Abstract
We identified a short RNA fragment, complementary to the Escherichia coli 23S rRNA segment comprising nucleotides 735 to 766 (in domain II), which when expressed in vivo results in the suppression of UGA nonsense mutations in two reporter genes. Neither UAA nor UAG mutations, examined at the same codon positions, were suppressed by the expression of this antisense rRNA fragment. Our results suggest that a stable phylogenetically conserved hairpin at nucleotides 736 to 760 in 23S rRNA, which is situated close to the peptidyl transferase center, may participate in one or more specific interactions during peptide chain termination.
A variety of experimental approaches have indicated sites of rRNA likely to directly participate in ribosomal functions or to be involved in interactions with ribosomal ligands. These include cross-linking studies (7, 38, 39), antibiotic binding (15), chemical footprinting (33), hydroxyl radical probing (47), oligonucleotide probing (27), electron microscopy (1, 41), and mutational studies (34, 50). However, little information is available concerning regions of rRNA important for the termination of translation. Mutations affecting the efficiency of termination are clustered in a few regions of the RNAs of both the small and large ribosomal subunits (2, 3, 29–32). Nevertheless, little is known about the possible involvement of other rRNA segments in the termination of translation.
Recently, a novel approach was developed for the identification of functionally important regions of rRNA (44). In this technique, random fragments of rRNA are expressed in vivo either in the direct orientation or in the complementary (antisense) orientation, and clones expressing RNA fragments affecting different aspects of protein synthesis can be selected by functional screening. Previously, this technique allowed the identification of a fragment of domain I of 23S rRNA that, when expressed in either the direct or antisense orientation, caused a read-through of the UGA termination codon (4). Here we report the identification of a novel RNA fragment with suppressor activity. In contrast to the previously identified fragment, the new suppressor RNA is complementary to a segment of domain II of 23S rRNA, and it induces a stop codon read-through only when present in the antisense orientation.
MATERIALS AND METHODS
Strains, plasmids, and materials.
All bacterial strains used were derivatives of Escherichia coli K-12. The JM109 strain was used for most of the cloning experiments (49). GI724 cells (24) were used for analysis of the chloramphenicol acetyltransferase (CAT) protein produced as the result of suppression of nonsense mutations in its gene, cat. The construction of the RNA fragment expression library in the pGEX-derived pPOT1 vector (44), growth media, and genetic procedures were described previously (4, 36). Enzymes were obtained from Promega or New England Biolabs. Chemicals and antibiotics were obtained from Fisher or Sigma.
Construction of reporter plasmids.
The reporter plasmids pCAT101, pCAT102, and pCAT103 (Table 1) were derivatives of pACYC184 (14) with a UGA, UAA, or UAG stop codon, respectively, replacing the Arg codon at position 74 of the 220-codon CAT gene. For constructing these plasmids, a segment of cat in pACYC184 was PCR amplified with the primer CGGAATTCCGGATGAGCATT, complementary to the 3′ end of cat, and three primers differing only in the sequence of the termination triplet (underlined) were introduced instead of Arg codon 74 in cat, namely, CGGAATTCTGAATGGCAATGAAAGACGGT, CGGAATTCTAAATGGCAATGAAAGACGGT, and CGGAATTCTAGATGGCAATGAAAGACGGT. The resulting PCR products were cut with EcoRI and introduced into the unique EcoRI site located in the cat gene on pACYC184, which restored the integrity of cat except for the presence of the engineered nonsense codons. The presence of each nonsense codon at codon position 74 and the lack of other mutations in the cat gene in pCAT101, pCAT102, and pCAT103 were verified by DNA sequence analysis.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pPOT1 | pGEX-derived plasmid used to clone the library of rRNA gene fragments | 44; transcription of the RNA fragment (from Ptac) is IPTG inducible |
| pA750 | pPOT1, from the library, containing the 32-nucleotide rDNA fragment whose transcript is complementary to nucleotides 735 to 766 of 23S rRNA | This work |
| pS750 | pPOT1 with the A750 insert in the opposite transcriptional orientation, yielding a transcript corresponding to the direct sequence of nucleotides 735 to 766 of 23S rRNA | This work |
| pACYC184 | Contains the wild-type cat gene | 14 |
| pCAT101 | pACYC184 derivative containing the cat gene with a UGA nonsense mutation at codon 74 | This work |
| pCAT102 | pACYC184 derivative containing the cat gene with a UAA nonsense mutation at codon 74 | This work |
| pCAT103 | pACYC184 derivative containing the cat gene with a UAG nonsense mutation at codon 74 | This work |
| pNC160 | Randomly chosen plasmid from the rRNA fragment library which contains a 160-nucleotide rRNA gene fragment whose transcript is complementary to nucleotides 377 to 537 of 23S rRNA | This work |
Screening of the random rRNA fragment library for suppressor RNA fragments.
CAT, the cat gene product, renders cells resistant to chloramphenicol. The mutant cat gene in pCAT101 has a UGA nonsense mutation at codon position 74. Consequently, cells transformed with pCAT101 remain sensitive to chloramphenicol. Cells carrying pCAT101 were transformed with the rRNA fragment library, and 106 transformed cells were plated on Luria-Bertani (LB) agar medium containing selective antibiotics for pCAT101 and pPOT1, namely, tetracycline (15 μg/ml) and ampicillin (100 μg/ml), as well as 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and chloramphenicol (20 μg/ml). Of the 24 Camr transformants obtained, only one exhibited the properties of the sought-after RNA fragment-induced nonsense suppression, namely, IPTG-dependent chloramphenicol resistance cotransferable with the plasmid. The pPOT1-based library plasmid isolated from this clone contained a segment of 23S rRNA gene, corresponding to positions 735 to 766 of 23S rRNA, inserted in the complementary (antisense) orientation relative to the Ptac promoter. The plasmid did not confer chloramphenicol resistance on its own, but in the presence of IPTG, an inducer of the Ptac promoter, it rendered cells carrying pCAT101 resistant to chloramphenicol. These results supported the hypothesis that the resistance was produced by a read-through of the mutant termination codon in cat. To be sure that no pPOT1 vector mutations constituted or contributed to the suppressor activity, the rRNA gene fragment from the selected library plasmid was subcloned into the original vector, pPOT1, in both orientations. Plasmids with each orientation were named pA750 (antisense; corresponding to the original, selected library plasmid) and pS750 (sense) (Table 1) and were tested for the ability to suppress nonsense mutations (see Results).
In vivo tests of suppression specificity.
The specificity of nonsense suppression was tested with termination codons in two reporter systems. First, plasmids pCAT101, pCAT102, and pCAT103 (respectively, UGA, UAA, and UAG; Table 1) were examined for the ability to confer resistance to chloramphenicol upon in vivo expression of the rRNA fragment encoded in the pA750 plasmid. Second, E. coli strains with all three stop codons at each of five codon positions, 15, 102, 115, 211, and 243, in trpA (4, 28, 35) were tested for a fragment-generated read-through resulting in an active trpA-encoded protein. Such nonsense suppression would be indicated by a Trp+ phenotype, that is, the ability of the cells to grow on glucose minimal medium in the absence of tryptophan or indole. All the indicated nonsense mutations in trpA could be suppressed by rRNA suppressor mutants or a variety of tRNA suppressors (36). The mutant trpA genes were contained in the cysB-trp-tonB region of the chromosome carried by the Fredericq episome, a conjugative plasmid (17). Mutations in genes of the trp operon were also used to test the UGA-suppressing RNA fragment for the ability to suppress frameshift mutations, namely, two +1 mutations, trpA8 (45) and trpE9777 (10), and a −1 mutation, trpE91 (6). The method for the detection of read-through and frameshifting on plates has been described elsewhere (11, 36, 45).
Expression and analysis of the CAT polypeptide.
E. coli cells of strain GI724 carrying pCAT101, pCAT102, or pCAT103 were analyzed for the production of the complete CAT protein in response to expression of the rRNA fragment encoded in pA750. For each, liquid LB medium containing ampicillin (100 μg/ml) and tetracycline (20 μg/ml) was inoculated from overnight cultures. Bacteria were grown to an optical density (A650) of 1 in the presence and absence of 1 mM IPTG. Cells from 30 μl of each culture were pelleted and resuspended in 20 μl of sample loading buffer, incubated at 96°C for 5 min, and loaded onto a 16% sodium dodecyl sulfate (SDS) gel (22). Proteins were transferred to nitrocellulose membranes (Hybond-C Pure; Amersham) and probed with anti-CAT antibodies (5 Prime → 3 Prime, Inc.) according to the manufacturer’s protocol.
RESULTS
Selection for suppressor rRNA fragments.
An expression rRNA fragment library was constructed previously by the cloning of random fragments of the E. coli rrnB operon into the pPOT1 vector behind the IPTG-inducible Ptac promoter (44). The induction of the promoter results in production in the cell of specific rRNA fragments in a direct (sense) or complementary (antisense) orientation. The RNA fragments that are produced are flanked by hairpins formed by plasmid-derived sequences corresponding to the lac operator and the trpL terminator. To screen the library for RNA fragments that when expressed in vivo can suppress nonsense mutations, the plasmid pCAT101 was constructed, which is compatible with pPOT1 and has a mutant cat gene containing a UGA nonsense mutation at codon position 74. Cells carrying the pCAT101 plasmid were transformed with the random rRNA fragment library, and a library plasmid was selected that conferred chloramphenicol resistance to the cells carrying the cat gene with a UGA nonsense mutation (see Materials and Methods for details).
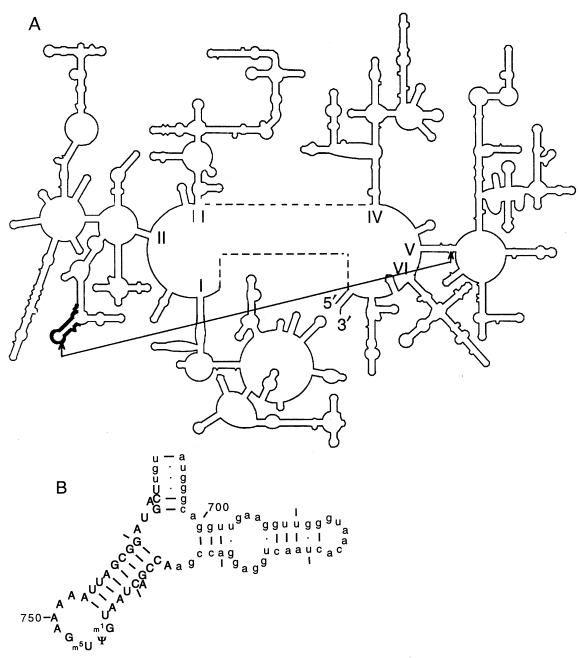
Sequence analysis of the ribosomal DNA (rDNA) fragment present in the isolated library plasmid identified a short 32-nucleotide sequence corresponding to nucleotides 735 to 766 of the 23S rRNA gene (Fig. 1). The rRNA gene segment in the selected plasmid was in the antisense orientation relative to the Ptac promoter, so that the transcribed RNA was complementary to the corresponding region of 23S rRNA. Northern hybridization analysis demonstrated that the plasmid-encoded RNA of the expected size is indeed produced in the cell upon IPTG induction (data not shown). The rRNA gene fragment from the selected library plasmid was subcloned into the original vector, pPOT1, in both orientations to produce two plasmids, pA750 (antisense; corresponding to the original, selected library plasmid) and pS750 (sense) (Table 1). Only pA750 could confer chloramphenicol resistance on cells carrying pCAT101. The chloramphenicol resistance of the clones containing pCAT101 and pA750 depended on the expression of the RNA fragment from pA750 because (i) the chloramphenicol resistance phenotype was cotransferable with pA750 and (ii) it was evident only in the presence of IPTG, an inducer of the Ptac promoter, which in pA750 controlled transcription of the cloned rRNA gene segment (Fig. 2). pA750 did not confer chloramphenicol resistance in the absence of pCAT101, indicating that production in vivo of the RNA fragment complementary to the 23S rRNA segment from nucleotides 735 to 766 in some way suppresses the nonsense mutation in pCAT101, resulting in the production of a functional CAT protein.
FIG. 1.
Location in E. coli 23S rRNA of the sequence complementary to the suppressor fragment isolated from the rRNA gene fragment library. (A) The secondary structure of 23S rRNA, with the segment from nucleotides 735 to 766 shown in boldface. Also indicated is a cross-link (two arrows connected by a line) between the hairpin 750 loop and the peptidyl transferase region (8). (B) Detail of the region of 23S rRNA which includes hairpin 750 (nucleotides 736 to 760). Known modified nucleotides are indicated. The sequence to which the suppressor fragment is complementary (nucleotides 735 to 766) is shown in uppercase and boldface.
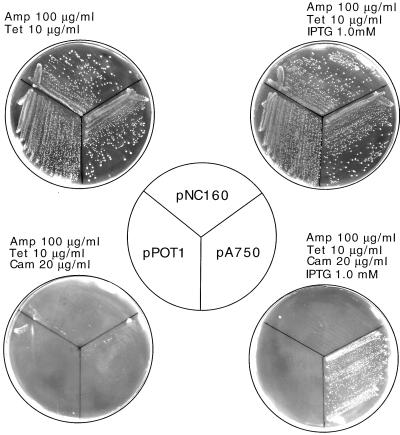
FIG. 2.
Suppression of a UGA nonsense mutation caused by the in vivo expression of an RNA fragment complementary to the E. coli 23S rRNA sequence comprising nucleotides 735 to 766. E. coli cells, carrying pCAT101 plasmid containing the cat gene with a nonsense UGA mutation, were transformed with either the pPOT1 vector, pA750, which encodes the suppressor RNA fragment, or a control plasmid, pNC160, which contained a 160-bp DNA fragment corresponding to nucleotides 377 to 537 of 23S rRNA inserted into pPOT1. After single-colony isolations, the cells were streaked on LB agar medium containing ampicillin (Amp) and tetracycline (Tet), with or without IPTG and chloramphenicol (Cam), as shown.
Specificity of nonsense suppression induced by the selected fragment.
The RNA fragment encoded in pA750 caused the suppression of a UGA mutation. To test whether it can also suppress the other nonsense codons besides UGA, pA750 was introduced into cells carrying pCAT102 (UAA) or pCAT103 (UAG) (Table 1). The resultant transformants remained chloramphenicol sensitive in the presence of IPTG, indicating the lack of suppression of the UAA or UAG nonsense mutations. pS750, in which a corresponding fragment of rRNA was expressed in the sense orientation, did not cause the suppression of any of the nonsense codons in the three cat reporter constructs (pCAT101, pCAT102, and pCAT103). To verify the UGA specificity of the fragment-mediated suppression in a second reporter gene and to examine the possible influence of the mRNA nucleotide sequence context of the stop codon on the suppression (11, 37), cells harboring any one of the three nonsense codon mutations at each of several trpA codon positions (15, 102, 115, 211, and 243) were transformed with pA750. The transformants were tested for the ability to grow on glucose minimal medium lacking Trp, in the presence and absence of IPTG. The result of this experiment (Table 2) was that the rRNA fragment encoded by pA750 suppressed UGA (but not UAA or UAG) mutations at positions 15 and 115 but did not suppress any of the nonsense codons at position 102, 211, or 243. This confirmed the observation made with the cat reporter that the pA750-encoded RNA fragment causes UGA-specific suppression and indicated in addition that the suppression was context dependent.
TABLE 2.
IPTG-dependent suppression of trpA nonsense mutations in the presence of pA750a
| UGA codon position in trpAb | Suppressionc with growth at:
|
|||||
|---|---|---|---|---|---|---|
| 31°C
|
37°C
|
41°C
|
||||
| Control | IPTG | Control | IPTG | Control | IPTG | |
| 15 | − | + | − | + | − | − |
| 102 | − | − | − | − | − | − |
| 115 | − | +++ | − | +++ | − | − |
| 211 | − | − | − | − | − | − |
| 243 | − | − | − | − | − | − |
The growth of replica-plated patches of cells on agar plates with glucose minimal medium, relative to suppression-independent general growth on glucose minimal medium supplemented with indole or tryptophan (3, 23), was evaluated. No suppression was observed with strains transformed with either the pPOT1 vector or pS750 (Table 1).
The UAA and UAG mutations at each codon position were also examined, but no suppression was observed.
Agar medium contained or did not contain (control) 1 mM IPTG. −, no growth up to 6 days; + to +++, increasing amounts of growth over control. “Failure” implies only the failure to grow on glucose minimal medium (that is, lack of suppression); the pA750 strains grow well (with or without IPTG) at all three temperatures on glucose minimal medium supplemented with either tryptophan or indole.
We also tested the ability of the pA750-encoded RNA fragment to stimulate frameshifting. For that, E. coli strains were used that contain trpA genes with either +1 or −1 frameshift mutations (see Materials and Methods). None of the frameshift mutations tested was suppressed by the pA750-encoded RNA fragment (data not shown).
The RNA fragment encoded by pA750 causes specific read-through of the UGA stop codon and production of full-length CAT protein from the mutant gene.
Western blot analysis was used to test whether expression in the cell of the pA750-encoded RNA fragment caused a read-through of the UGA nonsense codon in the mutant cat gene and production of the full-length CAT protein. Cells transformed with pA750 and either pCAT101, pCAT102, or pCAT103 were grown in the presence or absence of IPTG and, after separation of the total cellular protein in an SDS gel, the CAT protein was detected with antibodies. Figure 3 shows that an apparently full-size CAT protein was produced only in the cells carrying pCAT101 (the UGA mutation) when the expression of the pA750-encoded RNA fragment was induced by IPTG. This result strongly supports the conclusion that expression of the RNA fragment complementary to 23S rRNA nucleotides 735 to 766 allowed ribosomes to specifically read through the UGA nonsense codon.
FIG. 3.
Influence of the expression of a short RNA fragment, complementary to the E. coli 23S rRNA sequence from nucleotides 735 to 766, on a translational read-through at UGA, UAA, and UAG nonsense codons in the cat gene. Cells carrying the cat gene with one of the three nonsense mutations were transformed with either pPOT1 vector or pA750. Cultures were grown with (+) or without (−) IPTG, total cellular proteins were separated in SDS gels and blotted, and CAT protein was detected with CAT-specific antibodies (see text for details). The pACYC184 lane shows CAT protein expressed in cells transformed with pACYC184 plasmid (wild-type cat); the CAT lane shows purified CAT protein.
DISCUSSION
We described here the selection, from a random rRNA fragment library, of an RNA fragment, complementary to 23S rRNA nucleotides 735 to 766, that allows a read-through of the UGA termination codon in certain mRNA sequence contexts.
An agent such as an RNA fragment could cause a termination codon read-through in one of two general ways. First, it could decrease translation accuracy, allowing certain tRNAs to misread codons that differ from their own. Second, it could interfere with the function of the termination apparatus, thereby shifting the balance between termination and natural low-frequency stop codon read-through. However, it has been reasoned that rRNA mutations that lead to codon-specific nonsense suppression (that is, the read-through of only specific termination codons) are more likely to be primarily defective in chain termination than to represent increased misreading per se by tRNAs (30–32). That reasoning was supported recently by in vitro experiments in which the ribosomes from two UGA-specific rRNA suppressor mutants were shown to be defective in termination, exhibiting major defects preferentially in termination depending on release factor 2 (RF2), which works specifically at UGA (2, 3). It is reasonable, therefore, to suggest that the antisense fragment characterized here as a UGA-specific nonsense suppressor affects RF2-dependent peptide chain termination.
Since nonsense suppression was observed only with the antisense rRNA fragment (one that can base-pair with 23S rRNA) but not with the same fragment in the sense orientation, it is possible that suppression required the complementary interaction of the selected RNA fragment with 23S rRNA in the ribosome. Several observations support this suggestion. The rRNA region corresponding to the selected fragment forms a hairpin structure (hairpin 750; Fig. 1), which is highly conserved and is present even in the minimalist rRNA of animal mitochondria (20). Nucleotides in the loop of hairpin 750 can be modified in the ribosome by various chemical reagents, indicating that this rRNA sequence may be accessible for the binding of an antisense rRNA fragment (21, 26, 48). The nucleotide sequence of the hairpin loop exhibits a high degree of conservation among the three domains of organisms, eucarya, archaea, and bacteria (16). In the tertiary structure of rRNA in the ribosome, hairpin 750 is located in the immediate vicinity of the peptidyl transferase center (near the central loop of domain V of 23S rRNA). Evidence for this includes cross-linking of the two regions (42, 43), simultaneous protection by antibiotics of nucleotides in hairpin 750 and in the central loop of domain V (21, 26, 48), and the possible existence of a base triple involving Ψ 746, G2057, and C2611 proposed on the basis of covariation analysis (19) (Fig. 1). The proximity of hairpin 750 to the central loop of domain V suggests that the hairpin may be involved in functions of the peptidyl transferase center. Since there is evidence that the ribosomal peptidyl transferase center is directly involved in the hydrolysis of ribosome-bound peptidyl tRNA at termination (5, 12, 13, 46) the distortion of peptidyl transferase structure induced by binding of the antisense fragment to hairpin 750 may result in the inhibition of peptidyl tRNA hydrolysis during RF2-dependent termination.
A less direct mechanism for the action of the antisense fragment can also be suggested. Modified nucleotides, which are clustered in the vicinity of functional centers in the 23S rRNA, may be important for ribosome functioning (9, 23, 25, 40). There are three modified nucleotides in the hairpin 750 region of E. coli 23S rRNA, nucleotides 745 (m1G), 746 (Ψ), and 747 (m5U) (9, 25) (Fig. 1B), and at least one of these modifications was shown to be important for normal protein synthesis (18). Consequently, binding of the antisense rRNA fragments to E. coli 23S rRNA may inhibit the modification of nucleotides in the loop of hairpin 750, resulting in altered ribosomal activity and the suppression of UGA nonsense mutations.
Recently, another RNA fragment with suppressor activity was isolated from the same random rRNA fragment expression library using a trpA (UGA115) reporter (4). The identified rRNA fragment corresponded to the E. coli 23S rRNA sequence from nucleotides 74 to 136, in domain I, and could suppress UGA nonsense mutations when expressed in both (sense and antisense) orientations. In contrast to that finding, the RNA fragment described here exhibits suppressor activity only when expressed in the antisense orientation, that is, when the expressed RNA is complementary to the ribosomal RNA target sequence. The reason for such a difference is not clear, but it is likely that the fragments identified in the two studies act by different mechanisms.
ACKNOWLEDGMENTS
We thank P. Kloss for technical assistance, T. Tenson and A. Neyfakh for stimulating discussions, and W. J. Pagel for editorial consultation.
This work was supported by a grant to A.S.M. from the National Science Foundation (MCB9420768) and a grant to E.J.M. from the National Institute of General Medical Sciences (GM21499).
REFERENCES
- 1.Agrawal R K, Penczek P, Grassucci R A, Frank J. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc Natl Acad Sci USA. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arkov A L, Freistroffer D V, Ehrenberg M, Murgola E J. Mutations in RNAs of both ribosomal subunits cause defects in translation termination. EMBO J. 1998;17:1507–1514. doi: 10.1093/emboj/17.5.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arkov, A. L., D. V. Freistroffer, M. Y. Pavlov, M. Ehrenberg, and E. J. Murgola. Mutations in conserved regions of ribosomal RNAs decrease the productive association of peptide-chain release factors with the ribosome during translation termination. Submitted for publication. [DOI] [PubMed]
- 4.Arkov A L, Mankin A, Murgola E J. An rRNA fragment and its antisense can alter decoding of genetic information. J Bacteriol. 1998;180:2744–2748. doi: 10.1128/jb.180.10.2744-2748.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arkov, A. L., and E. J. Murgola. Unpublished results.
- 6.Atkins J F, Nichols B P, Thompson S. The nucleotide sequence of the first externally suppressible −1 frameshift mutant, and of some nearby leaky frameshift mutants. EMBO J. 1983;2:1345–1350. doi: 10.1002/j.1460-2075.1983.tb01590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barta A, Steiner G, Brosius J, Noller H F, Kuechler E. Identification of a site on 23S ribosomal RNA located at the peptidyl transferase center. Proc Natl Acad Sci USA. 1984;81:3607–3611. doi: 10.1073/pnas.81.12.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brimacombe R, Mitchell P, Müller F. The organization of rRNA, tRNA, and mRNA in the ribosome. In: Zimmermann R A, Dahlberg A E, editors. Ribosomal RNA: structure, evolution, processing, and function in protein biosynthesis. Boca Raton, Fla: CRC Press; 1996. pp. 129–147. [Google Scholar]
- 9.Brimacombe R, Mitchell P, Osswald M, Stade K, Bochkariov D. Clustering of modified nucleotides at the functional center of bacterial ribosomal RNA. FASEB J. 1993;7:161–167. doi: 10.1096/fasebj.7.1.8422963. [DOI] [PubMed] [Google Scholar]
- 10.Bronson M F, Yanofsky C. Characterization of mutations in the tryptophan operon of Escherichia coli by RNA nucleotide sequencing. J Mol Biol. 1974;88:913–916. doi: 10.1016/0022-2836(74)90407-0. [DOI] [PubMed] [Google Scholar]
- 11.Buckingham R H, Sörensen P, Pagel F T, Hijazi K A, Mims B H, Brechemier-Baey D, Murgola E J. Third position base changes in codons 5′ and 3′ adjacent UGA codons affect UGA suppression in vivo. Biochim Biophys Acta. 1990;1050:259–262. doi: 10.1016/0167-4781(90)90177-4. [DOI] [PubMed] [Google Scholar]
- 12.Caskey C T. Peptide chain termination. Trends Biochem Sci. 1980;5:234–237. [Google Scholar]
- 13.Caskey C T, Beaudet A L, Scolnick E M, Rosman M. Hydrolysis of fMet-tRNA by peptidyl transferase. Proc Natl Acad Sci USA. 1971;68:3163–3167. doi: 10.1073/pnas.68.12.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cundliffe E. Recognition sites for antibiotics within rRNA. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function, and evolution. Washington, D.C: American Society for Microbiology; 1990. pp. 479–490. [Google Scholar]
- 16.Egebjerg J, Larsen N, Garrett R A. Structural map of 23S rRNA. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function, and evolution. Washington, D.C: American Society for Microbiology; 1990. pp. 168–179. [Google Scholar]
- 17.Fredericq P. The recombination of colicinogenic factors with other episomes and plasmids. In: Wolstenholme G E, O’Connor M, editors. Bacterial plasmids and episomes. Boston, Mass: Little, Brown & Co.; 1969. pp. 163–174. [Google Scholar]
- 18.Gustafsson C, Persson B C. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J Bacteriol. 1998;180:359–365. doi: 10.1128/jb.180.2.359-365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutell R R. Comparative sequence analysis and the structure of 16S and 23S rRNA. In: Zimmermann R A, Dahlberg A E, editors. Ribosomal RNA: structure, evolution, processing, and function in protein biosynthesis. Boca Raton, Fla: CRC Press; 1996. pp. 111–128. [Google Scholar]
- 20.Gutell R R, Gray M W, Schnare M N. A compilation of large subunit (23S and 23S-like) ribosomal RNA structures: 1993. Nucleic Acids Res. 1993;21:3055–3074. doi: 10.1093/nar/21.13.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen L H, Mauvais P, Douthwaite S. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol Microbiol. 1999;31:623–632. doi: 10.1046/j.1365-2958.1999.01202.x. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lane B G, Ofengand J, Gray M W. Pseudouridine in the large-subunit (23 S-like) ribosomal RNA. The site of peptidyl transfer in the ribosome? FEBS Lett. 1992;302:1–4. doi: 10.1016/0014-5793(92)80269-m. [DOI] [PubMed] [Google Scholar]
- 24.LaVallie E R, DiBlasio E A, Kovacic S, Grant K L, Schendel P F, McCoy J M. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Bio/Technology. 1993;11:187–193. doi: 10.1038/nbt0293-187. [DOI] [PubMed] [Google Scholar]
- 25.McCloskey J A, Crain P F. The RNA modification database—1998. Nucleic Acids Res. 1998;26:196–197. doi: 10.1093/nar/26.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moazed D, Noller H F. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- 27.Muralikrishna P, Cooperman B S. A photolabile oligodeoxyribonucleotide probe of the peptidyltransferase center: identification of neighboring ribosomal components. Biochemistry. 1991;30:5421–5428. doi: 10.1021/bi00236a014. [DOI] [PubMed] [Google Scholar]
- 28.Murgola E J. tRNA, suppression, and the code. Annu Rev Genet. 1985;19:57–80. doi: 10.1146/annurev.ge.19.120185.000421. [DOI] [PubMed] [Google Scholar]
- 29.Murgola E J. Ribosomal RNA in peptide chain termination. In: Zimmermann R A, Dahlberg A E, editors. Ribosomal RNA: structure, evolution, processing, and function in protein biosynthesis. Boca Raton, Fla: CRC Press; 1996. pp. 357–372. [Google Scholar]
- 30.Murgola E J, Göringer H U, Dahlberg A E, Hijazi K A. Ribosomal RNA and UGA-dependent peptide chain termination. In: Cech T, editor. Molecular biology of RNA. New York, N.Y: Alan R. Liss; 1989. pp. 221–229. [Google Scholar]
- 31.Murgola E J, Hijazi K A, Göringer H U, Dahlberg A E. Mutant 16S ribosomal RNA: a codon-specific translational suppressor. Proc Natl Acad Sci USA. 1988;85:4162–4165. doi: 10.1073/pnas.85.12.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murgola E J, Pagel F T, Hijazi K A, Arkov A L, Xu W, Zhao S Q. Variety of nonsense suppressor phenotypes associated with mutational changes at conserved sites in Escherichia coli ribosomal RNA. Biochem Cell Biol. 1995;73:925–931. doi: 10.1139/o95-100. [DOI] [PubMed] [Google Scholar]
- 33.Noller H F, Moazed D, Stern S, Powers T, Allen P N, Robertson J M, Weiser B, Triman K. Structure of rRNA and its functional interactions in translation. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function, and evolution. Washington, D.C: American Society for Microbiology; 1990. pp. 73–92. [Google Scholar]
- 34.O’Connor M, Brunelli C A, Firpo M A, Gregory S T, Lieberman K R, Lodmell J S, Moine H, Van Ryk D I, Dahlberg A E. Genetic probes of ribosomal RNA function. Biochem Cell Biol. 1995;73:859–868. doi: 10.1139/o95-093. [DOI] [PubMed] [Google Scholar]
- 35.Pagel, F. T., and E. J. Murgola. Unpublished results.
- 36.Pagel F T, Zhao S Q, Hijazi K A, Murgola E J. Phenotypic heterogeneity of mutational changes at a conserved nucleotide in 16 S ribosomal RNA. J Mol Biol. 1997;267:1113–1123. doi: 10.1006/jmbi.1997.0943. [DOI] [PubMed] [Google Scholar]
- 37.Poole E S, Brown C M, Tate W P. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995;14:151–158. doi: 10.1002/j.1460-2075.1995.tb06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prince J B, Taylor B H, Thurlow D L, Ofengand J, Zimmermann R A. Covalent crosslinking of tRNAVal to 16S RNA at the ribosomal P site: identification of crosslinked residues. Proc Natl Acad Sci USA. 1982;79:5450–5454. doi: 10.1073/pnas.79.18.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rinke-Appel J, Junke N, Osswald M, Brimacombe R. The ribosomal environment of tRNA: crosslinks to rRNA from positions 8 and 20:1 in the central fold of tRNA located at the A, P, or E site. RNA. 1995;1:1018–1028. [PMC free article] [PubMed] [Google Scholar]
- 40.Sirum-Connolly K, Peltier J M, Crain P F, McCloskey J A, Mason T L. Implications of a functional large ribosomal RNA with only three modified nucleotides. Biochimie. 1995;77:30–39. doi: 10.1016/0300-9084(96)88101-6. [DOI] [PubMed] [Google Scholar]
- 41.Stark H, Rodnina M V, Rinke-Appel J, Brimacombe R, Wintermeyer W, Van Heel M. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature. 1997;389:403–406. doi: 10.1038/38770. [DOI] [PubMed] [Google Scholar]
- 42.Stiege W, Atmadja J, Zobawa M, Brimacombe R. Investigation of the tertiary folding of Escherichia coli ribosomal RNA by intra-RNA cross-linking in vivo. J Mol Biol. 1986;191:135–138. doi: 10.1016/0022-2836(86)90429-8. [DOI] [PubMed] [Google Scholar]
- 43.Stiege W, Glotz C, Brimacombe R. Localisation of a series of intra-RNA cross-links in the secondary and tertiary structure of 23S RNA, induced by ultraviolet irradiation of Escherichia coli 50S ribosomal subunits. Nucleic Acids Res. 1983;11:1687–1706. doi: 10.1093/nar/11.6.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tenson T, DeBlasio A, Mankin A. A functional peptide encoded in the Escherichia coli 23S rRNA. Proc Natl Acad Sci USA. 1996;93:5641–5646. doi: 10.1073/pnas.93.11.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tucker S D, Murgola E J, Pagel F T. Missense and nonsense suppressors can correct frameshift mutations. Biochimie. 1989;71:729–739. doi: 10.1016/0300-9084(89)90089-8. [DOI] [PubMed] [Google Scholar]
- 46.Vogel Z, Zamir A, Elson D. The possible involvement of peptidyl transferase in the termination step of protein biosynthesis. Biochemistry. 1969;8:5161–5168. doi: 10.1021/bi00840a070. [DOI] [PubMed] [Google Scholar]
- 47.Wilson K S, Noller H F. Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell. 1998;92:131–139. doi: 10.1016/s0092-8674(00)80905-8. [DOI] [PubMed] [Google Scholar]
- 48.Xiong L, Shah S, Mauvais P, Mankin A S. A ketolide resistance mutation in domain II of 23S rRNA reveals proximity of hairpin 35 to the peptidyl transferase centre. Mol Microbiol. 1999;31:633–639. doi: 10.1046/j.1365-2958.1999.01203.x. [DOI] [PubMed] [Google Scholar]
- 49.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 50.Zimmermann R A. The decoding domain. In: Zimmermann R A, Dahlberg A E, editors. Ribosomal RNA: structure, evolution, processing, and function in protein biosynthesis. Boca Raton, Fla: CRC Press; 1996. pp. 277–309. [Google Scholar]