Abstract
Large bone defects remain an unsolved clinical challenge because of the lack of effective vascularization in newly formed bone tissue. 3D bioprinting is a fabrication technology with the potential to create vascularized bone grafts with biological activity for repairing bone defects. In this study, vascular endothelial cells laden with thermosensitive bio-ink were bioprinted in situ on the inner surfaces of interconnected tubular channels of bone mesenchymal stem cell-laden 3D-bioprinted scaffolds. Endothelial cells exhibited a more uniform distribution and greater seeding efficiency throughout the channels. In vitro, the in situ bioprinted endothelial cells can form a vascular network through proliferation and migration. The in situ vascularized tissue-engineered bone also resulted in a coupling effect between angiogenesis and osteogenesis. Moreover, RNA sequencing analysis revealed that the expression of genes related to osteogenesis and angiogenesis is upregulated in biological processes. The in vivo 3D-bioprinted in situ vascularized scaffolds exhibited excellent performance in promoting new bone formation in rat calvarial critical-sized defect models. Consequently, in situ vascularized tissue-engineered bones constructed using 3D bioprinting technology have a potential of being used as bone grafts for repairing large bone defects, with a possible clinical application in the future.
Keywords: 3D bioprinting, In situ vascularization, RNA sequencing Analysis, Large segmental bone defects, Tissue engineering
Abbreviations: Alkaline phosphatase, (ALP); Alizarin red S, (ARS); analysis of variance, (ANOVA); bone mesenchymal stem cells, (BMSCs); bone mineral density, (BMD); bone volume to tissue volume, (BV/TV); complementary DNA, (cDNA); 4′,6-diamidino-2-phenylindole, (DAPI); differentially expressed genes, (DEGs); Dulbecco's modified Eagle's medium, (DMEM); Dulbecco's phosphate-buffered saline, (DPBS); ethylenediamine tetraacetic acid, (EDTA); endothelial cells, (ECs); extracellular matrix, (ECM); fetal bovine serum, (FBS); Fourier-transform infrared, (FTIR); 3D printed GelMA hydrogel scaffold, (G); 3D dual-extrusion bioprinted GelMA hydrogel and RAOECs-laden 3P hydrogel scaffold, (G-3PR); 3D bioprinted BMSCs-laden GelMA hydrogel scaffold, (GB); 3D dual-extrusion bioprinted BMSCs-laden GelMA hydrogel and RAOECs-laden 3P hydrogel scaffold, (GB-3PR); gene ontology, (GO); gelatin methacryloyl, (GelMA); green fluorescent protein, (GFP); glyceraldehyde-3-phosphate dehydrogenase, (GAPDH); hematoxylin and eosin, (H&E); lithium phenyl-2,4,6-trimethylbenzoylphosphinate, (LAP); micro-computed tomography, (micro-CT); nuclear magnetic resonance, (NMR); optical density, (OD); paraformaldehyde, (PFA); phosphate-buffered saline, (PBS); polyethylene glycol, (PEG); polylactic acid, (PLA); PLA-PEG-PLA, (3P); polyvinylidene fluoride, (PVDF); radioimmunoprecipitation assay, (RIPA); rat aortic endothelial cells, (RAOECs); real-time polymerase chain reaction, (RT-PCR); standard deviation, (SD); tissue-engineered bone, (TEB); tris buffered saline with Tween-20, (TBST)
Graphical abstract
Highlights
-
•
3D bioprinting was used to fabricate in situ vascularized tissue engineered bone.
-
•
In situ bioprinted endothelial cells exhibited uniform distribution and greater seeding efficiency.
-
•
3D-bioprinted scaffold produced coupling between angiogenesis and osteogenesis.
1. Introduction
The majority of segmental bone defects result from congenital defects, trauma, and bone tumors [[1], [2], [3]]. Bone defects that exceed 50% of the bone diameter or over 2 cm in length are called large segmental bone defects [4] and result in delayed union or nonunion during the rehabilitation period. Current clinical strategies for repairing and regenerating bone defects mainly use autografts, allografts, xenografts, and inorganic grafts [5,6]. However, these methods have limitations, such as low donor mass, poor osteoinductive activity, risk of infection, host immunoreaction, and poor biological activity, imposing restrictions on practical application [1,6,7]. Hence, the clinical healing of large segmental bone defects remains a big challenge for surgeons, and synthetic bone grafts for promoting bone healing have become a focus of medical research. Compared with traditional therapies, tissue engineering is a promising approach for bone defect repair that can overcome the aforementioned limitations and act as an effective tool for application [8].
With the development of biomaterials [9], stem cells [10], nanomedicine [11] and tissue engineering technologies [12], bone replacement materials have been updated through scientific research. Whereas the first generation of inert materials such as alloys mainly play the role of “substitution support,” the third generation of biodegradable materials now exhibit biological activity. In addition to meeting the basic requirements of mechanical support, bone repair materials need to have excellent bone induction activity and vascular induction activity to promote rapid regeneration of bone tissue. Autografts are presently considered as the “Gold Standard” [13,14] for segmental bone defect repair and reconstruction because autogenous bone contains abundant blood vessels, and vascularization is a crucial process during the growth and development of bone [15]. Vascularization plays an important role in bone defect repair, which is important for provide nutrients and factors to support the biological processes of related cells of bone repair and avoid bone tissue necrosis [16]. By contrast, the lack of vascularization within the constructs of tissue-engineered bone (TEB) has been an obstacle to its in vivo application [17]. Angiogenesis occurs prior to mineralized bone formation during fracture repair [18], whereas vascular regeneration and remodeling play an important role in the repair of bone defects, enabling the delivery of nutrients and oxygen to cells in vivo and elimination of metabolic waste in vitro [19,20]. Neovascularization can be induced by the injection of specific growth factors, bioinspired surface modification of implants, and implantation of prevascularized TEB during the bone repair process [[20], [21], [22]]. However, spontaneous angiogenesis cannot be effectively achieved because of the decomposition of bioactive factors, modified surface wear particles, and prevascular necrosis during tissue development and repair. According to the anatomical characteristics of bone, both vasculogenic and osteogenic cells are arranged in an osteon that contains a central channel and numerous transversely oriented channels, in which various types of blood vessels form the vascular network of bone tissues [16,[23], [24], [25]]. Because of the complexity of endoskeletal vessels, traditional manufacturing technology may fail to recapitulate vascular physiology [20]. Thus, the fabrication of vascularized TEB as a bone replacement material has become a new possibility for large segmental bone defect healing in the future. Three-dimensional (3D) bioprinting provides a viable solution for achieving precise positioning of cells, tunable mechanical properties, and accurate control of the internal and external architecture, to establish and facilitate the clinical translation of vascularized tissue-engineered bone grafts.
3D bioprinting uses computer-assisted accumulation manufacturing to accurately control the positions of tissue cells, extracellular matrix (ECM), and biomolecules in the overall 3D structure, to enable biological activity and ultimately achieve living functionalities to fulfill the requirements of tissue replacement and organ transplantation [26,27]. The application of 3D bioprinting may enable the design of scaffold materials to be optimized for personalized and accurate repair and for the slow and controlled release of drugs or bioactive factors through a combination of gel and microsphere methods [28]. The 3D bioprinting technology utilizes cell-laden bio-inks to fabricate 3D vascularized constructs, offering a potential for vascularized TEBs. The osteon-mimetic scaffolds were fabricated by using 3D bioprinting technology and satisfactory biocompatibility and promotion of angiogenesis and osteogenesis in vitro and prompted the new blood vessels and new bone formation in vivo [16]. Coaxial 3D bioprinting can directly print straight vascular channels using a coaxial nozzle that simultaneously extrudes the outer hydrogel material and inner sacrificial material. However, it is difficult for coaxial 3D bioprinting to form vascular networks and to bioprint submicron-sized capillaries [29]. By contrast, Folkman reported that capillary endothelial cells (ECs) could form capillary networks when cultured in a tumor-conditioned medium [30]. Thus, it appears that ECs can develop an entire vascular network in vitro. Subsequently, it has been conceived that ECs can be seeded on the surfaces of 3D printed porous scaffolds to form complex vascular networks. However, this strategy is not widely accepted because it results in a non-uniform distribution of ECs throughout the channels [31]. Thus, the use of 3D bioprinting to fabricate scaffolds with effective internal vascularization and osteoinductive bioactivity for bone defect repair has become a medical research hotspot.
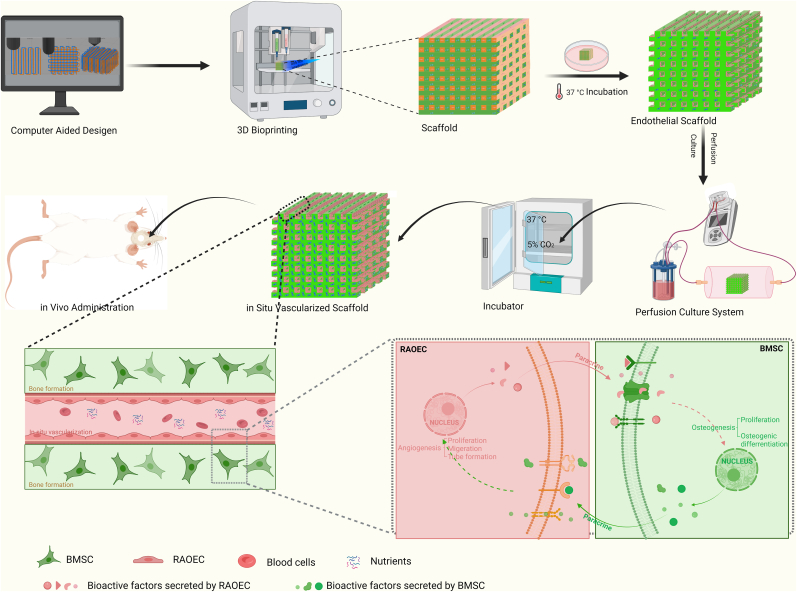
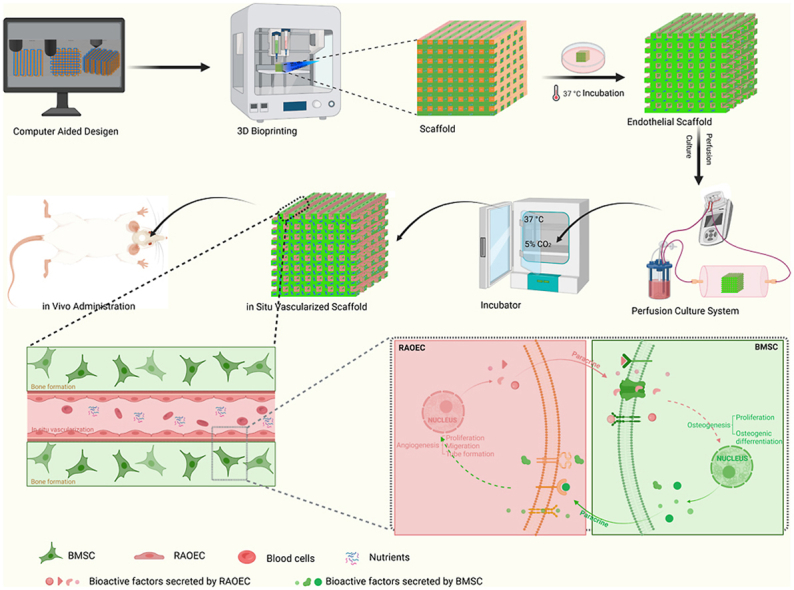
In this study, we developed a bioprinting strategy that enables ECs to form in situ vascular networks within a bone tissue-engineering scaffold. Two types of bio-inks are used to print the TEB: one, which is used as a matrix bio-ink, is a photocrosslinked extracellular matrix hydrogel supplemented with bone mesenchymal stem cells for osteogenesis, and the other, which is used as a templating bio-ink, is a thermosensitive hydrogel supplemented with ECs for angiogenesis. The templating bio-ink is printed line by line alongside the matrix bio-ink in alternate parallel lines, forming a one-layer structure, and then the 3D bioprinting is repeated layer by layer to create the construct structures. After the scaffolds are photocrosslinked, the ECs adhere onto the surfaces of the interconnected tubular channels by sacrificing the templating bio-ink. This in situ EC distribution via 3D bioprinting ensures a far greater cell seeding uniformity than that achieved using a conventional postseeding approach. In vitro experiments were then conducted to evaluate whether the 3D-bioprinted in situ vascularized tissue-engineered scaffolds had excellent osteogenesis and angiogenesis performance. RNA sequencing was used to analyze the expression of genes related to osteogenesis and angiogenesis. Finally, we applied scaffolds to a rat critical-size calvarial defect model to observe bone regeneration. This in situ vascularized tissue-engineered bone constructed using 3D bioprinting has a potential of being used as a bone graft for repairing large bone defects, with a possible clinical application in the future (Scheme 1).
Scheme 1.
Schematic representation of 3D bioprinting of in situ vascularized tissue engineered bone construction and application in repairing bone defects.
2. Materials and methods
2.1. Materials
Gelatin methacryloyl (GelMA), lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP), and a portable lamp that emits 405 nm irradiation were purchased from Engineering for Life (Suzhou, China). Polylactic acid (PLA)–polyethylene glycol (PEG)–PLA (3P) thermo-sensitive hydrogel was synthesized as a triblock polymer in accordance with a method in a previous study [32], with molecular weights of 3300–3500 Da for the PEG segment, and 1300–1500 Da for the PLA segment. The 3P temperature-sensitive hydrogel was synthesized as follows: i) PEG was added into a flask to remove water via melting and vacuum. ii) After cooling, monomer DL-lactide, solvent anhydrous toluene, and catalyst stannous caprylate were added. After the nitrogen was replaced, the system was heated to 200 °C and stirred for 4 h to facilitate a reflux reaction. iii) After the reaction, the product was dissolved in dichloromethane, and then added to n-hexane for precipitation and washing. The precipitated product was melted at a high temperature and vacuumed to remove the residual monomer and solvent before use. iv) The synthesized triblock polymer was prepared with water into a 10 wt% aqueous solution, and then sterilized using a 0.22 μm filter to obtain a temperature-sensitive hydrogel.
2.2. Characterization of thermo-sensitive hydrogel
Physical characterization of the 3P temperature-sensitive hydrogel was performed via nuclear magnetic resonance (NMR) and Fourier-transform infrared (FTIR) spectroscopy. In the NMR spectroscopy experiment, PLA–PEG–PLA was dissolved in D2O and stirred for 2 h. The 1H NMR spectra were recorded at room temperature using a Bruker-400 NMR at 400 MHz. Chemical shifts are reported in ppm, based on deuteroxide as an internal standard. In the FTIR spectroscopy experiment, the structure of PLA–PEG–PLA was analyzed using FTIR (Vertex 70v; Bruker, Germany). The hydrogels were first frozen at −80 °C, then lyophilized, pulverized, and mixed with KBr at a ratio of 1:100 (w/w). The samples were then made into disks using a tablet press. The analysis was performed using pulverized samples ranging from 500 to 4000 cm−1 with a resolution of 2 cm−1. Each sample was scanned 140 times to acquire the FTIR spectrum based on the average of all the scans.
2.3. Cell culture
2.3.1. BMSC extraction and cell culture conditions
Bone mesenchymal stem cells (BMSCs) were isolated from the femora and tibias of 3-week-old Sprague–Dawley rats, as described in a previous study [22]. The Sprague–Dawley rats were sacrificed via cervical dislocation and sterilized in 75% ethyl alcohol for 10 min. The femora and tibias were separated without attached soft tissue using sterile instruments in a biosafety cabinet. To expose the marrow cavity, both ends of the femora and tibias were removed. The bone marrow was rinsed using 5 mL low-glucose Dulbecco's modified Eagle's medium (DMEM; Gibco, USA) supplemented with 15% v/v fetal bovine serum (FBS) and 1% penicillin–streptomycin. The flushing fluid was filtered through a 100 μm sterile cell strainer. The cell suspension was centrifuged at 1000 rpm for 10 min. The centrifugal precipitate was suspended in cell culture medium and cultured in a 25 cm2 cell culture flask, which was incubated in a cell incubator (37 °C, 5% CO2). The medium was changed at 12 h for semi-quantitative medium exchange, at 24 h for whole medium exchange, and once three days thereafter. The BMSCs were passaged when the cell density reached approximately 90%.
2.3.2. Flow cytometric identification of BMSCs
The third-generation BMSCs were digested, centrifuged, and resuspended in Dulbecco's phosphate-buffered saline (DPBS). The samples were incubated in CD29-PE, CD31-PE, CD90-PE, CD44-APC, and CD-45APC for 30 min. BD FACSCanto was used to detect the BMSC surface molecules via flow cytometry. FlowJo software (version 10.5.2) was used for data analysis.
2.3.3. RAOEC lines and cell culture conditions
Rat aortic endothelial cell (RAOEC) lines were obtained from Cell Applications (USA). The RAOECs were cultured in DMEM supplemented with 10% v/v FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C with 5% CO2 in a cell incubator. The RAOECs were passaged when the cell density reached approximately 90%.
2.3.4. Generation of GFP-BMSCs and mCherry-RAOECs
Self-fluorescence-expressing cells were constructed via lentivirus infection as described in a previous study [33]. pLVX-EF1α-IRES-ZsGreen1 lentiviral vectors and packaging plasmids (PsPAX2 and pMD 2. G) co-transfected HEK293T cells to produce lentiviruses carrying ZsGreen1 fluorescent protein (GFP). Lentiviruses carrying mCherry fluorescent protein were purchased from GENE (Shanghai, China). BMSCs at passage 3 and RAOECs were seeded in 6-well plates at a density of 0.8 × 106 cells/well and cultured at 37 °C in 5% CO2 incubator. After the cells were grown to 60% confluence, lentiviruses with different fluorescent reporters were added to cell culture media containing 5 μg/mL polybrene. The cells were then incubated for 72 h to harvest self-fluorescent cells. ZsGreen1-BMSCs (GFP-BMSCs) and mCherry-RAOECs were sorted via BD Aria flow cytometry. The GFP-BMSCs and mCherry-RAOECs cultured in their respective culture media were used for the subsequent experiments.
2.4. Preparation of bio-inks
2.4.1. 5 wt% GelMA bio-ink
Freeze-dried GelMA was dissolved in DMEM at a weight fraction of 5 wt% with 0.25% (w/v) LAP, while light heating was avoided, in a water bath at 50 °C for 20 min. The 5 wt% GelMA solution was immediately filtered using a 0.22 μm sterile cell strainer for sterilization.
2.4.2. 5 wt% GelMA-BMSC bio-ink
After digestion with trypsin, the centrifuged BMSCs, which had a density of 5.0 × 106 cells/mL [34], were gently mixed with the prepared 5 wt% GelMA solution. The GelMA-BMSC solution was then placed in an assorted 10 cc cartridge (Nordson EFD, USA) at a 4 °C refrigerator for 10 min to undergo a sol–gel transition.
2.4.3. 10 wt% 3P-RAOEC bio-ink
The synthesized thermosensitive hydrogel (3P) was also dissolved in DMEM at a weight fraction of 10 wt% with heating and stirring in a water bath at 50 °C for 1 h. The 3P solution was then filtered through a 0.22 μm sterile cell strainer for sterilization. After the temperature dropped to 37 °C, the 3P solution was supplemented with 5.0 × 106 RAOECs/mL [31]. Then, the 3P solution was placed in an assorted 10 cc cartridge in a 4 °C refrigerator for 10 min to undergo a sol–gel transition.
2.5. Technological parameters
SolidWorks software was used to build the two experimental design models via digital modeling. Both slice processes were performed on Simplify3D software, with the parameters set to 0.2 mm extrusion width, 0.2 mm layer height, 20% interior fill percentage, and 600 mm/min printing speed. The nozzle for 5 wt% GelMA or GelMA-BMSC bio-ink was set to have a 22 °C temperature with a 0.05–0.075 MPa extrusion pressure, whereas the nozzle for 10 wt% 3P-RAOEC bio-ink was set to have a 25 °C temperature with a 0.07–0.09 MPa extrusion pressure. Additionally, the print bed was set to 5 °C to maintain the shapes of the gels after printing.
2.6. 3D bioprinting tissue-engineered scaffolds
The 3D-bioprinted tissue-engineered scaffolds were prepared using an extrusion-based 3D bioprinter (Arnold3, Starlight Material Technology Ltd., China) equipped with two temperature-controlled nozzles. Both files of the slice model (length × width × height of 10 × 10 × 2.5 mm3, slice thickness of 0.2 mm) were imported into the bio-printer. After the distance between the nozzles and print bed was adjusted, the prepared bio-inks were placed in a barrel equipped with 27G needles (Nordson EFD, USA). The GelMA scaffolds (G group) and GelMA (BMSC) scaffolds (GB group) were printed using GelMA bio-ink and BMSC-loaded GelMA bio-ink via one print nozzle. By contrast, the GelMA-3P (RAOEC) scaffolds (G-3PR group) were printed using GelMA bio-ink and RAOEC-loaded 3P bio-ink via two print nozzles. The GelMA (BMSC)–3P (RAOEC) scaffolds (GB-3PR group) were printed using BMSC-loaded GelMA bio-ink and RAOEC-loaded 3P bio-ink via two print nozzles. The hydrogel scaffolds were then exposed to 405 nm blue light (25 mW/cm2) for 0.5 min to form covalently crosslinked GelMA hydrogel scaffolds. The G scaffolds and GB scaffolds were immersed in a 5.0 × 106 RAOEC suspension to produce GelMA–RAOEC scaffolds (G-R group) and GelMA (BMSC)–RAOEC scaffolds (GB-R group). The different scaffold groups were summarized in Table 1. The corresponding printing process for GB-3PR is shown in Movie S1 and Movie S2 in the Supporting Information. Finally, the scaffolds were incubated with the culture medium in a cell incubator at 37 °C and 5% CO2.
Table 1.
3D bioprinting tissue-engineered scaffolds.
| Group | Bio-ink | Number of nozzles |
|---|---|---|
| G | 5 wt% GelMA | One |
| GB | 5 wt% GelMA-BMSC | One |
| G-3PR | 5 wt% GelMA +10 wt% 3P-RAOEC bio-ink | Two |
| GB-3PR | 5 wt% GelMA-BMSC + 10 wt% 3P-RAOEC bio-ink | Two |
| G-R | 5 wt% GelMA + RAOEC suspension | One |
| GB-R | 5 wt% GelMA-BMSC + RAOEC suspension | One |
2.7. Biomechanical properties
To test the biomechanical properties of the tissue-engineered scaffolds, uniaxial compression and tensile experiments were performed using an electronic dynamic fatigue testing machine (Linear Testing System, ZwickRoell, Germany). All the test samples were placed on a detection platform. The instrument was preloaded with 0.2 N to ensure that the two compressed plates were completely in contact with the samples. Stress–strain relation measurements were performed at a compression rate of 0.2 × 10−3 mm/s until the samples were ruptured. Measurements were obtained from at least three samples from each group. The compressive modulus was calculated as the slope of the linear region from 0 to 20% strain.
2.8. Cell proliferation
To determine the proliferation of BMSCs and RAOECs in hydrogels bioprinted in different ways, the 3D-bioprinted scaffolds (GB, GB-R, and GB-3PR) were cultured in DMEM supplemented with 10% v/v fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C with 5% CO2 in a cell incubator for 3, 5, and 7 days. At each measurement time point, GFP-BMSCs and mCherry-RAOECs were visualized and captured using a confocal fluorescence microscope (Nikon, Japan). The scaffolds were then dissolved in GelMA-Lysate reagent (0.3 mg/mL) to produce a cell suspension. GFP-BMSCs and mCherry-RAOECs were harvested and counted via flow cytometry.
2.9. In vitro osteogenesis assays
2.9.1. Alkaline phosphatase (ALP) activity and staining
The 3D-bioprinted scaffolds (G, GB, GB-R, and GB-3PR) were cultured in osteogenic differentiation medium (Cyagen, USA) for 7 and 14 days. For the detection of ALP activity, partial scaffolds (G, GB, GB-R, and GB-3PR) were dissolved using GelMA-Lysate reagent to produce a cell suspension. BMSCs were sorted from the cell mixture via flow cytometry. After centrifugation at 2000 rpm for 10 min, the cell pellet was lysed to release ALP protein using 0.3% Triton X-100 (Sigma-Aldrich, USA). The supernatants were incubated using an ALP assay kit (Beyotime, China) for 10 min. The ALP activity of each group was then quantified using the Tecan Sunrise absorbance reader (Sunrise, Switzerland) at 405 nm and normalized against total protein content. For the evaluation of ALP staining, the partial scaffolds of each group were stained with an ALP staining kit (Beyotime, China). After the samples were washed with phosphate-buffered saline (PBS), images were captured using a digital camera. The ALP-stained scaffolds were then dissolved using GelMA-Lysate reagent and cultured in 24-well plates for 24 h. Representative images were acquired using a microscope.
2.9.2. Alizarin red S (ARS) staining
Mineral deposits of the scaffolds were detected via ARS staining. For osteogenic induction for 7 and 14 days, the scaffolds (G, GB, GB-R, and GB-3PR) were stained with alizarin red dye solution (Cyagen, USA). After the samples were washed with PBS, the images were captured using a digital camera. The ARS-stained scaffolds were then dissolved in GelMA-Lysate reagent and cultured in 24-well plates for 24 h. Representative images were acquired using a microscope. Afterward, the mineral deposits were dissolved in 10 mM cetylpyridinium chloride and quantified (Macklin, China), and the optical density (OD) was measured using the Tecan Sunrise absorbance reader at 570 nm.
2.10. In vitro angiogenesis analysis
2.10.1. In situ angiogenesis assay
The perfusion culture system (MedPrin, China) consisted of a peristaltic pump, silicone tubes, a petri dish with a shelf, and a bottle containing the culture fluids (Fig. S1, Supporting Information). The 3D-bioprinted scaffolds (G-R, GB-R, G-3PR, and GB-3PR) were perfused continuously at a 300 mL/min speed during incubation in a cell incubator (37 °C, 5% CO2) for 3, 7, and 12 days (Movie S3, Supporting Information). The medium was replaced when the pH was below 7.1, and glucose consumption was greater than 30%. Vascular morphology was analyzed using a Leica TCS SP5 confocal microscope.
2.10.2. Migration assay
A scratch wound assay was used to quantify RAOEC migration. This study tested the ability of RAOECs to fill an area with a confluent monolayer of ECs after injury with a pipette tip. After 3 days of perfusion culture, the scaffolds (G-R, GB-R, G-3PR, and GB-3PR) were dissolved using GelMA-Lysate reagent. All cells were collected from miscible liquids that were processed via centrifugation at 1000 rpm for 5 min, after which the supernatant was discarded, and complete medium was added for repeated washing and centrifugation. The RAOECs were sorted via flow cytometry. Extractive RAOECs from the four groups were seeded at a density of 2 × 105 cells/cm2 in 6-well plates and then incubated in a cell incubator. After the cells reached approximately 100% confluence, pipette tips were used to create a line through the middle of each well of the monolayer. After 0 and 24 h of incubation, images of the wells were captured using a microscope. In accordance with a method described in previous study, ImageJ software (Rawak Software Inc., Stuttgart, Germany) was used to determine overall gap closure in the images [20].
2.10.3. Tube formation assay
Herein, 96-well plates were precoated with 50 μL of Matrigel (Corning, USA), which was stored in a 4 °C refrigerator. Matrigel exhibited a natural sol–gel transition for 30 min at 37 °C. Extractive RAOECs as migration assay were seeded at a density of 2 × 104 cells/well onto the coated 96-well plates, which were then incubated in a cell incubator for 6 h. Images of the wells were captured using a microscope to record the formation of tube-like structures. The number of meshes was then analyzed using ImageJ and the Angiogenesis Analyzer plugin.
2.11. RNA-sequencing analysis
For in vitro osteogenesis, the 3D-bioprinted scaffolds (GB and GB-3PR) were cultured in osteogenic differentiation medium for 7 days. For in vitro angiogenesis, the 3D-bioprinted scaffolds (GB-R and GB-3PR) were incubated in a perfusion culture system for 7 days. These scaffolds were dissolved using the GelMA-Lysate reagent. BMSCs and RAOECs were sorted via flow cytometry. The total RNAs of the BMSCs and RAOECs were extracted using the FastPure Cell/Tissue Total RNA Isolation Kit V2 (Vazyme, China). RNA quality and integrity were tested using the Agilent 2100 RNA Nano 6000 assay kit (Agilent Technologies, CA, USA). Transcriptomic sequencing service was provided by Annoroad Gene Technology, Annoroad Inc. (Annoroad, China), and was performed using the Illumina sequencing platform. In filtering out differentially expressed genes (DEGs), |log2 fold-change| ≥ 1, p value < 0.05, and Q value < 0.05 were used as cut-offs. For further analysis, volcano maps and gene ontology (GO) statistics bar charts of the DEGs were plotted. DEGs involved in osteogenesis and angiogenesis are present in the KEGG enrichment of biological processes.
2.12. In vitro real-time polymerase chain reaction (RT-PCR)
The expression of osteogenesis and angiogenesis genes was measured via RT-PCR. For in vitro osteogenesis, 3D-bioprinted scaffolds (GB, GB-R, and GB-3PR) were cultured in osteogenic differentiation medium for 7 and 14 days. For in vitro angiogenesis, the 3D-bioprinted scaffolds (G-R, GB-R, G-3PR, and GB-3PR) were incubated in a perfusion culture system for 7 and 14 days. After the scaffolds were dissolved, the BMSCs and RAOECs were sorted via flow cytometry. The total RNAs of BMSCs and RAOECs were extracted using the FastPure Cell/Tissue Total RNA Isolation Kit V2. The RNA concentration was measured using an ultramicrobiological detector (Thermo, USA). The RNA was reverse transcribed into complementary DNA (cDNA) using a reverse transcription kit (Vazyme, China). Mixtures of the synthesized cDNA, gene primers (forward primers + reverse primers), and Taq Pro Universal SYBR qPCR Master Mix (Vazyme, China) were placed into PCR microplates. RT-PCR was performed using the StepOnePlus Real-Time PCR system (Biosystems, USA). The osteogenesis and angiogenesis gene expression levels were normalized to the expression level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The RT-PCR experiments were repeated three times for each group. The osteogenesis and angiogenic gene primers that were used are listed in Table 2.
Table 2.
Primers for real-time PCR.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| RUNX2 | CATGGCCGGGAATGATGAG | TGTGAAGACCGTTATGGTCAAAGTG |
| OPN | GCCGAGGTGATAGCTTGGCTTA | TTGATAGCCTCATCGGACTCCTG |
| Oxterix | TGACTGCCTGCCTAGTGTCTACA | TGGATGCCCGCCTTGT |
| ALP | CACGTTGACTGTGGTTACTGCTGA | CCTTGTAACCAGGCCCGTTG |
| Col 1a1 CD31 VEGF |
GACATGTTCAGCTTTGTGGACCTC GAACAAACTTGCAAGGAGCAGGAA GCTCTCTTGGGTGCACTGGA |
GGGACCCTTAGGCCATTGTGTA CACGGAGCAAGAAAGACTCTGA CACCGCCTTGGCTTGTCACA |
| PDGF | TGGCTCGAAGTCAGATCCACA | TTCTCGGGCACATGGTTAATG |
| HIF1a | GTCCCAGCTACGAAGTTACAGC | CAGTGCAGGATACACAAGGTTT |
| GAPDH | GGCACAGTCAAGGCTGAGAATG | ATGGTGGTGAAGACGCCAGTA |
2.13. Western blot
Western blotting was performed to analyze the expression of related proteins, as follows: Cell lysis of the BMSCs and RAOECs harvested via RT-PCR was induced using a radioimmunoprecipitation assay (RIPA) lysis buffer (Solarbio, China) to produce proteins. Samples were placed in a 100 °C metal bath for 15 min, leading to protein denaturation. The extracted proteins were quantified using a protein quantification kit (Abbkine, China). Proteins (20 μg) from each group were electrophoresed on SDS-PAGE gels (GenScript, China). Afterward, the proteins on the gel were transferred onto pre-activated polyvinylidene fluoride (PVDF) membranes (Millipore, USA). After being blocked for 2 h with 5% skim milk, the membranes were trimmed according to molecular weight and then incubated with primary antibodies against GAPDH (Abbkine, Cat #: A01020, 1:2000), RUNX2 (Abbkine, Cat #: ABP53087, 1:2000), and HIF-1a (Abbkine, Cat #: ABP51513, 1:2000) overnight on a shaker at 4 °C. The next day, the membranes were rewarmed to room temperature for 30 min, washed three times for 5 min each in tris buffered saline with Tween-20 (TBST) (Sangon Biotech, China), and then incubated with the secondary antibodies anti-mouse IgG (Abcam, Cat #: ab6728, 1:3000) and anti-rabbit IgG (Abcam, Cat #: ab6721, 1:3000) for 2 h. After washing, the protein bands were visualized using a chemiluminescence kit (Abbkine, China) on an automatic chemiluminescence system (Sage Creation, China). Finally, the relative intensity of the protein bands was analyzed using ImageJ software.
2.14. Surgical procedure
The in vivo bone repair effect was monitored in a rat critical-size calvarial defect model [3]. All animal experiments followed the instructions of the Animal Experiment Center and acquired permission from the Ethics Committee of Southern University of Science and Technology (Resolution Number: SUSTech-JY202108003) and the Laboratory Animal Guide for Ethical Review of Animal Welfare (GB/T 35,892–2018). Forty-eight Sprague–Dawley rats (male, 8 weeks old, weighing approximately 250 g) were purchased from the Laboratory Animal Center. Rats were divided into four groups according to the type of scaffold implantation: GB, GB-R, G-3PR, and GB-3PR. To reduce intraoperative excretion, the rats were made to fast for 12 h and restricted for 4 h preoperatively. After successful isoflurane inhalation anesthesia, the rats were immobilized in the prone position with subcutaneous injection of meloxicam (0.1 mg/kg). The hair on the head was removed with a razor, and the exposed skin was disinfected with iodophor. A 10 mm midline incision was made, and anatomical exposure of the skull was performed using subperiosteal dissection of the soft tissue. A 5 mm critical calvarial defect in the skull was produced using a cranial drill with a 5 mm diameter drill bit. The drill bit was cooled via flushing with sterile saline during drilling. After the cranial defect tissue was removed, 5 mm in diameter cylindrical scaffolds cropped from 3D-bioprinted scaffolds which were cultured in an incubator for one day were inserted into the defect. The periosteal layer was sutured well with 5–0 absorbable suture material, and the skin wound was closed with 4–0 silk suture. All the procedures were performed by the same surgeon and assistants. During postoperative care, prophylactic penicillin sodium antibiotic (4 IU/kg, intramuscular) and meloxicam (0.1 mg/kg, subcutaneous) were administered for 3 days.
2.15. Micro-computed tomography (micro-CT) evaluation
Three rats in each group were euthanized with carbon dioxide in the 1st, 4th, 8th, and 12th weeks after surgery. Calvarial specimens were obtained and fixed in 4% paraformaldehyde (PFA) for 48 h. The calvarial specimens were then scanned at a voltage of 60 kV and an electric current of 100 mA via micro-CT (SkyScan 1276, Bruker, Belgium). NRecon, CTAn, and CTvol of the supplied software were used to process the data and reconstitute the 3D image. The bone mineral density (BMD) and the percentage of bone volume to tissue volume (BV/TV) were analyzed in the 4th, 8th, and 12th week.
2.16. Histologic and immunofluorescence analysis
After fixation in 4% PFA for approximately 48 h, the harvested calvarial specimens were decalcified for approximately six weeks in ethylenediamine tetraacetic acid (EDTA)-decalcified fluid (OKA, China). Afterward, parts of the samples and the fixed hearts, livers, spleens, lungs, and kidneys were completely dehydrated in a tissue dehydrator and embedded in paraffin wax. Paraffin wax slices of 5 μm thicknesses were prepared using a microtome. Longitudinal sections were subjected to hematoxylin and eosin (H&E) staining and Masson staining in accordance with standard protocols. After the samples were sealed with nail polish, digital images were captured using a scanner (Leica, Germany). The newly generated bone tissues and the biosafety in vivo were analyzed for each group. The quantitative analysis of newly formed tissue by HE and Masson staining was performed by Image-Pro Plus.
In addition, other decalcified samples were dehydrated in 30% sucrose for three days at 4 °C and then embedded in OCT (Tissue-Tek, USA). For immunofluorescence analysis, 10-μm-thick sections were prepared using a freezing microtome. The sections were permeabilized with 0.5% Triton-X for 30 min at room temperature. The samples were washed 3 times in PBS for 5 min, after which they were blocked with 5% normal goat serum for 30 min. The samples were then incubated with primary antibodies, i.e., anti-vWF (Servicebio, China) or anti-CD31 (Servicebio, China), overnight at 4 °C in a lucifugal chamber. To remove unbound primary antibodies, the slides were washed three times in PBS for 5 min, followed by 1 h incubation with fluorescent secondary antibodies (A594 goat anti-rabbit or A647 goat anti-mouse) at room temperature. The samples were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 5 min and washed in PBS, after which they were sealed with an anti-fluorescence quencher. The samples were then observed, and their images collected using a fluorescence microscope (Leica, Germany). Neovascularization in each group was analyzed.
2.17. Statistical analysis
All data are expressed as mean ± standard deviation (SD), and statistical analyses were performed using GraphPad Prism software (version 6.0; La Jolla, CA, USA). Student's t-test was used to compare the statistical significance of the differences between the two groups. Statistically significant differences among multiple groups were analyzed via one-way analysis of variance (ANOVA) with a post-hoc Tukey's test for multiple comparisons. For all tests, a p-value <0.05 was considered statistically significant.
3. Results and discussion
3.1. Synthesis and characterization of 3P and GelMA hydrogels
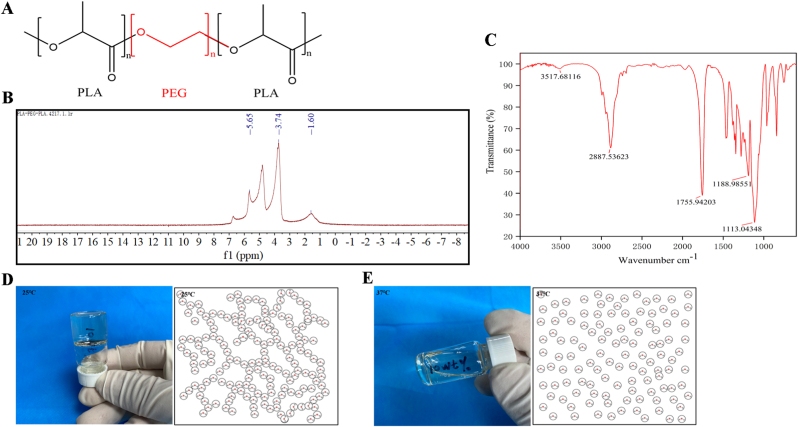
The PLA–PEG–PLA tri-block copolymer was synthesized, and its conjugation was confirmed via NMR and FTIR spectroscopy [32] in Fig. 1. The chemical structure (Fig. 1A) was analyzed via 1H NMR (Fig. 1B). The PLA contained protons of methyl and methine, which were observed at approximately 5.65 and 1.60 ppm, respectively. The presence of methylene protons in PEG was observed at approximately 3.74 ppm. By contrast, from the FTIR results (Fig. 1C), we observed sharp and intense bands at 1755.94 cm−1 and 1113.04 cm−1, which were attributed to the presence of carboxylic ester (C O) and ether (C–O) groups, respectively, indicating the formation of the PLA–PEG–PLA copolymer. In addition, the physical characteristics of the 3P thermo-sensitive hydrogel revealed a natural gel–sol transition. As shown in Fig. 1D and E, the synthesized 3P molecules were linked together, forming a gel state at 25 °C, as determined through macro and microscopic observations (Fig. 1D). Meanwhile, the 3P molecules were uniformly scattered and presented a sol state at 37 °C (Fig. 1E). Thus, the characteristics of the copolymer were confirmed, in line with the widespread use of PLA–PEG–PLA in many applications due to its superior compatibility and biodegradability [[35], [36], [37]].
Fig. 1.
(A) Chemical structure of synthetic PLA–PEG–PLA copolymer. (B) 1H NMR spectrum of PLA–PEG–PLA copolymer in D2O. (C) FTIR spectrum of PLA–PEG–PLA copolymer. (D) Macro and microscopic observations of morphology of PLA–PEG–PLA hydrogel at 25 °C. (E) Macro and microscopic observations of morphology of PLA–PEG–PLA hydrogel at 37 °C.
By contrast, GelMA is a photosensitive biomaterial that can be rapidly crosslinked and cured under blue or ultraviolet light to form a three-dimensional structure with a certain strength [38]. It supports cell proliferation and migration by loading tumor cells, cardiomyocytes, chondrocytes, and other cells. The mechanical properties of crosslinked hydrogels can be adjusted via changes in the degree of substitution and the concentration of the GelMA material. GelMA has good biocompatibility and is used mainly in tissue engineering, 2D/3D cell culture, and other tissue procedures. Similar to the 3P thermo-sensitive hydrogel, GelMA also exhibits the physical property of sol–gel transition [31].
3.2. Obtaining GFP-BMSCs and mCherry-RAOECs
The suspended cells rushed from the bone marrow were subcultured for 7 days. A colony of cells was formed at the bottom of the petri dish under the microscope and distributed in clusters that looked like whirlpools (Fig. S2A, Supporting Information). At present, BMSC surface antigens are considered to be non-monoclonal, expressing a series of surface markers of mesenchymal, endothelial, and epidermal cells, such as SH2, CD29, CD44, CD71, CD90, and other surface proteins, but do not express CD14, CD31, and CD45 of hematopoietic cells [39,40]. In this study, representative antigen markers CD29, CD31, CD44, CD45, and CD90 were selected to identify the extracted BMSCs. The results of flow cytometry show that the homogeneity of the third-generation BMSCs was above 90%. The proportions of CD31–CD45 cells in the BMSCs were 0.22% and 39.71%, respectively. The proportions of CD29+CD44+CD90+ cells in the BMSCs were 79.66%, 91.63%, and 95.12%, respectively (Fig. S2A, Supporting Information). Self-fluorescence-expressing BMSCs and RAOECs were transfected with lentiviruses. The lentivirus expression vector additionally expressed green and red fluorescent proteins to assess infection efficiency. The purity of the sorted populations was confirmed via GFP-BMSC and mCherry-RAOEC sorting strategies via flow cytometry, with values of 99.2% (Fig. S2B, Supporting Information) and 87.8% (Fig. S2C, Supporting Information), respectively. Ultimately, this method provides cell sources for the preparation of bio-inks.
3.3. Printability of hydrogel bio-ink
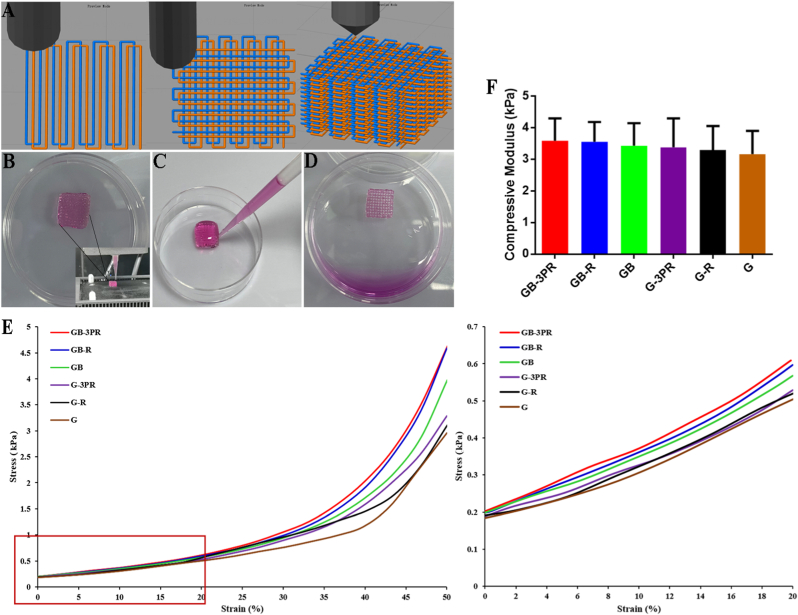
Because both GelMA and 3P hydrogels have the thermal properties of sol–gel transition, the 3D-bioprinted scaffolds were fabricated using an extrusion-based 3D bioprinter equipped with two temperature-controlled nozzles. After the bio-ink was prepared, the nozzles were warmed to a set temperature that allowed smooth filament extrusion, while the print bed was set to 5 °C to enable the extruded bio-ink to change quickly from sol to gel. To demonstrate the printing process, a slice of the cube model (length × width × height of 10 × 10 × 2.5 mm3, slice thickness of 0.2 mm) was accessed by the bio-printer. As shown in Movie S1 and Movie S2, the printing process based on two nozzles was continuous, and the motion trajectories were consistent with the technological parameters of the CAD design (Fig. 2A). We fabricated a 10 × 10 × 2.5 mm3 cuboid scaffold of hydrogel by stacking materials layer by layer. These results indicate that the proposed approach has excellent repeatability and accuracy. In this study, the GelMA bio-ink loaded with BMSCs was printed to form a scaffold with complex 3D interconnected networks after photocrosslinking, whereas the 3P bio-ink loaded with RAOECs was printed in a manner similar to the extruded lines of GelMA loaded with BMSCs, i.e., in alternate parallel lines. Subsequently, the scaffolds were structurally stable on the cooled print bed, exhibiting a void-free 3D shape (Fig. 2B). After the scaffolds were incubated at 37 °C for 1 h (Fig. 2C), the 3P hydrogel flowed out from the scaffold channels and then formed a porous scaffold construction with a about 500 μm pore and cross-sectional diameter (Fig. 2D), which is important for the diffusion of nutrients and waste products for the viability of hydrogel-loaded cells. During this incubation process, some RAOECs attached in situ to the surfaces of the scaffold channels.
Fig. 2.
(A) Path planning and design of 3D printed in situ vascularized tissue engineering bone model. (B) In situ vascularized scaffold printed using BMSC-loaded GelMA bio-ink and RAOEC-loaded 3P bio-ink via two print nozzles, presenting a void-free 3D shape. (C) Cell culture medium was added to surface of void-free scaffold at 37 °C. (D) 3P hydrogel flowed out from scaffold channels and then formed porous scaffold construction after incubation at 37 °C for 1 h. Mechanical properties of 3D-bioprinted scaffolds: (E) stress–strain curves of hydrogel scaffolds, and (F) compressive modulus analysis of hydrogel scaffolds. (n = 3, each group).
3.4. Mechanical properties
The mechanical properties of the 3D-bioprinted scaffolds were evaluated via uniaxial compression and tensile testing. The stress–strain curves of the hydrogel samples are shown in Fig. 2E. It was determined that the GB-3PR scaffold had an increasing trend in stress with respect to increasing strain that was similar to that exhibited by other hydrogel scaffolds. Moreover, the GB-3PR scaffold had no significant difference in term of compressive modulus (4.7 ± 1.90 kPa) compared to those of the GB-R (4.5 ± 1.84 kPa), GB (4.0 ± 1.39 kPa), G-3PR (4.2 ± 2.09 kPa), G-R (4.0 ± 1.71 kPa), and G (3.4 ± 1.55 kPa) scaffolds (Fig. 2F). These scaffolds had sufficient mechanical properties for maintaining their spatial structures in non-weight-bearing areas. The hydrogel scaffolds have good permeability, and the cells inside the bioprinted scaffolds can exchange substances to obtain necessary nutrients. However, as reported in a past study, although the mechanical properties of high-strength scaffolds can maintain structural integrity, they also restrain the biological activity of cells [41]. Excessive strength and stiffness of the scaffolds will damage the surrounding bone tissue and the stability of the bone-to-implant interface. To achieve bioprinted tissues with both proper structural fidelity and biofunctionality, a scaffold should possess appropriate mechanical and biological properties. Recent investigations have shown that GelMA hydrogels are more suitable cell-laden bioinks because of their high cell stability and viability, and have adequate mechanical strength to allow the scaffold to retain its initial geometry with a concentration (i.e., ≥ 5w/t %) [34,42]. Hydrogel scaffolds are beneficial for cell biological behavior and cellular interactions [43]. Hence, in situ bioprinting not only provides seed cells for vascularization but also satisfies the mechanical strength of TEBs required for bone defect sites.
3.5. In situ vascularized scaffold enhances osteogenic differentiation in vitro
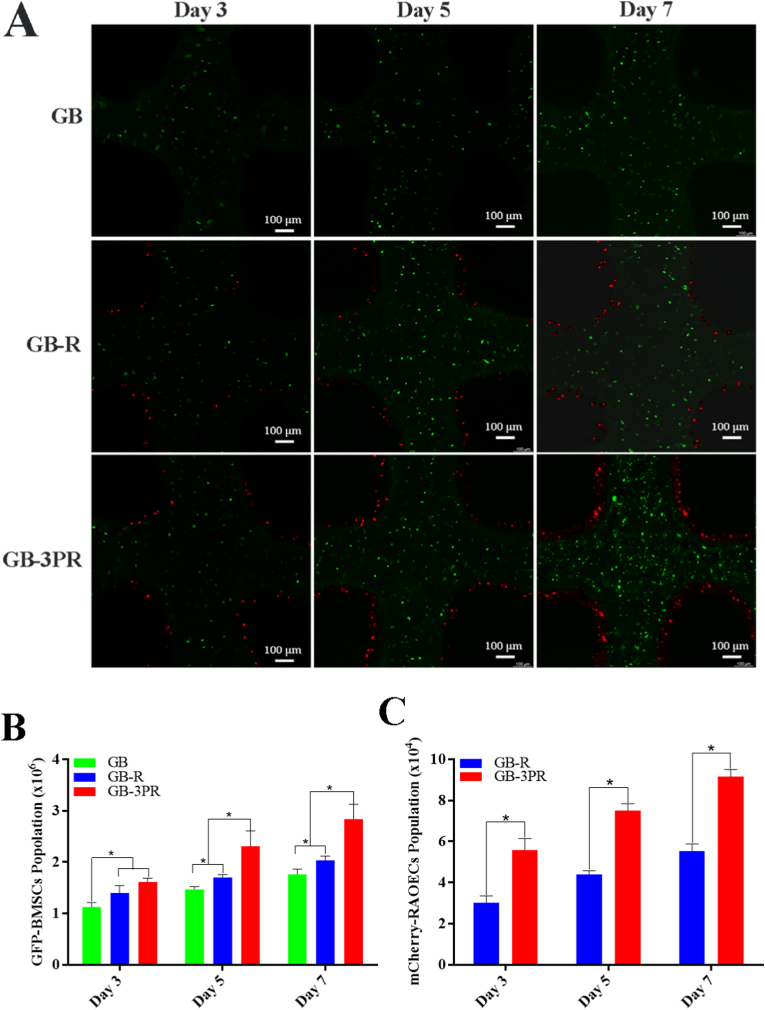
The proliferation of BMSCs and RAOECs was evaluated at the 3rd, 5th, and 7th days, as shown in Table 3 and Fig. 3. Compared to the GB and GB-R groups, the GB-3PR group clearly promoted the proliferation of BMSCs (Fig. 3A and B). Excellent cell proliferation provides the basis for osteogenic differentiation [44,45].
Table 3.
GFP-BMSCs proliferation on hydrogel scaffolds.
| Time | Index | GB | Group GB-R | GB-3PR | p (GB vs. GB-R) | p (GB vs. GB-3PR) | p (GB-R vs. GB-3PR) |
|---|---|---|---|---|---|---|---|
| Day 3 | Population (×106) | 1.1 ± 0.07 | 1.4 ± 0.12 | 1.6 ± 0.06 | 0.047 | 0.033 | 0.179 |
| Day 5 | Population (×106) | 1.5 ± 0.04 | 1.7 ± 0.05 | 2.3 ± 0.24 | 0.006 | 0.008 | 0.025 |
| Day 7 | Population (×106) | 1.8 ± 0.08 | 2.0 ± 0.07 | 2.8 ± 0.24 | 0.021 | 0.004 | 0.010 |
Fig. 3.
In vitro cell proliferation on hydrogel scaffolds. (A) Representative confocal fluorescence microscopy images of GFP-BMSC and mCherry-RAOEC proliferation after 3, 5, and 7 days of cultivation. (B) and (C) Quantitative analysis of cell proliferation. Calculated GFP-BMSC and mCherry-RAOEC population of each sample by flow cytometry. (n = 3, each group). ∗p < 0.05.
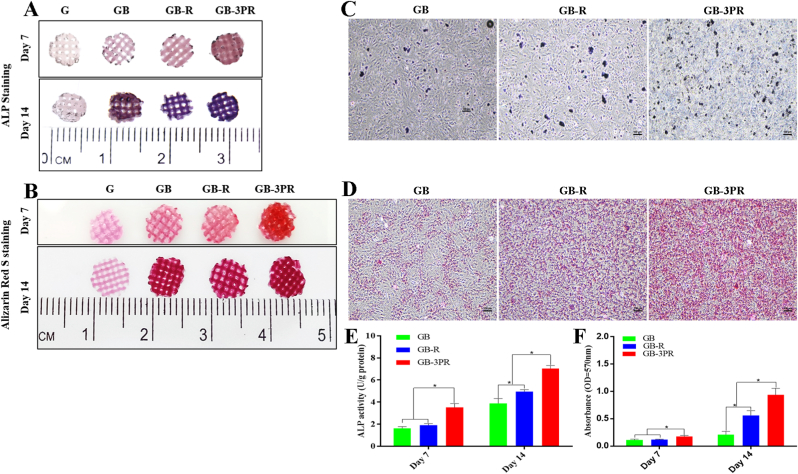
In our previous study, a tissue-engineered bone graft was prevascularized via the insertion of a femoral vascular bundle and implantation to treat large bone defects, and demonstrated a significantly higher volume of regenerated bone [22]. Considering the potential effects of vascular endothelial cells on the osteogenic ability of tissue-engineered bone, we designed a method for in situ 3D bioprinting of vascularized TEB to induce osteogenesis. The in vitro evaluation of osteogenic differentiation of BMSCs is shown in Fig. 4. ALP and ARS staining were performed to assess osteogenic differentiation of the scaffolds at the 7th and 14th days (Fig. 4A–D). The results show that the blue and red color intensities of the GB-3PR scaffold were visibly stronger than those of the GB-R and GB scaffolds. Moreover, for each group, the stained color for osteogenic induction at the 14th day was higher than that at the 7th day. Alkaline phosphatase was an early enzyme marker of the osteogenic differentiation of BMSCs [46]. According to the quantitative assays, the GB-3PR scaffold exhibited significantly higher ALP activity (3.5 ± 0.08 U/g, 7.1 ± 0.04 U/g) than those exhibited by the GB-R scaffold (1.9 ± 0.01 U/g, 5.0 ± 0.02 U/g) and GB scaffold (1.6 ± 0.01 U/g, 3.9 ± 0.1 U/g), at the 7th and 14th days (Fig. 4E). The osteogenic ability of the embedded BMSCs co-cultured with RAOECs may be attributed to their osteoinductive characteristics. The formation of inorganic calcium during osteogenic differentiation is a basic biological characteristic of the bone-repair process [47]. Deposition degrees of lysed calcium also supported the inference of better osteogenesis in the GB-3PR (0.2 ± 0.01, 0.9 ± 0.02) scaffold than in the GB-R scaffold (0.1 ± 0.01, 0.6 ± 0.2) and GB scaffold (0.1 ± 0.01, 0.2 ± 0.01) at the 7th and 14th days (Fig. 4F). The in situ vascularized TEB showed excellent osteogenic mineralization. In addition, the GB-R scaffold demonstrated significant differences with the GB scaffold after 14 days of osteogenic induction. Overall, the results indicate that in situ vascularization promoted the osteogenic differentiation of BMSCs.
Fig. 4.
In vitro evaluation of osteogenic differentiation of BMSCs in 3D-bioprinted scaffolds. (A) Alkaline phosphatase staining of hydrogel scaffolds induced by osteogenesis for 7 and 14 days. (B) Alizarin red S staining of hydrogel scaffolds induced by osteogenesis for 7 and 14 days. (C) Alkaline phosphatase staining of dissolved hydrogel scaffolds induced by osteogenesis for 14 days under microscopic observation. (D) Alizarin red S staining of dissolved hydrogel scaffolds induced by osteogenesis for 14 days under microscopic observation. (E) Quantitative analysis of alkaline phosphatase activity. (n = 4, each group). (F) Quantitative analysis of mineralized nodules. (n = 4, each group). ∗p < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. 3D in situ bioprinting promotes angiogenesis
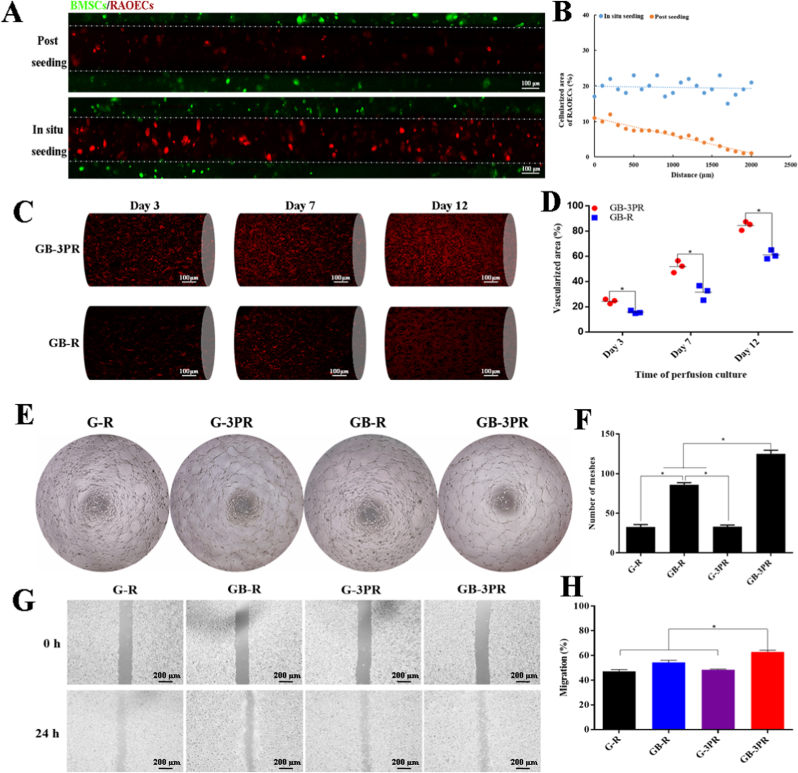
The GB-3PR group showed a remarkably uniform distribution of RAOECs adhering to the wall of GelMA loaded with BMSCs after the release of thermo-sensitive hydrogel at 37 °C. By contrast, the GB-R group showed an uneven gradient distribution as the distance increased (Fig. 5A). In addition, the GB-3PR group had a higher acellularized area of RAOECs than that in the GB-R group (Fig. 5B). These results indicate that the 3D-bioprinted in situ seeding approach could achieve a better cell seeding efficiency compared to that of the conventional postseeding approach. For in situ vascularization, the bioprinted scaffolds were perfused continuously during incubation in a cell incubator. Previous studies have shown that a tubular perfusion system bioreactor increases cell viability in 3D printed vascular networks [48]. The GB-3PR group exhibited obvious development of vascularization with the extension of incubation time (Fig. 5C). Simultaneously, quantitative analysis of the vascularized area showed statistical differences between the GB-3PR and GB-R groups (Fig. 5D). These scaffolds with post-seeded vascular endothelial cells lacked inherent vasculature and developed a necrotic core, where no nutrient exchange occurred as long as they were implanted. Hence, this 3D-bioprinted in situ vascularization method can be effectively used to produce engineered bone graft substitutes.
Fig. 5.
In vitro angiogenesis assays. (A) Representative confocal fluorescence microscopy images of mCherry-RAOECs, comparing postseeding method with 3D-bioprinted in situ seeding approach. (B) Assessment of uniformity of mCherry-RAOEC seeding. (C) Confocal fluorescence microscopy images assessing vasculogenic capacity of mCherry-RAOECs after 3, 7, and 12 days of perfusion culture. (D) Quantitative analysis of vascularized area. (E) and (F) Tube formation assay of RAOECs and quantitative analysis of formed meshes. (n = 3, each group). (G) and (H) Migration assay and quantitative analysis of RAOEC migration. (n = 3, each group). ∗p < 0.05.
Angiogenesis is a complicated and multi-step process that involves vascular endothelial cell proliferation, migration, and formation of structures [49,50]. As shown in Table 4 and Fig. 3A and C, the GB-3PR group exhibited better proliferation of RAOECs than those exhibited by the other groups, providing conditions for angiogenesis. Tube formation assays were then used to investigate the angiogenic process based on the formation of capillary-like structures. After three days of perfusion culture, the RAOECs were isolated from the 3D-bioprinted scaffolds. The results presented in Fig. 5E suggest that RAOECs isolated from the GB-3PR group formed more capillary-like networks with closed structures than those in the other groups. Quantification of the number of meshes revealed that the value for the GB-3PR group (125 ± 4.58) was significantly higher than the values for the G-R (32 ± 3.06), GB-R (86 ± 2.65), and G-3PR (33 ± 2.01) groups (Fig. 5F). In addition, postseeded RAOECs in the GB-R group were more easily able to self-assemble and elongate, forming capillary-like networks, than those in the G-R and G-3PR groups. Sorted RAOECs were also used to detect migration, revealing that the GB-3PR group had significant migratory activity compared to those exhibited by the other groups (Fig. 5G). Quantification of the scratch area indicated that wound closure for the GB-3PR group (62.8 ± 1.13%) was significantly faster than that the values for the G-R (47.2 ± 1.17%), GB-R (54.5 ± 1.34%), and G-3PR (48.5 ± 0.46%) groups (Fig. 5H). The RAOECs had a relatively better angiogenesis performance via coculture with BMSCs in the GB-3PR and GB-R groups than via lone cultures in the G-R and G-3PR groups, confirming that BMSC/RAOEC coculture promoted angiogenesis [51]. However, the GB-3PR group demonstrated markedly increased angiogenesis compared to that exhibited by the GB-R group when the BMSC/RAOEC coculture was used. 3D bioprinting of in situ vascularized tissue-engineered bone might form a symbiotic niche exerting an impact on intercellular interactions, which assisted RAOECs in forming new tube structures and were linked to the initial steps of angiogenesis. Hence, 3D in situ bioprinting provides a valuable tool for angiogenesis inside scaffolds as a result of endothelial cell proliferation, migration, branching, and lumen formation.
Table 4.
mCherry-RAOECs proliferation on hydrogel scaffolds.
| Time |
Index |
Group GB-R GB-3PR |
p (GB vs. GB-3R) |
|
|---|---|---|---|---|
| Day 3 | Population ( × 104) | 3.0 ± 0.27 | 5.6 ± 0.46 | 0.002 |
| Day 5 | Population (×104) | 4.4 ± 0.15 | 7.5 ± 0.27 | <0.001 |
| Day 7 | Population (×104) | 5.5 ± 0.27 | 9.2 ± 0.28 | <0.001 |
3.7. RNA-sequencing analysis
However, although 3D-printed scaffolds have been shown to be superior in promoting bone defect repair, the cell-level specific regulatory network had not yet been revealed, limiting their further application. In recent years, RNA-sequencing analysis has been developed as an effective analytical method and is widely used in biology and medicine [52,53]. In this study, 3D bioprinting of an in situ vascularized scaffold was performed, and RNA-sequencing analysis was used to explore the regulatory effect on both BMSCs and RAOECs compared with that for a control group.
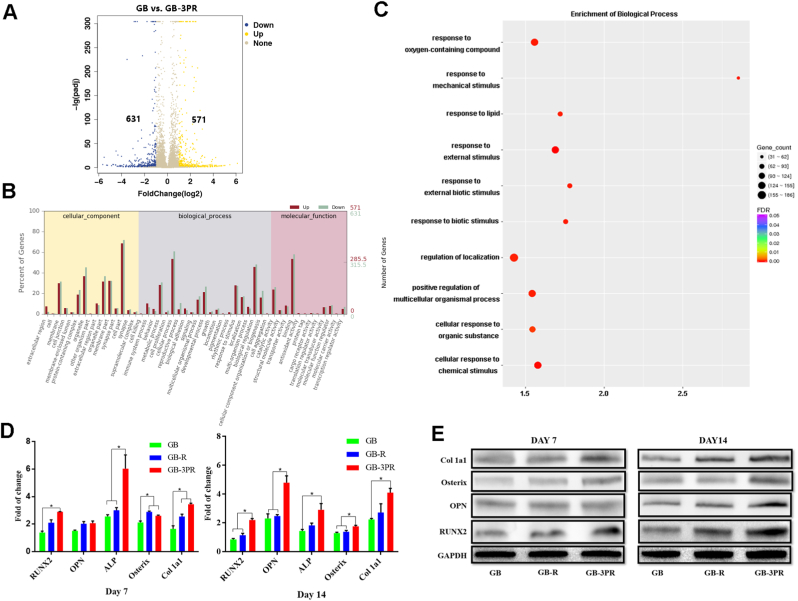
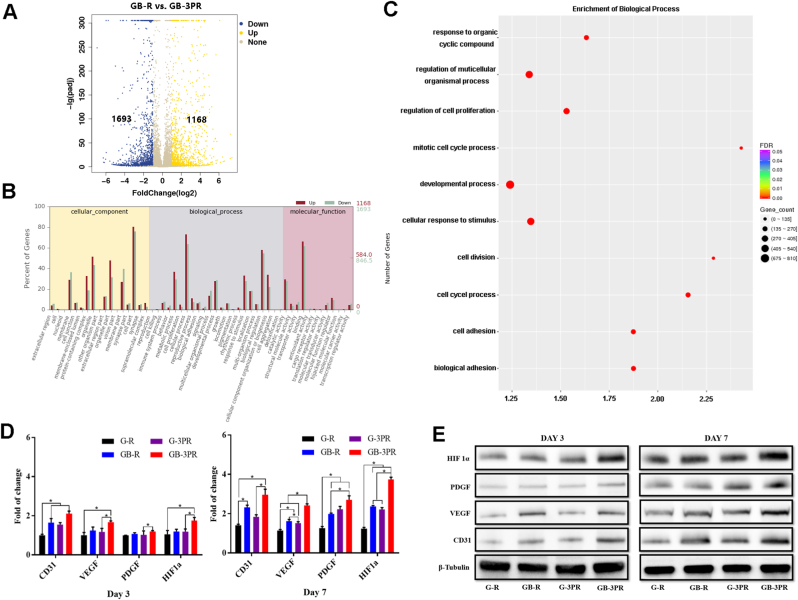
To validate the DEGs, both the osteogenic induction of obtained BMSCs and the angiogenesis of obtained RAOECs were analyzed via RNA sequencing [54], as shown in Fig. 6 and Fig. 7. The results show that the GB-3PR group had 571 upregulated DEGs relative to the GB group (Figs. 6A), and 1168 upregulated DEGs relative to the GB-R group (Fig. 7A). Although the osteogenic differentiation of BMSCs and angiogenesis of RAOECs in the GB-3PR group were better promoted than those in the other groups (Figs. 4 and 5), the function and relationship of DEGs in osteogenic induction and vascularization were not clear. GO enrichment was then used to analyze the functions of DEGs in cellular components, biological processes, and molecular functions. Compared with those in the GB group, the in vitro osteogenesis, gene expression for cell proliferation, biological adhesion, and biological regulation in biological processes were upregulated in the GB-3PR group (Fig. 6B). Moreover, in vitro angiogenesis and gene expression in the GB-3PR group were upregulated in cell proliferation, biological adhesion, and biological regulation in biological processes compared with those in the GB-R group (Fig. 7B). In this study, GO enrichment was used to analyze the functions of DEGs in biological processes. Compared to those of the GB group, the response to oxygen-containing compounds, response to external stimuli, and positive regulation of multicellular organismal processes were upregulated in the GB-3PR group (Fig. 6C), suggesting that GB-3PR could promote osteogenic differentiation of BMSCs. Furthermore, compared to those of the GB-R group, the regulation of multicellular organismal processes, regulation of cell proliferation, developmental processes, cellular responses to stimuli, cell adhesion, and biological adhesion were upregulated in the GB-3PR group (Fig. 7C), indicating that GB-3PR promoted RAOEC proliferation and adhesion ability, providing conditions for angiogenesis. In vitro real-time PCR analysis showed that the relative osteogenic mRNA levels of RUNX2, OPN, ALP, Osterix, and Col1a1 in the GB-3PR group were upregulated compared with those in the GB and GB-R groups (Table 5 and Fig. 6D), and that the relative angiogenic mRNA levels of CD31, VEGF, PDGF, and HIF1a in the GB-3PR group were upregulated compared with those in the G-R, G-3PR, and GB-R groups (Table 6 and Fig. 7D), which were consistent with the results of RNA sequencing analysis. Western blotting also showed higher osteogenic and angiogenic relative protein levels (Fig. 6, Fig. 7E). Therefore, 3D bioprinting of the in situ vascularized scaffold ultimately influenced the functions of BMSCs (proliferation and osteogenic differentiation) and RAOECs (proliferation and migration) via the upregulation of osteogenesis-related genes (RUNX2, OPN, ALP, Osterix, and Col1a1) and vascular-related genes (CD31, VEGF, PDGF, and HIF1a) of BMSC–RAOEC interactions at the transcriptional level, which is of great importance for their utilization in and guidance of the process of bone regeneration. In addition, 3D bioprinting of in situ vascularized scaffolds could be used to regulate the osteogenic ability of BMSCs and vascular capacity of RAOECs in biological processes and to adjust the molecular network in a symbiotic microenvironment to reduce barriers to bone regeneration. The potential biological mechanisms identified by transcriptome sequencing will be provided in further research.
Fig. 6.
(A) Volcano map of BMSC differentially expressed genes in GB and GB-3PR groups. (B) GO statistical histogram of differentially expressed genes in GB and GB-3PR groups. (C) Q-value enrichment map of GB-3PR in biological process. Enrichment ratio was calculated as (differentially expressed genes in this pathway/all differentially expressed genes)/(genes annotated to this pathway/all annotated genes). In filtering out differentially expressed genes, |log2 fold-change| ≥ 1, p-value < 0.05, and Q-value< 0.05 were used as cut-offs. (D) mRNA levels of osteogenesis-related genes RUNX2, OPN, ALP, Osterix, and Col1a1, as determined by real-time PCR. (n = 3, each group). (E) Protein levels of osteogenesis-related proteins RUNX2, OPN, Osterix, and Col1a1, as measured by western blotting. ∗p < 0.05.
Fig. 7.
(A) Volcano map of RAOEC differentially expressed genes in GB-R and GB-3PR groups. (B) GO statistical histogram of differentially expressed genes in GB-R and GB-3PR groups. (C) Q-value enrichment map of GB-3PR in biological process. Enrichment ratio was calculated as (differentially expressed genes in this pathway/all differentially expressed genes)/(genes annotated to this pathway/all annotated genes). In filtering out differentially expressed genes, |log2 fold-change| ≥ 1, p-value < 0.05, and Q-value< 0.05 were used as cut-offs. (D) mRNA levels of angiogenesis-related genes CD31, VEGF, PDGF, and HIF1a, as determined by real-time PCR. (n = 3, each group). (E) Protein levels of angiogenesis-related proteins CD31, VEGF, PDGF, and HIF1a, as measured by western blotting. ∗p < 0.05.
Table 5.
Fold change in mRNA levels of osteogenesis-related genes.
| Time | Gene | GB | Group GB-R | GB-3PR | p (GB vs. GB-R) | p (GB vs. GB-3PR) | p (GB-R vs. GB-3PR) |
|---|---|---|---|---|---|---|---|
| Day 7 | RUNX2 | 1.4 ± 0.15 | 2.1 ± 0.29 | 2.9 ± 0.02 | 0.097 | <0.001 | 0.003 |
| OPN | 1.5 ± 0.07 | 2.0 ± 0.22 | 2.1 ± 0.22 | 0.078 | 0.059 | 0.871 | |
| ALP | 2.6 ± 0.17 | 3.0 ± 0.27 | 6.0 ± 1.41 | 0.123 | 0.026 | 0.041 | |
| Osterix | 2.1 ± 0.16 | 2.9 ± 0.08 | 2.6 ± 0.07 | <0.001 | 0.016 | 0.051 | |
| Col 1a1 | 1.7 ± 0.32 | 2.6 ± 0.25 | 3.4 ± 0.11 | 0.035 | 0.002 | 0.009 | |
| Day 14 | RUNX2 | 0.8 ± 0.09 | 1.1 ± 0.17 | 2.2 ± 0.15 | 0.097 | <0.001 | 0.003 |
| OPN | 2.3 ± 0.45 | 2.5 ± 0.14 | 4.8 ± 0.67 | 0.648 | 0.012 | 0.009 | |
| ALP | 1.4 ± 0.16 | 1.8 ± 0.23 | 2.9 ± 0.61 | 0.114 | 0.029 | 0.078 | |
| Osterix | 1.3 ± 0.08 | 1.4 ± 0.12 | 1.7 ± 0.11 | 0.375 | 0.008 | 0.033 | |
| Col 1a1 | 2.2 ± 0.09 | 2.7 ± 0.87 | 4.1 ± 0.42 | 0.480 | 0.004 | 0.112 |
Table 6.
Fold change in mRNA levels of angiogenesis-related genes.
| Time | Gene | G-R | Group GB-R | G-3PR | GB-3PR | p (G-R vs. GB-3PR) | p (GB-R vs. GB-3PR) | p (G-3PR vs. GB-3PR) |
|---|---|---|---|---|---|---|---|---|
| Day 3 | CD31 | 1.0 ± 0.047 | 1.7 ± 0.172 | 1.6 ± 0.078 | 2.1 ± 0.182 | 0.001 | 0.060 | 0.016 |
| VEGF | 1.0 ± 0.116 | 1.2 ± 0.147 | 1.2 ± 0.148 | 1.7 ± 0.088 | 0.003 | 0.024 | 0.015 | |
| PDGF | 1.0 ± 0.008 | 1.1 ± 0.049 | 1.0 ± 0.164 | 1.2 ± 0.057 | 0.008 | 0.042 | 0.226 | |
| HIF1α | 1.1 ± 0.159 | 1.2 ± 0.086 | 1.2 ± 0.114 | 1.8 ± 0.226 | 0.022 | 0.032 | 0.033 | |
| Day 7 | CD31 | 1.4 ± 0.072 | 2.3 ± 0.156 | 1.8 ± 0.149 | 2.9 ± 0.393 | 0.005 | 0.099 | 0.019 |
| VEGF | 1.1 ± 0.078 | 1.6 ± 0.122 | 1.5 ± 0.103 | 2.4 ± 0.115 | <0.001 | 0.003 | 0.001 | |
| PDGF | 1.3 ± 0.116 | 1.9 ± 0.066 | 2.2 ± 0.204 | 2.7 ± 0.286 | 0.003 | 0.026 | 0.124 | |
| HIF1α | 1.2 ± 0.088 | 2.4 ± 0.059 | 2.2 ± 0.123 | 3.7 ± 0.153 | <0.001 | <0.001 | <0.001 |
3.8. In situ vascularized scaffold promotes bone defect repair in vivo
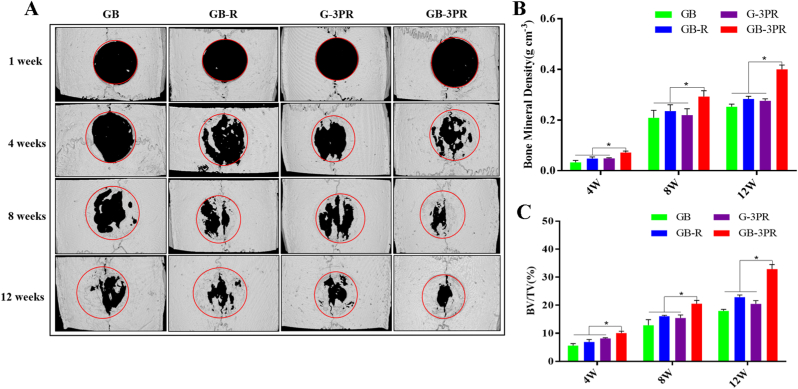
Given the results from the in vitro studies, the in vivo bone repair effect was monitored in a rat critical-size calvarial defect model (Fig. 8) and the bone regenerative capacity was evaluated via micro-CT in the 4th, 8th, and 12th weeks (Fig. 9). Neither infection nor weight loss was observed during the experimental period, and the rats remained active after the operation. Fig. 9A shows three-dimensional (3D) reconstructed images of the bone regeneration. The new bone tissue is marked by a red circle (5 mm in diameter), representing the defect area. Bone regeneration tends to grow from the edge of the defect to the center. Micro-CT images revealed that, by the 4th week, the GB, GB-R, and G-3PR groups had only limited new bone formation, whereas GB-3PR TEB promoted significant new bone formation. As the bone repair time increased, the GB-3PR group exhibited better bone formation than those exhibited by the GB, GB-R, and G-3PR groups. By contrast, the newly formed bone tissue in the GB-R group grew significantly more than those in the GB and G-3PR groups. This result indicates that in situ vascularized TEB is more likely to promote bone repair, significantly enhancing the connectivity of new bone tissue. In addition, the coculture of vascular endothelial cells in osteogenesis repair is conducive to bone regeneration [55]. The micro-CT parameters BMD (Fig. 9B) and BV/TV (Fig. 9C) for quantitative analysis were also obtained to further evaluate the observed results. The overall trend was aligned with the 3D reconstructed images. BMD and BV/TV measurements were successfully applied to the evaluation of the bone-healing process. The results indicated that the BMD and BV/TV ratios in GB-3PR group were higher than that in GB, GB-R and G-3PR groups in the 4th, 8th, and 12th weeks, respectively (Table 7). Interestingly, the symbiotic niche of BMSCs and RAOECs in the GB-R and GB-3PR groups had a better osteogenic effect than that of BMSCs in the GB group or RAOECs in the G-3PR group. Because the growth of blood vessels in bone and osteogenesis are coupled [25], bone-substitute materials with insufficient vascularization lead to poor bone regeneration in bone defect repairs [56]. By contrast, in situ vascularized TEB promotes bone repair by inducing blood vessel formation.
Fig. 8.
Construction of bone defect model and exhibition of scaffold implant operation process. (A) 5 mm diameter implanted bioprinted scaffolds. (B) 5 mm diameter defective cranial tissue. (C) Isolating surrounding soft tissue to expose skull and creating 5 mm diameter defect in rat critical-size calvarial model. (D) Inserting scaffolds into calvarial defect. (E) and (F) Suturing subcutaneous tissue and skin wounds.
Fig. 9.
Micro-CT evaluation of bone defect repair. (A) Three-dimensional reconstructed micro-CT images at 1, 4, 8, and 12 weeks after surgery. (B) and (C) Quantitative analysis of bone mineral density and bone volume/tissue volume (BV/TV) ratio at 4, 8, and 12 weeks after surgery. (n = 3, each group). ∗p < 0.05.
Table 7.
Quantitative analysis of the micro-CT parameters BMD and BV/TV.
| Time | Index | GB | Group GB-R |
G-3PR | GB-3PR | p (GB vs. GB-3PR) | p (GB-R vs. GB-3PR) | p (G-3PR vs. GB-3PR) |
|---|---|---|---|---|---|---|---|---|
| 4 weeks | BMD (g/cm3) | 0.03 ± 0.006 | 0.05 ± 0.005 | 0.05 ± 0.002 | 0.07 ± 0.004 | 0.002 | 0.008 | 0.003 |
| BV/TV (%) | 5.59 ± 0.571 | 6.89 ± 0.706 | 8.14 ± 0.198 | 10.06 ± 0.529 | 0.001 | 0.007 | 0.008 | |
| 8 weeks | BMD (g/cm3) | 0.21 ± 0.024 | 0.24 ± 0.020 | 0.22 ± 0.020 | 0.29 ± 0.019 | 0.018 | 0.043 | 0.020 |
| BV/TV (%) | 12.85 ± 1.64 | 16.09 ± 0.235 | 15.46 ± 0.871 | 20.53 ± 0.954 | 0.005 | 0.003 | 0.005 | |
| 12 weeks | BMD (g/cm3) | 0.25 ± 0.008 | 0.28 ± 0.009 | 0.28 ± 0.006 | 0.40 ± 0.014 | <0.001 | <0.001 | <0.001 |
| BV/TV (%) | 17.99 ± 0.423 | 22.85 ± 0.664 | 20.19 ± 0.944 | 32.85 ± 1.317 | <0.001 | <0.001 | <0.001 |
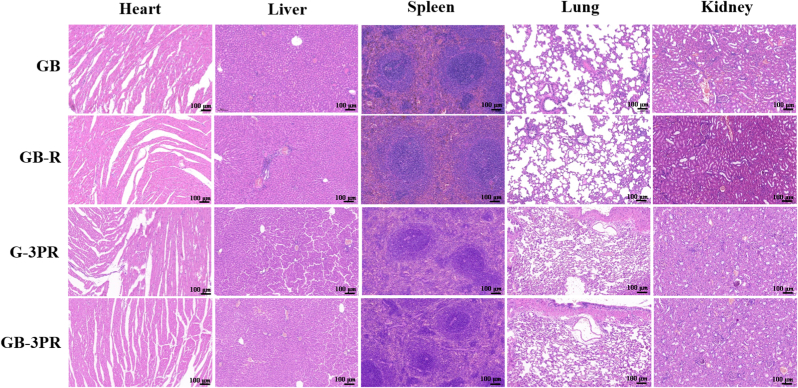
In treating bone defects using 3D-bioprinted TEBs, it was necessary to evaluate biosafety in vivo [57]. The main organs, including the heart, liver, spleen, lungs, and kidneys, were observed via hematoxylin and eosin (H&E) staining (Fig. 10). The results show no significant histopathological abnormalities in any of the groups, suggesting that these 3D-bioprinted TEBs had good biocompatibility without systemic toxicity.
Fig. 10.
Biosafety was evaluated by H&E staining of the heart, liver, spleen, lung, and kidney.
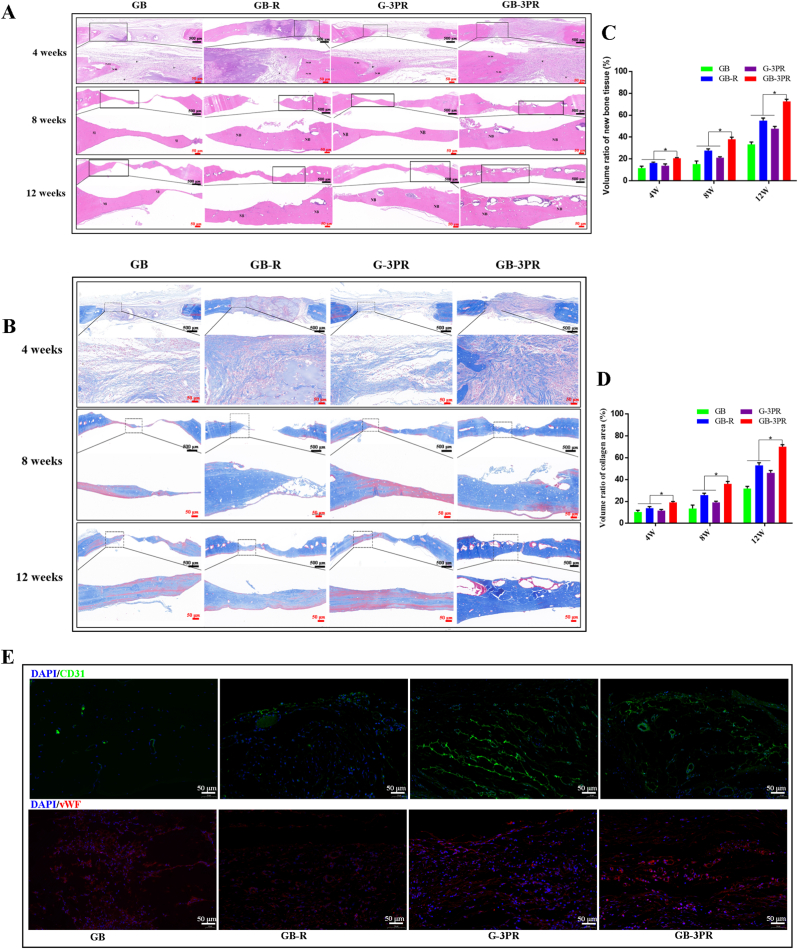
To further confirm the osteogenic induction effect, we performed H&E and Masson staining of the new bone tissue in the 4th, 8th, and 12th weeks. Fig. 11A and B shows fibrous tissue and new bone, respectively. No obvious inflammatory response was observed in the sections between the groups. In the 4th week, more fibrous tissues were observed in the GB-3PR group than in the GB, GB-R, and G-3PR groups. Mature bone tissue was observed in the 8th week. In agreement with the results of the micro-CT analysis, the results of quantitative analysis of HE and Masson staining showed greater evidence of new bone formation in the GB-3PR group than in the other groups (Fig. 11C and D). Moreover, the repaired tissues treated with GB-3PR TEB were much closer to those of normal bone tissue. To investigate the angiogenic activity of in situ vascularized TEB, immunofluorescence staining against CD31 and vWF was performed (Fig. 11E). The results showed that more CD31 and vWF were expressed in the GB-3PR group than in the GB, GB-R, or G-3PR group. In other words, the GB-3PR scaffolds resulted in an increase in blood vessel density and new bone thickness in vivo. This revealed that 3D-bioprinted in situ vascular endothelial cells easily formed vascular tissue networks that regulate bone regeneration.
Fig. 11.
Histomorphological analysis of newly formed tissue by hematoxylin–eosin (HE) staining (A) and Masson staining (B) at 4, 8, and 12 weeks after surgery. (C) and (D) Quantitative analysis of new bone tissue in the defect area. (n = 3, each group). (E) Immunofluorescence images of endogenous cells (CD31, green, and vWF, red) and DAPI (blue) in new tissue sections at 12 weeks after operation. ∗p < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In this study, the in situ vascularized hydrogel scaffold was implanted into a bone defect model, the niche provided by scaffolds promoted cell proliferation and adhesion, extracellular matrix adhesion, and new tissue formation. The symbiosis niche formed by scaffolds, cells, and regenerated tissues accelerated bone healing processes by influencing genes expression, signal transduction, and cellular function in the microenvironment. In addition, the in situ vascularized scaffold performed excellent biocompatibility in vitro and in vivo to promote the bone healing processes. Collectively, these results indicate that, among the given TEBs, the 3D-bioprinted in situ vascularized TEB had the best osteogenic and angiogenic effects for repairing bone defects.
4. Conclusion
In this study, we successfully fabricated vascularized tissue-engineered bone with biological activity to repair bone defects using 3D in situ bioprinting technology. Endothelial cells printed in situ exhibited more uniform distribution and greater seeding efficiency throughout the channels, promoting tube formation and forming a vascular network through culture in perfusion bioreactors. In vitro, the in situ vascularized scaffold exhibited a coupling effect between angiogenesis and osteogenesis. In addition, enrichment analysis of RNA sequencing revealed that 3D bioprinting of in situ vascularized scaffolds promoted osteogenesis and angiogenesis by enhancing gene expression in related biological processes. Furthermore, the in vivo 3D-bioprinted in situ vascularized scaffolds demonstrated excellent performance in promoting new bone formation in rat calvarial critical-sized defects. Whereas long-term research will be necessary, our study provides early evidence that 3D in situ bioprinting technology can control and direct in situ angiogenesis and subsequent bone regeneration, for improving clinical outcomes from bone grafting procedures in the future.
Credit author statement
Mingkui Shen: Conceptualization, Methodology, Material synthesis, Software, Validation, Formal analysis, Writing - original draft, Visualization. Lulu Wang: Conceptualization, Methodology, Validation, Investigation, Writing - review & editing, Validation. Yi Gao: Resources, Investigation, Resources. Li Feng: Validation, Formal analysis, Investigation, Resources. Chuangye Xu: Software, Validation, Investigation. Sijing Li: Formal analysis. Xiaohu Wang: Validation, Resources. Yulan Wu: Resources. Yao Guo: Investigation, Resources, Data curation. Guoxian Pei: Conceptualization, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgements
The authors would like to acknowledge the Southern University of Science and Technology for providing excellent testing equipment. This study was supported by the Research Startup Fund of the Southern University of Science and Technology (Y01416214).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2022.100382.
Contributor Information
Yao Guo, Email: guoy3@sustech.edu.cn.
Guoxian Pei, Email: nfperry@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhou K., Yu P., Shi X., Ling T., Zeng W., Chen A., Yang W., Zhou Z. Hierarchically porous hydroxyapatite hybrid scaffold incorporated with reduced graphene oxide for rapid bone ingrowth and repair. ACS Nano. 2019;13(8):9595–9606. doi: 10.1021/acsnano.9b04723. [DOI] [PubMed] [Google Scholar]
- 2.Wang J., Wu D., Zhang Z., Li J., Shen Y., Wang Z., Li Y., Zhang Z.Y., Sun J. Biomimetically ornamented rapid prototyping fabrication of an apatite-collagen-polycaprolactone composite construct with nano-micro-macro hierarchical structure for large bone defect treatment. ACS Appl. Mater. Interfaces. 2015;7(47):26244–26256. doi: 10.1021/acsami.5b08534. [DOI] [PubMed] [Google Scholar]
- 3.Spicer P.P., Kretlow J.D., Young S., Jansen J.A., Kasper F.K., Mikos A.G. Evaluation of bone regeneration using the rat critical size calvarial defect. Nat. Protoc. 2012;7(10):1918–1929. doi: 10.1038/nprot.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mauffrey C., Barlow B.T., Smith W. Management of segmental bone defects. J. Am. Acad. Orthop. Surg. 2015;23(3):143–153. doi: 10.5435/JAAOS-D-14-00018. [DOI] [PubMed] [Google Scholar]
- 5.Agarwal R., García A.J. Biomaterial strategies for engineering implants for enhanced osseointegration and bone repair. Adv. Drug Deliv. Rev. 2015;94:53–62. doi: 10.1016/j.addr.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho-Shui-Ling A., Bolander J., Rustom L.E., Johnson A.W., Luyten F.P., Picart C. Bone regeneration strategies: engineered scaffolds, bioactive molecules and stem cells current stage and future perspectives. Biomaterials. 2018;180:143–162. doi: 10.1016/j.biomaterials.2018.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu C., Ashok D., Nisbet D.R., Gautam V. Bioinspired surface modification of orthopedic implants for bone tissue engineering. Biomaterials. 2019;219 doi: 10.1016/j.biomaterials.2019.119366. [DOI] [PubMed] [Google Scholar]
- 8.Guan G., Qizhuang L., Liu S., Jiang Z., Zhou C., Liao W. 3D-bioprinted peptide coupling patches for wound healing. Materials today Bio. 2022;13 doi: 10.1016/j.mtbio.2021.100188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X.M., Wu K., Gao L., Wang L.P., Shi X.T. Biomaterial strategies for the application of reproductive tissue engineering. Bioact. Mater. 2022;14:86–96. doi: 10.1016/j.bioactmat.2021.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bacakova L., Zarubova J., Travnickova M., Musilkova J., Pajorova J., Slepicka P., Kasalkova N.S., Svorcik V., Kolska Z., Motarjemi H., et al. Stem cells: their source, potency and use in regenerative therapies with focus on adipose-derived stem cells - a review. Biotechnol. Adv. 2018;36(4):1111–1126. doi: 10.1016/j.biotechadv.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Chen X., Zhang Y.S., Zhang X.P., Liu C.S. Organ-on-a-chip platforms for accelerating the evaluation of nanomedicine. Bioact. Mater. 2021;6(4):1012–1027. doi: 10.1016/j.bioactmat.2020.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C., Huang W., Zhou Y., He L.B., He Z., Chen Z.L., He X., Tian S., Liao J.M., Lu B.H., et al. 3D printing of bone tissue engineering scaffolds. Bioact. Mater. 2020;5(1):82–91. doi: 10.1016/j.bioactmat.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang T., Wei Q., Zhou H., Jing Z., Liu X., Zheng Y., Cai H., Wei F., Jiang L., Yu M., et al. Three-dimensional-printed individualized porous implants: a new "implant-bone" interface fusion concept for large bone defect treatment. Bioact. Mater. 2021;6(11):3659–3670. doi: 10.1016/j.bioactmat.2021.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi K., Yanagisawa T., Shimabukuro M., Kishida R., Ishikawa K. Granular honeycomb scaffolds composed of carbonate apatite for simultaneous intra- and inter-granular osteogenesis and angiogenesis. Materials today Bio. 2022;14 doi: 10.1016/j.mtbio.2022.100247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Towler D.A. The osteogenic-angiogenic interface: novel insights into the biology of bone formation and fracture repair. Curr. Osteoporos. Rep. 2008;6(2):67–71. doi: 10.1007/s11914-008-0012-x. [DOI] [PubMed] [Google Scholar]
- 16.Sun X., Jiao X., Yang X., Ma J., Wang T., Jin W., Li W., Yang H., Mao Y., Gan Y., et al. 3D bioprinting of osteon-mimetic scaffolds with hierarchical microchannels for vascularized bone tissue regeneration. Biofabrication. 2022;14(3) doi: 10.1088/1758-5090/ac6700. [DOI] [PubMed] [Google Scholar]
- 17.Chiesa I., De Maria C., Lapomarda A., Fortunato G.M., Montemurro F., Di Gesù R., Tuan R.S., Vozzi G., Gottardi R. Endothelial cells support osteogenesis in an in vitro vascularized bone model developed by 3D bioprinting. Biofabrication. 2020;12(2) doi: 10.1088/1758-5090/ab6a1d. [DOI] [PubMed] [Google Scholar]
- 18.Li Z., Meyers C.A., Chang L., Lee S., Li Z., Tomlinson R., Hoke A., Clemens T.L., James A.W. Fracture repair requires TrkA signaling by skeletal sensory nerves. J. Clin. Invest. 2019;129(12):5137–5150. doi: 10.1172/JCI128428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xing F., Xiang Z., Rommens P.M., Ritz U. 3D bioprinting for vascularized tissue-engineered bone fabrication. Materials. 2020;13(10) doi: 10.3390/ma13102278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P., Sun Y., Shi X., Shen H., Ning H., Liu H. 3D printing of tissue engineering scaffolds: a focus on vascular regeneration. Bio-design and manufacturing. 2021:1–35. doi: 10.1007/s42242-020-00109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrabba M., Madeddu P. Current strategies for the manufacture of small size tissue engineering vascular grafts. Front. Bioeng. Biotechnol. 2018;6:41. doi: 10.3389/fbioe.2018.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng P., Li D., Gao Y., Cao T., Jiang H., Wang J., Li J., Zhang S., Song Y., Liu B., et al. Prevascularization promotes endogenous cell-mediated angiogenesis by upregulating the expression of fibrinogen and connective tissue growth factor in tissue-engineered bone grafts. Stem Cell Res. Ther. 2018;9(1):176. doi: 10.1186/s13287-018-0925-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez-Yague M.A., Abbah S.A., McNamara L., Zeugolis D.I., Pandit A., Biggs M.J. Biomimetic approaches in bone tissue engineering: integrating biological and physicomechanical strategies. Adv. Drug Deliv. Rev. 2015;84:1–29. doi: 10.1016/j.addr.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Andronowski J.M., Pratt I.V., Cooper D.M.L. Occurrence of osteon banding in adult human cortical bone. Am. J. Phys. Anthropol. 2017;164(3):635–642. doi: 10.1002/ajpa.23297. [DOI] [PubMed] [Google Scholar]
- 25.Kusumbe A.P., Ramasamy S.K., Adams R.H. Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature. 2014;507(7492):323–328. doi: 10.1038/nature13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vijayavenkataraman S., Yan W.C., Lu W.F., Wang C.H., Fuh J.Y.H. 3D bioprinting of tissues and organs for regenerative medicine. Adv. Drug Deliv. Rev. 2018;132:296–332. doi: 10.1016/j.addr.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Murphy S.V., Atala A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014;32(8):773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 28.Derakhshanfar S., Mbeleck R., Xu K.G., Zhang X.Y., Zhong W., Xing M. 3D bioprinting for biomedical devices and tissue engineering: a review of recent trends and advances. Bioact. Mater. 2018;3(2):144–156. doi: 10.1016/j.bioactmat.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shao L., Gao Q., Xie C., Fu J., Xiang M., He Y. Directly coaxial 3D bioprinting of large-scale vascularized tissue constructs. Biofabrication. 2020;12(3) doi: 10.1088/1758-5090/ab7e76. [DOI] [PubMed] [Google Scholar]
- 30.Folkman J., Haudenschild C. Angiogenesis in vitro. Nature. 1980;288(5791):551–556. doi: 10.1038/288551a0. [DOI] [PubMed] [Google Scholar]
- 31.Ouyang L., Armstrong J.P.K., Chen Q., Lin Y., Stevens M.M. Void-free 3D bioprinting for in-situ endothelialization and microfluidic perfusion. Adv. Funct. Mater. 2020;30(1) doi: 10.1002/adfm.201908349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danafar H., Rostamizadeh K., Davaran S., Hamidi M. PLA-PEG-PLA copolymer-based polymersomes as nanocarriers for delivery of hydrophilic and hydrophobic drugs: preparation and evaluation with atorvastatin and lisinopril. Drug Dev. Ind. Pharm. 2014;40(10):1411–1420. doi: 10.3109/03639045.2013.828223. [DOI] [PubMed] [Google Scholar]
- 33.Liu J., Yang S., Cao B., Zhou G., Zhang F., Wang Y., Wang R., Zhu L., Meng Y., Hu C., et al. Targeting B7-H3 via chimeric antigen receptor T cells and bispecific killer cell engagers augments antitumor response of cytotoxic lymphocytes. J. Hematol. Oncol. 2021;14(1):21. doi: 10.1186/s13045-020-01024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin J., Yan M., Wang Y., Fu J., Suo H. 3D bioprinting of low-concentration cell-laden gelatin methacrylate (GelMA) bioinks with a two-step cross-linking strategy. ACS Appl. Mater. Interfaces. 2018;10(8):6849–6857. doi: 10.1021/acsami.7b16059. [DOI] [PubMed] [Google Scholar]
- 35.He G., Ma L.L., Pan J., Venkatraman S. ABA and BAB type triblock copolymers of PEG and PLA: a comparative study of drug release properties and "stealth" particle characteristics. Int. J. Pharm. 2007;334(1–2):48–55. doi: 10.1016/j.ijpharm.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 36.Massadeh S., Almohammed I., Barhoush E., Omer M., Aldhawi N., Almalik A., Alaamery M. Development of epirubicin-loaded biocompatible polymer PLA-PEG-PLA nanoparticles: synthesis, characterization, stability, and in vitro anticancerous assessment. Polymers. 2021;13(8) doi: 10.3390/polym13081212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Girard E., Chagnon G., Moreau-Gaudry A., Letoublon C., Favier D., Dejean S., Trilling B., Nottelet B. Evaluation of a biodegradable PLA-PEG-PLA internal biliary stent for liver transplantation: in vitro degradation and mechanical properties. J. Biomed. Mater. Res. B Appl. Biomater. 2021;109(3):410–419. doi: 10.1002/jbm.b.34709. [DOI] [PubMed] [Google Scholar]
- 38.Kurian A.G., Singh R.K., Patel K.D., Lee J.H., Kim H.W. Multifunctional GelMA platforms with nanomaterials for advanced tissue therapeutics. Bioact. Mater. 2022;8:267–295. doi: 10.1016/j.bioactmat.2021.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Ugarte D.A., Alfonso Z., Zuk P.A., Elbarbary A., Zhu M., Ashjian P., Benhaim P., Hedrick M.H., Fraser J.K. Differential expression of stem cell mobilization-associated molecules on multi-lineage cells from adipose tissue and bone marrow. Immunol. Lett. 2003;89(2–3):267–270. doi: 10.1016/s0165-2478(03)00108-1. [DOI] [PubMed] [Google Scholar]
- 40.Luo X., Li H., Sun X., Zuo Q., Li B., Zhu Y., Wei W., Gu X. Promotion of differentiating bone marrow mesenchymal stromal cells (BMSCs) into cardiomyocytes via HCN2 and HCN4 cotransfection. BioMed Res. Int. 2021;2021 doi: 10.1155/2021/5529276. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Lee H.P., Gu L., Mooney D.J., Levenston M.E., Chaudhuri O. Mechanical confinement regulates cartilage matrix formation by chondrocytes. Nat. Mater. 2017;16(12):1243–1251. doi: 10.1038/nmat4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colosi C., Shin S.R., Manoharan V., Massa S., Costantini M., Barbetta A., Dokmeci M.R., Dentini M., Khademhosseini A. Microfluidic bioprinting of heterogeneous 3D tissue constructs using low-viscosity bioink. Adv. Mater. 2016;28(4):677–684. doi: 10.1002/adma.201503310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheikhi A., de Rutte J., Haghniaz R., Akouissi O., Sohrabi A., Di Carlo D., Khademhosseini A. Microfluidic-enabled bottom-up hydrogels from annealable naturally-derived protein microbeads. Biomaterials. 2019;192:560–568. doi: 10.1016/j.biomaterials.2018.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mehrasa M., Asadollahi M.A., Nasri-Nasrabadi B., Ghaedi K., Salehi H., Dolatshahi-Pirouz A., Arpanaei A. Incorporation of mesoporous silica nanoparticles into random electrospun PLGA and PLGA/gelatin nanofibrous scaffolds enhances mechanical and cell proliferation properties. Materials science & engineering C, Materials for biological applications. 2016;66:25–32. doi: 10.1016/j.msec.2016.04.031. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Q., Qin M., Zhou X., Nie W., Wang W., Li L., He C. Porous nanofibrous scaffold incorporated with S1P loaded mesoporous silica nanoparticles and BMP-2 encapsulated PLGA microspheres for enhancing angiogenesis and osteogenesis. J. Mater. Chem. B. 2018;6(42):6731–6743. doi: 10.1039/c8tb02138d. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura T., Nakamura-Takahashi A., Kasahara M., Yamaguchi A., Azuma T. Tissue-nonspecific alkaline phosphatase promotes the osteogenic differentiation of osteoprogenitor cells. Biochem. Biophys. Res. Commun. 2020;524(3):702–709. doi: 10.1016/j.bbrc.2020.01.136. [DOI] [PubMed] [Google Scholar]
- 47.Pan F.F., Shao J., Shi C.J., Li Z.P., Fu W.M., Zhang J.F. Apigenin promotes osteogenic differentiation of mesenchymal stem cells and accelerates bone fracture healing via activating Wnt/β-catenin signaling. Am. J. Physiol. Endocrinol. Metabol. 2021;320(4):E760–e771. doi: 10.1152/ajpendo.00543.2019. [DOI] [PubMed] [Google Scholar]
- 48.Ball O., Nguyen B.B., Placone J.K., Fisher J.P. 3D printed vascular networks enhance viability in high-volume perfusion bioreactor. Ann. Biomed. Eng. 2016;44(12):3435–3445. doi: 10.1007/s10439-016-1662-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piard C., Jeyaram A., Liu Y., Caccamese J., Jay S.M., Chen Y., Fisher J. 3D printed HUVECs/MSCs cocultures impact cellular interactions and angiogenesis depending on cell-cell distance. Biomaterials. 2019;222 doi: 10.1016/j.biomaterials.2019.119423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eilken H.M., Adams R.H. Dynamics of endothelial cell behavior in sprouting angiogenesis. Curr. Opin. Cell Biol. 2010;22(5):617–625. doi: 10.1016/j.ceb.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 51.Mayer H., Bertram H., Lindenmaier W., Korff T., Weber H., Weich H. Vascular endothelial growth factor (VEGF-A) expression in human mesenchymal stem cells: autocrine and paracrine role on osteoblastic and endothelial differentiation. J. Cell. Biochem. 2005;95(4):827–839. doi: 10.1002/jcb.20462. [DOI] [PubMed] [Google Scholar]
- 52.Ji C., Qiu M., Ruan H., Li C., Cheng L., Wang J., Li C., Qi J., Cui W., Deng L. Transcriptome analysis revealed the symbiosis niche of 3D scaffolds to accelerate bone defect healing. Adv. Sci. 2022;9(8) doi: 10.1002/advs.202105194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ayturk U. RNA-Seq in skeletal biology. Curr. Osteoporos. Rep. 2019;17(4):178–185. doi: 10.1007/s11914-019-00517-x. [DOI] [PubMed] [Google Scholar]
- 54.Stark R., Grzelak M., Hadfield J. RNA sequencing: the teenage years. Nat. Rev. Genet. 2019;20(11):631–656. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- 55.Anada T., Pan C.C., Stahl A.M., Mori S., Fukuda J., Suzuki O., Yang Y. Vascularized bone-mimetic hydrogel constructs by 3D bioprinting to promote osteogenesis and angiogenesis. Int. J. Mol. Sci. 2019;20(5) doi: 10.3390/ijms20051096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klenke F.M., Liu Y., Yuan H., Hunziker E.B., Siebenrock K.A., Hofstetter W. Impact of pore size on the vascularization and osseointegration of ceramic bone substitutes in vivo. J. Biomed. Mater. Res., Part A. 2008;85(3):777–786. doi: 10.1002/jbm.a.31559. [DOI] [PubMed] [Google Scholar]
- 57.Liu B., Li J., Lei X., Cheng P., Song Y., Gao Y., Hu J., Wang C., Zhang S., Li D., et al. 3D-bioprinted functional and biomimetic hydrogel scaffolds incorporated with nanosilicates to promote bone healing in rat calvarial defect model. Materials science & engineering C, Materials for biological applications. 2020;112 doi: 10.1016/j.msec.2020.110905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.