Summary
The presence of anti-citrullinated protein autoantibodies (ACPA) is a hallmark feature of rheumatoid arthritis (RA), which causes chronic joint destruction and systemic inflammation. Based on ACPA status, RA patients can be sub-grouped into two major subsets: ACPA-positive RA (ACPA+ RA) and ACPA-negative RA (ACPA– RA). Accumulating evidence have suggested that ACPA+ RA and ACPA– RA are two distinct disease entities with different underlying pathophysiology. In contrast to the well-characterized pathogenic mechanisms of ACPA+ RA, the etiology of ACPA– RA remains largely unknown. In this review, we summarized current knowledge about the primary drivers of ACPA– RA, particularly focusing on the serological, cellular, and molecular aspects of immune mechanisms. A better understanding of the immunopathogenesis in ACPA– RA will help in designing more precisely targeting strategies, and paving the road to personalized treatment. In addition, identification of novel biomarkers in ACPA– RA will substantially promote early treatment and improve the outcomes.
Keywords: Rheumatoid arthritis, Anti-citrullinated protein autoantibodies (ACPA)-negative, Immunopathogenesis, Biomarkers
Introduction
Rheumatoid arthritis (RA) is a chronic and systemic inflammatory autoimmune disorder characterized by synovial inflammation, progressive erosive arthritis and extra-articular involvements.1 The presence of anti-citrullinated protein autoantibodies (ACPA) represents a hallmark feature of RA. Based on ACPA status, RA patients can be sub-grouped into two major subsets: ACPA-positive RA (ACPA+ RA) and ACPA-negative RA (ACPA– RA).1 ACPA can be detected in circulation years before the onset of clinical overt symptoms, and the presence of ACPA usually associates with more aggressive bone and joint destruction, suggesting the citrulline-specific immune response is critical in disease initiation and evolution in ACPA+ RA.1 In contrast, the etiology of ACPA– RA remains largely unknown. Genetic studies have implied that ACPA+ RA and ACPA– RA are two distinct disease entities with differential underlying pathophysiology.2 There also lies some differences in clinical characteristics between the two subsets. It's showed that ACPA– RA had more tender joints, more difficulty in clenching fists and shorter symptom duration at the time of first presentation with arthralgia compared to ACPA+ RA, but ACPA+ RA progressed to arthritis more quickly thereafter.3 As for extra-articular manifestations, a lower probability was reported in ACPA– RA4 (Figure 1). A better understanding of the immunopathogenesis, may shed lights on developing novel targeting interventions in ACPA– RA. In this review, we outline the current knowledge on the primary drivers of ACPA– RA, particularly focusing on the serological, cellular, and molecular mechanisms of the immune response.
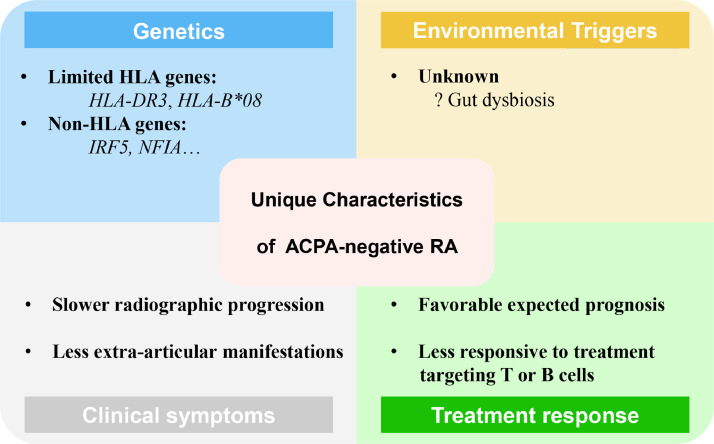
Figure 1.
Presumed unique characteristics of ACPA-negative RA. Based on limited evidence and current knowledge, unique features of ACPA-negative RA on genetics, environmental triggers, clinical symptoms and treatment response are presented with omission of shared common features with ACPA-positive RA.
Genetic and environmental factors in ACPA– RA
RA affects 0.5-1% of the population worldwide. ACPA+ RA constitutes approximately two-thirds of all RA patients. A recent epidemiological study showed a significant increase in the incidence of rheumatoid factor (RF)-negative RA whereas a decrease in RF-positive RA in 2005-2014 compared with previous decades.5 Generally, the development of RA starts from a preclinical phase, followed by a transition phase and then to clinical RA.1 The transition is considered as a result of a complex interplay between predisposing genes and environmental triggers. Interestingly, ACPA+ RA and ACPA– RA differ in genetic and environmental risk factors. The influence of HLA genes is more limited in ACPA– RA compared to ACPA+ counterpart. The genetic risk factors in ACPA+ RA is mainly on human leukocyte antigen (HLA) class II molecules.6 For example, HLA-DRB1 shared epitope alleles strongly associate with ACPA+ RA.2 In contrast, it has been shown that DR3 allotype is associated with ACPA– RA, but not with ACPA+ RA.7 Further, a recent study revealed that HLA-B*08 group of alleles and the related amino acid variant Asp-9 were associated with an ACPA– RA subset positive for anti-carbamylated protein (anti-CarP) antibodies.8 The differential involvement of HLA-II molecules versus HLA-I molecules between ACPA+ RA and ACPA– RA suggest different pathogenetic mechanisms underlying the two subsets. In non-HLA genetic polymorphisms, markers at ANKRD55, STAT4, C5orf30, PTPN22, ELMO1, RUNX1 and BLK are associated with both ACPA+ RA and ACPA– RA,2,9, 10, 12 while SPP1, IRF5, PRL and NFIA are preferentially associated with ACPA– RA.10,11,13 These genetic risk variants may contribute to disease pathogenesis in ACPA– RA.
Gut dysbiosis is associated with immune alterations, serving as an important environmental factor that promote RA pathogenesis.14 So far, the impact of dysbiosis on ACPA– RA remains unclear. Findings from K/BxN mice, a mouse model of inflammatory arthritis, have shown that segmented filamentous bacteria (SFB) colonization exacerbates disease severity via Th17 induction.15 Subsequent studies using IL1rn knockout (IL1rn−/−) mice also support a role of dysbiosis in disease pathogenesis, as spontaneous onset of arthritis was associated with the alterations of gut microbiome and arthritis was attenuated in IL1rn−/− mice under germ-free (GF) condition.16,17 Of interest, a recent study indicated that ACPA+ RA patients had higher proportions of Blautia, Akkermansia, and Clostridiales than ACPA– RA patients.18 And butyrate-metabolizing species were implicated in determining ACPA status by another recent study.19 The authors found that butyrate-consuming species were preferentially enriched in ACPA+ RA patients, whereas butyrate-producing species were preferentially enriched in ACPA– RA patients.19 Further studies on dissecting the role of dysbiosis in ACPA– RA patients will be of great importance to better understand the involvement of environmental factor in the pathogenesis of ACPA– RA.
Immunopathogenesis in RA
Immune dysregulation in the pathogenesis of ACPA– RA
Two different subsets of macrophages reside in synovial lining: MHCII−CX3CR1+ and MHCII+CX3CR1lo populations.20 The former populates the synovial tissue early during embryonic development and has a protective role in maintaining joint integrity, while the latter originates from bone marrow and requires a contribution from circulating monocytes.20 In RA, circulating monocytes are recruited to the joint, where they differentiate into classically activated macrophages, producing pro-inflammatory mediators, such as IL-1β to drive the pathology of synovitis.20,21 Single-cell RNA sequencing (scRNA-seq) analysis revealed that synovial macrophages displayed an inflammatory phenotype (M1), characterized by upregulated expression of IL1B gene and absent gene expression of TGFB1 and CD36 (phagocytic) in ACPA– RA in comparison to ACPA+ RA.22 Furthermore, the gene expression of CCL13, CCL18 and MMP3 were significantly upregulated, while HLA-DRB5 was absent in ACPA– RA synovial macrophage subsets.22 Similar to synovial macrophages, in synovial dendritic cell (DC) subsets of ACPA– RA, the gene expression levels of CCL13, CCL18 and MMP3 were also significantly upregulated, and HLA-DRB5 was absent.22 Aberrantly elevated levels of CCL13 and CCL18 in the serum and synovial tissues from RA patients contribute to immune cell infiltration, synovial cell proliferation and angiogenesis.23, 24, 25 MMP3 has been shown as an early and reliable marker of disease activity and joint and bone damage.26 HLA-DRB5 is identified on haplotypes with HLA-DRB1*15, 16 alleles.27 The precise role of HLA-DRB5 remains unclear, but may be associated with antigen processing and presentation activity.22 Elevated expression of HLA-DRB5 transcripts and increased frequency of the HLA-DRB5*01:05 allele was observed in scleroderma patients with interstitial lung disease.28 In contrast, the HLA-DRB5*null individuals were at increased risk for developing secondary progressive multiple sclerosis.29 Further mechanistic studies are needed to dissect the role of HLA-DRB5 in the pathogenesis of RA, in particular how the loss of HLA-DRB5 expression modulates the immune response in ACPA– RA.
Granulocyte macrophage-colony stimulating factor (GM-CSF) is an important driver of tissue inflammation and arthritic pain, and targeting GM-CSF pathway is effective in both preclinical models and RA clinical trials.30 GM-CSF is produced by a variety of cells, including Th17 cells, type 2 innate lymphoid cells (ILC2), synovial natural killer (NK) cells, synovial stromal cells and CD163+ sublining macrophages.31, 32, 33 GM-CSF skews synovial tissue macrophage towards pro-inflammatory phenotype, characterized by over-expression of activin A, MMP12 and TNFα.33 CCL21 were also elevated in the synovial tissue in RA compared to controls.34 Functionally, CCL21 attracts circulating monocytes into the joint and differentiates them to M1 macrophages, which promotes osteoclastogenesis.35
Genetic variants of IRF5 have been shown involved in a unique disease etiology and pathogenesis in ACPA– RA.13 IRF5 deficiency reduces arthritis severity in K/BxN model, which is probably attributed to the elimination of the role of IRF5 in mediating proinflammatory cytokine production downstream of TLR7 and TLR3.36 Neutrophils represent the most abundant cell population in RA synovial fluid, contributing to joint inflammation and tissue damage. Deficiency in IRF5 also impairs the production of neutrophil chemo-attractants in macrophages and reduces neutrophil trafficking to the joint.37
Taken together, innate immune dysregulation in ACPA– RA is characterized by upregulation of IL-1β, CCL13, CCL18, and MMP3 in synovial macrophages and DCs. Genetic variants of ACPA– RA-risk genes may also modulate myeloid cell functions and promote pro-inflammatory phenotype, leading to osteoclastogenesis and progressive joint destruction (Figure 2).
Figure 2.
Dysregulation of immune cells mainly involved in the pathogenesis of ACPA-negative RA. The dysregulation of innate and adaptive immunity, as well as synovial stromal cells, were depicted in ACPA-negative RA. Synovial macrophages are skewed to pro-inflammatory M1 phenotype with up-regulated gene expression of IL1B, CCL13, CCL18, MMP3 and down-regulated expression of TGFB1 and CD36. FLS and fibroblasts are also important drivers in the pathogenesis of ACPA-negative RA. Adaptive immune dysregulation is mainly associated with IL-6 driven CD4+ T cells dysregulation, Treg/Tfh/Tph cells and abnormal class-switching of B cell subsets. DC, dendritic cells; NK, natural killer; ILC, innate lymphoid cells; FLS, fibroblast-like synoviocyte; MMP, matrix metalloproteinase; Tfh, follicular helper T; Tph, peripheral helper T.
Dysregulated T cell responses are implicated in RA pathogenesis, irrespective of autoantibody status. Gene expression profiling of circulating CD4+ T cells revealed that genes involved in T cell differentiation were over-represented in both ACPA+ and ACPA– RA, compared to inflammatory non-RA arthritis and osteoarthritis (OA).38 STAT4 is a key transcription factor in the differentiation of Th1 cells, which plays a pathogenic role in the development of RA. SNPs within STAT4 loci confer the risks of both ACPA+ and ACPA– RA.2,39 Impaired induction of PTPN22 could also trigger a molecular signature of preclinical RA in both ACPA+ and ACPA– subset. The hypercitrullination, caused by attenuated activity of PTPN22, is responsible for aberrant production of Th2 and Th17 cytokines.40 RUNX1 is a transcription factor critical for Th17 differentiation by inducing RORγt expression.41 We previously showed that the expression of enhancer of zeste homolog 2 (EZH2) was decreased in CD4+ T cells from RA patients, which downregulated RUNX1 and upregulated SMAD7, and ultimately suppressed Tregs differentiation.42 SNPs within RUNX1 loci may relate to Treg/Th17 imbalance, thus confers risk in RA. Additionally, SNPs within ANKRD55 loci are one of the most compelling non-HLA risk alleles associated with both ACPA+ and ACPA– RA.10 ANKRD55 is highly expressed in CD4+ T cells,43 and its polymorphisms may affect glycoprotein 130 (gp130) signaling44 which is critical for downstream cascade of a number of cytokines, including IL-6. Of interest, serum IL-6 levels from ACPA– RA were the highest compared with ACPA+ RA, inflammatory non-RA controls and OA.38 Consistently, basal pSTAT3 in CD4+ T cells was significantly higher in patients with ACPA– RA than in inflammatory non-RA controls and non-inflammatory arthritis controls.45 IL-6-driven CD4+ T cell activation via STAT3 is an early feature in RA, particularly in ACPA– RA disease.38,45
ScRNA-seq analysis revealed that S100A8hiGZMB+ effector CD4+ T (Teff) cells expressing cytotoxic genes (including GNLY and GZMB) were significantly elevated in ACPA– RA compared to healthy controls (HCs).22 Furthermore, the abundance of GZMK+ IFN-act central memory T (TCM) cells expressing central memory CD4+ T cell and interferon pathway genes (including GZMK, ISG15, IFI6, CCR7, GPR183, SELL, TCF7, and IL7R) was also higher in RA patients, especially in ACPA- RA, compared to HCs.22 Of interest, the CD4+ T cells in synovial tissue from ACPA– RA displayed an increased proinflammatory cytokine profile with a significant increase in TNF-expressing capacity and marked polyfunctionality compared to ACPA+ RA.46 ScRNA-seq analysis also identified a distinct high expression of MMP3 in synovial tissue T cells in ACPA– RA.22
Treg dysregulation in ACPA– RA remains poorly defined. Using IL1rn−/− mice, Levescot et al. showed IL-1β induced the osteoclastogenic capacity in synovial Tregs which displayed preserved suppressive capacity and aerobic metabolism but aberrant expression of RANKL and a striking capacity to drive RANKL-dependent osteoclast differentiation.47 Tregs in ACPA– RA synovium may have similar characteristics considering scRNA-seq data found that ACPA– RA synovial macrophages preferentially produced IL-1β. Besides, gene signatures of S100A8hiGZMB+ Teff cells in ACPA– RA were also enriched for “osteoclast differentiation”, as suggested by the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) enrichment analyses.22
Follicular helper T (Tfh) cells provide critical help for B cells development as well as autoantibodies production. Treatment-naïve ACPA– RA patients displayed significantly higher frequency of Tfh cells compared to patients undergoing treatment.46 Peripheral helper T (Tph) (CXCR5–PD1+) cells are a novel CD4+ subset with enhanced migratory capacity towards sites of inflammation and enhanced ability to promote antibody and proinflammatory cytokine production.48,49 Intriguingly, levels of circulating Tph cells were similar between ACPA– RA and ACPA+ RA.46 These findings suggest that Tfh/Tph cells are also involved in the pathogenesis of ACPA– RA.
Dysregulated circulating plasma cell was revealed in ACPA– RA by scRNA-seq analysis, characterized by deficient HLA-DRB5+ plasma cells and enriched IGHG4+ plasma cells.22 A lack of HLA-DRB5 expression and impaired “antigen processing and presentation activity” were also observed in synovial memory B cells of ACPA– RA.22 Of note, comparing to HCs, naïve B cells from ACPA– RA patients displayed an additional differentiation pathway from naïve B cells to IGHG4+ plasma cells due to the lack of an “intermediate” HLA-DRB5+ plasma B.22 The functional relevance of IGHG4+ plasma cells remains unclear, but was suggested to be associated with “osteoclast differentiation” by the KEGG pathway and GO enrichment analyses.22 In addition, circulating plasma cells from ACPA– RA displayed a higher expression of IGHM and lower expressions of IGHG3 and IGHA1 in comparison to those in ACPA+ RA.22 Of interest, a recent study showed that the CD138+ plasma cells in the synovial tissue were more enriched in ACPA– RA compared to psoriatic arthritis, highlighting a critical role of B cells and autoantibodies in the pathogenesis of ACPA– RA.50 Thus, the absence of an “intermediate” HLA-DRB5+ plasma cell, together with higher expression of IGHM and IGHG4, and lower expressions of IGHG3 in circulating B cells suggested the presence of an abnormal class-switching of B cell subsets in ACPA– RA.
Immune-related adverse events associated inflammatory arthritis (irAE-IA) is a newly recognized IA with the utilization of immune checkpoint inhibitors (ICIs), which immunologically resembles seronegative RA.51 ICIs, such as anti-cytotoxic T-lymphocyte antigen-4 (CTLA-4) or anti-programmed death 1 (PD-1) antibodies, activate autoreactive T cells by targeting the negative regulatory molecules.52 Exploration of the shared underlying mechanisms between irAE-IA and ACPA– RA will also help dissect the role of autoreactive T cells in ACPA– RA.
Collectively, these findings suggest that adaptive immune dysregulation in ACPA– RA may be associated with IL-6 mediated CD4+ T cells dysregulation, increased levels of “osteoclast differentiation”-associated CD4+ Teff cells and Tfh/Tph cells, as well as aberrant class-switching in B cell subsets (Figure 2).
Synovial stromal cell dysregulation in ACPA– RA
The synovial pathotypes in RA have been characterized into three types based on the presence of stromal cells and infiltrating leukocytes: lympho-myeloid (the presence of B cells with myeloid cells), diffuse-myeloid (myeloid predominance with little B cells), and pauci-immune (fibroblast-like synoviocytes (FLS) predominance with little immune cell infiltration).53,54 ScRNA-seq on synovial tissue recently revealed lower gene expression levels of markers of plasma cells, exhausted T cells and cytotoxic T/NK cells in the synovial tissue from ACPA– RA compared to ACPA+ RA, suggesting that ACPA– RA are more likely clustered into diffuse-myeloid or pauci-immune category.22 Similarly, van Oosterhout et al also found more fibrosis and synovial lining thickening with less lymphocyte infiltration in synovial tissue from ACPA– RA.55 Of interest, these findings are consistent with those from Humby et al. who revealed that diffuse-myeloid or pauci-immune RA were less likely to develop joint damage progression than lympho-myeloid pathotype.54 Thus, synovial stromal cells may play a more important role in the immunopathogenesis in ACPA– RA compared to ACPA+ RA (Figure 3).
Figure 3.
Patterns of immune cells involvement in the synovial tissue of ACPA-positive and ACPA-negative RA. Both innate and adaptive immune cells are involved in the pathogenesis of RA in synovial tissues, but different patterns may exist in ACPA-positive RA and ACPA-negative RA. Compared with ACPA-positive RA with rich immune cells infiltration and ACPA mediated cytotoxic effects, ACPA-negative RA is characterized by less lymphocyte infiltration but increased proinflammatory cytokine profile in CD4+ T cells. By producing robust MMPs and pro-inflammatory cytokines and chemokines (e.g., IL-6, CXCL12, CCL2), synovial stromal cells may play a more important role in ACPA-negative RA.
Over production of MMPs by FLS contributes to the destruction of the collagen-rich structures in the joint tissues, and enables FLS invasion.56 FLS also secrete a repertoire of cytokines (e.g., IL-6), chemokines (e.g., CCL2, CXCL12) and pro-angiogenic factors to recruit immune cells to perpetuate joint inflammation.56,57 Further, by stimulating the expression of adhesion molecules on endothelial cells, FLS indirectly promote the immune cell infiltration to the joint.56 A recent study revealed that the combination of ferroptosis inducers and TNF inhibitors could result in fibroblast death and attenuate arthritis severity in collagen-induced arthritis model, implicating a potential target for therapy.58
Several studies have proposed that FLS or fibroblasts can be sub-grouped into at least two main pathogenic subsets: the THY1+ subset and THY1– subset.56,57,59, 60, 61, 62 The THY1+ subset, which locates in sublining layer, represents the immune effector subset. In RA, this fibroblast subset is substantially expanded and correlated with disease activity.59 A recent study showed that NOTCH3 signaling contributes to THY1+ fibroblast differentiation, which is also required for the development of inflammatory arthritis.63 The THY1– subset, on the other hand, locates in lining layer. As the source of the MMP13 and MMP3, this subset is proposed as joint-destructive subset.56,57,59, 60, 61, 62 Both subsets are involved in the pathogenesis of ACPA– RA.56,57,59, 60, 61, 62
Inflammatory tissue priming is critical for the transitioning from acute self-limiting inflammation to chronic persistent inflammation in RA.64 A recent study has shown that synovial fibroblasts mediate the inflammatory tissue priming, which depends on intracellular complement C3 and C3a receptor expression. The C3-C3a-C3aR axis drives metabolic reprogramming of fibroblasts, leading to functional changes of THY1+ fibroblast.65 Since this mechanism is mainly observed in leukocyte-rich inflammatory phenotype of arthritis, further study is needed to investigate whether a similar mechanism also exists in ACPA– RA.
C5orf30 polymorphisms is associated with ACPA– RA.10 C5orf30 is predominately expressed by synovial fibroblasts and synovial macrophages in the lining and sublining layer. C5orf30 is highly expressed in the synovium of RA patients compared to controls. Mice with C5orf30 deficiency displayed markedly accentuated joint inflammation and tissue damage. Mechanistically, loss of C5orf30 in fibroblasts led to an auto-aggressive phenotype characterized by increased cell migration and invasion.66 As C5orf30 is not expressed at significant levels in lymphoid cell lines or RA synovial lymphocytes,66 this phenotype may highlight the important role of synovial fibroblast in the joint inflammation and damage (Figure 2).
Novel biomarkers in ACPA– RA
The presence of autoantibodies targeting post-translational modification (PTM) represents a hallmark feature of RA. Although ACPA is not present in ACPA– RA, autoantibodies targeting other PTMs or non-modified antigens, including anti-CarP antibodies, anti-acetylated protein antibodies (AAPA), anti-malondialdehyde antibodies (anti-MDA), anti-malondialdehyde-acetaldehyde antibodies (anti-MAA), anti-homocysteinylated (Hcy)-alpha 1 antitrypsin (A1AT) antibodies, anti-PAD4 and anti-BRAF antibodies could possibly be detected in ACPA– RA.67, 68, 69, 70, 71, 72 These autoantibodies may be implicated in the pathogenesis of ACPA– RA. For example, anti-CarP antibody can predict radiographic progression in patients with ACPA– RA.67 In addition, anti-MDA antibodies significantly correlated with disease activity and inflammatory markers, and some robustly enhanced osteoclastogenesis in vitro.69
Utilizing high-density protein microarray, we recently identified two novel autoantibodies (anti-PTX3 and anti-DUSP11 autoantibody) in RA. Different from other autoantibodies that are predominantly present in ACPA+ RA (i.e. anti-PTMs), the prevalence of anti-PTX3 and anti-DUSP11 autoantibodies are similar between ACPA+ RA and ACPA– RA.73 The levels of anti-PTX3 antibody also positively correlated with disease activity and inflammatory markers.73 Notably, the serum levels of anti-PTX3 antibodies decreased dramatically in RA patients after effective treatment, suggesting that anti-PTX3 antibodies may assist early diagnosis and treatment monitoring in ACPA– RA.73 Of interest, we previously showed that increased circulating PTX3 in RA could synergize with complement C1q to activate inflammasome and pyroptosis.74 Thus, it would be interesting to determine whether the binding of anti-PTX3 antibody to PTX3 inhibit its pathogenic activity or not.
Another recent study identified increased serum levels of soluble scavenger receptor-A (sSR-A) in patients with RA which correlate with clinical and immunological features of the disease.75 Importantly, higher sSR-A can be detected in 63% patients with early RA and 66% established RA patients, including 50% of ACPA– RA.75 Although the pathogenic mechanisms of sSR-A in RA remains unclear, mice deficient in SR-A or administered with SR-A inhibitor displayed less disease severity, suggesting that targeting this molecule may have a therapeutic potential in RA.
Conclusions and future perspective
At present, the etiology and disease progression of ACPA– RA remains largely unknown. It may involve a complex interplay between the innate and adaptive arms of the immune response and the synovial stromal cells. Specifically, synovial macrophages and DCs displayed elevated gene expression levels of CCL13, CCL18 and MMP3, and synovial memory B cells displayed a lack of HLA-DRB5 expression and impaired “antigen processing and presentation activity”. In periphery, IL-6 mediated CD4+ T cells dysregulation and elevated IGHG4+ plasma cells may be implicated in the disease pathogenesis. Remarkably, the scRNA-seq and synovial histology data suggest that the immunopathology of synovitis in ACPA– RA is greatly driven by synovial stromal and myeloid cells, with minor involvement of adaptive immune cells. Also, different responses to targeting therapies between the ACPA+ RA patients and ACPA– RA patients implied differential underlying pathogenesis. ACPA+RA patients displayed significant improvement in outcomes with abatacept or rituximab treatments, whereas no difference or a better clinical response was observed in ACPA– RA patients treated with tocilizumab or tofacitinib or TNF-α inhibitors.76,77 However, the involvement of adaptive immunity should not be ignored in ACPA– RA, especially the specific pathogenic adaptive immune cell subsets as well as newly identified autoantibodies.
It should be pointed out that multiple key challenges remain to be addressed. The first one is the immune triggers in ACPA– RA at temporal and spatial levels. Compared to ACPA+ RA, the immune triggers in ACPA– RA remains largely unknown. Second, most of the studies reviewed in this article are based on general RA population. It is unclear whether these mechanisms are indeed present in ACPA– RA. Although these studies claimed that both ACPA+ RA and ACPA– RA patients were included, they did not specify whether the positive findings were from the ACPA+ RA patients or from ACPA– RA patients. It will be of great importance to make substantial comparisons between ACPA+ RA and ACPA– RA patients in the further studies. In addition, many of the mechanistic assumptions in this review are from the genome-wide association studies (GWAS) on genetic risk variants in ACPA– RA. Although a number of genetic risk variants for ACPA– RA have been identified, the precise pathogenic mechanisms associated with the causal variants remain to be explored. Further, some of the mechanistic explanations for ACPA– RA have been developed on the basis of animal models, lack of epidemiological evidence. It is necessary to verify these findings in patients with ACPA– RA in real world studies. Lastly, ACPA– RA is usually regarded as a milder disease compared to ACPA+ counterpart, but it is probably more heterogeneous with more divergent long-term outcome and a more complex etiopathology.78 Thus, further stratify ACPA– RA patients into different subsets using reliable biomarkers and dissect the underlying mechanisms in different subgroups will be of great clinical relevance.
A better understanding of the immune mechanisms in ACPA– RA will facilitate the designing of more precise targeting therapies against putative pathogenic subsets, and provide the promise for a personalized treatment strategy. In addition, identification of novel biomarkers for the early recognition of ACPA– RA will substantially promote early treatment and ultimately improve the outcomes.
Outstanding questions
Most studies focused on the general RA population without comparing the potential difference between ACPA+ and ACPA– RA. The specific immune triggers and mechanisms in ACPA– RA remains to be elucidated. Further explorations are needed to discover discriminative biomarkers and crucial mechanisms in this subset, so as to facilitate prompt diagnosis and precise therapy.
Search strategy and selection criteria
We conducted data search and collection in PubMed, using the terms “rheumatoid arthritis”, “anti-citrullinated protein autoantibodies”, “ACPA-positive RA”, “ACPA-negative RA”, “biomarkers”, “genetics”, “gut microbiome”, “adaptive immunity”, “innate immunity”, “synovial cells” and “treatment”. Articles written in English and published between 2000 and 2022 were included.
Contributors
X.Z. and Y.L. conceived the study and supervised the work. Y.L., K.L. and M.W. were participated in data collection, interpretation and manuscript writing. K.L. and Y.L. designed and completed the figures. L.Z. and X.Z. revised the final manuscript.
Declaration of interests
All the authors have nothing to disclose.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (81788101, 81630044, 81971521, 82171778), the CAMS Innovation Fund for Medical Sciences (CIFMS) (2021-I2M-1-017, 2021-I2M-1-047), and Capital's Funds for Health Improvement and Research (2020-2-4019). Figures were created with BioRender.com.
Contributor Information
Yudong Liu, Email: yudongliu1983@126.com.
Xuan Zhang, Email: zxpumch2003@sina.com.
References
- 1.Smolen JS, Aletaha D, Barton A, et al. Rheumatoid arthritis. Nat Rev Dis Primers. 2018;4:18001. doi: 10.1038/nrdp.2018.1. [DOI] [PubMed] [Google Scholar]
- 2.Padyukov L, Seielstad M, Ong RT, et al. A genome-wide association study suggests contrasting associations in ACPA-positive versus ACPA-negative rheumatoid arthritis. Ann Rheum Dis. 2011;70(2):259–265. doi: 10.1136/ard.2009.126821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgers LE, van Steenbergen HW, Ten Brinck RM, Huizinga TW, van der Helm-van Mil AH. Differences in the symptomatic phase preceding ACPA-positive and ACPA-negative RA: a longitudinal study in arthralgia during progression to clinical arthritis. Ann Rheum Dis. 2017;76(10):1751–1754. doi: 10.1136/annrheumdis-2017-211325. [DOI] [PubMed] [Google Scholar]
- 4.Turesson C, Jacobsson LT, Sturfelt G, Matteson EL, Mathsson L, Rönnelid J. Rheumatoid factor and antibodies to cyclic citrullinated peptides are associated with severe extra-articular manifestations in rheumatoid arthritis. Ann Rheum Dis. 2007;66(1):59–64. doi: 10.1136/ard.2006.054445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myasoedova E, Davis J, Matteson EL, Crowson CS. Is the epidemiology of rheumatoid arthritis changing? Results from a population-based incidence study, 1985-2014. Ann Rheum Dis. 2020;79(4):440–444. doi: 10.1136/annrheumdis-2019-216694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kampstra ASB, Toes REM. HLA class II and rheumatoid arthritis: the bumpy road of revelation. Immunogenetics. 2017;69(8-9):597–603. doi: 10.1007/s00251-017-0987-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verpoort KN, van Gaalen FA, van der Helm-van Mil AH, et al. Association of HLA-DR3 with anti-cyclic citrullinated peptide antibody-negative rheumatoid arthritis. Arthritis Rheum. 2005;52(10):3058–3062. doi: 10.1002/art.21302. [DOI] [PubMed] [Google Scholar]
- 8.Regueiro C, Casares-Marfil D, Lundberg K, et al. HLA-B*08 identified as the most prominently associated major histocompatibility complex locus for anti-carbamylated protein antibody-positive/anti-cyclic citrullinated peptide-negative rheumatoid arthritis. Arthritis Rheumatol. 2021;73(6):963–969. doi: 10.1002/art.41630. [DOI] [PubMed] [Google Scholar]
- 9.Eyre S, Bowes J, Diogo D, et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012;44(12):1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viatte S, Massey J, Bowes J, et al. Replication of associations of genetic loci outside the HLA region with susceptibility to anti-cyclic citrullinated peptide-negative rheumatoid arthritis. Arthritis Rheumatol. 2016;68(7):1603–1613. doi: 10.1002/art.39619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gazal S, Sacre K, Allanore Y, et al. Identification of secreted phosphoprotein 1 gene as a new rheumatoid arthritis susceptibility gene. Ann Rheum Dis. 2015;74(3):e19. doi: 10.1136/annrheumdis-2013-204581. [DOI] [PubMed] [Google Scholar]
- 12.Arandjelovic S, Perry JSA, Lucas CD, et al. A noncanonical role for the engulfment gene ELMO1 in neutrophils that promotes inflammatory arthritis. Nat Immunol. 2019;20(2):141–151. doi: 10.1038/s41590-018-0293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sigurdsson S, Padyukov L, Kurreeman FA, et al. Association of a haplotype in the promoter region of the interferon regulatory factor 5 gene with rheumatoid arthritis. Arthritis Rheum. 2007;56(7):2202–2210. doi: 10.1002/art.22704. [DOI] [PubMed] [Google Scholar]
- 14.Jiao Y, Wu L, Huntington ND, Zhang X. Crosstalk between gut microbiota and innate immunity and its implication in autoimmune diseases. Front Immunol. 2020;11:282. doi: 10.3389/fimmu.2020.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu HJ, Ivanov II, Darce J, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32(6):815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdollahi-Roodsaz S, Joosten LA, Koenders MI, et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J Clin Invest. 2008;118(1):205–216. doi: 10.1172/JCI32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogier R, Ederveen THA, Boekhorst J, et al. Aberrant intestinal microbiota due to IL-1 receptor antagonist deficiency promotes IL-17- and TLR4-dependent arthritis. Microbiome. 2017;5(1):63. doi: 10.1186/s40168-017-0278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang HI, Li JR, Liu CC, et al. An association of gut microbiota with different phenotypes in chinese patients with rheumatoid arthritis. J Clin Med. 2019;8(11):1770. doi: 10.3390/jcm8111770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He J, Chu Y, Li J, Meng Q, Liu Y, Jin J, et al. Intestinal butyrate-metabolizing species contribute to autoantibody production and bone erosion in rheumatoid arthritis. Sci Adv. 2022;8(6):eabm1511. doi: 10.1126/sciadv.abm1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misharin AV, Cuda CM, Saber R, et al. Nonclassical Ly6C(-) monocytes drive the development of inflammatory arthritis in mice. Cell Rep. 2014;9(2):591–604. doi: 10.1016/j.celrep.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Udalova IA, Mantovani A, Feldmann M. Macrophage heterogeneity in the context of rheumatoid arthritis. Nat Rev Rheumatol. 2016;12(8):472–485. doi: 10.1038/nrrheum.2016.91. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, Liu Y, Jin S, et al. Single-cell sequencing of immune cells from anticitrullinated peptide antibody positive and negative rheumatoid arthritis. Nat Commun. 2021;12(1):4977. doi: 10.1038/s41467-021-25246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwamoto T, Okamoto H, Iikuni N, et al. Monocyte chemoattractant protein-4 (MCP-4)/CCL13 is highly expressed in cartilage from patients with rheumatoid arthritis. Rheumatology (Oxford) 2006;45(4):421–424. doi: 10.1093/rheumatology/kei209. [DOI] [PubMed] [Google Scholar]
- 24.Iwamoto T, Okamoto H, Toyama Y, Momohara S. Molecular aspects of rheumatoid arthritis: chemokines in the joints of patients. FEBS J. 2008;275(18):4448–4455. doi: 10.1111/j.1742-4658.2008.06580.x. [DOI] [PubMed] [Google Scholar]
- 25.van Lieshout AW, Fransen J, Flendrie M, et al. Circulating levels of the chemokine CCL18 but not CXCL16 are elevated and correlate with disease activity in rheumatoid arthritis. Ann Rheum Dis. 2007;66(10):1334–1338. doi: 10.1136/ard.2006.066084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lerner A, Neidhofer S, Reuter S, Matthias T. MMP3 is a reliable marker for disease activity, radiological monitoring, disease outcome predictability, and therapeutic response in rheumatoid arthritis. Best Pract Res Clin Rheumatol. 2018;32(4):550–562. doi: 10.1016/j.berh.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Creary LE, Sacchi N, Mazzocco M, et al. High-resolution HLA allele and haplotype frequencies in several unrelated populations determined by next generation sequencing: 17th International HLA and Immunogenetics Workshop joint report. Hum Immunol. 2021;82(7):505–522. doi: 10.1016/j.humimm.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odani T, Yasuda S, Ota Y, et al. Up-regulated expression of HLA-DRB5 transcripts and high frequency of the HLA-DRB5*01:05 allele in scleroderma patients with interstitial lung disease. Rheumatology (Oxford) 2012;51(10):1765–1774. doi: 10.1093/rheumatology/kes149. [DOI] [PubMed] [Google Scholar]
- 29.Caillier SJ, Briggs F, Cree BA, et al. Uncoupling the roles of HLA-DRB1 and HLA-DRB5 genes in multiple sclerosis. J Immunol. 2008;181(8):5473–5480. doi: 10.4049/jimmunol.181.8.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burmester GR, McInnes IB, Kremer J, et al. A randomised phase IIb study of mavrilimumab, a novel GM-CSF receptor alpha monoclonal antibody, in the treatment of rheumatoid arthritis. Ann Rheum Dis. 2017;76(6):1020–1030. doi: 10.1136/annrheumdis-2016-210624. [DOI] [PubMed] [Google Scholar]
- 31.Hirota K, Hashimoto M, Ito Y, et al. Autoimmune Th17 cells induced synovial stromal and innate lymphoid cell secretion of the cytokine GM-CSF to initiate and augment autoimmune arthritis. Immunity. 2018;48(6):1220–1232. doi: 10.1016/j.immuni.2018.04.009. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Louis C, Souza-Fonseca-Guimaraes F, Yang Y, et al. NK cell-derived GM-CSF potentiates inflammatory arthritis and is negatively regulated by CIS. J Exp Med. 2020;217(5):e20191421. doi: 10.1084/jem.20191421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuentelsaz-Romero S, Cuervo A, Estrada-Capetillo L, et al. GM-CSF expression and macrophage polarization in joints of undifferentiated arthritis patients evolving to rheumatoid arthritis or psoriatic arthritis. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.613975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pickens SR, Chamberlain ND, Volin MV, Pope RM, Mandelin AM, 2nd, Shahrara S. Characterization of CCL19 and CCL21 in rheumatoid arthritis. Arthritis Rheum. 2011;63(4):914–922. doi: 10.1002/art.30232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Raemdonck K, Umar S, Palasiewicz K, et al. CCL21/CCR7 signaling in macrophages promotes joint inflammation and Th17-mediated osteoclast formation in rheumatoid arthritis. Cell Mol Life Sci. 2020;77(7):1387–1399. doi: 10.1007/s00018-019-03235-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duffau P, Menn-Josephy H, Cuda CM, et al. Promotion of inflammatory arthritis by interferon regulatory factor 5 in a mouse model. Arthritis Rheumatol. 2015;67(12):3146–3157. doi: 10.1002/art.39321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss M, Byrne AJ, Blazek K, et al. IRF5 controls both acute and chronic inflammation. Proc Natl Acad Sci USA. 2015;112(35):11001–11006. doi: 10.1073/pnas.1506254112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pratt AG, Swan DC, Richardson S, et al. A CD4 T cell gene signature for early rheumatoid arthritis implicates interleukin 6-mediated STAT3 signalling, particularly in anti-citrullinated peptide antibody-negative disease. Ann Rheum Dis. 2012;71(8):1374–1381. doi: 10.1136/annrheumdis-2011-200968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seddighzadeh M, Gonzalez A, Ding B, et al. Variants within STAT genes reveal association with anticitrullinated protein antibody-negative rheumatoid arthritis in 2 European populations. J Rheumatol. 2012;39(8):1509–1516. doi: 10.3899/jrheum.111284. [DOI] [PubMed] [Google Scholar]
- 40.Chang HH, Liu GY, Dwivedi N, et al. A molecular signature of preclinical rheumatoid arthritis triggered by dysregulated PTPN22. JCI Insight. 2016;1(17):e90045. doi: 10.1172/jci.insight.90045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9(11):1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao XY, Li YT, Jiang X, et al. EZH2 deficiency attenuates Treg differentiation in rheumatoid arthritis. J Autoimmun. 2020;108 doi: 10.1016/j.jaut.2020.102404. [DOI] [PubMed] [Google Scholar]
- 43.Hu X, Kim H, Stahl E, Plenge R, Daly M, Raychaudhuri S. Integrating autoimmune risk loci with gene-expression data identifies specific pathogenic immune cell subsets. Am J Hum Genet. 2011;89(4):496–506. doi: 10.1016/j.ajhg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lill CM, Schjeide BM, Graetz C, et al. Genome-wide significant association of ANKRD55 rs6859219 and multiple sclerosis risk. J Med Genet. 2013;50(3):140–143. doi: 10.1136/jmedgenet-2012-101411. [DOI] [PubMed] [Google Scholar]
- 45.Anderson AE, Pratt AG, Sedhom MA, et al. IL-6-driven STAT signalling in circulating CD4+ lymphocytes is a marker for early anticitrullinated peptide antibody-negative rheumatoid arthritis. Ann Rheum Dis. 2016;75(2):466–473. doi: 10.1136/annrheumdis-2014-205850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Floudas A, Canavan M, McGarry T, et al. ACPA status correlates with differential immune profile in patients with rheumatoid arthritis. Cells. 2021;10(3):647. doi: 10.3390/cells10030647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levescot A, Chang MH, Schnell J, et al. IL-1beta-driven osteoclastogenic Tregs accelerate bone erosion in arthritis. J Clin Invest. 2021;131(18) doi: 10.1172/JCI141008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao DA, Gurish MF, Marshall JL, et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature. 2017;542(7639):110–114. doi: 10.1038/nature20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakuragi T, Yamada H, Haraguchi A, et al. Autoreactivity of peripheral helper T cells in the joints of rheumatoid arthritis. J Immunol. 2021;206(9):2045–2051. doi: 10.4049/jimmunol.2000783. [DOI] [PubMed] [Google Scholar]
- 50.Alivernini S, Bruno D, Tolusso B, et al. Differential synovial tissue biomarkers among psoriatic arthritis and rheumatoid factor/anti-citrulline antibody-negative rheumatoid arthritis. Arthritis Res Ther. 2019;21(1):116. doi: 10.1186/s13075-019-1898-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Jaquith JM, McCarthy-Fruin K, et al. Immune checkpoint inhibitor-induced inflammatory arthritis: a novel clinical entity with striking similarities to seronegative rheumatoid arthritis. Clin Rheumatol. 2020;39(12):3631–3637. doi: 10.1007/s10067-020-05162-9. [DOI] [PubMed] [Google Scholar]
- 52.Calabrese LH, Calabrese C, Cappelli LC. Rheumatic immune-related adverse events from cancer immunotherapy. Nat Rev Rheumatol. 2018;14(10):569–579. doi: 10.1038/s41584-018-0074-9. [DOI] [PubMed] [Google Scholar]
- 53.Lewis MJ, Barnes MR, Blighe K, et al. Molecular portraits of early rheumatoid arthritis identify clinical and treatment response phenotypes. Cell Rep. 2019;28(9):2455–2470. doi: 10.1016/j.celrep.2019.07.091. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Humby F, Lewis M, Ramamoorthi N, et al. Synovial cellular and molecular signatures stratify clinical response to csDMARD therapy and predict radiographic progression in early rheumatoid arthritis patients. Ann Rheum Dis. 2019;78(6):761–772. doi: 10.1136/annrheumdis-2018-214539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Oosterhout M, Bajema I, Levarht EW, Toes RE, Huizinga TW, van Laar JM. Differences in synovial tissue infiltrates between anti-cyclic citrullinated peptide-positive rheumatoid arthritis and anti-cyclic citrullinated peptide-negative rheumatoid arthritis. Arthritis Rheum. 2008;58(1):53–60. doi: 10.1002/art.23148. [DOI] [PubMed] [Google Scholar]
- 56.Nygaard G, Firestein GS. Restoring synovial homeostasis in rheumatoid arthritis by targeting fibroblast-like synoviocytes. Nat Rev Rheumatol. 2020;16(6):316–333. doi: 10.1038/s41584-020-0413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei K, Nguyen HN, Brenner MB. Fibroblast pathology in inflammatory diseases. J Clin Invest. 2021;131(20) doi: 10.1172/JCI149538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu J, Feng Z, Chen L, et al. TNF antagonist sensitizes synovial fibroblasts to ferroptotic cell death in collagen-induced arthritis mouse models. Nat Commun. 2022;13(1):676. doi: 10.1038/s41467-021-27948-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Croft AP, Campos J, Jansen K, et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature. 2019;570(7760):246–251. doi: 10.1038/s41586-019-1263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang F, Wei K, Slowikowski K, et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol. 2019;20(7):928–942. doi: 10.1038/s41590-019-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mizoguchi F, Slowikowski K, Wei K, et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat Commun. 2018;9(1):789. doi: 10.1038/s41467-018-02892-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stephenson W, Donlin LT, Butler A, et al. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat Commun. 2018;9(1):791. doi: 10.1038/s41467-017-02659-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei K, Korsunsky I, Marshall JL, et al. Notch signalling drives synovial fibroblast identity and arthritis pathology. Nature. 2020;582(7811):259–264. doi: 10.1038/s41586-020-2222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schett G, Neurath MF. Resolution of chronic inflammatory disease: universal and tissue-specific concepts. Nat Commun. 2018;9(1):3261. doi: 10.1038/s41467-018-05800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Friscic J, Bottcher M, Reinwald C, et al. The complement system drives local inflammatory tissue priming by metabolic reprogramming of synovial fibroblasts. Immunity. 2021;54(5):1002–1021. doi: 10.1016/j.immuni.2021.03.003. e10. [DOI] [PubMed] [Google Scholar]
- 66.Muthana M, Hawtree S, Wilshaw A, et al. C5orf30 is a negative regulator of tissue damage in rheumatoid arthritis. Proc Natl Acad Sci U S A. 2015;112(37):11618–11623. doi: 10.1073/pnas.1501947112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ajeganova S, van Steenbergen HW, Verheul MK, et al. The association between anti-carbamylated protein (anti-CarP) antibodies and radiographic progression in early rheumatoid arthritis: a study exploring replication and the added value to ACPA and rheumatoid factor. Ann Rheum Dis. 2017;76(1):112–118. doi: 10.1136/annrheumdis-2015-208870. [DOI] [PubMed] [Google Scholar]
- 68.Thiele GM, Duryee MJ, Anderson DR, et al. Malondialdehyde-acetaldehyde adducts and anti-malondialdehyde-acetaldehyde antibodies in rheumatoid arthritis. Arthritis Rheumatol. 2015;67(3):645–655. doi: 10.1002/art.38969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gronwall C, Amara K, Hardt U, et al. Autoreactivity to malondialdehyde-modifications in rheumatoid arthritis is linked to disease activity and synovial pathogenesis. J Autoimmun. 2017;84:29–45. doi: 10.1016/j.jaut.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 70.Juarez M, Bang H, Hammar F, et al. Identification of novel antiacetylated vimentin antibodies in patients with early inflammatory arthritis. Ann Rheum Dis. 2016;75(6):1099–1107. doi: 10.1136/annrheumdis-2014-206785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Colasanti T, Sabatinelli D, Mancone C, et al. Homocysteinylated alpha 1 antitrypsin as an antigenic target of autoantibodies in seronegative rheumatoid arthritis patients. J Autoimmun. 2020;113 doi: 10.1016/j.jaut.2020.102470. [DOI] [PubMed] [Google Scholar]
- 72.Auger I, Balandraud N, Rak J, Lambert N, Martin M, Roudier J. New autoantigens in rheumatoid arthritis (RA): screening 8268 protein arrays with sera from patients with RA. Ann Rheum Dis. 2009;68(4):591–594. doi: 10.1136/ard.2008.096917. [DOI] [PubMed] [Google Scholar]
- 73.Li K, Mo W, Wu L, et al. Novel autoantibodies identified in ACPA-negative rheumatoid arthritis. Ann Rheum Dis. 2021;80(6):739–747. doi: 10.1136/annrheumdis-2020-218460. [DOI] [PubMed] [Google Scholar]
- 74.Wu XY, Li KT, Yang HX, et al. Complement C1q synergizes with PTX3 in promoting NLRP3 inflammasome over-activation and pyroptosis in rheumatoid arthritis. J Autoimmun. 2020;106 doi: 10.1016/j.jaut.2019.102336. [DOI] [PubMed] [Google Scholar]
- 75.Hu F, Jiang X, Guo C, et al. Scavenger receptor-A is a biomarker and effector of rheumatoid arthritis: a large-scale multicenter study. Nat Commun. 2020;11(1):1911. doi: 10.1038/s41467-020-15700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harrold LR, Connolly SE, Wittstock K, et al. Baseline anti-citrullinated protein antibody status and response to abatacept or non-TNFi biologic/targeted-synthetic DMARDs: US observational study of patients with RA. Rheumatol Ther. 2022;9(2):465–480. doi: 10.1007/s40744-021-00401-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin CT, Huang WN, Tsai WC, et al. Predictors of drug survival for biologic and targeted synthetic DMARDs in rheumatoid arthritis: analysis from the TRA Clinical Electronic Registry. PLoS One. 2021;16(4) doi: 10.1371/journal.pone.0250877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matthijssen XME, Niemantsverdriet E, Huizinga TWJ, van der Helm-van Mil AHM. Enhanced treatment strategies and distinct disease outcomes among autoantibody-positive and -negative rheumatoid arthritis patients over 25 years: a longitudinal cohort study in the Netherlands. PLoS Med. 2020;17(9) doi: 10.1371/journal.pmed.1003296. [DOI] [PMC free article] [PubMed] [Google Scholar]