Abstract
Based on the lung adenocarcinoma (LUAD) gene expression data from the cancer genome atlas (TCGA) database, the Stromal score, Immune score and Estimate score in tumor microenvironment (TME) were computed by the Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data (ESTIMATE) algorithm. And gene modules significantly related to the three scores were identified by weighted gene co-expression network analysis (WGCNA). Based on the correlation coefficients and P values, 899 key genes affecting tumor microenvironment were obtained by selecting the two most correlated modules. It was suggested through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis that these key genes were significantly involved in immune-related or cancer-related terms. Through univariate cox regression and elastic network analysis, genes associated with prognosis of the LUAD patients were screened out and their prognostic values were further verified by the survival analysis and the University of ALabama at Birmingham CANcer (UALCAN) database. The results indicated that eight genes were significantly related to the overall survival of LUAD. Among them, six genes were found differentially expressed between tumor and control samples. And immune infiltration analysis further verified that all the six genes were significantly related to tumor purity and immune cells. Therefore, these genes were used eventually for constructing a Naive Bayes projection model of LUAD. The model was verified by the receiver operating characteristic (ROC) curve where the area under curve (AUC) reached 92.03%, which suggested that the model could discriminate the tumor samples from the normal accurately. Our study provided an effective model for LUAD projection which improved the clinical diagnosis and cure of LUAD. The result also confirmed that the six genes in the model construction could be the potential prognostic biomarkers of LUAD.
Keywords: Naive Bayes model, Tumor microenvironment, Lung adenocarcinoma, Weighted gene co-expression network analysis, Prognostic biomarkers
1. Introduction
Lung cancer is a common malignant tumor in the world. It is the main reason of cancer morbidity and death which accounts for 11.6% of the total cancer cases and 18.4% of deaths caused by cancer (Bray et al., 2018). LUAD is the most common histological type of lung cancer, which is often caused by tumor cells driving oncogene aberrations (Saito et al., 2016). Recently, the incidence of LUAD has raised year by year. Though great progress has been made in immunotherapy and molecular targeted therapy (Osmani et al., 2017), the recurrence and mortality rates of LUAD remain high. Therefore, exploring new biomarkers and potential therapeutic targets is crucial for the diagnosis, cure and prognosis of LUAD.
In the past, cancer was considered as a heterogeneous disease of abnormal mutation in tumor cells. However, increasing researches have proved that tumor progression is the result of a variety of complex signaling pathways caused by tumor cells in their microenvironment (Hanahan & Coussens, 2012; Hui & Chen, 2015). TME is a complicated biological system compounded from proliferating tumor cells, stromal cells, immune cells, infiltrating inflammatory cells and extracellular matrix molecules (Whiteside, 2008). Some studies suggested that TME had important effect on tumor invasion, angiogenesis and infinite proliferation, and could significantly affect the treatment and clinical outcomes of cancer patients (Jain, 2013; Quail & Joyce, 2013). Immune cell and stromal cell are two principal nontumor ingredients in TME. The degrees of infiltration of the two cells have been reported to have significant impacts on the prognosis (Apollonio et al., 2021; Cheng et al., 2019). Therefore, evaluating the two kinds of cells in TME is helpful for better diagnose of tumor patients and their prognosis. To effectively measure the proportion of stromal cells and immune cells in TME, Yoshihara et al. used the ESTIMATE algorithm to infer the tumor purity and the infiltration fraction of stromal cells and immune cells of tumor tissues through gene expression level (Yoshihara et al., 2013). Previous studies have shown that TME-related biomarkers based on ESTIMATE algorithm were efficient for tumor diagnosis and prognosis. For example, Chen et al. found that CD2 could be a new biomarker of breast cancer, which can be used as a supplement to TNM (Tumor, Node, Metastasis) staging to improve the clinical prognosis of patients (Chen et al., 2021). Qi et al. found that thbs1 could be a biomarker of glioma, and its overexpression was related to patients’ poor prognosis (Qi et al., 2021). Recently, TME has received extensive attention due to its significant impact on the progression of lung cancer (Altorki et al., 2019; Wood et al., 2014). Bi et al. found that BTK had potential to be a prognostic factor for lung adenocarcinoma and an indicator for tumor microenvironment (Bi et al., 2020). Jiang et al. established immune-related gene pair signature to predict lung adenocarcinoma prognosis (Jiang et al., 2020). Zhang et al. identified the key genes and characterizations of tumor immune microenvironment in lung adenocarcinoma and lung squamous cell carcinoma (Zhang et al., 2020). Bischoff et al. revealed distinct tumor microenvironmental patterns in lung adenocarcinoma based on single-cell RNA sequencing (Bischoff et al., 2021). Nevertheless, most of the studies on the TME of LUAD were to find the differential genes between normal and cancer samples in different stages or between immune score and matrix score. There is a lack of research on the prognostic genes of lung adenocarcinoma tumor microenvironment from the perspective of constructing gene co-expression network.
In this study, we calculated the Estimate score, Immune score and Stromal score of every LUAD sample using the ESTIMATE algorithm and identified the gene modules related to TME through WGCNA. Univariate cox regression analysis and elastic net analysis were performed to select the key genes correlated with the prognosis of LUAD, and UACLAN database was applied to collect the genes differentially expressed between cancer and normal tissues. Then, immune infiltration analysis was conducted to verify the selected prognostic genes. Finally, a Naive Bayes model was constructed based on the prognostic genes for LUAD projection.
2. Materials and methods
2.1. Data collection and preprocessing
The gene expression data (526 cancer samples and 56 normal samples of LUAD) and the clinical data were obtained from TCGA database (https://xenabrowser.net). The age, gender, stage, TNM, survival time and survival status of the patients were collected from the clinical data, and the clinical information for 513 samples were extracted after integration with the gene expression data.
2.2. Calculation of the three scores
ESTIMATE is a tool to predict tumor purity, which can calculate the infiltration level of immune cells and stromal cells in tumor tissues using gene expression data. Based on ssGSEA algorithm, ESTIMATE calculates Immune score and Stromal score through the proportion of immune cells and stromal cells in tumor microenvironment, and adds the two scores together to obtain the Estimate score that predicts the purity of the tumor sample.
2.3. Correlation between the three scores and the clinical characteristics
The samples were classified as the low and high groups based on the median of the three scores, and Kaplan Meier survival analysis was conducted to test the association between the overall survival of LUAD patients and the high or low groups. The Kruskal test method was used to study the distribution differences of the three scores for different stages, genders, ages and TNM stages. 0.05 was taken as the threshold of P value.
2.4. Construction of weighted gene co-expression network
WGCNA can cluster the highly related genes into the same gene module according to gene expressions. We used WGCNA package in R to structure the co-expression network to obtain gene co-expression modules. Firstly, the top-quartile genes with the greatest variance were collected to calculate Pearson correlation and construct similarity matrix. And an appropriate soft threshold (parameter β) was applied to construct the adjacency matrix. Next, the adjacency matrix was converted into a topological overlap matrix according to the similarity of topological overlap matrix. Then, the Dynamic Tree Cut algorithm was used to classify the genes according to their expressions, and the modules with similarity greater than 0.75 were merged. Finally, the relationships between the gene modules and the three scores were calculated to explore the modules highly correlated with LUAD and identify the key genes in the modules.
2.5. Enrichment analysis
GO function and KEGG pathway enrichment analyses were performed through DAVID database (https://david.ncifcrf.gov/) (Huang et al., 2009a, 2009b) to better understand the potential biological functions of the key genes. The enrichment analysis results were imported into R and the results with P < 0.05 were screened out.
2.6. Screening of the prognostic genes
The selected key genes were inspected by univariate cox regression analysis and Elastic net analysis in R to screen survival related genes of LUAD. Elastic net is a regularization method which combines the advantages of Lasso and Ridge regression and performs feature selection without losing more variables. The common genes of the screening results of the two methods were ultimately considered as the prognostic factors that affected the survival of the patients with LUAD, where 0.05 was taken as the threshold of P value.
2.7. Verification of the prognosis genes
Survival package in R was applied to conduct survival analysis of the prognostic genes and explore their prognostic significance for the patients with LUAD. UALCAN (http://ualcan.path.uab.edu/index.html) (Chandrashekar et al., 2017) is an online resource to analyze cancer transcriptome data based on TCGA database, which can perform biomarker identification, expression spectrum analysis, survival analysis and so on. To ensure the accuracy of the results, gene prognosis value in LUAD was further verified through UALCAN database.
2.8. Differential expression analysis
Wilcox test was applied to examine the difference of the expressions of the genes between cancer and control samples, and the boxplot of each gene expression was drawn. Similarly, the UALCAN database was applied to further verify gene expression differences between cancer and control samples.
2.9. Immune infiltration analysis
The infiltration levels of six immune cells (B cells, CD4+ T cells, CD8+ T cells, Neutprphils, Macrophases and Dedritic cells) in tumor tissues could be detected by TIMER database (https://cistrome.shinyapps.io/timer/) (Li et al., 2016, 2017). In our study, to further confirm the selected prognostic genes, the correlation between the genes and the infiltration levels of six immune cells were analyzed by TIMER database.
2.10. Construction of Naive Bayes model for LUAD projection
Naive Bayes is a simplified technique based on Bayesian algorithm for constructing classifier which is essentially a conditional probability model. Naive Bayes model has stable classification efficiency which performs well on small-scale data sets and can handle multiple classification tasks. In theory, Naive Bayes model has the smallest error rate compared with other classification algorithms. In this study, the genes related to prognosis of LUAD screened from the above steps were used for constructing the Naive Bayes model by R software to discriminate the tumor samples from the normal. The accuracy of the model was verified by AUC of the ROC curve.
3. Results
3.1. Scores calculation and clinical correlation analysis
The Stromal score (−1761.58–2077.13), Immune score (−927.38–3412.03) and Estimate sore (−2317.18–4847.22) were obtained by ESTIMATE algorithm. As shown in Fig. 1, all the three scores were significantly associated with the LUAD patients’ survival. As shown in Fig. 2, Stromal score was significantly different in sex, age and M stage. Immune score was significantly different in sex, age, stage and T stage. And Estimate score showed significant differences in sex, age, stage, T stage and N stage (Supplementary Fig. 1). The results indicated that the proportion of stromal cells, immune cells and tumor purity in TME were associated with the progression of LUAD, and the scores were helpful for evaluating the prognosis for the patients of LUAD.
Fig. 1.
Correlation between the three kinds of scores and survival of LUAD patients.
Fig. 2.
Correlation between clinical features and Stromal score or Immune score.
3.2. Construction of gene co-expression network
The soft threshold β was selected as 4 (Fig. 3A and B) to guarantee the scale-free network when constructing the gene co-expression network. In the network analysis, 15 gene modules were detected (Fig. 3C). The p-values for the correlations between each module and the three scores were calculated (Fig. 3D). Considering the correlation coefficient and P value, the blue module and the red module were highly correlated with the three scores which were selected as the key modules. The blue module contained 717 genes and the red module contained 182 genes. Altogether, there were 899 genes considered as the key genes.
Fig. 3.
Construction and module analysis of co-expression network.
3.3. Enrichment analysis
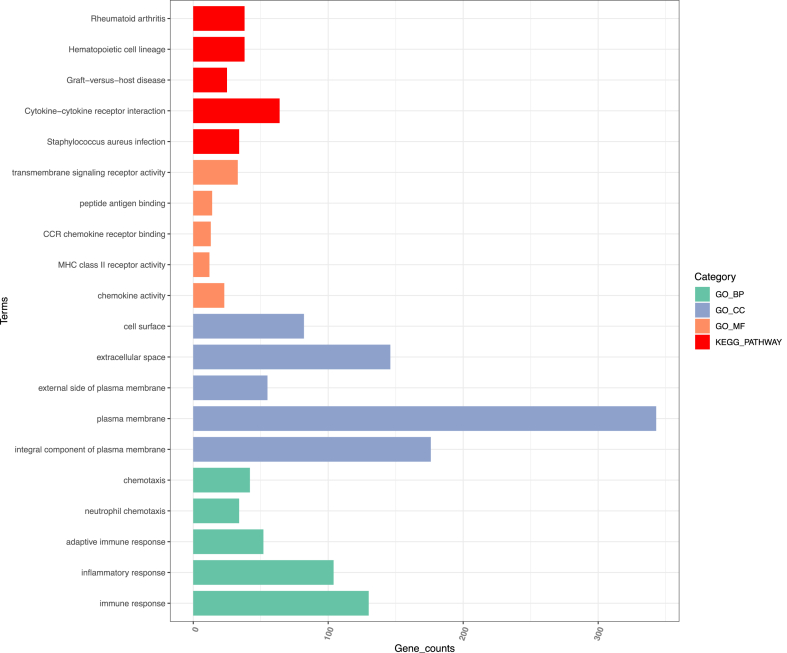
In the results of enrichment analysis on the 899 key genes, 51 KEGG pathways and 488 GO results were identified (P < 0.05). The first 5 KEGG pathways and the first 15 GO results including 5 biological processes, 5 cell components and 5 molecular functions were extracted respectively (Fig. 4). It could be seen from the results that the most enriched KEGG pathway terms were related to Staphylococcus aureus infection, Hematopoietic cell lineage, Graft-versus-host disease, Cytokine-cytokine receptor interaction and Rheumatoid arthritis. The most enriched GO terms were related to immune response, adaptive immune response, neutrophil chemotaxis, inflammatory response, chemotaxis, integral component of plasma membrane, plasma membrane, external side of plasma membrane, extracellular space, cell surface, MHC class II receptor activity, CCR chemokine receptor binding, chemokine activity, peptide antigen binding and transmembrane signaling receptor activity.
Fig. 4.
Results of enrichment analysis of the 899 key genes.
3.4. Screening of the prognostic genes
There were 146 genes screened out by univariate cox regression analysis (Supplementary Table 1) and 23 genes screened by elastic net regression analysis (Supplementary Table 2) significantly associated with the survival of LUAD patients. Finally, 13 prognostic genes (ARG2, CCL3L3, CCR2, CD74, CDCP1, CTSL, ERO1B, FURIN, HLA-DRB55, KCTD12, MERTK, MS4A7, PIK3CG) were identified from the intersection of the two methods.
3.5. Verification of the prognosis genes
The survival analysis results indicated that all the 13 genes were significantly related to the survival of LUAD patients (Fig. 5). While the results of survival analysis in UALCAN database showed that eight genes (CCR2, CD74, CDCP1, FURIN, HLA-DRB5, KCTD12, MERTK and PIK3CG) were significantly related to the survival of LUAD (Supplementary Fig. 2), 3 genes (MS4A7, CCL3L3, ARG2) did not pass the survival analysis and 2 genes (CTSL and ERO1B) were not found in the UACLAN database. Therefore, the results implied that the eight genes (CCR2, CD74, CDCP1, FURIN, HLA-DRB5, KCTD12, MERTK and PIK3CG) had prognostic value for the survival of the patients with LUAD.
Fig. 5.
Survival analysis results of the 13 selected genes (ARG2, CCL3L3, CCR2, CD74, CDCP1, CTSL, ERO1B, FURIN, HLA-DRB5, KCTD12, MERTK, MS4A7 and PIK3CG).
3.6. Differential expression analysis
Wilcox test method was applied to test the expression differences of the eight genes between LUAD and control samples. Boxplots revealed significant differences in the expressions of 7 genes except for MERTK (Fig. 6A). While the results of differential expression analysis on UACLAN database showed that six genes expressed differentially between cancer and control samples except CCR2 and MERTK (Fig. 6B). Therefore, the results suggested that the six genes including CD74, CDCP1, FURIN, HLA-DRB5, KCTD12 and PIK3CG could be the important genes that significantly affected the process of LUAD.
Fig. 6.
Differential expression analysis of the prognostic genes based on TCGA database (A) and UACLAN database (B).
3.7. Immune infiltration analysis
The immune infiltration levels of the six genes (CD74, CDCP1, FURIN, HLA-DRB5, KCTD12 and PIK3CG) were analyzed by timer database (Fig. 7). It could be seen from the results that five of these genes were negatively related to tumor purity except FURIN which was positively related to tumor purity and negatively related to all the immune cells. Besides, genes CD74, CDCP1, HLA-DRB5, KCTD12 and PIK3CG were positively related to most of the six immune cells except that CDCP1 was negatively related to B cell.
Fig. 7.
Immune infiltration analysis of CD74, CDCP1, FURIN, HLA-DRB5, KCTD12 and PIK3CG.
3.8. Construction of Naive Bayes model for LUAD projection
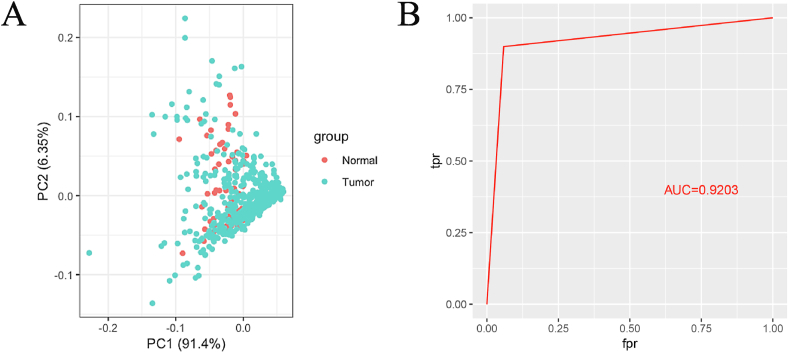
Principal component analysis (PCA) revealed that (Fig. 8A) most of the tumor and normal samples of the dataset overlapped each other, making it difficult to distinguish. The six genes related to prognosis of LUAD were used for constructing a Naive Bayes model. The result of the ROC curve showed that the model could discriminate the tumor samples accurately from the normal where the AUC reached 92.03% (Fig. 8B). Therefore, this model could be used for LUAD projection which could improve the clinical diagnosis level effectively. Moreover, the high projection accuracy of the model confirmed the importance of the six selected genes on LUAD which could be considered as the prognosis biomarkers and therapy targets.
Fig. 8.
PCA result of the two kinds of samples of the dataset (panel A) and ROC curve of the Naive Bayes model with AUC (panel B).
4. Discussion
Tumor microenvironment can affect the tumor cells proliferation and accelerate cell invasion and metastasis, while the existence of tumor cells will change the composition of TME in turn. The interactivity between tumor cells and their microenvironment has turned into a hot spot of recent research which provided potential biomarkers and therapeutic targets for clinical research (Genova et al., 2017).
In this study, the Stromal score, Immune score and Estimate score of each sample of LUAD were output using ESTIMATE algorithm. The results of correlation analysis suggested that the three scores were significantly related to the prognosis and clinical characteristics of LUAD, which further proved that evaluation of infiltration levels of the two kinds of cells was of great value in the diagnosis and prognosis of LUAD. Weighted gene co-expression network analysis is an effective tool of detecting tumor-related modules and genes. Establishing the co-expression network promoted the network-based gene screening method which could be applied to detect therapeutic targets or biomarkers (Langfelder & Horvath, 2008). By constructing co-expression network, we identified 899 key genes that influenced the microenvironment of LUAD by selecting two modules that were the most correlated with the three kinds of scores. GO and KEGG enrichment analyses indicated that the selected genes were significantly involved in cancer and immune related pathways. For example, MK2 promoted colon cancer growth by regulating the activity and recruitment of Macrophage chemokine (Phinney et al., 2018). Staphylococcus aureus was a toxin that promoted opportunistic bacterial lung infections (Cohen et al., 2016). Both univariate cox regression and elastic net regression were used to search the prognostic genes that affected the survival of patients with LUAD. There were 13 prognostic genes obtained, six of them (CD74, CDCP1, FURIN, HLA-DRB5, KCTD12 and PIK3CG) were found differentially expressed between cancer and normal samples by R package and UALCAN database. Immune infiltration analysis showed that the six genes were all related to tumor purity and six kinds of immune cells. Therefore, the six genes were used for modelling. A Naive Bayes model was constructed consequently where the AUC reached 92.03%. The high projection accuracy of the model provided us new effect method for the diagnosis and treatment of LUAD. At the same time, it discovered the importance of the six selected prognostic genes on LUAD.
Literatures suggested that CD74, CDCP1 and FURIN were associated with prognosis and treatment of lung adenocarcinoma. CD74 (MHC class II invariant chain) is a nonpolymorphic type II transmembrane glycoprotein which not only plays an important role in liver fibrosis and Alzheimer's disease but also participates in tumor progression (Su et al., 2017). It was shown that CD74, as an interferon gamma response gene, was a prognostic risk marker related to immune infiltration of LUAD (Yao et al., 2021). CD74 interacted with CD44 and enhanced the occurrence and metastasis of breast cancer through RHOA-mediated cofilin phosphorylation (Liu et al., 2016). The CUB domain-containing protein 1 (CDCP1) is a transmembrane protein that is phosphorylated after activation and may be associated with the proliferation and apoptosis of tumor cells (Uekita et al., 2014). At present, CDCP1 has been confirmed to abnormally overexpress in colorectal cancer and lung adenocarcinoma, and is a prognostic factor of LUAD (Ikeda et al., 2009; Scherl-Mostageer et al., 2001). In certain gastrointestinal cancers, CDCP1 was also a potential prognostic and therapeutic target, regulating peritoneal diffusion of gastric cancer (Uekita et al., 2008). FURIN is an important component of protoprotein processing enzyme family. It is highly expressed in many tumors. Studies have reported that FURIN inhibitor could inhibit proliferation and metastasis of A549 cells of LUAD by down regulating FURIN activity (Bassi et al., 2001). Meanwhile, inhibition of FURIN could lead to the loss or reduction of the invasiveness and tumorigenicity of human cancers, indicating that FURIN is a prospective target for tumor treatment (Ma et al., 2014).
Besides, the other three genes (HLA-DRB5, KCTD12 and PIK3CG) have been proven to be therapeutic targets for some other cancers. HLA-DRB5 has been proven to be a candidate gene for idiopathic pulmonary fibrosis (Gong et al., 2020) and a prognostic gene in the immune microenvironment of endometrial cancer (Ma et al., 2020), and has predictive value for survival of patients with gastric cancer (Hang et al., 2018). Potassium channel tetramerization domain containing 12 (KCTD12) has been shown to promote G1/S transition in breast cancer by sensitizing the AKT/FOXO1 signaling (Ye et al., 2020), and exert its inhibitory actions in esophageal squamous cell carcinoma by inhibiting the growth of stem cells and chromatin remodeling (Abbaszadegan et al., 2018). Class I phosphoinositide 3-kinase (PI3K) signaling pathway regulates many key cell functions such as cell growth, movement, survival and division (Hawkins & Stephens, 2007). In the PI3K family, PIK3CG (PI3Kgamma) has functions to regulate cell immunity and inflammation (Kaneda et al., 2016). It has been discovered that PIK3CG is a possible therapeutic target for breast cancer. Inhibiting PIK3CG activation can enhance the therapeutic efficacy of paclitaxel on breast cancer (Chang et al., 2020).
5. Conclusions
In the current work, we carried out a variety of bioinformatics analysis on lung adenocarcinoma based on the tumor microenvironment and weighted gene co-expression network analysis and identified six key genes (CD74, CDCP1, FURIN, HLA-DRB5, KCTD12 and PIK3CG) which could act as the potential prognostic biomarkers and therapeutic targets of LUAD. With the six genes, a Naive Bayes model was constructed which could project LUAD accurately. Our study proposed new perspectives for the diagnosis and treatment of lung adenocarcinoma. However, our study is based on bioinformatics analysis which mainly rely on data from open access databases. It is expecting that further experimentations are conducted to explore the function of the model and roles of these screened genes in LUAD.
Authors' contributions
All authors designed the research, performed the experiments, analyzed the results and wrote and revised the manuscript.
Declaration of competing interest
The authors declare there is no conflict of interest.
Acknowledgments
Our deepest gratitude goes to the editors and anonymous reviewers for their careful work and thoughtful suggestions that have helped to improve this paper substantially. The work was supported by the National Natural Science Foundation of China (No. 12071382), the Bowang scholar youth talent program (Zhiqiang Ye) of Chongqing Normal University, the Natural Science and Engineering Research Council of Canada, and the Canada Research Chair Program (JWu).
Handling editor: Dr HE DAIHAI HE
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.idm.2022.07.009.
Contributor Information
Xu Zhang, Email: zhangxu1107@163.com.
Jianhong Wu, Email: wujh@mathstat.yorku.ca, wujhhida@gmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abbaszadegan M.R., Taghehchian N., Li L., et al. Contribution of KCTD12 to esophageal squamous cell carcinoma. BMC Cancer. 2018;18(1):853. doi: 10.1186/s12885-018-4765-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altorki N.K., Markowitz G.J., Gao D., et al. The lung microenvironment: An important regulator of tumour growth and metastasis. Nature Reviews Cancer. 2019;19(1):9–31. doi: 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apollonio B., Ioannou N., Papazoglou D., et al. Understanding the immune-stroma microenvironment in B cell malignancies for effective immunotherapy. Frontiers Oncology. 2021;11 doi: 10.3389/fonc.2021.626818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi D.E., Lopez De Cicco R., Mahloogi H., et al. Furin inhibition results in absent or decreased invasiveness and tumorigenicity of human cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(18):10326–10331. doi: 10.1073/pnas.191199198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff P., Trinks A., Obermayer B., et al. Single-cell RNA sequencing reveals distinct tumor microenvironmental patterns in lung adenocarcinoma. Oncogene. 2021;40(50):6748–6758. doi: 10.1038/s41388-021-02054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi K.W., Wei X.G., Qin X.X., et al. BTK has potential to Be a prognostic factor for lung adenocarcinoma and an indicator for tumor microenvironment remodeling: A study based on TCGA data mining. Frontiers Oncology. 2020;10:424. doi: 10.3389/fonc.2020.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., et al. Ualcan: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Hong L., Liu Y., et al. Targeting PIK3CG in combination with paclitaxel as a potential therapeutic regimen in claudin-low breast cancer. Cancer Management and Research. 2020;12:2641–2651. doi: 10.2147/CMAR.S250171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H.S., Lee J.X.T., Wahli W., et al. Exploiting vulnerabilities of cancer by targeting nuclear receptors of stromal cells in tumor microenvironment. Molecular Cancer. 2019;18(1):51. doi: 10.1186/s12943-019-0971-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Meng Z., Zhang L., et al. CD2 is a novel immune-related prognostic biomarker of invasive breast carcinoma that modulates the tumor microenvironment. Frontiers in Immunology. 2021;12 doi: 10.3389/fimmu.2021.664845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T.S., Hilliard J.J., Jones-Nelson O., et al. Staphylococcus aureus α toxin potentiates opportunistic bacterial lung infections. Science Translational Medicine. 2016;8(329):329ra31. doi: 10.1126/scitranslmed.aad9922. [DOI] [PubMed] [Google Scholar]
- Genova C., Rijavec E., Grossi F. Tumor microenvironment as a potential source of clinical biomarkers in non-small cell lung cancer: Can we use enemy territory at our advantage? Journal of Thoracic Disease. 2017;9(11):4300–4304. doi: 10.21037/jtd.2017.10.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W., Guo P., Liu L., et al. Integrative analysis of transcriptome-wide association study and mRNA expression profiles identifies candidate genes associated with idiopathic pulmonary fibrosis. Frontiers in Genetics. 2020;11 doi: 10.3389/fgene.2020.604324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Coussens L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21(3):309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- Hang X., Li D., Wang J., et al. Prognostic significance of microsatellite instability associated pathways and genes in gastric cancer. International Journal of Molecular Medicine. 2018;42(1):149–160. doi: 10.3892/ijmm.2018.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins P.T., Stephens L.R. PI3Kgamma is a key regulator of inflammatory responses and cardiovascular homeostasis. Science. 2007;318(5847):64–66. doi: 10.1126/science.1145420. [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui L., Chen Y. Tumor microenvironment: Sanctuary of the devil. Cancer Letters. 2015;368(1):7–13. doi: 10.1016/j.canlet.2015.07.039. [DOI] [PubMed] [Google Scholar]
- Ikeda J., Oda T., Inoue M., et al. Expression of CUB domain containing protein (CDCP1) is correlated with prognosis and survival of patients with adenocarcinoma of lung. Cancer Science. 2009;100(3):429–433. doi: 10.1111/j.1349-7006.2008.01066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R.K. Normalizing tumor microenvironment to treat cancer: Bench to bedside to biomarkers. Journal of Clinical Oncology. 2013;31(17):2205–2218. doi: 10.1200/JCO.2012.46.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Gao Y., Zhang N., et al. Establishment of immune-related gene pair signature to predict lung adenocarcinoma prognosis. Cell Transplantation. 2020;29 doi: 10.1177/0963689720977131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M.M., Messer K.S., Ralainirina N., et al. PI3Kγ is a molecular switch that controls immune suppression. Nature. 2016;539(7629):437–442. doi: 10.1038/nature19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. Wgcna: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Fan J., Wang B., et al. Timer: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Research. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Severson E., Pignon J.-C., et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biology. 2016;17(1):174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Chu S., Yao S., Li Y., Fan S., Sun X., Su L., Liu X. CD74 interacts with CD44 and enhances tumorigenesis and metastasis via RHOA-mediated cofilin phosphorylation in human breast cancer cells. Oncotarget. 2016;7(42):68303–68313. doi: 10.18632/oncotarget.11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.C., Fan W.J., Rao S.M., et al. Effect of Furin inhibitor on lung adenocarcinoma cell growth and metastasis. Cancer Cell International. 2014;14:43. doi: 10.1186/1475-2867-14-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Zhang J.K., Yang D., et al. Identification of novel prognosis-related genes in the endometrial cancer immune microenvironment. Aging (Albany NY) 2020;12(21):22152–22173. doi: 10.18632/aging.104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani L., Askin F., Gabrielson E., et al. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): Moving from targeted therapy to immunotherapy. Seminars in Cancer Biology. 2017;68:103–109. doi: 10.1016/j.semcancer.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phinney B.B., Ray A.L., Peretti A.S., et al. MK2 regulates macrophage chemokine activity and recruitment to promote colon tumor growth. Frontiers in Immunology. 2018;9:1857. doi: 10.3389/fimmu.2018.01857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi C., Lei L., Hu J., et al. Thrombospondin-1 is a prognostic biomarker and is correlated with tumor immune microenvironment in glioblastoma. Oncology Letters. 2021;21(1):22. doi: 10.3892/ol.2020.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nature Medicine. 2013;19(11):1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M., Shiraishi K., Kunitoh H., et al. Gene aberrations for precision medicine against lung adenocarcinoma. Cancer Science. 2016;107:713–720. doi: 10.1111/cas.12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl-Mostageer M., Sommergruber W., Abseher R., et al. Identification of a novel gene, CDCP1, overexpressed in human colorectal cancer. Oncogene. 2001;20(32):4402–4408. doi: 10.1038/sj.onc.1204566. [DOI] [PubMed] [Google Scholar]
- Su H., Na N., Zhang X., et al. The biological function and significance of CD74 in immune diseases. Inflammation Research. 2017;66(3):209–216. doi: 10.1007/s00011-016-0995-1. [DOI] [PubMed] [Google Scholar]
- Uekita T., Fujii S., Miyazawa Y., et al. Oncogenic Ras/ERK signaling activates CDCP1 to promote tumor invasion and metastasis. Molecular Cancer Research. 2014;12(10):1449–1459. doi: 10.1158/1541-7786.MCR-13-0587. [DOI] [PubMed] [Google Scholar]
- Uekita T., Tanaka M., Takigahira M., et al. CUB-domain-containing protein 1 regulates peritoneal dissemination of gastric scirrhous carcinoma. American Journal Of Pathology. 2008;172(6):1729–1739. doi: 10.2353/ajpath.2008.070981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside T.L. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27(45):5904–5912. doi: 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood S.L., Pernemalm M., Crosbie P.A., et al. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treatment Reviews. 2014;40(4):558–566. doi: 10.1016/j.ctrv.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Yao B., Wang L., Wang H., et al. Seven interferon gamma response genes serve as a prognostic risk signature that correlates with immune infiltration in lung adenocarcinoma. Aging (Albany NY) 2021;13(8):11381–11410. doi: 10.18632/aging.202831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R.Y., Kuang X.Y., Zeng H.J., et al. KCTD12 promotes G1/S transition of breast cancer cell through activating the AKT/FOXO1 signaling. Journal of Clinical Laboratory Analysis. 2020;34(8) doi: 10.1002/jcla.23315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K., Shahmoradgoli M., Martínez E., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature Communications. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.M., Chen J.H., Cheng T.L., et al. Identification of the key genes and characterizations of tumor immune microenvironment in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) Journal of Cancer. 2020;11(17):4965–4979. doi: 10.7150/jca.42531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.