Abstract
The eut operon of Salmonella typhimurium encodes proteins involved in the cobalamin-dependent degradation of ethanolamine. Previous genetic analysis revealed six eut genes that are needed for aerobic use of ethanolamine; one (eutR), encodes a positive regulator which mediates induction of the operon by vitamin B12 plus ethanolamine. The DNA sequence of the eut operon included 17 genes, suggesting a more complex pathway than that revealed genetically. We have correlated an open reading frame in the sequence with each of the previously identified genes. Nonpolar insertion and deletion mutations made with the Tn10-derived transposable element T-POP showed that at least 10 of the 11 previously undetected eut genes have no Eut phenotype under the conditions tested. Of the dispensable eut genes, five encode apparent homologues of proteins that serve (in other organisms) as shell proteins of the carboxysome. This bacterial organelle, found in photosynthetic and sulfur-oxidizing bacteria, may contribute to CO2 fixation by concentrating CO2 and excluding oxygen. The presence of these homologues in the eut operon of Salmonella suggests that CO2 fixation may be a feature of ethanolamine catabolism in Salmonella.
Under aerobic conditions, Salmonella typhimurium can use ethanolamine as a sole source of carbon, nitrogen, and energy (44, 47). However, this growth depends on exogenous cobalamin, a required cofactor that Salmonella cannot synthesize in the presence of oxygen. Under anaerobic conditions, vitamin B12 is made, but Salmonella cannot use ethanolamine as a carbon or energy source, even with the alternative electron acceptor nitrate or fumarate. Recently this paradox has been resolved by the finding that the anaerobic electron acceptor tetrathionate allows Salmonella to use endogenous B12 to support anaerobic degradation of ethanolamine as a sole source of nitrogen, carbon, and energy (12). Anaerobic use of ethanolamine may be important to Salmonella, since this carbon source is a constituent of an abundant class of lipids which would be provided to anaerobic gut inhabitants as part of the host’s dietary intake.
The initial genetic analysis of the eut operon was done with mutants defective in aerobic degradation of ethanolamine on medium including cobalamin. A large set of mutations were sorted into six complementation groups (eutABCDER) and ordered by deletion mapping (44, 45). More recent genetic tests have identified a seventh complementation group, eutT (54, 67).
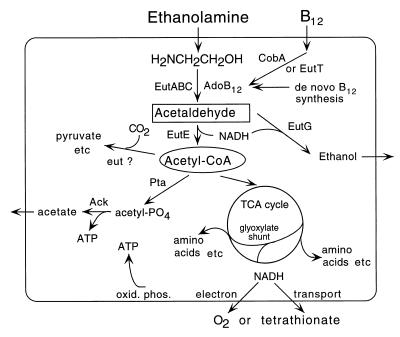
The standard reactions in ethanolamine utilization are diagrammed in Fig. 1, with proposed roles for several Eut proteins. One previously identified gene, eutR, encodes a positive regulatory protein which mediates induction of the operon by ethanolamine plus cobalamin (46, 53). Two genes (eutBC) encode subunits of the cobalamin-dependent ethanolamine ammonia lyase (27, 45), which converts ethanolamine to acetaldehyde and ammonia (13, 50). The eutE gene encodes the second enzyme in the pathway, acetaldehyde dehydrogenase, which forms acetyl-coenzyme A (CoA) (45). As expected, bacteria with mutations in this gene can use ethanolamine as a source of nitrogen but not carbon [a Eut(N+ C−) phenotype]. The EutT enzyme appears to be an adenosyl transferase, converting CNB12 to AdoB12, and the EutA protein appears to protect the lyase (EutBC) from inhibition by CNB12 (54). We propose below that the eutG gene encodes an alcohol dehydrogenase. No function has been assigned to the eutD gene, whose mutants have a Eut(N+ C−) phenotype.
FIG. 1.
Suggested pathway for metabolism of ethanolamine. Gene assignments are based on direct assays (EutBCE and CobA), on mutant phenotypes (EutA, EutT, Ack, Pta, and glyoxylate shunt), or on sequence similarity (EutG). The homologues of carboxysome shell proteins suggest the possibility of CO2 fixation, which has not been demonstrated. In the diagram, the outer boundary is the cell membrane; the role of carboxysomes in this pathway is unknown.
We report here the complete DNA sequence of the eut operon and adjacent regions, including about 7 kb of new sequence and several corrections of previously reported data. Previously sequenced parts of the operon include the eutB and eutC genes (27) and a nonoverlapping 8-kb fragment (60). Surprisingly, the operon includes 17 open reading frames, suggesting that 11 eut genes escaped detection by the initial genetic analysis. Here we correlate the genetic and physical maps of the operon and analyze available information on the function of each of the 17 genes. Using insertions of a new transposon (T-POP) and derived deletion mutations, we provide evidence that at least 10 of the 11 extra genes are not needed for aerobic ethanolamine metabolism. Five of the extra eut genes encode homologues of three families of proteins that serve in other prokaryotes as shell proteins of the carboxysome, an organelle which stimulates CO2 fixation and has been suggested to concentrate CO2 (3, 25, 29, 57). We propose that a similar organelle forms in Salmonella and supports catabolism of ethanolamine by a route that involves CO2 fixation.
MATERIALS AND METHODS
Bacterial strains.
All strains used in this study are derivatives of S. typhimurium; in view of the large number of strains used, strain numbers are listed only in data tables and in the text. Isolation of all zfa insertions (near the eut operon) and all eut mutations with allele numbers below 205 was described previously (44–46). Transposon Tn10dTc is a transposition-defective derivative of transposon Tn10 (68). The T-POP transposon, derived from Tn10dTc, directs tetracycline-inducible promoters into genes adjacent to its insertion site (42). MudA and MudJ elements are transposition-defective derivatives of phage Mu (15, 16). MudP and MudQ (MudP22 elements) have the ends of phage Mu but include a chloramphenicol resistance determinant and a P22 prophage that cannot excise when induced but packages a limited region of the chromosome adjacent to the MudP or -Q insertion site (70). Strains carrying MudP or MudQ insertions near the eut operon were induced in order to obtain template DNA for sequencing the eut operon.
Media, chemicals, and enzymes.
The rich medium was Luria-Bertani broth. The carbon-free minimal medium was NCE (5), and the carbon- and nitrogen-free minimal medium was NCN (43). Ethanolamine hydrochloride (3) at 0.4% was used as a carbon source in the last two media. MacConkey agar base (Difco) was used as a colorimetric indicator of acid production and was prepared according to the manufacturer’s specifications.
When used, antibiotics were present at the following concentrations: ampicillin, 50 μg/ml; tetracycline, 20 μg/ml (selection) or 2 μg/ml (T-POP transcription); chlortetracycline, 10 μg/ml (T-POP transcription); kanamycin, 50 μg/ml; and chloramphenicol, 20 μg/ml. Chlortetracycline was activated by autoclaving it with the medium. The chromogenic β-galactosidase substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside; Diagnostic Chemicals) was used at a final concentration of 0.01%. Cyanocobalamin (CN-B12; Sigma) was used at a final concentration of 200 nM.
Crystalline bovine serum, Ficoll (type 400), and cresol red were from Sigma. Premixed deoxynucleoside triphosphates were from Pharmacia. Isotopically labelled nucleotides (32P and 33P) were from Dupont, New England Nuclear. Hexadecyltrimethylammonium bromide (CTAB) was from Aldrich. Taq polymerase was purchased from Promega, TaqStart antibody was from Clontech, and proteinase K was from Gibco-BRL.
Genetic techniques.
Transduction crosses were mediated by the high-frequency generalized transducing phage P22 HT105/1 int-201 (51). Transductants were freed of phage by streaking them on green indicator plates (17). Cells were cross-streaked with the P22 clear plaque mutant H5 to verify phage sensitivity.
Selecting insertions of T-POP in the eut operon.
The T-POP derivative of transposon Tn10 directs tetracycline-induced promoters out of each end (42). In this cross, the donor (TT18797) carried the T-POP insertion on an Escherichia coli F′, plasmid; the lack of homology prevents recombination between the transduced T-POP region and the recipient chromosome. For some crosses, the recipient (TT17428) carried a standard Tn10 transposase (plasmid pZT380); in other cases, the recipient (TT17437) expressed a mutant form of IS10 transposase (plasmid pNK2881) that allows transposition with relaxed target site specificity (4). Selected tetracycline-resistant clones inherited T-POP by transposition into random sites in the chromosome. A large collection (>10,000) of random-insertion clones were pooled to create the T-POP pool.
Transducing phage prepared on the T-POP pool were used to transduce a recipient that carried a eutR::MudJ insertion; the lacZ gene of this recipient is not expressed, since it lacks the EutR protein required for operon induction. Clones were sought which formed red (Lac+) colonies on MacConkey agar-lactose-tetracycline plates (due to the T-POP promoter) and white colonies without tetracycline (when the T-POP promoter is repressed).
Making deletion mutations by using insertions of T-POP.
Phenotypically Eut− insertion mutants were subjected to selection for aerobic growth on ethanolamine plus vitamin B12. Some surviving clones carried a deletion that removed the inserted material and extended into adjacent regions of the eut operon that are not essential to the Eut+ phenotype. Four different in-frame Eut+ deletions that lie between the promoter and the eutR gene were isolated; each was made from a different parental eut insertion.
Preparation of chromosomal DNA.
Crude template DNA for rapid PCR mapping was prepared by resuspending a 50-μl cell pellet of an overnight culture in Tris-EDTA (TE) buffer, holding it at 95°C for 3 min, spinning out cell debris, and using the supernatant directly. These preparations lost template efficiency with repeated freezing and thawing or storage on ice and were unsatisfactory for sequencing.
Chromosomal DNA preparations for sequencing were prepared as suggested by Knut Jahreis (personal communication). A fresh overnight cell culture (1.5 ml) was centrifuged and resuspended in 567 μl of TE buffer (10 mM Tris, pH 8.3, 1 mM EDTA). Sodium dodecylsulfate (15 μl of a 20% solution) and proteinase K (3 μl of a 20-mg/ml solution) were added, and the suspension was incubated for 1 h at 37°C. NaCl (100 μl of a 5 M solution) was then added with gentle but complete mixing. CTAB was then added (80 μl of a solution of 41 mg of NaCl and 100 mg of CTAB in 1 ml of H2O) with gentle mixing. After 10 min at 65°C, the mixture was extracted with 1 volume of CIA (chloroform-isoamyl alcohol [24:1]). The aqueous phase was saved and drawn repeatedly through a 22-gauge syringe needle to fragment the DNA. The preparation was then extracted twice with phenol-CIA (1:1), and the final aqueous phase was extracted with 1 volume of 1-butanol. DNA was precipitated by addition of 1 volume of isopropanol and was recovered by centrifugation. The pellet was washed once with 70% ethanol, placed under vacuum until nearly (but not completely) dry, resuspended in 100 μl of H2O, and stored at −20°C.
PCR methods. (i) Standard amplification techniques.
All PCRs were done in glass capillaries with an AirCycler thermal cycler (Idaho Technology). The buffers and conditions were as described in protocols provided by the company, with the following modifications: cresol red was used as the indicator dye, and magnesium was used at 1, 2, or 3 mM. Products for sequencing were purified with Wizard PCR purification kits (Promega). The two methods described below were used to amplify unknown sequence adjacent to a single known region.
(ii) Semirandom amplification.
At sufficiently low stringency, a primer will often misprime close enough to its correct binding site that amplification of the intervening DNA will occur with a single primer (P1). The most stringent conditions of magnesium and annealing temperature which still allow one or a small number of misprimed bands to form are determined. These bands are excised and used as templates in a reamplification reaction at high stringency, using P1 with a nested primer oriented in the same direction (P2) at a 1:100 molar ratio of P1 to P2. P1 will continue to initiate at the unknown end, but P2 will dominate priming at the known end, leading to amplification of a fragment differing in size from the original product by the distance between the ends of P1 and P2. This difference is diagnostic of a product derived from the known region. The technique generates template which can be sequenced from either end by using P1 or P2.
(iii) Nested amplification.
The second method uses one primer in known sequence (K primer) and an ambiguous N primer, which is designed to misprime at nearby sites. The amplified region is between the known K primer and all of the sites at which the N primer acts. This method is sensitive to initial template complexity. It works well on DNA extracted from MudP22 heads or on large PCR products.
The four N primers had the sequence ACTTCTCAACAACTCAGGACGAACA(N)10XCAGC, where X is replaced by G, A, T, or C, yielding primers NG, NA, NT, and NC. The reamplification primer (P primer) is identical in sequence to the common portion of the initial oligonucleotide preceding the run of 10 ambiguous bases in the N primers.
Four initial primer extensions were done with NX primers at extremely low stringency (annealing temperature of 40°C). Wizard PCR columns were used to remove the primers and most of the large template DNA, which binds irreversibly to the columns. Extension times were less than 1 min, so most products are short enough to be easily eluted.
The extensions were then used as templates in standard amplification reactions containing a known primer and the shorter P primer, which recognizes the outside end of all NX-primed products. Reamplification with a nested known primer can be used to identify correctly anchored fragments. Optimally, amplification with a nested known primer and the P primer yielded a series of products separated by an average of 256 bases. The unknown ends of all products can be sequenced with the P primer.
Sequencing the eut operon and identifying insertion sites.
Sequencing templates were PCR-amplified genomic regions between genetically mapped Tn10 and Mud insertions or between one such insertion and previously determined eut sequence. The approximate positions of insertions were judged by the size of the fragment; the precise position was determined by sequencing the junctions between the element and adjacent chromosomal sequence. To amplify regions resistant to PCR, MudP22-packaged DNA was sequenced directly; this DNA was obtained from phage particles released after inducing one of the MudP or MudQ lysogens (TT14884, TT15254, or TT15632). MudP and MudQ elements are described above.
Sequencing was done according to the method of Sanger et al (49) with variations described in manganese reagent Sequenase or ThermoSequenase kits (Amersham Life Science) and in protocols for dye-terminator sequencing (Applied Biosystems). The latter was carried out at the University of Utah Health Sciences DNA Sequencing Facility, headed by Margaret Robertson. Primers were synthesized by Robert Schackmann at the University of Utah Health Sciences DNA/Peptide Synthesis Facility.
Nucleotide sequence accession number.
The sequence described here has GenBank accession no. AF093749.
RESULTS
Catabolism of ethanolamine.
A current view of ethanolamine catabolism is diagrammed in Fig. 1. This scheme is consistent with previous genetic analyses and includes some of the gene assignments proposed here. Acetyl-CoA is formed by the sequential activity of the vitamin B12-dependent lyase and the dehydrogenase whose genes were identified genetically (eutBC and eutE). Acetyl-CoA can be converted to acetyl phosphate and excreted as acetate, yielding one molecule of ATP; these reactions (Pta and Ack functions) are required for aerobic growth on ethanolamine (28). Acetyl-CoA can enter the tricarboxylic acid (TCA) cycle and provide both a carbon and an energy source by respiration of oxygen. Under anaerobic conditions, tetrathionate can be used as an alternative electron acceptor, but other alternative acceptors, including nitrate and fumarate, do not support anaerobic growth on ethanolamine (12). The TCA cycle is thought to be essential, since mutants blocked in the glyoxalate shunt fail to use ethanolamine (12). If NADH generated by the TCA cycle exceeds that which can be removed by respiration, acetaldehyde may serve as an electron sink by being reduced to ethanol (Fig. 1). The energy yield from ethanolamine by these pathways might be expected to exceed that provided by acetate, because ethanolamine can enter cells by diffusion and be converted to acetyl-CoA by a dehydratase with no energetic cost. In contrast, acetate must be transported and converted to acetyl-CoA at the cost of at least one ATP.
Since the vitamin B12 cofactor of ethanolamine ammonia lyase is only made anaerobically, we suspect that under natural conditions a major use of ethanolamine may occur in the absence of oxygen. In the absence of any electron acceptor, conversion of ethanolamine to excreted acetate, catalyzed by the Pta and Ack activities, provides a source of energy (ATP) but not of carbon; this use of ethanolamine is detected as a stimulation of anaerobic growth on dilute casamino acids (12). When tetrathionate is provided as the alternative electron acceptor, ethanolamine can serve anaerobically as a nitrogen, carbon, and energy source, using endogenous vitamin B12. Anaerobic growth on ethanolamine (or propanediol) with tetrathionate as an electron acceptor are the only conditions known to us under which vitamin B12 synthesis is required for growth of wild-type cells. We propose that many of the extra Eut enzymes may be involved in CO2 fixation. This fixation may be required because so much carbon is lost as excreted acetate and ethanol (Fig. 1).
Sequence of the eut operon.
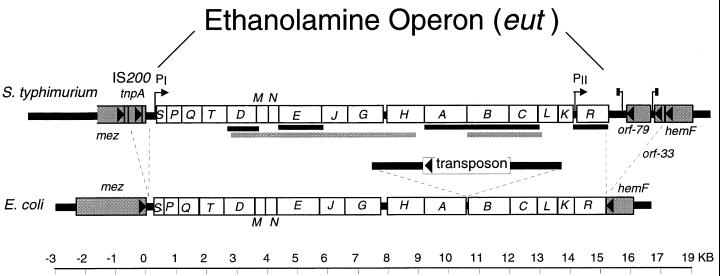
The eut operon sequence was completed and is diagrammed in Fig. 2. The portions determined previously are indicated (27, 60). The operon includes 17 genes. This was surprising, because only six genes were identified genetically (eutD, -E, -A, -B, -C, and -R). Features of the sequence are listed in Table 1, and selected alignments with other genes are given in Table 2. A copy of the transposable element IS200 was found upstream of the eut operon. Nucleotides are numbered with respect to the first base to the right of this IS200 copy (Fig. 2). Sequence downstream of eut structural genes includes a probable transcription terminator and the nearby hemF gene. The sequence of the homologous region from E. coli is indicated for comparison and will be described later.
FIG. 2.
Diagram of the eut operon sequence. The genes underlined in black were discovered by genetic characterization of mutants defective for aerobic use of ethanolamine. The regions underlined in gray were sequenced previously (27, 60). The transposon shown in the E. coli sequence was found in one isolate of E. coli (8) but not in another (69).
TABLE 1.
Features of the eut locus sequence
| Namea | Gene organizationb
|
Codonsc
|
Protein
|
Comment | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | Stop | Sense | Overlap | GC | 3d.GC | CAI | χ2 | Length (aa) | MW | pI | ||
| ′maeB | −1221 | −766 | → | − | 0.55 | 0.60 | 0.292 | 0.817 | >151 | >1,610 | 6.9 | One of two malic enzymes |
| IS200 | −721 | 0 | → | IS200 element; copy V | ||||||||
| tnpA | −578 | −120 | → | − | 0.46 | 0.56 | 0.250 | 0.572 | 152 | 17,958 | 10.1 | IS200 transposase (Tnp) |
| orf′ | 1 | 57 | ← | − | 0.58 | 0.79 | 0.192 | 1.926 | 19 | 1,907 | 7.5 | atsB-like fragment |
| eutS | 361 | 696 | → | − | 0.51 | 0.54 | 0.291 | 1.128 | 111 | 11,673 | 5.7 | Carboxysome structural protein? |
| eutP | 709 | 1188 | → | + | 0.52 | 0.54 | 0.243 | 0.645 | 159 | 17,726 | 5.9 | Unknown function; ATP binding motif |
| eutQ | 1166 | 1855 | → | + | 0.57 | 0.68 | 0.398 | 1.691 | 229 | 24,992 | 4.7 | Unknown function |
| eutT | 1852 | 2655 | → | + | 0.59 | 0.68 | 0.298 | 1.155 | 267 | 30,239 | 6.4 | Cobalamin adenosyl transferase |
| eutD | 2652 | 3668 | → | − | 0.62 | 0.68 | 0.336 | 1.373 | 338 | 36,266 | 7.5 | Resembles substrate domain of Pta |
| eutM | 3709 | 3999 | → | − | 0.60 | 0.67 | 0.411 | 2.002 | 96 | 9,843 | 6.5 | Carboxysome structural protein? |
| eutN | 4099 | 4398 | → | − | 0.57 | 0.65 | 0.315 | 2.094 | 99 | 10,351 | 5.1 | Carboxysome structural protein? |
| eutE | 4410 | 5813 | → | − | 0.60 | 0.69 | 0.354 | 1.360 | 467 | 49,259 | 6.9 | Aldehyde oxidoreductase |
| eutJ | 5824 | 6663 | → | + | 0.61 | 0.72 | 0.342 | 1.105 | 279 | 30,018 | 4.9 | Possible chaperonin |
| eutG | 6653 | 7828 | → | − | 0.57 | 0.71 | 0.320 | 1.429 | 391 | 40,570 | 7.2 | Alcohol dehydrogenase |
| eutH | 7948 | 9195 | → | + | 0.57 | 0.73 | 0.361 | 1.489 | 372 | 39,053 | 5.8 | Membrane protein |
| eutA | 9192 | 10595 | → | − | 0.61 | 0.71 | 0.297 | 0.899 | 467 | 49,527 | 5.0 | Possible chaperonin |
| eutB | 10607 | 11968 | → | + | 0.57 | 0.73 | 0.425 | 1.265 | 453 | 49,449 | 4.7 | ET/NH4d subunit |
| eutC | 11987 | 12883 | → | − | 0.61 | 0.70 | 0.364 | 1.475 | 298 | 32,137 | 5.9 | ET/NH4 subunit |
| eutL | 12893 | 13552 | → | − | 0.60 | 0.64 | 0.320 | 0.841 | 219 | 22,696 | 4.5 | Carboxysome structural protein? |
| eutK | 13565 | 14059 | → | − | 0.60 | 0.61 | 0.289 | 1.092 | 164 | 17,421 | 7.5 | Carboxysome structural protein? |
| PR | 14078 | → | Constitutive internal promoter | |||||||||
| eutR | 14107 | 15159 | → | − | 0.54 | 0.61 | 0.280 | 0.815 | 350 | 40,055 | 7.2 | AraC family positive regulator |
| term | 15677 | → | Probable terminator | |||||||||
| orf79 | 15828 | 16619 | ← | − | 0.53 | 0.54 | 0.297 | 0.382 | 263 | 30,021 | 4.8 | Unknown |
| term | 16673 | ← | Probable terminator | |||||||||
| orf33 | 16751 | 17077 | ← | − | 0.43 | 0.45 | 0.201 | 0.788 | 109 | 12,020 | 8.4 | Unknown |
| hemF | 17098 | 17997 | ← | − | 0.57 | 0.65 | 0.353 | 1.213 | 299 | 34,430 | 6.4 | Coproporphyrinogen oxidase |
| 18000 | 18869 | ← | 0.51 | 0.54 | 0.310 | 0.555 | 289 | 31,659 | 11.0 | N-acetylmuramylalanine amidase | ||
Component of the eut operon. The vertical line indicates the elements included in the eut operon.
The column headed “sense” gives the transcriptional direction, with right-facing arrows indicating counterclockwise on the standard Salmonella map; “overlap” indicates whether or not (+ and −, respectively) the stop codon overlaps the initiation codon of the next gene, implying potential translational coupling or interference (40a). Start and stop codon positions are in nucleotides numbered from the end of IS200.
GC, fractional G+C content of the gene; 3d.GC, G+C content of codon third positions; CAI, codon adaptation index of Sharp and Li (52a); χ2, measure of codon usage bias (55a).
ET/NH4, ethanolamine ammonia lyase.
TABLE 2.
Key alignments of Eut proteins
| Target | Similar protein(s) found | Organism | P(N)a |
|---|---|---|---|
| EutS | PduB (organelle structural?) | Salmonella typhimurium | 0.10 |
| EutP | None | ||
| EutQ | None | ||
| EutT | None | ||
| EutD | Pta (phosphate acetyltransferase) | Paracoccus denitrificans | 2.0e-85 |
| EutM | PduA (organelle structural?) | Salmonella typhimurium | 1.1e-37 |
| CcmK (carboxysome structural) | Synechocystis sp. strain PCC6803 | 7.6e-31 | |
| EutK | Escherichia coli | 5.3e-22 | |
| EutN | CcmL (carboxysome structural) | Synechococcus | 3.3e-19 |
| EutE | SucD (succinate-semialdehyde dehydrogenase) | Clostridium kluyveri | 2.0e-36 |
| EutJ | FtsA (cell division) | Bacillus subtilis | 6.1e-07 |
| DnaK (heat shock chaperonin) | Synechocystis sp. strain PCC6803 | 2.2e-05 | |
| EutG | FucO (lactaldehyde reductase on propanediol oxidoreductase) | Escherichia coli | 6.7e-66 |
| EutH | YxeR (unknown) | Bacillus subtilis | 2.2e-105 |
| MTCY04C12.24c (ABC sulphate transporter?) | Mycobacterium tuberculosis | 0.064 | |
| EutA | FtsA (cell division) | Bacillus subtilis | 0.25 |
| DnaK (heat shock chaperonin) | Thermomicrobium roseum | 0.73 | |
| EutB | EutB (ethanolamine ammonia lyase large subunit) | Rhodococcus erythropolis | 1.5e-140 |
| EutC | EutB (ethanolamine ammonia lyase small subunit) | Rhodococcus erythropolis | 7.6e-26 |
| EutL | PduB (organelle structural?) | Salmonella typhimurium | 1.4e-07 |
| EutK | EutM | Escherichia coli | 4.4e-22 |
| PduA (organelle structural?) | Salmonella typhimurium | 4.0e-21 | |
| CcmK (carboxysome structural) | Synechococcus sp. strain PCC7942 | 2.1e-17 | |
| EutR | OxoS (putative regulatory protein) | Pseudomonas putida | 9.4e-18 |
| ThcR (probable regulatory protein) | Rhodococcus NI86/21 | 5.0e-14 | |
| XylS (transcriptional activator of XylDLEGF operon) | Pseudomonas putida | 1.9e-12 | |
| TmbS (positive regulator of tmb-meta operon) | Pseudomonas putida | 9.4e-12 | |
| HrpB (positive regulation of hypersensitive response) | Burkholderia solanacearum | 7.7e-09 | |
| CbdS (positive regulator of cbd operon) | Pseudomonas sp. strain pBAH1 | 9.2e-09 | |
| HrpXv (positive regulator of hrp cluster) | Xanthomonas campestris | 1.6e-06 | |
| PocR (positive regulator of pdu operon) | Salmonella typhimurium | 0.084 |
This score, given by the BLAST program, is the probability that the observed resemblance between entire sequences could occur by chance. For those alignments with high P(N) scores, the relationship is based on shared motifs discussed in the text.
By investigating the function of each of the 11 extra genes, we hoped to gain a better understanding of ethanolamine metabolism. A series of eut mutations were characterized to demonstrate the mutant phenotype of each gene and to correlate the genetic and physical maps of the region. The mutant phenotypes explain why so many genes were missed in the initial genetic analysis of aerobic Eut− mutants.
Correlating the genetic and physical maps of the eut operon.
The physical locations of many genetically mapped insertions, deletions, and point mutations were determined from the sizes of PCR fragments or by sequencing. Table 3 lists the positions of insertion mutations. Correlation of these sites with the sites of genetically mapped deletion endpoints validates the genetic map (44) and supports the gene assignments listed below.
TABLE 3.
Positions of eut insertion mutations
| Locusa | Genotypeb | Strain | Gene positionc | Operon positiond |
|---|---|---|---|---|
| tktB | zfa-3649::Tn10 | TT13441 | ∼1330 | ∼−5640 |
| tnpA | zfa-3646::Tn10 (A) | TT13438 | 10 | −566 |
| zfa-3645::Tn10 | TT11568 | ∼340 | ∼−240 | |
| tnpA/eutS | zfa-3644::Tn10 | TT11567 | ∼60 | |
| eutS | eut-334::TPOP (A) | TT20356 | 43 | 403 |
| eutP | eut-171::Mu dJ (B) | TT13752 | 21 | 730 |
| eut-272::TPOP (A) | TT19099 | ∼140 | ∼850 | |
| eut-273::TPOP (B) | TT19100 | ∼140 | ∼850 | |
| eut-274::TPOP (A) | TT19101 | ∼240 | ∼950 | |
| eut-275::TPOP (B) | TT19102 | ∼240 | ∼950 | |
| eut-267::TPOP (A) | TT18814 | 259 | 968 | |
| eut-276::TPOP (A) | TT19103 | ∼340 | ∼1050 | |
| eutQ | eut-18::Mu dA (B) | TT10271 | 304 | 1470 |
| eutT | eut-11::Mu dA (B) | TT10647 | 156 | 2008 |
| eut-17::Mu dA (B) | TT10653 | 703 | 2555 | |
| eut-184::Mu dA (A) | TT13764 | 733 | 2585 | |
| eutD | eut-277::TPOP (B) | TT19104 | ∼200 | ∼2850 |
| eut-172::Mu dJ (A) | TT13753 | 409 | 3061 | |
| eutD/eutM | eut-168::Mu dJ (A) | TT13750 | 3701 | |
| eutM | eut-278::TPOP (B) | TT19105 | ∼140 | ∼3850 |
| eut-271::TPOP (A) | TT19098 | ∼190 | ∼3900 | |
| eutN/eutE | eut-6::Mu dA (B) | TT10642 | 4407 | |
| eutE | eut-24::Mu dA (B) | TT10660 | ∼50 | ∼4460 |
| eut-12::Mu dA (B) | TT10648 | ∼100 | ∼4510 | |
| eut-10::Mu dA (B) | TT10646 | ∼450 | ∼4860 | |
| eut-163::Mu dJ | TT13745 | ∼700 | ∼5110 | |
| eut-279::TPOP (A) | TT19106 | ∼940 | ∼5350 | |
| eut-181::Mu dJ (B) | TT13762 | ∼1020 | ∼5430 | |
| eutJ | eut-269::TPOP (A) | TT19096 | ∼280 | ∼6100 |
| eutG | eut-3::Mu dA (B) | TT10639 | ∼480 | ∼7130 |
| eut-178::Mu dJ | TT13759 | ∼590 | ∼7240 | |
| eut-4::Mu dA (B) | TT10640 | ∼780 | ∼7430 | |
| eut-26::Mu dA (B) | TT10662 | ∼960 | ∼7610 | |
| eut-270::TPOP (A) | TT19097 | 976 | 7628 | |
| eut-173::Mu dJ (B) | TT13754 | ∼1040 | ∼7690 | |
| eut-20::Mu dA (B) | TT10656 | ∼1080 | ∼7730 | |
| eutG/eutH | eut-160::Mu dJ (B) | TT13742 | ∼7890 | |
| eut-154::Mu dJ (B) | TT13737 | ∼7890 | ||
| eutH | eut-9::Mu dA (B) | TT10645 | ∼80 | ∼8030 |
| eut-203::Mu dJ (B) | TT13778 | ∼140 | ∼8090 | |
| eut-25::Mu dA (B) | TT10661 | 508 | 8456 | |
| eut-21::Mu dA (B) | TT10657 | ∼680 | ∼8630 | |
| eut-22::Mu dA (B) | TT10658 | ∼680 | ∼8630 | |
| eut-192::Mu dJ (B) | TT13769 | ∼1130 | ∼9080 | |
| eutA | eut-208::Tn10dTc (B) | TT10644 | ∼5 | ∼9200 |
| eut-176::Mu dJ (A) | TT13757 | ∼350 | ∼9540 | |
| eut-1::Mu dA (B) | TT10637 | ∼690 | ∼9880 | |
| eut-183::Mu dJ (B) | TT13763 | ∼690 | ∼9880 | |
| eut-280::TPOP (A) | TT19107 | ∼960 | ∼10150 | |
| eutB | eut-5::Mu dA (B) | TT10641 | ∼10 | ∼10610 |
| eut-8::Mu dA (B) | TT10644 | ∼710 | ∼11310 | |
| eut-281::TPOP (A) | TT19108 | ∼940 | ∼11550 | |
| eut-282::TPOP (A) | TT19109 | ∼1040 | ∼11650 | |
| eutC | eut-2::Mu dA (B) | TT10638 | 48 | 12035 |
| eut-283::TPOP (B) | TT19110 | ∼460 | ∼12450 | |
| eutL | eut-284::TPOP (A) | TT19111 | ∼60 | ∼12950 |
| eut-15::Mu dA (B) | TT10651 | 64 | 12957 | |
| eut-23::Mu dA (B) | TT10659 | 73 | 12966 | |
| eut-177::Mu dA (B) | TT13758 | 108 | 13001 | |
| eut-34::Mu dA (B) | TT10670 | 117 | 13010 | |
| eutK | eut-268::TPOP (A) | TT18828 | ∼340 | ∼13900 |
| eut-285::TPOP (B) | TT19112 | ∼340 | ∼13900 | |
| eut-286::TPOP (A) | TT19113 | ∼390 | ∼13950 | |
| eutR | eut-156::Mu dJ (B) | TT13738 | 28 | 14135 |
| eut-205::Tn10 | TT13893 | 862 | 14969 | |
| eut trailer | eut-38::Mu dA (B) | TT10674 | 15364 | |
| Outside eut operon | zfa-3648::Tn10 | TT13440 | 15930 |
Insertions with two gene designations (X/Y) are in the interval between the indicated genes.
“(B)” after a Mu-derived element indicates that the eut operon was fused to the lac operon of the element; “(A)” indicates the opposite orientation. An “(A)” after a Tn10-derived element indicates that tetracycline-regulated expression of downstream eut genes is from the tetA promoter of the T-POP element; “(B)” implies the opposite orientation. For both Mu- and Tn10-derived elements, no symbol means that the orientation was not determined.
Nucleotide position within the affected gene. ∼, positions were determined by sizing PCR fragments and are approximate; other positions were exactly determined by sequencing.
Distances are from the proximal end of the IS200 insertion at the left end of the operon (Fig. 1).
Deletion mutations (Table 4) were made from insertion mutations in two ways. Four deletion mutants (eutPQTD, eutDM, eutJG, and eutGH) were selected, each as a spontaneous Eut+ derivative of a different eut::T-POP insertion; all four deletions are in frame and should not cause a polar effect on expression of downstream genes. Additional deletions were made by recombination between eut::T-POP insertions in the same orientation; these constructed deletions have a T-POP element at the deletion join point which allows induced expression of genes distal to the deletion (see below). Point mutations initially classified by complementation tests and genetic mapping were later sequenced to provide a cross-reference between the genetic and physical maps (Table 5).
TABLE 4.
Deletions
| Deletion | Nucleotidesa | Gene(s) | Strain | Source |
|---|---|---|---|---|
| Del913 | (−)566 … | All of eut-cysA | TT14526 | Tn10 × Tn10 |
| Del1955 | (−)566–15930 | All of eut | TT20606 | Tn10 × Tn10 |
| Del763 | 730 … | eutS171–cysA | TT11734 | Mu dA × Mu dA |
| Del744 | 1470 … | eutQ18–cysA | TT11715 | Mu dA × Mu dA |
| Del739 | 2008 … | eutT11–cysA | TT11710 | Mu dA × mu dA |
| Del743 | 2555 … | eutT17–cysA | TT11714 | Mu dA × Mu dA |
| Del762 | 2585 … | eutT184–cysA | TT11733 | Mu dA × Mu dA |
| Del734 | 4407 … | eut6–cysA | TT11705 | Mu dA × Mu dA |
| Del752 | 8456 … | eutH25–cysA | TT11723 | Mu dA × Mu dA |
| Del730 | 12035 … | eutC2–cysA | TT11701 | Mu dA × Mu dA |
| Del741 | 12957 … | eutL15–cysA | TT11712 | Mu dA × Mu dA |
| Del750 | 12966 … | eutL23–cysA | TT11721 | Mu dA × Mu dA |
| Del756 | 13001 … | eutL177–cysA | TT11727 | Mu dA × Mu dA |
| Del747 | 13010 … | eutL34–cysA | TT11718 | Mu dA × Mu dA |
| Del754 | 14135 … | eutR156–cysA | TT11725 | Mu dA × Mu dA |
| Del863 | 15364 … | eut-38–cysA | TT13783 | Mu dA × Mu dA |
| eut-333 | 750–2919 | eutP–eutD | TT20581 | In-frame deletion |
| eut-302 | 2692–3994 | eutD–eutM | TT19189 | In-frame deletion |
| eut-300 | 6646–7438 | eutJ–eutG | TT19187 | In-frame deletion |
| eut-301 | 6945–8362 | eutG–eutH | TT19168 | In-frame deletion |
| eut-339 | 403–∼850 | eutS–eutP | TT20586 | T-POP × T-POP |
| eut-340 | 403–∼5350 | eutS–eutE | TT20587 | T-POP × T-POP |
| eut-336 | 403–∼13950 | eutS–eutK | TT20583 | T-POP × T-POP |
| eut-338 | ∼5350–∼13950 | eutE–eutK | TT20585 | T-POP × T-POP |
| eut-337 | ∼6100–∼13950 | eutJ–eutK | TT20584 | T-POP × T-POP |
| eut-335 | ∼10150–∼13950 | eutA–eutK | TT20582 | T-POP × T-POP |
All positions are relative to the base immediately distal to the IS200 insertion (Fig. 2); numbers preceded with (−) are to the left of that reference point. ∼, numbers are approximate positions determined by the sizes of PCR fragments or fine-structure mapping.
TABLE 5.
Point mutations
| Allele | Mutation
|
Strain | Phenotype | |
|---|---|---|---|---|
| Amino acida | Codon | |||
| eutT77 | Q61Am | CAG→TAG | TT11494 | eut(N)− |
| eutT62b | Q62Am | CAG→TAG | TT11479 | eut(N)− |
| eutT86 | P76L,Q77Am | CCACAG→CTATA | TT11503 | eut(N)− |
| eutT67 | Q77Am | CAG→TAG | TT11484 | eut(N)+ |
| eutT10 | W125Op | TGG→TGA | TT11518 | eut(N)+ |
| eutT75 | Q127Oc | CAA→TAA | TT11492 | eut(N)+ |
| eutT78 | Q134Op | TGG→TGA | TT11406 | eut(N)+ |
| eutT74 | Q177Am | CAG→TAG | TT11491 | eut(N)+ |
| eutD64 | Q18Am | CAG→TAG | TT11481 | eut(N)+ |
| eutD53 | Q35Oc | CAA→TAA | TT11470 | eut(N)− |
| eutD10 | Q156Am | CAG→TAG | TT11515 | eut(N)+ |
| eutD12 | R173Op | CGA→TGA | TT11538 | eut(N)+ |
Original amino acid (left) and substitution (right) flank the position of the affected codon in the gene. Nonsense codons are abbreviated as follows: Am, amber (TAG); Op, opal (TGA); Oc, ocher (TAA).
Unlisted mutations eutT88 (TT11505) and eutT90 (TT11507) are recurrences of the same mutation as listed mutation eutT62, i.e., Q184Am.
Use of T-POP insertions to define gene functions.
The transposable element T-POP was derived from transposon Tn10dTc (42). Weak tetracycline-inducible transcripts emerge from both ends of the parent transposon Tn10dTc (63). Stronger regulated outward transcription is seen for the derived T-POP element because internal transcription terminators have been deleted. When no tetracycline is provided, a T-POP insertion has a strong polar effect on expression of distal genes in an operon, allowing detection of insertions that prevent expression of genes required for a Eut+ phenotype. Tetracycline induces expression of downstream genes and, in effect, abolishes the polarity effect of the insertion. In the presence of tetracycline, a T-POP insertion is defective only for the gene in which it inserts. Genes with no mutant phenotype can be identified because their T-POP insertions cause a Eut− phenotype (by a polar effect on distal eut genes) that is corrected by addition of tetracycline. This correction is not seen if the target gene is essential to a Eut+ phenotype.
The eutS, -P, -Q, -T, -D, -M, -J, -G, -H, -L, and -K genes are not essential for aerobic ethanolamine degradation.
Available insertions of T-POP in many genes (eutSPDMJGJK) cause a Eut− phenotype that is corrected by addition of tetracycline (Table 6). In some cases the correction is incomplete, suggesting that the T-POP promoters may not be sufficiently strong to provide a wild-type Eut+ phenotype; this is frequently true for insertions in orientation B, which directs the weaker tetR promoter downstream. Alternatively, the target gene may encode a protein that makes a minor contribution but is not essential to ethanolamine degradation. The phenotypes scored (Table 6) were aerobic use of ethanolamine as a sole carbon and energy source (tested on minimal ethanolamine-vitamin B12 plates), and acid production on MacConkey medium containing ethanolamine and vitamin B12.
TABLE 6.
Phenotypes of eut::T-POP insertions
| Gene | Allelea | Operon locationb | Strain no. | Growth on NCE ethanolamine B12c
|
Color on MacConkey ethanolamine B12d
|
||
|---|---|---|---|---|---|---|---|
| −Tc | +Tc | −Tc | +Tc | ||||
| eutS | eut-334 (A) | 403 | TT20357 | 0 | 3 | 0 | 4 |
| eutP | eut-272 (A) | ∼850 | TT19099 | 0 | 3 | 0 | 4 |
| eut-273 (B) | ∼850 | TT19100 | 0 | 0 | 0 | 0 | |
| eut-274 (A) | ∼950 | TT19101 | 0 | 0 | 0 | 3 | |
| eut-275 (B) | ∼950 | TT19102 | 0 | 1 | 0 | 0 | |
| eut-267 (A) | 968 | TT18814 | 0 | 1 | 0 | 4 | |
| eut-276 (A) | ∼1050 | TT19103 | 0 | 1 | 0 | 4 | |
| eutD | eut-277 (B) | ∼2850 | TT19104 | 0 | 0 | 0 | 2 |
| eutM | eut-278 (B) | ∼3850 | TT19105 | 0 | 0 | 0 | 3 |
| eut-271 (A) | ∼3900 | TT19098 | 0 | 0 | 0 | 4 | |
| eutE | eut-279 (A) | ∼5350 | TT19106 | 0 | 0 | 0 | 0 |
| eutJ | eut-269 (A) | ∼6100 | TT19096 | 0 | 3 | 0 | 3 |
| eutG | eut-270 (A) | 7628 | TT19097 | 0 | 3 | 0 | 3 |
| eutA | eut-280 (A) | ∼10150 | TT19107 | 0e | 0e | 0 | 1e |
| eutB | eut-281 (A) | ∼11550 | TT19108 | 0f | 0f | 0 | 0f |
| eut-282 (A) | ∼11650 | TT19109 | 0f | 0f | 0 | 0f | |
| eutC | eut-283 (B) | ∼12450 | TT19110 | 0f | 0f | 0 | 0f |
| eutL | eut-284 (A) | ∼12950 | TT19111 | 1 | 3 | 2 | 3 |
| eutK | eut-268 (A) | ∼13900 | TT18828 | 0g | 3 | 3 | 4 |
| eut-285 (B) | ∼13900 | TT19112 | 0g | 3 | 3 | 4 | |
| eut-286 (A) | ∼13950 | TT19113 | 0g | 3 | 3 | 4 | |
A, induced transcription is from the tetA promoter of the T-POP element; B, T-POP insertion is in the opposite orientation.
Nucleotide position is distance from the first nucleotide at the left of IS200V. ∼, approximate location determined by PCR; other positions are by direct sequence determination.
Growth is scored on solid minimal medium with ethanolamine as sole carbon source. 0, no growth; 1, slow growth; 2, moderate growth; 3, growth as strong as wild type.
The Eut phenotype can be scored by acid production, which produces a red color on this medium. 0, white; 1, pale pink; 2, pink; 3, red; 4, dark red.
In eutA mutants, the EutBC lyase is inhibited by CNB12 at the concentration used here; thus, eutA mutants show weak growth similar to that of eutBC mutants.
Strains that lack the eutBC (lyase) but still express the eutE gene are scored here as 0; they show extremely weak but reliably scorable growth that is due to a minor (EutE-dependent) secondary pathway of ethanolamine degradation.
The “0” response of eutK insertions without tetracycline may reflect blockage of eutR transcription from a weak internal promoter between the eutL and eutK insertions used here.
In-frame spontaneous deletions and constructed deletions with a T-POP insertion at the join point showed phenotypes that helped determine the importance of eut genes (Table 7). In-frame deletions should have no polarity effect, and the constructed deletions with T-POP at the junction point have a polarity effect that is corrected by addition of tetracycline. Note that strains lacking the lyase (EutBC) protein show very slight growth on ethanolamine as long as they express the eutE gene. This peculiarity reflects a minor secondary route for degradation of ethanolamine that is currently under investigation (see Discussion).
TABLE 7.
Aerobic phenotypes of eut deletions
| Classa | Deletion | Removed | Strain | Growthb
|
|
|---|---|---|---|---|---|
| −Tc | +Tc | ||||
| 1 | eut+ (Tcr) | TT13440 | 3 | ||
| Del1955 | All eut | TT20606 | 0 | ||
| Δeut-300 | eutJ–eutG | TT19187 | 3 | ||
| Δeut-301 | eutG–eutH | TT19188 | 3 | ||
| Δeut-302 | eutD–eutM | TT19189 | 3 | ||
| Δeut0333 | eutP–eutD | TT20582 | 3 | ||
| 2 | eut+ (Tcr) | TT13440 | 3 | 3 | |
| Δeut-338 (T-POP) | ΔeutE–eutK | TT20585 | 0c | 0 | |
| Δeut–335 (T-POP) | ΔeutA–eutK | TT20582 | 0 | 0 | |
| Δeut-336 (T-POP) | eutS–eutK | TT20583 | 0 | 0 | |
| Δeut-337 (T-POP) | eutJ–eutK | TT20584 | 0 | 0 | |
| Δeut-339 (T-POP) | eutS–eutP | TT20586 | 0 | 2 | |
| Δeut-340 (T-POP) | eutS–eutE | TT20587 | 0 | 0 | |
| Δeut-302, Δeut-337 (T-POP) | eutD–eutM, eutJ–eutK | TT20589 | 0 | 0 | |
| Δeut-333 Δeut-337 (T-POP) | eutP–eutD, eutJ–eutK | TT20588 | 0 | 0 | |
Class 1 deletions are in-frame deletions derived from T-POP insertions. They retain no Tn10 material and show no polar effect on downstream eut genes. Class 2 deletions were constructed by recombination between T-POP insertions in different eut genes. They carry a T-POP insertion at the deletion junction point and are strongly polar but express distal eut genes in the presence of tetracycline.
Growth responses are scored as follows: 0, no growth; 1, very weak; 2, intermediate; 3, like wild type. Strains that express EutE but not EutBC (lyase) are listed as 0; they show extremely weak (EutE-dependent) growth that is due to a secondary minor route for conversion of ethanolamine to acetaldehyde (see Discussion). Tc, tetracycline.
Strains that lack the eutBC (lyase) gene but still express the eutE gene are scored here as 0; they show extremely weak but reliably scorable growth that is due to a minor (EutE-dependent) secondary pathway of ethanolamine degradation.
These results confirm the earlier genetic studies showing that the eutABCE and -R genes are needed for the aerobic Eut+ phenotype; all other eut genes tested have no aerobic mutant phenotype. No appropriate insertions or deletions were available for testing the phenotype of a eutN defect, but since mutations in this gene were not detected in the original genetic analysis, we presume that eutN, like the other extra genes, has no aerobic phenotype. The eutD gene was initially identified by mutations with a Eut− phenotype, but multigene deletions that remove the eutD gene are phenotypically Eut+; this suggests that the phenotype of EutD point mutations was due to polar effects on other genes. This will be discussed later.
Homologues of carboxysome proteins.
Five genes (eutS, -M, -N, -L, and K) encode small proteins similar to the shell proteins of the carboxysome, an organelle found in photosynthetic and sulfur-oxidizing bacteria (57). These genes also resemble the pduA and -B genes of in the Salmonella pdu operon, which encode enzymes needed for vitamin B12-dependent degradation of propanediol (10, 11, 18, 47). The eutM and -N genes were identified previously, and their sequence similarity to carboxysome proteins was noted (60); these genes were originally designated cchA and cchB and have been renamed, since it is now clear that they are part of the ethanolamine (eut) operon.
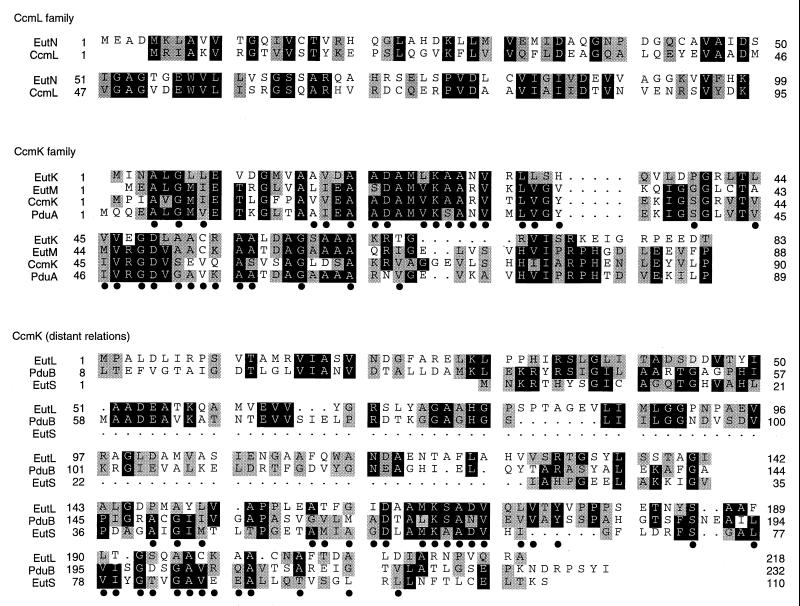
The EutN protein is very similar to the CcmL protein of Synechococcus (Fig. 3). The EutM, EutK, and PduA proteins are clearly homologous to the CcmK protein of Synechococcus. The longer EutL, EutS, and PduB proteins are clearly similar to each other and are less obviously related to the others; their distant similarity to the CcmK family (EutMK and PduA) was inferred from a shared multiple-alignment profile (24). The C-terminal portions of the EutL, EutS, and PduB proteins are most similar to that of the PduA protein. Sequence features that the three proteins share with PduA are indicated in Fig. 3; these were identified with the profile program of the Genetics Computer Group sequence analysis software package.
FIG. 3.
Alignment of carboxysome shell protein homologues. The top panel shows the alignment of the EutN protein with the CcmL protein of Synechococcus. The middle panel aligns the EutK and EutM proteins with the CcmK protein of Synechococcus and the PduA protein of the Salmonella pdu (propanediol utilization) operon. The bottom panel aligns the EutL and EutS proteins with the PduB protein from the Salmonella pdu operon. The EutLS family is most similar to the PduA protein of the EutK, EutN, CcmK class. The sequence features shared by the EutLS-PduB family and the PduA protein are indicated by the black dots below the sequences.
The eutP and eutQ genes.
The predicted EutP and EutQ proteins do not resemble others in current databases. Neither gene has a Eut− mutant phenotype when mutants are tested aerobically in an otherwise-wild-type background.
The EutP protein has an ATP-GTP binding motif of the P-loop class (Prosite motif PS00017). In addition, a second motif found in EutP was shared with a subset of the proteins containing the first motif. The extended motif is not represented in the Prosite database. Proteins sharing the entire motif either were components of ABC transporters, contributed to antibiotic resistance, or supported bacteriocin pumping, suggesting a transport function.
The eutT gene.
Point mutations in the eutT gene were included in the original set of Eut mutations but appeared to owe their phenotype to polarity on distal genes. This is supported by the fact that all are nonsense mutations (Table 5). This interpretation is consistent with the observation that the most upstream eutT point mutations cause a Eut(N− C−) phenotype and distal mutations allow the use of ethanolamine as a nitrogen source [Eut(N+ C−)] (Table 5); this might reflect position-dependent variation in polarity effects. More support for this possibility came from the finding that rho polarity suppressors corrected the Eut(C−) phenotype of eutT nonsense mutations (67) and that in-frame eutT deletions have no Eut phenotype (see above).
Recently, it was found that lack of EutT function causes a Eut− phenotype in a cobA mutant strain (54) which lacks the general cobalamin adenosyl transferase (26, 61, 62). The eutT gene appears to encode a second cobalamin adenosyl transferase, which converts CNB12 to AdoB12 (the lyase cofactor) (54). The pdu operon of Salmonella also appears to encode an adenosyl transferase (pduG) (1, 9, 66) which is very similar in sequence to a demonstrated adenosyl transferase (OrfZ) in the diol dehydratase operon of Citrobacter (22, 52). Surprisingly, the amino acid sequence of the EutT protein shows no similarity to that of the CobA adenosyl transferase (20, 23, 61) or to that of the adenosyl transferase PduG/OrfZ associated with diol dehydratase operons in Salmonella and Citrobacter (9, 52). Thus, it appears that three extremely different enzymes are able to catalyze adenosylation of cobalamin—EutT, CobA, and PduG/OrfZ.
The eutD gene.
Some point mutations in the eutD gene have a Eut(N− C−) phenotype, and others are Eut(N+C−) (44, 45). Point mutations in this gene constituted a clear complementation group in the original genetic tests; they complemented mutants in all other genes and did not cause a measurable decrease in the level of ethanolamine ammonia lyase or acetaldehyde dehydrogenase, encoded by distal genes (45). These point mutations were assigned to an open reading frame by correlating map positions with the physical locations of insertion sites determined by PCR; their location was confirmed by sequencing (Table 5). However the finding that all of the eutD point mutations are nonsense types made it reasonable that their phenotype might have been due to polarity effects.
A eutD::T-POP insertion mutant remains phenotypically Eut(C−) (on minimal medium) even when tetracycline is added to induce downstream genes, but tetracycline restores the ability to produce acid, suggesting partial correction (Table 6). Unfortunately, the only available eutD::T-POP insertion is in the B orientation, which provides only weak induction of distal functions. These results make it difficult to decide whether the phenotypes seen for eutD mutations are due to polarity effects or an inherent lack of EutD function. However, the nonpolar deletion mutations (eutPQTD and eutDM), which remove both the eutD gene and additional adjacent material, are Eut+ aerobically. The simplest interpretation is that eutD point mutations owe their phenotype to polarity effects on multiple downstream genes and a simple EutD defect causes no aerobic phenotype.
The predicted EutD protein sequence is very similar to the C-terminal half of Pta (phosphotransacetylase) and MeaB (NADP-dependent malate oxidoreductase, or malic enzyme). The Pta enzyme catalyzes conversion of acetyl-CoA to acetyl phosphate, and MeaB catalyzes the conversion of malate to pyruvate with release of CO2.
The function of the domain shared by these three proteins is not known, but we suspect that it may provide substrate specificity rather than catalytic activity. Several malate oxidoreductases align only with the N-terminal domains of Mez and Pta and share no similarity with EutD protein (e.g., Streptococcus bovis [GenBank accession no. U35659]); the substrate specificities of these single-domain proteins are reportedly relaxed. Similarly, several malate-decarboxylating enzymes, malic enzymes (which produce pyruvate), and malolactic enzymes (which produce lactate) resemble the N-terminal domain of Pta but lack the C-terminal domain that is homologous to the EutD sequence. Because the two classes of homologues of Pta and Mez enzymes seem to have catalytic domains which are not similar to EutD, we suspect that EutD is not an independent catalyst but may serve as a subunit of a larger complex, perhaps one involved in CO2 fixation.
The eutE and eutG genes (an aldehyde dehydrogenase and an alcohol dehydrogenase).
The eutE gene was initially identified in mutants which could use ethanolamine as a source of nitrogen but not carbon (45). Direct assay revealed that these mutants lack acetaldehyde dehydrogenase, which converts acetaldehyde to acetyl-CoA (44). The gene was initially sequenced by Stojiljkovic et al. (60), who noted that the predicted amino acid sequence of the protein was strikingly similar to that of the aldehyde oxidoreductase domain of the AdhE family of alcohol dehydrogenases-aldehyde oxidoreductases. Sequencing of mutations in the eutE complementation group demonstrated that they affect this open reading frame. The EutE sequence most closely resembled that of NADP-dependent succinate-semialdehyde dehydrogenase of Clostridium kluyveri, which catalyzes formation of succinyl-CoA (59).
The EutG protein appears to be an alcohol dehydrogenase (aldehyde reductase) (60). The best BLAST alignment was with lactaldehyde reductase (1,2-propanediol oxidoreductase) of E. coli. The EutE and EutG sequences aligned in tandem without overlap along the E. coli AdhE sequence, with EutE resembling the C-terminal aldehyde oxidoreductase domain and EutG resembling the N-terminal alcohol dehydrogenase domain. The AdhE protein is known to catalyze reduction of acetyl-CoA to acetaldehyde and further to ethanol. We propose that the EutE and EutG proteins together catalyze the same reactions as AdhE. During growth on ethanolamine, EutE catalyzes formation of acetyl-CoA (as shown previously) and EutG may help to maintain redox balance by reducing some aldehyde to ethanol. Mutants of the eutG gene have no Eut phenotype under the conditions tested, presumably because NADH+ can be recycled via respiratory enzymes or other alcohol dehydrogenases (Fig. 1). In the eut operon, the tandem arrangement of the eutE and eutG genes is interrupted by the eutJ gene.
The eutJ and eutA genes may encode chaperonins.
The eutJ gene had no mutant phenotype. The inferred amino acid sequence of the EutJ protein showed similarity to that of members of the DnaK family of heat shock chaperonins (60). A comparison of the conserved cores of EutJ, EutA, and the E. coli DnaK protein is shown in Fig. 4.
FIG. 4.
Sequence motif that EutJ and EutA proteins share with the chaperonin DnaK. Only EutJ shows significant similarity to DnaK over its entire length; EutJ and EutA are not significantly similar to each other, but they share the motif mentioned above. The central DIGGT motif is part of a nucleotide binding loop in the DnaK protein (55).
Mutations in the eutA gene cause a distinct Eut(N− C−) phenotype under aerobic conditions with CNB12 and defined one of the original eut complementation groups (45). These mutants became phenotypically Eut(N+ C−) when AdoB12 was provided instead of CNB12. A eutA mutant shows normal induction of the operon by CNB12 or AdoB12, demonstrating that it is not defective for cobalamin adenosylation (54). Recent results suggest that EutA protects the lyase from inhibition by CNB12 (54). It is important to remember that eutA mutants retain their Eut(C−) phenotype even when AdoB12 is provided, suggesting that the protein plays some additional role.
The EutA sequence is weakly related to the same group of proteins that show similarity with EutJ (Table 2 and Fig. 4). A motif common to EutJ, EutA, and the DnaK family proteins was the tract DIGGGT. This sequence pattern is part of the nucleotide binding loop in the crystal structure of DnaK protein (55).
The EutJ and EutA proteins may be important in assembling the carboxysome or in refolding lyase. The adenosyl moiety of AdoB12 is cleaved during catalysis (2) and may be subject to occasional loss from the enzyme or destruction by inappropriate reactions (65). Cobalamins without adenosine bind strongly to the enzyme and inhibit its activity in vitro (7). Replacement of damaged AdoB12 may require removal by refolding lyase. The ability to remove inhibitory forms of vitamin B12 from lyase may contribute to the ability of EutA to protect lyase from inhibition. A function of this sort has been reported for the vitamin B12-dependent enzyme propanediol dehydratase (65).
The eutH gene encodes a membrane protein of unknown function.
The eutH gene had no mutant phenotype. The deduced EutH amino acid sequence suggests 11 membrane-spanning segments capped at their ends with short tracts of polar residues. Although a role in ethanolamine transport has been suggested for the EutH protein (60), genetic data indicate that no ethanolamine transport functions are encoded within the operon (45). However, if sufficient ethanolamine enters cells by other means, this gene could encode a transporter with a very slight mutant phenotype. This is true for the propanediol diffusion facilitator PduF, which makes only a minimal contribution to the ability of cells to grow on that carbon source (18, 19). Unlinked mutations previously thought to affect ethanolamine transport have recently been shown to affect vitamin B12 uptake (41, 64). The EutH protein has no resemblance to a reported ethanolamine transporter, EutP, from Rhodococcus (GenBank accession no. U17129). Other possibilities are that the EutH protein increases uptake of vitamin B12 or facilitates efflux of acetaldehyde or acetate produced during ethanolamine catabolism.
The eutBC genes encode ethanolamine ammonia lyase.
The assignment of ethanolamine ammonia lyase to the eutBC genes was initially based on enzyme assays of mutants for these two genes (44). The cloned sequence that complemented these two mutant types provided the first sequence for an ethanolamine ammonia lyase (27). The sequence data reported here contain several corrections of the originally reported sequence. Use of the improved sequence may help identify cobalamin binding motifs (38). The only other described homologue of lyase is from Rhodococcus (GenBank accession no. L24492), whose eutB and eutC homologues are adjacent but do not appear to be part of a larger operon.
The EutR protein is a positive regulatory protein of the AraC family.
The eutR gene was identified in mutants with a Eut− phenotype that were unable to induce the operon in response to the regulatory effectors, ethanolamine and vitamin B12 (45, 46). The EutR protein is encoded within the operon and thus positively controls its own synthesis. This autocatalytic cycle is essential for full operon induction. Coinduction of lyase (EutBC) and EutR may serve to equalize their competition for a small pool of AdoB12, allowing operon control to remain sensitive to cofactor levels over a wide range of vitamin B12 concentrations (53).
The EutR protein is similar in amino acid sequence to a variety of known regulatory proteins in the AraC family. As is typical for this family, the similarity is restricted to the C-terminal helix-turn-helix domain. Since operon transcription is induced only in the presence of both ethanolamine and vitamin B12, the EutR protein may bind both effectors. This has not been demonstrated experimentally and places heavy demands on the EutR protein to recognize two effectors, a DNA binding site and components of the transcription apparatus. It would simplify matters if the requirement for vitamin B12 induction were to help convert ethanolamine to acetaldehyde, which served as sole inducer. However, mutants that lack lyase show normal operon induction by vitamin B12 plus ethanolamine, consistent with direct recognition of the two effectors (46).
The region between the eut operon and the hemF gene.
The region between the eut operon and the hemF gene includes a sequence resembling a Rho-independent transcription terminator located 519 bases from the end of the eutR gene (Fig. 2 and Table 1). A heavily exploited Mud-lac insertion mutant (eut-38::MudA) lies between the last gene in the operon (eutR) and this proposed terminator (Table 3). Strains with this insertion are phenotypically Eut+ but show β-galactosidase induction in response to eut operon regulatory effectors (46). This insertion lies within the transcribed region of the operon but promoter distal to all structural genes.
In Salmonella, two open reading frames (Orf79 and Orf33) are found between the eut operon and the nearby hemF gene. The orientation of the hemF gene, Orf33, and Orf79 is opposite to that of the eut operon. In E. coli, only 5 nucleotides separate the eutR and hemF coding sequences (Fig. 2); each transcript appears to be terminated by a rho-dependent terminator located within the coding sequence of the neighboring gene. A potential transcription terminator for Orf33 was found between Orf33 and Orf79. No significant alignments were found between the translated product of Orf79 or Orf33 and proteins in the database.
The region upstream of the eut operon.
The 1,200 bases upstream of the first gene of the eut operon (eutS) includes one of the six IS200 elements found in the chromosome of S. typhimurium LT2 (32, 35, 48). The element is flanked by pairs of A residues, as seen in other examples of IS200 insertions (31). Upstream of the insertion sequence is the meaB gene, encoding NADP-dependent malic enzyme (malate → pyruvate) (39, 40). The meaB gene and the IS200 element are separated by 42 bases. To the left of meaB are the genes (tktB and talA) for transketolase and transaldolase, enzymes which act in the pentose-phosphate shunt. They form an apparent operon whose orientation is opposite to that of the eut and meaB genes.
The eut operon has a main promoter and a minor internal promoter.
The main regulated promoter (PI) is activated by EutR when both ethanolamine and AdoB12 are present and requires Crp protein as a global regulator (46). A good potential ς70 binding site was found 83 nucleotides before the start of the eutS gene. This lies within a noncoding region well conserved between Salmonella and E. coli. We assume, but have not yet demonstrated, that the EutR regulator binds a site within this region to stimulate transcription. Although the operon is subject to catabolite repression (46), we have found no likely Crp-binding site in the sequence in this region.
The second promoter (PII) lies adjacent to the eutR gene and appears to provide a low constitutive level of EutR regulator sufficient to initiate induction of the main promoter (46, 53). Location of the PII promoter in the Salmonella operon was determined by mRNA runoff extension primed by oligonucleotides complementary to mRNA sequence within the eutR gene (data not shown). This message starts 29 bases before the beginning of eutR in the eutKR interval. The existence of this promoter does not preclude the existence of additional weak promoters further upstream which might contribute to the basal level of EutR protein. No obvious ς70 consensus is associated with PII.
Comparing the eut operons of S. typhimurium and E. coli.
Initial biochemical work on ethanolamine degradation was done for E. coli, with little parallel genetic analysis. Both S. typhimurium and E. coli use the same degradative pathway, and both sets of enzymes are induced by the presence of ethanolamine plus vitamin B12 (6, 7, 33, 34). As diagrammed in Fig. 2, the E. coli operon sequence encodes close homologues of the 17 genes described above for Salmonella (8). The presence of a eut operon in E. coli is surprising in that E. coli does not make the needed vitamin B12 cofactor de novo (36, 37). Furthermore, E. coli cannot reduce tetrathionate, a process that seems essential for anaerobic ethanolamine degradation by Salmonella. For both organisms, the eut operon has a rather high G+C content, suggesting acquisition by horizontal transfer. However, the two sequences differ at only 17% of aligned positions, a degree of conservation expected for genes that have been inherited vertically from the common ancestor of Salmonella and E. coli. A surprising feature of the E. coli eut operon sequenced by Blattner and coworkers (8) is the presence of an insertion element between the eutA and eutB genes which does not damage either of the flanking genes (Fig. 2). This element is not found in other K-12 genomes (69).
DISCUSSION
The complete sequence of the eut operon includes 17 genes, of which only 6 are required for aerobic use of ethanolamine as a carbon or nitrogen source. The functions encoded by the extra genes may be needed for ethanolamine use under unknown conditions, or they may make a slight contribution that escaped our detection. We initially expected that the extra genes would be required for anaerobic growth. This has recently been tested, since it was found that wild-type strains can grow anaerobically on ethanolamine if the electron acceptor tetrathionate is provided (12). However, the extra eut genes tested thus far are also nonessential for anaerobic growth with tetrathionate (28). It seems that the lack of mutant phenotypes for the extra genes is due to an alternative pathway for ethanolamine degradation that can supply some of the Eut functions and prevent the detection of eut mutations in some genes. In the presence of mutations that appear to block this alternative pathway, all genes in the eut operon have a Eut− phenotype aerobically and anaerobically (28).
The five Eut proteins that are similar to carboxysome components suggest that the ethanolamine pathway may involve fixation of CO2. In photosynthetic bacteria (Synechococcus) and in sulfur oxidizers (Thiobacillus), this protein-bounded organelle is thought to concentrate CO2 and exclude O2; this supports activity of RUBISCO, the enzyme directly involved in CO2 fixation (14, 25, 29, 30, 56, 58). In Salmonella, structures resembling carboxysomes have recently been observed by electron microscope following induction of the pdu (9) or eut (21) operon, but fixation of CO2 has not yet been shown to accompany growth on ethanolamine.
ACKNOWLEDGMENTS
This work was supported in part by NIH grant GM34804.
We thank Tom Fazzio and David Sheppard for helpful discussions and sharing unpublished results during the course of this work.
REFERENCES
- 1.Ailion M, Roth J R. Repression of the cob operon of Salmonella typhimurium by adenosylcobalamin is influenced by mutations in the pdu operon. J Bacteriol. 1996;179:6084–6091. doi: 10.1128/jb.179.19.6084-6091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babior B M, Carty T J, Abeles R H. The mechanism of action of ethanolamine ammonia-lyase, a B12-dependent enzyme. J Biol Chem. 1974;249:1689–1695. [PubMed] [Google Scholar]
- 3.Baker S H, Jin S, Aldrich H C, Howard G T, Shively J M. Insertion mutation of the form I cbbL gene encoding ribulose bisphosphate carboxylase/oxygenase (RuBisCO) in Thiobacillus neapolitanus results in expression of form II RuBisCO, loss of carboxysomes, and an increased CO2 requirement for growth. J Bacteriol. 1998;180:4133–4139. doi: 10.1128/jb.180.16.4133-4139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender J, Kleckner N. IS10 transposase mutations that specifically alter target site recognition. EMBO J. 1992;11:741–750. doi: 10.1002/j.1460-2075.1992.tb05107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz D, Hushon J M, Whitfield H J, Jr, Roth J R, Ames B N. Procedure for identifying nonsense mutations. J Bacteriol. 1968;96:215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackwell C M, Scarlett F A, Turner J M. Microbial metabolism of amino alcohols: control of formation and stability of partially purified enthanolamine ammonia-lyase in Escherichia coli. J Gen Microbiol. 1977;98:133–139. doi: 10.1099/00221287-98-1-133. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell C M, Turner J M. Microbial metabolism of amino alcohols: purification and properties of coenzyme B12-dependent ethanolamine ammonia-lyase of Escherichia coli. Biochem J. 1978;175:555–563. doi: 10.1042/bj1750555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner F R, Plunkett G R, Bloch C A, Perna N T, Burland V, Riley M, Collado V J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 9.Bobik, T. Personal communication.
- 10.Bobik T, Xu Y, Jeter R, Otto K, Roth J R. Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase. J Bacteriol. 1997;179:6633–6639. doi: 10.1128/jb.179.21.6633-6639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bobik T A, Ailion M, Roth J R. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J Bacteriol. 1992;174:2253–2266. doi: 10.1128/jb.174.7.2253-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bobik, T. A., J. Tingey, and J. R. Roth. Unpublished data.
- 13.Bradbeer C. The clostridial fermentations of choline and ethanolamine. I. Preparation and properties of cell-free extracts. J Biol Chem. 1965;240:4669–4674. [PubMed] [Google Scholar]
- 14.Cannon G C, English R S, Shively J M. In situ assay of ribulose-1,5-bisphosphate carboxylase/oxygenase in Thiobacillus neapolitanus. J Bacteriol. 1991;173:1565–1568. doi: 10.1128/jb.173.4.1565-1568.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casadaban M J, Cohen S N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castilho B A, Olfson P, Casadaban M J. Plasmid insertion mutagenesis and lac gene fusion with mini-Mu bacteriophage transposons. J Bacteriol. 1984;158:488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan R K, Botstein D, Watanabe T, Ogata Y. Specialized transduction of tetracycline by phage P22 in Salmonella typhimurium. II. Properties of a high frequency transducing lysate. Virology. 1972;50:883–898. doi: 10.1016/0042-6822(72)90442-4. [DOI] [PubMed] [Google Scholar]
- 18.Chen P, Andersson D I, Roth J R. The control region of the pdu/cob regulon in Salmonella typhimurium. J Bacteriol. 1994;176:5474–5482. doi: 10.1128/jb.176.17.5474-5482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, P., and J. R. Roth. Unpublished results.
- 20.Crouzet J, Levy-Schil S, Cameron B, Cauchois L, Rigault S, Rouyez M-C, Blanche F, Debussche L, Thibaut D. Nucleotide sequence and genetic analysis of a 13.1-kilobase-pair Pseudomonas denitrificans DNA fragment containing five cob genes and identification of structural genes encoding cob(I)alamin adenosyltransferase, cobyric acid synthase, and bifunctional cobinamide kinase-cobinamide phosphate guanylyltransferase. J Bacteriol. 1991;173:6074–6087. doi: 10.1128/jb.173.19.6074-6087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Czymmek, K. Unpublished results.
- 22.Daniel, R., and G. Gottschalk. Personal communication.
- 23.Debussche L, Couder M, Thibaut D, Cameron B, Crouzet J, Blanche F. Purification and partial characterization of cob(I)alamin adenosyltransferase from Pseudomonas denitrificans. J Bacteriol. 1991;173:6300–6302. doi: 10.1128/jb.173.19.6300-6302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devereux J. Profile analysis: associating distantly related proteins and finding structural motifs. In: Devereux J, editor. Program manual: sequence analysis software package. Madison, Wis: Genetics Computer Group; 1989. pp. 7.3–7.5. [Google Scholar]
- 25.English R S, Lorbach S C, Qin X, Shively J M. Isolation and characterization of a carboxysome shell gene from Thiobacillus neapolitanus. Mol Microbiol. 1994;12:647–654. doi: 10.1111/j.1365-2958.1994.tb01052.x. [DOI] [PubMed] [Google Scholar]
- 26.Escalante-Semerena J C, Suh S-J, Roth J R. cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J Bacteriol. 1990;172:273–280. doi: 10.1128/jb.172.1.273-280.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faust L P, Connor J A, Roof D M, Hoch J A, Babior B M. Cloning, sequencing and expression of the genes encoding the adenosylcobalamin-dependent ethanolamine ammonia-lyase of Salmonella typhimurium. J Biol Chem. 1990;265:12462–12466. [PubMed] [Google Scholar]
- 28.Fazzio, T. Unpublished results.
- 29.Friedberg D, Jager K M, Kessel M, Silman N J, Bergman B. Rubisco but not Rubisco activase is clustered in the carboxysomes of the cyanobacterium Synechococcus sp. PCC 7942: Mud-induced carboxysomeless mutants. Mol Microbiol. 1993;9:1193–1201. doi: 10.1111/j.1365-2958.1993.tb01248.x. [DOI] [PubMed] [Google Scholar]
- 30.Friedberg D, Kaplan A, Ariel R, Kessel M, Seijffers J. The 5′-flanking region of the gene encoding the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase is crucial for growth of the cyanobacterium Synechococcus sp. strain PCC 7942 at the level of CO2 in air. J Bacteriol. 1989;171:6069–6076. doi: 10.1128/jb.171.11.6069-6076.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haack, K., and J. Roth. Unpublished.
- 32.Haack K, Roth J R. Recombination between chromosomal IS200 elements supports frequent duplication formation in Salmonella typhimurium. Genetics. 1995;141:1245–1252. doi: 10.1093/genetics/141.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones P W, Turner J M. Interrelationships between the enzymes of ethanolamine metabolism in Escherichia coli. J Gen Microbiol. 1984;130:299–308. doi: 10.1099/00221287-130-2-299. [DOI] [PubMed] [Google Scholar]
- 34.Jones P W, Turner J M. A model for common control of enzymes of ethanolamine metabolism in Escherichia coli. J Gen Microbiol. 1984;130:849–860. doi: 10.1099/00221287-130-4-849. [DOI] [PubMed] [Google Scholar]
- 35.Lam S, Roth J R. Genetic mapping of insertion sequence IS200 copies in Salmonella typhimurium strain LT2. Genetics. 1983;105:801–812. doi: 10.1093/genetics/105.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence J G, Roth J R. The cobalamin (coenzyme B12) biosynthetic genes of Escherichia coli. J Bacteriol. 1995;177:6371–6380. doi: 10.1128/jb.177.22.6371-6380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence J G, Roth J R. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics. 1995;142:11–24. doi: 10.1093/genetics/142.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludwig M, Matthews R. Structure-based perspectives on B12-dependent enzymes. Annu Rev Biochem. 1997;66:269–313. doi: 10.1146/annurev.biochem.66.1.269. [DOI] [PubMed] [Google Scholar]
- 39.Murai T, Tokushige M, Nagai J, Katsuki H. Physiological functions of NAD- and NADP-linked malic enzymes in Escherichia coli. Biochem Biophys Res Commun. 1971;43:875–881. doi: 10.1016/0006-291x(71)90698-x. [DOI] [PubMed] [Google Scholar]
- 40.Murai T, Tokushige M, Nagai J, Katsuki H. Studies on regulatory functions of malic enzymes. I. Metabolic functions of NAD- and NADP-linked malic enzymes in Escherichia coli. J Biochem (Tokyo) 1972;71:1015–1028. doi: 10.1093/oxfordjournals.jbchem.a129850. [DOI] [PubMed] [Google Scholar]
- 40a.Oppenheim D S, Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980;95:785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Toole G A, Escalante-Semerena J C. Identification and initial characterization of the eutF locus of Salmonella typhimurium. J Bacteriol. 1991;173:5168–5172. doi: 10.1128/jb.173.16.5168-5172.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rappleye C A, Roth J R. A Tn10 derivative (“T-POP”) for isolation of insertions A Tn10 derivative (“T-POP”) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J Bacteriol. 1997;179:5827–5834. doi: 10.1128/jb.179.18.5827-5834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ratzkin B, Roth J. Cluster of genes controlling proline degradation in Salmonella typhimurium. J Bacteriol. 1978;133:744–754. doi: 10.1128/jb.133.2.744-754.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roof D M, Roth J R. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1988;170:3855–3863. doi: 10.1128/jb.170.9.3855-3863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roof D M, Roth J R. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1989;171:3316–3323. doi: 10.1128/jb.171.6.3316-3323.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roof D M, Roth J R. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J Bacteriol. 1992;174:6634–6643. doi: 10.1128/jb.174.20.6634-6643.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roth J R, Lawrence J G, Bobik T A. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 48.Sanderson K E, Sciore P, Liu S, Hessel A. Location of IS200 on the genomic map of Salmonella typhimurium LT2. J Bacteriol. 1993;175:7624–7628. doi: 10.1128/jb.175.23.7624-7628.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scarlett F A, Turner J M. Microbial metabolism of amino alcohols. Ethanolamine catabolism mediated by coenzyme B12-dependent ethanolamine ammonia-lyase in Escherichia coli and Klebsiella aerogenes. J Gen Microbiol. 1976;95:173–176. doi: 10.1099/00221287-95-1-173. [DOI] [PubMed] [Google Scholar]
- 51.Schmieger H. A method for detection of phage mutants with altered transducing ability. Mol Gen Genet. 1971;110:378–381. doi: 10.1007/BF00438281. [DOI] [PubMed] [Google Scholar]
- 52.Seyfried M, Daniel R, Gottschalk G. Cloning, sequencing, and overexpression of the genes encoding coenzyme B12-dependent glycerol dehydratase of Citrobacter freundii. J Bacteriol. 1996;178:5793–5796. doi: 10.1128/jb.178.19.5793-5796.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52a.Sharp P M, Li W H. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheppard D E, Roth J R. A rationale for autoinduction of a transcriptional activator: ethanolamine ammonia-lyase (EutBC) and the operon activator (EutR) compete for adenosyl-cobalamin in Salmonella typhimurium. J Bacteriol. 1994;176:1287–1296. doi: 10.1128/jb.176.5.1287-1296.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheppard, D. E., and J. R. Roth. Vitamin B12 adenosylation functions in the eut operon of Salmonella typhimurium. Submitted for publication.
- 55.Sheppard, P. Personal communication.
- 55a.Shields D C, Sharp P M, Higgins D G, Wright F. “Silent” sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol Biol Evol. 1998;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- 56.Shively J M. Inclusion bodies of prokaryotes. Annu Rev Microbiol. 1974;28:167–187. doi: 10.1146/annurev.mi.28.100174.001123. [DOI] [PubMed] [Google Scholar]
- 57.Shively J M, Ball F, Brown D H, Saunders R E. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- 58.Shively J M, Bock E, Westphal K, Cannon G C. Icosahedral inclusions (carboxysomes) of Nitrobacter agilis. J Bacteriol. 1977;132:673–675. doi: 10.1128/jb.132.2.673-675.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sohling B, Gottschalk G. Molecular analysis of the anaerobic succinate degradation pathway in Clostridium kluyveri. J Bacteriol. 1996;178:871–880. doi: 10.1128/jb.178.3.871-880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stojiljkovic I, Bäumler A J, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suh S-J, Escalante-Semerena J C. Cloning, sequencing and overexpression of cobA, which encodes ATP:corrinoid adenosyltransferase in Salmonella typhimurium. Gene. 1993;129:93–97. doi: 10.1016/0378-1119(93)90701-4. [DOI] [PubMed] [Google Scholar]
- 62.Suh S-J, Escalante-Semerena J C. Purification and initial characterization of the ATP:corrinoid adenosyltransferase encoded by the cobA gene of Salmonella typhimurium. J Bacteriol. 1995;177:921–925. doi: 10.1128/jb.177.4.921-925.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takiff H E, Baker T, Copeland T, Chen S, Court D L. Locating essential Escherichia coli genes by using mini-Tn10 transposons: the pdxJ operon. J Bacteriol. 1992;174:1544–1553. doi: 10.1128/jb.174.5.1544-1553.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas M G, O’Toole G A, Escalante-Semerena J C. Molecular characterization of eutF mutants of Salmonella typhimurium LT2 identifies eutF lesions as partial-loss-of-function tonB alleles. J Bacteriol. 1999;181:368–374. doi: 10.1128/jb.181.2.368-374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toraya T, Mori K. A reactivating factor for coenzyme B12-dependent diol dehydratase. J Biol Chem. 1999;274:3372–3377. doi: 10.1074/jbc.274.6.3372. [DOI] [PubMed] [Google Scholar]
- 66.Walter D, Ailion M, Roth J R. Genetic characterization of the pdu operon: use of 1,2 propanediol in Salmonella typhimurium. J Bacteriol. 1996;179:1013–1022. doi: 10.1128/jb.179.4.1013-1022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Warnick, L. Unpublished results.
- 68.Way J C, Davis M A, Morisato D, Roberts D E, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 69.Yamamoto Y, Aiba H, Baba T, Hayashi K, Inada T, Isono K, Itoh T, Kimura S, Kitagawa M, Makino K, Miki T, Mitsuhashi N, Mizobuchi K, Mori H, Nakade S, Nakamura Y, Nashimoto H, Oshima T, Oyama S, Saito N, Sampei G, Satoh Y, Sivasundaram S, Tagami H, Takahashi H, Takeda J, Takemoto K, Uehara K, Wada C, Yamagata S, Horiuchi T. Construction of a contiguous 874 kb sequence of the Escherichia coli K-12 genome corresponding to 50.0-68.8 min region on the linkage map and analysis of its sequence features. DNA Res. 1997;4:91–113. doi: 10.1093/dnares/4.2.91. [DOI] [PubMed] [Google Scholar]
- 70.Youdarian P, Sugiono P, Brewer K L, Higgins N P, Elliot T. Packaging specific segments of the Salmonella chromosome with locked-in Mud-P22 prophages. Genetics. 1988;118:581–592. doi: 10.1093/genetics/118.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]