Abstract
The increasing host range of canine morbillivirus (CDV) affecting important wildlife species such as Lions, Leopard, and Red Pandas has raised the concern. Canine distemper is a pathogen of dogs affecting the respiratory, gastrointestinal, and nervous systems. Seventeen lineages of CDV are reported, and the eighteenth lineage was proposed in 2019 from India. Marked genomic differences in the genome of wild-type virus and vaccine strain are also reported.The variations at the epitope level can be differentiated using specific monoclonal antibodies in neutralization tests. Keeping in mind the current status of the emergence of CDV, genetic and molecular study of circulating strains of the specific geographical region are the essential components of the disease control strategy. New target-based diagnostics and vaccines are in need to counter the effects of the emerging virus population. Control of CDV is necessary to save the endangered, vulnerable, and many other wildlife species to maintain balance in the ecological system. This review provides an overview on emergence reported in CDV, diagnostics developed till today, and a perspective on the disease control strategy, keeping wildlife in consideration.
Keywords: Canine morbillivirus (CDV), Emergence, Wildlife, Diagnosis, Monoclonal antibodies (mAb)
Introduction
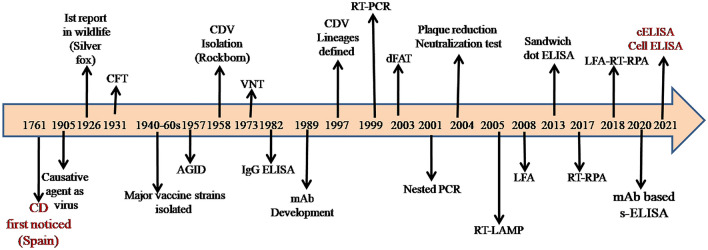
Canine distemper (CD) is primarily a viral disease of dogs and can infect several wildlife species. The disease was first reported in Spain (1761) and from there is believed to spread across the world [16, 17]. Edward Jenner was the first person to mention the disease name “canine distemper” and Carre first studied its etiological agent in 1905 [4]. The important milestones in the CDV research timeline are presented in Fig. 1.The disease is caused by canine morbillivirus or canine distemper virus (CDV), a member of the genus Morbillivirus and the family Paramyxoviridae [49]. The other members of genus like Rinderpest virus (RPV) and Measles virus (MV) are known to cause devastating diseases in both animals and humans from ages [132]. The host range of CDV includes members of Canidae, Procyonidae, Mustelidae, Ursidae, Viverridae, Felidae, Ailruidae, Ursidae, Hyaenidae, Tayassuidae, Cercopithecidae, and so on [72, 73]. Collectively, more than 20 families of Carnivores and non-carnivores are reported to be affected [34]. CD is a multisystemic disease that affects the respiratory, gastrointestinal, and nervous systems of the animal [81]. The incubation period varies from 1 to 4 weeks and the disease manifests as acute systemic and chronic encephalitic forms [101]. Virions are highly contagious, transmitted through aerosolized nasal, oral, and ocular fluid [4]. Real-time reverse-transcription polymerase chain reaction (RT-qPCR) based kinetics study demonstrated the virus shedding from the rectum (4 to 20th days post-infection/dpi), nose (2 to 20th dpi), blood (2 to 12th dpi), with the peak of virus titre at 10th, 12th, and 6th dpi respectively [110]. Infected animals shed virus in urine up to 60–90 days after recovery from the acute phase of the disease [42]. The virus causes severe immunosuppression and the affected animals show the clinical signs as biphasic fever, cough, conjunctivitis, diarrhea, and anorexia. The first peak of fever is seen after infection, followed by the second peak, after 6–9 days [95]. In some cases, the chronic stage of the disease occurs, manifested as seizures, tremors, twitching movements, and hard pads, referred to as “Old dog Encephalitis” [135].
Fig. 1.
The important milestones in the CDV research timeline
CDV is a non-segmented negative-sense RNA virus of 15,960 nucleotides, wrapped over by a lipoprotein envelope. The genome comprises six structural proteins viz. nucleocapsid protein (N), phosphoprotein (P), matrix protein (M), fusion protein (F), haemagglutinin protein (H), and the large protein (L) [80]. Non-structural protein (C) is encoded from the gene sequence of P protein byan overlapping open reading frame [118]. The study of detailed genetic sequence and the identification of conserved regions help to find targets for the specific identification of any pathogen. With the advancement in molecular studies after the 1990s, different genetic lineages of MeV [133], RPV [21], PPRV [113], and CDV [19] were identified, which are now one of the criteria for characterizing viral isolates based on their geographic origin. Till 2014 nine genetic lineages were reportedbased on H protein sequence identity (more than 95%) [107]. The number of genetic lineages has reached seventeen, namely America 1 to America 5, Asia 1 to Asia 4, Europe/ South America 1, South America 2 and 3, Europe wildlife, Arctic, Rockborn-like, Africa 1 and Africa 2 [33, 57]. In 2019, the eighteenth lineage was proposed (India-1/Asia-5) by our laboratory, which represents the strains circulating in India [13].
Vaccines are the most important component of a viral disease control programs. Formalin-inactivated virus vaccine strains were used earlier to provide active immunity to dogs. The inactivated vaccine induces less antibody titre and is found to be less efficacious [4]. Ferret-passaged and egg-based vaccines of CDV have also been used for vaccination. The cell-culture attenuated strains, Ondersteepoort, Synder Hill, Lederle, Convac, and Rockborn are widely used as live vaccines [25]. These vaccine strains provide effective immunity up to 4.4 years in dogs [50]. Rockborn strain was found to possess the residual virulence causing post-vaccination encephalitis, therefore its use was withdrawn in the mid-1990s [74]. The details of well-known strains of CDV are compiled in Table 1. Canarypoxvirus and Equine Herpesvirus type-1 (EHV-1) viral vector-based CDV vaccines are also found to protect dogs as well as wildlife species [91, 104, 144]. Many CDV strains have emerged in past few decades with different pathogenic characteristics and out of them apathogenic/naturally attenuated as a vaccine candidate isalso reported [131].
Table 1.
Important (well-characterized) strains of CDV
| Viral strain | Lineage | Available gene sequences | Accession no | Reference |
|---|---|---|---|---|
| Convac | America-1 | H gene | Z35493 | [63] |
| Ondersteepoort | America-1 | Complete genome | AF014953 | [122] |
| SynderHill | America-1 | H gene | AF259552 | [45] |
| Lederle | America-1 | H gene | EF418782 | Not available |
| Rockborn | America-1 | H gene | GU810819 | [74] |
| Dog/Bly/Ind/2018 | India-1/ Asia 5 | H gene | MF964178 | [13] |
This review provides a piece of comprehensive information on the emergence of CDV resulting in expansion of its host range, the current status of the disease, diagnostic tools developed till today, concluding with a perspective on strategic advancement needed for effective prevention and control of the disease.
Current status and the emergence of the disease
CD is an endemic disease with a high frequency of outbreaks reported from all parts of the world. The serological survey has shown the high prevalence of the virus in the free-ranging dog population in many countries [23, 66, 119]. In India, several reports on isolation and characterization of CDV in dogs as well as wildlife from different geographical regions are present [10, 52, 62, 75, 128]. The dogs are the most common source of transmission of CDV to wildlife. Infection in endangered species like Siberian tigers, Ethiopian wolves, Red Panda [154], and other vulnerable wildlife species like Cheetah and Lions [5, 79, 117] shows the severe impact of CDV. From the very first report in silver fox [4], the list of wild animals is continuously increasing with the increased surveys, for example, Tamandua tetradactyla [73], mesocarnivores like Mink, Skunk, Racoon [83]. Cheetah, domestic cats, and Asian Elephants are reported to be infected with the virus as indicated by the presence of specific antibodies but do not show any clinical signs [80]. The Phocine distemper virus (PDV) is also considered to be derived from CDV as a result of species jumping, selection pressure, and host adaptation [58]. CDV is now an emerging viral risk for several wildlife species [111], some of which are given in Table 2.
Table 2.
Host range of CDV
| Family | Affected animals | Scientific names | Reference |
|---|---|---|---|
| Canidae | Silver fox | Vulpes vulpes | [4] |
| Ethiopion wolf | Canis simensis | [41] | |
| Coyotes | Canis latrans | [44] | |
| Jackal | Canis aureus | [4] | |
| Ursidae | Giant pandas | Ailuropoda melanoleuca | [36, 98] |
| Black bears | Ursus Americana | [26] | |
| Ailuridae | Red Panda | Ailurus fulgens | [20, 154] |
| Felidae | Lion | Panthera leo | [93] |
| Leopard | Panthera pardus | [8] | |
| Siberian tiger (Amur tiger) | Panthera tigrisaltaica | [99] | |
| Cercopthecidae | Rhesus monkey | Macaca mulata | [29, 97] |
| Cynomologus Macaques | Macaca fascicularis | [105, 127] | |
| Japanese Monkey | Macaca fusata | [152] | |
| Procyonidae | Racoons | Procyon lotor | [4] |
| Hyaenidae | Hyaena | Crocuta crocuta | [45] |
| Mustelidae | Ferrets | Mustela putorius furo | [122] |
| Minks | Neovison vison | [4] | |
| Viveridae | Civets | Pagumalarvata | [4] |
| Myrmecophagidae | Collared anteater | Tamandua tetradactyla | [73] |
| Giant anteater | Myrmecophaga tridactyla | [30] | |
| Elephantidae | Asian Elephant | Elephas maximus | [86] |
The H protein of the virus is immunodominant and is responsible for interacting with host cellular receptors—SLAM (Signaling lymphocytic activation molecule) and nectin-4. The H protein possesses a high percentage of mutations which undergoes positive selection to adapt to its new host. Therefore, the H protein is considered to be main force behind virus affinity towards different cells and acquiring the ability to infect a new host [34].The interaction of H protein with SLAM and nectin-4 receptors has been depicted in Fig. 2. From the study of host cell receptors, the SLAM and nectin-4 receptors of dogs, felids, and small ruminants show some conserved residues and motifs which represent the specificity to particular morbillivirus [108, 145]. Nectin-4 receptor is comparatively conserved and is responsible for cell-to-cell spread within the host [76, 149]. SLAM receptor is the entry receptor for the virus and possesses mutations at several amino acid residues among the different hosts. Along with many other amino acids, the 76th position of the SLAM receptor plays a key role in host cell recognition and virus binding [85]. Based on the computational analysis, it was revealed that the Histidine residues at 28th position of N-terminal of SLAM receptors of Macaca are responsible for the stable interaction with H protein of CDV [150]. In our one of the unpublished data, most of the cases of wildlife population outbreaks showed changes at residue 549 of viral H protein.It was further confirmed that the Y549H mutation was positively selected by selection pressure. And the mutation at this particular residue might be a key determinant in adaption of virus in wildlife population.Also the mutation Y549H in H protein has been reported to beinvolved inthe interaction of virus with the V domain of mink SLAM receptors [38].
Fig. 2.
CDV tropism vis-à-vis receptor fidelity in domestic and wildlife animal population
With the ability of the virus to infect primates and adapt to the new host, few in-vitro studies were conducted manipulating human cell receptors (SLAM) and the H protein of the CDV [106]. These studies showed that the CDV has the fair possibility to emerge as a human pathogen with few mutations [16, 130]. As MeV shows cross-protective immunity with CDV, only the unimmunized population for MeV might be susceptible to new CDV [98]. The CDV isolates are classified as old CDV (vaccine strains) and new CDV based on the differences in their antigenic characteristic in neutralization tests [78]. Even different biotypes of different physiological properties and pathogenic patterns are reported [87, 126]. This variation is reflected in increasing cases of outbreaks of CD in vaccinated populations [2, 102, 136]. The widely used vaccine strains belong to the America-1 lineage, which possesses significant genetic variation from the circulating CDV strains of other lineages in different geographical areas [13].
Co-circulation of different lineages may result in homologous recombination and the emergence of viral variants or sub-lineages [46, 39, 94, 153]. The possibility of recombination of old vaccine strains with new viral variants has been suggested as the reason for the evolution of CDV and disease in vaccinated animals [28]. These are anticipated explanations, as the evolutionary analysis and emergence of CDV is not clearly studied and described yet. Being a pathogen of such a wide host range and the significance of wildlife in ecological balance, the monitoring and research of CDV require utmost priority without any critical gaps.
Available diagnostic tools for CDV
Ferrets were used as an experimental animal for diagnosis in earlier days [32]. Later, virus isolation started ascommonlaboratory practice for diagnosis using ferret kidney cells, dog alveolar macrophage culture, and Vero cells [4].Primarily, the diagnosis is done based on the history of animals and the clinical signs indicating multi-systemic affection. Tissue samples like the external epithelium of affected linings viz. hard pad, urothelium, uvea contain the persistent virus [54]. The sign and symptoms of the disease are not very noticeable at the early stage, especially in wildlife. Therefore, the correct and rapid diagnosis is the key to the management and control of the disease. Today’s requirement of an in-depth genetic study of pathogens puts forth the demand for highly sensitive and specific diagnostic assays. There are several diagnostic tests with varied sensitivity, specificity, required time, and different levels of skills to perform the test [106]. The diagnostic tools developed for CDV detection are included in Table 3.
Table 3.
Diagnostic tools for routine diagnosis of CDV in laboratory and field
| Diagnostic tool | Target | Application | References | |
|---|---|---|---|---|
|
1. Virus detection a. Virus isolation |
(MDCK, Vero cells-SLAM B95a, etc) | Virus |
Gold standard test Highly sensitive, helpful for generating virus repository (Require live virus titre, specific cell line and cell culture facilities) |
[7] [107] [120] |
| Direct ELISA | CDV antigen | Detects antigen in serum | [119] | |
| b. Antigen detection | Sandwich ELISA | H protein |
High specificity Detection and quantitation |
[91] |
| F protein | Efficient in field application with fecal and serum samples | [148] | ||
| Sandwich dot ELISA | Virus | Epidemiological surveillance | [65] | |
| LFA | F protein | Practically applicable in the field for quick diagnosis | [1] | |
| c. Nucleic acid detection | RT-PCR | N gene | Standard laboratory test | [39] |
| One-step nested-RT-PCR | N gene | 100-fold sensitivity than RT-PCR and nested PCR | [56] | |
| Double step real time-RT-PCR | N gene |
Highly sensitive and specific Quantitate viral load in clinical samples |
[34] | |
| One-step real-time RT-PCR | -- | To study viral replication and kinetics of viral RNA load in infection | [105] | |
| RT-LAMP assay | H gene |
100-times sensitive than RT-PCR Only 1 hour reaction |
[66] | |
| 2. Virus-specific antibody detection | ELISA | IgG Antibody |
Detect within 6 days of infection Sensitive as SNT |
[11] |
| Dot blot assay | N-protein specific IgM | Detecting recent infections | [10, 17] | |
| Capture sandwich ELISA | N-protein specific IgG & IgM Antibody | No Cross-reactivity with other Morbilliviruses | [131, 60, 61] |
*Diagnostic sensitivity (Dsn), Diagnostic specificity (Dsp)
NA –Information not available
Virus detection
Virus isolation
Virus isolation is the “gold standard” test for the diagnosis of viral diseases. CDV presents a low success rate in virus isolation due to its high sensitivity of virus to light and temperature [4]. CDV isolation often requires supplementation with canine or ferret pulmonary macrophages [7], co-cultivation with mitogen-stimulated lymphocytes derived from healthy dogs’ and ferret’s blood. Stable expression of canine SLAM receptors (CD150) on Vero cells results in enhanced isolation of field isolates of CDV, especially from dogs [112]. A virulent CDV was first isolated and adapted in dog kidney cells by Rockborn [103]. MDCK (Madin-Darby canine kidney), MV1 Lu (Mink lung epithelial cells), and Vero cells are commonly used for the primary isolation of CDV, but with a low success rate [67]. B95a, a marmoset lymphoblastoid cell line is efficient in CDV isolation similar to other morbilliviruses—RPV and PPRV [53, 61, 125].
After several blind passages, the virus adapts to the cell line and shows the visible changes appreciated under the light microscope. Cytopathic effect for CDV is similar to other morbilliviruses, especially measles virus, characterized by degenerated cells, granular appearance, vacuolization, and multi-nucleated giant cells (syncytia) [67]. The onset of CPE usually varies with strain of the virus, type of cell line used, incubation, and media condition for cells. The vaccine and wild-type strains can be differentiated by the type of CPE, as the level of attenuation of the strain is directly proportional to the fusion efficiency of H protein [139]. The CPE of attenuated strains is differentiated by the size of syncytia i.e. large patches of fused cells visible under the light microscope.
Nucleic acid detection
The antigen and nucleic acid detection do not require the live virus in the samples, as required in virus isolation. Molecular assays present the advantage of sensitive and specific detection in both the antemortem and post-mortem samples. RT-PCR is the widely used molecular diagnostic test and detects CDV in whole blood, serum, and cerebrospinal fluid [40]. Nested PCR combined with conventional RT-PCR gives comparatively high specificity [60]. Also, nested-PCR is more sensitive than RT-PCR and Immunofluorescence assay for CDV diagnosis of clinical samples like urine, blood, and saliva [51, 116]. RT-qPCR using the TaqMan probe based on CDV-N and P genes is highly sensitive and specific over other tests [35, 109]. For rapid identification, the sequencing of the whole genome by nanopore technology is used [92].
Many commonly used molecular tests are modified to differentiate wild-type and vaccine strains based on the variations in their genomic sequences (Table 4). For example, firstly the conserved sequences are targeted to amplify the both, followed by primer specific to either wild-type or vaccine strains. Also, the RFLP results in different number of fragments in both the strains. These significant variations might be responsible for the outbreaks in vaccinated populations as well.
Table 4.
Molecular tests developed to differentiate wild-type and vaccine CDV strains
Monoclonal antibodies in CD diagnosis
Orvell et al. first developed the mAbs against N, F, H, and P protein of CDV [89]. The mAbshold the potential to be used in immunodiagnostics as well as immunotherapy of CDV [14, 15, 48]. Recently, two different studies shows the role of mAbs in identifying the linear B-cell epitopes on P and H protein [68, 115]. Sandwich ELISA based on monoclonal antibodies (mAb) against H protein can detect the CDV in conjunctiva, nasal swab, and lungs [96]. mAb-based sandwich dot ELISA is rapid in detecting the CDV for epidemiological surveillance [69]. A sandwich ELISA using two mAbs against different epitopes of F protein was also developed to detect CDV in serum and fecal samples [155]. Monoclonal antibodies provide high specificity to the diagnostic assays [1, 96]. MAbs developed against H and N proteins of PPRV has been proved to be irreplaceable components of c-ELISA and s-ELISA, respectively [120, 121]. These diagnostic assays have contributed significantly to the PPRV diagnosis and its control program in India. The learnings from research in PPRV, the sibling member of CDV, can guidefuture research of CDV diagnostics and vaccines in many ways.
CDV—antibody detection
Dogs show lifelong protection from the disease through induction of high titre neutralizing antibodies (1:100) after infection, which usually achieves a peak in 2–3 weeks [6]. Most of the studies have reported that antibody titre of 1:32 is indicative of borderline protective titre against CDV infection [9], while some found it to be in the range of 1:80 to 1:160 [129]. An increase in IgG titer greater than fourfold within 14 days indicates infection in even recently vaccinated animals [55]. Although the virus neutralization test (VNT) is the gold standard and OIE accepted test, it is time-taking and requires conventional virology skills to perform the test and the handling of live viruses. This makes it less preferable over other immunological assays like ELISA, IFT, etc. ELISA is widely used to detect IgM and IgG antibodies till three months of infection in both dogs and non-dog hosts [43] as well as gives consistent specificity and sensitivity as serum neutralization test (SNT) [84]. The plaque reduction neutralization test is also used to evaluate the pre and post-vaccination status of the animal [90]. Immunoperoxidase plaque staining for CDV detection is another specific and sensitive technique for serological study [123].
There are cross-reactive epitopes among CDV, MV, PPRV, and RPV and should be considered while testing serological samples [114]. M protein shares the highest number of cross-reactive epitopes among CDV and MV [88]. F protein also shares the homology in the cleavage site (six amino acids) for furin protease and is replaceable between CDV and MV [134, 138]. F protein shares the high heterotypic cross-reactive epitopes among all structural proteins of the genus, showing the evolutionary relationship [116]. Therefore, the recombinant F protein-based vaccines provide heterotypic butincomplete protection. Recent antigenic profiling of morbilliviruses (CDV, PPRV & MV) from our laboratory using a panel of in-house developed six mAbs has indicated that PPRV possesses a more close antigenic relationship to CDV as compared to MV (unpublished information).
Point of care diagnostic tests for CDV
Field diagnostic tests are the need of the hour to detect the infection on-site and control the spread of disease. RT-loop-mediated isothermal amplification (RT-LAMP) has been developed with high sensitivity (100%), specificity (93.3%), and 100 fold lower detection limits as compared to conventional RT-PCR [24, 148]. RT-recombinase polymerase assay (RT-RPA) having a portable, user-friendly tube scanner has been developed [142]. A Microfluidic paper-based test device has also been developed using nanoparticle-coated polyclonal antibodies raised in rabbits. The test also presents a potential for field diagnostic tests as suggested by studying clinical data [77]. An incubation-free LFA-RT-RPA assay is also developed for CDV detection using a gold particle-conjugated anti-FAM antibody but presents the limitation of nucleic acid extraction requirement [143]. One-tube reverse transcription-insulated isothermal polymerase chain reaction has also been developed as another point-of-care diagnostic which presents comparable results with real time RT-PCR [146].
Point-of-care diagnostic test should be simple enough to be easily performed by a layman with mere instructions; hence the lateral flow assay (LFA) holds the advantage over all other tests. Also, the problem of sample collection in wildlife and the transport-related damage raises the need for specific, easy, and on-site diagnostic assays like LFA to diagnose the disease at an early stage. CDV-LFA was developed in the sandwich assay format using two monoclonal antibodies (mAbs) of IgG1 isotype against two different epitopes of F protein viz. 9D3 as a bio label and 7B2 in the ‘test’ line, resulting in sensitive and specific detection [1]. Different commercial LFA kits are also available from different countries like Rapigen Inc. CDV Antigen test, Lilif™ CDV Rapid Antigen kit, etc. Recently, a mAb based LFA is developed in our laboratory for CDV antigen detection in sandwich mode (unpublished) and antibody detection in competitive mode [56].
Differential diagnosis
The clinical signs of CD are similar to other viral diseases caused by Canine parvovirus (CPV), Canine coronavirus (CCoV), canine adenovirus (CAV), Rabies virus, influenza virus, and some bacterial and parasitic diseases [59]. According to a study conducted in selected areas of Madhya Pradesh (India) CDV, CPV and CAV are more prevalent than Rabies but are comparatively less in focus due to unnoticeable signs in dogs and zoonotic unimportance [23, 100]. Feral dogs harbor the CDV, CPV, and CAV very often [47, 82]. Even in serological surveys, other wild carnivores have reported the presence of antibodies against CDV and CPV altogether [132]. Therefore, there is also a need for diagnostic tests for differential detection of the virus and virus-specific antibodies. Important to today’s scenario, one-step duplex PCR was also developed for identification and differential diagnosis between CDV and Canine Coronavirus (CCoV) targeting the CDV-H gene and CCoV-M gene [141]. Similarly, a one-step triplex PCR detecting CDV, CPV, and Canine Kobu virus was developed, for differential diagnosis as well as surveillance [70].
Conclusion
CD is an endemic disease of dogs and was under control until some time ago due to the availability of effective vaccines. The strains Ondersteepoort, Convac, Synder Hill, Rockborn, were isolated and came in use around 1960s. In the past few decades, a large number of new cases are constantly reported in the vaccinated population of dogs and a large number of wildlife species, which has raised the concern towards disease control and wildlife protection. Some constant mutations have been found in H protein like Y549H which are responsible for the host range expansion of the virus. The difference in circulating strains and the emergence of new lineages confirms the continuous mutations in virus isolates. Yet more studies on recombination and evolution of CDV are required.
Control of CDV needs specific diagnostics, vaccines improvement to make it effective in dog population as well as wildlife, and proper serological surveillance to prevent unexpected outbreaks. Therefore, there is a need for novel targets, for both the modified diagnostics as well as vaccines to counteract the effect of circulating virus. Our laboratory isolated “CDV (Dog)/Bly/Ind/2018”, which represents the currently circulating CDV strains of India [13]. This strain of the virus has been extensively characterized and has been proposed as an attenuated vaccine candidate (Indian Patent application number 202011057169, dated 30/12/2020). A panel of monoclonal antibodies to this strain has been developed and characterized (unpublished information).Conventional virology techniques like virus isolation and virus neutralization test are still in use for laboratory diagnostics because of their high specificity. Molecular assays like RT-PCR, RT-qPCR, and LAMP are used when the rapid diagnosis is required. For the serological study, ELISA based on IgM and IgG antibodies are commonly used to detect recent infection and later phases of the disease, respectively. Point-of-care diagnostics are easy to perform and give a quick accurate diagnosis, hold an important place in disease diagnosis and management plan.
Widely used conventional cell culture adapted vaccines sometimes do not provide complete protection and presents residual virulence/retained pathogenicity, especially in cross-species vaccination in wildlife. To overcome the disadvantages presented by attenuated vaccines, recombinant, and other new-generation vaccines can be developed [22]. Looking at the wide host range and poor accessibility to the affected wildlife species, a focus on the development of an oral vaccine against canine morbillivirus may provide some solution. Control of disease needs to be planned at the root level i.e. control in feral dogs and strictly defined boundaries of protected areas to block the dog-wildlife interface and prevent spillover of disease to wildlife. Apart from dogs, ferrets, minks, and raccoons also serve as a reservoir of CDV and are responsible for transmission and co-circulation of different lineages of overlapping geographical regions between dogs and wildlife [3, 148]. Therefore, vaccination of these species is another important component for the control of CDV [50, 147]. Keeping in mind the role of dogs in virus transmission to wildlife, immediate mitigation strategies, and their execution is the need of the hour [27]. The broad host range of CD makes it difficult to eradicate, but with strategic research and applicability at the field level, the increasing cases and outbreaks can be controlled.
Funding
The authors are thankful to the Director and Joint Directors ICAR-IVRI for providing the necessary facility. The authors would like to acknowledge the funding agency Science and Engineering Research Board-Department of Science and Technology (SERB-DST) Government of India (Grant No EEQ/2018/000823).
Declarations
Conflict of interest
The authors declared no potential conflicts of interest concerning research, authorship, and/or publication of this article.
Consent for publication
The authors confirm that the work described here has not been published or under consideration in any other journal. The authors agree to publication in the journal of virus disease.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kaushal Kishor Rajak, Email: kaushalvirol@gmail.com.
Rabindra Prasad Singh, Email: Rabindra.Singh@icar.gov.in.
References:
- 1.An DJ, Kim TY, Song DS, Kang BK, Park BK. An immunochromatography assay for rapid antemortem diagnosis of dogs suspected to have canine distemper. J Virol methods. 2008;147(2):244–249. doi: 10.1016/j.jviromet.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anis E, Holford AL, Galyon GD, Wilkes RP. Antigenic analysis of genetic variants of canine distemper virus. Vet Microbiol. 2018;219:154–160. doi: 10.1016/j.vetmic.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 3.Anis E, Needle DB, Stevens B, Yan L, Wilkes RP. Genetic characteristics of canine distemper viruses circulating in wildlife in the united states. J Zoo Wildl Med. 2020;50(4):790–797. doi: 10.1638/2019-0052. [DOI] [PubMed] [Google Scholar]
- 4.Appel M, Robson DS. A microneutralization test for canine distemper virus. Am J Vet Res. 1973;34(11):1459–1463. [PubMed] [Google Scholar]
- 5.Appel MJ, Yates RA, Foley GL, Bernstein JJ, Santinelli S, Spelman LH, Miller LD, Arp LH, Anderson M, Barr M, Summers BA. Canine distemper epizootic in lions, tigers, and leopards in North America. J Vet Diagn Invest. 1994;6(3):277–288. doi: 10.1177/104063879400600301. [DOI] [PubMed] [Google Scholar]
- 6.Appel MJ. Pathogenesis of canine distemper. Am J Vet Res. 1969;30:1167–1182. [PubMed] [Google Scholar]
- 7.Appel MJG, Gillespie JH. Canine Distemper Virus. In: Canine Distemper Virus. 1972
- 8.Appel MJG, Shek WR, Shesberadaran H, Norrby E. Measles virus and inactivated canine distemper virus induce incomplete immunity to canine distemper. Arch Virol. 1984;82(1):73–82. doi: 10.1007/BF01309369. [DOI] [PubMed] [Google Scholar]
- 9.Appel MJ, Pearce-Kelling S, Summers BA. Dog lymphocyte cultures facilitate the isolation and growth of virulent canine distemper virus. J Vet Diagn Invest. 1992;4(3):258–263. doi: 10.1177/104063879200400306. [DOI] [PubMed] [Google Scholar]
- 10.Ashmi JM, Thangavelu A, Senthilkumar TMA, Manimaran K. Molecular characterization of canine distemper virus from Tamil Nadu, India. Indian J Anim Sci. 2017;87:1062–1067. [Google Scholar]
- 11.Barben G, Stettler M, Jaggy A, Vandevelde M, Zurbriggen A. Detection of IgM antibodies against a recombinant nucleocapsid protein of canine distemper virus in dog sera using a dot-blot assay. J Vet Med. 1999;46(2):115–122. doi: 10.1046/j.1439-0442.1999.00198.x. [DOI] [PubMed] [Google Scholar]
- 12.Bernard SL, Shen DT, Gorham JR. Antigen requirements and specificity of enzyme-linked immunosorbent assay for detection of canine IgG against canine distemper viral antigens. Am J Vet Res. 1982;43(12):2266–2269. [PubMed] [Google Scholar]
- 13.Bhatt M, Rajak KK, Chakravarti S, Yadav AK, Kumar A, Gupta V, Chander V, Mathesh K, Chandramohan S, Sharma AK, Mahendran K. Phylogenetic analysis of haemagglutinin gene deciphering a new genetically distinct lineage of canine distemper virus circulating among domestic dogs in India. Transbound Emerg Dis. 2019;66(3):1252–1267. doi: 10.1111/tbed.13142. [DOI] [PubMed] [Google Scholar]
- 14.Bi Z, Wang Y, Pan Q, Xia X, Xu L. Development of CDV-specific monoclonal antibodies for differentiation of variable epitopes of nucleocapsid protein. Vet Microbial. 2017;211:84–91. doi: 10.1016/j.vetmic.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 15.Bi Z, Xia X, Wang Y, Mei Y. Development and characterization of neutralizing monoclonal antibodies against canine distemper virus hemagglutinin protein. Microbiol Immunol. 2015;59(4):202–208. doi: 10.1111/1348-0421.12238. [DOI] [PubMed] [Google Scholar]
- 16.Bieringer M, Han JW, Kendl S, Khosravi M, Plattet P, Schneider-Schaulies J. Experimental adaptation of wild-type canine distemper virus (CDV) to the human entry receptor CD150. PLoS ONE. 2013;8(3):e57488. doi: 10.1371/journal.pone.0057488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blancou J. Dog distemper: imported into Europe from South America? His Med Vet. 2004;29(2):35–41. [PubMed] [Google Scholar]
- 18.Blixenkrone-Moller M, Pedersen IR, Appel MJ, Griot C. Detection of IgM antibodies against canine distemper virus in dog and mink sera employing enzyme-linked immunosorbent assay (ELISA) J Vet Diagn Invest. 1991;3(1):3–9. doi: 10.1177/104063879100300102. [DOI] [PubMed] [Google Scholar]
- 19.Bolt G, Jensen TD, Gottschalck E, Arctander P, Appel MJ, Buckland R, Blixenkrone-M M. Genetic diversity of the attachment (H) protein gene of current field isolates of canine distemper virus. J Gen Virol. 1997;78(2):367–372. doi: 10.1099/0022-1317-78-2-367. [DOI] [PubMed] [Google Scholar]
- 20.Bush RM, Roberts MS. Distemper in captive red pandas. International Zoo Yearbook. 1977.
- 21.Chamberlain RW, Wamwayi HM, Hockley E, Shaila MS, Goatley L, Knowles NJ, Barrett T. Evidence for different lineages of rinderpest virus reflecting their geographic isolation. J Gen Virol. 1993;74(12):2775–2780. doi: 10.1099/0022-1317-74-12-2775. [DOI] [PubMed] [Google Scholar]
- 22.Chappuis G. Control of canine distemper. Vet Microbiol. 1995;44(2–4):351–358. doi: 10.1016/0378-1135(95)00028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhary V. Threats of disease spillover from domestic dogs to wild carnivores in the Kanha tiger reserve, India. 2016.
- 24.Cho HS, Park NY. Detection of Canine distemper virus in blood samples by reverse transcription loop-mediated isothermal amplification. J Vet Med. 2005;52(9):410–413. doi: 10.1111/j.1439-0450.2005.00886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Confer AW, Kahn DE, Koestner A, Krakowka S. Biological properties of a canine distemper virus isolate associated with demyelinating encephalomyelitis. Infect Immun. 1975;11(4):835–844. doi: 10.1128/iai.11.4.835-844.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cottrell WO, Keel MK, Brooks JW, Mead DG, Phillips JE. First report of clinical disease associated with canine distemper virus infection in a wild black bear (Ursus americana) J Wildl Dis. 2013;49(4):1024–1027. doi: 10.7589/2013-02-027. [DOI] [PubMed] [Google Scholar]
- 27.da Costa VG, Saivish MV, Rodrigues RL, de Lima Silva RF, Moreli ML, Krüger RH. Molecular and serological surveys of canine distemper virus: a meta-analysis of cross-sectional studies. PLoS ONE. 2019;14(5):e0217594. doi: 10.1371/journal.pone.0217594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.da FontouraBudaszewski R, Streck AF, Weber MN, Siqueira FM, Guedes RLM, Canal CW. Influence of vaccine strains on the evolution of canine distemper virus. Infect Gen Evol. 2016;41:262–269. doi: 10.1016/j.meegid.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 29.Dalldorf G, Douglass M, Robinson HE. Canine distemper in the rhesus monkey (Macaca mulatta) J Exp Med. 1938;67(2):323. doi: 10.1084/jem.67.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Debesa Belizario Granjeiro M, Lima Kavasaki M, Morgado TO, Avelino Dandolini Pavelegini L, Alvesde Barros M, Fontana C, de Assis Bianchini M, de Oliveira Souza A, Goncalves Lima Oliveira Santos AR, Lunardi M, Jorge Mendonça A. First report of a canine morbillivirus infection in a giant anteater (Myrmecophaga tridactyla) in Brazil. Vet Med Sci. 2020;6(3):606–611. doi: 10.1002/vms3.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong XY, Li WH, Zhu JL, Liu WJ, Zhao MQ, Luo YW, Chen JD. Detection and differentiation of wild-type and vaccine strains of canine distemper virus by a duplex reverse transcription polymerase chain reaction. Iranian J Vet Res. 2015;16(2):172. [PMC free article] [PubMed] [Google Scholar]
- 32.Dunkin GW, Laidlaw pp. Studies in dog distemper. II. Experimental distemper in the dog. J Compo Path. 1926;39:213–221. doi: 10.1016/S0368-1742(26)80021-9. [DOI] [Google Scholar]
- 33.Duque-Valencia J, Forero-Muñoz NR, Díaz FJ, Martins E, Barato P, Ruiz-Saenz J. Phylogenetic evidence of the intercontinental circulation of a Canine distemper virus lineage in the Americas. Sci rep. 2019;9:1–15. doi: 10.1038/s41598-019-52345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duque-Valencia J, Sarute N, Olarte-Castillo XA, Ruíz-Sáenz J. Evolution and interspecies transmission of canine distemper virus—An outlook of the diverse evolutionary landscapes of a multi-host virus. Viruses. 2019;11(7):582. doi: 10.3390/v11070582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elia G, Decaro N, Martella V, Cirone F, Lucente MS, Lorusso E, Trani LD, Buonavoglia C. Detection of canine distemper virus in dogs by real-time RT-PCR. J Virol Met. 2006;136(1–2):171–176. doi: 10.1016/j.jviromet.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Feng N, Yu Y, Wang T, Wilker P, Wang J, Li Y, Sun Z, Gao Y, Xia X. Fatal canine distemper virus infection of giant pandas in China. Sci Rep. 2016;6(1):1–7. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer CD, Gräf T, Ikuta N, Lehmann FK, Passos DT, Makiejczuk A, Silveira MA, Jr, Fonseca AS, Canal CW, Lunge VR. Phylogenetic analysis of canine distemper virus in South America clade 1 reveals unique molecular signatures of the local epidemic. Infect Gen Evol. 2016;41:135–141. doi: 10.1016/j.meegid.2016.03.029. [DOI] [PubMed] [Google Scholar]
- 38.Fischer CDB, Ikuta N, Canal CW, Makiejczuk A, da Costa AM, Cardoso C, Lehmann FK, Fonseca ASK, Lunge VR. Detection and differentiation of field and vaccine strains of canine distemper virus using reverse transcription followed by nested real time PCR (RT-nqPCR) and RFLP analysis. J Virol Met. 2013;194(1–2):39–45. doi: 10.1016/j.jviromet.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freitas LA, Leme RA, Saporiti V, Alfieri AA, Alfieri AF. Molecular analysis of the full-length F gene of Brazilian strains of canine distemper virus shows lineage co-circulation and variability between field and vaccine strains. Virus Res. 2019;264:8–15. doi: 10.1016/j.virusres.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Frisk AL, König M, Moritz A, Baumgärtner W. Detection of canine distemper virus nucleoprotein RNA by reverse transcription-PCR using serum, whole blood, and cerebrospinal fluid from dogs with distemper. J Clin Microbiol. 1999;37(11):3634–3643. doi: 10.1128/JCM.37.11.3634-3643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gordon CH, Banyard AC, Hussein A, Laurenson MK, Malcolm JR, Marino J, Regassa F, Stewart AME, Fooks AR, Sillero-Zubiri C. Canine distemper in endangered Ethiopian wolves. Emerg Infect Dis. 2015;21(5):824. doi: 10.3201/eid2105.141920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greene CE, Appel MJ. Canine distemper. In: Clinical microbiology and infectious disease of the dog and cat. Edited by Greene, C. E. Saunders, Elsevier. 1984.
- 43.Greene CE. Infectious diseases of the dog and cat. 4. Philadelphia, Amsterdam: WB Saunders\Elsevier Science; 2006. [Google Scholar]
- 44.Guo W, Evermann JF, Foreyt WJ, Knowlton FF, Windberg LA. Canine distemper virus in coyotes: a serologic survey. J Am Vet Med Assoc. 1986;189(9):1099–1100. [PubMed] [Google Scholar]
- 45.Haas L, Hofer H, East M, Wohlsein P, Liess B, Barrett T. Canine distemper virus infection spotted hyaenas in Serengeti. Vet Microbiol. 1996;49:147–152. doi: 10.1016/0378-1135(95)00180-8. [DOI] [PubMed] [Google Scholar]
- 46.Haas L, Martens W, Greiser-Wilke I, Mamaev L, Butina T, Maack D, Barrett T. Analysis of the haemagglutinin gene of current wild-type canine distemper virus isolates from Germany. Virus Res. 1997;48(2):165–171. doi: 10.1016/S0168-1702(97)01449-4. [DOI] [PubMed] [Google Scholar]
- 47.Headley SA, Oliveira TE, Pereira AH, Moreira JR, Michelazzo MM, Pires BG, Marutani VHB, Xavier AA, Di Santis GW, Garcia JL, Alfieri AA. Canine morbillivirus (canine distemper virus) with concomitant canine adenovirus, canine parvovirus-2, and Neospora caninum in puppies: a retrospective immunohistochemical study. Sci Rep. 2018;8(1):1–16. doi: 10.1038/s41598-018-31540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirayama N, Senda M, Nakashima N, Takagi M, Sugiyama M, Yoshikawa Y, Yamanouchi K. Protective effects of monoclonal antibodies against lethal canine distemper virus infection in mice. J Gen Virol. 1991;72(11):2827–2830. doi: 10.1099/0022-1317-72-11-2827. [DOI] [PubMed] [Google Scholar]
- 49.“International Committee on Taxonomy of Viruses (ICTV): https://ictv.global/taxonomy/”
- 50.Jensen TH, Nielsen L, Aasted B, Pertoldi C, Blixenkrone-Moller M. Canine distemper virus DNA vaccination of mink can overcome interference by maternal antibodies. Vaccine. 2015;33(11):1375–1381. doi: 10.1016/j.vaccine.2015.01.029. [DOI] [PubMed] [Google Scholar]
- 51.Jozwik A, Frymus T. Comparison of the immunofluorescence assay with RT-PCR and nested PCR in the diagnosis of canine distemper. Vet Res Commun. 2005;29(4):347–359. doi: 10.1023/B:VERC.0000048528.76429.8b. [DOI] [PubMed] [Google Scholar]
- 52.Kadam RG, Karikalan M, Mohan C, Varshney R, Singh R, War ZA, Pawde AM, Ghosh M, Beena V, Singh KP, Sharma AK. Molecular and pathological screening of canine distemper virus in Asiatic tigers, lions, leopards, snow leopards, clouded leopards, leopard cats, jungle cats, civet cats, fishing cat, and jaguar of different states. India Infect Gene Evol. 2022;98:105211. doi: 10.1016/j.meegid.2022.105211. [DOI] [PubMed] [Google Scholar]
- 53.Kai C, Ochikubo F, Okita M, Linuma T, Mikami T, Kobnne F, Yamanouchi K. Use of B95a cells for isolation of canine distemper virus from clinical cases. J Vet Med Sci. 1993;55(6):1067–1070. doi: 10.1292/jvms.55.1067. [DOI] [PubMed] [Google Scholar]
- 54.Kapil S, Neel T. Canine distemper virus antigen detection in external epithelia of recently vaccinated, sick dogs by fluorescence microscopy is a valuable prognostic indicator. J Clin Microbiol. 2015;53(2):687–691. doi: 10.1128/JCM.02741-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kapil S, Yeary TJ. Canine distemper spillover in domestic dogs from urban wildlife. Vet Clin Small Anim. 2011;41(6):1069–1086. doi: 10.1016/j.cvsm.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karki M, Rajak KK, Singh P, Fayaz A, Kiran Yadav AK, Bhatt M, Rai V, Einstein C, Singh R. P. Optimization of competitive lateral flow assay for detection of canine distemper virus antibody. The Pharma innovation J. 2022
- 57.Ke GM, Ho CH, Chiang MJ, Sanno-Duanda B, Chung CS, Lin MY, Shi YY, Yang MH, Tyan YC, Liao PC, Chu PY. Phylodynamic analysis of the canine distemper virus hemagglutinin gene. BMC Vet Res. 2015;11(1):1–15. doi: 10.1186/s12917-015-0491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kennedy JM, Earle JA, Omar S, Abdullah HA, Nielsen O, Roelke-Parker ME, Cosby SL. Canine and phocine distemper viruses: Global spread and genetic basis of jumping species barriers. Viruses. 2019;11(10):944. doi: 10.3390/v11100944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim HH, Yang DK, Seo BH, Cho IS. Serosurvey of rabies virus, canine distemper virus, parvovirus, and influenza virus in military working dogs in Korea. J Vet Med Sci. 2018;80:18–0012. doi: 10.1292/jvms.18-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim YH, Cho KW, Youn HY, Yoo HS, Han HR. Detection of canine distemper virus (CDV) through one step RT-PCR combined with nested PCR. J Vet Sci. 2001;2(1):59–63. doi: 10.4142/jvs.2001.2.1.59. [DOI] [PubMed] [Google Scholar]
- 61.Kobune F, Sakata H, Sugiyama M, Sugiura A. B95a, a marmoset lymphoblastoid cell line, as a sensitive host for rinderpest virus. J Gen Virol. 1991;72(3):687–692. doi: 10.1099/0022-1317-72-3-687. [DOI] [PubMed] [Google Scholar]
- 62.Kodi H, Putty K, Ganji VK, Bhagyalakshmi B, Reddy YN, Satish K, Prakash MG. H gene-based molecular characterization of field isolates of canine distemper virus from cases of canine gastroenteritis. Indian J Ani Res. 2021;55(5):561–567. [Google Scholar]
- 63.Kovamees J, Blixenkrone-Moller M, Norrby E. The nucleotide and predicted amino acid sequence of the attachment protein of canine distemper virus. Virus res. 1991;19(2–3):223–233. doi: 10.1016/0168-1702(91)90048-Z. [DOI] [PubMed] [Google Scholar]
- 64.Latha D, Geetha M, Ramadass P, Narayanan RB. Development of recombinant nucleocapsid protein based IgM-ELISA for the early detection of distemper infection in dogs. Vet Immunol Immunop. 2007;119(3–4):278–286. doi: 10.1016/j.vetimm.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 65.Latha D, Geetha M, Ramadass P, Narayanan RB. Evaluation of ELISA based on the conserved and functional middle region of nucleocapsid protein to detect distemper infection in dogs. Vet Microbiol. 2007;120(3–4):251–260. doi: 10.1016/j.vetmic.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 66.Latha D, Srinivasan SR, Thirunavukkarasu PS, Gunaselan L, Ramadass P, Narayanan R B. Assessment of canine distemper virus infection in vaccinated and unvaccinated dogs. Indian J Biotechnol. 2007c
- 67.Lednicky JA, Meehan TP, Kinsel MJ, Dubach J, Hungerford LL, Sarich NA, Witecki KE, Braid MD, Pedrak C, Houde CM. Effective primary isolation of wild-type canine distemper virus in MDCK, MV1 Lu and Vero cells without nucleotide sequence changes within the entire haemagglutinin protein gene and in subgenomic sections of the fusion and phospho protein genes. J Virol Met. 2004;118(2):147–157. doi: 10.1016/j.jviromet.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 68.Li S, Yi L, Cao Z, Cheng Y, Tong M, Wang J, Lin P, Cheng S. Identification of linear B-cell epitopes on the phosphoprotein of canine distemper virus using four monoclonal antibodies. Virus Res. 2018;257:52–56. doi: 10.1016/j.virusres.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 69.Li Z, Zhang Y, Wang H, Jin J, Li W. Sandwich-dot enzyme-linked immunosorbent assay for the detection of canine distemper virus. Can J Vet Res. 2013;77(4):303–308. [PMC free article] [PubMed] [Google Scholar]
- 70.Liu D, Liu F, Guo D, Hu X, Li Z, Li Z, Ma J, Liu C. One–step triplex PCR/RT–PCR to detect canine distemper virus, canine parvovirus, and canine kobuvirus. J Vet Med Sci. 2018;81:17–0442. doi: 10.1292/jvms.17-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu DF, Liu CG, Tian J, Jiang YT, Zhang XZ, Chai HL, Yang TK, Yin XC, Zhang HY, Liu M, Hua YP. Establishment of reverse transcription loop-mediated isothermal amplification for rapid detection and differentiation of canine distemper virus infected and vaccinated animals. Infect Gen Evol. 2015;32:102–106. doi: 10.1016/j.meegid.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loots AK, Mokgokong PS, Mitchell E, Venter EH, Kotze A, Dalton DL. Phylogenetic analysis of canine distemper virus in South African wildlife. PLoS ONE. 2018;13(7):e0199993. doi: 10.1371/journal.pone.0199993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lunardi M, Darold GM, Amude AM, Headley SA, Sonne L, Yamauchi KCI, Boabaid FM, Alfieri AF, Alfieri AA. Canine distemper virus active infection in order Pilosa, family Myrmecophagidae, species Tamandua tetradactyla. Vet Microbiol. 2018;220:7–11. doi: 10.1016/j.vetmic.2018.04.030. [DOI] [PubMed] [Google Scholar]
- 74.Martella V, Blixenkrone-Møller M, Elia G, Lucente MS, Cirone F, Decaro N, Nielsen L, Banyai K, Carmichael LE, Buonavoglia C. Lights and shades on an historical vaccine canine distemper virus, the Rockborn strain. Vaccine. 2011;29(6):1222–1227. doi: 10.1016/j.vaccine.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 75.Martella V, Elia G, Buonavoglia C. Canine distemper virus. Vet Clin N Am Small AnimPract. 2008;38(4):787–797. doi: 10.1016/j.cvsm.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 76.Mateo M, Navaratnarajah CK, Willenbring RC, Maroun JW, Iankov I, Lopez M, Sinn PL, Cattaneo R. Different roles of the three loops forming the adhesive interface of nectin-4 in measles virus binding and cell entry, nectin-4 homodimerization, and heterodimerization with nectin-1. J Virol. 2014;88:14161–14171. doi: 10.1128/JVI.02379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mazzu-Nascimento T, Donofrio FC, Bianchi BC, Travensolo RDF, Souza Junior JB, Moraes DA, Varanda LC, Carrilho E. Based Microfluidics Immunoassay for Detection of Canine Distemper Virus. Braz Arch Biol Tech. 2017; 60.
- 78.Mochizuki M, Hashimoto M, Hagiwara S, Yoshida Y, Ishiguro S. Genotypes of canine distemper virus determined by analysis of the hemagglutinin genes of recent isolates from dogs in Japan. J Clin Microbiol. 1999;37(9):2936–2942. doi: 10.1128/JCM.37.9.2936-2942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mourya DT, Yadav PD, Mohandas S, Kadiwar RF, Vala MK, Saxena AK, Shete-Aich A, Gupta N, Purushothama P, Sahay RR, Gangakhedkar RR. Canine distemper virus in asiatic Lions of Gujarat State, India. Emerg Infect Dis. 2019;25(11):2128. doi: 10.3201/eid2511.190120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Munson L, Marker L, Dubovi E, Spencer JA, Evermann JF, O'Brien SJ. Serosurvey of viral infections in free-ranging Namibian cheetahs (Acinonyx jubatus) J Wildl Dis. 2004;40(1):23–31. doi: 10.7589/0090-3558-40.1.23. [DOI] [PubMed] [Google Scholar]
- 81.Murphy FA, Gibbs EPJ, Horzinek MC, Studdert MJ. Vet. Virol. 3rded. San Diego, California Academic Press. 1999.
- 82.Nayak RM, Jadav KK, Rajput N, Gupta S, Rokde A, Singh KP. Surveillance of Major Canine Pathogens in Feral Dogs and Big Cats at the Domestic-Wildlife Interface in Panna Tiger Reserve. India J Anim Res. 2020;10(2):303–308. [Google Scholar]
- 83.Needle DB, Burnell VC, Forzán MJ, Dubovi EJ, Schuler KL, Bernier C, Hollingshead NA, Ellis JC, Stevens BA, Tate P, Anis E. Infection of eight mesocarnivores in New Hampshire and Vermont with a distinct clade of canine distemper virus in 2016–2017. J Vet Diagn Invest. 2019;31(4):562–567. doi: 10.1177/1040638719847510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Noon KF, Rogul M, Binn LN, Keefe TJ, Marchwicki RH, Appel MJ. Enzyme-linked immunosorbent assay for evaluation of antibody to canine distemper virus. Am J Vet Res. 1980;41(4):605–609. [PubMed] [Google Scholar]
- 85.Ohishi K, Suzuki R, Maeda T, Tsuda M, Abe E, Yoshida T, Endo Y, Okamura M, Nagamine T, Yamamoto H, Maruyama T. Recent host range expansion of canine distemper virus and variation in its receptor, the signaling lymphocyte activation molecule, in carnivores. J Wildl Dis. 2014;50(3):596–606. doi: 10.7589/2013-09-228. [DOI] [PubMed] [Google Scholar]
- 86.Oni O, Wajjwalku W, Boodde O, Chumsing W. Canine distemper virus antibodies in the Asian elephant (Elaphas maximus) Vet Rec. 2006;159(13):420. doi: 10.1136/vr.159.13.420. [DOI] [PubMed] [Google Scholar]
- 87.Origgi FC, Plattet P, Sattler U, Robert N, Casaubon J, Mavrot F, Pewsner M, Wu N, Giovannini S, Oevermann A, Stoffel MH. Emergence of canine distemper virus strains with modified molecular signature and enhanced neuronal tropism leading to high mortality in wild carnivores. Vet Pathol. 2012;49(6):913–929. doi: 10.1177/0300985812436743. [DOI] [PubMed] [Google Scholar]
- 88.Orvell C, Norrby E. Immunological relationships between homologous structural polypeptides of measles and canine distemper virus. J Gen Virol. 1980;50(2):231–245. doi: 10.1099/0022-1317-50-2-231. [DOI] [PubMed] [Google Scholar]
- 89.Orvell C, Sheshberadaran H, Norrby E. Preparation and characterization of monoclonal antibodies directed against four structural components of canine distemper virus. J Gen Virol. 1985;66(3):443–456. doi: 10.1099/0022-1317-66-3-443. [DOI] [PubMed] [Google Scholar]
- 90.Oyedele OI, Oluwayelu DO, Cadmus SIB, Odemuyiwa SO, Adu FD. Protective levels of canine distemper virus antibody in an urban dog population using plaque reduction neutralization test. Onderstepoort J Vet Res. 2004;71(3):227–230. doi: 10.4102/ojvr.v71i3.264. [DOI] [PubMed] [Google Scholar]
- 91.Pan Z, Liu J, Ma J, Jin Q, Yao H, Osterrieder N. The recombinant EHV-1 vector producing CDV hemagglutinin as potential vaccine against canine distemper. MicrobPathog. 2017;111:388–394. doi: 10.1016/j.micpath.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 92.Peserico A, Marcacci M, Malatesta D, Di Domenico M, Pratelli A, Mangone I, D’Alterio N, Pizzurro F, Cirone F, Zaccaria G, Cammà C. Diagnosis and characterization of canine distemper virus through sequencing by MinION nanopore technology. Sci Rep. 2019;9(1):1–9. doi: 10.1038/s41598-018-37497-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Piat BL. Susceptibility of young lions to dog distemper. Bull. Serv. Elev. In-dustr. anim. AOF, 3. 1950.
- 94.Piewbang C, Radtanakatikanon A, Puenpa J, Poovorawan Y, Techangamsuwan S. Genetic and evolutionary analysis of a new Asia-4 lineage and naturally recombinant canine distemper virus strains from Thailand. Sci Rep. 2019;9(1):1–8. doi: 10.1038/s41598-018-37186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pope JP, Miller DL, Riley MC, Anis E, Wilkes RP. Characterization of a novel canine distemper virus causing disease in wildlife. J Vet Diagn Invest. 2016;28(5):506–513. doi: 10.1177/1040638716656025. [DOI] [PubMed] [Google Scholar]
- 96.Potgieter LN, Ajidagba PA. Quantitation of canine distemper virus and antibodies by enzyme-linked immunosorbent assays using protein A and monoclonal antibody capture. J Vet Diagn Invest. 1989;1(2):110–115. doi: 10.1177/104063878900100203. [DOI] [PubMed] [Google Scholar]
- 97.Qiu W, Zheng Y, Zhang S, Fan Q, Liu H, Zhang F, Wang W, Liao G, Hu R. Canine distemper outbreak in rhesus monkeys, China. Emerg Infect Dis. 2011;17(8):1541. doi: 10.3201/eid1708.101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qiu X, Mainka SA (1993) Review of mortality of the giant panda (Ailuropoda melanoleuca). J Zoo Wildl Med.; 425–429.
- 99.Quigley KS, Evermann JF, Leathers CW, Armstrong DL, Goodrich J, Duncan NM, Miquelle DG. Morbillivirus infection in a wild Siberian tiger in the Russian Far East. J Wildl Dis. 2010;46(4):1252–1256. doi: 10.7589/0090-3558-46.4.1252. [DOI] [PubMed] [Google Scholar]
- 100.Quintero-Gil C, Rendon-Marin S, Martinez-Gutierrez M, Ruiz-Saenz J. Origin of canine distemper virus: consolidating evidence to understand potential zoonoses. Front Microbial. 2019;10:1982. doi: 10.3389/fmicb.2019.01982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rendon-Marin S, Budaszewski RF, Canal CW, Ruiz-Saenz J. Tropism and molecular pathogenesis of canine distemper virus. Virol J. 2019;16(1):1–15. doi: 10.1186/s12985-019-1136-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Riley MC, Wilkes RP. Sequencing of emerging canine distemper virus strain reveals new distinct genetic lineage in the United States associated with disease in wildlife and domestic canine populations. Virol J. 2015;12(1):1–10. doi: 10.1186/s12985-015-0445-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rockborn G. Canine distemper virus in tissue culture. Archiv für die gesamteVirusforschung. 1958;8(4):485–492. doi: 10.1007/BF01243067. [DOI] [Google Scholar]
- 104.Sadler RA, Ramsay E, McAloose D, Rush R, Wilkes RP. Evaluation of two canine distemper virus vaccines in captive tigers (Panthera tigris) J Zoo Wildlife Med. 2016;47(2):558–563. doi: 10.1638/2015-0223.1. [DOI] [PubMed] [Google Scholar]
- 105.Sakai K, Nagata N, Ami Y, Seki F, Suzaki Y, Iwata-Yoshikawa N, Suzuki T, Fukushi S, Mizutani T, Yoshikawa T, Morikawa S. Lethal canine distemper virus outbreak in cynomolgus monkeys in Japan in 2008. J Virol. 2013;87(2):1105–1114. doi: 10.1128/JVI.02419-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Santhamani R, Singh RP, Njeumi F. Peste des petits ruminants diagnosis and diagnostic tools at a glance: perspectives on global control and eradication. Arch Virol. 2016;161(11):2953–2967. doi: 10.1007/s00705-016-3009-2. [DOI] [PubMed] [Google Scholar]
- 107.Sarute N, Delgado MV, Carrau L, Benech A, Francia L, Pérez R, Panzera Y. First genome sequence of a canine distemper virus strain from South America. Genome Announc. 2014;2(5):e01009–e1014. doi: 10.1128/genomeA.01009-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sawatsky B, Cattaneo R, von Messling V. Canine distemper virus spread and transmission to naive ferrets: selective pressure on signaling lymphocyte activation molecule-dependent entry. J Virol. 2018;92(15):e00669–e718. doi: 10.1128/JVI.00669-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scagliarini A, dal Pozzo F, Gallina L, Vaccari F, Morganti L. Taq-Man based real time PCR for the quantification of canine distemper virus. Vet Res Commun. 2007;31:261. doi: 10.1007/s11259-007-0020-9. [DOI] [PubMed] [Google Scholar]
- 110.Sehata G, Sato H, Ito T, Imaizumi Y, Noro T, Oishi E. Use of quantitative real-time RT-PCR to investigate the correlation between viremia and viral shedding of canine distemper virus, and infection outcomes in experimentally infected dogs. J Vet Med Sci. 2015;77(7):851–855. doi: 10.1292/jvms.14-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Seimon TA, Miquelle DG, Chang TY. Canine distemper virus: an emerging disease in wild endangered. MBio. 2013;4(4):e00410–e413. doi: 10.1128/mBio.00410-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Seki F, Ono N, Yamaguchi R, Yanagi Y. Efficient isolation of wild strains of canine distemper virus in Vero cells expressing canine SLAM (CD150) and their adaptability to marmoset B95a cells. J Virol. 2003;77(18):9943–9950. doi: 10.1128/JVI.77.18.9943-9950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shaila MS, Shamaki D, Forsyth MA, Diallo A, Goatley L, Kitching RP, Barrett T. Geographic distribution and epidemiology of peste des petits ruminants viruses. Virus Res. 1996;43(2):149–153. doi: 10.1016/0168-1702(96)01312-3. [DOI] [PubMed] [Google Scholar]
- 114.Sheshberadaran H, Norrby E, McCullough KC, Carpenter WC, Örvell C. The antigenic relationship between measles, canine distemper and rinderpest viruses studied with monoclonal antibodies. J Gen Virol. 1986;67(7):1381–1392. doi: 10.1099/0022-1317-67-7-1381. [DOI] [PubMed] [Google Scholar]
- 115.Shi P, Cao Z, Cheng Y, Cheng S, Yi L. Identification of Linear B-Cell Epitopes on Hemagglutinin Protein of Canine Distemper Virus Using Two Monoclonal Antibodies. Front Vet Sci. 2020;7:47. doi: 10.3389/fvets.2020.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shin YJ, Cho KO, Cho HS, Kang SK, Kim HJ, Kim YH, Park HS, Park NY. Comparison of one-step RT-PCR and a nested PCR for the detection of canine distemper virus in clinical samples. Aust Vet J. 2004;82(1–2):83–86. doi: 10.1111/j.1751-0813.2004.tb14651.x. [DOI] [PubMed] [Google Scholar]
- 117.Si W, Zhou S, Wang Z, Cui SJ. A multiplex reverse transcription-nested polymerase chain reaction for detection and differentiation of wild-type and vaccine strains of canine distemper virus. Virol J. 2010;7(1):1–6. doi: 10.1186/1743-422X-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sidhu MS, Husar W, Cook SD, Dowling PC, Udem SA. Canine Distemper terminal and intergenic non-protein coding nucleotide sequences: completion of the entire CDV genome sequence. Virology. 1993;193(1):66–72. doi: 10.1006/viro.1993.1103. [DOI] [PubMed] [Google Scholar]
- 119.Sindhu N, Borah J, Shah S, Rajput N, Jadav KK. Is canine distemper virus (CDV) a lurking threat to large carnivores? A case study from Ranthambhore landscape in Rajasthan. India JoTT. 2019;11(9):14220–14223. [Google Scholar]
- 120.Singh RP, Sreenivasa BP, Dhar P, Bandyopadhyay SK. A sandwich-ELISA for the diagnosis of Peste des petits ruminants (PPR) infection in small ruminants using anti-nucleocapsid protein monoclonal antibody. Arch Virol. 2004;149(11):2155–2170. doi: 10.1007/s00705-004-0366-z. [DOI] [PubMed] [Google Scholar]
- 121.Singh RP, Sreenivasa BP, Dhar P, Shah LC, Bandyopadhyay SK. Development of a monoclonal antibody based competitive-ELISA for detection and titration of antibodies to peste des petits ruminants (PPR) virus. Vet Microbial. 2004;98(1):3–15. doi: 10.1016/j.vetmic.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 122.Slanetz CA, Smetana H. An epizootic disease of ferrets caused by a filterable virus. J Exp Med. 1937;66(6):653. doi: 10.1084/jem.66.6.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Soma T, Ishii H, Hara M, Ohe K, Hagimori I, Ishikawa Y, Taneno A. Detection of canine distemper virus antigen in canine serum and its application to diagnosis. Vet Rec. 2003;153(16):499–501. doi: 10.1136/vr.153.16.499. [DOI] [PubMed] [Google Scholar]
- 124.Soma T, Ishii H, Hara M, Yamamoto S, Yoshida T, Kinoshita T, Nomura K. Comparison of immunoperoxidase plaque staining and neutralizing tests for canine distemper virus. Vet Res Commun. 2001;25(4):311–325. doi: 10.1023/A:1010682726245. [DOI] [PubMed] [Google Scholar]
- 125.Sreenivasa BP, Singh RP, Mondal B, Dhar P, Bandyopadhyay SK. Marmoset B95a cells: a sensitive system for cultivation of peste des petits ruminants (PPR) virus. Vet Res Commun. 2006;30(1):103–108. doi: 10.1007/s11259-005-3200-5. [DOI] [PubMed] [Google Scholar]
- 126.Summers BA, Greisen HA, Appel MJG. Canine distemper encephalomyelitis: variation with virus strain. J Comp Pathol. 1984;94(1):65–75. doi: 10.1016/0021-9975(84)90009-4. [DOI] [PubMed] [Google Scholar]
- 127.Sun Z, Li A, Ye H, Shi Y, Hu Z, Zeng L. Natural infection with canine distemper virus in hand-feeding rhesus monkeys in China. Vet Microbiol. 2010;141(3–4):374–378. doi: 10.1016/j.vetmic.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 128.Swati D, D., Uppal, S. K., Verma, R. Isolation and phylogenetic characterization of Canine distemper virus from India. Virus Dis. 2015;26(3):133–140. doi: 10.1007/s13337-015-0256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Taguchi M, Namikawa K, Maruo T, Orito K, Lynch J, Sahara H. Antibody titers for canine parvovirus type-2, canine distemper virus, and canine adenovirus type-1 in adult household dogs. Can Vet J. 2011;52(9):983. [PMC free article] [PubMed] [Google Scholar]
- 130.Takeda M, Seki F, Yamamoto Y, Nao N, Tokiwa H. Animal morbilliviruses and their cross-species transmission potential. Curr Opin Virol. 2020;41:38–45. doi: 10.1016/j.coviro.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 131.Takenaka A, Yoneda M, Seki T, Uema M, Kooriyama T, Nishi T, Fujita K, Miura R, Tsukiyama-Kohara K, Sato H, Kai C. Characterization of two recent Japanese field isolates of canine distemper virus and examination of the avirulent strain utility as an attenuated vaccine. Vet Microbial. 2014;174(3–4):372–381. doi: 10.1016/j.vetmic.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 132.Taques II, Morgado TO, Braga ÍA, Paz RC, Corrêa SH, Fritzen JT, Alfieri AA, Aguiar DM. Antibodies against canine distemper virus, parvovirus and Ehrlichia spp. in wild captive carnivores in midwestern Brazil. Pesq Vet Bras. 2018;38:1681–1684. doi: 10.1590/1678-5150-pvb-5333. [DOI] [Google Scholar]
- 133.Taylor MJ, Godfrey E, Baczko K, Ter Meulen V, Wild TF, Rima BK. Identification of several different lineages of measles virus. J Gen Virol. 1991;72(1):83–88. doi: 10.1099/0022-1317-72-1-83. [DOI] [PubMed] [Google Scholar]
- 134.Uhl EW, Kelderhouse C, Buikstra J, Blick JP, Bolon B, Hogan RJ. New world origin of canine distemper: interdisciplinary insights. Int J Paleopathol. 2019;72(1):83–88. doi: 10.1016/j.ijpp.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 135.Vandevelde M, Kristensen B, Braund KG, Greene CE, Swango LJ, Hoerlein BF. Chronic canine distemper virus encephalitis in mature dogs. Vet Pathol. 1980;17(1):17–29. doi: 10.1177/030098588001700102. [DOI] [PubMed] [Google Scholar]
- 136.Vergara-Wilson V, Hidalgo-Hermoso E, Sanchez CR, Abarca MJ, Navarro C, Celis-Diez S, Soto-Guerrero P, Diaz-Ayala N, Zordan M, Cifuentes-Ramos F, Cabello-Stom J. Canine distemper outbreak by natural infection in a group of vaccinated maned wolves in captivity. Pathog. 2021;10(1):51. doi: 10.3390/pathogens10010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Von Messling V, Harder TC, Moennig V, Rautenberg P, Nolte I, Haas L. Rapid and sensitive detection of immunoglobulin M (IgM) and IgG antibodies against canine distemper virus by a new recombinant nucleocapsid protein-based enzyme-linked immunosorbent assay. J Clin Microbiol. 1999;37(4):1049–1056. doi: 10.1128/JCM.37.4.1049-1056.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Von Messling V, Milosevic D, Devaux P, Cattaneo R. Canine distemper virus and measles virus fusion glycoprotein trimers: partial membrane-proximal ectodomain cleavage enhances function. J Virol. 2004;78(15):7894–7903. doi: 10.1128/JVI.78.15.7894-7903.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Von Messling V, Zimmer G, Herrler G, Haas L, Cattaneo R. The hemagglutinin of canine distemper virus determines tropism and cytopathogenicity. J virol. 2001;75(14):6418–6427. doi: 10.1128/JVI.75.14.6418-6427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang F, Yan X, Chai X, Zhang H, Zhao J, Wen Y, Wu W. Differentiation of canine distemper virus isolates in fur animals from various vaccine strains by reverse transcription-polymerase chain reaction-restriction fragment length polymorphism according to phylogenetic relations in china. Virol J. 2011;8(1):1–8. doi: 10.1186/1743-422X-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang J, Luo Y, Liang L, Li J, Cui S. A fast and simple one-step duplex PCR assay for canine distemper virus (CDV) and canine coronavirus (CCoV) detection. Arch Virol. 2018;163(12):3345–3349. doi: 10.1007/s00705-018-3982-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang J, Wang J, Li R, Liu L, Yuan W. Rapid and sensitive detection of canine distemper virus by real-time reverse transcription recombinase polymerase amplification. BMC Vet Res. 2017;13(1):1–7. doi: 10.1186/s12917-017-1180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang J, Wang J, Li R, Shi R, Liu L, Yuan W. Evaluation of an incubation instrument-free reverse transcription recombinase polymerase amplification assay for rapid and point-of-need detection of canine distemper virus. J Virol Met. 2018;260:56–61. doi: 10.1016/j.jviromet.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Welter J, Taylor J, Tartaglia J, Paoletti E, Stephensen CB. Vaccination against canine distemper virus infection in infant ferrets with and without maternal antibody protection, using recombinant attenuated poxvirus vaccines. J Virol. 2000;74(14):6358–6367. doi: 10.1128/JVI.74.14.6358-6367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wilkes RP, Sanchez E, Riley MC, Kennedy MA. Real-time reverse transcription polymerase chain reaction method for detection of canine distemper virus modified live vaccine shedding for differentiation from infection with wild-type strains. J Vet Diag Invest. 2014;26(1):27–34. doi: 10.1177/1040638713517232. [DOI] [PubMed] [Google Scholar]
- 146.Wilkes RP, Tsai YL, Lee PY, Lee FC, Chang HFG, Wang HTT. Rapid and sensitive detection of canine distemper virus by one-tube reverse transcription-insulated isothermal polymerase chain reaction. BMC Vet Res. 2014;10(1):1–8. doi: 10.1186/s12917-014-0213-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Williams ES, Anderson SL, Cavender J, Lynn C, List K, Hearn C, Appel MJG. Vaccination of black-footed ferret (Mustela nigripes)× Siberian polecat (M. eversmanni) hybrids and domestic ferrets (M. putorius furo) against canine distemper. J Wildl Dis. 1996;32(3):417–423. doi: 10.7589/0090-3558-32.3.417. [DOI] [PubMed] [Google Scholar]
- 148.Wostenberg DJ, Walker N, Fox KA, Spraker TR, Piaggio AJ, Gilbert A. Evidence of two cocirculating canine distemper virus strains in mesocarnivores from northern Colorado, USA. J Wildl Dis. 2018;54(3):534–543. doi: 10.7589/2017-09-238. [DOI] [PubMed] [Google Scholar]
- 149.Yadav AK, Rajak KK, Bhatt M, Kumar A, Chakravarti S, Sankar M, Muthuchelvan D, Kumar R, Khulape S, Singh RP, Singh RK. Comparative sequence analysis of morbillivirus receptors and its implication in host range expansion. Can J Microbial. 2019;65(11):783–794. doi: 10.1139/cjm-2019-0008. [DOI] [PubMed] [Google Scholar]
- 150.Yamamoto Y, Nakano S, Seki F, Shigeta Y, Ito S, Tokiwa H, Takeda M. Computational analysis reveals a critical point mutation in the n-terminal region of the signaling lymphocytic activation molecule responsible for the cross-species infection with canine distemper virus. Molecules. 2021;26(5):1262. doi: 10.3390/molecules26051262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yi L, Cheng S, Xu H, Wang J, Cheng Y, Yang S, Luo B. Development of a combined canine distemper virus specific RT-PCR protocol for the differentiation of infected and vaccinated animals (DIVA) and genetic characterization of the hemagglutinin gene of seven Chinese strains demonstrated in dogs. J Virol Met. 2012;179:281–287. doi: 10.1016/j.jviromet.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yoshikawa Y, Ochikubo F, Matsubara Y, Tsuruoka H, Ishii M, Shirota K, Nomura Y, Sugiyama M, Yamanouchi K. Natural infection with canine distemper virus in a Japanese monkey (Macaca fuscata) Vet Microbial. 1989;20(3):193–205. doi: 10.1016/0378-1135(89)90043-6. [DOI] [PubMed] [Google Scholar]
- 153.Yuan C, Liu W, Wang Y, Hou J, Zhang L, Wang G. Homologous recombination is a force in the evolution of canine distemper virus. PLoS ONE. 2017;12:e0175416. doi: 10.1371/journal.pone.0175416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang H, Shan F, Zhou X, Li B, Zhai JQ, Zou SZ, Wu MF, Chen W, Zhai SL, Luo ML. Outbreak and genotyping of canine distemper virus in captive Siberian tigers and red pandas. Sci Rep. 2017;7(1):1–7. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang Y, Xu G, Zhang L, Zhao J, Ji P, Li Y, Liu B, Zhang J, Zhao Q, Sun Y, Zhou EM. Development of a double monoclonal antibody–based sandwich enzyme-linked immunosorbent assay for detecting canine distemper virus. Appl Microbiol Biotechnol. 2020;104(24):10725–10735. doi: 10.1007/s00253-020-10997-y. [DOI] [PMC free article] [PubMed] [Google Scholar]