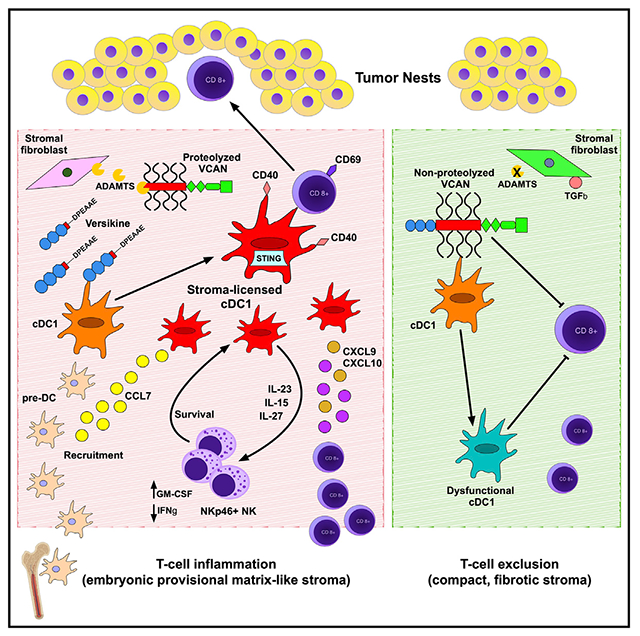
SUMMARY
Stimulatory type 1 conventional dendritic cells (cDC1s) engage in productive interactions with CD8+ effectors along tumor-stroma boundaries. The paradoxical accumulation of “poised” cDC1s within stromal sheets is unlikely to simply reflect passive exclusion from tumor cores. Drawing parallels with embryonic morphogenesis, we hypothesized that invasive margin stromal remodeling generates developmentally conserved cell fate cues that regulate cDC1 behavior. We find that, in human T cell-inflamed tumors, CD8+ T cells penetrate tumor nests, whereas cDC1s are confined within adjacent stroma that recurrently displays site-specific proteolysis of the matrix proteoglycan versican (VCAN), an essential organ-sculpting modification in development. VCAN is necessary, and its proteolytic fragment (matrikine) versikine is sufficient for cDC1 accumulation. Versikine does not influence tumor-seeding pre-DC differentiation; rather, it orchestrates a distinctive cDC1 activation program conferring exquisite sensitivity to DNA sensing, supported by atypical innate lymphoid cells. Thus, peritumoral stroma mimicking embryonic provisional matrix remodeling regulates cDC1 abundance and activity to elicit T cell-inflamed tumor microenvironments.
Graphical abstract
In brief
T cell-inflamed tumor microenvironments are a prerequisite for immunotherapy efficacy; however, why some tumors are inflamed and others not remains poorly understood. Papadas et al. link stromal reaction dynamics with T cell-induced inflammation. Peritumoral stroma emulating embryonic provisional matrix remodeling regulates cDC1-NK-CD8+ crosstalk to promote T cell repriming and penetration into tumor nests.
INTRODUCTION
Tumor antigen cross-presentation and CD8+ T cell effector priming by stimulatory type 1 conventional dendritic cells (cDC1s) is integral to spontaneous and therapeutic anti-tumor immunity (Binnewies et al., 2018; Broz et al., 2014; Gajewski, 2015; Hildner et al., 2008; Salmon et al., 2016). In addition to effector priming in the lymph node and in the tumor microenvironment (TME) (Thompson et al., 2010), cDC1s regulate effector cell influx into the TME (Spranger et al., 2017). From a translational perspective, cDC1s are crucial for responses to vaccination strategies, immune checkpoint inhibitors (Oba et al., 2020; Salmon et al., 2016; Sanchez-Paulete et al., 2016), and engineered immune effector cells (e.g., chimeric antigen receptor T [CAR-T] cells) (Kuhn et al., 2020).
Several key studies have shown that stimulatory cDC1s are excluded from interdigitating tumor nestlets and locate in peritumoral stroma (Bell et al., 1999; Broz et al., 2014; Hubert et al., 2020; Lavin et al., 2017; Mattiuz et al., 2021; Spranger et al., 2015). However, the mechanisms that retain cDC1s at the tumor periphery remain poorly understood. To understand this paradoxical localization, we drew parallels with embryonic development, where provisional matrix remodeling signals generate powerful cues that regulate cell fate, phenotype, and behavior essential for morphogenesis (Nandadasa et al., 2014).
The large aggregating extracellular matrix proteoglycan versican (VCAN) is a central component of the embryonic provisional matrix, playing key non-redundant roles in development of the cardiovascular system and limbs (Islam and Watanabe, 2020; Nandadasa et al., 2014; Papadas et al., 2020; Wight et al., 2020; Zhang et al., 2012). Vcan-null mice die in utero by embryonic day 10.5 because of defects along the anterior-posterior cardiac axis (Mjaatvedt et al., 1998). VCAN proteolysis by ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) proteases at the Glu441-Ala442 bond (V1 isoform enumeration) is an essential requirement that acts in part through the specific neoactivities of the released bioactive N-terminal fragment (matrikine) versikine (McCulloch et al., 2009; Nandadasa et al., 2014; Timms and Maurice, 2020). Disruption of the Glu441-Ala442 proteolytic site that generates versikine leads to developmental abnormalities (Islam et al., 2020; Nandadasa et al., 2021).
VCAN proteolysis at the versikine C-terminal Glu441-Ala442 bond correlates with CD8+ T cell infiltration in solid and hematopoietic human tumors (Emmerich et al., 2020; Hope et al., 2014, 2017). We demonstrated that recombinant versikine elicits immunomodulatory transcripts in myeloid cells in vitro (Hope et al., 2016), which was later confirmed by others (Han et al., 2020; Islam et al., 2022). Versikine also promotes cDC1 generation from FLT3L-treated bone marrow (BM) in vitro (Hope et al., 2017). These immunostimulatory in vitro activities of the proteolytic fragment versikine appear to be at odds with the immunosuppressive actions of parental non-proteolyzed VCAN (Tang et al., 2015). Here we provide compelling in vivo evidence connecting tumor stroma remodeling and matrikine activity with cDC1 abundance and function.
RESULTS
cDC1s localize in peritumoral stroma and are regulated by the VCAN pathway
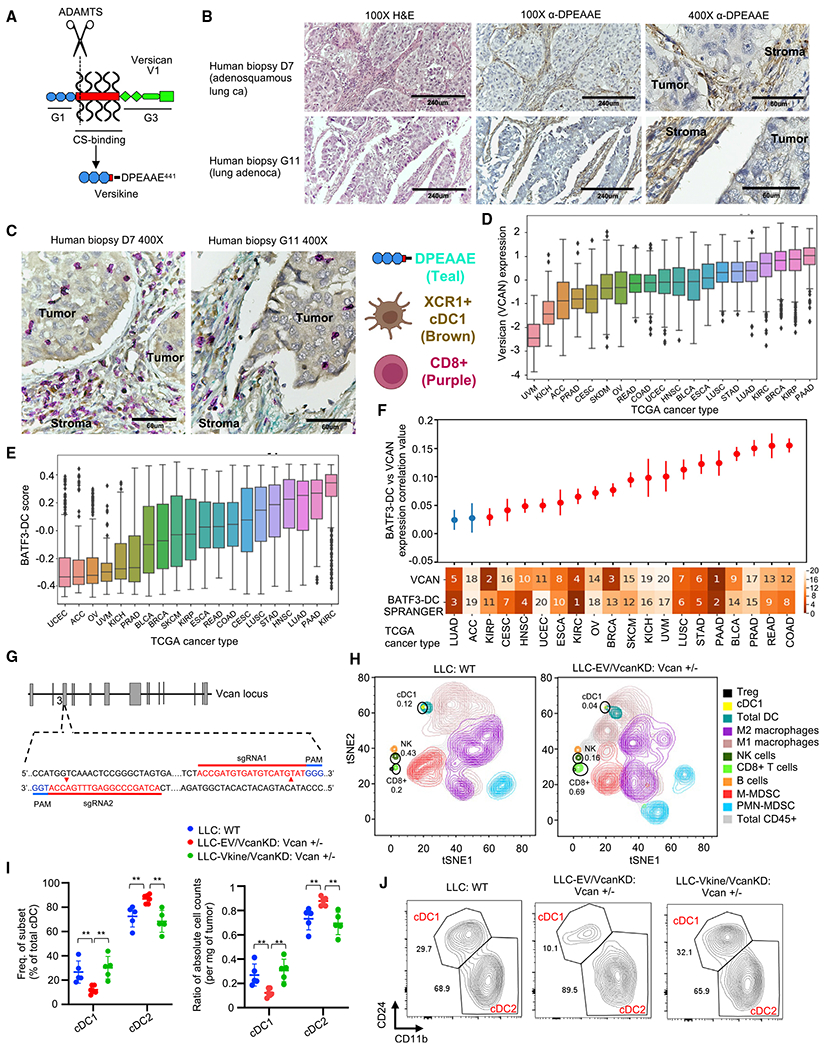
VCAN and its binding partner hyaluronan are cardinal components of the provisional extracellular matrix in development, wound healing, and cancer (Wight, 2017). In tumors, stromal provisional matrix is thought to coordinate critical pro-tumor functions (e.g., angiogenesis) and prime conversion toward collagen-rich, desmoplastic stroma (Yamauchi et al., 2018). For deposition into stromal matrix, VCAN is sourced from stromal mesenchymal cells, immune infiltrating cells (particularly myeloid cells, such as macrophages), and, in some cases (such as in lung carcinomas), the tumor cells themselves (Papadas and Asimakopoulos, 2020). However, VCAN proteolytic processing is located primarily in stroma because of the local activity of stromal fibroblast-derived ADAMTS VCANases (Hope et al., 2014). Using an immunohistochemistry (IHC)-validated antibody against DPEAAE, a neoepitope generated through VCAN proteolysis at Glu441-Ala442 (V1 isoform) (Figure 1A), we observed VCAN proteolysis signal in approximately 83% of human lung cancer cases in a stromal distribution (Figure 1B; Table S1). To determine the location of cDC1s relative to sites of stromal VCAN proteolysis, we performed multiplex IHC with antibodies detecting the cDC1 lineage marker XCR1 and CD8. Remarkably, even in T cell-inflamed tumors demonstrating intra-epithelial CD8+ T cell penetration, XCR1+ cells were confined within stromal sheets recurrently undergoing VCAN proteolysis (Figure 1C; Table S1). The antibody against XCR1 has been previously validated (Porciuncula et al., 2021), and we confirmed close correlation between XCR1 staining and the signal for the cDC1 lineage marker CLEC9A in human tonsils (Tullett et al., 2014; Figure S1A).
Figure 1. The VCAN pathway regulates tumor cDC1s.
For a Figure360 author presentation of this figure, see https://doi.org/10.1016/j.celrep.2022.111201.
(A) Schematic showing versican (VCAN)-V1 functional domains and site-specific proteolysis to generate versikine (scissors represent ADAMTS proteolytic cleavage). CS, chondroitin sulphate.
(B) Stromal distribution of anti-DPEAAE IHC staining in human lung cancers. DPEAAE constitutes the C terminus of versikine (chromogen, DAB; counterstain, hematoxylin). 10× objective: scale bars, 240 μm; 40× objective: scale bars, 60 μm.
(C) Triple IHC staining of human lung cancers (DPEAAE, teal; XCR1, brown; CD8, purple).
(D) Distribution of VCAN expression across TCGA carcinomas (gdc.cancer.gov), ordered on the horizontal axis by median VCAN expression.
(E) Distribution of cDC1 (BATF3-DC) score across TCGA carcinomas, ordered on the horizontal axis by median measured cDC1 score.
(F) Levels of correlation between cDC1 (BATF3-DC) score and VCAN expression across TCGA carcinomas. The ranked median of VCAN expression and measured cDC1 (BATF3-DC) score is shown across the x axis (1, highest; 20, lowest). Significant (q < 0.1) correlations after multiple hypothesis correction are colored red. Error bars represent the standard error of the correlation coefficient measured using Python statsmodels.
(G) Generation of Vcan+/− mice through CRISPR-Cas9-based targeting of Vcan exon 3.
(H) Mass cytometry of CD45+ cells from WT (LLC implanted into WT recipients, left) and Vcan-depleted tumors (LLCVcanKD tumor cells implanted into Vcan+/− recipients, right).
(I) Quantification of frequency (left) and absolute count ratios (cDC1/cDC1+cDC2 and cDC2/cDC1+cDC2) in WT, Vcan-depleted (LLC-EVVcanKD: Vcan+/−), and versikine (Vkine)-rescued (LLC-VkineVcanKD: Vcan+/−) tumors. Data are presented as mean ± SEM. n = 5 for each group. *p < 0.05, **p < 0.01, ***p < 0.001.
(J) Representative flow cytometry plots showing cDC1 and cDC2 frequency in WT, Vcan-depleted (LLC-EVVcanKD: Vcan+/−), and Vkine-rescued (LLC-VkineVcanKD: Vcan+/−) tumors (gating according to Figure S1F).
In vitro experiments were performed in technical triplicates. In vivo cohort sizes are shown in individual panels. All experiments were reproduced independently at least twice.
Matrikines (such as versikine) have been defined as “peptides liberated by partial proteolysis of extracellular matrix macromolecules which are able to regulate cell activities not triggered by their full-size parent macromolecules” (Gaggar and Weathington, 2016). Notwithstanding its distinct neo-activity, versikine ultimately derives from parental VCAN through ADAMTS proteolysis (Figure 1A); therefore, we hypothesized that VCAN expression (the substrate for versikine) and cDC1 abundance correlate in human cancer. We compared VCAN gene expression and cDC1 signature scores (Spranger et al., 2017) across 7,591 samples from 20 The Cancer Genome Atlas (TCGA) cancer types (Figures 1D and 1E). A significantly positive correlation between VCAN expression and cDC1 signature scores was observed in several human carcinomas (Figure 1F), suggesting that the VCAN pathway broadly regulates cDC1s.
To dissect the relevant mechanisms, we generated novel Vcan-targeted models that disrupt exons coding for Vcan’s N terminus (Vcan exons 2–6). The widely used Vcanhdf null mutant (hdf, heart defect) targeting exon 7 is embryonic lethal in homozygosity (Mjaatvedt et al., 1998). Vcanhdf hemizygosity demonstrates functional haploinsufficiency for CD8+-mediated control of viral infection (McMahon et al., 2016). We used CRISPR-Cas9-mediated mutagenesis to disrupt exon 3 sequences, abolishing transcription of all Vcan isoforms (and, consequently, generation of versikine) (Figure 1G). We derived two founders (Vcan1053 and Vcan1058) bearing Vcan exon 3 deletions (16 bp and 47 bp, respectively) (Figures S1B and S1C). We confirmed defective Vcan message induction after stimulation of BM-derived macrophages (BMDM) with the Toll-like receptor (TLR)-4 agonist lipopolysaccharide (LPS) (Figure S1D). BMDMs stimulated with LPS preferentially transcribe the V1 isoform, the precursor to versikine (Chang et al., 2014). The Vcan1053 transgenic line demonstrated the most severe defect in Vcan message induction (hereafter designated Vcan+/−).
Lewis lung carcinoma (LLC) cells produce Vcan cell autonomously (Kim et al., 2009). We knocked down endogenous Vcan expression in LLC cells using short hairpin RNA (shRNA) targeting Vcan exon 8 (encoding for the glycosaminoglycan (GAG)β domain in the VCAN- V1 isoform, the major isoform produced in LLC; Figure 1A), hereafter referred to as LLCVcanKD. We validated the reduced transcription of Vcan in LLCVcanKD cells using 5′ and -3′ Vcan primers, as shown in Figure S1E. We characterized the intratumoral immune contexture in LLCVcanKD tumors implanted in Vcan+/− mice through mass cytometry and compared it with wild-type (WT) controls (Figure 1H; Table S2). Vcan depletion resulted in expansion of CD8+ T cells, consistent with the known role of non-proteolyzed VCAN in T cell exclusion (Evanko et al., 2012; Gorter et al., 2010; McMahon et al., 2016). In keeping with our hypothesis, we observed cDC1 loss in Vcan-depleted tumors (Figure 1H). To corroborate the mass cytometry findings, we delineated intratumoral DCs through 9-color flow cytometry (Laoui et al., 2016; see Figure S1F for the gating strategy). cDC1s were depleted in LLCVcanKD tumors implanted into Vcan+/− mice (Figures 1I and 1J). In contrast, steady-state splenic cDC1s were not reduced (in fact, they were mildly increased) in Vcan+/− mice (Figure S1G). Ectopic expression of versikine in LLCVcanKD cells restored near-physiological cDC1 abundance (Figures 1I and 1J). Intratumoral DC absolute count ratios corroborated the cell frequency findings (Figures 1I and 1J) despite fluctuations in total cDC counts using the collagenase/hyaluronidase tumor dissociation protocol delineated in the STAR Methods (Figure S1H). Vcan depletion and versikine add-back did not affect tumor growth rates (Figure S1I). These results demonstrate that VCAN is necessary and that its proteolytic product versikine is sufficient for cDC1 abundance in the TME.
The VCAN matrikine versikine promotes cDC1 abundance in vivo
VCAN proteolysis is a composite event that produces two simultaneous, coupled consequences: first, parental VCAN clearance (Islam et al., 2022), and second, the novel activities of the released matrikine versikine. To uncouple versikine’s activity from the effects of parental VCAN depletion, we generated LLC cells stably expressing hemagglutinin (HA)-tagged versikine in the WT background (Figure 2A). Expression of versikine in LLC cells did not result in grossly visible increases in angiogenesis or hemorrhagic propensity (Figure 2B). Ectopically expressed versikine was readily detectable by anti-HA tag western blotting in murine tumor lysates at approximately 75 kDa (Figures 2C and S2A).
Figure 2. The VCAN-matrikine versikine promotes cDC1 abundance in vivo.
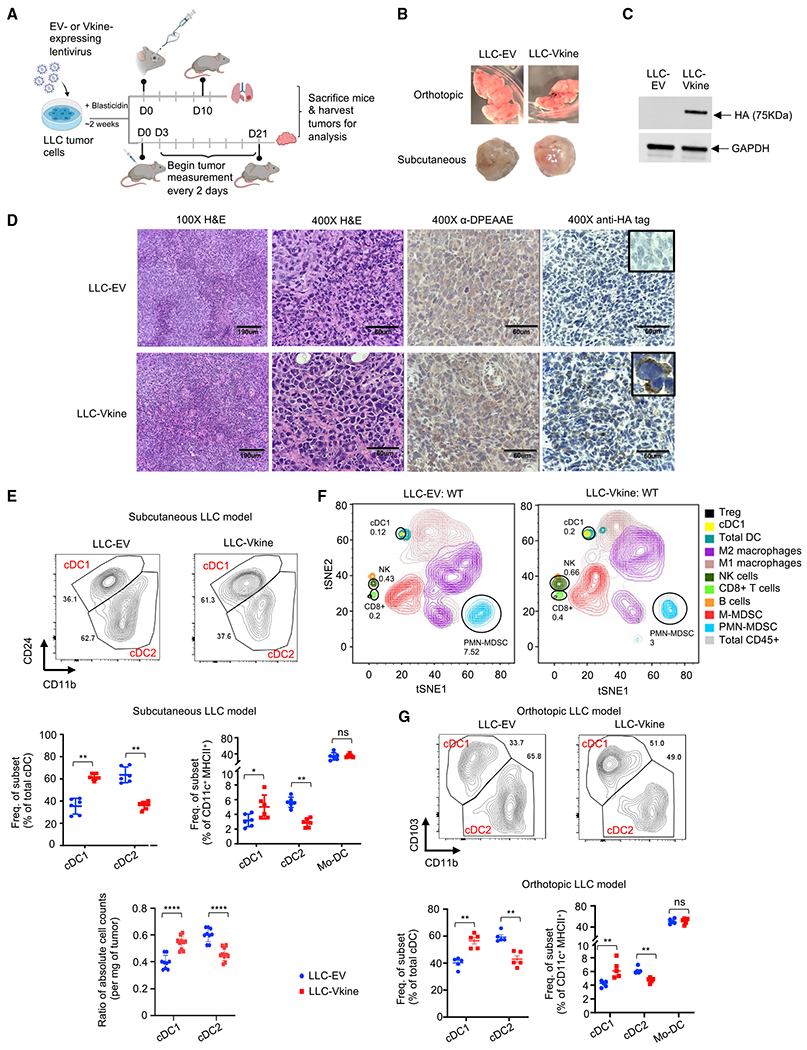
(A) Schematic of the experiment. LLC tumor cells were engineered to express hemagglutinin (HA)-tagged versikine (LLC-Vkine) or empty vector controls (LLC-EV) and injected subcutaneously (s.c.) on the flank or intravenously using a retro-orbital approach.
(B) Gross morphology of orthotopic (top) and s.c. (bottom) LLC-EV and LLC-Vkine tumors.
(C) Anti-HA tag western blotting detects a 75-kDa band in LLC-Vkine tumor lysates, consistent with versikine. See the full blot in Figure S2A.
(D) Representative immunohistochemistry (IHC) images showing α-DPEAAE and HA tag staining of LLC-EV and LLC-Vkine tumors. Endogenous DPEAAE proteolysis is low level and similar between LLC-EV and LLC-Vkine. Anti-HA staining localizes in a membranous distribution in LLC-Vkine cells (inset, larger magnification).
(E) Flow cytometric analysis of cDC subsets in s.c. LLC-EV and LLC-Vkine tumors (gating strategy according to Figure S1F) (Laoui et al., 2016) and quantification of cDC and tumor-associated DC (TADC) frequency (top) and absolute count ratios (cDC1/cDC1+cDC2 and cDC2/cDC1+cDC2) (bottom).
(F) Comparison of immune contexture (CD45+ fraction) in LLC-EV versus LLC-Vkine tumors by 31-marker mass cytometry.
(G) Flow cytometry analysis of cDC subsets in orthotopic LLC-EV and LLC-Vkine tumors (lung metastases induced by intravenous injection). A summary of cDC and TADC subset frequencies is depicted on the right.
Data are presented as mean ± SEM and are from one of three independent experiments with n = 5 or 6 for each group. *p < 0.05, **p < 0.01, ***p < 0.001. In vitro experiments were performed in technical triplicates. In vivo cohort sizes are shown in individual panels. All experiments were reproduced independently at least twice.
Murine implantable tumor models do not recapitulate the human architecture of epithelial nests and stromal sheets; this limitation has been attributed to acquisition of mesenchymal features through successive passaging (Guerin et al., 2020; Figure 2D). However, the LLC model does retain physiological relevance because of its tumor-intrinsic production of Vcan that regulates myeloid cells in the TME (Kim et al., 2009). In this regard, LLC mimics a subset of human lung cancers with detectable VCAN production and processing in stromal and epithelial compartments (Figure S2B; negative controls in Figure S2C). Ectopically expressed versikine was detected in a membranous distribution consistent with its accumulation in the pericellular glycocalyx (Figure 2D), a physiological site of VCAN cleavage (Hattori et al., 2011). Membranous localization of ectopic versikine was also seen in B16 melanoma tumor cells that transcribe a very low to undetectable endogenous Vcan message (Figure S2D).
There were no differences in the growth rates between LLC-empty vector (EV) and LLC-versikine (LLC-Vkine) tumors (Figure S2E). We analyzed cDC populations by conventional flow cytometry (Figure S1F). We confirmed cDC1 expansion in LLC-Vkine tumors (Figure 2E), notably the opposite phenotype to that of Vcan depletion (Figure 1J), as well as unchanged monocytic-derived dendritic cell (Mo-DC) frequency (Figure 2E). In the later stages of our study, optimized tumor cell dissociation protocols permitted determination of absolute DC counts; only cDC1s expanded in absolute terms (Figure S2F). By mass cytometry, in addition to cDC1 accumulation, we observed expansion of an innate lymphoid NK1.1+ NKp46+ population, an increase in intratumoral CD8+ T cells, as well as polymorphonuclear myeloid-derived suppressor cell (PMN-MDSC) depletion (Figure 2F), the latter being consistent with IRF8 regulation by versikine (Hope et al., 2016; Waight et al., 2013). To confirm cDC1 expansion in the orthotopic lung milieu, we injected EV- and versikine (Vkine)-expressing LLC cells intravenously to seed the relevant lung microenvironment and harvested lungs bearing metastatic deposits on day 10 (Figure 2G). LLC-Vkine tumors within the relevant lung microenvironment also showed enhanced cDC1s (Figure 2G). As with prior reports, we found CD103 to be more consistent compared with CD24 for cDC1 enumeration in native lung tissue (Cabeza-Cabrerizo et al., 2021; Misharin et al., 2013).
Versikine promoted cDC1s in the BALB/c-derived, orthotopic 4T1 mammary carcinoma model (Figure S2G). Growth rates of 4T1 versikine-replete tumors did not differ from their EV counterparts (Figure S2H). Earlier we reported a role of VCAN proteolysis in shaping the human BM myeloma immune microenvironment (Hope et al., 2016). More recently, we developed the first Ras-driven immunocompetent myeloma model, VQ (Wen et al., 2021). Versikine-replete VQ myeloma tumors demonstrated enhanced cDC1s (Figure S2I). Myeloma clinical progression was unaffected by versikine (Figure S2J). Therefore, versikine promotes cDC1 abundance in solid and hematopoietic cancers across murine genetic backgrounds.
Pre-DC differentiation is unaffected by versikine
To explain how VCAN proteolytic products promote tumor cDC1 density, we first determined whether this occurred through uncommitted tumor-seeding pre-DC differentiation (Diao et al., 2010). Our rationale was based on our prior observation that recombinant versikine promoted cDC1 generation from mouse BM treated with FLT3L in vitro (Hope et al., 2017). cDC1 “signature” transcripts (Irf8, Batf3, Cxcl9, and Cxcl10) were increased in the bulk transcriptome of versikine-replete (LLC-Vkine) tumors (Figure 3A). Irf8 is a “terminal selector” for the cDC1 lineage (Sichien et al., 2016). The Batf3 transcript increase corroborates versikine-induced cDC1 abundance because the Batf3 expression range is very narrow (Figure S3A; Table S3). Id2 transcripts did not differ between versikine-replete and control tumors, but Id2 is expressed more broadly and not highly expressed in cDC1s (Figure S3A; Table S3).
Figure 3. Versikine selectively activates cDC1 in vivo.
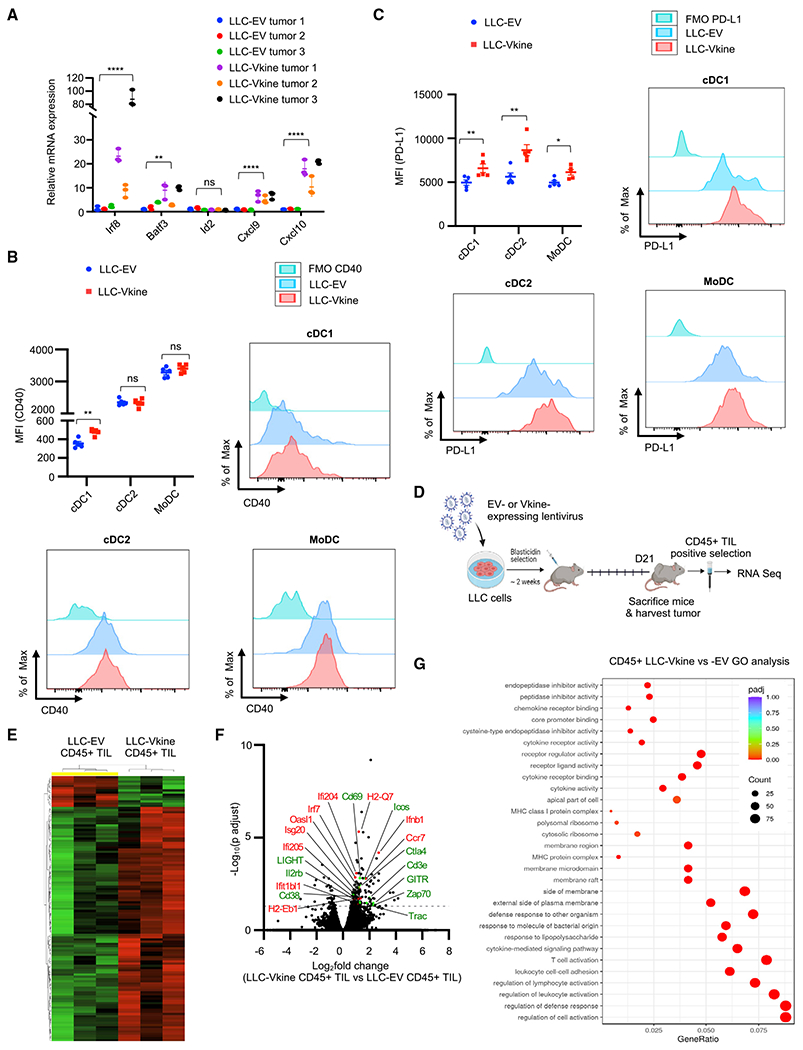
(A) RT-PCR analysis for cDC1 “signature” transcripts in bulk LLC-EV and LLC-Vkine tumor mRNA. Data are presented as mean ± SEM.
(B) Summary of CD40 staining intensity (MFI, mean fluorescence intensity) in DC subsets from LLC-EV and LLC-Vkine tumors (experiment 1). A second, independent experiment (experiment 2) is depicted in Figure S3F. Examples of individual histogram plots for each DC subset are shown.
(C) Summary of PD-L1 staining intensity in DC subsets from LLC-EV and LLC-Vkine tumors. Examples of individual histogram plots for each DC subset are shown.
(D) Layout of the experiment to compare transcriptomic profiles in LLC-EV versus LLC-Vkine tumor immune infiltrates.
(E) Hierarchical clustering of transcriptomic profiles by RNA sequencing (RNA-seq) analysis of CD45+ tumor-infiltrating leukocytes (TILs) extracted from LLC-EV versus LLC-Vkine tumors.
(F) Volcano plot highlighting key differentially expressed genes in CD45+ TILs from LLC-Vkine tumors compared with LLC-EV tumors. Genes whose overexpression has been linked to APC activation are shown in red and genes whose overexpression has been linked to T cell activation in green.
(G) Gene Ontology (GO) analysis of pathways enriched in CD45+ fractions from LLC-Vkine versus LLC-EV tumors.
ns, non-significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Data are presented as mean ± SEM. In vitro experiments were performed in technical triplicates. In vivo cohort sizes are shown in individual panels. All experiments were reproduced independently at least twice.
To test our differentiation hypothesis, we sorted CD45.2+ pre-DC precursors from the BM of Flt3l in-vivo-mobilized donor mice (Figures S3B and S3C). Donor mice were implanted with Flt3l-secreting B16 tumor cells to provide a continuous source of circulating Flt3l, as described previously (Vremec, 2016). CD45.2+ pre-DCs were adoptively transferred intratumorally into subcutaneous LLC-EV and LLC-Vkine tumors implanted into CD45.1+ recipients. 72 h after adoptive transfer, tumors were dissociated, and CD45.2+ as well as endogenous CD45.1+ DC fractions were enumerated by flow cytometry. The CD45.1+ endogenous cDC composition served as an internal control. As expected, CD45.1+ endogenous cDC1s were increased in LLC-Vkine tumors (Figure S3D). In contrast, CD45.2+ cDC1s and cDC2s did not differ between LLC-Vkine and -EV controls (Figure S3E). Thus, stromal signals did not affect pre-DC differentiation into cDC1s versus cDC2s within the time frame of our differentiation assay and given the assay’s limitations (Limitations of the study).
Versikine selectively activates cDC1s in vivo
Because pre-DC differentiation could not explain cDC1 accumulation, we tested the hypothesis that versikine regulates the cDC1 activation-survival cycle (Fuertes Marraco et al., 2011). Our rationale was supported by the association between VCAN proteolysis and T cell infiltration (Hope et al., 2016, 2017), suggesting a role of versikine in cDC1 activation. We were particularly interested in CD40 expression because of recent work showing that cDC1s cross-prime CD4+ T cells and are licensed through CD40 back-signaling to augment CD8+ T cell priming and anti-tumor responses (Ferris et al., 2020) as well as the reported anti-apoptotic functions of CD40 in cDC1s (Lin et al., 2020). Flow cytometry analysis of cDC subsets from LLC-EV and LLC-Vkine tumors demonstrated that versikine selectively upregulates CD40 expression in tumor cDC1s but not tumor cDC2s or Mo-DCs (Figure 3B and S3F). CD40 expression is weakest at baseline in intratumoral cDC1s. In contrast, all 3 intratumoral DC subsets upregulate PD-L1 when exposed to versikine in vivo (Figure 3C).
CD40 induction in cDC1s would be expected to promote T cell activation in the TME. To test this hypothesis, we compared the transcriptomic profiles of CD45+ tumor-infiltrating leukocytes (TILs) isolated from versikine-replete versus control LLC tumors (Figures 3D–3G; Table S4). We detected a compelling T cell costimulation and activation signature (Cd69, Ctla-4, Icos, Zap70, Il2rb, Cd38, Light, and Gitr) (Figures 3F and 3G) as well as a significant increase in T cell-specific transcripts (CD3e and T cell receptor [TCR] genes), consistent with the CD8+ T cell expansion seen by mass cytometry (Figure 2F). We detected hallmarks of antigen-presenting cell (APC) activation (upregulation of MHCII, Ccr7, Ifnb1, Irf7, and several interferon-responsive genes) (Figure 3F).
cDC1 activation by versikine is cell autonomous
To elucidate whether cDC1 activation by versikine was cell autonomous, we took advantage of the cDC1 cell model MutuDC1940 (Fuertes Marraco et al., 2012). To explore steady-state changes in the MutuDC1940 transcriptome in the presence of versikine, we generated stable MutuDC1940-Vkine cell lines through lentiviral transduction (Figure 4A). MutuDC1940-Vkine cells had a slightly more developed dendritic appearance compared with EV (Figure 4B).
Figure 4. cDC1 activation by versikine is cell autonomous.
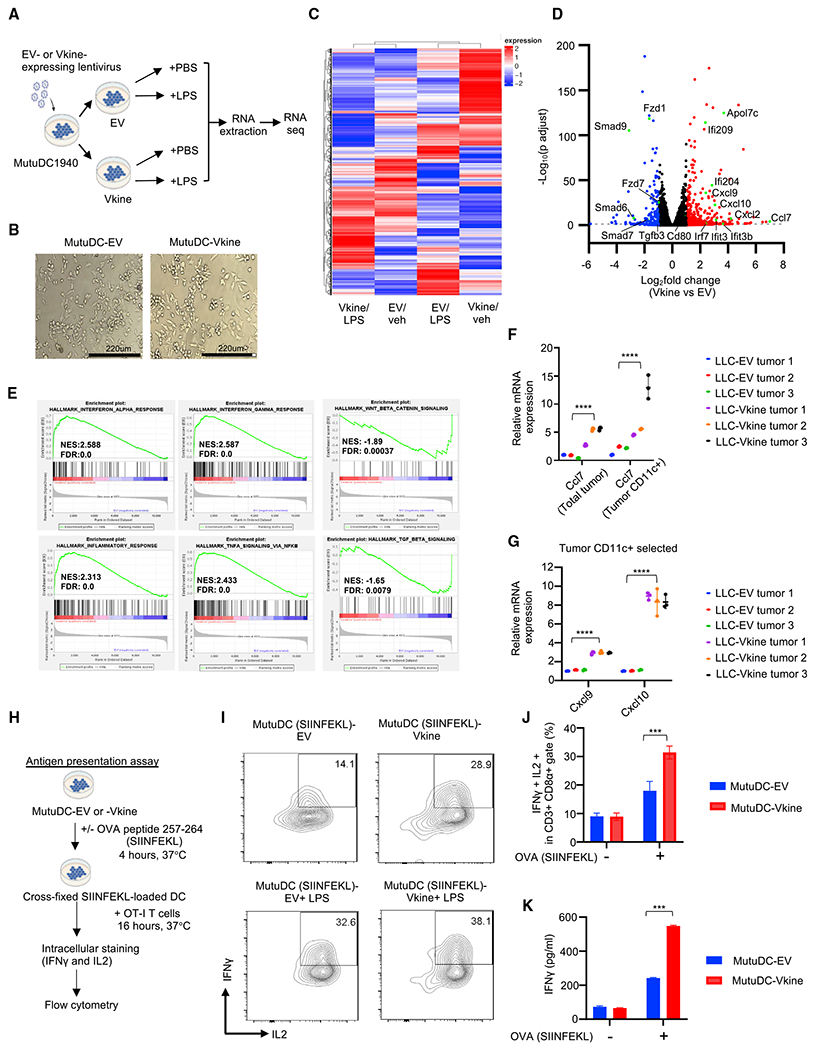
(A) Schematic layout of the experiment. MutuDC1940-EV or -Vkine cells were stimulated for 4 h with vehicle (PBS) or the TLR4 agonist lipopolysaccharide (LPS) (100 ng/mL) before RNA extraction.
(B) Gross morphology of MutuDC1940 cells engineered to express versikine (Vkine) or empty vector (EV). Phase contrast, 100× magnification; scale bar, 220 μm.
(C) Hierarchical clustering of MutuDC1940 transcriptomic profiles expressing EV or versikine (Vkine) and stimulated with the TLR4 agonist LPS or vehicle (PBS).
(D) Volcano plot highlighting key differentially expressed genes in MutuDC1940-Vkine versus -EV cells (without LPS).
(E) Gene set enrichment analysis (GSEA) of significantly upregulated (left and center) and downregulated (right) pathways in MutuDC1940-Vkine versus -EV cells (without LPS).
(F) Ccl7 RT-PCR using LLC-EV and LLC-Vkine tumor bulk mRNA (left) and CD11c+ magnetically separated fraction mRNA (right).
(G) Cxcl9 and Cxcl10 RT-PCR using CD11c+ magnetically separated fractions from LLC-EV or LLC-Vkine tumors.
(H) Schematic of the antigen presentation experiment.
(I) Flow cytometry for endogenous IFN-γ and IL-2 of OT-I CD8+ T cells co-cultured with SIINFEKL peptide-loaded MutuDC1940 cells, EV- or Vkine-expressing, with or without LPS.
(J) Quantitation of OT-I flow cytometry analysis of the antigen presentation assay.
(K) IFN-γ by ELISA in supernatants from OT-I and MutuDC1940:SIINFEKL co-cultures in the antigen presentation assay.
Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. In vitro experiments were performed in technical triplicates. In vivo cohort sizes are shown in individual panels. All experiments were reproduced independently at least twice.
Versikine elicited a co-stimulatory transcriptional program in MutuDC1940 cells distinct from the transcriptional program elicited by the TLR4 agonist LPS (Figure 4C and S4A). The combination of versikine and LPS elicited a transcriptional signature distinct from either stimulus alone (Figure 4C). Versikine-upregulated genes involved in DC maturation (interferon-stimulated genes such as Ifi209 and Ifi204), chemokines (Ccl7, Ccl2, Cxcl9, and Cxcl10), and co-stimulatory signals (Cd80 and Cd40) (Figure 4D; Table S5). Downregulated genes included components of transforming growth factor β (TGF-β) and Wnt pathways, both associated with immunosuppression (Conejo-Garcia et al., 2016). Gene set enrichment analysis (GSEA) confirmed upregulation of immune activation gene sets (e.g., interferon [IFN]-α response, IFN-γ signaling, nuclear factor κB [NF-κB]-induced tumor necrosis factor [TNF] signaling, and inflammation) and downregulation of immunosuppressive Wnt-β-catenin and TGF-β signaling (Figure 4E). Several of the top hits were confirmed by RT-PCR and, at the protein level, by ELISA (Figures S4B–S4E). Key hits were confirmed after exposure of MutuDC1940 cells to supernatants from HEK293 cells secreting versikine (Figure S4F). One of the top hits, Ccl7, has been shown recently to act as a cDC1 chemoattractant (Zhang et al., 2020). LLC-Vkine tumors expressed high bulk Ccl7 transcripts (Figure 4F), as did immunomagnetically separated CD11c+ cells from LLC-Vkine tumors (Figure 4F).
We then interrogated the functional consequences of versikine’s transcriptional program. To test versikine’s effect on T cell priming by cDC1s, we carried out antigen presentation assays using the OVA (ovalbumin) antigen system in conjunction with TCR-engineered OT-IT cells (Figure 4H). Vkine- and EV-MutuDC1940 cells were pulsed with SIINFEKL peptides and co-cultured with OT-I cells (Figures 4I–4K and S4G–S4I). Versikine alone more than doubled the percentage of primed OT-I cells secreting IFN-γ and interleukin-2 (IL-2) by flow cytometry, confirmed through IFN-γ ELISA of culture supernatants. Combining versikine with LPS further augmented T cell priming. Thus, versikine synergized with classical “danger” signals to maximize stimulatory DC antigen presentation and T cell priming.
cDC1 accumulation requires atypical innate lymphoid support
Although cDC1 activation by versikine was cell autonomous, cDC1 accumulation might still require supporting actors, such as natural killer (NK) cells (Bodder et al., 2020). To determine whether versikine regulates cDC1/NK cell cross-talk in vivo, we characterized CD11c+ DCs from primary versikine-replete versus EV tumors. Freshly explanted CD11c+ cells from LLC-Vkine tumors expressed higher levels of the NK regulators IL-23 (α subunit), IL-27 (p28 and EBI3 subunits), and IL-15 (Figure 5A). Therefore, versikine’s cDC1 activation program incorporated an NK cell-activating module. Versikine-exposed DCs demonstrated low IL-12α(p35) and IL-12β(p40) subunit expression but very high IL-23α expression. We speculate that IL-23α could combine with IL-12β(p40) secreted by other activated myeloid cells (e.g., macrophages) to form bioactive IL-23 heterodimers, similar to extracellular IL-12p70 generation (Gerber et al., 2021).
Figure 5. cDC1 accumulation requires innate lymphoid support.
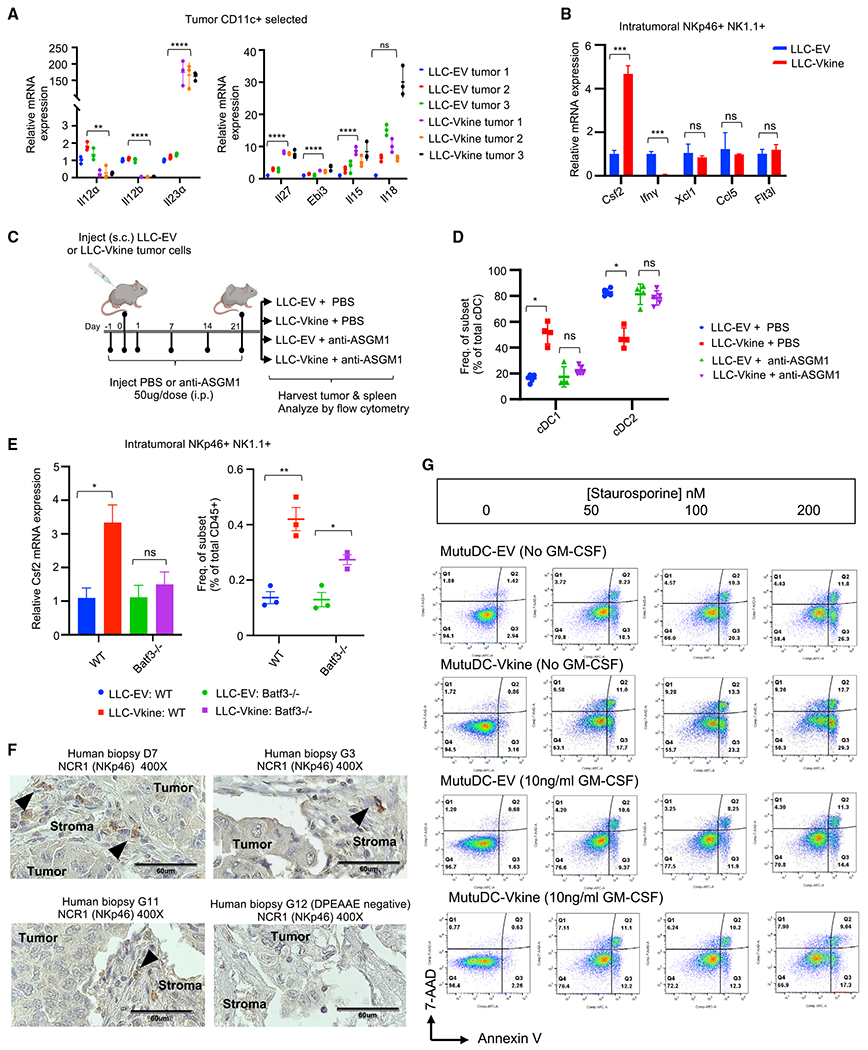
(A) RT-PCR for NK cell-activating cytokine transcripts expressed by ex vivo magnetically separated CD11c+ cells from LLC-EV and LLC-Vkine tumors.
(B) RT-PCR profile of NKp46+ NK1.1+ cells flow-sorted from LLC-EV and LLC-Vkine tumors. Data are presented as mean ± SEM, n = 3 for each group.
(C) Schematic of the NK cell depletion experiment.
(D) Summary of cDC subset frequency by flow cytometric analysis in LLC-EV versus LLC-Vkine tumors after treatment with NK cell-depleting antibody (anti-ASGM1) or vehicle (PBS).
(E) Csf2 (GM-CSF) RT-PCR of RNA extracted from NKp46+ NK1.1+ cells flow-sorted from LLC-EV and LLC-Vkine tumors growing in WT or Batf3−/− hosts.
(F) Stromal localization of NCR1+ (NKp46+) cells in human lung cancers (chromogen, DAB; counterstain, hematoxylin). 40× objective: scale bar, 60 μm.
(G) Annexin V/7-AAD apoptosis assay of MutuDC1940-EV or -Vkine dendritic cells exposed to graded staurosporine concentrations with or without murine GM-CSF.
Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. In vitro experiments were performed in technical triplicates. In vivo cohort sizes are shown in individual panels. All experiments were reproduced independently at least twice.
We hypothesized that versikine may trigger a distinct stroma-specific pattern of cDC1-NK cell communication (Peterson and Barry, 2020). Previous studies have implicated the NK cell-derived differentiation/survival mediator FLT3L as well as the chemo-attractants XCL1 and CCL5 in cDC1 support (Barry et al., 2018; Bottcher et al., 2018). NK cell-derived IFN-γ has been shown recently to induce the cDC1 “terminal selector” IRF8 (Lopez-Yglesias et al., 2021). NKp46+ NK1.1+ cells from LLC-Vkine tumors were potent expressors of Csf2 (granulocyte-macrophage colony stimulating factor [GM-CSF]) and relatively weak expressors of IFN-γ compared with the LLC-EV-derived counterparts (Figure 5B), whereas expression of Xcl1, Flt3l, and Ccl5 remained unchanged (Figure 5B). Versikine causes induction/expansion of an atypical NK cell subset expressing low IFN-γ despite high cytotoxicity receptor (NKp46) expression (Glasner et al., 2018) and robust Csf2 (GM-CSF), an essential survival factor for cDC1s (Greter et al., 2012). These results suggest that versikine engages innate lymphoid cells through a previously unreported mechanism in tumors. Versikine-induced NK cells are reminiscent of a recently reported spleen-resident ILC1-like subset that nurses cDC1s and promotes CD8+ T cell priming in viral infection (Flommersfeld et al., 2021).
To determine whether versikine - requires innate lymphoid cells to support cDC1 abundance in the TME, we used an anti-asialo-GM1 antibody (anti-ASGM1) for in vivo NK cell depletion (Figure 5C). Anti-ASGM1 antibody treatment (Figure S5A) completely abrogated versikine-mediated enhancement of cDC1s in the TME (Figures 5D and S5B).
Versikine regulates cDC1-NK cell cross-talk through cDC1s
The results so far suggested that versikine regulates cDC1s and NK cells in a virtuous circle but left open the question of whether versikine’s primary target is cDC1s or NK cells. To answer this question, we repeated the experiment in Batf3−/− mice (Hildner et al., 2008), which lack intratumoral cDC1s (Figure S5E). In Batf3−/− recipients, versikine was unable to induce Csf2 (GM-CSF) in intratumoral NKp46+ NK1.1+ cells despite a modest increase in NK cell frequency (Figure 5E). This result suggested that the primary target of versikine in the cDC1-NK cell cycle was cDC1s. This model would pre-suppose proximity of cDC1s and NKp46+ cells in human tumor stroma. NCR1 (NKp46+) cells in human tumors were almost exclusively stromal in their location (Figure 5F).
The evidence so far supports a model where versikine acts directly on stromal cDC1s to activate a pro-immunogenic program. Like other known cDC1 activators (e.g., TLR ligands), versikine-mediated activation simultaneously triggers a homeostatic propensity for apoptosis (Fuertes Marraco et al., 2011). This pro-apoptotic propensity is mitigated/rescued by NK cell-derived GM-CSF. The end result is accumulation of activated cDC1s (which we called stroma-licensed cDC1s) in stroma undergoing VCAN proteolysis. Consistent with this model, versikine’s MutuDC1940 signature incorporates a pro-apoptotic module (Figure S4A). Experimentally, we confirmed that versikine-expressing MutuDC1940 cells are sensitive to apoptotic signals and that this sensitivity is mitigated/rescued by exogenous murine GM-CSF (Figure 5G). In the TME, NK cells are dominant GM-CSF producers compared with sparser GM-CSF expressors, such as basophils (Figures S5C and S5D).
TLR2 and CD44 are dispensable for cDC1 accumulation in response to versikine
Our data are consistent with versikine signaling through a DC receptor whose identity is unknown. Although cDC1s do not robustly express TLR2, non-proteolyzed VCAN is thought to act through TLR2 expressed broadly by DC subsets (Kim et al., 2009; Tang et al., 2015). However, Tlr2 loss had no effect on versikine-induced cDC1 accumulation (Figures S5F–S5H).
We also considered the possibility that versikine may activate hyaluronan-dependent signaling pathways in vivo. VCAN’s N-terminal link domains bind hyaluronan (Figure 1A). We previously demonstrated recombinant versikine bioactivity in the absence of bound hyaluronan in vitro (Hope et al., 2016). Loss of Cd44, encoding the major hyaluronan receptor (Misra et al., 2015), did not affect versikine-induced cDC1 accumulation in vivo (Figures S5I–S5K).
Stroma-licensed cDC1s are “poised” and hypersensitive to nucleic acid sensing in vivo
Previous studies have highlighted the paradox of immunogenic DC accumulation along the tumor rim (Pai et al., 2020), but no compelling mechanism has emerged. We hypothesized that peritumoral stroma-licensed DCs are “poised” to respond to physiological maturation signals arising from necrotic tumor cells. Thus, versikine-induced CD40 expression and “stroma licensing” could serve to amplify these innate cancer-sensing signals (Borst et al., 2018). The cGAS/STING pathway, a sensor of exogenous double-stranded DNA, has emerged as a central mediator of innate sensing of tumors (Corrales et al., 2015). We reasoned that stroma-licensed DCs may respond to very low doses of a STING agonist (Figure 6A). Corrales et al. (2015) established dose-response relationships for the intratumorally administered murine STING agonist DMXAA: the maximum tolerated dose (MTD) was 500 μg; unacceptable toxicity was observed at higher doses (Corrales et al., 2015). LLC-EV and LLC-Vkine tumors were challenged with subtherapeutic doses of the STING agonist DMXAA (150–200 μg) or vehicle (NaHCO3). Tumor response curves are shown in Figure 6B and survival plots (Kaplan-Meier) in Figure 6C. EV tumors did not appreciably respond to vehicle or subtherapeutic doses of DMXAA (DMXAA200). In contrast, versikine lowered the response threshold to DMXAA so that versikine-replete tumors demonstrated a consistent response to single subtherapeutic DMXAA doses. Versikine-replete tumors routinely developed necrotic eschars by 24 h after subtherapeutic DMXAA injection (Figure 6D). In contrast, none of the control mice developed eschars within this time frame. To determine whether versikine - reduced the therapeutic threshold through a classic type I IFN response to DMXAA, we harvested tumors for RNA extraction 2 h after DMXAA administration. Versikine-replete tumors demonstrated a several-fold increase in IFN-α transcripts (particularly IFN-α2 and IFN-α4) and, to a lesser degree, IFN-β1 transcripts (Figure 6E). A list of up- and down-regulated genes is provided in Table S6. These results demonstrate that versikine - lowered the threshold for a classic type I IFN-mediated STING agonist response.
Figure 6. Stroma-licensed cDC1s are “poised” and hypersensitive to nucleic acid sensing in vivo.
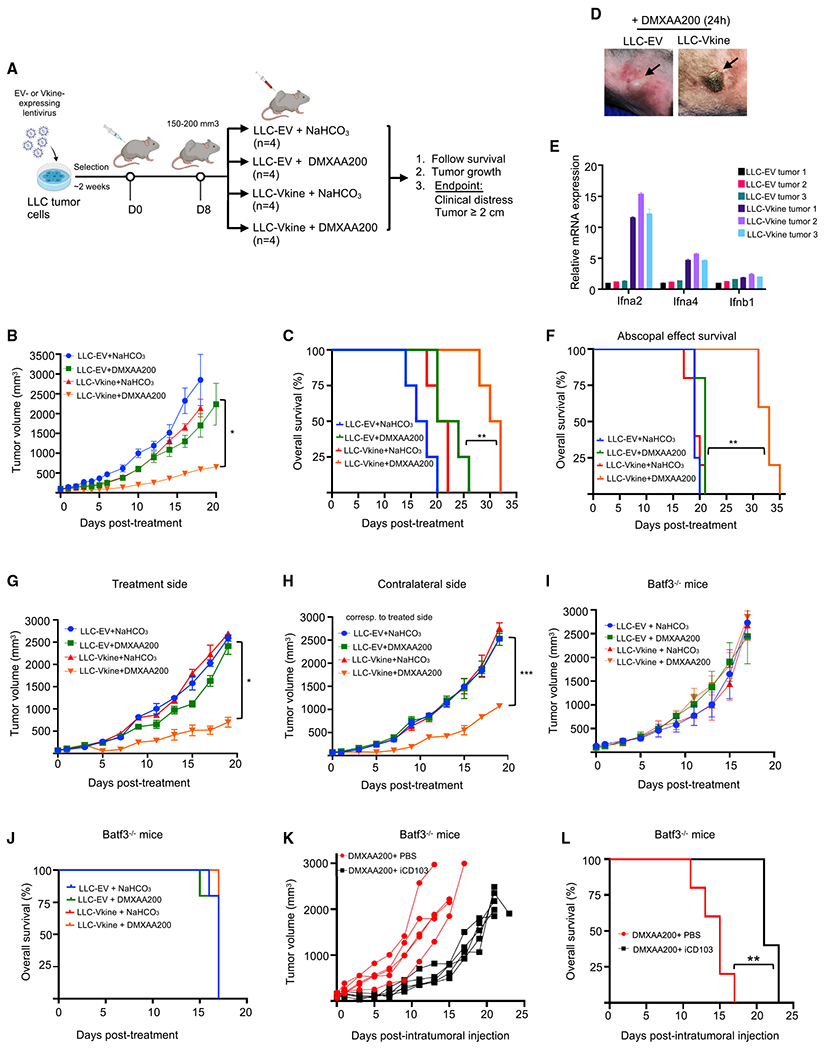
(A) Schematic of the experiment.
(B) Growth curves of LLC-EV and LLC-Vkine tumors challenged with a single subtherapeutic dose (200 μg) of intratumoral (IT) DMXAA (DMXAA200) or vehicle (NaHCO3) on day 0.
(C) Kaplan-Meier survival curves for the experiment in (B); **p < 0.01 by log rank test.
(D) Representative images showing development of hemorrhagic necrosis and a necrotic eschar in LLC-Vkine but not LLC-EV tumors 24 h after IT DMXAA200 administration.
(E) Transcriptomic profile of LLC-EV and LLC-Vkine tumors harvested 2 h after IT DMXAA200 (Table S6).
(F) Versikine -DMXAA synergy generates an abscopal effect in LLC tumors that produces a survival advantage. **p < 0.01 by log rank test.
(G) Growth curves of treatment-side LLC-EV and LLC-Vkine tumors challenged with a single subtherapeutic dose (200 μg) of IT DMXAA (DMXAA200) or vehicle (NaHCO3) on day 0.
(H) Growth curves of contralateral side unmanipulated LLC tumors; treated side as in (G).
(I) Response to DMXAA200 is lost in Batf3−/− recipients. Shown are growth curves of LLC-EV and LLC-Vkine tumors challenged with a single subtherapeutic dose (200 μg) of IT DMXAA (DMXAA200) or vehicle (NaHCO3) on day 0 in Batf3−/− recipients.
(J) Batf3 loss abrogates the survival advantage seen in the WT (C).
(K) Efficacy of DMXAA200 in LLC-Vkine tumors implanted into Batf3−/− recipients is restored after adoptive transfer of iCD103 (Figures S6D and S6E).
(L) Adoptive transfer of iCD103 in LLC-Vkine tumors implanted into Batf3−/− recipients restores the survival advantage of mice treated with DMXAA200. **p < 0.01 by log rank test.
Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. In vitro experiments were performed in technical triplicates. In vivo cohort sizes are shown in individual panels. All experiments were reproduced independently at least twice.
To determine whether low doses of the STING agonist could generate an abscopal effect, we examined mice bearing tumors implanted in both flanks. The treated side was inoculated with EV- or versikine-expressing LLC cells; the contralateral, non-treated side was inoculated with unmanipulated (untransduced) LLC cells. Ectopic versikine is bound in the pericellular halo (glycocalyx) (Figure 2D) and probably does not circulate to an appreciable degree. We observed a consistent abscopal effect when versikine-replete tumors were injected with 200 μg DMXAA (Figures 6F–6H). EV tumors treated with the same subtherapeutic dose failed to elicit any response on the treatment or contralateral side. STING agonist hypersensitivity produced consistent primary tumor and abscopal effects across genetic backgrounds; e.g., in the orthotopic 4T1 mammary carcinoma model (Figures S6A–S6C).
Our hypothesis that stromal matrikines render cDC1 hypersensitive to nucleic acid sensing in vivo predicts that DMXAA200 would be ineffective in the absence of cDC1s. Thus, we repeated the experiment delineated in Figure 6A in Batf3−/− recipients. DMXAA200 was globally ineffective in the Batf3-null background, and all survival benefit was lost (Figures 6I and 6J). To confirm that the responsible actors were cDC1s (rather than another Batf3-expressing lineage), we attempted to rescue the null phenotype with intratumoral adoptive transfer of iCD103, BM-derived primary cDC1-like cells generated in culture (Mayer et al., 2014; Figure S6D). MutuDC1940 cells cannot be used for adoptive transfer experiments because of their immunogenicity. We confirmed a consistent cDC1-like phenotype in iCD103 cells (Figure S6E). Adoptive transfer of iCD103 cells restored subtherapeutic STING agonist efficacy (Figure 6K) and survival benefits (Figure 6L). To confirm the findings in a different C57BL6/J model, we chose the B16 melanoma model. B16 tumors responded to subtherapeutic doses of DMXAA in the presence of versikine but not EV (Figures S6F and S6G). Efficacy was lost in the Batf3-null background (Figure S6H), but response to subtherapeutic doses of the STING agonists was restored, at least in a subset of mice, upon iCD103 adoptive transfer (iCD103 “take” in B16, an “immune-cold” tumor, was less efficient than in LLC, an “immune-hot” tumor; Lechner et al., 2013; Figure S6I). Therefore, stroma-licensed cDC1s are “poised” and hypersensitive to nucleic acid sensing in vivo.
Versikine promotes antigen-specific CD8+ T cell responses in vivo
We wanted to determine whether hypersensitivity of stroma-licensed cDC1s to DNA sensing translates into enhanced antigen-specific effector responses in vivo. To this end, we employed the OVA system as an in vivo model antigen (Figure 7A). EV- and versikine-expressing LLC cells were additionally engineered to express full-length OVA (LLC-OVA). EV- or versikine-replete LLC-OVA tumors were challenged with therapeutic DMXAA doses (500 μg). Five days after challenge, spleens were harvested and analyzed by flow cytometry for antigen-specific effector responses using an antigen-specific tetramer assay. Versikine more than doubled the magnitude of the antigen-specific response in the CD8+ T cell compartment, as determined by MHCI:SIINFEKL-tetramer staining (Figure 7B). Spleens contained a larger proportion of CD8+CD62L+CD44+ cells with a central memory phenotype (Figure S7A). The results demonstrate that stromal matrikines enhance antigen-specific CD8+ T cell effector responses in vivo.
Figure 7. Versikine promotes CD8+ responses and overcomes resistance to anti-PD1 inhibitors in vivo.
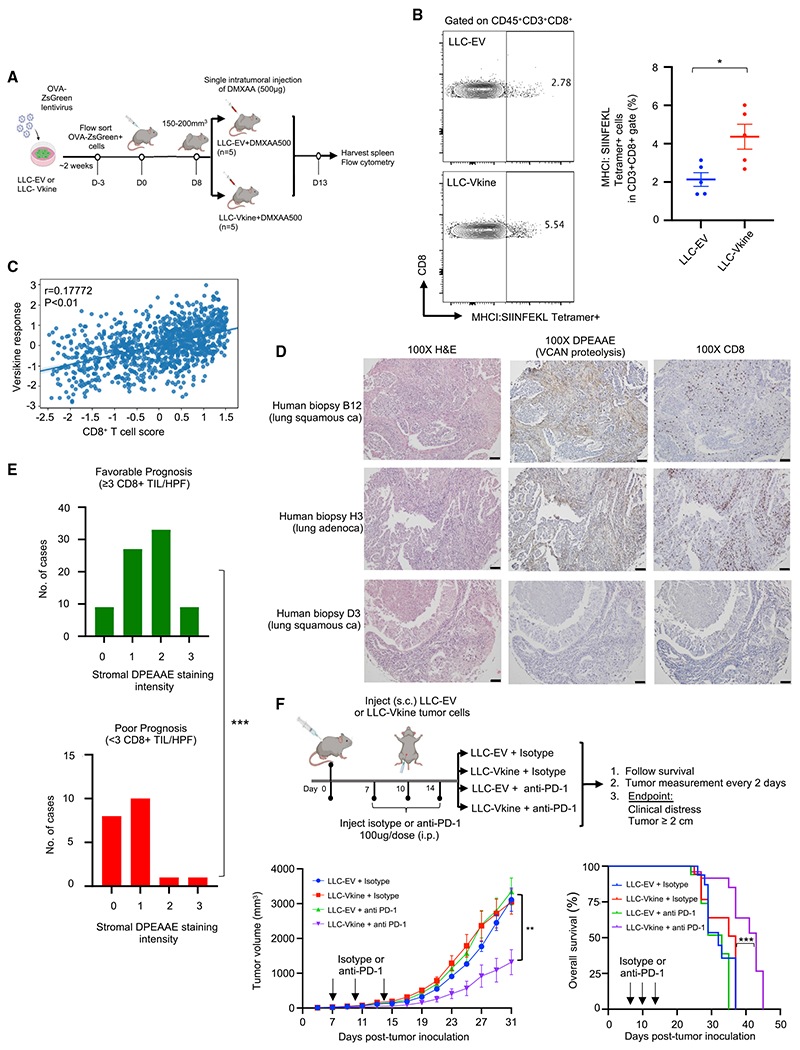
(A) Schematic of the experiment.
(B) Frequency of MHCI:SIINFEKL tetramer+CD8+ splenocytes in mice bearing LLC-EV versus LLC-Vkine tumors 5 days after challenge with a therapeutic dose of a STING agonist (DMXAA500). Data are presented as mean ± SEM.
(C) Correlation between in vitro versikine signature and CD8+ T cell scores across TCGA human lung cancers. Significance was measured using a linear model while accounting for total immune infiltration.
(D) DPEAAE staining in human lung cancers and associated CD8+ infiltration. 10× objective: scale bar, 50 μm.
(E) Distribution of DPEAAE stromal staining intensity across lung cancer prognostic subgroups (pauci-immune [poor prognosis] and immune-rich [favorable prognosis] at cutoff 3 CD8+ TILs/HPF). p < 0.001 by two-tailed Mann-Whitney test.
(F) Top: schematic of the experiment. Bottom: tumor growth rates and survival curves of LLC-EV and LLC-Vkine-bearing animals treated with 3 doses of anti-PD1 antibody or isotype control.
*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. In vitro experiments were performed in technical triplicates. In vivo cohort sizes are shown in individual panels. All experiments were reproduced independently at least twice.
The stroma-licensed cDC1 signature correlates with CD8+ T cell scores in human lung cancer
To correlate stromal cDC1 licensing with CD8+ T cell density in human cancers, we generated a unique cDC1 response signature from 200 genes whose expression was significantly altered in versikine-activated MutuDC1940 cells (Figures 4C and 4D). We then correlated this stroma-cDC1 signature with CD8+ T cell scores estimated in TCGA expression data for 1,017 lung cancers (STAR Methods). The results are shown in Figures 7C and S7B. We observed a significant but weak correlation between stroma-licensed cDC1 transcriptional profiles and CD8+ T cell scores. There were obvious limitations in this analysis (application of an in vitro murine cultured cell-generated signature to primary bulk human tumor data), but the results did support a connection between stromal cDC1 licensing and CD8+ T cell density in human cancer.
Stromal VCAN proteolysis correlates with CD8+ infiltration in human lung cancer
CD8+ T cell infiltration has prognostic significance in human lung cancer (Zeng et al., 2016) as well as predictive significance for efficacy of checkpoint inhibitor-based immunotherapy (Fumet et al., 2018). A cutoff of 3–5 CD8+ T cells/HPF has been used in some studies to designate CD8+ TIL-rich versus -poor tumors bearing a favorable and unfavorable prognosis, respectively (see individual studies referenced in a meta-analysis; Zeng et al., 2016). To determine whether stromal VCAN proteolysis was associated with prognostic immune infiltration groups in humans, we subdivided 98 non-small cell lung cancer (NSCLC) biopsies in our tissue microarray (TMA) into pauci-immune (0–2 CD8+ TILs/HPF in stromal and epithelial compartments, n = 26) and immune-rich (≥3 CD8+ TILs/HPF in stromal or epithelial compartments, n = 72). The distribution of stromal DPEAAE staining intensity (0, 1, 2, or 3, assessed by pathologist K.A.M.) was compared between the groups. The results are shown in Figures 7D, 7E, S7C, and Table S1. We found a statistically significant association between stromal VCAN proteolysis intensity and CD8+ T cell infiltration in human NSCLC.
The VCAN matrikine versikine overcomes resistance to anti-PD1 checkpoint inhibition immunotherapy in vivo
cDC1s are critical for responses to checkpoint inhibitors (Sanchez-Paulete et al., 2016). Thus, we tested the effect of versikine on anti-PD1 responses in the refractory LLC model (Figures 7F and S7D). Versikine sensitized at least a subset of LLC tumors to a short course of anti-PD1 checkpoint inhibition-based immunotherapy, resulting in enhanced animal survival.
DISCUSSION
Our data address several unsettled conundrums in tumor DC biology: first, the persistence of a rare subset of stimulatory DCs within the highly immune-suppressive TME of established tumors, characterized by abundant regulatory myeloid cells; second, the enigmatic topography of cDC1s along tumor margins distal to tumor nests; third, the mechanisms regulating T cell infiltration versus the T cell-excluded phenotypes, characterized by stalling of immune effectors within the peritumoral stroma; fourth, the clinical impetus to increase cDC1 density within tumors and promote infiltration by natural and engineered T cell and other immune effectors.
Activated cDC1s are preferentially located in the peritumoral matrix, distal to digitating tumor nestlets, where they are poised to interact with transiting CD8+ T cell effectors (Broz et al., 2014; Hubert et al., 2020; Mattiuz et al., 2021). The accumulation and retention of immunogenic DCs along the tumor rim presents a paradox that cannot be explained by simply invoking passive cDC1 displacement from tumor nests. The existing literature does not adequately explain why cDC1s are preferentially retained within peritumoral stroma and how they become “poised” to interact with T cells in that location. The mere persistence of stimulatory cDC1s in established tumors has been recognized as “counterintuitive” (Balan et al., 2020). Therefore, we took a tumor-extrinsic viewpoint and reasoned that the accumulation, persistence, and function of stimulatory DCs at the tumor periphery may be regulated by signals emanating from invasive margin matrix remodeling. In embryonic development, provisional matrix remodeling signals regulate cellular phenotypes and fates to guide tissue plane forging (Wight, 2017). This consideration brings matrix proteoglycans into sharp focus, in particular VCAN, a key player in tissue plane specification in embryonic and adult tissues (Fava et al., 2018; Islam and Watanabe, 2020; Nandadasa et al., 2014).
Our findings suggest that conserved, provisional matrix-remodeling signals exert homeostatic control of nascent tumors through regulation of peritumoral cDC1 abundance and activity. This notion challenges the general concept of tumor provisional matrix as pro-tumor (Yamauchi et al., 2018). Stroma-licensed peritumoral cDC1s promote T cell infiltration into tumor nests through re-priming, activation, and expansion of chemo-attracted T cell effectors within invasive margins. In small nascent tumors surrounded by stroma rich in remodeling provisional matrix signals (e.g., versikine), the process may well end in tumor elimination through cDC1 activation and T cell-mediated destruction (immunoediting stage: “elimination”; Mittal et al., 2014). Tumors that survive with persisting stromal-DC signaling may display T cell inflammation (immunoediting stage: “equilibrium”). Attenuation of stromal provisional matrix remodeling signals (e.g., TGF-β-mediated downregulation of ADAMTS proteases; Cross et al., 2005), progressive accumulation of non-proteolyzed VCAN (Tang et al., 2015), and eventual transition into fibrotic stroma (Mariathasan et al., 2018; Yamauchi et al., 2018) redresses the balance and results in effector cell stalling within the tumoral border and immune exclusion (immunoediting stage: “escape”).
The prevailing view in the literature casts peritumoral stroma overwhelmingly in the role of immune “barrier” (Joyce and Fearon, 2015), but our data support a more dynamic and fluid perspective. Thus, a nuanced distinction between “stimulatory stroma” (mimicking embryonic or wound healing provisional matrix remodeling) and “regulatory stroma” (akin to compact, fibrotic, collagen-rich matrix in development and wound healing) may permit design of accurate personalized immunotherapy approaches. Early evidence from the clinic demonstrates superior responses to checkpoint inhibition by tumors displaying robust VCAN proteolysis (Deming et al., 2020). Therapeutic repletion and redistribution of matrix-DC activation signals to the tumor core may be exploited to generate or potentiate a “hot” immune TME that sensitizes immune-evasive tumors to immunotherapy to promote antitumor responses and/or overcome resistance.
Limitations of the study
Murine implantable models do not replicate the orderly architecture of stromal sheets dividing epithelial nests found in human solid cancers because of acquisition of mesenchymal features from continuous passage (Guerin et al., 2020). Our data do not entirely discriminate between survival and recruitment as the mechanism of cDC1 support by NK cells in versikine-replete tumors, although we hypothesize that survival is dominant. Interpretation of the pre-DC differentiation assay (Figure S3) may be limited by harvesting of pre-DCs from B16-Flt3l tumor-bearing donors, a fact that could influence the differentiation potential of adoptively transferred pre-DCs. A fuller understanding of how versikine selectively targets cDC1s awaits identification of the putative versikine receptor.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Fotis Asimakopoulos (fotis@health.ucsd.edu).
Materials availability
Mouse lines and all other reagents reported in this paper are available from the lead contact upon execution of a Materials Transfer Agreement (MTA).
Data and code availability
RNAseq data have been deposited at GEO and are publicly available (accession GEO: GSE199938). Original western blot images appear in Supplementary Data. Microscopy data will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal strains and regulatory approval
C57BL/6J (JAX stock 000664), BALB/cJ (JAX stock 000651), B6.129 (Cg)-Cd44tm1Hbg/J (Cd44−/− JAX stock 005085), B6.129S(C)-Batf3tm1Kmm/J (Batf3−/−, JAX stock 013755), B6.129-Tlr2tm1Kir/J (Tlr2−/−, JAX stock 004650), C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-I, JAX stock 003831) and VQ mice (Wen et al., 2021) were housed, cared for, and used in accordance with the Guide for Care and Use of Laboratory Animals (NIH Publication 86-23) under IACUC-approved protocols #M5476 and #S19109 in the University of Wisconsin-Madison and University of California, San Diego respectively.
Generation of Vcan +/− mice using CRISPR-Cas9 gene editing
A mixture of two gRNA (25ng each, sgRNA#1 5′-ACTAGCCCGGAGTTTGACCA-3′, sgRNA#2 5′-ACCGATGTGATGTCATGTAT-3′) targeting mouse Vcan exon 3 and Cas9 protein (40ng; PNABio) was injected into the pronucleus of one-cell fertilized embryos from C57LB/6J females. Injected embryos were transferred into pseudo-pregnant females. Tail samples were taken at weaning, and the targeted region was characterized using targeted ultradeep sequencing. Briefly, the targeted region was PCR amplified using the following primers:
>207A.VCAN.ex2.F.1.6N.ILTS.1
acactctttccctacacgacgctcttccgatctNNNNNNACTGTCTTGGTGGCCCAGAAC.
>207A.VCAN.ex2.R.1.ILTS.1
gtgactggagttcagacgtgtgctcttccgatctTCTCTGGTACCATGCTGCCTTTC.
Samples were indexed & pooled, and the pool was sequenced on a MiSeq 2×250 Nano. Resultant sequences were quality filtered, trimmed, and analyzed with CRISPResso (Pinello et al., 2016). Founders were backcrossed to C57LB/6J mates, and F1s were characterized similarly.
For genotyping, DNA was extracted from mouse tail using genomic DNA extraction kit (Promega Wizard SV Genomic DNA Purification System, Catalog #: A2360), according to the manufacturer’s protocol. For PCR, Promega 2X GoTaq Master Mix (Catalog #: M7123), 1ul of template DNA and 10uM of each primer were used. The PCR conditions were 1min at 95°C followed by 35 cycles of 15 s at 95°C, 15 s at 60°C, 30 s at 72°C and a final extension of 1 min at 72°C. Primers for target sequences are listed on Table S7.
Cell lines and primary cell culture
Lewis Lung Carcinoma (LLC, ATCC CRL-1642) and B16-F10 melanoma (B16, ATCC CRL-6322) were cultured in complete DMEM medium (10-013 CV Corning DMEM with 10% fetal calf serum, 50μM 2-mercaptoethanol, 100U/mL Penicillin, 100μg/mL Streptomycin, 292ng/mL L-Glutamine). 4T1 mammary carcinoma cell line (4T1, ATCC CRL-2359) was cultured in complete RPMI (10-040 CV Corning RPMI 1640 with 10% fetal calf serum, 50μM 2-mercaptoethanol, 100U/mL Penicillin, 100μg/mL streptomycin, 10mM non-essential amino acids and 1M HEPES buffer). VQ4935 cells (Wen et al., 2021) were cultured in suspension in Iscove’s DMEM medium [10-016-CV Corning Iscove’s DMEM supplemented with 10% Fetal calf serum, 50μM 2-mercaptoethanol, 100U/mL penicillin, 100μg/mL streptomycin, 10mM non-essential amino acids and 10ng/mL IL-6 (Peprotech)]. Immortalized mouse dendritic cells (MutuDC1940, Applied Biological Materials Inc. #T0528) were cultured in Iscove’s DMEM medium supplemented with 10% fetal calf serum, 292ng/mL L-glutamine, 50μM 2-mercaptoethanol, 1% of 7.5% sodium bicarbonate (w/v). iCD103 in vitro differentiation was performed using bone marrow cells from female C57BL/6J mice at 6-12 weeks of age according to the protocol by Mayer and colleagues (Mayer et al., 2014). 15-16 days after start of the culture, DC were harvested, immunophenotyped and used for experiments.
Constructs
pLenti6-UbC-VKine-HA and pLenti6-UbC-VKine-Myc has been previously described (Hope et al., 2016; McCulloch et al., 2009). Ovalbumin (OVA) amplicon was PCR amplified from pcDNA3-OVA (Addgene #64599) and cloned into pHIV-Luc-ZsGreen backbone (Addgene #39196). All lentiviral constructs were transformed into NEB 5-alpha competent cells (#C2987U) for propagation of plasmid DNA. All plasmids were prepped and purified using Macharey-Nagel NucleoBond Xtra Maxi kit (# 740414.50).
Human lung cancer TMA
Human lung cancer TMA was commercially obtained from US Biomax (BC041115e). Fresh cuts were obtained and used for immunohistochemistry analysis.
METHOD DETAILS
Lentiviral transduction
HEK293T cells were transfected with a mixture of ps-PAX2 (packaging plasmid) and pVSV-G (envelope plasmid), and transfer plasmids encoding respective open reading frames or empty control. On Day 2 post-transfection, pseudotype virus-containing culture medium was harvested, filtered, supplemented with 7.5 μg/mL polybrene (Sigma-Aldrich), and immediately applied to target cells for spinfection (120min, 2500xg at 32C). After spinfection, the medium was exchanged for fresh complete RPMI1640 medium. Target cells were passaged at least three times after retroviral transduction.
Generation of HA-tagged versikine- and OVA-ZsGreen-expressing cell lines
LLC, 4T1, B16-F10 melanoma and MutuDC1940 cells were transduced with HA tagged versikine (Vkine) - or empty vector (EV)- containing lentivirus as detailed above. The cells were selected with 10μg/mL blasticidin for 2 weeks. HA-tagged versikine expression was confirmed by western blotting using anti-HA antibody (clone: C29F4, Cell Signaling). LLC- EV or -Vkine cell lines were transduced with pHIV-Luc-OVA-ZsGreen lentivirus. LLC-OVA expressing cells were FACS-sorted based on ZsGreen expression to ensure comparable transduction rates between different cell lines.
shRNA mediated VCAN knockdown
The lentiviral shRNA vector set targeting mouse Vcan (NM_019389.2) and scrambled control were purchased from GeneCopoeia (#MSH080253-LVRU6H and #CSHCTR001-LVRU6H). In brief, 2×105 LLC cells were plated per well in a 6-well plate and incubated overnight. Next day, 2mL freshly harvested lentiviral supernatant (expressing either shambled control, Vcan shRNA#1, #2 or #3), 1 mL of culture medium and 7.5μg/mL polybrene was added per well. The plate was centrifuged at 800g for 2h at 37°C and returned to CO2 incubator. After 72h, 200μg/mL Hygromycin B was added and the cells were under antibiotic selection for 2 weeks. Vcan knockdown was confirmed by RT-PCR as shown in Figure S1.
Tumor cell inoculations and tumor growth measurement
Cells were harvested by trypsinization and washed in PBS. Mice were under isoflurane anesthesia during tumor injections. 5x105 LLC cells were injected subcutaneously (s.c.) in 100μL endotoxin-free PBS on the flank of recipient mice. 105 4T1 cells were injected orthotopically in the mammary fat pad of the mice. Tumor growth was measured using a digital caliper. Tumor volumes were measured biweekly and estimated by using the formula: Tumor volume = length x (width) 2 divided by 2, where length represented the largest tumor diameter and width represented the perpendicular tumor diameter. Intratumoral injections were performed using a 28G insuilin syringe, when tumors had reached 100-150 mm3, using surgical forceps to hold the tumor constantly. For intravenous (i.v.) inoculations, we adopted a retro-orbital approach. Mice were anesthetized using inhaled isoflurane in a chamber. The eyeball was partially protruded from the socket by applying downward pressure to the skin dorsal and ventral to the eye. Injections were performed by placing the needle, bevel face-down, in order to decrease the likelihood of damaging the eyeball. Once the injection was complete, the needle was slowly and smoothly withdrawn. Triple antibiotic ophthalmic ointment was then applied to the eye.
Intraperitoneal injection was performed using a 28.5G insulin-syringe with the head tilted down. The needle was inserted at a 30° angle in the lower left or right quadrant. Transplantation of myeloma VQ4935 cells was performed via intracardiac injection after the 6-8 week old C57BL/6J recipient mice were sub-lethally irradiated at 6.0 Gy using an X-RAD 320 Irradiator. Intracardiac injection was performed by placement of needle in the fourth intercostal space and into the left ventricle. The needle was inserted at a 90° angle in the middle of the imaginary line connecting the sternal notch and xyphoid process serving as anatomical landmarks, and the needle was inserted slightly left of the sternum.
Anti-PD1 treatments
Recipient syngeneic mice (10 per arm) were injected with LLC-EV or LLC-Vkine cells (5 × 10^5 cells per inoculum). Antibody treatments were with 100 ug of antibody in 100 uL of volume each (aPD1: Bio X Cell InVivoPlus, rat IgG2a, clone RPM1-14, Cat# BP0146, Lot# 806321J2B; Isotype control: Bio X Cell InVivoPlus, rat IgG2a, clone 2A3, Cat# BE0089, Lot# 796721M2). Treatments were administered on days 7, 10, and 14 post-inoculation. Tumor burden was tracked by measuring tumors with an electronic caliper every two days, beginning with Day 5 post-inoculation. Mice reached endpoint when they were found dead, were in clear distress, or when tumors reached 20 mm in any dimension. Animals found dead were considered to have reached endpoint on the off-day of measurements.
Processing of tumor tissue
Unless stated otherwise, tumors were excised 21 days after transplantation. For subsequent analysis by flow cytometry, tumors were cut into pieces and digested with either Collagenase Ia (1mg/mL) C2674 Sigma Aldrich and Hyaluronidase V (0.1mg/mL) H6254 Sigma Aldrich for 40min at 37°C or with a mouse tumor dissociation kit (Miltenyi Biotec #130-096-730) using gentle MACS dissociator. Tissue was passed through a 70μm cell strainer (Falcon) and washed with FACS buffer (PBS with 1% FCS) before proceeding with antibody staining. For RNA isolation, homogenization was performed in RLT buffer (QIAGEN) facilitated by a closed tissue grinder system (Fisher brand #02-542-09, 15mL).
Mass cytometry
Tumor tissue was harvested and processed for mass cytometry analyses using the protocol described above for flow cytometry. After single cell suspensions were acquired, cells were washed with PBS, centrifuged at 300-400g for 5 minutes and supernatant was discarded by aspiration. Cells were resuspended in PBS and Cell-ID Cisplatin (Fluidigm, #201064) was added to a concentration of 5uM. After rigorous mixing, cells were incubated at room temperature for 5 minutes. Cells were then quench stained with MaxPar Cell Staining Buffer (Fluidigm, #201068) using 5× the volume of the cell suspension, centrifuged and supernatant was discarded by aspiration. The process was continued with surface staining. 50ul of the antibody cocktail was added to each tube so the total staining volume was 100ul (50ul of cell suspension+ 50ul antibody cocktail). Cells were stained for an hour at room temperature. All antibodies used for staining were either bought pre-conjugated to metal isotopes or were conjugated using the Maxpar Antibody Labelling Kit (Fluidigm, 201160B) (Table S7). Following incubation, cells were washed by adding 2mL Maxpar Cell Staining Buffer to each tube, then centrifuged at 300xg for 5 minutes and supernatant was removed by aspiration. This step was repeated for a total of 2 washes, and cells were resuspended in residual volume by gently vortexing after final wash/aspiration. Cells were then fixed with 1.6% FA solution and incubated at room temperature for 10 minutes. Finally, cells were labelled with Cell-ID Intercalator-Ir (Fluidigm, #201192A) at a final concentration of 125nM, incubated for an hour at room temperature and then analyzed on a Helios instrument (WB injector). All samples were resuspended in sufficient volume of 0.1 EQ beads (Fluidigm, #201078 by diluting one part beads to 9 parts Maxpar Cell Acquisition (CAS) solution.
Analysis of mass cytometry data using viSNE
To visualize the immune contexture, the immune milieu of the tumor (CD45+) was enriched by manual gating among single events, equally subsampled to 6,000 events, then run through a Barnes Hut implementation of the t-SNE algorithm, viSNE, in the R package ‘Rtsne’, using optimized parameters (iterations:1000, perplexity:30, learning rate:455). All markers listed in Table S2 to characterize the myeloid and lymphoid linages were selected for viSNE, excluding CD45.
Flow cytometry and fluorescence-activated cell sorting
Flow cytometric analyses were performed using an LSR II and/or LSR Fortessa X20. Data were analyzed using FlowJo (Tree Star). DAPI (0.5 mg/mL, Sigma-Aldrich) or a Live/Dead fixable cell stain (Ghost 780 Tonbo Biosciences) was used to exclude dead cells in all experiments, and anti-CD16/CD32 antibody (2.4G2) was used to block non-specific binding of antibodies via Fc-receptors. Flow cytometry antibodies are listed in Table S7. Quantification of total cell numbers by flow cytometry was done using fluorescent beads (Biolegend Precision beads). For intracellular staining of IFNγ and IL-2 in vitro, cells were treated with Golgi Plug (Brefeldin A 500×) and were collected 4h later. Intracellular staining was performed in permeabilization buffer (eBioscience) for 30min and cells were subsequently analyzed by flow cytometry. Sorting of tumor cells after retroviral transduction was done using a BD FACSAria or a BD FACSAria Fusion. Purity of cell populations was determined by re-analysis of a fraction of sorted cell samples.
Generation of iCD103 in vitro
1.5x106 BM cells were cultured in 10mL RPMI1640 medium supplemented with 10% heat-inactivated FCS (Biochrom), penicillin/streptomycin and 50μM β-mercaptoethanol. Recombinant human FLT3L (300-19, Peprotech) and recombinant murine GM-CSF (315-03, Peprotech) were added at day 0 of the culture. 5mL complete medium was added between day 5 and day 6 to minimize apoptosis. Non-adherent cells were harvested on day 9, counted and re-plated at 3x106cells in 10mL complete medium supplemented with FLT3L and GMCSF as on day 0. Non-adherent iCD103 were harvested on days 15-16. Cells were then validated by assaying for CD103, CD24, Clec9A, and CD11c by flow cytometry.
ELISA
MutuDC1940 cells were left unstimulated or were in vitro stimulated with LPS for 8 or 24 hours at 37°. Cell-free supernatant was assessed for CXCL9 (R&D Quantikine mouse CXCL9 #MCX900) and IL27p28 (R&D Quantikine mouse IL27p28 #M2728) protein levels by ELISA according to the manufacturer’s instructions (R&D). For the antigen-presentation assay, cell-free supernatants were collected and assessed for IFNγ levels (R&D Quantikine mouse IFNγ #P233156).
Immunoblotting
Whole-cell lysates were prepared by boiling cells in Laemmli Sample Buffer (Bio-Rad) supplemented with 100 mM DTT for 10 min at a final concentration of 107 cells per milliliter. A total of 105 cells or 20 mg protein was resolved by SDS-PAGE and transferred to Immobilon-P PVDF membranes (Millipore). Membranes were blocked in 5% milk in TBS-T (25 mM Tris-HCl [pH 7.4], 0.13 M NaCl, 2.7 mM KCl). Primary antibodies (anti-HA [C29F4; Cell Signaling Technologies], anti-DPEAAE [PA1-1748A; Thermo]) were diluted in 5% milk-TBS-T, and membranes were incubated overnight at 4°C. Secondary Ab–HRP conjugate, as well as anti-GAPDH–HRP conjugate (A00192; GenScript), incubations were carried out for 1 h at room temperature. Signal detection was achieved using Amersham ECL.
Immunohistochemistry
Paraffin-embedded murine tumor sections and unstained 4-5 μm-thick human lung carcinoma TMA (US Biomax Inc., BC041115e) sections were deparaffinized and rehydrated using standard methods. Antigen retrieval was carried out in citrate buffer, pH 6.0 (Vector Laboratories, #H-3300) for DPEAAE and HA; and pH 8.0 for XCR1 and CD8 (Abcam, ab93680). Primary antibodies included αDPEAAE (PA1-1748A, Thermo Fisher), anti-HA (C29F4, Cell Signaling Technology), anti-XCR1 (D2F8T, Cell Signaling Technology), anti-CLEC9A (ab223188, Abcam), anti-NCR1 (NKp46) (MAB1850, R&D) and anti-CD8 (C8/144B, Ebioscience). The αDPEAAE neoepitope antibody has been previously validated (Foulcer et al., 2015). Stained slides were examined using an Echo Revolve microscope with attached digital camera. αDPEAAE immunostaining score was assessed (by pathologist KAM) by scoring staining intensity (0 for no staining, 1 for low/weak staining, 2 for moderate staining and 3 for strong/intense staining) as previously described (Hope et al., 2017).
Dual staining XCR1/CLEC9A on human tonsil was performed by the UW TRIP lab as follows: The experiment was run on Roche Ventana Medical System’s Discovery Ultra Automated platform. Deparaffinization was carried out on the instrument, as was heat-induced epitope retrieval in the form of “cell conditioning” with CC1 buffer (Ventana #950-500), a Tris based buffer pH 8.4 for approximately 56 minutes at 95°C. Slide was incubated with the first primary antibody XCR1 diluted 1:40 in DaVinci Green antibody diluent (BioCare Medical #PD900H) for 60 min at 37°. Slide was rinsed with Reaction Buffer (Ventana #950-300) for the preset time and duration. Discovery OmniMap anti-Rabbit HRP (Ventana #760-4311) was applied for 16 min at 37°. Slide was rinsed with Reaction Buffer (Ventana #950-300) for the preset time and duration. Discovery ChromoMap DAB detection (Ventana #760-159) was applied for the preset time. Denaturing agent in the form of Discovery Inhibitor (Ventana #760-4840) was applied for the preset time. Slide was incubated with the second primary antibody CLEC9A diluted 1:50 in Ventana antibody diluent with casein (Ventana #760-219) for 60 min at 37°. Slide was rinsed with Reaction Buffer (Ventana #950-300) for the preset time and duration. Discovery OmniMap anti-Rabbit HRP (Ventana #760-4311) was applied for 16 min at 37°. Slide was rinsed with Reaction Buffer (Ventana #950-300) for the preset time and duration. Discovery Teal HRP detection kit (Ventana #760-247) was applied for 32 minutes. Slide was removed from the instrument and rinsed with dawn dishsoap and warm tap water followed by rinsing with dH2O. Slide was counterstained with Harris hematoxylin (1:5 diluted in dH2O) for 45 seconds. Slide was rinsed with dH2O. Slide was dehydrated in the oven (60 degrees C) followed by dipping in Xylene. Slide was coversliped.
Triple staining DPEAAE/XCR1/CD8+ on lung cancer TMA was performed by the UW TRIP lab as follows: The experiment was run on Roche Ventana Medical System’s Discovery Ultra Automated platform. Deparaffinization was carried out on the instrument, as was heat-induced epitope retrieval in the form of “cell conditioning” with CC1 buffer (Ventana #950-500), a Tris based buffer pH 8.4 for approximately 56 minutes at 95°C. Slide was incubated with the first primary antibody XCR1 diluted 1:25 in DaVinci Green antibody diluent (BioCare Medical #PD900H) for 60 min at 37°. Slide was rinsed with Reaction Buffer (Ventana #950-300) for the preset time and duration. Discovery OmniMap anti-Rabbit HRP (Ventana #760-4311) was applied for 16 min at 37 degrees. Slide was rinsed with Reaction Buffer (Ventana #950-300) for the preset time and duration. Discovery ChromoMap DAB detection (Ventana #760-159) was applied for the preset time. Denaturing agent in the form of Discovery Inhibitor (Ventana #760-4840) was applied for the preset time. Slide was incubated with the second primary antibody DPEAAE diluted 1:800 in Ventana antibody diluent with casein (Ventana #760-219) for 28 min at 37 degrees. Slide was rinsed with Reaction Buffer (Ventana #950-300) for the preset time and duration. Discovery OmniMap anti-Rabbit HRP (Ventana #760-4311) was applied for 16 min at 37 degrees. Slide was rinsed with Reaction Buffer (Ventana #950-300) for the preset time and duration. Discovery Teal HRP detection kit (Ventana #760-247) was applied for 32 minutes. Denaturing agent in the form of Discovery Inhibitor (Ventana #760-4840) was applied for the preset time. Slide was incubated with the third primary antibody CD8 (pre-diluted ready to use) for 16 min at 37 degrees. Slide was rinsed with Reaction Buffer (Ventana #950-300) for the preset time and duration. Discovery OmniMap anti-Rabbit HRP (Ventana #760-4311) was applied for 16 min at 37 degrees. Slide was rinsed with Reaction Buffer (Ventana #950-300) for the preset time and duration. Discovery Purple HRP detection kit (Ventana #760-229) was applied for 16 minutes. Slide was removed from the instrument and rinsed with dawn dishsoap and warm tap water followed by rinsing with dH2O. Slide was counterstained with Harris hematoxylin (1:5 diluted in dH2O) for 45 seconds. Slide was rinsed with dH2O. Slide was dehydrated in the oven (60 degrees C) followed by dipping in Xylene. Slide was coversliped.
An Olympus BX43 microscope with 40X objective (400X magnification) was used for data points reported as per “high power field”.
Imaging and morphometric analysis of XCR1+CLEC9A colocalization
Imaging of a human tonsil slide was performed on a Vectra 2 multispectral scanner (Akoya Biosciences). The stained slide was then loaded onto the instrument and 24 8-bit Bright Field 20X images were acquired for analysis. Antibodies against human XCR1 and CLEC9A are listed in the Key Resources Table. A customized spectral library algorithm for all chromogens and counterstain was created using Nuance Software version 3.0.2 (PerkinElmer). The inForm software version 2.4.7 was used to segment tissue subcellular compartments (nucleus, cytoplasm or membrane), and to measure biomarker expression. The double positivity algorithm was used to measure colocalization of cells expressing XCR1 and CLEC9A as double-positive percentage rate.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| HA-Tag (C29F4) | Cell Signaling | 3724S; RRID:AB_1549585 |
| DPEAAE | Thermo Fisher | PA1-1748A; RRID:AB_2304324 |
| XCR1 (D2F8T) | Cell Signaling | 44665S; RRID:AB_2799269 |
| CLEC9A | Abcam | ab223188;RRID:AB_2884022 |
| CD8 (C8/144B) | eBioscience | 14-0085-80; RRID:AB_11151339 |
| c-myc (9E10)-chimeric | Novus Bio | NBP2-52636; RRID:AB_962987 |
| GAPDH– HRP | GenScript | A00192 |
| aPD-1 rat IgG2a (RPM1-14) | Bio X Cell InVivoPlus | BP0146; RRID:AB_10949053 |
| Isotype control (2A3) | Bio X Cell InVivoPlus | BE0089; RRID:AB_1107769 |
| NKp46/NCR1 | R&D | MAB1850; RRID:AB_2149153 |
| Anti-Asialo-GM1 | Wako Chemicals | NC1345696 |
| Flow cytometry antibodies are listed in Table S7 (sheet 1) | Table S7 (sheet 1) | Table S7 (sheet 1) |
| CyTOF antibodies are listed in Table S7 (sheet 3) | Table S7 (sheet 3) | Table S7 (sheet 3) |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| pcDNA3-OVA | Addgene | 64599 |
| pHIV-Luc-ZsGreen | Addgene | 39196 |
| 5-alpha competent cells | New England Biolabs | C2987U |
| psPAX2 | Addgene | 12260 |
| pCMV-VSVg | Addgene | 8454 |
| shRNA clone against mouse Vcan | GeneCopoeia | MSH080253-LVRU6H |
| shRNA scrambled control clone for psi-LVRU6H | GeneCopoeia | CSHCTR001-LVRU6H |
|
| ||
| Biological samples | ||
|
| ||
| Human Lung Cancer TMA | US Biomax | BC041115e |
| Mouse subcutaneous tumor | This manuscript | N/A |
| Mouse lung | This manuscript | N/A |
| Mouse spine | This manuscript | N/A |
| Mouse long bones | This manuscript | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Ghost Dye 780 | Tonbo Biosciences | 13-0865 |
| DAPI | Sigma-Aldrich | D9542 |
| Collagenase Ia | Sigma-Aldrich | C2674 |
| Hyaluronidase V | Sigma-Aldrich | H6254 |
| Recombinant murine IL-6 | Peprotech | 216-16 |
| Recombinant murine GM-CSF | Peprotech | 315-03 |
| Recombinant human FLT3L | Peprotech | 300-19 |
| Chondroitinase ABC from Proteus vulgaris | Sigma-Aldrich | C2905 |
| Methanol | Fisher Scientific | A452-4 |
| TBS 10x | Corning | 46-012-CM |
| Tween20 | Sigma-Aldrich | P1379 |
| Clear-Rite 3 | Epredia | 6901 |
| 10x Tris/Glycine Buffer | Biorad | 1610734 |
| 10x Tris/Glycine/SDS Buffer | Biorad | 1610732 |
| Citrate-based Antigen Unmasking Solution | Vector Laboratories | H-3300 |
| Isoflurane USP | Midwest Vet Supply | NDC 13985-528-60 |
| 2-mercaptoethanol | Gibco | 21985-023 |
| DTT | Thermo Fisher | R0861 |
| 16% Formaldehyde Solution(w/v) | Thermo Scientific | TD264520 |
| Polybrene | Sigma-Aldrich | TR1003 |
| Hygromycin B | InvivoGen | ant-hg-1 |
| Blasticidin | InvivoGen | ant-bl-1 |
| LPS, E.Coli 0111:B4 | Sigma-Aldrich | LPS25 |
| DMXAA | InvivoGen | tlrl-dmx |
| Cell-ID Cisplatin | Fluidigm | 201064 |
| Cell-ID Intercalator-Ir | Fluidigm | 201192A |
| EQ beads | Fluidigm | 201078 |
| Staurosporine | Selleckchem | S1421 |
| OVA peptide 257-264 (SIINFEKL) | GenScript | RP10611 |
| Glycine 1M Solution | Sigma-Aldrich | 67419-1ML-F |
| UltraPure 0.5M EDTA | Invitrogen | 15575-038 |
| ECL Western Blotting Detection Reagents | Amersham | RPN2106 |
| Glutaraldehyde EM Grade 8% | Electron Microscopy Sciences | 2912.19.5000 |
| Cell Activation Cocktail with Brefeldin A | Biolegend | 423303 |
| Blotting grade blocker | Biorad | 1706404 |
| Bovine Serum Albumin (BSA) | Fisher bioreagents | BP 9703-100 |
| Isopropyl Alcohol | Sigma-Aldrich | PX1835-2 |
| DMSO | Sigma-Aldrich | D2650 |
| Phusion High Fidelity Polymerase | New England Biolabs | M0530S |
|
| ||
| Critical commercial assays | ||
|
| ||
| Wizard SV Genomic DNA Purification System | Promega | A2360 |
| GoTaq Master Mix | Promega | M7123 |
| RNAeasy Mini Kit | Qiagen | 74104 |
| Tissue Grinder System | Fisher Brand | 02-542-09 |
| NucleoBond Xtra Maxi kit | Macherey-Nagel | 740414.50 |
| Maxpar Antibody Labelling Kit | Fluidigm | 201160B |
| APC Annexin V Apoptosis Detection Kit with 7AAD | Biolegend | 640930 |
| Intracellular Fixation and Permeabilization Buffer | eBioscience | 88-8824-00 |
| AMPure XP | Beckman Coulter | A63880 |
| NEBNext® Ultra™ RNA Library Prep Kit for Illumina® | New England Biolabs | E7530L |
| NEBNext® RNA First Strand Synthesis Module | New England Biolabs | E7771 |
| Mouse Tumor Dissociation Kit | Miltenyi Biotec | 130-096-730 |
| Precision Count Beads | Biolegend | 424902 |
| RT2 Profiler PCR Array | Qiagen | PAMM-021Z |
| RT2 SYBR® Green qPCR Mastermix | Qiagen | 330529 |
| iSCript Reverse Transcription Supermix | Biorad | 1708840 |
| RT2 SYBR® Green qPCR Mastermix | Biorad | 330529 |
| SsoAdvanced Universal SYBR Green Supermix | Biorad | 1725272 |
| Quantikine mouse CXCL9 | R&D | MCX900 |
| Quantikine mouse IL27p28 | R&D | M2728 |
| Quantikine mouse IFNγ | R&D | P233156 |
| ToxinSensor Gel Clot Endotoxin Assay Kit | Genscript | L00351 |
| USER® Enzyme | New England Biolabs | M5508 |
| TruSeq PE Cluster Kit v3-cBot-HS | Illumina | PE-401-3001 |
|
| ||
| Deposited data | ||
|
| ||
| Raw and analyzed data | This manuscript | GEO: GSE199938 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Lewis Lung Carcinoma (LLC) | ATCC | CRL-1642 |
| B16F10 Melanoma | ATCC | CRL-6322 |
| 4T1 Mammary carcinoma | ATCC | CRL-2359 |
| MutuDC1940 | Applied Biological Materials | T0528 |
| VQ4935 | Wen et al., 2021 | N/A |
| HEK293T myc-versikine cells | Suneel S. Apte gift | N/A |
| HEK293T cells | ATCC | CRL-3216 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6J | The Jackson Laboratory | 000664 |
| Mouse: BALB/cJ | The Jackson Laboratory | 000651 |
| Mouse: B6.129 (Cg)-Cd44tm1Hbg/J | The Jackson Laboratory | 005085 |
| Mouse: B6.129S(C)-Batf3tm1Kmm/J | The Jackson Laboratory | 013755 |
| Mouse: B6.129-Tlr2tm1Kir/J | The Jackson Laboratory | 004650 |
| C57BL/6J-Tg(TcraTcrb), (OT-I) | The Jackson Laboratory | 003831 |
| VQ mice | Wen et al., 2021 | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| Oligonucleotide qPCR primers are listed in Table S7 (sheet 2) | Table S7 (sheet 2) | Table S7 (sheet 2) |
| Oligonucleotide genotyping primers are listed in Table S7 (sheet 4) | Table S7 (sheet 4) | Table S7 (sheet 4) |
|
| ||
| Recombinant DNA | ||
|
| ||
| pLenti6-UbC-VKine-HA | Hope et al., 2016 | N/A |
| pLenti6-UbC-VKine-Myc | McCulloch et al., 2009 | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo 10.7.1 | Tree Star | N/A |
| Graph Pad Prism Version:9.3.1 (350) | Dotmatics | N/A |
| Biorender | Biorender | N/A |
| SnapGene 6.0 | Dotmatics | N/A |
| OmniGraffle 7 | Omni Group | N/A |
| RT2 Profiler Data analysis | Qiagen | N/A |
|
| ||
| Other | ||
|
| ||
| Iscove’s DMEM medium | Corning | 15-016-CV |
| DMEM medium | Corning | 10-013-CV |
| RPMI 1640 medium | Corning | 10-040-CV |
| PBS | Corning | 21-040-CV |
| HEPES buffer 1M | Gibco | 15630-080 |
| Fetal Calf Serum One Shot | Gibco | A38400-01 |
| Antibiotic-antimycotic | Corning | 30-004-CI |
| Sodium Bicarbonate 7.5% (w/v) | Corning | 25-035-CI |
| MEM Non-essential amino acids | Gibco | 11140-050 |
| Glutamax | Gibco | 35050-061 |
| 0.25% Trypsin | Corning | 25-053-CI |
| BD Insulin Syringe | BD Insulin Syringe | 329461 |
| 5mL Polystyrene Round-Bottom Tube | Falcon | 352235 |
| Dual Chamber Cell Counting Slides | Biorad | 145-0011 |
| gentleMACS C Tubes | Miltenyi Biotec | 130-093-237 |
| MaxPar Cell Staining Buffer | Fluidigm | 201068 |
| MaxPar Fix and Perm Buffer | Fluidigm | 201067 |
| MaxPar Cell Acquisition Solution | Fluidigm | 201244 |
| Trypan Blue Stain (0.4%) | Gibco | 15250-061 |
| Red Blood Cell Lysing Buffer Hybri-Max | Sigma-Aldrich | R7757 |
| Mini-PROTEAN TGX Stain-Free Gels | Biorad | 4568096 |
| 2x Laemmli Sample Buffer | Biorad | 1610737 |
| Immobilon®-P PVDF Membrane | Millipore Sigma | IPVH15150 |
| PES 0.45 μm Filter | Cell Treat | 229748 |
| Sartorius Vivaspin 20 | Sartorius | VS2001 |
| Precision Plus Protein Dual Color Standards | Biorad | 161-0374 |
| Brilliant Stain Buffer | BD Biosciences | 563794 |
| UltraComp eBeads | Invitrogen | 01-2222-41 |
RNA isolation and quantitative real-time PCR
RNA was isolated using QIAGEN RNeasy Mini Kit and cDNA was synthesized using the iScript Reverse Transcription Supermix (Biorad). Quantitative real-time (qRT-PCR) analysis was performed using SsoAdvanced Universal SYBR Green Supermix (Biorad #1725272) according to the manufacturer’s instructions on an CFX96 Touch Real Time PCR detection (Biorad) using the relative standard curve method. PCR conditions were 2min at 50°C, 10min at 95°C followed by 40 2-step cycles of 15 s at 95°C and 1 min at 60°C. Primers for the targets listed in Table S7 as well as SDHA for normalization control, were used to assess relative gene expression.
For DMXAA-response analysis, RT2 Profiler PCR Array (QIAGEN, Cat. no. PAMM-021Z) was used. In brief, RNA was isolated from tumors and was reverse transcribed using kits mentioned above. cDNA was mixed with RT2 SYBR® Green qPCR Mastermix (Cat. no. 330529). The mixture was aliquoted across the RT2 Profiler PCR Array (in 96-well format) and was run on the Real Time PCR machine. Data analysis was performed using the manufacturer’s online platform for RT2 Profiler Data analysis software.
Generation of versikine cell culture supernatant
5x106 HEK293 and HEK-Vkine expressing cells (a kind gift of Dr. Suneel S. Apte, Cleveland Clinic Lerner Research Institute) were seeded in T-175 cell culture flasks and cultured in DMEM 10% FBS media. After 75 to 80% confluency, the cell media was changed to DMEM 1% FBS media. Subsequently, media supernatant was collected after 48 hours of incubation. The collected supernatant was centrifuged to remove debris and filtered with 0.45μ filter. The filtered supernatant was then concentrated 30 times to the initial volume using Sartorius Vivaspin 20, 10,000 MWCO PES concentrator (Cat. No. VS2001). Endotoxin assay was performed using Genscript ToxinSensor Gel Clot Endotoxin Assay Kit (Cat. No. L00351) according to the manufacturer’s instructions to rule out contamination. The presence of versikine in concentrated supernatant was confirmed using western blot using c-Myc Antibody (Novus Bio-c-Myc Antibody (9E10) - Chimeric NBP2-52636). Concentrated supernatant containing versikine was then used to treat MutuDC1940 cells. 2 × 105 MutuDC1940 cells per well were seeded in 12 well plate. The following day, 10% supernatant was added to the plate media. Cells were then incubated for 72 hours. After incubation, total RNA was extracted from MutuDC1940 cells.
Library preparation for RNA-seq
A total amount of 1 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 150–200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then 3 μL USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using PE Cluster Kit cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina platform and paired-end reads were generated.
RNA-seq data analysis
Raw data (raw reads) of FASTQ format were firstly processed through fastp. In this step, clean data (clean reads) were obtained by removing reads containing adapter and poly-N sequences and reads with low quality from raw data. At the same time, Q20, Q30 and GC content of the clean data were calculated. All the downstream analyses were based on the clean data with high quality. Reference genome and gene model annotation files (GRCm38) were downloaded from genome website browser (NCBI/UCSC/Ensembl) directly. Paired-end clean reads were aligned to the reference genome using the Spliced Transcripts Alignment to a Reference (STAR) software (v2.6.1d). FeatureCounts (v1.5.0-p3) was used to count the read numbers mapped of each gene. RPKM of each gene was calculated based on the length of the gene and reads count mapped to this gene. Differential expression analysis between two conditions/groups (three biological replicates per condition) was performed using DESeq2 R package (v1.20.0). DESeq2 provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting p values were adjusted using the Benjamini and Hochberg’s approach for controlling the False Discovery Rate (FDR). Genes with an adjusted p value <0.05 found by DESeq2 were assigned as differentially expressed.
Gene set enrichment analysis
Gene Set Enrichment Analysis (Subramanian et al., 2005) was performed by comparing MutuDC1940-Vkine (treated with PBS, 4h) RNA-seq data to the corresponding MutuDC1940-EV sample. 4736 differentially expressed gene features for each condition were ranked by the signal to noise metric of GSEA and the analysis was performed using the standard weighted enrichment statistic against human gene sets contained in the Molecular Signatures Database (MSigDB v7.4) that included all (H) Hallmark gene sets, (C2) curated gene sets, and (C3) motif gene sets. The normalized enrichment score (NES) was calculated using 1000 gene set permutations.
NK cell depletion in vivo
For depletion of NK cells, mice were injected i.p. with 50 ug of anti-asialoGM1 (Wako Pure Chemical Industries, 100μL/mouse) on days −1, 0, 7, 14 around tumor inoculation.
MutuDC1940 apoptosis assay
2×106 MutuDC1940-EV or -Vkine cells were plated per well in a 6-well plate and allowed to attach overnight at 37C in 5% CO2 incubator. Next day, cells were pretreated with 10ng/mL murine GM-CSF (Peprotech, catalog no # 315-03) for 6 h, followed by staurosporine [(AM-2282), Selleckchem, Catalog no. S1421] treatment at indicated doses for 24h. Next day, for apoptosis assay, cells and medium were harvested, washed twice with cold PBS and then resuspended in Annexin-V binding buffer. The cell suspension was stained with APC Annexin V and 7-AAD viability dye according to manufacturer’s instructions (APC Annexin V Apoptosis Detection Kit with 7-AAD, Biolegend, catalog #640930). Each sample was analyzed by flow cytometry with proper machine settings.
Antigen presentation assay
MutuDC1940 were cultured and treated with LPS or PBS control respectively overnight. Next day, the cells were harvested and plated on 96-well round bottom plates at a density of 100,000 cells per plate. DC were then loaded with OVA peptide 257-264 (SIINFEKL) (3ng/mL) and incubated for 4 hours at 37°C. MutuDC1940 were then washed with 0.1% PBS-BSA and centrifuged at 800 × g and were fixed with 50μL per well of freshly made PBS-glutaraldehyde (GTA) 0.008% (vol/vol) and incubated for 5 minutes on ice. 50μL of PBS-glycine 0.4M was added to the PBS-GTA 0.008% solution and cells were centrifuged at 800 × g for 2 minutes at 4°C. Plates were subsequently flicked. Finally, 100 μL of PBS-glycine 0.2M was added to each well and centrifugation of the plates at 800 × g for 2 min at 4°C followed. Fixed DC were then washed twice with 200 μL/well of T cell culture medium (RPMI 1640 containing 10% heat inactivated FBS, 100 IU/mL penicillin, 100μg/mL streptomycin, 2mM glutamax, 50μM β-mercaptoethanol, 1xMEM non-essential amino acids, 1× sodium pyruvate) before being resuspended in 100 ul/well of the same medium. 100,000 OT-I T cells per well in 100μL T cell culture medium were added (to a final volume of 200μL). The co-cultured OT-I T cells with the cross-fixed DCs were incubated for 18 h at 37°C. Cell activation cocktail with brefeldin A (PMA/Ionomycin and Brefeldin A Biolegend, #423303) was added to the wells 4 hours before harvesting. At the time of the harvest, plates were spun down at 800 x g for 2 minutes at 4°C and supernatant was kept for subsequent cytokine analysis. They were then washed with 100μL of 0.1% PBS-BSA before proceeding with live/dead staining with fixable viability Ghost 780 dye (Tonbo Biosciences) for 30 min at 4°C in PBS. Cells were then washed with 0.1% PBS-BSA and stained with a cocktail of 70μL/well of the surface markers (CD8a and CD3) for 40 minutes at 4°C. They were then fixed and permeabilized using the eBiosciences fixation and permeabilization buffer set (eBioscience 88-8824-00) according to the manufacturer’s instructions followed by intracellular staining of IFNγ and IL-2 in permeabilization buffer. Finally, cells were washed 2 times with 0.1% PBS-BSA, centrifuged and resuspended in 100μL/well of PBS-BSA and then analyzed by flow cytometry.
cDC1 and VCAN landscape analysis of TCGA datasets
Level 4 gene expression data were downloaded from the TCGA Data Portal and filtered to retain only cancer types of known epithelial origin for a total of 7591 samples across 20 different cancer types. Single-sample Gene Set Enrichment Analysis (ssGSEA) was performed as described in Barbie and co-authors (Barbie et al., 2009) to measure the signature of gene sets designed to measure overall immune infiltration (Yoshihara et al., 2013), cDC1 density and CD8+ T cell density, as previously published (Spranger et al., 2017). TCGA samples were grouped by cancer type and sorted based on median expression of versican (VCAN) and median cDC1 signature. To measure cancer-specific relationships between VCAN expression and the cDC1 signature an ordinary least squares linear model was fit on these two variables to measure their relationship within each cancer type. Nominal p values from these 20 different models were corrected for multiple hypothesis testing using the Benjamini-Hochberg method and q-values less than 0.1 were considered statistically significant.
Computational modeling of CD8+ T cell density and versikine response signature
Differential expression using DESeq2 was performed to identify genes that were differentially expressed between the PBS-EV and PBS-versikine conditions (Figures 4A and 4C). Genes with a q-value of less than 0.1 were considered significant. We define two gene sets to measure the response to versikine by selecting the 100 genes more significantly induced (versikine-up) and most significantly repressed (versikine-down). These mouse genes were then mapped to their human orthologues using the HGNC Comparison of Orthology Predictions (HCOP) tool (Eyre et al., 2007). ssGSEA (Barbie et al., 2009) was then used to measure the signature of these two gene sets in the 1017 lung samples in the TCGA cohort and overall versikine response level was summarized as the difference between versikine-up and versikine-down signature levels. This versikine response signature was then compared to the CD8 effector T cell signature using an ordinary least squares linear model including the overall immune infiltration signature as a covariate. p-values of less than 0.05 were considered significant.
Graphics
Graphics and diagrams were created using BioRender and Omnigraffle.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using GraphPad Prism software (GraphPad version 9.0.0) or Python version 3.6.6 with stats models version 0.10.0. Statistical significance was determined using unpaired two-tailed Student’s t-test, unpaired non-parametric Mann-Whitney test and non-parametric two-way ANOVA, as indicated in figure legends. The log rank (Mantel-Cox) test was used to determine statistical significance for overall survival in in vivo experiments. Data are shown as mean ± SEM. Significance was assumed with *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001.
Supplementary Material
Highlights.
Tumor stroma remodeling generates cDC1 survival, recruitment, and activation cues
Stroma-licensed cDC1s overexpress CD40 and are hypersensitive to dsDNA sensing
Stromal remodeling promotes atypical NK cells that are GM-CSFhi IFNγlo
T cell repriming by stroma-licensed cDC1s may overcome exclusion at tumor margins
ACKNOWLEDGMENTS
This work was supported by the NIH/National Cancer Institute (R01CA252937, R01CA230275, and U01CA196406), the American Cancer Society (127508-RSG-15-045-01-LIB), the Leukemia and Lymphoma Society (6551-18), the UW Trillium Myeloma Fund, and the Robert J. Shillman Foundation. A. Papadas was supported by the A.G. Leventis, Gerondelis, and Mentzelopoulos Foundations. A.O. was supported by NLM (T15LM011271). We thank Cheryl Kim, Denise Hinz, and Chris Dillingham (LJI Flow Cytometry); Dagna Sheerar (UWCCC Flow Cytometry); Jesus Olvera and Cody Fine (UCSD Human Embryonic Stem Cell Core); Dustin Rubinstein and Kathy Krentz (UW Biotechnology Center); Vishal Chanana and the UW TRIP lab, supported by P30CA014520; Elsa Molina (UCSD Stem Cell Genomics); and the Emory NIH Tetramer Core. We thank Suneel S. Apte (Cleveland Clinic) for the kind gift of versikine-expressing HEK293 cells; Emanuela Romano, Marine Gros, and Amigorena lab members (Institut Curie, Paris, France) for help with antigen presentation assays; Damya Laoui (Free University, Brussels, Belgium) for help with DC gating strategies; and Sasha Rakhmilevich, Paul Sondel, Mario Otto, Peiman Hematti, Silvio Gutkind, and their teams for steadfast support.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111201.
DECLARATION OF INTERESTS
F.A. and C.H. are listed as inventors on US patent US20170258898A1: “Versikine for inducing or potentiating an immune response.”
REFERENCES
- Balan S, Radford KJ, and Bhardwaj N (2020). Unexplored horizons of cDC1 in immunity and tolerance. Adv. Immunol 148, 49–91. [DOI] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. (2009). Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry KC, Hsu J, Broz ML, Cueto FJ, Binnewies M, Combes AJ, Nelson AE, Loo K, Kumar R, Rosenblum MD, et al. (2018). A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor micro-environments. Nat. Med 24, 1178–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell D, Chomarat P, Broyles D, Netto G, Harb GM, Lebecque S, Valladeau J, Davoust J, Palucka KA, and Banchereau J (1999). In breast carcinoma tissue, immature dendritic cells reside within the tumor, whereas mature dendritic cells are located in peritumoral areas. J. Exp. Med 190, 1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, et al. (2018). Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med 24, 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bödder J, Zahan T, van Slooten R, Schreibelt G, de Vries IJM, and Flórez-Grau G (2020). Harnessing the cDC1-NK cross-talk in the tumor microenvironment to battle cancer. Front. Immunol 11, 631713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst J, Ahrends T, Bąbała N, Melief CJM, and Kastenmüller W (2018). CD4(+) T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol 18, 635–647. [DOI] [PubMed] [Google Scholar]
- Böttcher JP, Bonavita E, Chakravarty P, Blees H, Cabeza-Cabrerizo M, Sammicheli S, Rogers NC, Sahai E, Zelenay S, and Reis e Sousa C (2018). NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell 172, 1022–1037.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz ML, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, Barczak A, Rosenblum MD, Daud A, Barber DL, et al. (2014). Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell 26, 938–952. [DOI] [PubMed] [Google Scholar]
- Cabeza-Cabrerizo M, Cardoso A, Minutti CM, Pereira da Costa M, and Reis e Sousa C (2021). Dendritic cells revisited. Annu. Rev. Immunol 39, 131–166. [DOI] [PubMed] [Google Scholar]
- Chang MY, Tanino Y, Vidova V, Kinsella MG, Chan CK, Johnson PY, Wight TN, and Frevert CW (2014).A rapid increase in macrophage-derived versican and hyaluronan in infectious lung disease. Matrix Biol. 34, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conejo-Garcia JR, Rutkowski MR, and Cubillos-Ruiz JR (2016). State-of-the-art of regulatory dendritic cells in cancer. Pharmacol. Ther 164, 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, Woo SR, Lemmens E, Banda T, Leong JJ, et al. (2015). Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11, 1018–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross NA, Chandrasekharan S, Jokonya N, Fowles A, Hamdy FC, Buttle DJ, and Eaton CL (2005). The expression and regulation of ADAMTS-1, −4, −5, −9, and −15, and TIMP-3 by TGFbeta1 in prostate cells: relevance to the accumulation of versican. Prostate 63, 269–275. [DOI] [PubMed] [Google Scholar]
- Deming DA, Emmerich P, Turk AA, Lubner SJ, Uboha NV, LoConte NK, Mulkerin D, Kim DH, Matkowskyj KA, Weber SM, et al. (2020). Pembrolizumab (Pem) in combination with stereotactic body radiotherapy (SBRT) for resectable liver oligometastatic MSS/MMR proficient colorectal cancer (CRC). J. Clin. Oncol 38, 4046. [Google Scholar]
- Diao J, Zhao J, Winter E, and Cattral MS (2010). Recruitment and differentiation of conventional dendritic cell precursors in tumors. J. Immunol 184, 1261–1267. [DOI] [PubMed] [Google Scholar]
- Emmerich P, Matkowskyj KA, McGregor S, Kraus S, Bischel K, Qyli T, Buehler D, Pasch C, Babiarz C, Depke M, et al. (2020). VCAN accumulation and proteolysis as predictors of T lymphocyte-excluded and permissive tumor microenvironments. J. Clin. Oncol 38, 3127. [Google Scholar]
- Evanko SP, Potter-Perigo S, Bollyky PL, Nepom GT, and Wight TN (2012). Hyaluronan and versican in the control of human T-lymphocyte adhesion and migration. Matrix Biol. 31, 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre TA, Wright MW, Lush MJ, and Bruford EA (2007). HCOP: a searchable database of human orthology predictions. Brief. Bioinform 8, 2–5. [DOI] [PubMed] [Google Scholar]
- Fava M, Barallobre-Barreiro J, Mayr U, Lu R, Didangelos A, Baig F, Lynch M, Catibog N, Joshi A, Barwari T, et al. (2018). Role of ADAMTS-5 in aortic dilatation and extracellular matrix remodeling. Arterioscler. Thromb. Vasc. Biol 38, 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris ST, Durai V, Wu R, Theisen DJ, Ward JP, Bern MD, Davidson JT 4th, Bagadia P, Liu T, Briseño CG, et al. (2020). cDC1 prime and are licensed by CD4(+) T cells to induce anti-tumour immunity. Nature 584, 624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flommersfeld S, Böttcher JP, Ersching J, Flossdorf M, Meiser P, Pachmayr LO, Leube J, Hensel I, Jarosch S, Zhang Q, et al. (2021). Fate mapping of single NK cells identifies a type 1 innate lymphoid-like lineage that bridges innate and adaptive recognition of viral infection. Immunity 54, 2288–2304.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulcer SJ, Day AJ, and Apte SS (2015). Isolation and purification of versican and analysis of versican proteolysis. Methods Mol. Biol 1229, 587–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes Marraco SA, Grosjean F, Duval A, Rosa M, Lavanchy C, Ashok D, Haller S, Otten LA, Steiner QG, Descombes P, et al. (2012). Novel murine dendritic cell lines: a powerful auxiliary tool for dendritic cell research. Front. Immunol 3, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes Marraco SA, Scott CL, Bouillet P, Ives A, Masina S, Vremec D, Jansen ES, O’Reilly LA, Schneider P, Fasel N, et al. (2011). Type I interferon drives dendritic cell apoptosis via multiple BH3-only proteins following activation by PolyIC in vivo. PLoS One 6, e20189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumet JD, Richard C, Ledys F, Klopfenstein Q, Joubert P, Routy B, Truntzer C, Gagné A, Hamel MA, Guimaraes CF, et al. (2018). Prognostic and predictive role of CD8 and PD-L1 determination in lung tumor tissue of patients under anti-PD-1 therapy. Br. J. Cancer 119, 950–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggar A, and Weathington N (2016). Bioactive extracellular matrix fragments in lung health and disease. J. Clin. Invest 126, 3176–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski TF (2015). The next hurdle in cancer immunotherapy: overcoming the non-T-cell-inflamed tumor microenvironment. Semin. Oncol 42, 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AN, Abdi K, and Singh NJ (2021).The subunits of IL-12, originating from two distinct cells, can functionally synergize to protect against pathogen dissemination in vivo. Cell Rep. 37, 109816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasner A, Levi A, Enk J, Isaacson B, Viukov S, Orlanski S, Scope A, Neuman T, Enk CD, Hanna JH, et al. (2018). NKp46 receptor-mediated interferon-gamma production by natural killer cells increases fibronectin 1 to alter tumor architecture and control metastasis. Immunity 48, 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorter A, Zijlmans HJ, van Gent H, Trimbos JB, Fleuren GJ, and Jordanova ES (2010). Versican expression is associated with tumor-infiltrating CD8-positive T cells and infiltration depth in cervical cancer. Mod. Pathol 23, 1605–1615. [DOI] [PubMed] [Google Scholar]
- Greter M, Helft J, Chow A, Hashimoto D, Mortha A, Agudo-Cantero J, Bogunovic M, Gautier EL, Miller J, Leboeuf M, et al. (2012). GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity 36,1031–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin MV, Finisguerra V, Van den Eynde BJ, Bercovici N, and Trautmann A (2020). Preclinical murine tumor models: a structural and functional perspective. Elife 9, e50740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han CY, Kang I, Harten IA, Gebe JA, Chan CK, Omer M, Alonge KM, den Hartigh LJ, Gomes Kjerulf D, Goodspeed L, et al. (2020). Adipocyte-derived versican and macrophage-derived biglycan control adipose tissue inflammation in obesity. Cell Rep. 31, 107818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori N, Carrino DA, Lauer ME, Vasanji A, Wylie JD, Nelson CM, and Apte SS (2011). Pericellular versican regulates the fibroblast-myofibroblast transition: a role for ADAMTS5 protease-mediated proteolysis. J. Biol. Chem 286, 34298–34310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, et al. (2008). Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope C, Emmerich PB, Papadas A, Pagenkopf A, Matkowskyj KA, Van De Hey DR, Payne SN, Clipson L, Callander NS, Hematti P, et al. (2017). Versican-derived matrikines regulate batf3-dendritic cell differentiation and promote T cell infiltration in colorectal cancer. J. Immunol 199, 1933–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope C, Foulcer S, Jagodinsky J, Chen SX, Jensen JL, Patel S, Leith C, Maroulakou I, Callander N, Miyamoto S, et al. (2016). Immunoregulatory roles of versican proteolysis in the myeloma microenvironment. Blood 128, 680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope C, Ollar SJ, Heninger E, Hebron E, Jensen JL, Kim J, Maroulakou I, Miyamoto S, Leith C, Yang DT, et al. (2014). TPL2 kinase regulates the inflammatory milieu of the myeloma niche. Blood 123, 3305–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert M, Gobbini E, Couillault C, Manh TPV, Doffin AC, Berthet J, Rodriguez C, Ollion V, Kielbassa J, Sajous C, et al. (2020). IFN-III is selectively produced by cDC1 and predicts good clinical outcome in breast cancer. Sci. Immunol 5, eaav3942. [DOI] [PubMed] [Google Scholar]
- Islam S, Chuensirikulchai K, Khummuang S, Keratibumrungpong T, Kongtawelert P, Kasinrerk W, Hatano S, Nagamachi A, Honda H, and Watanabe H (2020). Accumulation of versican facilitates wound healing: Implication of its initial ADAMTS-cleavage site. Matrix Biol. 87, 77–93. [DOI] [PubMed] [Google Scholar]
- Islam S, Jahan N, Shahida A, Karnan S, and Watanabe H (2022). Accumulation of versican and lack of versikine ameliorate acute colitis. Matrix Biol. 107, 59–76. [DOI] [PubMed] [Google Scholar]
- Islam S, and Watanabe H (2020). Versican: a dynamic regulator of the extracellular matrix. J. Histochem. Cytochem 68, 763–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce JA, and Fearon DT (2015). T cell exclusion, immune privilege, and the tumor microenvironment. Science 348, 74–80. [DOI] [PubMed] [Google Scholar]
- Kim S, Takahashi H, Lin WW, Descargues P, Grivennikov S, Kim Y, Luo JL, and Karin M (2009). Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature 457, 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn NF, Lopez AV, Li X, Cai W, Daniyan AF, and Brentjens RJ (2020). CD103(+) cDC1 and endogenous CD8(+) T cells are necessary for improved CD40L-overexpressing CAR T cell antitumor function. Nat. Commun 11, 6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoui D, Keirsse J, Morias Y, Van Overmeire E, Geeraerts X, Elkrim Y, Kiss M, Bolli E, Lahmar Q, Sichien D, et al. (2016). The tumour microenvironment harbours ontogenically distinct dendritic cell populations with opposing effects on tumour immunity. Nat. Commun 7, 13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Kobayashi S, Leader A, Amir EAD, Elefant N, Bigenwald C, Remark R, Sweeney R, Becker CD, Levine JH, et al. (2017). Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell 169, 750–765.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner MG, Karimi SS, Barry-Holson K, Angell TE, Murphy KA, Church CH, Ohlfest JR, Hu P, and Epstein AL (2013). Immunogenicity of murine solid tumor models as a defining feature of in vivo behavior and response to immunotherapy. J. Immunother 36, 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Huffman AP, Wattenberg MM, Walter DM, Carpenter EL, Feldser DM, Beatty GL, Furth EE, and Vonderheide RH (2020). Type 1 conventional dendritic cells are systemically dysregulated early in pancreatic carcinogenesis. J. Exp. Med 217, e20190673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Yglesias AH, Burger E, Camanzo E, Martin AT, Araujo AM, Kwok SF, and Yarovinsky F (2021). T-bet-dependent ILC1- and NK cell-derived IFN-gamma mediates cDC1-dependent host resistance against Toxoplasma gondii. PLoS Pathog. 17, e1008299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, Kadel EE III, Koeppen H, Astarita JL, Cubas R, et al. (2018). TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiuz R, Brousse C, Ambrosini M, Cancel JC, Bessou G, Mussard J, Sanlaville A, Caux C, Bendriss-Vermare N, Valladeau-Guilemond J, et al. (2021). Type 1 conventional dendritic cells and interferons are required for spontaneous CD4(+) and CD8(+) T-cell protective responses to breast cancer. Clin. Transl. Immunology 10, e1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer CT, Ghorbani P, Nandan A, Dudek M, Arnold-Schrauf C, Hesse C, Berod L, Stüve P, Puttur F, Merad M, and Sparwasser T (2014). Selective and efficient generation of functional Batf3-dependent CD103+ dendritic cells from mouse bone marrow. Blood 124, 3081–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch DR, Nelson CM, Dixon LJ, Silver DL, Wylie JD, Lindner V, Sasaki T, Cooley MA, Argraves WS, and Apte SS (2009). ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Dev. Cell 17, 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Ye S, Izzard L, Dlugolenski D, Tripp RA, Bean AGD, McCulloch DR, and Stambas J (2016). ADAMTS5 is a critical regulator of virus-specific T cell immunity. PLoS Biol. 14, e1002580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GRS, and Perlman H (2013). Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am. J. Respir. Cell Mol. Biol 49, 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Hascall VC, Markwald RR, and Ghatak S (2015). Interactions between hyaluronan and its receptors (CD44, RHAMM) regulate the activities of inflammation and cancer. Front. Immunol 6, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal D, Gubin MM, Schreiber RD, and Smyth MJ (2014). New insights into cancer immunoediting and its three component phases–elimination, equilibrium and escape. Curr. Opin. Immunol 27, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjaatvedt CH, Yamamura H, Capehart AA, Turner D, and Markwald RR (1998). The Cspg2 gene, disrupted in the hdf mutant, is required for right cardiac chamber and endocardial cushion formation. Dev. Biol 202, 56–66. [DOI] [PubMed] [Google Scholar]
- Nandadasa S, Burin des Roziers C, Koch C, Tran-Lundmark K, Dours-Zimmermann MT, Zimmermann DR, Valleix S, and Apte SS (2021). A new mouse mutant with cleavage-resistant versican and isoform-specific versican mutants demonstrate that proteolysis at the Glu(441)-Ala(442) peptide bond in the V1 isoform is essential for interdigital web regression. Matrix Biol. 10, 100064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandadasa S, Foulcer S, and Apte SS (2014). The multiple, complex roles of versican and its proteolytic turn over by ADAMTS proteases during embryogenesis. Matrix Biol. 35, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba T, Long MD, Keler T, Marsh HC, Minderman H, Abrams SI, Liu S, and Ito F (2020). Overcoming primary and acquired resistance to anti-PD-L1 therapy by induction and activation of tumor-residing cDC1s. Nat. Commun 11, 5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai SI, Cesano A, and Marincola FM (2020). The paradox of cancer immune exclusion: immune oncology next frontier. Cancer Treat Res. 180, 173–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadas A, Arauz G, Cicala A, Wiesner J, and Asimakopoulos F (2020). Versican and versican-matrikines in cancer progression, inflammation, and immunity. J. Histochem. Cytochem 68, 871–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadas A, and Asimakopoulos F (2020). Versican in the tumor microenvironment. Adv. Exp. Med. Biol 1272, 55–72. [DOI] [PubMed] [Google Scholar]
- Peterson EE, and Barry KC (2020). The natural killer-dendritic cell immune Axis in anti-cancer immunity and immunotherapy. Front. Immunol 11, 621254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinello L, Canver MC, Hoban MD, Orkin SH, Kohn DB, Bauer DE, and Yuan GC (2016). Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol 34, 695–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porciuncula A, Morgado M, Gupta R, Syrigos K, Meehan R, Zacharek SJ, Frederick JP, and Schalper KA (2021). Spatial mapping and immunomodulatory role of the OX40/OX40L pathway in human non-small cell lung cancer. Clin. Cancer Res 27, 6174–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon H, Idoyaga J, Rahman A, Leboeuf M, Remark R, Jordan S, Casanova-Acebes M, Khudoynazarova M, Agudo J, Tung N, et al. (2016). Expansion and activation of CD103(+) dendritic cell progenitors at the tumor site enhances tumor responses to therapeutic PD-L1 and BRAF inhibition. Immunity 44, 924–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Paulete AR, Cueto FJ, Martínez-López M, Labiano S, Morales-Kastresana A, Rodríguez-Ruiz ME, Jure-Kunkel M, Azpilikueta A, Aznar MA, Quetglas JI, et al. (2016). Cancer immunotherapy with immunomodulatory anti-CD137 and anti-PD-1 Monoclonal antibodies requires BATF3-dependent dendritic cells. Cancer Discov. 6, 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichien D, Scott CL, Martens L, Vanderkerken M, Van Gassen S, Plantinga M, Joeris T, De Prijck S, Vanhoutte L, Vanheerswynghels M, et al. (2016). IRF8 transcription factor controls survival and function of terminally differentiated conventional and plasmacytoid dendritic cells, respectively. Immunity 45, 626–640. [DOI] [PubMed] [Google Scholar]
- Spranger S, Bao R, and Gajewski TF (2015). Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235. [DOI] [PubMed] [Google Scholar]
- Spranger S, Dai D, Horton B, and Gajewski TF (2017). Tumor-Residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell 31, 711–723.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Diao J, Gu H, Khatri I, Zhao J, and Cattral MS (2015). Toll-like receptor 2 activation promotes tumor dendritic cell dysfunction by regulating IL-6 and IL-10 receptor signaling. Cell Rep. 13, 2851–2864. [DOI] [PubMed] [Google Scholar]
- Thompson ED, Enriquez HL, Fu YX, and Engelhard VH (2010). Tumor masses support naive T cell infiltration, activation, and differentiation into effectors. J. Exp. Med 207, 1791–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms KP, and Maurice SB (2020). Context-dependent bioactivity of versican fragments. Glycobiology 30, 365–373. [DOI] [PubMed] [Google Scholar]
- Tullett KM, Lahoud MH, and Radford KJ (2014). Harnessing human cross-presenting CLEC9A(+)XCR1(+) dendritic cells for immunotherapy. Front. Immunol 5, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vremec D (2016). The isolation and enrichment of large numbers of highly purified mouse spleen dendritic cell populations and their in vitro equivalents. Methods Mol. Biol 1423, 61–87. [DOI] [PubMed] [Google Scholar]
- Waight JD, Netherby C, Hensen ML, Miller A, Hu Q, Liu S, Bogner PN, Farren MR, Lee KP, Liu K, and Abrams SI (2013). Myeloid-derived suppress or cell development isregulated by a STAT/IRF-8 axis. J. Clin. Invest 123, 4464–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Rajagopalan A, Flietner ED, Yun G, Chesi M, Furumo Q, Burns RT, Papadas A, Ranheim EA, Pagenkopf AC, et al. (2021). Expression of NrasQ61R and MYC transgene in germinal center B cells induces a highly malignant multiple myeloma in mice. Blood 137, 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wight TN (2017). Provisional matrix: a role for versican and hyaluronan. Matrix Biol. 60-61, 38–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wight TN, Kang I, Evanko SP, Harten IA, Chang MY, Pearce OMT, Allen CE, and Frevert CW (2020). Versican-A critical extracellular matrix regulator of immunity and inflammation. Front. Immunol 11, 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi M, Barker TH, Gibbons DL, and Kurie JM (2018). The fibrotic tumor stroma. J. Clin. Invest 128, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, et al. (2013). Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun 4, 2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng DQ, Yu YF, Ou QY, Li XY, Zhong RZ, Xie CM, and Hu QG (2016). Prognostic and predictive value of tumor-infiltrating lymphocytes for clinical therapeutic research in patients with non-small cell lung cancer. Oncotarget 7, 13765–13781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Yang W, Wang P, Deng Y, Dong YT, Liu FF, Huang R, Zhang P, Duan YQ, Liu XD, et al. (2020). CCL7 recruits cDC1 to promote antitumor immunity and facilitate checkpoint immunotherapy to non-small cell lung cancer. Nat. Commun 11, 6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Miao L, and Wang L (2012). Inflammation amplification by Versican: the first mediator. Int. J. Mol. Sci 13, 6873–6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data have been deposited at GEO and are publicly available (accession GEO: GSE199938). Original western blot images appear in Supplementary Data. Microscopy data will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data is available from the lead contact upon request.


