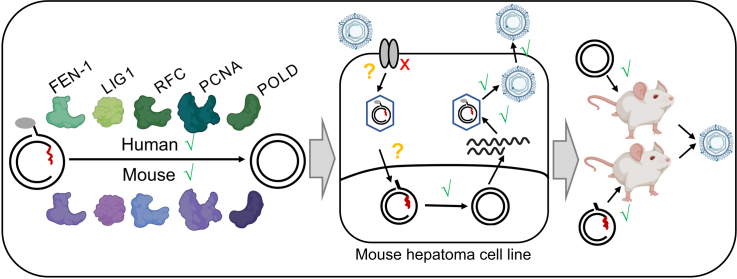
Abstract
Background & Aims
HBV has a narrow host restriction, with humans and chimpanzees representing the only known natural hosts. The molecular correlates of resistance in species that are commonly used in biomedical research, such as mice, are currently incompletely understood. Expression of human NTCP (hNTCP) in mouse hepatocytes enables HBV entry, but subsequently covalently closed circular (cccDNA) does not form in most murine cells. It is unknown if this blockade in cccDNA formation is due to deficiency in repair of relaxed circular DNA (rcDNA) to cccDNA.
Methods
Here, we deployed both in vivo and in vitro virological and biochemical approaches to investigate if murine cells contain a complete set of repair factors capable of converting HBV rcDNA to cccDNA.
Results
We demonstrate that HBV cccDNA does form in murine cell culture or in mice when recombinant rcDNA without a protein adduct is directly introduced into cells. We further show that the murine orthologues of core components in DNA lagging strand synthesis, required for the repair of rcDNA to cccDNA in human cells, can support this crucial step in the HBV life cycle. It is worth noting that recombinant HBV rcDNA substrates, either without a protein adduct or containing neutravidin to mimic HBV polymerase, were used in our study; it remains unclear if the HBV polymerase removal processes are the same in mouse and human cells.
Conclusions
Collectively, our data suggest that the HBV life cycle is blocked post entry and likely before the repair stage in mouse cells, which yields critical insights that will aid in the construction of a mouse model with inbred susceptibility to HBV infection.
Lay summary
Hepatitis B virus (HBV) is only known to infect humans and chimpanzees in nature. Mouse models are often used in modeling disease pathogenesis and preclinical research to assess the efficacy and safety of interventions before they are then tested in human participants. However, because mice are not susceptible to HBV infection it is difficult to accurately model human infection (and test potential treatments) in mouse models. Herein, we have shown that mice are able to perform a key step in the HBV life cycle, tightening the net around the possible reason why HBV can not efficiently infect and replicate in mice.
Keywords: viral hepatitis, hepatitis B virus, species tropism, animal model
Abbreviations: cccDNA, covalently closed circular DNA; FEN-1, flap endonuclease 1; HCC, hepatocellular carcinoma; HDD, hydrodynamic delivery; hNTCP, human sodium taurocholate co-transporting polypeptide; LIG1, DNA ligase 1; NA-RrcDNA, neutravidin-recombinant relaxed circular DNA; PCNA, proliferating cell nuclear antigen; POLδ, DNA polymerase delta; rcDNA, relaxed circular DNA; RFC, replication factor C; RrcDNA, recombinant relaxed circular DNA; ssDNA, single-stranded DNA
Graphical abstract
Highlights
-
•
Mouse RFC, POLD, LIG1, PCNA, FEN1 can catalyze formation of cccDNA.
-
•
rcDNA to cccDNA conversion can be supported in mouse cell lines in vitro and in vivo.
-
•
rcDNA (+) and (-) strands can be repaired similarly by mouse and human host factors in vitro.
-
•
Block in the HBV life-cycle in mouse cells is likely at (a) step(s) prior to intranuclear replication.
Introduction
HBV causes chronic infection in an estimated 257-400 million individuals worldwide.[1], [2], [3] Chronic infection can lead to a broad spectrum of disease outcomes, including cirrhosis and/or hepatocellular carcinoma (HCC). Although hepatitis B can be effectively prevented with a prophylactic vaccine, and viremia can be suppressed with antiviral therapy, there is rarely a cure for infected patients. Efforts to devise improved – and potentially curative – antiviral therapies have been reignited by new insights into the viral life cycle and HBV’s interactions with the host cell. However, the scarcity of animal models faithfully recapitulating infection and the clinical features associated with chronic hepatitis B poses significant challenges for systematic testing of approaches to cure HBV infection.4
HBV is a partially double-stranded DNA virus of the Hepadnaviridae family, genus Orthohepadnavirus. Although viruses genetically similar to HBV have been identified in a variety of species, the etiologic agent of hepatitis B in humans has a remarkably narrow tissue and host range, limited to hepatocytes in humans and chimpanzees.5 The mechanistic basis for this highly restricted tropism has not been fully deciphered, and consequently it has proven difficult to establish the entire viral life cycle in traditionally non-permissive species.
Although chimpanzees were historically the most important animal model for HBV research, their use as an experimental model has been restricted by their limited availability, high associated costs, and considerable ethical concerns.4,[6], [7], [8], [9] Hepadnaviruses related to HBV have been discovered in other species, such as woodchucks (WHBV), domestic ducks (DHBV), Beechey ground squirrels (GSHBV), and woolly monkeys[5], [6], [7], [8], [9], [10], [11], [12], [13]; however, while these surrogate models have played an important role in deciphering the molecular virology of HBV, there are distinct genetic differences that limit their utility.6,[10], [11], [12], [13]
Rodent models would undoubtedly be the most convenient due to their low cost, rapid propagation, and the prevalence of experimental tools for their genetic manipulation; however, mice and rats do not support HBV infection. Numerous complementary efforts have been undertaken to overcome the apparent resistance,[5], [6], [7], [8], [9] and transient or stable transgenic expression of the HBV genome in mice has enabled the study of HBV immunity and pathogenesis.[14], [15], [16], [17], [18], [19], [20], [21], [22] However, these models do not recapitulate faithfully the infectious cycle of HBV, thus limiting their use. In efforts to break the species barriers, humanized mice, i.e. mice transplanted with human hepatocytes, have been created that are fully susceptible to HBV infection.[23], [24], [25], [26], [27], [28], [29] These mice have a highly immunodeficient status, however, which is one of the major concerns about this model.[23], [24], [25], [26], [27], [28], [29]
The discovery of the human sodium taurocholate co-transporting polypeptide (hNTCP, or SLC10A) as the receptor for HBV and its satellite hepatitis delta virus raised hopes that murine models with inbred susceptibility to HBV infection could be generated.[30], [31], [32] However, while HBV can enter hNTCP-expressing hepatocytes in mice, blocks in subsequent steps of the viral life cycle hinder further progression of infection.[30], [31], [32], [33] Following entry into human cells, the HBV genome is uncoated and the relaxed circular genome (rcDNA) is transported into the nucleus. Here, covalently closed circular DNA (cccDNA), which serves as the transcriptional template for all viral gene products, is formed through mechanisms that rely on the host cell DNA repair machinery.34 Currently, there is insufficient experimental evidence that murine cells can support these steps. In mice stably expressing the HBV genome, pre-genomic HBV RNAs are transcribed from the transgenic integrant but cccDNA is hardly detectable.35 Yet, HBV transgenic mice produce virions that were shown to be infectious in chimpanzees,36 suggesting that assembly and release of HBV is supported in mouse cells. Conceivably, human-specific factors may be missing or rodent-specific factors may limit formation and/or maintenance of cccDNA. In vitro evidence suggests that cccDNA formation could be detected in a clone derived from the murine hepatoma cell line AML12,37 which is consistent with the fact that the minimal factors required for this step are highly conserved across species.38Furthermore, it was shown that in HBV transgenic mice lacking hepatocyte nuclear factor 1α, low levels of cccDNA could be detected,39 suggesting that the block in the HBV life cycle in murine cells is not absolute. Heterokaryons formed from mouse hepatoma cells expressing hNTCP and human hepatoma cells are susceptible to HBV infection,33 suggesting that there are possible missing human-specific factors critical for the HBV life cycle in mouse hepatocytes.
It is still a mystery why cccDNA does not form in murine cells even after successful viral entry. The answer to this question would enable the generation of immunocompetent mouse models that are fully susceptible to HBV infection, which would be valuable tools to study HBV pathogenicity, immune response, and tumorigenesis. Several steps take place after viral entry that lead to cccDNA formation, including nucleocapsid transport, capsid disassembly, and rcDNA repair.34 Defects in any of these steps can cause failure of cccDNA formation. We have recently developed biochemical reconstitution assays to directly study rcDNA repair in human cells.38,40 In this study, we aimed to investigate if the rcDNA repair step is supported in murine cells. Our findings are consistent with the notion that there is a step(s) post HBV entry, but before rcDNA conversion to cccDNA, that is the block(s) in cccDNA biogenesis in mouse cells.
Materials and methods
For detailed materials and methods please refer to the supplementary information.
Results
Murine hepatoma cell lines support repair of recombinant rcDNA
HBV rcDNA harbors 4 individual lesions: 1) a protein adduct (HBV polymerase) covalently linked to the 5’ end of the minus strand; 2) a 10 nt DNA flap on the minus strand; 3) an RNA primer covalently linked to the 5’ terminus of the plus strand; and 4) a single-stranded DNA (ssDNA) gap on the plus strand (Fig. S1). Formation of cccDNA requires successful repair of all these lesions. Hepa1.6 is a commonly used murine hepatoma cell line that does not support cccDNA formation, while clones from another murine hepatoma cell line (AML12) have been shown to support limited cccDNA formation.33,37 We first investigated if both Hepa 1.6 and AML12 cell lines were proficient in the rcDNA repair step. Because it is very challenging and laborious to purify HBV rcDNA from the virion, we took advantage of our recently developed protocol to recombinantly generate HBV rcDNA, which shares critical features with authentic rcDNA38: RrcDNA (mimics the deproteinated virion-derived rcDNA) and neutravidin-RrcDNA complex (NA-RrcDNA, mimics the authentic rcDNA with the attachment of HBV polymerase). Our recombinant substrates differ from native HBV rcDNA in 2 main aspects. First, the RNA primers in RrcDNA and NA-RrcDNA are not capped. Second, authentic HBV rcDNA contains the HBV polymerase covalently linked to a 5’ flap via a tyrosyl-phosphodiester bond. NA-RrcDNA contains neutravidin attached to the 10-nt flap of the minus strand through a non-covalent NA-biotin interaction, which does not contain a tyrosyl-phosphodiester bond.
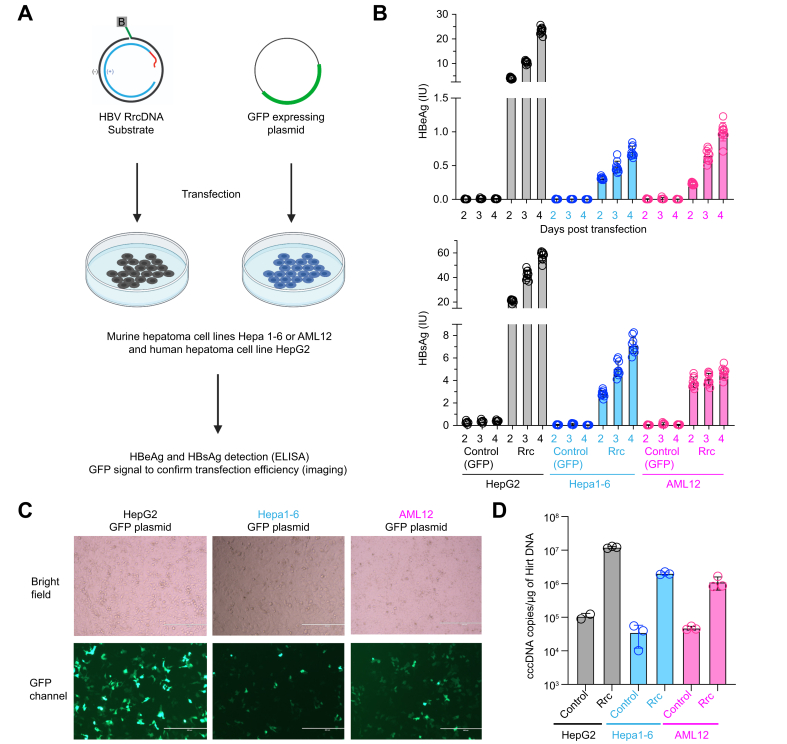
Transfection of recombinant HBV rcDNA (RrcDNA) into these 2 cell lines and the human hepatoma cell line HepG2 bypasses the viral entry and capsid disassembly steps, and thus allows examination of the rcDNA repair step in these cell lines. Post transfection, all cell lines expressed HBeAg, which is a surrogate marker for HBV cccDNA formation, and HBsAg (Fig. 1A-B). The kinetics of HBeAg and HBsAg levels are similar among these cell lines, with continuously increased levels from day 2 to day 4 post transfection (Fig. 1B). It is worth noting that the HBeAg and HBsAg reached higher levels in the human HepG2 cell line, likely due to higher transfection efficiency of the HepG2 cell line in these experiments (Fig. 1B-C). Other factors such as growth rate, transcription and translation levels may also contribute. Consistent with these results, cccDNA levels can also be detected in all 3 cell lines after transfection of recombinant rcDNA (Fig. 1D). These findings indicate that both murine cell lines can support certain levels of rcDNA repair.
Fig. 1.
Murine hepatoma cell lines support repair of reporter rcDNA.
(A) HBV RrcDNA or a GFP-expressing plasmid are transfected into murine Hepa1.6, AML12 or human HepG2 cell lines. (B) HBeAg and HBsAg levels in the supernatant as detected by ELISA. Supernatants from samples transfected with recombinant rcDNA (n = 9) or control (n = 6) were collected at day 2, 3 and 4. The height of the boxes indicates the mean of all measurements, error bars represent SD. Levels of significance (multiple unpaired t tests): p <0.000001 for HBeAg and HBsAg levels between control and RrcDNA transfection at day 2, 3, and 4, for HepG2, Hepa1-6, and AML. (C) The transfection efficiency of RrcDNA or GFP-expressing plasmid (48 hours post transfection) are evaluated by fluorescence imaging. (D) cccDNA levels from HepG2, Hepa1-6, and AML12 cells after 3 days of transfection of recombinant rcDNA were evaluated by qPCR. Levels of significance (multiple unpaired t tests): p = 0.000278, 0.000064, and 0.017174 between control and RrcDNA for HepG2, Hepa1-6, and AML, respectively. DNA were extracted from these cells via Hirt’s extraction method. Hirt DNA was treated with T5 exonuclease before being subjected to qPCR.
Hydrodynamic delivery of HBV rcDNA into mouse hepatocytes in vivo results in HBV replication
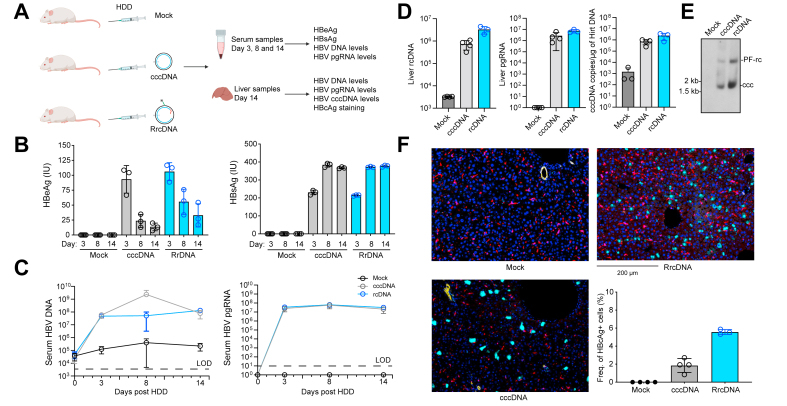
To directly test whether cccDNA can be formed in vivo in murine hepatocytes, we hydrodynamically injected RrcDNA or cccDNA into immunodeficient non-obese diabetic (NOD) recombinase activating gene 1 (Rag1-/-) and interleukin 2 receptor γ-chain (IL2γRNULL) deficient mice (NRG, Fig. 2A). NRG mice were chosen since they lack functional B, T and natural killer cells and were previously shown to sustain high levels of HBsAg, HBeAg and HBV DNA over several weeks following hydrodynamic delivery (HDD) of a 1.3x HBV genome or recombinant cccDNA.29 HDD of either RrcDNA or cccDNA resulted in similarly high levels of HBeAg, HBsAg (Fig. 2B), and HBV DNA and pregenomic RNA (Fig. 2C) in circulation. To further corroborate these data, we quantified intrahepatic replication intermediates and viral protein expression. Both RrcDNA and cccDNA, but not mock injected cohorts, had equivalently high copy numbers of HBV DNA, pregenomic RNA and cccDNA in liver tissues (Fig. 2D-E). It is worth noting that 2 major bands are observed on Southern blot (Fig. 2E), the band above the cccDNA band is either protein-free rcDNA or nicked cccDNA. Likewise, the frequencies of HBcAg-bearing cells were 1-3% and 5-6% for the cccDNA- and rcRNA-injected groups, respectively, while there was no detectable HBcAg signal in PBS-injected mice (Fig. 2F). It is not clear why rcDNA-injected groups had a higher frequency of HBcAg+ cells, one possibility is that the uptake of recombinant rcDNA is more efficient than cccDNA via the HDD procedure. Collectively, these data demonstrate that HBV cccDNA can form in vivo if the direct precursor, recombinant protein-free rcDNA, is delivered into cells, which provides additional evidence that mouse hepatocytes support the intranuclear and subsequent steps of the HBV replication cycle.
Fig. 2.
HDD of recombinant HBV rcDNA and cccDNA into mouse liver leads to robust expression of HBc protein.
(A) Three cohorts of mice (n = 3-4 in each cohorts), and PBS control, cccDNA, RrcDNA were hydrodynamically delivered into each cohort via tail vain injection. HBeAg, HBsAg levels were tested in mouse serum at day 3, 8, 14. HBV DNA, pgRNA, and HBcAg expression was examined in liver tissue extracted on day 14 post HDD. (B) Serum HBeAg and HBsAg levels at day 3, 8, and 14 were tested by ELISA. The height of the boxes indicates the mean of all measurements, error bars represent SD. Levels of significance (multiple unpaired t tests): p = 0.000074, 0.002305, 0.007249 for HBeAg levels between mock and cccDNA at day 3, 8, and 14 respectively; p = 0.000003, 0.000782, 0.017366 for HBsAg levels between mock and cccDNA at day 3, 8, and 14, respectively. (C) Serum HBV DNA and pgRNA levels at day 3, 8, and 14 were tested by qPCR or RT-qPCR respectively. (D) Liver HBV DNA, pgRNA, and cccDNA levels at day 14 were tested by qPCR or RT-qPCR. (E) Liver HBV cccDNA levels at day 14 were detected by Southern blot. PF-rc, protein-free rcDNA; ccc, cccDNA. Levels of significance between cccDNA and RrcDNA (multiple unpaired t tests): p = 0.009166 (liver rcDNA), 0.060249 (liver pgRNA), 0.074832 (liver cccDNA). (F) Liver tissues of all mice were stained via multiplex fluorescent immunohistochemistry targeting HBcAg-positive cells (cyan), DAPI-nuclei (dark blue), CK17/19-ductal epithelial cells (yellow), and CD68-macrophages (red). The percentage of HBcAg-positive, CK17/19- and CD68-negative cells are calculated and plotted (lower right panel). Levels of significance (multiple unpaired t tests): p = 0.001594 (mock vs. cccDNA), 0.000535 (mock vs. RrcDNA), 0.004692 (cccDNA vs. RrcDNA).
Murine hepatoma cell extracts fully support the conversion of rcDNA to cccDNA
We have previously established a biochemical reconstitution system to directly examine the repair process of rcDNA using yeast and human hepatoma cell nuclear extracts.38 We have previously generated RrcDNA and NA-RrcDNA substrates38 (Fig. 3B). The former mimics deproteinated HBV rcDNA intermediate, while the latter contains RrcDNA in complex with neutravidin via a biotin moiety on RrcDNA, mimicking authentic HBV rcDNA. NA-RrcDNA could be repaired to form cccDNA by nuclear extracts of either Hepa1.6 or AML12 cells (Fig. S2). We also confirmed that nuclear extracts derived from Hepa1-6 cells can convert virion-derived deproteinated HBV rcDNA into cccDNA (Fig. S3). These results indicate that murine cell nuclear extracts of Hepa1.6 and AML12 contain a complete set of factors to convert rcDNA into cccDNA.
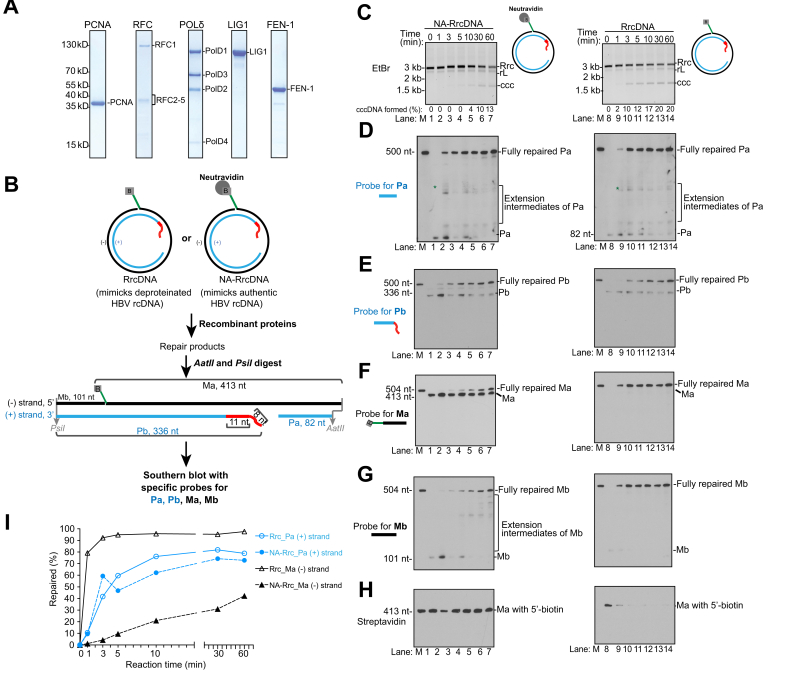
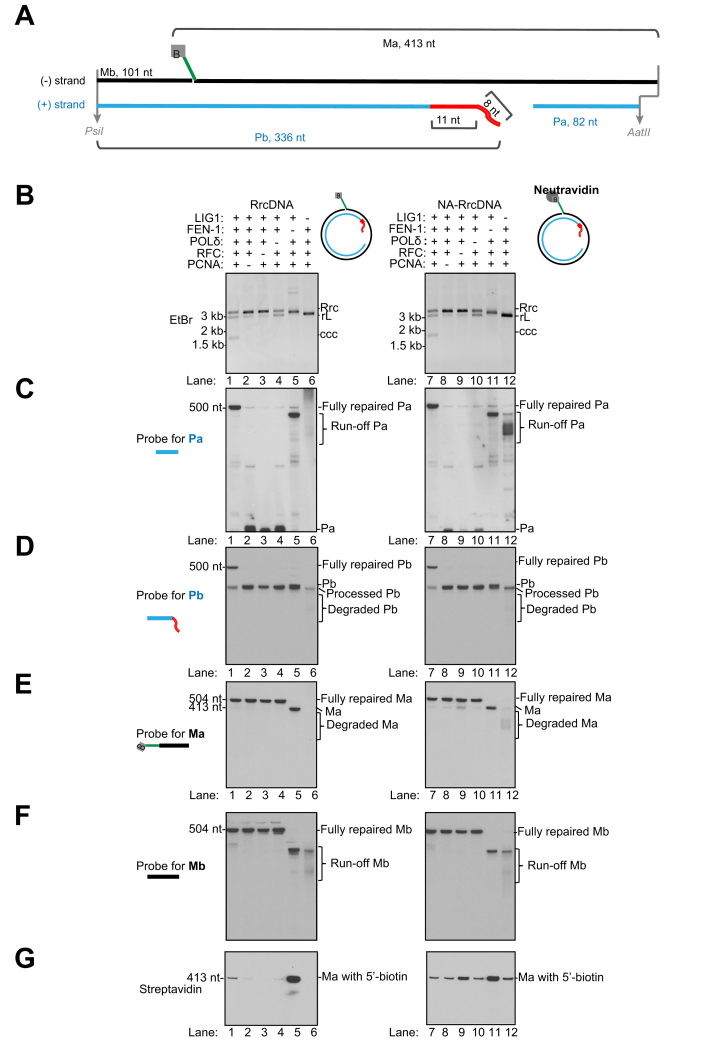
Fig. 3.
Five purified murine factors reconstitute repair of rcDNA to form cccDNA.
(A) Coomassie blue staining of purified murine recombinant proteins PCNA, RFC, POLδ, FEN-1 and LIG1. (B) Schematics showing a biochemical assay that monitors the repair of all individual lesions of HBV rcDNA substrates RrcDNA and NA-RrcDNA. The repair products are digested to generate 4 fragments of interest: Pa- and Pb-containing lesions on the plus strand; Ma- and Mb-harboring lesions on the minus strand. The fates of these fragments can be examined via Southern blot. Green line, biotinylated flap; B, biotin; red line, RNA primer. (C) Time course cccDNA formation assays with RrcDNA (lanes 1–7) or NA-RrcDNA (lanes 8–14) and 5 purified murine factors, as depicted in (A). “% cccDNA formed” was calculated by dividing the intensity of the ccc band by the sum of the intensities of the RrcDNA, linear RrcDNA and cccDNA bands. The absolute values are displayed above each lane number. (D) The plus-strand Pa fragment repair is monitored by Southern blot. “∗” indicates extension products of Pa that reach the 5′ end of Pb. (E) The repair of the plus-strand Pb fragment is monitored by Southern blot. (F-G) Repair of minus-strand Ma and Mb fragments are monitored by Southern blot. (H) Removal of biotin-harboring flap in Ma is detected by Streptavidin blot. (I) The repair efficiencies of plus and minus strands are calculated from (E) and (F) and plotted. “% repaired” is calculated by dividing the band intensities of fully repaired Pa or Ma by the sum of the band intensities of unrepaired, intermediate, and fully repaired Pa or Ma. M, marker; Rrc, RrcDNA; rL, recombinant linear RrcDNA; ccc, cccDNA.
Five core factors of the murine DNA lagging strand synthesis machinery fully reconstitute HBV cccDNA formation in biochemical assays
We have previously shown that 5 human factors involved in DNA lagging strand synthesis, namely proliferating cell nuclear antigen (PCNA), replication factor C (RFC) complex, DNA polymerase delta (POLδ), flap endonuclease 1 (FEN-1), and DNA ligase 1 (LIG1), are necessary and sufficient to convert rcDNA into cccDNA in biochemical reconstitution assays.38,40 Since these 5 factors are highly conserved in mice, with over 90% amino acid sequence identity and similarity (Table 1), we hypothesized that the murine orthologues would be sufficient to repair rcDNA to form cccDNA in murine cells. We purified these 5 recombinant murine factors to near homogeneity (Fig. 3A) and found that these 5 factors can indeed convert NA-RrcDNA and RrcDNA into cccDNA (Fig. 3C). The kinetics of repair of NA-RrcDNA were slower compared to those of RrcDNA (Fig. 3C, compare lanes 1-7 to lanes 8-14), indicating that the protein adduct on the minus strand retarded the repair of NA-RrcDNA. These observations are consistent with the repair of RrcDNA and NA-RrcDNA with 5 recombinant human factors at similar concentrations in biochemical assays (Fig. S4 and Fig. S5B). Notably, the repair kinetics of NA-RrcDNA, which mimic the authentic HBV rcDNA, are very similar in repair systems containing either purified human or murine factors (Fig. S5C). However, the repair of RrcDNA, a substrate mimicking the deproteinated repair intermediate of HBV rcDNA, is slower with murine factors than human factors (Fig. S5C). Since the repair for rcDNA to form cccDNA involves several distinct steps, including removal of the protein adduct on the minus strand and the repair of the remaining lesions on both strands, the rates of repair are most likely different between human and murine factors.
Table 1.
Similarity and identity of murine factors.
| Protein | Identity | Similarity |
|---|---|---|
| mPCNA | 96.95% | 98.47% |
| mFEN-1 | 96.58% | 97.89% |
| mLIG1 | 84.10% | 89.43% |
| mPOLD1 | 89.53% | 94.22% |
| mPOLD2 | 94.26% | 97.23% |
| mPOLD3 | 84.40% | 90.81% |
| mPOLD4 | 83.33% | 88.89% |
| mRFC1 | 82.02% | 89.14% |
| mRFC2 | 91.55% | 95.21% |
| mRFC3 | 97.20% | 98.04% |
| mRFC4 | 90.96% | 95.34% |
| mRFC5 | 94.13% | 97.36% |
Repair of rcDNA plus and minus strands requires different sets of murine factors
We next examined the mechanisms of repair of individual lesions on rcDNA with purified murine factors. We have previously developed an assay to monitor the repair of each lesion via Southern blot.40 In this assay, the repair products are digested with restriction enzymes to generate 4 HBV ssDNA fragments containing individual lesions (Fig. 3B). Pa and Pb fragments are on the plus strand, while Ma and Mb fragments are on the minus strand. The fate of these fragments and the repair process could be detected by Southern blot using sequence-specific probes (Fig. 3B). Using this assay, we previously examined the repair process of individual lesions using 5 recombinant human factors and 2 recombinant substrates, NA-RrcDNA and RrcDNA.40 We have shown that: 1) repair of the plus strand requires all 5 factors, while minus strand repair requires FEN-1 and LIG1; and 2) the repair process is similar to the maturation of the Okazaki fragments.
Now using these 5 recombinant murine factors, we found that the repair process is remarkably similar to human repair systems. Omission of any of these 5 factors abrogated cccDNA formation and drastically reduced the amount of fully repaired Pa and Pb fragments of the plus strand (Fig. 4A-D). However, only the omission of FEN-1 and LIG1 abrogated the formation of fully repaired Ma and Mb fragments (Fig. 4E-F). These results indicate that repair of the rcDNA plus strand requires all 5 murine factors, while the repair of the minus strand only requires FEN-1 and LIG1.
Fig. 4.
Examination of repair intermediates generated when individual murine repair factors are omitted.
(A) Schematic depicting the 4 fragments in unrepaired RrcDNA digested by AatII/PsiI. (B) Repair products of RrcDNA (lanes 1–6) or NA-RrcDNA (lanes 7–12) (with 5 purified murine factors) was examined by agarose gel electrophoresis stained by ethidium bromide. Omitted factors are indicated by “−”. (C) Repair products from (B) were digested with AatII/PsiI and the plus-strand Pa fragment was monitored by Southern blot as in Fig. 3D. (D) The plus strand Pb fragment was detected by Southern blot as in Fig. 3E. (E-F) Minus-strand Ma and Mb fragments were examined by Southern blot as in Fig. 3F-G. (G) Removal of the biotin-containing flap of Ma is evaluated by Streptavidin blot as in Fig. 3H. Rrc, RrcDNA; rL, recombinant linear RrcDNA; ccc, cccDNA.
The repair of plus strand by murine factors
We have previously shown that the repair of the plus strand with the human factors resembles the maturation of Okazaki fragments (Fig. S6A). In this model, the free 3′-OH containing Pa fragment on the plus strand is equivalent to a primer and can be recognized by the RFC complex, which recruits and loads PCNA.41 PCNA interacts with POLδ and the PCNA-POLδ complex can extend Pa until it reaches the 5′ terminus of Pb and gradually displaces the RNA primer (Fig. S6A, step +i). Displacement of the RNA primer leads to the formation of an RNA flap structure on Pb that can be recognized and processed by FEN-1, leading to degradation and shortening of Pb (Fig. S6A, step +ii). The processed Pa and Pb fragments are then ligated by LIG1 to form the fully repaired plus strand (Fig. S6A, step +iii).
Consistent with this repair model, in the murine 5-protein repair system, the ssDNA gap on the plus strand is filled by extension of the 5’ terminus of the Pa fragment, leading to formation of a specific repair intermediate at 1 min (Fig. 3D, asterisk). This extension of the Pa fragment is dependent on PCNA, RFC, and POLδ, since omission of any of these factors led to accumulation of unprocessed Pa fragment (Fig. 4B-C, lanes 2-4, 8-10, Fig. S7A-B). In contrast, omission of FEN-1 and LIG1 still led to extension of Pa (Fig. 4C, lanes 5-6, 11-12). It is noteworthy that omission of FEN-1 and LIG1 led to different extension intermediates of Pa. When FEN-1 is omitted, the Pa fragment is extended until it reaches the end of its template, Ma, generating a specific run-off Pa fragment (Fig. S7C). However, when LIG1 is omitted, FEN-1 can degrade the un-ligated template Ma fragment, thus leading to heterogeneous run-off Pa fragments (Fig. S7D).
The Pb fragment contains an RNA primer that blocks the ligation reaction, and thus needs to be fully removed to generate a DNA terminus suitable for ligation. The processing and shortening of Pb is best observed when ligation is prevented by omission of LIG1 (Fig. 4D, lane 6 and 12, Fig. S7D). These results are consistent with the model that the efficient processing of Pb requires extension of Pa to displace the 5’ end of the RNA-containing fragment, generating a 5’ flap that could be recognized and removed by FEN-1 (Fig. S6A, step +ii). Subsequent ligation of Pa and Pb leads to formation of fully repaired products (Fig. 3D-E). The repair efficiencies of Pa and Pb are comparable when either RrcNA or NA-RrcDNA are used as substrates (Fig. 3I), indicating that the protein adduct on the minus strand does not affect the repair of the plus strand. These results are similar to our findings using purified human factors (Fig. S5A-E, I) and are consistent with a model that the repair of the plus strand lesions resembles the maturation process of Okazaki fragments (Fig. S6A).
The repair of minus strand by murine factors
We next evaluated the repair kinetics of the minus-strand Ma and Mb fragments in our purified murine protein system (Fig. 3F,G). For the NA-RrcDNA substrate, the Ma fragment containing the flap and protein adduct persisted, and fully repaired Ma and Mb fragments only gradually accumulated (Fig. 3F, lanes 8-14; 3I). The slow removal of the flap was also confirmed by the persistence of the biotin moiety at the 5′ terminus of the flap, which binds to neutravidin (Fig. 3H, lanes 8-14). The slow kinetics of NA-RrcDNA minus-strand repair also mirrored those of cccDNA formation (Fig. 3C, lanes 8-14), indicating that minus-strand repair is rate-limiting for cccDNA formation in the presence of the protein adduct. Remarkably, without the protein adduct, the repair of the minus strand of RrcDNA was robust, with close to 80% completion within 1 min (Fig. 3F, G, lanes 1-7; 3I). In our repair system, only FEN-1 contains endonuclease activity that could result in flap removal, so we confirmed that omission of FEN-1 drastically reduced the removal of flap (Fig. 4G, lane 5). The slow kinetics of removal of the flap in the presence of the protein adduct is most likely due to inhibition of FEN-1 endonuclease activity by bulky protein adducts on the 5’ end of the substrates.42
Like Pa, the Mb fragment contains a free 3′-OH and can be extended by POLδ-PCNA. When FEN-1 was inhibited by the 5′ protein adduct, Mb extension by POLδ-PCNA was evident (Fig. 3G, lanes 8-14). However, when the protein adduct was absent, Mb extension product was minimal and was repaired quickly (Fig. 3G, lanes 1-7), indicating that the removal of the flap and ligation of Ma and Mb were coordinated and fast, leading to reduced Mb extension events.
In addition, our observation that the 5’ protein adduct affects the repair kinetics of the minus strand but not the plus strand indicates that the repair of the plus and minus strands are generally independent events using the recombinant rcDNA substrates in our biochemical assays. It is worth noting that the presence of neutravidin instead of HBV polymerase in the substrate may alter the repair process. These observations are consistent with our findings using purified human factors40 (Fig. S5F-I) and are consistent with a model that the repair of the minus strand lesions could be accomplished by FEN-1-mediated removal of the DNA flap and the protein adduct, followed by ligation of Ma and Mb fragments (Fig. S6B). It is worth noting that the repair kinetics of the NA-RrcDNA minus strand appear to be faster in the murine repair system than in the human repair system, which is most likely due to the intrinsic differences in enzymatic activities between human and murine orthologues.
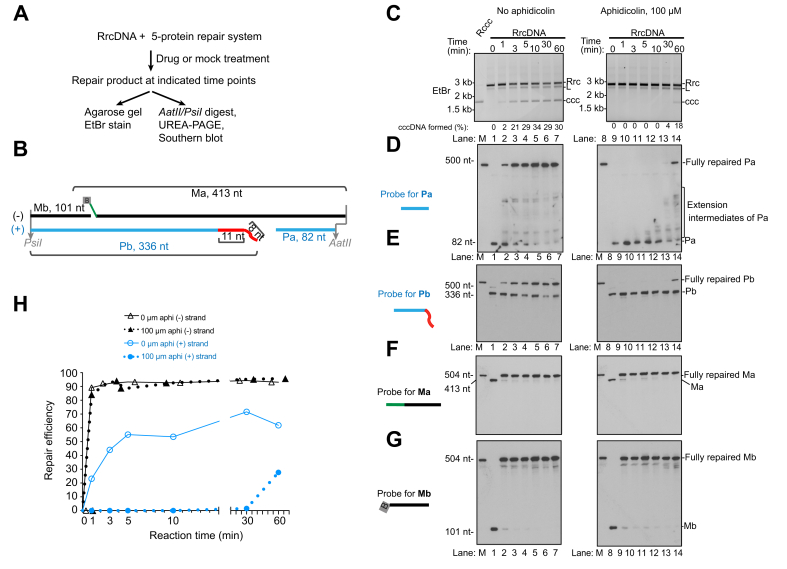
Aphidicolin specifically blocks the plus strand repair of HBV rcDNA in murine biochemical repair systems
The findings shown above indicate that the same set of 5 factors in both humans and mice are necessary and sufficient to convert HBV rcDNA into cccDNA, and the repair mechanisms are similar between humans and mice. Since we have previously shown that DNA polymerase inhibitor aphidicolin could specifically delay plus strand repair in biochemical systems using human factors,40 we test whether this is also true in the murine 5-protein repair system with RrcDNA as the substrate. Indeed, aphidicolin treatment drastically delayed the formation of cccDNA (Fig. 5A-C, compare lanes 1-7 and 8-14). This delay resulted from the slow rate of RFC-PCNA-POLD-mediated extension of the Pa fragment of the plus strand (Fig. 5D-E, compare lanes 1-7 to lanes 8-14). In contrast, the repair of the minus strand was not affected (Fig. 5F-G, 5H). Consistent with these biochemical findings, treatment of Hepa1-6 and AML12 cells with aphidicolin reduced HBeAg, HBsAg, and cccDNA levels post transfection of RrcDNA (Fig. S8). These results are also consistent with our findings that the 5 murine and human repair factors convert HBV rcDNA into cccDNA by similar mechanisms.
Fig. 5.
DNA polymerase inhibitor aphidicolin treatment specifically inhibits the repair of the rcDNA plus strand.
(A) Schematic of a time course assay to examine the effects of aphidicolin on repair of HBV RrcDNA plus and minus strands. (B) A simplified schematic depicting the 4 fragments in unrepaired RrcDNA digested by AatII/PsiI. (C) Repair of the RrcDNA substrate under treatments of no aphidicolin (1% DMSO, lanes 1–7) or aphidicolin (100 μM in 1% DMSO, lanes 8–14) was examined by agarose gel stained by ethidium bromide. “% cccDNA formed” was calculated as described in Fig. 3C. (D-G) Repair of plus-strand fragments Pa, Pb, and minus-strand fragments Ma, Mb was monitored by Southern blot as in Fig. 3D-G. (H) Repair efficiency of plus strand (E) and minus strand (F) was calculated and plotted as in Fig. 3I. M, marker; Rrc, RrcDNA; rL, recombinant linear RrcDNA; ccc, cccDNA.
Discussion
HBV cccDNA biogenesis is key for establishing HBV infection and is a complex process consisting of multiple steps including viral entry, capsid transport, and disassembly, as well as repair of rcDNA.34,[43], [44], [45] HBV cccDNA biogenesis is defective in most murine cells and thus is a barrier to the development of an immunocompetent mouse model that can be used to study HBV pathogenesis and immune response. It is currently not known which step(s) in HBV cccDNA biogenesis are defective in mice. A comprehensive examination of each step in cccDNA biogenesis in mouse cells is required to identify the defective steps and breach the cccDNA formation barrier in mouse cells. Herein, we have thoroughly examined the rcDNA repair step in murine cells via a combination of in vitro and in vivo approaches.
We provide 5 lines of evidence that mouse cells contain the required factors to support rcDNA repair: 1) transfection of recombinant rcDNA directly into 2 mouse hepatoma cell lines Hepa1.6 and AML12 led to HBeAg and HBsAg expression. Although the levels of HBeAg and HBsAg were lower compared to human HepG2 cells, this could be due to higher transfection efficiency in HepG2 cells; 2) HDD of recombinant rcDNA or cccDNA into mouse livers led to similar levels of HBeAg, HBsAg and HBcAg expression, as well as viral replication; 3) nuclear extracts of Hepa1.6 and AML12 cell lines supported repair of rcDNA to form cccDNA; 4) 5 purified recombinant murine factors (PCNA, POLD, RFC, FEN-1, and LIG1) were necessary and sufficient to convert rcDNA into cccDNA in biochemical reconstitution assays; and 5) the repair mechanisms of rcDNA via 5 murine factors resembled those via human factors.
These 5 factors are highly conserved in humans and mice, and to a lesser extent in humans and yeast (Table S1), and we have previously shown that yeast factors can convert recombinant rcDNA into cccDNA, therefore it is not surprising that these murine factors can repair rcDNA to form cccDNA like their human counterparts. Mechanistically, there are similarities and differences between these 2 sets of factors in rcDNA repair. The similarities include: 1) the repair of the minus strand only requires FEN-1 and LIG1 in both human and murine repair systems, which is consistent with the hypothesis that minus strand repair is accomplished by FEN-1-mediated removal of the 10 nt flap and the protein adduct followed by LIG1-mediated ligation; 2) the repair of the plus strand requires all 5 factors in both systems, and the repair process also resembles the maturation of Okazaki fragments, where the ssDNA gap is filled by RFC-PCNA-POLD-mediated extension, which displaces the 5’ RNA primer-containing strand. Subsequent FEN-1-mediated trimming and LIG1-mediated ligation lead to full repair of the plus strand; 3) the DNA polymerase inhibitor aphidicolin specifically inhibits repair of the plus strand but not the minus strand; 4) the presence of the 5’ protein adduct inhibits repair of the minus strand by retarding the removal of the flap. However, there are also differences between the murine and human repair systems. The repair of rcDNA requires removal of the protein adduct and subsequent repair of the remaining lesions, and we found that the repair of the NA-RrcDNA minus strand seems to be faster when using murine factors (Fig. 3I and Fig. S5I), which indicates that the removal of the protein adduct and flap is likely faster in the mouse repair system. However, the kinetics of repair of the deproteinated rcDNA mimetic RrcDNA are slower when 5 murine factors are used (Fig. S5C). These results indicate that human and mouse factors exhibit different repair rates during different steps of rcDNA repair, even though overall cccDNA formation rates from NA-RrcDNA substrates are comparable in mouse and human repair systems using recombinant proteins at similar concentrations. It is worth noting that although biochemical data suggest that these 5 murine factors can repair rcDNA like their human counterparts, it would be interesting to test if these murine factors can replace human ones to allow cccDNA formation in cells. In addition, the NA-RrcDNA substrate used in this study contains neutravidin attached to the minus strand through a non-covalent NA-biotin interaction, which does not contain a tyrosyl-phosphodiester bond. Therefore, the removal of HBV polymerase via some factors specific for HBV polymerase or tyrosyl-phosphodiester bond could not be evaluated using NA-RrcDNA, and it is not clear if murine cells have similar ability to remove HBV polymerase from rcDNA as human cells.
Our findings indicate that among viral entry, nucleocapsid transport, capsid disassembly, and repair of rcDNA in cccDNA biogenesis, the repair of rcDNA can be supported to a certain level in murine cells, so it is likely that at least 1 other step is blocked in murine cells. It has been shown that HBV entry is permitted by expressing human HBV entry factor NTCP in mouse cells, without leading to cccDNA biogenesis.29,31,33 In addition, steps of the viral life cycle following/downstream of cccDNA formation are unlikely to be responsible for the block of HBV infection in murine cells, since transfection or HDD of cccDNA into mouse cells can support viral mRNA transcription and HBV replication22,46 and infectious HBV is assembled and released.36 These lines of evidence and our findings suggest that a step between viral entry and rcDNA repair is defective in mouse cells, and this step may be nucleocapsid transport and/or capsid disassembly. These steps are complex processes that require many viral and host factors. It has been reported that importins that interact with the HBV capsids, host kinase- and phosphatase-mediated phosphorylation/dephosphorylation of capsids, as well as the HBV polymerase could be involved in these processes.34 It remains to be determined if murine cells are defective in any of these host-viral interactions or if other unknown mechanisms are involved. In addition, it is also unclear if mouse and human cells have comparable ability to remove HBV polymerase from rcDNA. Future studies are required to comprehensively examine these steps in cccDNA biogenesis in murine cells.
Financial support
This study was funded in part by grants from the National Institutes of Health (R01 AI138797, R01AI107301, R01AI146917, R01AI153236, R01AI168048 all to A.P.), a Research Scholar Award from the American Cancer Society (RSG-15-048-01-MPC to A.P.), a Burroughs Wellcome Fund Award for Investigators in Pathogenesis (101539), and funds from the US Department of Defense (W81XWH1810237) and Princeton University. This work also utilized an instrument acquired from a NIH SIG grant (S10OD026983-01). L.W. is a recipient of a postdoctoral fellowship award from the New Jersey Commission on Cancer Research (NJCCR, DFHS17PPC011). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authors’ contributions
The project was conceived by L.W. and A.P. Experiments were performed by all authors. Data were analyzed by L.W., A.T., N.C. and A.P. The manuscript was written by L.W. and A.P with input from all authors.
Data availability statement
All data are included in this study. Additional information are available upon request.
Conflict of interest
The authors do not have any conflict of interest pertaining to this study.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgments
We thank all members of the Ploss lab for critical discussion of the data and manuscript. This study was funded in part by grants from the National Institutes of Health (R01 AI138797, R01AI107301, R01AI146917, R01AI153236, R01AI168048 all to A.P.), a Research Scholar Award from the American Cancer Society (RSG-15-048-01-MPC to A.P.), a Burroughs Wellcome Fund Award for Investigators in Pathogenesis (101539), and funds from the US Department of Defense (W81XWH1810237) and Princeton University. This work also utilized an instrument acquired from a NIH SIG grant (S10OD026983-01). L.W. is a recipient of a postdoctoral fellowship award from the New Jersey Commission on Cancer Research (NJCCR, DFHS17PPC011).
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2022.100534.
Contributor Information
Lei Wei, Email: weilei@westlake.edu.cn.
Alexander Ploss, Email: aploss@princeton.edu.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Revill P.A., Chisari F.V., Block J.M., Dandri M., Gehring A.J., Guo H., et al. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol Hepatol. 2019;4:545–558. doi: 10.1016/S2468-1253(19)30119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schweitzer A., Horn J., Mikolajczyk R.T., Krause G., Ott J.J. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546–1555. doi: 10.1016/S0140-6736(15)61412-X. [DOI] [PubMed] [Google Scholar]
- 3.Yuen M.F., Chen D.S., Dusheiko G.M., Janssen H.L.A., Lau D.T.Y., Locarnini S.A., et al. Hepatitis B virus infection. Nat Rev Dis Primers. 2018;4:18035. doi: 10.1038/nrdp.2018.35. [DOI] [PubMed] [Google Scholar]
- 4.Ploss A., Strick-Marchand H., Li W. Animal models for hepatitis B: does the supply meet the demand? Gastroenterology. 2021;160:1437–1442. doi: 10.1053/j.gastro.2020.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winer B.Y., Ploss A. Determinants of hepatitis B and delta virus host tropism. Curr Opin Virol. 2015;13:109–116. doi: 10.1016/j.coviro.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y., Maya S., Ploss A. Animal models of hepatitis B virus infection-success, challenges, and future directions. Viruses. 2021;13:777. doi: 10.3390/v13050777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason W.S. Animal models and the molecular biology of hepadnavirus infection. Cold Spring Harb Perspect Med. 2015;5:a021352. doi: 10.1101/cshperspect.a021352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burwitz B.J., Zhou Z., Li W. Animal models for the study of human hepatitis B and D virus infection: new insights and progress. Antivir Res. 2020;182:104898. doi: 10.1016/j.antiviral.2020.104898. [DOI] [PubMed] [Google Scholar]
- 9.Guo W.N., Zhu B., Ai L., Yang D.L., Wang B.J. Animal models for the study of hepatitis B virus infection. Zool Res. 2018;39:25–31. doi: 10.24272/j.issn.2095-8137.2018.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Littlejohn M., Locarnini S., Yuen L. Origins and evolution of hepatitis B virus and hepatitis D virus. Cold Spring Harb Perspect Med. 2016;6:a021360. doi: 10.1101/cshperspect.a021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Summers J., Smolec J.M., Snyder R. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc Natl Acad Sci U S A. 1978;75:4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason W.S., Seal G., Summers J. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J Virol. 1980;36:829–836. doi: 10.1128/jvi.36.3.829-836.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanford R.E., Chavez D., Brasky K.M., Burns R.B., 3rd, Rico-Hesse R. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc Natl Acad Sci U S A. 1998;95:5757–5761. doi: 10.1073/pnas.95.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Araki K., Miyazaki J., Hino O., Tomita N., Chisaka O., Matsubara K., et al. Expression and replication of hepatitis B virus genome in transgenic mice. Proc Natl Acad Sci U S A. 1989;86:207–211. doi: 10.1073/pnas.86.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farza H., Hadchouel M., Scotto J., Tiollais P., Babinet C., Pourcel C. Replication and gene expression of hepatitis B virus in a transgenic mouse that contains the complete viral genome. J Virol. 1988;62:4144–4152. doi: 10.1128/jvi.62.11.4144-4152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang P.L., Althage A., Chung J., Chisari F.V. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc Natl Acad Sci U S A. 2002;99:13825–13830. doi: 10.1073/pnas.202398599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chisari F.V., Pinkert C.A., Milich D.R., Filippi P., McLachlan A., Palmiter R.D., et al. A transgenic mouse model of the chronic hepatitis B surface antigen carrier state. Science. 1985;230:1157–1160. doi: 10.1126/science.3865369. [DOI] [PubMed] [Google Scholar]
- 18.Huang L.R., Gabel Y.A., Graf S., Arzberger S., Kurts C., Heikenwalder M., et al. Transfer of HBV genomes using low doses of adenovirus vectors leads to persistent infection in immune competent mice. Gastroenterology. 2012;142:1447–1450 e1443. doi: 10.1053/j.gastro.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Dion S., Bourgine M., Godon O., Levillayer F., Michel M.L. Adeno-associated virus-mediated gene transfer leads to persistent hepatitis B virus replication in mice expressing HLA-A2 and HLA-DR1 molecules. J Virol. 2013;87:5554–5563. doi: 10.1128/JVI.03134-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang D., Liu L., Zhu D., Peng H., Su L., Fu Y.X., et al. A mouse model for HBV immunotolerance and immunotherapy. Cell Mol Immunol. 2014;11:71–78. doi: 10.1038/cmi.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye L., Yu H., Li C., Hirsch M.L., Zhang L., Samulski R.J., et al. Adeno-associated virus vector mediated delivery of the HBV genome induces chronic hepatitis B virus infection and liver fibrosis in mice. PLoS One. 2015;10:e0130052. doi: 10.1371/journal.pone.0130052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li G., Zhu Y., Shao D., Chang H., Zhang X., Zhou D., et al. Recombinant covalently closed circular DNA of hepatitis B virus induces long-term viral persistence with chronic hepatitis in a mouse model. Hepatology. 2018;67:56–70. doi: 10.1002/hep.29406. [DOI] [PubMed] [Google Scholar]
- 23.Dandri M., Burda M.R., Torok E., Pollok J.M., Iwanska A., Sommer G., et al. Repopulation of mouse liver with human hepatocytes and in vivo infection with hepatitis B virus. Hepatology. 2001;33:981–988. doi: 10.1053/jhep.2001.23314. [DOI] [PubMed] [Google Scholar]
- 24.Meuleman P., Libbrecht L., De Vos R., de Hemptinne B., Gevaert K., Vandekerckhove J., et al. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology. 2005;41:847–856. doi: 10.1002/hep.20657. [DOI] [PubMed] [Google Scholar]
- 25.Bissig K.D., Wieland S.F., Tran P., Isogawa M., Le T.T., Chisari F.V., et al. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J Clin Invest. 2010;120:924–930. doi: 10.1172/JCI40094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Billerbeck E., Mommersteeg M.C., Shlomai A., Xiao J.W., Andrus L., Bhatta A., et al. Humanized mice efficiently engrafted with fetal hepatoblasts and syngeneic immune cells develop human monocytes and NK cells. J Hepatol. 2016;65:334–343. doi: 10.1016/j.jhep.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strick-Marchand H., Dusseaux M., Darche S., Huntington N.D., Legrand N., Masse-Ranson G., et al. A novel mouse model for stable engraftment of a human immune system and human hepatocytes. PLoS One. 2015;10:e0119820. doi: 10.1371/journal.pone.0119820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bility M.T., Cheng L., Zhang Z., Luan Y., Li F., Chi L., et al. Hepatitis B virus infection and immunopathogenesis in a humanized mouse model: induction of human-specific liver fibrosis and M2-like macrophages. Plos Pathog. 2014;10:e1004032. doi: 10.1371/journal.ppat.1004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winer B.Y., Shirvani-Dastgerdi E., Bram Y., Sellau J., Low B.E., Johnson H., et al. Preclinical assessment of antiviral combination therapy in a genetically humanized mouse model for hepatitis delta virus infection. Sci Transl Med. 2018;10:9328. doi: 10.1126/scitranslmed.aap9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z., et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan H., Peng B., He W., Zhong G., Qi Y., Ren B., et al. Molecular determinants of hepatitis B and D virus entry restriction in mouse sodium taurocholate cotransporting polypeptide. J Virol. 2013;87:7977–7991. doi: 10.1128/JVI.03540-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni Y., Lempp F.A., Mehrle S., Nkongolo S., Kaufman C., Falth M., et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 33.Lempp F.A., Mutz P., Lipps C., Wirth D., Bartenschlager R., Urban S. Evidence that hepatitis B virus replication in mouse cells is limited by the lack of a host cell dependency factor. J Hepatol. 2016;64:556–564. doi: 10.1016/j.jhep.2015.10.030. [DOI] [PubMed] [Google Scholar]
- 34.Wei L., Ploss A. Mechanism of hepatitis B virus cccDNA formation. Viruses. 2021;13:1463. doi: 10.3390/v13081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guidotti L.G., Matzke B., Schaller H., Chisari F.V. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chisari F.V. Rous-Whipple Award Lecture. Viruses, immunity, and cancer: lessons from hepatitis B. Am J Pathol. 2000;156:1117–1132. doi: 10.1016/s0002-9440(10)64980-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui X., Guo J.T., Hu J. Hepatitis B virus covalently closed circular DNA formation in immortalized mouse hepatocytes associated with nucleocapsid destabilization. J Virol. 2015;89:9021–9028. doi: 10.1128/JVI.01261-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei L., Ploss A. Core components of DNA lagging strand synthesis machinery are essential for hepatitis B virus cccDNA formation. Nat Microbiol. 2020;5:715–726. doi: 10.1038/s41564-020-0678-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raney A.K., Eggers C.M., Kline E.F., Guidotti L.G., Pontoglio M., Yaniv M., et al. Nuclear covalently closed circular viral genomic DNA in the liver of hepatocyte nuclear factor 1 alpha-null hepatitis B virus transgenic mice. J Virol. 2001;75:2900–2911. doi: 10.1128/JVI.75.6.2900-2911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei L., Ploss A. Hepatitis B virus cccDNA is formed through distinct repair processes of each strand. Nat Commun. 2021;12:1591. doi: 10.1038/s41467-021-21850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majka J., Burgers P.M. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol. 2004;78:227–260. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- 42.Gloor J.W., Balakrishnan L., Bambara R.A. Flap endonuclease 1 mechanism analysis indicates flap base binding prior to threading. J Biol Chem. 2010;285:34922–34931. doi: 10.1074/jbc.M110.165902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia Y., Guo H. Hepatitis B virus cccDNA: formation, regulation and therapeutic potential. Antivir Res. 2020;180:104824. doi: 10.1016/j.antiviral.2020.104824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marchetti A.L., Guo H. New insights on molecular mechanism of hepatitis B virus covalently closed circular DNA formation. Cells. 2020;9:2430. doi: 10.3390/cells9112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 46.Yan Z., Zeng J., Yu Y., Xiang K., Hu H., Zhou X., et al. HBVcircle: a novel tool to investigate hepatitis B virus covalently closed circular DNA. J Hepatol. 2017;66:1149–1157. doi: 10.1016/j.jhep.2017.02.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in this study. Additional information are available upon request.