Abstract
The genetic basis for the structural diversity of capsule polysaccharide (CPS) in Streptococcus pneumoniae serogroup 19 (consisting of types 19F, 19A, 19B, and 19C) has been determined for the first time. In this study, the genetic basis for the 19A and 19C serotypes is described, and the structures of all four serogroup 19 cps loci and their flanking sequences are compared. Transformation studies show that the structural difference between the 19A and 19F CPSs is likely to be a consequence of differences between their respective polysaccharide polymerase genes (cps19aI and cps19fI). The CPS of type 19C differs from that of type 19B by the addition of glucose. We have identified a single gene difference between the two cps loci (cps19cS), which is likely to encode a glucosyl transferase. The arrangement of the genes within the cps19 loci is highly conserved, with 13 genes (cps19A to -H and cps19K to -O) common to all four serogroup 19 members. These cps genes encode functions required for the synthesis of the shared trisaccharide component of the group 19 CPS repeat unit structures. Furthermore, the genetic differences between the group 19 cps loci identified are consistent with the CPS structures of the individual serotypes. Functions have been assigned to nearly all of the cps19 gene products, based on either gene complementation or similarity to other proteins with known functions, and putative biosynthetic pathways for production of all four group 19 CPSs have been proposed.
Streptococcus pneumoniae (the pneumococcus) is an important cause of invasive disease in human populations throughout the world, resulting in high morbidity and mortality. Control of pneumococcal disease is being complicated by the increasing prevalence of antibiotic-resistant strains and the suboptimal clinical efficacy of existing vaccines. S. pneumoniae produces a polysaccharide capsule, which is essential for virulence because it protects the pneumococcus from the nonspecific immune defenses of the host during an infection (2). All fresh isolates from patients with pneumococcal infection are encapsulated, and spontaneous nonencapsulated (rough) derivatives of such strains are almost completely avirulent.
There are now 90 recognized serotypes of S. pneumoniae (18), each of which produces a structurally distinct capsular polysaccharide (CPS). Classical genetic studies carried out by Austrian et al. (3) demonstrated that the S. pneumoniae genes required for biosynthesis and expression of CPS are closely linked on the pneumococcal chromosome. This fact enabled us to clone and sequence the capsule locus from S. pneumoniae type 19F (designated cps19f) (15, 31). Our studies were initially concentrated on S. pneumoniae type 19F because it is one of the commonest causes of invasive disease in children and because the type 19F CPS is one of the poorest immunogens in this group (11). We have since characterized the type 19B capsule locus (designated cps19b) and the 5′ portion of the type 19A capsule locus (designated cps19a) (32, 33). The immuno-cross-reactive types 19F, 19A, 19B, and 19C are all members of group 19. In one study, group 19 pneumococci accounted for 7% of isolates from cases of invasive disease (40). Of these, 65% were caused by type 19F, 34% were caused by type 19A, and 1% were caused by type 19B; type 19C was a very rare cause of disease in this study.
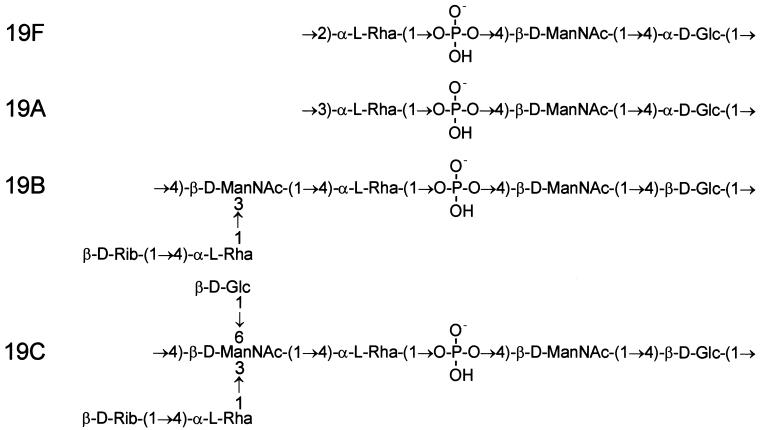
The CPS structures of types 19F and 19A are quite similar, as are those for types 19B and 19C (Fig. 1). However, the latter two have an extra sugar in the backbone and a disaccharide side chain. When compared with cps19f, the cps19b locus has been shown to contain extra genes required for biosynthesis of the more complicated type 19B CPS repeat unit, as well as a different polysaccharide repeat unit transporter and polysaccharide polymerase (32).
FIG. 1.
Biological repeat units of pneumococcal type 19F, 19A, 19B, and 19C CPSs. The orders of the sugars in the repeat units have been altered compared to the published chemical structures for 19F (36), 19A (21), and 19B and 19C (6), reflecting the fact that glucose is the first sugar in the biological repeat unit. d-Glc, glucose; d-ManNAc, N-acetylmannosamine; l-Rha, rhamnose; d-Rib, ribose.
Analysis of purified type 19A CPS has yielded two distinct putative structures. One is the same as type 19F except for a 1→3 linkage (rather than 1→2) between Glc and Rha (Fig. 1) (21). This difference would necessitate an alteration only in the specificity of the polysaccharide polymerase (Cps19fI). The alternative structure involves the same trisaccharide backbone and interunit linkage as type 19F but with additional β-d-GlcpNAc-(1→3)-β-d-Galp-(1-PO4−→2) and α-l-Fucp-(1-PO4−→3) side chains attached to the Glc and Rha, respectively (24). This would necessitate a number of additional enzyme activities not found for the cps locus of type 19F strains. Interestingly, individual type 19A strains were subsequently reported to be capable of producing either structural type, depending on the growth conditions (25). Analysis of the 5′ portion of the cps19a locus revealed that it is similar to cps19f, with the first seven genes arranged in the same order. However, many of these genes have only 70 to 80% nucleotide sequence identity with their cps19f counterpart, suggesting either that the two loci diverged long ago or that portions of these loci have separate origins (33).
The last member of serogroup 19 is type 19C. The cps19c locus is predicted to contain both the extra genes present in cps19b (32) and an additional gene to encode an additional transferase required for the glucose side chain present in the type 19C CPS.
In this study, DNA sequence analysis for both the remainder of the type 19A cps locus and the cps19c locus was undertaken, and in conjunction with transformation studies, the identities of the type 19A- and 19C-specific genes were determined. These data complete the characterization of the genetic loci for all members of S. pneumoniae group 19. The data explain the genetic mechanisms used by S. pneumoniae to generate diversity in CPS structure and are of relevance to the evolution of other S. pneumoniae cps loci.
MATERIALS AND METHODS
Bacterial strains and plasmids.
S. pneumoniae Rx1-19F-I, an unencapsulated insertion-duplication mutant (in which the cps19fI gene has been interrupted) of Rx1-19F (a derivative of Rx1 expressing type 19F capsule), was constructed as described elsewhere (31). A clinical isolate of S. pneumoniae type 19A, strain 1777/39, was obtained from Jorgen Henrichsen, Statens Seruminstitüt, Copenhagen, Denmark, and was designated 19A1. Clinical isolates of S. pneumoniae type 19A (designated 19A2) and 19C were obtained from Chi-Jen Lee, Center for Biologics, Food and Drug Administration, Bethesda, Md. Six Australian clinical isolates of S. pneumoniae type 19A were obtained from Mike Gratten, Acute Respiratory Infections Research and Reference Unit, Centre for Public Health Sciences, Queensland Health, Brisbane, Australia. All other clinical isolates were from the Women’s and Children’s Hospital, Adelaide, South Australia, Australia. Pneumococci were routinely grown in Todd-Hewitt broth with 0.5% yeast extract or on blood agar. Where appropriate, erythromycin was added to media at a concentration of 0.2 μg/ml.
Escherichia coli K-12 DH5α (Bethesda Research Laboratories, Gaithersburg, Md.) was grown in Luria-Bertani broth (27) with or without 1.5% (wt/vol) Bacto-agar (Difco Laboratories, Detroit, Mich.). Where appropriate, ampicillin was added to the growth medium at a concentration of 50 μg/ml.
The vector pBluescript KS(+) was obtained from Stratagene, La Jolla, Calif.
Bacterial transformation.
Transformation of E. coli with plasmid DNA was carried out as described by Brown et al. (7). The unencapsulated S. pneumoniae strain Rx1-19F-I was transformed as described previously for strain D39 (5).
Assessment of encapsulation.
Production of capsule by pneumococci was assessed by the quellung reaction, using factor-specific antisera obtained from Statens Seruminstitüt, Copenhagen, Denmark. This was performed by Mike Gratten.
DNA manipulations.
S. pneumoniae chromosomal DNA was extracted and purified by using the Wizard genomic DNA purification kit (Promega Corporation, Madison, Wis.). Chromosomal DNA was purified according to the manufacturer’s instructions except that cell lysis was induced by the addition of 0.1% (wt/vol) deoxycholate followed by incubation at 37°C for 10 min. Plasmid DNA was isolated from E. coli by the alkaline lysis method (28). Analysis of recombinant plasmids was carried out by digestion of DNA with one or more restriction enzymes under the conditions recommended by the supplier. Restricted DNA was electrophoresed in 0.8 to 1.5% agarose gels with a Tris-borate-EDTA buffer system as described by Maniatis et al. (27).
Long-range PCR.
The Expand Long Template PCR System (Boehringer, Mannheim, Germany) was used for long-range PCR according to the manufacturer’s instructions.
Southern hybridization analysis.
Chromosomal DNA (2.5 μg) was digested with appropriate restriction enzymes, and the digests were electrophoresed on agarose gels in Tris-borate-EDTA buffer. DNA was then transferred to a positively charged nylon membrane (Hybond N+; Amersham, Amersham, England) as described by Southern (39), hybridized to digoxigenin (DIG)-labelled probe DNA, washed, and then developed with anti-DIG–alkaline phosphatase conjugate (Boehringer) and 4-nitroblue tetrazolium–X-phosphate substrate according to the manufacturer’s instructions. DIG-labelled lambda DNA, restricted with HindIII, was used as a DNA molecular size marker.
DNA sequencing and analysis.
DNA sequencing of various PCR products was carried out by using dye terminator chemistry with specifically designed primers on an Applied Biosystems model 373A automated DNA sequencer. Nested deletions of pJCP484, which contains the type 19C cps locus, were constructed by the method of Henikoff (17) with an Erase-a-Base kit (Promega). This DNA was transformed into E. coli DH5α, and the resulting plasmid DNA was characterized by restriction analysis. Double-stranded template DNA for sequencing was prepared as recommended in the Applied Biosystems sequencing manual. The sequences of both strands were then determined by using dye-labelled primers on an Applied Biosystems model 373A automated DNA sequencer. The sequence was analyzed by using DNASIS and PROSIS version 7.0 software (Hitachi Software Engineering, South San Francisco, Calif.). The program BLASTX 2.0 (1) was used to translate DNA sequences and conduct homology searches of the protein databases available at the National Center for Biotechnology Information, Bethesda, Md. The program PROFILEGRAPH (19) was used to align hydropathy plots generated by the method of Kyte and Doolittle (23).
Nucleotide sequence accession numbers.
The nucleotide sequences for the cps19a locus and the 5′ intergenic region in S. pneumoniae 19A1 have been deposited with GenBank under accession no. AF094575. The nucleotide sequences for the cps19a2 and cps19c loci have been deposited with GenBank under accession no. AF105113 and AF105116, respectively. The nucleotide sequences for the 5′ intergenic regions from strains 19A2, 19B, and 19C have been deposited with GenBank under accession no. AF105112, AF105114, and AF105115, respectively.
RESULTS AND DISCUSSION
PCR amplification of the 3′ region of the type 19A cps locus.
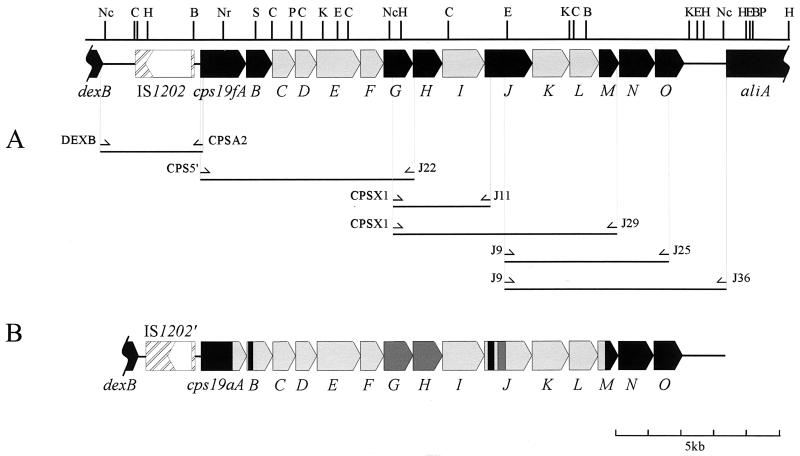
The number and arrangement of the genes in the 5′ portion of the cps19a locus were found to be identical to those in the cps19f locus (33). It was assumed that the arrangement of the genes in the remainder of the two loci would also be similar. Thus, a series of overlapping DNA fragments containing type 19A-specific genes flanked by conserved sequences was generated by long-range PCR with primers based on the cps19f sequence. A map of the PCR products spanning the entire cps19a locus is shown in Fig. 2A. DNAs from two different type 19A clinical isolates (19A1 and 19A2) were used as templates. Interestingly, the PCR products amplified from regions between cps19A and cps19J were identical in size for both type 19A isolates and type 19F, but the 19A2 isolate differed in the 3′ region of the cps19a locus; the PCR products obtained from the cps19J-to-cps19O region were either smaller or absent from 19A2. This suggests that part of this region of the cps19a locus from this strain may have been deleted.
FIG. 2.
Schematic representation of the PCR products amplified by using cps19f primers. The cps19f genes which hybridized (at high stringency) to type 19A chromosomal DNA are shown in black, and the remainder of the locus is shown in grey. (A) Primers used to amplify the cps19a locus. (B) The cps19a locus. Regions with >95% nucleotide sequence identity with the cps19f locus are shown in black. Regions with 90 to 95% identity are shown in dark grey, and regions with 70 to 90% identity are shown in light grey. Restriction sites are as follows: Nc, NcoI; C, ClaI; H, HindIII; B, BAMHI; Nr, NruI; S, SphI; P, PstI; K, KpnI; E, EcoRI.
Sequence analysis of the cps19a locus.
The sequences of the PCR products from 19A1 were determined by using specifically designed primers. Analysis of the compiled sequence revealed that the entire cps19f and cps19a loci are indeed very closely related. The cps19a locus has the same number of open reading frames (ORFs) organized in an order identical to those in cps19f, with homologies to the cps19f genes ranging from 70.1 to 99.4% identity (Fig. 2B). The sequences and properties of the cps19aA to -G genes have been reported previously (33). The sizes and percent identities of the cps19aH to -O and cps19fH to -O protein products are shown in Table 1.
TABLE 1.
Comparison of the cps19a and cps19f ORFs
| cps19a ORF | % G+Ca | Predicted size of protein product
|
cps19f ORF | % G+C | Predicted size of protein product
|
% Identity between cps19a and cps19f ORFs
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Da | No. of aab | Da | No. of aa | DNA | aa | ||||||
| cps19aH | 32.2 | 34,455 | 292 | cps19fH | 30.3 | 34,474 | 292 | 90.8 | 95.2 | ||
| cps19aI | 32.7 | 51,604 | 444 | cps19fI | 29.7 | 51,734 | 445 | 78.5 | 80.7 | ||
| cps19aJ | 33.0 | 54,650 | 474 | cps19fJ | 29.7 | 55,055 | 473 | 82.3 | 83.3 | ||
| cps19aK | 36.9 | 40,749 | 362 | cps19fK | 35.2 | 40,950 | 362 | 85.2 | 92.8 | ||
| cps19aL | 43.3 | 32,242 | 289 | cps19fL | 42.3 | 32,215 | 289 | 79.9 | 92.4 | ||
| cps19aM | 41.2 | 22,408 | 198 | cps19fM | 41.5 | 22,379 | 198 | 87.6 | 94.4 | ||
| cps19aN | 42.4 | 39,086 | 349 | cps19fN | 42.1 | 39,053 | 349 | 98.2 | 99.1 | ||
| cps19aO | 41.3 | 32,330 | 283 | cps19fO | 41.5 | 32,330 | 283 | 99.4 | 99.3 | ||
Percent guanine plus cytosine in coding region.
aa, amino acids.
Notwithstanding the overall similarity between the cps19a and cps19f loci, several interesting differences between the two loci were noted. The intergenic gaps between the cps19a genes and the cps19f genes are all similar, except for that between cps19aK and cps19aL, which is much larger (152 nucleotides) than that between cps19fK and cps19fL (38 nucleotides). The largest variation between the cps19a and cps19f loci occurs in the 5′ intergenic region. This region in the 19A1 strain has several deletions compared to the same region in type 19F, but the 3′ intergenic regions of types 19F and 19A1 are almost identical (96.7% identity). The differences in the 5′ intergenic region are discussed below.
A distinct crossover point was identified at the 3′ end of the locus within the cps19M gene; the first 348 nucleotides of cps19aM have 80.3% identity to cps19fM, whereas the remainder of cps19aM is 98% identical to cps19fM (Fig. 2B). The remainder of the cps19a and cps19f loci and the intergenic region preceding aliA are >99% identical. However, no such distinct point of divergence has been identified at the 5′ ends of the loci. Instead, the cps19aAB genes present a mosaic pattern with small regions of various degrees of identity to the cps19fAB genes, ranging from 76.6 to 100% (33).
The overall identity between cps19fJ and cps19aJ is only 82%, which is insufficient for the cps19fJ probe to hybridize to the cps19aJ gene under high-stringency conditions. However, on closer examination of the sequences, two small regions (nucleotides 10605 to 10784 and 10910 to 11116) at the 5′ end of cps19aJ were found to have >90% DNA sequence identity (97.6 and 93.2%, respectively) to cps19fJ, which presumably accounts for the Southern hybridization data obtained previously (31). It is tempting to speculate that these highly conserved regions (with >90% identity) may be important for the function of Cps19aJ and Cps19fJ, which putatively transport the same trisaccharide repeat unit across the plasma membrane.
Comparison of Cps19aI and Cps19fI.
The putative polysaccharide polymerases, Cps19aI and Cps19fI, are predicted to form different glycosidic linkages in type 19A (α1→3) and type 19F (α1→2) CPS, respectively. As these two proteins are 80.7% identical, their amino acid sequences were examined to identify any potentially significant differences between them. A cluster of nonconservative amino acid substitutions is located in the region between amino acids 290 and 320. No such clustering of nonconservative amino acid substitutions was observed when either Cps19fH and Cps19aH or Cps19fJ and Cps19aJ were compared. The region where these clustered substitutions occur is predicted to be in a loop located on the outer surface of the cytoplasmic membrane, based on the topology of the O-antigen polymerase (Rfc/Wzy) from Shigella flexneri (10). The CPS repeat units are predicted to be transported across the cytoplasmic membrane prior to polymerization (41). Thus, the external location of the nonconserved regions in Cps19aI and Cps19fI is consistent with that of the putative catalytic site in these proteins.
Capsule transformation from type 19F to 19A.
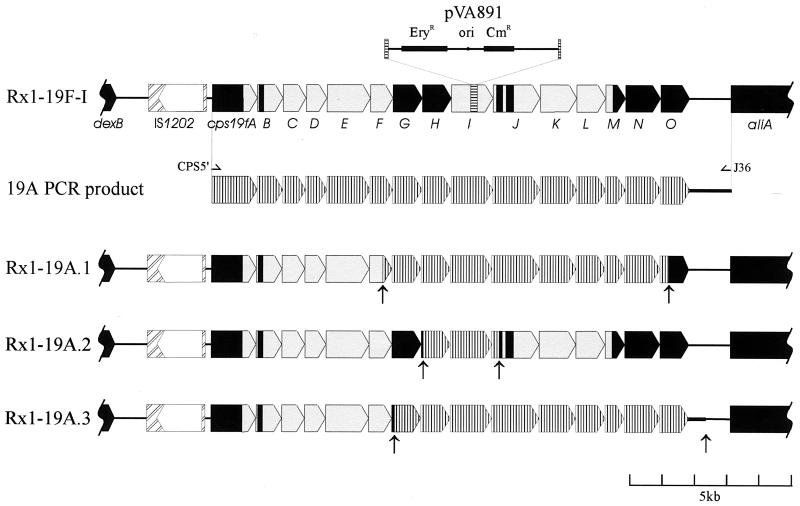
To confirm that the cps19a locus was sufficient for type 19A CPS biosynthesis, a 16.5-kb PCR product from the 5′ end of cps19aA to the 5′ end of aliA was amplified by using the primers CPS5′ and J36 (Fig. 1). This DNA product was used to transform Rx1-19F-I, an unencapsulated, erythromycin-resistant derivative of Rx1-19F in which the cps19fI gene had been disrupted by insertion-duplication mutagenesis with pVA891, as described previously (31). Several smooth transformants were checked for erythromycin sensitivity, indicating loss of the pVA891 sequence. Southern hybridization analysis confirmed the absence of both pVA891 and the cps19fI gene and the presence of the cps19aI gene. The presence or absence of both the cps19aC (located in the 5′ region of the cps19a locus) and cps19aK (located in the 3′ region of the cps19a locus) genes in the three individual transformants was also investigated to determine the sites of recombinational exchange between the cps19f locus and the type 19A PCR product (data not shown). The production of a type 19A capsule by these three smooth transformants, designated Rx1-19A.1 to -3, was then confirmed by the quelling reaction.
Based on the Southern hybridization data, the crossover points between the cps19f locus and the type 19A PCR product were then identified by sequencing the regions where recombination was predicted to have occurred. A diagrammatic representation indicating the positions of the recombination points is shown in Fig. 3. Two transformants (Rx1-19A.1 and -3) were similar, resulting in the exchange of a large region of the cps19f locus, from cps19fG to cps19fN (including cps19fO in Rx1-19A.3) for the homologous region from cps19a. On the other hand, Rx1-19A.2 was derived from exchange of a much smaller region of the cps19f locus, involving only cps19H and cps19I (Fig. 3). The cps19aH gene has 90.8% nucleotide identity to cps19fH, and the encoded highly conserved putative rhamnosyl transferases (95.2% amino acid identity) are predicted to be functionally identical in both type 19F and 19A CPS biosynthesis. The cps19aI and cps19fI genes are less conserved, with only 78.5% nucleotide identity, and the encoded putative polysaccharide polymerases (80.7% amino acid identity) are predicted to form different glycosidic linkages (as described above). Thus, these data show that it is possible to alter capsule production from type 19F to type 19A by replacing no more than two genes in the cps19f locus and that the presence of the cps19aI gene probably determines the 19A serotype.
FIG. 3.
Diagrammatic representation of the crossover points in the Rx1-19A transformants. The cps19f locus is shown in solid colors, where black represents >90% identity between the cps19f and cps19a loci and grey indicates regions with 70 to 80% identity. The horizontally hatched region within cps19fI indicates the site of pVA891 insertion. The cps19a locus is vertically hatched. The arrows indicate the points from which the sequence becomes cps19a specific.
Characterization of a gene rearrangement in the cps locus from strain 19A2.
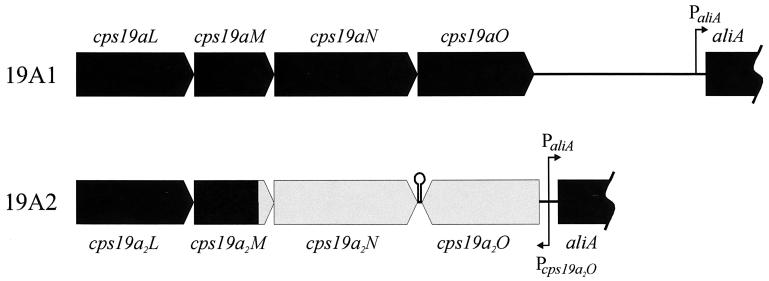
The 19A2 PCR product obtained by using primers J9 and J36 (Fig. 1), which amplified the 3′ region of the cps19a locus containing the dTDP-Rha biosynthesis genes (cps19L to -O), was smaller than those from both Rx1-19F and 19A1. To identify the deletion present in 19A2, this PCR product was sequenced, and this region of the locus was designated cps19a2.
Analysis of the sequence identified a gene rearrangement in the 3′ region of the cps19a2 locus, as well as a deletion of 1.4 kb of DNA between the end of cps19aO and the start of aliA (Fig. 4). The first 3,347 nucleotides of the cps19a2 sequence have 99.8% identity to cps19a, followed by 1,185 nucleotides with 80% identity to cps19a and 84% identity to cps19f. The remainder of the sequence then diverges until the final 94 nucleotides, which are 90% identical to the same region in cps19a. The conserved regions contain the genes cps19a2JKLMN, with a recombination point approximately 120 nucleotides from the end of cps19a2M. The next 1.1 kb of DNA contains an inverted copy of cps19a2O (Fig. 4) with 76.4% identity to cps19aO and cps19fO. A potential promoter was identified upstream of cps19a2O in the same region (but on the opposite DNA strand) as that for aliA. There are 61 nucleotides between the stop codons of cps19a2N and cps19a2O, and a stem-loop structure which could be a transcription terminator (ΔG = −30.5 kcal/mol) was identified in this region (Fig. 4).
FIG. 4.
Diagrammatic representation of the cps19aL-aliA regions of cps19a and cps19a2. The black region of cps19a2 is >90% identical to the same region in cps19a, and the grey region exhibits 75 to 80% identity to the equivalent regions in cps19a. The positions of the potential promoter sequences and the putative stem-loop terminator are also indicated.
When the 3′ regions of the cps loci from six Australian type 19A isolates were examined by PCR, none were found to contain the same rearrangement as seen in strain 19A2. However, two of the type 19A isolates did appear to contain extra DNA at the 3′ end of the locus, which has not been investigated further and may indicate the presence of yet another insertion sequence (IS) element in the 3′ intergenic region. The occurrence of IS elements in the intergenic regions flanking the cps loci in different S. pneumoniae strains is common and has been previously reported for several different serotypes (13, 15, 22, 34).
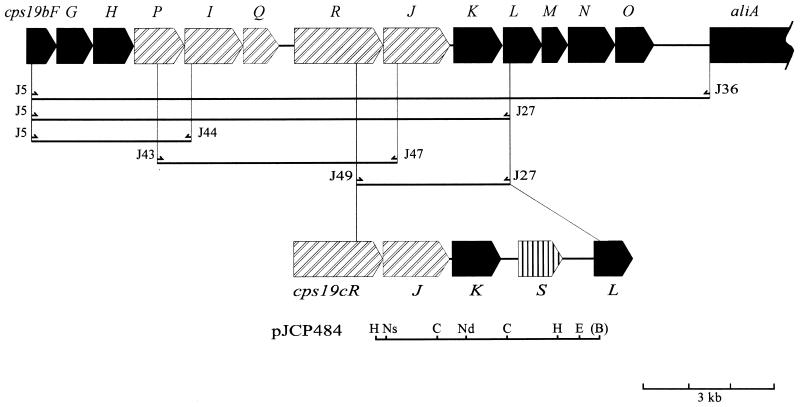
Isolation of the type 19C-specific cps region.
The exact location of the extra gene predicted to be present in the type 19C cps locus was investigated by using long-range PCR with a variety of primer pairs (Fig. 5). PCR products obtained with the type 19C template appeared to be approximately 2 kb larger than respective type 19B PCR products with any primer combination that spanned the cps19K-L region. Accordingly, the PCR product amplified from type 19C DNA by using primers J49 and J27 was purified and cloned into pBluescript KS(+), generating pJCP484 (Fig. 5). A map of the 5.3-kb PCR product was also constructed by using the restriction enzymes BamHI, ClaI, HindIII, NsiI, NdeI, and EcoRI (Fig. 5).
FIG. 5.
Physical map of part of the cps19c locus. Boxed arrows represent potential ORFs. Gene designations are indicated below the map; cps19cB to -S are abbreviated as B to S, respectively. Restriction sites are as follows: B, BamHI; C, ClaI; E, EcoRI; H, HindIII; Nd, NdeI; Ns, NsiI. The region of DNA subcloned into pBluescript KS(+) is shown below the map. The BamHI restriction site is bracketed because it is generated by the J27 primer and is not present in the chromosome of S. pneumoniae type 19C.
Both strands of the pneumococcal DNA insert, and nested derivatives thereof, were subjected to sequence analysis in order to compile the sequence of this portion of the cps19c locus. Examination of the compiled sequence revealed, as expected, that the first 2.9 kb of sequence at the 5′ end has a high degree of similarity to the cps19b sequence. This region contains the homologues of cps19bR, cps19bJ, and cps19bK (cps19cR, cps19cJ, and cps19cK), which exhibited 98.5, 99.7, and 94.9% identity, respectively. The sequence then diverges (at nucleotide 2906 of the cps19c sequence) just prior to the end of cps19cK; the sequence of nucleotides 2954 to 3155 exhibits 74.8% identity to the 5′ region of cps19bL, but does not contain an ORF, and then diverges from the cps19b sequence. An additional potential ORF, designated cps19cS (Fig. 5), is located between cps19cK and cps19cL and has a TTG start codon, which is preceded by a ribosome binding site. The closest potential ATG start codon is located 138 nucleotides downstream, but it is not preceded by a ribosome binding site. As predicted, the 3′ end of the cps19c sequence again shows similarity to the cps19b sequence, starting from nucleotide 5017; this is immediately before the start of the cps19cL gene (Fig. 5), which has 90.6% identity to cps19bL. There are potentially significant intergenic gaps immediately before and after the cps19cS gene, of 370 and 633 nucleotides, respectively. However, no potential stem-loop structures or obvious promoter sequences were found in these intergenic regions.
Characterization of Cps19cS.
The type 19C-specific ORF cps19cS is located between cps19cK and cps19cL in the cps19c locus (nucleotides 3276 to 4385) and encodes a putative 43.2-kDa protein containing 343 amino acids. This hydrophilic protein has a hydrophobicity index (according to Kyte and Doolittle [23]) of −0.23 and a predicted pI of 5.18. The region from the 3′ end of cps19cK to the 5′ end of cps19cL has a G+C content of 30.4%, increasing slightly to 31.4% for the cps19cS coding region. This is lower than the G+C contents of the two flanking genes cps19cK (35.3%) and cps19cL (42.6%).
Database searches with Cps19cS found significant similarity to the C termini of various known or putative glycosyl transferases (Table 2). Interestingly, one of these glycosyl transferases, CpoA, is possibly involved in teichoic acid biosynthesis in S. pneumoniae (14). Cps19cS exhibits 21% identity along its entire length to WaaG (Table 2), a proven α(1→3) glucosyl transferase involved in lipopolysaccharide core biosynthesis in E. coli and Salmonella enterica serovar Typhimurium (16). Thus, Cps19cS could function as the glucosyl transferase required for the addition of the β(1→6)-linked Glc side chain to the backbone in type 19C CPS biosynthesis.
TABLE 2.
Similarity of Cps19cS to other proteins
| Proteina | % Identityb to:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| MjRfbU | AaMtfC | AfGal | SaCapM | YeTrsD | VcRfbV | EaAmsD | StWaaG | SpCpoA | |
| Cps19cS | 26 (127) | 28.5 (158) | 22.9 (223) | 32.3 (93) | 23 (222) | 26.8 (142) | 22.2 (221) | 21 (300) | 28.7 (94) |
| MjRfbU | 100 | 30.3 (195) | 34.2 (190) | 25.4 (244) | 26.3 (179) | 25.4 (213) | 26.9 (119) | 30.4 (161) | 24.3 (173) |
| AaMtfC | 100 | 24.8 (311) | 23.7 (359) | 21.6 (361) | 34.6 (373) | 23 (183) | 21.9 (233) | 19.5 (246) | |
| AfGal | 100 | 25.1 (175) | 20.9 (344) | 24.5 (372) | 25.6 (203) | 27.9 (183) | 21.5 (251) | ||
| SaCapM | 100 | 25.6 (164) | 20.9 (349) | 17.7 (220) | 13.9 (230) | 15.2 (230) | |||
| YeTrsD | 100 | 30.3 (165) | 33.1 (362) | 20.7 (227) | 25.4 (114) | ||||
| VcRfbV | 100 | 26.3 (152) | 22.8 (136) | 22.4 (250) | |||||
| EaAmsD | 100 | 19.8 (177) | 25.2 (111) | ||||||
| StWaaG | 100 | 21.6 (180) | |||||||
| SpCpoA | 100 | ||||||||
Cps19cS, S. pneumoniae Cps19cS; MjRfbU, Methanococcus jannaschii RfbU (GenBank accession no. F64500); AaMtfC, Aquifex aeolicus MtfC (GenBank accession no. AE000693); AfGal, Archaeoglobus fulgidus galactosyl transferase (GenBank accession no. AE000983); SaCapM, Staphylococcus aureus CapM (26); YeTrsD, Yersinia enterocolitica TrsD (38); VcRfbV, Vibrio cholerae RfbV (12); EaAmsD, Erwinia amylovora AmsD (8); StWaaG, S. enterica serovar Typhimurium WaaG (16); SpCpoA, S. pneumoniae CpoA (14).
Percentage of identical amino acids determined with FASTA as implemented in PROSIS. Numbers in parentheses indicate the number of amino acids over which the identity occurs.
Serotype distribution of cps19cS.
To examine the relationship between cps19cS and encapsulation loci of other S. pneumoniae serotypes, a SacI-HindIII DNA fragment from a nested deletion derivative of pJCP484 corresponding to nucleotides 3160 to 4269 of the cps19c sequence was labelled with DIG and used to probe (at high stringency) Southern blots of restricted chromosomal DNAs from representative pneumococci belonging to serotypes and serogroups 2, 3, 4, 6, 7F, 7B, 8, 9N, 9V, 12, 14, 16, 17, 18, 19F, 19A, 19B, 20, 22, 23F, and 24. None of these serotypes had a high-stringency homologue to cps19cS (result not shown). However, this is not surprising when the structures for their CPSs are examined, because none contain a Glc side chain with a β(1→6) linkage (40).
Transformation of S. pneumoniae type 19F to type 19C.
We have previously demonstrated that capsule production was altered from type 19F to type 19B by replacing cps19fIJ with the central region of cps19b, which contains the cps19bPIQRJ genes and determines the 19B serotype (32). A similar approach was taken to determine whether cps19cS is indeed the gene responsible for the additional Glc side chain which distinguishes type 19C CPS. A large PCR product of the cps19c region between cps19cF and aliA was amplified by using primers J5 and J36 (Fig. 5) and transformed into Rx1-19F-I (as described above). The resultant transformant, expressing type 19C CPS, would be predicted to contain the cps19cPIQRJ genes required for both type 19B and 19C CPS biosynthesis as well as cps19cS. The cps19cK gene, which is located between cps19cJ and cps19cL, would also replace the almost identical cps19fK gene (94.9% identity). However, the encoded UDP-GlcNAc-2-epimerase, while essential for CPS biosynthesis in all group 19 members, is not serotype determining (31).
A smooth transformant was found to be erythromycin sensitive, indicating loss of the pVA891 sequence. Southern hybridization confirmed the absence of both pVA891 and the cps19fI gene and the presence of the cps19cP, -J, and -S genes (data not shown). The production of a type 19C capsule by the transformant, designated Rx1-19C, was then confirmed by the quellung reaction. This result showed that it is possible to alter capsule production from type 19F to type 19C by replacing cps19fIJ with the cps19cPIQRJ genes (required for both type 19B and 19C CPS biosynthesis) and the cps19cS gene. Hence, cps19cS determines the 19C serotype.
Sequence variation in the 5′ intergenic region of serogroup 19.
The PCR products from the 5′ intergenic regions of 19A1, 19A2, Rx1-19F, 19B, and 19C, amplified by using the DEXB and CPSA2 primers (Fig. 2), were highly variable in size, ranging from approximately 1.2 kb for 19A2 to 2 kb for 19A1 and 19B and 4 kb for Rx1-19F and 19C. The PCR products from 19A1, 19A2, 19B, and 19C were sequenced by using specific primers, and the 5′ intergenic regions were compared to that from Rx1-19F.
Interestingly, the 5′ intergenic regions of 19A1, 19B, and 19C are all almost identical, with three conserved deletions compared to 19F, of 58, 742, and 321 bp, respectively. These three deletions remove all but 150 nucleotides of the 1-kb intergenic region between dexB and IS1202, as well as the 3′ end of IS1202 (up to the stop codon of the putative transposase). Another mutation at nucleotide position 2151 introduces a stop codon which interrupts the putative transposase in 19A1, 19B, and 19C. The 5′ intergenic region of the S. pneumoniae type 23F Mexican drug-resistant strain Him18 (37) is almost identical to that of 19A1, 19B, and 19C, suggesting that these strains may have a shared clonal origin.
The larger size of the PCR product obtained from 19C is due to the presence of an additional IS element, designated IS19C. This 1.2-kb IS element is inserted into the inverted repeat of IS1202, adjacent to the cps19c locus, and is flanked at both ends by a 13-bp direct repeat, followed by 14 bp of unique DNA and then a 14-bp inverted repeat. The ORF which encodes the putative transposase in IS19C lies in the same orientation as that for IS1202 and opposite to that of the cps19c genes. This putative transposase has 67.5% amino acid identity to the transposase encoded by IS1239 from Streptococcus pyogenes, but at the DNA sequence level these IS elements exhibit negligible similarity. The transposases from these two IS elements also have 28 to 36% amino acid similarity to other transposases found in several different bacterial species, including IS30 from E. coli.
Analysis of the sequences indicated that the 5′ intergenic region of 19A2 is almost identical to that of 19F, except that it does not contain a copy of IS1202 in the 5′ intergenic region, although Southern hybridization data have previously shown that this type 19A strain does contain a copy of IS1202 in its chromosome (29). When PCR products from the 5′ intergenic regions from the six Australian type 19A isolates were examined by electrophoresis, they were all the same size as that from 19A2 (data not shown). A type 19F strain which lacks IS1202 has also been previously reported (29). The 5′ intergenic regions of four different S. pneumoniae strains belonging to serotypes 2, 3, 14, and 23F were also found to be almost identical to that from 19A2 (13, 20, 22, 30).
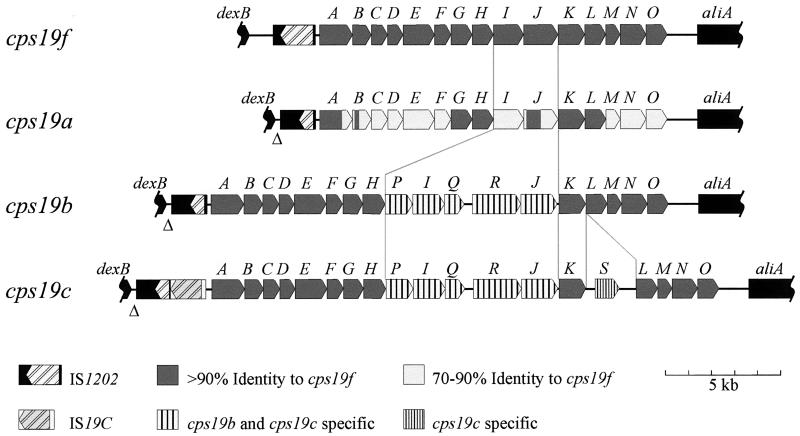
Conclusions.
S. pneumoniae group 19 is the first group for which the cps loci from all of the members (19F, 19A, 19B, and 19C) have been completely characterized (Fig. 6). Functions have been assigned to the majority of the cps19 gene products, based on either gene complementation or similarity to other proteins with known functions (15, 31, 32). The ability of PCR products containing either complete or partial cps loci to transform pneumococci from one serotype to another demonstrated that the cps19 loci contain sufficient genetic information for expression of type-specific CPSs.
FIG. 6.
Comparison of the cps loci of S. pneumoniae group 19.
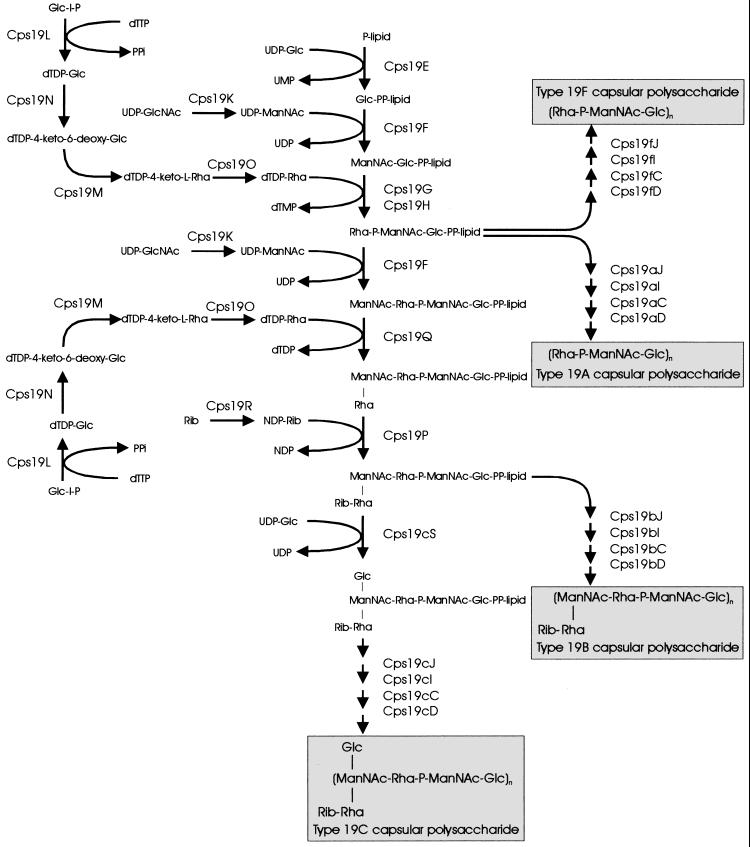
The structural similarities between the CPS repeat units from all four members of serogroup 19 are reflected in the highly conserved arrangement of their cps loci, with 13 genes (cps19A to -H, and cpsK to -O) common to all four serogroup members, as shown in Fig. 6. These 13 common genes encode functions required for the synthesis of the shared trisaccharide component of the group 19 CPS structures. Furthermore, the genetic differences between the group 19 cps loci identified are consistent with the differences in the CPS structures of the individual serotypes. This information has been used to propose biosynthetic pathways for each of the serotypes (Fig. 7) by a mechanism analogous to that proposed for Rol/Cld/Wzz- and Rfc/Wzy-dependent O-antigen assembly in S. enterica serogroups B and E (41).
FIG. 7.
Putative biosynthetic pathways for 19F, 19A, 19B, and 19C.
Transformation studies have shown that the genes which are present in the cps19a locus are functionally homologous to their cps19f counterparts and are sufficient for type 19A CPS biosynthesis and hence that the biosynthetic pathway for type 19A CPS is essentially identical to that proposed for type 19F CPS (Fig. 7). This is consistent with the fact that according to the structure proposed by Lee and Fraser (24), type 19A CPS differs from type 19F only by the type of glycosidic linkage between identical trisaccharide repeat units.
No additional genes, which might be involved in type 19A CPS biosynthesis, were identified either in or adjacent to the cps19a locus. Thus, the extra genes required to synthesize the side chains in the alternative type 19A CPS structure proposed by Lee et al. (25) must be located elsewhere on the S. pneumoniae chromosome. It is not known if these extra putative genes (if they exist at all) are present in all pneumococci or are specific to type 19A strains. The ability to alter CPS production from type 19F to type 19A (as judged by the quellung reaction with factor-specific sera) by exchange of no more than two cps19 genes, including the putative polysaccharide polymerase gene (cps19I), suggests that the nature of the glycosidic linkage formed by cps19I (joining the repeat units) is serotype determining for types 19F and 19A. Furthermore, this would be inconsistent with the formation of the alternative type 19A CPS structure, in which the repeat units are joined via the same glycosidic linkage as in type 19F.
The cps19c locus is almost identical to the cps19b locus except that an extra gene (cps19cS) has inserted between cps19cK and cps19cL. This gene is most likely to encode the glucosyl transferase required for the addition of the Glc side chain in the type 19C repeat unit. Interestingly, all three putative transferases involved in the addition of side chains to type 19B and/or 19C CPS, Cps19cS, Cps19P, and Cps19Q, appear to be cytoplasmic enzymes, as they lack both a leader sequence for export to the cell surface and a hydrophobic transmembrane sequence which could anchor them to the cell membrane. Thus, the Rha-Rib disaccharide side chain present in both type 19B and 19C CPSs and the Glc side chain specific to type 19C CPS are most probably added to the repeat units in the cytoplasm, before translocation to the outer surface by Cps19J and subsequent polymerization by Cps19I. It is interesting that the Glc side chain does not appear to interfere with the function of either the putative repeat unit transporter (Cps19bJ and Cps19cJ) or the putative polysaccharide polymerase (Cps19bI and Cps19cI), as the proteins encoded by cps19b and cps19c are almost identical (99.7 and >95%, respectively) and are able to function in the biosynthesis of both the type 19B and type 19C CPSs. The type 19C cps locus contains 19 genes, and at 21 kb, it is the largest pneumococcal capsule gene cluster characterized to date.
Comparison of the serogroup 19 cps loci shows that CPS structural diversity has evolved from genetic exchange in the central region of the cps locus. Recombinational exchange of small DNA fragments within the cps locus has been previously reported for S. pneumoniae (33). This mechanism could facilitate the generation of novel serotypes by the addition and/or replacement of specific transferases, the polysaccharide polymerase, and/or the repeat unit transporter in the cps locus, altering the structure of the CPS expressed. Interestingly, serotypes 2 and 23F, which, like all of the members of group 19, contain Rha in their CPS, have a similar arrangement of their cps genes (20, 30, 37). All contain the conserved cpsA to -E genes at the 5′ end of the cps locus and the genes involved in dTDP-Rha biosynthesis at the 3′ end of the locus, whereas the central serotype-determining regions of these loci are unique. Thus, these serotypes could have a common ancestor and result from recombinational exchanges within the cps locus. This mechanism, on a larger scale, has been shown to result in the replacement of the entire cps locus of antibiotic-resistant clones of S. pneumoniae, thus altering the expressed serotype (4, 9, 35).
ACKNOWLEDGMENT
This work was supported by a grant from the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austrian R. Some observations on the pneumococcus and on the current status of pneumococcal disease and its prevention. Rev Infect Dis. 1981;3(Suppl.):S1–S17. doi: 10.1093/clinids/3.supplement_1.s1. [DOI] [PubMed] [Google Scholar]
- 3.Austrian R, Bernheimer H P, Smith E E B, Mills G T. Simultaneous production of two capsular polysaccharides by pneumococcus. II. The genetic and biochemical bases of binary capsulation. J Exp Med. 1959;110:585–602. doi: 10.1084/jem.110.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes D M, Whittier S, Gilligan P H, Soares S, Tomasz A, Henderson F W. Transmission of multidrug-resistant serotype 23F Streptococcus pneumoniae in group day care: evidence suggesting capsular transformation of the resistant strain in vivo. J Infect Dis. 1995;171:890–896. doi: 10.1093/infdis/171.4.890. [DOI] [PubMed] [Google Scholar]
- 5.Berry A M, Yother J, Briles D E, Hansman D, Paton J C. Reduced virulence of a defined pneumolysin-negative mutant of Streptococcus pneumoniae. Infect Immun. 1989;57:2324–2330. doi: 10.1128/iai.57.7.2037-2042.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beynon L M, Richards J C, Perry M B, Kniskern P J. Antigenic and structural relationships within group 19 Streptococcus pneumoniae: chemical characterization of the specific capsular polysaccharides of type 19B and 19C. Can J Chem. 1991;70:131–137. [Google Scholar]
- 7.Brown M C M, Weston A, Saunders J R, Humphreys G O. Transformation of E. coli C600 by plasmid DNA at different phases of growth. FEMS Microbiol Lett. 1979;5:219–222. [Google Scholar]
- 8.Bugert P, Geider K. Molecular analysis of the ams operon required for exopolysaccharide synthesis of Erwinia amylovora. Mol Microbiol. 1995;15:917–933. doi: 10.1111/j.1365-2958.1995.tb02361.x. [DOI] [PubMed] [Google Scholar]
- 9.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 10.Daniels C, Vindurampulle C J, Morona R. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy) Mol Microbiol. 1998;28:1211–1222. doi: 10.1046/j.1365-2958.1998.00884.x. [DOI] [PubMed] [Google Scholar]
- 11.Douglas R M, Paton J C, Duncan S J, Hansman D. Antibody response to pneumococcal vaccination in children younger than five years of age. J Infect Dis. 1983;148:131–137. doi: 10.1093/infdis/148.1.131. [DOI] [PubMed] [Google Scholar]
- 12.Fallarino A, Mavrangelos C, Stroeher U H, Manning P A. Identification of additional genes required for O-antigen biosynthesis in Vibrio cholerae O1. J Bacteriol. 1997;179:2147–2153. doi: 10.1128/jb.179.7.2147-2153.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García E, López R. Molecular biology of the capsular genes of Streptococcus pneumoniae. FEMS Microbiol Lett. 1997;149:1–10. doi: 10.1111/j.1574-6968.1997.tb10300.x. [DOI] [PubMed] [Google Scholar]
- 14.Grebe T, Paik J, Hakenbeck R. A novel resistance mechanism against beta-lactams in Streptococcus pneumoniae involves CpoA, a putative glycosyl transferase. J Bacteriol. 1997;179:3342–3349. doi: 10.1128/jb.179.10.3342-3349.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guidolin A, Morona J K, Morona R, Hansman D, Paton J C. Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 19F. Infect Immun. 1994;62:5384–5396. doi: 10.1128/iai.62.12.5384-5396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinrichs D E, Monteiro M A, Perry M B, Whitfield C. The assembly system for the lipopolysaccharide R2 core-type of Escherichia coli is a hybrid of those found in Escherichia coli K-12 and Salmonella enterica. Structure and function of the R2 WaaK and WaaL homologues. J Biol Chem. 1998;23:8849–8859. doi: 10.1074/jbc.273.15.8849. [DOI] [PubMed] [Google Scholar]
- 17.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 18.Henrichsen J. Six newly recognized types of Streptococcus pneumoniae. J Clin Microbiol. 1995;33:2759–2762. doi: 10.1128/jcm.33.10.2759-2762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann K, Stöffel W. PROFILEGRAPH: an interactive graphical tool for protein sequence analysis. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/8.4.331. [DOI] [PubMed] [Google Scholar]
- 20.Iannelli F, Pearce B J, Pozzi G. The type 2 capsule locus of Streptococcus pneumoniae. J Bacteriol. 1999;181:2652–2654. doi: 10.1128/jb.181.8.2652-2654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katzenellenbogen E, Jennings H J. Structural determination of the capsular polysaccharide of Streptococcus pneumoniae type 19A 57. Carbohydr Res. 1983;124:235–245. doi: 10.1016/0008-6215(83)88459-6. [DOI] [PubMed] [Google Scholar]
- 22.Kolkman M A B, Wakarchuk W, Nuijten P J M, van der Zeijst B A M. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyl transferases required for the biosynthesis of the tetrasaccharide subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 23.Kyte J, Doolittle R F. A simple method for displaying the hydrophobic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 24.Lee C-J, Fraser B A. The structures of the cross-reactive types 19 (19F) and 57 (19A) pneumococcal polysaccharides. J Biol Chem. 1980;255:6847–6853. [PubMed] [Google Scholar]
- 25.Lee C-J, Fraser B A, Boykins R A, Li J P. Effect of culture conditions on the structure of Streptococcus pneumoniae type 19A 57 capsular polysaccharide. Infect Immun. 1987;55:1819–1823. doi: 10.1128/iai.55.8.1819-1823.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin W S, Cunneen T, Lee C Y. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J Bacteriol. 1994;176:7005–7016. doi: 10.1128/jb.176.22.7005-7016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 28.Morelle G. A plasmid extraction procedure on a miniprep scale. Focus. 1989;11.1:7–8. [Google Scholar]
- 29.Morona J K, Guidolin A, Morona R, Hansman D, Paton J C. Isolation, characterization, and nucleotide sequence of IS1202, an insertion sequence of Streptococcus pneumoniae. J Bacteriol. 1994;176:4437–4443. doi: 10.1128/jb.176.14.4437-4443.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morona J K, Miller D C, Coffey T J, Vindurampulle C J, Spratt B G, Morona R, Paton J C. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniae type 23F. Microbiology. 1999;145:781–789. doi: 10.1099/13500872-145-4-781. [DOI] [PubMed] [Google Scholar]
- 31.Morona J K, Morona R, Paton J C. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol. 1997;23:751–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- 32.Morona J K, Morona R, Paton J C. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniae type 19B. J Bacteriol. 1997;179:4953–4958. doi: 10.1128/jb.179.15.4953-4958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morona J K, Morona R, Paton J C. Analysis of the 5′ portion of the type 19A capsule locus identifies two classes of cpsC, cpsD, and cpsE genes in Streptococcus pneumoniae. J Bacteriol. 1999;181:3599–3605. doi: 10.1128/jb.181.11.3599-3605.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muñoz R, Mollerach M, López R, García E. Molecular organisation of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol Microbiol. 1997;25:79–92. doi: 10.1046/j.1365-2958.1997.4341801.x. [DOI] [PubMed] [Google Scholar]
- 35.Nesin M, Ramirez M, Tomasz A. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J Infect Dis. 1998;177:707–713. doi: 10.1086/514242. [DOI] [PubMed] [Google Scholar]
- 36.Ohno N, Yadomae T, Miyazaki T. The structure of the type specific polysaccharide of pneumococcus type XIX. Carbohydr Res. 1980;80:297–304. doi: 10.1016/s0008-6215(00)84638-8. [DOI] [PubMed] [Google Scholar]
- 37.Ramirez M, Tomasz A. Molecular characterization of the complete 23F capsular polysaccharide locus of Streptococcus pneumoniae. J Bacteriol. 1998;180:5273–5278. doi: 10.1128/jb.180.19.5273-5278.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skurnik M, Venho R, Toivanen P, al-Hendy A. A novel locus of Yersinia enterocolitica serotype O:3 involved in lipopolysaccharide outer core biosynthesis. Mol Microbiol. 1995;17:575–594. doi: 10.1111/j.1365-2958.1995.mmi_17030575.x. [DOI] [PubMed] [Google Scholar]
- 39.Southern E. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 40.van Dam J E G, Fleer A, Snippe H. Immunogenicity and immunochemistry of Streptococcus pneumoniae capsular polysaccharide. Antonie van Leeuwenhoek. 1990;58:1–47. doi: 10.1007/BF02388078. [DOI] [PubMed] [Google Scholar]
- 41.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]