Summary
The All of Us Research Program seeks to engage at least one million diverse participants to advance precision medicine and improve human health. We describe here the cloud-based Researcher Workbench that uses a data passport model to democratize access to analytical tools and participant information including survey, physical measurement, and electronic health record (EHR) data. We also present validation study findings for several common complex diseases to demonstrate use of this novel platform in 315,000 participants, 78% of whom are from groups historically underrepresented in biomedical research, including 49% self-reporting non-White races. Replication findings include medication usage pattern differences by race in depression and type 2 diabetes, validation of known cancer associations with smoking, and calculation of cardiovascular risk scores by reported race effects. The cloud-based Researcher Workbench represents an important advance in enabling secure access for a broad range of researchers to this large resource and analytical tools.
Keywords: precision medicine, All of Us Research Program, electronic health records, cohort study, cloud-based analytics
Highlights
-
•
The All of Us Research Program has released data for over 315,000 participants
-
•
Demonstration projects support the utility and validity of the All of Us dataset
-
•
The cloud-based Researcher Workbench provides secure, low-cost compute power
The bigger picture
The engagement of participants in the research process and broad availability of data to diverse researchers are essential elements in building precision medicine equitably available for all. The NIH has established the ambitious All of Us Research Program to build one of the most diverse health databases in history with tools to support research to improve human health. Here, we present the initial launch of the Researcher Workbench with data types including surveys, physical measurements, and electronic health record data with validation studies to support researcher use of this novel platform. Broad access for researchers to data like these is a critical step in returning value to participants seeking to support the advancement of precision medicine and improved health for all.
The initial release of the All of Us Research Program data reflects diverse participants with broad information, reproduces known associations, and provides rich opportunities for research. The dataset and tools form a strong foundation for cohort growth and future research, advancing the program mission to improve human health and advance precision medicine.
Introduction
The NIH’s All of Us Research Program (All of Us) is a longitudinal cohort study aimed at advancing precision medicine and improving human health through partnering with one million or more diverse participants across the United States.1 Informed by the success of prospective longitudinal cohorts and the more recent research use of electronic health records (EHRs), All of Us combines participant-derived information from surveys (participant-provided information [PPI]) and physical measurements (PMs), EHRs, biospecimens, wearables, and planned links to external data sources to allow for both active and passive data collection; participants may also consent to recontact.2 Whereas a conventional biorepository design delivers data to investigators, the All of Us program has adopted a different infrastructure, described here, to “bring researchers to the data” in a cloud-based environment.3 This approach should both enhance data storage and security, as well as provide facile access to data and analysis tools to a broad range of researchers including those in computationally underdeveloped environments. This infrastructure will enable both hypothesis-generating approaches as well as traditional hypothesis testing by researchers with diverse interests and capabilities, with the ultimate goal of improving individualized care and outcomes.
All of Us launched national recruitment in May 2018 and as of June 2021 had enrolled over 387,000 participants, of whom 295,000 had provided biospecimens and survey data. Recruitment is accomplished by a large multi-disciplinary consortium, with enrollment centers in varied settings including health provider organizations and community partners. Specific emphasis in the program has been placed on recruiting participants from groups that have been historically underrepresented in biomedical research (UBR), and as of May 2021, over 75% of participants are identified as UBR including racial and ethnic groups, income levels, educational attainment, rural living area, sexual and gender minorities, and individuals with disabilities.4 All of Us is committed to engaging participants longitudinally, ensuring access to their own data and to results of research, including support for a participant partnership program to inform the direction of the program and research processes.1
The cloud-based Researcher Workbench5 described here has been developed to democratize access for researchers by eliminating requirements for large local infrastructure and to enhance data security by minimizing individual data copies.6 The platform is designed to meet the FAIR principles of research—Findable, Accessible, Interoperable, and Reusable—developed to address concerns about the reuse of scholarly data on behalf of a diverse set of stakeholders representing academia, industry, funding agencies, and scholarly publishers.7 Additionally, All of Us has developed policies to lower barriers to data access necessitated by human subjects research review by removing known identifiers and applying privacy preserving methodology enabling a “passport model” that grants broad access to the non-human subjects research dataset that was approved by the program institutional review board instead of burdening researchers with completing the conventional project-by-project mode of review. Currently, researchers are approved from institutions that have signed a data-use agreement and after completing ethics training by using their eRA Commons ID.
All of Us has adopted a philosophy of early, iterative data release and the establishment of demonstration projects with the goal of evaluating the quality, usefulness, validity, and diversity of the research dataset and platform.1,8 A particular challenge for All of Us, compared with other large cohorts such as the UK Biobank and the Million Veterans Program, is harmonizing many data sources, necessitating demonstration of data validity and utility.9,10 A core value of All of Us is to ensure equal access to the data by researchers; therefore, demonstration projects presented here were not designed to make significant biological discoveries but rather to describe the cohort and validate the Researcher Workbench structure by replicating previous findings. Here, we describe the demographics of the first 315,007 participants and the results of demonstration projects investigating treatment pathways of diabetes and depression medication, the relation of smoking to cancer, and calculation of baseline cardiovascular disease risk scores using the data and tools in the All of Us Researcher Workbench. We also estimated compute costs for these analyses. All methods, cohorts used, and relevant analytical code have been made available in the Researcher Workbench for replication and reuse.
Results
Demographics of the dataset
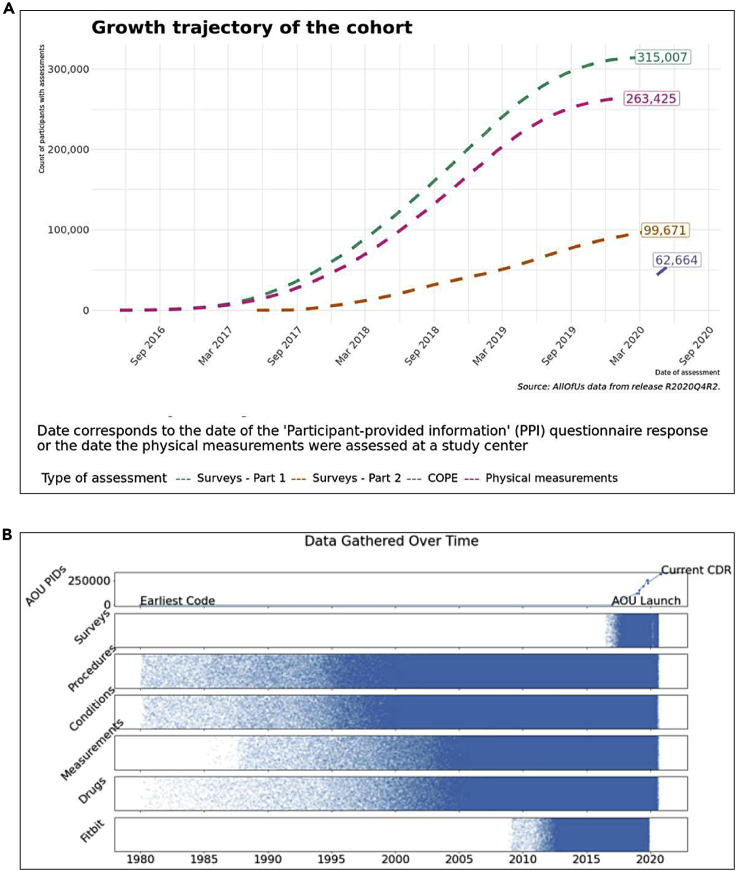
The beta launch of the Researcher Workbench includes data from 315,007 total participants. Figure 1A displays an overview of the data types available, including PMs obtained in person, surveys completed electronically, and EHRs from enrolling partners. In the dataset analyzed, personal identifiers were removed, and a random backward date shift (1–365 days) was introduced; researchers may access non-deidentified datasets only with specific approvals. The date shift causes some survey data to appear before the start of program enrollment.
Figure 1.
Overview of data types included in the beta-release curated data repository
(A) Growth trajectory of participant data types after enrollment. Survey part 1 (green) includes “the Basics,” “Lifestyle,” and “Overall Health” surveys; survey part 2 (pink) includes “Personal Medical History,” “Health Care Access and Utilization,” and “Family Medical History.” Physical measurement accrual is shown in red, and the COVID-19 Participant Experience (COPE) survey is shown in purple. Note that the flattening is artificial due to the random date shift introduced to protect participant privacy.
(B) Historical availability of participants’ electronic health record (EHR), survey, and device data.
By design, all participants have data from the first of the part 1 surveys, “the Basics” survey, which must be completed before participants are eligible to complete other steps in the All of Us protocol. The second two surveys, “Overall Health” and “Lifestyle,” have 307,756 and 306,316 participant responses, respectively. The part 2 surveys distributed 90 days after enrollment, “Healthcare Access & Utilization,” “Family History,” and “Personal Medical History,” have 98,541, 91,695, and 89,261 participant responses, respectively. The most recently launched survey, COVID-19 Participant Experience (COPE), has 62,664 responses, and 8,435 participants have data from a Fitbit device. Of those with any survey response, 263,425 have at least one PM recorded, and 203,813 have any EHR data included. The total number of participants who have any survey response, PM, and EHR data is 196,709. Additional breakdowns of individual data types are shown in Figure S1. Figure 1B shows the historical availability of EHR and Fitbit data by structured domains of information.
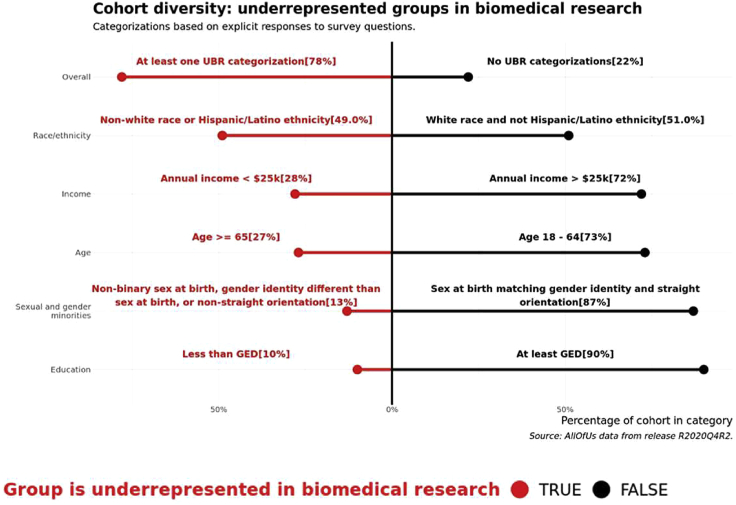
Demographic information included in the dataset is extracted from “the Basics” survey response. Figure 2 details additional survey responses into the program definitions of participant status as UBR. Notably, 49% of participants identified with a population other than White alone, and 13% of participants identified with a sexual or gender minority group. Overall, 78% of participants were included in at least one UBR category. Additional breakdowns are shown in Figure S2.
Figure 2.
UBR metrics
Depiction of the proportion of participants that are underrepresented in biomedical research (UBR) based on program definitions. A participant is included in the overall category if they meet at least one criterion among the race/ethnicity, income, age, sexual orientation, education, gender identity, and sex at birth designations. The sexual and gender minorities category shows aggregates of any participant with a UBR response to questions on sexual orientation or gender identity or sex at birth. GED, General Education Development (i.e., high school diploma or equivalent).
Treatment-pathway visualization
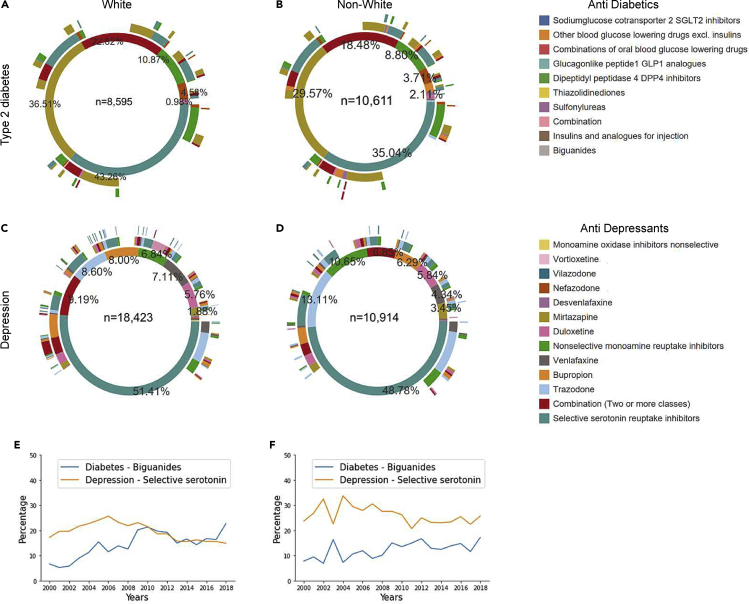
Depression and type 2 diabetes (T2D) are common diseases for which multiple medications are used. The order of treatment(s) prescribed after common disease diagnosis was determined to demonstrate medication mapping and use of hierarchies in the dataset. The numbers of participants meeting inclusion criteria to map treatment pathways were 19,206 total participants with T2D and 29,337 with depression. The number of participants contributed by individual consortium EHR sites are shown in Table S1. The treatment-pathway visualizations are shown in Figures 3A–3D, and the percentage of usage of most common medications by year is in Figures 3E and 3F, with separate counts for White and non-White participants. The innermost circle represents the first medication class prescribed, and the circles expanding outward are the second and third medication classes occurring in the EHR after diagnosis. The most common first medication classes were biguanides for T2D and selective serotonin-reuptake inhibitors for depression in both White and non-White participants. However, the proportion of those treated first with the most common medication prescribed for both T2D and depression differed between White and non-White participants (p < 0.01), and the order of subsequent medication use differs as well. These results replicated published analyses.11
Figure 3.
Medication sequencing for participants who have diabetes and depression grouped by race
(A) Anti-diabetic medication sequences for White participants.
(B) Anti-diabetic medication sequences for non-White participants.
(C) Antidepressant medication sequences for White participants.
(D) Antidepressant medication sequences for non-White participants.
(E) Percentage of White participants who were prescribed one medication that is the most common one from years 2000–2018.
(F) Percentage of non-White participants who were prescribed one medication that is the most common one from years 2000–2018. The difference in counts of first anti-diabetic in (A) and (B) and the counts of first antidepressants in (C) and (D) for each medication between White and non-White participants was significant (p by chi-square was <0.05).
Cancer phenome-wide association with smoking study
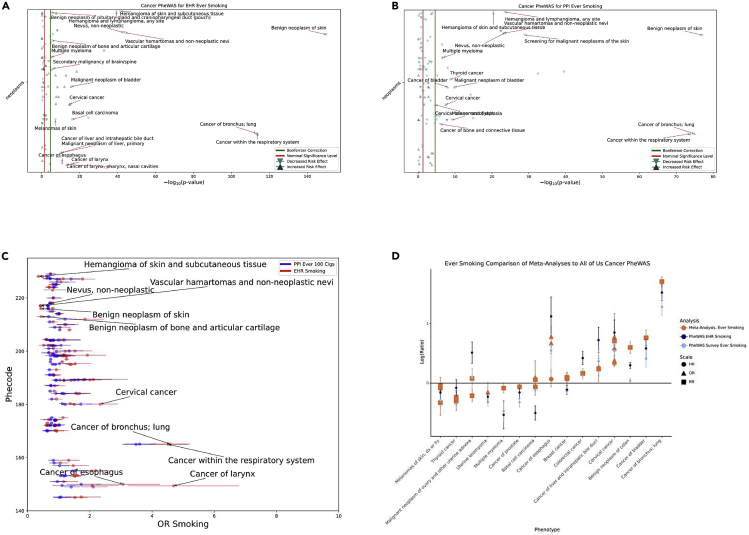
A cancer phenome-wide association study (PheWAS) was performed to determine whether known associations with smoking could be replicated and to compare effect sizes of smoking exposure gathered from EHR billing codes with smoking exposure determined from survey data.12,13 A total of 32,755 participants were identified as EHR ever smokers and 145,844 as EHR never smokers using billing codes. The survey responses identified 122,524 participants as ever smokers, 55,986 participants as current smokers, and 175,809 never smokers. In both analyses using PPI data, the PPI never-smoking group was used as the comparison group. The overlap of these participants is shown in Table S2. The results of the cancer phenome-wide associations for EHR ever smoking and survey ever smoking are shown in Figures 4A and 4B. Effect sizes for the results of the top five EHR non-protective and protective phenome-wide significant cancer associations matched to their phenome-wide significant result from the survey ever-smoking cancer PheWAS are shown in Table S3. An expanded results list for each is included in Table S8. The top cancer phenotypes for which ever smoking was a risk include respiratory cancers and cervical cancer, both known associations. Smoking was protective against cutaneous-related neoplastic outcomes, which has also been previously reported.14, 15, 16 A comparison between the effect sizes seen in EHR results versus survey ever smokers is shown in Figure 4C. 20% of the EHR effect sizes were statistically significantly higher than the ever-smoking effects. The comparison of effect sizes seen in All of Us data with literature is shown in Figure 4D.17 For 12 out of 15 of the phenotypes, there was at least one cancer PheWAS result whose confidence interval overlapped with the confidence interval reported in the corresponding meta-analysis.
Figure 4.
Cancer PheWAS ever-smoking EHR and survey comparison
(A) Manhattan plot for Cancer PheWAS using EHR ever smoking as exposure. Results are the −log10 (p value) of the corresponding logistic regression adjusted for age at last relevant EHR code, sex at birth, race and ethnicity from surveys, EHR length, and number of unique billing codes per record. Up arrows indicate non-protective associations, and down arrows indicate protective ones. Phenotype labels are given to the top ten phenotypes based on magnitude of effect size for both protective and non-protective effects.
(B) Manhattan plot for cancer PheWAS using survey ever smoking as exposure, with the same presentations as (A).
(C) Comparison of survey smoking-regression estimates (colored in blue) to EHR smoking-regression estimates (colored in red) for cancer outcomes.
(D) PheWAS EHR ever-smoking (dark blue) and survey ever-smoking (light blue) effect sizes and confidence intervals compared with published meta-analyses (orange). Estimates are presented on the natural log ratio scale (odds ratio [OR] or risk ratio [RR]). Estimates below the horizontal line represent protective effects, and estimates above the line represent non-protective effects. Each meta-analysis plot point shape represents whether the effect size from the literature was an OR, HR, or an RR, recognizing that RR and ORs are not directly comparable except in the case of rare disease.
Cardiovascular disease risk calculation
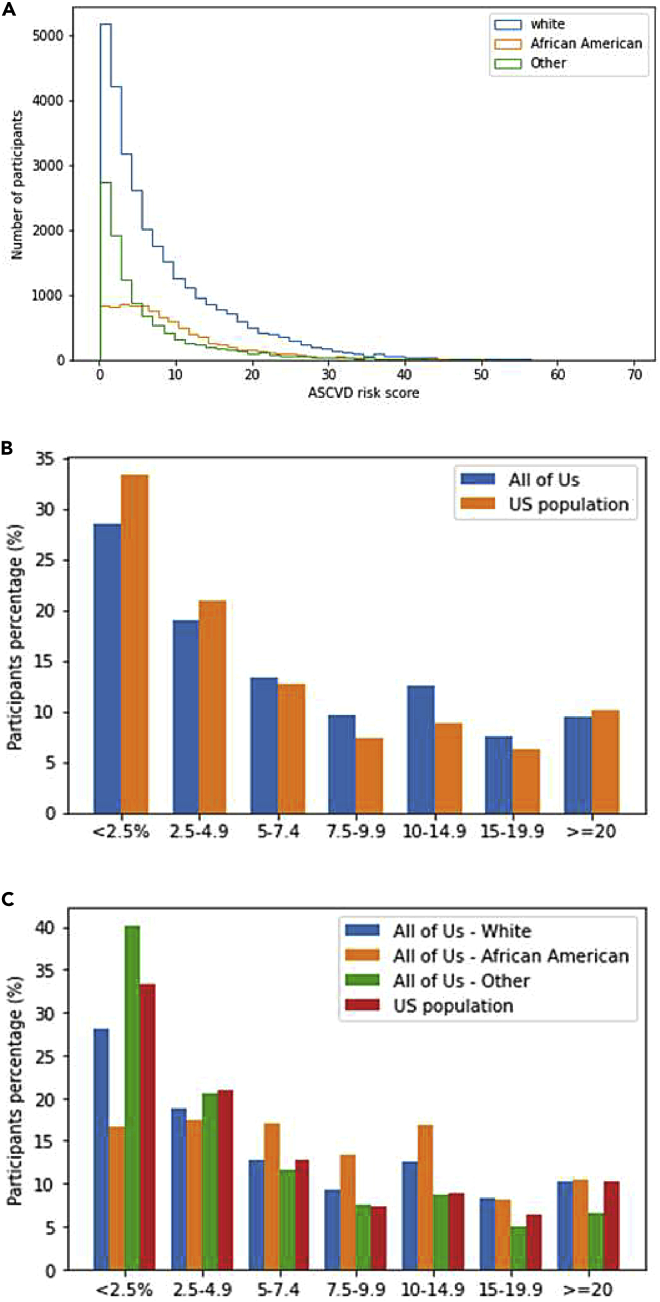
A number of tools have been developed to calculate risk for atherosclerotic cardiovascular disease (ASCVD). Many of these incorporate race in their risk estimates. In this analysis, we estimated ASCVD risk using the American College of Cardiology/American Heart Association (ACC/AHA) 2013 Pooled Cohort Equations.18 Participants were included if aged 40–79, and the following model parameters were available from the EHR data: total cholesterol (TC), high-density lipoprotein cholesterol (HDL), systolic blood pressure (SBP), and hypertension treatment status, diabetes status, and no evidence of existing ASCVD on enrollment. There were 49,982 participants with all parameters necessary for calculation of the ASCVD risk score prior to observation of any cardiovascular event. Among these participants, 32,148 (64.3%) were assigned female sex at birth, 9,331 (18.7%) were African American, 10,564 (21.1%) had other a single or two or more races assigned to “other” for score calculation, 30,087 (60.1%) were White, and 6,603 (13.2%) were smokers. Table S4 summarizes demographic information for All of Us participants, participants who have any EHR data, and participants with sufficient data to calculate risk scores. The mean age of participants with calculated scores was 57.3 (SD ± 9.9) years, and the mean SBP was 127.4 (SD ± 14.0). There were 8,821 (16.4%) participants who had the onset of new CVD within 10 years of measurement.
Across all three groups, scores were significantly different (p < 0.001) by race (Figure 5A; Tables S5 and S6). In pairwise comparison, risk scores for White and other race participants were lower than for African American race (p < 0.001) in both scores calculated at any time and those calculated within a year of enrollment. Other race participant scores were significantly lower than White participants in the scores at any time overall and within a year of enrollment (p < 0.001). We compared the percentage of All of Us participants at each ASCVD risk threshold to the US population scores as estimated previously.18 The trend in the percentages of participants by risk and race groups in seven ASCVD score groups is similar in both studies, as shown in Figures 5B and 5C.
Figure 5.
Baseline cardiovascular disease risk calculations
(A) Baseline 10 year ASCVD cardiovascular disease risk calculations (%). A histogram of the cardiovascular disease risk score for participants with necessary measurements grouped by race group into White, African American, and other. The difference among the cardiovascular risk scores across the three race groups was statistically significant (p value for the Kruskal-Wallis H test was 0). Mann-Whitney p value was <0.001 when comparing the risk scores for White versus African American participants, other versus African American, and White versus other.
(B) Comparing the percentage of All of Us participants to the US population in each ASCVD risk category as published in ACC/AHA guidelines. The risk score for US population was calculated by applying the pooled cohort equations (i.e., ASCVD score) to the National Health and Nutrition Examinations Surveys.
(C) Comparing the percentage of All of Us participants in each race group with the US population in each ASCVD risk category as published in ACC/AHA guidelines. The risk score for US population was calculated by applying the pooled cohort equations (i.e., ASCVD score) to the National Health and Nutrition Examinations Surveys.
Cost and sharing of analytic methods and code
The total compute cost for all analyses, from the beginning to the submission of this paper, was approximately $96. Table S7 indicates the rates and cost of individual analyses. All analyses presented have been made available within the Featured Workspaces section of the Researcher Workbench that permits researchers to view and duplicate code for replication of analyses or adaptation for their own use.
Discussion
A core principle of the All of Us program is to provide data to researchers quickly, in a manner that is transparent to participants.1 The initial launch of the cloud-based Researcher Workbench was in May 2020, 2 years after the national launch of participant enrollment, and includes robust security practices for participant protection.19 Currently, access is provided to all researchers whose institutions sign a data-access agreement; the approvals are at US academic institutions and other health research non-profits, with future plans to expand access to researchers in industry and the international community in 2022 as well as to create paths for citizen scientists in the future. The cloud-based analysis platform not only enhances data security but also enables ready sharing and reproduction of research findings; all code of demonstrations described here can be copied and replicated from the Featured Workspaces in the Researcher Workbench. Additionally, the use of the Observational Medical Outcomes Partnership (OMOP) Common Data Model, supported by a broad coalition of users, sets a foundation for interoperability with other cohorts made realistic by cloud-based sharing of code for replication and reuse.20 By making computational tools available with the data, All of Us expands researcher access to those who do not have resources to store and compute on large datasets and provides a foundation for the future addition of storage-intensive data types such as whole-genome sequences and digital health data streams. To increase transparency of research using the All of Us data to the public, including study participants, the public ResearchHub website includes a description of each workspace within the Researcher Workbench and a directory of all researchers approved for access. All of Us is also committed to the direct return of results to participants, including engagement with participant ambassadors in the policy process21 who found that the need for demonstration of responsible curation and research use was important to earn and retain participants’ trust and show how their data might be used to further health research.
The demonstration projects described here, together with the availability of the analysis code for researcher reuse, show the potential of the cohort for a variety of research purposes. For example, the description of medication sequencing in common complex diseases such as T2D and depression speaks to the validity of the data aggregated from over 30 individual healthcare facilities in showing expected treatment patterns. As with all these examples, the code needed to reproduce these results is provided, giving researchers the foundation to extract medications from the data model and extrapolate them to classes using a common medication ontology. Other discoveries may be advanced in this growing dataset, with the entire PheWAS package now available in the Researcher Workbench for researcher reuse and new hypothesis generation. Finally, the calculation of the ASCVD pooled risk scores also shows the feasibility of detailed derivation of multiple data elements required for this estimation. While this ASCVD calculation, which included historical EHR data, likely demonstrates survivor bias in those included, the replication of known race relationships to established risk models and ongoing ability to monitor should provide valuable baseline data for decades to come.
The projects described here aim to replicate prior findings and show how the dataset can be used, without preempting significant discoveries. We provide example visualizations of general cohort characteristics to illustrate the heterogeneity of available data types and the diversity of the cohort. EHR data are currently available for roughly two-thirds of the cohort at this time; we expect that this proportion will grow over time. All surveys are now available at enrollment, and implementation trials are underway to increase response rates further. The program has committed to improving survey completion rates including development of a reassessment module to monitor outcomes over time. Nearly 100% of participants with data in the Researcher Workbench have a biospecimen available, and generation of genomic data on this cohort has begun. Thus, given the high proportion (over 75%) of participants included from groups traditionally UBR, this cohort will provide the foundation for genome-based studies in minority populations as well as with ongoing collection of EHR outcomes data be uniquely well suited to studying health disparities.22
These replication projects highlight the value of EHR data obtained from many healthcare partners merged with direct participant data from measurements and surveys and made available in a privacy-sensitive, secure, powerful compute environment. Researcher Workbench support services also include an interactive monitored user forum to communicate with the program and other researchers, as well as an integrated help desk ticketing system to support researchers and gather feedback for the program. The code for the projects presented here and others completed will also be made directly available to researchers in the Featured Workspaces section of the Researcher Workbench for replication and reuse,5 benefitting researchers by saving work to generate new code when existing approaches can be adapted, as well as returning value to participants in fulfilling FAIR principles and elevating reproducibility and validity of findings.
Because participants contribute data in many different ways, the cohort enables prospective, retrospective, cross-sectional, and nested case-control analyses. The coronavirus 2019 (COVID-19) crisis occurring concurrently with the planned launch of the Researcher Workbench provided All of Us the opportunity to rapidly adapt and serve the emergent need for COVID-19-relevant research. In-person enrollment of participants paused, and the consortium pivoted to use biospecimens for antibody testing to localize early spread of the virus, developed new surveys that align with other cohorts to gather data directly from participants, and worked to ensure appropriate capture of COVID-19-relevant EHR data.23 While the current curation timeline and model did not allow real-time provision of pandemic-related data to researchers, both the COVID-19 survey data and COVID-19 serology data are now available in the Researcher Workbench for retrospective analyses of the health outcomes in the cohort less than 6 months from generation.
Limitations of the beta-launch platform presented here include access that is more restricted than the planned full release with expanded access options planned in 2022 to reach industry and international researchers. Finding the balance of privacy, security, and sharing is ongoing, and to fulfill the pledge to protect participants, this limitation has given the program an opportunity to learn how to share widely and wisely. Additional risk in this limited launch is paucity of data specific for researchers focused on health disparities and minority health given the generalizations required for privacy at this time and a lack of a comprehensive assessment of social determinants of health, which will be captured in the next survey that will be offered to participants. The program also plans to release these data in 2022, including more granular demographic information and the exact dates of events without the date shift included in the current release. The requirements of knowledge of Python 3 or R to perform analyses in the Researcher Workbench may exclude some researchers unfamiliar with these methods. Currently, batch workflow is not available, and computational ability to deal with larger datasets will be required when genomics and other wearable data expand to enable deep-learning techniques; these capabilities are also slated for release in 2022. Questions asked during the researcher registration process and workspace descriptions are providing valuable data to the program regarding diversity of research topics as well as of the researcher community. Because we have prioritized early, iterative data release and the speed of sharing this dataset, some data types are limited. Also, the date shift introduced to decrease identifiability of participants by disallowing comparison of rare events found with actual dates in publicly available reporting makes seasonal and cross-sectional research in relation to major events impossible. The response rate of later-release surveys is low but increasing with earlier delivery of all surveys and focused efforts at maintaining ongoing engagement with the program. Heterogeneity of EHR data, including needs for harmonization and data missingness, can hinder studies, and specific efforts are focused on improving conformance to the data model and completeness from existing sites, as well as exploring newer direct links such as Apple Health Record linkage, Sync for Science, and other Fast Health Interoperability Resource (FHIR)-based efforts allowing for participant health record data donation.24, 25, 26 Additionally, the calculations to balance reidentification risk will be updated as the cohort grows, which will likely allow for fewer generalizations in future data releases, allowing more granular inspection of groups traditionally underrepresented in research. Finally, as many other large cohorts are developing, including the UK Biobank and the Million Veteran Program, learning to interoperate and jointly analyze data will be paramount. Notably, All of Us, unlike other large resources, has an explicit commitment to return data to participants. The opportunities to develop new methodologies to handle data at scale are greater than ever, and the low-cost, secure Researcher Workbench platform fulfills a great unmet need to advance precision medicine research including future implementation of machine-learning approaches.27,28
While significant progress has been done to allow for the safe sharing of All of Us data with the research community, many challenges lie ahead in navigating the future of All of Us research, including ensuring ongoing engagement with diverse participants, reduction of data missingness, and rapid expansion of data types including digital health technology, genomics, and external data linkages. The beta launch of the Researcher Workbench begins a process of iterative improvement, fulfilling the goal of providing data to researchers early and often. The All of Us Research Program looks forward to incorporating feedback from the research community on this initial release of data and tools.
The initial release of the All of Us Research Program data reflects diverse participants with broad information, reproduces known associations, and provides rich opportunities for research. The dataset and tools form a strong foundation for cohort growth and future research advancing the program mission to improve human health and advance precision medicine.
Experimental procedures
Resource availability
Lead contact
Requests for additional information about the findings presented in this study may be directed to the lead author, Andrea Ramirez (andrea.ramirez@nih.gov). Requests for information about the All of Us Researcher Workbench platform, including access, may be directed to the All of Us Researcher Workbench support team (support@researchallofus.org). For more information about the All of Us Research Program data and tools, please visit https://www.researchallofus.org/.
Materials availability
Study materials are made available through the Researcher Workbench at https://researchallofus.org.
Methods
Protocol
The goals, recruitment methods and sites, and scientific rationale for All of Us have been described previously.1 Participants consent to the study and authorize the sharing of EHRs through an online portal or smartphone application, after which they can answer health surveys, share digital health data (such as any Fitbit model and Apple HealthKit), and can view their study information. Through in-person visits, participants are invited to contribute biospecimens and undergo PMs including systolic and diastolic blood pressure, height, weight, heart rate, waist and hip measurement, wheelchair use, and current pregnancy status. Structured EHR data are transferred from enrolling sites at least once per quarter.
Data curation and privacy methodology
Surveys, PM, and EHR are mapped to the OMOP common data model v.5.2 maintained by the Observational Health Data Sciences and Informatics (OHDSI) collaborative.20 Where the model does not support necessary concepts, custom concepts are added in collaboration with the OHDSI community, linked to existing concepts where possible, and documented in the open-source Athena resource, a repository of vocabularies used in OMOP and supported by Odysseus Data Services.29 Participants were included in the beta-launch curated data repository (CDR) if they responded to at least the first survey, “the Basics.” To protect participant privacy, a series of data transformations were applied including data suppression of codes with a high risk of identification such as military status; generalization of categories including age, sex at birth, gender identity, sexual orientation, and race; and date shifting by a random number of days from 1 to 365 implemented consistently across each participant record, causing some data to appear to have accrued before program start. General conformance rules are applied to meet the standard conventions of the OMOP data model including dropping invalid dates and extreme values, resulting in a base version of the CDR. Additional cleaning steps for selected lab data (from EHRs) and PMs were performed to standardize units and values resulting in the processed CDR, on which the analyses presented here were performed. Documentation on privacy implementation and creation of the CDR is available in the Research Hub at www.researchallofus.org and in the All of Us Registered Tier CDR Data Dictionary.30
Platform
The dataset was accessed through the All of Us Researcher Workbench, a cloud-based analytic platform custom built by the program for approved researchers. The Workbench is built on top of the Terra platform (terra.bio), which is also utilized for a number of other NIH-funded studies including the National Cancer Institute (NCI Cloud Resources), the National Heart, Lung, and Blood (NHLBI) BioData Catalyst, and the National Human Genome Research Institute (NHGRI) AnVIL. The Workbench exceeds Federal Information Security Management Act (FISMA) moderate security standards and undergoes routine security testing.31 The All of Us Researcher Workbench platform includes project-specific spaces, termed workspaces, featuring a description of the project and permitting sharing among teams of collaborators. Workspaces include access to graphical “point and click” interface tools to select participants (a “cohort builder”) using a variety of Boolean criteria across data types and selection of data elements for analysis. Analyses are currently performed using Jupyter Notebooks.32 The notebooks currently enable use of saved datasets and direct query using R and Python 3 programming languages. Access to the Researcher Workbench and data are free. Compute and storage accrue usage cost. The Researcher Workbench uses Google Compute Engine for computational resources in the cloud and Google Cloud Storage for storage in the cloud.
Access
All researchers who access the data for analyses are currently authorized and approved via a 6-step process that includes registration, affiliation with an institution that has completed a Data Use and Registration Agreement, identity verification via login.gov, completion of ethics training, and attestation to a data use agreement. Approval to use the dataset for the specified demonstration projects was obtained from the All of Us Institutional Review Board. Results reported are in compliance with the All of Us Data and Statistics Dissemination Policy disallowing disclosure of group counts under 20 to protect participant privacy.33
Descriptive visualizations
The age displayed reflects the age when the CDR version used in this report was generated in the summer of 2020. Presence of a data-type survey, PM, or EHR was counted if at least one observation was present within each category. To assess race and ethnicity, participants were asked “Which categories describe you? Select all that apply. Note, you may select more than one group” in the “the Basics” survey. Responses were mapped to the race variable in the OMOP Person table directly for the responses White, Asian, and Black, African American, or African, and responses Middle Eastern or North African and Native Hawaiian or other Pacific Islander were generalized to “Other single population”.
Currently, all participants responding American Indian or Alaska Native have been removed from the CDR, as All of Us goes through official consultation with tribal leaders on the research use of data. Participants choosing any two of the categories were labeled “More than one population.” Skipped questions were omitted, and the responses “None of these fully describe me” and “I prefer not to answer” or non-responses to these categories were individually mapped in the data model and grouped as “Not specified” for visualization for the analyses presented here. The “Not specified” group included participants who chose Hispanic, Latino, or Spanish. This response was mapped to the ethnicity variable, allowing reflection of both race and Hispanic ethnicity. Program designations of status as UBR were adapted to data available in the CDR.34
Treatment-pathway visualization
The order of treatment prescribed after common disease diagnosis was determined for T2D and depression to demonstrate medication mapping and use of hierarchies in the OMOP common data model. For each condition, the time of earliest diagnosis was identified, and medications were extracted using the OMOP hierarchy as in the previously published work. Medications were then grouped into generalized classes based on their main ingredient using the anatomical therapeutic chemical (ATC) classification.35 Participants were included if their first medication related to the disease was prescribed after the earliest diagnosis code for that disease, they had two or more diagnosis codes for the disease, and they had at least 3 years of medication records with at least a single structured occurrence of the drug. We determined the number of participants whose monotherapy was the most common first medication in any given year between 2000 and 2016. Each of these analyses was performed separately on the participants identified as White and compared with those included in any non-White response. A chi-square test was used to compare medication sequences between races.
Phenome-wide association of cancer with smoking study
To define ever-smoking exposure from EHR data (EHR ever smoker), we identified all participants with at least two instances on separate calendar days of ICD-9-CM codes 305.1∗ (tobacco use disorder), 649.0∗ (tobacco use disorder complicating pregnancy, childbirth, or the puerperium), V15.82 (history of tobacco use), and 989.84 (toxic effect of tobacco) or ICD-10-CM codes Z72.0 (tobacco Use), Z71.6 (tobacco abuse counseling), O99.33∗ (tobacco use disorder complicating pregnancy, childbirth, and the puerperium), Z87.891 (personal history of nicotine dependence), F17.2∗ (nicotine dependence) excluding F17.22∗ (nicotine Dependence, chewing tobacco), and T65.2∗ (Toxic effect of tobacco and nicotine) excluding T65.21∗ (toxic effect of chewing tobacco). To define never smokers from EHR data (EHR never smoker), zero occurrences of the defining codes above and at least one other ICD-9-CM or ICD-10-CM code that was not T65.21∗ or F17.22∗ was required. To define smoking exposure from survey data (survey ever smoker), the “Lifestyle” survey responses were used. Specifically, the response to “Have you smoked at least 100 cigarettes in your entire life?” was used to include participants as a survey ever smoker, and the branching logic question “Do you now smoke cigarettes every day, some days, or not at all?” with the response “Every day” was used to designate current smokers. Conversely, participants answering “No” to the 100-cigarettes question were included as survey never smokers, and participants skipping the question were excluded. The logistic regression model used in the PheWAS analyses was implemented using the statsmodels Python module, optimized for compute efficiency, and made flexible for reuse with variable inputs. The analysis was corrected for age at last code occurrence, sex at birth, race and ethnicity as generalized from survey responses, EHR length as reflected by time between first and last billing code, and unique billing codes per record.
To compare the phenome-wide associated effects found in the respective PheWAS analyses to prior results, we searched PubMed for meta-analyses that produced effects comparable to the odds ratios produced by the logistic regressions. We first searched PubMed for all meta-analyses related to smoking using the R package easyPubMed and query "(tobacco[TI] OR smoking[TI]) AND meta-analysis[All Fields]") and found 1,840 results. Computable restrictions were then applied including limitation to active smoking-exposed individuals and excluding genetic and smoking-cessation studies, resulting in 538 studies. Manual review then included only those titles with phenotypes represented in the PheCode ontology.36, 37, 38 We then restricted to only those meta-analyses that could be matched with a phenome-wide significant result from at least one of the PheWAS analyses and a comparable effect, finding 51 ever-smoking meta-analyses across 38 unique phenotypes, of which 15 had a phenome-wide significant result that was related to an oncologic outcome with ever-smoking exposure, among phenotypes where there was a PheWAS result in both EHR and PPI. Results were plotted to compare with All of Us results for EHR and survey ever-smoking phenotypes.
Cardiovascular disease risk calculation
Ten-year ASCVD risk was calculated according to the 2013 Pooled Cohort Equations.18 Participants were included if aged 40–79, and the following EHR data were available: TC, HDL, SBP, and treatment status, diabetes status, and no evidence of existing ASCVD. We used the gender that was assigned to the participants at birth. The codes used to identify ASCVD outcome, diabetes, hypertension, and treatment are presented in Table S4. We removed values outside the valid ranges for the scores: TC 130–320, HDL 20–100, and SBP 90–200 mm Hg. Measurements were included within 1 year of the most frequently available variable, SBP. If multiple measures were in the window, the median was used. Current smoking status was taken from the participants’ survey response within “Lifestyle” branching logic to age started smoking and age stopped smoking to determine if smoking occurred during the score calculation. The beta coefficients for African American race were used if the participant selected only Black, African American, or African and White if the participant selected only White in “the Basics” survey. Any other response, multiple responses, or skip was designated as other here, which also uses the White-race beta coefficients in the ASCVD model. To compare risk scores among race groups, the Kruskal-Wallis H and Mann-Whitney U non-parametric tests were used. To calculate the optimal risk, we used 170 for TC, 50 for HDL, 110 for SBP, and status as non-smoker, non-hypertensive, and non-diabetic.
Acknowledgments
The All of Us Research Program is supported (or funded) by grants through the National Institutes of Health, Office of the Director: Regional Medical Centers: 1 OT2 OD026549, 1 OT2 OD026554, 1 OT2 OD026557, 1 OT2 OD026556, 1 OT2 OD026550, 1 OT2 OD 026552, 1 OT2 OD026553, 1 OT2 OD026548, 1 OT2 OD026551, and 1 OT2 OD026555; IAA #: AOD 16037; Federally Qualified Health Centers: HHSN 263201600085U; Data and Research Center: 5 U2C OD023196; Biobank: 1 U24 OD023121; The Participant Center: U24 OD023176; Participant Technology Systems Center: 1 U24 OD023163; Communications and Engagement: 3 OT2 OD023205 and 3 OT2 OD023206; and Community Partners: 1 OT2 OD025277, 3 OT2 OD025315, 1 OT2 OD025337, and 1 OT2 OD025276. In addition to the funded partners, the All of Us Research Program would not be possible without the contributions made by its participants. See Document S1 for additional information on All of Us Research program members and their affiliations.
Author contributions
Conceptualization, A.H.R, M.B., C.W., H.A.-C., E.B., M.C., C.R.C., E.C., L.O.-M., B.K.A., M.A., R.M.C., C.O., M.F., D.B.G., P.G., S.J.H., E.W.K., P.K., B.K., J.W.S., S.D.S., J.W., J.C.D., R.J.C., D.G., P.A.H., G.H., A.P., and D.M.R.; methodology, A.H.R., L.S., D.J.S., and F.R.; formal analysis, L.S., D.J.S., A.H., J.Q., and F.R.; investigation, A.H.R., K.M., J.C.D., and D.M.R.; writing – original draft, A.H.R., L.S., D.J.S., and K.M.; writing – review & editing, A.H.R., J.Q., F.R., R.L., K.M., K.N., A.K., H.X., C.W., H.A.-C., E.B., M.C., C.R.C., E.C., L.O.-M., S.S., B.K.A., M.A., R.M.C., C.O., M.F., D.B.G., P.G., S.J.H., E.W.K., P.K., B.K., J.W.S., S.D.S., J.W., J.H., S.M., C.L., S.A.D., K.G., J.C.D., R.J.C., D.G., P.A.H., G.H., A.P., and D.M.R.; data curation, K.N., A.K., H.X., and R.J.C.; visualization, L.S., D.J.S., A.H., and F.R.; resources, N.D. and K.N.M.; project administration, A.H.R., R.L., K.G., J.C.D., D.G., P.A.H., A.P., and D.M.R.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. We worked to ensure that the study questionnaires were prepared in an inclusive way. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: August 12, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2022.100570.
Contributor Information
Andrea H. Ramirez, Email: andrea.ramirez@nih.gov.
Dan M. Roden, Email: dan.roden@vumc.org.
the All of Us Research Program:
Brian Ahmedani, Christine D. Cole Johnson, Habib Ahsan, Donna Antoine-LaVigne, Glendora Singleton, Hoda Anton-Culver, Eric Topol, Katie Baca-Motes, Steven Steinhubl, James Wade, Mark Begale, Praduman Jain, Scott Sutherland, Beth Lewis, Bruce Korf, Melissa Behringer, Ali G. Gharavi, David B. Goldstein, George Hripcsak, Louise Bier, Eric Boerwinkle, Murray H. Brilliant, Narayana Murali, Scott Joseph Hebbring, Dorothy Farrar-Edwards, Elizabeth Burnside, Marc K. Drezner, Amy Taylor, Veena Channamsetty, Wanda Montalvo, Yashoda Sharma, Carmen Chinea, Nancy Jenks, Mine Cicek, Steve Thibodeau, Beverly Wilson Holmes, Eric Schlueter, Ever Collier, Joyce Winkler, John Corcoran, Nick D’Addezio, Martha Daviglus, Robert Winn, Consuelo Wilkins, Dan Roden, Joshua Denny, Kim Doheny, Debbie Nickerson, Evan Eichler, Gail Jarvik, Gretchen Funk, Anthony Philippakis, Heidi Rehm, Niall Lennon, Sekar Kathiresan, Stacey Gabriel, Richard Gibbs, Edgar M. Gil Rico, David Glazer, Joannie Grand, Philip Greenland, Paul Harris, Elizabeth Shenkman, William R. Hogan, Priscilla Igho-Pemu, Cliff Pollan, Milena Jorge, Sally Okun, Elizabeth W. Karlson, Jordan Smoller, Shawn N. Murphy, Margaret Elizabeth Ross, Rainu Kaushal, Eboni Winford, Febe Wallace, Parinda Khatri, Vik Kheterpal, Akinlolu Ojo, Francisco A. Moreno, Irving Kron, Rachele Peterson, Usha Menon, Patricia Watkins Lattimore, Noga Leviner, Juno Obedin-Maliver, Mitchell Lunn, Lynda Malik-Gagnon, Lara Mangravite, Adria Marallo, Oscar Marroquin, Shyam Visweswaran, Steven Reis, Gailen Marshall, Jr., Patrick McGovern, Deb Mignucci, John Moore, Fatima Munoz, Gregory Talavera, George T. O'Connor, Christopher O'Donnell, Lucila Ohno-Machado, Greg Orr, Fornessa Randal, Andreas A. Theodorou, Eric Reiman, Mercedita Roxas-Murray, Louisa Stark, Ronnie Tepp, Alicia Zhou, Scott Topper, Rhonda Trousdale, Phil Tsao, Lisa Weidman, Scott T. Weiss, David Wellis, Jeffrey Whittle, Amanda Wilson, Stephan Zuchner, and Michael E. Zwick
Supplemental information
Data and code availability
Data and code used in this study are available as a featured workspace to registered researchers of the All of Us Researcher Workbench. For information about access, please visit https://www.researchallofus.org/.
References
- 1.Rutter J.L., Philippakis A., Jenkins G., Smoller J.W., Jenkins G., Dishman E., All of Us Research Program Investigators The “All of Us” Research Program. N. Engl. J. Med. 2019;381:668–676. doi: 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cronin R.M., Jerome R.N., Mapes B., Andrade R., Johnston R., Ayala J., Schlundt D., Bonnet K., Kripalani S., Goggins K., et al. Development of the initial surveys for the all of us research program. Epidemiology. 2019;30:597–608. doi: 10.1097/EDE.0000000000001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramirez A.H., Gebo K.A., Harris P.A. Progress with the all of us research program: opening access for researchers. JAMA. 2021;325:2441–2442. doi: 10.1001/jama.2021.7702. [DOI] [PubMed] [Google Scholar]
- 4.Devaney S. All of us. Nature. 2019;576:S14–S17. doi: 10.1038/d41586-019-03717-8. [DOI] [Google Scholar]
- 5.All of Us Research Hub . 2020. Researcher Workbench.https://www.researchallofus.org/workbench/ [Google Scholar]
- 6.Paten B. A Data Biosphere for Biomedical Research. Medium. 2017 https://medium.com/@benedictpaten/a-data-biosphere-for-biomedical-research-d212bbfae95d [Google Scholar]
- 7.Wilkinson M.D., Dumontier M., Aalbersberg I.J.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.W., Da Silva Santos L.B., Bourne P.E., et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denny J.C., Devaney S.A., Gebo K.A. The “all of us” research program. N. Engl. J. Med. 2019;381:1884–1885. doi: 10.1056/NEJMc1912496. [DOI] [PubMed] [Google Scholar]
- 9.Gaziano J.M., Concato J., Brophy M., Fiore L., Pyarajan S., Breeling J., Whitbourne S., Deen J., Shannon C., Humphries D., et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 10.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M., et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hripcsak G., Ryan P.B., Duke J.D., Shah N.H., Park R.W., Huser V., Suchard M.A., Schuemie M.J., DeFalco F.J., Perotte A., et al. Characterizing treatment pathways at scale using the OHDSI network. Proc. Natl. Acad. Sci. USA. 2016;113:7329–7336. doi: 10.1073/pnas.1510502113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denny J.C., Ritchie M.D., Basford M.A., Pulley J.M., Bastarache L., Brown-Gentry K., Wang D., Masys D.R., Roden D.M., Crawford D.C. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denny J.C., Bastarache L., Ritchie M.D., Carroll R.J., Zink R., Mosley J.D., Field J.R., Pulley J.M., Ramirez A.H., Bowton E., et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 2013;31:1102–1110. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z., Wang Z., Yu Y., Zhang H., Chen L. Smoking is inversely related to cutaneous malignant melanoma: results of a meta-analysis. Br. J. Dermatol. 2015;173:1540–1543. doi: 10.1111/bjd.13998. [DOI] [PubMed] [Google Scholar]
- 15.Pirie K., Beral V., Heath A.K., Green J., Reeves G.K., Peto R., McBride P., Olsen C.M., Green A.C. Heterogeneous relationships of squamous and basal cell carcinomas of the skin with smoking: the UK Million Women Study and meta-analysis of prospective studies. Br. J. Cancer. 2018;119:114–120. doi: 10.1038/s41416-018-0105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song F., Qureshi A.A., Gao X., Li T., Han J. Smoking and risk of skin cancer: a prospective analysis and a meta-analysis. Int. J. Epidemiol. 2012;41:1694–1705. doi: 10.1093/ije/dys146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hripcsak G., Duke J.D., Shah N.H., Reich C.G., Huser V., Schuemie M.J., Suchard M.A., Park R.W., Wong I.C.K., Rijnbeek P.R., et al. Observational health data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud. Health Technol. Inf. 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- 18.Goff D.C., Lloyd-Jones D.M., Bennett G., Coady S., D’Agostino R.B., Gibbons R., Greenland P., Lackland D.T., Levy D., O’Donnell C.J., et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk. Circulation. 2014;129:S49–S73. doi: 10.1161/01.cir.0000437741.48606.98. [DOI] [PubMed] [Google Scholar]
- 19.National Institute of Health Precision Medicine Initiative: Privacy and Trust Principles. 2020. https://allofus.nih.gov/protecting-data-and-privacy/precision-medicine-initiative-privacy-and-trust-principles
- 20.OHDSI OMOP Common Data Model. 2021. https://www.ohdsi.org/data-standardization/the-common-data-model/
- 21.National Institutes of Health All of Us Participant Partners. 2020. https://allofus.nih.gov/about/who-we-are/all-us-participant-partners
- 22.Chakravarthy R., Stallings S.C., Williams M., Hollister M., Davidson M., Canedo J., Wilkins C.H. Factors influencing precision medicine knowledge and attitudes. PLoS One. 2020;15:e0234833. doi: 10.1371/journal.pone.0234833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Althoff K.N., Schlueter D.J., Anton-Culver H., Cherry J., Denny J.C., Thomsen I., et al. Antibodies to SARS-CoV-2 in all of us research program participants, January 2-March 18, 2020. Clin. Infect. Dis. 2021;74:584–590. doi: 10.1093/cid/ciab519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gettinger A., Zayas-Cabán T. HITECH to 21st century cures: clinician burden and evolving health IT policy. J. Am. Med. Inf. Assoc. 2021;28:1022–1025. doi: 10.1093/jamia/ocaa330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Apple Developer HealthKit. 2021. https://developer.apple.com/documentation/healthkit
- 26.HealthIT.gov Sync for Science. 2021. https://www.healthit.gov/topic/sync-science
- 27.Johnson K.B., Wei W.Q., Weeraratne D., Frisse M.E., Misulis K., Rhee K., Zhao J., Snowdon J.L. Precision medicine, AI, and the future of personalized health care. Clin. Transl. Sci. 2021;14:86–93. doi: 10.1111/cts.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denny J.C., Collins F.S. Precision medicine in 2030-seven ways to transform healthcare. Cell. 2021;184:1415–1419. doi: 10.1016/j.cell.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Athena 2021. https://athena.ohdsi.org/search-terms/start
- 30.OHDSI All of us registered tier CDR data dictionary v3 (R2019Q4R3) 2021. https://docs.google.com/spreadsheets/d/1dsvJV8B7EXQj5EWa2XG-KAhs-l7FsQnyJSSFMstLF2U/edit#gid=183931508
- 31.Cybersecurity and Infrastructure Security Agency Federal Information Security Modernization Act. 2021. https://www.cisa.gov/federal-information-security-modernization-act
- 32.Jupyter Project Jupyter. 2021. https://www.jupyter.org
- 33.All of Us Research Hub Data Access and Use. 2021. https://www.researchallofus.org/data-tools/data-access/
- 34.Mapes B.M., Foster C.S., Kusnoor S.V., Epelbaum M.I., AuYoung M., Jenkins G., Lopez-Class M., Richardson-Heron D., Elmi A., Surkan K., et al. Diversity and inclusion for the All of Us research program: a scoping review. PLoS One. 2020;15:e0234962. doi: 10.1371/journal.pone.0234962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.WHOCC ATC/DDD Index. 2021. https://www.whocc.no/atc_ddd_index/
- 36.Carroll R.J., Bastarache L., Denny J.C. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30:2375–2376. doi: 10.1093/bioinformatics/btu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei W.-Q., Bastarache L.A., Carroll R.J., Marlo J.E., Osterman T.J., Gamazon E.R., Cox N.J., Roden D.M., Denny J.C. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One. 2017;12:e0175508. doi: 10.1371/journal.pone.0175508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu P., Gifford A., Meng X., Li X., Campbell H., Varley T., Zhao J., Carroll R., Bastarache L., Denny J.C., et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med. Inform. 2019;7:e14325. doi: 10.2196/14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and code used in this study are available as a featured workspace to registered researchers of the All of Us Researcher Workbench. For information about access, please visit https://www.researchallofus.org/.