Abstract
Corynebacterium diphtheriae, the causative agent of diphtheria, utilizes various host compounds to acquire iron. The C. diphtheriae hmuO gene encodes a heme oxygenase that is involved in the utilization of heme and hemoglobin as iron sources. Transcription of the hmuO gene in C. diphtheriae is controlled under a dual regulatory mechanism in which the diphtheria toxin repressor protein (DtxR) and iron repress expression while either heme or hemoglobin is needed to activate transcription. In this study, two clones isolated from a C. diphtheriae chromosomal library were shown to activate transcription from the hmuO promoter in Escherichia coli. Sequence analysis revealed that these activator clones each carried distinct genes whose products had significant homology to response regulators of two-component signal transduction systems. Located upstream from each of these response regulator homologs are partial open reading frames that are predicted to encode the C-terminal portions of sensor kinases. The full-length sensor kinase gene for each of these systems was cloned from the C. diphtheriae chromosome, and constructs each carrying one complete sensor kinase gene and its cognate response regulator were constructed. One of these constructs, pTSB20, which carried the response regulator (chrA) and its cognate sensor kinase (chrS), was shown to strongly activate transcription from the hmuO promoter in a heme-dependent manner in E. coli. A mutation in chrA (chrAD50N), which changed a conserved aspartic acid residue at position 50, the presumed site of phosphorylation by ChrS, to an asparagine, abolished heme-dependent activation. These findings suggest that the sensor kinase ChrS is involved in the detection of heme and the transduction of this signal, via a phosphotransfer mechanism, to the response regulator ChrA, which then activates transcription of the hmuO promoter. This is the first report of a bacterial two-component signal transduction system that controls gene expression through a heme-responsive mechanism.
Corynebacterium diphtheriae is a gram-positive, nonsporulating bacterium that is the causative agent of diphtheria. The primary virulence determinant in C. diphtheriae is the diphtheria toxin (DT), a 58,000-Da secreted protein which has been extensively studied (31). The tox gene, which encodes DT, is regulated at the transcriptional level by the diphtheria toxin repressor protein (DtxR) and iron (2, 38). DtxR, which is functionally similar to the Escherichia coli ferric uptake repressor protein (Fur) (13), is a global iron-dependent repressor that regulates the expression of at least eight genes in C. diphtheriae (2, 23, 36, 38, 40, 41, 45). The importance of iron in the regulation of bacterial virulence determinants has been well established, and the ability to acquire sufficient iron during infection has been shown to be important for a number of bacterial pathogens to be fully virulent (10, 24, 49).
Systems involved in the acquisition of iron by bacteria include high-affinity siderophore transport systems (3) and siderophore-independent mechanisms in which bacterial pathogens utilize iron from various host sources, such as transferrin, lactoferrin, heme, or hemoglobin (22, 26). The molecular mechanism involved in the transport of heme and its subsequent utilization as an iron source has been examined in several gram-negative pathogens (15, 18, 22, 28, 30, 43, 44, 46, 51). These systems include a heme-specific outer membrane receptor, which is required for the uptake of heme into the periplasm, and an ATP-binding cassette transporter complex that is involved in the transport of heme through the cytoplasmic membrane. It was proposed that these bacteria contain a heme oxygenase-like enzyme that functions in the removal of the heme-bound iron (22, 44). However, proteins with a heme-degrading activity have not been identified in any of these gram-negative species, and the mechanism involved in the extraction of iron from heme remains to be determined.
In C. diphtheriae, the ability to utilize iron from transferrin was shown to be siderophore dependent, while the use of iron from heme and hemoglobin was independent of the siderophore uptake system (36). Mutants of C. diphtheriae and Corynebacterium ulcerans that were unable to utilize heme and hemoglobin as iron sources have been isolated and characterized (36). Clones carrying the C. diphtheriae hmuO gene were shown to complement several of the Corynebacterium heme utilization mutants. The product of the hmuO gene has significant amino acid homology to eukaryotic heme oxygenases. Heme oxygenases, which had not been previously identified in bacteria but are well known in eukaryotic systems, are involved in the oxidative degradation of heme through the cleavage of the heme porphyrin ring and the subsequent production of CO, iron, and biliverdin (25). The HmuO protein from C. diphtheriae was purified and shown to have an enzymatic activity that is similar to that observed for eukaryotic heme oxygenases (50). It is proposed that the role of HmuO in the utilization of heme as an iron source in C. diphtheriae is in the degradation of heme and the subsequent release of the heme-bound iron. It is believed that Corynebacterium mutants deficient in HmuO activity are unable to extract the iron from heme and, therefore, defective in their ability to use heme as an iron source. Bacterial heme oxygenases have recently been identified in species of Cyanobacterium (5).
In gram-negative bacteria, most of the systems involved in the transport and utilization of heme-bound iron are repressed in high-iron environments; this repression is mediated through the Fur protein (22, 30, 51). In pathogenic species of Haemophilus, the expression of the transferrin and hemoglobin receptors is repressed by heme (8, 19, 29). The mechanism involved in this regulation has not been determined. Expression studies with the C. diphtheriae hmuO gene revealed that transcription from the hmuO promoter was under a dual regulatory mechanism, which involved repression by DtxR and iron and activation by heme (37). DNase I footprinting experiments showed that purified DtxR, in the presence of a divalent metal, bound to an approximately 30-bp region that overlapped the hmuO promoter. Expression of the hmuO promoter from a promoter-probe plasmid in C. diphtheriae revealed that only low levels of transcription were observed unless a heme source, either heme or hemoglobin, was added to the growth medium. Northern blot analysis and primer extension studies provided additional evidence that transcription of the hmuO gene was activated by heme (37). Genes that are activated by heme or other heme-containing compounds have not been previously reported for bacteria; however, several genes in eukaryotic systems, including those encoding certain heme oxygenases, are regulated at the transcriptional level by heme (52).
In this study, the mechanism involved in heme activation of the hmuO promoter was investigated. Two independent clones from a C. diphtheriae library were shown to activate expression of an hmuO promoter-lacZ fusion construct in E. coli. The recombinant plasmids were shown to encode the genes designated chrA and cstA, whose predicted products are homologous to response regulators of two-component signal transduction systems. Immediately upstream from chrA and cstA are open reading frames that are predicted to encode the cognate sensor kinase genes, which have been designated chrS and cstS, respectively. A construct carrying the entire coding region for chrS and its cognate response regulator chrA was shown to activate expression of the hmuO promoter in E. coli in the presence of heme. This is the first report of a two-component signal transduction system in which transcriptional activation is mediated through heme.
MATERIALS AND METHODS
Bacterial strains and media.
E. coli DH5α (Bethesda Research Laboratories, Gaithersburg, Md.) was used throughout this study in the analysis of the hmuO promoter-lacZ reporter fusion constructs (PhmuO-lac) and for routine plasmid isolation. C. diphtheriae C7(−) (17) was originally obtained from the strain collection of Randall K. Holmes. Luria-Bertani (LB) medium (27) was used for culturing of E. coli, while heart infusion broth (Difco, Detroit, Mich.) containing 0.2% Tween 80 (HIBTW) was used for growth of C. diphtheriae C7(−). Permanent stocks of bacterial strains were maintained in 20% glycerol at −70°C. When needed, antibiotics were added to LB medium for E. coli as follows: 10 μg of tetracycline/ml, 34 μg of chloramphenicol/ml, and 100 μg of ampicillin/ml. Chloramphenicol at 2 μg/ml was added to HIBTW for growth of C. diphtheriae strains which harbor plasmids. LB medium and HIBTW were made low iron by the addition of ethylenediamine di(o-hydroxyphenylacetic acid) (EDDA) which was deferrated by the method of Rogers (33). EDDA was added to HIBTW-containing media at 50 μg/ml and to LB media at 2.5 μg/ml. Hemin (bovine) was added to C. diphtheriae cultures at 25 μg/ml and to E. coli cultures at 100 μg/ml, and hemoglobin (human) was added to cultures of both types at 10 μM. Isopropyl-β-d-thiogalactopyranoside (IPTG) (Bethesda Research Laboratories) was used at 0.5 mM. Antibiotics, EDDA, Tween 80, hemin, and hemoglobin were purchased from Sigma Chemical Co. (St. Louis, Mo.).
Plasmid construction and DNA manipulation.
The promoter probe vector pCM502 (Cmr) (37), which contains a promoterless lacZ gene, was used for the construction of the six PhmuO-lac fusion plasmids (see Fig. 1). The DNA inserts present in the six PhmuO-lac fusion plasmids were generated by using PCR and seven different oligonucleotide primers. Six of the primers, designated PO-1, PO-2, PO-3, PO-4, PO-5, and PO-6, are 32 nucleotides (nt) in length: the 17-nt sequences at their 3′ ends are complementary to unique sequences upstream of the hmuO promoter (sense strand), and the 15-nt sequences at the 5′ ends contain a SalI site. The primer on the antisense strand, designated PO-Bam, was used in PCRs with each of the six primers described above to generate the inserts in the various PhmuO-lac fusions. The primer PO-Bam contains sequences at its 3′ region that are complementary to a sequence 240 bp downstream from the hmuO promoter, and it contains a unique BamHI site in a 5′ tail region. The six PCR products each have a unique SalI site in the region upstream of the hmuO promoter and a unique BamHI site in the downstream sequence. The six fragments were digested with BamHI and SalI and then ligated into the corresponding sites in pCM502 to generate the PhmuO-lac fusions for which maps are shown in Fig. 1. All of the fragments terminated at different locations upstream of the hmuO promoter but shared a common downstream terminus. The DNA sequences of the inserts in the six promoter fusion plasmids were determined, and it was confirmed that no sequence changes had occurred during construction of these plasmids.
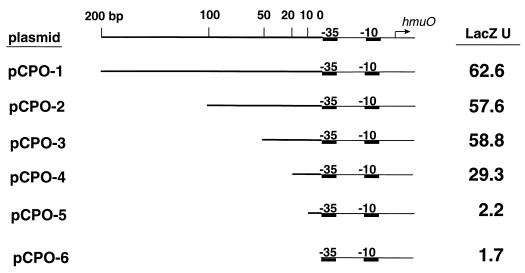
FIG. 1.
Linear maps of the hmuO promoter region present on the six promoter-lac fusions. The hmuO promoter is indicated by the −10 and −35 elements. The top line indicates the distance from the −35 sequence in base pairs. The arrow indicates the start site of transcription of the hmuO gene (transcription begins at an A residue 40 bp downstream from the 5′ end of the −35 sequence [37]). LacZ activity (LacZ U) for each fusion was determined in C. diphtheriae C7(−) grown in low-iron medium in the presence of 25 μg of hemin/ml. LacZ units were determined as previously described (39). Values are means of three independent experiments, and standard deviations did not vary by greater than 15% from the mean.
Plasmids pWKS30 and pWSK29, which carry ampicillin resistance determinants, are low-copy-number plasmids that contain the pSC101 origin of replication (47) and were used in the construction of the plasmids pTSB20, pTSB50, and pTSB20-50. The plasmid pTSB20 was constructed in a two-step process, as follows: the 1.5-kb BamHI-HindIII fragment was excised from pTSB2.B (Fig. 2) and ligated into the BamHI-HindIII site of pWKS30 to generate plasmid pW2-1.5. In the second step, the 1.5-kb HindIII fragment in pKSH51 (identified from the colony hybridization; see below) was ligated into the HindIII site of pW2-1.5 to create pTSB20. The plasmid pTSB50 was also generated in a two-step process as follows: the 2.4-kb PstI-EcoRI insert in pTSB5.R (Fig. 3) was ligated into the PstI-EcoRI site of pWSK29 to produce plasmid pW5-2.4. In the second step, a 1.2-kb PstI fragment in pR5-38 (obtained from the colony hybridization; see below) was ligated to the PstI site in pW5-2.4 to create pTSB50. Plasmid pTSB20-50 was constructed by ligating the 3.5-kb insert of pTSB20 (present on a PvuII fragment) into a HindIII site of pTSB50. The HindIII site, which is present in vector sequences, was made blunt prior to ligation. All of the genes present on pTSB20, pTSB50, and pTSB20-50 are oriented such that they are predicted to be under transcriptional control of the lac promoter on the vector.
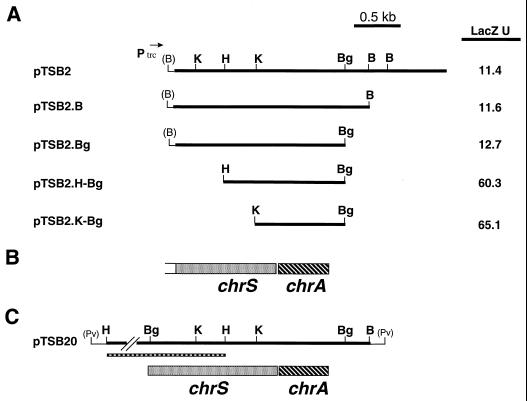
FIG. 2.
(A) Restriction maps of plasmid pTSB2 and various subclones. The direction of transcription from the plasmid-encoded trc promoter is indicated by the arrow. LacZ assays were done with E. coli DH5α/pCPO-1 that carried the various activator clones. Bacteria were grown in LB medium in the presence of 0.5 mM IPTG, and LacZ units were determined by the method of Miller (27). Values are means of three independent experiments, and standard deviations did not vary by greater than 15% from the mean. (B) Genetic map of the chrS and chrA genes present on plasmid pTSB2. The genetic map is aligned with the restriction maps shown in panel A. (C) Restriction and genetic maps of plasmid pTSB20. The thin boxed region below the restriction map indicates the location of the 1.5-kb HindIII fragment that is present in plasmid pKSH51 and contains the 5′ portion of the chrS gene. Only a portion of the HindIII fragment is shown. The restriction and genetic maps are aligned with each other and with the maps shown in panels A and B. Restriction sites are as follows: B, BamHI; Bg, BglII; H, HindIII; K, KpnI; Pv, PvuII. Sites shown in parentheses indicate restriction sites present in vector sequences.
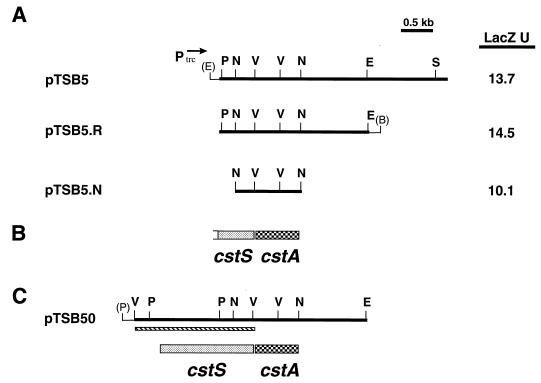
FIG. 3.
(A) Restriction maps of plasmid pTSB5 and various subclones. Direction of transcription from the plasmid encoded trc promoter is indicated by arrow. LacZ assays were done with E. coli DH5α/pCPO-1 that carried the various activator clones. Bacteria were grown in LB medium in the presence of 0.5 mM IPTG, and LacZ units (U) were determined by the method of Miller (27). Values are means of three independent experiments, and standard deviations did not vary by greater than 15% from the mean. (B) Genetic map of the cstS and cstA genes present on plasmid pTSB5. The genetic map is aligned with the restriction maps shown in panel A. (C) Restriction and genetic maps of plasmid pTSB50. The thin boxed region below the restriction map indicates the location of the 1.8-kb EcoRV fragment that is present in plasmid pR5-38 and contains the cstS gene. The restriction and genetic maps are aligned with each other and to the maps shown in panels A and B. Restriction sites are as follows: B, BamHI; E, EcoRI; N, NruI; P, PstI; S, SalI; V, EcoRV. Sites shown in parentheses indicate restriction sites present in vector sequences.
The plasmid pTSB2-5 was constructed by ligating the 2-kb BamHI fragment from pTSB2.B into the BamHI site of pTSB5.R. The 2-kb BamHI fragments in pTSB2-5 and in pTSB2.B are in the same orientation relative to the trc promoter present on plasmid pTrc99A. The pSHU9 plasmid (Tcr, pAT153 replicon) was a gift from Shelley M. Payne, University of Texas at Austin, and it contains a 9-kb insert that carries genes for the heme transport system from Shigella dysenteriae (28). To optimize expression of the heme transport genes on pSHU9, EDDA was added to the growth medium at 2.5 μg/ml. The plasmid pBluescript KS (Stratagene, La Jolla, Calif.) was used for routine cloning experiments and for preparing DNA for sequencing. Plasmids were transformed into C. diphtheriae by electroporation (14) and into E. coli as previously described (12).
Construction of the C. diphtheriae chromosomal library.
The expression vector pTrc99A (Ampr) (Pharmacia, Milwaukee, Wis.), which was used in the construction of the C. diphtheriae library, contains the pBR322 origin of replication and the lacIq gene. The plasmid library was constructed as follows. C. diphtheriae C7(−) chromosomal DNA was isolated as previously described (42) and then partially digested with Sau3AI. DNA fragments of 3 to 7 kb were excised from a 1% agarose gel and purified by using a Gene Clean Spin kit from Bio 101 (Vista, Calif.). The DNA fragments were ligated into the BamHI site of the pTrc99A vector that had been treated with shrimp alkaline phosphatase (Amersham, Cleveland, Ohio).
Identification and cloning of fragments from the C7(−) chromosome that carry the sensor kinase genes.
Since plasmids pTSB2 and pTSB5 carry only the 3′ portions of the coding regions for the cstS and chrS genes, additional restriction mapping analysis of the C. diphtheriae chromosome was done to identify restriction fragments that might carry the 5′ regions of these two genes. 32P-labeled DNA fragments obtained from either plasmid pTSB2 or pTSB5 were used as probes to hybridize to a chromosomal digest of C. diphtheriae C7(−) DNA (21, 34). Hybridization studies using the pTSB2 probe identified a 1.5-kb HindIII fragment that was predicted to contain the 5′ region of the sensor kinase gene chrS. Similarly, hybridization analysis with a pTSB5 probe indicated that a 1.8-kb EcoRV fragment present in the C. diphtheriae chromosome contains the 5′ portion of the cstS gene. Both the 1.5-kb HindIII fragment and the 1.8-kb EcoRV fragment contain approximately 500-bp sequences that are present on the cloned sequences of pTSB2 and pTSB5, respectively. The 1.5-kb HindIII fragment and the 1.8-kb EcoRV fragment were cloned into the pBluescript KS vector as follows. C. diphtheriae chromosomal DNAs were digested separately with either HindIII or EcoRV, and DNA fragments in the size range of 1 to 3 kb were excised from a 1% agarose gel, purified, and ligated into the appropriate restriction sites in the KS vector. The recombinants were transformed into DH5α, and then clones that carried the insert of interest were identified by colony hybridization (34). Plasmid pKSH51 carried the 1.5-kb HindIII insert and plasmid pR5-38 carried the 1.8-kb EcoRV insert.
DNA sequence analysis.
The DNA sequences for both strands of a 2,021-bp region that includes the complete sequence of the chrS and chrA genes were determined. Double-stranded DNA templates were sequenced by the chain termination method of Sanger et al. (35) by using a DNA sequencing kit from Amersham. The complete DNA sequences for both strands of a 2,404-bp region containing the cstS and cstA genes were also determined. Sequences were compiled and analyzed by using the Genetics Computer Group (GCG) program (Madison, Wis.). Amino acid homologies were identified by using a BLAST search of the SwissProt protein database. Amino acid alignments were done by the GAP program (GCG), and putative transmembrane regions were identified by the TMpred program (16).
LacZ assays.
Cultures (18 h) of E. coli and C. diphtheriae were used to inoculate fresh medium at a 1:100 dilution which was then grown for 16 to 18 h at 37°C with shaking. Supplements were added to the medium as indicated. LacZ activities were determined for E. coli by the method of Miller (27) and for C. diphtheriae as previously described (39).
Mutant construction.
A point mutation was introduced into the chrA gene by using inverse PCR and utilizing the useful properties of the class II restriction enzyme BsaI (New England BioLabs, Beverly, Mass.). The mutation results in replacement of the Asp residue at position 50 (D50) in the wild-type gene product (ChrA) with an Asn (N50) in the mutant gene product (ChrAD50N). The mutagenesis procedure utilized two 33-bp oligonucleotide primers, MUTN50T, 5′-CGCGGTCTCACCAACATCCAAATGCCAGGCACC-3′ (sense strand), and MUTN50B, 5′-CGCGGTCTCGTTGGTGACAACAACGTCGATGCC-3′ (antisense strand), which each contain a unique BsaI recognition site (underlined) and a single base change from the wild-type chrA sequence (boldface type; G to A for MUTN50T and C to T for MUTN50B). The 24 nt sequences at the 3′ ends of the two primers are complementary to sequences on opposite strands of the chrA gene, while the 9 nt at the 5′ end contain noncomplementary sequences and include a BsaI site. The primers are designed to anneal to circular template DNA containing the cloned chrA gene in a tail-to-tail inverted manner, such that there exists a 6-bp complementary overlap between the primers in the region that is immediately 3′ to the BsaI site. The template DNA used for the PCR was the plasmid pKBH1.2, which contains the 1.2-kb BglII-HindIII fragment from pTSB2 (chrA+) ligated into the BamHI-HindIII sites of pBluescript KS. Inverse PCR was done by using Vent polymerase (New England BioLabs), and the reaction mixture contained 10 ng of template DNA, 0.5 μg of each primer, and 300 μM deoxynucleoside triphosphates in 1× Vent polymerase buffer (New England BioLabs). The reaction was run under the following conditions: 94°C for 30 s, 55°C for 30 s, and 72°C for 6 min for 28 cycles and a final cycle of 72°C for 10 min. The 4-kb linear PCR product was digested with BsaI, which generated two large fragments, each of approximately 2 kb (BsaI cuts within the ampicillin resistance gene on the vector and also in the 5′ tail regions of the primers). The two fragments were ligated, and Ampr transformants were isolated. Since BsaI has a cut site that is adjacent to, but does not overlap, its recognition sequence, digestion of the PCR product with BsaI followed by ligation will result in the removal of the BsaI recognition sequence from the 5′ tail regions of the primers without affecting any of the sequences 3′ of the recognition site and will generate complementary overhangs. Therefore, BsaI digestion of the PCR product followed by ligation of the two fragments is predicted to reconstitute the sequence of the original plasmid, pKBH1.2, with the incorporation of a single nucleotide substitution. Plasmid DNA was obtained from one of the transformants, and the DNA sequence of the 1.2-kb insert was determined, confirming the presence of the point mutation within the chrA gene and further showing that no other sequence changes had occurred. The resulting plasmid was designated pKBH1.2-D50N.
The plasmids pWBH20 and pWBH20-D50N were constructed by using a two-step process that was similar to that used for the construction of pTSB20. In the first step, the inserts in plasmids pKBH1.2 and pKBH1.2-D50N were excised with PvuII and ligated into the EcoRV site of the low-copy-number vector pWKS30 to produce pWBH1.5 and pWBH1.5-D50N, respectively. In the second step, 1.5-kb HindIII fragments from pKSH51 were ligated into the HindIII sites of pWBH1.5 and pWBH1.5-D50N to generate plasmids pWBH20 and pWBH20-D50N, respectively. The Ptrc99A expression vector was used to construct plasmids pPBH2 and pPBH2-D50N, which contain the 1.2-kb inserts from pKBH1.2 and pKBH1.2-D50N, respectively.
Nucleotide sequence accession numbers.
The sequences of the 2,021-bp region containing chrS and chrA and the 2,404-bp region containing cstS and cstA were assigned GenBank accession no. AF161327 and AF161328, respectively.
RESULTS
Sequences required for heme activation of the hmuO promoter.
To identify sequences needed for the heme-dependent activation of the hmuO promoter, six hmuO promoter-lacZ transcriptional fusion constructs (PhmuO-lac) that contained various amounts of C. diphtheriae DNA sequences upstream of the hmuO promoter were created. Plasmid pCPO-1 contained 200 bp of upstream sequences, while plasmids pCPO-2 through pCPO-6 contained decreasing amounts of the native C. diphtheriae sequences upstream of the hmuO promoter (Fig. 1). The six PhmuO-lac fusion plasmids were examined for transcriptional activity in C. diphtheriae C7(−) that was grown in low-iron medium in the presence of heme. Expression of the PhmuO-lac fusion on plasmids pCPO-1, pCPO-2, and pCPO-3 resulted in similar levels of LacZ activity (Fig. 1). However, the PhmuO-lac fusion on plasmid pCPO-4, which contained only 20 bp of upstream sequence, exhibited a twofold decrease in expression relative to the fully induced levels seen with pCPO-1 (Fig. 1). The plasmids pCPO-5 and pCPO-6 exhibited little if any heme-dependent activation. C7(−) carrying pCPO-1 gave 2.3 U of LacZ activity in low-iron medium in the absence of heme, and similar LacZ levels in the same medium were observed for DH5α carrying pCPO-2 through pCPO-6 (data not shown). Hemoglobin also activated the expression of the hmuO promoter in a manner similar to that seen in the presence of heme (data not shown). These findings indicate that sequences upstream of the hmuO promoter are required for heme induction and further suggest that the upstream region may contain a binding site for a factor involved in the heme-responsive activation.
Identification of genes involved in the activation of the hmuO promoter.
In an earlier study, it was shown that the hmuO promoter is poorly expressed in E. coli DH5α (37). Consistent with this earlier finding, the PhmuO-lac fusion on pCPO-1 is also expressed at low levels in DH5α (Table 1), and colonies of DH5α carrying pCPO-1 plated onto LB agar medium containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) were white, which is indicative of low-level LacZ expression (data not shown). To identify the gene(s) that activates the expression of the hmuO promoter, a C. diphtheriae chromosomal library, which was constructed by using the expression plasmid pTrc99A, was transformed into DH5α carrying the plasmid pCPO-1. Transformants were plated onto LB agar medium containing X-Gal and IPTG (IPTG was used to induce the trc promoter on pTrc99A). Since DH5α carrying pCPO-1 produces white colonies on LB medium containing X-Gal and IPTG, blue colonies should represent transformants that contain recombinant clones capable of activating the expression of the hmuO promoter present on pCPO-1. Four blue colonies were identified after screening of approximately 10,000 library transformants. The four unique clones were designated pTSB1, pTSB2, pTSB3, and pTSB5 and had inserts of different sizes that ranged from 2.8 kb for pTSB2 to 5.6 kb for pTSB1. Restriction mapping analysis of these clones indicated that the entire 2.8-kb insert of the plasmid pTSB2 was contained within the larger inserts of the plasmids pTSB1 and pTSB3 (Fig. 2A and data not shown). Additionally, the inserts in pTSB1, pTSB2, and pTSB3 all shared the same left end terminus relative to the map of pTSB2 shown in Fig. 2A. Restriction analysis of the 3.7-kb insert in the plasmid pTSB5 indicated that it did not share sequences with the other three plasmids (Fig. 3A). DH5α carrying pCPO-1 and carrying each of the four putative activator clones produced blue colonies on X-Gal-containing medium only in the presence of IPTG (data not shown). This indicated that expression from the IPTG-inducible trc promoter, present on pTrc99A, was essential for each of these clones to activate the hmuO promoter. The dependence on the trc promoter indicated that either the putative activator gene(s) on the four clones lacked their native C. diphtheriae promoter or the native promoter was inadequately active in DH5α.
TABLE 1.
Effect of activator clones on expression of Phmuo-lac fusions in DH5α
| Plasmid(s) | IPTGa | LacZ Ub |
|---|---|---|
| pCPO-1 (PhmuO-lac) | + | 0.8 |
| pCPO-1, pTSB2 | −c | 0.9 |
| pCPO-1, pTSB2 | + | 11.4 |
| pCPO-1, pTSB2, pSHU9 (+He)d | + | 9.3 |
| pCPO-4, pTSB2 | + | 4.3 |
| pCPO-5, pTSB2 | + | 0.7 |
| pCPO-1, pTSB5 | − | 0.9 |
| pCPO-1, pTSB5 | + | 13.7 |
| pCPO-1, pTSB5, pSHU9 (+He)d | + | 10.4 |
| pCPO-4, pTSB5 | + | 5.6 |
| pCPO-5, pTSB5 | + | 0.6 |
| pCPO-1, pTSB2-5 | − | 55.7 |
| pCPO-1, pTSB2-5 | + | 352.1 |
| pCPO-1, pTSB2-5, pSHU9 (+He)d | + | 323.7 |
IPTG was added as indicated (+) at 0.5 mM.
LacZ units (U) were determined by the method of Miller (27). Values are means of three independent experiments, and standard deviations did not vary by greater than 15% from the mean.
pCPO-1, pCPO-4, and pCPO-5 showed similar LacZ activities in the absence of IPTG.
pSHU9 enables DH5α to transport hemin; hemin (+He) was added at 100 μg/ml.
Effect of the activator clones on expression of the hmuO promoter.
Expression of the hmuO promoter on pCPO-1, pCPO-4, and pCPO-5, in the presence of the various activator clones, was quantitated in liquid culture medium by measuring β-galactosidase levels (27). Only low levels of promoter activity were seen with the four clones in the absence of IPTG (Table 1 and data not shown). Greater than 10-fold induction of LacZ activity was seen when DH5α carried both pCPO-1 and either pTSB2 or pTSB5 and was grown in the presence of IPTG. Similar LacZ levels were seen with the clones pTSB1 (13.2 U) and pTSB3 (9.6 U) in the presence of IPTG. The plasmids pTSB1 and pTSB3 were not characterized further since these results suggested that all of the sequences needed for activation of the hmuO promoter reside in pTSB2. The PhmuO-lac fusion on pCPO-4 was less responsive than that on pCPO-1 to IPTG induction in the presence of plasmids pTSB2 and pTSB5; the LacZ levels were two- to threefold lower than those observed with pCPO-1 (Table 1). The plasmids pTSB2 and pTSB5 did not induce the expression of the PhmuO-lac fusion on pCPO-5. These results showed that there exists a similar trend for the heme activation obtained with pCPO-1, pCPO-4, and pCPO-5 in C. diphtheriae (Fig. 1) and for the induction caused by pTSB2 and pTSB5 in E. coli.
Subclones of the plasmids pTSB2 and pTSB5 were constructed in pTrc99A to identify the smallest region that would maintain induction of the hmuO promoter on pCPO-1. The smallest such subclone from pTSB2 identified was pTSB2.K-Bg, which contained a 900-bp KpnI-BglII fragment (Fig. 2A). Subclones of pTSB2 which contained deletions at the left end had a five- to sixfold increase in LacZ expression of the PhmuO-lac fusion on pCPO-1 (Fig. 2A, plasmids pTSB2.H-Bg and pTSB2.K-Bg). The reason for this increase in activation is not clear, although it may be due either to the loss of a repressor function or to the more proximal location of the putative activator gene to the trc promoter present on the vector. The smallest subclone of pTSB5 maintaining induction was an approximately 1-kb NruI fragment on pTSB5.N (Fig. 3A). No significant differences in the activation of the hmuO promoter were observed for any of the pTSB5 subclones.
To assess the effect that both activator genes together would have on the expression of the hmuO promoter in E. coli, DNA fragments from pTSB2 and pTSB5 were placed in tandem onto the pTrc99A vector to generate plasmid pTSB2-5. The presence of pTSB2-5 in DH5α carrying pCPO-1 resulted in a high level of induction (352.1 U) of the hmuO promoter in the presence of IPTG (Table 1). This level of expression was 25-fold higher than the LacZ activity seen with either of the activators alone: 11.4 U for pTSB2 and 13.7 U for pTSB5. This finding suggested that the presence of both activators on the same plasmid caused a synergistic effect on the activation of the hmuO promoter. Relatively high levels of LacZ activity (55.7 U) were also detected with pTSB2-5 even in the absence of IPTG induction (Table 1), which may have been caused by low-level expression from the “leaky” trc promoter.
Almost all laboratory strains of E. coli, including DH5α, are unable to transport heme through their outer membranes unless a heme transport system, such as those present in certain bacterial pathogens, is provided in trans. The activation of the hmuO promoter in DH5α carrying pCPO-1 and either pTSB2, pTSB5, or pTSB2-5 was not affected by the addition of heme to the medium even in the presence of the plasmid pSHU9 (28), which encodes the S. dysenteriae heme transport system and enables DH5α to transport heme (Table 1).
Sequence analysis of the activator genes on pTSB2 and pTSB5.
To identify the genes present on pTSB2 and pTSB5 that are required for activation of the hmuO promoter, the DNA sequences of the 1.7-kb BamHI-BglII insert on pTSB2.Bg and the 1.2-kb left end region of plasmid pTSB5.R (Fig. 2A and Fig. 3A, respectively) were determined. DNA sequence analysis indicated that a single open reading frame was present in the KpnI-BglII fragment from pTSB2.Bg (the smallest subcloned region that maintains activation). This open reading frame was designated chrA (Corynebacterium heme-responsive activator) (Fig. 2B) and is predicted to encode a product of 199 amino acids that has significant homology to response regulators of two-component signal transduction systems. Immediately upstream from chrA is a partial open reading frame for a gene that is designated chrS (Corynebacterium heme-responsive sensor), whose product has homology to the sensor kinase component of two-component signal transduction systems (Fig. 2B). The chrS open reading frame is predicted to encode a product containing 340 amino acids; however, the 5′ portion of the chrS coding region is not present on pTSB2 or the other two related clones, pTSB1 and pTSB3. The chrS termination codon TGA overlaps the ATG start codon for chrA, suggesting that the genes are organized as an operon similar to other genes encoding two-component systems. Since chrA is the only complete gene present on the KpnI-BglII fragment on plasmid pTSB2.K-Bg, this suggests that only the response regulator is needed for activation of the hmuO promoter in E. coli and this effect is observed only when the IPTG-inducible trc promoter is active.
Sequence analysis of the 1.2-kb region of pTSB5.R revealed that a single open reading frame was present on the 1-kb NruI fragment, and this open reading frame, designated cstA for Corynebacterium signal transduction activator, also has significant homology to response regulators of two-component systems (Fig. 3B). Upstream from cstA is a partial open reading frame for a gene designated cstS, which is predicted to encode the C-terminal 137 amino acids of a sensor kinase. The predicted amino acid sequences of CstA and ChrA show the highest homology to proteins in the NarL and NarP family of response regulators (both ChrA and CstA show homologies to proteins in this family that range from 30 to 40% amino acid identity over the entire length of the protein). Proteins in this family of response regulators are known to bind DNA and function as transcriptional activators (32). ChrA and CstA are greater than 40% identical to each other at the amino acid level and share numerous residues that are conserved among all response regulators within the NarL and NarP family (Fig. 4) (1). The conserved amino acids include an aspartate residue (Asp50 for ChrA and Asp54 for CstA), which is the site of phosphorylation in other response regulators (32).
FIG. 4.
Amino acid sequence alignment of ChrA and CstA. Amino acid residues that are highly conserved among other response regulators within the NarP and NarL family (1) are shown in boldface. The conserved Asp (D) residues marked with an asterisk (D50 in ChrA and D54 in CstA) are known to be the sites of phosphorylation in other response regulators.
Cloning and sequence analysis of the sensor kinase homologs encoded by chrS and cstS.
Although the chrA and cstA genes were able to activate the expression of the hmuO promoter in E. coli, the heme-dependent activation of hmuO observed in C. diphtheriae could not be reconstituted in E. coli with either of these genes. To determine if the sensor kinase genes, chrS and cstS, are needed for heme-dependent activation in E. coli, the 5′ portions of these genes along with upstream regions were cloned from the chromosome of C. diphtheriae. A 1.5-kb HindIII fragment, which carries the 5′ region of the chrS gene, was used to construct the plasmid pTSB20, which contains the complete coding region for chrS and chrA (Fig. 2C). Similarly, a 1.8-kb EcoRV fragment that was cloned from the chromosome of C. diphtheriae was used to construct the plasmid pTSB50, which contains the complete coding region for cstS and cstA (Fig. 3C). The plasmids pTSB20 and pTSB50 contain the pSC101 origin of replication and replicate at a low copy number in E. coli. Low-copy-number plasmids were used in the analysis of the chrS and cstS genes since the predicted products of these genes are presumed to be membrane associated and could be deleterious at high levels.
Sequence analysis of the chrS and cstS genes indicated that they are predicted to encode proteins of 417 and 408 amino acids, respectively. Both proteins showed the highest homology (approximately 30% identity) in their C-terminal halves to the UhpB and DegS sensor kinases (data not shown). The putative sensor domain of CstS, located at the N-terminal region, had no significant homology with any proteins in the GenBank database. The N-terminal sensor domain of ChrS had 28% identity with the N-terminal region of the SenR protein from Streptomyces reticuli (GenBank accession no. Y14336). The ChrS and SenR proteins are 36% identical at the amino acid level over their entire sequences. A specific function for SenR has not been reported, although it is proposed to function as the sensor component in a two-component system. Analysis of the amino acid sequences for both ChrS and CstS, using the TMpred program, predicts both proteins to have multiple transmembrane helices in their N-terminal 200 amino acids (data not shown).
Effect of chr and cst operons on the expression of the hmuO promoter.
Plasmid pTSB20 (chrS+, chrA+) and plasmid pTSB50 (cstS+, cstA+) were transformed into DH5α carrying pCPO-1 to determine what effect these genes had on the expression of the PhmuO-lac fusions in the presence and absence of heme. Plasmid pTSB20 in DH5α carrying pCPO-1 showed relatively low LacZ activity regardless of the presence heme (Table 2). However, when pSHU9, which carries a heme transport system, was moved into this strain, greater than 20-fold induction was seen in the presence of heme (the LacZ activity was 4.6 U in the absence of heme and 102.5 U in the presence of heme [Table 2]). This high level of heme induction was dependent on the presence of a functional heme transport system supplied by the pSHU9 plasmid. Heme activation in E. coli also required the presence of both the chrS and chrA genes, since clones (constructed on the same low-copy-number plasmids) containing only the chrA gene did not exhibit heme induction (data not shown). Furthermore, the presence of pTSB20 also conferred heme induction on the PhmuO-lac fusion on pCPO-4, but only very low LacZ activity was seen with pCPO-5 (Table 2). The relative levels of heme-induced expression from the PhmuO-lac fusions on pCPO-1, pCPO-4, and pCPO-5 by pTSB20 (chrA+, chrS+) in E. coli are similar to those observed for the same PhmuO-lac fusions in C. diphtheriae (Fig. 1). The plasmid pTSB50 transformed into DH5α carrying pCPO-1 and pSHU9 showed only a low level of LacZ activity that was not affected by the presence of heme (Table 2). Plasmid pTSB20-50, which contained the inserts of both pTSB20 and pTSB50 on the same low-copy-number plasmid, showed a level of heme-induced LacZ activity in DH5α carrying pCPO-1 and pSHU9 (96 U) that was similar to that observed for pTSB20.
TABLE 2.
Effects of sensor and/or activator clones on the expression of PhmuO-lac fusions in the presence and absence of heme in DH5α
| Plasmids | Hemea | LacZ Ub |
|---|---|---|
| pTSB20, pCPO-1 (PhmuO-lac) | − | 5.3 |
| pTSB20, pCPO-1 | + | 6.7 |
| pTSB20, pCPO-1, pSHU9 (He+)c | + | 102.5 |
| pTSB20, pCPO-1, pSHU9 (He+) | − | 4.6 |
| pTSB20, pCPO-4, pSHU9 (He+) | + | 52.3 |
| pTSB20, pCPO-4, pSHU9 (He+) | − | 7.3 |
| pTSB20, pCPO-5, pSHU9 (He+) | + | 3.4 |
| pTSB20, pCPO-5, pSHU9 (He+) | − | 0.8 |
| pTSB50, pCPO-1, pSHU9 (He+) | + | 1.7 |
| pTSB50, pCPO-1, pSHU9 (He+) | − | 1.3 |
Hemin was added to cultures (+) at 100 μg/ml.
LacZ units (U) were determined by the method of Miller (27). Values are means of three independent experiments, and standard deviations did not vary by greater than 15% from the mean.
Plasmid pSHU9 (He+) enables DH5α to transport heme.
Effect of the chrAD50N mutant allele on expression of the hmuO promoter.
Site-directed mutagenesis was used to introduce a point mutation into the chrA gene which resulted in the replacement of the aspartate residue at position 50 with an asparagine. Based on sequence homologies with other response regulators, the Asp50 residue in ChrA is the presumed site of phosphorylation by its cognate sensor kinase ChrS. The plasmid pWBH20-D50N, which carries both the mutant chrA allele (chrAD50N) and the wild-type chrS gene on the low-copy-number vector pWKS30, was examined to determine the effect of the chrAD50N mutation on the heme-dependent activation of the hmuO promoter. DH5α carrying both pCPO-1 and pWBH20-D50N showed only low LacZ activity and very weak heme-dependent activation of the PhmuO-lac fusion present on pCPO-1 (Table 3). Plasmid pWBH20, which carries the wild-type copies of the chrA and chrS genes on pWKS30, showed levels of heme-dependent activation of the PhmuO-lac fusion that were greater than 18-fold higher than those observed for the chrAD50N mutant allele (the LacZ activities were 66.4 and 3.6 U, respectively [Table 3]). These results indicated that replacement of the Asp residue at position 50 with an Asn in the chrAD50N gene product virtually abolished all heme-responsive activation when chrAD50N was expressed from a low-copy-number plasmid.
TABLE 3.
Effect of the chrAD50N mutation on the expression of a PhmuO-lac fusion in DH5α
| Plasmids | Hemea | IPTG | LacZ Ub |
|---|---|---|---|
| pWBH20-D50N, pCPO-1, pSHU9 | − | − | 1.1 |
| pWBH20-D50N, pCPO-1, pSHU9 | + | − | 3.6 |
| pWBH20, pCPO-1, pSHU9c | − | − | 8.1 |
| pWBH20, pCPO-1, pSHU9 | + | − | 66.4 |
| pPBH2-D50N, pCPO-1 | − | − | 1.4 |
| pPBH2-D50N, pCPO-1 | − | + | 32.4 |
| pPBH2, pCPO-1 | − | − | 1.5 |
| pPBH2, pCPO-1 | − | + | 61.8 |
Hemin was added to cultures (+) at 100 μg/ml.
LacZ units (U) were determined by the method of Miller (27). Values are means of three independent experiments, and standard deviations did not vary by greater than 15% from the mean.
Plasmid pSHU9 enables DH5α to transport heme.
However, when the chrAD50N allele was present on the higher-copy-number vector pTrc99A (pPBH2-D50N) and expressed from the strong IPTG-inducible trc promoter, the chrAD50N gene product exhibited a capacity to activate the hmuO promoter that was only twofold lower than the activation observed for the wild-type chrA gene on plasmid pPBH2 (the activities were 32.4 and 61.8 U, respectively [Table 3]). Activation of the PhmuO-lac fusion by both the wild-type chrA gene and the chrAD50N allele required the presence of IPTG, and this activation occurred in the absence of the chrS gene, since chrS is not present on the plasmid pPBH2-D50N or pPBH2. This finding indicates that the chrAD50N gene product has the capability to activate transcription, although at reduced levels relative to those induced by the wild-type gene product, and that the Asp50 residue is not essential for transcriptional activation if the gene is expressed at high levels.
DISCUSSION
Bacteria utilize a variety of mechanisms to adapt to and interact with the environment, including the well-characterized two-component signal transduction systems (for a review see reference 32). Two-component regulatory systems typically consist of a sensor protein that monitors the environment and a cognate response regulator which is involved in controlling gene expression. In this study, the mechanism involved in the heme-responsive activation of the hmuO promoter has been reconstituted in E. coli. The genes, chrS and chrA, that encode homologs of two-component signal transduction systems were shown to activate the expression of a PhmuO-lac fusion construct in E. coli. The activation required heme or hemoglobin in the growth medium and a functional heme transport system that was provided in trans. Although this system has been reconstituted in an E. coli background, it very closely mimics the heme-dependent activation observed in C. diphtheriae, which strongly suggests that chrS and chrA are the relevant activators in C. diphtheriae.
The hmuO gene is the only bacterial gene whose expression is known to be activated by heme, and this is the first report in which heme has been identified as the environmental stimulus for a two-component regulatory system. Relatively little is known about heme-regulated gene expression in other bacterial systems. Numerous bacterial pathogens can transport and utilize heme as an iron source, and most of the genes that encode products mediating these heme transport functions are regulated by iron (22, 30, 51). However, the expression of certain proteins involved in the transport of heme and iron by pathogenic species of Haemophilus (8, 19, 29) and Porphyromonas (9, 20) has been shown to be heme repressible. The mechanisms involved in the regulation of genes encoding heme-repressed proteins in these organisms have not been described.
Sequences upstream from the hmuO promoter were shown to be important for heme-dependent transcription both in C. diphtheriae and in E. coli. The PhmuO-lac fusion on plasmid pCPO-3, which contained only 50 bp of C. diphtheriae-specific sequences upstream of the −35 sequence (which is 90 bp upstream from the start of transcription [37]), was fully induced by heme. The PhmuO-lac fusions on pCPO-4 and pCPO-5, which had fewer upstream sequences than pCPO-3, showed significantly reduced heme induction. These findings indicate that sequences within the 90-bp region upstream from the start site of transcription from the hmuO promoter, and most likely upstream of the −35 element, are required for heme induction and may contain the binding site for the response regulator encoded by the chrA gene. In support of this possibility, the related response regulators, NarP, NarL, and UhpA, bind to sequences within 80 bp of the transcriptional start sites of some of the genes that they regulate (6, 7).
The activator clones pTSB2 and pTSB5, which contain the chrA and cstA genes, respectively, were isolated from a plasmid library due to their ability to activate the expression of the PhmuO-lac fusion on pCPO-1 in E. coli. The cloned chrA and cstA genes in E. coli are able to activate the transcription of the hmuO promoter in the absence of their cognate sensor kinase genes, chrS and cstS, respectively. Several factors may contribute to the capacity of the chrA and cstA genes to activate transcription in the absence of their cognate sensor kinases. These include (i) low-level phosphorylation of ChrA and CstA by endogenous nonspecific kinases in E. coli, (ii) cross talk with other two-component systems, and (iii) overexpression of the products of the chrA and cstA genes due to the presence of these genes on high-copy-number plasmids. The expression of these genes is further enhanced since they are under the transcriptional control of the IPTG-induced trc promoter. Other investigators have made observations similar to those described here showing that multicopy plasmids carrying genes encoding response regulators are able to activate transcription in the absence of their cognate sensor kinases (11, 48).
The deletion constructs pTSB2.H-Bg and pTSB2.K-Bg, which carry the chrA gene and a portion of the 3′ region of chrS, exhibited an enhanced activity relative to the activity of the parent clone, pTSB2 (Fig. 2A). This enhanced activity may be due to the more proximal location of the chrA gene to the trc promoter or to the deletion of sequences containing the chrS gene. The truncated chrS gene present on either pTSB2.H-Bg or pTSB2.K-Bg lacks the codon for the conserved histidine (H215), which is proposed to be the site of phosphorylation and is predicted to be essential for the activities of sensor kinases. The truncated chrS gene on the parent clone pTSB2 contains this conserved histidine (H215)-encoding codon, and it is possible that any putative peptide produced from this truncated chrS gene (either initiating from a weak internal start codon or present as part of a translational fusion with upstream vector sequences) could potentially have activity. Since sensor kinases are known to have both positive and negative effects on the activities of their cognate response regulators (32), it is possible that a truncated ChrS product produced from pTSB2 may exert a repressor effect on the ChrA activator. However, when the conserved histidine codon is removed from the chrS sequence, as in the two-deletion constructs, the repressor effect is alleviated and enhanced expression is observed.
The chrAD50N mutant allele, when expressed at high levels, was also able to activate transcription of the hmuO promoter in the absence of its cognate sensor kinase. Similarly, other investigators have shown that a mutation in the gene encoding the E. coli UhpA response regulator, in which the Asp54 residue (the site of phosphorylation by its cognate sensor kinase) is replaced with an Asn, results in the loss of all activity when the gene is present in low copy number but that transcriptional activity is maintained when the gene is expressed from a multicopy plasmid (48). The presumed DNA binding domain for ChrA, based on sequence homologies with other response regulators in the NarP and NarL family, is predicted to be located in the C-terminal portion of the protein and would not be directly affected by the D50N mutation. The results obtained with the ChrAD50N and UhpAD54N response regulators indicate that phosphorylation at the conserved Asp residue is not essential for activating transcription when these proteins are strongly expressed.
The findings in this study indicate that the two genes chrA and cstA are able to activate expression at the hmuO promoter. Additionally, both genes showed similar levels of activation when expressed in the various promoter deletion constructs (Table 1), which suggests that the products of chrA and cstA may have DNA binding sites that are very near each other. Plasmid pTSB2-5, which carries both the chrA and cstA genes under control of the trc promoter, activates transcription of the PhmuO-lac fusion on pCPO-1 to levels greater than 25-fold higher than those produced by either pTSB2 or pTSB5, each of which carries only one of the activators (Table 1). Evaluation of the results for the tandem construct is difficult since protein levels are not known. While it is clear that additional studies are required to fully understand the mechanism of this enhanced activity of the chrA and cstA genes on pTSB2-5, one possible mechanism by which the expression may be increased could involve the formation of mixed dimers or multimers of the ChrA and CstA proteins. Since the DNA binding sites of ChrA and CstA may be close to each other, the presence of mixed multimers may result in either greater stability of a protein-DNA complex or an alteration of the conformation of the proteins so that there exists a more optimal interaction with RNA polymerase, which results in the enhanced transcription.
The presence of both response regulators, chrA and cstA, and their cognate sensors, chrS and cstS, together on plasmid pTSB20-50 did not result in a synergistic effect similar to that seen for pTSB2-5, which contained only the response regulator genes. Since the presence of cstA and cstS on the same plasmid, pTSB50, failed to activate the expression of the hmuO promoter, it is unclear what role, if any, the cstA and cstS genes have in the regulation of the hmuO promoter in C. diphtheriae. Since cstA did not activate the hmuO promoter in the presence of the cstS gene but did in the absence of cstS, it is possible that the product of cstS may repress the activity of cstA. It is possible that an environmental factor other than heme may be required for the cstA-cstS system to activate transcription at the hmuO promoter. Additional studies are needed to define the role of the cstA and cstS genes in C. diphtheriae.
It is clear from the results of this study that heme-dependent activation of the hmuO promoter in E. coli requires the presence of both the chrA and chrS genes and a functional heme transport system, which serves to transport heme through the outer membrane. The evidence strongly suggests that the putative sensor kinase encoded by chrS is involved in the detection of heme, which is presumed to be the environmental signal. Alternatively, it is possible that the actual environmental stimulus is not heme but is a factor that is produced in response to the presence of heme in the medium. While additional studies are required to determine the mechanism by which ChrS detects heme (or other signals), it is likely that sequences in the N-terminal portion or sensor domain of ChrS are involved in the detection of the environmental stimulus. Amino acid sequence analysis using the TMpred program predicts that there are at least four transmembrane helices in the N-terminal 180 amino acids of ChrS. Extracytoplasmic loop regions between transmembrane helices have been proposed to be involved in the detection of environmental stimuli by other sensor kinases (4, 32), and it is plausible that regions between the putative membrane-spanning regions in ChrS may have a role in the detection of heme. In the reconstituted system in E. coli, it is proposed that the ChrS protein resides in the cytoplasmic membrane and that the extracytoplasmic loop regions are involved in the detection of heme that has been transported into the periplasm by means of the heme transport system encoded by the genes present on the plasmid pSHU9. In the gram-positive organism C. diphtheriae, ChrS is also predicted to reside in the cytoplasmic membrane; however, the extracytoplasmic loop regions would be involved in detecting extracellular heme. If the activity of ChrS is like that of other related sensor kinases, it is presumed that detection of an environmental stimulus by ChrS should result in autophosphorylation at the conserved histidine residue, H215. The phosphoryl group on H215 could then be transferred to the conserved Asp residue (D50) on ChrA, which would allow ChrA to activate transcription at the hmuO promoter. The findings from this study indicate that the D50 residue in ChrA is needed for heme-responsive activation of the hmuO promoter, since the ChrAD50N protein had little if any capacity to activate the hmuO promoter in the presence of heme (Table 3). This observation supports the sequence homology data which predicts that the D50 residue of ChrA functions as the site of phosphorylation by ChrS and, therefore, should have a direct role in a heme-dependent signal transduction mechanism. A model depicting how this phosphotransfer signaling mechanism may function to control hmuO transcription in C. diphtheriae is presented in Fig. 5.
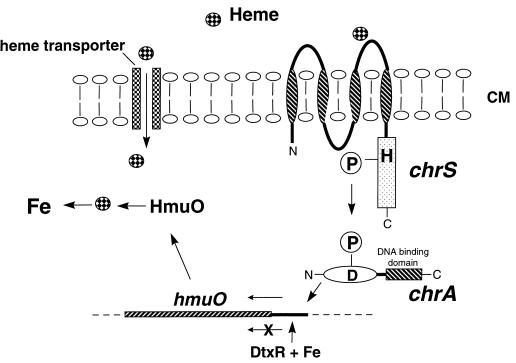
FIG. 5.
Proposed mechanism of heme-responsive activation at the hmuO promoter in C. diphtheriae. The sensor kinase ChrS is proposed to detect extracellular heme at its N-terminal sensor domain, which is predicted to contain at least two transmembrane helices (indicated by striped ovals) and two extracellular loop regions. The detection of heme by ChrS is proposed to result in autophosphorylation at a conserved histidine (H, H215) that is located in the cytosolic kinase domain of ChrS (boxed region). The phosphoryl group (indicated by a circled P) is then transferred to a conserved Asp residue (D, D50) on ChrA. Phosphorylation is proposed to activate the DNA binding ability of ChrA and allows ChrA to bind upstream of the hmuO promoter and activate transcription. Transcription of the hmuO promoter can also be repressed by DtxR during growth in the presence of iron. The hmuO gene, therefore, is optimally expressed in low-iron environments in the presence of heme. Additionally, a C. diphtheriae heme-specific transporter has been proposed to be involved in the transport of heme into the cytosol (7a), where the HmuO protein is proposed to degrade the cytosolic heme and liberate the heme-bound iron.
While the construction of defined mutations in the chrS and/or chrA genes in the chromosome of C. diphtheriae should provide additional evidence for the function of these genes in C. diphtheriae, the capability to perform allelic replacement or transposon mutagenesis in C. diphtheriae is not yet available due to the lack of genetic tools. The findings in this study expand our knowledge as to the variety of environmental factors that can function as stimuli for two-component systems. The regulatory systems reported in this study are the first two-component signal transduction systems described for the genus Corynebacterium, and future research will focus on identifying additional genes controlled by these regulatory systems and the characterization of functional domains present in these regulatory proteins.
ACKNOWLEDGMENTS
I thank Scott Stibitz for advice on the mutagenesis technique and for his helpful comments on the manuscript. I also thank Clare Schmitt and Sue Drazek for helpful comments and discussions.
REFERENCES
- 1.Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. Structure of the E. coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 2.Boyd J M, Manish O N, Murphy J R. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc Natl Acad Sci USA. 1990;87:5968–5972. doi: 10.1073/pnas.87.15.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun V, Hantke K, Koster W. Bacterial iron transport: mechanisms, genetics, and regulation. Met Ions Biol Syst. 1998;35:67–145. [PubMed] [Google Scholar]
- 4.Chiang R C, Cavicchiol R, Gunsalus R P. “Locked-on” and “locked-off” signal transduction mutations in the periplasmic domain of the Escherichia coli NarQ and NarX sensors affect nitrate- and nitrite-dependent regulation by NarL and NarP. Mol Microbiol. 1997;24:1049–1060. doi: 10.1046/j.1365-2958.1997.4131779.x. [DOI] [PubMed] [Google Scholar]
- 5.Cornejo J, Willows R D, Beale S I. Phytobilin biosynthesis: cloning and expression of a gene encoding soluble ferredoxin-dependent heme oxygenase from Synechocystis sp. PCC 6803. Plant J. 1998;15:99–107. doi: 10.1046/j.1365-313x.1998.00186.x. [DOI] [PubMed] [Google Scholar]
- 6.Dahl J L, Wei B-Y, Kadner R J. Protein phosphorylation affects binding of the Escherichia coli transcription activator UhpA to the uhpT promoter. J Biol Chem. 1997;272:1910–1919. doi: 10.1074/jbc.272.3.1910. [DOI] [PubMed] [Google Scholar]
- 7.Darwin A J, Tyson K L, Busby S J W, Stewart V. Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K-12 depends on DNA binding site arrangement. Mol Microbiol. 1997;25:583–595. doi: 10.1046/j.1365-2958.1997.4971855.x. [DOI] [PubMed] [Google Scholar]
- 7a.Drazek, E. S., et al. Submitted for publication.
- 8.Elkins C. Identification and purification of a conserved heme-regulated hemoglobin-binding outer membrane protein from Haemophilus ducreyi. Infect Immun. 1995;63:1241–1245. doi: 10.1128/iai.63.4.1241-1245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genco C A, Odusanya B M, Brown G. Binding and accumulation of hemin in Porphyromonas gingivalis are induced by hemin. Infect Immun. 1994;62:2885–2892. doi: 10.1128/iai.62.7.2885-2892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths E. Iron and bacterial virulence—a brief overview. Biol Met. 1991;4:7–13. doi: 10.1007/BF01135551. [DOI] [PubMed] [Google Scholar]
- 11.Haldimann A, Fisher S L, Daniels L L, Walsh C T, Wanner B L. Transcriptional regulation of the Enterococcus faecium BM4147 vancomycin resistance gene cluster by the VanS-VanR two-component regulatory system in Escherichia coli K-12. J Bacteriol. 1997;179:5903–5913. doi: 10.1128/jb.179.18.5903-5913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 13.Hantke K. Cloning of the repressor protein gene of iron regulated systems in Escherichia coli K-12. Mol Gen Genet. 1984;197:337–341. doi: 10.1007/BF00330982. [DOI] [PubMed] [Google Scholar]
- 14.Haynes J A, Britz M L. Electrotransformation of Brevibacterium lactofermentum and Corynebacterium glutamicum: growth in Tween 80 increases transformation frequencies. FEMS Microbiol Lett. 1989;61:329–334. [Google Scholar]
- 15.Henderson D P, Payne S M. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J Bacteriol. 1994;176:3269–3277. doi: 10.1128/jb.176.11.3269-3277.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann D, Stoffel W. Tmbase—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:116. [Google Scholar]
- 17.Holmes R K, Barksdale L. Genetic analysis of tox+ and tox− bacteriophages of Corynebacterium diphtheriae. J Virol. 1969;3:586–598. doi: 10.1128/jvi.3.6.586-598.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornung J M, Jones H A, Perry R D. The hmu locus of Yersinia pestis is essential for utilization of free haemin and haem-protein complexes as iron sources. Mol Microbiol. 1996;20:725–739. doi: 10.1111/j.1365-2958.1996.tb02512.x. [DOI] [PubMed] [Google Scholar]
- 19.Jin H, Ren Z, Pozsgay J M, Elkins C, Whitby P W, Morton D J, Stull T L. Cloning of a DNA fragment encoding a heme-repressible hemoglobin-binding outer membrane protein from Haemophilus influenzae. Infect Immun. 1996;64:3134–3141. doi: 10.1128/iai.64.8.3134-3141.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karunakaran T, Maden T, Kuramitsu H. Isolation and characterization of a hemin-regulated gene, hemR, from Porphyromonas gingivalis. J Bacteriol. 1997;179:1898–1908. doi: 10.1128/jb.179.6.1898-1908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kidd F J. In-situ hybridization to agarose gels. Focus. 1983;6:3–4. [Google Scholar]
- 22.Lee C B. Quelling the red menace: heme capture by bacteria. Mol Microbiol. 1995;18:383–390. doi: 10.1111/j.1365-2958.1995.mmi_18030383.x. [DOI] [PubMed] [Google Scholar]
- 23.Lee J H, Wang T, Ault K, Liu J, Schmitt M P, Holmes R K. Identification and characterization of three new promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. Infect Immun. 1997;65:4273–4280. doi: 10.1128/iai.65.10.4273-4280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Litwin C M, Calderwood S B. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–149. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maines M D. Characterization and regulation of heme oxygenase isozymes. In: Maines M D, editor. Heme oxygenase: clinical applications and functions. Boca Raton, Fla: CRC Press; 1992. pp. 109–144. [Google Scholar]
- 26.Mietzner T A, Morse S A. The role of iron-binding proteins in the survival of pathogenic bacteria. Annu Rev Nutr. 1994;14:471–493. doi: 10.1146/annurev.nu.14.070194.002351. [DOI] [PubMed] [Google Scholar]
- 27.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 28.Mills M, Payne S M. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J Bacteriol. 1995;177:3004–3009. doi: 10.1128/jb.177.11.3004-3009.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morton D J, Musser J M, Stull T L. Expression of the Haemophilus influenzae transferrin receptor is repressible by hemin but not elemental iron alone. Infect Immun. 1993;61:4033–4037. doi: 10.1128/iai.61.10.4033-4037.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Occhino D A, Wycoff E E, Henderson D P, Wrona T J, Payne S M. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, and exbD genes. Mol Microbiol. 1998;29:1493–1507. doi: 10.1046/j.1365-2958.1998.01034.x. [DOI] [PubMed] [Google Scholar]
- 31.Pappenheimer A M., Jr Diphtheria toxin. Annu Rev Biochem. 1977;46:69–94. doi: 10.1146/annurev.bi.46.070177.000441. [DOI] [PubMed] [Google Scholar]
- 32.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 33.Rogers H J. Iron-binding catechols and virulence in Escherichia coli. Infect Immun. 1973;7:445–458. doi: 10.1128/iai.7.3.445-456.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt M P. Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenaese and is required for acquisition of iron from heme and hemoglobin. J Bacteriol. 1997;179:838–845. doi: 10.1128/jb.179.3.838-845.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt M P. Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infect Immun. 1997;65:4634–4641. doi: 10.1128/iai.65.11.4634-4641.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt M P, Holmes R K. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect Immun. 1991;59:1899–1904. doi: 10.1128/iai.59.6.1899-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt M P, Holmes R K. Characterization of a defective diphtheria toxin repressor (dtxR) allele and analysis of dtxR transcription in wild-type and mutant strains of Corynebacterium diphtheriae. Infect Immun. 1991;59:3903–3908. doi: 10.1128/iai.59.11.3903-3908.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt M P, Holmes R K. Cloning, sequence, and footprint analysis of two promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. J Bacteriol. 1994;176:1141–1149. doi: 10.1128/jb.176.4.1141-1149.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitt M P, Talley B G, Holmes R K. Characterization of lipoprotein IRP1 from Corynebacterium diphtheriae, which is regulated by the diphtheria toxin repressor (DtxR) and iron. Infect Immun. 1997;65:5364–5367. doi: 10.1128/iai.65.12.5364-5367.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiller J, Groman B, Coyle M. Plasmids in Corynebacterium diphtheriae and diphtheroids mediating erythromycin resistance. Antimicrob Agents Chemother. 1980;18:814–821. doi: 10.1128/aac.18.5.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stojiljkovic I, Hantke K. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram negative bacteria. EMBO J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stojiljkovic I, Hantke K. Transport of hemin across the cytoplasmic membrane through a hemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol Microbiol. 1994;13:719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 45.Tao X, Nikolaus S, Zeng H, Ringe D, Murphy J R. Iron, DtxR and the regulation of diphtheria toxin expression. Mol Microbiol. 1994;14:191–197. doi: 10.1111/j.1365-2958.1994.tb01280.x. [DOI] [PubMed] [Google Scholar]
- 46.Torres A G, Payne S M. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1997;23:825–833. doi: 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- 47.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 48.Webber C A, Kadner R J. Action of receiver and activator modules of UhA in transcriptional control of the Escherichia coli sugar phosphate transport system. Mol Microbiol. 1995;15:883–893. doi: 10.1111/j.1365-2958.1995.tb02358.x. [DOI] [PubMed] [Google Scholar]
- 49.Weinberg E D. Iron and infection. Microbiol Rev. 1978;42:45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilks A, Schmitt M P. Expression and characterization of a heme oxygenase (HmuO) from Corynebacterium diphtheriae. J Biol Chem. 1998;273:837–841. doi: 10.1074/jbc.273.2.837. [DOI] [PubMed] [Google Scholar]
- 51.Wyckoff E E, Duncan D, Torres A G, Mills M, Maase K, Payne S M. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol Microbiol. 1998;28:1139–1152. doi: 10.1046/j.1365-2958.1998.00873.x. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida T, Biro P, Cohen T, Muller R M, Shibahara S. Human heme oxygenase cDNA and induction of its mRNA by hemin. Eur J Biochem. 1988;171:457. doi: 10.1111/j.1432-1033.1988.tb13811.x. [DOI] [PubMed] [Google Scholar]