Abstract
Objective
Osteoarthritis (OA) is characterized by the chronic and progressive deterioration of articular cartilage. Chondrocyte senescence could lead to a shift in the balance between extracellular matrix (ECM) component synthesis and degradation. Small noncoding RNAs (sncRNAs), including microRNAs (miRNAs), P-element-induced wimpy testis-(PIWI-) interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), and repeat-associated siRNAs (rasiRNAs), are a class of important epigenetic molecules. We aimed to gain insights into the changes and roles of sncRNA in chondrocyte senescence.
Design
Healthy mouse postnatal chondrocytes were isolated, and a replicative aging model was constructed. We used small RNA sequencing (small RNA-seq) to generate extensive small RNA data. We identified differentially expressed sncRNAs and performed tissue-specific analysis using real-time quantitative polymerase chain reaction (qRT-PCR). β-galactosidase staining was used to detect chondrocyte senescence. The results showed that the expression profiles of sncRNA in passage 5 chondrocytes were significantly different from those in passage 0 chondrocytes. The expression of sncRNA was tissue specific. We found that 40 miRNAs were upregulated and 70 miRNAs were downregulated during chondrocyte senescence, and that miR-132-5p expression inhibition prevented chondrocyte senescence. We found that 8 piRNAs were upregulated and 17 piRNAs were downregulated during chondrocyte senescence, and that piRNA piR_025576 overexpression delayed chondrocyte senescence. We found that 24 snoRNAs were upregulated and 28 snoRNAs were downregulated during chondrocyte senescence, and that snoRNA ENSMUSG00000087935 overexpression delayed chondrocyte senescence. We found that 5 snRNAs were upregulated and 6 snRNAs were downregulated during chondrocyte senescence, and that snRNA ENSMUSG00000064682 overexpression delayed chondrocyte senescence. We found that 1 rasiRNA was upregulated and 4 rasiRNAs were downregulated during chondrocyte senescence.
Conclusions
These findings might provide novel insights into OA pathogenesis and contribute to the development of candidates for targeted therapeutics in OA.
Keywords: chondrocyte senescence, miRNA, piRNA, small nuclear RNAs, small nucleolar RNAs, RasiRNAs
Introduction
Osteoarthritis (OA) is a prevalent multiple factor-associated disorder that is characterized by chronic and progressive deterioration of articular cartilage. Several risk factors for the development of OA exist, including prior joint injury, obesity, diabetes mellitus, hyperglycemia, and anatomical factors related to joint shape and alignment; however, the most prevalent risk factor is increasing age.1-3 Cellular senescence, one of the hallmarks of aging, is a form of irreversible cellular arrest characterized by senescence-associated beta-galactosidase (SA-β-Gal) activity and the release of harmful proinflammatory molecules, growth factors, and matrix metalloproteinases as part of the senescence-associated secretory phenotype (SASP). 4 As the accumulation of senescent cells reduces cellular proliferation and disordered tissue function and metabolism, senescence has been implicated in the pathogenesis and/or progression of a variety of aging-associated diseases, including OA.3,5
Although OA has recently been regarded as a whole joint disease rather than merely dysfunctional cartilage, chondrocytes are primarily known to have a major role in the pathology of OA.5-8 Chondrocytes, the unique resident cell type in articular cartilage, regulate anabolic and catabolic pathways to maintain cartilage homeostasis. The senescence of chondrocytes is expected to lead to a shift in the balance between extracellular matrix (ECM) component synthesis and degradation through metalloproteinase components (i.e., MMP1, MMP2, MMP3, MMP10, MMP13, and more) of the SASP response.5,9 There was a causal relationship between the occurrence and development of OA, which was significantly correlated with age and chondrocyte aging, but the more specific pathological mechanism has not been fully clarified. Studies have also confirmed that the expression of SA-β-Gal in articular cartilage is positively correlated with the severity of OA. 10 The increase in the positive rate of aging chondrocytes reduced the repair ability of cartilage, aggravated cartilage degeneration, and led to an increase in the incidence rate of OA. Understanding the molecular mechanism of chondrocyte aging is of great significance for the treatment of OA and the development of new therapies.
The exact mechanisms through which senescence can affect cartilage health remain elusive, although it is believed to be caused by multiple molecular mechanisms rather than a single etiology. Recent studies have shown that small noncoding RNAs (sncRNAs), a class of important epigenetic regulatory molecules, play an important role in regulating cellular senescence in OA.11-14 SncRNAs are short RNA species, typically fewer than 200 nucleotides in length, that are not translated into proteins but have other structural or regulatory biological roles. 15 These include microRNAs (miRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), P-element-induced wimpy testis (PIWI-)-interacting RNAs (piRNAs), small interfering RNAs (siRNAs), transfer RNAs (tRNAs), and repeat-associated siRNAs (rasiRNAs). SncRNAs are promising candidates for targeted therapeutics in OA due to their small size, and their activity can be modulated via small molecules and biological delivery systems.15,16
In the present study, primary mouse chondrocytes were isolated and subjected to 5 additional passages to simulate chondrocyte replicative senescence in vitro. To gain insights into sncRNA changes in chondrocyte senescence, we used small RNA sequencing (small RNA-seq) to generate extensive small RNA data. We identified differentially expressed sncRNAs and performed tissue-specific analysis. These findings might provide novel insight into OA pathogenesis and contribute to the development of candidates for targeted therapeutics in OA.
Materials and Methods
Cell Isolation and Culture
Mouse primary articular chondrocytes were isolated from the femoral heads, femoral condyles, and tibial plateaus of mice at postnatal day 5 as previously described. 17 The animal study was reviewed and approved by the Ethics Committee of Xinhua Hospital affiliated with Shanghai Jiao Tong University School of Medicine. Cartilage was minced into small pieces. After digestion with 0.25% trypsin and collagenase D solution at 0.5 mg/ml, chondrocytes were collected and cultured in Dulbecco’s modified Eagle medium DMEM (1 g glucose per ml; Sigma, cat. no. D5546) supplemented with 10% fetal bovine serum (FBS) (Gibco, Thermo Fisher Scientific, USA) and 1% penicillin-streptomycin. Chondrocytes at passages 0 to 5 were used in all experiments.
SA-β-Gal Staining
The Senescence Detection Kit (BioVision, San Francisco, CA, USA) was used to stain SA-β-Gal. Chondrocytes at passages 1 and 5 were rinsed in phosphate-buffered saline (PBS). After fixation with fixative solution at room temperature for 15 minutes, the cells were washed twice with PBS. Next, the cells were added to a staining solution mixture and incubated overnight at 37°C. Chondrocytes were then washed twice with PBS, and the senescent cells stained blue were analyzed under a light microscope.
qRT-PCR Analysis
Total RNA was extracted from cells using an RNeasy Mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions, and cDNA was synthesized from total RNA (1 µg) using reverse transcriptase (TaKaRa Biotechnology, Otsu, Japan). The complementary DNA (cDNA; 10 ng) products were used as the template for amplification using SYBR Green PCR Master Mix (TaKaRa Biotechnology). Furthermore, mRNA expression was detected by an ABI 7500 sequencing detection system (Applied Biosystems, Foster City, CA, USA). The primer lists are shown in Supplementary Material S1.
Small RNA-Seq
The original image data file obtained by high-throughput sequencing was transformed into the original sequenced reads by base calling analysis. The results are stored in FASTQ (FQ) file format, which contains the sequence information of sequenced reads and their corresponding sequencing quality information. First, the original data were preprocessed, joint sequences were removed, low-quality sequences were filtered, and then the FASTQ format was converted to a FASTA format. After data preprocessing, we used Bowtie software to compare the sequences to the reference genome, and only the sequences aligned to the reference genome were selected for subsequent analysis. To comprehensively predict noncoding RNAs of different lengths, we first extracted the sequencing sequences aligned to the reference genome in each sample and then clustered the sequences of all samples using cd-hit to remove the same or similar sequences. Then, we used infernal software to search the RFAM database. After searching, we matched the small RNA sequence in the RFAM database and then compared the sequencing data of each sample to count the read count of small RNA of each sample. The read count was additionally subjected to TPM standardization to calculate the proportion of each million reads in the comparison. For the experiments with biological repetition, we used DESeq for the analysis. For experiments without biological duplication, we used edger for the analysis. Finally, the genes with Padj less than 0.05 and a difference multiple greater than or equal to 2 were selected as small RNAs with significant differential expression.
Statistical Analysis
Statistical analysis was performed using IBM SPSS Statistics 24, and graphs were generated by GraphPad Prism 9. Independent sample t tests were used to analyze the mRNA expression results from the qPCR assays. P values less than 0.05 were considered significant.
Results
Elevation of Cell Senescence with Increasing Passages
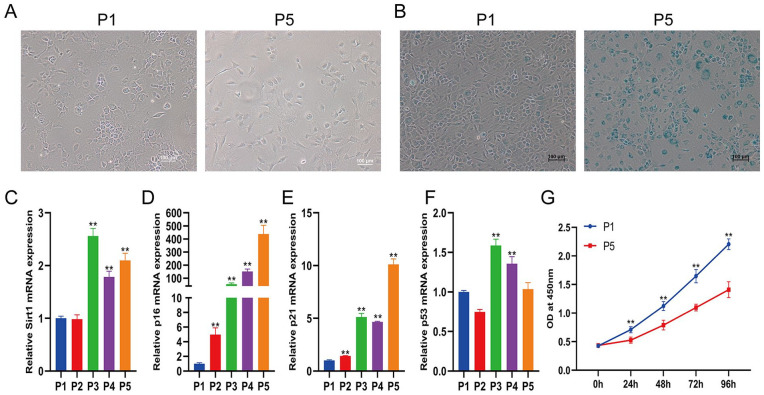
First, the morphological changes in the chondrocytes were recorded. Mouse cartilage chondrocytes were isolated and continuously subcultured to passage 5. At passage 1, chondrocytes had a characteristic tetris-shaped, smooth cell surface, and grew regularly. Unlike the cells at passage 1, almost all chondrocytes at passage 5 exhibited irregular shapes with pseudopods and became larger and flatter ( Fig. 1A ). The SA-β-Gal activity test is usually performed for the identification of senescent cells. The senescence status of chondrocytes at passage 5 was confirmed by the SA-β-Gal staining assay, which showed increased lysosomal β-galactosidase activity specifically in senescent cells ( Fig. 1B ). To further evaluate senescence at the molecular level, the mRNA expression levels of the cellular senescence-related proteins Sirt1, p16, p21, and p53 were detected at different passages. The mRNA expression of Sirt1, p16, p21, and p53 also showed increasing trends as the passage number increased ( Fig. 1C-F ). The proliferation rate of chondrocytes at passage 5 was significantly decreased compared with that of chondrocytes at passage 1 ( Fig. 1G ). These results suggest that chondrocytes undergo replicative senescence during in vitro amplification.
Figure 1.
Elevation of cell senescence with increasing passages. (A) The morphology of chondrocytes at passages 1 and 5. The scale bar represents 100 μm. (B) β-Gal staining of chondrocytes at passages 1 and 5. The scale bar represents 100 μm. Real-time quantitative polymerase chain reaction analysis of the mRNA expression of Sirt1 (C), p16 (D), p21 (E), and p53 (F) in chondrocytes from passages 1 to 5. (C) CCK-8 detected the cell viability of chondrocytes at passages 1 and 5. CCK-8 = Cell Counting Kit-8. ** indicates P < 0.01 compared to passage 1.
Identification of Differentially Expressed miRNAs and Tissue-Specific Analysis
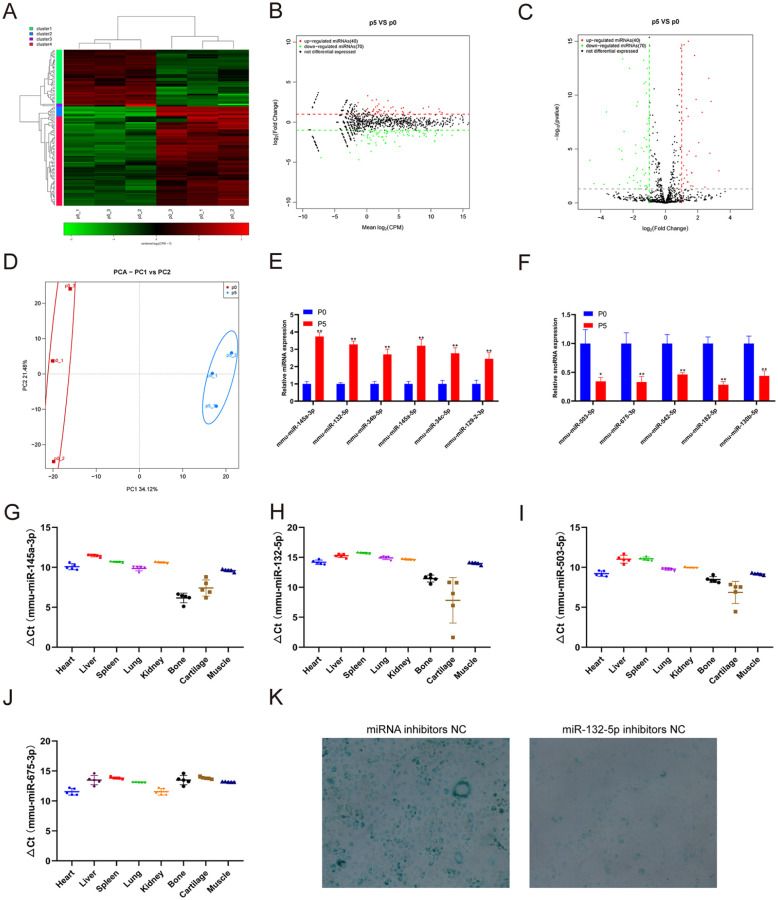
As important epigenetic regulators, miRNAs bind to specific regions of target mRNAs, resulting in reduced protein expression and thus participating in many cellular processes. We performed small RNA-seq to analyze the miRNA expression profiling change in replicative senescence chondrocytes. The small RNA-seq results showed that 40 miRNAs were upregulated and 70 miRNAs were downregulated in chondrocytes at passage 5 compared with those at passage 0 ( Fig. 2A-D ). Then, we further verified the expression of several miRNAs with the greatest difference in the sequencing results using qRT-PCR. Consistent with the sequencing results, the qRT-PCR results showed that the expression of miR-145a-3p, miR-132-5p, miR-34b-5p, miR-145a-5p, miR-34c-5p and miR-129-3p was increased in chondrocytes at passage 5 compared with that at passage 0 ( Fig. 2E ). Moreover, the expression of miR-503-5p, miR-675-3p, miR-542-5p, miR-182-5p, and miR-130b-5p was decreased in chondrocytes at passage 5 compared with that at passage 0 ( Fig. 2F ). We also detected the expression of miR-145a-3p, miR-132-5p, miR-503-5p, and miR-675-3p in different mouse tissues ( Fig. 2G-J ). β-galactosidase staining showed that inhibiting the expression of miR-132-5p in chondrocytes could effectively inhibit the aging of chondrocytes ( Fig. 2K ).
Figure 2.
Identification of differentially expressed miRNAs and tissue-specific analysis. (A) Heatmap showing the differentially expressed miRNAs between chondrocytes at passages 1 and 5. (B) MA showing the differentially expressed miRNAs between chondrocytes at passages 1 and 5. (C) Volcano plot showing the differentially expressed miRNAs between chondrocytes at passages 1 and 5. (D) Principal component analysis showing the differentially expressed miRNAs between chondrocytes at passage 1 and passage 5. (E) qRT-PCR analysis of miRNA expression in chondrocytes at passage 1 compared to that at passage 5. (F) qRT-PCR analysis of miRNA expression in chondrocytes within passage 1 compared to that at passage 5. qRT-PCR analysis of (G) mmu-miR-145a-3p, (H) mmu-miR-132-5p, (I) mmu-miR-503-5p, and (J) mmu-miR-675-3p in different tissues. (K) β-Gal staining of chondrocytes at passage 5 with miRNA inhibitor NC or miR-132-5p inhibitor. qRT-PCR = real-time quantitative polymerase chain reaction; MA = minus-average; NC = negative control; CPM = count per million; PCA = principle component Analysis. **indicates P < 0.01 compared to passage 1.
Identification of Differentially Expressed piRNAs and Tissue-Specific Analysis
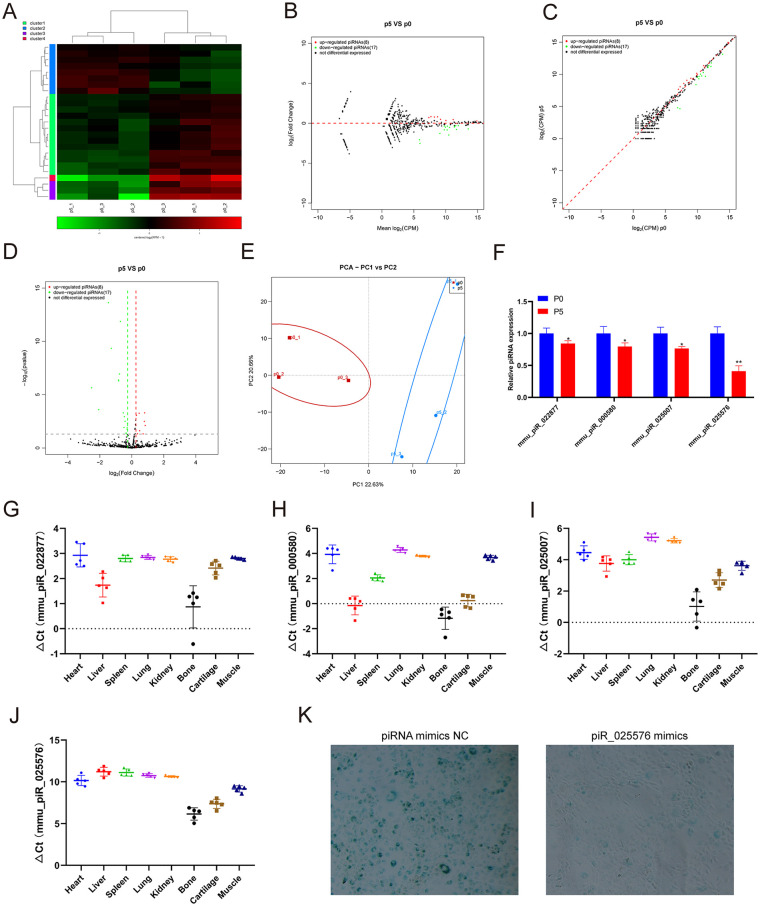
Compared with miRNAs, piRNAs are a relatively new type of sncRNA and have attracted increasing attention in recent years. piRNAs, which have a length of 24-31 nucleotides, play regulatory roles, such as silencing the transcriptional gene process, regulating mRNA stability, interacting with multiple proteins, and maintaining stem cell functions. 18 We performed small RNA-seq to analyze piRNA expression profiling changes in chondrocytes undergoing replicative senescence. The small RNA-seq results showed that 8 piRNAs were upregulated and 17 piRNAs were downregulated in chondrocytes at passage 5 compared with those at passage 0 ( Fig. 3A-D ). Principal components analysis (PCA) showed that the aging of chondrocytes was closely related to the expression of piRNAs ( Fig. 3E ). Then, we further verified the expression of the differentially expressed piRNAs using qRT-PCR. Consistent with the sequencing results, the qRT-PCR results showed that the expression of piR_022877, piR_000580, piR_025007, and piR_025576 was decreased in chondrocytes at passage 5 compared with those at passage 0 ( Fig. 3F ). However, no piRNA was verified to be upregulated in chondrocytes at passage 5 using qRT-PCR. We also detected the expression of piR_022877, piR_000580, piR_025007, and piR_025576 in different mouse tissues ( Fig. 3G-J ). β-galactosidase staining showed that piRNA piR_025578 overexpression in chondrocytes could effectively inhibit the aging of chondrocytes ( Fig. 3K ).
Figure 3.
Identification of differentially expressed piRNAs and tissue-specific analysis. (A) Heatmap showing the differentially expressed piRNAs between chondrocytes at passages 1 and 5. (B) MA showing the differentially expressed piRNAs between chondrocytes at passages 1 and passage 5. (C) Scatter plot showing the differentially expressed piRNAs between chondrocytes at passages 1 and 5. (D) Volcano plot showing the differentially expressed piRNAs between chondrocytes at passages 1 and 5. (E) Principal components analysis showing the differentially expressed piRNAs between chondrocytes at passages 1 and 5. (F) qRT-PCR analysis of piRNA expression in chondrocytes within passage 1 compared to that at passage 5. qRT-PCR analysis of (G) mmu_piR_022877, (H) mmu_piR_000580, (I) mmu_piR_025007, and (J) mmu_piR_025576 in different tissues. (K) β-Gal staining of chondrocytes at passage 5 with piRNA mimics NC or piR_025576 mimics. qRT-PCR = real-time quantitative polymerase chain reaction; MA = minus-average; pLVX = plasmid lentiviral vector X; CPM = count per million; PCA = principle component analysis. * indicates P < 0.05 and ** indicates P < 0.01 compared to passage 1.
Identification of Differentially Expressed snoRNAs and Tissue-Specific Analysis
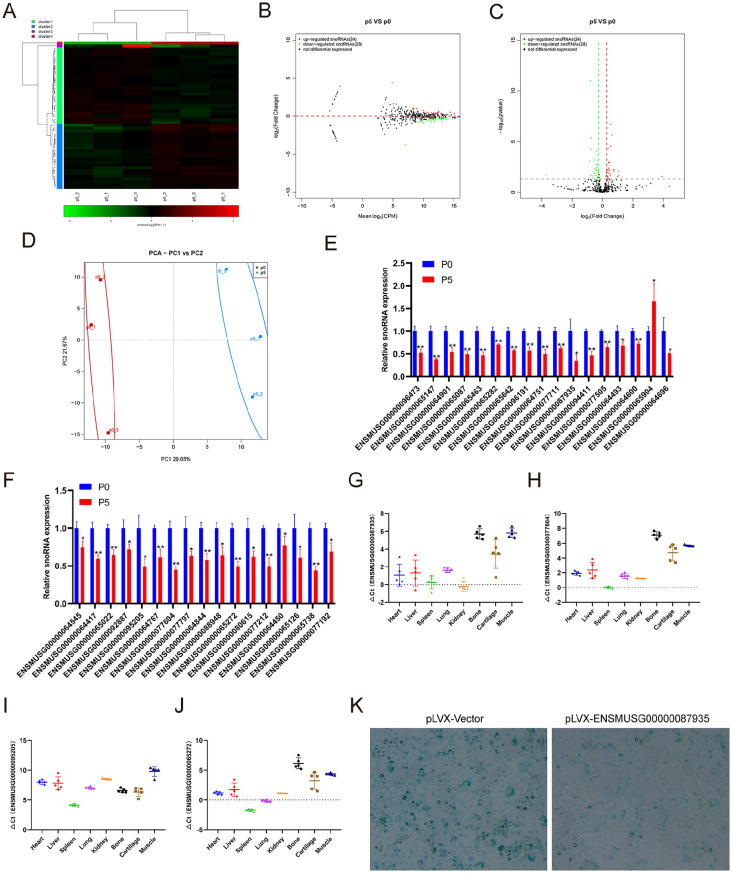
SnoRNAs direct chemical modification of other RNA substrates to fine-tune spliceosomal splicing and are mainly involved in endoribonucleolytic pre-rRNA processing. 19 The role of snoRNAs in cartilage aging has attracted increasing attention. 12 We performed small RNA-seq to analyze the snoRNA expression profiling change in replicative senescent chondrocytes. The small RNA-seq results showed that 24 snoRNAs were upregulated and 28 snoRNAs were downregulated in chondrocytes at passage 5 compared with those at passage 0 ( Fig. 4A-C ). PCA showed that the aging of chondrocytes was closely related to the expression of snoRNA ( Fig. 4D ). Then, we further verified the expression of several snoRNAs with significant differences in the sequencing results using qRT-PCR. Consistent with the sequencing results, the qRT-PCR results showed that the downregulated snoRNAs were decreased in chondrocytes at passage 5 compared with those at passage 0 ( Fig. 4E ). However, only ENSMUSG00000065904 was verified to be upregulated, which was consistent with the sequencing results. The other 16 snoRNAs with upregulated sequencing results were verified to have decreased expression ( Fig. 4F ). We also detected the expression of ENSMUSG00000087935, ENSMUSG00000077604, ENSMUSG00000095205, and ENSMUSG00000065272 in different mouse tissues ( Fig. 4G-J ). β-galactosidase staining showed that overexpression of the snoRNA ENSMUSG00000087935 in chondrocytes effectively inhibited the aging of chondrocytes ( Fig. 4K ).
Figure 4.
Identification of differentially expressed snoRNAs and tissue-specific analysis. (A) Heatmap showing the differentially expressed snoRNAs between chondrocytes at passages 1 and 5. (B) MA showing the differentially expressed snoRNAs between chondrocytes at passages 1 and 5. (C) Volcano plot showing the differentially expressed snoRNAs between chondrocytes at passages 1 and 5. (D) Principal component analysis showing the differentially expressed snoRNAs between chondrocytes at passages 1 and 5. (E) qRT-PCR analysis of snoRNA expression in chondrocytes at passage 1 compared with that at passage 5. (F) qRT-PCR analysis of snoRNA expression in chondrocytes at passage 1 compared with that at passage 5. qRT-PCR analysis of (G) ENSMUSG00000087935, (H) ENSMUSG00000077604, (I) ENSMUSG00000095205, and (J) ENSMUSG00000065272 in different tissues. (K) β-Gal staining of chondrocytes at passage 5 with pLVX-Vecotor or pLVX-ENSMUSG00000087935. qRT-PCR = real-time quantitative polymerase chain reaction; MA = minus-average; pLVX = plasmid lentiviral vector X; CPM = count per million; PCA = principle component analysis. * indicates P < 0.05 and ** indicates P < 0.01 compared with passage 1.
Identification of Differentially Expressed snRNAs and Tissue-Specific Analysis
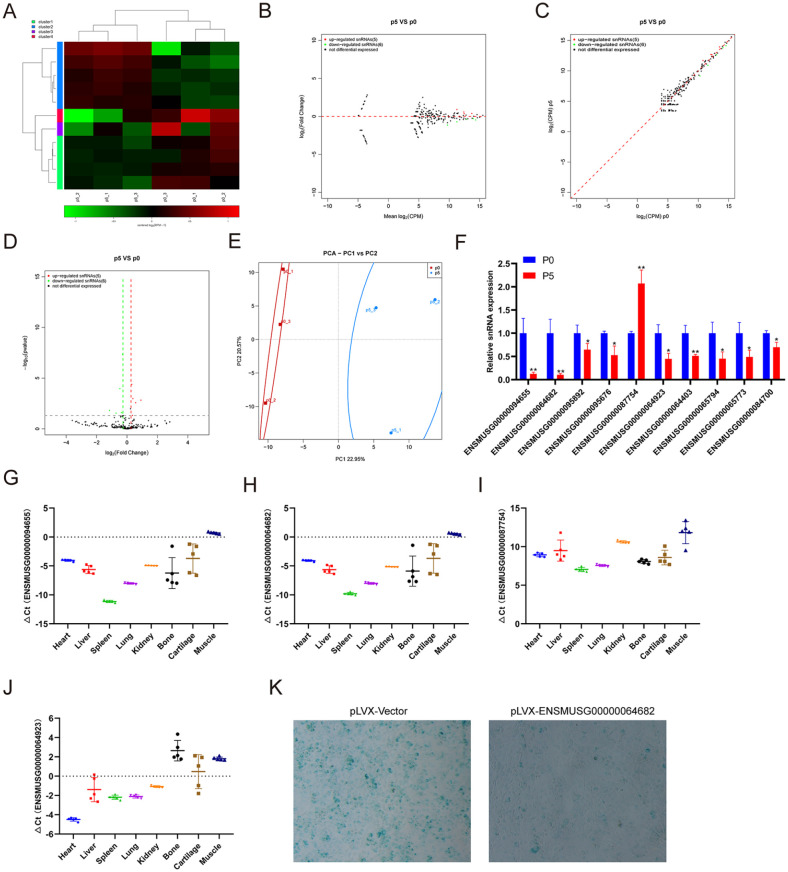
SnRNA is the main component of RNA spliceosomes in the posttranscriptional processing of eukaryotes, with a length of 100-215 nucleotides. At present, there is no literature on snRNAs directly related to chondrocytes or cartilage. We performed small RNA-seq to analyze the snRNA expression profiling change in chondrocytes undergoing replicative senescence. The small RNA-seq results showed that 5 snRNAs were upregulated and 6 snRNAs were downregulated in chondrocytes at passage 5 compared with those at passage 0 ( Fig. 5A-D ). PCA showed that the aging of chondrocytes was closely related to the expression of snoRNA ( Fig. 5E ). Then, we further verified the expression of several snRNAs with significant differences in the sequencing results using qRT-PCR. Consistent with the sequencing results, the qRT-PCR results showed that the expression of nine snRNAs was decreased and the expression of one snRNA was increased in chondrocytes at passage 5 compared with those at passage 0 ( Fig. 5F ). The expression of ENSMUSG00000064923, ENSMUSG00000064403, ENSMUSG00000065794, and ENSMUSG00000065773 was consistent with the sequencing results. We also detected the expression of ENSMUSG00000094655, ENSMUSG00000064682, ENSMUSG00000087754, and ENSMUSG00000064923 in different mouse tissues ( Fig. 5G-J ). β-galactosidase staining showed that snRNA ENSMUSG00000064682 overexpression in chondrocytes could effectively inhibit the aging of chondrocytes ( Fig. 5K ).
Figure 5.
Identification of differentially expressed snRNAs and tissue-specific analysis. (A) Heatmap showing the differentially expressed snRNAs between chondrocytes at passages 1 and 5. (B) MA showing the differentially expressed snRNAs between chondrocytes at passages 1 and 5. (C) Scatter plot showing the differentially expressed snRNAs between chondrocytes at passages 1 and 5. (D) Volcano plot showing the differentially expressed snRNAs between chondrocytes at passages 1 and 5. (E) Principal components analysis showing the differentially expressed snRNAs between chondrocytes at passages 1 and 5. (F) qRT-PCR analysis of snRNA expression in chondrocytes at passage 1 compared with that at passage 5. qRT-PCR analysis of (G) ENSMUSG00000094655, (H) ENSMUSG00000064682, (I) ENSMUSG00000087754, and (J) ENSMUSG00000064923 in different tissues. (K) β-Gal staining of chondrocytes at passage 5 with pLVX-Vecotor or pLVX-ENSMUSG000000648682. qRT-PCR = real-time quantitative polymerase chain reaction. * indicates P < 0.05 and ** indicates P < 0.01 compared with passage 1.
Identification of Differentially Expressed rasiRNAs
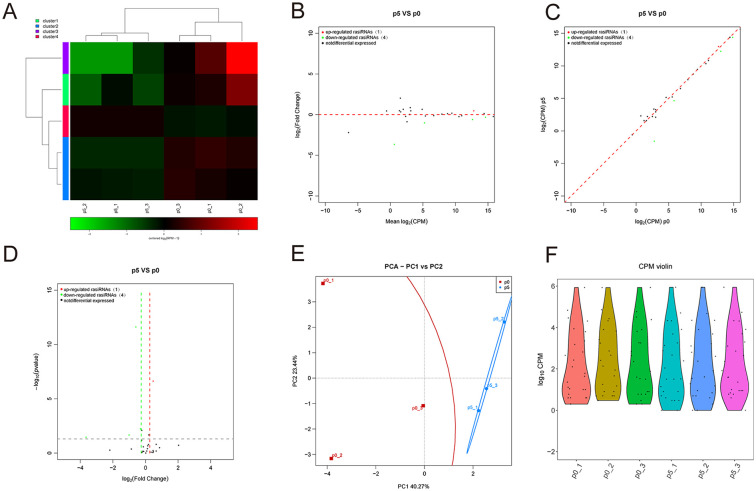
RasiRNAs of 24-28 nucleotides in length may assume a role in sperm confrontation and consolidation. 20 Unfortunately, there are no studies in the literature focusing on repeat RNAs that are directly related to chondrocytes or cartilage. We performed small RNA-seq to analyze the repeat RNA expression profiling change in replicative senescent chondrocytes. The small RNA-seq results showed that only one repeat RNA was upregulated, and 4 repeat RNAs were downregulated in chondrocytes at passage 5 compared with passage 0 ( Fig. 6A-D ). PCA showed that the aging of chondrocytes was closely related to the expression of rasiRNA ( Fig. 6E ). There was no significant difference in total rasiRNA count per million (CPM) in senescent chondrocytes ( Fig. 6F ).
Figure 6.
Identification of differentially expressed rasiRNAs and tissue-specific analysis. (A) Heatmap showing the differentially expressed rasiRNAs between chondrocytes at passages 1 and 5. (B) MA showing the differentially expressed rasiRNAs between chondrocytes at passages 1 and 5. (C) Scatter plot showing the differentially expressed rasiRNAs between chondrocytes at passages 1 and 5. (D) Volcano plot showing the differentially expressed rasiRNAs between chondrocytes at passages 1 and 5. (E) Principal components analysis showing the differentially expressed rasiRNAs between chondrocytes at passages 1 and 5. (F) CPM violin plot showing the differentially expressed rasiRNAs between chondrocytes at passages 1 and 5. MA =Minus-Average, NC = Negative control, CPM = Count per million, PCA = Principle Component Analysis.
Discussion
Cellular senescence is a driver of various aging-associated disorders, including OA. OA chondrocytes exhibit features that are similar to senescent cells, such as specific senescent markers, telomere shortening, and increased SA-β Gal activity.21,22 Exploring anti-cellular senescence strategies may be a promising approach to prevent or cure OA. The exact mechanism of chondrocyte senescence remains elusive, although two different mechanisms of senescence have been suggested in chondrocytes, including stress-induced premature senescence (SIPS) and replicative senescence.23,24 It was concluded that replicative senescence contributes to either the development or the progression of OA. 21
SncRNAs, a large class of regulatory molecules, are involved in organism development and coordination of biological processes, including metabolism, maintaining genome integrity, and immune and stress responses. 25 Recent studies have suggested that sncRNAs play an important role in regulating cellular senescence in OA.11-14 The aim of this study was to systematically investigate the sncRNA profile changes in senescent chondrocytes after long-term expansion in vitro. We found that mouse chondrocytes at passage 5 showed obvious senescence characteristics, including increased SA-β Gal activity and senescence-related gene expression. Moreover, the expression profiles of miRNAs, snRNAs, snoRNAs, piRNAs, and rasiRNAs in passage 5 chondrocytes were significantly different from those in passage 0 chondrocytes.
MiRNAs, the most studied sncRNAs, exert their negative regulatory functions by partially or fully binding complementary sequences in the 3ʹ untranslated region of their messenger RNA (mRNA) targets, thus inhibiting mRNA translation. Our results showed that 6 miRNAs were upregulated and 5 miRNAs were downregulated ( Fig. 2G and H). Some differentially expressed miRNAs have been reported to be related to chondrocyte metabolism or cellular senescence. For example, miR-145a-3p and miR-145a-5p were upregulated at passage 5. It has been reported that miR-145 contributes to impaired ECM in OA cartilage in part by targeting Smad3 26 and negatively regulates tumor necrosis factor (TNF)-α-mediated signaling activation and the induction of cartilage matrix degradation mechanically through the MKK4-JNK/p38-c-Jun/ATF2 axis during OA pathogenesis. 27 In addition, miR-145 plays a role in regulating cellular senescence; for example, miR-145 induces the senescence of activated hepatic stellate cells through the activation of the p53 pathway by ZEB2. 28 The expression of miR-145a-3p in the saliva of patients with Parkinson’s disease, which is an aging-related disease, was significantly increased. 29 The expression of miR-34b-5p in aged skin was significantly increased, and the overexpression of miR-34b-5p in skin fibroblasts resulted in cell cycle arrest. 30 MiR-34b-5p plays an important role in the process of mouse vascular aging. 31 On the other hand, miR-34b-5p may be a biomarker for poststroke social isolation of aged mice. 32 MiR-34c-5p promoted ultraviolet irradiation-induced senescence of epidermal fibroblasts. 33 MiR-34c-5p inhibits the progression of leukemia by inducing the senescence of myeloid leukemia stem cells. 34 MiR-145a-3p, miR-34b-5p, and miR-34c-5p were also highly expressed in aging chondrocytes. A recent study suggested that miR-503-5p was downregulated in the cartilage tissues of OA patients compared with normal people and that HDAC2 could promote OA through the miR-503-5p/SGK1 axis, while overexpression of miR-503-5p resulted in promoted proliferation and reduced apoptosis of rat primary chondrocytes. 35 In addition, miR-675-3p expression levels were significantly decreased in OA patient cartilage and interleukin (IL)-1β-treated chondrocytes, while overexpression of miR-675-3p inhibited IL-1β-stimulated apoptosis, matrix degradation and inflammation in chondrocytes by targeting GNG5. 36 The results of the present study showed that the expression of miR-503-5p decreased in senescent chondrocytes, which implies that miR-503-5p might play an important role in regulating chondrocyte senescence. Since the function of the other differentially expressed miRNAs in senescent chondrocytes remains unclear, further studies are needed.
PiRNAs are a type of small RNA with a length of approximately 24-13 nucleotides. The precursors of piRNAs are derived from tandem repeat sequences called piRNA clusters and form a mature piRNA/PIWI complex via two route-dependent or independent “Ping-Pong” amplification pathways. 18 In the past, piRNAs were thought to exist only in the reproductive system to regulate the growth and development of germ cells. Recently, it was reported that piRNAs are also expressed in several other human tissues with tissue specificity. 13 PiRNAs play roles in transcriptional or posttranscriptional gene silencing pathways by combining Piwi subfamily proteins to form piRNA complexes. At present, piRNA research mainly focuses on its regulation of the occurrence and development of cancer, including colorectal cancer, breast cancer and lung cancer. 13 To our knowledge, there are no reports on the relationship between piRNAs and OA or cartilage, and the role of piRNAs in chondrocytes is unknown. Our results showed that the expression of piR_022877, piR_000580, piR_025007, and piR_025576 was decreased in chondrocytes at passage 5 compared with passage 0, while no piRNA was verified to upregulate piR_022877, piR_000580, piR_025007, and piR_025576 ( Fig. 3G ). An increasing number of epigenetic studies have shown that epigenetic regulation of gene expression plays an important role in the pathological mechanism of chondrocyte aging and OA. 37 The aging process of OA chondrocytes involves an increase in the expression of a variety of matrix-degrading enzymes. The demethylation of the promoter sites of various matrix-degrading enzymes, such as MMP-3, MMP-9, and ADAMTS-4, is related to their enhanced expression in OA. 38 PiRNA is involved in the regulation of DNA methylation. 39 PiRNA-823 promotes the proliferation of myeloma by regulating DNA methylation. 40 PiRNA-6426 promotes the methylation of SOAT1 and heart repair. 41 We also detected piRNAs in multiple tissues or organs ( Fig. 3H-K ), although their specific functions need to be further elucidated. The results showed that piR_000580 and piR_025576 were highly expressed in cartilage and may be involved in the regulation of DNA methylation modification in chondrocytes.
SnoRNAs, a class of evolutionarily conserved noncoding small guide RNAs with lengths of 60-300 nucleotides, are extensively studied noncoding RNAs that primarily accumulate in the nucleoli. 42 SnoRNAs are involved in the direct chemical modification of other RNA substrates and the regulation of alternative splicing and posttranscriptional modification of snRNA, tRNA, and mRNA, while others exhibit miR-like activity. 43 Recently, snoRNAs have been increasingly studied in aging and OA. A previous mouse study demonstrated alterations in the snoRNA profile of young compared with old and OA compared with healthy controls in joints and serum, highlighting the potential of snoRNAs to be used as novel markers for OA and aging. 44 In human cartilage, snoRNAs are differentially expressed due to aging (including SNORD96A and SNORD44) and OA (including SNORD26 and SNORD116). 12 Altering SNORD26 or SNORD96A expression resulted in changes in chondrogenic-, hypertrophic-, rRNA- and OA-related gene expression. 12 U3, one of the most abundant snoRNAs playing a key role in ribosome biogenesis, was found to have significantly lower expression in human OA cartilage and chondrocytes. The protein translational capacity of chondrocyte cultures with diminished U3 snoRNA expression was significantly reduced, while ectopic induction of U3 snoRNA expression resulted in increased translational capacity. 45 In the present study, we detected 17 downregulated snoRNAs in chondrocytes at passage 5 compared with passage 0 ( Fig. 4H ). However, only ENSMUSG00000065904 was verified to be upregulated, which was consistent with the sequencing results. The other 16 snoRNAs with upregulated sequencing results were verified to have decreased expression.
SnRNA is the main component of the RNA spliceosome in the posttranscriptional processing of eukaryotes and participates in splicing pre-mRNA and noncoding transcripts in cells. In humans, its length is approximately 60-330 nucleotides, and it can be divided into 14 categories. U1-U7 are rich in uridylate. U3 snRNA is related to the maturation of 28S rRNA in the nucleolus, while U1 is related to the splicing and processing of precursor mRNA in the nucleus. Rn7SK is a highly conserved snRNA transcribed by RNA polymerase III and is a regulator of Pol II activity through inhibiting the function of positive transcriptional elongation factor b. A recent study demonstrated that Rn7SK participates in the regulation of cellular senescence, with the transient knockdown of Rn7SK in mesenchymal stem cells (MSCs) leading to delayed senescence, while its overexpression shows the opposite effects. 46 However, the role of snRNA in chondrocytes and its relationship with OA have not yet been reported. We found that the expression of snRNA ENSMUSG00000087754 increased and the other 9 snRNAs decreased at passage 5 ( Fig. 5G ). The function and potential targets of these snRNAs in chondrocytes will be explored and verified in our subsequent studies. However, many sequencing results for snoRNA and snRNA are inconsistent with the expression trends observed via qRT-PCR. We speculate that there may be many stem-loop structures in snoRNA and snRNA, which will affect the sequencing or qRT-PCR results.
At present, the treatment of OA is still a clinical challenge, and the understanding of the molecular mechanism is still unclear. SncRNAs are promising candidates for targeted therapeutics in OA due to their small size, and their activity can be modulated via small molecules and biological delivery systems.15,16 However, the study of sncRNAs in the aging process of chondrocytes is not sufficient. Anti-senescence therapy is a frequently discussed clinical issue, and research into this topic involves new drug research, drug-target protein detection, and disease mechanism exploration. Through screening, we will be able to verify the target genes of the above differentially expressed sncRNAs in subsequent in vivo and in vitro studies. The sequencing results showed that rasiRNA was differentially expressed in senescent chondrocytes. However, due to the scarcity of research on rasiRNA and its short length, it is difficult to verify the sequencing results by conventional real-time PCR. More studies are needed to explore the role of rasiRNA in chondrocyte aging.
Conclusion
In this study, we detected the expression changes in miRNAs, piRNAs, snoRNAs, snRNAs and repeat RNAs during chondrocyte aging by high-throughput small RNA sequencing and detected the expression of miRNAs, piRNAs, snoRNAs, and snRNAs during chondrocyte aging by qRT-PCR. We found that the expression of miRNA, piRNA, snoRNA, snRNA, and repeat RNA changes significantly during chondrocyte aging and is tissue specific.
Supplemental Material
Supplemental material, sj-xlsx-1-car-10.1177_19476035221118165 for Changes in Small Noncoding RNA Expression during Chondrocyte Senescence by Fei Xiao, Chenglong Wang, Jianping Peng, Xing Zhou, Ding Ma, Yu Wang, Yanpeng Li, Xiaodong Chen and Chuandong Wang in CARTILAGE
Footnotes
Author Contributions: Conceptualization: Xiaodong Chen and Chuandong Wang; methodology: Fei Xiao, Chenglong Wang and Jianping Peng; software: Yanpeng Li, Xing Zhou, Ding Ma and Yu Wang; data curation: Chuandong Wang and Fei Xiao; writing, review, and editing: Fei Xiao and Chao Shen, Junfeng Zhu, Chuandong Wang.
Acknowledgments and Funding: This work is supported by Shanghai Sailing Program (20YF1429100), National Natural Science Foundation of China (82172473, 81802191, and 82072462), Natural Science Foundation of Shanghai (19ZR1433100), Interdisciplinary of Medicine and Engineering Foundation of Shanghai Jiao Tong University (YG2019ZDA22), and Biomedical Technology Support Program of Shanghai (20S31900200). Natural Science Foundation of Shandong Province (ZR2019PH068)
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: This study was conducted under the standard approved by the Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine.
ORCID iD: Chuandong Wang  https://orcid.org/0000-0003-0528-4105
https://orcid.org/0000-0003-0528-4105
Supplementary material for this article is available on the Cartilage website at http://cart.sagepub.com/supplemental.
References
- 1. Loeser RF, Collins JA, Diekman BO. Ageing and the pathogenesis of osteoarthritis. Nat Rev Rheumatol. 2016;12:412-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rahmati M, Nalesso G, Mobasheri A, Mozafari M. Aging and osteoarthritis: central role of the extracellular matrix. Ageing Res Rev. 2017;40:20-30. [DOI] [PubMed] [Google Scholar]
- 3. Coryell P, Diekman B, Loeser R. Mechanisms and therapeutic implications of cellular senescence in osteoarthritis. Nat Rev Rheumatol. 2021;17:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. López-Otín C, Blasco M, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McCulloch K, Litherland G, Rai T. Cellular senescence in osteoarthritis pathology. Aging Cell. 2017;16:210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hwang H, Kim H. Chondrocyte apoptosis in the pathogenesis of osteoarthritis. Int J Mol Sci. 2015;16:26035-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peffers M, Liu X, Clegg P. Transcriptomic signatures in cartilage ageing. Arthritis Res Ther. 2013;15:R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Au M, Liu Z, Rong L, Zheng Y, Wen C. Endothelin-1 induces chondrocyte senescence and cartilage damage via endothelin receptor type B in a post-traumatic osteoarthritis mouse model. Osteoarthritis Cartilage. 2020;28:1559-71. [DOI] [PubMed] [Google Scholar]
- 9. Greene MA, Loeser RF. Aging-related inflammation in osteoarthritis. Osteoarthritis Cartilage. 2015;23:1966-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao SG, Zeng C, Li LJ, Luo W, Zhang FJ, Tian J, et al. Correlation between senescence-associated beta-galactosidase expression in articular cartilage and disease severity of patients with knee osteoarthritis. Int J Rheum Dis. 2016;19:226-32. [DOI] [PubMed] [Google Scholar]
- 11. Ma X, Zheng Q, Zhao G, Yuan W, Liu W. Regulation of cellular senescence by microRNAs. Mech Ageing Dev. 2020;189:111264. [DOI] [PubMed] [Google Scholar]
- 12. Peffers M, Chabronova A, Balaskas P, Fang Y, Dyer P, Cremers A, et al. SnoRNA signatures in cartilage ageing and osteoarthritis. Sci Rep. 2020;10:10641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang K, Wang T, Gao XQ, Chen XZ, Wang F, Zhou LY. Emerging functions of piwi-interacting RNAs in diseases. J Cell Mol Med. 2021;25:4893-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zacharjasz J, Mleczko A, Bąkowski P, Piontek T, Bąkowska-Żywicka K. Small noncoding RNAs in knee osteoarthritis: the role of MicroRNAs and tRNA-derived fragments. Int J Mol Sci. 2021;22:5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balaskas P, Green J, Haqqi T, Dyer P, Kharaz Y, Fang Y, et al. Small non-coding RNAome of ageing chondrocytes. Int J Mol Sci. 2020;21:5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ali SA, Peffers MJ, Ormseth MJ, Jurisica I, Kapoor M. The non-coding RNA interactome in joint health and disease. Nat Rev Rheumatol. 2021;17:692-705. [DOI] [PubMed] [Google Scholar]
- 17. Gosset M, Berenbaum F, Thirion S, Jacques C. Primary culture and phenotyping of murine chondrocytes. Nat Protoc. 2008;3:1253-60. [DOI] [PubMed] [Google Scholar]
- 18. Chen S, Ben S, Xin J, Li S, Zheng R, Wang H, et al. The biogenesis and biological function of PIWI-interacting RNA in cancer. J Hematol Oncol. 2021;14:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peffers M, Cremers A, Welting TJM. Small nucleolar RNA expression profiling in cartilage. Methods Mol Biol. 2021;2245:135-49. [DOI] [PubMed] [Google Scholar]
- 20. Krawetz S, Kruger A, Lalancette C, Tagett R, Anton E, Draghici S, et al. A survey of small RNAs in human sperm. Hum Reprod. 2011;26:3401-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parsch D, Brümmendorf T, Richter W, Fellenberg J. Replicative aging of human articular chondrocytes during ex vivo expansion. Arthritis Rheum. 2002;46:2911-6. [DOI] [PubMed] [Google Scholar]
- 22. Zhang X, He SH, Liang X, Li W, Li T, Li D. Aging, cell senescence, the pathogenesis and targeted therapies of osteoarthritis. Front Pharmacol. 2021;12:728100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ashraf S, Cha BH, Kim JS, Ahn J, Han I, Park H, et al. Regulation of senescence associated signaling mechanisms in chondrocytes for cartilage tissue regeneration. Osteoarthritis Cartilage. 2016;24:196-205. [DOI] [PubMed] [Google Scholar]
- 24. Rim Y, Nam Y, Ju J. The role of chondrocyte hypertrophy and senescence in osteoarthritis initiation and progression. Int J Mol Sci. 2020;21:2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Proshkina E, Solovev I, Koval L, Moskalev A. The critical impacts of small RNA biogenesis proteins on aging, longevity and age-related diseases. Ageing Res Rev. 2020;62:101087. [DOI] [PubMed] [Google Scholar]
- 26. Yang B, Kang X, Xing Y, Dou C, Kang F, Li J, et al. Effect of microRNA-145 on IL-1β-induced cartilage degradation in human chondrocytes. FEBS Lett. 2014;588:2344-52. [DOI] [PubMed] [Google Scholar]
- 27. Hu G, Zhao X, Wang C, Geng Y, Zhao J, Xu J, et al. MicroRNA-145 attenuates TNF-α-driven cartilage matrix degradation in osteoarthritis via direct suppression of MKK4. Cell Death Dis. 2017;8:e3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang J, Lu Y, Yang P, Chen Q, Wang Y, Ding Q, et al. MicroRNA-145 induces the senescence of activated hepatic stellate cells through the activation of p53 pathway by ZEB2. J Cell Physiol. 2019;234:7587-99. [DOI] [PubMed] [Google Scholar]
- 29. Chen Y, Zheng J, Su L, Chen F, Zhu R, Chen X, et al. Increased salivary microRNAs that regulate DJ-1 gene expression as potential markers for Parkinson’s disease. Front Aging Neurosci. 2020;12:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li T, Yan X, Jiang M, Xiang L. The comparison of microRNA profile of the dermis between the young and elderly. J Dermatol Sci. 2016;82:75-83. [DOI] [PubMed] [Google Scholar]
- 31. Li H, Wang X, Lu X, Zhu H, Li S, Duan S, et al. Co-expression network analysis identified hub genes critical to triglyceride and free fatty acid metabolism as key regulators of age-related vascular dysfunction in mice. Aging (Albany NY). 2019;11:7620-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Banerjee A, Chokkalla AK, Shi JJ, Lee J, Venna VR, Vemuganti R, et al. Microarray profiling reveals distinct circulating miRNAs in aged male and female mice subjected to post-stroke social isolation. Neuromolecular Med. 2021;23:305-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou BR, Guo XF, Zhang JA, Xu Y, Li W, Wu D, et al. Elevated miR-34c-5p mediates dermal fibroblast senescence by ultraviolet irradiation. Int J Biol Sci. 2013;9:743-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peng D, Wang H, Li L, Ma X, Chen Y, Zhou H, et al. MiR-34c-5p promotes eradication of acute myeloid leukemia stem cells by inducing senescence through selective RAB27B targeting to inhibit exosome shedding. Leukemia. 2018;32:1180-8. [DOI] [PubMed] [Google Scholar]
- 35. Wang Z, Zhou N, Wang W, Yu Y, Xia L, Li N. HDAC2 interacts with microRNA-503-5p to regulate SGK1 in osteoarthritis. Arthritis Res Ther. 2021;23:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shen X, Cheng Y, Dong Q, Zheng M. MicroRNA-675-3p regulates IL-1β-stimulated human chondrocyte apoptosis and cartilage degradation by targeting GNG5. Biochem Biophys Res Commun. 2020;527:458-65. [DOI] [PubMed] [Google Scholar]
- 37. Kim S, Wyckoff J, Morris AT, Succop A, Avery A, Duncan GE, et al. DNA methylation associated with healthy aging of elderly twins. Geroscience. 2018;40:469-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roach HI, Yamada N, Cheung KS, Tilley S, Clarke NM, Oreffo RO, et al. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. 2005;52:3110-24. [DOI] [PubMed] [Google Scholar]
- 39. Zoch A, Auchynnikava T, Berrens RV, Kabayama Y, Schopp T, Heep M, et al. SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation. Nature. 2020;584:635-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang L, et al. PiRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29:196-206. [DOI] [PubMed] [Google Scholar]
- 41. Zhong N, Nong X, Diao J, Yang G. PiRNA-6426 increases DNMT3B-mediated SOAT1 methylation and improves heart failure. Aging (Albany NY). 2022;14:2678-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liang J, Wen J, Huang Z, Chen XP, Zhang BX, Chu L. Small nucleolar RNAs: insight into their function in cancer. Front Oncol. 2019;9:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stepanov GA, Filippova JA, Komissarov AB, Kuligina EV, Richter VA, Semenov DV. Regulatory role of small nucleolar RNAs in human diseases. Biomed Res Int. 2015;2015:206849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Steinbusch M, Fang Y, Milner P, Clegg P, Young D, Welting T, et al. Serum snoRNAs as biomarkers for joint ageing and post traumatic osteoarthritis. Sci Rep. 2017;7:43558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ripmeester E, Caron M, van den Akker G, Surtel D, Cremers A, Balaskas P, et al. Impaired chondrocyte U3 snoRNA expression in osteoarthritis impacts the chondrocyte protein translation apparatus. Sci Rep. 2020;10:13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Musavi M, Kohram F, Abasi M, Bolandi Z, Ajoudanian M, Mohammadi-Yeganeh S, et al. Rn7SK small nuclear RNA is involved in cellular senescence. J Cell Physiol. 2019;234:14234-45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xlsx-1-car-10.1177_19476035221118165 for Changes in Small Noncoding RNA Expression during Chondrocyte Senescence by Fei Xiao, Chenglong Wang, Jianping Peng, Xing Zhou, Ding Ma, Yu Wang, Yanpeng Li, Xiaodong Chen and Chuandong Wang in CARTILAGE