Abstract
Objective
Aberrant activity of androgen receptor (AR) is the primary cause underlying development and progression of prostate cancer (PCa) and castration-resistant PCa (CRPC). Androgen signaling regulates gene transcription and lipid metabolism, facilitating tumor growth and therapy resistance in early and advanced PCa. Although direct AR signaling inhibitors exist, AR expression and function can also be epigenetically regulated. Specifically, lysine (K)-specific demethylases (KDMs), which are often overexpressed in PCa and CRPC phenotypes, regulate the AR transcriptional program.
Methods
We investigated LSD1/UTX inhibition, two KDMs, in PCa and CRPC using a multi-omics approach. We first performed a mitochondrial stress test to evaluate respiratory capacity after treatment with MC3324, a dual KDM-inhibitor, and then carried out lipidomic, proteomic, and metabolic analyses. We also investigated mechanical cellular properties with acoustic force spectroscopy.
Results
MC3324 induced a global increase in H3K4me2 and H3K27me3 accompanied by significant growth arrest and apoptosis in androgen-responsive and -unresponsive PCa systems. LSD1/UTX inhibition downregulated AR at both transcriptional and non-transcriptional level, showing cancer selectivity, indicating its potential use in resistance to androgen deprivation therapy. Since MC3324 impaired metabolic activity, by modifying the protein and lipid content in PCa and CRPC cell lines. Epigenetic inhibition of LSD1/UTX disrupted mitochondrial ATP production and mediated lipid plasticity, which affected the phosphocholine class, an important structural element for the cell membrane in PCa and CRPC associated with changes in physical and mechanical properties of cancer cells.
Conclusions
Our data suggest a network in which epigenetics, hormone signaling, metabolite availability, lipid content, and mechano-metabolic process are closely related. This network may be able to identify additional hotspots for pharmacological intervention and underscores the key role of KDM-mediated epigenetic modulation in PCa and CRPC.
Keywords: PCa, KDMs, AR, Metabolism, Lipid, Cell stiffness
Abbreviations: AR, androgen receptor; CRPC, castration resistant prostate cancer; FASN, fatty acid synthase; KDM, lysine demethylase; LSD1, lysine-specific demethylase 1; PCa, prostate cancer; PCs, phosphatidylcholines; UTX, lysine-specific demethylase 6A
Highlights
-
•
KDMs inhibition promotes increases H3K4me2 and H3K27me3 in PCa and CRPC, which causes cancer selective pro-apoptotic pathways.
-
•
KDMs regulate AR expression in PCa and CRPC, reducing ATP production, mitochondrial respiration and intermediate metabolites availability.
-
•
Epigenetic controls metabolic pathways and redirects lipid metabolic cascade.
-
•
KDMs inhibition alters lipid distribution and composition, impacting on physical and mechanical properties of PCa and CRPC.
1. Introduction
Prostate cancer (PCa) is the second leading cause of cancer death in men worldwide [1], and androgen receptor (AR) is a key driving force in the tumorigenesis process. AR supports gene transcription involved in primary tumor formation, progression, and metastasis [2]. The standard pharmacological treatment for PCa is based on the use of androgen deprivation therapy (ADT) and AR ligand inhibitors (e.g., enzalutamide and second-generation derivatives) to block hormone signaling via sequestration of AR, thus preventing receptor nuclear translocation and the subsequent activation of AR target genes [3]. However, 20–40% of PCa patients do not respond to androgen antagonist therapies and develop castration-resistant PCa (CRPC) [4] as a consequence of long-term adaptation [5,6]. An alternative therapeutic strategy, independent of AR presence and androgen antagonist sensitivity, therefore remains an urgent and unmet need for CRPC patients. Intriguingly, unlike other types of cancer that often exhibit the Warburg effect, PCa and CRPC present a “lipid-based metabolic phenotype”, mainly supported directly and indirectly by AR signaling [7]. Indeed, AR promotes the transcription of genes involved in lipid synthesis such as SREBF1 [8], which promotes fatty acid synthesis via fatty acid synthase (FASN) and acetyl-CoA carboxylase (ACC), making lipids the primary source of ATP supporting cancer growth and progression [9,10]. In addition, cellular mechanics and metabolism are tightly interconnected. Thus, the resulting changes in metabolism, in turn, affect the regulation of every level of cell biology, including the physical and mechanical properties of cells and tissues [11]. To execute its function, AR transcriptionally associates with epigenetic enzymes in multi-complexes [12]. Lysine (K)-specific demethylases (KDMs) physically interact with AR [13], and methylation of lysines is associated with a repressive or permissive chromatin environment for AR-mediated transcription. Specifically, LSD1 (KDM1A) and UTX (KDM6A) promote AR expression [14] and contribute to its transcriptional activity, while inhibition of both epigenetic enzymes is associated with a reduction in AR-mediated transcriptionalsignaling [[15], [16], [17]]. We previously reported the link between KDM epigenetic modulation and estrogen receptor alpha (ERα) in breast cancer (BC) [18]. Given the similarity between BC and PCa, here we translated the anticancer effects of a dual KDM inhibitor of LSD1/UTX (MC3324) to prostate models and provide data supporting the efficacy of epigenetic treatment in both PCa and CRPC cell systems. In PCa and CRPC, LSD1 and UTX inhibition i) globally increases methylation on H3K4me2 and H3K27me3 residues, ii) downregulates AR independently of androgen antagonist sensitivity, iii) perturbs ATP production, iv) alters intermediate metabolite availability, v) blocks lipid metabolic cascade, changing the amount and subclasses of cellular lipids, and vi) alters physical and mechanical properties of cells. Inhibition of LSD1/UTX changes the levels of ATP and metabolites in PCa and CRPC by downregulating glycolysis and lipogenesis/lipolysis enzymes. As a result, cancer growth is slowed. Metabolic substrates are fundamental in determining lipid content and accelerating PCa cell proliferation, growth, and plasticity, contributing to mechanical features of tumor cells Here, the traditional paradigm of the histone code as being regulated by epigenetic readers, writers, and erasers (e.g., LSD1, UTX) working in association with transcriptional factors (e.g., AR) is shifting to one in which the “ink” (metabolites and energy precursors) is required to sustain PCa and CRPC. AR signaling and metabolite-producing enzymes can be epigenetically regulated to weaken bioenergy pathways, thus reducing proliferation index and overcoming cell death resistance in PCa and CRPC, causing a disruption of the tumor balance via alterations in mechano-metabolic properties. The direct link between lipid metabolism and PCa is corroborated by preclinical and clinical trials investigating the effect of FASN inhibitors in the management of CRPC patients [19,20], (https://clinicaltrials.gov/ct2/show/NCT04337580NCT04337580). Here, we show that upstream epigenetic regulation of lipid metabolism via LSD1/UTX inhibition may represent an additional approach for interfering with hormone and metabolic signaling to overcome resistance mechanisms associated with CRPC.
2. Methods
2.1. Chemicals
Tranylcypromine (TCP, #13492-01-8) and GSK-J4 (#1373423-53-0) were purchased from Sigma–Aldrich (St Louis, MO, USA) and were used at a final concentration of 100 μM and 5 μM, respectively. MC3324 was synthesized by Prof. Mai's group (“Sapienza” University of Rome), as reported in [21] describing MC3324 structure. The compound was obtained by fusing TCP and IOX1 KDM inhibitors and was used in each experiment at a final concentration of 25 μM.
2.2. Cell culture
LNCaP, C4-2, and C4–2 B cell lines were purchased from ATCC (Milan, Italy). 22RV1, DU145, and PC3 cells lines were kindly provided by Prof. Carmen Jeronimo (Institute of Biomedical Sciences Abel Salazar [ICBAS] University of Porto). PNT2 cell line was kindly provided by Prof. Antimo Migliaccio (University of Campania “Luigi Vanvitelli”). Cells were grown in Roswell Park Memorial Institute culture medium (RPMI; EuroClone, Milan, Italy, ECB9006L), supplemented with 10% heat-inactivated fetal bovine serum (FBS; Sigma–Aldrich, F7524), antimicrobials (100 U/mL penicillin, 100 μg/mL streptomycin [EuroClone, ECB3001D], 250 ng/mL amphotericin B [EuroClone, ECM0009D], 2 mM l-glutamine [EuroClone, ECB3000D]), and 1% essential amino acids solution (MEM; EuroClone, ECB3054D). All cell lines were cultivated at 37 °C with 5% CO2 and were checked for mycoplasma contamination using EZ-PCR Mycoplasma Test Kit (Biological Industries; #20-700-20).
2.3. Histone extraction
Histones were extracted as reported in [22]. Briefly, after treatment with the indicated compounds, cells were collected and washed two times with PBS. Then, cell pellets were re-suspended in triton extraction buffer (TEB; PBS containing 0.5% Triton X 100 [v/v], 2 mmol/L PMSF, 0.02% [w/v] NaN3); lysis was performed for 10 min with stirring at 4 °C, and samples were centrifuged at 2000 rpm for 10 min at 4 °C. After the wash step, samples were then precipitated in 0.2 N HCl overnight at 4 °C for acid histone extraction. The day after, the supernatant was recovered and protein concentration quantified by Bradford assay (Bio-Rad Protein Assay Dye Reagent Concentrate, #5000006) (Bio-Rad, California, U.S.A.). H3K4me2, H3K27me3, H3K9me2 (Diagenode, Ougrée, Belgium, pAB-035-050, C15410069, C15200154), and H4 (Abcam, Cambridge, UK, ab17036) were used according to the manufacturer's instructions.
2.4. Western blot analysis
Cell pellets were suspended in lysis buffer (50 mmol/L Tris–HCl pH 7.4, 150 mmol/L NaCl, 1% NP40, 10 mmol/L NaF, 1 mmol/L PMSF, and protease inhibitor cocktail). Next, the lysis reaction was carried out for 15 min at 4 °C, samples were centrifuged at 13,000 rpm for 30 min at 4 °C, and protein concentration quantified by Bradford assay (Bio-Rad Protein Assay Dye Reagent Concentrate, #5000006). A total of 50 μg of each sample was loaded on 8%, 10%, or 15% polyacrylamide gels and electro-blotted on nitrocellulose membrane. Immunoreactive signals were detected with a horseradish peroxidase-conjugated secondary antibody (Bio-Rad, #1705046, #1706464). Primary antibodies were: AR (sc-52309), P53 (sc-126), tubulin (sc-5286), and SQSTM1 (sc-48402) purchased from Santa Cruz Biotechnology (Dallas, USA); BAX (ab53154), caspase 9 (ab9502), caspase 8 (ab9746), BCL2 (ab28725), p-eNOS (ab184154), eNOS (ab5589), p-AKT (ab81283) and LSD1 (ab17721) from Abcam. Fatty Acid and Lipid Metabolism Antibody Sampler Kit (#8335), GLUT1 (D3J3 A) and GAPDH (D16H11) were purchased from Cell Signaling Technology (Danvers, MA, USA); acetylated-tubulin (T7451) from Sigma–Aldrich. All antibodies were used according to the manufacturer's instructions. Semi-quantitative analysis was performed using ImageJ software (version 1.44), and the relative intensities are reported in figures.
2.5. RNA isolation and real-time PCR
Total RNA was purified as previously described in [23]. RNA samples were quantified using NanoDrop 1000 Spectrophotometer V3.8 (Thermo Fisher Scientific), and their quality was checked using an Agilent RNA 6000 Nano Kit Guide (Agilent Technologies, Santa Clara, U.S.A.). VILO cDNA Synthesis Kit (Invitrogen, Monza and Brianza, Italy, #11754050) was used to convert RNA into cDNA. Then, 50 ng of cDNA was added with 1X SYBR Green PCR Master Mix (Bio-Rad #1708880), according to the manufacturer's instructions. Primers used are listed in Supplementary File Table 4.
2.6. Cell cycle and cell death analysis
For cell cycle analysis and PI evaluation, PC3 cells were plated (2 × 105 cells/mL) and after treatment with MC3324 at 25 μM for 6 h, 24 h, and 48 h were processed as described in [24]. The results were acquired on a BD Accuri TM C6 flow cytometer system (BD Biosciences, New Jersey, U.S.A.).
2.7. shLSD1 transfection
shLSD1 vector (Santa Cruz, sc-60970) and the empty vector shSCR (Santa Cruz, sc-108060) were used. 1 μg of each vector was transfected into C4-2 cells using an Amaxa Nucleofector (Lonza), according to the manufacturer's protocol. shLSD1 and shSCR were maintained in RPMI medium (EuroClone) with 10% heatinactivated FBS (Sigma–Aldrich), 1% glutamine (EuroClone), 1% penicillin/streptomycin (EuroClone), 0.1% gentamycin (EuroClone), 500 μg/mL G418 (Gibco) and 1% essential amino acids solution (EuroClone) at 37 °C in air containing 5% CO2. Downregulation of LSD1 was confirmed by Western blot.
2.8. Cellular mitochondrial stress and ATP production
Metabolic status was investigated on a Seahorse XF24 Analyzer (Agilent Technologies) with standard 24-well Seahorse microplates. A Mito Stress Test Kit (Agilent Technologies, #103015) was used to assess oxygen consumption ratio (OCR) and extracellular acidification rate (ECAR) after MC3324 treatment. In brief, 2 × 104 cells were seeded into plates 12 h prior to analysis. The medium was then replaced with 175 μL of non-buffered RPMI containing 10 mM glucose, 2 mM glutamine, and 1 mM pyruvate. The cells were then treated with 25 μM MC3324 for 3 h at 37 °C and incubated in a CO2-free incubator at 37 °C for 1 h to allow for temperature and pH equilibration before being loaded into the XF24 Analyzer. The injection sequence was programmed as follows: 1st, oligomycin (1 μM at final concentration); 2nd, carbonyl cyanide m-chlorophenylhydrazone (FCCP; 1 μM at final concentration); 3rd, rotenone and antimycin A (1 μM and 0.5 μM at final concentrations, respectively). Data were analyzed with Wave software (version 2.2.0, Seahorse Bioscience, Agilent Technologies, Santa Clara, CA, USA). Experiments were performed in triplicates. P-values were calculated using t-test. Statistical significance is expressed as ∗ p-value <0.05. Standard deviations are reported as error bars.
2.8.1. Sample preparation and untargeted lipidomic analysis
After treatment for 6 h with MC3324 at 25 μM, PCa cells were harvested. Lipids were extracted by adding to the cell pellet 1.5 mL chloroform: MeOH (2:1 v/v), 0.5 mL ultrapure water, vortexed for 1 min, and centrifuged at 3000 rpm for 10 min at 4 °C. The lower phase was then transferred to a new tube for evaporation, and dried under nitrogen. The dried extract was re-suspended with 200 μL isopropyl alcohol: MeOH (1:1 v/v); 5 μL LPC (12:0) was added for internal standard. Each sample was then centrifuged at 12,000 rpm for 10 min at 4 °C, and the supernatant was transferred for liquid chromatography–mass spectrometry (LC–MS) analysis. Separation was performed by Ultra Performance Liquid Chromatography UPLC (Thermo Fisher Scientific, Ultimate 3000LC). The LC system comprised an ACQUITY UPLC BEH C18 column (100 mm × 2.1 mm, 1.7 μm). The mobile phase was composed of solvent A (60% ACN + 40% H2O + 10 mM HCOONH4) and solvent B (10% ACN + 90% isopropyl alcohol + 10 mM HCOONH4) with a gradient elution (0–10.5 min, 30–100% B; 10.5–12.5 min, 100% B; 12.5–12.51 min, 100−30% B; 12.51–16 min, 30% B). The flow rate of the mobile phase was 0.3 mL/min. The column temperature was maintained at 40 °C and the sample manager temperature was set at 4 °C. Mass spectrometry parameters in ESI + mode were: Heater Temp, 300 °C; Sheath Gas Flow Rate, 45 arb; Aux Gas Flow Rate, 15 arb; Sweep Gas Flow Rate, 1 arb; Spray Voltage, 3.0 kV; Capillary Temp, 350 °C; S-Lens RF Level, 30%. Mass spectrometry parameters in ESI- mode were: Heater Temp, 300 °C; Sheath Gas Flow Rate, 45 arb; Aux Gas Flow Rate, 15 arb; Sweep Gas Flow Rate, 1 arb; Spray Voltage, 3.2 kV; Capillary Temp, 350 °C; S-Lens RF Level, 60%. Five quality control (QC) samples were run to avoid small changes in both chromatographic retention time and signal intensity.
2.8.2. Statistical analysis and software tool analysis
Raw data were acquired and aligned using Lipid Search software version 5.0 (Thermo Fisher Scientific) based on the m/z value and retention time of the ion signals. Ions from both ESI+ and ESI- were merged and imported into SIMCA-P multivariate data analysis software (version 14.1) for multivariate analysis. Principal component analysis (PCA) was first used as an unsupervised method for data visualization and outlier identification. Supervised regression modeling was then performed on the dataset using partial least squares discriminant analysis (PLS-DA) or orthogonal PLS-DA (OPLS-DA) to identify potential biomarkers. Biomarkers were filtered and confirmed by combining the results of variable importance in projection (VIP) values (VIP >1.0), t-test (p < 0.05), FC > 2. A combination of databases was used to visualize the pathway analysis: Lipid Maps (https://www.lipidmaps.org/) and Lipid Pathway Enrichment Analysis (LIPEA; https://lipea.biotec.tu-dresden.de/home) for the lipid species present in treated and untreated conditions; lipidomeR Integrative Visualizations of the Lipidome software was used to identify lipidomic content as R package. Lipids were categorized by lipid classes and presented on two-dimensional maps organized by lipid size and level of saturation. Experiments were performed in triplicates for each condition. P-values were calculated using t-test.
2.9. Co-immunoprecipitation
After induction of MC3324 at 25 μM for 6 h in C4-2 cells, co-immunoprecipitation (Co-IP) of endogenously expressed AR protein was performed using whole cell lysate (800 μg) in Co-IP buffer (10 mM Tris pH 7.5, 50 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM DTT, 10 mM sodium molybdate, 0.2 mM PMSF, 1X Roche protease inhibitor cocktail). Cell lysis was obtained with sonication using Diagenode's Bioruptor (Diagenode, B01020001). Protein A/G Plus Agarose (Santa Cruz, sc-2003) was coated with AR antibody and IgG (Santa Cruz, sc-2025) and mixed gently for 2 h at 4 °C for immunoprecipitation, in triplicate. A fraction of the resulting complexes was washed three times with Wash Buffer 1 (10 mM Tris pH 7.5, 50 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM DTT, 10 mM sodium molybdate, 0.2 mM PMSF, 1X Roche protease inhibitor cocktail), three times with Wash Buffer 2 (10 mM Tris pH 7.5, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 10 mM sodium molybdate, 0.2 mM PMSF, 1X Roche protease inhibitor cocktail), then denatured and eluted in 2X bromophenol blue as a control for IP (Supp. Figure 17). The remaining IP fraction was then processed for downstream MS analysis. First, proteins were digested and eluted from Ab-bead complexes by adding trypsin (Promega, Trypsin/Lys-C Mix Mass Spec Grade, V5071) and incubating for 30 min at room temperature. Eluates were then washed with 2 M urea, 50 mM Tris pH 7.5, and 5 mM chloroacetamide, and then fully digested overnight at room temperature [25]. Digestion was stopped by adding trifluoroacetic acid TFA to a final concentration of 1% v/v, and peptide cleanup was performed by dual C18 Stage tip and SP3. Briefly, three C18 disks were stacked on top of each other and transferred to a pipette tip. Tips were conditioned with methanol and 80% acetonitrile–0.5% acetic acid in LCMS-grade H2O (Buffer B) and equilibrated with 0.5% acetic acid in LCMS-grade H2O (Buffer A). Samples were loaded and washed with Buffer A, then eluted with Buffer B. Peptides were then dried and kept at −80 °C until use. Any remaining detergents from the IP protocol were removed using the SP3 protocol [26]. Dry peptides from the C18 cleanup were incubated with 2 μL of a 50:50 mixture of SeraMag-A and SeraMag-B (Sigma–Aldrich, GE29343057) beads and 200 μL acetonitrile. Beads were then washed once more with pure acetonitrile and eluted by incubation with 2% DMSO in LCMS-grade H2O. Peptides were then dried and re-suspended in a solution containing 0.1% formic acid in LCMS-grade H2O. Data are available at the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD029249.
2.9.1. Proteomic analysis: cell lysis and protein digestion
PCa cells were treated for 24 h with MC3324 at 25 μM and were then washed twice with PBS and harvested by centrifuge at 1200 rpm for 5 min. Next, the cell pellets were re-suspended in RIPA buffer supplemented with protease inhibitors (Thermo Fisher Scientific, Halt Protease Inhibitor; 0.5 M EDTA). Cells were lysed in a Bioruptor sonicator bath (Diagenode) for 20 min (20 cycles: 30 s ON, 30 s OFF), and lysates were centrifuged at 12,000 RPM for 30 min at 4 °C. Protein-containing supernatants were collected for downstream digestion. Protein concentration of whole cell lysates was measured by bicinchoninic acid assay (Thermo Fisher Scientific), while AR IP samples were measured on a DS-11-FX (DeNovix, DE, USA) at 280 nm. A total of 100 μg of proteins was digested for downstream proteome analyses. Solubilized proteins were precipitated using ice-cold methanol and centrifuged down at 12,000 rpm for 20 min at 4 °C. Protein pellets were re-suspended in 100 mM Tris buffer containing 100 mM dithiothreitol and 4% w/v sodium dodecyl sulphate (SDS, pH 8.0), heated at 95 °C for 30 min at 600 rpm. A total of 8 M urea in 100 mM Tris buffer pH 8.0 solution was added to dilute SDS, samples were loaded on 30 KDa molecular filters (Sigma–Aldrich) and centrifuged at 12,000 rpm for 20 min. Filters were washed twice with 8 M urea buffer and incubated in 50 mM iodoacetamide in 8 M urea buffer for 30 min (in the dark). Filters were washed four times (2 × 8 M urea buffer, 2 × 50 mM triethylammonium bicarbonate buffer pH 8.0), and proteins were digested with trypsin (enzyme–protein ratio 1:50) at 37 °C for 16 h under agitation (650 rpm). Filters were then centrifuged at 12,000 rpm for 20 min to extract tryptic peptides.
2.9.2. Strong anion exchange peptide fractionation
Tryptic peptide mixtures were fractionated for downstream MS analysis using strong anion exchange (SAX). Digested peptides were dried, then re-suspended in Britton and Robinson Universal Buffer (BRUB; 20 mM phosphoric acid, 20 mM boric acid, and 20 mM acetic acid in ultrapure water) pH 11 and loaded on SAX Stage tips (Sigma–Aldrich) combined with C18 filters (Sigma–Aldrich). SAX filter-containing tips were used to elute peptides onto C18 tips (Sigma–Aldrich) using BRUB at decreasing pH: 8, 6, 5, 4, and 3. C18 tips were then washed with 0.1% formic acid solution, and peptides eluted with 0.1% formic acid and 80% acetonitrile in ultrapure water. SP3 peptide purification was subsequently performed on dried eluates. SP3 beads (Thermo Fisher Scientific) were added to peptides, peptides were captured by adding 200 μL acetonitrile and eluted with 2% DMSO in water. Supernatants were dried and stored at −80 °C until MS analysis.
2.10. High-resolution mass spectrometry for AR IP and whole proteome analysis
Digested peptides derived from AR IP and whole proteome digests were re-suspended in 3% acetonitrile and 0.1% FA in ultrapure water and then analyzed on a Q-Exactive HF-X mass spectrometer coupled to a Proxeon EASY 1200 nano-liquid chromatography system (Thermo Fisher Scientific). For AR IP and whole fractionated proteome samples, a volume corresponding to 1 ug of digested peptides was analyzed. Digested peptide mixtures were injected into a reverse phase EasySpray (Thermo Fisher Scientific) analytical column (ID 75 μm × 50 cm C18 2 μm 100 Å particle size). Gradient was run using LCMS-grade water with 0.1% FA (solvent A) and 80% acetonitrile with 0.1% FA (solvent B) for 120 min. Gradient was run as follows over a 350 μL/min flow-rate: 90 min10–30% solvent B, 20 min 30–45% solvent B, 1 min 45–95% solvent B, and 9.5 min 95% solvent B. Eluting peptides were subjected to a 1.8 kV spray voltage. Full scans were acquired at 60,000 resolution and the 15 most intense ions were fragmented using high-energy induced collision dissociation, whose spectra were collected at 15,00 resolution. Precursor ions with charge 1 and > 6 and with intensities lower than 1.7 × E4 were excluded from triggering fragmentation. Ion accumulation time was set to 60 msec. Automatic gain control was set to 1xE5. Dynamic exclusion was enabled and set to 20 s. Thermo RAW files were acquired using Xcalibur software (version 4.1).
2.11. Analysis of mass spectrometry data
MS analysis-derived RAW files were analyzed using MaxQuant (version 1.6.14.0) and MS spectra searched using the Andromeda search engine. The Uniprot-Swissprot human proteome database (version released: 2020.02.24) was used for database searches. Selected protease was trypsin. Carbamidomethylation of Cys residues was selected as fixed modification. Met oxidation and acetylation of N-terminal residues were selected as variable modifications. The Label-free Quantification (LFQ) option was selected. Identification of peptides resulting from missed cleavages was allowed. Precursor ion tolerance was 20 ppm and 4.5 ppm for first and main searches, respectively. Match-between-run option was enabled, and settings left to default. MaxQuant search-derived protein intensities were used for statistical analyses. Protein tables were filtered for protein q-value (<0.01), contaminant (excluded), reverse sequences (excluded), and unique peptides (at least 1).
2.11.1. Statistical analysis
AR IP and proteome tables were filtered for missing data (cutoff: 30%), and log2 transformed. Pseudocount (value: 0.1) was applied to the protein table prior to log2 transformation. Differential expression was assessed by Welch-corrected t-test (AR IP) and DeqMS (peptide identification-adjusted linear model; proteome). Pathway enrichment between conditions (treated vs untreated) was performed using Gene Set Enrichment Analysis (GSEA) against the Hallmarks database (version 5.2). Settings were as follows: permutation type, gene set; scoring, classic; metric, t test. Other parameters were kept to default settings. False discovery rate cutoff to call significant pathways was set to 0.25. Normalized enrichment scores were plotted to define enrichment levels. All data were analyzed in R (version 3.6). Data are available at the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD029525.
2.11.2. Metabolomic analysis: metabolite extraction for nuclear magnetic resonance (NMR) analysis
PCa cells were treated for 6 h with MC3324 at 25 μM and were washed twice with PBS and then harvested by centrifuge at 1200 rpm for 5 min. A dual phase extraction procedure introduced by Bligh and Dyer in 1959 [27] was employed, with slight modifications, to extract intracellular metabolites, as already reported elsewhere [28]. Briefly, 6 mL cold methanol (−20 °C) and 6 mL chloroform were added to the original aqueous solution (5.4 mL) containing quenched cells to obtain a mixture of water, methanol, and chloroform in a volume ratio of 0.9:1:1, corresponding to a total volume of 17.4 mL. The mixture was then incubated for 20 min on ice and repeatedly vortexed to facilitate the extraction. Next, centrifugation at 4000g at 4 °C was performed for 20 min to obtain a two-phase extract made of an upper phase containing water-soluble intracellular metabolites and an organic lower phase including non-polar metabolites such as lipids. The skin-like layer between the two phases entrapped proteins and macromolecules. The upper and lower phases were then separated and carefully transferred into different falcon tubes. Solvents were completely removed from both fractions using a vacuum concentrator (hydrophilic phase) and under a gentle flow of N2 gas (organic phase).
2.11.3. NMR spectroscopy
Aqueous cell extracts were dissolved in 600 μL D2O, briefly vortexed, and transferred into 5-mm NMR tubes. All one-dimensional 1H NMR spectra were acquired at 300 K on a Bruker Avance NEO 600 MHz spectrometer (Bruker BioSpin, Rheinstetten, Germany) equipped with a QCI cryo-probe set for 5-mm sample tubes and a cooled SampleJet autosampler. The 1H NMR spectra of hydrophilic cell extracts were acquired with Topspin 4.1 (Bruker BioSpin) using the ‘noesygppr1d’ pulse sequence allowing for a quantitative evaluation even closer to the water signal [29], which was presaturated at 4.698 ppm. All the experiments were performed with an acquisition time of 3.67 s, a relaxation delay of 4 s, receiver gain of 101, 128 scans, 4 dummy scans, and a spectral width of 17,857 Hz (29.755 ppm). All samples were automatically tuned, matched, and shimmed. Prior to Fourier transformation, free induction decays were multiplied by an exponential function equivalent to a 0.3-Hz line-broadening factor. The transformed spectra were then automatically corrected for phase and baseline distortions and calibrated using TopSpin built-in processing tools.
2.11.4. Metabolite identification
Assignment of the hydrophilic metabolites was achieved by i) analysis of literature data [28,30], ii) comparison with the chemical shifts of metabolites in the Human Metabolome Database (HMDB), and iii) peak fitting routine within the spectral database in Chenomx NMR Suite 5.0 software package (Chenomx, Edmonton, AB, Canada). A representative 1H NMR with the relative assignment is reported in Supp. Figure 21.
2.11.5. NMR data reduction and processing
NMR spectra were imported into MATLAB (Mathworks, Natick, MA, USA, R2015b), where spectral regions above 10 ppm and below 0 ppm were removed because they contained only noise. To correct for spectral misalignment, an interval-based alignment step was carried out using the icoshift algorithm [31] and choosing the alanine doublet at 1.49 ppm as reference signal. Then, in order to reduce the model complexity, peak areas of the well-separated resonances of 20 selected metabolites were manually integrated and submitted to data analysis as a data matrix made of 18 rows (samples) × 20 columns (metabolites). This data matrix was then submitted to the PLS toolbox version 8.6.1 (Eigenvector Research, Manson, WA, USA), where it was normalized according to the total area (1-norm) and then autoscaled. Autoscaling employs standard deviation as a scaling factor, thus giving all metabolites the same chance to affect the model and the mean-centering, which is needed to compute the Principal Component Analysis (PCA).
2.12. Bulk RNA sequencing analysis
PCa, advanced PCa, benign prostatic hyperplasia (BPH) and CRPC RNA sequencing data were downloaded from the NCBI repository GSE80609 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80609). Statistical analyses were performed by using R (version 3.6.4). Fragments Per Kilobase of exon per Million mapped reads (FPKM) was normalized by the quantile method, log2 transformed, and median-centered across genes and samples. To compare sample subgroups in terms of gene expression, the DESeq package that utilizes a negative binomial model was used to detect differentially expressed genes from count data matrix. Expression differences in genes were considered statistically significant if the p-value was <0.05 and the fold difference in expression between two sample groups was ≥1.
2.12.1. Acoustic force spectroscopy cell seeding and measurements
Cell suspensions were harvested and centrifuged at 1200 rpm for 5 min, and the supernatant was discarded. Cells were re-suspended in RPMI at an average concentration of 1.3 × 106 cells ml−1 and transferred into a 1.5 mL Eppendorf tube. Then, 40 μL of 7.9 μm-diameter silica beads were washed and re-suspended in 80 μL medium. The beads were mixed with the cell suspension at a cell-to-bead ratio of 1:5, and the mixture was injected into the flow channel of the AFS chip up to a desired cell confluence of 60–70% and incubated at 37 °C with 5% CO2 for 3 h to allow the cells to attach to the bottom of the AFS channels under static conditions. After incubation, the AFS chip was connected to a peristaltic pump for growth media perfusion. Flow rate was set to 30 μL/min, until culture achieved 90–100% confluency. All perfusion experiments were performed within a dry incubator to avoid damaging the AFS chip. Upon reaching full confluency, the chip was placed on the AFS microscope stage, and the temperature controller was set to the physiological temperature of 37 °C. To measure the non-linear creep response of cells, a constant force (ranging from 1.026 nN to 1.467 nN), was applied for 20 s. The displacement at each step was fitted by a creep-compliance model (see section below: 2.16.2). To measure the effect of MC3324 on PC3 cell mechanics, creep responses were measured at 3 h, 6 h, and 24 h of incubation. All measurements were performed at 37 °C with a peak-to-peak driving voltage of 30 Vpp at 14.51 MHz frequency. Images were acquired with a bright-field inverted microscope equipped with a 1.3 MP camera recording at 60 Hz (IDS, UI-324CP) in combination with an air 20 × 0.75 NA objective (Nikon, CFI Plan APO, VC 20×, MRD70200). The bead z-position was determined using a predefined look-up table (LUT), a library of radial profiles as a function of z position with 100 nm steps, created from a series of microsphere images prior to application of the acoustic force.
2.12.2. Acoustic force calibration
To determine the acoustic radiation force, we performed a force-balance on acoustically driven beads. The acoustic force results from the standing acoustic wave and pushes the bead toward the acoustic node. The acoustic wave amplitude is proportional to the voltage applied to the piezo-element, which excites the wave according to:
| (1) |
where F is the acoustic force, V the voltage applied to the piezo-element, and c the conversion factor.
The conversion factor is found by a setup calibration. During calibration (i.e., without a cell), a single bead is subjected to gravitational, acoustic, and drag forces according to:
| (2) |
| (3) |
with
| (4) |
where g is gravity, r is the bead radius, ρbead is the density of the silica bead, ρmedium is the density of PBS, ubead is the bead velocity, γBrenner is the correction factor for Stokes drag coefficient, η is the viscosity of the medium, and h is the height of the bead center to the surface. Brenner's drag coefficient was determined by measuring the viscosity of the bulk fluid and directly inserting it into Equation [6]. In order to find the experimental local viscosity, η, and to use it for the determination of the effective drag coefficient, γBrenner, a method based on the terminal velocity of the bead, was used as previously published [32]. Briefly, the terminal velocity of the bead is tracked as it settles from the acoustic node toward the bottom.
2.12.3. AFS data fitting
To quantify the viscoelastic properties of PC3 cell line, the standard linear liquid (SLL) model was utilized, where the creep compliance of the cell is captured by:
| (5) |
where Ea is the elasticity associated with the cytoskeleton, ηa the viscosity associated with the cytoskeleton, and ηc is the background viscosity. Each force step (i.e., z-height versus time) was first converted to J(t) according to:
| (6) |
where z(t) is the extension curve in z-direction obtained by pulling on the cell, F is the applied force, and r is the particle radius. J(t) was then plotted as a function of time and Equation [1] was fitted to the resultant curves.
3. Results
3.1. LSD1/UTX inhibition induces AR signaling downregulation and antiproliferative effects
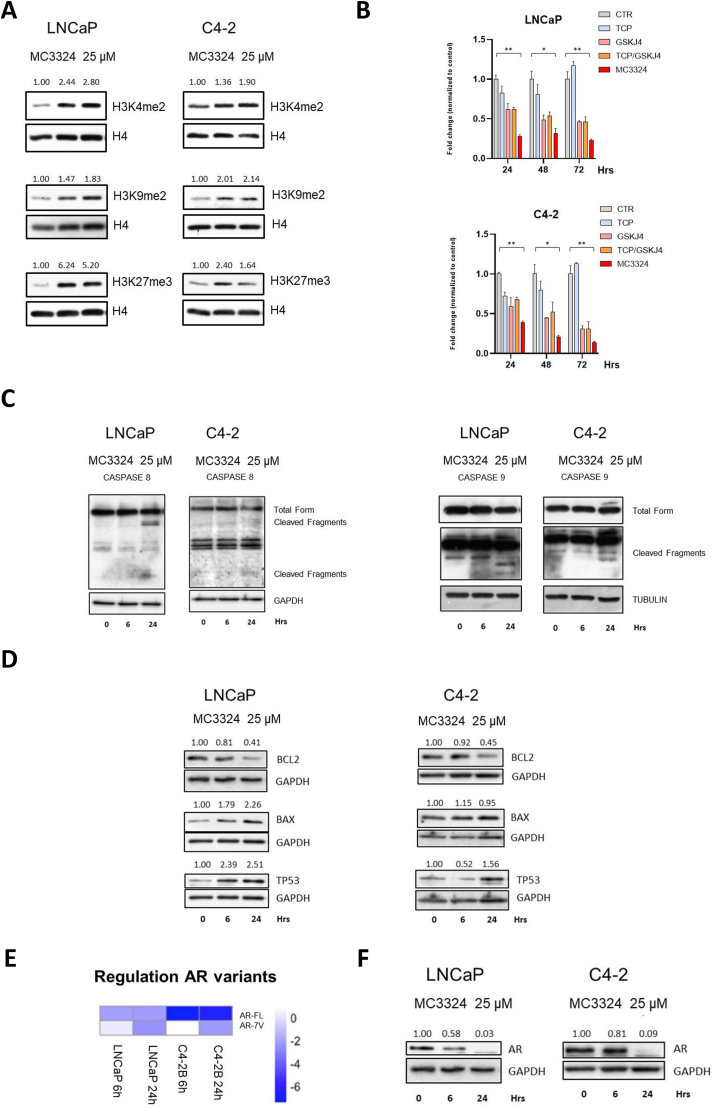
MC3324 globally increased dimethylation of histone H3 at lysines 4 and 9 (H3K4me2, H3K9me2), two LSD1 targets, and trimethylation of histone H3 at lysine 27 (H3K27me3), a UTX target, in both PCa (LNCaP) and CRPC (C4-2) cell lines (Figure 1A). Epigenetic rebalance was coupled with a strong antiproliferative effect and increased anticancer efficacy compared to TCP and GSK-J4 TCP, commercially available inhibitors of LSD1 and UTX, respectively, used either alone or in combination (1:4 ratio) (Figure 1B). Among all available LSD1 inhibitors, TCP was chosen because MC3324 is constituted with its active scaffold. Cell growth was impaired in both PCa and CRPC models, suggesting that dual KDM inhibition may be a promising interfering strategy in endocrine-resistant or AR-null PCa (Supp. Figure 1). Orthogonal assays have been applied to PC3 cell line to confirm cell death induction and cell cycle arrest (Supp. Figure 2). The antiproliferative effect was associated with the induction of pro-apoptotic proteins after treatment (Figure 1C and Supp. Figure 3A-B). Furthermore, we observed an overexpression of TP53 (Figure 1D and Supp. Figure 3C), a major tumor suppressor, whose upregulation is associated with remodulation of cancer signaling and reduced oncogenic activity [33]. MC3324 induced time-dependent downregulation of AR at protein and mRNA level (Figure 1E–F). AR downregulation was also confirmed in C4–2 B cells (Supp. Figure 3D), CRPC bone metastasis model androgen-independent. By inhibiting the epigenetic modifiers LSD1 and UTX, treatment with MC3324 reduced cell survival and activated cell death mechanisms. MC3324 cancer selectivity was shown in the normal prostate PNT2 cell line. Although a reduction in proliferation was observed (Supp. Figure 4A), H3k4me2 and H3k27me3 were not increased (Supp. Figure 4B), caspases were not cleaved (Supp. Figure 4C), and, interestingly, AR expression levels were not downregulated (Supp. Figure 4D). Taken together, these findings suggest the epigenetic modulation of LSD1/UTX as an alternative/additional therapeutic approach in PCa and CRPC phenotypes.
Figure 1.
Epi-downregulation in androgenic signaling and apoptotic triggering. A) Western blot analysis of histone methylation levels after MC3324 treatment (25 μM) in LNCaP and C4-2 cells. Increase in H3K4me2, H3K9me2, and H3K27me3 levels was evaluated after 6 h and 24 h of MC3324 treatment. The relative increase was quantified with ImageJ software (1.46r, NIH, USA) and reported as normalized to control intensity. Experiments were performed in triplicates. B) MTT assay results expressed as fold change in treated cells normalized to the mean of control in LNCaP and C4-2 cells after induction with MC3324 (25 μM), tranylcypromine (TCP; 100 μM) and GSK-J4 (25 μM) for 24 h, 48 h, and 72 h. Experiments were performed in triplicates. Standard deviations are reported as error bars, while p-values were calculated using t-test. Statistical significance is expressed as p-value ∗ <0.05, ∗∗ <0.01, ∗∗∗ <0.001. C) Western blot analysis of extrinsic and intrinsic apoptotic proteins after MC3324 treatment (25 μM) after 6 h and 24 h in LNCaP and C4-2 cells. Experiments were performed in triplicates. D) Western blot analysis of BCL2 and BAX acting as anti-apoptotic and pro-apoptotic proteins, respectively, after MC3324 treatment (25 μM) after 6 h and 24 h in LNCaP and C4-2 cells; relative expression of TP53 after treatment is also shown. Experiments were performed in triplicates. E) Real-time PCR data, in triplicates, showing AR downregulation by MC3324 treatment (25 μM) in LNCaP and C4-2 cells after 6 h and 24 h. F) Western blot analysis showing AR downregulation in LNCaP and C4-2 cells after MC3324 treatment (25 μM) for 6 h and 24 h.
3.2. LSD1/UTX inhibition affects maximal respiration and ATP production
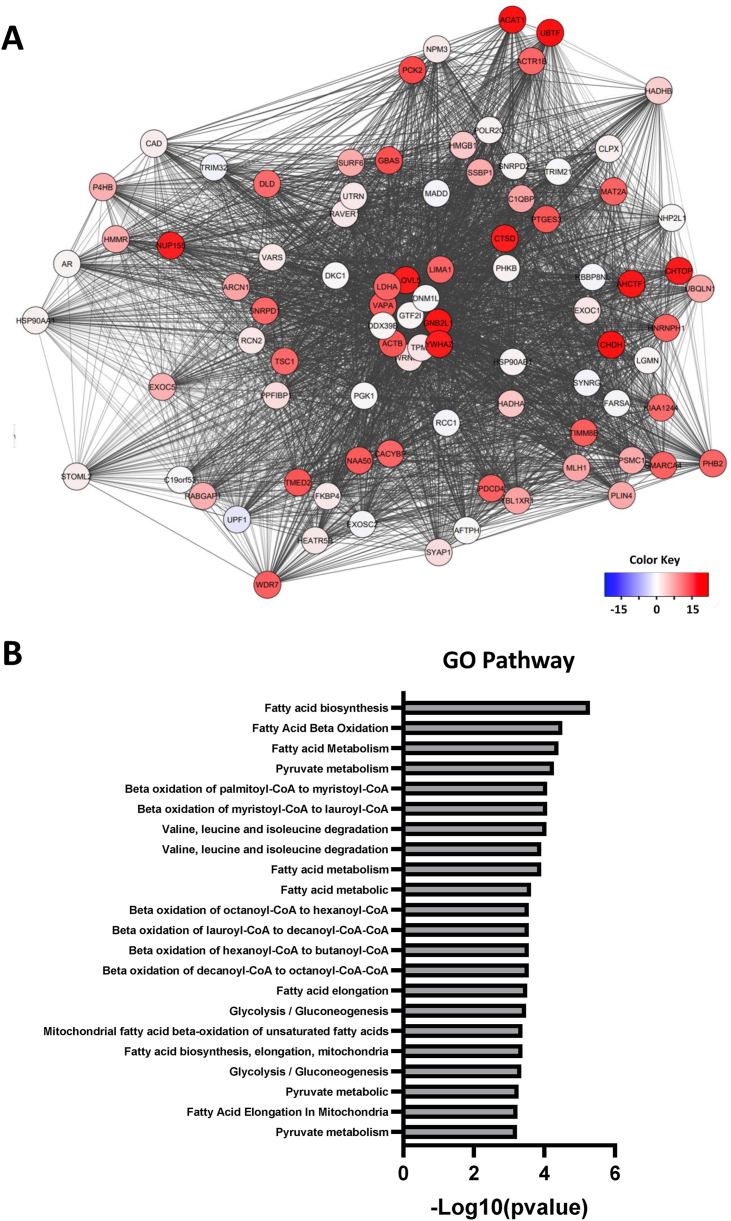
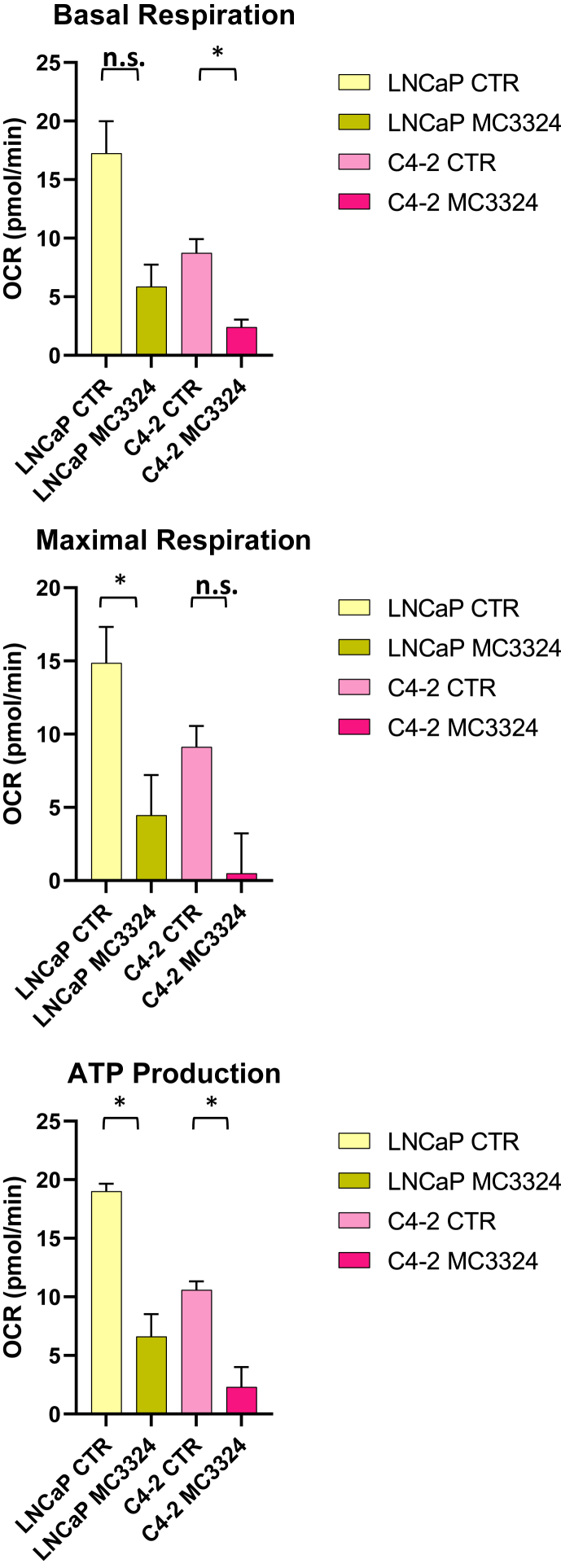
Metabolic evolution is recognized as a fundamental hallmark of cancer, and AR is known to orchestrate metabolism and biosynthesis at key regulatory steps in PCa and CRPC [9]. The AR interactome of a CRPC cell line was analyzed following short-term MC3324 treatment, and AR interactors were identified and grouped by biological pathway (Figure 2A–B). The results returned proteins active in metabolism and metabolic pathways such as ACAT1 and ELOVL5, both involved in fatty acid synthesis, LDHA, active in the conversion of pyruvate to l-lactate, and DLD, a mitochondrial enzyme that plays a vital role in energy production [34]. The displacement of AR metabolic interactors, following MC3324 treatment, was coupled with the alteration of pathways involved in cellular energy activity (glycolysis, lipolysis, and glutaminolysis) in PCa and CRPC. Globally, MC3324 treatment at 25 μM for 3 h reduced ATP production, impairing maximal mitochondrial respiration capacity in LNCaP and C4-2 cells (Figure 3). Lower mitochondrial efficiency was also observed in the other CRPC and AR-null cell lines (Supp. Figure 5), suggesting that LSD1/UTX inhibition shuts down prostate metabolism independently of AR expression. These findings suggest an impairment in mitochondrial functions with a block of energetic processes that disrupts the onco-metabolic balance supporting uncontrolled proliferation of PCa and CRPC cells.
Figure 2.
Interactome of AR. A) AR interaction network of the C4-2 CRPC cell line. Proteins identified by AR pulldown assay after treatment with MC3324 (25 μM) were annotated and clustered. B) GO term annotation for metabolic functions associated with AR interactors.
Figure 3.
Mitochondrial stress test. Metabolic flux analysis reported as oxygen consumption rate (OCR) measurements performed in MC3324-treated and untreated (control) LNCaP and C4-2 cell lines. P-values were calculated using t-test based on triplicate values. Statistical significance is expressed as ∗ p-value <0.05. Standard deviations are reported as error bars.
3.3. MC3324 treatment alters metabolic gene expression profiles
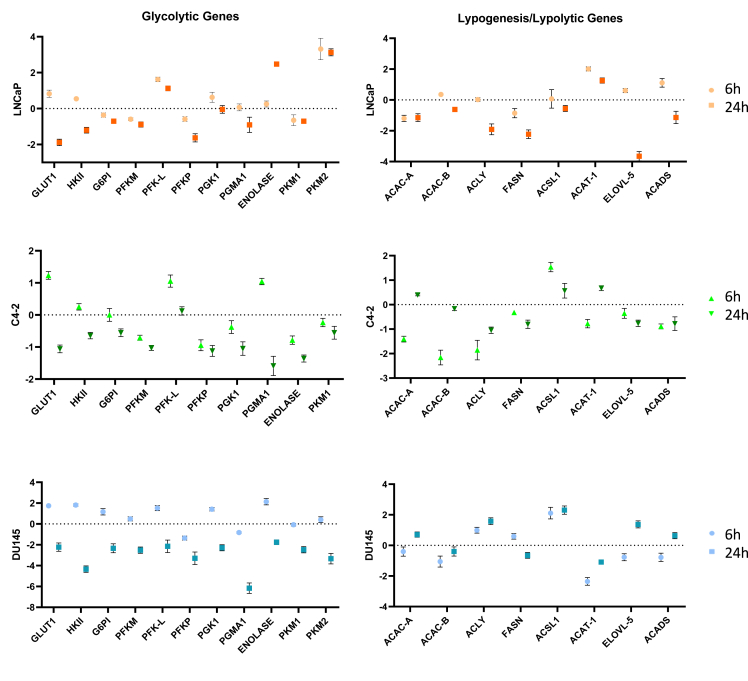
Reduced mitochondrial activity leads to the impairment of cancer cell homeostasis. The reduction in ATP production is due to the alteration of genes involved in glycolysis and lipid pathways following MC3324 treatment. We investigated the differential expression of enzymes catalyzing key reaction steps in glucose transport and conversion and in beta oxidation of fatty acids. MC3324 treatment at 6 h induced GLUT1 upregulation in LNCaP, PCa cell line, C4-2, and DU145 CRPC cells (Figure 4), probably as a protective mechanism in response to ATP production impairment. A similar trend was observed for HKII, the second enzyme involved in glycolysis; complete downregulation of GLU1 and HKII in this pathway was detected at 24 h of MC3324 treatment. Because PCa and CRPC are lipid-enriched cancers (Supp. Figure 6), we also investigated ATP citrate lyase (ACLY), which acts as a bridge between glycolysis and lipogenesis [35]. Following MC3324 treatment, ACLY mRNA expression was reduced in AR-positive and androgen-sensitive LNCaP cells, in AR-positive and androgen-insensitive C4-2 cells, and in AR-null DU145 cells. The different response to MC3324 treatment was also observed for other metabolic genes (Figure 4), highlighting PCa heterogeneity. Interestingly, the lipid metabolic gene FASN, encoding the key enzyme in fatty acid synthesis, was found downregulated after MC3324 treatment at 24 h in PCa and CRPC models. FASN plays a pivotal role in the acquisition of tumor phenotype and directly favors PCa progression [36], as confirmed by its overexpression in PCa and CRPC associated with poor outcomes [37]. Bulk RNA sequencing re-analysis (GSE80609) of patients with different degrees of disease severity (PCa = 16, advanced PCa = 9, CRPC = 12) compared to those with benign prostatic hyperplasia (BPH = 8) revealed FASN overexpression in all cancer stages (Supp. Figure 7), further strengthening the correlation between PCa/CRPC and the “lipid phenotype” acquired in oncogenic onset [38]. To further confirm that KDMs play a key role in AR expression and metabolic control of prostate cancer we silenced LSD1 with shRNA. The downregulation of LSD1 (Supp. Figure 8A), similarly to its pharmacological inhibition, induces AR downregulation together to key metabolic proteins downregulation (Supp. Figure 8B). Although the involvement of LSD1 in the regulation of lipogenic genes is already well known [39], the role of UTX is less clear [40,41]. These findings suggest a potential role for KDM enzymes in cancer treatment through repression of the metabolic oncogene FASN. Our results show that metabolic downregulation via LSD1/UTX inhibition may represent a novel strategy to modulate metabolic pathways in PCa and CRPC, overcoming limitations related to AR expression and androgen sensitivity.
Figure 4.
Glycolytic and lipogenesis/lipolysis gene expression. Graphs from real-time PCR data showing different expression levels of genes involved in glycolysis and in lipogenesis/lipolysis processes in LNCaP, C4-2, and DU145 cell lines after 6 h and 24 h of MC3324 treatment at 25 μM. Standard deviations are reported as error bars based on triplicate values. Primers used are available in Supplementary File Table 4.
3.4. LSD1/UTX inhibition disrupts lipid metabolism
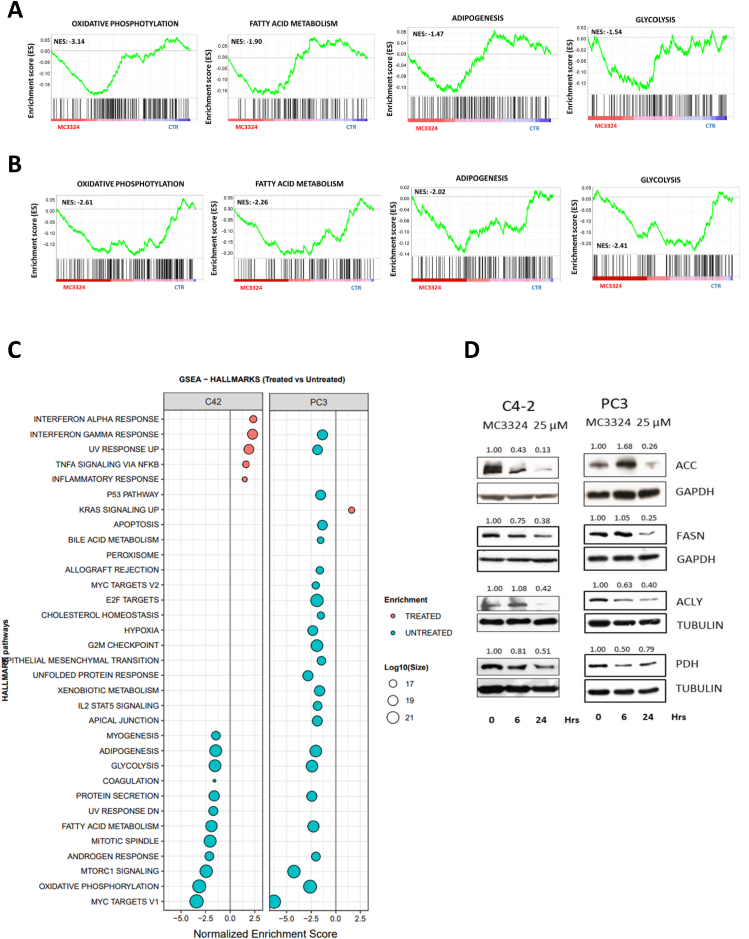
The differential mRNA expression observed in key steps of energy processes may account for the reduced metabolic capacity of PCa and CRPC following MC3324 treatment. To further confirm this hypothesis, we performed proteomic analyses to investigate the relative abundance of enzymes involved in metabolism. Specifically, we selected two CRPC cell lines recognized as having accelerated lipid metabolism: AR-positive and androgen-insensitive C4-2 cells and AR-null PC3 cells. The Pearson correlation and hierarchical clustering on the proteomic dataset indicated a good correlation (Supp. Figure 9), and normalized enrichment score showed a reduction, after MC3324 treatment, in oxidative phosphorylation, fatty acid metabolism, adipogenesis, and glycolysis (Figure 5 A-B-C), all pathways directly connected to metabolic processes. To further validate these findings, we investigated the expression level of proteins in key steps of lipid metabolism in both systems (Figure 5 D). We observed a downregulation of ACC and FASN, pivotal proteins in lipogenesis, as well as downregulation of ACLY and PDH, involved in carbohydrate and lipid communication, and in acetyl-CoA procurement, respectively. These findings indicate an epigenetic regulation of metabolic signaling, highlighting the existence of a druggable axis between LSD1/UTX inhibition and metabolic circuits promoting the more aggressive phenotype. We also observed a downregulation of pathways equally important in supporting tumor growth and progression (Supp. Figure 10), indicating a depletion of androgen response signaling, accompanied by reduced protein secretion and a reduction of the pathway driven by MYC, a known oncogene overexpressed in cancer (Supp. Figure 11). While it is not completely clear at which level the cascade of events begins, the pleiotropic effect observed after LSD1/UTX inhibition culminated in proliferative arrest and cell death. Indeed, metabolic signaling is not only related to energy processes, but mediates and enhances other cellular activities with multiple effects such as duplication and cellular trafficking. Interestingly, mTORC1 pathway was also reduced after MC3324 treatment (Supp. Figure 11). Since mTORC1 is widely associated with autophagic activity, we investigated the main actors in autophagy to further validate whole proteome findings. In C4-2 cells, Western blot analysis showed an increase in phospho-AKT and upregulation/phosphorylation of eNOS, an AKT downstream target, as well as downregulation of acetylated tubulin and upregulation of SQSTM1 (Supp. Figure 12A), with upregulation of phosphatidylinositol species (Supp. Figure 12B). Autophagy impairment affects fatty acid recycling [42], thus showing a further block of autophagic branch (Supp. Figure 12C). All these findings indicate that LSD1/UTX inhibition switches off oncogenic signaling and lipid metabolism, acting as the driving energy source in CRPC systems. The autophagy axis also seems to be disrupted, suppressing further energy supply.
Figure 5.
Switch-off of metabolic pathways. Enrichment plot for C4-2 (A) and for PC3 (B) cells showing that MC3324 treatment downregulates key metabolic- and lipid-related pathways. C) GSEA enrichment of modified pathways after MC3324 treatment in C4-2 and PC3 cell lines. Experiments were performed in triplicates for each condition. D) Western blot analysis in C4-2 and PC3 cells after MC3324 treatment (25 μM) for 6 h and 24 h in ACC, FASN, ACLY, and PDH genes involved in lipid metabolism. Experiments were performed in triplicates. The relative increase was quantified with ImageJ software (1.46r, NIH, USA). Images were normalized using gapdh/tubulin as loading control.
3.5. MC3324 treatment alters untargeted lipidomic contents
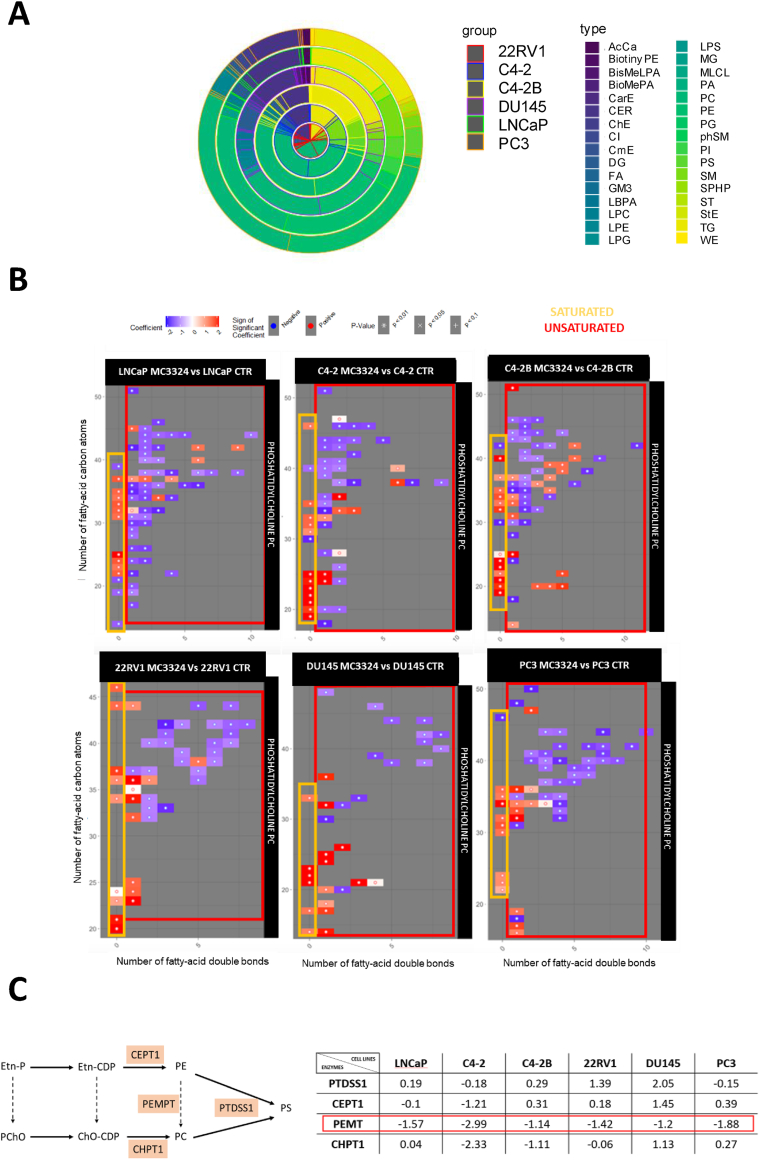
Having observed mitochondrial ATP production impairment with a modulation of metabolic genes following LSD1/UTX inhibition in PCa and CRPC models, we explored the evolution of lipid species after MC3324 treatment. We investigated lipid content by carrying out an untargeted lipidomic analysis comparing MC3324-treated (6 h) and untreated PCa cells. The untargeted lipidomic analysis was performed using liquid chromatography high-resolution mass spectrometry (LC–MS/MS) (Supp. Figure 13A-B) (Supplementary File Tables 2–3). In total, we identified more than 15 different lipid classes modulated after MC3324 treatment, and more than 300 lipid species were quantified for each prostate cell line, detected in electron spray positive-ion and negative-ion mode. The classes identified included: phosphatidylcholines (PCs), phosphatidylethanolamines, triacylglycerols, ceramides, phosphatidylserines, sphingomyelins, phosphatidylglycerols, cholesteryl esters, and diacylglycerols (Figure 6A). Heatmaps show two panels of differentially expressed lipid species (Supp. Figure 14) after MC3324 treatment. In our analysis, differential expression of PCs indicated an increase in saturated species in all cells, but an overall reduction in unsaturated classes, regardless of AR expression or androgen sensitivity (Figure 6B). PCs can be synthetized from phosphatidylethanolamines by a process of trimethylation on the primary amine in a dense network of enzymatic activities [43,44]. Real-time PCR analysis showed downregulation of phosphatidylethanolamine N methyltransferase (PEMT) mRNA levels after 24 h of MC3324 induction in both PCa and CRPC models, suggesting the impairment in PC production, which can affect tumorigenic progression to the extent of inducing an arrest in proliferation and cell death (Figure 6C). Furthermore, metabolomic analysis of selected PCa and CRPC cell lines (Supp. Figure 15) orthogonally confirmed the statistically significant reduction in PC lipid species upon MC3324 treatment (Supp. Figure 16). PCa is reported to modulate PC content by increasing the production of unsaturated fatty acids [45]. The study described three unsaturated lipids, PC(38:5), PC(40:3), and PC(42:4), which when overexpressed are associated with PCa. Our lipidomic analysis revealed downregulation of all these three lipid species after MC3324 treatment, when expressed (Supp. Figure 17). Taken together, our results show that LSD1/UTX inhibition regulates PC expression in PCa and CRPC cell lines by inducing metabolic plasticity in lipid acquisition. Dual targeting of lipid supply may be a valid treatment approach to overcome therapeutic resistance associated with AR expression or androgen sensitivity.
Figure 6.
Lipid content alteration. A) Multiple pie chart showing relative lipid class content in prostate cell lines. The six rings correspond to different PCa cell lines: 22RV1 (red), C4-2 (blue), C4–2 B (yellow), DU145 (violet), LNCaP (green), PC3 (orange); for each cell line, relative lipid class subgroups are highlighted with different colors inside the circle. B) Phosphatidylcholine (PC) species based on saturation degree (x axis) and length of fatty acid (y axis) grouped by saturated (yellow) and unsaturated (red) matrix. C) Enzymes involved in phosphatidylserine (PS) synthesis from phosphatidylethanolamines (PE) or phosphatidylcholines (PC). Relative expression of PTDSS1, CEPT1, PEMT, and CHPT1 genes from real-time PCR data after MC3324 treatment for 24 h in the indicated PCa cell lines. Experiments were performed in triplicates.
3.6. MC3324 treatment modulates lipid classes associated with cell death
Acyl carnitine is reported to interfere with key functional properties of progression and angiogenesis in PCa both in vitro and in vivo [46]. This lipid class was shown to decrease acetyl-CoA inside mitochondria, with reduced ATP production in the tricarboxylic acid cycle, reducing cell proliferation through apoptosis and suggesting a protective role against PCa. In our lipidomic dataset, acyl carnitine was found upregulated after MC3324 treatment in LNCaP, DU145, and PC3 cell lines (Supp. Figure 18). Several studies describe a strong interconnection between lipids, mitochondrial activity, and cell death. The sphingolipid class is known to play a role in cell death, cell survival, and therapy resistance in many cancers, including PCa [47,48], and ceramide, a key molecule in sphingolipid metabolism, is generally associated with antiproliferative effects and mitochondrial-mediated apoptosis. In our dataset, the sphingomyelin class was mainly upregulated in all six cell lines after MC3324 treatment (Supp. Figure 19). In addition, we observed a substantial difference in the number of species found differentially expressed. AR-positive cell lines showed a large number of sphingomyelins (LNCaP = 33, C4-2 = 23, C4–2 B = 22, 22RV1 = 11) compared to the small number found in AR-null cell lines (DU145 = 3, PC3 = 4) (Supp. Figure 19). Other members of the sphingolipid class were also altered, suggesting that their modulation could contribute to mitochondrial degeneration and favor cell death.
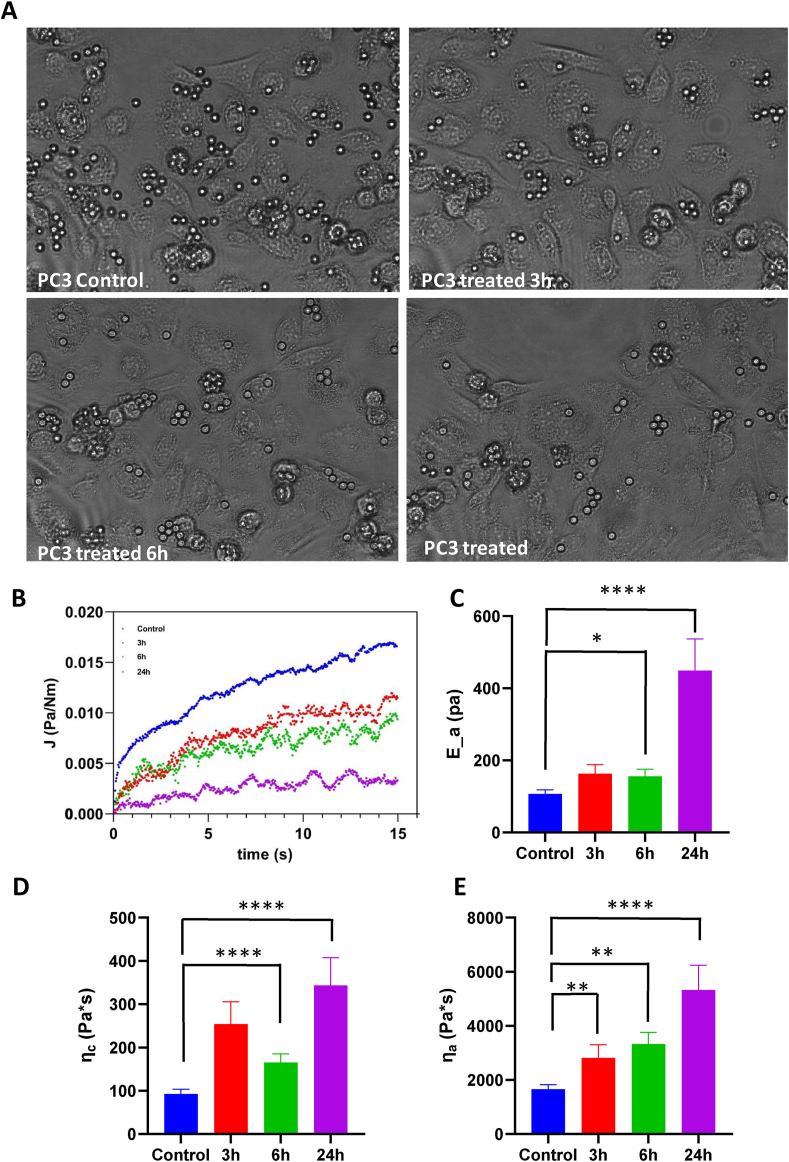
3.7. MC3324 treatment stiffens PC3 cells
It is widely acknowledged that cells can be described as mechanical systems with properties orchestrated by biochemical cues [49]. Tumorigenesis induces substantial alterations in both structure and composition of the actin network [50,51] resulting in a general loss of cell rigidity, which, in turn, provides critical advantages for matrix infiltration and migration [52,53]. We therefore investigated whether MC3324 is able to increase the stiffness of PCa cells, potentially lowering their metastatic potential. We measured the mechanical properties of PC3 cells using acoustic force spectroscopy, a high-throughput force spectroscopy technique that has only recently been adapted to probe single-cell mechanics [54,55]. PC3 cells were confined between silica beads (7.9 μm in diameter) and the glass surface of the AFS microfluidic chip. Acoustic forces were applied via a piezo element, which generates standing acoustic waves that push the beads up toward the acoustic node, instantaneously stretching the cells. From the resulting stress–strain curves, the creep compliance (the total load strain per unit of stress) was determined, from which the viscoelastic properties of the PC3 cells were obtained. Cells were measured before the addition of MC3324 followed by rapid solution exchange for exposure to the compound. Upon MC3324 exposure, cells underwent a drastic change in morphology, taking on a distinctly flatter appearance (Figure 7A). Example traces for creep compliance at each time point of MC3324 exposure are shown in Figure 7B. MC3324 caused significant stiffening of PC3 cells in a time-dependent manner (Figure 7C). Cell stiffness is associated with the apoptotic response of cells [56,57], which our data revealed to be upregulated after addition of MC3324. Inhibition of ACC activity with soraphen A is reported to increase the saturation of fatty acids in LNCaP cells [58], and we also found upregulated saturated PC levels in PCa after KDM inhibition, which caused an increase in the elastic modulus of the lipid bilayer. The viscous modulus associated with the cytoskeleton also showed a significant time-dependent increase (Figure 7D). The increase in cell stiffness was accompanied by a significant time-dependent increase in cytoplasmic viscosity (Figure 7E). Cytoplasm rheology critically depends on the spatial and temporal scale of the deformation. In our experimental conditions, the cytoplasm response was more similar to that of a viscous fluid, which was previously linked to the effect of ATP depletion [59], a process also induced by MC3324. Taken together, these results indicate that PCa mechanics and metabolism can be molecularly and epigenetically targeted.
Figure 7.
Mechanical and physical properties. MC3324 alters the viscoelastic properties of PC3 cells. A) PC3 culture inside the AFS chip at different time points of MC3324 exposure. B) Example traces of PC3 creep compliance at different time points of MC3324 exposure. C) Graph showing that MC3324 significantly increases the average elastic modules Ea in a time-dependent manner (p < 0.0001). D) Graph showing that MC3324 significantly increases the average viscosity ηa in a time-dependent manner (p < 0.0001). E) Graph showing that the average background viscosity ηc of PC3 cells significantly increases upon stimulation by MC3324 in a time-dependent manner (p < 0.0001).
4. Discussion
CRPC continues to be a major burden for public healthcare systems, and overcoming treatment resistance requires the identification of alternative therapeutic targets/strategies. In PCa and CRPC, the KDMs LSD1 and UTX are overexpressed and co-localize with both AR and promoters of its transcription, triggering a self-sustaining loop [[60], [61], [62]]. This functional relation suggests a potential role for epigenetic compounds in modulating oncogenic signaling and highlights their possible use in breaking this loop. The present study reveals that the indirect downregulation of AR through LSD1/UTX inhibition induces histone methylation on residues H3K4me2, H3K9me2, and H3K27me3, thereby promoting cell death and a metabolic shift in PCa and CRPC, as well as an alteration in cellular mechanics. We investigated the effect of the LSD1/UTX inhibitor MC3324 at metabolomic, proteomic, lipidomic, and phenotypic level, showing that epigenetic code, receptor signaling, lipids, and mechano-metabolic features are tightly connected and pathologically compromised in PCa and CRPC. Regardless of androgen sensitivity and AR expression, our data indicate a correlation between epigenetic activity and tumorigenic processes, suggesting that LSD1 and UTX play a fundamental role in tumor growth in PCa and CRPC and that both epi-enzymes can be considered promising targets for therapy. AR is one of the major drivers of both cancer types and is supported by a plethora of partners [63], while its downregulation is associated with a positive patient outcome [61]. Although inhibitors of LSD1 and UTX were previously reported to limit PCa advancement [64], this is to the best of our knowledge the first study to show that simultaneous LSD1/UTX inhibition is able to modulate androgen signaling in PCa and CRPC. Our findings suggest that this approach may overcome ADT resistance associated with the aggressive phenotype. A reduction in AR transcription leads to a block of proliferation with induction of cell death in forms of PCa such as AR-positive CRPC. The absence of AR impacts with an avalanche-like effect on the transcription of thousands of genes that directly or indirectly regulate cell proliferation and death, affecting the entire cellular organization. However, in our study LSD1/UTX inhibition also reduced cancer growth in AR-null CRPC. The dual inhibition of these enzymes induced a block of ATP production through downregulation of the main actors in glycolysis, such as GLUT1, the first enzyme responsible for glucose transport inside cells reported to be overexpressed in different types of human cancers [65], and in oxidative phosphorylation in PCa and CRPC phenotype, as shown by proteomic analysis, thereby disrupting pivotal energy pathways recognized as drivers of CRPC, including those involved in fatty acid metabolism. Lipids are essential molecules for cellular structure, energy, and communication in lipid-enriched tumors, such as PCa [66]. Bioactive lipids are in fact also used as physiological signaling molecules involved in PCa progression until onset of the CRPC phenotype [35]. Recent reports revealed that epigenetic modifications play a pivotal role in energy metabolism [67] through the regulation of lipid and other metabolic pathways. Metabolism and epigenetic processes regulate each other in an interacting system to create a complex network promoting malignant growth and conferring treatment resistance [68]. Our lipidomic analysis reveals changes in the architecture of tumor membrane leading to tumor cell death. Lipid species share one or more fatty acids in their chemical structures linked to a scaffold characterizing each class, and PCs are one of the main components of the bilayer cell membrane [43]. Saturation degree classifies fatty acids as either saturated (without double bonds) or unsaturated (with one or more double bonds). The presence/absence of double bonds influences PC spatial arrangement and geometry, substantially modifying the biological function of the molecule [69]. In unsaturated PCs, the double bond forces the fatty acid tail to fold, giving it a spatial conformation able to increase membrane fluidity due to the space created by tails [70]. This conformation makes the membrane more elastic and more easily prone to communication with other cells and with microenvironment, thus facilitating vesicle trafficking and the cellular duplication phase [71]. In contrast, a membrane constituted mostly of saturated fatty acid species loses this fluidity because PCs are more compact and thus loses all the advantages described above. Maintaining physiological cell membrane fluidity is required for its correct function and is associated with cell viability [72]. Our lipidomic and metabolomic results indicate an increase in saturated PCs and their downregulation in the whole class, respectively, suggesting a membrane structure evolution from fluid to more rigid with the loss of mechanical properties for cancer cells following LSD1/UTX inhibition. Indeed, our data reveal a stiffening of PCa cells following LSD1/UTX inhibition. By linking biometabolic functions with physical–mechanical properties, we show that PCa mechanics and metabolism can be epigenetically regulated. Membrane functionality is related to lipid composition, and interfering in cancer progression through modulation of cell membrane components may therefore represent a valid strategy to avoid cell communication and trafficking within the tumor microenvironment. In CRPC, inactivation of the tumor suppressor phosphatase and tensin homolog (PTEN) by deletion or mutation [65] is a common genomic aberration and leads to an alteration of the PI3K–AKT axis directly involved in controlling mTORC1 signaling and the related autophagy process, with adverse oncological outcomes [73]. AKT regulates the phosphorylation state of eNOS, whose activity is linked to autophagic block via SQSTM1 accumulation, compromising clearance of misfolded proteins [74]. In addition, the acetylated form of tubulin is required for fusion of autophagosomes with lysosomes, driving autophagolysosome formation and degradation [75]. Downregulation of mTORC1 pathway observed in our proteomic analysis of the CRPC cell lines and in the further analysis of a selected CRPC cell line, C4-2, suggests that LSD1/UTX blocking may also disrupt this alternative energy path. This finding was supported by combining proteomic data with lipidomic analysis, which highlighted a variation in the phosphatidylinositol species regulating upstream the autophagic axis. CRPC progression is driven by an accelerated metabolism and the acquisition of a lipid metabolic phenotype where the radically altered lipogenesis/lipolysis cycle supports proliferation and increases resistance to other forms of treatment, such as radiotherapy. The strong dependence on an enriched lipid compartment was already identified as a possible targetable weakness for this type of tumor [76]. FASN inhibition results in cancer cell death, a reduction in tumor size [19], and sensitization of androgen-dependent and -independent CRPC to radiotherapy [77,78]. Furthermore, FASN inhibitor compounds have reached clinical studies in solid tumor patients, including a phase 2 trial that very recently started in 2021 investigating taxane therapy in combination with FASN inhibition in CRPC patients (NCT04337580). The purpose of the trial is to study this class of pharmacological agents for the treatment of specific tumors via disruption of cancer metabolism. Identifying tumor types susceptible to FASN inhibitors will represent a crucial step toward their possible therapeutic use and the identification of criteria for patient eligibility. In line with this new strategy for PCa therapy, the multi-omics approach we used here shows that simultaneous inhibition of LSD1 and UTX could be beneficial in the treatment of PCa and CRCP phenotypes by directly and indirectly triggering a biological cascade that epigenetically blocks energy production, thus interfering with metabolism, lipid content, and cellular mechanics.
5. Conclusions
Exploiting an epigenetic approach to control cancer mechanics and metabolism and to interrupt the accelerated lipid evolution observed in CRPC may represent an alternative strategy to treat this highly aggressive disease.
Funding
This research was funded by V:ALERE 2019 EPI-MS; V:ALERE CIRCE; Campania Regional Government Technology Platform “Lotta alle Patologie Oncologiche”: iCURE; Campania Regional Government FASE2: IDEAL; MIUR, Proof of Concept POC01_00043; POR Campania FSE 2014–2020 ASSE III. PON RI 2014/2020 “Dottorati Innovativi con Caratterizzazione Industriale”. Nuovi Farmaci e Biomarkers di Risposta e Resistenza Farmacologica nel Cancro del ColonRetto – NABUCCO no. 1682, MISE. TMJE and AM acknowledge support from Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWA-IDG: NWA.1228.192.309).
Authors' contributions
Conceptualization, U.C., C.P.; formal analysis, U.C., C.P., T.M.J.E., M.B.; funding acquisition, R.B., L.A. and A.M.; investigation, U.C., C.P., T.M.J.E., T.D.M., V.C. and N.I.; methodology, E.P., A.T., M.M.N., N.D.G., S.C., T.M.J.E., A.M.; project administration, R.B., L.A., and A.M.; resources, L.A., A.M.; software, M.B., T.D.M., E.N., D.R. and A.M.; writing—original draft, U.C., C.P, T.M.J.E.; writing—review and editing, R.B., L.A, and A.M.
Availability of data and materials
Data are available on request from the authors. The data that support the findings of this study are available from the corresponding author, (RB), upon reasonable request. MS/MS data are available at the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD029249 (interactome) and PXD029525 (whole proteomics).
Ethics approval and consent to participate
Not applicable
Consent to participate
Not applicable
Consent to publish
Each author has approved the submitted version of the manuscript.
Acknowledgments
We thank C. Fisher for English language editing and Gregorio Favale for help with FACS analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101561.
Contributor Information
Ugo Chianese, Email: ugo.chianese@unicampania.it.
Chiara Papulino, Email: chiara.papulino@unicampania.it.
Eugenia Passaro, Email: eugenia.passaro@unicampania.it.
Tom MJ. Evers, Email: t.m.j.evers@lacdr.leidenuniv.nl.
Mehrad Babaei, Email: m.babaei@lacdr.leidenuniv.nl.
Antonella Toraldo, Email: antonella.toraldo@unicampania.it.
Tommaso De Marchi, Email: tommaso.de_marchi@med.lu.se.
Emma Niméus, Email: emma.nimeus@med.lu.se.
Vincenzo Carafa, Email: vincenzo.carafa@unicampania.it.
Maria Maddalena Nicoletti, Email: mariamaddalena.nicoletti@unicampania.it.
Nunzio Del Gaudio, Email: nunzio.delgaudio@unicampania.it.
Nunzia Iaccarino, Email: nunzia.iaccarino@unina.it.
Antonio Randazzo, Email: antonio.randazzo@unina.it.
Dante Rotili, Email: dante.rotili@uniroma1.it.
Antonello Mai, Email: antonello.mai@uniroma1.it.
Salvatore Cappabianca, Email: salvatore.cappabianca@unicampania.it.
Alireza Mashaghi, Email: a.mashaghi.tabari@lacdr.leidenuniv.nl.
Fortunato Ciardiello, Email: fortunato.ciardiello@unicampania.it.
Lucia Altucci, Email: lucia.altucci@unicampania.it.
Rosaria Benedetti, Email: rosaria.benedetti@unicampania.it.
Conflict of interest
The authors declare that they have no competing interests.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Augello M.A., Den R.B., Knudsen K.E. AR function in promoting metastatic prostate cancer. Cancer & Metastasis Reviews. 2014;33(2–3):399–411. doi: 10.1007/s10555-013-9471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawahara T., Inoue S., Kashiwagi E., Chen J., Ide H., Mizushima T., et al. Enzalutamide as an androgen receptor inhibitor prevents urothelial tumorigenesis. Am J Cancer Res. 2017;7(10):2041–2050. [PMC free article] [PubMed] [Google Scholar]
- 4.Dong L., Zieren R.C., Xue W., de Reijke T.M., Pienta K.J. Metastatic prostate cancer remains incurable, why? Asian J Urol. 2019;6(1):26–41. doi: 10.1016/j.ajur.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fizazi K., Scher H.I., Miller K., Basch E., Sternberg C.N., Cella D., et al. Effect of enzalutamide on time to first skeletal-related event, pain, and quality of life in men with castration-resistant prostate cancer: results from the randomised, phase 3 AFFIRM trial. The Lancet Oncology. 2014;15(10):1147–1156. doi: 10.1016/S1470-2045(14)70303-1. [DOI] [PubMed] [Google Scholar]
- 6.Nelson P.S. Molecular states underlying androgen receptor activation: a framework for therapeutics targeting androgen signaling in prostate cancer. Journal of Clinical Oncology. 2012;30(6):644–646. doi: 10.1200/JCO.2011.39.1300. [DOI] [PubMed] [Google Scholar]
- 7.Mah C.Y., Nassar Z.D., Swinnen J.V., Butler L.M. Lipogenic effects of androgen signaling in normal and malignant prostate. Asian J Urol. 2020;7(3):258–270. doi: 10.1016/j.ajur.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Audet-Walsh E., Vernier M., Yee T., Laflamme C., Li S., Chen Y., et al. SREBF1 activity is regulated by an AR/mTOR nuclear Axis in prostate cancer. Molecular Cancer Research. 2018;16(9):1396–1405. doi: 10.1158/1541-7786.MCR-17-0410. [DOI] [PubMed] [Google Scholar]
- 9.Butler L.M., Centenera M.M., Swinnen J.V. Androgen control of lipid metabolism in prostate cancer: novel insights and future applications. Endocrine-Related Cancer. 2016;23(5):R219–R227. doi: 10.1530/ERC-15-0556. [DOI] [PubMed] [Google Scholar]
- 10.Rice M.A., Malhotra S.V., Stoyanova T. Second-generation antiandrogens: from discovery to standard of care in castration resistant prostate cancer. Frontiers in Oncology. 2019;9:801. doi: 10.3389/fonc.2019.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evers T.M.J., Holt L.J., Alberti S., Mashaghi A. Publisher Correction: reciprocal regulation of cellular mechanics and metabolism. Nature Metabolism. 2021;3(6):876–877. doi: 10.1038/s42255-021-00423-6. [DOI] [PubMed] [Google Scholar]
- 12.Cai C., Yuan X., Balk S.P. Androgen receptor epigenetics. Translational Andrology and Urology. 2013;2(3):148–157. doi: 10.3978/j.issn.2223-4683.2013.09.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffey K., Rogerson L., Ryan-Munden C., Alkharaif D., Stockley J., Heer R., et al. The lysine demethylase, KDM4B, is a key molecule in androgen receptor signalling and turnover. Nucleic Acids Research. 2013;41(8):4433–4446. doi: 10.1093/nar/gkt106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morozov V.M., Li Y., Clowers M.M., Ishov A.M. Inhibitor of H3K27 demethylase JMJD3/UTX GSK-J4 is a potential therapeutic option for castration resistant prostate cancer. Oncotarget. 2017;8(37):62131–62142. doi: 10.18632/oncotarget.19100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo J., Attard G., Balk S.P., Bevan C., Burnstein K., Cato L., et al. Role of androgen receptor variants in prostate cancer: report from the 2017 mission androgen receptor variants meeting. European Urology. 2018;73(5):715–723. doi: 10.1016/j.eururo.2017.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan L., Rai G., Roggero C., Zhang Q.J., Wei Q., Ma S.H., et al. KDM4/JMJD2 histone demethylase inhibitors block prostate tumor growth by suppressing the expression of AR and BMYB-regulated genes. Chemistry & Biology. 2015;22(9):1185–1196. doi: 10.1016/j.chembiol.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson S., Fan L., Sahgal N., Qi J., Filipp F.V. The histone demethylase KDM3A regulates the transcriptional program of the androgen receptor in prostate cancer cells. Oncotarget. 2017;8(18):30328–30343. doi: 10.18632/oncotarget.15681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benedetti R., Dell'Aversana C., De Marchi T., Rotili D., Liu N.Q., Novakovic B., et al. Inhibition of histone demethylases LSD1 and UTX regulates ERalpha signaling in breast cancer. Cancers. 2019;11(12) doi: 10.3390/cancers11122027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H.W., Chang Y.F., Chuang H.Y., Tai W.T., Hwang J.J. Targeted therapy with fatty acid synthase inhibitors in a human prostate carcinoma LNCaP/tk-luc-bearing animal model. Prostate Cancer and Prostatic Diseases. 2012;15(3):260–264. doi: 10.1038/pcan.2012.15. [DOI] [PubMed] [Google Scholar]
- 20.Zadra G., Ribeiro C.F., Chetta P., Ho Y., Cacciatore S., Gao X., et al. Inhibition of de novo lipogenesis targets androgen receptor signaling in castration-resistant prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(2):631–640. doi: 10.1073/pnas.1808834116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rotili D., Tomassi S., Conte M., Benedetti R., Tortorici M., Ciossani G., et al. Pan-histone demethylase inhibitors simultaneously targeting Jumonji C and lysine-specific demethylases display high anticancer activities. Journal of Medicinal Chemistry. 2014;57(1):42–55. doi: 10.1021/jm4012802. [DOI] [PubMed] [Google Scholar]
- 22.Carafa V., Nebbioso A., Cuomo F., Rotili D., Cobellis G., Bontempo P., et al. RIP1-HAT1-SIRT complex identification and targeting in treatment and prevention of cancer. Clinical Cancer Research. 2018;24(12):2886–2900. doi: 10.1158/1078-0432.CCR-17-3081. [DOI] [PubMed] [Google Scholar]
- 23.Dell'Aversana C., Giorgio C., D'Amato L., Lania G., Matarese F., Saeed S., et al. miR-194-5p/BCLAF1 deregulation in AML tumorigenesis. Leukemia. 2018;32(2):573. doi: 10.1038/leu.2017.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carafa V., Poziello A., Della Torre L., Giovannelli P., Di Donato M., Safadeh E., et al. Enzymatic and biological characterization of novel sirtuin modulators against cancer. International Journal of Molecular Sciences. 2019;20(22) doi: 10.3390/ijms20225654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubner N.C., Mann M. Extracting gene function from protein-protein interactions using Quantitative BAC InteraCtomics (QUBIC) Methods. 2011;53(4):453–459. doi: 10.1016/j.ymeth.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 26.Hughes C.S., Foehr S., Garfield D.A., Furlong E.E., Steinmetz L.M., Krijgsveld J. Ultrasensitive proteome analysis using paramagnetic bead technology. Molecular Systems Biology. 2014;10:757. doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 28.Lauri I., Savorani F., Iaccarino N., Zizza P., Pavone L.M., Novellino E., et al. Development of an optimized protocol for NMR metabolomics studies of human colon cancer cell lines and first insight from testing of the protocol using DNA G-quadruplex ligands as novel anti-cancer drugs. Metabolites. 2016;6(1) doi: 10.3390/metabo6010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iaccarino N., Varming C., Agerlin Petersen M., Viereck N., Schutz B., Toldam-Andersen T.B., et al. Ancient Danish apple cultivars-A comprehensive metabolite and sensory profiling of apple juices. Metabolites. 2019;9(7) doi: 10.3390/metabo9070139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schepkens C., Dallons M., Dehairs J., Talebi A., Jeandriens J., Drossart L.M., et al. A new classification method of metastatic cancers using a (1)H-NMR-Based approach: a study case of melanoma, breast, and prostate cancer cell lines. Metabolites. 2019;9(11) doi: 10.3390/metabo9110281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savorani F., Tomasi G., Engelsen S.B. icoshift: a versatile tool for the rapid alignment of 1D NMR spectra. Journal of Magnetic Resonance. 2010;202(2):190–202. doi: 10.1016/j.jmr.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Evers T.M.J., Sheikhhassani V., Haks M.C., Storm C., Ottenhoff T.H.M., Mashaghi A. Single-cell analysis reveals chemokine-mediated differential regulation of monocyte mechanics. iScience. 2022;25(1):103555. doi: 10.1016/j.isci.2021.103555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chopra H., Khan Z., Contreras J., Wang H., Sedrak A., Zhu Y. Activation of p53 and destabilization of androgen receptor by combinatorial inhibition of MDM2 and MDMX in prostate cancer cells. Oncotarget. 2018;9(5):6270–6281. doi: 10.18632/oncotarget.23569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yumnam S., Kang M.C., Oh S.H., Kwon H.C., Kim J.C., Jung E.S., et al. Downregulation of dihydrolipoyl dehydrogenase by UVA suppresses melanoma progression via triggering oxidative stress and altering energy metabolism. Free Radical Biology and Medicine. 2021;162:77–87. doi: 10.1016/j.freeradbiomed.2020.11.037. [DOI] [PubMed] [Google Scholar]
- 35.Dang Q., Chen Y.A., Hsieh J.T. The dysfunctional lipids in prostate cancer. American Journal of Clinical and Experimental Urology. 2019;7(4):273–280. [PMC free article] [PubMed] [Google Scholar]
- 36.Buckley D., Duke G., Heuer T.S., O'Farrell M., Wagman A.S., McCulloch W., et al. Fatty acid synthase - modern tumor cell biology insights into a classical oncology target. Pharmacology & Therapeutics. 2017;177:23–31. doi: 10.1016/j.pharmthera.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 37.Cao Z., Xu Y., Guo F., Chen X., Ji J., Xu H., et al. FASN protein overexpression indicates poor biochemical recurrence-free survival in prostate cancer. Disease Markers. 2020;2020:3904947. doi: 10.1155/2020/3904947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yun S.J., Kim S.K., Kim J., Cha E.J., Kim J.S., Kim S.J., et al. Transcriptomic features of primary prostate cancer and their prognostic relevance to castration-resistant prostate cancer. Oncotarget. 2017;8(70):114845–114855. doi: 10.18632/oncotarget.22296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdulla A., Zhang Y., Hsu F.N., Xiaoli A.M., Zhao X., Yang E.S., et al. Regulation of lipogenic gene expression by lysine-specific histone demethylase-1 (LSD1) Journal of Biological Chemistry. 2014;289(43):29937–29947. doi: 10.1074/jbc.M114.573659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ota K., Komuro A., Amano H., Kanai A., Ge K., Ueda T., et al. High fat diet triggers a reduction in body fat mass in female mice deficient for utx demethylase. Scientific Reports. 2019;9(1):10036. doi: 10.1038/s41598-019-46445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang D., He J., Dai Y., Zhou H., Zhang C., Leng Q., et al. Targeting KDM6A suppresses SREBP1c-dependent lipid metabolism and prostate tumorigenesis. Cancer Research. 2021 doi: 10.1158/0008-5472.CAN-21-1825. [DOI] [PubMed] [Google Scholar]
- 42.Butler D.E., Marlein C., Walker H.F., Frame F.M., Mann V.M., Simms M.S., et al. Inhibition of the PI3K/AKT/mTOR pathway activates autophagy and compensatory Ras/Raf/MEK/ERK signalling in prostate cancer. Oncotarget. 2017;8(34):56698–56713. doi: 10.18632/oncotarget.18082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nature Reviews Molecular Cell Biology. 2008;9(2):112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Veen J.N., Kennelly J.P., Wan S., Vance J.E., Vance D.E., Jacobs R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2017;1859(9 Pt B):1558–1572. doi: 10.1016/j.bbamem.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 45.Barfeld S.J., Itkonen H.M., Urbanucci A., Mills I.G. Androgen-regulated metabolism and biosynthesis in prostate cancer. Endocrine-Related Cancer. 2014;21(4):T57–T66. doi: 10.1530/ERC-13-0515. [DOI] [PubMed] [Google Scholar]
- 46.Baci D., Bruno A., Cascini C., Gallazzi M., Mortara L., Sessa F., et al. Acetyl-L-Carnitine downregulates invasion (CXCR4/CXCL12, MMP-9) and angiogenesis (VEGF, CXCL8) pathways in prostate cancer cells: rationale for prevention and interception strategies. Journal of Experimental & Clinical Cancer Research. 2019;38(1):464. doi: 10.1186/s13046-019-1461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voelkel-Johnson C., Norris J.S., White-Gilbertson S. Interdiction of sphingolipid metabolism revisited: focus on prostate cancer. Advances in Cancer Research. 2018;140:265–293. doi: 10.1016/bs.acr.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guha P., Kaptan E., Gade P., Kalvakolanu D.V., Ahmed H. Tunicamycin induced endoplasmic reticulum stress promotes apoptosis of prostate cancer cells by activating mTORC1. Oncotarget. 2017;8(40):68191–68207. doi: 10.18632/oncotarget.19277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levayer R. Solid stress, competition for space and cancer: the opposing roles of mechanical cell competition in tumour initiation and growth. Seminars in Cancer Biology. 2020;63:69–80. doi: 10.1016/j.semcancer.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Izdebska M., Zielinska W., Halas-Wisniewska M., Grzanka A. Involvement of actin and actin-binding proteins in carcinogenesis. Cells. 2020;9(10) doi: 10.3390/cells9102245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aseervatham J. Cytoskeletal remodeling in cancer. Biology. 2020;9(11) doi: 10.3390/biology9110385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cross S.E., Jin Y.S., Rao J., Gimzewski J.K. Nanomechanical analysis of cells from cancer patients. Nature Nanotechnology. 2007;2(12):780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 53.Katira P., Zaman M.H., Bonnecaze R.T. How changes in cell mechanical properties induce cancerous behavior. Physical Review Letters. 2012;108(2):28103. doi: 10.1103/PhysRevLett.108.028103. [DOI] [PubMed] [Google Scholar]
- 54.Nguyen A., Brandt M., Muenker T.M., Betz T. Multi-oscillation microrheology via acoustic force spectroscopy enables frequency-dependent measurements on endothelial cells at high-throughput. Lab on a Chip. 2021;21(10):1929–1947. doi: 10.1039/d0lc01135e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silvani G., Romanov V., Cox C.D., Martinac B. Biomechanical characterization of endothelial cells exposed to shear stress using acoustic force spectroscopy. Frontiers in Bioengineering and Biotechnology. 2021;9:612151. doi: 10.3389/fbioe.2021.612151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lam W.A., Rosenbluth M.J., Fletcher D.A. Chemotherapy exposure increases leukemia cell stiffness. Blood. 2007;109(8):3505–3508. doi: 10.1182/blood-2006-08-043570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lulevich V., Zink T., Chen H.Y., Liu F.T., Liu G.Y. Cell mechanics using atomic force microscopy-based single-cell compression. Langmuir. 2006;22(19):8151–8155. doi: 10.1021/la060561p. [DOI] [PubMed] [Google Scholar]
- 58.Jump D.B., Torres-Gonzalez M., Olson L.K. Soraphen A, an inhibitor of acetyl CoA carboxylase activity, interferes with fatty acid elongation. Biochemical Pharmacology. 2011;81(5):649–660. doi: 10.1016/j.bcp.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yubero M.L., Kosaka P.M., San Paulo A., Malumbres M., Calleja M., Tamayo J. Effects of energy metabolism on the mechanical properties of breast cancer cells. Commun Biol. 2020;3(1):590. doi: 10.1038/s42003-020-01330-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu D., Zhang C., Shen Y., Nephew K.P., Wang Q. Androgen receptor-driven chromatin looping in prostate cancer. Trends in Endocrinology and Metabolism. 2011;22(12):474–480. doi: 10.1016/j.tem.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crea F., Sun L., Mai A., Chiang Y.T., Farrar W.L., Danesi R., et al. The emerging role of histone lysine demethylases in prostate cancer. Molecular Cancer. 2012;11:52. doi: 10.1186/1476-4598-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hua C., Chen J., Li S., Zhou J., Fu J., Sun W., et al. KDM6 demethylases and their roles in human cancers. Frontiers in Oncology. 2021;11:779918. doi: 10.3389/fonc.2021.779918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Regufe da Mota S., Bailey S., Strivens R.A., Hayden A.L., Douglas L.R., Duriez P.J., et al. LSD1 inhibition attenuates androgen receptor V7 splice variant activation in castration resistant prostate cancer models. Cancer Cell International. 2018;18:71. doi: 10.1186/s12935-018-0568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fang Y., Liao G., Yu B. LSD1/KDM1A inhibitors in clinical trials: advances and prospects. Journal of Hematology & Oncology. 2019;12(1):129. doi: 10.1186/s13045-019-0811-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J., Ye C., Chen C., Xiong H., Xie B., Zhou J., et al. Glucose transporter GLUT1 expression and clinical outcome in solid tumors: a systematic review and meta-analysis. Oncotarget. 2017;8(10):16875–16886. doi: 10.18632/oncotarget.15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stoykova G.E., Schlaepfer I.R. Lipid metabolism and endocrine resistance in prostate cancer, and new opportunities for therapy. International Journal of Molecular Sciences. 2019;20(11) doi: 10.3390/ijms20112626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miranda-Goncalves V., Lameirinhas A., Henrique R., Jeronimo C. Metabolism and epigenetic interplay in cancer: regulation and putative therapeutic targets. Frontiers in Genetics. 2018;9:427. doi: 10.3389/fgene.2018.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ravindran Menon D., Hammerlindl H., Torrano J., Schaider H., Fujita M. Epigenetics and metabolism at the crossroads of stress-induced plasticity, stemness and therapeutic resistance in cancer. Theranostics. 2020;10(14):6261–6277. doi: 10.7150/thno.42523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Szlasa W., Zendran I., Zalesinska A., Tarek M., Kulbacka J. Lipid composition of the cancer cell membrane. Journal of Bioenergetics and Biomembranes. 2020;52(5):321–342. doi: 10.1007/s10863-020-09846-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Casares D., Escriba P.V., Rossello C.A. Membrane lipid composition: effect on membrane and organelle structure, function and compartmentalization and therapeutic avenues. International Journal of Molecular Sciences. 2019;20(9) doi: 10.3390/ijms20092167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu X., Daniels G., Lee P., Monaco M.E. Lipid metabolism in prostate cancer. American Journal of Clinical and Experimental Urology. 2014;2(2):111–120. [PMC free article] [PubMed] [Google Scholar]
- 72.Los D.A., Murata N. Membrane fluidity and its roles in the perception of environmental signals. Biochimica et Biophysica Acta. 2004;1666(1–2):142–157. doi: 10.1016/j.bbamem.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 73.Jamaspishvili T., Berman D.M., Ross A.E., Scher H.I., De Marzo A.M., Squire J.A., et al. Clinical implications of PTEN loss in prostate cancer. Nature Reviews Urology. 2018;15(4):222–234. doi: 10.1038/nrurol.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sahani M.H., Itakura E., Mizushima N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy. 2014;10(3):431–441. doi: 10.4161/auto.27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xie R., Nguyen S., McKeehan W.L., Liu L. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biology. 2010;11:89. doi: 10.1186/1471-2121-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schlaepfer I.R., Rider L., Rodrigues L.U., Gijon M.A., Pac C.T., Romero L., et al. Lipid catabolism via CPT1 as a therapeutic target for prostate cancer. Molecular Cancer Therapeutics. 2014;13(10):2361–2371. doi: 10.1158/1535-7163.MCT-14-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chuang H.Y., Lee Y.P., Lin W.C., Lin Y.H., Hwang J.J. Fatty acid inhibition sensitizes androgen-dependent and -independent prostate cancer to radiotherapy via FASN/NF-kappaB pathway. Scientific Reports. 2019;9(1):13284. doi: 10.1038/s41598-019-49486-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kant S., Kumar A., Singh S.M. Tumor growth retardation and chemosensitizing action of fatty acid synthase inhibitor orlistat on T cell lymphoma: implication of reconstituted tumor microenvironment and multidrug resistance phenotype. Biochimica et Biophysica Acta. 2014;1840(1):294–302. doi: 10.1016/j.bbagen.2013.09.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on request from the authors. The data that support the findings of this study are available from the corresponding author, (RB), upon reasonable request. MS/MS data are available at the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD029249 (interactome) and PXD029525 (whole proteomics).
Data will be made available on request.