Abstract
Background
Tuberculosis (TB) had been the leading lethal infectious disease worldwide for a long time (2014–2019) until the COVID-19 global pandemic, and it is still one of the top 10 death causes worldwide. One important reason why there are so many TB patients and death cases in the world is because of the difficulties in precise diagnosis of TB using common detection methods, especially for some smear-negative pulmonary tuberculosis (SNPT) cases. The rapid development of metabolome and machine learning offers a great opportunity for precision diagnosis of TB. However, the metabolite biomarkers for the precision diagnosis of smear-positive and smear-negative pulmonary tuberculosis (SPPT/SNPT) remain to be uncovered. In this study, we combined metabolomics and clinical indicators with machine learning to screen out newly diagnostic biomarkers for the precise identification of SPPT and SNPT patients.
Methods
Untargeted plasma metabolomic profiling was performed for 27 SPPT patients, 37 SNPT patients and controls. The orthogonal partial least squares-discriminant analysis (OPLS-DA) was then conducted to screen differential metabolites among the three groups. Metabolite enriched pathways, random forest (RF), support vector machines (SVM) and multilayer perceptron neural network (MLP) were performed using Metaboanalyst 5.0, “caret” R package, “e1071” R package and “Tensorflow” Python package, respectively.
Results
Metabolomic analysis revealed significant enrichment of fatty acid and amino acid metabolites in the plasma of SPPT and SNPT patients, where SPPT samples showed a more serious dysfunction in fatty acid and amino acid metabolisms. Further RF analysis revealed four optimized diagnostic biomarker combinations including ten features (two lipid/lipid-like molecules and seven organic acids/derivatives, and one clinical indicator) for the identification of SPPT, SNPT patients and controls with high accuracy (83–93%), which were further verified by SVM and MLP. Among them, MLP displayed the best classification performance on simultaneously precise identification of the three groups (94.74%), suggesting the advantage of MLP over RF/SVM to some extent.
Conclusions
Our findings reveal plasma metabolomic characteristics of SPPT and SNPT patients, provide some novel promising diagnostic markers for precision diagnosis of various types of TB, and show the potential of machine learning in screening out biomarkers from big data.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-022-07694-8.
Keywords: Tuberculosis (TB), Mycobacterium tuberculosis (Mtb), Smear-positive/negative pulmonary tuberculosis, Diagnostic biomarkers, Random forest, Machine learning, Metabolome, Metabolite
Background
According to WHO reports, tuberculosis (TB) caused by mycobacterium tuberculosis (Mtb) had been the leading lethal infectious disease worldwide for a long time (2014–2019) until the COVID-19 global pandemic (2020–2021) [1], and there were ∼ 10 million new TB cases every year [2, 3]. According to the data collected from the National Notifiable Disease Reporting System (NNDRS), the annual incidence of Xinjiang is 169.05/100,000 and the mean annual rate of reported PTB (pulmonary tuberculosis) in Kashgar was 450.91/100,000 from 2011 to 2020 [4]. Why are there so many TB patients and death cases around the world? One reason is because of the difficulties in precise diagnosis of TB, especially for some smear-negative pulmonary tuberculosis (SNPT) cases that usually show similar symptoms to other lung diseases [5, 6]. In some countries/regions, SNPT patients even account for more than 50% of all TB cases [7].
At present, although three common methods (sputum-smear microscopy, sputum culture tests and Xpert MTB/RIF assays) can achieve relatively precise diagnosis for most TB patients, they still have some disadvantages (such as relatively low sensitivity for sputum-smear microscopy, time-consuming for sputum culture, and relatively high cost for Xpert), further leading to some false negative/positive cases [1, 6, 8–10]. The failure diagnosis may result in delayed treatment, poor therapeutic effect and higher treatment costs [11, 12]. Nowadays, how to timely and accurately detect various types of TB remains a substantial challenge for global TB control.
The rapid development of various omics technologies offers a great opportunity for precision diagnosis of various types of diseases [13–16]. Among them, metabolome has been widely applied in biomarker discovery for the detection, diagnosis and treatment of various diseases, since they have been reported to be closely associated with disease genotypes and phenotypes [17]. In the TB research field, Deng et al. reported significantly changed glutathione and histamine in the urine of active TB patients, which could distinguish them from latent tuberculosis infected patients [18]; Huang et al.provided some potential plasma metabolite biomarkers (Xanthine, 4-Pyridoxate, and d-glutamic acid) for TB diagnosis [19]; Sun et al.revealed some potential metabolite biomarkers for pediatric TB diagnosis by l-valine, pyruvic acid and betaine in plasma [20]. However, the metabolite biomarkers for precision diagnosis of smear-positive and smear-negative tuberculosis (SPPT and SNPT) remain to be uncovered.
In our study, we performed plasma metabolomic analyses from 27 SPPT patients, 37 SNPT patients and 36 controls. Metabolomic profiling revealed dysfunctional fatty acid and amino acid metabolisms in SPPT and SNPT patients. Four optimized diagnostic biomarker combinations (two lipid/lipid-like molecules and seven organic acids/derivatives, and one clinical indicator) were then screened out for precise diagnosis of SPPT and SNPT patients and controls through the random forest (RF). The classification performance of the four combinations was further verified by other two machine learning methods: support vector machines (SVM) and multilayer perceptron neural network (MLP). Our findings revealed the metabolomic characteristics of SPPT and SNPT patients, provided some promising diagnostic markers for precision diagnosis of various types of TB patients, and showed the potential of machine learning in the detection of diagnostic biomarkers.
Methods
Study participates
In our study, all the TB patients (including 27 SPPT and 37 SNPT patients) were recruited from the Tuberculosis Prevention and Treatment Institute of Kashgar, the Second People’s Hospital of Aksu, and the Kuqa County Infectious Disease Hospital during October 2017 to October 2018. 36 control people (Ctrl) without TB infection from the First Affiliated Hospital of Xinjiang Medical University were also enrolled (Table 1, Additional file 1: Fig. S1). The diagnosis of TB was based on clinical symptoms and microbiological evidence according to Diagnosis for Pulmonary Tuberculosis (WS 288-2017). SPPT patients were diagnosed when one of the following microbiological evidence was obtained: (1) positive stain for acid-fast bacilli, (2) positive culture for Mtb, (3) positive Xpert test. SNPT patients were diagnosed based on the classical clinical symptoms although acid-fast bacilli were negative. The exclusion criteria included: (1) the TB patients in treatment period; (2) the TB patients with other chronic or acute diseases such as pregnancy complications, cardiac dysfunction, renal disease, psychiatric disease, gastrointestinal disease, uncontrolled hypertension, and some severe stress states (including cardiovascular and cerebrovascular events, severe infection, traumatic surgery, and severe wasting diseases). This study was approved by the Ethical Committee of First Affiliated Hospital of Xinjiang Medical University (20171123-06-1908A).
Table 1.
Baseline characteristics of SPPT and SNPT patients
| Total (N = 100) | SPPT (N = 27) | SNPT (N = 37) | Control (N = 36) | *Adjusted p-value (SPPT/Ctrl) | *Adjusted p-value (SNPT/Ctrl) | *Adjusted p-value (SPPT/SNPT) | χ2, F or H value (SPPT/SNPT/Ctrl) | p-value (SPPT/SNPT/Ctrl) | |
|---|---|---|---|---|---|---|---|---|---|
| Gender (%) | 0.162 | 0.43 | 0.449 | 4.868$ | 0.088 | ||||
| Male | 52 (52.0) | 18 (66.7) | 20 (54.1) | 14 (38.9) | |||||
| Female | 48 (48.0) | 9 (33.3) | 17 (45.9) | 22 (61.1) | |||||
| Age (years, median [Q1–Q3]) | 53.50 (35.00–67.25) | 51.00 (32.50–71.00) | 60.00 (49.00–71.00) | 43.50 (34.00–59.25) | 0.087 | 0.011 | 0.315 | 6.369 | 0.041 |
| Occupations (%) | – | – | 0.934 | – | – | ||||
| Farmer | 54 (54.0) | 23 (85.2) | 31 (83.8) | 0 (0.0) | |||||
| Retiree | 11 (11.0) | 1 (3.7) | 1 (2.7) | 9 (25.0) | |||||
| Student | 4 (4.0) | 1 (3.7) | 3 (8.1) | 0 (0.0) | |||||
| Other | 6 (6.0) | 2 (7.4) | 2 (5.4) | 2 (5.6) | |||||
| (Missing value) | 25 (25.0) | 0 (0.0) | 0 (0.0) | 25 (69.4) | |||||
| Marital status (%) | – | – | 0.183 | – | – | ||||
| Single | 7 (7.0) | 5 (18.5) | 2 (5.4) | 0 (0.0) | |||||
| Married | 92 (92.0) | 22 (81.5) | 35 (94.6) | 35 (97.2) | |||||
| (Missing value) | 1 (1.0) | 0 (0.0) | 0 (0.0) | 1 (2.8) | |||||
| BMI (kg/m2, mean[SD]) | 23.19 (4.44) | 20.22 (3.95) | 22.82 (3.93) | 25.33 (4.13) | < 0.001 | 0.022 | 0.027 | 10.02# | < 0.001 |
| Smoking status (%) | – | – | – | – | – | ||||
| Never | 79 (79.0) | 24 (88.9) | 34 (91.9) | 21 (58.3) | |||||
| Current | 11 (11.0) | 3 (11.1) | 2 (5.4) | 6 (16.7) | |||||
| Former | 1 (1.0) | 0 (0.0) | 1 (2.7) | 0 (0.0) | |||||
| (Missing value) | 9 (9.0) | 0 (0.0) | 0 (0.0) | 9 (25.0) | |||||
| Drinking status (%) | – | – | – | – | – | ||||
| Never | 80 (80.0) | 25 (92.6) | 37 (100.0) | 18 (50.0) | |||||
| Current | 11 (11.0) | 2 (7.4) | 0 (0.0) | 9 (25.0) | |||||
| Former | 1 (1.0) | 0 (0.0) | 0 (0.0) | 1 (2.8) | |||||
| (Missing value) | 8 (8.0) | 0 (0.0) | 0 (0.0) | 8 (22.2) | |||||
| TB contact (%) | – | – | < 0.001 | – | – | ||||
| Yes | 44 (68.8) | 8 (29.6) | 36 (97.3) | – | |||||
| No | 16 (25.0) | 16 (59.3) | 0 (0.0) | – | |||||
| (Missing value) | 4 (6.3) | 3 (11.1) | 1 (2.7) | ||||||
| TB treatment (%) | – | – | 0.836 | – | – | ||||
| New cases of TB | 17 (26.6) | 7 (25.9) | 10 (27.0) | – | |||||
| Previously treated | 41 (64.1) | 14 (51.9) | 27 (73.0) | – | |||||
| (Missing value) | 6 (9.4) | 6 (22.2) | 0 (0.0) | ||||||
| Cavitary pulmonary TB (%) | 33 (51.6) | 19 (70.4) | 14 (37.8) | – | – | – | 0.02 | – | |
| Symptoms (%) | |||||||||
| Cough | 60 (92.3) | 22 (81.5) | 37 (100.0) | – | – | – | 0.024 | – | – |
| Expectoration | 60 (92.3) | 22 (81.5) | 37 (100.0) | – | – | – | 0.024 | – | – |
| Dyspnea | 35 (54.7) | 6 (22.2) | 29 (78.4) | – | – | – | < 0.001 | – | – |
| Chest discomfort | 13 (20.3) | 5 (18.5) | 8 (21.6) | – | – | – | 1 | – | – |
| Fever | 5 (7.8) | 3 (11.1) | 2 (5.4) | – | – | – | 0.713 | – | – |
| Hemoptysis | 2 (3.1) | 1 (3.7) | 1 (2.7) | – | – | – | 1 | – | – |
| Chest pain | 2 (3.1) | 1 (3.7) | 1 (2.7) | – | – | – | 1 | – | – |
| Nausea | 1 (1.6) | 0 (0.0) | 1 (2.7) | – | – | – | - | – | – |
| Fatigue | 2 (3.1) | 2 (7.4) | 0 (0.0) | – | – | – | 0.174 | – | – |
| Night sweats | 1 (1.6) | 0 (0.0) | 1 (2.7) | – | – | – | – | – | – |
| Short of breath | 3 (4.7) | 3 (11.1) | 0 (0.0) | – | – | – | – | – | – |
BMI body mass index, Data are shown as n (%), mean (SD) or median (Q1–Q3). p-values are calculated after exclusion of missing data for that variable; *Adjusted p-value for multiple comparisons using Bonferroni-Holm correction. SD: standard deviation; (Q1–Q3): 25th Quartile–75th Quartile. $Chi aquare test; #One Way ANOVA;
Plasma sample preparation
A total of 0.5–1 mL of the whole blood sample from each participant was collected by cubital vein phlebotomy using a heparin anticoagulation collection tube. The blood samples were then centrifuged for 10 min (1500 rpm/min, 4 °C) to remove the blood cells, and the supernatants were immediately frozen in liquid nitrogen and stored at − 80 °C until use. Frozen plasma samples were slowly thawed at 4 °C, and each 100 μL aliquot was mixed with 400 μL of pre-cooled methanol/acetonitrile (1:1, v/v) solution. After the vortex, the mixture was incubated at − 20 °C for 10 min, and then centrifuged for 15 min (14,000 rcf, 4 °C). The supernatants were freeze-dried and reconstituted in 100 μL acetonitrile/water (1:1, v/v) solution for LC–MS/MS analysis (Shanghai Applied protein technology Co., Ltd, Shanghai, China).
Metabolite measurement
Metabolites were extracted from plasma samples. Untargeted metabolomics analysis was conducted by using ultra-high-performance liquid chromatography (UHPLC, 1290 Infinity LC, Agilent Technologies, Palo Alto, CA, USA) and a quadrupole time-of-flight mass spectrometer (TripleTOF 6600; AB Sciex, Framingham, MA, USA). The separation was performed using a 2.1 mm × 100 mm ACQUITY UPLC BEH 1.7 μm column (Waters, Wexford, Ireland). The mobile phase consisted of A. 25 mM ammonium acetate with 25 mM ammonium hydroxide; B. acetonitrile. Gradient elution was performed as follows: 95% B for 0.5 min, and was reduced linearly to 65% in 7 min, next, the gradient was reduced to 40% in 2 min, increased to 95% in 0.1 min, then with a re-equilibration period employed for 3 min. The flow rate was set to 0.3 mL min−1, column temperature at 25 °C and injection volume of 2 µL. The ESI conditions were as follows: Ion Source Gas1(Gas1): 40 psi; Ion Source Gas2 (Gas2): 80 psi; curtain gas (CUR): 30 psi; source temperature: 650℃; IonSpray Voltage Floating (ISVF) ± 5500 V. The raw data were converted to MzXML by MSconventer (ProteoWizard, Palo Alto, CA, USA), and imported into XCMS software (Scripps Research Institute, La Jolla, CA, USA) for alignment, feature detection, retention time correction, and data filtering.
Bioinformatics analysis
Multivariable analysis was conducted using SIMCA-P software (version 14.1 Umetrics, Umea, Sweden). The orthogonal partial least squares-discriminant analysis (OPLS-DA, Umetrics, Umea, Sweden) was then performed to screen the differential metabolites, and the robustness of the OPLS-DA model was evaluated by using the sevenfold cross-validation and response permutation testing. Differentially abundant metabolites (DAMs) were confirmed based on variable importance in projection (VIP) > 1 obtained from the OPLS-DA model and Student’s t-test p values (p < 0.05). The chemical taxonomy of DAMs was determined according to “The Human Metabolome Database (HMDB)” (https://hmdb.ca/). Metabolite enriched pathway analysis was implemented with the online software of Metaboanalyst 5.0 [21].
Data preprocessing
After removing the indicators with a large proportion of missing values (≥ 20%, for details see Additional file 1: Table S1), 24 remaining clinical indicators and 96 DAMs were included to screen out potential diagnostic biomarkers. Categorical variables were then coded with dummy variables. A total of 100 individuals (27 SPPT patients, 37 SNPT patients and 36 controls) were then randomly separated into a training set (n = 81) and a test set (n = 19) using createDataPartition function in R caret package (http://topepo.github.io/caret/data-splitting.html). Further K-Nearest Neighbor was adopted to impute the missing values of the remaining indicators [22]. Specifically, a KNN model (http://topepo.github.io/caret/pre-processing.html) was created based on the training set, which was then applied to predict the missing values in the test set. As a result, the standardized data sets were obtained. Principal component analysis (PCA) was then applied to detect global clinical indicators and metabolic alterations among different samples [23]. Pearson correlation coefficients among the clinical indicators and DAMs were calculated by the findCorrelation function in R software (https://github.com/topepo/caret/blob/master/pkg/caret/R/findCorrelation.R). The features with high mean absolute correlations (≥ 0.7) were excluded (Additional file 2).
Biomarker detection and verification using three machine learning methods (RF, SVM and MLP)
First, the pre-select 20 clinical indicators and 58 identified DAMs (78 features, defined as F0 set) were included for the classification of SPPT/Ctrl, SNPT/Ctrl, SPPT/SNPT and SPPT/SNPT/Ctrl groups. RF was then adopted to evaluate the classification performance of the F0 set. AUCs were calculated by receiver operating characteristic (ROC) analysis using the roc () function of pROC package in R [24].
We then used recursive feature elimination (R package caret) to decrease the number of features in the RF model (parameter use "rfFuncs” and “cv”) [25]. Mean decrease in Gini coefficient (MDG) was further used for measuring variable importance, and the combinations of important features with accuracy over 90% were finally selected for machine learning. Here, the selected features in SPPT/Ctrl, SNPT/Ctrl, SPPT/SNPT and SPPT/SNPT/Ctrl groups were defined as F1, F2, F3 and F4, respectively. Ultimately, the classification accuracies of the above four feature sets were verified by other two machine learning methods: SVM and MLP. The SVM was realized using “e1071” R package. The MLP classification algorithm including two hidden layers (each layer consists of 15 nodes) was completed using the “Tensorflow” package of Python [26]. To avoid overfitting, tenfold cross-validation (CV) was employed on the train set, which was further randomly split into 90% for “actual train set” and 10% for “validation set” for ten times. Ultimately, the test sets were used to evaluate the accuracy, sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of each trained model. The codes were deposited on GitHub (https://github.com/ChenF-Lab/SPPT.git).
Statistical analysis
The continuous variables were described using mean (standard deviation), median and interquartile ranges (Q1–Q3). The categorical variables were described as frequency rates and percentages. Independent samples t-test was used for comparing means of normally distributed variables while Mann Whitney U test for not normally distributed variables. One-Way ANOVA or Kruskal Wallis test were used to compare variables among three groups. Categorical variables were compared using the chi-square test. Bonferroni-Holm correction was applied to obtain the corrected p-value for multiple comparisons. All the statistical analyses were performed using R software (version 4.0.2; an open-source free software). Two-sided p values of less than 0.05 were considered statistically significant.
Results
Demographics and clinical characteristics of the SPPT and SNPT patients
In our study, 64 TB patients, including 27 SPPT patients and 37 SNPT patients, were enrolled to identify the biomarker candidates for tuberculosis diagnosis. 36 non-TB individuals were also included as controls. Here, the majority of TB patients are males (59.4%), and more than 80% of TB patients are farmers. The median age of SNPT patients was 60.0 years old (Q1–Q3: 49.00–71.00), which was significantly higher than that of SPPT patients (51.0 years old, Q1–Q3: 32.50–71.00) and controls (43.5 years old, Q1–Q3: 34.00–59.25). The mean BMIs of SPPT and SNPT patients were 20.22 kg/m2 (SD: 3.95) and 22.82 kg/m2 (SD: 3.93), respectively, which were significantly lower than controls (p < 0.001). The common symptoms were cough (92.3%) and expectoration (92.3%), followed by dyspnea (54.7%) and chest discomfort (20.3%). Notably, 70.4% of SPPT patients belong to cavitary pulmonary TB which has been previously demonstrated to be associated with higher bacterial load [27] (Table 1).
Clinical characteristic analysis showed significantly decreased albumin and serum creatinine, and increased erythrocyte sedimentation rate (ESR) for the TB patients (Table 2). Here, the albumin of SPPT patients was significantly lower than that of SNPT patients (SPPT: 35.30 g/L; SNPT: 39.20 g/L; adjusted p = 0.002), indicating more serious chronic inflammation/malnutrition for the SPPT patients [28, 29]; the serum creatinine was significantly lower in TB patients compared with controls, but showed no difference between SPPT and SNPT patients, suggesting renal injury induced by tuberculous drugs; the ESR of SPPT patients (67.50 mm/h) was significantly higher than that of SNPT patients (43.00 mm/h), and ESR had been reported to identify active tuberculosis and differentiate pulmonary tuberculosis from bacterial community-acquired pneumonia [30].
Table 2.
Clinical indicators of SPPT and SNPT patients
| Normal range | SPPT | SNPT | Control | *Adjusted p-value (SPPT/Ctrl) | *Adjusted p-value (SNPT/Ctrl) | *Adjusted p-value (SPPT/SNPT) | H value (SPPT/SNPT/Ctrl) | p-value (SPPT/SNPT/Ctrl) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | Median (Q1–Q3) | Patients | Median (Q1–Q3) | Patients | Median (Q1–Q3) | |||||||
| Blood routine | ||||||||||||
| Leucocytes, × 109/L | 3.5–9.5 | 27 | 7.20 (5.89–8.61) | 37 | 7.65 (6.32–8.74) | 36 | 6.49 (5.66–7.54) | 0.009 | 0.032 | 0.413 | 6.86 | 0.032 |
| Neutrophils, × 109/L | 1.8–6.3 | 10 | 7.83 (4.62–11.61) | 37 | 5.10 (3.92–5.94) | 36 | 3.82 (3.18–4.51) | < 0.001 | < 0.001 | 0.068 | 19.6 | < 0.001 |
| Erythrocytes, × 1012/L | 4.3–5.8 | 27 | 4.28 (3.88–4.66) | 37 | 4.78 (4.35–4.98) | 36 | 4.77 (4.46–5.25) | < 0.001 | 0.682 | 0.003 | 14.06 | 0.001 |
| Hemoglobin, g/L | 130–175 | 27 | 117.00 (104.50–134.00) | 37 | 139.00 (132.00–151.00) | 36 | 140.00 (132.00–151.50) | < 0.001 | 0.961 | < 0.001 | 29.01 | < 0.001 |
| Platelets, × 109/L | 125–300 | 26 | 305.50 (238.50–371.75) | 37 | 225.00 (201.00–279.00) | 36 | 265.00 (234.50–348.25) | 0.15 | 0.003 | 0.003 | 10.31 | 0.006 |
| Eosinophils, × 109/L | 0.02–0.52 | 27 | 0.12 (0.04–0.34) | 37 | 0.15 (0.10–0.22) | 36 | 0.11 (0.07–0.24) | 0.12 | 0.174 | 0.12 | 0.61 | 0.739 |
| Basophils, × 109/L | 0–0.06 | 27 | 0.02 (0.00—0.07) | 37 | 0.02 (0.01–0.05) | 36 | 0.02 (0.01—0.02) | < 0.001 | 0.005 | < 0.001 | 0.73 | 0.693 |
| Blood biochemistry | ||||||||||||
| Total protein, g/L | 65–85 | 27 | 66.50 (58.75–0.30) | 37 | 66.30 (62.70–68.70) | 34 | 73.40 (70.93–77.18) | < 0.001 | < 0.001 | 0.402 | 31.68 | < 0.001 |
| Albumin, g/L | 40–55 | 27 | 35.30 (31.00–38.75) | 37 | 39.20 (36.00–43.00) | 34 | 44.59 (42.73–45.66) | < 0.001 | < 0.001 | 0.002 | 47.21 | < 0.001 |
| Globulin, g/L | 20–40 | 16 | 30.65 (26.00–35.75) | 37 | 26.40 (24.00–29.30) | 34 | 28.53 (26.36–32.54) | 0.189 | 0.087 | 0.043 | 7.63 | 0.022 |
| Triglyceride, mmol/L | 0.5–1.9 | 25 | 0.99 (0.80–1.22) | 35 | 1.14 (0.89–1.60) | 35 | 1.08 (0.76–1.63) | 0.83 | 0.696 | 0.696 | 1.8 | 0.408 |
| Total cholesterol, mmol/L | 2.3–5.2 | 25 | 3.45 (2.87–3.69) | 35 | 3.63 (3.27–4.58) | 35 | 4.15 (3.76–4.98) | < 0.001 | 0.048 | 0.302 | 12.12 | 0.002 |
| ASP, IU/L | 9–60 | 27 | 23.00 (18.00–36.50) | 37 | 21.00 (18.00–31.00) | 35 | 19.80 (17.65–22.30) | 0.014 | 0.043 | 0.508 | 3.75 | 0.153 |
| ALT, IU/L | 9–50 | 27 | 21.50 (12.55–43.15) | 37 | 19.00 (15.00–29.00) | 35 | 24.20 (15.20–30.40) | 0.269 | 0.269 | 0.586 | 0.51 | 0.776 |
| AKP, IU/L | 45–125 | 27 | 68.00 (53.65–102.50) | 37 | 98.00 (74.00–129.00) | 32 | 76.85 (65.80–89.80) | 0.155 | 0.005 | 0.005 | 10.13 | 0.006 |
| γ-GT, IU/L | 10–60 | 27 | 33.70 (22.00–55.00) | 37 | 26.00 (20.00–59.00) | 32 | 21.50 (15.75–33.50) | 0.189 | 0.189 | 0.559 | 3.79 | 0.15 |
| Creatinine, μmol/L | 57–97 | 27 | 55.20 (44.80–70.70) | 37 | 54.00 (47.00–72.00) | 36 | 65.77 (60.15–74.81) | 0.025 | 0.022 | 0.961 | 7.93 | 0.019 |
| Total bilirubin, μmol/L | 0–26 | 27 | 9.60 (7.90–11.85) | 37 | 11.93 (9.20–19.60) | 34 | 12.13 (10.27–13.95) | 0.042 | 0.123 | 0.042 | 6.61 | 0.037 |
| Direct bilirubin, μmol/L | 0–8 | 27 | 3.39 (2.30–4.50) | 37 | 2.18 (0.30–3.30) | 6 | 2.31 (2.12–3.82) | 0.421 | < 0.001 | < 0.001 | 6.48 | 0.039 |
| Indirect bilirubin, μmol/L | 0–14 | 15 | 6.30 (4.23—8.37) | 37 | 7.90 (6.11–12.15) | 6 | 6.40 (6.20–6.55) | 0.402 | 0.115 | 0.115 | 4.53 | 0.104 |
| Inflammatory-related biomarkers | ||||||||||||
| ESR, mm/h | 0–15 | 26 | 67.50 (43.75–94.25) | 35 | 43.00 (14.00–62.00) | 0 | – | – | – | 0.003 | – | |
| C-reaction protein, mg/L | 0–4 | 27 | 16.53 (9.69–65.08) | 33 | 1.67 (0.80–3.67) | 0 | – | – | – | < 0.001 | – | |
| Procalcitonin, ng/mL | 0–0.05 | 15 | 0.10 (0.07–0.36) | 34 | 0.02 (0.01—0.11) | 0 | – | – | – | 0.013 | – | |
Data are shown as median (Q1–Q3); Missing data of variables are omitted here and showed in the Additional file 1: Table S1; *Adjusted p-value for multiple comparisons using Bonferroni-Holm correction. SD: standard deviation; (Q1–Q3): 25th Quartile—75th Quartile. ASP: aspartate aminotransferase; ALT: alanine aminotransferase; AKP: Alkaline phosphatase; γ-GT: γ-glutamyl transpeptidase; ESR: Erythrocyte sedimentation rate
Additionally, neutrophils, C-reactive protein and procalcitonin were significantly upregulated in SPPT patients than in SNPT ones, while the hemoglobin of SPPT patients was significantly downregulated than that of SNPT ones. These indicators were all in the normal range for the SNPT patients, reflecting stronger immune and inflammatory reactions of SPPT patients.
Plasma metabolomic analysis showing dysfunctional fatty acid and amino acid metabolisms in SPPT and SNPT patients
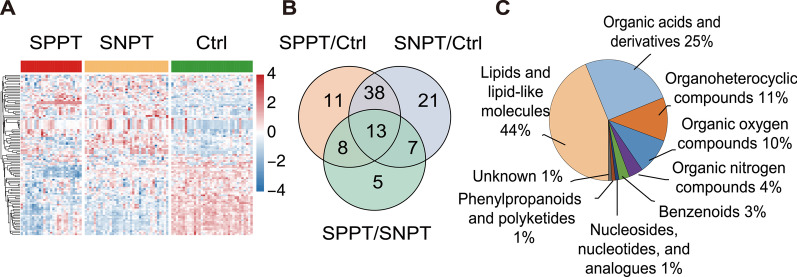
Metabolome analysis was performed on the plasma samples from SPPT, SNPT and Ctrl groups, and a total of 103 DAMs were identified (Fig. 1A, B and Additional files 3, 4, 5). The heatmap showed the DAM expression profiles for the three groups, and the metabolomic profiling of SPPT patients was more similar to that of SNPT patients rather than controls (Fig. 1A). We then classified all DAMs into nine categories based on their chemical taxonomy according to “The Human Metabolome Database” (https://hmdb.ca/), including “Lipids and lipid-like molecules” (~ 44%), “Organic acids and derivatives” (~ 25%), “Organoheterocyclic compounds” (12%) and “Organic oxygen compounds” (~ 10%) (Fig. 1C).
Fig. 1.
Plasma metabolomic analysis for the SPPT patients, SNPT patients and controls. A Heatmap showing 103 differential abundant metabolites (DAMs, VIP > 1, p < 0.05) among the three groups. The colored bar above the heatmap represent the SPPT (red), SNPT (orange) and Ctrl (green) samples. The color key indicates the scaled expression levels of the 103 metabolites for the three groups. B Venn diagram showing the differential metabolites among the three groups. C Pie chart showing the chemical classification of the 103 significantly differentially abundant metabolites according to the HMDB database
In the SPPT/Ctrl group, 70 DAMs were identified, most of which were lipids/lipid-like molecules (31) and organic acids/derivatives (16) (Additional file 3). Compared with controls, 77% (24/31) of the lipids/lipid-like molecules (19 fatty acyls, 3 glycerophospholipids, etc.) and 81.5% (13/16) of the organic acids/derivatives showed significantly down-regulated trend (FC < 1, p < 0.05) in the SPPT group, indicating the dysfunctional lipid and amino acid metabolisms in the SPPT patients as previously reported [31, 32].
In the SNPT/Ctrl group, 79 DAMs were obtained, most of which also belonged to lipid/lipid-like molecules (37, top-1) and organic acids/derivatives (16, top-2) (Additional file 4). Compared to controls, 73% (27/37) of lipids/lipid-like molecules and 56% (9/16) organic acids/derivatives showed significantly down-regulated trend in the SNPT samples, also indicating the dysfunctional lipid and amino acid metabolisms in the SNPT patients.
In the SPPT/SNPT group, 33 DAMs were identified, most of which also belonged to lipid/lipid-like molecules (17) and organic acids/derivatives (10) (Additional file 5): 53% (9/17) of the lipid/lipid-like molecules were significantly downregulated (4 fatty acyls, 2 glycerophospholipids, 2 prenol lipids, etc.), and 47% (8/17) of them were significantly up-regulated (5 fatty acyls and 3 steroids/steroid derivatives); 90% (9/10) of the organic acids/derivatives were significantly down-regulated (eight carboxylic acids and derivatives and one organic carbonic acid/derivative), and only one was significantly up-regulated (hydroxy acid/derivative).
In all, the three groups (SPPT/Ctrl, SPPT/Ctrl and SPPT/SNPT) showed significant enrichments in lipids/lipid-like molecules (top-1) and organic acids/derivatives (top-2).
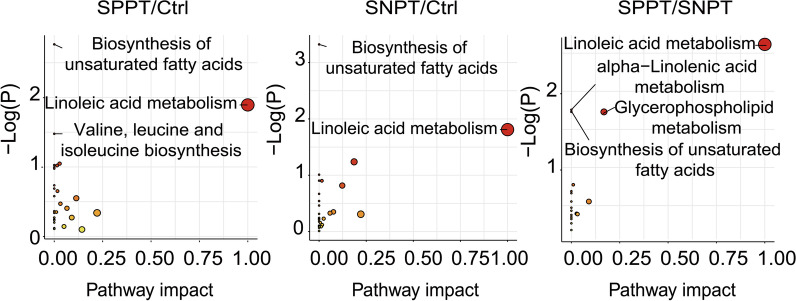
To evaluate the metabolic characteristics of the three groups, we further performed the pathway analysis for these DAMs using MetaboAnalyst 5.0. The results showed significantly differential enrichment of lipid and amino acid metabolism related pathways among the three groups (Fig. 2 and Additional file 1: Table S2–S4). In the SPPT/Ctrl group, the DAMs were significantly enriched in two fatty acid metabolism related pathways (“Biosynthesis of unsaturated fatty acids pathway”, “Linoleic acid metabolism pathway”) and one amino acid metabolism related pathway (“Valine, leucine and isoleucine biosynthesis pathway”), indicating significantly unregulated unsaturated fatty acid and amino acid metabolisms in the SPPT samples as previously reported [32–35]. In the SNPT/Ctrl group, the DAMs were significantly enriched in the same two fatty acid-related pathways as those in the SPPT/Ctrl group. In the SPPT/SNPT group, four lipid-related metabolic pathways, including “Linoleic acid metabolism pathway”, “Glycerophospholipid metabolism pathway”, “alpha-Linolenic acid metabolism pathway” and “Biosynthesis of unsaturated fatty acids pathway”, were significantly enriched, indicating more serious dysfunction of fatty acid metabolisms in the SPPT patients than in the SNPT patients. Overall, the two significant enrichment unsaturated fatty acid metabolism related pathways were shared by the three groups (SPPT/Ctrl, SNPT/Ctrl and SPPT/SNPT), indicting similar dysfunctional fatty acid metabolisms among the three groups; they should be associated with disease progress of TB.
Fig. 2.
Scatter plots showing the significantly enriched metabolic pathways among the three groups. The size and color of circles indicate the impact score and p-value of the enriched pathways, respectively
Taken together, the above results showed the dysfunctions of fatty acid and amino acid metabolisms in the SPPT and SNPT patients, where these dysfunctions in the SPPT patients were more serious than those in the SNPT patients.
Precise classification among the three groups using DAMs and clinical indicators
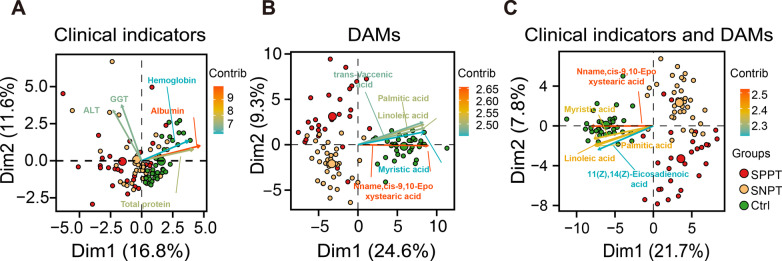
We then investigated the classification effect for the three groups (SPPT, SNPT patients and controls) using all the practicable clinical laboratory indicators (24) and DAMs (96). Here, seven drug-related metabolites (Dehydroabietic acid, Dyphylline, EDTA, Levofloxacin, Norethindrone Acetate, Sunitinib and Thioetheramide-PC) were excluded to increase the general applicability of classification according to HMDB database [36, 37]. PCA analysis was first applied to explore whether clinical indicators and DAMs could be used to distinguish the SPPT, SNPT and Control samples (Fig. 3): DAMs displayed obvious separation while clinical indicators showed poor separation among the three groups; clinical indicators combined with DAMs showed the best classification performance among the three groups. Here the top ten contributed variables of PC1 and PC2 are all belong to DAMs, indicating a greater contribution of DAMs than clinical indicators (Additional file 1: Fig. S2).
Fig. 3.
Principal component analyses of clinical indicators (A), DAMs (B) and their combination (C) among SPPT, SNPT and controls. The color key indicates the contribution of the top 5 variables from high (reddish arrows) to low contribution (bluish arrows)
All the 120 features (24 clinical indicators and 96 DAMs) were further calculated for correlation coefficients between pairwise features (Additional file 2). 42 features were excluded due to their higher mean absolute correlation coefficients (≥ 0.7), and the remaining 78 features were denoted as F0 set for classification analysis among the three groups. RF and ROC analyses were then used to evaluate the classification performance of the 78 features for the SPPT/Ctrl, SNPT/Ctrl, SPPT/SNPT and SPPT/SNPT/Ctrl groups. The results showed the tenfold cross-validation average accuracy of 98% (SD: 0.06), 100% (SD: 0.00) and 92% (SD: 0.09) for the binary classifications of the SPPT/Ctrl, SNPT/Ctrl and SPPT/SNPT groups in validation sets, respectively (Additional file 1: Table S5). Further, 100% accuracy (AUC: 1.00) was obtained for all the binary classifications of in test sets (Additional file 1: Fig. S3). For the three-class classification of SPPT/SNPT/Ctrl group, the 78 features also showed good classification performance in validation sets (average accuracy: 95% (SD: 0.09) and test set (accuracy: 94.74%; sensitivity: 80%, 100% and 100% for SPPT, SNPT and control groups; and specificity: 100%, 91.67%, and 100% for SPPT, SNPT and control groups; PPV: 100%, 87.50% and 100% for SPPT, SNPT and control groups; NPV: 93.33%, 100% and 100% for SPPT, SNPT and control groups;). These indicated the precise classification among the SPPT and SNPT patients and controls using the combination of clinical indicators and DAMs (F0).
Selecting the optimized biomarker combinations to precisely identify any one of the SPPT and SNPT patients and controls.
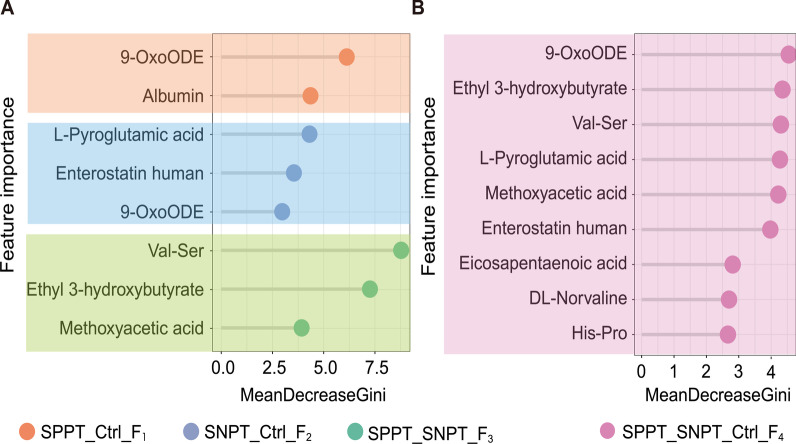
To explore the optimized diagnostic biomarker combinations, we then evaluated the contribution of features to the classification using random forest algorithm. The results revealed the optimized biomarker combinations with higher accuracy (> 0.9, Additional file 1: Fig. S4) for precision binary and three-class classifications among the three groups in training sets, including a two biomarker combination (albumin and 9-OxoODE, defined as “F1 set”) for precisely distinguishing SPPT from controls, a three biomarker combination (L-Pyroglutamic acid (PGA), Enterostatin human and 9-OxoODE, defined as “F2 set”) for precisely differentiating SNPT from controls, a three biomarker combination (Val-Ser, Methoxyacetic acid (MAA) and Ethyl 3-hydroxybutyrate, defined as “F3 set”) for precisely distinguishing SPPT from SNPT, and a nine biomarker combination (9-OxoODE, PGA, Val-Ser, Ethyl 3-hydroxybutyrate, MAA, Enterostatin human, DL-Norvaline, His-Pro and Eicosapentaenoic acid (EPA), defined as “F4 set”) for simultaneously precise identification of SPPT and SNPT patients and controls (Fig. 4, Additional file 1: Table S6).
Fig. 4.
Importance of the screened features for identifying SPPT, SNPT patients from controls. A Importance of the clinical and metabolic features from different optimized combinations for precisely binary classification of SPPT/Ctrl, SNPT/Ctrl and SPPT/SNPT groups (from top to bottom) using random forest model. B Importance of the clinical and metabolic features from the four optimized combinations for simultaneous classification of SPPT, SNPT and Ctrl groups
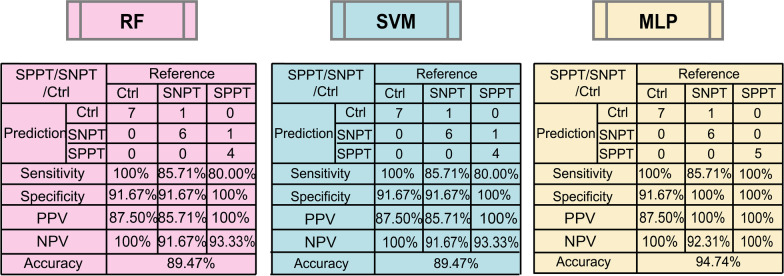
The binary classification performance of the above biomarker combinations (F1, F2 and F3) was further verified in test sets with high accuracy, sensitivity and specificity (accuracy: 83.33% for SPPT/Ctrl classifier, 92.86% for SNPT/Ctrl classifier, 83.33% for SPPT/SNPT classifier; sensitivity: 80.00% for SPPT/Ctrl classifier, 85.71% for SNPT/Ctrl classifier, 80.00% for SPPT/SNPT classifier; specificity: 85.71% for SPPT/Ctrl classifier, 100% for SNPT/Ctrl classifier, 85.71% for SPPT/SNPT classifier; PPV: 80.00% for SPPT/Ctrl classifier, 100% for SNPT/Ctrl classifier, 80.00% for SPPT/SNPT classifier; NPV: 85.71% for SPPT/Ctrl classifier, 87.50% for SNPT/Ctrl classifier, 85.71% for SPPT/SNPT classifier; Table 3). In the SPPT/SNPT/Ctrl group, the optimized biomarker combination (F4: 9 features) could achieve higher three-class classification accuracy (89.47%), sensitivity (80%, 85.71% and 100% for SPPT, SNPT and control groups), specificity (100%, 91.67%, and 91.67% for SPPT, SNPT and control groups), PPV (100%, 85.71% and 87.50% for SPPT, SNPT and control groups) and NPV (93.33%, 91.67% and 100% for SPPT, SNPT and control groups) (Fig. 5). These results demonstrated good performance of the four feature sets (F1–F4) for precise identification of any one of the SPPT and SNPT patients and controls.
Table 3.
Classification performance of binary classifications with selected feature combinations on test sets
| Accuracy | Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|---|
| RF | |||||
| SPPT/Ctrl (F1) | 83.33% | 80.00% | 85.71% | 80.00% | 85.71% |
| SNPT/Ctrl (F2) | 92.86% | 85.71% | 100% | 100% | 87.50% |
| SPPT/SNPT (F3) | 83.33% | 80.00% | 85.71% | 80.00% | 85.71% |
| SVM | |||||
| SPPT/Ctrl (F1) | 91.67% | 80.00% | 100% | 100% | 87.50% |
| SNPT/Ctrl (F2) | 92.86% | 85.71% | 100% | 100% | 87.50% |
| SPPT/SNPT (F3) | 91.67% | 80.00% | 100% | 100% | 87.50% |
| MLP | |||||
| SPPT/Ctrl (F1) | 83.33% | 60.00% | 100% | 100% | 77.78% |
| SNPT/Ctrl (F2) | 92.86% | 85.71% | 100% | 100% | 87.50% |
| SPPT/SNPT (F3) | 91.67% | 80.00% | 100% | 100% | 87.50% |
F1 set: albumin and 9-OxoODE; F2 set: l-Pyroglutamic acid, Enterostatin human and 9-OxoODE; F3 set: Val-Ser, Methoxyacetic acid and Ethyl 3-hydroxybutyrate
Fig. 5.
Confusion matrixes for discriminating SPPT, SNPT and controls with F4 set in the test sets. Confusion matrixes from left to right show the classification performance of SPPT/SNPT/Ctrl groups in the test sets using RF, SVM and MLP models, respectively. F4 set: 9-OxoODE, PGA, Val-Ser, Ethyl 3-hydroxybutyrate, MAA, Enterostatin human, DL-Norvaline, His-Pro and Eicosapentaenoic acid
The other two machine learning methods (SVM and MLP) were further adopted to verify the classification performance of the above-mentioned four biomarker combinations. As expected, the above four biomarker combinations showed high classification accuracy in SVM and MLP methods as that in RF method (Table 3, Fig. 5). Especially, compared with RF and SVM methods, MLP displayed the best classification performance (accuracy: 94.74%; sensitivity: 100%, 85.71% and 100% for SPPT, SNPT and control groups, specificity: 100%, 91.67%, and 100% for SPPT, SNPT and control groups, PPV: 100%, 100% and 87.50% for SPPT, SNPT and control groups and NPV: 100%, 92.31% and 100% for SPPT, SNPT and control groups) for simultaneously discriminating the SPPT and SNPT patients and controls (Fig. 5), indicating the potential in disease classification/diagnosis for MLP.
Discussion
Our study revealed significantly enrichment of lipid/lipid-like molecules and organic acids/derivatives in the SPPT and SNPT patients, indicating the dysfunctional fatty acid and amino acid metabolisms, which is in agreement with previous reports [32–35]. Here, the SPPT samples showed a more serious dysfunction in fatty acid and amino acid metabolisms. Further, four promising diagnostic marker combinations (including nine lipid/lipid-like and organic acids/derivatives molecules and one clinical indicator) were screened out for precise classification of SPPT patients, SNPT patients and controls with high accuracy (83.33–92.86%): a lipid-like molecule combined with a clinical indicator (albumin and 9-OxoODE) could precisely differentiate SPPT patients from controls (accuracy: 83.33%); two lipid/lipid-like and one organic acid molecules (PGA, Enterostatin human and 9-OxoODE) could precisely distinguish SNPT patients from controls (accuracy: 92.86%); three organic acid molecules (Val-Ser, MAA and Ethyl 3-hydroxybutyrate) could precisely classify SPPT and SNPT patients (accuracy: 83.33%); two lipid/lipid-like and seven organic acid molecules (9-OxoODE, PGA, Val-Ser, Ethyl 3-hydroxybutyrate, MAA, Enterostatin human, DL-Norvaline, His-Pro and EPA) could simultaneously precise identify SPPT patients, SNPT patients and controls (accuracy: 89.47%).
As we know, lipids/lipid-like molecules are a type of important structural material of Mtb, especially in the bacterial cell wall [38], which possesses a rich repository of lipid remodeling enzymes to utilize host fatty acids for their survival in the harsh hypoxic microenvironment [39], further, resulting in serious dysfunctional lipid metabolism in TB patients [40]. For amino acid metabolism, since TB is a chronic consumptive disease, various types of amino acids and proteins are essential for Mtb to survive in the human body, thus leading to the dysfunctional amino acid metabolism for TB patients [32]. As expected, our study identified some significantly differential (up-/down-regulated) lipid and amino acid metabolites to precisely discriminate SPPT patients, SNPT patients and controls through machine learning methods. Certainly, these markers and panels warrant further confirmation and optimization with larger sample size studies.
The nine lipid/lipid-like and organic acids/derivatives molecules from four potential diagnostic biomarker combinations include two lipid/lipid-like molecules (9-OxoODE and EPA), and seven organic acids/derivatives (PGA, DL-Norvaline, MAA, His-Pro, Val-Ser, Ethyl 3-hydroxybutyrate and Enterostatin human) (Additional files 3, 4, 5).
First, the two lipid/lipid-like molecules show significant downregulation/inhibition in the SPPT and SNPT patients (Additional file 1: Fig. S5). Here, 9-OxoODE ranks the first, the first and the third in the classification biomarkers for SPPT/Ctrl, SPPT/SNPT/Ctrl and SNPT/Ctrl groups, respectively (Fig. 4). A previous study has shown that the significantly inhibited 9-OxoODE also reflects a negative regulation for lipolysis induced inflammatory response in SPPT and SNPT patients, since 9-OxoODE (metabolite of linoleic acid) can activate the lipogenic machinery as a ligand nuclear receptor in PPAR-α and PPAR-γ [41–44]. Another lipid/lipid-like molecule, EPA ranks seventh in the classification biomarkers for SPPT/SNPT/Ctrl (Fig. 4). Previous studies have reported that significantly downregulated EPA can result in dysfunctional inflammatory responses in TB patients by downregulating the pro-inflammatory cytokines and upregulating lipid synthesis of immune cells [45].
For the abovementioned seven organic acids/derivatives as potential classification biomarkers, compared with controls, three ones (PGA, MAA and DL-Norvaline) show significant downregulation and His-Pro shows significant upregulation in both SPPT and SNPT patients (Additional file 1: Fig. S5). Here, PGA ranks the first and fourth for the classification biomarkers for SNPT/Ctrl and SPPT/SNPT/Ctrl groups, respectively (Fig. 4). Significantly downregulated PGA has been reported to improve the Mtb growth by inhibiting the biosynthesis of glutathione in human bodies [46–49]. MAA ranks third and fifth among the classification biomarkers for SPPT/SNPT and SPPT/SNPT/Ctrl groups, respectively (Fig. 4). Significantly downregulated MAA could result in a poor inhibition of mPTPB essential for the survival of Mtb, since it has been shown to catalyze the formation of an inhibitor of a Mycobacterium protein (tyrosine phosphatase B: mPTPB) [50]. In addition, DL-Norvaline and His-Pro rank the eighth and ninth among the classification biomarkers for SPPT/SNPT/Ctrl group (Fig. 4), both of which showed similar expressed trends, suggesting the dysfunction in both SPPT and SNPT patients.
The remaining three organic acid biomarker molecules (Val-Ser, Ethyl 3-hydroxybutyrate and Enterostatin human) show differential enrichment between the SPPT and SNPT patients. Here Val-Ser and Ethyl 3-hydroxybutyrate show specifically downregulated and upregulated in SPPT patients, respectively (Additional file 1: Fig. S5). They rank the first and second among the features for the differentiation of SPPT/SNPT group, and rank the third and second among the features for the differentiation of SPPT/SNPT/Ctrl group, respectively (Fig. 4). “Enterostatin human” was specifically upregulated in SNPT patients, and ranks the second and sixth among the selected features for the differentiation of SNPT/Ctrl and SPPT/SNPT/Ctrl groups, respectively (Fig. 4). The three organic acids/derivatives with specific changes in only one group display unique feature for the classification of various types of TB patients.
In addition, a clinical indicator of albumin ranks second in the feature set for the differentiation of SPPT/Ctrl group, indicating the better precision diagnosis of SPPT patients through combining metabolome and clinical indicators (Fig. 4). Previous reports have indicated a prognostic marker of TB patients for albumin, which is a critical nutrient and inflammation related protein marker [51].
Our finding further shows the potential of machine learning in the precise diagnosis of SPPT and SNPT patients. Machine learning is becoming ubiquitous for analyzing multi-dimensional big data, and has been widely applied to many biological/medical fields, including diagnostic biomarker identification [52], therapeutic targets detection [53], disease progression prediction [54], and causal relationship between phenotype and genotype [55]. In our study, three machine learning methods are used to screen out potential biomarkers for precise classification of various types of TB from multidimensional data. RF was first adopted to screen out precise classification biomarkers, since it has been widely applied to classification and feature selection for big data; we then obtained some important classification features according to the ranks of variables and their predictive importance. Previous studies have also demonstrated the good performance of RF method for discriminating TB from Non-TB [56]. The other two machine learning methods (SVM and MLP) were further used to verify the classification accuracy of the biomarker combinations. SVM is an ensemble machine learning to improve classification performance compared with a single classifier, which has also been applied in the prediction of disease progression such as breast cancer [57]. MLP is very famous for its autonomic learning capacity without the requirement of previous knowledge, which has also been used in the diagnosis of TB [58] and assessment of prognostic risk for SNPT patients [59]. Our research indicated the best classification performance of MLP for simultaneously identifying the SPPT, SNPT, and controls, with the highest accuracy of 94.74%, suggesting the advantage of MLP over RF and SVM to some extent.
There are also some limitations in our study. Although we have included all the TB patients meeting the inclusion and exclusion criteria in the three hospitals during 2017–2018 (the Tuberculosis Prevention and Treatment Institute of Kashgar, the Second People’s Hospital of Aksu, and the Kuqa County Infectious Disease Hospital), this is indeed a limitation of our study for not calculating the needed sample size as epidemiological survey. The relatively small training and test sets may decrease the statistical power of the results, and this point warrants further confirmation and optimization with larger sample size studies in the future. In addition, we do not observe the impact of the demographic factors (age, occupation, BMI, etc.) on the metabolomic profiles (data not shown), but further confirmation with larger samples is also warranted. Certainly, to translate our classification model into clinical practice, many standardized works about data/workflow/sampling are still required. Overall, all binary and three-class classifiers obtained from our study showed good performance for precisely identifying SPPT, SNPT and Ctrl groups in spite of some limitations, and some classification biomarkers have also been reported to be closely associated with TB [45, 49, 50].
Conclusions
Our current study not only screens out four biomarker combinations for precise detection of SPPT and SNPT patients through combining plasma metabolites with clinical indicators, but also shows promising application of machine learning on the identification of diagnostic biomarkers from multi-dimensional big data.
Over recent decades, despite the rapid advancement of various diagnostic technologies, diagnostic errors (missed, delayed, or wrong diagnoses) are still the most common problems for many important diseases, such as lung cancer [52]. Multi-omics and machine learning provide powerful tools for solving these problems, and researchers can achieve precise classifications/diagnoses for the misdiagnosed diseases through integrating multi-omics data with machine learning [15, 18, 52]. Our research presents a successful attempt to precisely detect various types of TBs by integrating multi-omics data with machine learning, and further provides a good example and workflow for future studies on the precision diagnosis of various misdiagnosed diseases.
Supplementary Information
Additional file 2. Correlation matrix of 120 features.
Additional file 3. Detailed information of the 70 DAMs between the SPPT patients and controls.
Additional file 4. Detailed information of the 79 DAMs between the SNPT patients and controls.
Additional file 5. Detailed information of the 33 DAMs between the SPPT and SNPT patients.
Acknowledgements
The authors wish to acknowledge all the study participants who contributed to this work, as well as the clinical research staff of the participating institutions, who made this research possible.
Abbreviations
- Ctrl
Control people
- CV
Cross validation
- DAMs
Differentially abundant metabolites
- EPA
Eicosapentaenoic acid
- ESR
Erythrocyte sedimentation rate
- IQR
Interquartile range
- KNN
K-nearest neighbor
- MAA
Methoxyacetic acid
- MDG
Mean decrease in Gini coefficient
- MLP
Multilayer perceptron neural network
- Mtb
Mycobacterium tuberculosis
- OPLS-DA
Orthogonal partial least squares-discriminant analysis
- PCA
Principal component analysis
- PGA
L-pyroglutamic acid
- RF
Random forest
- SNPT
Smear-negative pulmonary tuberculosis
- ROC
Receiver operating characteristic
- SPPT
Smear-positive pulmonary tuberculosis
- SVM
Support vector machines
- TB
Tuberculosis
- VIP
Variable importance in projection
Author contributions
FC, JingW, HW, and WBZ, contributed to the conception and design of the study JieW, JYJ and XLZ, performed the bioinformatics analyses XH, BT, YL, YFY and CBC collected blood samples XH, JieW, JYJ, XLZ, CDL and YLY drew the figures FC, JingW, XH, JieW, JYJ, XLZ and QW wrote the manuscript All authors read and approved the final manuscript.
Funding
This research was funded by State Key Laboratory of Pathogenesis, Prevention and Treatment of High Incidence Diseases in Central Asia, Xinjiang Medical University (Grant No. SKL-HIDCA-2021-JH10, SKL-HIDCA-2020-38, SKL-HIDCA-2020-36 and SKL-HIDCA-2020-35), Major Science and Technology Special Project in Xinjiang Uygur Autonomous Region (Grant No. 2017A03006-2), National Natural Science Foundation of China (NSFC) (Grant No. 82060609), Funds for International Cooperation and Exchange of the National Natural Science Foundation of China (Grant No. 32061143024), Key research and development project in Hainan Province (ZDYF2021SHFZ228).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files, further inquiries can be directed to the corresponding authors. Metabolomics data have been deposited to the EMBL-EBI MetaboLights database with the identifier MTBLS3787 [60]. The data and code used for the analysis in this study are available on GitHub (https://github.com/ChenF-Lab/SPPT.git).
Declarations
Ethics approval and consent to participate
This study was approved by the Ethical Committee of First Affiliated Hospital of Xinjiang Medical University (Record number 20171123-06-1908A) and project supported by Hainan Province Clinical Medical Center. All enrolled subjects provided written informed consent. All methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xin Hu, Jie Wang, Yingjiao Ju and Xiuli Zhang contributed equally to this work
Contributor Information
Jing Wang, Email: tlfwj@163.com.
Fei Chen, Email: chenfei@big.ac.cn.
References
- 1.World Health Organization. Global tuberculosis report 2021. Geneva: World Health Organization; 2021. https://www.who.int/teams/global-tuberculosis-programme/data.
- 2.Bussi C, Gutierrez MG. Mycobacterium tuberculosis infection of host cells in space and time. FEMS Microbiol Rev. 2019;43(4):341–361. doi: 10.1093/femsre/fuz006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang H, Ding N, Yang T, Li C, Jia X, Wang G, et al. Cross-sectional Whole-genome sequencing and epidemiological study of multidrug-resistant Mycobacterium tuberculosis in China. Clin Infect Dis. 2019;69(3):405–413. doi: 10.1093/cid/ciy883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tusun D, Abulimiti M, Mamuti X, Liu Z, Xu D, Li G, et al. The epidemiological characteristics of pulmonary tuberculosis—Kashgar Prefecture, Xinjiang Uygur Autonomous Region, China, 2011–2020. China CDC Wkly. 2021;3(26):557–561. doi: 10.46234/ccdcw2021.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lv L, Li C, Zhang X, Ding N, Cao T, Jia X, et al. RNA Profiling analysis of the serum exosomes derived from patients with active and latent Mycobacterium tuberculosis infection. Front Microbiol. 2017;8:1051. doi: 10.3389/fmicb.2017.01051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang G, Zhang L, Zhang M, Pan L, Wang F, Huang J, et al. Screening and assessing 11 Mycobacterium tuberculosis proteins as potential serodiagnostical markers for discriminating TB patients from BCG vaccinees. Genom Proteom Bioinf. 2009;7(3):107–115. doi: 10.1016/S1672-0229(08)60039-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campos LC, Rocha MV, Willers DM, Silva DR. Characteristics of patients with smear-negative pulmonary tuberculosis (TB) in a Region with High TB and HIV Prevalence. PLoS ONE. 2016;11(1):e0147933. doi: 10.1371/journal.pone.0147933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steingart KR, Ng V, Henry M, Hopewell PC, Ramsay A, Cunningham J, et al. Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis. 2006;6(10):664–674. doi: 10.1016/S1473-3099(06)70602-8. [DOI] [PubMed] [Google Scholar]
- 9.Chakaya J, Khan M, Ntoumi F, Aklillu E, Fatima R, Mwaba P, et al. Global tuberculosis report 2020—reflections on the global TB burden, treatment and prevention efforts. Int J Infect Dis. 2021;113:S7. doi: 10.1016/j.ijid.2021.02.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorman SE, Schumacher SG, Alland D, Nabeta P, Armstrong DT, King B, et al. Xpert MTB/RIF ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. Lancet Infect Dis. 2018;18(1):76–84. doi: 10.1016/S1473-3099(17)30691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Getahun H, Harrington M, O'Brien R, Nunn P. Diagnosis of smear-negative pulmonary tuberculosis in people with HIV infection or AIDS in resource-constrained settings: informing urgent policy changes. Lancet. 2007;369(9578):2042–2049. doi: 10.1016/S0140-6736(07)60284-0. [DOI] [PubMed] [Google Scholar]
- 12.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363(11):1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olivier M, Asmis R, Hawkins GA, Howard TD, Cox LA. The need for multi-omics biomarker signatures in precision medicine. Int J Mol Sci. 2019;20(19):4781. doi: 10.3390/ijms20194781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang E, Cho WCS, Wong SCC, Liu S. Disease biomarkers for precision medicine: challenges and future opportunities. Genom Proteom Bioinf. 2017;15(2):57–58. doi: 10.1016/j.gpb.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L, Wu J, Shi M, Wang F, Lu H, Liu J et al. New metabolic alterations and predictive marker pipecolic acid in sera for esophageal squamous cell carcinoma. Genom Proteom Bioinf. 2022. [DOI] [PMC free article] [PubMed]
- 16.Li Y, Chen L. Big biological data: challenges and opportunities. Genom Proteom Bioinf. 2014;12(5):187–189. doi: 10.1016/j.gpb.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.German JB, Bauman DE, Burrin DG, Failla ML, Freake HC, King JC, et al. Metabolomics in the opening decade of the 21st century: building the roads to individualized health. J Nutr. 2004;134(10):2729–2732. doi: 10.1093/jn/134.10.2729. [DOI] [PubMed] [Google Scholar]
- 18.Deng J, Liu L, Yang Q, Wei C, Zhang H, Xin H, et al. Urinary metabolomic analysis to identify potential markers for the diagnosis of tuberculosis and latent tuberculosis. Arch Biochem Biophys. 2021;704:108876. doi: 10.1016/j.abb.2021.108876. [DOI] [PubMed] [Google Scholar]
- 19.Huang H, Shi LY, Wei LL, Han YS, Yi WJ, Pan ZW, et al. Plasma metabolites Xanthine, 4-Pyridoxate, and d-glutamic acid as novel potential biomarkers for pulmonary tuberculosis. Clin Chim Acta. 2019;498:135–142. doi: 10.1016/j.cca.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 20.Sun L, Li JQ, Ren N, Qi H, Dong F, Xiao J, et al. Utility of novel plasma metabolic markers in the diagnosis of pediatric tuberculosis: a classification and regression tree analysis approach. J Proteome Res. 2016;15(9):3118–3125. doi: 10.1021/acs.jproteome.6b00228. [DOI] [PubMed] [Google Scholar]
- 21.Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49(W1):W388–W396. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan R, Yang T, Cao J, Lu K, Zhang ZC, et al. Missing data imputation by K nearest neighbours based on grey relational structure and mutual information. Appl Intell. 2015;43:614–632. doi: 10.1007/s10489-015-0666-x. [DOI] [Google Scholar]
- 23.Abdi H, Williams LJ. Principal component analysis. Wiley Interdiscip Rev Comput Stat. 2010;2:433–459. doi: 10.1002/wics.101. [DOI] [Google Scholar]
- 24.Bewick V, Cheek L, Ball J. Statistics review 13: receiver operating characteristic curves. Crit Care. 2004;8:508. doi: 10.1186/cc3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breiman L. Random forests. Mach Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 26.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. Tensor flow: large-scale machine learning on heterogeneous systems. 2015. Available online at: tensorflow.org.
- 27.Palaci M, Dietze R, Hadad DJ, Ribeiro FK, Peres RL, Vinhas SA, et al. Cavitary disease and quantitative sputum bacillary load in cases of pulmonary tuberculosis. J Clin Microbiol. 2007;45(12):4064–4066. doi: 10.1128/JCM.01780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang W, Wu M, Yang K, Ertai A, Wu S, Geng S, et al. Factors associated with negative T-SPOT.TB results among smear-negative tuberculosis patients in China. Sci Rep. 2018;8(1):4236. doi: 10.1038/s41598-018-22495-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakao M, Muramatsu H, Arakawa S, Sakai Y, Suzuki Y, Fujita K, et al. Immunonutritional status and pulmonary cavitation in patients with tuberculosis: a revisit with an assessment of neutrophil/lymphocyte ratio. Respir Investig. 2019;57(1):60–66. doi: 10.1016/j.resinv.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Berhane M, Melku M, Amsalu A, Enawgaw B, Getaneh Z, Asrie F. The role of neutrophil to lymphocyte count ratio in the differential diagnosis of pulmonary tuberculosis and bacterial community-acquired pneumonia: a cross-sectional study at Ayder and Mekelle Hospitals, Ethiopia. Clin Lab 2019, 65(4). [DOI] [PubMed]
- 31.Shvets OM, Shevchenko OS, Todoriko LD, Shevchenko RS, Yakimets VV, Choporova OI, et al. Carbohydrate and lipid metabolic profiles of tuberculosis patients with bilateral pulmonary lesions and mycobacteria excretion. Wiad Lek. 2020;73(7):1373–1376. doi: 10.36740/WLek202007113. [DOI] [PubMed] [Google Scholar]
- 32.Zhang P, Zhang W, Lang Y, Qu Y, Chen J, Cui L. 1H nuclear magnetic resonance-based metabolic profiling of cerebrospinal fluid to identify metabolic features and markers for tuberculosis meningitis. Infect Genet Evol. 2019;68:253–264. doi: 10.1016/j.meegid.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Collins JM, Walker DI, Jones DP, Tukvadze N, Liu KH, Tran VT, et al. High-resolution plasma metabolomics analysis to detect Mycobacterium tuberculosis-associated metabolites that distinguish active pulmonary tuberculosis in humans. PLoS ONE. 2018;13(10):e0205398. doi: 10.1371/journal.pone.0205398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frediani JK, Jones DP, Tukvadze N, Uppal K, Sanikidze E, Kipiani M, et al. Plasma metabolomics in human pulmonary tuberculosis disease: a pilot study. PLoS ONE. 2014;9(10):e108854. doi: 10.1371/journal.pone.0108854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou A, Ni J, Xu Z, Wang Y, Lu S, Sha W, et al. Application of (1)h NMR spectroscopy-based metabolomics to sera of tuberculosis patients. J Proteome Res. 2013;12(10):4642–4649. doi: 10.1021/pr4007359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim E, Kang YG, Kim YJ, Lee TR, Yoo BC, Jo M, et al. Dehydroabietic acid suppresses inflammatory response via suppression of Src-, Syk-, and TAK1-mediated pathways. Int J Mol Sci. 2019;20(7):1593. doi: 10.3390/ijms20071593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kartha S, Yan L, Ita ME, Amirshaghaghi A, Luo L, Wei Y, et al. Phospholipase A2 inhibitor-loaded phospholipid micelles abolish neuropathic pain. ACS Nano. 2020;14(7):8103–8115. doi: 10.1021/acsnano.0c00999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jankute M, Cox JA, Harrison J, Besra GS. Assembly of the mycobacterial cell Wall. Annu Rev Microbiol. 2015;69:405–423. doi: 10.1146/annurev-micro-091014-104121. [DOI] [PubMed] [Google Scholar]
- 39.Srivastava S, Chaudhary S, Thukral L, Shi C, Gupta RD, Gupta R, et al. Unsaturated lipid assimilation by mycobacteria requires auxiliary cis-trans enoyl CoA isomerase. Chem Biol. 2015;22(12):1577–1587. doi: 10.1016/j.chembiol.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 40.Mu J, Yang Y, Chen J, Cheng K, Li Q, Wei Y, et al. Elevated host lipid metabolism revealed by iTRAQ-based quantitative proteomic analysis of cerebrospinal fluid of tuberculous meningitis patients. Biochem Biophys Res Commun. 2015;466(4):689–695. doi: 10.1016/j.bbrc.2015.08.036. [DOI] [PubMed] [Google Scholar]
- 41.Goto T, Lee JY, Teraminami A, Kim YI, Hirai S, Uemura T, et al. Activation of peroxisome proliferator-activated receptor-alpha stimulates both differentiation and fatty acid oxidation in adipocytes. J Lipid Res. 2011;52(5):873–884. doi: 10.1194/jlr.M011320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andres Contreras G, De Koster J, de Souza J, Laguna J, Mavangira V, Nelli RK, et al. Lipolysis modulates the biosynthesis of inflammatory lipid mediators derived from linoleic acid in adipose tissue of periparturient dairy cows. J Dairy Sci. 2020;103(2):1944–1955. doi: 10.3168/jds.2019-17256. [DOI] [PubMed] [Google Scholar]
- 43.Armstrong MM, Diaz G, Kenyon V, Holman TR. Inhibitory and mechanistic investigations of oxo-lipids with human lipoxygenase isozymes. Bioorg Med Chem. 2014;22(15):4293–4297. doi: 10.1016/j.bmc.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattmiller SA, Carlson BA, Gandy JC, Sordillo LM. Reduced macrophage selenoprotein expression alters oxidized lipid metabolite biosynthesis from arachidonic and linoleic acid. J Nutr Biochem. 2014;25(6):647–654. doi: 10.1016/j.jnutbio.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Nienaber A, Baumgartner J, Dolman RC, Ozturk M, Zandberg L, Hayford FEA, et al. Omega-3 fatty acid and iron supplementation alone, but not in combination, lower inflammation and anemia of infection in Mycobacterium tuberculosis-infected mice. Nutrients. 2020;12(9):2897. doi: 10.3390/nu12092897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orlowski M, Meister A. The gamma-glutamyl cycle: a possible transport system for amino acids. Proc Natl Acad Sci U S A. 1970;67(3):1248–1255. doi: 10.1073/pnas.67.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gamarra Y, Santiago FC, Molina-Lopez J, Castano J, Herrera-Quintana L, Dominguez A, et al. Pyroglutamic acidosis by glutathione regeneration blockage in critical patients with septic shock. Crit Care. 2019;23(1):162. doi: 10.1186/s13054-019-2450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balazy M, Kaminski PM, Mao K, Tan J, Wolin MS. S-Nitroglutathione, a product of the reaction between peroxynitrite and glutathione that generates nitric oxide. J Biol Chem. 1998;273(48):32009–32015. doi: 10.1074/jbc.273.48.32009. [DOI] [PubMed] [Google Scholar]
- 49.Ly J, Lagman M, Saing T, Singh MK, Tudela EV, Morris D, et al. Liposomal glutathione supplementation restores TH1 cytokine response to Mycobacterium tuberculosis infection in HIV-infected individuals. J Interferon Cytokine Res. 2015;35(11):875–887. doi: 10.1089/jir.2014.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He R, Zeng LF, He Y, Wu L, Gunawan AM, Zhang ZY. Organocatalytic multicomponent reaction for the acquisition of a selective inhibitor of mPTPB, a virulence factor of tuberculosis. Chem Commun (Camb) 2013;49(20):2064–2066. doi: 10.1039/c3cc38961h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu YR, Yi ZJ, Guan SZ, Zhang SY, Li M. Proteomic analysis of sputum in patients with active pulmonary tuberculosis. Clin Microbiol Infect. 2012;18(12):1241–1247. doi: 10.1111/j.1469-0691.2012.03824.x. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J, Han X, Gao C, Xing Y, Qi Z, Liu R, et al. 5-Hydroxymethylome in circulating cell-free DNA as a potential biomarker for non-small-cell lung Cancer. Genom Proteom Bioinf. 2018;16(3):187–199. doi: 10.1016/j.gpb.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riniker S, Wang Y, Jenkins JL, Landrum GA. Using information from historical high-throughput screens to predict active compounds. J Chem Inf Model. 2014;54(7):1880–1891. doi: 10.1021/ci500190p. [DOI] [PubMed] [Google Scholar]
- 54.Wang J, Xie X, Shi J, He W, Chen Q, Chen L, et al. Denoising autoencoder, a deep learning algorithm, aids the identification of a novel molecular signature of lung adenocarcinoma. Genom Proteom Bioinf. 2020;18(4):468–480. doi: 10.1016/j.gpb.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akkasi A, Moens MF. Causal relationship extraction from biomedical text using deep neural models: a comprehensive survey. J Biomed Inform. 2021;119:103820. doi: 10.1016/j.jbi.2021.103820. [DOI] [PubMed] [Google Scholar]
- 56.Yang Q, Chen Q, Zhang M, Cai Y, Yang F, Zhang J, et al. Identification of eight-protein biosignature for diagnosis of tuberculosis. Thorax. 2020;75(7):576–583. doi: 10.1136/thoraxjnl-2018-213021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang MW, Chen CW, Lin WC, Ke SW, Tsai CF. SVM and SVM ensembles in breast cancer prediction. PLoS ONE. 2017;12(1):e0161501. doi: 10.1371/journal.pone.0161501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Er O, Temurtas F, Tanrikulu AC. Tuberculosis disease diagnosis using artificial neural networks. J Med Syst. 2010;34(3):299–302. doi: 10.1007/s10916-008-9241-x. [DOI] [PubMed] [Google Scholar]
- 59.de Souza Filho JBO, de Seixas JM, Galliez R, de Braganca Pereira B, de Mello FCQ, Dos Santos AM, et al. A screening system for smear-negative pulmonary tuberculosis using artificial neural networks. Int J Infect Dis. 2016;49:33–39. doi: 10.1016/j.ijid.2016.05.019. [DOI] [PubMed] [Google Scholar]
- 60.Haug K, Cochrane K, Nainala VC, Williams M, Chang J, Jayaseelan KV, et al. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020;48(D1):D440–D444. doi: 10.1093/nar/gkz1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 2. Correlation matrix of 120 features.
Additional file 3. Detailed information of the 70 DAMs between the SPPT patients and controls.
Additional file 4. Detailed information of the 79 DAMs between the SNPT patients and controls.
Additional file 5. Detailed information of the 33 DAMs between the SPPT and SNPT patients.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files, further inquiries can be directed to the corresponding authors. Metabolomics data have been deposited to the EMBL-EBI MetaboLights database with the identifier MTBLS3787 [60]. The data and code used for the analysis in this study are available on GitHub (https://github.com/ChenF-Lab/SPPT.git).