Abstract
Transcription of mga, encoding the multiple virulence gene regulator of the group A streptococcus, is positively autoregulated. This regulation requires a DNA region (Pmga) that contains both a promoter proximal to mga (P2) and a promoter located further upstream (P1). To determine if Mga has a direct role in this process, its ability to bind to specific sequences within Pmga was tested. A purified fusion of Mga to the C-terminal end of maltose-binding protein (MBP-Mga), encoded by malE-mga, was shown previously to bind to the promoter regions of Mga-regulated genes, including scpA and emm. We report here that MBP-Mga can function in vivo to regulate emm and mga. Electrophoretic mobility shift assays and DNase I footprinting were used to demonstrate specific binding of MBP-Mga to two ca. 59-bp binding sites in Pmga centered around bases −108 and −180 from the major P2 start of transcription. Mga binding sites from Pemm and PscpA were shown to compete for binding at the two Pmga sites, suggesting that the same domain of Mga interacts at all of these promoter targets. Deletion of the distal Pmga binding site (site I) in vivo resulted in loss of Mga-dependent transcription from the P2 start. However, the same lesion resulted in an increase in P1 transcription that was independent of Mga. This suggests the existence of a repressor of mga transcription with a binding site overlapping those of Mga.
The group A streptococcus (GAS), Streptococcus pyogenes, is a human pathogen that elicits a wide array of suppurative diseases of the skin and throat, including pharyngitis and impetigo. In some cases, GAS infections can lead to serious nonsuppurative sequelae such as rheumatic fever and glomerulonephritis (4). Serious invasive streptococcal diseases, such as necrotizing fasciitis, myositis, and streptococcal toxic shock syndrome, are characterized by infections within normally sterile sites of the human body and can often lead to death in typically healthy individuals (2). The ability of GAS to adapt and grow within the different niches of the human host that it encounters during the course of these infections suggests an environmentally responsive control of its virulence determinants.
Mga is a multiple-gene regulator of GAS that responds to environmental signals by activating transcription of several virulence genes. These include the genes encoding the antiphagocytic M protein (emm), C5a peptidase (scpA), and in strains where these are present, M-like proteins (e.g., fcrA, enn, and sph) and serum opacity factor (sof) (5, 8, 10, 17, 24, 28). Mga functions as a DNA-binding protein that interacts directly with sequences in the promoters of two Mga-regulated genes, emm and scpA (22). Binding of Mga occurs at a 45-bp site within each of the two promoters centered 52 bp upstream of the start of transcription, which may allow Mga to interact with RNA polymerase and stimulate transcription initiation (22).
Mga is also required for its own positive autoregulation, and this requires 473 bp of DNA located directly upstream of its coding sequence (25). This extended Pmga region contains two distinct promoters: a distal start of transcription, P1, and a start site located proximal to mga, P2 (12, 25). Studies on an M type 6 GAS strain have determined that the P2 promoter represents the major start site for mga transcription. It exhibits about 10-fold more activity than the P1 promoter (25). The use of dual promoters (P1 and P2) within Pmga has been demonstrated for other GAS serotypes as well (e.g., M types 12 and 49) and most likely represents a general mechanism of mga expression (3, 27). Currently, very little is known about the specifics of mga regulation and the role of Mga in this process.
Many bacterial transcription factors regulate their own expression by binding directly to their own promoters (14, 19). To aid in the identification of other Mga-regulated promoters in GAS, a consensus Mga binding sequence was derived from Pemm and PscpA binding studies (22). Inspection of the Pmga region revealed no significant sequence homology with the consensus Mga binding element found upstream of Pemm and PscpA. This suggests that Mga does not bind directly to Pmga as it does to the other promoters. However, the Mga protein possesses two distinct N-terminal domains that show predicted helix-turn-helix DNA-binding motifs (26). Therefore, it is possible that Mga binding to Pmga involves different residues of the protein than those that bind to Pemm and PscpA. To investigate a possible direct role of Mga in autoregulation of mga expression, the ability of Mga to bind specifically within the Pmga regulatory region was investigated.
MATERIALS AND METHODS
Bacterial strains and media.
S. pyogenes JRS4 is a streptomycin-resistant derivative of serotype M6 strain D471 (33). GAS strains JRS14 (mga-1) (8) and JRS519 (mga-10) (23) are Mga− derivatives of JRS4. The pVIT GAS strain RTG229 was derived from JRS4 and contains the transposon Tn916-J4 (12).
Escherichia coli DH5α (hsdR17 recA1 gyrA endA1 relA1) was used as the host for plasmid constructions. E. coli was grown in Luria-Bertani broth (32), and S. pyogenes was grown in Todd-Hewitt medium supplemented with 0.2% yeast extract. Antibiotics were used at the following concentrations: ampicillin at 100 μg/ml for E. coli, chloramphenicol at 30 μg/ml for E. coli, erythromycin at 750 μg/ml for E. coli and 0.1 μg/ml for S. pyogenes, kanamycin at 50 μg/ml for E. coli and 300 μg/ml for S. pyogenes, and spectinomycin at 100 μg/ml for E. coli and S. pyogenes.
DNA manipulations.
Plasmid DNA was isolated from E. coli using the Wizard Maxiprep and Miniprep systems (Promega). Genomic DNA was isolated from GAS by the method of Chassy (9). DNA fragments were isolated from agarose gels by electroelution (20). Colony hybridization was performed with the DIG system (Boehringer Mannheim) using PCR-labeled DNA probes. DNA sequencing was done by the method of Sanger et al. (31) using Sequenase (Bethesda Research Laboratories). PCR for cloning and promoter probes was performed using Pfu high-fidelity DNA polymerase (Stratagene). PCR for diagnostic assays was performed using Taq DNA polymerase (Sigma).
Construction of the Pmga-malE-mga plasmid pJRS2019.
To place the malE-mga fusion allele of pJRS2016 (22) downstream of the mga promoter, 698 bp of DNA upstream of the mga gene was amplified using the template pJRS180 (26) and the mutagenic primers Pmga-HIII and Pmga-Pst (Table 1) to introduce HindIII (5′) and PstI (3′) restriction sites, respectively. The resulting Pmga fragment was inserted into HindIII-PstI-digested pBluescript II KS− (Stratagene) to produce pB-Pmga. A 2.9-kb fragment containing a promoterless malE-mga (MBP-Mga) fusion allele was amplified from pJRS2016 using the mutagenic primers MalMga-Pst and MalMga-Bam (Table 1) to introduce PstI (5′) and BamHI (3′) restriction sites, respectively. The malE-mga fragment was cloned into PstI-BamHI-digested pB-Pmga to produce pB-2019. To allow replication in GAS, the 6.6-kb HindIII Pmga-malE-mga fragment from pB-2019 was cloned into HindIII-digested pLZ12-Spec (15) to produce pJRS2019.
TABLE 1.
PCR primers used in this study
| Gene and primer | Sequence (5′–3′)a |
|---|---|
| mga | |
| MgaL3_Bam | caggatccGGATTTTAATGAAAGAATTT |
| MalMga-Pst | gcgctgcagATGAAAATCGAAGAAGGTAAAC |
| MalMga-Bam | cgcggatccCGACGTTGTAAAACGACGGC |
| Mga-s1dL | gcggaattcTTTTGTCACTAACTTAATTAGGTTTTTT |
| Mga-s1dR | gcgaattcggtaccGCATAAATGGCATAAAA |
| OYR-1 | AGAGCATAAATGGCATA |
| OYR-2 | ACCTAATTAAGTTAGTGAC |
| OYR-3 | GATATAGGTCTTAC |
| OYR-4 | GTACCATCAACATTGCG |
| OYR-5 | CCTCAATCTCAGCGTCACC |
| OYR-22 | TAGACCCCAAATTCCCGT |
| OYR-23 | AATTGACTGAAGTATGATAGAAT |
| OYL-1 | TTATGCCATTTATGCTCT |
| OYL-2 | TCTTGTTATTTCGTGAG |
| OYL-12 | CATGTTAGTGAGACATGT |
| OYL-13 | GACGGCAGAGTATCCCTTGT |
| OYL-14 | GTCACTAACTTAATTAGGT |
| OYL-15 | CAGTCACGATCACGCAAT |
| OYL-24 | GCGATGAAAGTCCAAGGG |
| Pmga_HIII | gcgccaagcttCAATCTGCGAGATTAGAGTAA |
| Pmga_Pst | ggcctgcagGTATACCCTTCTTTTTAATT |
| Pmga_Bam | cgggatccTAAGTTAACCAGTTCACAA |
| Pmga_Xho | ggctcgagACCTTGTATACCCTTCTTTT |
| Pmga2-PE | TTAGTTCTCTCCACTGTTGAC |
| scpA | |
| C5up346-L | AAGAATGAGATTAAGGAGGTCACA |
| C5up346-R | GCGCAATGGCAAGTTTGTC |
| emm | |
| M6up236-L | GCACGCCCCCCCCTC |
| M6up236-R | TAGTGTCTATTCGTG |
| OM6-19 | GAGTGTAATAGGGGCAGGA |
| OM6-30 | AAGGGCACCAATGTCTTGT |
| cat86* | |
| CAT86-L2 | ATATGACGTTAACCCGATGCT |
| CAT86-R2 | TGTCAATAAGCCACCTCAAA |
| CAT86-PE | GGTGAAAGTGCTCTTTTCGC |
| recA | |
| SRA-L | GCGTTCAGGAAGTCTAGCTC |
| SRA-R | CTGATGCTACTGCCATAGCAG |
The sequences in boldface are introduced restriction sites.
Expression and purification of MBP-Mga from E. coli.
MBP-Mga was purified from E. coli as described previously (22). Briefly, cultures of E. coli SA2817 containing pJRS2016 (Ptac-malE-mga) were grown at 30°C in Luria-Bertani broth plus ampicillin (100 μg/ml) and expression of the protein was induced by addition of 6 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were harvested by passage through a prechilled French pressure cell, and MBP-Mga was purified over an amylose column (New England Biochemicals [NEB]). Protein concentrations were determined using the Bio-Rad protein assay kit.
Electrophoretic mobility shift assays.
Promoter probes were generated by PCR amplification using the plasmid template pJRS180 (26) and primers described in Table 1. PCR fragments were end labeled with [γ-32P]ATP using T4 polynucleotide kinase (NEB). Labeled fragments were excised from a 5% polyacrylamide gel, extracted by crush-and-soak elution, and purified using the QIAquick PCR purification system (Qiagen).
Mobility shift assays were performed as described previously (22) in a total reaction volume of 25 μl containing 12 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 1 mM EDTA, 0.6 mM dithiothreitol, 60 mM KCl, 5 mM MgCl2, and 40 ng of poly(dI-dC) per μl. A constant amount of labeled promoter probe DNA (ca. 20 to 40 pM) and various concentrations of MBP-Mga (10 to 320 nM) were used in each reaction. Competition experiments were performed by inclusion of cold probes prior to protein addition. Gels were processed as described below. Gels were dried under vacuum at 80°C for 1 h and autoradiographed using phosphor exposure screens. Images were then scanned at 176-μm resolution using a Molecular Dynamics PhosphorImager 445SI or Storm 860, and the resulting data were analyzed with the ImageQuaNT software (version 4.2a).
DNase I protection assays.
Substrate fragments for nuclease protection were prepared in two different ways. (i) PCR-generated promoter fragment 23-15 was amplified from pJRS180 (26) using the primers OYR23 and OYL15 (Table 1). The product was cloned by blunt-end ligation into the HincII site of pNEB193 (NEB) to produce pJRS503, and the orientation of the insert was verified by PCR analysis. A strand-specific 3′-labeled insert from pJRS503 was produced using the Klenow fragment of DNA polymerase as described elsewhere (18). (ii) Strand-specific labeled PCR-derived promoter fragments were generated using either a sense or an antisense primer that had been end labeled as described above. Labeled promoter probes were purified through polyacrylamide gels as described above.
DNase I protection assays were performed as described previously in a total reaction volume of 100 μl (22). Various amounts of unlabeled competitor DNAs were added, when appropriate, prior to addition of protein. Reactions were stopped by addition of DNase I inhibitor solution (750 mM ammonium acetate, 75 mM EDTA, 33 μg of tRNA per ml), concentrated by ethanol precipitation, and separated on a 6% sequencing gel. The method of Maxam and Gilbert (21) was used to sequence the labeled fragments. Gels were processed as described above.
Construction of pVIT GAS strains to study Pmga deletions.
The pVIT system (7) was used as described previously (12) with some modifications (described below). In this system, designed for in vivo analysis of activity of Pmga in single copy in GAS, a small region of Tn916 is included in a pVIT vector (vectors for integration into Tn916) which can then integrate into a single copy of Tn916 in the chromosome of GAS strain RTG229. Following the procedures of Geist et al. (12), different Pmga promoter fragments were fused to a promoterless cat86* (cat) reporter gene in the plasmid pVIT164 (12) and integrated via vector homology into a resident Tn916 transposon located outside the mga locus in the GAS chromosome in strain RTG229. Thus, determination of cat-specific transcription in these strains allows analysis of the introduced Pmga constructs in single copy in either a wild-type mga background or an mga mutant background.
Plasmids for chromosomal integration were constructed in the vector pVIT164 as follows. The positive control plasmid pJRS530 contains a 493-bp PCR fragment containing the minimal Pmga regulatory region (25) amplified from pJRS180 (26) using primers Pmga_Bam and Pmga_Xho (Table 1) and cloned into BamHI-XhoI-digested pVIT164. A deletion of the Mga binding site I (mga-12) was constructed as follows. pJRS528 contains the 493-bp PCR fragment from pJRS530 cloned into BamHI-XhoI-digested pBluescript II KS− (Stratagene). The mga-12 allele was created by inverse PCR on pJRS528 using the diverging primers Mga-s1dR and Mga-s1dL (Table 1), resulting in a deletion of 63 bp of GAS DNA including Mga binding site I and the introduction of an EcoRI restriction site. The inverse PCR product was digested with EcoRI and religated to create pMga-s1d. The 410-bp BamHI-XhoI mga-12 fragment from pMga-s1d was cloned into BamHI-XhoI-digested pVIT164 to produce pJRS5301. The plasmid pJRS578 contains a 139-bp PCR fragment including only the Pmga2 promoter amplified from pJRS2050 (1) using primers MgaL3_Bam and Pmga_Xho (Table 1) and cloned into BamHI-XhoI-digested pVIT164.
The pVIT strains were constructed in the GAS strain RTG229 as described previously (12). Briefly, integrational plasmids were linearized with PvuII and RTG229 was transformed with ca. 200 μg of the linear DNA. The promoter fusions to cat along with an ΩKm-2 marker on the linear plasmids recombine into a resident Tn916 transposon via flanking homology and result in the replacement of an erythromycin resistance gene. Resulting pVIT strains will be erythromycin sensitive, kanamycin resistant, and wild type for mga. Negative control strain JRS529 was generated by introduction of the pVIT164 vector alone into RTG229. Introduction of the plasmids pJRS530, pJRS5301, and pJRS578 resulted in the pVIT strains JRS530, JRS5301 (mga-12), and JRS578 (mga-21), respectively.
Inactivation of mga in pVIT GAS strains.
The plasmid pJRS586 contains a 456-bp PCR fragment internal to the mga coding sequence amplified from pJRS2050 using primers OYR4 and OYL13 (Table 1) and cloned blunt into HincII-digested pUC-Spec (15). Since pUC-Spec cannot replicate in GAS, spectinomycin-resistant transformants of GAS result from integration into mga via homologous recombination and insertional inactivation. The pVIT GAS strains JRS529, JRS530, and JRS5301 were transformed with ca. 300 μg of pJRS586, and spectinomycin-resistant transformants were isolated after growth at 37°C for 2 days. Potential mga mutant strains were verified both by using PCR across the mga gene and by RNA slot blot analysis as described below (data not shown). The resulting mutant strains were called JRS1529 (from JRS529), JRS1530 (from JRS530), and JRS15301 (from JRS5301).
Primer extension analysis.
Total RNA was extracted from samples in mid-exponential phase as described previously (23). Growth was assayed by measuring absorbance using a Klett-Summerson photoelectric colorimeter. Primer extension was performed as described previously (11). Specific primers for cat (cat86-PE) and mga (Pmga2-PE) are shown in Table 1. In primer extension experiments requiring quantitation, both labeled primers were included in the same reaction. Levels of cat-specific product were normalized to levels of the Pmga2-PE product within the same reaction. The primer extension products were run on a 6% denaturing acrylamide gel (Ameresco) along with the appropriate sequencing reactions. The SequiTherm Excel II DNA sequencing kit (Epicentre Technologies) was used according to the manufacturer’s protocol to sequence either pJRS530 or pJRS5301 using the labeled cat86-PE primer (Table 1). Gels were processed as described above.
RNA slot blot analyses.
Total RNA purified as described above was transferred to a Zeta-probe membrane (Bio-Rad) and hybridized with a labeled DNA probe as previously described (34). Labeled membranes were autoradiographed by using Phosphor Imager exposure screens (Molecular Dynamics) and imaged as described above. Normalized units were defined as the band intensity (pixel volume) for any experimental probe at a particular RNA concentration divided by that of a specific control probe (either recA or wild-type mga) at the same RNA concentration.
DNA probes internal to the coding region for each gene were generated by PCR amplification from GAS strain JRS4 genomic DNA using the following primer pairs (Table 1): mga 5′, OYR4 and OYL13; mga 3′, OYR5 and OYL24; cat86, CAT86-L2 and CAT86-R2; emm, OM6-30 and OM6-19. Resulting fragments were radiolabeled with [α-32P]dATP by random priming (20).
RESULTS
The MBP-Mga fusion protein activates transcription in vivo.
Previous studies showed that a fusion of the E. coli maltose-binding protein to the amino terminus of Mga (MBP-Mga) bound specifically to the promoters of Mga-regulated genes from the serotype M6 GAS strain JRS4 (22). However, the ability of MBP-Mga to function in vivo to autoregulate and activate other Mga-regulated genes in a GAS strain lacking wild-type Mga was never demonstrated.
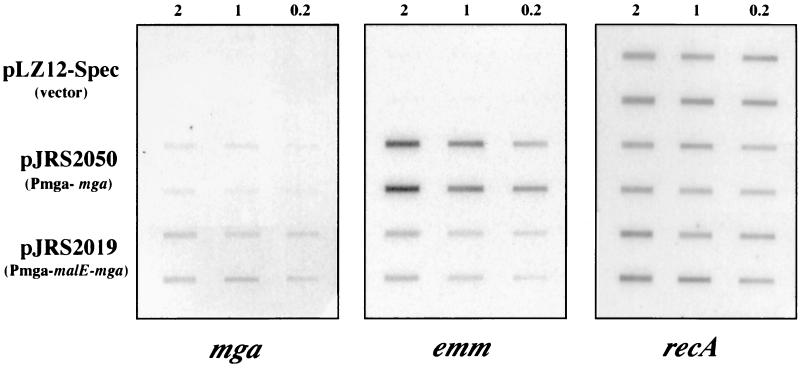
The plasmid pJRS2050 (1), containing the wild-type mga gene from GAS strain D471 (serotype M6) with its native promoter, has been shown previously to complement several mga mutations in different GAS strains (1, 23). To test the in vivo activity of MBP-Mga, JRS14, a GAS of serotype M6 containing a Tn916 insertion within the mga promoter region (mga1) that results in loss of Mga activity (6), was used. A promoterless malE-mga allele, encoding MBP-Mga, was cloned downstream of the native mga regulatory region (Pmga region) in the vector pLZ12-Spec to produce pJRS2019 (Materials and Methods), which was introduced by electroporation into JRS14. Total RNA was isolated at mid-exponential phase and assayed by hybridization for transcripts from mga, emm, and, as a control for equal loading of gels, recA.
Compared to the recA control, no transcripts for mga or emm were detectable in JRS14 containing pLZ12-Spec, the vector without an insert (Fig. 1). As previously observed (1), JRS14/pJRS2050, which contains the wild-type mga allele under the control of its own promoter, produced both mga and emm transcripts (Fig. 1). JRS14/pJRS2019 (carrying Pmga-malE-mga) also produced both mga and emm transcripts (Fig. 1). This is consistent with the previous finding that MBP-Mga positively autoregulates its own expression. However, unexpectedly, pJRS2019 resulted in more mga transcript than did pJRS2050 and less emm transcript (Fig. 1). Thus, it appears that the ability of MBP-Mga to positively autoregulate mga transcription is actually enhanced slightly over that of Mga. The unexpected decrease in transcripts of the Mga-regulated gene emm could be a consequence of either less-efficient activation by the fusion protein MBP-Mga or the overexpression of this protein.
FIG. 1.
Ability of the malE-mga allele encoding MBP-Mga to activate transcription in vivo. Total RNA for slot blot analysis was isolated from JRS14 containing either pLZ12-Spec (vector), pJRS2050 (wild-type mga), or pJRS2019 (Pmga-malE-mga) at mid-exponential growth, and 2, 1, or 0.2 μg of RNA was loaded per slot in duplicate. Samples were transferred to nitrocellulose, and membranes were reacted with specific DNA probes internal to the coding regions of mga, emm, and recA as shown.
Mga binds to specific sequences upstream of mga.
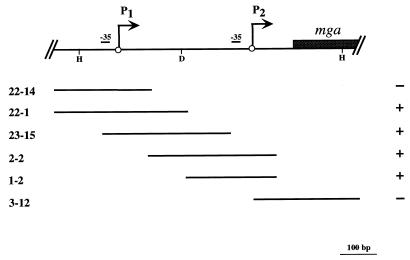
In the M6 GAS strain JRS4, a 473-bp region directly upstream of mga, which includes the distal P1 and the proximal P2 starts of transcription, is required for maximal mga expression (25). To begin a study on the autoregulation of mga in GAS, the ability of Mga to bind to sequences within this Pmga region was determined. Six overlapping promoter probes encompassing this region were amplified by PCR, end labeled with [γ-32P]ATP, and tested for binding to the MBP-Mga fusion protein by electrophoretic mobility shift assay.
Increasing amounts of MBP-Mga resulted in decreased mobility of four overlapping Pmga region probes: 22-1, 23-15, 2-2, and 1-2 (Fig. 2 and data not shown). Addition of the respective unlabeled probe to each reaction mixture prevented formation of the complexed species, whereas addition of a nonspecific unlabeled probe did not (data not shown). No detectable effect on the mobility of the flanking probes 22-14 and 3-12 was ever observed, even with protein concentrations of up to 350 nM, indicating that MBP-Mga does not bind to these sequences.
FIG. 2.
Summary of Pmga promoter probes bound by MBP-Mga in electrophoretic mobility shift assays. The diagram at the top shows the chromosomal region containing the regulatory sequences upstream of mga (Pmga) from serotype M6 GAS strain JRS4. The 5′ end of the mga coding sequence is represented by the shaded box, and important restriction sites are shown (H, HpaI; D, DraI). Two starts of transcription, P1 and P2, reported by Okada et al. (25) are shown with their respective −35 hexamers. Solid lines designate PCR-derived promoter probes using the following primer pairs found in Table 1: 22-14, OYR22 and OYL14; 22-1, OYR22 and OYL1; 23-15, OYR23 and OYL15; 2-2, OYR2 and OYL2; 1-2, OYR1 and OYL2; 3-12, OYR3 and OYL12. Results from mobility shift assays with MBP-Mga are indicated as positive (+) or negative (−) at the right.
DNase I protection assays define two MBP-Mga binding sites upstream of the mga P2 promoter.
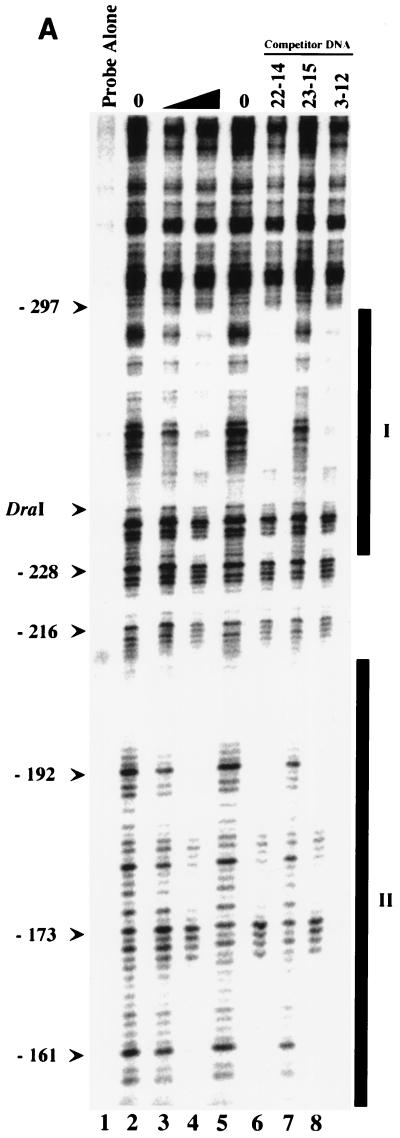
To identify the sequences within the Pmga region that interact with MBP-Mga, protection of this region from DNase I digestion was determined in the presence of the fusion protein. Probe 23-15 (Fig. 2) was labeled on the sense strand and incubated with increasing amounts of MBP-Mga (Materials and Methods). Two regions were protected from DNase I digestion; these define a distal binding site, I, of ca. 61 bp and a proximal binding site, II, of ca. 58 bp (Fig. 3A and B). DNase I analysis of probe 23-15 labeled on the antisense strand protected regions almost identical to those described above and did not reveal any additional binding sites (Fig. 4). Additional DNase I analysis of probes 22-1, 2-2, and 1-2 (Fig. 2) verified the results reported above (data not shown). Since probes 22-1 and 1-2 contain only a single binding site, these data indicate that Mga can bind either site I or II in the absence of the other site.
FIG. 3.
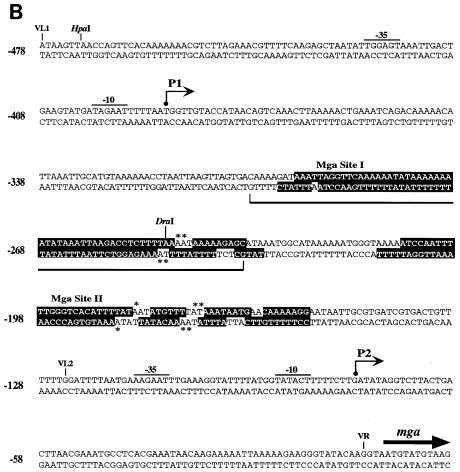
DNase I protection analysis of MBP-Mga binding to Pmga. (A) The promoter probe 23-15 was labeled on the sense strand (Materials and Methods). Labeled probe (2 pM) was incubated with increasing amounts of MBP-Mga as follows: lane 1, probe alone; lanes 2 and 5, probe plus DNase I; lane 3, probe plus 120 nM MBP-Mga and DNase I; lane 4, probe plus 240 nM MBP-Mga and DNase I. Unlabeled competitor sequences (40 nM) were added prior to addition of MBP-Mga as follows: lane 6, lane 4 reaction mixture plus 22-14; lane 7, lane 4 reaction mixture plus 23-15; lane 8, lane 4 reaction mixture plus 3-12. Thick vertical bars designate positions of Mga binding sites I and II. The DraI restriction site is shown; nucleotide positions are shown relative to the start of mga translation (see panel B). Base pair positions are shown on the left. (B) Location of Mga binding sites within Pmga. Shown is 478 bp of sequence upstream of the mga coding region (thick arrow). The P1 and P2 starts of transcription (thin arrows) were determined by primer extension (this study) and differ slightly from those reported by Okada et al. (25). The −10 and −35 hexamers for both starts are designated by overlines. Nucleotides protected from DNase I by MBP-Mga binding are shaded, while those hypersensitive to DNase I digestion are indicated by asterisks. The region deleted in mga-12 GAS strain JRS5301 is shown by the bracket below the sequence. The DraI restriction site corresponding to the mga-1 Tn916 insertion site (8) is indicated. The VL1, VL2, and VR sites are described in Fig. 5. Nucleotides are numbered relative to the start of mga translation (+1).
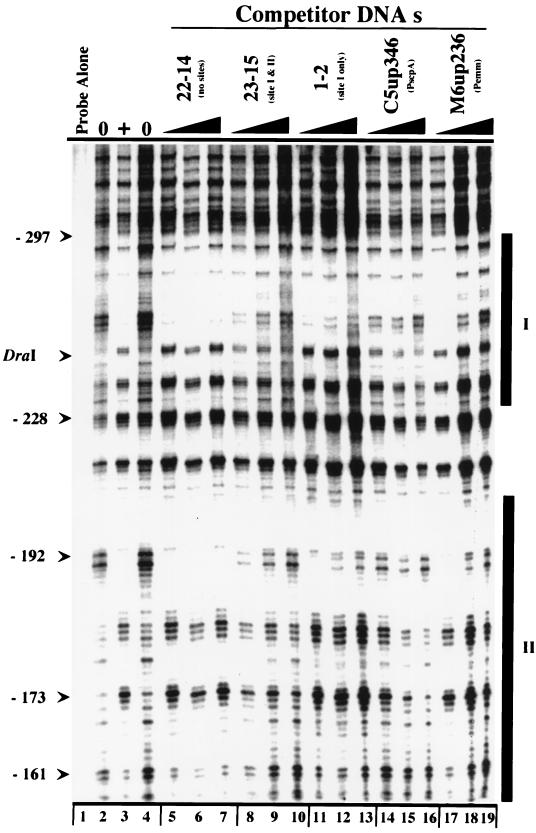
FIG. 4.
Comparison of Mga binding sites from different promoters by competition DNase I protection analysis. The promoter probe 23-15 was labeled on the antisense strand (Materials and Methods). Labeled probe (80 pM) was incubated with a constant amount of MBP-Mga (84 nM). Increasing concentrations of various unlabeled competitor sequences (final concentrations, 5, 20, and 40 nM) were added prior to incubation with protein. Lanes: 1, probe alone; 2 and 4, probe plus DNase I; 3, probe plus MBP-Mga and DNase I; 5 to 7, lane 3 contents plus increasing 22-14; 8 to 10, lane 3 contents plus increasing 23-15 containing sites I and II; 11 to 13, lane 3 contents plus increasing 1-2 containing site II only; 14 to 16, lane 3 contents plus increasing C5up346 (PscpA; 22); 17 to 19, lane 3 contents plus increasing M6up246 (Pemm; 22). Thick vertical bars designate positions of Mga binding sites I and II. The DraI restriction site is shown; nucleotide positions are shown relative to the start of mga translation (Fig. 3B).
The sequences in site I protected by MBP-Mga extend from base −153 to base −211 upstream of the mga open reading frame. This places the center of the site at base −104 from the start of transcription at the P2 promoter, which is the major promoter used in vivo (25) (Fig. 3A and B). In binding site II (Fig. 3A), the protected sequences extend from base −234 to base −295 upstream of the mga coding sequence and place the center of the site at base −185 from the P2 transcript start (Fig. 3B). Binding of MBP-Mga in this region suggests that Mga acts directly to activate the major P2 transcriptional start of mga and thus to positively autoregulate its expression.
Addition of unlabeled probe 23-15 inhibited the binding to both protected regions, while probes 22-14 and 3-12, which did not show binding to MBP-Mga by mobility shift assays (Fig. 2), also failed to prevent protection in DNase I assays (Fig. 3A and 4). DNase I footprint analysis also showed that MBP-Mga occupies both sites I and II simultaneously, even at the lowest protein concentration used (Fig. 3A and 4). These data demonstrate that binding of MBP-Mga is specific at binding sites I and II and that MBP-Mga appears to have similar affinities for both of the sites within the Pmga region.
Comparison of Mga binding to different Mga-regulated promoters.
Mga has been shown to bind directly to the promoters of the Mga-activated genes emm (Pemm) and scpA (PscpA) (22), in addition to binding to the Pmga region. Since the consensus sequence recognized by Mga at Pemm and PscpA is not apparent at Pmga, it seemed possible that different domains of the 62-kDa Mga protein are involved in binding to these promoters. To investigate this, competition DNase I protection assays were performed. The Pmga promoter probe 23-15 (Fig. 2) was labeled at the 5′ end of the antisense strand with [γ-32P]ATP, and increasing concentrations of various unlabeled promoter probes (Fig. 2) were added prior to incubation with MBP-Mga (Materials and Methods). The negative control probe 22-14 had no effect on MBP-Mga binding (Fig. 4, lanes 5 to 7), whereas two Pmga probes, 23-15 and 1-2, inhibited binding of Mga in a concentration-dependent fashion (Fig. 4, lanes 8 to 13). Addition of either the PscpA fragment C5up346 or the Pemm fragment M6up236 at concentrations similar to those of the Pmga-specific probes resulted in loss of DNase I protection by MBP-Mga (Fig. 4, lanes 14 to 19). These results are consistent with the possibility that the same domain of the Mga protein binds to its own promoter and to other Mga-regulated promoters.
Mga binding site I is involved in autoregulation by Mga.
Since Mga binds in vitro within the Pmga promoter region, Mga may function directly in autoregulation in vivo. To test this, a deletion of Mga binding site I (mga-12) was constructed and assayed for its effect on transcription from the Pmga promoter. The pVIT system was used to provide a single chromosomal copy of the wild-type or mutant form of Pmga transcribing the reporter cat gene, while Mga protein is supplied from its unlinked normal chromosomal location (Materials and Methods).
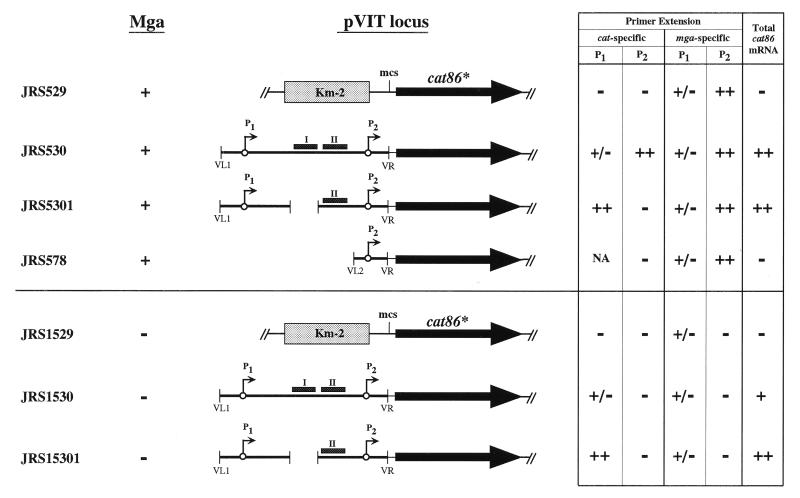
Four pVIT-derived strains were compared (Fig. 5): strain JRS529, containing the integrated pVIT164 vector with no Pmga insert driving cat transcription; strain JRS530, which has the wild-type Pmga region cloned in front of cat; strain JRS5301, which contains a 64-bp deletion that removes Mga binding site I (mga-12) cloned upstream of cat; and strain JRS578, which contains a deletion of the DNA upstream of P2 (including the P1 promoter and both Mga binding sites) cloned upstream of cat.
FIG. 5.
Summary of in vivo analyses of Mga binding site I. All pVIT strains shown were constructed for this study as described previously (12; Materials and Methods). Presence (+) or absence (−) of wild-type Mga protein is indicated for each strain background. Transcriptional fusions to a promoterless cat86* gene (thick arrow) are inserted at the multiple cloning site (mcs), recombined at the pVIT locus, and are shown schematically for each strain as follows. Control strains JRS529 and JRS1529 contain promoterless cat only, JRS530 and JRS1530 contain the full-length Pmga fragment cloned upstream of cat, JRS5301 and JRS15301 contain the mga-12 deletion of Mga binding site I cloned upstream of cat, and JRS578 has the mga-21 deletion allele containing only the P2 promoter cloned upstream of cat. Restriction sites introduced by PCR for cloning are shown as VL1, VL2, and VR (Fig. 3B). Pmga promoters (thin arrows), Mga binding sites I and II (small shaded boxes), and the ΩKm-2 interposon (large shaded box) are indicated. Results from transcriptional analyses of each strain are summarized at the right (−, no detectable transcript; +/−, barely detectable transcript; +, detectable transcript; ++, more transcript; NA, not applicable).
Specific initiation of transcription from the P1 and P2 mga promoters was determined for each strain by extension of two different primers, of which one was specific for cat (cat86-PE; Fig. 6A and Table 1), to assay transcription at the pVIT locus, and one was specific for mga (Pmga2-PE; Fig. 6A and Table 1), to check transcription from the wild-type mga locus. From the mga-specific primer, all four strains produced a strong P2 product and a barely detectable P1 product (Fig. 5, mga specific, and 6B). This is in agreement with published data on Pmga (25, 27) and provides an internal control for direct comparison of transcription at the pVIT locus.
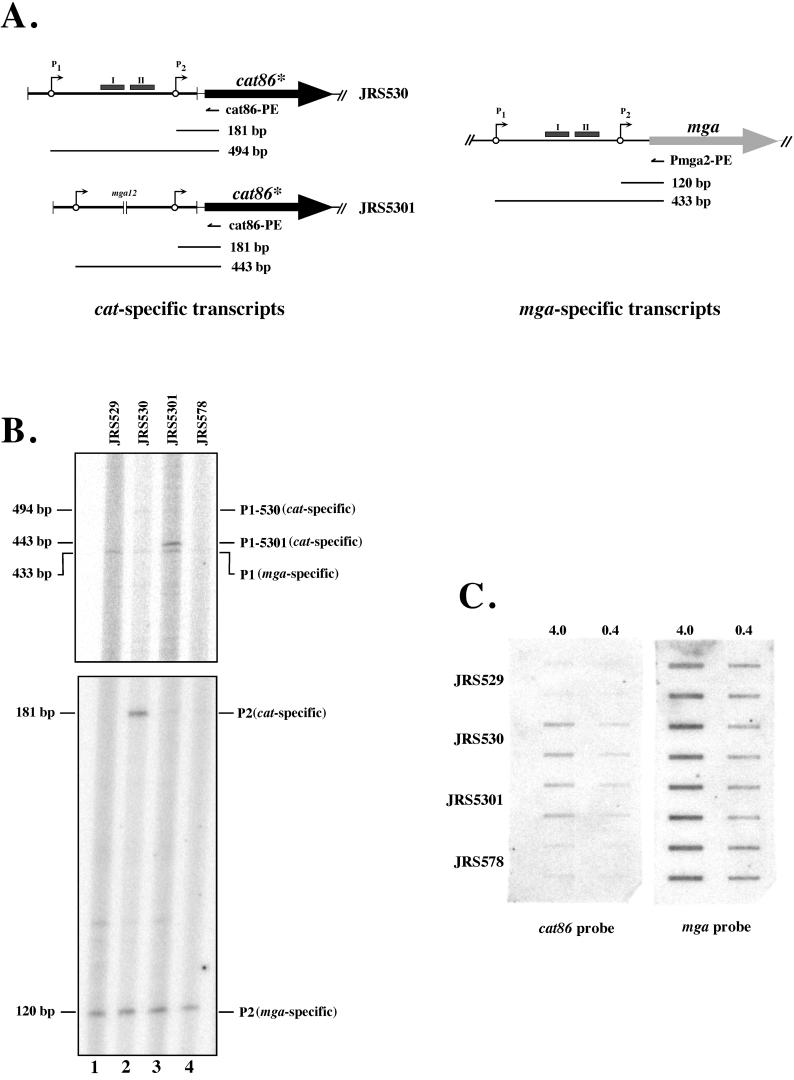
FIG. 6.
Transcriptional studies of Mga binding site I deletion. Whole-cell RNA was isolated from pVIT strains JRS529 (control), JRS530 (wild-type Pmga), JRS5301 (Mga binding site I deletion allele mga-12), and JRS578 (P2-only allele mga-21) at mid-exponential growth (Materials and Methods). (A) Diagram of expected results of primer extension analysis. Primers used for extension reactions (small arrows) and expected sizes of extension products (solid bars) are shown. The diagrammed cat-specific transcripts represent pVIT strains JRS530 and JRS530, while the mga-specific transcripts shown are present in all four strains tested. Pmga transcriptional start sites (circles with arrows), Mga binding sites (shaded boxes), deletions (parallel vertical bars), the cat86* gene (solid heavy arrow), and the mga gene (shaded heavy arrow) are indicated. (B) Analysis of P1 and P2 transcriptional starts using primer extension. The extension products were labeled based on appropriate sequencing reactions run concurrently (data not shown) and comparison to the Pmga sequence shown in Fig. 3B. Primer specific for the wild-type mga locus (Pmga2-PE; Table 1) was included as an internal control in each reaction mixture along with the experimental cat-specific primer (cat86-PE; Table 1). Primer-specific products are identified at the right as either derived from cat86-PE (cat specific) or derived from Pmga2-PE (mga specific). The locations on the gel of P1-530 (cat specific) and P1-5301 (cat specific) differ because of the mga-12 deletion downstream of P1 in these strains. (C) Total cat86* transcript in different pVIT strains identified by slot blot analysis. Total RNA isolated as described above was loaded in duplicate (vertical rows) at 4 and 0.4 μg of RNA per slot. Samples were transferred to nitrocellulose, and membranes were reacted with specific DNA probes internal to the coding regions of cat86* and mga.
As expected, JRS529 (promoterless cat) produced no detectable cat-specific Pmga promoter transcripts from its pVIT locus (Fig. 6B) and JRS530 (wild-type Pmga-cat) generated a cat-specific Pmga profile almost identical to the wild-type Pmga pattern described above (Fig. 5 and 6B). The JRS578 strain, which lacks both Mga binding sites, as well as P1 (mga-21), showed no detectable P2 activity from the cat-specific primer (Fig. 5 and 6B). This supports previous observations (3, 12, 25) suggesting that P2 transcription requires sequences upstream of the deletion in this strain.
At the pVIT locus of JRS5301, which has a deletion in Mga binding site I (mga-12), there was a complete loss of cat-specific P2 initiation and a corresponding strong increase in the level of cat-specific P1-derived transcripts (Fig. 5 and 6B). These data suggest that the region encompassing Mga binding site I is important for both the activation of the downstream P2 promoter and repression of the upstream transcript initiated at P1. However, the overall effect of the loss of Mga binding site I on expression of mga was not obvious since the amount of cat transcript (assayed by hybridization) in JRS5301 (mga-12) was almost the same as that in wild-type strain JRS530 (Fig. 5 and 6C).
Mga binding at site I is required for P2 activation.
To determine whether the requirement for binding site I for transcription from P2 is dependent on Mga, the wild-type mga gene was insertionally inactivated in the pVIT strain JRS530 to produce JRS1530 (Materials and Methods). As expected, no mga-specific transcript was detectable by hybridization in JRS1530 (data not shown). In the absence of Mga in strain JRS1530, which has an intact Pmga region at the pVIT locus, no cat-specific P2 product was detected in primer extension studies (Fig. 5; data not shown). Since this transcript was present in JRS530, which differs from JRS1530 only by having an intact mga gene, this supports the interpretation from mga-12 binding site I deletion strain JRS5301 (above) that Mga binding is needed to activate transcription from the P2 promoter.
Repression of P1 requires binding site I but not Mga.
At the pVIT locus of JRS5301, which has a deletion in binding site I, there is a strong increase in the amount of cat-specific P1 transcript compared to JRS530, which has an intact Pmga region (Fig. 5 and 6B). Although there is no detectable cat-specific transcription from P2 in JRS5301 (see above), the total amount of cat transcript in this strain was almost the same as for wild-type strain JRS530 (Fig. 5 and 6C). Thus, it appears that binding site I is needed not only for activation of the P2 promoter but also for repression of the P1 promoter in the Pmga region.
To determine whether this repression is dependent on Mga, the wild-type mga gene was insertionally inactivated in strain JRS529 to produce JRS1529 and in strain JRS5301 to produce JRS15301. P1 in strain JRS1530, the Mga− strain, still produced a detectable mga-specific P1 transcript, as shown by primer extension analysis (data not shown). Therefore, transcription from P1 does not appear to require Mga.
Surprisingly, the absence of Mga had no effect on cat-specific P1 promoter activity in strain JRS15301, compared with its Mga+ parent, JRS5301 (Fig. 5; data not shown). Both JRS530 and JRS1530 produce barely detectable amounts of a cat-specific transcript that initiates at P1, while JRS5301 and JRS15301, with binding site I deleted, produce significantly higher amounts of cat-specific P1 transcripts. Therefore, it can be deduced that repression of the P1 promoter requires binding site I (see above) and that this repression does not involve Mga. These data suggest the existence of a protein other than Mga that binds at Mga binding site I to inhibit transcription from the P1 promoter.
DISCUSSION
Mga binds to two sites within Pmga in vitro.
Expression of mga is positively autoregulated, and this control requires the entire 473-bp regulatory region (Pmga) located upstream of mga (12, 25). Because Mga is a DNA-binding protein that binds within the promoters of the Mga-regulated emm and scpA genes, it seemed possible that autoregulation by Mga was also direct. To address this, an MBP-Mga fusion protein was used in DNase I footprinting and gel retardation experiments to assay specific Mga binding within the Pmga region. The Pmga region contains two promoters, a major one located proximal to the coding region (P2) and one located further upstream that produces much less transcript (Fig. 3A and 4). We have shown here that Mga binds to two 59-bp sites upstream of the major P2 transcriptional start site within Pmga (sites I and II). Mga also appears to bind to both sites with equal affinity since a DNA fragment containing only site II (fragment 1-2) competed equally well for binding of MBP-Mga to both sites on separate DNA fragments.
It is anticipated from these in vitro binding studies that in strain JRS5301, which has binding site I deleted, Mga will still be bound to site II. With Mga bound there, it might be expected to repress transcription initiating from upstream promoter P1. However, this does not occur (Fig. 5 and 6). This suggests either that Mga binds weakly to a single site and is displaced during transcription or that binding in vivo differs from that in vitro, perhaps due to supercoiling.
No transcription initiating from P2 was detected in JRS5301, although Mga should be bound at site II and would be expected to activate this nearby downstream promoter. This suggests the possibility that Mga must occupy both binding sites I and II to activate P2, perhaps to achieve an appropriate polymerization state for association with RNA polymerase (RNAP).
What defines an Mga recognition site within different promoters?
Although Mga has two helix-turn-helix motifs in its amino-terminal end that may be directly involved in protein-DNA interactions, DNA fragments containing the Pemm or the PscpA binding sites competed for MBP-Mga binding in DNase I footprinting experiments at both Pmga sites even though the sites were on different DNA fragments. Thus, Mga may use the same domain to interact with the sites found in Pmga, Pemm, and PscpA. Alternatively, Mga may utilize a separate domain to bind to Pemm and PscpA, producing a conformational change that would affect binding to Pmga.
If a single domain of Mga interacts with all three sites, binding might involve recognition of similar DNA sequences in all cases. However, the two regions within Pmga protected by MBP-Mga show very little DNA sequence similarity to the consensus Mga binding site previously derived from the Pemm and PscpA promoters (22). Using sequence alignment, the four individual Mga binding sites were found to have less than 50% sequence similarity to each other and attempts to generate a more accurate consensus using the four known sites were not successful. It should be remembered that DNase I footprinting analysis of regions protected by a protein are likely to include many nucleotides that are not directly contacted by that protein. This may be even more likely with larger proteins such as the 62-kDa Mga protein, which is at least twice as large as most bacterial DNA-binding proteins. Furthermore, the high AT content of the sequence in the regions bound by Mga makes it even more difficult to identify a possible recognition consensus in the absence of experimental knowledge of the nucleotides contacted.
Both of the Pmga binding sites that are protected by Mga are much larger than the sites protected within Pemm and PscpA. MBP-Mga protects a 45-bp region upstream of emm and scpA, while the two Mga-protected regions within Pmga are both 59 bp long. Since the competition experiments indicated that the same domain of Mga is involved in the DNA-protein interactions at all four sites, the disparity in size may reflect the number of Mga molecules that are bound at the different sites. Some bacterial regulators bind to their DNA target sequences as homomultimeric species, such as dimers or tetramers, and it seems possible that a larger Mga species is required for binding in the Pmga region than in Pemm or PscpA.
Distances of the Mga binding sites from their target promoters.
Direct interaction between a regulator protein and RNAP bound at a promoter is the primary mode of activation of genes in bacteria (13, 14, 30). Transcriptional activators of bacteria bind upstream of the promoter that they regulate in close proximity to RNAP. This allows the factor to interact with subunits of RNAP (either α or ς) to stabilize the initiation complex and thus activate transcription (16, 30). For both Pemm and PscpA, Mga binds to a single site centered around base −52 from the start of transcription and slightly overlapping the −35 hexamer (22). Although the exact mechanism of Mga activation of emm and scpA expression has not been determined, the physical location of the Mga binding site for each of these promoters suggests that Mga acts via a direct interaction with the adjacent RNAP.
The location of Mga binding sites within Pmga, centered around bases −104 (site I) and −185 (site II) from the P2 promoter, is much further upstream from the target promoter than the sites within Pemm and PscpA. The greater distance of these sites from P2 within Pmga (over 100 bp upstream) suggests that Mga functions by a different mechanism of activation in this promoter than that which it utilizes at Pemm and PscpA.
Several different models may be suggested to explain the ability of Mga to activate the P2 promoter from distant sites. Bacterial transcriptional activators that use remote sites often interact with promoters that use RNAP associated with alternate sigma factors (14). Although this may be the case for Mga activation of P2, the presence of alternative sigma factors in GAS has yet to be investigated. Alternatively, Mga may interact with an additional factor that binds to a site adjacent to RNAP at the P2 promoter. An example of this can be found in E. coli, where some activator proteins bound at remote sites interact with the global regulator Crp, which binds close to the promoter to activate transcription (14). Finally, an additional Mga binding site located proximal to P2 may exist. In this case, DNA looping might cause Mga to interact with RNAP. However, such a third site must exhibit a lower affinity for Mga than the two defined here since no additional sites were detected by DNase I analysis at the levels of protein used in our study.
Evidence of a repressor of P1 activity.
Even though the role of the P1 promoter in determining appropriate mga expression remains unclear, the observation that P1 activity is regulated suggests that it is important. If synthesis of the P1 repressor is itself regulated, it may allow P1 to respond differently to environmental signals than P2. This will enable the organism to be attuned more sensitively to environmental changes. In agreement with this, transcription from the P1 promoter does not appear to require Mga (Fig. 5 and 6). Although a previous study reported that P1 was dependent on Mga for activity (3), those results were based on a spontaneous mutant of an M12 GAS strain that may differ from the strain used here.
Expression of mga is positively autoregulated, which implies that some form of negative regulation exists to limit exponentially increasing Mga synthesis. We have provided the first suggestion of such a negative element in the M6 strain studied. Deletion of the sequences containing Mga binding site I resulted in an increase in transcriptional initiation from the P1 promoter, even in the absence of Mga. This suggests that this lesion removes a cis-acting element necessary for repression of transcription from the P1 start site. Thus, while deletion of site I, which is upstream of P2, prevents Mga-dependent activation of the P2 promoter, it also leads to derepression of P1 transcription (Fig. 5 and 6). This might occur by binding of a classical repressor that blocks RNA polymerase from progressing through site I, which is 80 bp downstream of P1. Although Podbielski et al. recently identified a global negative regulator that may affect mga expression in an M type 49 GAS strain (29), they reported that this gene is absent from the M6 strain used in this study. Thus, the negative regulator implied by our work remains to be discovered.
The possible involvement of a second trans-acting molecule in the regulation of mga expression indicates that control of mga is more complex than originally thought. Since the recognition site for this new regulator overlaps a binding site for Mga, one could imagine that these two factors might compete for access to these sites. This would create a new level of regulation that may involve both binding affinities and protein-protein interactions. Since all of the different strains studied so far (3, 25, 27) are similar in Pmga architecture, our current and subsequent findings may be applicable to other serotypes of GAS and thus provide a general theme of mga regulation. Only with further investigation of mga regulation will we be able to propose a more detailed model of this intricate process that will lead us to a better understanding of this important human pathogen.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant R37-AI20723, and K.S.M. was supported in part by National Research Service award AI09460 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Andersson G, McIver K S, Heden L, Scott J R. Functional equivalence of divergent mga genes of group A streptococcus. Gene. 1995;175:77–81. doi: 10.1016/0378-1119(96)00124-2. [DOI] [PubMed] [Google Scholar]
- 2.Bisno A L. Group A streptococcal infections and acute rheumatic fever. N Engl J Med. 1991;325:783–793. doi: 10.1056/NEJM199109123251106. [DOI] [PubMed] [Google Scholar]
- 3.Bormann N E, Cleary P P. Transcriptional analysis of mga, a regulatory gene in Streptococcus pyogenes: identification of monocistronic and bicistronic transcripts that phase vary. Gene. 1997;200:125–134. doi: 10.1016/s0378-1119(97)00392-2. [DOI] [PubMed] [Google Scholar]
- 4.Bronze M S, Dale J B. The reemergence of serious group A streptococcal infections and acute rheumatic fever. Am J Med Sci. 1996;311:41–54. doi: 10.1097/00000441-199601000-00008. [DOI] [PubMed] [Google Scholar]
- 5.Caparon M G, Geist R T, Perez-Casal J, Scott J R. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J Bacteriol. 1992;174:5693–5701. doi: 10.1128/jb.174.17.5693-5701.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caparon M G, Scott J R. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell. 1989;59:1027–1034. doi: 10.1016/0092-8674(89)90759-9. [DOI] [PubMed] [Google Scholar]
- 7.Caparon M G, Scott J R. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 1991;204:556–586. doi: 10.1016/0076-6879(91)04028-m. [DOI] [PubMed] [Google Scholar]
- 8.Caparon M G, Scott J R. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci USA. 1987;84:8677–8681. doi: 10.1073/pnas.84.23.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chassy B M. A gentle method for the lysis of oral streptococci. Biochem Biophys Res Commun. 1976;68:603–608. doi: 10.1016/0006-291x(76)91188-8. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Bormann N, Cleary P P. VirR and Mry are homologous trans-acting regulators of M protein and C5a peptidase expression in group A streptococci. Mol Gen Genet. 1993;241:685–693. doi: 10.1007/BF00279912. [DOI] [PubMed] [Google Scholar]
- 11.Froehlich B, Husmann L, Caron J, Scott J R. Regulation of rns, a positive regulatory factor for pili of enterotoxigenic Escherichia coli. J Bacteriol. 1994;176:5385–5392. doi: 10.1128/jb.176.17.5385-5392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geist R T, Okada N, Caparon M G. Analysis of Streptococcus pyogenes promoters by using novel Tn916-based shuttle vectors for the construction of transcriptional fusions to chloramphenicol acetyltransferase. J Bacteriol. 1993;175:7561–7570. doi: 10.1128/jb.175.23.7561-7570.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gralla J D. Activation and repression of E. coli promoters. Curr Opin Genet Dev. 1996;6:526–530. doi: 10.1016/s0959-437x(96)80079-7. [DOI] [PubMed] [Google Scholar]
- 14.Gralla J D, Collado-Vides J. Organization and function of transcription regulatory elements. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1232–1245. [Google Scholar]
- 15.Husmann L K, Scott J R, Lindahl G, Stenberg L. Expression of protein Arp, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect Immun. 1995;63:345–348. doi: 10.1128/iai.63.1.345-348.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kihlberg B M, Cooney J, Caparon M G, Olsen A, Bjork L. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb Pathog. 1995;19:299–315. doi: 10.1016/s0882-4010(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 18.Lu F, Churchward G. Conjugative transposition: Tn916 integrase contains two independent DNA binding domains that recognize different DNA sequences. EMBO J. 1994;13:1541–1548. doi: 10.1002/j.1460-2075.1994.tb06416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maloy S, Stewart V. Autogenous regulation of gene expression. J Bacteriol. 1993;175:307–316. doi: 10.1128/jb.175.2.307-316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 21.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 22.McIver K S, Heath A S, Green B D, Scott J R. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A streptococcus. J Bacteriol. 1995;177:6619–6624. doi: 10.1128/jb.177.22.6619-6624.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McIver K S, Scott J R. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J Bacteriol. 1997;179:5178–5187. doi: 10.1128/jb.179.16.5178-5187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLandsborough L A, Cleary P P. Insertional inactivation of virR in Streptococcus pyogenes M49 demonstrates that VirR functions as a positive regulator of streptococcal C5a peptidase and M protein in OF+ strains. Dev Biol Stand. 1995;85:149–152. [PubMed] [Google Scholar]
- 25.Okada N, Geist R T, Caparon M G. Positive transcriptional control of mry regulates virulence in the group A streptococcus. Mol Microbiol. 1993;7:893–903. doi: 10.1111/j.1365-2958.1993.tb01180.x. [DOI] [PubMed] [Google Scholar]
- 26.Perez-Casal J, Caparon M G, Scott J R. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Podbielski A, Flosdorff A, Weber-Heynemann J. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect Immun. 1995;63:9–20. doi: 10.1128/iai.63.1.9-20.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Podbielski A, Peterson J A, Cleary P. Surface protein-CAT reporter fusions demonstrate differential gene expression in the vir regulon of Streptococcus pyogenes. Mol Microbiol. 1992;6:2253–2265. doi: 10.1111/j.1365-2958.1992.tb01401.x. [DOI] [PubMed] [Google Scholar]
- 29.Podbielski A, Woischnik M, Leonard B, Schmidt K. Characterization of nra, a global negative regulator gene in group A streptococci. Mol Microbiol. 1999;31:1051–1064. doi: 10.1046/j.1365-2958.1999.01241.x. [DOI] [PubMed] [Google Scholar]
- 30.Rhodius V A, Busby S J W. Positive activation of gene expression. Curr Opin Microbiol. 1998;1:152–159. doi: 10.1016/s1369-5274(98)80005-2. [DOI] [PubMed] [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott J R. A new gene controlling lysogeny in phage P1. Virology. 1972;48:282–283. doi: 10.1016/0042-6822(72)90139-0. [DOI] [PubMed] [Google Scholar]
- 33.Scott J R, Guenthner P C, Malone L M, Fischetti V A. Conversion of an M− group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J Exp Med. 1986;164:1641–1651. doi: 10.1084/jem.164.5.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas P S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci USA. 1980;9:5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]