Abstract
The outer membrane (OM) of Gram-negative bacteria is an evolving antibiotic barrier composed of a glycerophospholipid (GP) inner leaflet and a lipopolysaccharide (LPS) outer leaflet. The two-component regulatory system CrrAB has only recently been reported to confer high-level polymyxin resistance and virulence in Klebsiella pneumoniae. Mutations in crrB have been shown to lead to the modification of the lipid A moiety of LPS through CrrAB activation. However, functions of CrrAB activation in the regulation of other lipids are unclear. Work here demonstrates that CrrAB activation not only stimulates LPS modification but also regulates synthesis of acyl-glycerophosphoglycerols (acyl-PGs), a lipid species with undefined functions and biosynthesis. Among all possible modulators of acyl-PG identified from proteomic data, we found expression of lipid A palmitoyltransferase (PagP) was significantly upregulated in the crrB mutant. Furthermore, comparative lipidomics showed that most of the increasing acyl-PG activated by CrrAB was decreased after pagP knockout with CRISPR-Cas9. These results suggest that PagP also transfers a palmitate chain from GPs to PGs, generating acyl-PGs. Further investigation revealed that PagP mainly regulates the GP contents within the OM, leading to an increased ratio of acyl-PG to PG species and improving OM hydrophobicity, which may contribute to resistance against certain cationic antimicrobial peptides resistance upon LPS modification. Taken together, this work suggests that CrrAB regulates the palmitoylation of PGs and lipid A within the OM through upregulated PagP, which functions together to form an outer membrane barrier critical for bacterial survival.
Supplementary key words: lipidomics, glycerophospholipids (GP), glycerophosphoglycerols (PG), palmitoylation, phospholipids metabolism, proteomics, two-component system CrrAB, palmitoyltransferase PagP, lipopolysaccharide, cationic antimicrobial peptides (CAMP)
Abbreviations: acyl-PG, acyl-glycerophosphoglycerols; Ara4N, 4-amino-4-deoxy-L-arabinose; CAMP, cationic antimicrobial peptide; CL, cardiolipin; IM, inner membrane; IPA, isopropanol; LPS, lipopolysaccharide; MeOH, methanol; MIC, minimum inhibitory concentration; MRM, multiple reaction monitoring; OM, outer membrane; PE, glycerophosphoethanolamines; PG, glycerophosphoglycerols; GP, glycerophospholipid
Graphical abstract
Gram-negative bacteria have a complex cell envelope consisting of an inner membrane (IM), a thin peptidoglycan layer, and an outer membrane (OM) (1). The OM exhibits an asymmetric lipid bilayer with lipopolysaccharide (LPS) in the outer leaflet and phospholipids (GP) in the inner leaflet. The specific structure serves as an effective and selective permeability barrier that protects the cells from toxic agents such as antibiotics, bile salts, and detergents, allowing bacterial survival under adverse conditions (2).
Klebsiella pneumoniae is a type of Gram-negative bacteria, which is best known as a pathogen of the human respiratory system that causes pneumonia. The emergence of multidrug resistance especially polymyxin resistance in K. pneumoniae necessitates a critical need for new treatments (3, 4). The common mechanisms of polymyxin resistance involve LPS modification, which is mediated by arnBCADTEF operon activated by two-component regulatory systems of PhoPQ, PmrAB, CrrAB in K. pneumoniae (5, 6). LPS consists of lipid A, core oligosaccharide, and O-specific antigen. Gram-negative bacteria mainly decorate the structure of lipid A, the bioactive component of LPS, in response to polymyxin treatments to promote antimicrobial resistance and interfere with the ability of host recognition. The general lipid A remodeling contains 4-amino-4-deoxy-L-arabinose (L-Ara4N), phosphoethanolamine addition, and palmitoylation, which would lead to perturbation of OM permeability (7). As major constituents of the OM, GP contents may vary with the lipid A alterations to maintain a healthy balance between membrane integrity and lipid A modification. Given that the maintenance of OM is essential for bacterial survival, understanding the GP regulation of OM may empower the development of new therapeutics.
In Escherichia coli, the cell membranes are composed of three major GP species including ∼75% glycerophosphoethanolamines (PEs), ∼20% glycerophosphoglycerols (PGs), and 5% cardiolipin (CL) (8, 9). Like E. coli, the membrane glycerophospholipids of K. pneumoniae mainly consist of PE, PG, and CL. Each phospholipid class comprises a multitude of molecular species defined by acyl chain compositions. GPs are synthesized in the cytoplasm or inner membrane and transported to the OM by specialized transporters. Unlike LPS transport, GP transport between the IM and the OM is bidirectional, which still requires thorough investigation (10). It has been demonstrated that the Mla system and the Tol-Pal complex were important for retrograde transport of GP (11, 12). Recently, AsmA-like proteins YhdP, TamB, and YdbH were reported to be necessary for proper anterograde transport of GP (13, 14).
To prevent uncontrolled breakdown resulting from surrounding mislocalized GP, Gram-negative bacteria regulate the GP contents from the outer leaflet through several mechanisms. PagP is an OM palmitoyltransferase that removes sn-1 palmitate residue (C16:0) from GP and transfers it to lipid A or PG, producing hepta-acylated Lipid A or palmitoylated PG with lysophospholipid as by-products (15, 16, 17). PagP is expressed at a low level in unstressed cells but can be induced by PhoPQ or membrane damage (18). Unlike PagP, another OM phospholipase PldA removes both sn-1 and sn-2 fatty acid side chains from GP and lysophospholipid that have miscompartmentalized to the outer leaflet of the OM (19). Moreover, it is known that accumulated GP in the OM can be translocated back to the IM by retrograde (OM to IM) GP transport to maintain the OM homeostasis. The Mla system consisting of an ABC transporter MlaFEDB, a periplasmic protein MlaC, and an OM lipoprotein MlaA has been shown to be a phospholipid transport system that is important for maintaining OM asymmetry (12, 20).
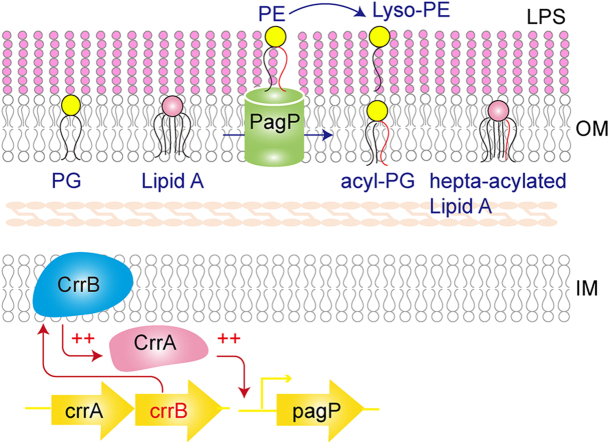
Studies on two-component systems including PhoPQ, PmrAB, and CrrAB, regulators of polymyxin resistance, mainly focused on LPS modification (21). However, our understanding of whether GP alterations vary with lipid A modification is still lacking. Mutations in crrB have only recently been reported to regulate high-level polymyxin resistance through CrrAB activation (22). Here we analyzed the GP alterations regulated by CrrAB using lipidomic analysis and found that CrrAB activated PagP, which appears to act as a bifunctional palmitoyltransferase that transfers palmitate chain from GP to both lipid A and PG within the OM, generating palmitoylated lipid A and acyl-PG in K. pneumoniae. The altered outer membrane may attenuate lipopolysaccharide activation of TLR4 signal transduction pathway and contribute to selective cationic antimicrobial peptide (CAMP) resistance. Our work presented that the phospholipid regulation varies with lipopolysaccharide modification, which provides insight into possible antimicrobial strategies that target the OM barrier.
Materials and methods
Bacterial strains and culture
Chromosomal deletions of pagP, mlaC were performed in the crrBP151S mutant, which was a mutant of K. pneumoniae ATCC BAA 2146. K. pneumoniae cells were grown in LB broth at 37°C with shaking until the exponential phase (A600 0.6–0.8). Minimum inhibitory concentrations (MICs) were determined in 96-well plates using a standard 2-fold microdilution method as described before (22).
Gene editing with CRISPR-Cas9
Gene editing in K. pneumoniae was performed with CRISPR-Cas9 as previously described with some modifications (23). Wild-type sequences were analyzed via the CRISPOR website to identify appropriate N20 sequences (24). The N20 sequence was assembled into plasmid pSGKP-spe (Addgene, catalog#117234) by Golden gate assembly to form a gRNA cassette. The gRNA cassette and ssDNA template were electroporated into the competent cells with plasmid pCas-apr (Addgene, catalog#117231) for gene editing. All the primers used were shown in supplemental Table S1.
Lipid A characterization with ESI-TOF-MS
Lipid A was isolated using mild acid hydrolysis as described before (22). The dried lipids were redissolved in chloroform/IPA/MeOH/H2O (1:1:1:0.5, v/v) and characterized by an U3000 HPLC system (Thermo Fisher) coupled to a Triple TOF 5600 mass spectrometer (AB Sciex) on negative ion mode with a mass range from 1,100 to 2,400 m/z. A volume of 2 μl of each sample was separated with ZORBAX RRHD 300 Å SB C8 (Agilent, 4.6x150 mm,5 μm) using a gradient consisting of mobile phase A (MeOH/acetonitrile/IPA, 1:1:1, 5 mM NH4OAC) and mobile phase B (MeOH/IPA, 1:4, 5 mM NH4OAC). The gradient started from 10% B to 60% B over 5 min, then to 100% B at 18 min, followed by a wash with 100% B for another 2 min, and re-equilibration for 5 min with 10% B at a flow rate of 1 ml/min. Data were acquired using an ion spray voltage of 4.5 kV, curtain gas of 35 PSI, nebulizer gas of 60 PSI, and an interface heater temperature of 600°C.
The outer and inner membrane isolation
Outer and inner membrane fractions were isolated using a lysozyme/EDTA spheroplasting method and homogenization (25). Cells were resuspended in 8 ml of 0.5 M sucrose, 10 mM Tris pH 7.5. Lysozyme (20 μg/ml), phenylmethylsulfonyl (2 mM), and DNase I were added to the suspension, followed by the addition of 8 ml of 5 mM EDTA. Cells were homogenized by French Press (GLEN MILLS, America). Total membranes were isolated by centrifugation using an MLA-55 rotor in Optima MAX-XP Ultracentrifuge (Beckman) at 185,000 g for 1 h and resuspended in 20% (w/v) sucrose (1 mM EDTA, 1 mM Tris, pH 7.5). The inner and outer membranes were separated by the use of a defined 73%-53%–20% sucrose gradient through ultracentrifugation with an MLS-50 rotor (250,000 g, 16 h, 4°C). Western blots for OmpA (OM) and NADH dehydrogenase assay (IM) were used to assess the purity of the membranes.
Lipidomic analysis
All experiments were conducted with four technical replicates for three biological replicates. Lipid extraction was performed with methyl-tert-butyl ether as described before with some modifications (26). Cell pellets were resuspended in 1×PBS and disrupted by ultrasonication (4 s/burst, ∼6 s between bursts, 2 min in total). An aliquot equal to 500 μg protein was used for lipid extraction after protein determination with a Pierce BCA protein assay kit (Thermo Fisher). Lipids were extracted with a double-phase solution (methyl-tert-butyl ether/MeOH/H2O, 10:3:2.5, v/v/v). Commercial 16:0-d31-18:1 PE, 16:0-d31-18:1 PC, 16:0-d31-18:1 PG, 16:0-d31-18:1 PS, C17 Ceramide (d18:1/17:0), 1,3-17:0 D5 diacylglycerol, D33-FFA 17:0 from Avanti lipids were added to the samples as internal standards for untargeted lipidomics, and 16:0-d31-18:1 PE, 16:0-d31-18:1 PG, 16:0 LPE-d9 were used as internal standards for targeted lipidomics. The upper phase was collected, dried under nitrogen, and redissolved in chloroform/MeOH (1:1, 10 mM NH4OAC) for LC/MS/MS. The pooled QC samples were injected during the batch analysis.
Untargeted lipidomic analysis was performed on Triple TOF 5600 (AB Sciex) equipped with a U3000 system (Thermo Fisher). Samples were separated with ACQUITY UPLC HSS T3 (Waters, 2.1×100 mm, 1.8 μm) using a multistep gradient that consisted of mobile phase A (MeOH/acetonitrile/H2O, 1:1:1, v/v, 5 mM NH4OAC) and mobile phase B (MeOH/IPA, 1:4, v/v, 5 mM NH4OAC). The gradient is from 10% B to 10% B within 1 min, 1 min–6 min to 60% B, 6 min–18 min to 100% B, then keep at 100% B for 2 min, followed by equilibration with 10% B for 5 min at a flow rate of 250 μl/min. Samples were analyzed by a full scan followed by 10 MS/MS scan from 50 to 1,250 m/z on positive and negative ion modes. The electrospray voltage was set as 5.5 kV in positive mode and 4.5 kV in negative ion mode. Nebulizer gas (GS1), heater gas (GS2), curtain gas (CUR) were separately set to 60, 60, 35 PSI, and interface heater temperature was set to 600°C. The collision energy was set to 35 ± 15 eV.
Targeted lipidomic analysis of PE, PG, acyl-PG, Lyso-PL was conducted on API 6500 Qtrap (AB Sciex) with multiple reaction monitoring (MRM) in negative ion mode. The LC condition was the same as described above. The Q1/Q3 transitions of the identified lipids were based on the MS2 spectra from Triple TOF 5600, which were described in supplemental Table S2.
Data processing for lipidomics
Untargeted lipidomic analyses were performed through commercial software lipidview 1.3, peakview 2.1, and multiquant 3.0 (AB Sciex). Raw data of a pooled QC sample were input to lipidview 1.3 for lipid identification. Precursor and fragment mass tolerance was set to 10 ppm, and minimum signal-to-noise ratio of TOF peaks was set to 3. The identification result from lipidview 1.3 was checked with peakview 2.1 based on accurate mass, retention time, isotopic pattern, and MS2 spectrum. Samples was quantified based on the identification result with multiquant 3.0 using peak area and normalized to nM lipids/mg protein with internal standards. Lipids with a fold change above 1.5 and P-value below 0.05 were thought to be altered. Statistical analysis was performed with Student’s t test or one-way ANOVA with Tukey multiple comparisons on R software.
Results
CrrAB activation increases the level of acyl-glycerophosphoglycerols with long acyl chains
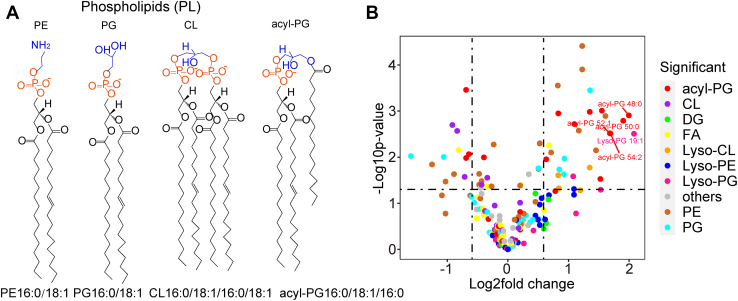
Mutations in the CrrAB two-component regulatory systems confer a high level of polymyxins resistance (27). K. pneumoniae ATCC BAA 2146 and its paired crrB mutant crrBP151S, which were reported previously, were employed to study the lipid alterations regulated by CrrAB activation (22). Untargeted lipidomic analysis of K. pneumoniae based on ESI-TOF-MS was built for lipid identification and quantification. The volcano plot showed that four of the top five altered lipids belonged to acyl-PG species, including acyl-PG 48:0 (3.99-fold), acyl-PG 50:0 (3.74-fold), acyl-PG 54:2 (3.23-fold), and acyl-PG 52:1 (3.21-fold) (Fig. 1B). Acyl-PG, modified PG molecules, are acylated with fatty acyl chains at the sn-3 position on the glycerol backbone of PG (Fig. 1A). Unlike predominant PE and PG in the membrane, the biosynthesis and function of acyl-PG have not been defined. In the study, we identified 22 kinds of acyl-PG in the sum composition level, which may contain different types of isomers differing in acyl chain compositions based on the fragmentation patterns in both positive and negative ion modes (Table 1 and supplemental Fig. S1). The alterations of acyl-PG showed that crrB mutant tended to synthesize acyl-PG with long acyl chains. For the saturated acyl-PG, the level of short-chain acyl-PG (C42-C44) was reducing or unvarying, while the level of longer chain acyl-PG (C46-C50) was increased at the sum composition level in the crrB mutant compared with wild type. Likewise, the relative abundance of acyl-PG with one, two, or three of double bonds in the attached fatty acids presented a similar tendency as the increasing number of carbons (Table 1).
Fig. 1.
Lipidomic analysis of WT and crrB mutant based on ESI-TOF-MS. A: The representative structure of PE, PG, CL, and acyl-PG. B: The volcano plot indicated that the level of acyl-PG species, especially acyl-PG 48:0, acyl-PG 50:0, acyl-PG 54:2, and acyl-PG 52:1, were highly elevated in the crrB mutant in comparison with control. The experiment was repeated three times with four technical replicates each, and the data are given in supplemental File S2.
Table 1.
The alterations of acyl-PG species annotated based on ESI-TOF-MS
| Sum Composition | Molecular lipid Information | FCa | P-value |
|---|---|---|---|
| acyl-PG 42:0 | acyl-PG 14:0_16:0/12:0 | 0.80 | 2.20E-01 |
| acyl-PG 44:0 | acyl-PG 14:0_16:0/14:0 + acyl-PG 16:0_16:0/12:0 + acyl-PG 12:0_16:0/16:0 | 1.09 | 6.33E-01 |
| acyl-PG 46:0 | acyl-PG 14:0_16:0/16:0 | 2.14 ↑ | 1.91E-03 |
| acyl-PG 48:0 | acyl-PG 16:0_16:0/16:0 | 3.99 ↑ | 1.24E-03 |
| acyl-PG 50:0 | acyl-PG 16:0_18:0/16:0 | 3.74 ↑ | 1.62E-03 |
| acyl-PG 42:1 | acyl-PG 14:0_16:1/12:0 | 1.15 | 1.54E-01 |
| acyl-PG 44:1 | acyl-PG 16:0_16:1/12:0 | 0.62 ↓ | 3.47E-04 |
| acyl-PG 46:1 | acyl-PG 16:0_16:1/14:0 + acyl-PG 16:0_18:1/12:0 + acyl 14:0_16:1/16:0 + acyl 14:0_16:0/16:1 | 0.90 | 3.16E-01 |
| acyl-PG 48:1 | acyl-PG 16:0_16:1/16:0 | 1.78 ↑ | 1.12E-03 |
| acyl-PG 50:1 | acyl-PG 16:0_18:1/16:0 | 2.93 ↑ | 9.80E-04 |
| acyl-PG 52:1 | acyl-PG 18:0_18:1/16:0 | 3.21 ↑ | 3.03E-03 |
| acyl-PG 44:2 | acyl-PG 14:0_14:1/16:1 + acyl-PG 12:0_16:1/16:1 + acyl-PG 12:0_14:1/18:1 | 0.76 | 1.01E-02 |
| acyl-PG 46:2 | acyl-PG 16:1_18:1/12:0 + acyl-PG 14:0_18:1/14:1 + acyl-PG 14:0_16:1/16:1 | 0.64 ↓ | 8.60E-03 |
| acyl-PG 48:2 | acyl-PG 16:0_16:1/16:1 | 0.94 | 6.60E-01 |
| acyl-PG 50:2 | acyl-PG 16:0_18:1/16:1 + acyl-PG 16:1_18:1/16:0 + acyl-PG 16:0_16:1/18:1 | 1.55 ↑ | 1.11E-02 |
| acyl-PG 52:2 | acyl-PG 16:0_18:1/18:1 + acyl-PG 18:1_18:1/16:0 | 2.55 ↑ | 1.04E-03 |
| acyl-PG 54:2 | acyl-PG 18:1_18:0/18:1 | 3.23 ↑ | 3.06E-03 |
| acyl-PG 46:3 | acyl-PG 16:1_16:1/14:1 | 0.62 ↓ | 1.04E-02 |
| acyl-PG 48:3 | acyl-PG 16:1_18:1/14:1 + acyl-PG 16:1_16:1/16:1 | 0.88 | 4.74E-01 |
| acyl-PG 50:3 | acyl-PG 16:1_18:1/16:1 | 1.15 | 3.73E-01 |
| acyl-PG 52:3 | acyl-PG 16:1_18:1/18:1 + acyl-PG 18:1_18:1/16:1 | 1.72 | 5.44E-02 |
| acyl-PG 54:3 | acyl-PG 18:1_18:1/18:1 | 2.88 ↑ | 2.97E-02 |
The fold change of the lipid levels in the crrB mutant compared with the wild type. Data are given in supplemental File S2.
CrrAB activates the expression of palmitoyltransferase PagP and Mla system proteins
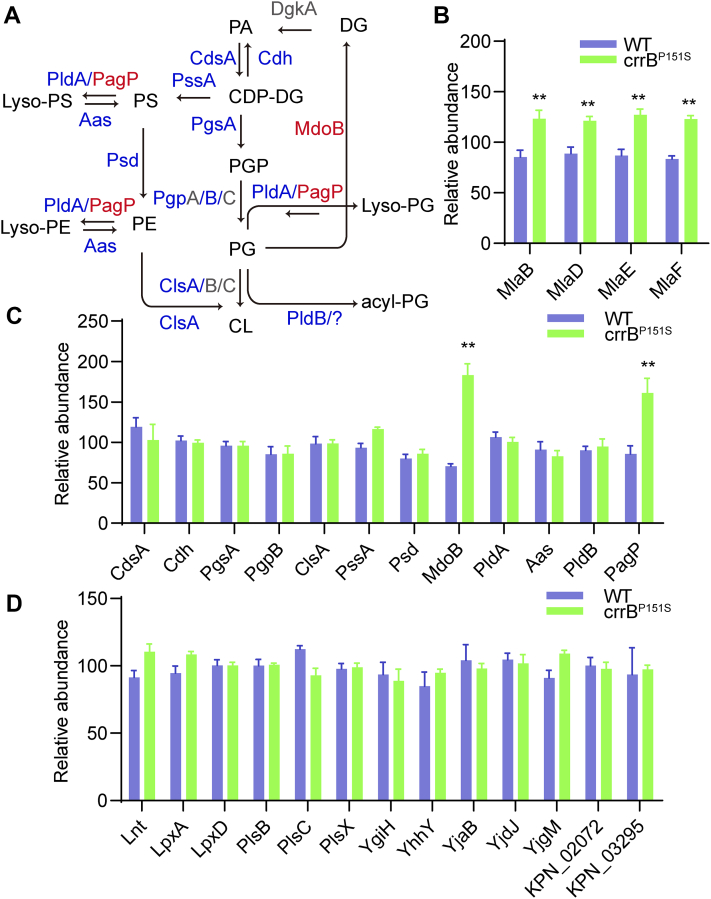
CrrAB activation leads to an increase of long-chain acyl-PG, which must be associated with the synthesis and metabolism of GP. In order to uncover the determinant, we screened the identified enzymes that mediate the synthesis and metabolism of GP in our previous proteomic data (Fig. 2A, C) (22). Considering that the synthesis of acyl-PG is not fully understood, we also sorted out the acyltransferases identified in K. pneumoniae including Lnt, LpxA, LpxD, PlsB, PlsC, PlsX, YgiH, YhhY, YjaB, YjdJ, YjdM, KPN_02072, and KPN_03295 that were hypothesized to transfer an acyl chain from GP to PG, forming acyl-PG (Fig. 2D). Of all enzymes mentioned above, only palmitoyltransferase PagP (1.92-fold) and phosphoglycerol transferase I MdoB (2.60-fold) were upregulated in the crrB mutant compared with the wild type (Fig. 2C). MdoB has no correlation with the production of acyl-PG. Unlike MdoB, PagP has been reported as a lipid A palmitoyltransferase, and we hypothesized that the PagP may transfer an acyl chain to PG to produce acyl-PG, leading to accumulated acyl-PG. Besides, the expression of Mla proteins including MlaB (1.40-fold), MlaD (1.35-fold), MlaE (1.47-fold), MlaF (1.47-fold), which were thought to be involved in lipid trafficking, were upregulated in the crrB mutant (Fig. 2B) (28). As GP is synthesized on IM, and then transported to OM, upregulated Mla system complex may play a role in acyl-PG generation as GP transporter.
Fig. 2.
CrrAB activation led to an increased level of palmitoyltransferase PagP and Mla system proteins. A: The identified biosynthetic and metabolic pathway of bacterial phospholipid. Proteins in blue, red, and gray, respectively, represented proteins that were unaltered, upregulated, and unidentified in the crrB mutant compared with the wild type. B: The expression of Mla system proteins including MlaB, MlaD, MlaE, and MlaF were upregulated in the crrB mutant. C: The expression of enzymes that mediate the synthesis and metabolism of phospholipids in K. pneumoniae. D: The expression of acyltransferases identified in the K. pneumoniae that were hypothesized to mediate the synthesis of acyl-PG had no difference between WT and crrB mutant. The experiment was repeated three times with two technical replicates each and the proteomic data are available via ProteomeXchange with identifier PXD018528.
Elevated long-chain acyl-PG contents attributed to the upregulated palmitoyltransferase PagP in the crrB mutant
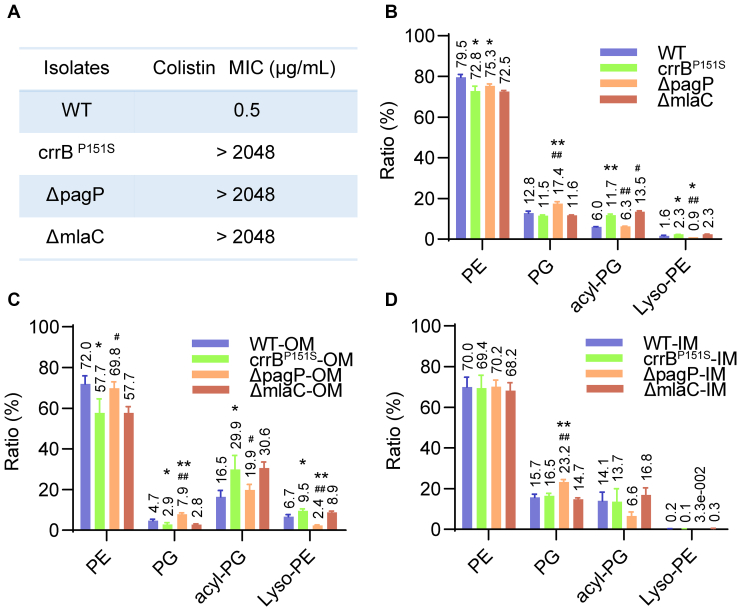
To explore it, we knocked out the pagP and mlaC gene of the Mla system in the crrB mutant by CRISPR-Cas9, respectively. PagP and MlaC were not the determinants of polymyxin resistance since the colistin MIC showed no difference before and after the gene knockout in the crrB mutant (Fig. 3A). Targeted lipidomic analysis of acyl-PG, PE, and PG from strains including WT, crrB mutant, ΔpagP, and ΔmlaC based on MRM was conducted according to the results of untargeted lipidomics. The results showed that the acyl-PG percentage of the total amount was significantly reduced after pagP knockout. Simultaneously, the ratio of PG species was elevated and that of lyso-PE was sharply diminished. Unlike PagP, the removal of mlaC brought minor changes to GP species (Fig. 3B).
Fig. 3.
The alterations of the relative abundance among each GP species. A: The colistin MIC of strains. B: Each GP percentage of the total lipid amount detected by ESI-MRM-MS. C: Each GP percentage of total lipid amount within the OM. D: Each GP percentage of total lipid amount within the IM. ∗(P-value<0.05) or ∗∗(P-value<0.01) indicates a significance of crrBP151S, ΔpagP, or ΔmlaC compared with WT. #(P-value<0.05) or ##(P-value<0.01) shows a significance of ΔpagP or ΔmlaC in comparison with crrBP151S. The pagP and mlaC deletions were constructed in the crrBP151S mutant.
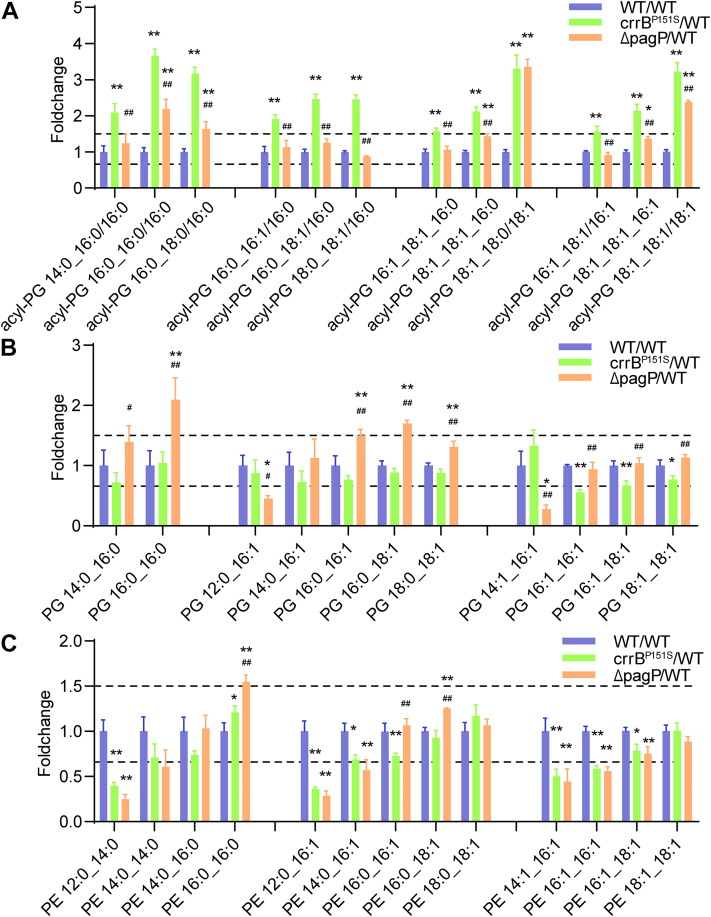
PagP is a palmitoyltransferase, which was reported to transfer a palmitate chain to lipid A (29). Interestingly, most of the elevated acyl-PG with longer acyl chains in the crrB mutant owned C16:0 acyl moiety at sn-3. Acyl-PG 16:1_18:1_16:0, acyl 18:1_18:1_16:0, and acyl-PG 18:1_18:1_16:1 had several isomers differing in acyl chain of sn-3 position (Fig. 4A and Table 1). After pagP knockout, all of the acyl-PG with C16:0 or C16:1 at the sn-3 position appeared to be diminished, while acyl-PG (18:1_18:1)_18:0 (1.02-fold) and acyl-PG (18:1_18:1)_18:1 (0.74-fold) showed a minor difference (Fig. 4A). At the same time, the removal of pagP brought an increase in most of the PG, which may regard as acyl chain acceptors to producing acyl-PG. However, the level of PG 12:0_16:1 and PG 14:1_16:1 were reduced after pagP knockout, which was in accordance with the results that no acyl-PG using PG 12:0_16:1 and PG 14:1_16:1 as acyl chain acceptor were identified (Fig. 4B and Table 1). Furthermore, the crrB mutant showed a reduction in the level of PE with short acyl chains like PE 12:0_14:0, PE 12:0_16:1, PE 14:0_16:1, PE 16:0_16:1, PE 14:1_16:1, PE 16:1_16:1, PE 16:1_18:1. PagP inactivation in the crrB mutant had a minor impact on nonpalmitoylated PE contents, but lead to an increase of palmitoylated PE like PE 16:0_16:0, PE 16:0_16:1, and PE 16:0_18:1 (Fig. 4C). Thus, PagP may transfer palmitate from PE to PG, resulting in accumulated acyl-PG in the crrB mutant.
Fig. 4.
The alterations of acyl-PG, PG, and PE of whole-cell lysates. A: Of the 12 increased acyl-PG in the crrB mutant, 11 were decreased after pagP knockout. The level of acyl-PG 18:1_18:0/18:1 and acyl-PG 18:1_18:0/18:1 showed a relatively minor difference before and after pagP knockout. B: PG alterations in the crrB mutant before and after pagP knockout. C: The alterations of PE with and without a palmitate chain in the crrB mutant before and after pagP knockout. Dashed lines marked the fold change threshold of 1.5. The experiment was repeated three times with four technical replicates each, and the data are given in supplemental File S3. ∗(P-value<0.05) or ∗∗(P-value<0.01) indicates a significance of crrBP151S or ΔpagP compared with WT. #(P-value<0.05) or ##(P-value<0.01) shows a significance of ΔpagP in comparison with crrABP151S.
CrrAB remodels the lipid components within the OM through PagP
As we know, PagP is an outer membrane protein. To further explore the extent of variations in the IM and OM content, lipidomic analyses of IM and OM contents based on MRM were conducted. The purity of the IM and OM were assessed by NADH dehydrogenase assay (IM) and Western blot of OmpA (OM), suggesting that the IM and OM were separated successfully (supplemental Fig. S2). The inner and outer membranes exhibit different GP distributions. As we can see, the level of lyso-PE and the ratio of acyl-PG to PG on the OM was much higher than that on the IM (Fig. 3C, D). The variations of GP caused by CrrAB activation mainly focused on the OM, which could be recovered after pagP knockout (Fig. 3C, D). However, the removal of mlaC brought a minor difference on the IM and OM in the crrB mutant (Fig. 3C, D).
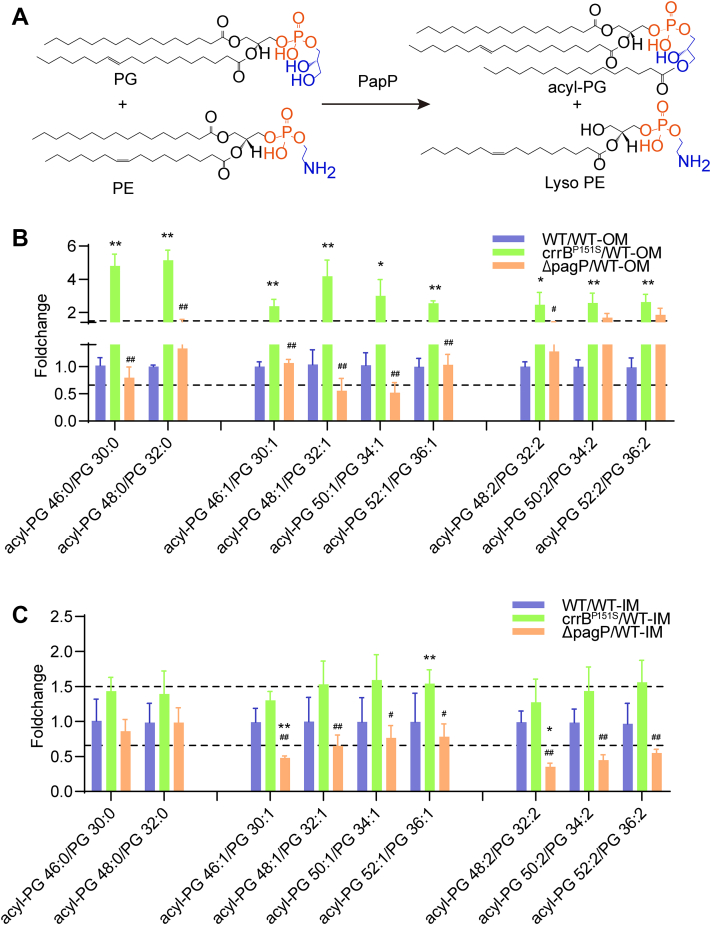
As the results mentioned above, we hypothesized that PGs are the acceptor substrates of PagP, which produce acyl-PG after receiving a palmitate chain (C16:0) (Fig. 5A). To reduce the deviation from the amount of membrane protein caused by CrrAB activation and PagP removal, the ratio of products to substrates like acyl-PG 48:1 (acyl-PG 16:0_16:1/16:0) to PG 32:1 (PG 16:0_16:1) was employed to assess the effect of PagP on the IM and OM. If the expression of PagP was upregulated, the ratio of products to substrates would be increased; otherwise, it would be decreased. CrrAB activation had a minor impact on the ratio of acyl-PG to PG, but PagP defect significantly reduced the ratio of acyl-PG to PG within the IM (Fig. 5C). Distinct from the IM, the ratio of OM acyl-PG to its accordingly PG substrate in the crrB mutant, like predominant acyl-PG 48:1 to PG 32:1 and acyl-PG 50:1 to PG 34:1, was sharply increased compared with the wild type. When the pagP gene was knocked out, most of them were reduced toward the level of the wild type (Fig. 5B). The alterations of OM PE were similar to that of the whole cells, while that of IM had minor variations (supplemental Fig. S3). Therefore, OM palmitoylated PE may be the donor substrates of the palmitate chains. Our results showed that CrrAB activation mainly remodels the GP contents within the OM other than IM, which depends on the PagP functioning on the OM.
Fig. 5.
The alterations of lipid contents within the outer and inner membranes. A: The hypothesis of the generation of acyl-PG mediated by PagP mediator. B: The ratio of OM acyl-PG to its accordingly PG was significantly increased in the crrB mutant compared with the wild type, and decreased after pagP knockout within the outer membrane. C: The ratio of IM acyl-PG to its accordingly PG substrate showed a minor variance of crrB mutant compared with the WT, but significantly decreased in the pagP knockout group. The experiment was repeated three times with four technical replicates each, and the data are given in supplemental File S4. ∗(P-value<0.05) or ∗∗(P-value<0.01) indicates a significance of crrBP151S or ΔpagP compared with WT. #(P-value<0.05) or ##(P-value<0.01) shows a significance of ΔpagP in comparison with crrABP151S.
PagP is responsible for the lipid A palmitoylation
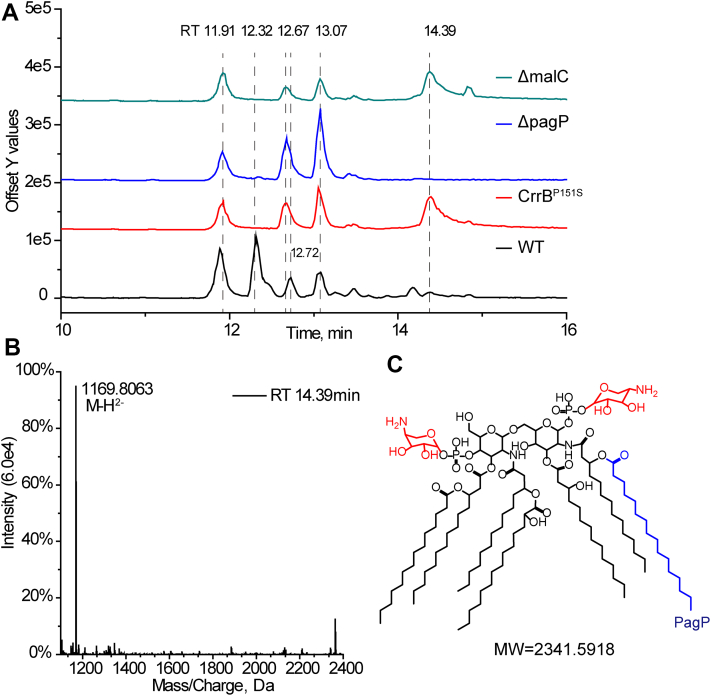
CrrAB activation induces lipid A modification with L-Ara4N and palmitoylation (22). In the study, extracted lipid A in the wild type was separated into four peaks on reversed-phase HPLC with retention times of 11.91, 12.32, 12.72, and 13.07 min, while that of the crrB mutant was separated into four peaks with retention times of 11.91, 12.67, 13.07, and 14.39 min (Fig. 6A). Specific fractions of the wild type at 12.32 and 12.72 min, which presented ions of m/z 1760.1693 and 1744.1822 in mass spectrum, respectively, correspond to the lipid A without modification (supplemental Fig. S4). Specific fractions of the crrB mutant at 12.67 and 14.39 min, whose corresponding ions were at m/z 2102.3389 and 1169.8063, respectively, represented lipid A with two L-Ara4N molecules and lipid A modified with two L-Ara4N and a palmitoylation (Figs. 6 and S4). Compared with the base peak of crrB mutant with pagP, the pagP knockout strain lost the peak at 14.39 min, which revealed that lipid A dropped the palmitate chain, corresponding to the previous study that PagP transfers a palmitate chain from GP to lipid A (29). Moreover, the lipid A profile of the crrB mutant showed little difference before and after the mlaC gene knockout (Fig. 6A). Therefore, the palmitoylation of lipid A in the crrB mutant was indeed mediated by pagP, which did not rely on the Mla system.
Fig. 6.
Lipid A analysis based on LC/MS. A: The base peak of lipid A in each strain. B: The corresponding mass spectrum at the retention time of 14.39 min. The ion at m/z 1169.8063 was doubly deprotonated. C: The corresponding lipid A structure at the retention time of 14.39 min represented lipid A with two L-Ara4N and a palmitoylation.
Discussion
Recent advances have revealed that CrrAB activates LPS modification including L-Ara4N and phosphoethanolamine, leading to polymyxin resistance (22, 27, 30). Here we investigated that CrrAB activation not only stimulates LPS remodeling but also regulates the GP components especially acyl-PG species to maintain a balance within the OM.
It is of note that, although LPS was modified by L-Ara4N and palmitoylation via elevated expression of arnBCADTEF and pagP, CrrAB activation appeared to have a minor impact on the LPS biosynthesis and degradation pathway since the expression of proteins linked to LPS biosynthesis (LpxACDHBK/WaaA/LpxL), transport (LptACDE), and degradation (YciM/FtsH) remained stable compared with the wild type (data not shown) (31, 32). As a consequence, CrrAB activation regulated the ratio of modified LPS to unmodified LPS but presumably did not alter the total LPS level. Since lipid A modified with two L-Ara4N and a palmitoylation was the last-eluted on reversed-phase HPLC, the production of palmitoylated lipid A increased the hydrophobicity of the outer leaflet in the OM. Meanwhile, the ratio of acyl-PG species to PE and PG species in the crrB mutant was significantly elevated with the OM (∼2-fold), which increased the hydrophobicity of the inner leaflet in the OM. Interestingly, GP contents within the IM were almost invariable. Thus, the increased acyl-PG within the OM was thought to cooperate with the palmitoylated lipid A to maintain the balance of OM. The alteration of GP in the inner leaflet and the modified LPS in the outer leaflet increases the hydrophobicity of the OM. Previous studies have reported that elevated hydrophobicity could block surface association and membrane intercalation of amphipathic alpha-helical CAMP, promoting resistance to various CAMPs such as C18G (7, 33). According to this, elevated acyl-PG accompanied with palmitoylated lipid A in the crrB mutant may cocontribute to certain CAMP resistance. It is of note that the palmitoylation of lipid A caused by PagP was not the determinant of polymyxin resistance since the polymyxin MIC was still above 2048 μg/ml after blocking the palmitoylation of lipid A and PG by pagP knockout in the crrB mutant. PagP is required for Bordetella bronchiseptica to persist in the mouse respiratory tract (34) and to resist antibody-mediated complement lysis (35). Previous studies reported that PagP-mediated lipid A palmitoylation can attenuate LPS activation of the TLR4/MD2/CD14 innate immune receptor, conferring resistance to host immune defenses (36, 37, 38), which was in accordance with that crrB mutants display an increased in vivo virulence (27).
Acyl-PG, also known as hemi bis(monoacylglycero) phosphate, is a specific lipid species with undefined biosynthesis and function (39). Our results showed that acyl-PG species are enriched in membrane especially OM in K. pneumoniae (Fig. 3C, D). Combined with the untargeted and targeted lipidomics, we presented the acyl-PG profiles of K. pneumoniae, including 22 kinds in bond levels with more fatty acyl compositions by fragmentation patterns in both positive and negative ion modes (Table 1). The fragmentation patterns of acyl-PG in negative ion mode were already described by Hsu FF et al. and Coulon D et al., which offered the fragment ion of the fatty acid chains at sn-1, sn-2, or sn-3 (40, 41). However, the corresponding position of fatty acid chains was difficult to determine. Here we found that the molecular ion [M+Na+] of acyl-PG fragmented at the linkage between phosphate and a carbon atom on both sides, producing ions [R3COOCH2CH(OH)CH2OPO3HNa]+ and [R1COOR2COOCH2CH(OH)CH2OPO3HNa]+, offering the information of fatty acid chain at sn-3 position (supplemental Fig. S1). The increased level of acyl-PG in the crrB mutant backing toward the wild type after pagP knockout revealed that PagP is one of the enzymes responsible for acyl-PG generation, which corresponds to a previous study that purified PagP transfers palmitate to PG (16). PagP is a lipid A palmitoyltransferase possessing precise catalyzation activated by PhoPQ or OM damage (17). The different alterations of palmitoylated and nonpalmitoylated PE before and after the removal of PE are corresponding to the results that PagP can distinguish palmitate from other acyl chains found in GP (29) (Fig. 4C). Considering that PagP was an OM protein remodeling the distribution of OM lipids, acyl-PG was thought to be synthesized in the OM (42). The decreased acyl-PG ratio and increased PG ratio within the IM after pagP knockout suggested that enriched OM acyl-PG may translocate to IM and PG synthesized in the IM may transfer to OM as substrates when PagP was activated (Fig. 3D). To date, the pathways for anterograde and retrograde transport of GP are less understood. The Mla system has been implicated in both retrograde and anterograde GP transport (43, 44). Mla proteins including MlaB, MlaD, MlaE, MlaF were overexpressed in the crrB mutant (Fig. 2B). However, the GP contents including PE, PG, acyl-PG, Lyso-PE within the OM and IM in the crrB mutant lack significant changes in the defect of the Mla system. Thus, we suspected that the Mla pathway targets a relatively minor population of OM GP molecules or there were unknown lipid trafficking systems offsetting the function of the Mla system in the process of CrrAB activation. Previous studies have reported that the MsbA-dependent flip of lipids from the IM to the OM is required for lipid A palmitoylation in vivo (18) and it has been demonstrated to possess phospholipid flippase activity in vitro using fluorescent phospholipid derivatives (45). Since a previous study reported that the active site of PagP was located on the outer leaflet of the OM, phospholipids like PG and PE, normally found in the inner leaflet, may translocate to the outer leaflet as substrates, and acyl-PG products were transported to the inner leaflet when CrrAB was activated (42).
Except for acyl-PG, phospholipids including PE, PG, lyso-PE altered in the crrB mutant to some extent. Interestingly, the levels of acyl-PG (18:1_18:1)_18:0 and acyl-PG (18:1_18:1)_18:1 that should not be affected by the expression of PagP were elevated in the crrB mutant. Furthermore, the level of PE without palmitate like PE 12:0_14:0, PE 12:0_16:1, PE 14:0_16:1, PE 14:1_16:1, PE 16:1_16:1, PE 16:1_18:1 that should not affected by PagP was diminished in the crrB mutant. However, the expression of proteins belonging to de novo pathway of PE, PG, and CL like CdsA, PssA, Psd, PgsA, PgpA/B/C, ClsA, PldA showed no difference between the crrB mutant and the wild type. Hence, in order to keep a balance between membrane integrity and lipid A modification, CrrAB activation may modulate the GP contents through other pathways.
Our study demonstrates that the CrrAB two-component system regulated lipid A palmitoylation and the outer membrane GP through elevated palmitoyltransferase PagP in K. pneumoniae. Although studies about PagP are extensive, most studies focused on lipid A palmitoylation. Our study suggests that PagP acts as a bifunctional palmitoyltransferase to PG and lipid A, contributing to acyl-PG generation. The finding broadens our understanding of PL metabolism and regulation, assisting in new therapeutic strategies targeting on OM barrier.
Data availability
Proteomic data are available via ProteomeXchange with identifier PXD018528.
Supplemental data
This article contains supplemental data
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Professor Quanjiang Ji from Shanghai Tech University for providing plasmids for gene editing. This work was supported by grants from the National Natural Science Foundation of China (32141003, 82104249, and 82104248), and the CAMS Innovation Fund for Medical Science (2021-1-I2M-030).
Author contributions
L. S., F. Y. conceptualization; L. S., Y. Z., T. C., Z. X. methodology; L. S. investigation; Y. Z. resources; L. S. writing – original draft; F. Y., X. Y. writing – review & editing; X. L., N. L. supervision.
Contributor Information
Fuquan Yang, Email: fqyang@ibp.ac.cn.
Xuefu You, Email: xuefuyou@imb.pumc.edu.cn.
Supplemental data
Reference
- 1.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.May K.L., Grabowicz M. The bacterial outer membrane is an evolving antibiotic barrier. Proc. Natl. Acad. Sci. U. S. A. 2018;115:8852–8854. doi: 10.1073/pnas.1812779115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC) Centers for Disease Control and Prevention; Atlanta, GA: 2013. Antibiotic Resistance Threats in the United States, 2013. [Google Scholar]
- 4.Nang S.C., Azad M.A.K., Velkov T., Zhou Q.T., Li J. Rescuing the last-line polymyxins: achievements and challenges. Pharmacol. Rev. 2021;73:679–728. doi: 10.1124/pharmrev.120.000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trimble M.J., Mlynarcik P., Kolar M., Hancock R.E. Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 2016;6:a025288. doi: 10.1101/cshperspect.a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y.Y., Wang Y., Walsh T.R., Yi L.X., Zhang R., Spencer J., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 7.Needham B.D., Trent M.S. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 2013;11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raetz C.R., Dowhan W. Biosynthesis and function of phospholipids in Escherichia coli. J. Biol. Chem. 1990;265:1235–1238. [PubMed] [Google Scholar]
- 9.Lopez-Lara I.M., Geiger O. Bacterial lipid diversity. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:1287–1299. doi: 10.1016/j.bbalip.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Jones N.C., Osborn M.J. Translocation of phospholipids between the outer and inner membranes of Salmonella typhimurium. J. Biol. Chem. 1977;252:7405–7412. [PubMed] [Google Scholar]
- 11.Shrivastava R., Jiang X., Chng S.S. Outer membrane lipid homeostasis via retrograde phospholipid transport in Escherichia coli. Mol. Microbiol. 2017;106:395–408. doi: 10.1111/mmi.13772. [DOI] [PubMed] [Google Scholar]
- 12.Malinverni J.C., Silhavy T.J. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douglass M.V., McLean A.B., Trent M.S. Absence of YhdP, TamB, and YdbH leads to defects in glycerophospholipid transport and cell morphology in Gram-negative bacteria. PLoS Genet. 2022;18 doi: 10.1371/journal.pgen.1010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz N., Davis R.M., Kumar S. YhdP, TamB, and YdbH Are Redundant but Essential for Growth and Lipid Homeostasis of the Gram-Negative Outer Membrane. mBio. 2021;12 doi: 10.1128/mBio.02714-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalebroux Z.D., Miller S.I. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr. Opin. Microbiol. 2014;17:106–113. doi: 10.1016/j.mib.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalebroux Z.D., Matamouros S., Whittington D., Bishop R.E., Miller S.I. PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1963–1968. doi: 10.1073/pnas.1316901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bishop R.E., Gibbons H.S., Guina T., Trent M.S., Miller S.I., Raetz C.R. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia W., El Zoeiby A., Petruzziello T.N., Jayabalasingham B., Seyedirashti S., Bishop R.E. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J. Biol. Chem. 2004;279:44966–44975. doi: 10.1074/jbc.M404963200. [DOI] [PubMed] [Google Scholar]
- 19.Dekker N. Outer-membrane phospholipase A: known structure, unknown biological function. Mol. Microbiol. 2000;35:711–717. doi: 10.1046/j.1365-2958.2000.01775.x. [DOI] [PubMed] [Google Scholar]
- 20.Kamischke C., Fan J., Bergeron J., Kulasekara H.D., Dalebroux Z.D., Burrell A., et al. The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. Elife. 2019;8 doi: 10.7554/eLife.40171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhagirath A.Y., Li Y., Patidar R., Yerex K., Ma X., Kumar A., et al. Two Component Regulatory Systems and Antibiotic Resistance in Gram-Negative Pathogens. Int. J. Mol. Sci. 2019;20:E1781. doi: 10.3390/ijms20071781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun L., Rasmussen P.K., Bai Y., Chen X., Cai T., Wang J., et al. Proteomic changes of klebsiella pneumoniae in response to colistin treatment and crrB mutation-mediated colistin resistance. Antimicrob. Agents Chemother. 2020;64:e02200–02219. doi: 10.1128/AAC.02200-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., Wang S., Chen W., Song L., Zhang Y., Shen Z., et al. CRISPR-Cas9 and CRISPR-assisted cytidine deaminase enable precise and efficient genome editing in klebsiella pneumoniae. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.01834-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Concordet J.-P., Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucl. Acids Res. 2018;46:W242–W245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cian M.B., Giordano N.P., Mettlach J.A., Minor K.E., Dalebroux Z.D. Separation of the cell envelope for gram-negative bacteria into inner and outer membrane fractions with technical adjustments for acinetobacter baumannii. J. Vis. Exp. 2020 doi: 10.3791/60517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matyash V., Liebisch G., Kurzchalia T.V., Shevchenko A., Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McConville T.H., Annavajhala M.K., Giddins M.J., Macesic N., Herrera C.M., Rozenberg F.D., et al. CrrB positively regulates high-level polymyxin resistance and virulence in klebsiella pneumoniae. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shrivastava R., Chng S.S. Lipid trafficking across the gram-negative cell envelope. J. Biol. Chem. 2019;294:14175–14184. doi: 10.1074/jbc.AW119.008139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bishop R.E. The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Mol. Microbiol. 2005;57:900–912. doi: 10.1111/j.1365-2958.2005.04711.x. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Y.H., Lin T.L., Lin Y.T., Wang J.T. Amino acid substitutions of CrrB responsible for resistance to colistin through CrrC in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2016;60:3709–3716. doi: 10.1128/AAC.00009-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guest R.L., Rutherford S.T., Silhavy T.J. Border control: regulating LPS Biogenesis. Trends Microbiol. 2021;29:334–345. doi: 10.1016/j.tim.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fivenson E.M., Bernhardt T.G. An essential membrane protein modulates the proteolysis of LpxC to control lipopolysaccharide synthesis in Escherichia coli. mBio. 2020;11 doi: 10.1128/mBio.00939-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo L., Lim K.B., Poduje C.M., Daniel M., Gunn J.S., Hackett M., et al. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 34.Preston A., Maxim E., Toland E., Pishko E.J., Harvill E.T., Caroff M., et al. Bordetella bronchiseptica PagP is a Bvg-regulated lipid A palmitoyl transferase that is required for persistent colonization of the mouse respiratory tract. Mol. Microbiol. 2003;48:725–736. doi: 10.1046/j.1365-2958.2003.03484.x. [DOI] [PubMed] [Google Scholar]
- 35.Pilione M.R., Pishko E.J., Preston A., Maskell D.J., Harvill E.T. pagP is required for resistance to antibody-mediated complement lysis during Bordetella bronchiseptica respiratory infection. Infect. Immun. 2004;72:2837–2842. doi: 10.1128/IAI.72.5.2837-2842.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simpson B.W., Trent M.S. Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol. 2019;17:403–416. doi: 10.1038/s41579-019-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller S.I., Ernst R.K., Bader M.W. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 38.Kawasaki K., Ernst R.K., Miller S.I. Purification and characterization of deacylated and/or palmitoylated lipid A species unique to Salmonella enterica serovar Typhimurium. J. Endotoxin Res. 2005;11:57–61. doi: 10.1179/096805105225006696. [DOI] [PubMed] [Google Scholar]
- 39.Showalter M.R., Berg A.L., Nagourney A., Heil H., Carraway K.L., 3rd, Fiehn O. The emerging and diverse roles of Bis(monoacylglycero) phosphate lipids in cellular physiology and disease. Int. J. Mol. Sci. 2020;21:8067. doi: 10.3390/ijms21218067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coulon D., Bure C. Acylphosphatidylglycerol (acyl-PG) or N-acylphosphatidylethanolamine (NAPE)? J. Mass Spectrom. 2015;50:1318–1320. doi: 10.1002/jms.3701. [DOI] [PubMed] [Google Scholar]
- 41.Hsu F.F., Turk J., Shi Y., Groisman E.A. Characterization of acylphosphatidylglycerols from Salmonella typhimurium by tandem mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 2004;15:1–11. doi: 10.1016/j.jasms.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Hwang P.M., Choy W.Y., Lo E.I., Chen L., Forman-Kay J.D., Raetz C.R., et al. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powers M.J., Trent M.S. Intermembrane transport: glycerophospholipid homeostasis of the Gram-negative cell envelope. Proc. Natl. Acad. Sci. U. S. A. 2019;116:17147–17155. doi: 10.1073/pnas.1902026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughes G.W., Hall S.C.L., Laxton C.S., Sridhar P., Mahadi A.H., Hatton C., et al. Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat. Microbiol. 2019;4:1692–1705. doi: 10.1038/s41564-019-0481-y. [DOI] [PubMed] [Google Scholar]
- 45.Eckford P.D., Sharom F.J. The reconstituted Escherichia coli MsbA protein displays lipid flippase activity. Biochem. J. 2010;429:195–203. doi: 10.1042/BJ20100144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomic data are available via ProteomeXchange with identifier PXD018528.