Abstract
Acute hepatopancreatic necrosis disease (AHPND) is a contagious disease for the shrimp cultivation, thus early detection of disease is an unmet need. This present study documented for the first time a simple lateral flow immunoassay (LFIA) strip using polyclonal antibodies was created for the rapid detection both of PirAvp and PirBvp protein simultaneously. LFIA method based on the principle of sandwich format. The label is the colloidal gold. The polyclonal antibody was conjugated with the colloidal gold acting as biorecognition element and coated onto the conjugate pad. The rabbit anti-Pirvp, anti-PirBvp antibodies, and goat anti-rabbit IgG antibody were separately sprayed onto a nitrocellulose membrane to form two test lines and one control line, respectively. The appearance of red bands at the control line and the test line indicated a positive result. A single coloured band at control area indicated a negative result. The limit of detection of LFIA was found to be 125 ng, which could be visually detected by naked eye within 15 min. There was no cross-reactivity observed with VPnon-AHPND. Furthermore, the sensitivity and specificity of LFIA were 94.0% and 98.0%, respectively. The developed test strip could be a game changer for early and in situ diagnosis of AHPND.
Keywords: AHPND, Anti-PirAvp, Anti-PirBvp, Colloidal gold, Lateral flow immunoassay strip
Introduction
Acute hepatopancreatic necrosis disease (AHPND) is a newly emerging disease of cultured shrimp. AHPND was first appeared in China in 2009, Viet Nam in 2010, Malaysia in 2010, Thailand in 2011, Mexico in 2013, the Philippines in 2015, South American countries in 2016, Bangladesh and the United States of America in 2017, Taiwan in 2018, South Korea in 2019, and Okinawa Prefecture of Japan in 2020 (FAO 2020). It causes high mortality in two of the most commercially cultured shrimp species including Penaeus monodon and Penaeus vannamei in many countries. Economic losses caused by this disease were estimated over USD 7 billion each year (FAO 2020). The agent was known to lead to AHPND was strain of Vibrio parahaemolyticus, carrying a 69–70 kb plasmid that encoded two PirAvp and PirBvp toxins, which are similar to the Photorhabdus insect-related binary toxin (Lee et al. 2015; Prachumwat et al. 2018). PirAvp–PirBvp are secreted toxins that affect the hepatopancreas. The suggested role of PirAvp is as a receptor-binding part and the PirBvp is the main player in inducing hepatopancreatic necrosis, hence losing one of these toxins reduces or even completely abrogate the disease signs (Sirikharin et al. 2015; Lin et al. 2019). Typical signs of affected shrimp by AHPND include atrophied hepatopancreas, empty gut, and stomach. Histopathological examination shows the hepatopancreatic tubule epithelial cells sloughing into the lumen and hemocytic infiltration (Lightner 2012). AHPND is a contagious disease for the shrimp cultivation, thus early detection of disease is an unmet need. PCR-based multiplex or simultaneous amplification of the PirAvp–PirBvp toxin genes are presently used as a reliable diagnostic or detection tool for AHPND (Kumar et al. 2020; Mai-Hoang et al. 2021). However, many things make it difficult for this method to apply in farmed ponds in situ such as time-consuming, expensive instruments required, and skilled personnel (Kumar et al. 2020). Unlike PCR, loop-mediated isothermal amplification (LAMP) is an alternative to PCR assay for the detection of V. parahaemolyticus at 65 °C in very short time and requires only a water bath. LAMP may lead to a false positive test result because of non-specific amplicons products and cross contamination (Hu et al. 2021; Xu et al. 2022). Recently, the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) technology has a potential for diagnostics. CRISPR combines nucleic acid pre-amplification with CRISPR–Cas enzyme for specific recognition of target DNA or RNA (Kaminski et al. 2021; Li et al. 2021). This technique improves the sensitive and specific, but time-consuming and expensive due to its dependence on fluorescence equipment. In cases of limited resources, it is very inconvenient to use this method (Xu et al. 2022). Therefore, antibody-dependent methods, e.g. ELISA, Western blot, and immunochromatographic strip test, to quickly detect AHPND and offer easy-to-use on-site for farmer was developed recently (Wangman et al. 2019; Mai et al. 2020). In addition, this method can combine with LAMP, CRISPR/Cas to offer a simple visualization (Hu et al. 2021; Xu et al. 2022). In this present study, we aimed to fabricate a lateral flow immunoassay (LFIA) strip to monitor the risks and rapid detection potential impacts of aquatic disease, particularly AHPND. LFIA method based on the principle of sandwich format. Binding event at reaction lines occurs between colloidal gold–polyclonal antibody conjugates, PirAvp and/or PirBvp protein(s) from analyzed sample, and immobilized antibodies onto a nitrocellulose membrane. Therefore, LFIA can detect both of PirAvp and PirBvp proteins simultaneously.
Materials and methods
Materials
Bacterial strains
VPAHPND XN89 and VPnon-AHPND XN8 were given by National Center for Genetic Engineering and Biotechnology (BIOTEC), Thailand.
Reagents
Colloidal gold solution (CG-020), backing card (MIBA-020–60 mm) were purchased from DCNovations, USA. Membrane and pads (sample pad-81132250, conjugate pad-8133-2250, nitrocellulose membrane (NCM)—10547004, and absorbent pad—8116-2250), were provided from Cytiva Life Sciences, Sweden. Goat anti-rabbit IgG antibody (R5506) was purchased from Sigma, USA. Culture media were purchased from BD Difco. Other reagents were of analytical grade.
Polyclonal antibody preparation
The process of antibody production was published elsewhere (Duong et al. 2021; Nguyen-Phuoc et al. 2021). All animal experiments followed the regulations of the Directive 2010/63/EU guideline approved by The Animal Care and Use Committee of University of Science, VNU-HCM in Ho Chi Minh City (ethical code 12/18-0599-01). Briefly, anti-PirAvp and anti-PirBvp polyclonal antibodies were produced from rabbits inoculated with the purified recombinant PirAvp/PirBvp protein. Then, these antibodies were purified by Hitrap protein G HP column (Cytiva Life Sciences). The affinity-purified antibodies were dialysed against phosphate buffer saline (PBS), pH 7.2. The concentration of antibody was measured by Bradford method. Specificity of anti-PirAvp and anti-PirBvp polyclonal antibodies was checked by dot blot technique with other Vibrio species (Duong et al. 2021; Nguyen-Phuoc et al. 2021).
Parameter optimisation of colloidal gold–polyclonal antibody conjugate
Determination of the optimum pH for the antibody labelling with colloidal gold
Colloidal gold solutions (DCNovations, USA) was adjusted pH values from 6 to 11 with an increment of 0.5 unit. Then, antibody solution was added to obtain a concentration 10 µg/ml. The mixtures were maintained under orbital shaking for 30 min at ambient temperature. Then, NaCl solution was added to each sample at concentration 10%. Two hours later, the changes in absorption peak after conjugation were detected by ultraviolet–visible (UV/Vis) spectrophotometer at range of wavelengths from 400 to 700 nm (UV1800, Shimadzu). Data were recorded with UV Probe software. Peaks were automatically marked with 1 and the optimum condition was highlighted in blue.
Determination of the optimum polyclonal Ab concentration for the antibody labelling with colloidal gold
Different amounts of polyclonal antibody were added to 1 ml of colloidal gold solution (DCNovations, USA) at optimum pH value to give concentrations of 0, 1, 5, 10, 20 μg/ml. The mixtures were maintained under orbital shaking for 30 min at ambient temperature. Then, NaCl solution was added to each sample at concentration 10%. Two hours later, the changes in absorption peak after antibody conjugation were detected by an UV/Vis spectrophotometer at range of wavelengths from 400 to 700 nm (UV1800, Shimadzu) as above.
Preparation and characterization of stable colloidal gold with polyclonal antibody
One ml of colloidal gold solution (DCNovations, USA) was adjusted pH 9 using 0.2 M K2CO3. Aliquots of 75 μl of antibody solution with the concentration of 137 µg/ml were added the pH adjusted gold solution as instructed by manfacturer. The mixtures were maintained under orbital shaking for 30 min at ambient temperature. Then, BSA solution was added to the suspension at concentration 10% and incubated for 1 h at ambient temperature. After that, the conjugate solution was centrifuged for 20 min at 12,000 rpm. The pellet was resuspended in 5 mM Borate, 1% BSA, pH 9.0, and stored at 4 °C. The formation of colloidal gold–polyclonal antibody conjugate was recorded with UV/Vis spectrophotometer at range of wavelengths from 400 to 700 nm (UV1800, Shimadzu).
Parameter optimisation of lateral flow immunoassay strip (LFIA)
Membrane selection
The sample pads and nitrocellulose membrane (NCM) tested to improve the effectiveness of LFIA. The NCMs with different capillary flow rate included Whatman FF80HP (60–100 s/4 cm), Whatman FF120HP (90–150 s/4 cm), and Whatman FF170HP (140–200 s/4 cm) (Cytiva Life Sciences).
Sample pads included Whatman CF1 (176 µm thickness with capillary flow rate at 207.3 s/4 cm), and Whatman CF3 (322 µm thickness with capillary flow rate at 174.3 s/4 cm) (Cytiva Life Sciences).
Determination of the running buffer
Different running buffers were tested: buffer 1 (5 mM Tris–HCl, pH 7.2), buffer 2 (5 mM phosphate buffer, pH 7.2), buffer 3 (5 mM Tris–HCl, pH 7.2, 1% Triton X-100), buffer 4 (5 mM phosphate buffer, pH 7.2, 1% Triton X-100), buffer 5 (5 mM Tris–HCl, pH 7.2, 1% Tween 20), and buffer 6 (5 mM phosphate buffer, pH 7.2, 1% Tween 20).
Preparation of conjugate pad
The colloidal gold–polyclonal antibody conjugate solution was diluted in 2 mM borate buffer with 5% sucrose and dropped 5 μL on glass fiber pad, and the pad was desiccated at 37 °C for 2 h.
Preparation of nitrocellulose membrane
The rabbit anti-PirAvp (1 mg/ml), anti-PirBvp (1 mg/ml) antibody, and goat anti-rabbit IgG antibody (1 mg/ml) in PBS were automatically dispensed onto nitrocellulose membrane at a rate of 3.5 μl/cm to generate one control line and two test lines using the lateral flow reagent dispenser (ClaremontBio, USA). The control line and test lines were located about 5 mm apart. The membrane was desiccated at 37 °C for 2 h.
Assembly of the LFIAs
The nitrocellulose membrane was stuck in the middle of the adhesive plastic backing card (0.01” thick, 60 mm width, DCNovations). The absorbent pad covered a part of the nitrocellulose membrane about 3 mm. The conjugate pad covered a part of the nitrocellulose membrane about 2 mm. The sample pad covered a part of the conjugate pad about 2 mm. Then, the LFIA was cut into test strips of 5 mm in width using Programmable strip cutter (Shanghai Kinbio Tech Co., Ltd, China). The test strips were stored in a desiccator cabinet at 4 °C for further study.
LFIA procedure
Sample in the running buffer with a volume of 100 µL was transferred into the sample pad of the test strip. The result was evaluated after 10–15 min by the naked eye. The appearance of red bands at the test line and the control line indicated a positive result. A single coloured band at control area indicated a negative result.
Evaluating the limit of detection (LOD) of the LFIA
A mixture of the purified recombinant PirAvp and PirBvp proteins was spiked at 500–250–125–62.5 ng/ml in the running buffer and 100 µL was applied to the test strip. The spike of each concentration was observed in triplicate. Negative sample in the running buffer was also applied to the test strip in parallel. The result was read as instructed above.
Evaluating the sensitivity and specificity of the LFIA
To evaluate our method, the LFIA was applied to detect PirAvp and PirBvp proteins spiked in shrimp samples. VPAHPND-free shrimps were spiked with bacteria at 5 × 103 CFU/ml and incubated at 30 °C in 20 mL of TSB medium. For negative samples and positive samples, VPnon-AHPND strain XN8 and VPAHPND strain XN89 were added, respectively. After 18 h, shrimp heads were crushed in 5 mL of the running buffer and 100µL of the supernatant was applied to the test strip. The result was read as instructed above. In parallel, VPAHPND-spiked shrimp samples were verified using PCR with GMIF1-2 primers (Mai Hoang et al. 2021).
Sensitivity and specificity were calculated according to formula below:
Results
Optimisation of colloidal gold–polyclonal antibody conjugate
Antibody was added to the different colloidal gold solutions with pH range from 6 to 11. The spectrum of the colloidal gold solutions corresponds to the pH when NaCl was added at a concentration of 10% (Fig. 1). Data showed that the absorbance value of solution increased with increasing pH value. When the pH reached 9.0, the absorbance curve saturated and decreased when pH was greater than 9. Accordingly, the best pH of the colloidal gold was 9.0.
Fig. 1.
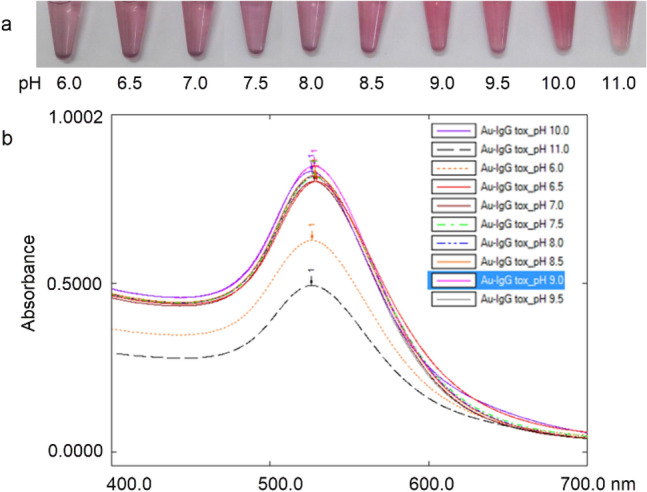
The pH optimisation of colloidal gold. a Photos of the resulting gold aggregation test at different pHs; b UV–Vis spectra of colloidal gold antibody conjugates at different pHs of colloidal gold corresponding to the photos in a
In Fig. 2, UV–Vis spectral absorbance with UV Probe software showed that the peak of the colloidal gold solutions changed in antibody concentration-dependent manner. When amount of antibody was insufficient, the spectral peak of colloidal gold solution was broadened and reduced in intensity. The absorbance of the solution increased as antibody concentration increased. After binding to antibody, colloidal gold remained stable throughout the procedure. There was no aggregation of colloidal gold at antibody concentration of 5, 10, and 20 µg/ml was observed at 520 nm. However, the absorbance value of antibody solution at 10 µg/ml was higher than the remaining two antibody solutions. Therefore, for economy’s sake this antibody concentration was used to label antibody with the colloidal gold nanoparticle solution because it maintained the stable conjugate.
Fig. 2.
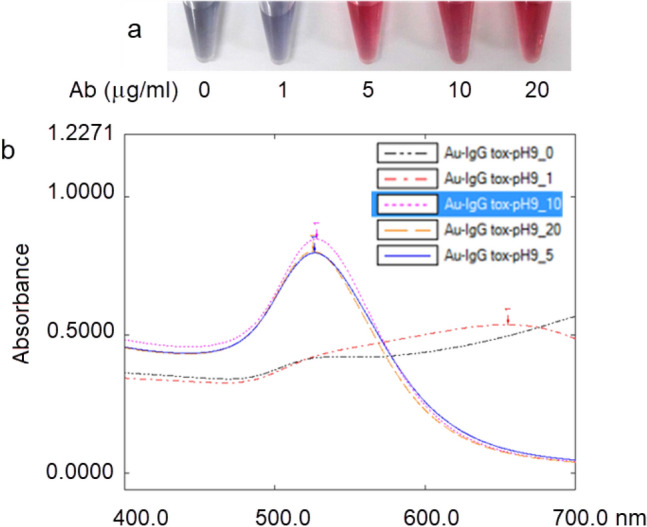
Optimal concentration of antibody. a Photos of the resulting gold aggregation test; b UV–Vis spectra of colloidal gold antibody conjugates after addition of different antibody concentration corresponding to the photos in a
The maximum absorption for colloidal gold solution was at wavelength of 523 nm and the solution was red in colour. After direct absorption of the antibody onto the colloidal gold particles, there was slight red shift in the adsorption peak to 526 nm due to the interaction between the colloidal gold and the antibodies compared to that of bare colloidal gold (Fig. 3). After a successful conjugation, there was a minor red-shifting of the absorbance peak position in the UV/Vis spectra, but the overall shape of the spectra remains the same before and after conjugation.
Fig. 3.
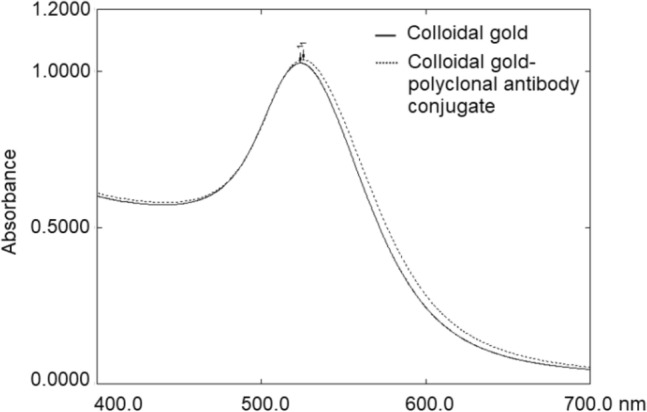
UV/Vis spectra of colloidal gold and the colloidal gold–polyclonal antibody conjugate
Optimisation of lateral flow immunoassay strip
Parameters were tested to improve the specificity and sensitivity of LFIA including types of nitrocellulose membrane, sample pad, and the running buffers. Corresponding to each parameter optimized, the used PirAvp or PirBvp protein concentration was 200 ng and applied to the test strip. For optimized conditions, the negative control was carried out in parallel. Digitally documented analysis showed Whatman FF170HP had slightly higher signal intensities than Whatman FF80HP, and FF120HP. In the case of sample pads, the Whatman CF3 performed better than Whatman CF1. The obtained intensity of test and control lines with buffer 4 (5 mM phosphate buffer, pH 7.2, 1% Triton X-100) were stable, that meant the flow front was not deformed. After optimizing LFIA, Whatman FF170HP NCM, Whatman CF3 sample pad and buffer 4 were used for further study (Fig. 4).
Fig. 4.
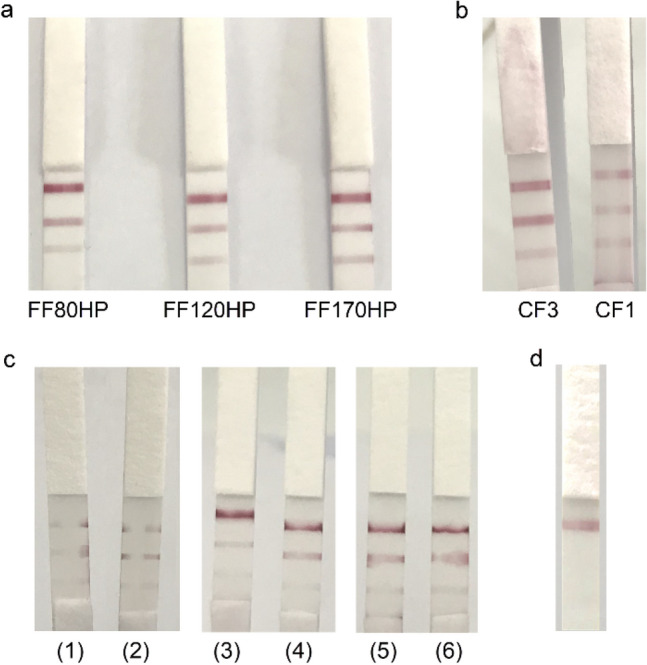
Parameter optimisation of LFIA. a Selecting the nitrocellulose membrane (FF80HP, FF120HP and FF170HP); b selecting the sample pad (CF1 and CF3); c selecting the running buffer. Buffer 1(5 mM Tris–HCl, pH 7.2), buffer 2 (5 mM phosphate buffer, pH 7.2), buffer 3 (5 mM Tris–HCl, pH 7.2, 1% Triton X-100), buffer 4 (5 mM phosphate buffer, pH 7.2, 1% Triton X-100), buffer 5 (5 mM Tris–HCl, pH 7.2, 1% Tween 20), and buffer 6 (5 mM phosphate buffer, pH 7.2, 1% Tween 20); d negative control. Data represent three independent repeats. Top band, control line; middle band, test line (rPirBvp); bottom band, test line (rPirAvp)
Limit of detection (LOD) of the LFIA
When using proteins from isolates of VPAHPND, the test strips gave positive result, while VPnon-AHPND gave a negative result (Fig. 5). Results indicated that LFIA had LOD for detecting rPirBvp protein at 62.5 ng. To a lesser extent, similar results were seen for rPirAvp protein at 125 ng.
Fig. 5.
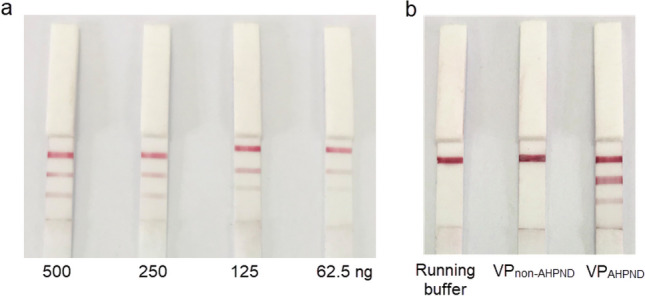
Limit of detection (LOD) and technical specificity of the LFIA. a LOD of the LFIA; b specificity of the LFIA. Data represent three independent repeats. Top band, control line; middle band, test line (rPirBvp); bottom band, test line (rPirAvp)
Evaluating the sensitivity and specificity of the LFIA
VPAHPND-free shrimp, VPnon-AHPND strain XN8, and VPAHPND strain XN89 were verified using PCR method (Fig. 6a). The performance of LFIAs was tested with sensitivity and specificity in VPAHPND (XN89)-spiked shrimp samples (Fig. 6b). A positive LFIA test result was found in 47 out of 50 positive spiked samples and 1 out of 50 negative spiked samples. As a result, the sensitivity and specificity of the LFIA diagnostic test were 94.0% and 98.0%, respectively (Table 1). In parallel, shrimp samples were verified using PCR with GMIF1-2 primers (Fig. 6c).
Fig. 6.
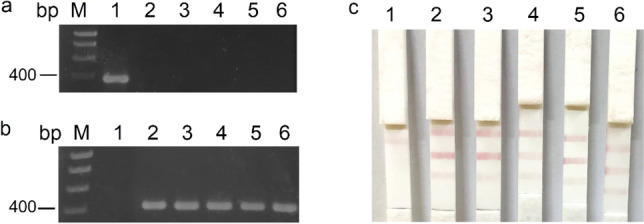
The performance of the LFIA and PCR. a PCR verification of VPAHPND status of input materials. M: ladder; 1: VPAHPND strain XN89; 2: VPnon-AHPND strain XN8, 3–6: VPAHPND-free shrimp samples. b The performance of the LFIA. 1: Negative spiked sample; 2–6: positive spiked samples. c PCR results corresponding to the photos in b. Data represent three independent repeats
Table 1.
Sensitivity and specificity of LFIA in spiked shrimp sample
| LFIA test | Total | ||
|---|---|---|---|
| Positive | Negative | ||
| Spiked sample | |||
| Positive | 47 | 3 | 50 |
| Negative | 1 | 49 | 50 |
| Total | 48 | 52 | 100 |
Discussion
Colloidal gold conjugated with antibody is an effective agent for diagnostic applications (Azzazy and Mansour 2009). The performance of immunoassay strip depends on quality of colloidal gold–antibody conjugate. In the present study, the antibodies are passively adsorbed onto colloidal golds based on a combination of electrostatic and hydrophobic interactions of the antibody and the particle surface (Thobhani et al. 2010). Therefore, pH and antibody concentration play a key role in the adsorption process to maintain the stability of colloidal gold–antibody conjugate. The best pH and the minimum amount of antibody was determined by salt-induced aggregation. If there was not enough amount of antibody that adsorbed onto colloidal gold surface, gold particles would aggregate in the presence of NaCl, and a colour change would be observed from red to blue in the solution by naked eye. This meant that the colloidal gold was unstable after the labelling process. Therefore, it was essential that the conjugate was stable before it was incorporated into a LFIA as an unstable conjugate would aggregate on the test strip and reduce assay performance. To perform a more accurate optimisation, an UV/Vis spectrophotometer was used as a detection system at range of wavelengths from 400 to 700 nm. This was a simple yet robust method for measurement and comparison the UV/Vis spectra before and after conjugation. After a successful conjugation, there was a minor red-shifting of the absorbance peak position in the UV/Vis spectra, but the overall shape of the spectra remains the same before and after conjugation (NanoComposix University 2021). In general, a red shift of 3–7 nm in peak absorbance was typical of protein binding on the colloidal gold (Yeo et al. 2015). In this study, mixtures of different pH and concentration were prepared. It was shown that the pH 9.0 and the antibody concentration of 10 µg/ml were the most optimized values. Colloidal gold remained stable after binding with antibody throughout the procedure.
Membrane and pads are the main physical components of LFIA. The sample pad is located at the beginning of the strip test. This component absorbs the sample, ensures an even flow, and filters out interfere particles from the sample solution. The choice of material and design for the sample pad could greatly affect the overall system (Millipore 2013; NanoComposix 2016). In this study, Whatman CF3 performed better than Whatman CF1 based on the thickness of the pad. The thickness of Whatman CF3 and CF1 was 322 µm and 176 µm, respectively. The thickness affects not only the bed volume but also the consistent flow. Because of thicker sample pad, the retention of the solution on the membrane is higher and the flow rate is relatively slow and constant (Parolo et al. 2020). A slower flow helps to increase the biological interaction between the target with the labelled conjugate. Therefore, it gives a higher sensitivity of the LFIA.
The membrane is the part of the LFIA where the signal is generated, therefore, it is apparently indispensable material to the system. Capillary flow rate is the most important parameter of the membrane (Millipore 2013). It affects the assay sensitivity and specificity. High capillary flow rate helps to promote the interaction between the target with the labelled conjugate, thus raising the sensitivity of the strip test. However, high capillary flow rate also increases the possibility of non-specific binding. Thus, it is important to determine the different capillary flow rates of the membranes during the development of an LFIA (Parolo et al. 2020). In this study, the NCMs with different capillary flow rate included Whatman FF80HP (60–100 s/4 cm), Whatman FF120HP (90–150 s/4 cm), and Whatman FF170HP (140–200 s/4 cm) were tested. Whatman FF170HP produced the highest signal to noise interaction.
The type and concentration of the running buffer affect the pH and ionic strength of the solution during the assay. This gives not only the same condition for different samples but also maintain interaction between the target and labelled conjugate. Moreover, the use of surfactant in the running buffer to minimize the non-specific binding and promote the flow of the detection reagents along the different pads and resolubilize of the conjugate (Parolo et al. 2020). Especially, the immobilization of a capture reagent solution in the nitrocellulose membrane, causing the reagent line to permeate the solution longer than the surrounding the untreated membrane area. When the sample solution passes, the flow front becomes deformed. To preclude this, an effective way is to add a small amount of surfactant to promote rewetting of the line (Millipore 2013). Therefore, buffer containing 5 mM phosphate, 1% Triton X-100, pH 7.4 was better than the same buffer containing 1% Tween 20.
AHPND is a newly emerging disease, spreading quickly without any treatment, causing great damage to the shrimp industry. Many studies showed that the presence of both proteins is required to cause AHPND (Sirikharin et al. 2015). Some mutant strains that have completely lost one of the two genes which coding PirAvp or PirBvp protein, do not cause AHPND; hence detecting both toxins confirms the presence of PirABvp that plays a major role in causing AHPND. Therefore, the main goal of this work is to simultaneously detect the AHPND-causing PirAvp and PirBvp proteins (100% mortality) in the shrimp.
Our developed LFIA could detect both of PirAvp and PirBvp protein with high sensitivity and specificity (94.0% and 98.0%, respectively), and low limit of detection (LOD) (125 ng for PirAvp and 62.5 ng for PirBvp). According to Wangman et al. (2019) research group have developed a monoclonal antibody-dependent famer-friendly strip test only for the detection of PirBvp with the LOD of the strip was around 625 ng/ml. In similar way, a study used PirA-specific polyclonal antibodies to develop flow-through assay test which able to detect 121 ng/ml of PirAvp protein (Hanumanthappa et al. 2020). Our LFIAs could detect two toxins simultaneously at 125 ng/ml. Meanwhile, the amount of PirAvp and PirBvp proteins require to cause 100% mortality with AHPND histopathology is 10 μg/g per shrimp (Sirikharin et al. 2015), meaning that this test strip can detect the disease early in the field. In the study, we only compared LFIA with the PCR method at the spiked bacteria concentration of 5 × 103 CFU/ml and the result indicated the LFIA was in line with PCR result. LFIA could detect PirAvp and PirBvp proteins in the solution of bacterial culture, but the corresponding concentration of these proteins had not been determined. In addition, LFIA readout combined with molecular assays such as LAMP, and RPA-CRISPR/Cas12a was also developed to detect AHPND. The detection limit of LAMP-LFIA was 2.1 × 10–4 ng/µl, corresponding to 630 fg per reaction and RPA-CRISPR/Cas12a-LFIA was 200 copies/μL (Hu et al. 2021; Li et al. 2021). However, such methods need 1–2 h to prepare samples and read results. Moreover, in the case of shrimp ponds with a Vibrio strain carrying the PirAvp–PirBvp toxin genes that are mutated or silenced for protein expression, gene-based AHPND detection methods may not be accurate. Meanwhile, our protein-based test strip method could determine whether the shrimp ponds really have AHPND disease. It is also more cost-effective because it does not require high expertise as well as associated equipment. Thus, the newly developed LFIA could be used for in-field diagnosis of AHPND.
In summary, we have developed for the first time a simple LFIAs for rapid detection both of PirAvp and PirBvp protein simultaneously using lab-made polyclonal antibodies. The developed test strip could be a game changer for early and in situ diagnosis of AHPND.
Acknowledgements
This work was funded by the Mekong Delta Program Office under project No: 19/2018/HĐ-KHCN-TNB.ĐT/14-19/C31. Hieu Tran-Van was a recipient of the ASEA-UNINET, (project no.: ASEA 2020-2021/Vet Med Uni/1).
Author contributions
HT-V, N-DD, and K-HN-P conceived and designed the research. N-DD, K-HN-P, K-YTD, T-DM-H, T-BH, and N-TTN conducted the experiments. HT-V, N-DD, K-HN-P, and TLT evaluated the data. HT-V, N-DD, and K-HN-P wrote the manuscript. All the authors read and approved the manuscript.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Footnotes
Ngoc-Diem Duong and Khai-Hoan Nguyen-Phuoc: First authorship shared.
References
- Azzazy HM, Mansour MM. In vitro diagnostic prospects of nanoparticles. Clin Chim Acta. 2009;403:1–8. doi: 10.1016/j.cca.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Duong ND, Nguyen-Phuoc KH, Do TKY, Nguyen TNT, Tran LT, Tran-Van H. Production of polyclonal antibody against the recombinant PirBvp protein of Vibrio parahaemolyticus. J Genet Eng Biotechnol. 2021;19(70):1–8. doi: 10.1186/s43141-021-00172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO Fisheries and Aquaculture Circular No. 1190 . Shrimp acute hepatopancreatic necrosis disease strategy manual. Rome: FAO; 2020. [Google Scholar]
- Hanumanthappa SK, Kumar BTN, Patil P, Poojary SR, Ballyaya AP, Ramesh KS, Shankar KM, Tyagi A, Holeyappa SA. Polyclonal antibody-based farmer-friendly flow-through test for the detection of acute hepatopancreatic necrosis disease in shrimp. Aquac Res. 2020;51(7):2863–2869. doi: 10.1111/are.14625. [DOI] [Google Scholar]
- Hu YQ, Huang XH, Guo LQ, Shen ZC, LV LX, Li FX, Zhou ZH, Zhang DF, Rapid and visual detection of Vibrio parahaemolyticus in aquatic foods using blaCARB-17 gene-based loop-mediated isothermal amplification with lateral flow dipstick (LAMP-LFD) J Microbiol Biotechnol. 2021;31(12):1672–1683. doi: 10.4014/jmb.2107.07022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski MM, Abudayyeh OO, Gootenberg JS, Zhang F, Collins JJ. CRISPR-based diagnostics. Nat Biomed Eng. 2021;5(7):643–656. doi: 10.1038/s41551-021-00760-7. [DOI] [PubMed] [Google Scholar]
- Kumar R, Ng TH, Wang HC. Acute hepatopancreatic necrosis disease in penaeid shrimp. Rev Aquac. 2020;12:1867–1880. doi: 10.1111/raq.12414. [DOI] [Google Scholar]
- Lee CT, Chen IT, Yang YT. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. PNAS. 2015;112(34):10798–10803. doi: 10.1073/pnas.1503129112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Lin N, Feng Z, Lin M, Guan B, Liang W, Wang Q, Li M, You Y, Chen K, Chen Q. CRISPR/Cas12a based rapid molecular detection of acute hepatopancreatic necrosis disease in shrimp. Front Vet Sci. 2021;8:1–10. doi: 10.3389/fvets.2021.819681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightner D (2012) Biology and pathology of early mortality syndrome of shrimp. Global outlook for aquaculture leadership, Bangkok, 40
- Lin SJ, Chen YF, Hsu KC, Chen YL, Ko TP, Lo CF, Wang HC, Wang HC. Structural insights to the heterotetrameric interaction between the Vibrio parahaemolyticus PirAvp and PirBvp toxins and activation of the Cry-like pore-forming domain. Toxins. 2019;11(233):1–15. doi: 10.3390/toxins11040233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai HN, Cruz-Flores R, Dhar AK. Development of an indirect enzyme linked immunoassay (iELISA) using monoclonal antibodies against Photorhabdus insect related toxins, PirAVp and PirBVp released from Vibrio spp. J Microbiol Methods. 2020;176:1–9. doi: 10.1016/j.mimet.2020.106002. [DOI] [PubMed] [Google Scholar]
- Mai-Hoang TD, Tien HL, Chau-Hoang HM, Nguyen-Phuoc KH, Pham QH, Tran LT, Tran-Van H. A novel PCR method for simultaneously detecting Acute hepatopancreatic necrosis disease (AHPND) and mutant-AHPND in shrimp. Aquac. 2021 doi: 10.1016/j.aquaculture.2020.736336. [DOI] [Google Scholar]
- Millipore EMD (2013) Rapid lateral flow test strips: considerations for product development. EMD Millipore Corporation: Billerica, MA, USA, 29, pp 702–707
- NanoComposix . Lateral flow assay development guide. San Diego: NanoComposix; 2016. pp. 1–37. [Google Scholar]
- NanoComposix University . Interpreting-particle-spectra-during-protein-conjugation. San Diego: NanoComposix University; 2021. [Google Scholar]
- Nguyen-Phuoc KH, Duong ND, Phan VT, Do TKY, Nguyen TNT, Tran LT, Tran-Van H. Generation and evaluation of polyclonal antibodies specific for ToxA from Vibrio parahaemolyticus causing acute hepatopancreatic necrosis disease (AHPND) in shrimp. Mol Biol Res Commun. 2021;10(1):23–32. doi: 10.22099/mbrc.2020.38774.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolo C, Sena-Torralba A, Bergua JF, Fuentes-Chust C, Hu L, Rivas L, Álvarez-Diduk R, Calucho E, Nguyen EP, Cinti S, Quesada-González D, Merkoçi A. Tutorial: design and fabrication of nanoparticle based lateral-flow immunoassays. Nat Protoc. 2020;15(12):3788–3816. doi: 10.1038/s41596-020-0357-x. [DOI] [PubMed] [Google Scholar]
- Prachumwat A, Taengchaiyaphum S, Mungkongwongsiri N. Update on early mortality syndrome/acute hepatopancreatic necrosis disease by April 2018. J World Aquac Soc. 2018;50:5–17. doi: 10.1111/jwas.12559. [DOI] [Google Scholar]
- Sirikharin R, Taengchaiyaphum S, Sanguanrut P, Chi TD, Mavichak R, Proespraiwong P, Sritunyalucksana K. Characterization and PCR detection of binary, Pir-like toxins from Vibrio parahaemolyticus isolates that cause acute hepatopancreatic necrosis disease (AHPND) in shrimp. PLoS ONE. 2015;10(5):1–16. doi: 10.1371/journal.pone.0126987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thobhani S, Attree S, Boyd R, Kumarswami N, Noble J. Bioconjugation and characterisation of gold colloid-labelled proteins. J Immunol Methods. 2010;356:60–69. doi: 10.1016/j.jim.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Wangman P, Chaivisuthangkura P, Taengchaiyaphum S, Pengsuk C, Sithigorngul P, Longyant S. Development of a rapid immunochromatographic strip test for the detection of Vibrio parahaemolyticus toxin B that cause acute hepatopancreatic necrosis disease. J Fish Dis. 2019;43(2):207–214. doi: 10.1111/jfd.13115. [DOI] [PubMed] [Google Scholar]
- Xu H, Tang H, Li R, Xia Z, Yang W, Zhu Y, Liu Z, Lu G, Ni S, Shen J. A new method based on LAMP-CRISPR–Cas12a-lateral flow immunochromatographic strip for detection. Infect Drug Resist. 2022;15:685–696. doi: 10.2147/IDR.S348456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo ELL, Yeo HY, Chua AJS, Ng ML, Parthasarathy K, Kah JCY. Understanding aggregation-based assays: nature of protein corona and number of epitopes on antigen matters. RSC Adv. 2015;5:14982–14993. doi: 10.1039/C4RA12089B. [DOI] [Google Scholar]


