Abstract
RNA-binding proteins play vital roles in regulating gene expression and cellular physiology in all organisms. Bacterial RNA-binding proteins can regulate transcription termination via attenuation or antitermination mechanisms, while others can repress or activate translation initiation by affecting ribosome binding. The RNA targets for these proteins include short repeated sequences, longer single-stranded sequences, RNA secondary or tertiary structure, and a combination of these features. The activity of these proteins can be influenced by binding of metabolites, small RNAs, or other proteins, as well as by phosphorylation events. Some of these proteins regulate specific genes, while others function as global regulators. As the regulatory mechanisms, components, targets, and signaling circuitry surrounding RNA-binding proteins have become better understood, in part through rapid advances provided by systems approaches, a sense of the true nature of biological complexity is becoming apparent, which we attempt to capture for the reader of this review.
Keywords: RNA-binding protein, gene regulation, attenuation, antitermination, translation, sRNA
INTRODUCTION
Bacteria have the capacity to rapidly alter metabolism, physiology, behavior, and morphology in response to changing environmental conditions by altering gene expression. Although transcription initiation plays a vital role in response to environmental stimuli, regulation that occurs after transcription initiation is now recognized as a common regulatory strategy in all organisms. A wide variety of posttranscriptional regulatory mechanisms have been identified, including regulated transcription termination (attenuation and antitermination), regulation of translation initiation, and changes in mRNA stability. The factors responsible for sensing and responding to changing environmental conditions include RNA-binding proteins and small RNAs (sRNAs). This review focuses on mechanisms that RNA-binding proteins utilize to control bacterial gene expression, although the role of sRNAs that affect the activity of certain RNA-binding proteins will also be discussed. Excellent reviews describing enzymes involved in mRNA turnover, as well as the mechanism by which the RNA chaperone Hfq mediates sRNA-mRNA base-pairing, have been published recently, and these are not reviewed here (38, 72, 88, 116).
Once transcription initiates, the transcription elongation complex is subject to regulatory events including RNA polymerase pausing and transcription termination. These processes are modulated by the general transcription elongation factors NusA and NusG (23, 90, 152). During transcription the nascent transcript can adopt alternative antiterminator and terminator structures that dictate whether RNA polymerase continues to transcribe into the downstream protein-coding sequences, or whether transcription terminates in the 5′ untranslated region (5′ UTR). NusA and NusG can stimulate RNA polymerase pausing, which can synchronize RNA polymerase position with RNA folding and/or binding of a regulatory factor to the nascent transcript (23, 152, 155). Transcription attenuation is defined as promotion of transcription termination in the 5′ UTR by the action of a regulatory molecule (translating ribosome, RNA-binding protein, RNA, metabolite), with the default being transcription read-through into the downstream structural gene(s). Antitermination is distinguished from attenuation in that the action of the regulatory molecule results in transcription read-through, with the default being termination (47). Mechanisms involving the RNA-binding proteins HutP and the Bgl-Sac family of proteins of Bacillus subtilis and Escherichia coli are used to illustrate well-characterized antitermination mechanisms, while the actions of trp RNA–binding attenuation protein (TRAP) and PyrR of B. subtilis are used to describe transcription attenuation.
RNA-binding proteins are also capable of regulating translation initiation. Although the vast majority of the known regulatory mechanisms repress translation initiation, a few examples of translational activation have been identified (14). RNA-binding proteins can promote the formation of alternative RNA structures that dictate the accessibility of 30S ribosomal subunits to the Shine-Dalgarno (SD) sequence, a critical component of the ribosome-binding site. Sequestration of the SD sequence in RNA structure represses translation, whereas destabilization of such structures can activate translation. Translation can also be repressed directly such that the RNA-binding protein directly occludes ribosome binding. Mechanisms of RNA-binding-protein-mediated translational control are illustrated by descriptions of TRAP, CsrA, and the E. coli cold shock protein CspA.
RNA-binding proteins throughout the biological world interact with noncoding RNAs that regulate gene expression either by altering the activity of the RNA-binding protein itself or by influencing the formation of sRNA-mRNA base-pairing interactions. The three-factor interactions thus generated can create diverse regulatory effects and patterns. The CsrA/RsmA family of bacterial proteins provides an example of RNA-binding proteins whose activity is regulated by binding to sRNAs containing many high-affinity CsrA-binding sites, which decrease the availability of these proteins to interact with mRNA targets with lower binding affinity. The global regulatory roles of CsrA/RsmA proteins and their sRNAs have attracted widespread attention (112, 143). A second RNA-binding protein, RapZ, interacts with two related sRNAs, affecting the base-pairing activity of one of these sRNAs while being sequestered and inhibited by the second sRNA, which acts as a decoy (50). The FinO/ProQ family of RNA-binding proteins offers a final example of protein-sRNA-mRNA interactions, only recently recognized for potentially diverse and pervasive regulatory roles (60, 95).
REGULATED TRANSCRIPTION TERMINATION: ATTENUATION AND ANTITERMINATION
Bgl-Sac Family of Antitermination Proteins of B. subtilis and E. coli
A common antitermination mechanism controls expression of genes involved in the utilization of β-glucosides, sucrose, and glucose in E. coli and/or B. subtilis (3, 129). The general antitermination mechanism was first described for BglG-mediated regulation of the E. coli bglGFBH operon (4, 5). Subsequent studies of the SacT, SacY, LicT, and GlcT antitermination proteins from B. subtilis contributed greatly to our understanding of this fascinating mechanism (e.g., 9, 20, 122, 131, 135, 136). Each antitermination protein consists of an N-terminal RNA-binding domain and two regulatory domains called PRD1 and PRD2. These regulatory domains are the sites of reversible phosphorylation at conserved histidine residues in response to the cognate carbon source (6, 84, 130). In the absence of the carbon source, these proteins are rendered inactive by phosphorylation of PRD1 by the cognate membrane-bound EII transporter of the phosphotransferase system (PTS) (54, 78). In the presence of the carbon source the EII transporter dephosphorylates PRD1, while HPr, a component of the PTS that is sugar-nonspecific, phosphorylates PRD2 (54, 78, 115, 130). When PRD1 is unphosphorylated and PRD2 is phosphorylated the antitermination protein forms an active homodimer (5, 115, 121). The active dimer then binds to its target transcripts and stabilizes an otherwise weak antiterminator structure in the 5′ UTR termed RAT, for RNA antiterminator (5, 12). As a consequence, an overlapping intrinsic terminator is unable to form, leading to transcription of the operon. In the absence of bound protein, the terminator hairpin forms and transcription halts upstream of the coding sequence (Figure 1). One notable exception to this antitermination mechanism is the apparent absence of an intrinsic terminator that overlaps the RAT in the 5′ UTR of the sacXY operon (136). The mechanism by which this operon is regulated by SacT and SacY has not been elucidated.
Figure 1.
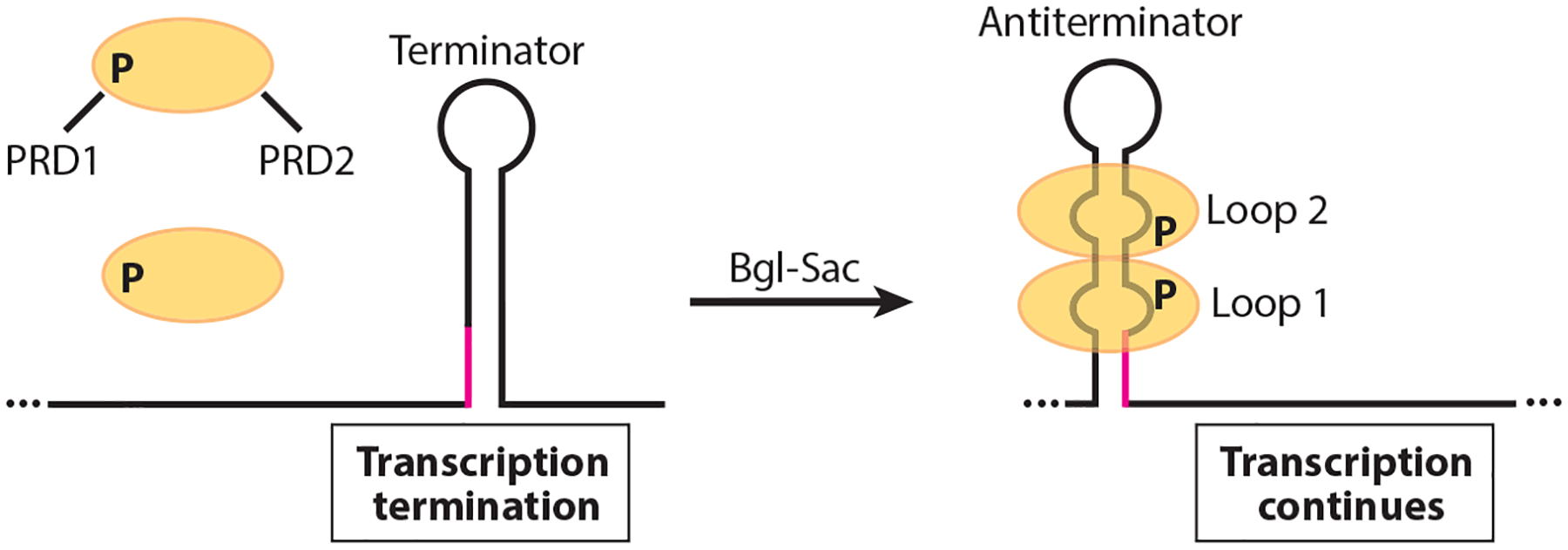
Model of the Bgl-Sac family antitermination mechanism. In the absence of the cognate carbon source the antitermination protein (yellow ovals) is rendered inactive by phosphorylation of PRD1. In the presence of the carbon source, PRD1 is dephosphorylated and PRD2 is phosphorylated, resulting in protein dimerization. The active dimer then binds to its cognate RNA target, thereby stabilizing an otherwise weak antiterminator structure. As a consequence, an overlapping intrinsic terminator is unable to form (overlap in magenta), leading to transcription of the operon involved in catabolism of the cognate carbon source. In the absence of bound protein, the terminator hairpin forms and transcription halts upstream of the coding sequence.
Structural studies from several Bgl-Sac family proteins confirmed that the RNA-binding domain forms a symmetrical homodimer, with each monomer forming a four-stranded β sheet (32, 84, 163). A solution structure of the LicT RNA-binding domain in complex with its cognate RAT showed that LicT interacts with two bulges within the RAT, and with the minor groove of the stem between these two loops (163). Despite their sequence and structural similarity, these antitermination proteins are highly specific for their cognate RNA targets. Substitution of residues in SacT, LicT, and SacY that contact their cognate RATs resulted in reduced binding specificity (61). A crucial arginine residue in LicT was also identified that is required for high affinity and binding specificity. Interestingly, introduction of this residue in SacY increased its RNA-binding affinity but reduced its specificity for its cognate RNA target. These results led to the suggestion that this family of antitermination proteins evolved to maintain specificity by compromising binding affinity for their cognate RNA targets (61).
HutP-Mediated Antitermination in B. subtilis
The B. subtilis hutPHUIGM operon encodes an RNA-binding protein that regulates expression of the operon (HutP), a histidine transporter (HisM), and enzymes for the utilization of histidine as a carbon and nitrogen source (73). HutP regulates expression of the hut operon by an antitermination mechanism in response to histidine. Under low histidine, an intrinsic terminator causes transcription to terminate in the hutP-hutH intercistronic region (150). However, under histidine excess, histidine-activated HutP binds to the nascent hut transcript and prevents formation of the intrinsic terminator such that transcription continues into the downstream genes (53, 93, 94). Thus, antitermination allows increased transport and utilization of histidine as a carbon and nitrogen source (Figure 2).
Figure 2.
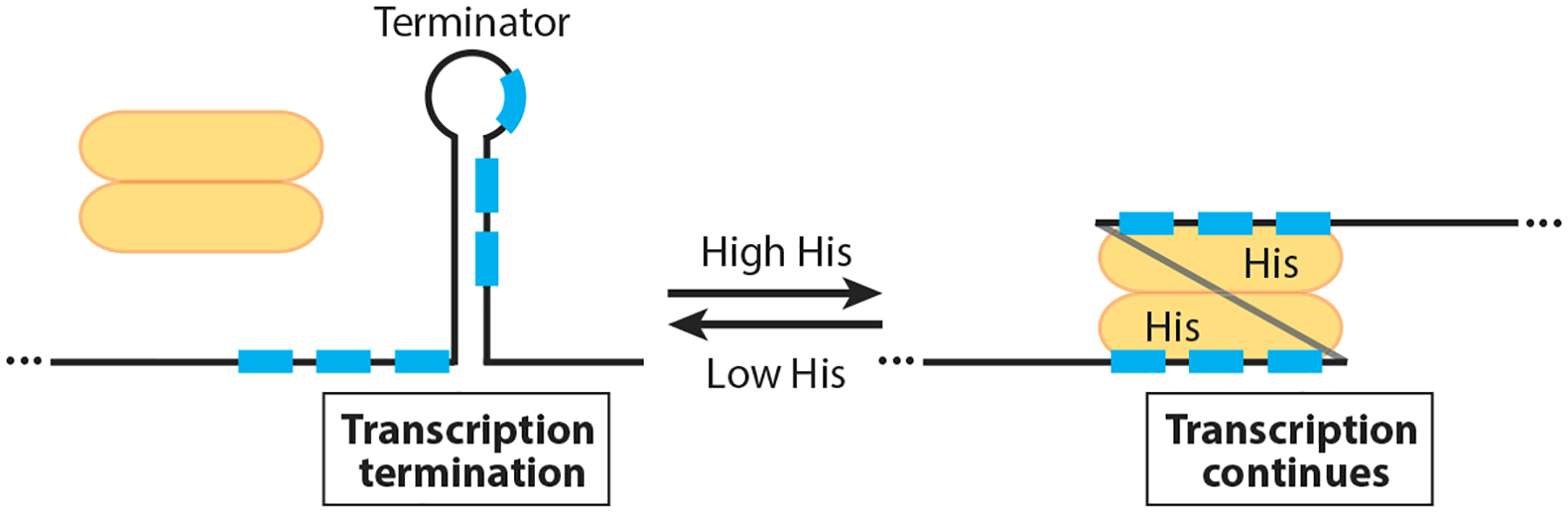
Model of the HutP antitermination mechanism. Under low histidine conditions, the HutP dimer (yellow) is inactive and an intrinsic terminator causes transcription to terminate in the hutP-hutH intercistronic region. In the presence of excess histidine, histidine-activated HutP binds to six NAG triplet repeats (blue boxes) in the nascent hut transcript, thereby preventing formation of the intrinsic terminator such that transcription continues into the downstream histidine utilization genes.
HutP functions as a homohexamer, and binding of both histidine and Mg2+ is required to activate its RNA-binding activity (94). A crystal structure of HutP demonstrated that each monomer consists of a four-stranded antiparallel β sheet, with two α helices located on each side of the β sheet (76). A structure of the activated HutP-histidine-Mg2+ complex demonstrated that histidine and Mg2+ interact with one another, that histidine contacts several amino acid residues in HutP, and that six molecules of both histidine and Mg2+ bind to each HutP hexamer (76). When activated by histidine and Mg2+, HutP binds to NAG triplet repeats that are separated by two to four nonconserved spacer residues (74, 93). A crystal structure of activated HutP in complex with an RNA containing three UAG repeats revealed that one RNA molecule bound to one surface of HutP and a second RNA molecule bound to an equivalent surface on the opposite side of HutP. The bound RNA molecules adopted a triangular shape on each HutP RNA-binding surface (75). The natural RNA target in the hutP-hutH intercistronic region contains two clusters of three NAG repeats, with the two clusters separated by 20 nucleotides. A crystal structure of HutP with an RNA molecule containing six UAG repeats that mimicked the natural RNA target demonstrated that one cluster bound to each HutP surface, with the long spacer separating each cluster wrapping around one side of the HutP hexamer (76).
PyrR-Mediated Attenuation in B. subtilis
The B. subtilis pyrRPBC-AA-AB-KDFE operon encodes an RNA-binding protein that regulates expression of the operon (PyrR), a uracil transporter (PyrP), and the enzymes for de novo biosynthesis of UMP (137). PyrR functions as a homodimer and is responsible for sensing both UMP and UTP in the cell (26, 81). Whereas UMP and UTP activate PyrR to bind to its RNA targets, GMP, GDP, and GTP bind to the same allosteric site of PyrR and compete with UMP/UTP binding (26, 29, 68).
PyrR regulates expression of the pyr operon by three essentially identical transcription attenuation mechanisms, each involving overlapping antiterminator and terminator structures (Figure 3). These attenuators are located in the pyr operon 5′ UTR, as well as in the intercistronic regions between pyrR and pyrP and between pyrP and pyrB (82, 83, 139). In each case, UMP-activated PyrR binds to an RNA structure called the binding loop. Since the binding loop includes nucleotides that are required for antiterminator formation, bound PyrR favors termination (82, 83, 139). Thus, the binding loop functions as an anti-antiterminator when bound by PyrR. Each of the three PyrR binding loops contains an identical ARUCCAGAGAGGYU sequence with the underlined residues residing in the loop of the hairpin. In addition to this binding element, a conserved sequence/structural motif is present closer to the base of the binding loop. Mutagenesis studies demonstrated that substitution of several of the residues in these two RNA elements inhibited PyrR binding (26). Mutational studies of PyrR further revealed that a basic concave surface is involved in RNA recognition (119). Further details of PyrR-RNA interaction will require a structure of this complex.
Figure 3.
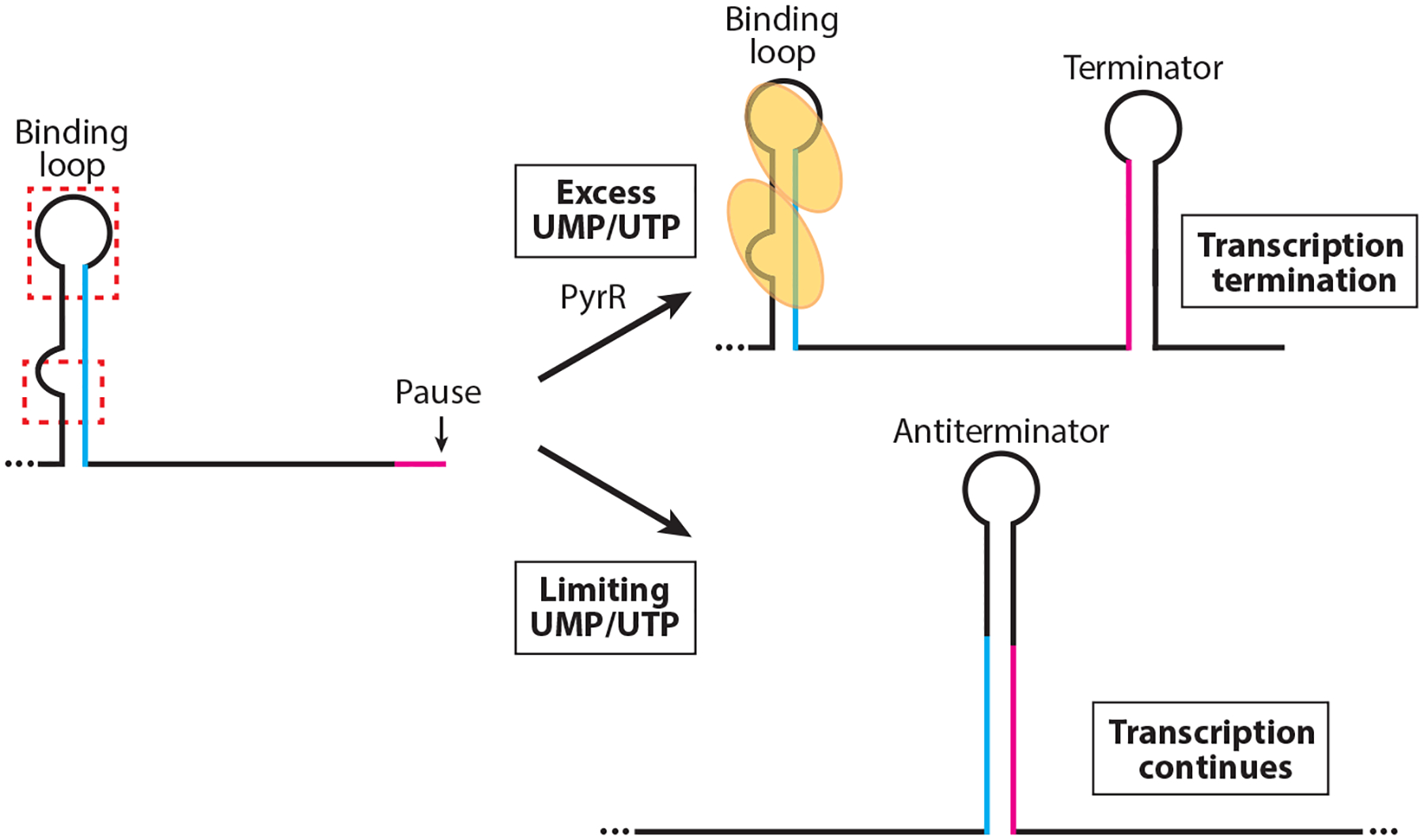
Model of the PyrR-mediated transcription attenuation mechanism. During transcription RNA polymerase pauses downstream of the PyrR-binding loop and within the overlap between the antiterminator and terminator structures (magenta). PyrR (yellow ovals) is not activated in limiting UMP/UTP conditions and does not bind to the binding loops in the 5′ UTR, or to the pyrR-pyrP and pyrP-pyrB intergenic regions. Once RNA polymerase resumes transcription, the antiterminator forms, which prevents formation of the mutually exclusive terminator hairpin (overlap in blue). In excess UMP/UTP conditions, UMP/UTP-activated PyrR binds to two regions of the binding loop (red dashed boxes). Pausing allows additional time for PyrR binding. Bound PyrR promotes transcription termination by preventing formation of the antiterminator structure.
NusA is a general transcription elongation factor that binds to RNA polymerase soon after sigma subunit release. RNA polymerase pausing appears to play a role in the three PyrR attenuation mechanisms. NusA stimulates pausing in vitro at positions that are present within the 3′ side of all three antiterminator structures (166). Each of these pause sites is positioned such that it could provide additional time for PyrR binding.
In addition to regulating pyr operon expression, PyrR is capable of catalyzing the uracil phosphoribosyltransferase (UPRTase) reaction involved in the uracil salvage pathway. However, the affinity for uracil is much lower than that of the UPRTase activity encoded by upp, suggesting that PyrR does not play a significant role in this biochemical pathway (134, 138). Rather, it was proposed that the UPRTase activity may have been its original function prior to evolving its RNA-binding activity (137).
TRAP-Mediated Attenuation in B. subtilis
The B. subtilis trpEDCFBA operon and trpG (pabA) encode the enzymes to convert chorismic acid, the common aromatic amino acid branch point, to tryptophan. mtrB encodes TRAP of B. subtilis. TRAP is composed of 11 identical subunits arranged in a single ring and is responsible for regulating tryptophan biosynthesis and transport in response to tryptophan (13). Tryptophan activates TRAP by binding between each adjacent subunit. Nine hydrogen bonds are formed between tryptophan and amino acids residing on adjacent subunits, with the indole ring buried in a hydrophobic pocket (7, 8). NMR studies indicate that tryptophan binding causes a structural change that is required for high-affinity TRAP-RNA interaction (86). Tryptophan-activated TRAP binds to NAG triplet repeats (GAG>UAG>AAG>CAG), with each repeat typically separated by two or three nonconserved residues (15, 17, 19). A crystal structure of the TRAP-RNA complex demonstrated that the RNA wraps around the perimeter of the TRAP ring and that each triplet repeat interacts with a KKR motif on the TRAP perimeter (7).
TRAP regulates expression of the trp operon by a transcription attenuation mechanism in which mutually exclusive antiterminator and terminator structures can form in the nascent trp transcript (Figure 4a). Under limiting tryptophan, TRAP is not activated and does not bind to RNA. In this case the antiterminator forms in the 5′ UTR such that transcription proceeds into the trp operon structural genes (18, 96). Under excess tryptophan, tryptophan-activated TRAP binds to 11 triplet repeats in the RNA, 7 of which are present within the sequence that forms the antiterminator structure. As a consequence, bound TRAP promotes termination in the 5′ UTR by preventing formation of the antiterminator (18). The general transcription elongation factors NusA and NusG stimulate pausing just upstream of the critical overlap between the antiterminator and terminator structures. Pausing at this position provides additional time for TRAP to bind to the nascent transcript in vitro, although pausing at this position has not been substantiated in vivo (89, 151, 156). TRAP also contributes to termination in the 5′ UTR by a mechanism that is independent of its role in preventing formation of the antiterminator. Although the mechanism has not been firmly established, it appears that bound TRAP promotes forward translocation of RNA polymerase, thereby increasing the termination efficiency (85, 105).
Figure 4.
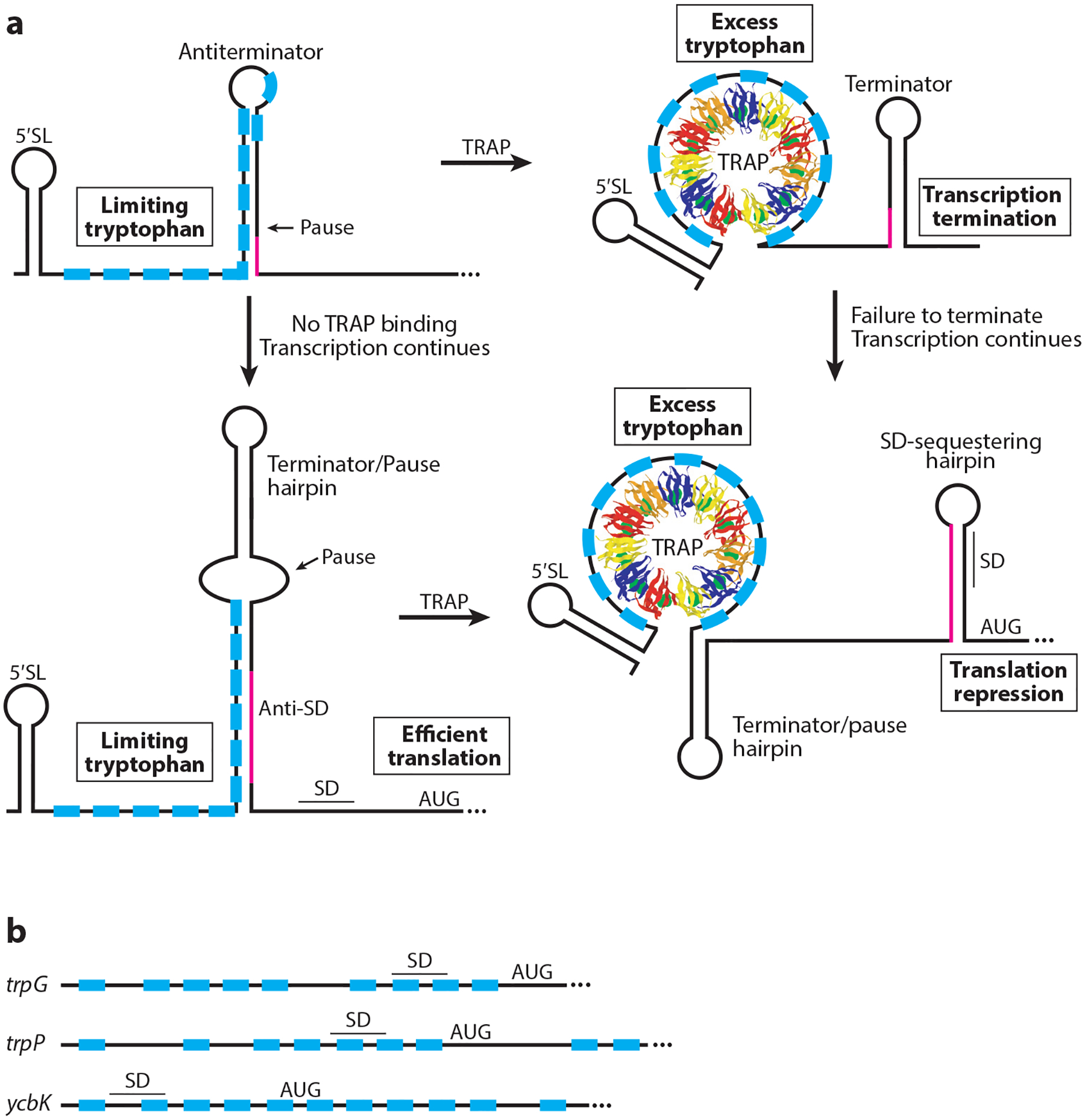
Models of TRAP-mediated transcription attenuation and translation repression mechanisms. The 11 TRAP subunits are shown in different colors, and tryptophan is shown as green spheres. (a, Top) Transcription attenuation mechanism. TRAP is not activated in limiting tryptophan conditions and does not bind to the RNA. Thus formation of the antiterminator results in transcription of the downstream genes. In excess tryptophan conditions, tryptophan-activated TRAP binds to the triplet repeats (blue boxes). Pausing may provide additional time for TRAP binding. Bound TRAP prevents formation of the antiterminator, resulting in transcription termination upstream of the coding sequences. Overlap between the antiterminator and terminator structures is shown in magenta. (a, Bottom) trpE translation control model. In tryptophan-limiting conditions the RNA adopts a structure such that the trpE SD sequence is single stranded and available for ribosome binding. When tryptophan is in excess, tryptophan-activated TRAP binds to its target sequence such that the trpE SD–sequestering hairpin forms, causing translational repression. RNA polymerase pausing provides additional time for TRAP binding. Overlap between the two alternative structures is shown in magenta. (b) TRAP-binding sites overlapping the trpG, trpP, and ycbK SD sequence and translation initiation regions. Bound TRAP represses translation of each gene. Abbreviations: 5′ SL, 5′ stem-loop; SD, Shine-Dalgarno; TRAP, trp RNA–binding attenuation protein. Adapted with permission from Reference 14.
In addition to the antiterminator and terminator structures, a 5′ stem-loop (5′ SL) structure that forms just upstream of the triplet-repeat region participates in the attenuation mechanism (35). Not only does TRAP-5′ SL interaction increase the affinity of TRAP for trp leader RNA, but this interaction also orients the downstream triplet repeats for interaction with the 11 KKR motifs that lie on the TRAP perimeter, thus increasing the likelihood that TRAP will bind in time to promote termination (87).
Another protein called anti-TRAP functions as a TRAP antagonist by binding to tryptophan-activated TRAP (144, 145). Binding of one trimeric anti-TRAP protein is sufficient to prevent TRAP-RNA interaction (124). Expression of rtpA, the gene encoding anti-TRAP, responds to the accumulation of uncharged tRNATrp via the T box riboswitch mechanism (56, 118). Thus, B. subtilis controls tryptophan metabolism by sensing the levels of both free tryptophan and uncharged tRNATrp.
REGULATION OF TRANSLATION INITIATION
TRAP-Mediated Translational Repression in B. subtilis
In addition to regulating expression of the trpEDCFBA operon by transcription attenuation, TRAP regulates translation initiation of trpE (Figure 4a). In the absence of bound TRAP, trp operon read-through transcripts adopt an RNA secondary structure in which the trpE SD sequence is available for ribosome binding. However, when TRAP is bound to a read-through transcript, an RNA structure forms that sequesters the trpE SD sequence such that ribosome binding is inhibited (33, 117). A NusG-dependent RNA polymerase pause site located 3–4 nucleotides downstream from the 5′ UTR termination sites participates in this translation control mechanism by providing additional time for TRAP binding (154, 155, 156). NusG-dependent pausing depends on sequence-specific contacts with unpaired T residues in the nontemplate DNA strand within the paused transcription bubble (154). Because the trpE and trpD coding regions overlap by 29 nucleotides, TRAP-dependent translational repression of trpE is extended to trpD via translational coupling, a process in which translation of the downstream cistron is at least partially dependent on translation of the cistron just upstream. In addition, TRAP-dependent formation of the trpE SD–sequestering hairpin allows Rho access to the nascent transcript, causing transcriptional polarity (158).
TRAP also represses translation initiation of the unlinked trpG (pabA) (34, 162), trpP (118, 161), and ycbK (160) genes in response to tryptophan (Figure 4b). trpG is the second gene in an operon primarily concerned with folate biosynthesis, trpP (tryptophan transport) is a single-gene operon, while ycbK (unknown function) is in the rtpA-ycbK operon; recall that rtpA encodes anti-TRAP. TRAP binds to a triplet-repeat region that overlaps the cognate SD sequence and translation initiation regions of trpG, trpP, and ycbK, directly occluding ribosome binding in each case (34, 160, 161). Thus, TRAP coordinately regulates tryptophan biosynthesis and transport using three distinct mechanisms: by transcription attenuation of the trpEDCFBA operon, by promoting formation of the trpE SD-sequestering hairpin, and by directly blocking ribosome access to the trpG and trpP SD sequences.
CspA-Mediated Cold Shock Response in E. coli
When E. coli cells growing at 37°C experience a temperature downshift to 10°C, cell growth enters a lag phase for approximately 4 h. During this cold shock acclimation phase, the accumulation of 17 cold shock–induced proteins was demonstrated by 2D gel electrophoresis (67). The major cold shock protein is CspA, constituting approximately 13% of the total cellular protein synthesis during acclimation (46). CspA acts as an RNA chaperone that destabilizes global mRNA structure at low temperatures by binding preferentially to pyrimidine-rich RNA sequences (66). Bound CspA destabilizes secondary structures to promote translation or alter mRNA turnover of downstream genes.
CspA is structurally related to the cold shock domain of Y-box protein in eukaryotes, which forms a β-barrel structure with a positively charged RNA-binding surface (120). Three aromatic amino acids on the RNA-binding surface promote binding and destabilization of the RNA secondary structure. Base-stacking interactions between these aromatic amino acids and nucleotide bases in the apical loop of a hairpin provide the energy requirement to destabilize hydrogen bonding of the first few bases in the hairpin stem (110).
Expression of cspA is regulated posttranscriptionally by a temperature-sensitive RNA switch in its 5 UTR that adopts one of two secondary structures that regulate translation initiation (44). At 37°C, the 5′ UTR forms a secondary structure that sequesters the SD sequence and translation initiation codon in a base-paired structure that inhibits ribosome binding. After a temperature shift to 10°C, the 5′ UTR of cspA adopts an entirely different secondary structure in which the SD sequence and AUG start codon are no longer sequestered in a stable structure. This thermosensitive switch controlling ribosome accessibility is the first example of an RNA structure than can promote gene expression at low temperatures (44).
CspA regulates its own translation in an autoregulatory feedback mechanism during cold shock acclimation. As the cellular concentration of CspA increases in response to cold shock, it binds to the 5 UTR of its own mRNA, forcing a conformational rearrangement (167). This rearranged structure in the cspA 5′ UTR resembles the structure of the mRNA at 37°C, where translation is blocked and the transcript is rapidly degraded. The structure adopted by the mRNA is dictated by long-distance base-pairing interactions that include a conserved cold-box sequence in the 5′ UTR (42). This base-pairing interaction is a key feature in the structural rearrangement of the mRNA at a temperature shift or in the presence of bound protein (167). Moreover, limited accessibility of the SD and AUG start codon when the mRNA adopts this structure leads to rapid turnover of the cspA transcript (21).
During the acclimation phase of cold shock, the rate of protein synthesis decreases dramatically (67). This decrease is a result of depleted ribosome density in the 5′ ends of open reading frames (ORFs) and increased secondary structure of global mRNAs (167). Recovery of translation is directly correlated with a decrease in ORF mRNA secondary structure mediated by the activity of CspA. A mutational analysis identified RNase R as a second cold shock protein involved in global translational recovery (167). RNase R is required for the turnover of cspA mRNA at low temperatures when CspA is bound so as to achieve the appropriate cellular CspA:mRNA ratio (167). RNase R also regulates global mRNA turnover during cold shock acclimation in cooperation with the activity of CspA. Degradation of global mRNAs facilitated by RNase R during cold shock allows the cell to retain the correct CspA:mRNA ratio for efficient RNA remodeling (167).
CsrA-Mediated Translational Regulation
E. coli CsrA and its homologs in other organisms (e.g., CsrA, RsmA, RsmE) have been shown to repress translation of numerous genes. In many instances, translational repression leads to rapid turnover of the mRNA. Translational repression typically involves CsrA binding to multiple sites in the target transcript, one of which overlaps the SD sequence of the mRNA target, such that bound CsrA prevents ribosome binding (112, 143) (Figure 5a). The first such repression mechanism was identified for E. coli glgC, which encodes an enzyme involved in glycogen biosynthesis (22). CsrA binds to four sites in the glgCAP 5′ UTR, one of which overlaps the glgC SD sequence while the other three are located further upstream. CsrA homodimers are capable of bridging a high-affinity site in the glgC 5′ UTR to a lower-affinity site overlapping the SD sequence (112). A structural study of a Pseudomonas fluorescens RsmE-RNA complex confirmed that protein dimers are capable of binding to two sites within the same transcript (39). Since CsrA-binding sites resemble SD sequences, dual site bridging appears to be a common mechanism to allow effective CsrA-mediated repression while at the same time maintaining a strong SD sequence for translation initiation.
Figure 5.
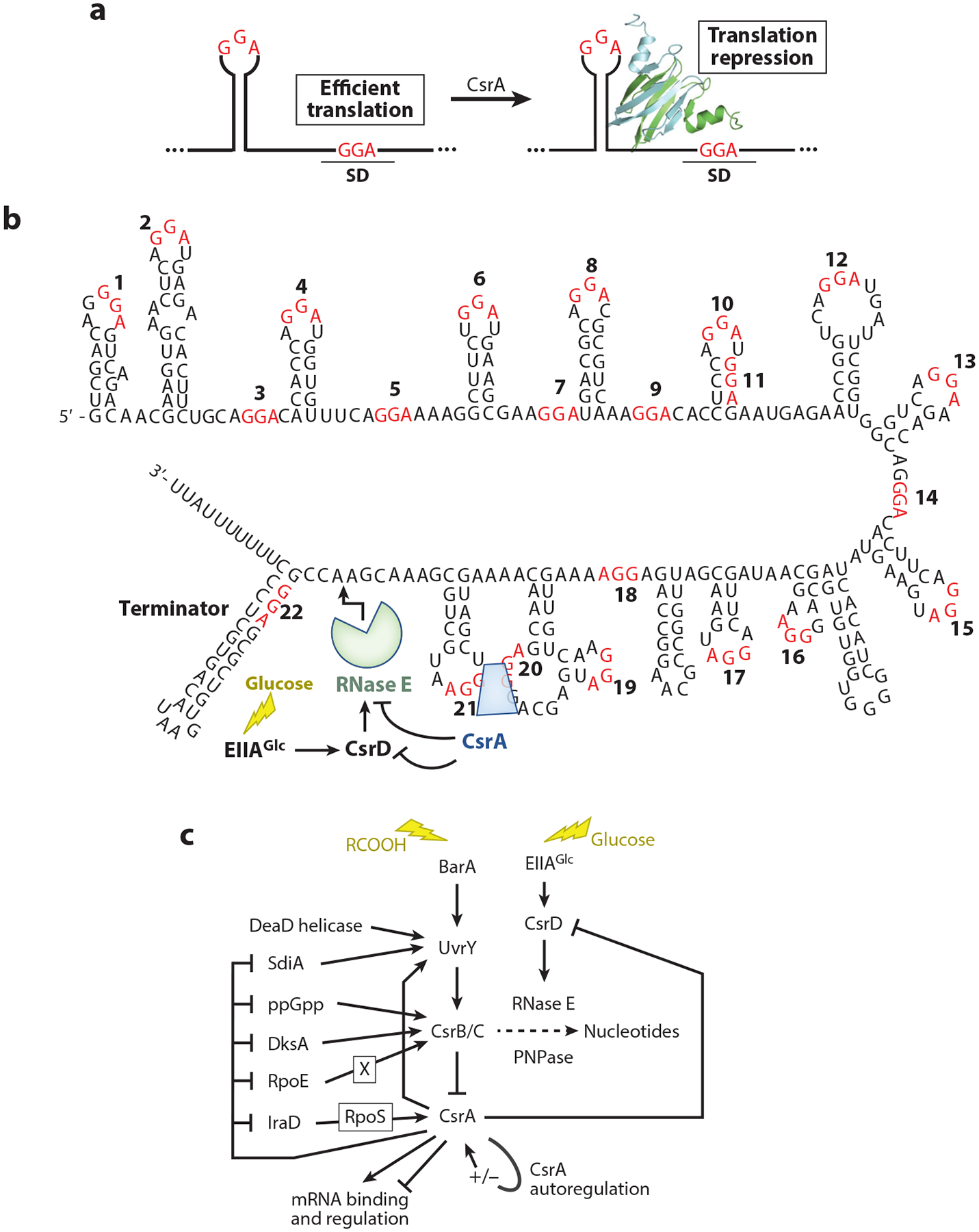
Csr-mediated regulatory pathways. (a) Common CsrA-mediated translation repression mechanism. CsrA dimers, depicted as green and blue ribbons, bind to two sites, one of which overlaps the Shine-Dalgarno (SD) sequence such that bound CsrA blocks ribosome binding. GGA motifs of CsrA-binding sites are in red. (b) CsrB sequence, secondary structure, and RNA-decay pathway. GGA motifs are numbered and shown in red. (c) Circuitry influencing the Csr system. The Csr system of Escherichia coli includes CsrA autoregulation, negative-feedback loops among the Csr components, and reciprocal interactions with other global regulatory systems. Autoregulation and feedback loops fine-tune CsrA activity and support robust regulatory responses, while interactions with other global regulatory systems integrate the Csr system into stress responses. The CsrA ribbon diagram in panel a is adapted with permission from Reference 112.
In addition to the general CsrA-mediated translational repression mechanism described above, there are several examples that deviate substantially from this mechanism. CsrA is capable of repressing translation of hfq by binding to a single site that overlaps its SD sequence (112). Since Hfq functions as an RNA chaperone that promotes numerous sRNA-mRNA base-pairing interactions (72, 116), the finding that CsrA represses hfq expression established a link between these two important global posttranscriptional regulatory systems. Another interesting example was described for the Pseudomonas aeruginosa psl transcript, required for biosynthesis of a biofilm matrix polysaccharide of this organism. RsmA binding to the psl 5′ UTR stabilizes an RNA structure that sequesters its SD sequence (62). Finally, CsrA represses translation of iraD, a gene encoding a protein that prevents targeted degradation of RpoS, the general stationary-phase and stress-response sigma factor of E. coli. Rather than directly repressing iraD translation, CsrA represses translation of a short leader peptide whose termination codon overlaps the iraD initiation codon. Since translation of the leader peptide and iraD is coupled, CsrA represses IraD synthesis entirely via translational coupling (99).
While translation repression is the most-studied mechanism by which CsrA controls gene expression, there are two reported examples of translational activation, although the detailed mechanisms have not been elucidated. P. aeruginosa RsmA activates translation of the phenazine biosynthetic gene cluster phz2, presumably by destabilizing an SD-sequestering hairpin (109). In addition, E. coli CsrA activates translation of moaA by altering the structure of the 5′ UTR, thereby promoting ribosome binding (101).
CsrA has also been shown to regulate gene expression via transcription attenuation and mRNA stabilization. The E. coli pgaABCD operon encodes proteins involved in the synthesis and secretion of a polysaccharide adhesin involved in biofilm formation (63, 148). In addition to CsrA-mediated repression of pgaA translation initiation (148), expression of the pga operon is regulated by a Rho-dependent transcription attenuation mechanism (43). In the absence of bound CsrA, an RNA secondary structure forms that prevents Rho from binding to the nascent transcript. However, when CsrA is bound to the pga 5′ UTR, this structure cannot form, thereby exposing a Rho-binding site, leading to Rho-dependent termination upstream of the pgaA coding sequence. Lastly, E. coli CsrA activates expression of the genes encoding a DNA binding activator of flagella biosynthesis and chemotaxis. CsrA binding to the 5′ end of the flhDC transcript stabilizes the mRNA by blocking the 5′ end–dependent cleavage pathway of RNase E (153). Since CsrA has been shown to bind hundreds of RNAs in E. coli and Salmonella enterica (59, 106), it is likely that future studies will reveal new CsrA-dependent regulatory mechanisms.
RNA-BINDING PROTEINS EXHIBITING COMPLEX INTERACTIONS WITH sRNAs
Csr (Rsm) Family Proteins
Competitive inhibition was introduced to the world of sRNAs and RNA-binding proteins when the global regulatory protein CsrA of E. coli was found to copurify in a large globular complex with a 369-nucleotide RNA molecule, CsrB, that antagonizes CsrA activity (79, 80, 113). CsrB RNA has 22 GGA-containing sequences, most of which are bound by CsrA (141) (Figure 5b). E. coli also expresses a second sRNA, CsrC, related structurally and functionally to CsrB (149). The GGA-containing binding sites of Csr family sRNAs are also closely related to mRNA consensus binding sites derived from SELEX and CLIP-seq studies (16, 36, 59, 106). Except for their repeated GGA-containing stem-loops, the Csr family sRNAs are not highly conserved (39, 64, 114, 143). In a well-studied example, the P. fluorescens RsmE:RsmZ complex was shown to be assembled via ordered and cooperative protein binding, although the structural diversity of Csr family sRNAs suggests that this result may not apply to all such interactions (39).
Phylogenetic examinations suggest that the CsrA-inhibitory sRNAs probably function throughout the Gammaproteobacteria. Species of this bacterial class are unique in expressing a conserved two-component signal transduction system, referred to as GacS-GacA, BarA-UvrY, BarA-SirA, etc., that is frequently, but not exclusively, dedicated to the transcription of Csr family sRNAs (27, 57, 165). Candidate CsrA-binding sRNAs have been identified by bioinformatics analyses in a variety of species (41), but to our knowledge no CsrA-sequestering sRNA has been confirmed outside of the Gammaproteobacteria.
Because Csr family sRNA levels control CsrA activity and widely influence physiology and virulence gene expression (112, 143), the syntheses of these RNAs have been investigated in several species, most intensively in E. coli. CsrB/C transcription responds to cues associated with limitation of nutrients, e.g., ppGpp, the elicitor of the stringent response; carboxylic acid–containing carbon metabolites such as formate, acetate, or TCA cycle intermediates; and the cAMP-Crp complex (28, 31, 40, 97, 123, 165). In addition, the expression of CsrB/C is subject to other conditions and regulatory factors, including quorum sensing (SdiA), envelope stress (σE), and DeaD-box RNA helicases (DeaD, SrmB), consistent with a far-reaching role of these sRNAs in monitoring and responding to cell physiology (133, 142, 157). The regulatory interactions between the Csr components themselves and other factors generate negative-feedback loops that support quick and potent responses to changing conditions (1, 97–99, 114, 132, 133, 143, 157) (Figure 5c). Numerous transcription factors are subject to CsrA regulation (106), including transcription factors that directly or indirectly regulate CsrB/C levels (Figure 5c). Such reciprocal regulatory circuitry implies that the Csr system can reinforce transcriptional regulation at the posttranscriptional level for genes that are regulated by both CsrA and an associated transcription factor (40). In addition, the integrated circuitry of systems in which a stress activates CsrB/C production, while CsrA represses one or more regulatory factors responding to that stress, predicts that the Csr system is involved in initiating and propagating stress responses, as well as restoring the basal activity of the stress response system when the consequences of stress have been resolved (e.g., 40, 99).
Turnover of the Csr RNAs in E. coli, Vibrio cholerae, and most likely other Enterobacteriaceae, Vibrionaceae, and Shewanellaceae, requires the activation of RNase E cleavage by a membrane-associated, nonnucleolytic protein that contains degenerate GGDEF and EAL domains (CsrD or MshH) (77, 132, 147). This role of CsrD allows it to impart global effects on gene expression (104). Because CsrD is activated upon its binding to the unphosphorylated form of EIIAGlc of the PTS pathway, degradation of CsrB/C is triggered by the presence of a preferred carbon source, such as glucose. Limited studies of Csr family sRNAs in species that lack a CsrD homolog find that their decay is directly inhibited by the binding of a Csr family protein (39, 146). In contrast, in csrD wild-type E. coli, CsrA does not affect CsrB decay (55). However, in strains deleted for csrD, CsrB is protected by CsrA against RNase E attack (141). In summary, CsrD is necessary for RNase E to cleave CsrB because this RNA is otherwise protected by CsrA binding, and the evolution of CsrD allowed CsrB/C decay in some species to become responsive to the availability of preferred carbon substrates (77, 141).
The classical Csr family sRNAs, largely composed of repeated GGA-containing stem-loop structures, perform no known function besides CsrA sequestration. However, it seems that other CsrA-binding RNAs sequester CsrA as a moonlighting function, in addition to performing their other roles in the cell (112). These include the abundant mRNA fimAICDHF of Salmonella, to which CsrA binding acts in a hierarchical fashion to prevent CsrA-activated expression of secondary fimbriae (128). The base-pairing sRNAs McaS and GadY seemingly use CsrA sequestration in regulating pgaABCD expression and biofilm formation (63, 69, 98, 100, 148). Such regulation is possible because the concentration of CsrA that is not bound to RNA in the cell is low (114), allowing tight-binding RNAs expressed at sufficient levels to compete for CsrA (RsmA) binding with lower-affinity transcripts.
Finally, CsrA-binding proteins have recently been found to serve as CsrA antagonists. One such protein, FliW, participates with CsrA in the regulation of motility in B. subtilis and likely functions widely as an inhibitor of CsrA in species outside of the Gammaproteobacteria (37, 92, 107, 159). Unlike the Csr family sRNAs, FliW inhibits CsrA activity using an allosteric mechanism (2, 91, 92). On the other hand, CsrA binding to the CesT protein, which also serves as a chaperone for type III secretion system (TTSS) effectors in enteropathogenic and enterohemorrhagic E. coli (EPEC and EHEC), inhibits CsrA activity by directly occluding its RNA-binding surfaces (71, 164). While their CsrA-inhibitory mechanisms differ, FliW and CesT both mediate regulation by binding to CsrA after their primary protein partners have been secreted through the pores of the flagellum or TTSS. Notably, EPEC and EHEC strains possess csrB and csrC genes as well as cesT, although their contributions to the regulation of CsrA activity remain to be elucidated (25).
RapZ
The enzyme glucosamine-6-phosphate synthase (GlmS) catalyzes the committed step of the de novo biosynthetic pathway for UDP-N-acetyl-d-glucosamine, an essential intermediary metabolite and precursor of peptidoglycan, lipopolysaccharide, enterobacterial common antigen, and the biofilm adhesin poly-β−1,6-N-acetylglucosamine (PGA) (24, 63, 70, 111, 127). The widely conserved protein RapZ (RNase adapter protein for sRNA GlmZ) and two sRNAs, GlmZ and GlmY, play a critical role in regulating the expression of glmS in E. coli (51, 103) (Figure 6a). In its un-cleaved form, GlmZ interacts with the RNA chaperone Hfq, which promotes GlmZ-glmS mRNA base-pairing. Bound GlmZ prevents formation of a glmS SD–sequestering hairpin, thus activating its translation (70, 140). RapZ binds to GlmZ sRNA and to the N-terminal catalytic domain of RNase E, triggering RNase E cleavage and inactivation of GlmZ, resulting in the loss of translational activation (52). GlmY sRNA is related in sequence and structure to GlmZ but does not bind with high affinity to Hfq or base-pair with glmS mRNA (51). RapZ binds to GlmY sRNA. However, unlike GlmZ, GlmY is not cleaved by RNase E upon binding to RapZ; it competes effectively with GlmZ for binding to RapZ (52). This results in stabilization of GlmZ when the active form of GlmY RNA is present. Consequently, conditions that favor GlmY accumulation, such as decreased glucosamine levels, prevent GlmZ cleavage and activate glmS translation (52, 108).
Figure 6.
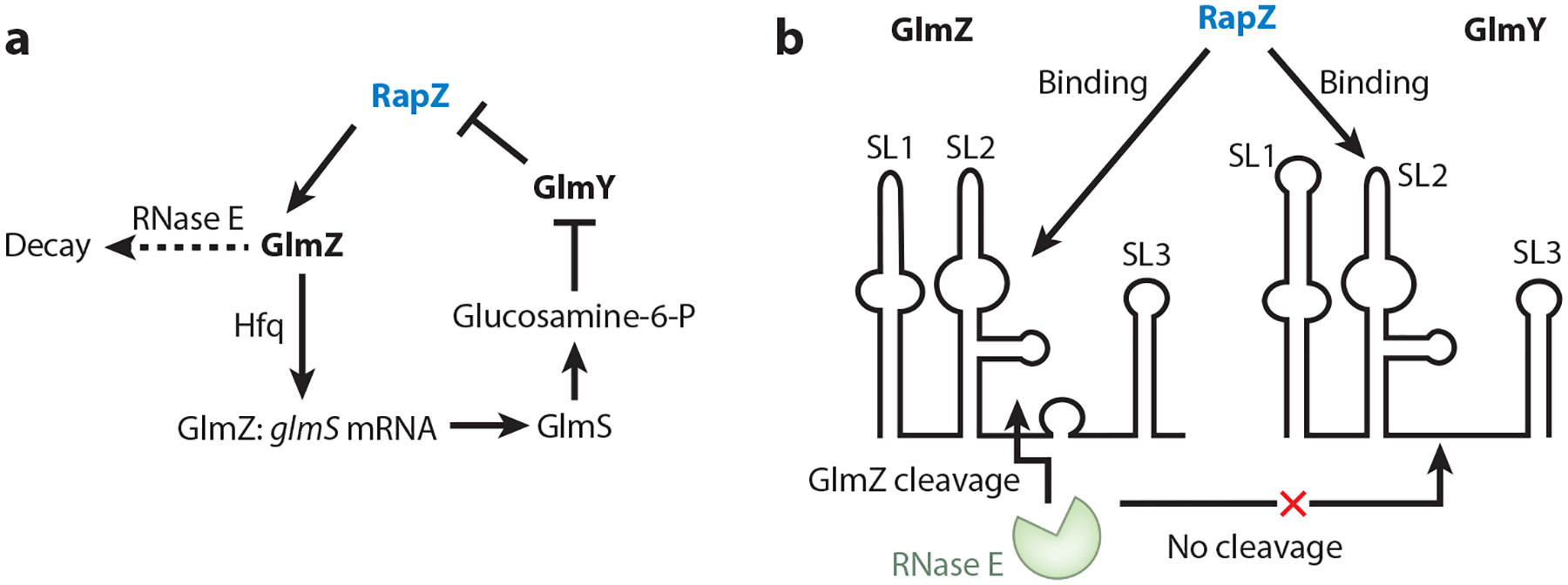
Regulation by the RNA-binding protein RapZ and its two related small RNA targets, GlmZ and GlmY. (a) Regulatory circuitry and (b) RapZ:GlmZ recognition involving stem-loop 2 (SL2) promote RNase E recognition and sequence-independent cleavage at the single-stranded base-pairing site of GlmZ. GlmY competes with GlmZ for RapZ binding without undergoing RapZ-mediated RNase E cleavage.
Despite their structural similarity, modest differences associated with stem-loop 2 (SL2) (Figure 6b) of these sRNAs permit RNase E to cleave GlmZ:RapZ, but not GlmY:RapZ. The discovery of an RNA aptamer (GlmZ SL1,2) that is recognized by an RNA-binding protein, thus rendering the RNA susceptible to RNase E cleavage, represented a new mechanism for initiating RNase E cleavage, distinct from the 5′ monophosphate recognition and the direct entry cleavage routes (51). The additional discovery that RNase E cleavage of GlmZ occurs independently of its nucleotide sequence suggested the adaptability of this system for designing regulatable RNA-decay mechanisms, useful in synthetic biology (51). Conceivably, the CsrD protein might invoke a related mechanism for triggering CsrB/C cleavage by RNase E (141), although this remains to be seen.
A crystal structure revealed that RapZ is a homotetrameric protein (48). The N-terminal domain of RapZ is related to kinases, such as adenylate kinase, and contains Walker A and B motifs, while the C-terminal domain is related to 6-phosphofructokinases. While RapZ is unrelated to other RNA-binding proteins, a high density of surface-exposed positive charges in the C terminus suggests this as a likely site for interaction with RNA. These findings have set the stage for the elucidation of the detailed structural biology of this fascinating RNA-binding protein.
ProQ/FinO Family Proteins
ProQ/FinO domain–containing proteins have gained attention as emerging regulatory RNA-binding proteins that possess RNA chaperone activities (58, 95, 125). E. coli FinO (fertility inhibition) was once considered a single member of its own class, until recent studies revealed a wider distribution of homologs on the chromosomes or plasmids of Proteobacteria (11). Thus far, three different RNA-binding proteins containing the ProQ/FinO domain have been studied (10, 11, 49, 125).
The F plasmid–encoded FinO protein promotes duplex formation between the complementary sequences of the cis-acting sRNA FinP and the 5′ UTR of traJ mRNA, thus stabilizing FinP against ribonuclease degradation, repressing translation initiation of traJ by blocking ribosome binding, and inhibiting plasmid conjugation (10, 45, 65) (Figure 7a). The chromosome-encoded RocC (repressor of competence) protein of Legionella pneumophila interacts with the trans-acting sRNA RocR to posttranscriptionally repress the expression of competence genes (comEA, comM, comEC, and comF) that are required for the DNA uptake by this bacterium (11) (Figure 7b). ProQ itself functions as a monomer and was initially identified as a regulator of the E. coli pro-line transporter ProP, although this regulatory mechanism remains to be determined (30). An approach designed to identify the RNA-protein partners in cellular complexes, Grad-seq, suggested the possibility of a global role for ProQ in the posttranscriptional regulatory networks of Salmonella, while CLIP-seq enabled the discovery of hundreds of RNA transcripts that directly interact with ProQ in S. enterica and E. coli. (58, 125). Although most of the currently identified ProQ-binding sRNAs remain uncharacterized, it is clear that only a few (21 out of 138) of its sRNA targets overlap with those of Hfq or CsrA, suggesting the existence of distinct features of ProQ-dependent sRNAs (58). Analysis of the sRNA targets of ProQ revealed that the interaction appears to be driven by recognition of RNA stem-loop structures rather than consensus sequence motifs, which is consistent with observations in FinO- and RocC-dependent sRNAs (11, 58, 65).
Figure 7.

Target small RNAs (sRNAs) of FinO/ProQ domain proteins. Putative ProQ-binding sites in yellow. Regions involved in base-pairing with target mRNAs are circled in red. Guanine-cytosine pairs are depicted by double bars, adenine-uracil pairs are depicted by single bars, and bars with circles represent guanine-uracil pairs. (a) cis-Acting FinP sRNA from Escherichia coli F plasmid, which is bound by FinO. (b) trans-Acting RocR sRNA from Legionella pneumophila, which is bound by RocC. (c) trans-Acting RaiZ sRNA from Salmonella enterica, which is dependent on ProQ.
One molecular mechanism for ProQ-mediated regulation was identified for the sRNA RaiZ, which interacts at high affinity with ProQ via its two 3′ -terminal stem-loops and the nucleotides in the 5′ double-stranded region (Figure 7c). RaiZ base-pairs with the 5′ UTR of hupA mRNA in a ProQ-dependent manner to prevent synthesis of HU-α by interfering with the formation of a translation initiation complex (126). Notwithstanding its importance as the first mechanistic example of ProQ regulation, this mechanism might not represent the predominant mode of action by ProQ. Indeed, of the ~500 RNAs recognized by ProQ, fewer than 8% were bound at mRNA 5′ UTRs. In contrast, ProQ bound >30% of RNAs at their 3′ ends (58).
An NMR structure of E. coli ProQ uncovered an extended architecture with globular N-terminal and C-terminal domains bridged by a flexible linker region (49). All three regions may be involved independently in RNA binding. Notably, the N-terminal domain represents the conserved ProQ/FinO domain, whereas the C-terminal domain not only shares partial similarity with Hfq but also resembles the Tudor domain superfamily of eukaryotic proteins, involved in RNA metabolism and recognition of modified amino acids in protein partners (49, 102). These advances will enable a broader and more detailed molecular understanding of ProQ regulation to be obtained in the future.
SUMMARY POINTS.
RNA-binding proteins can regulate transcription termination by attenuation (repression) or antitermination (activation) mechanisms.
RNA-binding proteins can regulate translation initiation by altering the accessibility of ribosome-binding sites by promoting or preventing formation of SD-sequestering hairpins and/or by directly occluding ribosome binding.
The interactions of RNA-binding proteins with target RNAs can regulate RNA decay by protecting a nuclease cleavage site or by facilitating nuclease recognition of an otherwise inaccessible site.
The activity of RNA-binding proteins can be altered by phosphorylation or by binding of effectors including metabolites, sRNAs, or other proteins.
RNA-binding proteins can regulate specific genes or function as global regulators by binding to hundreds of RNA targets.
RNA-binding proteins can be central to gene expression networks, in which they interact with transcription factors and other regulators to generate diverse and complex biological responses.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants GM059969 (T.R. and P.B.), GM098399 (P.B.), and GM066794 (T.R.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Adamson DN, Lim HN. 2013. Rapid and robust signaling in the CsrA cascade via RNA-protein interactions and feedback regulation. PNAS 110:13120–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altegoer F, Rensing SA, Bange G. 2016. Structural basis for the CsrA-dependent modulation of translation initiation by an ancient regulatory protein. PNAS 113:10168–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amster-Choder O 2005. The bgl sensory system: a transmembrane signaling pathway controlling transcriptional antitermination. Curr. Opin. Microbiol 8:127–34 [DOI] [PubMed] [Google Scholar]
- 4.Amster-Choder O, Houman F, Wright A. 1989. Protein phosphorylation regulates transcription of the β-glucoside utilization operon in E. coli. Cell 58:847–55 [DOI] [PubMed] [Google Scholar]
- 5.Amster-Choder O, Wright A. 1992. Modulation of the dimerization of a transcriptional antiterminator protein by phosphorylation. Science 257:1395–98 [DOI] [PubMed] [Google Scholar]
- 6.Amster-Choder O, Wright A. 1997. BglG, the response regulator of the Escherichia coli bgl operon, is phosphorylated on a histidine residue. J. Bacteriol 179:5621–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X, et al. 1999. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature 401:235–42 [DOI] [PubMed] [Google Scholar]
- 8.Antson AA, Otridge J, Brzozowski AM, Dodson EJ, Dodson GG, et al. 1995. The structure of the trp RNA attenuation protein. Nature 374:693–700 [DOI] [PubMed] [Google Scholar]
- 9.Arnaud M, Débarbouillé M, Rapoport G, Saier MH Jr., Reizer J. 1996. In vitro reconstitution of transcriptional antitermination by the SacT and SacY proteins of Bacillus subtilis. J. Biol. Chem 271:18966–72 [DOI] [PubMed] [Google Scholar]
- 10.Arthur DC, Edwards RA, Tsutakawa S, Tainer JA, Frost LS, Glover JM. 2011. Mapping interactions between the RNA chaperone FinO and its RNA targets. Nucleic Acids Res 39:4450–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Attaiech L, Boughammoura A, Brochier-Armanet C, Allatif O, Peillard-Fiorente F, et al. 2016. Silencing of natural transformation by an RNA chaperone and a multitarget small RNA. PNAS 113:8813–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aymerich S, Steinmetz M. 1992. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. PNAS 89:10410–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babitzke P 2004. Regulation of transcription attenuation and translation initiation by allosteric control of an RNA-binding protein: the Bacillus subtilis TRAP protein. Curr. Opin. Microbiol 7:132–39 [DOI] [PubMed] [Google Scholar]
- 14.Babitzke P, Baker CS, Romeo T. 2009. Regulation of translation initiation by RNA binding proteins. Annu. Rev. Microbiol 63:27–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babitzke P, Bear DG, Yanofsky C. 1995. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a toroid-shaped molecule that binds transcripts containing GAG or UAG repeats separated by two nucleotides. PNAS 92:7916–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babitzke P, Romeo T. 2007. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol 10:156–63 [DOI] [PubMed] [Google Scholar]
- 17.Babitzke P, Stults JT, Shire SJ, Yanofsky C. 1994. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J. Biol. Chem 269:16597–604 [PubMed] [Google Scholar]
- 18.Babitzke P, Yanofsky C. 1993. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. PNAS 90:133–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Babitzke P, Yealy J, Campanelli D. 1996. Interaction of the trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis with RNA: effects of the number of GAG repeats, the nucleotides separating adjacent repeats, and RNA secondary structure. J. Bacteriol 178:5159–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachem S, Stülke J. 1998. Regulation of the Bacillus subtilis GlcT antiterminator protein by components of the phosphotransferase system. J. Bacteriol 180:5319–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bae W, Jones PG, Inouye M. 1997. CspA, the major cold shock protein of Escherichia coli, negatively regulates its own gene expression. J. Bacteriol 179:7081–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P. 2002. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol 44:1599–610 [DOI] [PubMed] [Google Scholar]
- 23.Belogurov GA, Artsimovitch I. 2015. Regulation of transcript elongation. Annu. Rev. Microbiol 69:49–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertani B, Ruiz N. 2018. Function and biogenesis of lipopolysaccharides. EcoSal Plus 8. 10.1128/ecosalplus.ESP-0001-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhatt S, Edwards AN, Nguyen HT, Merlin D, Romeo T, Kalman D. 2009. The RNA binding protein CsrA is a pleiotropic regulator of the locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli. Infect. Immun 77:3552–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonner ER, D’Elia JN, Billips BK, Switzer RL. 2001. Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR. Nucleic Acids Res 29:4851–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, et al. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol 73:434–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camacho MI, Alvarez AF, Chavez RG, Romeo T, Merino E, Georgellis D. 2015. Effects of the global regulator CsrA on the BarA/UvrY two-component signaling system. J. Bacteriol 197:983–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chander P, Halbig KM, Miller JK, Fields CJ, Bonner HK, et al. 2005. Structure of the nucleotide complex of PyrR, the pyr attenuation protein from Bacillus caldolyticus, suggests dual regulation by pyrimidine and purine nucleotides. J. Bacteriol 187:1773–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaulk SG, Smith-Frieday MN, Arthur DC, Culham DE, Edwards RA, et al. 2011. ProQ is an RNA chaperone that controls ProP levels in Escherichia coli. Biochemistry 50:3095–106 [DOI] [PubMed] [Google Scholar]
- 31.Chavez RG, Alvarez AF, Romeo T, Georgellis D. 2010. The physiological stimulus for the BarA sensor kinase. J. Bacteriol 192:2009–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Declerck N, Vincent F, Hoh F, Aymerich S, van Tilbeurgh H. 1999. RNA recognition by transcriptional antiterminators of the BglG/SacY family: functional and structural comparison of the CAT domain from SacY and LicT. J. Mol. Biol 294:389–402 [DOI] [PubMed] [Google Scholar]
- 33.Du H, Babitzke P. 1998. trp-RNA binding attenuation protein-mediated long-distance RNA refolding regulates translation of trpE in Bacillus subtilis. J. Biol. Chem 273:20494–503 [DOI] [PubMed] [Google Scholar]
- 34.Du H, Tarpey R, Babitzke P. 1997. The trp-RNA binding attenuation protein regulates TrpG synthesis by binding to the trpG ribosome binding site of Bacillus subtilis. J. Bacteriol 179:2582–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du H, Yakhnin AV, Dharmaraj S, Babitzke P. 2000. trp RNA-binding attenuation protein-5′ stem-loop RNA interaction is required for proper transcription attenuation control of the Bacillus subtilis trpEDCFBA operon. J. Bacteriol 182:1819–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dubey AK, Baker CS, Romeo T, Babitzke P. 2005. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11:1579–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dugar G, Svensson SL, Bischler T, Wäldchen S, Reinhardt R, et al. 2016. The CsrA-FliW network controls polar localization of the dual-function flagellin mRNA in Campylobacter jejuni. Nat. Commun 7:11667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durand S, Condon C. 2018. RNases and helicases in gram-positive bacteria. Microbiol. Spectr 6:RWR-0003–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duss O, Michel E, Yulikov M, Schubert M, Jeschke G, Allain FH. 2014. Structural basis of the noncoding RNA RsmZ acting as a protein sponge. Nature 509:588–92 [DOI] [PubMed] [Google Scholar]
- 40.Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K, et al. 2011. Circuitry linking the Csr and stringent response global regulatory systems. Mol. Microbiol 80:1561–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fakhry CT, Kulkarni P, Chen P, Kulkarni R, Zarringhalam K. 2017. Prediction of bacterial small RNAs in the RsmA (CsrA) and ToxT pathways: a machine learning approach. BMC Genom 18:645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang L, Hou Y, Inouye M. 1998. Role of the cold-box region in the 5′ untranslated region of the cspA mRNA in its transient expression at low temperature in Escherichia coli. J. Bacteriol 180:90–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Figueroa-Bossi N, Schwartz A, Guillemardet B, D’Heygère F, Bossi L, Boudvillain M. 2014. RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes Dev 28:1239–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giuliodori AM, Di Pietro FD, Marzi S, Masquida B, Wagner R, et al. 2010. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell 37:21–33 [DOI] [PubMed] [Google Scholar]
- 45.Glover JM, Chaulk SG, Edwards RA, Arthur D, Lu J, Frost LS. 2015. The FinO family of bacterial RNA chaperones. Plasmid 78:79–87 [DOI] [PubMed] [Google Scholar]
- 46.Goldstein J, Pollitt NS, Inouye M. 1990. Major cold shock protein of Escherichia coli. PNAS 87:283–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gollnick P, Babitzke P. 2002. Transcription attenuation. Biochim. Biophys. Acta Gene Struct. Expr 1577:240–50 [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez GM, Durica-Mitic S, Hardwick SW, Moncrieffe MC, Resch M, et al. 2017. Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism. Nucleic Acids Res 45:10845–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gonzalez GM, Hardwick SW, Maslen SL, Skehel JM, Holmqvist E, et al. 2017. Structure of the Escherichia coli ProQ RNA-binding protein. RNA 23:696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Göpel Y, Khan MA, Görke B. 2014. Ménage à trois: post-transcriptional control of the key enzyme for cell envelope synthesis by a base-pairing small RNA, an RNase adaptor protein, and a small RNA mimic. RNA Biol 11:433–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Göpel Y, Khan MA, Görke B. 2016. Domain swapping between homologous bacterial small RNAs dissects processing and Hfq binding determinants and uncovers an aptamer for conditional RNase E cleavage. Nucleic Acids Res 44:824–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Göpel Y, Papenfort K, Reichenbach B, Vogel J, Görke B. 2013. Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA. Genes Dev 27:552–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gopinath SC, Balasundaresan D, Kumarevel T, Misono TS, Mizuno H, Kumar PK. 2008. Insights into anti-termination regulation of the hut operon in Bacillus subtilis: importance of the dual RNA-binding surfaces of HutP. Nucleic Acids Res 36:3463–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Görke B, Rak B. 1999. Catabolite control of Escherichia coli regulatory protein BglG activity by antagonistically acting phosphorylations. EMBO J 18:3370–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gudapaty S, Suzuki K, Wang X, Babitzke P, Romeo T. 2001. Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli. J. Bacteriol 183:6017–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henkin TM. 2014. The T box riboswitch: a novel regulatory RNA that utilizes tRNA as its ligand. Biochim. Biophys. Acta Gene Regul. Mech 1839:959–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heroven AK, Sest M, Pisano F, Scheb-Wetzel M, Steinmann R, et al. 2012. Crp induces switching of the CsrB and CsrC RNAs in Yersinia pseudotuberculosis and links nutritional status to virulence. Front. Cell. Infect. Microbiol 2:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holmqvist E, Li L, Bischler T, Barquist L, Vogel J. 2018. Global maps of ProQ binding in vivo reveal target recognition via RNA structure and stability control at mRNA 3′ ends. Mol. Cell 70:971–82 [DOI] [PubMed] [Google Scholar]
- 59.Holmqvist E, Wright PR, Li L, Bischler T, Barquist L, et al. 2016. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J 35:991–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hör J, Gorski SA, Vogel J. 2018. Bacterial RNA biology on a genome scale. Mol. Cell 70:785–99 [DOI] [PubMed] [Google Scholar]
- 61.Hübner S, Declerck N, Diethmaier C, Le Coq D, Aymerich S, Stülke J. 2011. Prevention of crosstalk in conserved regulatory systems: identification of specificity determinants in RNA-binding anti-termination proteins of the BglG family. Nucleic Acids Res 39:4360–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol 78:158–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, et al. 2008. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-β−1,6-N-acetyl-d-glucosamine. J. Bacteriol 190:3670–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Janssen KH, Diaz MR, Gode CJ, Wolfgang MC, Yahr TL. 2018. RsmV, a small noncoding regulatory RNA in Pseudomonas aeruginosa that sequesters RsmA and RsmF from target mRNAs. J. Bacteriol 200:e00277–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jerome LJ, Frost LS. 1999. In vitro analysis of the interaction between the FinO protein and FinP antisense RNA of F-like conjugative plasmids. J. Biol. Chem 274:10356–62 [DOI] [PubMed] [Google Scholar]
- 66.Jiang W, Hou Y, Inouye M. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem 272:196–202 [DOI] [PubMed] [Google Scholar]
- 67.Jones PG, VanBogelen RA, Neidhardt FC. 1987. Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol 169:2092–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jørgensen CM, Fields CJ, Chander P, Watt D, Burgner JW 2nd, et al. 2008. pyr RNA binding to the Bacillus caldolyticus PyrR attenuation protein—characterization and regulation by uridine and guanosine nucleotides. FEBS J 275:655–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jørgensen MG, Thomason MK, Havelund J, Valentin-Hansen P, Storz G. 2013. Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev 27:1132–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalamorz F, Reichenbach B, März W, Rak B, Görke B. 2007. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol. Microbiol 65:1518–33 [DOI] [PubMed] [Google Scholar]
- 71.Katsowich N, Elbaz N, Pal RR, Mills E, Kobi S, et al. 2017. Host cell attachment elicits posttranscriptional regulation in infecting enteropathogenic bacteria. Science 355:735–39 [DOI] [PubMed] [Google Scholar]
- 72.Kavita K, de Mets F, Gottesman S. 2018. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol 42:53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kumar PKR, Mizuno H. 2014. Metal ion-dependent anti-termination of transcriptional regulation of ribonucleoprotein complexes. Biophys. Rev 6:215–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumarevel T, Fujimoto Z, Karthe P, Oda M, Mizuno H, Kumar PK. 2004. Crystal structure of activated HutP: an RNA binding protein that regulates transcription of the hut operon in Bacillus subtilis. Structure 12:1269–80 [DOI] [PubMed] [Google Scholar]
- 75.Kumarevel T, Gopinath SC, Nishikawa S, Mizuno H, Kumar PK. 2004. Identification of important chemical groups of the hut mRNA for HutP interactions that regulate the hut operon in Bacillus subtilis. Nucleic Acids Res 32:3904–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kumarevel T, Mizuno H, Kumar PK. 2005. Structural basis of HutP-mediated anti-termination and roles of the Mg2+ ion and l-histidine ligand. Nature 434:183–91 [DOI] [PubMed] [Google Scholar]
- 77.Leng Y, Vakulskas CA, Zere TR, Pickering BS, Watnick PI, et al. 2016. Regulation of CsrB/C sRNA decay by EIIAGlc of the phosphoenolpyruvate: carbohydrate phosphotransferase system. Mol. Microbiol 99:627–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lindner C, Galinier A, Hecker M, Deutscher J. 1999. Regulation of the activity of the Bacillus subtilis antiterminator LicT by multiple PEP-dependent, enzyme I- and HPr-catalysed phosphorylation. Mol. Microbiol 31:995–1006 [DOI] [PubMed] [Google Scholar]
- 79.Liu MY, Gui G, Wei B, Preston JF 3rd, Oakford L, et al. 1997. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem 272:17502–10 [DOI] [PubMed] [Google Scholar]
- 80.Liu MY, Romeo T. 1997. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J. Bacteriol 179:4639–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu Y, Switzer RL. 1996. Transcriptional attenuation of the Bacillus subtilis pyr operon by the PyrR regulatory protein and uridine nucleotides in vitro. J. Bacteriol 178:7206–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu Y, Turner RJ, Switzer RL. 1995. Roles of the three transcriptional attenuators of the Bacillus subtilis pyrimidine biosynthetic operon in the regulation of its expression. J. Bacteriol 177:1315–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lu Y, Turner RJ, Switzer RL. 1996. Function of RNA secondary structures in transcriptional attenuation of the Bacillus subtilis pyr operon. PNAS 93:14462–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Manival X, Yang Y, Strub MP, Kochoyan M, Steinmetz M, Aymerich S. 1997. From genetic to structural characterization of a new class of RNA-binding domain within the SacY/BglG family of antiterminator proteins. EMBO J 16:5019–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McAdams NM, Gollnick P. 2014. The Bacillus subtilis TRAP protein can induce transcription termination in the leader region of the tryptophan biosynthetic (trp) operon independent of the trp attenuator RNA. PLOS ONE 9:e88097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McElroy C, Manfredo A, Wendt A, Gollnick P, Foster M. 2002. TROSY-NMR studies of the 91 kDa TRAP protein reveal allosteric control of a gene regulatory protein by ligand-altered flexibility. J. Mol. Biol 323:463–73 [DOI] [PubMed] [Google Scholar]
- 87.McGraw AP, Mokdad A, Major F, Bevilacqua PC, Babitzke P. 2009. Molecular basis of TRAP-5′ SL RNA interaction in the Bacillus subtilis trp operon transcription attenuation mechanism. RNA 15:55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mohanty BK, Kushner SR. 2018. Enzymes involved in posttranscriptional RNA metabolism in gram-negative bacteria. Microbiol. Spectr 6:RWR-0011–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mondal S, Yakhnin AV, Babitzke P. 2017. Modular organization of the NusA- and NusG-stimulated RNA polymerase pause signal that participates in the Bacillus subtilis trp operon attenuation mechanism. J. Bacteriol 199:e00223–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mondal S, Yakhnin AV, Sebastian A, Albert I, Babitzke P. 2016. NusA-dependent transcription termination prevents misregulation of global gene expression. Nat. Microbiol 1:15007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mukherjee S, Oshiro RT, Yakhnin H, Babitzke P, Kearns DB. 2016. FliW antagonizes CsrA RNA binding by a noncompetitive allosteric mechanism. PNAS 113:9870–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mukherjee S, Yakhnin H, Kysela D, Sokoloski J, Babitzke P, Kearns DB. 2011. CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol. Microbiol 82:447–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oda M, Kobayashi N, Fujita M, Miyazaki Y, Sadaie Y, et al. 2004. Analysis of HutP-dependent transcription antitermination in the Bacillus subtilis hut operon: identification of HutP binding sites on hut antiterminator RNA and the involvement of the N-terminus of HutP in binding of HutP to the antiterminator RNA. Mol. Microbiol 51:1155–68 [DOI] [PubMed] [Google Scholar]
- 94.Oda M, Kobayashi N, Ito A, Kurusu Y, Taira K. 2000. cis-Acting regulatory sequences for antitermination in the transcript of the Bacillus subtilis hut operon and histidine-dependent binding of HutP to the transcript containing the regulatory sequences. Mol. Microbiol 35:1244–54 [DOI] [PubMed] [Google Scholar]
- 95.Olejniczak M, Storz G. 2017. ProQ/FinO-domain proteins: another ubiquitous family of RNA match-makers? Mol. Microbiol 104:905–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Otridge J, Gollnick P. 1993. MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan dependent manner. PNAS 90:128–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pannuri A, Vakulskas CA, Zere T, McGibbon LC, Edwards AN, et al. 2016. Circuitry linking the catabolite repression and Csr global regulatory systems of Escherichia coli. J. Bacteriol 198:3000–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pannuri A, Yakhnin H, Vakulskas CA, Edwards AN, Babitzke P, Romeo T. 2012. Translational repression of NhaR, a novel pathway for multi-tier regulation of biofilm circuitry by CsrA. J. Bacteriol 194:79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Park H, McGibbon LC, Potts AH, Yakhnin H, Romeo T, Babitzke P. 2017. Translational repression of the RpoS antiadapter IraD by CsrA is mediated via translational coupling to a short upstream open reading frame. mBio 8:e01355–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Parker A, Cureoglu S, De Lay N, Majdalani N, Gottesman S. 2017. Alternative pathways for Escherichia coli biofilm formation revealed by sRNA overproduction. Mol. Microbiol 105:309–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Patterson-Fortin LM, Vakulskas CA, Yakhnin H, Babitzke P, Romeo T. 2013. Dual posttranscriptional regulation via a cofactor-responsive mRNA leader. J. Mol. Biol 425:3662–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pek JW, Anand A, Kai T. 2012. Tudor domain proteins in development. Development 139:2255–66 [DOI] [PubMed] [Google Scholar]
- 103.Pompeo F, Luciano J, Brochier-Armanet C, Galinier A. 2011. The GTPase function of YvcJ and its sub-cellular relocalization are dependent on growth conditions in Bacillus subtilis. J. Mol. Microbiol. Biotechnol 20:156–67 [DOI] [PubMed] [Google Scholar]
- 104.Potts AH, Leng Y, Babitzke P, Romeo T. 2018. Examination of Csr regulatory circuitry using epistasis analysis with RNA-seq (Epi-seq) confirms that CsrD affects gene expression via CsrA, CsrB and CsrC. Sci. Rep 8:5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Potter KD, Merlino NM, Jacobs T, Gollnick P. 2011. TRAP binding to the Bacillus subtilis trp leader region RNA causes efficient transcription termination at a weak intrinsic terminator. Nucleic Acids Res 39:2092–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Potts AH, Vakulskas CA, Pannuri A, Yakhnin H, Babitzke P, Romeo T. 2017. Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat. Commun 8:1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Radomska KA, Wösten MMSM, Ordoñez SR, Wagenaar JA, van Putten JPM. 2017. Importance of Campylobacter jejuni FliS and FliW in flagella biogenesis and flagellin secretion. Front. Microbiol 8:1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reichenbach B, Maes A, Kalamorz F, Hajnsdorf E, Görke B. 2008. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res 36:2570–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ren B, Shen H, Lu ZJ, Liu H, Xu Y. 2014. The phzA2-G2 transcript exhibits direct RsmA-mediated activation in Pseudomonas aeruginosa M18. PLOS ONE 9:e89653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rennella E, Sára T, Juen M, Wunderlich C, Imbert L, et al. 2017. RNA binding and chaperone activity of the E. coli cold-shock protein CspA. Nucleic Acids Res 45:4255–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rick PD, Silver RP. 1996. Enterobacterial common antigen and capsular polysaccharides. In Escherichia coli and Salmonella: Cellular and Molecular Biology, ed. Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, et al. , pp. 104–22. Washington, DC: ASM. 2nd ed. [Google Scholar]
- 112.Romeo T, Babitzke P. 2018. Global regulation by CsrA and its RNA antagonists. Microbiol. Spectr 6:RWR-0009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Romeo T, Gong M, Liu MY, Brun-Zinkernagel AM. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol 175:4744–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Romeo T, Vakulskas CA, Babitzke P. 2013. Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Env. Microbiol 15:313–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rothe FM, Bahr T, Stülke J, Rak B, Görke B. 2012. Activation of Escherichia coli antiterminator BglG requires its phosphorylation. PNAS 109:15906–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Santiago-Frangos A, Woodson SA. 2018. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip. Rev. RNA 9:e1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sarsero JP, Merino E, Yanofsky C. 2000. A Bacillus subtilis gene of previously unknown function, yhaG, is translationally regulated by tryptophan-activated TRAP and appears to be involved in tryptophan transport. J. Bacteriol 182:2329–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sarsero JP, Merino E, Yanofsky C. 2000. A Bacillus subtilis operon containing genes of unknown function senses tRNATrp charging and regulates expression of the genes of tryptophan biosynthesis. PNAS 97:2656–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Savacool HK, Switzer RL. 2002. Characterization of the interaction of Bacillus subtilis PyrR with pyr mRNA by site-directed mutagenesis of the protein. J. Bacteriol 184:2521–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schindelin H, Jiang W, Inouye M, Heinemann U. 1994. Crystal structure of CspA, the major cold shock protein of Escherichia coli. PNAS 91:5119–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schmalisch MH, Bachem S, Stülke J. 2003. Control of the Bacillus subtilis antiterminator protein GlcT by phosphorylation: elucidation of the phosphorylation chain leading to inactivation of GlcT. J. Biol. Chem 278:51108–15 [DOI] [PubMed] [Google Scholar]
- 122.Schnetz K, Stülke J, Gertz S, Krüger S, Krieg M, et al. 1996. LicT, a Bacillus subtilis transcriptional antiterminator protein of the BglG family. J. Bacteriol 178:1971–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Septer AN, Bose JL, Lipzen A, Martin J, Whistler C, Stabb EV. 2015. Bright luminescence of Vibrio fischeri aconitase mutants reveals a connection between citrate and the Gac/Csr regulatory system. Mol. Microbiol 95:283–96 [DOI] [PubMed] [Google Scholar]
- 124.Sharma S, Gollnick P. 2014. Modulating TRAP-mediated transcription termination by AT during transcription of the leader region of the Bacillus subtilis trp operon. Nucleic Acids Res 42:5543–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Smirnov A, Förstner KU, Holmqvist E, Otto A, Günster R, et al. 2016. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. PNAS 113:11591–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Smirnov A, Wang C, Drewry LL, Vogel J. 2017. Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J 36:1029–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Steiner S, Lori C, Boehm A, Jenal U. 2013. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J 32:354–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sterzenbach T, Nguyen KT, Nuccio SP, Winter MG, Vakulskas CA, et al. 2013. A novel CsrA titration mechanism regulates fimbrial gene expression in Salmonella typhimurium. EMBO J 32:2872–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stülke J 2002. Control of transcription termination in bacteria by RNA-binding proteins that modulate RNA structures. Arch. Microbiol 177:433–40 [DOI] [PubMed] [Google Scholar]
- 130.Stülke J, Arnaud M, Rapoport G, Martin-Verstraete I. 1998. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol 28:865–74 [DOI] [PubMed] [Google Scholar]
- 131.Stülke J, Martin-Verstraete I, Zagorec M, Rose M, Klier A, Rapoport G. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol 25:65–78 [DOI] [PubMed] [Google Scholar]
- 132.Suzuki K, Babitzke P, Kushner SR, Romeo T. 2006. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev 20:2605–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Suzuki K, Wang X, Weilbacher T, Pernestig AK, Melefors O, et al. 2002. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol 184:5130–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tomchick DR, Turner RJ, Switzer RL, Smith JL. 1998. Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase. Structure 6:337–50 [DOI] [PubMed] [Google Scholar]
- 135.Tortosa P, Declerck N, Dutartre H, Lindner C, Deutscher J, Le Coq D. 2001. Sites of positive and negative regulation in the Bacillus subtilis antiterminators LicT and SacY. Mol. Microbiol 41:1381–93 [DOI] [PubMed] [Google Scholar]
- 136.Tortosa P, Le Coq D. 1995. A ribonucleic antiterminator sequence (RAT) and a distant palindrome are both involved in sucrose induction of the Bacillus subtilis sacXY regulatory operon. Microbiology 141:2921–27 [DOI] [PubMed] [Google Scholar]
- 137.Turnbough CL Jr., Switzer RL. 2008. Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors. Microbiol. Mol. Biol. Rev 72:266–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Turner RJ, Bonner ER, Grabner GK, Switzer RL. 1998. Purification and characterization of Bacillus subtilis PyrR, a bifunctional pyr mRNA-binding attenuation protein/uracil phosphoribosyltransferase. J. Biol. Chem 273:5932–38 [DOI] [PubMed] [Google Scholar]
- 139.Turner RJ, Lu Y, Switzer RL. 1994. Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism. J. Bacteriol 176:3708–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Urban JH, Vogel J. 2008. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLOS Biol 6:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vakulskas CA, Leng Y, Abe H, Amaki T, Okayama A, et al. 2016. Antagonistic control of the turnover pathway for the global regulatory sRNA CsrB by the CsrA and CsrD proteins. Nucleic Acids Res 44:7896–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vakulskas CA, Pannuri A, Cortés-Selva D, Zere TR, Ahmer BM, et al. 2014. Global effects of the DEAD-box RNA helicase DeaD (CsdA) on gene expression over a broad range of temperatures. Mol. Microbiol 92:945–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vakulskas CA, Potts AH, Babitzke P, Ahmer BM, Romeo T. 2015. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev 79:193–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Valbuzzi A, Gollnick P, Babitzke P, Yanofsky C. 2002. The anti-trp RNA-binding attenuation protein (Anti-TRAP), AT, recognizes the tryptophan-activated RNA binding domain of the TRAP regulatory protein. J. Biol. Chem 277:10608–13 [DOI] [PubMed] [Google Scholar]
- 145.Valbuzzi A, Yanofsky C. 2001. Inhibition of TRAP, the B. subtilis trp RNA-binding attenuation protein, by the Anti-TRAP protein, AT. Science 293:2057–5911557884 [Google Scholar]
- 146.Valverde C, Lindell M, Wagner EG, Haas DA. 2004. A repeated GGA motif is critical for the activity and stability of the riboregulator RsmY of Pseudomonas fluorescens. J. Biol. Chem 279:25066–74 [DOI] [PubMed] [Google Scholar]
- 147.Vijayakumar V, Vanhove AS, Pickering BS, Liao J, Tierney BT, et al. 2018. Removal of a membrane anchor reveals the opposing regulatory functions of Vibrio cholerae glucose-specific enzyme IIA in biofilms and the mammalian intestine. mBio 9:e00858–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol 56:1648–63 [DOI] [PubMed] [Google Scholar]
- 149.Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, et al. 2003. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol 48:657–70 [DOI] [PubMed] [Google Scholar]
- 150.Wray LV Jr., Fisher SH. 1994. Analysis of Bacillus subtilis hut operon expression indicates that histidine-dependent induction is mediated primarily by transcriptional antitermination and that amino acid repression is mediated by two mechanisms: regulation of transcription initiation and inhibition of histidine transport. J. Bacteriol 176:5466–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yakhnin AV, Babitzke P. 2002. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism in vitro. PNAS 99:11067–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yakhnin AV, Babitzke P. 2014. NusG/Spt5: Are there common functions of this ubiquitous transcription elongation factor? Curr. Opin. Microbiol 18:68–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yakhnin AV, Baker CS, Vakulskas CA, Yakhnin H, Berezin I, et al. 2013. CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol. Microbiol 87:851–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yakhnin AV, Murakami KS, Babitzke P. 2016. NusG is a sequence-specific RNA polymerase pause factor that binds to the non-template DNA within the paused transcription bubble. J. Biol. Chem 291:5299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yakhnin AV, Yakhnin H, Babitzke P. 2006. RNA polymerase pausing regulates translation initiation by providing additional time for TRAP-RNA interaction. Mol. Cell 24:547–57 [DOI] [PubMed] [Google Scholar]
- 156.Yakhnin AV, Yakhnin H, Babitzke P. 2008. Function of the Bacillus subtilis transcription elongation factor NusG in hairpin-dependent RNA polymerase pausing in the trp leader. PNAS 105:16131–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yakhnin H, Aichele R, Ades SE, Romeo T, Babitzke P. 2017. Circuitry linking the global Csr and σE- dependent cell envelope stress response systems. J. Bacteriol 199:e00484–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yakhnin H, Babiarz JE, Yakhnin AV, Babitzke P. 2001. Expression of the Bacillus subtilis trpEDCFBA operon is influenced by translational coupling and Rho termination factor. J. Bacteriol 183:5918–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yakhnin H, Pandit P, Petty TJ, Baker CS, Romeo T, Babitzke P. 2007. CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol. Microbiol 64:1605–20 [DOI] [PubMed] [Google Scholar]
- 160.Yakhnin H, Yakhnin AV, Babitzke P. 2006. The trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis regulates translation initiation of ycbK, a gene encoding a putative efflux protein, by blocking ribosome binding. Mol. Microbiol 61:1252–66 [DOI] [PubMed] [Google Scholar]
- 161.Yakhnin H, Zhang H, Yakhnin AV, Babitzke P. 2004. The trp RNA-binding attenuation protein of Bacillus subtilis regulates translation of the tryptophan transport gene, trpP (yhaG), by blocking ribosome binding. J. Bacteriol 186:278–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yang M, de Saizieu A, van Loon AP, Gollnick P. 1995. Translation of trpG in Bacillus subtilis is regulated by the trp RNA-binding attenuation protein (TRAP). J. Bacteriol 177:4272–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yang Y, Declerck N, Manival X, Aymerich S, Kochoyan M. 2002. Solution structure of the LicT-RNA antitermination complex: CAT clamping RAT. EMBO J 21:1987–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ye F, Yang F, Yu R, Lin X, Qi J, et al. 2018. Molecular basis of binding between the global post-transcriptional regulator CsrA and the T3SS chaperone CesT. Nat. Commun 9:1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zere TR, Vakulskas CA, Leng Y, Pannuri A, Potts AH, et al. 2015. Genomic targets and features of BarA-UvrY (-SirA) signal transduction systems. PLOS ONE 10:e0145035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang H, Switzer RL. 2003. Transcriptional pausing in the Bacillus subtilis pyr operon in vitro: a role in transcriptional attenuation? J. Bacteriol 185:4764–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang Y, Burkhardt DH, Rouskin S, Li GW, Weissman JS, Gross CA. 2018. A stress response that monitors and regulates mRNA structure is central to cold shock adaptation. Mol. Cell 70:274–86 [DOI] [PMC free article] [PubMed] [Google Scholar]


