Abstract
Prokaryote genomics started in earnest in 1995, with the complete sequences of two small bacterial genomes, those of Haemophilus influenzae and Mycoplasma genitalium. During the next quarter century, the prokaryote genome database has been growing exponentially, with no saturation in sight. For most of these 25 years, genome sequencing remained limited to cultivable microbes. Together with next generation sequencing methods, advances of metagenomics and single cell genomics have lifted this limitation, providing for an increasingly unbiased characterization of the global prokaryote diversity. Advances in computational genomics followed the progress of genome sequencing, even if occasionally lagging behind. Several major new branches of bacteria and archaea were discovered including Asgard archaea, the apparent closest relatives of eukaryotes and expansive groups of bacteria and archaea with small genomes thought to be symbionts of other prokaryotes. Comparative analysis of numerous prokaryote genomes spanning a wide range of evolutionary distances changed the conceptual foundations of microbiology, supplanting the notion of species genomes with fixed gene sets with that of dynamic pangenomes and the notion of a single Tree of Life with a statistical tree-like trend among individual gene trees. Strides were also made towards a theory and quantitative laws of prokaryote genome evolution.
The birth of microbial genomics
In the fall of 1995, 25 years before the time of this writing, J. Craig Venter‟s research institute (then The Institute for Genome Research, TIGR) released the first two complete sequences of bacterial genomes, both from opportunisitc human pathogens, the 1.78 Mbp genome of Haemophilus influenzae [1] and the 0.57 Mbp genome of Mycoplasma genitalium [2]. Comparison of these two small bacterial genomes inspired an attempt to reconstruct the minimal genetic complement of life that was inferred to consist of about 250 genes [3]. The first sequenced genome of an archaeon, Methanococcus (currently, Methanocaldococcus) jannaschii, followed promptly [4], along with several additonal bacteria including the photosynthetic cyanobacterium Synechococcus sp [5]. Within 3–4 years, the exponential growth of the collection of complete bacterial and archaeal genomes has settled in [6] (Figure 1). The comprehensive comparative analysis of microbial genomes that started as soon as the first two genomes became available made it clear that 70–80% of the genes in each genome were highly conserved in evolution such that orthologs could be identified in distantly related bacteria and/or archaea [7]. Thus, the genomes of prokaryotes provided ample material for functional and evolutionary inferences conducive to experimental validation. The flourishing research field of microbial genomics was born. In this short review, we briefly address several achievements of microbial genomics that appear most impactful as well as the major challenges in this field.
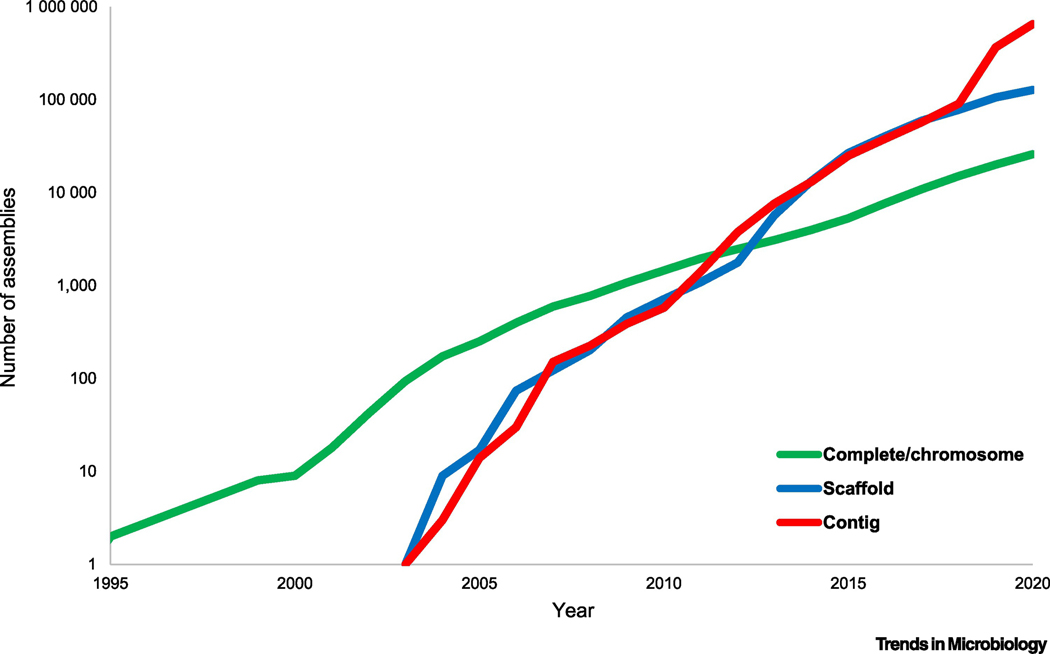
Figure 1. Exponential growth of the prokaryote genome database.
Number of genome assemblies at different assembly levels (ftp://ftp.ncbi.nih.gov/genomes/ASSEMBLY_REPORTS/) in the GenBank section of the NCBI Assembly database. The data points correspond to the end of the respective year; the X-axis starts with 1995, the year when the first two complete genomes were published.
Complete genome: all chromosomes are fully assembled with gaps not exceeding 10 ambiguous bases.
Chromosome: all chromosomes are fully assembled, but possibly containing gaps or unlocalized scaffolds.
Scaffold: sequence contigs are connected across gaps, but not placed on the chromosomes.
Contig: only unconnected contigs are available.
Advances and pitfalls of computational genomics
Genome sequences are useless unless adequate capabilities exist for the annotation and comparative analysis of genomes. At the time the first prokaryote genomes came along in the mid 1990s, the computational biology community was at best partially prepared for the genomic revolution, but developments followed quickly. Computational methods for gene prediction are essential in genomics but, in the case of prokaryotes, are relatively straightforward because of the dense coverage of these genomes by largely non-overlapping protein-coding genes [8–11]. Arguably, the two key developments in computational microbial genomics were the approaches for the identification of orthologous genes and the utilization of genome context information for functional inferences. Although the concept of orthology – evolutionary relationship between genes derived from the same ancestral gene in the most recent common ancestor of the compared organisms – was introduced by Walter Fitch as early as 1970 [12], orthology became an important concept only with the appearance of complete genome sequences because orthologous relationships cannot be reliably identified on incomplete gene sets [7, 13, 14]. Notwithstanding all the limitations imposed by the complexity of biological processes, the orthology conjecture, which posits that orthologous genes are responsible for equivalent functions in the respective organisms, generally, appears to hold [15]. Therefore, clusters of orthologous genes (COGs) can serve not only as the units for the reconstruction of microbial genome evolution but also as the most adequate platform for functional annotation of the genomes [7, 16–20]. As pointed out above, 70–80% of the protein-coding genes in a typical prokaryote genome are conserved across long evolutionary distances, and for most of these, biological function,at least, in general terms, can be reliably assigned by automatic comparison to position-specific scoring matrices or hidden Markov models derived from multiple alignments of protein sequences in well-curated collections of COGs. Superimposing the patterns of gene presence-absence in COGs onto phylogenetic trees for the most highly conserved genes (see below), the history of gene gain and loss in microbial genomes can be reconstructed using either maximum parsimony or the more robust maximum likelihood methods [21–25]. Given the highly dynamic character of the prokaryote evolution, such reconstuctions are an essential approach in microbial evolutionary genomics.
A substantial fraction of genes in any prokaryote genome are organized into operons, arrays of cotranscribed genes that are typically involved in the same pathway or process [26–28]. The operonic architectures are only partially conserved among distantly related prokaryotes, such that comparison of operons from diverse organisms yields networks of (potentially) functionally linked genes [29, 30, 31]. Hence the “guilt by association” approach that allows systematic prediction of the functions of “hypothetical” microbial genes based on their consistent linkage with functionally characterized genes [32–35]. With the growth of the collection of genomes across a broad range of evolutionary distances, the guilt by association approach has evolved into a powerful strategy for the discovery of new functional systems, particularly, those involved in highly variable functions, such as biological conflicts and signal transduction. The recent discovery of numerous, enormously diverse antivirus defense systems that tend to form distinct islands in prokaryote genomes may be considered the prime case in point for these approaches [36–39].
The progress in computational genomics over 25 years has been momentous, but serious problems persist and are even exacerbated by the rapid accumulation of genome sequences, many of them incompletely assembled. The most general and, apparently, most damning outstanding problem is that the exponential growth of genomic databases necessitates a near complete automation of genome analysis procedures to replace the combination of automatic and manual, case by case analyses, which was the most efficient approach in the early days of genomics. This results in notorious error propagation both in the construction of COGs and in the downstream genome annotation [40–42]. Identification of orthology is straightforward for highly conserved, single-copy genes with conserved domain architectures but remains an incompletely resolved challenge for faster evolving gene families with complex histories that include domain rearrangements, lineage-specific amplification of paralogous genes and gene loss as well as multiple horizontal gene transfers (HGT). Devising and implementing robust and efficient, phylogeny-based algorithms for this task and creating reliable, regularly updated databases of orthologous gene clusters remains a key task for computational genomics.
Metagenomics and single cell genomics usher in a new revolution in microbiology
The completion of the first microbial genome sequences was brought about by the perfection of whole genome shotgun (WGS) technique which remained the principal genome sequencing method for about a decade and a half since then. However, over the next few years, WGS has been nearly completely supplanted by Next Generation Sequencing (NGS) which brought to the table unprecedented sequencing depths but also hard problems with sequence assembly [43–45]. Once these difficulties have been largely overcome thanks to new, highly efficient assembly algorithms, such as Spades, assembly of numerous nearly complete genomes from metagenomics and single cell genomics data has become realistic [45–48].
Given the estimates indicating that less than 0.1% of prokaryotes represented in most environments can grow in culture [49, 50], metagenomics ushered in a revolution in microbiology by allowing unbiased, genome level surveys of microbial diversity. The Tara Ocean project that broadly explored the marine prokaryote diversity is a prime example of such a global survey [51, 52], and the exhaustive analysis of the human gut microbiome is another strong case in point [53]. In parallel, the advances of sing-cell genomics provide for partial sequencing of thousands of prokaryotic genomes across diverse habitats, allowing taxonomic assignment for the majority of metagenomic reads [53]. Perhaps, even more importantly, metagenomics and single-cell genomics brought about the discovery of entire major groups of uncultivable bacteria and archaea that shed new light on major aspects of microbial physiology, ecology and evolution as discussed in the next section.
All these remarkable advances notwithstanding, metagenomics changed the very notion of what a microbial genome sequence is because, in virtually all cases, it is impractical to assemble a closed circular chromosome sequence from metagenomic contigs. Thus, at present, when a recently sequenced “genome” of a bacterium or an archaeon is reported, by default, this implies a collection of contigs that have been placed in the same bin based on statistical properties of the contig sequences, such as oligonucleotide frequencies. In the current genome sequence databases, the number of such conditionally “complete” genomes at the scaffold or contig level exceeds the number of literally complete, closed genomes by orders of magnitude (Figure 1). This state of genome sequencing puts an extra onus on computational approaches to genome analysis and annotation. In particular, numerous contaminations to prokaryotic genomes ensue, stimulating the development of dedicated computational decontamination methods [54]. Evidently, it is highly desirable that, for each new group of prokaryotes that is discovered through metagenomics and single cell genomics approaches, at least a few truly complete, closed genomes were sequenced, to alleviate the concerns of possible incompleteness and contamination, and to obtain an example of the full gene repertoire for the given group of organisms.
Discovery of novel major groups of bacteria and archaea: impact on our understanding of microbial physiology, ecology and evolution
At least two momentous discoveries were enabled by the advances of metagenomics. First, metagenomic sequences allowed the delineation of two expansive groups of bacteria and archaea with small genomes, between 0.5–1 Mb that are thought to be symbionts (parasites, commensals or mutualists) of other prokaryotes [55–58]. Because of their apparent dependence on the respective hosts and despite their ubiquity in various environmental habitats, these microbes have been missed in the pre-metagenomic era (or detected only in surveys of the 16S rRNA diversity and not characterized in any detail). The only prominent exception was Nanoarchaeon equitans which was originally discovered through the observation of tiny cocci attached to the cell surface of the crenarchaeon Ignicoccus hospitalis and subsequently sequenced using the DNA isolated from a co-culture with I. hospitalis [59]. With the advent of metagenomics, it became apparent that N. equitans belonged to a large group of archaea with small genomes that form a clade in most phylogenetic trees and are known as the DPANN superphylum (named after the 5 constituent major groups of archaea: Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota and Nanohaloarchaeota), which includes several phylum-level branches [56, 60–62]. Parallel to the discovery of DPANN, an expansive branch of bacteria with similar genome sizes has been discovered and became known as Candidate Phyla Radiation (CPR), or later, Patescibacteria [60, 63]. Genome analysis indicates that both DPANN archaea and Patescibacteria encode a minimum of metabolic enzymes and therefore have to depend on other microbes for most metabolites [55, 57]. However, for most of these bacteria and archaea, it remains unclear whether their tiny cells are actually attached to host cells or are simply members of complex microbial communities with an extreme dependence on other members [57].
The second major discovery of metagenomics is the Asgard superphylum that includes a broad variety of archaea (all named after Norse deities) that encode numerous Eukaryotic Signature Proteins (ESPs), that is, homologs of proteins involved in characteristic eukaryotic functional systems, such as endomembranes and cytoskeleton. In most phylogenies of universal genes, eukaryotes cluster with or even within Asgard archaea, suggesting that these are the closest archaeal relatives of eukaryotes [64–66]. The first cultivation of an Asgard archaeon has been reported after a substantial diversity of Asgards has already been revealed by metagenomics, demonstrating synthrophy with Delta-proteobacteria and methanogenic archaea, consistent with the syntrophic scenarios for the origin of eukaryotes [67–69].
Taken together, the discoveries of DPANN and Asgard archaea, and Patescibacteria highlight the potential of metagenomics to reveal entire new continents in the microbial world. It is equally obvious, however, that genome analysis can take microbiologists only so far, and biological follow-up is essential.
The conceptual shift in microbial evolutionary genomics: horizontal genomics, statistical tree of life and dynamic pangenomes
Apart from the characterization of the microbial diversity and discoveries of new major groups of bacteria and archaea, genomics (and later, metagenomics) have transformed the most fundamental concepts of the structure and dynamics of the microbial world. This conceptual shift was triggered by the observations, made shortly after the first several complete genomes of bacteria and archaea were sequenced, that phylogenies of different conserved genes had different topologies. The differences were found to be so extensive that they could not be explained away by methodological artifacts, leading to the conclusion on extensive HGT being a key factor in the evolution of prokaryotes [5, 70–73]. Hence the idea of giving up the concept of the Tree of Life (ToL) for a net of life, devoid of any vertical component, gained much ground, leading to vivid debates on the extent of “horizontal genomics” [74–77]. Nevertheless, a comprehensive comparison of the topologies of the phylogenetic trees for individual conserved genes demonstrates the existence of a statistically significant consensus tree-like trend in the evolution of prokaryotes, even though the gene flow is quantitatively dominated by HGT [78–80]. Thus, the ToL survived the genomic revolution, but in a transformed version, as a statistical trend within the forest of individual gene trees, rather than a definitive representation of genome evolution [81].
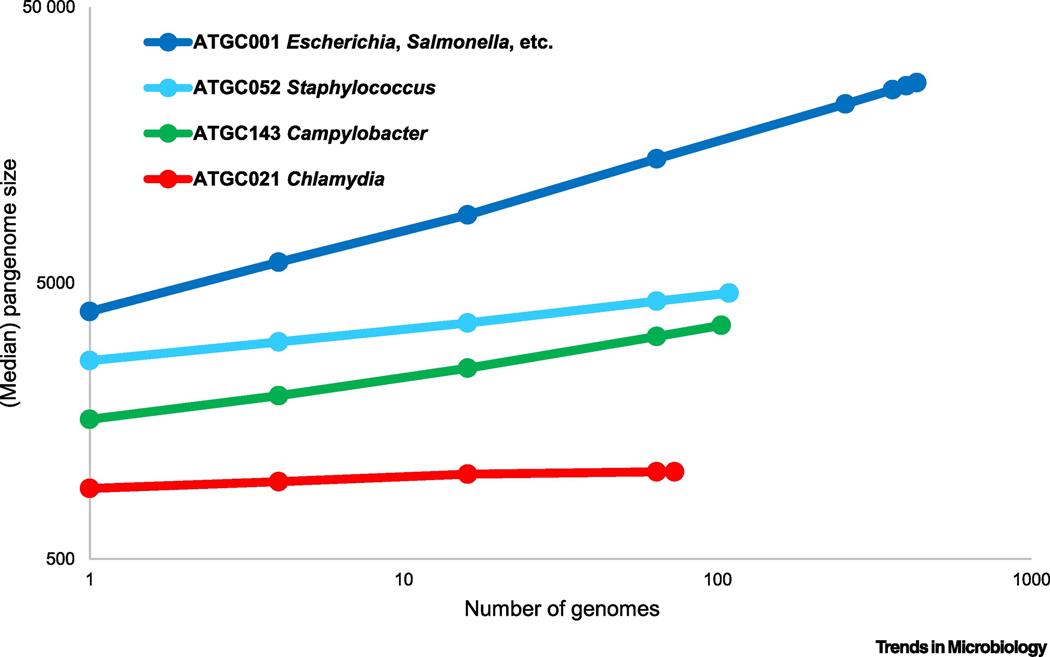
The second conceptual shift in microbiology is the emergence of the concept of the dynamic pangenome, that is, the entirety of the genes found in all representatatives of a prokaryotic species (notwithstanding the difficulty of defining the latter) [82–84]. The majority of the bacteria and archaea have open pangenomes, that is, sequencing of genomes of new isolates adds a set of new genes that were not detected in the previously available genomes from the same species, without obvious signs of saturation (Figure 2). This trend, certainly, does not imply that the pangenomes are infinite, but does indicate that most of them include orders of magnitude more genes than a typical individual genome [85]. Thus, in most bacteria and archaea, the relatively small, conserved core genome is associated with numerous accessory genes that comprise the bulk of the pangenome. However, the estimated size of pangenomes across the diversity of archaea and bacteria varies in within a broad range, with some having closed pangenomes that effectively saturate after a small number of isolates are sequenced (Figure 2).
Figure 2. Pangenomes of prokaryotes.
The plot (rarefaction curves; double logarithmic coordinates) show the increase in the total number of genes with the addition of new genomes for four clades of closely related bacteria. The points show the medians of the numbers of families of orthologous genes in 100 randomly sampled subsets of genomes within a clade. The clades represent four bacterial clusters from Alignable Tight Genome Clusters databse (ATGCs) [95]. ATGC001 is a cluster of 432 genomes from Escherichia, Salmonella, Enterobacter and other closely related families; ATGC052, 109 genomes of Staphylococcus aureus and S. argenteus; ATGC143, 103genomes of Campylobacter jejuni and C. coli; ATGC021, 73 genomes of Chlamydia trachomatis and C. muridarum.
Quantitative laws and theory of microbial genome evolution
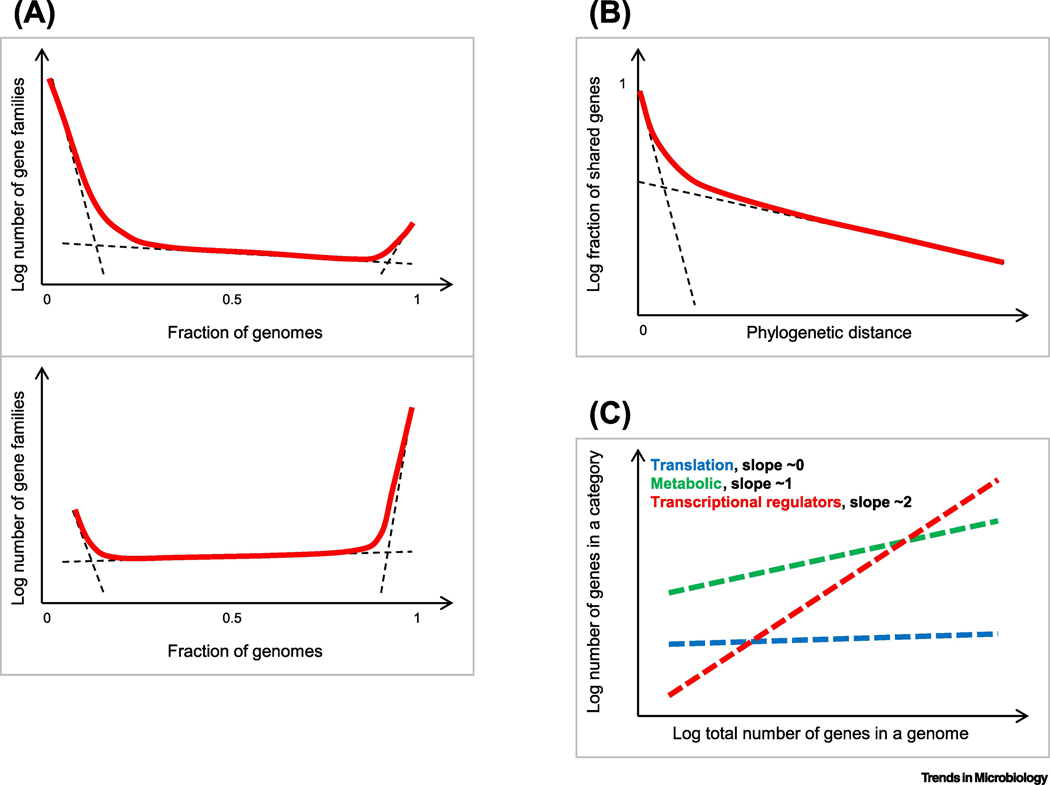
Comparative analysis of prokaryote genomes has yielded several quantitative regularities that can be construed as “laws” of evolutionary genomics (Figure 3) [86]. The crucial corollary of the discovery of pangenomes is that the key evolutionary process in prokaryotes is not point mutation but rather gene replacement via HGT and gene loss. A plot of the gene commonality (that is, sharing of orthologs) in any set of prokaryote genomes shows a universal, skewed U-shape, regardless of the evolutionary distances between the compared genomes (Figure 3a) [87]. This universal curve consists of three components that correspond to the small core of nearly universal genes, the much larger ‘shell’ of modetately conserved genes, and the huge ‘cloud’ of rare and unique genes (also known as ‘orphans’) (Figure 3a). The proportions between the components dramatically differ on the phylogenetic depth of the group; at the domain level (Bacteria or Archaea), the core consists of 100–200 genes, the shell contains thousands of genes, whereas the cloud of rare genes reaches into hundreds of thousands and, at the current state of sampling, is effectively unbounded, in line with the openness of most prokaryote pangenomes. The formation of this distinct plot shape can be accounted for by a stochastic model of genome evolution with non-uniformly distributed genes replacement rates, that is, differential effect of selection on different genes [88].
Figure 3. Quantitative laws of prokaryote genome evolution.
The schematic plots show: A. The universal gene commonality distribution. Top: prokaryote domains level (across multiple phyla); bottom: ATGC level (across closely related species or genera) [95]. Dashed lines show the approximate contributions of the individual components (cloud, shell and core); the solid line shows the observed combined distribution.
B. Two classes of prokaryote genes with slow and near-instantaneous replacement rates. Dashed lines show the exponential decay of the fast- and slow-decaying components; the solid line shows the observed combined fraction of gene families that are shared at different evolutionary distances.
C. Differential scaling of functional classes of prokaryote genes with the total number of genes in a genome
Mathematical modeling of prokaryote genome evolution by gene replacement shows that, to fit the observed dynamics of gene commonality decay during evolution, it is necessary to introduce two classes of genes [89]. The first class, combining the core and shell of the universal commonality distribution, includes about 90% of the genes in each genome that are replaced relatively slowly, whereas the second class (the “cloud”) consists of the remaining 10% or so of the genes that are replaced virtually instantaneously, in comparison (Figure 3b). A notable inference from this model is that the prokaryote genome universe consists of billions of distinct genes. In an indepndent line of analysis, it has been shown that the rate of gene trunover in prokaryotic genomes is proprotional to the standing nucleotide variation in the population, which is compatible with the notion that replacement of accessory genes is a predominantly neutral process [90].
Different functional classes of prokaryote genes show distinct scaling exponents with the total number of genes [87, 91–93]. Genes involved in information processing (replication, transcription, translation) are characterized by sublineal scaling, metabolic enzymes and transporters scale close to linearity, whereas regulatory and signal transduction genes scale superlinearly (Figure 3c). These notable regularities in the evolution of the prokaryote genome content can be accounted for by a model that includes two distinct parameters, selection coefficient, which defines the gene loss rate, and genome plasticity that reflects gene gain [94].
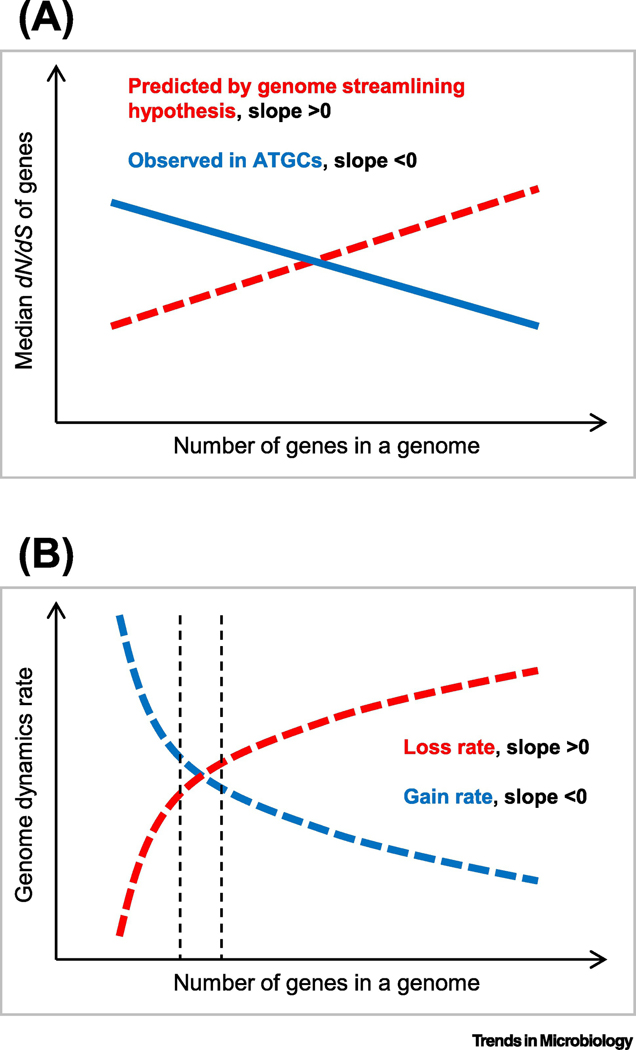
The availability of multiple groups of closely related bacterial and archaeal genomes [95], combined with the realization that the evolution of prokaryotes occurs, primarily, via gene replacements, provided for the development of a basic population-genetic theory of prokaryote genome and pangenome evolution. The general theoretical model of genome evolution developed by Lynch holds that genome evolution is shaped by the power of selection (primarily, purifying selection) that itself depends on the product of the selection coefficient (s) and effective population size (Ne) [96, 97]. In organisms with large Ne, selection is efficient such that features with even small negative s values can be eliminated. In contrast, in organisms with small Ne, genetic drift is a major contribution to the evolutionary process, such that even moderately deleterious features are often fixed in the population. Prokaryotes typically live in large populations (Ne up to 109), and so the theory predicts that, unlike the the genomes of multicellular organisms that accumulate large amounts of “junk” DNA, prokaryote should evolve under selection for streamlining because any piece of junk, even a small one, would be eliminated by purifying selection [98]. The dense packing of genes in prokaryote genomes appears to be compatible with this prediction. However, direct measurements of the strength of selection unexpectedly run afoul of the theoretical prediction. Selection in prokaryotes can be gauged by measuring the ratio of the non-synonymous to synonymous substitution rates (dN/dS) in multiple groups of closely related prokaryote genomes. Such measurements led to the unexpected conclusion on a highly significant negative correlation between dN/dS and the genome size in prokaryotes [99, 100] (Figure 4). In other words, in prokaryotes, the larger the genome, the stronger the protein-level selection. Stimulated by these findings, mathematical modeling of the evolution of prokaryote genomes by gene gain and loss has shown that the observed genome size distributions were best fit by models with positive, even if small, mean s values associated with gene gain [101, 102]. Thus, in the evolution of bacteria and archaea, the advantage of functional diversification conferred by the capture of new genes appears to often override the selection for genome streamlining, at least, up to a limit on the genome size.
Figure 4. Selection in the evolution of prokaryote genomes.
The schematic plots show:
A. The predicted and observed dependency of protein level selection, measured as dN/dS, on the total number of genes in a genome.
B. The utility hypothesis predicting that the gene gain rate decreases with the genome size because the relative impact on the fitness of a newly gained or lost gene is relatively smaller in larger genomes (diminishing return) whereas the intrinsic loss rate increases with the genome size because there are more genes to lose [101]. Vertical dashed lines around the equilibrium point indicate the range of the genome size expected to be observed in independently isolated genomes of the given species.
Along similar lines, capture of advantageous genes, in particular, those that ensure ability of microbes to explore new ecological niches, can lead to the expansion of pangenomes [103, 104]. Under this conceptual framework, organisms with large Ne tend to have larger pangenomes than those with small populations due to the strong positive selection driving fixation of acquired genes that confer even a slight fitness gain [105].
Conclusions
In the quarter century since its birth, microbial genomics evolved from a modest enterprise, where sequencing of each new genome was a feat in itself, to an expansive research field where discoveries stem from comparative analysis of hundreds or thousands of genomes. With the recent advances of metagenomics, the goal of completely charting the diversity of prokaryotes on earth, at least at coarse grain, might be within reach of the current generation of microbiology students. Above and beyond this striking quantitative progress, microbial genomics has transformed some of the fundamental concepts of evolutionary biology, replacing the notion of species genomes with fixed gene sets with that of dynamic pangenomes, and the single tree of life with the statistical tree-like trend in the forest of gene trees. Furthermore, comparative analysis of multiple genomes from many prokaryotic taxa provides for the discovery of quantitative laws of genome evolution and testing increasingly realistic theoretical models to explain the emergence of such laws. All these advances must not overshadow the hard challenges faced by microbial genomics, in particular, those associated with reliable automatic analysis of rapidly growing collections of genomes most of which in actuality are clusters of contigs rather than complete, closed genome sequences. Finally, we should never forget that genome analysis, however extensive, can only stimulate, augment and complement but by no means replace experimental microbiology.
How can we radically improve automated genome annotation?
How large is the diversity of bacteria and archaea on earth and how long will it take to chart it completely?
What is the size of a typical prokaryote pangenome and what determines it?
How common are host-parasite relationships between different prokaryotes?
Are there domain-level or superphylum-level groups of prokaryotes remaining to be discovered?
Highlights.
The database of prokaryote genomes has been growing exponentially for 25 years
There is no saturation of prokaryote diversity currently in sight
Most prokaryotes have dynamic, “open” pangenomes
Metagenomics makes key contributions to the study of prokaryote diversity
There is no single tree of life but a tree-like trend in evolution is discernible
Acknowledgements
The authors thank Dr. Purificacion Lopez-Garcia for critical reading of the manuscript. The authors’ research is supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine).
Footnotes
Competing interests
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fleischmann RD et al. (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd [see comments]. Science 269 (5223), 496–512. [DOI] [PubMed] [Google Scholar]
- 2.Fraser CM et al. (1995) The minimal gene complement of Mycoplasma genitalium. Science 270 (5235), 397–403. [DOI] [PubMed] [Google Scholar]
- 3.Mushegian AR and Koonin EV (1996) A minimal gene set for cellular life derived by comparison of complete bacterial genomes [see comments]. Proc Natl Acad Sci U S A 93 (19), 10268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bult CJ et al. (1996) Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii [see comments]. Science 273 (5278), 1058–73. [DOI] [PubMed] [Google Scholar]
- 5.Koonin EV and Galperin MY (1997) Prokaryotic genomes: the emerging paradigm of genome-based microbiology. Curr Opin Genet Dev 7 (6), 757–63. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Z. et al. (2020) Keeping up with the genomes: efficient learning of our increasing knowledge of the tree of life. BMC Bioinformatics 21 (1), 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tatusov RL et al. (1997) A genomic perspective on protein families. Science 278 (5338), 631–7. [DOI] [PubMed] [Google Scholar]
- 8.Borodovsky M. et al. (1994) Intrinsic and extrinsic approaches for detecting genes in a bacterial genome. Nucleic Acids Res 22 (22), 4756–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Besemer J. et al. (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29 (12), 2607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salzberg SL et al. (1998) Microbial gene identification using interpolated Markov models. Nucleic Acids Res 26 (2), 544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyatt D. et al. (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitch WM (1970) Distinguishing homologous from analogous proteins. Systematic Zoology 19, 99–106. [PubMed] [Google Scholar]
- 13.Fitch WM (2000) Homology a personal view on some of the problems. Trends Genet 16 (5), 227–31. [DOI] [PubMed] [Google Scholar]
- 14.Koonin EV (2005) Orthologs, Paralogs and Evolutionary Genomics. Annu Rev. Genet 39, 309–338. [DOI] [PubMed] [Google Scholar]
- 15.Gabaldon T. and Koonin EV (2013) Functional and evolutionary implications of gene orthology. Nat Rev Genet 14 (5), 360–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen LJ et al. (2008) eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res 36 (Database issue), D250–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen F. et al. (2006) OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res 34 (Database issue), D363–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trachana K. et al. (2011) Orthology prediction methods: a quality assessment using curated protein families. Bioessays 33 (10), 769–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huerta-Cepas J. et al. (2017) Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOGMapper. Mol Biol Evol 34 (8), 2115–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristensen DM et al. (2011) Computational methods for Gene Orthology inference. Brief Bioinform 12 (5), 379–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snel B. et al. (2002) Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res 12 (1), 17–25. [DOI] [PubMed] [Google Scholar]
- 22.Mirkin BG et al. (2003) Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol 3 (1), 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Csuros M. (2010) Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26 (15), 1910–2. [DOI] [PubMed] [Google Scholar]
- 24.Cohen O. and Pupko T. (2011) Inference of gain and loss events from phyletic patterns using stochastic mapping and maximum parsimony--a simulation study. Genome Biol Evol 3, 1265–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen O. et al. (2008) A likelihood framework to analyse phyletic patterns. Philos Trans R Soc Lond B Biol Sci 363 (1512), 3903–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacob F. and Monod J. (1961) Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol 3, 318–356. [DOI] [PubMed] [Google Scholar]
- 27.Salgado H. et al. (2000) Operons in Escherichia coli: genomic analyses and predictions. Proc Natl Acad Sci U S A 97 (12), 6652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolf YI et al. (2001) Genome alignment, evolution of prokaryotic genome organization and prediction of gene function using genomic context. Genome Res. 11, 356–372. [DOI] [PubMed] [Google Scholar]
- 29.Rogozin IB et al. (2002) Connected gene neighborhoods in prokaryotic genomes. Nucleic Acids Res 30 (10), 2212–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janga SC et al. (2005) Nebulon: a system for the inference of functional relationships of gene products from the rearrangement of predicted operons. Nucleic Acids Res 33 (8), 2521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Mering C. et al. (2007) STRING 7--recent developments in the integration and prediction of protein interactions. Nucleic Acids Res 35 (Database issue), D358–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aravind L. (2000) Guilt by association: contextual information in genome analysis. Genome Res 10 (8), 1074–7. [DOI] [PubMed] [Google Scholar]
- 33.Galperin MY and Koonin EV (2000) Who’s your neighbor? New computational approaches for functional genomics. Nat Biotechnol 18 (6), 609–613. [DOI] [PubMed] [Google Scholar]
- 34.Huynen M. et al. (2000) Exploitation of gene context. Curr Opin Struct Biol 10 (3), 366–70. [DOI] [PubMed] [Google Scholar]
- 35.Moreno-Hagelsieb G. and Collado-Vides J. (2002) A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics 18 Suppl 1, S329–36. [DOI] [PubMed] [Google Scholar]
- 36.Doron S. et al. (2018) Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359 (6379). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Millman A. et al. (2020) Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat Microbiol 5 (12), 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernheim A. and Sorek R. (2020) The pan-immune system of bacteria: antiviral defence as a community resource. Nat Rev Microbiol 18 (2), 113–119. [DOI] [PubMed] [Google Scholar]
- 39.Gao L. et al. (2020) Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 369 (6507), 1077–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen P. and Krogh A. (2005) Large-scale prokaryotic gene prediction and comparison to genome annotation. Bioinformatics 21 (24), 4322–9. [DOI] [PubMed] [Google Scholar]
- 41.Poptsova MS and Gogarten JP (2010) Using comparative genome analysis to identify problems in annotated microbial genomes. Microbiology (Reading) 156 (Pt 7), 1909–1917. [DOI] [PubMed] [Google Scholar]
- 42.Danchin A. et al. (2018) No wisdom in the crowd: genome annotation in the era of big data - current status and future prospects. Microb Biotechnol 11 (4), 588–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kisand V. and Lettieri T. (2013) Genome sequencing of bacteria: sequencing, de novo assembly and rapid analysis using open source tools. BMC Genomics 14, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forouzan E. et al. (2017) Evaluation of nine popular de novo assemblers in microbial genome assembly. J Microbiol Methods 143, 32–37. [DOI] [PubMed] [Google Scholar]
- 45.Giani AM et al. (2020) Long walk to genomics: History and current approaches to genome sequencing and assembly. Comput Struct Biotechnol J 18, 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sundquist A. et al. (2007) Whole-genome sequencing and assembly with high-throughput, short-read technologies. PLoS One 2 (5), e484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chitsaz H. et al. (2011) Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nat Biotechnol 29 (10), 915–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bankevich A. et al. (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19 (5), 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmeisser C. et al. (2007) Metagenomics, biotechnology with non-culturable microbes. Appl Microbiol Biotechnol 75 (5), 955–62. [DOI] [PubMed] [Google Scholar]
- 50.Teeling H. and Glockner FO (2012) Current opportunities and challenges in microbial metagenome analysis--a bioinformatic perspective. Brief Bioinform 13 (6), 728–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sunagawa S. et al. (2015) Ocean plankton. Structure and function of the global ocean microbiome. Science 348 (6237), 1261359. [DOI] [PubMed] [Google Scholar]
- 52.Ibarbalz FM et al. (2019) Global Trends in Marine Plankton Diversity across Kingdoms of Life. Cell 179 (5), 1084–1097 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt TSB et al. (2018) The Human Gut Microbiome: From Association to Modulation. Cell 172 (6), 11981215. [DOI] [PubMed] [Google Scholar]
- 54.Steinegger M. and Salzberg SL (2020) Terminating contamination: large-scale search identifies more than 2,000,000 contaminated entries in GenBank. Genome Biol 21 (1), 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castelle CJ et al. (2018) Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat Rev Microbiol 16 (10), 629–645. [DOI] [PubMed] [Google Scholar]
- 56.Castelle CJ and Banfield JF (2018) Major New Microbial Groups Expand Diversity and Alter our Understanding of the Tree of Life. Cell 172 (6), 1181–1197. [DOI] [PubMed] [Google Scholar]
- 57.Beam JP et al. (2020) Ancestral Absence of Electron Transport Chains in Patescibacteria and DPANN. Front Microbiol 11, 1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopez-Garcia P. and Moreira D. (2020) Physical connections: prokaryotes parasitizing their kin. Environ Microbiol Rep. [DOI] [PubMed] [Google Scholar]
- 59.Waters E. et al. (2003) The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci U S A 100 (22), 12984–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parks DH et al. (2017) Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol 2 (11), 1533–1542. [DOI] [PubMed] [Google Scholar]
- 61.Dombrowski N. et al. (2019) Genomic diversity, lifestyles and evolutionary origins of DPANN archaea. FEMS Microbiol Lett 366 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dombrowski N. et al. (2020) Undinarchaeota illuminate DPANN phylogeny and the impact of gene transfer on archaeal evolution. Nat Commun 11 (1), 3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown CT et al. (2015) Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523 (7559), 208–11. [DOI] [PubMed] [Google Scholar]
- 64.Spang A. et al. (2015) Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521 (7551), 173–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zaremba-Niedzwiedzka K. et al. (2017) Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541 (7637), 353–358. [DOI] [PubMed] [Google Scholar]
- 66.Eme L. et al. (2018) Archaea and the origin of eukaryotes. Nat Rev Microbiol 16 (2), 120. [DOI] [PubMed] [Google Scholar]
- 67.Imachi H. et al. (2020) Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 577 (7791), 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lopez-Garcia P. and Moreira D. (2020) Cultured Asgard Archaea Shed Light on Eukaryogenesis. Cell 181 (2), 232–235. [DOI] [PubMed] [Google Scholar]
- 69.Lopez-Garcia P. and Moreira D. (2020) The Syntrophy hypothesis for the origin of eukaryotes revisited. Nat Microbiol 5 (5), 655–667. [DOI] [PubMed] [Google Scholar]
- 70.Doolittle WF (1999) Phylogenetic classification and the universal tree. Science 284 (5423), 2124–9. [DOI] [PubMed] [Google Scholar]
- 71.Doolittle WF (1999) Lateral genomics. Trends Cell Biol 9 (12), M5–8. [PubMed] [Google Scholar]
- 72.Doolittle WF (2000) Uprooting the tree of life. Sci Am 282 (2), 90–5. [DOI] [PubMed] [Google Scholar]
- 73.Koonin EV et al. (2001) Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol 55, 709–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O’Malley MA and Boucher Y. (2005) Paradigm change in evolutionary microbiology. Stud Hist Philos Biol Biomed Sci 36 (1), 183–208. [DOI] [PubMed] [Google Scholar]
- 75.Bapteste E. et al. (2005) Do orthologous gene phylogenies really support tree-thinking? BMC Evol Biol 5 (1), 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doolittle WF and Bapteste E. (2007) Pattern pluralism and the Tree of Life hypothesis. Proc Natl Acad Sci U S A 104 (7), 2043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bapteste E. et al. (2009) Prokaryotic evolution and the tree of life are two different things. Biol Direct 4, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Puigbo P. et al. (2009) Search for a Tree of Life in the thicket of the phylogenetic forest. J. Biol 8, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Puigbo P. et al. (2010) The tree and net components of prokaryote evolution. Genome Biol Evol 2, 745–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Puigbo P. et al. (2013) Seeing the Tree of Life behind the phylogenetic forest. BMC Biol 11, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Malley MA and Koonin EV (2011) How stands the Tree of Life a century and a half after The Origin? Biol Direct 6, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Medini D. et al. (2005) The microbial pan-genome. Curr Opin Genet Dev 15 (6), 589–94. [DOI] [PubMed] [Google Scholar]
- 83.Medini D. et al. (2008) Microbiology in the post-genomic era. Nat Rev Microbiol 6 (6), 419–30. [DOI] [PubMed] [Google Scholar]
- 84.Medini D. et al. (2020) The Pangenome: A Data-Driven Discovery in Biology. In The Pangenome: Diversity, Dynamics and Evolution of Genomes (Tettelin H. and Medini D. eds), pp. 3–20. [Google Scholar]
- 85.Puigbo P. et al. (2014) Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes. BMC Biol 12, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koonin EV (2011) Are there laws of genome evolution? PLoS Comput Biol. 7, e1002173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koonin EV and Wolf YI (2008) Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 36 (21), 6688–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lobkovsky AE et al. (2013) Gene frequency distributions reject a neutral model of genome evolution. Genome Biol Evol 5 (1), 233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wolf YI et al. (2016) Two fundamentally different classes of microbial genes in a vast genomic universe. Nature Microbiology 2, 16208. [DOI] [PubMed] [Google Scholar]
- 90.Andreani NA et al. (2017) Prokaryote genome fluidity is dependent on effective population size. ISME J 11 (7), 1719–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van Nimwegen E. (2003) Scaling laws in the functional content of genomes. Trends Genet 19 (9), 479–84. [DOI] [PubMed] [Google Scholar]
- 92.Konstantinidis KT and Tiedje JM (2004) Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci U S A 101 (9), 3160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Molina N. and van Nimwegen E. (2009) Scaling laws in functional genome content across prokaryotic clades and lifestyles. Trends Genet 25 (6), 243–7. [DOI] [PubMed] [Google Scholar]
- 94.Sela I. et al. (2019) Selection and Genome Plasticity as the Key Factors in the Evolution of Bacteria. Phys. Rev. X 9, 031018. [Google Scholar]
- 95.Kristensen DM et al. (2017) ATGC database and ATGC-COGs: an updated resource for micro- and macro-evolutionary studies of prokaryotic genomes and protein family annotation. Nucleic Acids Res 45 (D1), D210–D218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lynch M. and Conery JS (2003) The origins of genome complexity. Science 302 (5649), 1401–4. [DOI] [PubMed] [Google Scholar]
- 97.Lynch M. (2007) The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci U S A 104 Suppl 1, 8597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lynch M. (2006) Streamlining and simplification of microbial genome architecture. Annu Rev Microbiol 60, 327–49. [DOI] [PubMed] [Google Scholar]
- 99.Novichkov PS et al. (2009) Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. J Bacteriol 191 (1), 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kuo CH et al. (2009) The consequences of genetic drift for bacterial genome complexity. Genome Res 19 (8), 14504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sela I. et al. (2016) Theory of prokaryotic genome evolution. Proc Natl Acad Sci U S A 113 (41), 11399–11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Iranzo J. et al. (2017) Disentangling the effects of selection and loss bias on gene dynamics. Proc Natl Acad Sci U S A 114, E616–E624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McInerney JO et al. (2017) Why prokaryotes have pangenomes. Nat Microbiol 2, 17040. [DOI] [PubMed] [Google Scholar]
- 104.McInerney JO et al. (2020) Pangenomes and Selection: The Public Goods Hypothesis. In The Pangenome: Diversity, Dynamics and Evolution of Genomes (Tettelin H. and Medini D. eds), pp. 151–167. [PubMed] [Google Scholar]
- 105.Bobay LM and Ochman H. (2018) Factors driving effective population size and pan-genome evolution in bacteria. BMC Evol Biol 18 (1), 153. [DOI] [PMC free article] [PubMed] [Google Scholar]