubMed] [Google Scholar]
- 15.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, et al. KRAS-dependent sorting of miRNA to exosomes. Elife. 2015;4:e07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu S, Zhang Y, Li Y, Luo L, Zhao Y, Yao Y. Extracellular vesicles in cardiovascular diseases. Cell Death Discov. 2020;6:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwok ZH, Wang C, Jin Y. Extracellular Vesicle Transportation and Uptake by Recipient Cells: A Critical Process to Regulate Human Diseases. Processes (Basel). 2021;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maas SLN, Breakefield XO, Weaver AM. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017;27(3):172–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xavier CPR, Caires HR, Barbosa MAG, Bergantim R, Guimaraes JE, Vasconcelos MH. The Role of Extracellular Vesicles in the Hallmarks of Cancer and Drug Resistance. Cells. 2020;9(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman DW, Noren Hooten N, Eitan E, Green J, Mode NA, Bodogai M, et al. Altered Extracellular Vesicle Concentration, Cargo, and Function in Diabetes. Diabetes. 2018;67(11):2377–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Povero D, Eguchi A, Li H, Johnson CD, Papouchado BG, Wree A, et al. Circulating extracellular vesicles with specific proteome and liver microRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PLoS One. 2014;9(12):e113651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eichner NZM, Gilbertson NM, Gaitan JM, Heiston EM, Musante L, LaSalvia S, et al. Low cardiorespiratory fitness is associated with higher extracellular vesicle counts in obese adults. Physiol Rep. 2018;6(10):e13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang TT, Liu CG, Gao SC, Zhang Y, Wang PC. The Serum Exosome Derived MicroRNA-135a, −193b, and −384 Were Potential Alzheimer’s Disease Biomarkers. Biomed Environ Sci. 2018;31(2):87–96. [DOI] [PubMed] [Google Scholar]
- 25.Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New Technologies for Analysis of Extracellular Vesicles. Chem Rev. 2018;118(4):1917–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamin RJ, McLaughlin LS. Plasma components: properties, differences, and uses. Transfusion. 2012;52 Suppl 1:9S–19S. [DOI] [PubMed] [Google Scholar]
- 27.Phillips W, Willms E, Hill AF. Understanding extracellular vesicle and nanoparticle heterogeneity: Novel methods and considerations. Proteomics. 2021;21(13–14):e2000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breitman TR, He RY. Combinations of retinoic acid with either sodium butyrate, dimethyl sulfoxide, or hexamethylene bisacetamide synergistically induce differentiation of the human myeloid leukemia cell line HL60. Cancer Res. 1990;50(19):6268–73. [PubMed] [Google Scholar]
- 29.Coumans FAW, Brisson AR, Buzas EI, Dignat-George F, Drees EEE, El-Andaloussi S, et al. Methodological Guidelines to Study Extracellular Vesicles. Circ Res. 2017;120(10):1632–48. [DOI] [PubMed] [Google Scholar]
- 30.Palviainen M, Saraswat M, Varga Z, Kitka D, Neuvonen M, Puhka M, et al. Extracellular vesicles from human plasma and serum are carriers of extravesicular cargo-Implications for biomarker discovery. PLoS One. 2020;15(8):e0236439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacroix R, Judicone C, Mooberry M, Boucekine M, Key NS, Dignat-George F, et al. Standardization of pre-analytical variables in plasma microparticle determination: results of the International Society on Thrombosis and Haemostasis SSC Collaborative workshop. J Thromb Haemost. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiam K, Mayne GC, Wang T, Watson DI, Irvine TS, Bright T, et al. Serum outperforms plasma in small extracellular vesicle microRNA biomarker studies of adenocarcinoma of the esophagus. World J Gastroenterol. 2020;26(20):2570–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zarovni N, Corrado A, Guazzi P, Zocco D, Lari E, Radano G, et al. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods. 2015;87:46–58. [DOI] [PubMed] [Google Scholar]
- 35.Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome Isolation Techniques. Theranostics. 2017;7(3):789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Bi J, Huang J, Tang Y, Du S, Li P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int J Nanomedicine. 2020;15:6917–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atha DH, Ingham KC. Mechanism of precipitation of proteins by polyethylene glycols. Analysis in terms of excluded volume. J Biol Chem. 1981;256(23):12108–17. [PubMed] [Google Scholar]
- 38.Yuana Y, Koning RI, Kuil ME, Rensen PC, Koster AJ, Bertina RM, et al. Cryo-electron microscopy of extracellular vesicles in fresh plasma. J Extracell Vesicles. 2013;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJ, Hole P, et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine. 2011;7(6):780–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welsh JA, Holloway JA, Wilkinson JS, Englyst NA. Extracellular Vesicle Flow Cytometry Analysis and Standardization. Front Cell Dev Biol. 2017;5:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel R, Willmott G, Kozak D, Roberts GS, Anderson W, Groenewegen L, et al. Quantitative sizing of nano/microparticles with a tunable elastomeric pore sensor. Anal Chem. 2011;83(9):3499–506. [DOI] [PubMed] [Google Scholar]
- 42.Akers JC, Ramakrishnan V, Nolan JP, Duggan E, Fu CC, Hochberg FH, et al. Comparative Analysis of Technologies for Quantifying Extracellular Vesicles (EVs) in Clinical Cerebrospinal Fluids (CSF). PLoS One. 2016;11(2):e0149866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip. 2014;14(11):1891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jorgensen M, Baek R, Pedersen S, Sondergaard EK, Kristensen SR, Varming K. Extracellular Vesicle (EV) Array: microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. J Extracell Vesicles. 2013;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jorgensen MM, Baek R, Varming K. Potentials and capabilities of the Extracellular Vesicle (EV) Array. J Extracell Vesicles. 2015;4:26048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koliha N, Wiencek Y, Heider U, Jungst C, Kladt N, Krauthauser S, et al. A novel multiplex bead-based platform highlights the diversity of extracellular vesicles. J Extracell Vesicles. 2016;5:29975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113(8):E968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin-Jaular L, Nevo N, Schessner JP, Tkach M, Jouve M, Dingli F, et al. Unbiased proteomic profiling of host cell extracellular vesicle composition and dynamics upon HIV-1 infection. EMBO J. 2021:e105492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skotland T, Sandvig K, Llorente A. Lipids in exosomes: Current knowledge and the way forward. Progress in Lipid Research. 2017;66:30–41. [DOI] [PubMed] [Google Scholar]
- 50.Skotland T, Sagini K, Sandvig K, Llorente A. An emerging focus on lipids in extracellular vesicles. Adv Drug Deliv Rev. 2020;159:308–21. [DOI] [PubMed] [Google Scholar]
- 51.Guduric-Fuchs J, O’Connor A, Camp B, O’Neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, He X, Li Q, Lai H, Zhang H, Hu Z, et al. EV-origin: Enumerating the tissue-cellular origin of circulating extracellular vesicles using exLR profile. Comput Struct Biotechnol J. 2020;18:2851–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clark J, Avula V, Ring C, Eaves LA, Howard T, Santos HP, et al. Comparing the Predictivity of Human Placental Gene, microRNA, and CpG Methylation Signatures in Relation to Perinatal Outcomes. Toxicol Sci. 2021;183(2):269–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ring C, Sipes NS, Hsieh JH, Carberry C, Koval LE, Klaren WD, et al. Predictive modeling of biological responses in the rat liver using in vitro Tox21 bioactivity: Benefits from high-throughput toxicokinetics. Comput Toxicol. 2021;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lage K, Hansen NT, Karlberg EO, Eklund AC, Roque FS, Donahoe PK, et al. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc Natl Acad Sci U S A. 2008;105(52):20870–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zavala J, Freedman AN, Szilagyi JT, Jaspers I, Wambaugh JF, Higuchi M, et al. New Approach Methods to Evaluate Health Risks of Air Pollutants: Critical Design Considerations for In Vitro Exposure Testing. Int J Environ Res Public Health. 2020;17(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rager JE, Bangma J, Carberry C, Chao A, Grossman J, Lu K, et al. Review of the environmental prenatal exposome and its relationship to maternal and fetal health. Reprod Toxicol. 2020;98:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clark J, Rager JE. Chapter 1. Epigenetics: An overview of CpG methylation, chromatin remodeling, and regulatory/non-coding RNAs. In: Fry RC, editor. Environmental Epigenetics in Toxicology and Public Health: Elsevier; 2020. p. 3–32. [Google Scholar]
- 59.Fry RC, Bangma J, Szilagyi J, Rager JE. Developing novel in vitro methods for the risk assessment of developmental and placental toxicants in the environment. Toxicol Appl Pharmacol. 2019;378:114635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aristizabal-Henao JJ, Ahmadireskety A, Griffin EK, Da Silva BF, Bowden JA. Lipidomics and environmental toxicology: Recent trend. Curr Opin Environ Sci Health. 2020;15:26–31. [Google Scholar]
- 61.Pathan M, Fonseka P, Chitti SV, Kang T, Sanwlani R, Van Deun J, et al. Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019;47(D1):D516–D9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vesiclepedia. Browse results for cell type/tissue: Plasma 2021. [cited 2021 May 15]. Available from: http://microvesicles.org/browse_results?org_name=&cont_type=&tissue=Plasma&gene_symbol=&ves_type=.
- 63.Kim J, Tan Z, Lubman DM. Exosome enrichment of human serum using multiple cycles of centrifugation. Electrophoresis. 2015;36(17):2017–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.LifeMapSciences. GeneCards THE HUMAN GENE DATABASE: MIR142 2021 [cited 2021 Nov 6]. Available from: https://www.genecards.org/cgi-bin/carddisp.pl?gene=MIR142&keywords=miR-142-3p.
- 65.LifeMapSciences. GeneCards THE HUMAN GENE DATABASE: MIR103A1 2021 [cited 2021 Nov 6]. Available from: https://www.genecards.org/cgi-bin/carddisp.pl?gene=MIR103A1.
- 66.LifeMapSciences. GeneCards THE HUMAN GENE DATABASE: MIRLET7A1 2021 [cited 2021 Nov 6]. Available from: https://www.genecards.org/cgi-bin/carddisp.pl?gene=MIRLET7A1&keywords=let-7a.
- 67.Endzelins E, Berger A, Melne V, Bajo-Santos C, Sobolevska K, Abols A, et al. Detection of circulating miRNAs: comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer. 2017;17(1):730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Catalano M, O’Driscoll L. Inhibiting extracellular vesicles formation and release: a review of EV inhibitors. J Extracell Vesicles. 2020;9(1):1703244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gong M, Yu B, Wang J, Wang Y, Liu M, Paul C, et al. Mesenchymal stem cells release exosomes that transfer miRNAs to endothelial cells and promote angiogenesis. Oncotarget. 2017;8(28):45200–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bobrie A, Krumeich S, Reyal F, Recchi C, Moita LF, Seabra MC, et al. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72(19):4920–30. [DOI] [PubMed] [Google Scholar]
- 71.Im EJ, Lee CH, Moon PG, Rangaswamy GG, Lee B, Lee JM, et al. Sulfisoxazole inhibits the secretion of small extracellular vesicles by targeting the endothelin receptor A. Nat Commun. 2019;10(1):1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chalmin F, Ladoire S, Mignot G, Vincent J, Bruchard M, Remy-Martin JP, et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest. 2010;120(2):457–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhuang X, Xiang X, Grizzle W, Sun D, Zhang S, Axtell RC, et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol Ther. 2011;19(10):1769–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McCann JV, Bischoff SR, Zhang Y, Cowley DO, Sanchez-Gonzalez V, Daaboul GD, et al. Reporter mice for isolating and auditing cell type-specific extracellular vesicles in vivo. Genesis. 2020;58(7):e23369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ryu AR, Kim DH, Kim E, Lee MY. The Potential Roles of Extracellular Vesicles in Cigarette Smoke-Associated Diseases. Oxid Med Cell Longev. 2018;2018:4692081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Comfort N, Smith C, Chillrud S, Yang Q, Baccarelli A, Jack D. Extracellular Vesicles in Saliva as Biomarkers of Exposure and Effect: A feasibility pilot in the context of the New York City Biking and Breathing Study. Environmental Epidemiology. 2019;3(80):doi: 10.1097/01.EE9.0000606548.23536.31. [DOI] [Google Scholar]
- 77.Martin PJ, Heliot A, Tremolet G, Landkocz Y, Dewaele D, Cazier F, et al. Cellular response and extracellular vesicles characterization of human macrophages exposed to fine atmospheric particulate matter. Environ Pollut. 2019;254(Pt A):112933. [DOI] [PubMed] [Google Scholar]
- 78.Le Goff M, Lagadic-Gossmann D, Latour R, Podechard N, Grova N, Gauffre F, et al. PAHs increase the production of extracellular vesicles both in vitro in endothelial cells and in vivo in urines from rats. Environ Pollut. 2019;255(Pt 1):113171. [DOI] [PubMed] [Google Scholar]
- 79.Hinzman CP, Baulch JE, Mehta KY, Girgis M, Bansal S, Gill K, et al. Plasma-derived extracellular vesicles yield predictive markers of cranial irradiation exposure in mice. Sci Rep. 2019;9(1):9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fares J, Kashyap R, Zimmermann P. Syntenin: Key player in cancer exosome biogenesis and uptake? Cell Adh Migr. 2017;11(2):124–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619–24. [DOI] [PubMed] [Google Scholar]
- 82.Di Vizio D, Morello M, Dudley AC, Schow PW, Adam RM, Morley S, et al. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am J Pathol. 2012;181(5):1573–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yan C, Hu J, Yang Y, Hu H, Zhou D, Ma M, et al. Plasma extracellular vesiclepackaged microRNAs as candidate diagnostic biomarkers for earlystage breast cancer. Mol Med Rep. 2019;20(5):3991–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Szajnik M, Derbis M, Lach M, Patalas P, Michalak M, Drzewiecka H, et al. Exosomes in Plasma of Patients with Ovarian Carcinoma: Potential Biomarkers of Tumor Progression and Response to Therapy. Gynecol Obstet (Sunnyvale). 2013;Suppl 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baran J, Baj-Krzyworzeka M, Weglarczyk K, Szatanek R, Zembala M, Barbasz J, et al. Circulating tumour-derived microvesicles in plasma of gastric cancer patients. Cancer Immunol Immunother. 2010;59(6):841–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim HK, Song KS, Park YS, Kang YH, Lee YJ, Lee KR, et al. Elevated levels of circulating platelet microparticles, VEGF, IL-6 and RANTES in patients with gastric cancer: possible role of a metastasis predictor. Eur J Cancer. 2003;39(2):184–91. [DOI] [PubMed] [Google Scholar]
- 88.Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015;40:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kupper N, Huppertz B. The endogenous exposome of the pregnant mother: Placental extracellular vesicles and their effect on the maternal system. Mol Aspects Med. 2021:100955. [DOI] [PubMed] [Google Scholar]
- 90.O’Reilly D, Dorodnykh D, Avdeenko NV, Nekliudov NA, Garssen J, Elolimy AA, et al. Perspective: The Role of Human Breast-Milk Extracellular Vesicles in Child Health and Disease. Adv Nutr. 2021;12(1):59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Merchant ML, Rood IM, Deegens JKJ, Klein JB. Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nat Rev Nephrol. 2017;13(12):731–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.UniProt. UniProtKB 2021 [cited 2021 May 15]. Available from: https://www.uniprot.org/uniprot/.
- 93.Laulagnier K, Javalet C, Hemming FJ, Chivet M, Lachenal G, Blot B, et al. Amyloid precursor protein products concentrate in a subset of exosomes specifically endocytosed by neurons. Cell Mol Life Sci. 2018;75(4):757–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liao Z, Jaular LM, Soueidi E, Jouve M, Muth DC, Schoyen TH, et al. Acetylcholinesterase is not a generic marker of extracellular vesicles. J Extracell Vesicles. 2019;8(1):1628592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wahlund CJE, Eklund A, Grunewald J, Gabrielsson S. Pulmonary Extracellular Vesicles as Mediators of Local and Systemic Inflammation. Front Cell Dev Biol. 2017;5:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Khalyfa A, Kheirandish-Gozal L, Gozal D. Exosome and Macrophage Crosstalk in Sleep-Disordered Breathing-Induced Metabolic Dysfunction. Int J Mol Sci. 2018;19(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schierer S, Ostalecki C, Zinser E, Lamprecht R, Plosnita B, Stich L, et al. Extracellular vesicles from mature dendritic cells (DC) differentiate monocytes into immature DC. Life Sci Alliance. 2018;1(6):e201800093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hough KP, Deshane JS. Cutting edge approaches for rapid characterization of airway exosomes. Methods. 2020;177:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Herrera Sanchez MB, Previdi S, Bruno S, Fonsato V, Deregibus MC, Kholia S, et al. Extracellular vesicles from human liver stem cells restore argininosuccinate synthase deficiency. Stem Cell Res Ther. 2017;8(1):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen W, Huang Y, Han J, Yu L, Li Y, Lu Z, et al. Immunomodulatory effects of mesenchymal stromal cells-derived exosome. Immunol Res. 2016;64(4):831–40. [DOI] [PubMed] [Google Scholar]
- 101.Oksvold MP, Kullmann A, Forfang L, Kierulf B, Li M, Brech A, et al. Expression of B-cell surface antigens in subpopulations of exosomes released from B-cell lymphoma cells. Clin Ther. 2014;36(6):847–62 e1. [DOI] [PubMed] [Google Scholar]
- 102.Multia E, Tear CJY, Palviainen M, Siljander P, Riekkola ML. Fast isolation of highly specific population of platelet-derived extracellular vesicles from blood plasma by affinity monolithic column, immobilized with anti-human CD61 antibody. Anal Chim Acta. 2019;1091:160–8. [DOI] [PubMed] [Google Scholar]
- 103.Muraoka S, Jedrychowski MP, Tatebe H, DeLeo AM, Ikezu S, Tokuda T, et al. Proteomic Profiling of Extracellular Vesicles Isolated From Cerebrospinal Fluid of Former National Football League Players at Risk for Chronic Traumatic Encephalopathy. Front Neurosci. 2019;13:1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jiang L, Shen Y, Guo D, Yang D, Liu J, Fei X, et al. EpCAM-dependent extracellular vesicles from intestinal epithelial cells maintain intestinal tract immune balance. Nat Commun. 2016;7:13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ravenhill BJ, Kanjee U, Ahouidi A, Nobre L, Williamson J, Goldberg JM, et al. Quantitative comparative analysis of human erythrocyte surface proteins between individuals from two genetically distinct populations. Commun Biol. 2019;2:350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Choi H, Kim Y, Mirzaaghasi A, Heo J, Kim YN, Shin JH, et al. Exosome-based delivery of super-repressor IkappaBalpha relieves sepsis-associated organ damage and mortality. Sci Adv. 2020;6(15):eaaz6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhu Z, Shen Y, Chen Y, Shi H, Shi Y. The exosome of platelet endothelial cell adhesion molecule-1 (PECAM1) protein: A potential risking star in high blood pressure patients (HBPP). Medicine (Baltimore). 2021;100(4):e21370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miranda J, Paules C, Nair S, Lai A, Palma C, Scholz-Romero K, et al. Placental exosomes profile in maternal and fetal circulation in intrauterine growth restriction - Liquid biopsies to monitoring fetal growth. Placenta. 2018;64:34–43. [DOI] [PubMed] [Google Scholar]
- 109.Mu W, Provaznik J, Hackert T, Zoller M. Tspan8-Tumor Extracellular Vesicle-Induced Endothelial Cell and Fibroblast Remodeling Relies on the Target Cell-Selective Response. Cells. 2020;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]


