Abstract
The hya operon of Escherichia coli is composed of the genes which synthesize uptake hydrogenase isoenzyme 1 (Hyd1). Although hya expression and Hyd1 synthesis occur only under anaerobic conditions, Hyd1 is not essential for growth. In this study we used a hya′-′lacZ fusion to characterize parameters of anaerobic growth that maximize hya expression in an attempt to further elucidate Hyd1 function. We found that the expression pattern of hya followed a decline of external pH. In buffered media where the pH value was set, the onset of hya expression initiated earlier in growth and reached a greater peak level in acidic than in alkaline medium. When cultures expressing hya were shifted from acidic to alkaline conditions, hya expression was arrested; shifting from alkaline to acidic conditions stimulated hya expression. Maximal expression of hya under all growth conditions required the sigma factor RpoS and transcriptional regulators AppY and ArcA. In the absence of RpoS or AppY, the response of hya expression onset to external pH was evident and maximal hya levels remained greater in acidic than in alkaline medium. However, the absence of ArcA led to a diminished response of expression onset to external pH and the loss of elevated expression at an acidic external pH. The fermentation end product formate slightly altered hya expression levels but was not required for hya to respond to external pH. In contrast to hya expression, the onset of hyb operon expression, encoding uptake hydrogenase isoenzyme 2, was constitutive with respect to external pH. However, external pH did affect hyb expression levels, which, in contrast to hya, were maximal in alkaline rather than acidic medium.
During anaerobic growth, Escherichia coli is capable of synthesizing two uptake hydrogenase isoenzymes, Hyd1 and Hyd2, which catalyze the oxidation of hydrogen gas (5, 32). Hyd1 and Hyd2 are αβ heterodimers, consisting of a small subunit and a nickel-containing catalytic large subunit (5, 32), that localize to the inner membrane facing the periplasmic space (28, 29). The H2 uptake-specific activity of purified Hyd2 is greater than that of Hyd1, while purified active Hyd1 is more resistant to denaturation by changes in pH and prolonged exposure to oxygen (5, 32). The pH optimum for Hyd1 ranges from 6 to 8, whereas that of Hyd2 is 8 (5, 32).
The genes encoding the Hyd1 small and large subunits, hyaA and hyaB, respectively, comprise part of the hya operon (24), while the genes encoding the Hyd2 small and large subunits, hybA and hybC, respectively, comprise part of the hyb operon (23). In addition to the structural genes, the hyaDEF and hybFG genes are required for posttranslational modification of nascent structural subunits (23, 25). Complete transcription of both operons is required for maximal expression of active Hyd1 and Hyd2 complexes (23–25).
The redundancy of uptake hydrogenase formation is partially explained by the differential regulation of hya and hyb operon expression and functions of Hyd1 and Hyd2 in anaerobic metabolism. Anaerobic induction of hyb expression requires the Fnr protein and is enhanced by exogenous fumarate (31, 40). Likewise, Hyd2 activity levels are maximal in cultures grown on hydrogen and fumarate (23, 31). An hyb mutant that lacks Hyd2 activity is unable to grow anaerobically on hydrogen and fumarate (23), which strongly suggests that Hyd2 functions in contributing electrons from hydrogen oxidation to fumarate reductase for the reduction of fumarate to succinate (5, 23, 31).
Hyd1 has no defined function in anaerobic metabolism, but it has been proposed to shuttle electrons from formate to fumarate during fumarate reduction (12) and/or to oxidize hydrogen gas and contribute electrons to the quinone pool (31). Transcription of hya is induced by anaerobiosis and repressed by nitrate (9) and requires the sigma factor RpoS (2) and the transcriptional regulators ArcA and AppY (9). Conflicting reports on the effects of carbon source on hya expression and Hyd1 synthesis during growth have been published. Sawers et al. reported that Hyd1 enzymatic activity was maximal in cultures grown on glucose and enhanced by the addition of exogenous formate (31). Formate also led to the recovery of Hyd1 activity in a pfl-1 mutant defective in formate production (17, 31). Furthermore, Bronsted and Atlung demonstrated that formate enhanced the level of hya transcription during fermentative growth on glucose (9). In contrast, Menon et al. have reported that the levels of active Hyd1 are essentially identical in cultures grown on either glucose or glucose plus formate (25).
Although the physiological function of Hyd1 has not yet been defined, the response of hya expression to carbon and phosphate starvation (2) and dependence on RpoS and AppY (2, 9) suggest that it is involved in response to stress. Here we report that during anaerobic growth, hya responds to the external pH (pHe). We also determined the effect of pHe on the expression of hyb to characterize the effect of pHe on anaerobic expression of both uptake hydrogenase operons.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in these experiments are listed in Table 1. All generalized transductions were conducted as described previously (34) with phage P1vir. To create strain JK22, the selectable marker zjj::Tn10 from donor strain ECL618 (14) was transduced into strain JK2. The resulting Apr Tetr transductants were then scored for sensitivity to toluidine blue, as described previously (15), to indicate inheritance of the arcA2 mutation.
TABLE 1.
E. coli strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| BW545 | ΔlacU169 rpsL | Laboratory collection |
| ECL618 | arcA2 zjj::Tn10 | 14a |
| LCB320 | F−thr-1 leu-6 thi-1 lacY tonA22 strA | 38b |
| LCB898 | Like LCB320 but pfl-1 | 38b |
| UM122 | HfrH thi-1 katF13::Tn10 | 21c |
| RW193 | ara-14 leuB6 secA206 lacY1 proC14 tsx-67 entA403 glnV44 λ− trpE38 rfbD1 rpsL109 xylA5 mtl-1 thi-1 | CGSCd |
| UT5600 | Like RW193 but Δ(ompT-fepC)266 | CGSCd |
| MC4100 | F−araD139 (arg-lac)U169 rpsL150 relA flb-5301 ptsF25 deoC1 | Laboratory collection |
| SE2065 | Like MC4100 but φhyb′-′lacZ, Kanr | 39e |
| JK2 | Like BW545 but attB::hya′-′lacZ, Apr | This study |
| JK1222 | Like JK2 but katF13::Tn10 | P1(UM122) × JK2 |
| JK22 | Like JK2 but arcA2 zjj::Tn10 | P1(ECL618) × JK2 |
| JK2320 | Like LCB320 but attB::hya′-′lacZ, Apr | P1(JK2) × LCB320 |
| JK2898 | Like LCB898 but attB::hya′-′lacZ, Apr | P1(JK2) × LCB898 |
| JK2193 | Like RW193 but attB::hya′-′lacZ, Apr | P1(JK2) × RW193 |
| JK256 | Like UT5600 but attB::hya′-′lacZ, Apr | P1(JK2) × UT5600 |
S. Iuchi (source of strain).
D. Clark.
P. Loewen.
Coli Genetic Stock Center (M. Berlyn, Curator).
K. T. Shanmugam.
Construction of the hya′-′lacZ chromosome fusion and strain JK2.
Plasmid pCL47 (24) containing the hya promoter and the entire hya operon was digested with EcoRI and BamHI. The 470-bp EcoRI-BamHI fragment containing the promoter region and first 164 bp of hyaA was isolated by agarose gel electrophoresis, purified, treated with EcoRI and BamHI, and ligated into EcoRI-BamHI-digested plasmid pMC1403 (Apr, ′lacZYA) (11). This created an in-frame fusion of codon 58 of hyaA with codon 8 of lacZ. Plasmid pAF11 was isolated by its ability to confer Lac+ Apr to BW545, and the presence of the hya′-′lacZ junction was confirmed by DNA sequencing.
The hya′-′lacZ fusion was recombined onto phage λRZ5 by previously described methods (34) to generate λAF11. A λAF11 lysate was used to infect BW545, and the Lac+ Apr single lysogen, JK2, was isolated. Strain JK2 is a merodiploid strain, since the hya locus is maintained when phage λAF11 integrates at the lambda attachment site on the chromosome.
Growth conditions.
Strains were routinely cultured at 37°C in liquid Luria broth (LB) (10% tryptone, 5% yeast extract, 10% NaCl) or on solid Luria agar medium (LB with 15% agar added). Carbon sources and supplements for growth in liquid media were used at the following concentrations: glucose, 0.4%; glycerol, 0.4%; sodium fumarate, 0.5%; sodium formate, 0.4%. Growth in liquid minimal media was performed with M63 medium (34) with 0.2% glucose as a carbon source. Antibiotics were used at the following concentrations: 100 μg of ampicillin per ml, 30 μg of kanamycin per ml, and 25 μg of tetracycline per ml. Cultures were grown aerobically by shaking 20-ml cultures in 250-ml flasks at 250 rpm on a rotary shaker. Anaerobic growth of cultures was carried out in either 15-, 20-, or 35-ml bottles fitted with a rubber stopper and capped (25).
For the screening of Lac+ lysogens on solid media, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside was added at 50 μg/ml (3). The appearance of blue colonies indicated expression of β-galactosidase activity.
Anaerobic expression of hya′-′lacZ and hyb′-′lacZ fusions.
To assay for either hya′-′lacZ or hyb′-′lacZ expression in cells, the appropriate strain was grown aerobically overnight in liquid medium and diluted 1:100 in 20 ml of LB or M63 medium (pH 7.2) containing either glucose (hya′-′lacZ expression assays) (25) or glycerol and fumarate (hyb′-′lacZ expression assays) (23). This culture was grown aerobically until mid-log phase (approximately 2 to 3 h). A 1-ml volume of cells was collected by centrifugation, washed, and resuspended 1:1 in fresh medium. These cells were then diluted 1:100 in 20 ml of either LB or M63 medium containing the appropriate carbon source and grown anaerobically as described above.
To assay for hya′-′lacZ or hyb′-′lacZ expression during anaerobic growth at an acidic pH, 2-(N-morpholino)ethanesulfonic acid (MES; pKa = 6.1) at 100 mM (35) was added to LB and the pH was adjusted to 5.5 using sodium hydroxide. To assay for expression during growth at basic pH, N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES; pKa = 7.4) was added at 100 mM to LB and the pH was adjusted to 8.5 with sodium hydroxide.
pHe shift experiments.
To measure the response of hya′-′lacZ expression to a shift from alkaline to acidic pHe, an aerobic culture of strain JK2 was prepared as above and used to inoculate 35 ml of LB containing glucose and buffered at pH 7.8 with 10 mM TES. This culture was grown anaerobically. Samples to determine pH, optical density at 600 nm (OD600), and β-galactosidase specific activity were taken at 15-min intervals for approximately two generations of growth (OD600 = 0.2). At this point, 15 ml of the culture was removed with a syringe and added to a prewarmed 15-ml stoppered bottle containing 1.0 M MES (pH 4.0) to give a final concentration of 63 mM and a pH of 5.8. Samples from both cultures were then taken at the time of the shift and then every 15 min for at least 2 h.
An identical procedure was used to determine the response of hya′-′lacZ expression in cells to a shift from acidic to alkaline pH. An anaerobic culture was prepared as above in 35 ml of LB containing glucose and buffered at pH 5.8 with 10 mM MES. The treated culture received 1.0 M TES (pH 8.5) to a final concentration of 150 mM to give a pH of 7.8. The shift in pH was timed to correspond to the time point of hya′-′lacZ induction.
β-Galactosidase activity assay, pHe determination, and measurement of optical density.
The cells from 1-ml culture samples were collected immediately by centrifugation, and the pH of the supernatant was determined. The cell pellet was resuspended in a 1-ml volume of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 · 7H2O, 50 mM β-mercaptoethanol; buffered at pH 7.0) (26) and kept at 4°C until assayed. Cell suspensions were assayed spectrophotometrically for the OD600. β-Galactosidase specific activity was assayed in duplicate as described previously (26) and measured spectrophotometrically by using the following calculation: 1 Miller unit = 1,000 × (OD420 − 1.75 OD550)/time (minutes) × 0.1 ml × OD600 (26). Specific activities are the average of the values obtained in the two assays. Each experiment was performed at least twice. All OD measurements were conducted with a Cary 2000 spectrophotometer; the culture pH was measured with a Radiometer PH62M pH meter.
RESULTS
Response of hya expression to pHe.
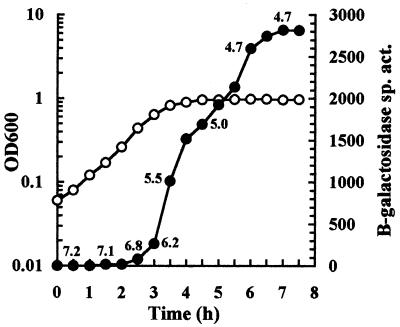
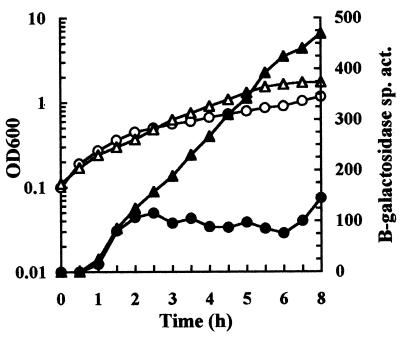
We initially cultured strain JK2 (attB::hya′-′lacZ) anaerobically in unbuffered LB containing glucose to monitor hya expression during growth (Fig. 1). The onset of hya expression was concurrent with the decline of the medium pH. Cultures began to express hya as the pHe began to decline. As cultures progressed in growth and the pHe continued to decline, hya expression continued to increase and eventually peaked in late stationary phase when the pHe had fallen to 4.7.
FIG. 1.
Time course of hya expression during anaerobic growth. Strain JK2 was cultured in unbuffered LB with glucose and assayed for β-galactosidase specific activity (Miller units) (solid circles) and OD600 (open circles). pHe values of selected samples are indicated.
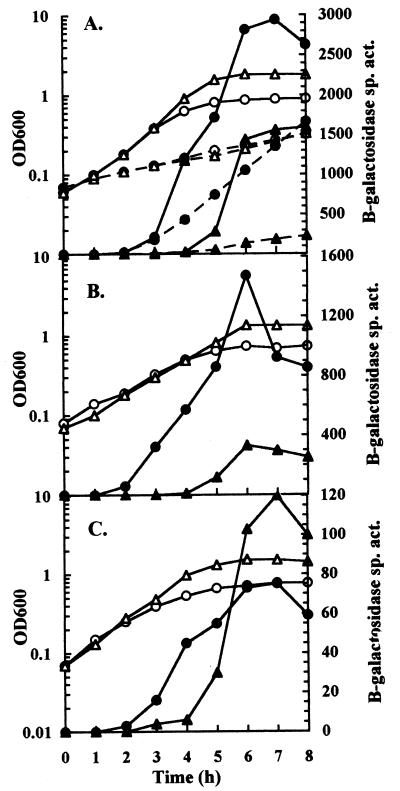
The apparent stimulation of hya expression by external acidification was tested by comparing expression in cultures grown in LB buffered at either an acidic or alkaline pH (Fig. 2A). The onset of hya expression occurred earlier during exponential growth in acidic LB than in alkaline LB, where the onset was delayed a further 3 h. After expression had initiated, a twofold-higher peak expression value was reached in acidic LB than in alkaline LB.
FIG. 2.
Effect of RpoS and ArcA on the response of hya expression to external pH. Cultures were grown in either LB (solid lines) or M63 medium (dashed lines) containing glucose and buffered at either pH 5.5 (circles) or pH 8.5 (triangles). Time point samples were assayed for β-galactosidase specific activity (Miller units) (solid symbols) and OD600 (open symbols). (A) Strain JK2 (hya′-′lacZ). (B) Strain JK1222 (rpoS::Tn10 hya′-′lacZ). (C) Strain JK22 (arcA2 hya′-′lacZ).
The above experiment was repeated in glucose minimal medium buffered at either acidic (pH 5.5) or alkaline (pH 8.5) pH. In minimal medium cultures, there was a reduction in both the growth rate and rate of hya expression compared to LB cultures (Fig. 2A). Although the expression rate was reduced approximately threefold in minimal medium, expression levels continuously increased during 10 h of growth (data not shown). Under acidic conditions, maximal expression was only 0.28-fold lower in minimal medium than in LB; however, under alkaline conditions expression was 7-fold lower in minimal medium than in LB. A comparison of pHe values of the media at the end of growth indicated a significant difference; in minimal medium the pHe was 8.2, but in LB it had fallen to a more neutral value of 7.2 (data not shown).
Effect of a pHe shift on hya expression.
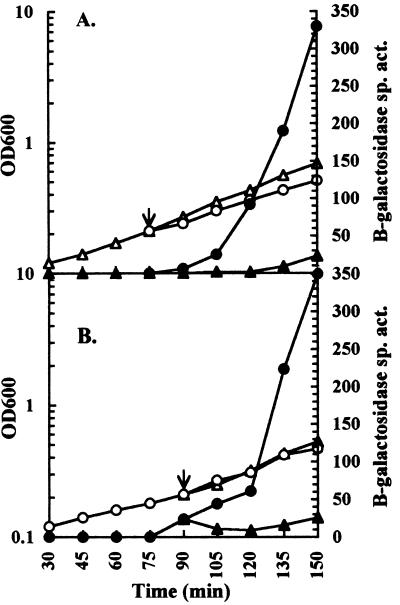
To test if the hya response to pHe could be initiated by a more rapid change in the pHe value, anaerobic cultures were grown to the point of early exponential phase and then the pHe value was shifted. As demonstrated in Fig. 3A, a shift to an acidic pHe value stimulated hya expression. The level of hya expressed in acidic versus untreated cultures was 6-, 13-, 70-, and 20-fold higher at 15, 30, 45, and 60 min postshift, respectively. A slight effect on the growth rate was observed in the treated culture (Fig. 3A).
FIG. 3.
Effect of a shift in pHe on hya expression. Growth of JK2 cultures was initiated in buffered LB containing glucose. At the point in growth indicated by the arrow, the cultures were split and buffer was added to one portion to achieve a shift in pH value. The cultures were monitored for β-galactosidase specific activity (Miller units) (solid symbols) and OD600 (open symbols). (A) Cultures grown in medium at pH 7.8 (10 mM TES) (triangles) and shifted to pH 5.8 (circles) by the addition of 1 M MES (pH 4). (B) Cultures grown in medium at pH 5.8 (10 mM MES) (circles) and shifted to pH 7.8 (triangles) by the addition of 1 M TES (pH 8).
In exponential-phase acidic cultures expressing hya, an abrupt shift of pHe to an alkaline value arrested expression within 15 min (Fig. 3B). Expression in the untreated control continued to increase throughout growth. As a result, the relative expression level in treated cultures was inhibited 4-, 7-, 14-, and 14-fold at 15, 30, 45, and 60 min, respectively, after the shift to an alkaline pHe.
Effect of Pfl activity and formate on hya expression.
In E. coli, the enzyme pyruvate formate-lyase (Pfl), encoded by the pfl gene, is responsible for the production of formate from pyruvate during fermentative growth (19). Initially, excess formate is excreted into the culture medium (7), and then, as the pHe declines, formate is transported back into the cell (30) and further metabolized (27).
Since formate enhances hya expression (9) and its uptake increases as pHe declines, we examined whether the buildup of extracellular formate was required for the response of hya expression to pHe. The expression of hya′-′lacZ in wild-type and Pfl− (38) strains was monitored in cultures grown in either acidic or alkaline medium (Table 2). Both strains expressed higher levels of hya in acidic than in alkaline medium. In Pfl− cultures, initial expression levels matched the levels in wild-type cultures. However, later in growth, levels in wild-type cultures surpassed levels in Pfl− cultures. In wild-type cultures, hya levels were 3-fold higher in acidic medium and 9.5-fold higher in alkaline medium than in Pfl− cultures. However, the overall magnitude of the pHe effect was larger in Pfl− cultures (33-fold) than in the wild-type cultures (10-fold). Most significantly, in the absence of Pfl activity and endogenous formate production, hya expression still responded to pHe.
TABLE 2.
Effect of formate and Pfl activity on hya′-′lacZ expression during growtha
| Strain | Formate | Growth phaseb
|
|||
|---|---|---|---|---|---|
| Exponential
|
Stationary
|
||||
| pHe | β-Galactosidase sp actc | pHe | β-Galactosidase sp act | ||
| JK2320d | − | 5 | 24 | 5 | 1,347 |
| − | 8 | 0.2 | 8 | 133 | |
| + | 8 | UDf | 8 | 22 | |
| JK2898 (pfl-1)d | − | 5 | 18 | 5 | 474 |
| − | 8 | UD | 8 | 14 | |
| JK2320e | − | 6 | 1 | 5 | 1,023 |
| + | 7 | 21 | 6 | 1,573 | |
| JK2898 (pfl-1)e | − | 7 | UD | 5 | 526 |
| + | 7 | 2 | 6 | 735 | |
Wild-type strain JK2320 and Pfl− derivative JK2898 (pfl-1) containing the hya′-′lacZ fusion were cultured anaerobically in LB with either 0.4% glucose or glucose plus 0.4% sodium formate as carbon sources.
Culture samples were taken in either the exponential or stationary phase of growth.
β-Galactosidase specific activity values are given in Miller units.
Cultures were grown in medium buffered at either pH 5.5 or pH 8.5.
Cultures were grown in unbuffered media.
UD, undetectable activity.
Since reduced hya expression in the Pfl− strain might have resulted from reduced formate production, we tested whether the addition of exogenous formate could rescue hya expression levels in Pfl− cultures (Table 2). Exogenous formate was found to stimulate hya expression, as previously reported (9), and had its greatest effect at the onset of expression. As expression levels peaked, the effect of formate diminished, and it could only partially rescue hya expression in the pfl mutant. Moreover, in alkaline medium, formate had no stimulatory affect on hya expression (Table 2). No significant difference was observed in the pHe value between wild-type and Pfl− cultures grown in unbuffered glucose medium (data not shown).
Response of hya expression to pHe in the absence of RpoS.
In E. coli, the levels of the sigma factor RpoS increase in response to conditions of stationary-phase growth (13), acidic pHe (6), and increasing concentrations of fermentation end products (33). We and others (2) have noted that RpoS is required for maximal expression of hya; thus, it was possible that the change in hya expression in response to pHe results from a change in RpoS-mediated expression. Therefore, we tested the response of hya to pHe in the absence of RpoS by culturing an RpoS− strain in either acidic or alkaline medium (Fig. 2B). In the absence of RpoS, the response of hya expression to pHe was not lost. The expression levels, however, were reduced compared to expression in wild-type cultures (compare Fig. 2B with Fig. 2A). In RpoS− cultures, maximal hya expression levels were achieved in early stationary phase and then rapidly declined, whereas in wild-type cultures, maximal expression was sustained in the stationary phase for a longer period (compare Fig. 2B with Fig. 2A). Growth of RpoS− cultures in acidic medium led to a fourfold increase in maximal expression levels over levels achieved in alkaline medium (Fig. 2B). There was no significant difference in pHe values during growth of RpoS− cultures compared to wild-type cultures (data not shown).
Response of hya expression to pHe in the absence of ArcA.
Like RpoS, ArcA is required for full expression of hya (9). A putative signal for ArcA activation is a change in proton motive force (Δp) (8, 16), and one component of Δp is pHe. Thus, the response of hya expression to pHe could involve ArcA. This possibility was tested by examining the hya response to pHe in the absence of ArcA (Fig. 2C). In both acidic and alkaline media, hya expression levels were reduced in ArcA− cultures compared to wild-type cultures (compare Fig. 2C with Fig. 2A). The difference in the onset of expression observed in wild-type cultures grown at the two pHe values was diminished in ArcA− cultures. Thereafter, as cultures progressed into stationary phase, the absence of ArcA abrogated elevated hya levels in acidic medium. Interestingly, ArcA− cultures expressed 1.6-fold higher levels in alkaline medium than in acidic medium. Again, no significant difference was observed in the values of pHe for ArcA− and wild-type cultures (data not shown).
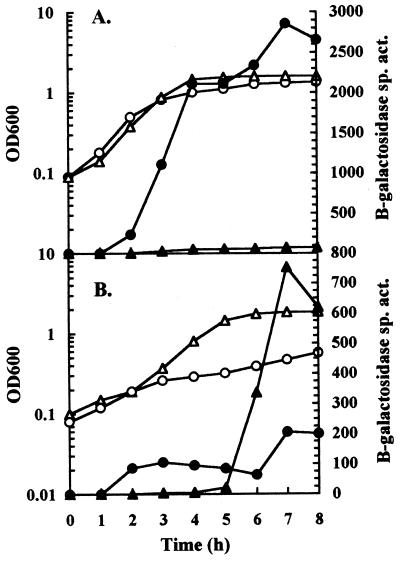
Expression of hya in response to pHe in the absence of AppY.
The transcriptional activator AppY is required for full expression of hya during anaerobic growth (2, 9). The response of hya expression to pHe was tested in the absence of AppY to determine whether AppY was involved in the response of hya to pHe (Fig. 4). The onset of hya expression in acidic medium occurred at the same point during growth of cultures without AppY as in wild-type cultures. However, in cultures without AppY, hya expression plateaued after only 1 h of growth whereas in wild-type cultures, hya expression continued to increase for an additional 6 h. In alkaline medium, the onset of hya expression in both strains was inhibited until entry into stationary phase. As above, wild-type cultures expressed higher levels in acidic than in alkaline medium whereas in AppY− cultures, maximal levels were 3.5-fold higher in alkaline than in acidic medium. In alkaline medium, the pHe value of AppY− cultures dropped to a neutral value 4 h later during growth than did the pHe value of wild-type cultures (data not shown); this was the same point in growth where hya expression in AppY− cultures reached maximal levels.
FIG. 4.
Effect of AppY on the response of hya expression to external pH. Cultures were grown in LB containing glucose buffered at pH 5.5 (circles) or 8.5 (triangles). Time point samples were assayed for β-galactosidase specific activity (Miller units) (solid symbols) and OD600 (open symbols). (A) Strain JK2193 (hya′-′lacZ). (B) Strain JK256 (ΔappY hya′-′lacZ).
Expression of hyb in response to pHe.
Anaerobic growth of E. coli cultures results in the synthesis of both Hyd1 and Hyd2 uptake hydrogenases. To further characterize anaerobic uptake hydrogenase operon expression in response to pHe, we studied the expression of a hyb′-′lacZ fusion in both acidic and alkaline media containing glycerol and fumarate (Fig. 5). The onset of hyb expression in cultures was constitutive with respect to the pHe value and occurred 30 min after the initiation of anaerobic growth. In contrast to hya expression, hyb expression levels in acidic medium achieved maximal values after only 2 h of growth whereas expression in alkaline medium continued to increase during the entire 8 h of growth. Consequently, maximal hyb expression levels attained in alkaline medium were fourfold higher than in acidic medium.
FIG. 5.
Response of hyb expression to external pH during growth. Strain SE2065 containing the hyb′-′lacZ chromosome fusion was cultured in LB containing glycerol and fumarate buffered at pH 5.5 (circles) or 8.5 (triangles). Time point samples were assayed for β-galactosidase specific activity (Miller units) (solid symbols) and OD600 (open symbols).
DISCUSSION
The difficulty in resolving the function of Hyd1 is partially due to the complexity of the way in which hya expression is regulated (2, 9). Anaerobic growth induces hya expression; however, E. coli does not require Hyd1 for anaerobic growth. Limitation of carbon and phosphate contributes to anaerobic expression (2), but the effect on Hyd1 synthesis is unknown. Another possible source of stress is acidification of the external environment (6), and as we have shown in this study, anaerobic expression of hya responds to pHe and is maximal at an acidic pHe. This response of hya was evident in both complex and minimal media, suggesting that components found in yeast extract or tryptone, such as amino acids, are not mediating the response. However, the level of expression achieved at alkaline pHe values was greater in complex medium than in minimal medium. This could reflect an nutritional enhancement and/or the greater acidification of alkaline LB cultures compared to alkaline minimal medium cultures. Regardless, in both minimal medium and LB, pHe exerts a significant effect on hya expression.
Formate is not required for hya response to pHe.
We found that exogenous formate enhances the level of hya expression, as previously reported (9), but that the degree of enhancement depended on the growth phase. Formate had a stronger influence during the initial stages of expression, but the influence diminished as expression became maximal. It is important to this study, however, that the response of hya expression to pHe was evident in the pfl mutant; thus, it does not require formate. While formate was not essential for the hya response to pHe, it was needed to achieve maximal expression levels. Formate is also essential for synthesis of active Hyd1 (31), since the expression of HydA and HypA, which are required to process Hyd1 precursors to active enzymes (17, 31), is formate dependent (21, 22). Thus, formate is essential for maximal hya expression and Hyd1 precursor processing but does not mediate the hya response to pHe.
RpoS is required to achieve and sustain maximal hya expression but does not mediate the response to pHe.
Although RpoS mediates changes in gene expression in response to pHe (6) and although maximal hya expression requires RpoS, the response of hya to pHe did not require RpoS. This is similar to the effect of RpoS on the hya response to phosphate starvation (2). The period of peak expression that occurred in stationary-phase growth in the absence of RpoS was shortened compared to that in wild-type cultures. Therefore, RpoS enhances and sustains hya expression but does not regulate its response to pHe. The effect of RpoS on hya during stationary-phase growth is in agreement with the increased level of RpoS in stationary-phase cells (13).
hya responds to pHe in the absence of AppY but at reduced levels.
The transcriptional regulator AppY, which is required for full anaerobic expression of hya (9), was not required for the early onset of hya expression in acidic medium but was needed to attain maximal expression levels. AppY had a greater influence on hya in the stationary phase of growth. This could reflect the elevated levels of appY transcription in stationary-phase cultures (10), which could presumably lead to increased AppY stimulation of hya expression.
ArcA is required for hya to respond to pHe and to achieve maximal expression.
Whereas maximal hya expression required AppY, RpoS, and ArcA, the response of hya to pHe was most strongly affected by ArcA. Without ArcA, hya expression appeared to respond primarily to changes in growth rate and not external pH. ArcA regulates gene expression in response to changes in the respiratory state of the cell (16) and may be activated by the accompanying decline in Δp (8, 16). Cultures that are growing anaerobically have a lower Δp value than aerobically growing cultures do (37). When cultures are grown in acidic medium or reach stationary phase, the Δp decreases further (18, 37). Since the conditions of growth that minimize Δp also maximize hya expression, it is plausible that the hya response to pHe is at least partially mediated by ArcA. As a result, anaerobic and pHe control of hya expression would be linked through ArcA.
hya and hyb respond differentially to pHe.
While both the hya and hyb operons responded to pHe, the hyb response reached maximal levels in alkaline medium and the hya response reached maximal levels in acidic medium. This expression pattern is consistent with the activity patterns of the two periplasmic uptake hydrogenases. Hyd2 has an alkaline pH optimum (5) and is at its highest levels during early growth phases, when the medium and periplasm are alkaline (23, 36). Hyd1 has an acidic optimum (32) and is at its highest levels during later stages of growth (25), when the pH of the medium and periplasm have declined (36) and the activity of Hyd2 has begun to diminish (23). Thus, the expression of hya and hyb is upregulated by pH conditions which favor the activity of the respective periplasmic enzymes. In support of this, active Hyd1 is present at severalfold higher levels in acidic cultures than in alkaline cultures throughout growth (data not shown). Expression of the hyb operon is also needed for maximal production of active Hyd1 (23), since one of the genes of hyb is required for the efficient processing of Hyd1 (23). Therefore, under acidic conditions when Hyd2 activity becomes less important, expression of hyb is maintained as a component of Hyd1 processing.
It is evident that hya expression achieved maximal levels in stationary phase under acidic conditions. Stationary-phase growth (13) and an acidic pHe are conditions of stress for E. coli and additionally they effect a reduction of Δp (18, 37). This suggests that the function of Hyd1 during anaerobic growth might be to maintain Δp in an energy-conserving manner (32). Indeed, Hyd1 hydrogen oxidation could contribute to Δp by vectorial electron transport, as in Desulfovibrio vulgaris (4), by releasing protons to the periplasm and donating electrons to an electron acceptor for the reduction of a cytoplasmic substrate. Utilization of hydrogen gas in this manner by Hyd1 could then promote or maintain Δp.
ACKNOWLEDGMENTS
We would like to thank D. P. Clark for strains LCB320 and LCB898, S. Iuchi for strain ECL618, P. C. Loewen for strain UM122, K. T. Shanmugam and J. C. Wendt for strain SE2065, and N. K. Menon and D. Gonzalez for helpful discussions.
This work was supported by NSF MCB 9005734 to A.E.P. and H. D. Peck, Jr., NIH GM34903-08 to A.E.P. and H. D. Peck, Jr., and DOE DE-FG09-89ER13614 to H. D. Peck, Jr., and A.E.P.
REFERENCES
- 1.Atlung T, Bronsted L. Role of the transcriptional activator AppY in regulation of the cyx appA operon of Escherichia coli by anaerobiosis, phosphate starvation, and growth phase. J Bacteriol. 1994;176:5414–5422. doi: 10.1128/jb.176.17.5414-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlung T, Knudsen K, Heerfordt L, Bronsted L. Effects of ςS and the transcriptional activator AppY on induction of the Escherichia coli hya and cbdAB-appA operons in response to carbon and phosphate starvation. J Bacteriol. 1997;179:2141–2146. doi: 10.1128/jb.179.7.2141-2146.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Siedman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 4.Badziong W, Thauer R K. Vectorial electron transport in Desulfovibrio vulgaris (Marburg) growing on hydrogen plus sulfate as sole energy source. Arch Microbiol. 1980;125:167–174. [Google Scholar]
- 5.Ballantine S P, Boxer D H. Isolation and characterisation of a soluble active fragment of hydrogenase isoenzyme 2 from the membranes of anaerobically grown Escherichia coli. Eur J Biochem. 1986;156:277–284. doi: 10.1111/j.1432-1033.1986.tb09578.x. [DOI] [PubMed] [Google Scholar]
- 6.Bearson S M D, Benjamin W H, Swords W E, Foster J W. Acid shock induction of RpoS is mediated by the mouse virulence gene mviA of Salmonella typhimurium. J Bacteriol. 1996;178:2572–2579. doi: 10.1128/jb.178.9.2572-2579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackwood A C, Neish A C, Ledingham G A. Dissimilation of glucose at controlled pH values by pigmented and nonpigmented strains of Escherichia coli. J Bacteriol. 1956;72:497–499. doi: 10.1128/jb.72.4.497-499.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogachev A V, Murtazina R A, Skulachev V P. Cytochrome d induction in Escherichia coli growing under unfavorable conditions. FEBS Lett. 1993;336:75–78. doi: 10.1016/0014-5793(93)81612-4. [DOI] [PubMed] [Google Scholar]
- 9.Bronsted L, Atlung T. Anaerobic regulation of the hydrogenase 1 (hya) operon of Escherichia coli. J Bacteriol. 1994;176:5423–5428. doi: 10.1128/jb.176.17.5423-5428.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bronsted L, Atlung T. Effect of growth conditions on expression of the acid phosphatase (cyx-appA) operon and the appY gene, which encodes a transcriptional activator of Escherichia coli. J Bacteriol. 1996;178:1556–1564. doi: 10.1128/jb.178.6.1556-1564.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casadaban M J, Chou J, Cohen S N. In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980;143:971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis K, Patel P, Wendt J C, Shanmugam K T. Purification and characterization of two forms of hydrogenase isoenzyme 1 from Escherichia coli. J Bacteriol. 1990;172:5750–5757. doi: 10.1128/jb.172.10.5750-5757.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hengge-Aronis R. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell. 1993;72:165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- 14.Iuchi S, Furlong D, Lin E C C. Differentiation of arcA, arcB, and cpxA mutant phenotypes of Escherichia coli by sex pilus formation and enzyme regulation. J Bacteriol. 1989;171:2889–2893. doi: 10.1128/jb.171.5.2889-2893.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iuchi S, Matsuda Z, Fujiwara T, Lin E C C. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol Microbiol. 1990;4:715–727. doi: 10.1111/j.1365-2958.1990.tb00642.x. [DOI] [PubMed] [Google Scholar]
- 16.Iuchi S, Weiner L. Cellular and molecular physiology of Escherichia coli in the adaptation to aerobic environments. J Biochem. 1996;120:1055–1063. doi: 10.1093/oxfordjournals.jbchem.a021519. [DOI] [PubMed] [Google Scholar]
- 17.Jacobi A, Rossman R, Bock A. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch Microbiol. 1992;158:444–451. doi: 10.1007/BF00276307. [DOI] [PubMed] [Google Scholar]
- 18.Kahsket E R. Stoichiometry of the H+-ATPase of Escherichia coli cells during anaerobic growth. FEBS Lett. 1983;154:343–346. doi: 10.1016/0014-5793(83)80179-3. [DOI] [PubMed] [Google Scholar]
- 19.Knappe J, Schacht J, Mockel W, Hopner T, Vetter H, Jr, Edenharder R. Pyruvate formate-lyase reaction in Escherichia coli. Eur J Biochem. 1969;11:316–327. doi: 10.1111/j.1432-1033.1969.tb00775.x. [DOI] [PubMed] [Google Scholar]
- 20.Loewen P C, Switala J, Triggs-Raine B L. Catalases HPI and HPII in Escherichia coli are induced independently. Arch Biochem Biophys. 1985;243:144–149. doi: 10.1016/0003-9861(85)90782-9. [DOI] [PubMed] [Google Scholar]
- 21.Lutz S, Bohm R, Beier A, Bock A. Characterization of divergent NtrA-dependent promoters in the anaerobically expressed gene cluster coding for hydrogenase 3 components of Escherichia coli. Mol Microbiol. 1990;4:13–20. doi: 10.1111/j.1365-2958.1990.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 22.Maier T, Binder U, Bock A. Analysis of the hydA locus of Escherichia coli: two genes (hydN and hypF) involved in formate and hydrogen metabolism. Arch Microbiol. 1996;165:333–341. doi: 10.1007/s002030050335. [DOI] [PubMed] [Google Scholar]
- 23.Menon N K, Chatelus C Y, DerVartanian M, Wendt J C, Shanmugam K T, Peck H D, Jr, Przybyla A E. Cloning, sequencing, and mutational analysis of the hyb operon encoding Escherichia coli hydrogenase 2. J Bacteriol. 1994;176:4416–4423. doi: 10.1128/jb.176.14.4416-4423.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menon N K, Robbins J, Peck H D, Jr, Chatelus C Y, Choi E S, Przybyla A E. Cloning and sequencing of a putative Escherichia coli [NiFe] hydrogenase-1 operon containing six open reading frames. J Bacteriol. 1990;172:1969–1977. doi: 10.1128/jb.172.4.1969-1977.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menon N K, Robbins J, Wendt J C, Shanmugam K T, Przybyla A E. Mutational analysis and characterization of the Escherichia coli hya operon, which encodes [NiFe] hydrogenase 1. J Bacteriol. 1991;173:4851–4861. doi: 10.1128/jb.173.15.4851-4861.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 27.Peck H D, Jr, Gest H. Formic dehydrogenase and the hydrogenlyase enzyme complex in coli-aerogenes bacteria. J Bacteriol. 1957;73:706–721. doi: 10.1128/jb.73.6.706-721.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhode M, Furstenau V, Meyer F, Przybyla A E, Peck Jr H D, Legall J, Choi E-S, Menon N K. Localization of membrane-associated (NiFe) and (NiFeSe) hydrogenases of Desulfovibrio vulgaris using immunoelectron microscopic procedures. Eur J Biochem. 1989;180:421–427. doi: 10.1111/j.1432-1033.1990.tb19134.x. [DOI] [PubMed] [Google Scholar]
- 29.Rodrigue A, Boxer D H, Mandrand-Berthelot M A, Wu L-F. Requirement for nickel of the transmembrane translocation of NiFe-hydrogenase 2 in Escherichia coli. FEBS Lett. 1996;392:81–86. doi: 10.1016/0014-5793(96)00788-0. [DOI] [PubMed] [Google Scholar]
- 30.Rossmann R, Sawers G, Bock A. Mechanism of regulation of the formate-hydrogenlyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol Microbiol. 1991;5:2807–2814. doi: 10.1111/j.1365-2958.1991.tb01989.x. [DOI] [PubMed] [Google Scholar]
- 31.Sawers R G, Ballantine S P, Boxer D H. Differential expression of hydrogenase isoenzymes in Escherichia coli K-12: evidence for a third isoenzyme. J Bacteriol. 1985;164:1324–1331. doi: 10.1128/jb.164.3.1324-1331.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawers R G, Boxer D H. Purification and properties of membrane-bound hydrogenase isoenzyme 1 from anaerobically grown Escherichia coli K12. Eur J Biochem. 1986;156:265–275. doi: 10.1111/j.1432-1033.1986.tb09577.x. [DOI] [PubMed] [Google Scholar]
- 33.Schellhorn H E, Stones V L. Regulation of katF and katE in Escherichia coli K-12 by weak acids. J Bacteriol. 1992;174:4769–4776. doi: 10.1128/jb.174.14.4769-4776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 35.Slonczweski J L, Gonzalez T N, Bartholomew F M, Holt N J. Mu d-directed lacZ fusions regulated by low pH in Escherichia coli. J Bacteriol. 1987;169:3001–3006. doi: 10.1128/jb.169.7.3001-3006.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stock J B, Rauch B, Roseman S. Periplasmic space in Salmonella typhimurium and Escherichia coli. J Biol Chem. 1977;252:7850–7861. [PubMed] [Google Scholar]
- 37.Tran Q H, Unden G. Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. Eur J Biochem. 1998;251:538–543. doi: 10.1046/j.1432-1327.1998.2510538.x. [DOI] [PubMed] [Google Scholar]
- 38.Varenne S, Casse F, Chippaux M, Pascal M C. A mutant of Escherichia coli deficient in pyruvate formate-lyase. Mol Gen Genet. 1975;141:181–184. doi: 10.1007/BF00267683. [DOI] [PubMed] [Google Scholar]
- 39.Wendt J C. Regulation of the hydrogen uptake (hup) gene of Escherichia coli. M.S. thesis. Gainesville: University of Florida; 1989. [Google Scholar]
- 40.Wendt J C, Maupin J A, Shanmugam K T. Abstracts of the 3rd International Conference on Molecular Biology of Hydrogenases 1991. 1991. Physiological and genetic regulation of dihydrogen metabolism in Escherichia coli; pp. 26–27. [Google Scholar]