Abstract
The ubihydroquinone-cytochrome c oxidoreductase (or the cytochrome bc1 complex) from Rhodobacter capsulatus is composed of the Fe-S protein, cytochrome b, and cytochrome c1 subunits encoded by petA(fbcF), petB(fbcB), and petC(fbcC) genes organized as an operon. In the work reported here, petB(fbcB) was split genetically into two cistrons, petB6 and petBIV, which encoded two polypeptides corresponding to the four amino-terminal and four carboxyl-terminal transmembrane helices of cytochrome b, respectively. These polypeptides resembled the cytochrome b6 and su IV subunits of chloroplast cytochrome b6f complexes, and together with the unmodified subunits of the cytochrome bc1 complex, they formed a novel enzyme, named cytochrome b6c1 complex. This membrane-bound multisubunit complex was functional, and despite its smaller amount, it was able to support the photosynthetic growth of R. capsulatus. Upon further mutagenesis, a mutant overproducing it, due to a C-to-T transition at the second base of the second codon of petBIV, was obtained. Biochemical analyses, including electron paramagnetic spectroscopy, with this mutant revealed that the properties of the cytochrome b6c1 complex were similar to those of the cytochrome bc1 complex. In particular, it was highly sensitive to inhibitors of the cytochrome bc1 complex, including antimycin A, and the redox properties of its b- and c-type heme prosthetic groups were unchanged. However, the optical absorption spectrum of its cytochrome bL heme was modified in a way reminiscent of that of a cytochrome b6f complex. Based on the work described here and that with Rhodobacter sphaeroides (R. Kuras, M. Guergova-Kuras, and A. R. Crofts, Biochemistry 37:16280–16288, 1998), it appears that neither the inhibitor resistance nor the redox potential differences observed between the bacterial (or mitochondrial) cytochrome bc1 complexes and the chloroplast cytochrome b6f complexes are direct consequences of splitting cytochrome b into two separate polypeptides. The overall findings also illustrate the possible evolutionary relationships among various cytochrome bc oxidoreductases.
Ubihydroquinone (UQH2)-cytochrome c (cytochrome bc1 complex) and plastohydroquinone (PQH2)-plastocyanine (PQ) (cytochrome b6f complex) oxidoreductases are major membrane protein complexes involved in electron transfer in bacteria and mitochondria and in chloroplasts, respectively. They contribute to the formation of membrane potential and proton gradient, which are coupled to ATP synthesis (for reviews see references 5, 14, 17, and 34). The cytochrome bc1 complex is located in the inner membrane of mitochondria (34) and the cytoplasmic membranes of bacteria (14), while the cytochrome b6f complex is located in the thylakoid membranes of chloroplasts (5) and the cytoplasmic membranes of cyanobacteria (17). In facultative phototrophic bacteria like Rhodobacter capsulatus, the cytochrome bc1 complex is part of the cyclic electron transfer around the photochemical reaction center and donates electrons to its physiological partners, the soluble cytochrome c2 and the membrane-associated cytochrome cy (14). In cyanobacteria, the cytochrome b6f complex is involved in the linear electron transfer between photosystem II and photosystem I, where PQ is the electron carrier (17).
The cytochrome bc1 complexes contain three catalytic subunits with redox-active prosthetic groups: cytochrome b is an integral membrane protein of 48 kDa, has eight transmembrane helices, and houses two noncovalently bound b-type hemes of low and high redox midpoint potentials (Em7) (cytochrome bL [Em7, around −90 mV) and cytochrome bH [Em7, around 50 mV]) (28); cytochrome c1 contains a covalently bound c-type heme, and the Fe-S protein subunit has a [2Fe-2S] cluster with a high Em value (14, 34). In R. capsulatus these subunits are encoded by petA (fbcF) (Fe-S protein), petB (fbcB) (cytochrome b), and petC (fbcC) (cytochrome c1) cistrons organized as an operon, petABC (fbcFBC) (14), which is essential for the photosynthetic (PS) growth of this species (6). The cytochrome b6f complexes, on the other hand, have four subunits: cytochrome b6 has four membrane-spanning helices and two b-type hemes (cytochrome bL [Em, around −170 mV] and cytochrome bH [Em, around −50 mV]), and su IV is formed from three membrane-spanning helices (5, 17). Together, these two subunits are highly homologous to the first seven transmembrane helices of cytochrome b. The remaining two subunits of the cytochrome b6f complexes are functional analogs of cytochrome c1 and the Fe-S protein subunits of cytochrome bc1 complexes and are called cytochrome f and Fe-S protein, respectively (5, 17). In the cytochrome bc1 complex, chemical catalysis of UQH2 to ubiquinone (UQ) follows the Q cycle model of Mitchell (25) and takes place in two distinct domains, Qo and Qi sites, located on the outer (near bL heme) and the inner (near bH heme) sides of the cytoplasmic membrane, respectively. A similar Q cycle scheme has also been proposed for the mechanism of function of the cytochrome b6f complexes (23).
Pronounced sequence homology is apparent between the cytochrome b and the cytochrome b6 plus the su IV subunits from various species (9). In particular, there is about 60% amino acid similarity between the amino-terminal four helices of cytochrome b and cytochrome b6 and between the remaining carboxyl-terminal part of cytochrome b and the su IV protein (12). This remarkable similarity has led to the proposal that the cytochrome b6 and su IV subunits may have originated from splitting of cytochrome b during evolution (36). Recently, a third kind of cytochrome bc complex, known as menahydroquinone-cytochrome c oxidoreductase, has been encountered among the gram-positive bacteria (33, 39). These cytochrome bc complexes contain a short cytochrome b6 subunit which is similar to those found in cytochrome b6f complexes but often associated with different variants of the remaining subunits. For example, in the case of Bacillus subtilis, the homolog of su IV is fused to the amino-terminal end of a cytochrome c1 homolog (40), and in the case of Heliobacillus mobilis, cytochrome c1 is an unusual diheme c-type cytochrome (38).
The structure of the mitochondrial bc1 complex is now available at atomic resolution (16, 37, 42), and the three-dimensional models built with X-ray diffraction data fully support previous findings from genetic, biochemical, and biophysical studies (3). In the case of the cytochrome b6f complex, although the structures of the soluble domains of cytochrome f (24) and the Fe-S protein (4) subunits are known, no high-resolution structure for a complete cytochrome b6f complex is yet available. In this work, using the available structural information and molecular genetic approaches, we have attempted to generate a novel variant of UQH2-cytochrome c oxidoreductase, named cytochrome b6c1 complex, by converting the three-cistronic petABC operon into a four-cistronic petAB6BIVC operon. For this purpose, we have split R. capsulatus cytochrome b genetically into two polypeptides analogous to the cytochrome b6 and su IV subunits of the cytochrome b6f complex. We found that these polypeptides assembled correctly with the Fe-S protein and cytochrome c1 subunits to form an active UQH2-cytochrome c oxidoreductase which was able to support PS growth of R. capsulatus. Similar, and in many aspects complementary, work has recently been reported by Kuras et al. (19), using R. sphaeroides cytochrome bc1 complex. Together, these studies demonstrate that it is feasible to convert a cytochrome bc1 complex into a four-subunit cytochrome b6f-like complex, and they illustrate evolutionary structural and functional similarities among the bacterial, mitochondrial, and chloroplast b- and c-type cytochrome-bearing redox-driven proton pumps.
MATERIALS AND METHODS
Growth conditions and strains used.
R. capsulatus and Escherichia coli (HB101, MV1190, and XL1-Red) strains were grown in MPYE and Luria-Bertani media, respectively, supplemented with appropriate antibiotics as needed (6, 8). PS growth rates and sensitivity to myxothiazol or stigmatellin were determined as described previously (7).
Molecular genetic techniques.
For construction of various mutants, site-directed mutagenesis was performed as described previously (1). The mutagenic oligonucleotide 5′GGG GTT GAA GTC CGC GCC GAA AAG GAC ACC3′ was used to delete the amino acid residues Arg231-Thr-Ser-Lys-Ala-Asp236 (located between base pair positions 2010 and 2038) of cytochrome b by using phage M13-mp10-BC1Smadel6 DNA as a template (1), yielding a petBΔ(231-236) mutant (13). The oligonucleotide 5′GTT GAA GTC CGC TGA GGG GAA AGA CAC ATG GCC GAA AAG GAC3′ (boldface letters indicate the termination and initiation codons, and underlined bases correspond to a putative ribosome binding site) was used for splitting petB into two cistrons, petB6 and petBIV, using petBΔ(231-236) as a template (Fig. 1). Following mutagenesis and screening by DNA sequence analysis, the SmaI-AsuII (SfuI) fragment containing the desired mutation was excised from the replicative form of the corresponding mutant phage and exchanged for its wild-type counterpart on plasmid pMTS1, which carries a wild-type copy of petABC and kanamycin resistance (15). The plasmid pSM1 thus obtained was introduced by triparental crosses into the R. capsulatus strain MT-RBC1, which carries a complete deletion of petABC, as described previously (1).
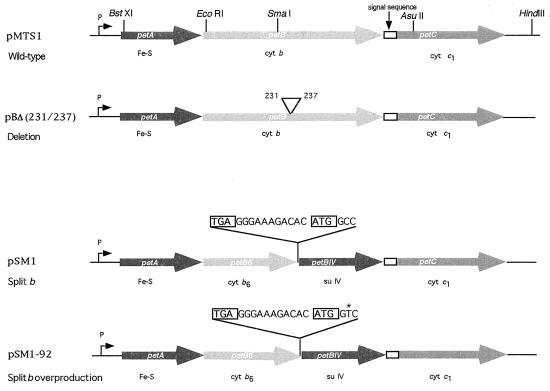
FIG. 1.
Genetic organization of petABC(fbcFBC) operon encoding the Fe-S, cytochrome b, and cytochrome c1 subunits of R. capsulatus cytochrome bc1 complex. Plasmids pMTS1 and pBΔ(231/237) refer to the wild-type petABC operon and to its variant carrying a deletion in cytochrome b extending from amino acid positions 231 to 237, respectively. Plasmids pSM1 and pSM1-92 correspond to petAB6BIVC derivatives encoding the cytochrome b6c1 complex, obtained after genetically splitting petB into two cistrons (petB6 and petBIV) by introduction of a stop (TGA [boxed]) codon, an intergenic (GGGAAAGACAC) sequence, and a start (ATG [boxed]) codon as indicated. pSM1-92 was derived from pSM1 and contains a single-base-pair substitution at the second base of the second codon of petBIV, as indicated by the asterisk. P and open rectangle, promoter of petABC and processed signal sequence of cytochrome c1, respectively.
Isolation of a mutant derivative of pSM1 overproducing cytochrome b6c1 complex.
To isolate a mutant derivative of pSM1 producing increased amounts of the cytochrome b6c1 complex, this plasmid was randomly mutagenized by passage via the E. coli mutator strain XL1-Red (Stratagene, Inc.) according to the protocol provided by the supplier. A mutagenized culture was used directly as a conjugal donor to MT-RBC1, and Kanr colonies were selected under respiratory growth conditions and tested for their PS growth ability. Transconjugants producing colonies larger than those of MT-RBC1/pSM1 were retained, and their abilities to grow on minimal medium A plates containing 10−7 M myxothiazol and 10−7 M stigmatellin were tested. Under these conditions, the parent strain, MT-RBC1/pSM1, failed to grow due to the small amount of the cytochrome b6c1 complex while MT-RBC1/pMTS1 overproducing the wild-type cytochrome bc1 complex formed regular-size colonies.
Biochemical and biophysical techniques.
Chromatophore membrane preparations were done in 50 mM MOPS (morpholinepropanesulfonic acid) buffer (pH 7.0) containing 100 mM KCl, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride with a French pressure cell as described previously (1). The bacteriochlorophyll concentration was determined spectroscopically using an absorption coefficient ɛ775 of 75 mM−1 cm−1, and the amount of protein was measured according to the method of Lowry et al., (21). Difference spectra, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblot analysis were performed as described previously (15, 31) with polyclonal and monoclonal antibodies against the subunits of R. capsulatus cytochrome bc1 complex. To determine the Em7 of the hemes bL and bH, optical potentiometric titrations from 300 to −200 mV were performed according to the method of Dutton (11) with a double-beam spectrophotometer (Biomedical Instrumentation Group, University of Pennsylvania) as described previously (31). Flash-activated (8-μs actinic light pulse) single-turnover cytochrome b reduction and cytochrome c rereduction kinetics were performed as described previously (31, 32) with a single-wavelength spectrophotometer (Biomedical Instrumentation Group).
For electron paramagnetic resonance (EPR) spectroscopy, chromatophore membranes were reduced with 20 mM Na ascorbate and kept frozen in liquid nitrogen until the spectra were recorded. The EPR spectroscopy conditions were as follows: modulation frequency, 100 kHz; modulation amplitude, 12.49 G; microwave frequency, 9.47 GHz; microwave power, 5 mW; temperature, 20 K (31, 32).
Chemicals.
N-Methyldibenzopyrazine sulfate (PMS), N-ethyldibenzopyrazine sulfate (PES), and antimycin A were purchased from Sigma (St. Louis, Mo). Myxothiazol and various restriction enzymes were obtained from Boehringer Mannheim Biochemicals, New England Biolabs, and other commercial suppliers. 2,3,5,6-Tetramethyl-p-phenylenediamine (diaminodurene) and 2-hydroxy-1,4-naphtho-quinone were from Aldrich, and stigmatellin was from Fluka. Pyocyanine was a gift from D. E. Robertson (University of Pennsylvania). All other chemicals were reagent grade and were purchased from commercial sources.
RESULTS
Elimination of residues 231 to 237 of R. capsulatus cytochrome b has no deleterious effect on cytochrome bc1 complex activity.
The work presented here was initiated in order to assess whether stepwise conversions of bacterial cytochrome bc1 complexes to their evolutionary relatives encountered in other organisms were feasible. As a first step toward this aim, a comparison of bacterial and mitochondrial cytochrome b sequences highlighted two insertions 6 and 14 amino acids long (positions 231 to 237 and 308 to 325 of R. capsulatus cytochrome b, respectively) that are present only in bacterial proteins (9). Thus, the roles of these insertions were probed by generating bacterial mutants lacking each of them separately [e.g., pBΔ(231/237) (Fig. 1)]. Deletion of these regions yielded mutant cytochrome bc1 complexes that were indistinguishable from a wild-type enzyme in their assembly, stability, and kinetic properties, as well as their abilities to support the PS growth of R. capsulatus (data not shown) (13). Therefore, it was concluded that neither of these regions had any deleterious effect on R. capsulatus cytochrome bc1 complex.
Characterization of MT-RBC1/pSM1 producing a cytochrome b6c1 complex.
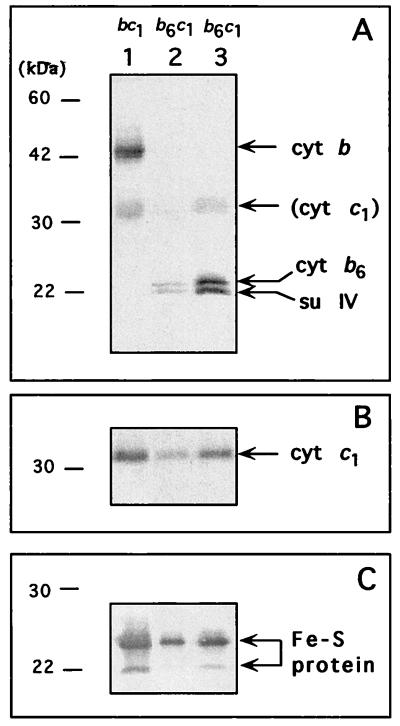
Next, the strain MT-RBC1/pSM1 producing a four-subunit decylbenzohydroquinone (DBH2):cytochrome c reductase was constructed by genetically splitting cytochrome b into two polypeptides, as described in Materials and Methods (Fig. 1). This mutant grew more slowly than its parent, MT-RBC1/pMTS1, in both enriched (MPYE) medium and minimal medium A under PS growth conditions (with a doubling time of 210 min versus 150 min for the parental strain in MPYE medium) and exhibited increased sensitivity to the Qo site inhibitors myxothiazol and stigmatellin (Table 1). Its PS growth on MPYE medium was completely inhibited in the presence of 10−7 M concentrations of these compounds, unlike its parent, which could readily tolerate these concentrations. Chromatophore membranes prepared with MT-RBC1/pSM1 contained smaller amounts of total b- and c-type cytochromes, as estimated by reduced minus oxidized optical difference spectra, and exhibited a cytochrome c reductase activity lower than that of a wild-type strain like MT-RBC1/pMTS1 (Table 1). SDS-PAGE and immunoblot analyses with polyclonal antibodies against R. capsulatus cytochrome b revealed that in MT-RBC1/pSM1 the native cytochrome b (48 kDa) was replaced by two smaller polypeptides similar in size to the cytochrome b6 and su IV subunits of the cytochrome b6f complexes (Fig. 2). Based on the locations of the stop and start codons introduced into petB, and hence the lengths of petB6 and petBIV, the upper (24-kDa) and lower (22-kDa) bands were tentatively assigned to the cytochrome b6 and su IV subunits of the cytochrome b6c1 complex. The gel data also confirmed that the four-subunit bc-type cytochrome complex, named cytochrome b6c1 complex, was present in smaller amounts than the wild-type bc1 complex.
TABLE 1.
Various properties of R. capsulatus strains harboring a cytochrome b6c1 complex or a cytochrome bc1 complex
| Strain | Growth (min)a | Inhibitor responseb | Cytochrome b (μM)c | DBH2-cytochrome c reductase (%)d |
Em7 (mV)
|
λmax (nm)
|
I50 (nM)e | Qo → bHf
|
Qo → cf
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bH | bL | bH | bL | Rate (s−1) | Eh (mV) | Rate (s−1) | Eh (mV) | ||||||
| MT-RBC1/pMTS1 (cytochrome bc1 complex) | 150 | Myxs | 1.97 | 100 | 49 | −125 | 560 | 558, 565 | 15 | 529 | 106 | 240 | 100 |
| Stgs | 118 | 208 | |||||||||||
| MT-RBC1/pSM1 (cytochrome b6c1 complex) | 210 | MyxHS | 0.44 | 7 | 56 | −110 | 560 | 560 | 7.6 | NDg | ND | ||
| StgHS | ND | ||||||||||||
| MT-RBC1/pSM1-92 (cytochrome b6c1 complex) | 180 | Myxs | 0.76 | 42 | 46 | −132 | 560 | 560 | 15 | 335 | 115 | 174 | 106 |
| Stgs | 112 | 200 | |||||||||||
Doubling time under PS growth conditions in MPYE medium.
Myx and Stg correspond to myxothiazol and stigmatellin, respectively. S and HS indicate sensitivity (inability to grow in the presence of 10−6 M) and hypersensitivity (inability to grow in the presence of 10−7 M) to these inhibitors, respectively.
Total amount of cytochrome b estimated from reduced minus oxidized optical difference spectra with an ɛ560–570 of 28 m−1 cm−1.
Cytochrome c reductase activity was determined as nanomoles of horse heart cytochrome c reduced per minute per milligram of total membrane proteins with DBH2 as an electron donor and was normalized for the amount of gy signal of the Fe-S protein as measured by EPR spectroscopy. In this instance, 100% corresponds to 7,099 nmol of cytochrome c reduced per min per mg of protein, normalized to the amplitude of the gy signal as shown in Fig. 5.
I50 refers to the concentration of antimycin A required to inhibit by 50% the cytochrome c reductase activity detected.
Qo → bH and Qo → c electron transfer rates were obtained by fitting the cytochrome b reduction and cytochrome c rereduction traces (not shown) to a single exponential equation, as described previously (31). Eh is the ambient potential at which the measurements were done, and c indicates the total amount of the cytochromes (c1 + c2 + cy).
ND, not done.
FIG. 2.
SDS-PAGE and immunoblot analysis of chromatophore membranes from appropriate R. capsulatus strains. Lanes 1, 2, and 3 correspond to MT-RBC1/pMTS1 (cytochrome bc1 complex), MT-RBC1/pSM1 (cytochrome b6c1 complex), and MT-RBC1/pSM1-92 (overproduced cytochrome b6c1 complex), respectively. In each case, 40 μg of membrane proteins per lane, heated at 37°C for 10 min, was used. At the end of electrophoresis, the gel was blotted to a nitrocellulose filter and probed with polyclonal antibodies specific for cytochrome b (A), cytochrome c1 (B), and Fe-S protein (C) of R. capsulatus cytochrome bc1 complex. Antibodies against cytochrome b were spiked with monoclonal antibodies against cytochrome c1 to better visualize the positions of cytochrome b6 and su IV relative to that of cytochrome c1. Molecular mass markers are indicated on the left in each case. Note that under the conditions used here the Fe-S protein runs as a doublet of approximately 24 and 22 kDa (28).
Overproduction of the cytochrome b6c1 complex.
To facilitate biochemical characterization of the cytochrome b6c1 complex, a derivative of MT-RBC1/pSM1 producing the complex in larger amounts was sought after random mutagenesis of plasmid pSM1 (see Materials and Methods). Following conjugation of a mutagenized pool of pSM1 into MT-RBC1, colonies which exhibited more vigorous PS growth than MT-RBC1/pSM1 on minimal medium A plates were chosen and tested for their abilities to grow by photosynthesis in the presence of 10−7 M myxothiazol and 10−7 M stigmatellin. Isolates as resistant to the Qo site inhibitors as MT-RBC1/pMTS1 were retained, and one of them (MT-RBC1/pSM1-92) was chosen for further studies. MT-RBC1/pSM1-92 contained in its chromatophore membranes substantially increased amounts of total b- and c-type cytochromes (Table 1) and exhibited higher cytochrome c reductase activity. This was approximately fivefold more and about two- to threefold less, than that found in MT-RBC1/pSM1 (cytochrome b6c1 complex) and MT-RBC1/pMTS1 (cytochrome bc1 complex), respectively (Table 1). SDS-PAGE and immunoblot analyses with antibodies against the subunits of the cytochrome bc1 complex confirmed these findings (Fig. 2). The data established the increased levels of the cytochrome b6 (24-kDa product of petB6) and su IV (22-kDa product of petBIV) subunits, as well as the absence of an intact cytochrome b (48-kDa product of petB, which usually runs as a 43- to 44-kDa band due to its hydrophobicity) in MT-RBC1/pSM1-92, which overproduced the cytochrome b6c1 complex (Fig. 2).
Molecular nature of the mutation in MT-RBC1/pSM1-92 leading to the overproduction of the cytochrome b6c1 complex.
To delimit the genetic location of the mutation(s) which conferred robust PS growth and increased cytochrome b6c1 complex activity, the SmaI-HindIII DNA fragment of pSM1-92 was exchanged in vitro for its counterpart in pSM1 (Fig. 1). The newly reconstructed plasmid also provided vigorous PS growth to MT-RBC1 on medium A plates, indicating that the desired mutation(s) was confined to the exchanged DNA fragment. Nucleotide sequence determination of the region extending from the SmaI site in petB to the 3′ end of petC revealed only a single C-to-T transition located at the second base of the second codon (underlined) (GCC to GTC) of petBIV (Fig. 1).
Biochemical characterizations of the cytochrome b6c1 complex.
Detailed analyses of the biochemical properties of the cytochrome b6c1 complex were undertaken, mainly with MT-RBC1/pSM1-92. Since one of the salient differences between the cytochrome bc1 and b6f complexes is their sensitivity and resistance, respectively, to the Qi site inhibitor antimycin A (5, 34), the response of the cytochrome b6c1 complex to this inhibitor was tested. The I50 value for antimycin A of the cytochrome b6c1 complex produced by MT-RBC1/pSM1-92 (approximately 15 nM) was identical to that of a wild-type cytochrome bc1 complex produced by MT-RBC1/pMTS1 (Table 1).
Next, the single-turnover kinetics exhibited by the cytochrome b6c1 complex were compared with those of a wild-type cytochrome bc1 complex by using light-activated, time-resolved kinetic spectroscopy (Table 1). Single-turnover cytochrome b reduction rates in the presence of antimycin A with MT-RBC1/pSM1-92 (335 s−1 at 115 mV and 112 s−1 at 200 mV) were similar to those observed with a wild-type strain like MT-RBC1/pMTS1 (529 s−1 at 106 mV and 118 s−1 at 208 mV). Further, a slightly lower cytochrome b reoxidation rate was also apparent in the case of the cytochrome b6c1 complex (data not shown), but this was not investigated further. Single-turnover cytochrome c rereduction rates exhibited by the cytochrome b6c1 complex in the absence of inhibitor were approximately 174 s−1 versus 240 s−1 for the wild-type cytochrome bc1 complex at comparable ambient potential (Eh) values (Table 1). These kinetics are not described in greater detail here, since extensive studies with similar findings have been reported in the case of R. sphaeroides cytochrome bc1 complex (19).
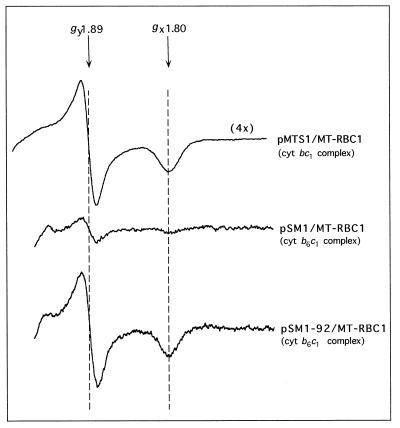
EPR spectroscopy was used to probe any possible effect of splitting cytochrome b into two peptides on the occupancy of the Qo site of the cytochrome b6c1 complex. The EPR spectrum of Na ascorbate-reduced chromatophore membranes of MT-RBC1/pSM1-92 was identical to that of a wild-type strain (MT-RBC1/pMTS1) and exhibited typical gx and gy signals at g values of 1.80 and 1.89, respectively (Fig. 3). Thus, in the case of the cytochrome b6c1 complex, the Qo site UQ/UQH2 occupancy, which is critical for fast enzyme kinetics (10), was very similar to that of a wild-type cytochrome bc1 complex. As expected, the amplitude of the signals in MT-RBC1/pSM1-92 and in MT-RBC1/pSM1 were about 33 and 5%, respectively, of those in MT-RBC1/pMTS1 (Figure 3), in agreement with the immunoblotting and optical difference spectrum data described above.
FIG. 3.
EPR spectra of chromatophore membranes from various R. capsulatus strains harboring either a cytochrome bc1 complex or a cytochrome b6c1 complex. The spectra were taken with 300 μl of samples containing 31.8 mg (MT-RBC1/pMTS1; cytochrome bc1 complex), 33.7 mg (MT-RBC1/pSM1; cytochrome b6c1 complex), and 25.2 mg (MT-RBC1/pSM1-92; overproduced cytochrome b6c1 complex) of membrane proteins per ml and reduced with 20 mM Na ascorbate. The EPR parameters were as described in Materials and Methods, and the gx and gy values are indicated by arrows. Note that a four-times-larger scale was used to record the spectrum of MT-RBC1/pMTS1, as indicated by the label.
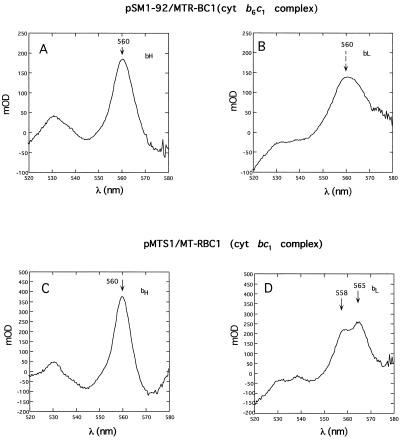
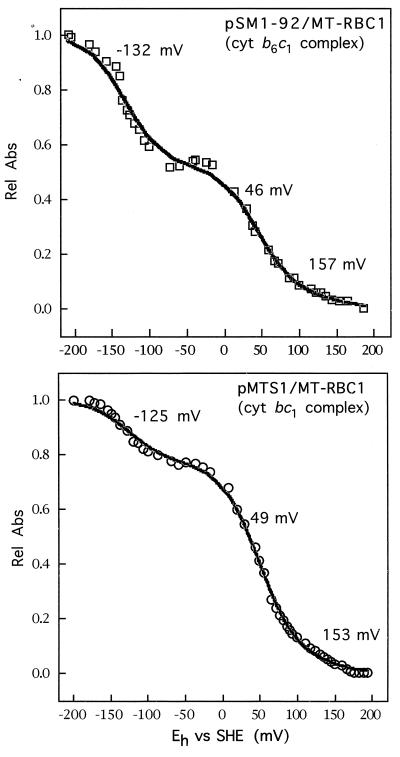
Lastly, the spectral and thermodynamic properties of the heme groups of cytochrome b6c1 complex were determined by dark-equilibrium optical redox titrations (Fig. 4 and 5). The data obtained indicated that the characteristics of hemes bH of the cytochrome b6c1 and cytochrome bc1 complexes were very similar. The λmax value of this heme group in MT-RBC1/pSM1-92 (also in MT-RBC1/pSM1 [not shown]) and MT-RBC1/pMTS1 was around 560 nm (Fig. 4A and C), and the corresponding Em7 values were about 46 mV (51 mV in MT-RBC1/pSM1 [not shown]) and 49 mV, respectively (Fig. 5). Likewise, the Em7 value of heme bL in the cytochrome b6c1 complex produced by MT-RBC1/pSM1-92 was around −132 mV (−131 mV for MT-RBC1/pSM1 [not shown]) and quite similar to that of the wild-type cytochrome bc1 complex (Em7, −125 mV) (Fig. 5). On the other hand, in the cytochrome b6c1 complex the unusual split α-peak (λmax, 558 and 565 nm) of heme bL, which is highly characteristic of this prosthetic group in the cytochrome bc1 complex, was modified to a nonsplit and broad form (λmax, approximately 560 nm) (Fig. 4B and D), like those found in the cytochrome b6f complexes.
FIG. 4.
Optical redox difference spectra of b-type cytochromes associated with the cytochrome bc1 complex and the cytochrome b6c1 complex of appropriate R. capsulatus strains. An amount of chromatophore membranes containing 80 and 50 μM bacteriochlorophyll was used for MTR-BC1/pSM1-92 (cytochrome b6c1 complex,) (A and B) and MT-RBC1/pMTS1 (cytochrome bc1 complex) (C and D), respectively. Panels A (spectrum taken at Eh of −6 mV minus that at Eh of 170 mV) and C (spectrum at −34 mV minus that at 160 mV) correspond to the cytochrome bH optical difference spectra, and panels B (spectrum at [−203 mV minus that at −63 mV) and D (spectrum at −197 mV minus that of −58 mV) correspond to the cytochrome bL optical difference spectra; the arrows indicate the corresponding λmax values observed in each case. Note the absence of a split α-peak for cytochrome bL of the cytochrome b6c1 complex (B). cyt, cytochrome; mOD, milli OD.
FIG. 5.
Dark potentiometric redox titrations of the b-type cytochromes of the cytochrome bc1 complex and the cytochrome b6c1 complex produced by appropriate R. capsulatus strains. The experimental conditions were as described in Materials and Methods, and the amounts of materials were as for Fig. 6. The data obtained were fitted to three n = 1 Nernst equations, and the Em7 values thus derived are indicated. Eh values were measured versus the standard hydrogen electrode (SHE). Rel Abs, relative absorbance.
DISCUSSION
In this work, large modifications of the bacterial UQH2-cytochrome c oxidoreductase were attempted by using the genetic system of R. capsulatus (1) and the structure of the cytochrome bc1 complex from mitochondria (16, 37, 42). It was hoped that such an attempt, if successful, would greatly help correlation of structural and functional similarities or differences observed among various cytochrome bc complexes (14). Encouraged by the nondeleterious effects of large deletions located in the de and ef loops of cytochrome b (13), we proceeded to split it into two peptides analogous to those found in chloroplast cytochrome b6f complexes. This construct generated a novel variant of UQH2-dependent cytochrome c reductase, named cytochrome b6c1 complex, which was fully functional to support the PS growth of R. capsulatus. Introduction of appropriate stop and start codons converted the three-cistronic petABC operon into a four-cistronic variant (petAB6BIVC) able to direct the synthesis of two separate polypeptides, cytochrome b6 and su IV, instead of cytochrome b (Fig. 1). These polypeptides assembled posttranslationally into a four-subunit cytochrome b6c1 complex (Fig. 6), resembling the cytochrome b6f complexes from chloroplasts (5, 17).
FIG. 6.
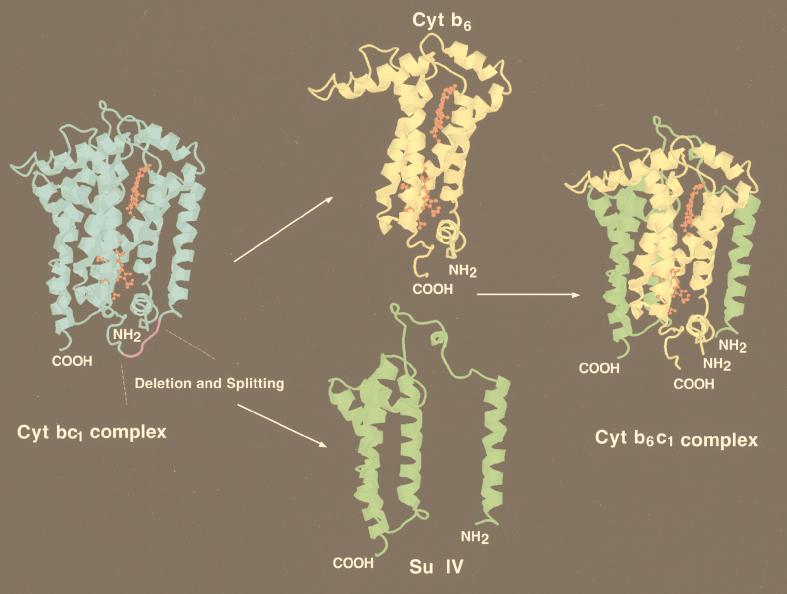
Three-dimensional depiction of cytochrome b6 and su IV subunits of the cytochrome b6c1 complex. These hypothetical models are intended to illustrate the consequences of splitting petB (cytochrome b [blue]) into petB6 (cytochrome b6 [yellow]) and petBIV (su IV [green]). Since the properties of the cytochrome b6c1 complex are similar to those of the cytochrome bc1 complex, it is assumed that the newly created cytochrome b6 and su IV subunits reassemble as in the native cytochrome b. X-ray coordinates from chicken mitochondrial cytochrome bc1 complex (41) were visualized with Rasmol, and the region of cytochrome b where splitting took place (purple) was modified by using homology between the chicken and R. capsulatus cytochrome b subunits. The heme bH and bL prosthetic groups are shown in red.
When cytochrome b was split, the steady-state amount of the cytochrome b6c1 complex dropped to about 1/10 that of a strain overproducing a wild-type cytochrome bc1 complex (MT-RBC1/pMTS1), compromising its ability to support growth. However, selective pressure for better growth readily led to a mutant (MT-RBC1/pSM1-92) producing roughly half the amount in MT-RBC1/pMTS1. The molecular mechanism underlying this overproduction was not determined, but the location and nature of the mutation found (a single C-to-T substitution at the second base of the second codon of petBIV) suggested that the translation efficiency of petBIV (su IV subunit) might have been improved (22). In any event, this increased production rendered possible detailed characterization of the properties of the cytochrome b6c1 complex. The analyses demonstrated that the amino-terminal four and the carboxyl-terminal four transmembrane helices of R. capsulatus cytochrome b could be synthesized separately and assembled correctly, as depicted in Fig. 6. Previously, several membrane proteins, like the E. coli lactose transporter (2, 41) the β2 subunit of the α2β2 adrenergic receptor (18), the tachykinin–neurokinin-1 receptor (NK1) (26), and various rhodopsins (20, 27), have been split into separate fragments and expressed independently. These studies have established that multispan transmembrane proteins can often be split into several independently expressed peptides that can self-assemble into functional multisubunit complexes in vivo. It appears that a major consequence of the splitting event is often a decrease in the steady-state amount of these membrane-bound complexes without complete loss of function. Then, as shown here, additional mutations increasing their amounts may be obtained.
The Qi site of the R. capsulatus cytochrome b6c1 complex was fully sensitive to antimycin A, suggesting that the resistance of the cytochrome b6f complexes to this inhibitor was not a consequence of splitting a long cytochrome b into two polypeptides. A consequence of the splitting event could be a decrease in the stability of the semiquinone species located near the Qi site, as implied by slightly slower cytochrome b reoxidation and decreased cytochrome c reduction kinetics seen in MT-RBC1/pSM1-92 (Table 1), and this remains to be studied in detail by EPR spectroscopy. In regard to the molecular basis of inhibitor resistance observed with the cytochrome b6f complexes, the presence of natural amino acid substitutions at specific positions of cytochrome b had been noted during our earlier studies (7).
The Em values of the cytochrome b hemes in the cytochrome b6f complexes are about 100 mV lower than those of their counterparts in the cytochrome bc1 complexes (5). This difference has sometimes been attributed to the cytochrome b6 and su IV subunits being separate polypeptides, unlike the bacterial or mitochondrial cytochrome b. The Em7 values of the cytochrome b hemes of R. capsulatus cytochrome b6c1 complex described here are very similar to those of a native cytochrome bc1 complex, indicating that splitting of cytochrome b is not the sole molecular basis of the different Em7 values found in the cytochrome b6f complexes. On the other hand, a clear consequence of splitting cytochrome b appears to be the spectral changes caused in cytochrome bL. Unlike various b-type cytochromes, cytochrome bL of the cytochrome bc1 complexes has a characteristic split α-band spectrum, attributed to its constrained axial ligands (30). In the cytochrome b6c1 complex this α-band is broader and similar to that of cytochrome bH, like that of the cytochrome b6f complexes (5, 17). This modification may be a consequence of the relaxation of this constraint upon splitting the cytochrome b polypeptide backbone (Fig. 4 and 6). Conversion of the split α-spectrum of cytochrome bL to a nonsplit form has also been observed during purification and storage of a reconstitutively active R. capsulatus cytochrome bc1 subcomplex (35). Interestingly, synthetic four-helix-bundle peptide models carrying two hemes, similar to those of cytochrome b, also exhibit identical and nonsplit α-band spectra with λmax of around 560 nm (29). Whether this modification affects the rate of electron transfer between the cytochrome bH and bL hemes remains to be seen.
Very recently, a R. sphaeroides cytochrome bc1 complex with a split cytochrome b subunit, similar to the cytochrome b6c1 complex described here, has been reported (19). R. sphaeroides and R. capsulatus are closely related species, and their cytochrome bc1 complexes are similar but not identical (14). In particular, the purified R. sphaeroides cytochrome bc1 complex contains an additional non-redox-active polypeptide (14 to 15 kDa) which is absent in other bacterial enzymes, including that of R. capsulatus (14). A comparison of the cytochrome b6c1 complex from R. capsulatus with the modified cytochrome bc1 complex of R. sphaeroides obtained by splitting its cytochrome b subunit (19) reveals that these engineered complexes exhibit different properties. While their electron transfer behaviors and responses to Qi and Qo site inhibitors appear comparable, their subunit compositions (four subunits in R. capsulatus versus five in R. sphaeroides) and several of their functional properties are different. In this respect, it is noteworthy that the spacer nucleotide sequences used to generate petB6 and petBIV are different in each case both in length and composition. In particular, while the R. capsulatus cytochrome b6c1 complex contains a deletion six amino acid residues long (position 231 to 237) encompassing the splitting site of cytochrome b, generating a situation highly analogous to those found in chloroplast cytochrome b6f complexes, that from R. sphaeroides contains no such deletion in the de loop of cytochrome b. Moreover, while R. capsulatus cytochrome b6c1 complex is produced in small amounts in the absence of any additional mutation improving its expression, the amount of the modified R. sphaeroides complex is apparently less affected (19). Thus, while R. capsulatus strains carrying a cytochrome b6c1 complex are more sensitive to Qo site inhibitors under PS growth conditions, no similar phenotype has been described in the case of R. sphaeroides. Finally, while the R. sphaeroides split cytochrome bc1 complex exhibits a modification of both the optical spectrum and the Em value (approximately −40 mV) of cytochrome bL, that of R. capsulatus cytochrome b6c1 complex has a wild-type-like Em value (−132 mV) despite its modified spectrum. It appears that seemingly similar genetic splittings of cytochrome b into two polypeptides do not lead to identical functional modifications. In any event, in both cases a clear outcome of splitting a long cytochrome b into two shorter polypeptides appears to be the relaxation of the constrained axial ligands of cytochrome bL heme, as revealed by its modified optical spectrum.
While further experiments with both R. capsulatus and R. sphaeroides are obviously required to better define the molecular basis of the similarities and differences highlighted by these studies, a clear conclusion is that genetic conversion of a bacterial cytochrome bc1 complex into a modified form reminiscent of that found in plants is indeed possible. Undoubtedly, future studies with similar approaches will further establish rigorous correlations between the structural and functional similarities and differences encountered among the various cytochrome bc oxidoreductases from bacteria, mitochondria, and chloroplasts.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM 38237.
We thank Kevin A. Gray for constructing the petBΔ(231-236) mutant, Saadettin Guner for determining the I50 values, and Brian R. Gibney for help in recording the EPR spectra.
REFERENCES
- 1.Atta-Asafo-Adjei E, Daldal F. Size of the amino-acid side-chain at position-158 of cytochrome b is critical for an active cytochrome bc1 complex and for photosynthetic growth of Rhodobacter capsulatus. Proc Natl Acad Sci USA. 1991;88:492–496. doi: 10.1073/pnas.88.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibi E, Kaback R H. Functional complementation of internal deletion mutants in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:4325–4329. doi: 10.1073/pnas.89.5.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brasseur G, Saribas A S, Daldal F. A compilation of mutations located in the cytochrome b subunit of the bacterial and mitochondrial bc1 complex. Biochim Biophys Acta. 1996;1275:61–69. doi: 10.1016/0005-2728(96)00051-5. [DOI] [PubMed] [Google Scholar]
- 4.Carrell C J, Zhang H, Cramer W A, Smith J L. Biological identity and diversity in photosynthesis and respiration: structure of the lumen-side domain of the chloroplast Rieske protein. Structure. 1997;5:1613–1625. doi: 10.1016/s0969-2126(97)00309-2. [DOI] [PubMed] [Google Scholar]
- 5.Cramer W A, Soriano G M, Ponomarev M, Huang D, Zhang H, Martinez S E, Smith J L. Some new structural aspects and old controversies concerning the cytochrome b6f complex of oxygenic photosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:447–508. doi: 10.1146/annurev.arplant.47.1.477. [DOI] [PubMed] [Google Scholar]
- 6.Daldal F, Davidson E, Cheng S. Isolation of the structural genes for the Rieske Fe-S Protein, cytochrome b and cytochrome c1, all compounds of the ubiquinol: cytochrome c2 oxidoreductase complex of Rhodopseudomonas capsulata. J Mol Biol. 1987;195:1–12. doi: 10.1016/0022-2836(87)90322-6. [DOI] [PubMed] [Google Scholar]
- 7.Daldal F, Tokito M K, Davidson E, Faham M. Mutations conferring resistance to quinol oxidation (Qz) inhibitors of the ubiquinol: cytochrome c2 oxidoreductase of Rhodobacter capsulatus. EMBO J. 1989;8:3951–3961. doi: 10.1002/j.1460-2075.1989.tb08578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson E, Ohnishi T, Tokito M K, Daldal F. R. capsulatus mutants lacking the Rieske FeS protein form a stable cytochrome bc1 subcomplex with an intact quinone reduction (Qi) site. Biochemistry. 1992;31:3351–3358. doi: 10.1021/bi00128a007. [DOI] [PubMed] [Google Scholar]
- 9.Degli-Esposti M D, De Vries S, Crimi M, Ghelli A, Patarnello T, Meyer A. Mitochondrial cytochrome b: evolution and structure of the protein. Biochim Biophys Acta. 1993;1143:243–271. doi: 10.1016/0005-2728(93)90197-n. [DOI] [PubMed] [Google Scholar]
- 10.Ding H, Moser C C, Robertson D E, Tokito M K, Daldal F, Dutton P L. Ubiquinone pair in the Qo site central to the primary energy conversion reactions of cytochrome bc1 complex. Biochemistry. 1995;34:15979–15996. doi: 10.1021/bi00049a012. [DOI] [PubMed] [Google Scholar]
- 11.Dutton P L. Redox potentiometry: determination of midpoint potentials of oxidation-reduction components of biological electron-transfer systems. Methods Enzymol. 1978;54:411–435. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- 12.Furbacher P N, Tae G-S, Cramer W A. Evolution and origins of the cytochrome bc1 and b6f complexes. In: Baltscheffsky H, editor. Origin and evolution of biological energy conversion. New York, N.Y: Wiley-VCH; 1996. pp. 221–253. [Google Scholar]
- 13.Gray, K. A., and F. Daldal. Unpublished data.
- 14.Gray K A, Daldal F. Mutational studies of the cytochrome bc1 complexes. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 747–774. [Google Scholar]
- 15.Gray K A, Dutton P L, Daldal F. Histidine 217 is necessary for efficient electron transfer between cytochrome bH and the quinone reductase (Qi) site in the cytochrome bc1 complex. Biochemistry. 1994;33:723–733. doi: 10.1021/bi00169a014. [DOI] [PubMed] [Google Scholar]
- 16.Iwata S, Lee J W, Okada K, Lee J K, Iwata M, Rasmussen B, Link T A, Ramaswamy S, Jap B K. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 17.Kallas T. The cytochrome b6f complex. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 259–317. [Google Scholar]
- 18.Kobilka B K, Kobilka T S, Daniel K, Regan J W, Caron M G, Lefkowitz R J. Chimeric alpha 2-beta 2-adrenergic receptors: delineation of domains involved in effector coupling and ligand binding specificity. Science. 1988;240:1310–1316. doi: 10.1126/science.2836950. [DOI] [PubMed] [Google Scholar]
- 19.Kuras R, Guergova-Kuras M, Crofts A R. Steps toward constructing a cytochrome b6f complex in the purple bacterium Rhodobacter sphaeroides: an example of the structural plasticity of a membrane cytochrome. Biochemistry. 1998;37:16280–16288. doi: 10.1021/bi9813476. [DOI] [PubMed] [Google Scholar]
- 20.Liao M-J, Huang K-S, Khorona H G. Regeneration of native bacteriorhodopsin structure from fragments. J Biol Chem. 1984;259:4200–4204. [PubMed] [Google Scholar]
- 21.Lowry O H, Rosenbrough N J, Farr A L, Randall R J. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 22.Makrides S C. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malkin R. Cytochrome bc1 and b6f complexes of photosynthetic membranes. Photosynth Res. 1992;33:121–136. doi: 10.1007/BF00039175. [DOI] [PubMed] [Google Scholar]
- 24.Martinez S E, Huang D, Szczepaniak A, Cramer W A, Smith J L. Crystal-structure of chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation. Structure. 1994;2:95–105. doi: 10.1016/s0969-2126(00)00012-5. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell P. The protonmotive Q cycle: a general formulation. FEBS Lett. 1976;59:137–139. doi: 10.1016/0014-5793(75)80359-0. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen S M, Elling C E, Schwartz T W. Split-receptors in the tachykinin neurokinin-1 system: mutational analysis of intracellular loop 3. Eur J Biochem. 1998;251:217–226. doi: 10.1046/j.1432-1327.1998.2510217.x. [DOI] [PubMed] [Google Scholar]
- 27.Ridge K D, Lee S S J, Yao L L. In vivo assembly of rhodopsin from expressed polypeptide fragments. Proc Natl Acad Sci USA. 1995;92:3204–3208. doi: 10.1073/pnas.92.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson D E, Ding H, Chelminski P R, Slaughter C, Hsu J, Moomaw C, Tokito M, Daldal F, Dutton P L. Hydroubiquinone cytochrome-c2 oxidoreductase from Rhodobacter capsulatus: definition of a minimal, functional isolated preparation. Biochemistry. 1993;32:1310–1317. doi: 10.1021/bi00056a016. [DOI] [PubMed] [Google Scholar]
- 29.Robertson D E, Farid R S, Moser C C, Urbauer J L, Mulholland S E, Pidikiti R, Lear J D, Wand A J, Degrado W F, Dutton P L. Design and synthesis of multi-heme proteins. Nature. 1994;368:425–431. doi: 10.1038/368425a0. [DOI] [PubMed] [Google Scholar]
- 30.Salerno J. Cytochrome electron spin resonance line shapes, ligand fields, and components stoichiometry, in ubiquinol-cytochrome c oxidoreductase. J Biol Chem. 1984;259:2331–2336. [PubMed] [Google Scholar]
- 31.Saribas A S, Ding H, Dutton P L, Daldal F. Tyrosine 147 of cytochrome b is required for efficient electron transfer at the ubihydroquinone oxidase site (Qo) of the cytochrome bc1 complex. Biochemistry. 1995;34:16004–16012. doi: 10.1021/bi00049a014. [DOI] [PubMed] [Google Scholar]
- 32.Saribas A S, Valkova-Valchonova M, Tokito M K, Zhang Z, Berry E A, Daldal F. Interactions between the cytochrome b, cytochrome c1, and Fe-S protein subunits at the ubihydroquinone oxidation site of the bc1 complex of Rhodobacter capsulatus. Biochemistry. 1998;37:8105–8115. doi: 10.1021/bi973146s. [DOI] [PubMed] [Google Scholar]
- 33.Sone N, Tsuchiya N, Inoue M, Noguchi S. Bacillus stearothermophilus qcr operon encoding Rieske FeS protein, cytochrome b6, and a novel-type cytochrome c1 of quinol-cytochrome c reductase. J Biol Chem. 1996;271:12457–12462. doi: 10.1074/jbc.271.21.12457. [DOI] [PubMed] [Google Scholar]
- 34.Trumpower B L, Gennis R B. Energy transduction by cytochrome complexes in mitochondrial and bacterial respiration: the enzymology of coupling electron transfer reactions to transmembrane proton translocation. Annu Rev Biochem. 1994;63:675–716. doi: 10.1146/annurev.bi.63.070194.003331. [DOI] [PubMed] [Google Scholar]
- 35.Valkova-Valchanova M B, Saribas A S, Gibney B R, Dutton P L, Daldal F. Isolation and characterization of a two-subunit cytochrome b-c1 subcomplex from Rhodobacter capsulatus and reconstitution of its ubihydroquinone oxidation (Qo) site with purified Fe-S protein subunit. Biochemistry. 1998;37:16242–16251. doi: 10.1021/bi981651z. [DOI] [PubMed] [Google Scholar]
- 36.Widger W R, Cramer W A, Herrman R G, Trebst A. Sequence homology and structural similarity between cytochrome b of mitochondrial complex III and the chloroplast b6-f complex: position of the cytochrome b hemes in the membrane. Proc Natl Acad Sci USA. 1984;81:674–678. doi: 10.1073/pnas.81.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia D, Yu C-A, Kim H, Xia J-Z, Kachurin A M, Zhang L, Yu L, Deisenhofer J. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong J, Inoue K, Bauer C E. Tracking molecular evolution of photosynthesis by characterization of a major photosynthesis gene cluster from Heliobacillus mobilis. Proc Natl Acad Sci USA. 1998;95:14851–14856. doi: 10.1073/pnas.95.25.14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu J, Hederstedt L, Piggot P J. The cytochrome bc complex (menaquinone:cytochrome c reductase) in Bacillus subtilis has a nontraditional subunit organization. J Bacteriol. 1995;177:6751–6760. doi: 10.1128/jb.177.23.6751-6760.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J, Le Brun N E. Studies of the cytochrome subunits of menaquinone:cytochrome c reductase (bc complex) of Bacillus subtilis: evidence for the covalent attachment of heme to the cytochrome b subunit. J Biol Chem. 1998;273:8860–8866. doi: 10.1074/jbc.273.15.8860. [DOI] [PubMed] [Google Scholar]
- 41.Zen K H, McKenna E, Bibi E, Hardy D, Kaback H R. Expression of lactose permease in contiguous fragments as a probe for membrane-spanning domains. Biochemistry. 1994;33:8198–8206. doi: 10.1021/bi00193a005. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z, Huang L, Shulmeister V M, Chi Y-I, Kim K K, Hung L-W, Crofts A R, Berry E A, Kim S-H. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]