Abstract
Bovine tuberculosis caused by Mycobacterium bovis, is a significant global pathogen causing economic loss in livestock and zoonotic TB in man. Several vaccine approaches are in development including reverse vaccinology which uses an unbiased approach to select open reading frames (ORF) of potential vaccine candidates, produce them as recombinant proteins and assesses their immunogenicity by direct immunization. To provide feasibility data for this approach we have cloned and expressed 123 ORFs from the M. bovis genome, using a mixture of E. coli and insect cell expression. We used a concatenated open reading frames design to reduce the number of clones required and single chain fusion proteins for protein pairs known to interact, such as the members of the PPE-PE family. Over 60% of clones showed soluble expression in one or the other host and most allowed rapid purification of the tagged bTB protein from the host cell background. The catalogue of recombinant proteins represents a resource that may be suitable for test immunisations in the development of an effective bTB vaccine.
Keywords: bovine tuberculosis, Mycobacterium bovis, genome, open reading frame, vaccine, expression, protein purification, PPE
Introduction
Bovine tuberculosis (bTB) is a major disease throughout the world and is particularly prominent in Africa and parts of Asia. While many developed countries have reduced or eliminated bTB from their cattle populations, major hotspots remain in Europe, America, Canada and New Zealand (Smith, 2012). Phylogenetic analysis of >1,000 non-redundant M. bovis genomes has suggested divergence from a last common ancestor with M. caprae ∼ 3,500 years ago with spread since aided by animal trade (Zimpel et al., 2020). Control is effected by large scale cattle testing and animals scoring positive are subsequently culled, with an acknowledged impact on sustainability and an increased risk of human infection (Chambers et al., 2014). Various control strategies have been implemented, including those with limited public appeal (Bielby et al., 2014), but it is generally accepted that control in the longer term will require the development of an efficacious cattle vaccine. Multiple potential vaccines have been described (Vordermeier and Hewinson, 2006; Kaufmann et al., 2017; McShane, 2019) but the lead contender for early use remains Bacillus Calmette-Guerin (BCG), the cornerstone of human vaccine programmes. However, many of the proteins present in the purified protein derivative (PPD) used in herd screening programs are also present in BCG leading to BCG immunised animals scoring positive, a complication if immunisation and screening occur concurrently. To overcome this, some protein markers, either missing or non-immunogenic in BCG, have been developed to differentiate between infected and vaccinated status (Whelan et al., 2010; Vordermeier et al., 2011; Srinivasan et al., 2019). An alternate approach, to develop a BCG based vaccine that does not lead to positivity in the screening test has also been investigated (Chandran et al., 2019). Although BCG derives from M. bovis, comparative genomics has revealed genomic losses in the variously passaged BCG stocks when compared to ancestral stocks (Zhang et al., 2013) suggesting that some BCG vaccine stocks may be incapable of providing suitable levels of protection unless supplemented with additional proteins representing the missing ORFs. A number of candidates have been investigated in this respect, including Ag85, ESAT-6, CFP-10 and members of the PPE family (Li et al., 2015; Dai et al., 2017; Tkachuk et al., 2017; Stylianou et al., 2018) although generally as single antigens rather than as a mixture of candidates. As a result, the case has also been made that current candidate vaccines lack diversity and that a broader range of proteins should be considered (Fletcher and Schrager, 2016).
Reverse vaccinology (Rappuoli, 2001; Serruto et al., 2012; Rappuoli et al., 2016) assumes that molecules on the cell surface of a target organism include those that may act as an effective vaccine even if they do not raise a significant serum response following natural infection. The availability of whole bacterial genomes, suitable bioinformatics and high throughput protein expression technologies allows this concept to be tested, with some notable successes (Muruato et al., 2017; Masignani et al., 2019). The feasibility of this approach is in part determined by how many and how easily bTB proteins can be expressed and purified. For Mycobacterium tuberculosis, progress in multiple protein expression has been made by the TB Structural Genomics Consortium, although the proteins selected were based primarily on their suitability as drug targets (Chim et al., 2011). To assess feasibility for proteins that might be targeted for vaccine use and to provide such proteins for testing we report a high throughput expression approach (Aricescu et al., 2006; Nettleship et al., 2010; Bird et al., 2014) to the expression of many full-length Mycobacterium bovis ORFs.
Materials and Methods
Selection of Vaccine Candidates
Candidate bTB proteins were selected by reference to lists derived from published bioinformatics approaches (e.g. (He et al., 2010; Monterrubio-López et al., 2015)) as well as current web sources (Kapopoulou et al., 2011; Dhanda et al., 2016). Throughout the study the genome of Mycobacterium bovis strain AF2122/97 was used as the reference strain. Proteins with a range of putative functions related to vaccine use were included, for example roles in virulence, predicted surface expression, previous use as a candidate vaccine and roles in the immune response and immune evasion.
Molecular Cloning
Sequences representing the candidate ORFs were ordered as custom designed double stranded DNA fragments (Integrated DNA Technologies, Belgium). Sequence predicted to encode transmembrane domains was removed and synthetic genes were codon optimised for Spodoptera frugiperda (Sf9) or E. coli to maximise expression in the relevant host. All DNA fragments were flanked with 18 base pairs of sequence homologous to the vector pTriEx 1.1 (EMD Biosciences), suitable for expression in E. coli under control of the T7 promoter or insect cells under control of the baculovirus very late P10 promoter. All clones were generated by Gibson assembly (Gibson et al., 2009). The sequence designs were such that translation initiated at the bTB ORF initiation codon and fused the C-terminus with the histidine tag present in the vector, except when otherwise indicated. The pTriEx-1.1 cloning vector was linearised by digestion with restriction enzymes XhoI and NcoI (Thermo Fisher Scientific, Paisley, United Kingdom) and 20 ng gene fragment was assembled into 50 ng linearized pTriEx-1.1 cloning vector using the NEBuilder HiFi DNA Assembly Protocol (NEB, United Kingdom). Clones were isolated following transformation of Stellar competent cells (Takara Bio, France) with confirmation by colony PCR followed by DNA sequencing.
E. Coli Expression
The default strain for protein expression in E. coli was SoluBL21 (Kim et al., 2015). Alternate hosts were E. coli T7 Express lysY (NEB) and LOBSTR (Andersen et al., 2013) which were used for low yielding constructs. Initial screening was achieved with cultures of ∼20 ml and scaled according to the outcome. Cells were grown at 37°C to an OD600 = 0.6 and induced by the addition of IPTG to 0.1 mM. After induction, growth was continued for 4 h at 37°C. Cultures were harvested by centrifugation at 4,000 × g for 15 min and disrupted for gel analysis or purification as required. Solubility was determined after lysis in 1% Triton-X detergent and gel analysis of the soluble and pellet fractions. Poor solubility was corrected, where possible, with low temperature induction (16°C) or by varying the IPTG concentration used for induction. Intractable expression was abandoned in favour of alternate constructs or the alternate host.
Recombinant Baculovirus Expression
Recombinant baculoviruses were produced by co-transfection of Sf9 cells with transfer vectors and linearised baculovirus DNA as described (Porta et al., 2013). Transfections were done in 6 well tissue culture plates at a monolayer confluency of 50%. Viruses were passaged when significant cytopathic effect was observed and viral stocks were amplified to high titre, typically 3 passages, prior to use for infection and protein detection. All viral stocks were produced in Sf9 cells. For protein expression, 100 ml suspension cultures of AoTni38 cells (Hashimoto et al., 2012) were infected at a density of 2 × 106 per ml with 10 ml of high titre (>107 pfu per ml) virus stock and the culture continued for 3 days. Cells were harvested by centrifugation and processed as described.
SDS PAGE
Protein samples were prepared using NuPAGE loading buffer (Fisher Scientific UK Ltd., United Kingdom) and heated to 98°C for 10 min before loading the gel. Proteins were separated by SDS-PAGE using 4–12% precast Tris-Glycine SDS polyacrylamide gels (Fisher Scientific UK Ltd., United Kingdom) for 30 min at 200 V. SDS-PAGE loading used the equivalent of 50 μL of bacterial culture or 5 × 104 insect cells per lane of a 10 lane, 10 cm gel. After electrophoresis, gels were subjected to either Coomassie Brilliant Blue R250 staining or transferred to a polyvinylidene difluoride (PVDF) membrane for western blot analysis.
Western Blot
Proteins were transferred electrophoretically to PVDF membranes (Immobilon®, Merck, Germany) by semi-dry transfer. Membranes were blocked with Protein-Free T20 (TBS) Blocking Buffer (Fisher Scientific UK Ltd., United Kingdom) for 1 h. All subsequent incubations and washes were done in Tris-buffer saline containing 0.1% Tween 80 (TBST). Membranes were incubated with monoclonal Anti-6X His tag® antibody conjugated to HRP (Abcam, United Kingdom) at 1:2000 dilution followed by three washes of 5 min each. The membrane was finally washed with TBS and antibody reactive bands were revealed using ECL Western Blotting Detection Reagent (Geneflow, United Kingdom) and imaged using a Syngene G-Box (Syngene, United Kingdom).
Purification
E.coli cell pellets were lysed in 10% of the original culture volume in a lysis buffer (500 mM NaCl, 20 mM Sodium phosphate buffer pH 7.4, 20 mM imidazole, 1% Triton X-100) supplemented with 1% lysozyme, 1 mM phenylmethanesulfonyl fluoride and 1 mM benzamidine hydrochloride and incubated for 10 min at 4°C. Samples were sonicated for 10 min on ice followed by centrifugation at 15,000 × g for 10 min. Infected insect cells were disrupted similarly but without lysozyme in the lysis buffer. Expressed proteins, with an affinity tag of six consecutive histidine residues, were purified by His-mag sepharose Ni beads (GE Life Sciences) using 200 µL bead slurry per preparation. Beads were incubated with the clarified lysate for a minimum of 2 h on a blood wheel before washing and elution according to the vendor’s instructions. For proteins that demonstrated a tendency to insolubility the lysis buffer also contained 34 mM SDS and the wash and elution buffers included 0.1% sodium sarkosyl (Schlager et al., 2012). For secreted proteins the clarified supernatant was adjusted to 0.5 mM nickel sulphate and used directly for IMAC pull-down. Constructs that were positive for expression but failed purification by one of these routes were abandoned.
Results
Identification and Generation of Sequences Encoding Putative Vaccine Candidates
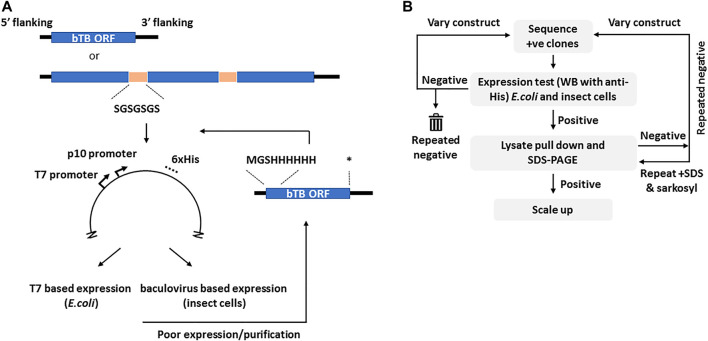
The selection of candidates for this study was based on one or more vaccine related properties, viz. their predicted biological function (cell surface protein, virulence or adhesion), predicted immunogenicity, previous use as a candidate vaccine or sequence conservation in multiple mycobacterial genomes. From these considerations the final ORFs selected were grouped into seven loose descriptive categories (Table 1). For ORFs representing complete proteins with a predicted molecular mass of greater than ∼15 kDa, synthetic DNA was produced to the entire coding region with flanking regions that ensured a unique initiator ATG downstream of the Shine-Dalgarno sequence and fusion with a sequence encoding polyhistidine at the carboxyl terminus of the expressed protein, both features of the vector used (Figure 1A). If a signal peptide was present in the original M. bovis ORF, assessed by routine submission to the SigP server (Nielsen et al., 2019), it was changed to that of honeybee melittin, known to be efficiently processed in insect cells (Tessier et al., 1991). Sequences predicted to encode transmembrane domains were deleted. In cases where expression of the recombinant protein was either poor or, more usually, insoluble, the design was changed to incorporate an N-terminal polyhistidine sequence after the initiator codon and a stop codon prior to the vector encoded polyhistidine sequence (Figure 1A).
TABLE 1.
The bTB ORFs selected for recombinant expression tests.
| Expression host | Soluble expression | Purification yield | ||
|---|---|---|---|---|
| Cell wall and cell processes | ||||
| Mb0923 | Outer membrane protein Omp A | E. coli & Insect | +++ | +++ |
| Mb0485 | Iron-regulated heparin binding hemagglutinin hbha | E. coli & Insect | +++ | +++ |
| Mb0419c | Probable glutamine-binding lipoprotein glnh (glnbp) | E. coli & Insect | ++ | + |
| Mb3840 | Exported repetitive protein precursor PirG | E. coli & Insect | +++ | + |
| Mb0348 | Isoniazid inductible gene protein iniB | E. coli & Insect | +++ | +++ |
| Mb2293 | Probable lipoprotein lppN | E. coli | +++ | +++ |
| Mb2807c | Probable lipoprotein lppU | Insect | +++ | +++ |
| Mb0598c | Probable lipoprotein lpqN | E. coli & Insect | +++ | +++ |
| Mb1260 | Probable lipoprotein lpqX | Insect | +++ | +++ |
| Mb1568c | Probable lipoprotein lprI | E. coli & Insect | +++ | +++ |
| Mb1891c | Alanine and proline rich secreted protein APA | E. coli & Insect | +++ | +++ |
| Mb3905 | 6 kda early secretory antigenic target esxa (Esat-6) | E. coli & Insect | +++ | +++ |
| Mb3904 | 10 kda culture filtrate antigen esxb (lhp) (cfp10) | E. coli & Insect | +++ | +++ |
| Mb0296 | Low molecular weight antigen 7 esxh (10 kda antigen) (cfp-7) | Insect | +++ | +++ |
| Mb0295 | Esat-6 like protein esxg | Insect | +++ | +++ |
| Mb1229 | Esat-6 like protein esxk (Esat-6 like protein 3) | E. coli & Insect | +++ | +++ |
| Mb1230 | Putative Esat-6 like protein esxi (Esat-6 like protein 4) | Insect | +++ | +++ |
| Mb1820 | Esat-6 like protein esxm | E. coli & Insect | +++ | +++ |
| Mb1821 | Putative Esat-6 like protein esxn (Esat-6 like protein 5) | Insect | +++ | ++ |
| Mb3475c | Esat-6 like protein esxu | E. coli & Insect | +++ | +++ |
| Mb3911c | Proteolytic substrate protein espb | E. coli & Insect | ++ | ++ |
| Mb0674 | Possible ribonucleotide-transport ATP-binding protein | E. coli & Insect | +++ | ++ |
| Mb1036 | Probable resuscitation-promoting factor rpfb | E. coli & Insect | ++ | - |
| Mb1445 | Aminoglycosides/tetracycline-transport integral membrane protein | E. coli & Insect | ++ | - |
| Mb3070 | Probable FeIII-dicitrate-binding periplasmic lipoprotein fecb | E. coli | ++ | ++ |
| Mb3338 | Acid phosphatase sapm | E. coli & Insect | ++ | ++ |
| Mb2653c | Probable conserved transmembrane protein | E. coli & Insect | - | - |
| Mb3646c | Esx-1 secretion-associated protein, espa | E. coli & Insect | +++ | + |
| Mb3645c | Esx-1 secretion-associated protein espc | E. coli & Insect | +++ | +++ |
| Mb3644c | ESX-1 secretion-associated protein espd | E. coli & Insect | - | - |
| Mb2002c | Immunogenic protein mpt64 (antigen mpt64/mpb64) | E. coli & Insect | +++ | +++ |
| Mb1762 | Probable conserved transmembrane protein | E. coli & Insect | 0 | ++ |
| Information pathway related proteins | ||||
| Mb0055 | Single-strand binding protein Ssb | E. coli & Insect | + | + |
| Mb0671 | 50s ribosomal protein l7/l12 rplL (sa1) | E. coli & Insect | +++ | +++ |
| Mb0704 | Probable iron-regulated elongation factor tu tuf | E. coli & Insect | ++ | + |
| Mb0670 | 50s ribosomal protein l10 rplJ | E. coli & Insect | +++ | ++ |
| Mb1656 | 30s ribosomal protein s1 rpsA | E. coli & Insect | +++ | +++ |
| Virulence related proteins | ||||
| Mb3441 | Possible antitoxin VapB47 | Insect | +++ | +++ |
| Mb2891 | Toxin RelG | E. coli & Insect | +++ | - |
| PE/PPE family proteins | ||||
| Mb2548 | PE family PE26 | Insect | + | + |
| Rv3622c | PE family protein PE32 | E. coli & Insect | +++ | +++ |
| Mb0293 | PE family protein PE5 | E.coli & Insect | +++ | +++ |
| Mb0294 | PE family protein PE4 | E.coli & Insect | +++ | +++ |
| Mb3902 | PE family-related protein PE35 | E.coli & Insect | +++ | +++ |
| Mb3922c | PE family protein PE36 | E.coli | +++ | +++ |
| Mb3504 | PE family protein PE31 | Insect | +++ | +++ |
| Mb0940c | PE family protein PE7 | Insect | +++ | +++ |
| Mb1835 | PE family protein PE20 | Insect | +++ | +++ |
| Mb1421 | PE family protein PE15 | E.coli & Insect | +++ | +++ |
| Mb3505 | PE family protein PPE 60 | E.coli & Insect | +++ | +++ |
| Mb1069c | PE family protein PE 8 | E.coli & Insect | +++ | +++ |
| Mb2457c | PE family protein PE 25 | E.coli & Insect | +++ | +++ |
| Mb1202c | PE family protein | E.coli & Insect | +++ | +++ |
| Mb3903 | PPE family protein PPE68 | E.coli & Insect | +++ | +++ |
| Mb3921c | PPE family-related protein PPE69 | E.coli & Insect | +++ | +++ |
| Mb0939c | PPE family protein PPE 14 | E.coli & Insect | +++ | +++ |
| Mb1836 | PPE family protein PPE 31 | E.coli & Insect | +++ | +++ |
| Mb1068c | PPE family protein PPE15 | E.coli & Insect | +++ | +++ |
| Mb1422 | PPE family protein PPE 20 | Insect | +++ | +++ |
| Mb2456c | PPE family protein PE41 | E.coli & Insect | +++ | +++ |
| Mb1228 | PPE family protein PPE 18 | E.coli & Insect | +++ | +++ |
| Mb1200c | PPE17 (part) | E.coli & Insect | +++ | +++ |
| Mb1201c | PPE 17 (part) | E.coli & Insect | +++ | +++ |
| Intermediary metabolism & respiration related proteins | ||||
| Mb3871 | Bacterioferritin BfrB | E.coli | +++ | +++ |
| Mb2981 | Possible glycosyl transferase | E.coli & Insect | ++ | + |
| Mb0130 | probable serine protease pepA | E.coli & Insect | ++ | ++ |
| Mb1272 | Probable malate dehydrogenase mdh | E.coli & Insect | ++ | - |
| Mb0977 | Probable succinyl-CoA synthetase (a chain) sucD | E.coli & Insect | ++ | ++ |
| Mb3652 | Inorganic pyrophosphatase PPA | E.coli & Insect | +++ | |
| Mb1129c | Fructose 1,6-bisphosphatase glpX | Insect | ++ | - |
| Mb3412 | Diterpene synthase | E.coli & Insect | ++ | ++ |
| Conserved hypotheticals & unknown proteins | ||||
| Mb2315c | Hypothetical protein | Insect | + | - |
| Mb3935c | Putative Esat-6 like protein esxf (Esat-6 like protein 13) | E. coli & Insect | +++ | +++ |
| Mb3920c | Possible Esat-6 like protein esxd | E. coli & Insect | +++ | +++ |
| Mb3934c | Putative Esat-6 like protein esxe (Esat-6 like protein 12) | E. coli & Insect | +++ | +++ |
| Mb1066c | Putative Esat-6 like protein esxI (Esat-6 like protein 1) | Insect | +++ | +++ |
| Mb1067c | Esat-6 like protein esxj | E. coli & Insect | +++ | +++ |
| Mb2375c | Putative Esat-6 like protein esx0 (Esat-6 like protein 6) | Insect | + | + |
| Mb3042c | Esat-6 like protein esxq (tb12.9) (Esat-6 like protein 8) | Insect | + | + |
| Mb3045c | Secreted Esat-6 like protein esxr (Esat-6 like protein 9) | E.coli & Insect | +++ | +++ |
| Mb3046c | Esat-6 like protein esxs | E.coli & Insect | +++ | +++ |
| Mb3474c | Putative Esat-6 like protein esxt | E.coli & Insect | +++ | +++ |
| Mb3919c | Esat-6 like protein esxc (Esat-6 like protein 11) | Insect | + | - |
| Mb0959 | Periplasmic phosphate-binding lipoprotein PstS1 | E. coli & Insect | +++ | +++ |
| Mb1858 | Conserved protein with fha domain, gara | E. coli & Insect | ++ | ++ |
| Mb1868c | Malate synthase G GlcB | E. coli & Insect | ++ | ++ |
| Mb1943c | Catalase-peroxidase-peroxynitritase T KatG | E. coli & Insect | ++ | ++ |
| Mb2006c | Probable cutinase precursor CFP21 | E. coli & Insect | ++ | +++ |
| Mb2057c | Stress protein induced by anoxia | E. coli & Insect | ++ | +++ |
| Mb2244 | Glutamine synthetase GlnA1 | E. coli & Insect | ++ | +++ |
| Mb2898 | Cell surface lipoprotein Mpt83 (lipoprotein P23) | E. coli & Insect | ++ | +++ |
| Mb2900 | Major secreted immunogenic protein Mpt70 | E. coli & Insect | ++ | ++ |
| Mb0169 | Conserved protein TB18.5 | E. coli & Insect | + | + |
| Mb0418c | Serine/threonine-protein kinase PknG | E. coli & Insect | +++ | +++ |
| Mb2477c | Probable resuscitation-promoting factor RpfE | E. coli & Insect | ++ | - |
| Mb1319 | Conserved protein | E. coli & Insect | ++ | ++ |
| Mb1596 | Involved in biotin biosynthesis | E. coli & Insect | ++ | ++ |
| Mb2656 | Universal stress protein family protein TB31.7 | E. coli & Insect | ++ | ++ |
| Mb3046c | Esat-6 like protein EsxS | E. coli & Insect | ++ | ++ |
| Mb2982c | Possible glycosyl transferase | E. coli & Insect | ++ | + |
| Mb0455c | Cyclopropane fatty acid synthase | E. coli & Insect | +++ | ++ |
| Mb2054c | pfkb | E. coli & Insect | +++ | +++ |
| Mb3157c | devR | E. coli & Insect | +++ | +++ |
| Mb2970c | Probable conserved lipoprotein LppX | E. coli & Insect | ++ | |
| Mb0891 | Possible resuscitation-promoting factor rpfA | E. coli & Insect | +++ | ++ |
| Mb1916 | Probable resuscitation-promoting factor rpfC | E. coli & Insect | ++ | ++ |
| Mb2410 | Probable resuscitation-promoting factor rpfD | E. coli & Insect | ++ | ++ |
| Mb3274c | Two component sensory transduction transcriptional regulatory protein mtrA | E. coli & Insect | +++ | +++ |
| Mb0463c | Conserved protein | E. coli & Insect | +++ | +++ |
| Mb3641 | Hypothetical arginine and proline rich protein | E. coli & Insect | ++ | - |
| Mb0062 | Hypothetical protein | E. coli & Insect | ++ | ++ |
| Mb1843 | Conserved protein | E. coli & Insect | ++ | ++ |
| Mb3743c | Conserved protein | E. coli & Insect | +++ | +++ |
| Mb1833c | Conserved protein | Insect | +++ | +++ |
| Mb0854c | Conserved protein | E. coli & Insect | +++ | +++ |
| Mb2058 | Conserved protein Acg | E. coli & Insect | ++ | - |
| Mb0337c | hypothetical protein | E. coli & Insect | ++ | ++ |
| Mb2660c | Conserved protein | E. coli & Insect | ++ | - |
| Mb 2659 | hypoxic response protein 1 hrp1 | E. coli | +++ | +++ |
| Mb3707 | Probable bifunctional membrane-associated penicillin-binding protein 1a/1b pona2 | E. coli & Insect | ++ | + |
| Mb0014c | Transmembrane Serine/therorine protein Kinase-B (pknb) | E. coli & Insect | ++ | +++ |
| Mb0979 | Probable conserved Transmembrane protein | E. coli & Insect | ++ | +++ |
| Mb0448 | GROEL protein-2 | E. coli & Insect | ++ | +++ |
Soluble expression levels: +++ - strongest band on SDS-PAGE, ++ - among the stronger bands, + - visible band, − no visible band. Purification yields: +++ ∼1 mg/L, ++ ∼0.1 mg/L, + <0.1 mg/L, - not purified. Greyed boxes required wash and elution buffers with 0.1% sodium sarkosyl.
FIGURE 1.
The cloning and selection strategies for the generation of the M. bovis recombinant protein atlas. Panel (A). Generic construct design showing the use of either complete or concatenated ORFs tagged at the C-terminus with polyhistidine as present in vector pTriEx1.1. In the event of poor expression the same ORFs were re-cloned with an N-terminal His tag as shown. The sequence of the flexible linker is indicated. Asterisk–stop codon. Panel (B). The protein expression and purification regimen showing the iterative nature of the process. The varied constructs are those with alternate His tags shown in (A). Initial screening was by Western blot for the His tag in all cases.
More than half of the expressed proteins that were problematic in recombinant form with a C-terminal polyhistidine were improved by this strategy, as described below. For proteins that were <15 kDa, or for proteins whose database entry did not contain an initiator methionine, for example some members of the esx family, sequences were concatenated before synthesis so that several ORFs were expressed together, connected by a Gly-Ser linker of 7 residues (Figure 1A). Similarly, for protein pairs such as the proline-proline-glutamate (PPE) and proline-glutamate (PE) complexes, the matching pair was synthesised as a single chain also connected by a flexible linker. The available crystal structures of such complexes show the C-terminus of the PPE partner lying close to the N-terminus of the paired PE (Strong et al., 2006), a distance easily spanned by the added linker. Some M. bovis ORFs were persistently difficult to express in either E. coli or insect cells and in these cases workaround strategies involved fusion with solubility tags including GFP, SUMO or the B1 domain of streptococcal protein G (GB1) (Lindhout et al., 2003; Pengelley et al., 2006; Lee et al., 2008). Some of these strategies improved expression or purification for some of the ORFs but none provided a universal route to improvement.
Protein Expression and Purification
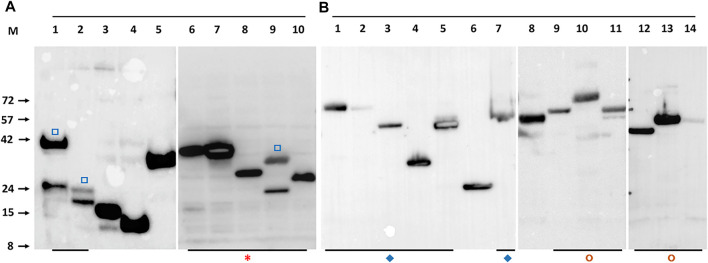
To maximise the possibility of efficient expression of a diverse range of selected M. bovis ORFs as soluble proteins, two different expression systems were used, E. coli and recombinant baculoviruses. Screening of the clones or recombinant viruses was done in small scale cultures by Western blot with a directly conjugated antibody recognizing the polyhistidine tag (Figure 1B). The default result was a band of the molecular mass predicted for the translated protein but occasionally the band identified by the Western blot had bands additional to the predicted mass (marked in Figure 2A). We assumed smaller sizes were the result of protein degradation, with the residual C-terminal His tagged fragment being detected, whereas larger proteins were post-translationally modified or oligomeric. Notably, the concatenated PPE-PE complexes gave rise to stable products of the predicted molecular mass (marked in Figure 2B) as did the Esx concatenates (marked in Figure 2A), some of which have been reported elsewhere (e.g., (Li et al., 2016; Mearns et al., 2017)). Concatenated constructs Mb1036_Mb1129c, Mb1272_Mb2477c, Mb3070_Mb3641 and Mb3338_Mb3644c also showed predominantly single bands at the predicted molecular mass when probed with an anti-His antibody (marked in Figure 2B).
FIGURE 2.
Example Western blot detection of recombinant M. bovis proteins with anti-His antibody. Panel (A). Detection of M. bovis ORFs expressed in induced E.coli. The lane identity and predicted molecular mass are: 1-Mb1228 (39 kDa), 2-Mb1916 (18 kDa), 3-Mb2410 (15 kDa), 4-Mb3441 (11 kDa), 5-Mb3412c (34 kDa), 6-EsxRST (32 kDa), 7-EsxNOQ (30 kDa), 8-EsxU (24 kDa), 9-EsxFDE (27 kDa), 10-EsxAB (21.2 kDa). Panel (B). Detection of M. bovis ORFs expressed in baculovirus infected insect cells. The lane identity and predicted molecular mass are: 1-PPE15_PE8 (66 kDa), 2-PPE20_PE15 (67 kDa), 3-PPE31_PE20 (51 kDa), 4-PPE41_PE25 (34 kDa), 5-PPE60_PE31 (53 kDa), 6-MTRA (28 kDa), 7-PPE14_PE7 (53 kDa), 8- Mb0463_Mb0674 (59 kDa), 9-Mb0854_Mb0977 (63 kDa), 10-Mb1036_Mb1129c (74 kDa), 11-Mb1272_Mb2477c (54 kDa), 12-Mb0891c (34 kDa), 13- Mb3070_Mb3641 (54 kDa), 14-Mb3338_Mb3644c (54 kDa). Open square (□) symbols indicate protein which show some breakdown as indicated by at least 2 His antibody reactive bands. Asterisk (*) indicates all Esx related proteins, most as concatenates. Diamonds (♦) indicate concatenated PPE-PE pairs. Circles (◘) other protein concatenates. M indicates the marker track, the molecular masses of which are given on the left of panel (A) in kilodaltons.
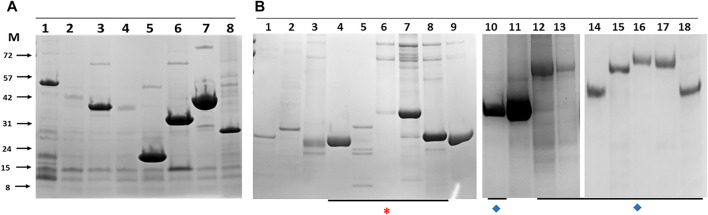
Clones positive for expression by western blot were assessed for purification by pull-down from detergent lysates using IMAC magnetic beads and the eluates assessed directly by SDS-PAGE and gel staining (Figure 3). The default outcome was a predominant band of the predicted molecular mass that also agreed with the mass identified by western blot with an anti-His antibody. The concatenated forms of the Esx proteins and many of the similarly linked PPE-PE fusions purified from the soluble fraction as single band products indicating little if any breakdown of the fusion protein at the linker junction (Figure 3B). For example, purified EsxRST and EsxU (Figure 3, panel B, lanes 7 and 8) match those blotted (Figure 2, panel A, lanes 6 and 8) and PPE-PE fusions PPE41_PE25, PPE14_PE7, PPE15_PE8 and PPE20_PE15 (Figure 3, panel B, lanes 10, 14, 16 and 17) matched the proteins identified by blot in crude lysates (Figure 2, panel B, lanes 4, 7, 1 and 2). Sequence concatenation as a viable strategy to reduce clone numbers and to produce single polypeptide versions of multimeric bTB protein complexes, therefore appears feasible, as has been shown for other proteins (reviewed in (Matsushima et al., 2008; van Rosmalen et al., 2017)). Overall, the number of recombinant proteins purified directly without any form of optimisation was ∼50% of the number of clones tested (Table 1), low expression levels or insolubility accounting for the majority of the failures.
FIGURE 3.
Example histidine-tagged M. bovis proteins purified by immobilised metal affinity chromatography pull down with magnetic beads from E.coli (A) and insect cells (B). The lane identity and predicted molecular masses are: Panel A, 1-Mb0704 (44 kDa), 2-Mb3070 (37 kDa), 3-Mb2054 (36 kDa), 4-Mb2002_Mb0062 (35 kDa), 5-Mb0671 (17 kDa), 6-Mb0854 (31 kDa), 7- Mb3157c_Mb3743c (37 kDa), 8-Mb0337 (26 kDa). Panel B, 1-PE5_PE32 (19.5 kDa), 2-LpqN (25 kDa), 3-LpqX (22 kDa), 4-EsxAB (21.2 kDa), 5-EsxFDE (27.3 kDa), 6-EsxNOQ (30 kDa), 7- EsxRST (32 kDa), 8-EsxU (24 kDa), 9- Mb0485 (21 kDa), 10-PPE41_PE25 (34 kDa), 11-Mb1228 (39 kDa), 12-PPE69_PE36 (46.6 kDa), 13- PPE68_PE35 (47.5 kDa), 14-PPE14_PE7 (51 kDa), 15-PPE4_PE5 (67 kDa), 16- PPE15_PE8 (66 kDa), 17- PPE20_PE15 (67 kDa), 18-PPE31_PE20 (51 kDa). Asterisk (*) indicates all Esx related proteins, most as concatenates. Diamonds (♦) indicate concatenated PPE-PE pairs. M indicates the marker track, the molecular masses of which are given on the left of panel (A) in kilodaltons.
Problematic Clones
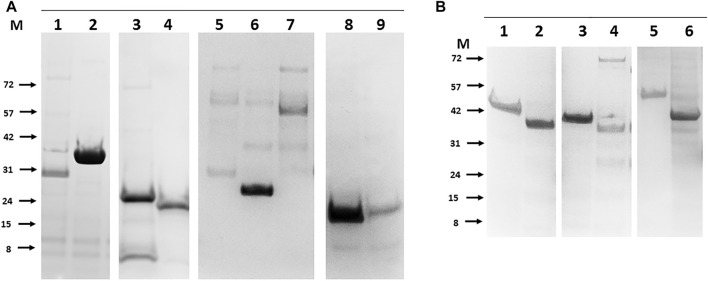
To improve the overall recombinant bTB protein recovery rate a limited number of ORF sequences were re-cloned as N-terminal His tagged variants and purification repeated. Cell wall proteins (Mb3338 and Mb3070), conserved hypothetical (Mb0854), ribosomal protein (Mb1656), three metabolic proteins (Mb0130, Mb3652 and Mb3871), and six proteins of unknown function (Mb2898, Mb2900, Mb2656, Mb1319, Mb1228 and Mb2244) were all rescued to purified soluble proteins by this approach (Figures 4A,B). Others, such as outer membrane proteins (Mb3644, Mb1445 and Mb2653) remained unworkable. These proteins may require folding partners to be co-expressed in order to be solubilised, as was the case for the original expression trials of PPE-PE family members (Strong et al., 2006).
FIGURE 4.
Rescue of soluble purified recombinant bTB proteins that were originally poorly expressed by alternate positioning of the His tag. Panel (A). Examples of N-terminal tagged proteins purified from E.coli. 1- Mb3338 (31 kDa), 2-Mb2656 (31 kDa), 3-Mb2898 (22 kDa), 4-Mb2900 (19 kDa), 5-Mb0130 (34 kDa), 6-Mb0854 (31 kDa), 7- Mb1656 (53 kDa), 8-Mb3652 (18 kDa), 9-Mb3871 (20 kDa). Panel (B). Examples of N-terminal tagged proteins purified from insect cells. 1- Mb0977_Mb3046c (43 kDa), 2-Mb0959 (39 kDa), 3-Mb1319 (44 kDa), 4-Mb3070 (37 kDa), 5-Mb2244 (53 kDa), 6-Mb1228 (39 kDa). Molecular weight markers (M) are labelled to the left of each panel and are in kilodaltons.
For some problematic proteins the case for their inclusion on grounds of likely candidacy as a vaccine component was such that several alternate forms of expression were assessed. EspA (Mb3646c) has been suggested to be part, with EspC, of the type VII secretion apparatus (discussed in (Ates and Brosch, 2017)) and an obvious candidate for inclusion (Chiliza et al., 2017). Recombinant EspC has been described (Lou et al., 2017; Salemi et al., 2020) and used diagnostically (Yan et al., 2018; Srinivasan et al., 2019; Villar-Hernández et al., 2020), but EspA has proven more difficult (Garces et al., 2010). EspA expression as either a C- or N-terminally His tagged protein was not successful and mutation of the single cysteine reported to drive dimer formation (Garces et al., 2010) had no effect. Fusion of EspA with EspC at either the N- or C- termini via a Gly-Ser linker did not improve expression but, of a number of enhancement tags investigated (GFP, SUMO and GB1), fusion of GFP to the N-terminus rescued detectable expression at the predicted molecular mass in both E. coli and insect cells and enabled modest levels to be purified (Figure 5A). Similarly, adding GB1 as an N- terminal fusion tag rescued expression of outer membrane protein Mb1762 and two unknown proteins, Mb1916 and Mb2410, allowing purification to reasonable levels, albeit with contaminating host derived proteins (Figure 5B). To assess if problematic expression was the result of any common feature in the proteins concerned, we inspected the AlphaFold predicted structures (Jumper et al., 2021) of the full length problematic proteins (Jumper et al., 2021) (Table 2). Of the 20 candidates analysed, only 4, Mb2244, Mb2656, Mb3652 and Mb3871 had available full length structures but in each case the proteins concerned were multimeric, some to a very high degree (Mb3871 is a 24mer), which may have limited expression here. Five proteins, Mb1656, Mb1916c, Mb2410, Mb2898 and Mb2900 had partial structures obtained with protein fragments suggesting that full length protein expression was not possible, and a further 5, Mb0130, Mb1228, Mb1319, Mb3070 and Mb3338, had fragment structures predicted on the basis of homology with other proteins whose structures have been solved. Of these 10 proteins, AlphaFold predictions suggested unstructured termini, predominantly at the N-terminus, as a likely limitation to stable protein expression. Unstructured termini, also with a predominance of unstructured N-termini, were associated with the 6 proteins (Mb0854, Mb1445, Mb1762, Mb2653, Mb3644 and Mb3646c) with no available structure, only 3 of which were recoverable here. Submission of the 20 candidate to the FuzDrop server (Hatos et al., 2022) showed a high score for likely aggregation and liquid-liquid phase separation associated with about half of the proteins for which there was either no structure or only a partial structure available (Table 2, indicated). Extensive protein engineering to reduce these scores might be necessary if these proteins were to be produced at a scale and purity required for a vaccine candidate.
FIGURE 5.
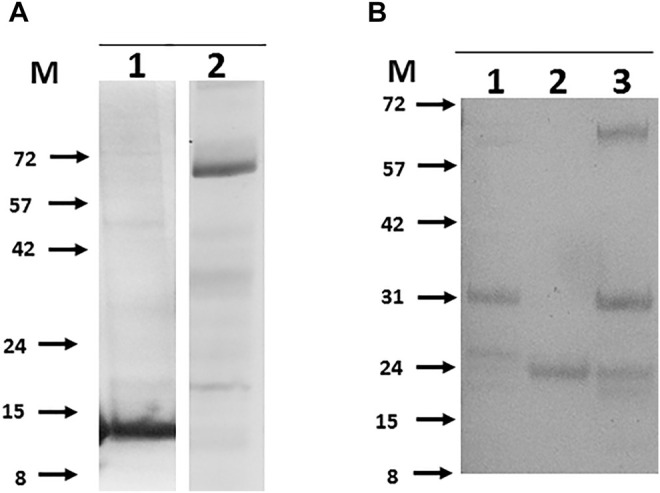
Rescued expression of problematic bTB proteins in E.coli by non-His fusion tags. Panel (A). The secretion accessory proteins EspC and EspA. 1- C-terminally His tagged EspC, 2- N-terminally GFP tagged EspA. Panel (B). 1-GB_Mb1762 (29 kDa), 2-GB_Mb1916 (25 kDa), 3-GB_Mb2410 (22 kDa) all expressed and purified as N-terminal fusions with the B1 domain of streptococcal protein G. Molecular weight markers (M) are labelled to left of each panel and are in kilodaltons.
TABLE 2.
Known and predicted structural features of difficult to express proteins.
| M.bovis | M. tuberculosis | Structure yes (Y), No (N), homology based (H) | Swiss-model | Alphafold Feature | LLPS Probability |
|---|---|---|---|---|---|
| Mb0130 | Rv1003 | H (2–230 of 285) | 3Kwp.1.B | None | 0.19 |
| Mb0854 | Rv0831c | N | Unstructured N-term | 0.2 | |
| Mb1228 | Rv1196 | H (2–175 of 391) | 5xfs.1.B | Unstructured C-term | 0.944 |
| Mb1319 | Rv1288 | H (164–447 of 456) | 6sx4.1.A | Burried N-term | 0.19 |
| Mb1445 | Rv1410c | N | Unstructured C-term | 0.136 | |
| Mb1656 | Rv1630 | Y (283–438 of 481) | 4NNI | Unstructured N-term; highly exteded structure | 0.225 |
| Mb1762 | Rv1733c | N | Unstructured N-term | 0.5 | |
| Mb1916c | Rv1884c | Y (68–153 of 176) | 4OW1 | Unstructured N-term | 0.31 |
| Mb2244 | Rv2220 | Y | 1HTQ | None | 0.19 |
| Mb2410 | Rv2389c | Y (50–127 of 154) | 4ow1.1.A | Unstructured N-term | 0.54 |
| Mb2653 | Rv2620c | N | None | 0.22 | |
| Mb2656 | Rv2623 | Y | 3CIS | None | 0.19 |
| Mb2898 | Rv2873 | Y (58–219 of 220) | 1nyo.1.A | Unstructured N-term | 0.63 |
| Mb2900 | Rv2875 | Y (31–193 of 193) | 1NYO | Unstructured N-term | 0.17 |
| Mb3070 | Rv3044 | H (68–352 of 359) | 3tny.1.A | Unstructured N-term | 0.76 |
| Mb3338 | Rv3310 | H (5–284 of 299) | 1e3c.1.B | Unstructured N-term | 0.2 |
| Mb3644 | Rv3614c | N | Unstructured N-term | 0.46 | |
| Mb3646c | Rv3616c | N | Unstructured C-term | 0.57 | |
| Mb3652 | Rv3628 | Y | 1.WCF | None | 0.18 |
| Mb3871 | Rv3841 | Y | 7O6E | None | 0.13 |
Discussion
Bovine tuberculosis (bTB) is a significant risk to animal health and welfare with estimates of >50 million cattle infected worldwide (Waters et al., 2012). Vaccination with M. bovis BCG, which is safe and inexpensive, shows varying efficacies (Francis, 1948; Berggren, 1981; Rodrigues and Smith, 1990) but can reduce spread in cattle indicating that control via vaccination is feasible (Hope et al., 2005; Lopez-Valencia et al., 2010; Waters et al., 2012). However, as up to 80% of BCG-vaccinated cattle may react positively to the tuberculin skin test at 6 months post-vaccination (Whelan et al., 2011), alternate vaccines including new attenuated mycobacterial vaccines (Khare et al., 2007; Waters et al., 2007; Blanco et al., 2013), DNA vaccines (Maue et al., 2004; Cai et al., 2005; Skinner et al., 2005; Maue et al., 2007) and virus-vectored vaccines (Vordermeier et al., 2009; Dean et al., 2014) have been investigated. The reverse vaccinology concept has provided a new serogroup B Neisseria meningitidis vaccine, Bexsero (Masignani et al., 2019) and has been used to select vaccine candidates for Staphylococcus aureus (Oprea and Antohe, 2013), Campylobacter jejuni (Muruato et al., 2017) and Streptococcus pneumonia (Argondizzo et al., 2015) in addition to Mycobacterium tuberculosis (Monterrubio-López et al., 2015). To test the feasibility for this approach we cloned and expressed >120 ORFs encoded by the reference M. bovis AF2122/97 genome and purified more than half of them using a combination of prokaryotic and eukaryotic expression. The list includes Esx protein family members, the PE and PPE families, a number of lipoproteins and others suggested to be potential vaccine candidates, including all of the targets predicted by the VaxiJen package (Monterrubio-López et al., 2015). A useful approach to reduce the number of clones required was to concatenate shorter ORFs or family partners together via flexible linkers. The successful expression and purification of many PPE-PE complexes (PPE4-PE5, PPE68-PE35, PPE69-PE36, PPE65-PE32, PPE41-PE25, PPE17-PE11, PPE15-PE8, PPE14-PE7, PPE31-PE20, PPE20-PE15 and PPE60-PE31) linked in this way was notable and was consistent with the PPE and PE partners folding to form a stable heterodimer (Strong et al., 2006). Many current TB vaccine candidates are fusions of several preselected ORFs (reviewed in (Dockrell, 2016)) and our data suggest that PPE-PE complexes, which have been noted as potential vaccine candidates (Stylianou et al., 2018), may be suitable additions. Currently, those candidate TB vaccines that contain PPE sequences, M72 and ID93, encode only fragments (Brennan, 2017). We did not investigate post-translational modification of any expressed protein as some of these are reportedly Mycobacteria specific (van Els et al., 2014) and may not occur in either of the expression hosts used although we noted no particular association with expression outcome. For example the heparin binding hemagglutinin (HBHA), which is methylated and toxic at high levels in Mycobacteria (Delogu et al., 2004), was well expression and purified from both E. coli and insect cells. We noted some trends in the outcome of expression, e.g. recovery of soluble rpf-like proteins (Kana et al., 2008; Mukamolova et al., 2010) was successful from the baculovirus system but not from E. coli where they were insoluble, and the combination of these two systems was beneficial overall, providing a success rate for the recovery of purified soluble protein of 67%, slightly higher than the 61% success rate reported during the development of Bexsero (Serruto et al., 2012; Masignani et al., 2019) and considerably higher than the ∼33% success rate previously reported for Mycobacterial protein expression using solely E. coli (Bashiri and Baker, 2015). An AlphaFold analysis of the proteins where re-cloning with N-terminal tags benefited expression or purification revealed that unstructured termini were a common feature. A tendency to aggregation was also noted by LLPS predication software (Hatos et al., 2022) with the regions identified located in the unstructured regions. Plausibly, the improvements in expression and recoverability for some the targets that were achieved by appending an N-terminal tag stabilised an otherwise disordered structure, reducing aggregation or degradation. A GFP fusion to the sequence encoding full length EspA, where the C-terminal 100 aa of the 319 aa EspA protein sequence has no predictable structure, uniquely allowed expression and purification of this vaccine candidate. A revised HTP scheme (Figure 1) might include an AlphaFold and FuzDrop screen for disorder prior to the selection of the endpoints for translation. That some proteins scored highly for LLPS is interesting given that such regions are associated with stress and lipid homeostasis (Vendruscolo, 2022), both of which are hallmarks of TB infection (Chow and Cox, 2011). Notwithstanding these considerations, our data suggest that the production of a large library of full length bTB proteins for candidate vaccine use is feasible. Recently the protection afforded by intravenous vaccination with BCG was shown to be correlate with the IgM component of the antibody response, although no antigen specificity was reported (Divangahi et al., 2021; Irvine et al., 2021). In addition, a monoclonal antibody to a single protein, PstS1 (Mb0959 in Table 1, purified in Figure 4B), was inhibitory in a whole blood Mtb growth inhibition assay (Watson et al., 2021). Antibodies from animals immunised with proteins from the range we describe here might be used to identify other targets relevant for vaccine development against Bovine TB or the related Mycobacterium tuberculosis and Mycobacterium leprae.
Acknowledgments
We thank Helen McShane, Elena Stylianou, Rachel Tanner, Martin Vordermeier and Bernardo Villarreal-Ramos for constructive comments. The work was funded by the Biotechnology and Biological Sciences Research Council.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
DP, MT, DR, AH, and AW did the experimental work. BC and IJ obtained the funding, supervised the work and wrote the first draft. All authors contributed to the final manuscript.
Funding
Funding for this research was from the BBSRC grant number BB/N004698/1.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Andersen K. R., Leksa N. C., Schwartz T. U. (2013). OptimizedE. Coliexpression Strain LOBSTR Eliminates Common Contaminants from His˗tag Purification. Proteins 81, 1857–1861. 10.1002/prot.24364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argondizzo A. P. C., da Mota F. F., Pestana C. P., Reis J. N., de Miranda A. B., Galler R., et al. (2015). Identification of Proteins in Streptococcus Pneumoniae by Reverse Vaccinology and Genetic Diversity of These Proteins in Clinical Isolates. Appl. Biochem. Biotechnol. 175, 2124–2165. 10.1007/s12010-014-1375-3 [DOI] [PubMed] [Google Scholar]
- Aricescu A. R., Assenberg R., Bill R. M., Busso D., Chang V. T., Davis S. J., et al. (2006). Eukaryotic Expression: Developments for Structural Proteomics. Acta Crystallogr. D. Biol. Cryst. 62, 1114–1124. 10.1107/s0907444906029805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ates L. S., Brosch R. (2017). Discovery of the Type VII ESX-1 Secretion Needle? Mol. Microbiol. 103, 7–12. 10.1111/mmi.13579 [DOI] [PubMed] [Google Scholar]
- Bashiri G., Baker E. N. (2015). Production of Recombinant Proteins inMycobacterium Smegmatisfor Structural and Functional Studies. Protein Sci. 24, 1–10. 10.1002/pro.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berggren S. A. (1981). Field Experiment with BCG Vaccine in Malawi. Br. Veterinary J. 137, 88–94. 10.1016/s0007-1935(17)31792-x [DOI] [PubMed] [Google Scholar]
- Bielby J., Donnelly C. A., Pope L. C., Burke T., Woodroffe R. (2014). Badger Responses to Small-Scale Culling May Compromise Targeted Control of Bovine Tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 111, 9193–9198. 10.1073/pnas.1401503111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird L. E., Rada H., Flanagan J., Diprose J. M., Gilbert R. J. C., Owens R. J. (2014). Application of In-Fusion Cloning for the Parallel Construction of E. coli Expression Vectors. Methods Mol. Biol. 1116, 209–234. 10.1007/978-1-62703-764-8_15 [DOI] [PubMed] [Google Scholar]
- Blanco F. C., Bianco M. V., Garbaccio S., Meikle V., Gravisaco M. J., Montenegro V., et al. (2013). Mycobacterium bovis Δmce2 Double Deletion Mutant Protects Cattle against Challenge with Virulent M. Bovis. Tuberculosis 93, 363–372. 10.1016/j.tube.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Brennan M. J. (2017). The Enigmatic PE/PPE Multigene Family of Mycobacteria and Tuberculosis Vaccination. Infect. Immun. 85. 10.1128/IAI.00969-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Tian X., Hu X. D., Li S. X., Yu D. H., Zhu Y. X. (2005). Combined DNA Vaccines Formulated Either in DDA or in Saline Protect Cattle from Mycobacterium bovis Infection. Vaccine 23, 3887–3895. 10.1016/j.vaccine.2005.03.025 [DOI] [PubMed] [Google Scholar]
- Chambers M. A., Carter S. P., Wilson G. J., Jones G., Brown E., Hewinson R. G., et al. (2014). Vaccination against Tuberculosis in Badgers and Cattle: an Overview of the Challenges, Developments and Current Research Priorities in Great Britain. Veterinary Rec. 175, 90–96. 10.1136/vr.102581 [DOI] [PubMed] [Google Scholar]
- Chandran A., Williams K., Mendum T., Stewart G., Clark S., Zadi S., et al. (2019). Development of a Diagnostic Compatible BCG Vaccine against Bovine Tuberculosis. Sci. Rep. 9, 17791. 10.1038/s41598-019-54108-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiliza T. E., Pillay M., Pillay B. (2017). Identification of Unique Essential Proteins from a Mycobacterium tuberculosis F15/LAM4/KZN Phage Secretome Library. Pathog. Dis. 75. 10.1093/femspd/ftx001 [DOI] [PubMed] [Google Scholar]
- Chim N., Habel J. E., Johnston J. M., Krieger I., Miallau L., Sankaranarayanan R., et al. (2011). The TB Structural Genomics Consortium: a Decade of Progress. Tuberculosis 91, 155–172. 10.1016/j.tube.2010.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow E. D., Cox J. S. (2011). TB Lipidomics-The Final Frontier. Chem. Biol. 18, 1517–1518. 10.1016/j.chembiol.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai F. Y., Wang J. F., Gong X. L., Bao L. (2017). Immunogenicity and Protective Efficacy of Recombinant Bacille Calmette-Guerin Strains Expressing mycobacterium Antigens Ag85A, CFP10, ESAT-6, GM-CSF and IL-12p70. Hum. Vaccin Immunother. 13, 1–8. 10.1080/21645515.2017.1279771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean G., Whelan A., Clifford D., Salguero F. J., Xing Z., Gilbert S., et al. (2014). Comparison of the Immunogenicity and Protection against Bovine Tuberculosis Following Immunization by BCG-Priming and Boosting with Adenovirus or Protein Based Vaccines. Vaccine 32, 1304–1310. 10.1016/j.vaccine.2013.11.045 [DOI] [PubMed] [Google Scholar]
- Delogu G., Bua A., Pusceddu C., Parra M., Fadda G., Brennan M. J., et al. (2004). Expression and Purification of Recombinant Methylated HBHA inMycobacterium Smegmatis. FEMS Microbiol. Lett. 239, 33–39. 10.1016/j.femsle.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Dhanda S. K., Vir P., Singla D., Gupta S., Kumar S., Raghava G. P. S. (2016). A Web-Based Platform for Designing Vaccines against Existing and Emerging Strains of Mycobacterium tuberculosis . PloS one 11, e0153771. 10.1371/journal.pone.0153771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divangahi M., Javid B., Kaufmann E. (2021). 100 Years of Antibody Solitude in TB. Nat. Immunol. 22 (2021), 1470–1471. 10.1038/s41590-021-01071-4 [DOI] [PubMed] [Google Scholar]
- Dockrell H. M. (2016). Towards New TB Vaccines: What Are the Challenges? Pathogens Dis. 74, ftw016. 10.1093/femspd/ftw016 [DOI] [PubMed] [Google Scholar]
- Fletcher H. A., Schrager L. (2016). TB Vaccine Development and the End TB Strategy: Importance and Current Status. Trans. R. Soc. Trop. Med. Hyg. 110, 212–218. 10.1093/trstmh/trw016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis J. (1948). Bovine Tuberculosis Including a Contrast with Human Tuberculosis. Indian Med. Gazette 83, 250–251. [Google Scholar]
- Garces A., Atmakuri K., Chase M. R., Woodworth J. S., Krastins B., Rothchild A. C., et al. (2010). EspA Acts as a Critical Mediator of ESX1-dependent Virulence in Mycobacterium tuberculosis by Affecting Bacterial Cell Wall Integrity. PLoS Pathog. 6, e1000957. 10.1371/journal.ppat.1000957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A., 3rd, Smith H. O. (2009). Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 6, 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Zhang S., Zhang S., Chen Y.-R., Blissard G. W. (2012). Correction: BTI-Tnao38, a New Cell Line Derived from Trichoplusia Ni, Is Permissive for AcMNPV Infection and Produces High Levels of Recombinant Proteins. BMC Biotechnol. 12, 12. 10.1186/1472-6750-12-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatos A., Tosatto S. C. E., Vendruscolo M., Fuxreiter M. (2022). FuzDrop on AlphaFold: Visualizing the Sequence-dependent Propensity of Liquid-Liquid Phase Separation and Aggregation of Proteins. Nucleic acids Res. 2022, gkac386. 10.1093/nar/gkac386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Xiang Z., Mobley H. L. (2010). Vaxign: the First Web-Based Vaccine Design Program for Reverse Vaccinology and Applications for Vaccine Development. J. Biomed. Biotechnol. 2010, 297505. 10.1155/2010/297505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope J. C., Thom M. L., Villarreal-Ramos B., Vordermeier H. M., Hewinson R. G., Howard C. J. (2005). Exposure to Mycobacterium avium Induces Low-Level Protection from Mycobacterium bovis Infection but Compromises Diagnosis of Disease in Cattle. Clin. Exp. Immunol. 141, 432–439. 10.1111/j.1365-2249.2005.02882.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine E. B., O’Neil A., Darrah P. A., Shin S., Choudhary A., Li W., et al. (2021). Robust IgM Responses Following Intravenous Vaccination with Bacille Calmette-Guérin Associate with Prevention of Mycobacterium tuberculosis Infection in Macaques. Nat. Immunol. 22, 1515–1523. 10.1038/s41590-021-01066-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly Accurate Protein Structure Prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana B. D., Gordhan B. G., Downing K. J., Sung N., Vostroktunova G., Machowski E. E., et al. (2008). The Resuscitation-Promoting Factors ofMycobacterium Tuberculosisare Required for Virulence and Resuscitation from Dormancy but Are Collectively Dispensable for Growthin Vitro. Mol. Microbiol. 67, 672–684. 10.1111/j.1365-2958.2007.06078.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapopoulou A., Lew J. M., Cole S. T. (2011). The MycoBrowser Portal: a Comprehensive and Manually Annotated Resource for Mycobacterial Genomes. Tuberculosis 91, 8–13. 10.1016/j.tube.2010.09.006 [DOI] [PubMed] [Google Scholar]
- Kaufmann S. H. E., Weiner J., von Reyn C. F. (2017). Novel Approaches to Tuberculosis Vaccine Development. Int. J. Infect. Dis. 56, 263–267. 10.1016/j.ijid.2016.10.018 [DOI] [PubMed] [Google Scholar]
- Khare S., Hondalus M. K., Nunes J., Bloom B. R., Garry Adams L. (2007). Mycobacterium bovis ΔleuD Auxotroph-Induced Protective Immunity against Tissue Colonization, Burden and Distribution in Cattle Intranasally Challenged with Mycobacterium bovis Ravenel S. Vaccine 25, 1743–1755. 10.1016/j.vaccine.2006.11.036 [DOI] [PubMed] [Google Scholar]
- Kim H.-W., Lee S.-Y., Park H., Jeon S.-J. (2015). Expression, Refolding, and Characterization of a Small Laccase from Thermus Thermophilus HJ6. Protein Expr. Purif. 114, 37–43. 10.1016/j.pep.2015.06.004 [DOI] [PubMed] [Google Scholar]
- Lee C.-D., Sun H.-C., Hu S.-M., Chiu C.-F., Homhuan A., Liang S.-M., et al. (2008). An Improved SUMO Fusion Protein System for Effective Production of Native Proteins. Protein Sci. 17, 1241–1248. 10.1110/ps.035188.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Xu M., Qin C., Xia L., Xiong Y., Xi X., et al. (2016). Recombinant Fusion ESAT6-CFP10 Immunogen as a Skin Test Reagent for Tuberculosis Diagnosis: an Open-Label, Randomized, Two-Centre Phase 2a Clinical Trial. Clin. Microbiol. Infect. 22, 889–e16. e16. 10.1016/j.cmi.2016.07.015 [DOI] [PubMed] [Google Scholar]
- Li W., Li M., Deng G., Zhao L., Liu X., Wang Y. (2015). Prime-boost Vaccination with Bacillus Calmette Guerin and a Recombinant Adenovirus Co-expressing CFP10, ESAT6, Ag85A and Ag85B of Mycobacterium tuberculosis Induces Robust Antigen-specific Immune Responses in Mice. Mol. Med. Rep. 12, 3073–3080. 10.3892/mmr.2015.3770 [DOI] [PubMed] [Google Scholar]
- Lindhout D. A., Thiessen A., Schieve D., Sykes B. D. (2003). High-yield Expression of Isotopically Labeled Peptides for Use in NMR Studies. Protein Sci. 12, 1786–1791. 10.1110/ps.0376003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Valencia G., Renteria-Evangelista T., Williams J. d. J., Licea-Navarro A., Mora-Valle A. D. l., Medina-Basulto G. (2010). Field Evaluation of the Protective Efficacy of Mycobacterium bovis BCG Vaccine against Bovine Tuberculosis. Res. veterinary Sci. 88, 44–49. 10.1016/j.rvsc.2009.05.022 [DOI] [PubMed] [Google Scholar]
- Lou Y., Rybniker J., Sala C., Cole S. T. (2017). EspC Forms a Filamentous Structure in the Cell Envelope ofMycobacterium Tuberculosisand Impacts ESX-1 Secretion. Mol. Microbiol. 103, 26–38. 10.1111/mmi.13575 [DOI] [PubMed] [Google Scholar]
- Masignani V., Pizza M., Moxon E. R. (2019). The Development of a Vaccine against Meningococcus B Using Reverse Vaccinology. Front. Immunol. 10, 751. 10.3389/fimmu.2019.00751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima N., Yoshida H., Kumaki Y., Kamiya M., Tanaka T., Izumi Y., et al. (2008). Flexible Structures and Ligand Interactions of Tandem Repeats Consisting of Proline, glycine, Asparagine, Serine, And/or Threonine Rich Oligopeptides in Proteins. Cpps 9, 591–610. 10.2174/138920308786733886 [DOI] [PubMed] [Google Scholar]
- Maue A. C., Waters W. R., Palmer M. V., Nonnecke B. J., Minion F. C., Brown W. C., et al. (2007). An ESAT-6:CFP10 DNA Vaccine Administered in Conjunction with Mycobacterium bovis BCG Confers Protection to Cattle Challenged with Virulent M. Bovis. Vaccine 25, 4735–4746. 10.1016/j.vaccine.2007.03.052 [DOI] [PubMed] [Google Scholar]
- Maue A. C., Waters W. R., Palmer M. V., Whipple D. L., Minion F. C., Brown W. C., et al. (2004). CD80 and CD86, but Not CD154, Augment DNA Vaccine-Induced Protection in Experimental Bovine Tuberculosis. Vaccine 23, 769–779. 10.1016/j.vaccine.2004.07.019 [DOI] [PubMed] [Google Scholar]
- McShane H. (2019). Insights and Challenges in Tuberculosis Vaccine Development. Lancet Respir. Med. 7, 810–819. 10.1016/s2213-2600(19)30274-7 [DOI] [PubMed] [Google Scholar]
- Mearns H., Geldenhuys H. D., Kagina B. M., Musvosvi M., Little F., Ratangee F., et al. (2017). H1:IC31 Vaccination Is Safe and Induces Long-Lived TNF-α+IL-2+CD4 T Cell Responses in M. tuberculosis Infected and Uninfected Adolescents: A Randomized Trial. Vaccine 35, 132–141. 10.1016/j.vaccine.2016.11.023 [DOI] [PubMed] [Google Scholar]
- Monterrubio-López G. P., González-Y-Merchand J. A., Ribas-Aparicio R. M. (2015). Identification of Novel Potential Vaccine Candidates against Tuberculosis Based on Reverse Vaccinology. Biomed. Res. Int. 2015, 483150. 10.1155/2015/483150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamolova G. V., Turapov O., Malkin J., Woltmann G., Barer M. R. (2010). Resuscitation-promoting Factors Reveal an Occult Population of Tubercle Bacilli in Sputum. Am. J. Respir. Crit. Care Med. 181, 174–180. 10.1164/rccm.200905-0661oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruato L. A., Tapia D., Hatcher C. L., Kalita M., Brett P. J., Gregory A. E., et al. (2017). Use of Reverse Vaccinology in the Design and Construction of Nanoglycoconjugate Vaccines against Burkholderia Pseudomallei. Clin. Vaccine Immunol. 24, 1–13. 10.1128/CVI.00206-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettleship J. E., Assenberg R., Diprose J. M., Rahman-Huq N., Owens R. J. (2010). Recent Advances in the Production of Proteins in Insect and Mammalian Cells for Structural Biology. J. Struct. Biol. 172, 55–65. 10.1016/j.jsb.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Nielsen H., Tsirigos K. D., Brunak S., von Heijne G. (2019). A Brief History of Protein Sorting Prediction. Protein J. 38, 200–216. 10.1007/s10930-019-09838-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprea M., Antohe F. (2013). Reverse-vaccinology Strategy for Designing T-Cell Epitope Candidates for Staphylococcus aureus Endocarditis Vaccine. Biologicals 41, 148–153. 10.1016/j.biologicals.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Pengelley S. C., Chapman D. C., Mark Abbott W., Lin H.-H., Huang W., Dalton K., et al. (2006). A Suite of Parallel Vectors for Baculovirus Expression. Protein Expr. Purif. 48, 173–181. 10.1016/j.pep.2006.04.016 [DOI] [PubMed] [Google Scholar]
- Porta C., Kotecha A., Burman A., Jackson T., Ren J., Loureiro S., et al. (2013). Rational Engineering of Recombinant Picornavirus Capsids to Produce Safe, Protective Vaccine Antigen. PLoS Pathog. 9, e1003255. 10.1371/journal.ppat.1003255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R., Bottomley M. J., D’Oro U., Finco O., De Gregorio E. (2016). Reverse Vaccinology 2.0: Human Immunology Instructs Vaccine Antigen Design. J. Exp. Med. 213, 469–481. 10.1084/jem.20151960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R. (2001). Reverse Vaccinology, a Genome-Based Approach to Vaccine Development. Vaccine 19, 2688–2691. 10.1016/s0264-410x(00)00554-5 [DOI] [PubMed] [Google Scholar]
- Rodrigues L. C., Smith P. G. (1990). Tuberculosis in Developing Countries and Methods for its Control. Trans. R. Soc. Trop. Med. Hyg. 84, 739–744. 10.1016/0035-9203(90)90172-b [DOI] [PubMed] [Google Scholar]
- Salemi O., Noormohammadi Z., Bahrami F., Siadat S. D., Ajdary S. (2020). Cloning, Expression and Purification of Espc, Espb and Espc/Espb Proteins of Mycobacterium tuberculosis ESX-1 Secretion System. Rep. Biochem. Mol. Biol. 8, 465–472. [PMC free article] [PubMed] [Google Scholar]
- Schlager B., Straessle A., Hafen E. (2012). Use of Anionic Denaturing Detergents to Purify Insoluble Proteins after Overexpression. BMC Biotechnol. 12, 95. 10.1186/1472-6750-12-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruto D., Bottomley M. J., Ram S., Giuliani M. M., Rappuoli R. (2012). The New Multicomponent Vaccine against Meningococcal Serogroup B, 4CMenB: Immunological, Functional and Structural Characterization of the Antigens. Vaccine 30 (2), B87–B97. 10.1016/j.vaccine.2012.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M. A., Wedlock D. N., de Lisle G. W., Cooke M. M., Tascon R. E., Ferraz J. C., et al. (2005). The Order of Prime-Boost Vaccination of Neonatal Calves with Mycobacterium bovis BCG and a DNA Vaccine Encoding Mycobacterial Proteins Hsp65, Hsp70, and Apa Is Not Critical for Enhancing Protection against Bovine Tuberculosis. Infect. Immun. 73, 4441–4444. 10.1128/iai.73.7.4441-4444.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. H. (2012). The Global Distribution and Phylogeography of Mycobacterium bovis Clonal Complexes. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 12, 857–865. 10.1016/j.meegid.2011.09.007 [DOI] [PubMed] [Google Scholar]
- Srinivasan S., Jones G., Veerasami M., Steinbach S., Holder T., Zewude A., et al. (2019). A Defined Antigen Skin Test for the Diagnosis of Bovine Tuberculosis. Sci. Adv. 5, eaax4899. 10.1126/sciadv.aax4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., Eisenberg D. (2006). Toward the Structural Genomics of Complexes: Crystal Structure of a PE/PPE Protein Complex from Mycobacterium tuberculosis . Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065. 10.1073/pnas.0602606103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylianou E., Harrington-Kandt R., Beglov J., Bull N., Pinpathomrat N., Swarbrick G. M., et al. (2018). Identification and Evaluation of Novel Protective Antigens for the Development of a Candidate Tuberculosis Subunit Vaccine. Infect. Immun. 86. 10.1128/IAI.00014-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier D. C., Thomas D. Y., Khouri H. E., Laliberié F., Vernet T. (1991). Enhanced Secretion from Insect Cells of a Foreign Protein Fused to the Honeybee Melittin Signal Peptide. Gene 98, 177–183. 10.1016/0378-1119(91)90171-7 [DOI] [PubMed] [Google Scholar]
- Tkachuk A. P., Gushchin V. A., Potapov V. D., Demidenko A. V., Lunin V. G., Gintsburg A. L. (2017). Multi-subunit BCG Booster Vaccine GamTBvac: Assessment of Immunogenicity and Protective Efficacy in Murine and guinea Pig TB Models. PloS one 12, e0176784. 10.1371/journal.pone.0176784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Els C. c. A. C. M., Corbière V. r., Smits K., van Gaans-van den Brink J. A. M., Poelen M. C. M., Mascart F., et al. (2014). Toward Understanding the Essence of Post-Translational Modifications for the Mycobacterium tuberculosis Immunoproteome. Front. Immunol. 5, 361. 10.3389/fimmu.2014.00361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rosmalen M., Krom M., Merkx M. (2017). Tuning the Flexibility of Glycine-Serine Linkers to Allow Rational Design of Multidomain Proteins. Biochemistry 56, 6565–6574. 10.1021/acs.biochem.7b00902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendruscolo M. (2022). Lipid Homeostasis and its Links with Protein Misfolding Diseases. Front. Mol. Neurosci. 15, 829291. 10.3389/fnmol.2022.829291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar-Hernández R., Blauenfeldt T., García-García E., Muriel-Moreno B., De Souza-Galvão M. L., Millet J. P., et al. (2020). Diagnostic Benefits of Adding EspC, EspF and Rv2348-B to the QuantiFERON Gold In-Tube Antigen Combination. Sci. Rep. 10, 13234. 10.1038/s41598-020-70204-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vordermeier M., Gordon S. V., Hewinson A. R. G. (2009). Antigen Mining to Define Mycobacterium bovis Antigens for the Differential Diagnosis of Vaccinated and Infected Animals: A VLA Perspective. Transbound. Emerg. Dis. 56, 240–247. 10.1111/j.1865-1682.2009.01070.x [DOI] [PubMed] [Google Scholar]
- Vordermeier M., Gordon S. V., Hewinson R. G. (2011). Mycobacterium bovis Antigens for the Differential Diagnosis of Vaccinated and Infected Cattle. Veterinary Microbiol. 151, 8–13. 10.1016/j.vetmic.2011.02.020 [DOI] [PubMed] [Google Scholar]
- Vordermeier M., Hewinson R. G. (2006). Development of Cattle TB Vaccines in the UK. Veterinary Immunol. Immunopathol. 112, 38–48. 10.1016/j.vetimm.2006.03.010 [DOI] [PubMed] [Google Scholar]
- Waters W. R., Palmer M. V., Buddle B. M., Vordermeier H. M. (2012). Bovine Tuberculosis Vaccine Research: Historical Perspectives and Recent Advances. Vaccine 30, 2611–2622. 10.1016/j.vaccine.2012.02.018 [DOI] [PubMed] [Google Scholar]
- Waters W. R., Palmer M. V., Nonnecke B. J., Thacker T. C., Scherer C. F. C., Estes D. M., et al. (2007). Failure of a Mycobacterium tuberculosis ΔRD1 ΔpanCD Double Deletion Mutant in a Neonatal Calf Aerosol M. Bovis Challenge Model: Comparisons to Responses Elicited by M. Bovis Bacille Calmette Guerin. Vaccine 25, 7832–7840. 10.1016/j.vaccine.2007.08.029 [DOI] [PubMed] [Google Scholar]
- Watson A., Li H., Ma B., Weiss R., Bendayan D., Abramovitz L., et al. (2021). Human Antibodies Targeting a Mycobacterium Transporter Protein Mediate Protection against Tuberculosis. Nat. Commun. 12, 602. 10.1038/s41467-021-20930-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan A. O., Clifford D., Upadhyay B., Breadon E. L., McNair J., Hewinson G. R., et al. (2010). Development of a Skin Test for Bovine Tuberculosis for Differentiating Infected from Vaccinated Animals. J. Clin. Microbiol. 48, 3176–3181. 10.1128/jcm.00420-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan A. O., Coad M., Upadhyay B. L., Clifford D. J., Hewinson R. G., Vordermeier H. M. (2011). Lack of Correlation between BCG-Induced Tuberculin Skin Test Sensitisation and Protective Immunity in Cattle. Vaccine 29, 5453–5458. 10.1016/j.vaccine.2011.05.057 [DOI] [PubMed] [Google Scholar]
- Yan Z. H., Yi L., Wei P. J., Jia H. Y., Wang J., Wang X. J., et al. (2018). Evaluation of Panels of Mycobacterium tuberculosis Antigens for Serodiagnosis of Tuberculosis. Int. J. Tuberc. Lung Dis. 22, 959–965. 10.5588/ijtld.18.0060 [DOI] [PubMed] [Google Scholar]
- Zhang W., Zhang Y., Zheng H., Pan Y., Liu H., Du P., et al. (2013). Genome Sequencing and Analysis of BCG Vaccine Strains. PloS one 8, e71243. 10.1371/journal.pone.0071243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimpel C. K., Patané J. S. L., Guedes A. C. P., de Souza R. F., Silva-Pereira T. T., Camargo N. C. S., et al. (2020). Global Distribution and Evolution of Mycobacterium bovis Lineages. Front. Microbiol. 11, 843. 10.3389/fmicb.2020.00843 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.