Abstract
Wnt signaling is essential to regulate embryonic development and adult tissue homeostasis 1–3. Aberrant Wnt signaling is frequently associated with cancers 4. Wnt proteins must undergo palmitoleoylation on their hairpin 2 by the endoplasmic reticulum (ER)-resident membrane-bound O-acyltransferase porcupine (PORCN) before acting as signaling molecules 5–7. This modification is indispensable for Wnt to bind its receptor Frizzled, thus triggering signaling 8. Here, we report four cryo-electron microscopy structures of human PORCN: 1) the complex with the palmitoleoyl-CoA substrate, 2) the complex with its inhibitor LGK974, an anti-cancer drug currently in clinical trials 9, 3) the complex with LGK974 and WNT3A hairpin 2 (denoted as WNT3Ap), and 4) the complex with a synthetic palmitoleoylated WNT3Ap analog. The structures reveal that the hairpin 2 of WNT3A, which is well conserved in all Wnt ligands, inserts into PORCN from the lumenal side and the palmitoleoyl-CoA accesses the enzyme from the cytosolic side. The catalytic histidine triggers the transfer of unsaturated palmitoleoyl group to the target serine on the Wnt hairpin 2 owing to the proximity of the two substrates. The inhibitor-bound structure shows that LGK974 occupies the palmitoleoyl-CoA binding site to prevent the reaction. Thus, this work provides a mechanism of Wnt acylation and advances the development of PORCN inhibitors for cancer treatment.
Introduction
Wnt signaling is critical for human development and stemness as one of the essential signal cascades1–3,5. Wnt signaling is triggered by the interactions between the secreted lipid-modified signaling glycoprotein Wnt and its receptor Frizzled 5,8. The endoplasmic reticulum (ER)-resident membrane-bound O-acyltransferase Porcupine (PORCN) transfers a palmitoleoyl moiety to a serine residue on Wnt ligands 5–7 (Extended Data Fig. 1a). This modification is essential for WNT signal activation and is akin to the necessity of the palmitoyl modification of the Hedgehog (HH) ligand in HH signal transduction 10,11. Structural studies showed that the lipid modifications on Wnt and Hedgehog are essential to recognize their receptors 8,12,13. The mutation of the acylation site on Wnt or the inhibition of PORCN activity abolishes the Wnt signal indicating the crucial role of PORCN-mediated palmitoleoylation in the Wnt signaling pathway 14. To date, nineteen human Wnt ligands have been identified that activate different signaling pathways through both paracrine and autocrine signaling 15. In contrast, PORCN is the only enzyme that catalyzes Wnt lipidation, implying a general principle of PORCN-mediated Wnt acylation 16.
PORCN is a member of the membrane-bound O-acyltransferase (MBOAT) family that transfers the acyl group from acyl-coenzyme or accessory protein to lipid or protein substrates. This family plays an important role in lipid metabolism and signal transduction 7,17. Previously, the structures of sterol O-acyltransferases (ACAT) and diglyceride O-acyltransferase (DGAT) revealed the mechanism of MBOAT-mediated lipid modification 18–23. The Hedgehog acyltransferase (HHAT) structures show that the flexible N-terminal peptide of Hedgehog inserts into HHAT, which adds a palmitoyl group to the main chain of the N-terminal cysteine on Hedgehog 24. In contrast, the lipid modification of Wnt occurs on the hydroxyl group of a serine residue that locates on the tip of Wnt hairpin 2. The sequence of Wnt hairpin 2 is highly conserved among different Wnts 25, which indicates this particular secondary structure is indispensable for PORCN to recognize its substrate.
The deficiency of PORCN and its mutations cause focal dermal hypoplasia (FDH) 26,27, a rare disease associated with abnormal development of the skin, hands, feet, and eyes. Notably, some small-molecule inhibitors, including LGK974 and ETC159, have been found to specifically disrupt PORCN’s enzymatic activity, hence abolishing Wnt signaling 28. This inhibition considerably reduces the size of Wnt-dependent tumors 29,30. LGK974 is currently in clinical trials for treating pancreatic cancer, melanoma, triple-negative breast cancer, and other malignancies 9. In this manuscript, we report the cryo-electron microscopy (cryo-EM) structures of 1) palmitoleoyl-CoA-bound PORCN, 2) LGK974-bound PORCN, 3) WNT3A hairpin 2 and LGK974 bound PORCN, and 4) synthetic palmitoleoylated WNT3A hairpin 2 analog bound PORCN. These structures elucidate the molecular mechanism of PORCN-mediated Wnt acylation and provide insights into the development of anti-cancer small molecules.
Overall Structures of PORCN
We purified human FLAG-tagged PORCN (isoform B, which lacks residues 235–239, compared with the canonical sequence) by anti-Flag M2 resin. The fluorescence-based acyltransferase assay was conducted to measure the activity of PORCN in vitro. Briefly, we incubated recombinant PORCN with palmitoleoyl-CoA and a WNT3A hairpin 2 peptide (residues 199–220, denoted as “WNT3Ap”), which can be recognized and palmitoleoylated by PORCN 25,31. PORCN-mediated palmitoleolylation will release free coenzyme A, which can form a fluorescent adduct with 7-diethylamino-3-(4-maleimidophenyl)-4-methylcoumarin (CPM). This assay shows that recombinant PORCN exhibits robust acyltransferase activity in vitro (Fig. 1a), while PORCN activity is inhibited by LGK974 (Fig. 1b). The liquid chromatography tandem mass spectrometry (LC-MS) analysis shows that >40% WNT3Ap was acylated, but less than 0.1% WNT3Ap was modified in the presence of LGK974 (Extended Data Fig. 1b and c). The WNT3Ap and the palmitoleoylated WNT3Ap have been verified by mass spectrometry analysis.
Fig. 1. Functional characterization and overall structures of human PORCN.
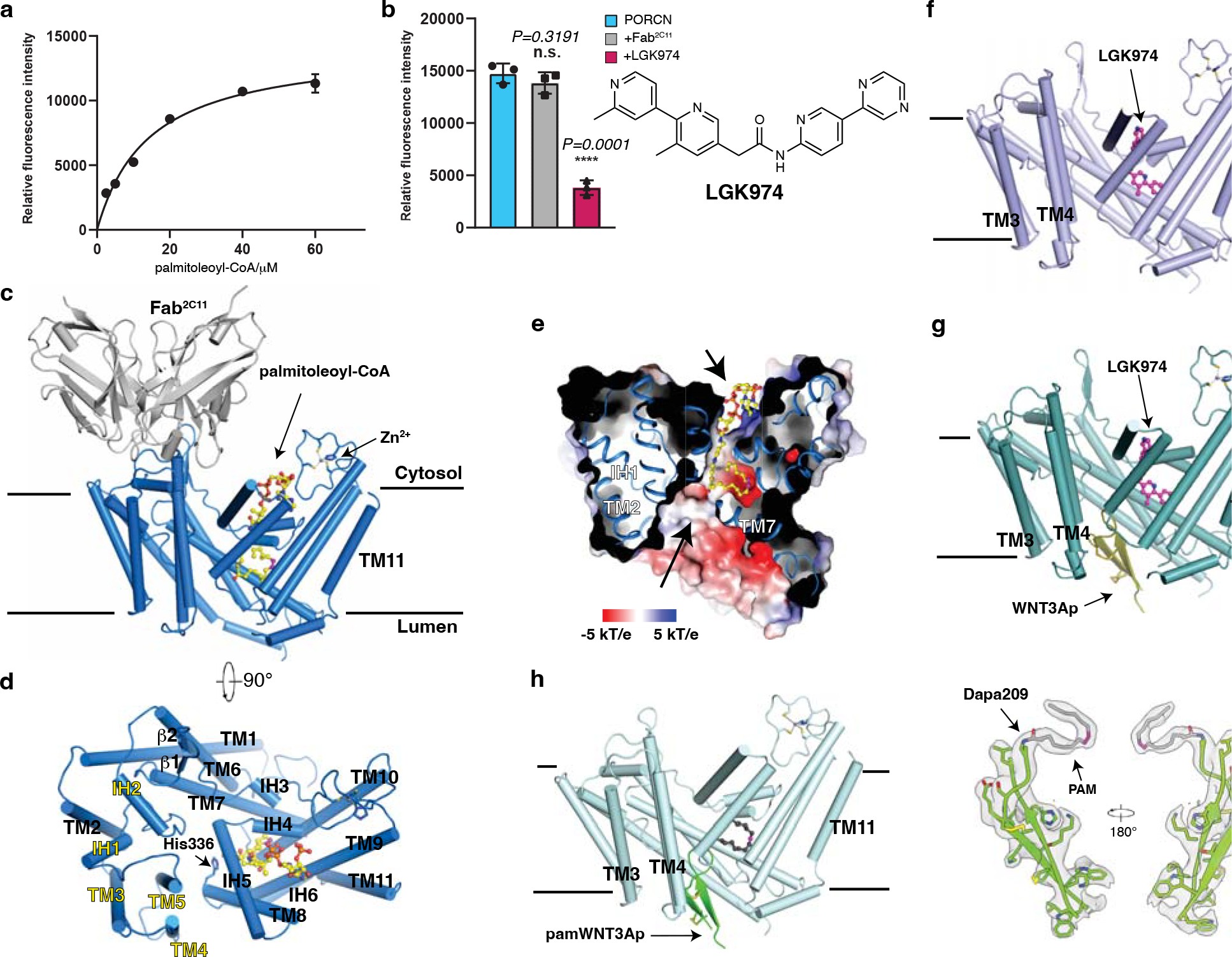
a, Concentration curve of the WNT3Ap acylation by PORCN with palmitoleoyl-CoA (Km=14.29 μM, fitted and calculated using GraphPad Prism 8). Data are mean ± s.d. (n=3 biologically independent experiments). b, LGK974 but not Fab2C11 inhibits the activity of PORCN in vitro. The chemical structure of LGK974 is shown on the right. Data are mean ± s.d. (n=3 biologically independent experiments). ****P ≤ 0.0001, two-tailed unpaired t-test using GraphPad Prism 8. c and d, Overall structure showing palmitoleoyl-CoA-bound PORCN viewed from the side of the membrane (c) or from the cytosol (d). The structural elements that bind to Fab2C11 are highlighted in yellow. e, Electrostatic surface representation of catalytic cavity viewed from the side of the membrane. The routes of substrates access are indicated by arrows. f, Overall structure showing LGK974-bound PORCN. g, Overall structure showing WNT3Ap-bound PORCN in the presence of LGK974. h, Overall structure showing pamWNT3Ap-bound PORCN. The cryo-EM map of pamWNT3Ap is shown. The palmitoleoyl-CoA is shown as sticks in yellow and LGK974 is shown as sticks in magenta. The cis double bond at C9 position of palmitoleoyl chain is colored in magenta. PAM, palmitoleic acid.
Because the molecular weight of PORCN is low and there is no notable feature outside of the detergent micelle, structural study by cryo-EM is challenging. Therefore, we generated a monoclonal antibody denoted 2C11 that explicitly binds to native PORCN. The binding of Fab2C11 to PORCN does not interfere with the activity of PORCN (Fig. 1b). Gel filtration reveals that PORCN and Fab2C11 form a complex and migrate as a single peak (Extended Data Fig. 2a).
To capture the substrate-bound state, we incubated recombinant human PORCN with palmitoleoyl-CoA prior to vitrification. The cryo-EM structure of PORCN with palmitoleoyl-CoA was determined at 3.1-Å resolution (Fig. 1c–e, Extended Data Figs. 2, 3, and Table 1). We also purified PORCN in the presence of LGK974 at a final concentration of 10 μM and determined the LGK974-bound structure at 3.1-Å (Fig. 1f, Extended Data Figs. 2, 3, and Table 1). The structure of the WNT3Ap-bound PORCN with LGK974 was determined at 2.9-Å resolution (Fig. 1g, Extended Data Figs. 4, 5, and Table 1). To capture the product-bound state, we incubated the WNT3Ap and palmitoleoyl-CoA with PORCN on ice for 1 hour. The resulting cryo-EM map revealed a weak covalent bond between Ser209 and the palmitoleoyl chain, indicating an incomplete acylation during the preparation, leading to the heterogeneity of the cryo-EM sample. To obtain a homogeneous and stable product analogue, we synthesized a WNT3Ap analog in which Ser209 is replaced by a (L)-2,3-diaminoproprionic acid (Dapa) residue, and this analogous serine residue is palmitoleoylated with an amide linkage (denoted as “pamWNT3Ap”, Extended Data Fig. 1d). We took advantage of this analog to determine the structure of pamWNT3Ap bound PORCN at 3.2-Å resolution. The covalent bond between Dapa209 and the palmitoleoyl chain can be well determined (Fig. 1h, Extended Data Figs. 4–5, and Table 1). The structurally observed binding mode of pamWNT3Ap is similar but may not be identical to that of the natural product owing to kinetic effects.
PORCN contains 11 transmembrane helices (TMs) with 6 intervening α-helices (IH) and 2 β-strands. Fab2C11 binds to the cytosolic region of TMs 3–5 and IHs 1–2 (Fig. 1d). There are two cavities on the opposite sides of PORCN, implying the routes of substrates access (Fig. 1e). TMs 7–10 create a cytosolic cavity to accommodate palmitoleoyl-CoA, while TMs 1, 2, 5, 6, and 7 generate the lumenal cavity to bind the Wnt ligand (Fig. 1e, g, and h). A previous study showed that PORCN can bind zinc at a 1:1 molar ratio 25. Consistent with this result, a zinc ion is coordinated by residues Cys370, Cys376, Cys380 and His382 in the cytosolic loop between TM9 and TM10 (Fig. 2a and b). Intriguingly, DHHC20, a palmitoyl S-acyltransferase, also employs a similar motif to engage the zinc ion 32, although it adopts a different fold with PORCN (Extended Data Fig. 6a).
Fig. 2. Overall structure of PORCN with palmitoleoyl-CoA.
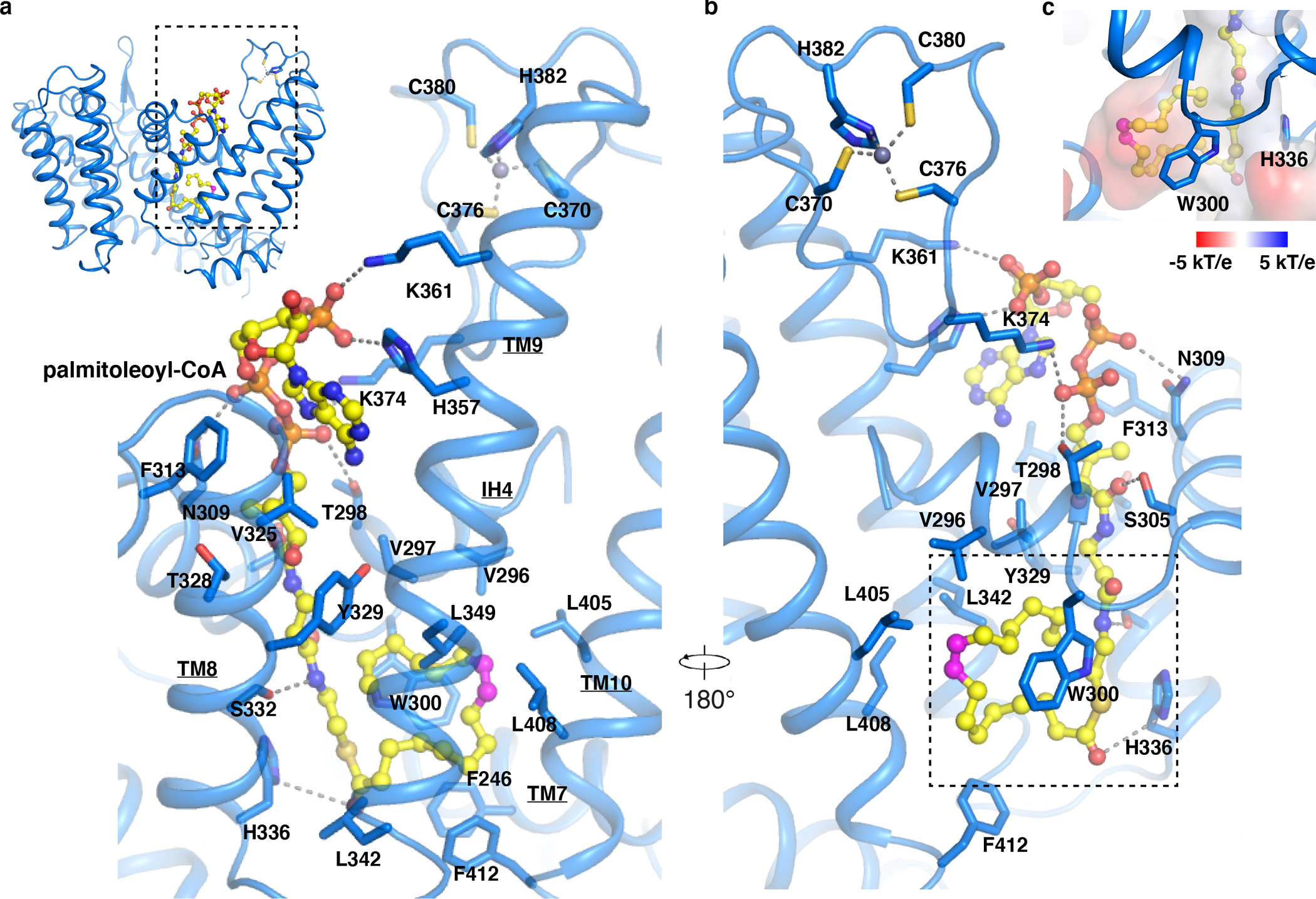
a and b, Overall view of the palmitoleoyl-CoA and zinc ion with the bound residues in PORCN. Dashed lines indicate the polar interactions. The cis double bond at C9 position of palmitoleoyl-CoA is colored in magenta. c, Electrostatic surface representation of palmitoleate bound cavity (dashed box in panel b).
ACATs and DGATs function as tetramers or dimers 18–23, while PORCN is a monomer similar to HHAT 24,33. A Dali search 34 shows that the overall structure of PORCN shares a similar fold with DltB (a bacterial MBOAT 35), HHAT, DGAT-1, and ACAT-1 with R.M.S.D. values of 3.4 Å, 3.7 Å, 4.4 Å, and 4.3 Å respectively (Extended Data Fig. 6b–e). The HHAT, DGAT, ACAT, and PORCN are conserved in acyl-CoA binding positions. Although the members of the MBOAT family have different numbers of TMs (9 TMs in ACATs and DGATs, 10 TMs in DltB, and 12 TMs in HHAT), all MBOATs employ six transmembrane helices (TMs 5–10 in PORCN) to form a catalytic core for the acyl-transfer reaction (Fig. 1d).
Palmitoleoyl-CoA binding site
The palmitoleoyl-CoA is observed in the cytosolic side (Fig. 2a). The cryo-EM map suggests that the palmitoleoyl group may adopt two conformations (curled-up and curled-down, Extended Data Fig. 3d). After 100 ns molecular dynamics (MD) simulations, the interaction of residues Trp300 and His357 to the palmitoleoyl-CoA in the curled-down conformation have been disrupted (Supplementary Videos 1 and 2). Calculated interaction energies (see Methods for details) of the palmitoleoyl-CoA ligand with surrounding amino acid side chains of PORCN indicate that the curled-up conformation is energetically more stable than the curled-down conformation (Extended Data Fig. 3d). Therefore, we modeled the palmitoleoyl-CoA in the curled-up position (Fig. 2a). The catalytic His336 is in spatial proximity to the carbonyl oxygen of the fatty acid, which is akin to a similar observation in substrate-bound HHAT 24. The unsaturated palmitoleoyl group curls in the center of the enzyme (Fig. 2a). Residues Thr298, Asn309, Tyr329, Ser332, His357, Lys361 and Lys374 engage with coenzyme A through polar interactions (Fig. 2a). Residues Phe246, Val296, Val297, Trp300, Leu342, Leu349, Leu405, Leu408 and Phe412 are responsible for accommodating the palmitoleate moiety (Fig. 2b and c).
Previous studies showed that PORCN is also highly selective for palmitoleoyl-CoA compared to other acyl-CoAs 25,31. Our structural observation illustrates that the center cavity of PORCN is a snug fit for palmitoleoyl chain, and differences in the carbon chain lengths and the double bond positions on acyl chain cause PORCN misrecognition, reducing the efficacy of the catalyzation 36. Moreover, residue Trp300 contributes to the stabilization of the palmitoleoyl-CoA acyl chain and provides the cavity with the stiffness necessary to particularly accommodate the palmitoleoyl chain (Fig. 2d).
LGK974-bound state
The structure of LGK974-bound PORCN reveals the interaction details between PORCN and its inhibitor. LGK974 adopts an “L” shape bound to the cytosolic pocket (Fig. 3a). Hydrophobic interactions with LGK974 are provided by residues Phe246, Val297, Trp300, Thr328, Tyr329, Leu349, and Leu408 (Fig. 3b). Residue Ser332 is responsible for engaging with the carbonyl oxygen of LGK974. Many potent PORCN inhibitors contain a carbonyl oxygen and adopt an “L” shape, suggesting a general binding mode between PORCN and its inhibitors 37. Structural comparison reveals that the binding site of LGK974 overlaps with the 4-phosphopantetheine and fatty-acid chain of palmitoleoyl-CoA (Fig. 3c), implying that the PORCN inhibitor functions as an acyl-CoA competitor to preclude its access, thereby preventing the reaction.
Fig. 3. Overall structure of LGK974-bound PORCN.
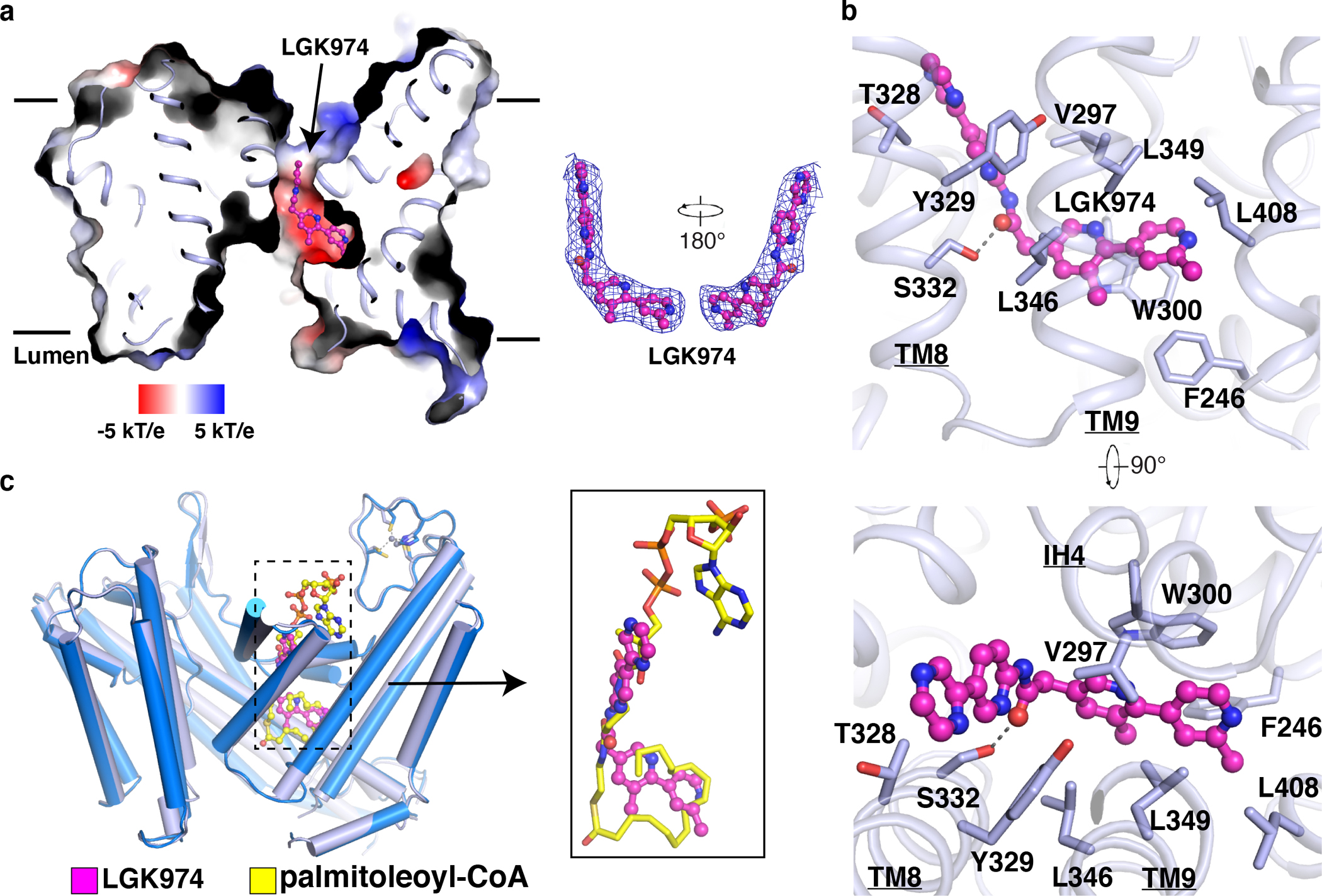
a, Electrostatic surface representation of LGK974-bound PORCN. LGK974 is shown as sticks in magenta. The cryo-EM map of LGK974 is shown at 5σ level. b, Interaction of LGK974 with cavity residues. c, The comparison of LGK974-bound structure with palmitoleoyl-CoA bound structure. The palmitoleoyl-CoA is shown as sticks in yellow and LGK974 is shown as sticks in magenta.
This mechanism is quite different from our previously reported mechanism for inhibiting ACAT-mediated cholesterol esterification. Specifically, inhibitors of ACATs, including nevanimibe and pyripyropene A, occupy the catalytic core of ACATs to prevent the proximity of cholesterol and acyl-CoA substrates (Extended Data Fig. 6e), but do not compete with either of them to bind the enzymes 19,23. The inhibitor-bound HHAT structure showed the inhibitor IMP1575 bound to the catalytic histidine of HHAT (Extended Data Fig. 6c) and introduces the conformational changes of the catalytic core 33. In contrast, the overall structures of LGK974 bound PORCN and palmitoleoyl-CoA bound PORCN share a similar conformation with a R.M.S.D. value of 0.4 Å (Fig. 3c), and the catalytic His336 does not bind to LGK974.
WNT3Ap-bound state
The WNT3Ap and the pamWNT3Ap are clearly resolved in the cryo-EM maps (Extended Data Fig. 5). Two pairs of disulfide bonds, which are widely conserved in all Wnt ligands (Fig. 4a), confine the peptide in a hairpin structure, allowing it to insert into the lumenal side of PORCN (Fig. 1g and h). Residue His206 of WNT3A forms a hydrophilic bond with the hydroxyl group of Ser249 of PORCN, and the main chains of residues Lys215 and Cys217 interact with residues Gln28 and Gln24 of PORCN, respectively (Fig. 4b). A previous biochemical assay showed that WNT1 peptide with a His221Ala mutation (equivalent to His206Ala of WNT3A) exhibits minimal acylation by PORCN 25 supporting our structural observation. Residues Phe67, Met128, Ile129, Val166, Phe167, Val204, Phe252, and Val253 of PORCN contribute hydrophobic contacts to bind WNT3Ap (Fig. 4b). The Cys17-Cys209 disulfide bond of PORCN confines the TM1 and TM6 in a conformation that hosts the WNT3Ap (Fig. 4b). Notably, residues Cys17, Gln24, Gln28, Cys209 and Ser249 are conserved across the different species of PORCN 25. Besides two disulfide bonds, His206 of WNT3A is also well conserved in WNT protein family (Fig. 4a). It is tempting to speculate that the hydrophilic interactions between different Wnt ligands and PORCN are consistent.
Fig. 4. Overall structures of PORCN with WNT3Ap and palmitoleoylated WNT3Ap.
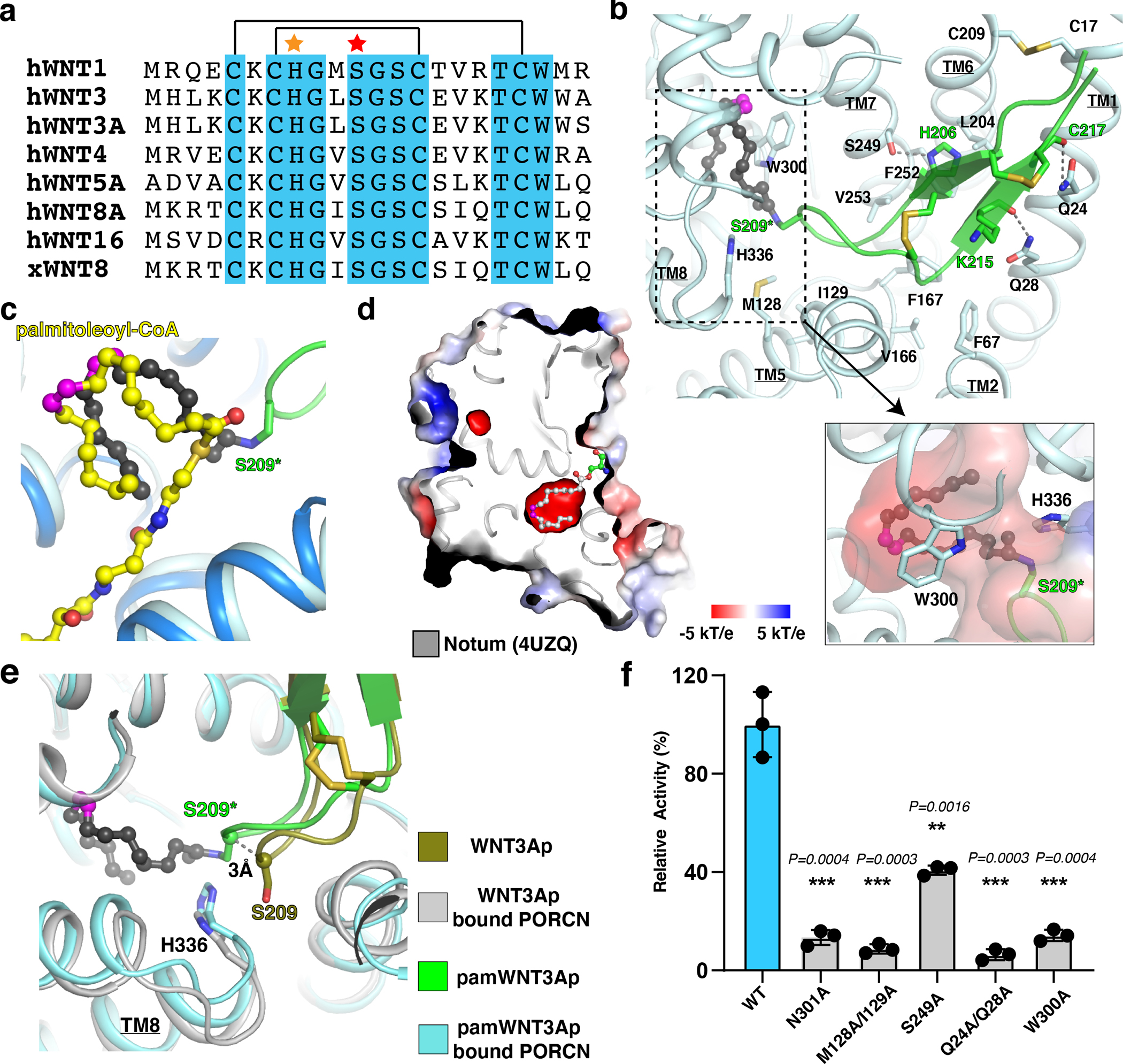
a, Sequence alignment of the hairpin 2 from the Wnt ligands. The conserved residues among Wnt ligands are highlighted in cyan. The disulfide bonds are indicated by black lines. The His206 is indicated by an orange star. The acylation serine of Wnt ligands is marked by a red star. b, Overall view of the pamWNT3Ap with the bound residues in PORCN. S209 in pamWNT3Ap is replaced by (L)-2,3-diaminoproprionic acid (S209*). PAM, palmitoleic acid (dark gray sticks). Electrostatic surface representation of palmitoleate bound cavity (dashed box) is shown. c, Comparison of palmitoleate moiety in the palmitoleoyl-CoA bound PORCN (yellow sticks) and pamWNT3Ap bound PORCN structures (black sticks). d, Electrostatic surface representation of Notum. The palmitoleate is shown as gray sticks. The cis double bond at C9 position of palmitoleoyl-CoA is colored in magenta. e, Structural comparison of unmodified WNT3Ap and pamWNT3Ap. f, Functional validation of residues Q24/Q28, M128/I129, S249, W300, and N301. Data are mean ± s.d. (n=3 biologically independent experiments). **P ≤ 0.01, ***P ≤ 0.001, two-tailed unpaired t-test using GraphPad Prism 8.
The center cavity of PORCN accommodates the palmitoleate moiety of the pamWNT3Ap (Fig. 4b), which is consistent with our observation in palmitoleoyl-CoA bound PORCN (Fig. 4c). The structure of palmitoleoyl-protein carboxylesterase Notum, which mediates depalmitoleoylation of Wnt ligands, reveals the molecular details of how palmitoleate binds to the center of Notum 38 (Fig. 4d). Structural comparison exhibits that the fatty acid chains in both Notum and PORCN, particularly the cis-Δ9-double bonds, share a similar position in the cavity, implying the conservative interaction mode between these two enzymes and palmitoleate. Compared to the WNT3Ap, there is a 3-Å shift of Ser209 of the pamWNT3Ap towards the catalytic core of PORCN (Fig. 4e), indicating the flexibility of the tip of Wnt hairpin 2 during the acylation. This shift may also be associated with the difference in the chemical structure of pamWNT3Ap and the natural product.
Four variants PORCNQ24A/Q28A, PORCNM128A/I129A, PORCNS249A, and PORCNW300A exhibit exemplary behavior in detergent solution (Extended Data Fig. 7) and present a considerably decreased enzymatic activity (Fig. 4f), which validates the essential role of these residues in Wnt lipidation. Previous mutagenesis confirmed that Tyr329 and Ser332 are required for PORCN activity 37,39. Moreover, mutations of Leu204, Val253, Ser332, Leu342, Leu349 and Phe412 lead to FDH (https://databases.lovd.nl/shared/genes/PORCN), highlighting the physiological importance of these residues in the biogenesis of Wnt signal.
Structural Model of WNT3A-bound PORCN
Owing to the conservation of Wnt hairpin 2, we superimposed our structure onto the structures of the full-length Wnt complex with cysteine-rich domain (CRD) of Frizzled 8 and the Wnt complex with its transporter Wntless (WLS) 40 (Extended Data Fig. 8a and b). Although the hairpin 2 of the Wnt ligands in all three structures are well aligned; the palmitoleate moiety positions are quite different, and the globular domain of full-length Wnt ligands in Wnt-CRD and Wnt-WLS complexes would clash with PORCN (Extended Data Fig. 8a and b).
We performed MD simulations to study how full-length Wnt ligand interacts with PORCN using the AlphaFold 41-predicted structure of full-length WNT3A and PORCN complex (Extended Data Fig. 8c). The predicted structure of PORCN is similar to our cryo-EM structure with an R.M.S.D value of 0.8 Å (Extended Data Fig. 8d). The MD simulations show that hairpin1 and hairpin 3 of WNT3A do not bind PORCN via stable interactions; while the hairpin 2 can undergo slightly conformational changes, consistent with our structural observation (Fig. 4e and Supplementary Video 3). It is possible that Wnt ligand may adopt various conformations when bound to PORCN, which is akin to our observation in the structure of the Hedgehog-HHAT complex. In the structure of the Sonic Hedgehog-N (SHH-N) complex with HHAT, the N-terminal peptide of SHH-N is resolved in the cryo-EM map; in contrast, the globular domain of SHH-N is not visible, implying that this domain adopts multiple conformations during the HHAT-mediated acylation (Extended Data Fig. 9).
Structural Comparison with HHAT
Previous remarkable work on HHAT revealed an archway in the TMs 8–10 may allow the acyl-CoA access to the enzyme 24; interestingly, our PORCN structure shows TM11 has blocked the putative entrance of the acyl-CoA archway (Fig. 5a). To test whether PORCN-TM11 moves in the membrane to allow the acyl-CoA access into the archway, as found in HHAT 24, we conducted an unbiased MD simulation (100 ns) of PORCN in a lipid environment (Supplementary Video 4). No large-scale movement of helices was observed on this time scale, suggesting the dynamics of PORCN-TM11 restricts acyl-CoA access into the archway. However, further investigations may be necessary to track dynamics over longer time periods.
Fig. 5. The structural comparison with HHAT.
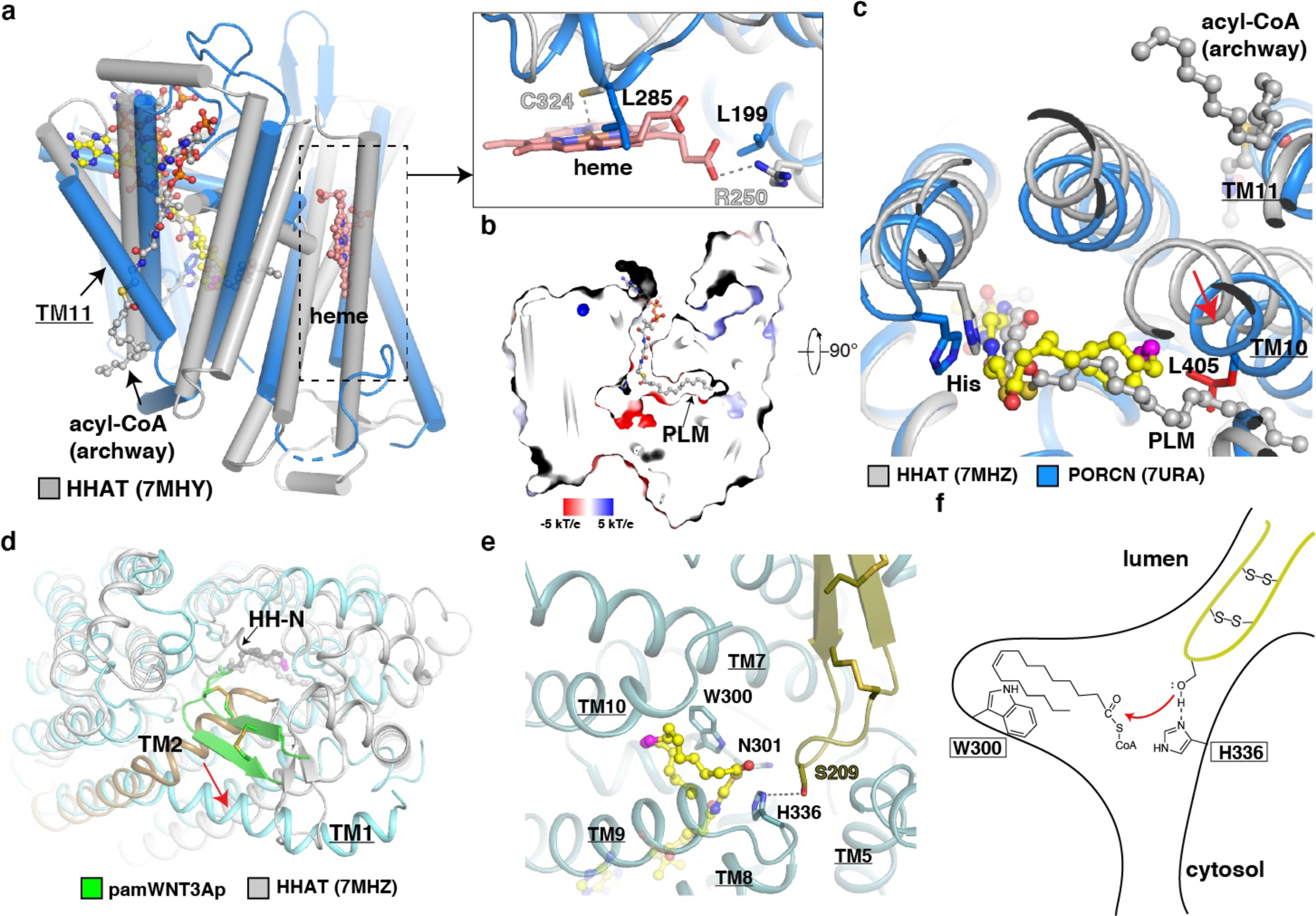
a, Membrane view of the structure of PORCN (blue) compared to that of acyl-CoA bound HHAT (gray). The acyl-CoA in the archway and heme of HHAT are indicated. The cis double bond at C9 position of palmitoleoyl-CoA is colored in magenta. A close-up view of the heme (salmon sticks) binding site in HHAT compared to that in PORCN. The dashed line indicates the interaction between HHAT residues and heme. b, Electrostatic surface representation of catalytic cavity of HHAT. The acyl-CoA in the cavity is shown in sticks. PLM, palmitic acid (gray sticks). c, Lumenal view of the structure of palmitoleoyl-CoA bound PORCN compared to that of acyl-CoA bound HHAT. The conformational difference of PORCN-TM10 is indicated by the red arrow. Residue Leu405 of PORCN is colored in red. TM11 of PORCN and the acyl-CoA in the archway are indicated. Palmitoleoyl-CoA is shown in yellow sticks. d, Lumenal view of the structure of pamWNT3Ap bound PORCN (light cyan) compared to that of HH product bound HHAT. The conformational difference between HHAT-TM2 (gold) and PORCN-TM1 is indicated by the red arrow. e, Docking palmitoleoyl-CoA into WNT3Ap/LGK974 bound PORCN (dark cyan). f, A proposed “one-step” working model.
PORCN and HHAT are required for modifying the Wnt and Hedgehog morphogens, respectively. Several differences between these two membrane-embedded enzymes should be noted. The lipid modification on Hedgehog involves a saturated fatty acid, while that of Wnt is unsaturated 36. The saturated fatty acid chain of acyl-CoA in HHAT adopts an extended state within the linear cavity, showing collinearity with the midline of the membrane (Fig. 5b); in contrast, the cis double bond at C9 position of palmitoleoyl moiety causes a kink of fatty acid chain to form a “C” shape in the catalytic core (Fig. 2c and 4b). A biochemical study confirmed that the catalytic core of PORCN enforces cis-Δ9 palmitoleoylation on Wnt proteins 42.
Moreover, a superimposition with the structure of HHAT shows that TM10 of PORCN shifts and its Leu405 clashes with the extended acyl-chain of acyl-CoA substrate in HHAT (Fig. 5c), indicating the cis-Δ9 ensures the palmitoleate in a proper conformation during the acyl transfer. HHAT recognizes the flexible N-terminal peptide of Hedgehog and transfers the acyl group to the N-terminus via N-acylation; in contrast, PORCN engages a well-folded hairpin and transfers the acyl group to the serine’s side chain via O-acylation. Compared to the PORCN-TM1, the N-terminus of HHAT-TM2 is located in the center of the HHAT, shrinking the diameter of the lumenal cavity (Fig. 5d). Therefore, only a flexible peptide can access the catalytic core; in contrast, the lumenal cavity of PORCN opens extensively, allowing the entire hairpin 2 of WNT to be inserted into the enzyme (Fig. 5d). In addition, the structural studies showed that HHAT protein binds a heme in the cytosolic leaflet (Fig. 5a), and this binding is essential for the HHAT activity 24,33. Although PORCN does not bind to heme, a previous study 25 along with our study shows that zinc ion binds to the cytosolic loop of PORCN (Fig. 2b). It remains unclear whether this zinc ion modulates the WNT acylation or functions to stabilize the structure.
Discussion
The distances between Ser209 of WNT3A and the catalytic His336 in the WNT3Ap/LGK974 bound PORCN structure is ~4-Å (Fig. 5e). Docking the palmitoleoyl-CoA into the WNT3Ap/LGK974 bound PORCN structure shows spatial proximity between the thioester bond of palmitoleoyl-CoA, the catalytic His336, and the hydroxyl group of WNT3Ap-Ser209. (Fig. 5e). Accoding to this observation, we propose a “one-step” working model for PORCN-mediated WNT acylation (Fig. 5f). The catalytic His336 deprotonates Ser209 of WNT3A first. The proximity of the deprotonated Ser209 on WNT3A and carbonyl group of palmitoleoyl-CoA allows the hydroxyl group of Ser209 to directly attack the thioester bond, forming the product. After the reaction, the free CoA is released into the cytosol. Trp300 and Asn301 may stabilize the palmitoleoyl-CoA since the mutations on these two residues weaken the enzymatic activity (Fig. 4f). The modified Wnt can either be released into the lumenal side or be delivered to its transporter WLS, and then the Wnt-WLS complex is transported through exocytic vesicles to the cell surface 37,40. Obviously, we could not exclude the working model of a “two-step” reaction in which an acyl-enzyme intermediate is generated during the reaction, like the previously proposed reaction mechanism of DHHC20 32. A following study on PORCN may distinguish the catalytic mechanism.
Pharmacological inhibition of PORCN is highly selective for the Wnt signaling and considerably reduces the growth of Wnt-dependent tumors, suggesting a promising role of PORCN inhibition in cancer treatment. Moreover, the hairpin 2 of Wnt was also shown to mediate interactions with its receptor Frizzled 8 and its transporter WLS 40. Together with our structural observation, these findings highlight the critical role of Wnt’s conserved hairpin 2 in signal transduction. Our work elucidates the mechanism of PORCN-mediated Wnt lipidation at the atomic level and provides the additional insights into the development of anti-cancer medicines targeting Wnt-related cancers.
Methods
Protein expression and purification
The cDNA encoding human PORCN isoform B (Horizon Discovery) and its mutants were cloned into pEG BacMam vector with an N-terminal FLAG tag. The cDNA encoding human WNT3A was cloned into pEG BacMam vector without tag. The coding region of each plasmid was sequenced to ensure the integrity of the construct. All proteins were expressed using baculovirus-mediated transduction of mammalian HEK-293S GnTI− cells (ATCC). 8 hours after adding the virus, 5 mM sodium butyrate was introduced, and cells were then incubated at 30°C to express proteins. 72 hours after infection, the cells were harvested and ready for protein purification.
For the purification of wild-type PORCN with inhibitor LGK974, the cells were infected with the PORCN-encoded virus only. The pellet from 2 liters of cells was resuspended in 40 ml lysis buffer (100 mM HEPES, 450 mM NaCl, 5 μg/ml leupeptin, 1 mM PMSF, 10 μM LGK974, 0.5 mM TCEP, pH 7.5) and disrupted by sonication. After low-speed centrifugation, the resulting supernatant was then incubated with 1% (w/v) lauryl maltose neopentyl glycol (LMNG, Anatrace) for 1 hour at 4°C. The insoluble fraction was removed by centrifugation (60,000 g, 4°C, 30 mins), and the supernatant was incubated for 1 hour with 1mL Anti-FLAG M2 resin (Sigma-Aldrich). The beads were then washed with 10 ml of high salt wash buffer (25 mM HEPES, 350 mM NaCl, 0.06% Digitonin, 10 μg/ml 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS, Anatrace), 10 μM LGK974, pH 7.5) and 10 mL of low salt wash buffer (25 mM HEPES, 250 mM NaCl, 0.06% Digitonin, 10 μg/ml POPS, 10μM LGK974, pH 7.5) by gravity flow. PORCN was eluted in 10mL elution buffer (20 mM HEPES, 150 mM NaCl, 0.06% Digitonin, 100 μg/mL 3x Flag peptide,10 μM LGK974, pH 7.5). The eluate was concentrated and further purified by size-exclusion chromatography (SEC, Superose-6 Increase, 10/300 GL column, GE Healthcare) in buffer (20 mM HEPES, 150 mM NaCl, 0.06% Digitonin, 10 μM LGK974, pH 7.5) prior to incubation with fabs for cryo-EM studies.
For purification of apo wild-type PORCN, the cells were infected with PORCN-encoded virus and WNT3A-encoded virus at a 4:1 ratio since PORCN alone does not have a good behaviour during purification. The pellet from 2 liters of cells was resuspended in 40 ml lysis buffer (100 mM HEPES, 450 mM NaCl, 5 μg/ml leupeptin, 1 mM PMSF, pH 7.5) and disrupted by sonication. The following step is the same as above, except excluding the LGK974 in all purification steps. The purifications of different mutants were similar to the purification of apo wild-type PORCN.
Antibody generation and purification of PORCN-Fab complexes
IgG-2C11, a mouse monoclonal anti-human PORCN antibody, was prepared by fusion of SP2-mIL6 mouse myeloma cells (ATCC no. CRL-2016) with splenic B lymphocytes obtained from mice (n = 1) at UT Southwestern with the approval of the Institutional Animal Care and Research Advisory Committee #2017-102391 as previously described 43. Briefly, 6–8 weeks old male New Zealand Black NZBWF1/J mice from Jackson Laboratory (strain #: 100008) were immunized with one primary and four boosts of purified recombinant human PORCN reconstituted with amphipols (50 μg) in phosphate buffered saline (PBS) combined with Sigma Adjuvant System. We used the combined techniques of ELISA, immunoblot analysis, and immunoprecipitation to identify antibodies that are preferentially bound to PORCN in its native but not SDS-denatured state. To clone IgG-2C11, total RNA was isolated from the hybridoma by RNA extraction kit (Qiagen) following the manufacturer’s protocol. Total RNA was subjected to reverse transcription reactions using Superscript III reverse transcription kit (Invitrogen), and the resultant cDNA was used as a template in PCR reactions with degenerate primers to amplify the variable regions. Sequences of the resulting PCR products were analyzed with the IMGT database (http://www.imgt.org/) to determine the variable regions of the light chain and heavy chain, which were then cloned into shuttle vectors for the light chain and the Fab region of heavy chain with a C-terminal 6xHis tag, respectively 44. The resulting constructs were co-transfected to HEK-293S GnTI− cells (ATCC) using PEI MAX transfection reagent (Polysciences) for expression at 37 °C. After 72 hours, the medium was harvested and applied to Ni-NTA gravity columns. Following several washes with buffer A (20 mM HEPES, 150 mM NaCl, pH 7.5) containing 20 mM imidazole, bound material was eluted in buffer A containing 250 mM imidazole. The eluate was then applied to a Superdex-200 Increase size-exclusion chromatography column (GE Healthcare) in buffer A, and peak fractions containing Fab2C11 were collected for complex assembly.
For the generation of PORCN-Fab2C11 complexes, purified Fab2C11 were combined with purified ~ 0.2 mg/ml PORCN (using a molar ratio of 1:1.5, PORCN:Fab) and incubated at 4°C for 1 hour. The samples were then concentrated and further purified by SEC (SEC, Superose-6 Increase, 10/300 GL column) in buffer (20 mM HEPES, 150 mM NaCl, 0.06% Digitonin, pH 7.5) with or without 10 μM LGK974.
Purification of HHAT–SHH-N–Fab3H02 complex
The cDNA encoding human HHAT (Horizon Discovery) containing a H379C mutation was cloned into pEG BacMam vector with a C-terminal FLAG tag. The cDNA encoding full-length human SHH was cloned into pEG BacMam vector without a tag. HHAT–SHH-N complex was expressed using baculovirus-mediated transduction of mammalian HEK-293S GnTI− cells (ATCC) by co-infection. 8 hours after adding the viruses, 10 mM sodium butyrate was introduced. 48 hours after infection, the cells were harvested for protein purification. The cell pellet was incubated with lysis-extraction buffer (20 mM HEPES, 300 mM NaCl, 1% LMNG, 5 μg/ml leupeptin, 1 mM PMSF, pH 7.5) for 2 hours at 4°C. The insoluble fraction was removed by centrifugation (60,000 g, 4°C, 30 mins), and the supernatant was incubated with Anti-FLAG-M2 resin (Sigma) for 1 hour. The beads were then washed with buffer (20 mM HEPES, 300 mM NaCl, 0.1% LMNG, 10 μg/ml POPS) by gravity flow. HHAT–SHH-N complex was eluted in buffer (20 mM HEPES, 150 mM NaCl, 0.1% LMNG, 10 μg/ml POPS, 25 μM 16:0 CoA, 100μg/mL 3x Flag peptide, 1 mM DTT, pH 7.5) and further purified by size-exclusion chromatography (SEC, Superose-6 Increase, 10/300 GL column, GE Healthcare) in buffer (20 mM HEPES, 150 mM NaCl, 0.06% Digitonin, pH 7.5).
The DNA sequences encoding the light chain and heavy chain of Fab3H02 were derived from PDB: 7MHZ and synthesized from Integrated DNA Technologies and cloned into shuttle vectors with a 6x His tag at the C-terminus of heavy chain. The expression and purification procedure are the same as that of Fab2C11. For the generation of HHAT–SHH-N–Fab3H02 complex, purified HHAT–SHH-N was incubated with Fab3H02 with a molar ratio of 1:1.5 (HHAT–SHH-N: Fab3H02) at 4°C for 1 hour. The mixture was then concentrated and purified by SEC (Superose-6 Increase, 10/300 GL column) in buffer (20 mM HEPES, 150 mM NaCl, 0.06% Digitonin, pH 7.5).
EM sample preparation and imaging for 300 kV Cryo-TEM
PORCN samples (~8 mg/ml) with 10 μM LGK974 and HHAT–SHH-N–Fab3H02 complex (~10mg/ml) were applied to Quantifoil R1.2/1.3 400 mesh Au holey carbon grids (Quantifoil), respectively. For the preparation of palmitoleoyl-CoA-bound PORCN samples, palmitoleoyl-CoA was added into the apo-PORCN sample at the final concentration of 1 mM before vitrification. For the preparation of LGK974/WNT3Ap-bound PORCN samples, WNT3Ap (MHLKCKCHGLSGSCEVKTCWWS, C5-C19, C7-C14, Biomatik) were mixed with LGK974-bound PORCN at a final concentration of 1 mM. For the preparation of product-bound PORCN, pamWNT3Ap was added into the apo-PORCN at the final concentration of 1.2 mM. The above two peptide mixtures were incubated on ice for 30 min before vitrification. The grids were blotted and plunged into liquid ethane for flash freezing using a Vitrobot Mark IV (FEI). The grids were imaged in a 300 kV Titan Krios (FEI) with a Gatan K3 Summit direct electron detector (Gatan). Data were collected using SerialEM 45 at 0.83 Å/pixel or 0.842 Å/pixel. Images were recorded for 5-second exposures in 50 subframes with a total dose of ~60 electrons per Å2.
Imaging processing and 3D reconstruction
Dark subtracted images were first normalized by gain reference. Motion correction was performed using MotionCor2 46 and performed in RELION-3.1 47. The contrast transfer function (CTF) was estimated using CTFFIND4 48. Auto picking was performed with crYOLO v1.7.6 using the general model 49 with the particle threshold of 0.1. About 1 million particles were extracted for each dataset. Subsequent 2D classification, multi-class Ab-Initio modeling, heterogenous 3D refinement, and non-uniform refinement of the best class were performed (Extended Data Figs. 2 and 4). For PORCN complexes, after a local CTF refinement, the final maps were obtained with a final local refinement in cryoSPARC. For HHAT–SHH-N–Fab3H02 complex, a subsequent 3D classification was performed in RELION-3.1 47 with the SHH-N region masked, followed by a 3D refinement and postprocessing for the best class.
Cryo-EM model building and refinement
For PORCN complexes, the initial model was built de novo based on an AlphaFold2 predicted structure and then manually adjusted using COOT 50. Residues 1–3, 223–233 and 415–424 were not built; residues 271–274 were built as poly-alanine due to limited local resolution. The model was refined in real space using PHENIX 51. For cross-validations, the final model was refined against one of the half maps generated by 3D auto-refine and the model vs. map FSC curves were generated in the comprehensive validation module in PHENIX. PHENIX and MolProbity 52 were used to validate the final model. Local resolutions were estimated using cyroSPARC local resolution estimation 53. For HHAT–SHH-N–Fab3H02 complex, structure of HHAT–Fab3H02 from PDB: 7MHZ and structure of SHH-N from PDB: 6E1H were docked into the cryo-EM map as initial models. The structure was built manually in COOT 50 and refined using PHENIX51. Structure Figures were generated using PyMOL (http://www.pymol.org), Chimera 54 and ChimeraX 55.
Fluorescence-based acylation assay
PORCN-mediated Wnt palmitoleoylation is measured by a fluorescence-based assay reported before to measure ACAT1 activity 19. Palmitoleoylation by PORCN results in the release of free CoA, which contains a sulfhydryl (−SH) group that reacts with CPM to produce a readily detectable fluorescent signal. The reaction buffer contains 25 mM HEPES pH 7.5, 150 mM NaCl, 4 mM Decyl Maltoside (DM) and 100 μg/mL POPS, which is reported as essential for PORCN activity 25. To measure the PORCN enzyme activity, the assay was carried out in a total volume of 10 μL with a final concentration of 1 μM protein, 50 μM WNT3Ap (home synthesized) and various concentrations (2.5 μM to 60 μM) of palmitoleoyl-CoA. The reaction is initiated by adding PORCN and incubated at 37°C for 30 mins (Fig. 1a). For the LGK974-inhibited group, PORCN enzyme was first treated with LGK974 with 1:50 molar ratio at 37 °C for 5 mins to suppress enzyme activity. The reaction was carried out in a similar way as apo-PORCN group but use LGK974 treated-PORCN instead of apo-PORCN. The reaction mixture was added to 90 μl of 50 μM CPM in CPM buffer (100 mM HEPES, 150 mM NaCl, pH 7.5) and incubated at room temperature for 15 mins. Relative fluorescence intensity was obtained by subtracting the background signal, which is the fluorescence intensity of the LGK974-inhibited group. The data were fitted to the Michaelis-Menten model in GraphPad Prism version 8, obtaining the kinetic parameters as well.
The Fab2C11 effect assay and LGK974 inhibition assay were carried out in a similar way. Apo-PORCN enzyme were either treated with Fab2C11 at a 1:5 molar ratio or treated with LGK974 at a 1:50 molar raio at 37°C for 30 mins. The assay was carried out in a total volume of 10 μL with a final concentration of 1 μM protein, 50 μM WNT3Ap and 50 μM palmitoleoyl-CoA. The reaction is initiated by adding PORCN and incubated at 37°C for 30 mins (Fig. 1b). The reaction mixture is then incubated with 90 μl of 50 μM CPM in CPM buffer at room temperature for 15 mins. Relative fluorescence intensity was obtained by subtracting the fluorescence intensity of the palmitoleoyl-CoA free reaction system.
The activity of PORCN mutants were measure in a similar way. Briefly, the reaction was carried out in 10 μL system with a final concentration of 1 μM wild type or mutant PORCN, 50 μM palmitoleoyl-CoA and 50 μM WNT3Ap. The reaction was initiated by adding PORCN and incubated at 37°C for 30 mins (Fig. 4e). The free-CoA release is measured as the same as above. Relative fluorescence intensity was obtained by subtracting the fluorescence intensity of the palmitoleoyl-CoA free reaction system.
All of the fluorescent signal is detected by BioTek Synergy Neo2 Hybrid Multi-Mode Reader (excitation 405 nm; emission 530 nm). The experiments in Figs. 1a, 1b, 4e and Extended Data Fig. 1b were repeated three times on different days with three independent group for each repeat. Similar results were obtained.
HPLC method for PORCN activity analysis
Solvents A (0.1% trifluoroacetic acid, TFA, in water) and B (0.1% TFA in 90% v/v acetonitrile in water) were used for peptide handling and HPLC. Reactions were stopped by addition of 9x volume of HPLC solvent (A:B = 70:30). 45 μL mixture was resolved on a HPLC column (DiscoveryBIO wide pore C5, 4.6×150mm, 5 μm) using a gradient in which percentage of solvent B increases from 20% to 80% in 6 minutes at 1.5 mL/min. Area of peaks of interest was automatically integrated from 280-nm chromatograms in OpenLab CDS ChemStation (Agilent) using the following parameters: slope sensitivity = 1.0; area reject = 0.2, height reject = 0.1. ESI-MS: m/z calculated for [M+3H]3+ 840.041, found 840.039, and m/z calculated for [palmitoleated WNT3Ap +3H]3+ 918.779, found 918.778.
Synthesis of WNT3Ap
WNT3Ap synthesis was modified from the previous protocol 25. Disulfide formation was monitored by resolving small aliquots on a HPLC column (Agilent Proshell 120 C18, 50×2.1 mm, 2.7 μm, 0.5 mL/min) using a gradient in which percentage of solvent B increases from 22% to 32% in 10 minutes. Eluate was analyzed using MSD (G6125, Agilent) for product identification. Peptide was quantified by absorbance measurement at 280 nm using an extinction coefficient of 11200 M−1cm−1.
Solid-phase peptide synthesis (SPPS) was performed on a Liberty Blue automated peptide synthesizer at 0.25mmol scale on H-Rink-Amide-ChemMatrix resin (Biotage 0.41 mmol/g). The target sequence, corresponding to WNT3A (residues 199–220) was H-MHLKC(Acm)KCHGLSGSCEVKTC(Acm)WWS-NH2. N-(9-fluorenyl)methoxycarbonyl (Fmoc)-protected amino acids (Novabiochem) were coupled using N,N-diisopropylcarbodiimide (DIC, Sigma-Aldrich) and Oxyma Pure (Novabiochem) in N,N-dimethylformamide (DMF, Sigma-Aldrich), and Fmoc deprotection was carried out using 20% (v/v) piperidine (Sigma-Aldrich) in DMF. Default settings of the synthesizer regarding reagent concentration, reagent usage for each coupling, vessel temperature, microwave and reaction time are followed. The peptide was then cleaved and deprotected by 4-hour treatment at room temperature with 20 mL cocktail containing TFA (Sigma-Aldrich) water, phenol (100% w/v in water, Fisher Scientific) and triisopropylsilane (TIPS, Sigma-Aldrich) in a volume ratio of 88:4:6:2. The peptide was precipitated with 180 mL ice-cold diethyl ether (Sigma-Aldrich), thoroughly washed (3×30 mL) with diethyl ether (3×30 mL) and then dissolved in 50 mL HPLC solvent (A:B = 80:20). Leftover resin was removed by filtration and the peptide was lyophilized.
Peptide was redissolved in 20 mL HPLC solvent (A: B = 80: 20) and was diluted in 800 mL buffer containing 50 mM Tris-HCl 8.0, 1 M guanidine hydrochloride (Gu-HCl, VWR), 5% (v/v) DMSO (Sigma Aldrich), and 10% (v/v) acetonitrile. This treatment forms the disulfide bond between Cys205 and Cys212. The mixture was degassed over a Schlenk line and stirred at RT overnight under N2 protection. The reaction was then acidified using TFA to pH = 2 and diluted with 1 liter of HPLC solvent (A: B = 80: 20). Upon passing through a 0.45-μm filter membrane, the mixture was applied to a preparative HPLC column (DiscoveryBIO wide pore C18, 21.2×250mm, 10 μm). Bound peptide was fractionated using a gradient in which percentage of solvent B increases from 20% to 70% in 150mL. Fractions containing the desired, one-disulfide intermediate was lyophilized.
The crude one-disulfide intermediate was dissolved in HPLC solvent B for at 1.0 mM total-peptide concentration. 5 mL of this solution was further diluted to 0.1 mM using acetonitrile containing 0.1% TFA. To deprotect the Acetaminomethyl (Acm) protection groups on Cys203 and Cys217 and form a disulfide between these residues, iodine was added to a final concentration of 0.5 mM. The mixture was incubated at 30°C for 30 minutes and then treated with ascorbic acid at a final concentration of 1.5 mM before subject to a rotary evaporator to remove organic solvents. The residual was taken up in 5 mL HPLC solvent (A: B = 90: 10) and applied to a Mono S 5/50 cation exchange column (Cytiva) operated using CEX Buffer A (100 mM Na-acetate 4.8 and 15% (v/v) acetonitrile) and CEX Buffer B (100 mM Na-acetate 4.8, 1 M NaCl and 15% (v/v) acetonitrile). Bound peptide was eluted using a gradient in which percentage of CEX Buffer B increases from 20% to 100% in 32 mL. Fractions containing WNT3Ap was applied to a semi-prep HPLC column (DiscoveryBIO wide pore C18, 10×250 mm, 5 μm). Bound peptide was fractionated using a gradient in which percentage of solvent B increases from 20% to 35% in 90 mL. Fractions containing pure WNT3Ap was lyophilized. ESI-MS: m/z calculated for [M+3H]3+ (C108H167N32O28S5) 840.041, found 840.039.
Synthesis of pamWNT3Ap
HPLC Solvents and LC-MSD conditions were the same to WNT3Ap synthesis except a gradient in which buffer B percentage increases from 40% to 60% in 10 minutes was used. SPPS was identical to WNT3Ap except Fmoc-Nβ-allyloxycarbonyl-L-2,3-diaminopropionic acid (Fmoc-Dapa(Alloc)-OH, Chempep) was incorporated in place of Fmoc-Ser(tBu)-OH at position 209, and that deprotection of the final Fmoc group was excluded from the automated program. To remove the side-chain Alloc protection group of Dapa209, the resin (0.05 mmol loading) was conditioned using dichloromethane (DCM, Sigma-Aldrich) and treated twice with 598 μL of phenylsilane (4.85 mmol) and 45.63 mg (39.5 μmol) of tetrakis(triphenylphosphine)palladium (Sigma-Aldrich) in 4 mL DCM at RT for 20 minutes. Then the resin was conditioned using a 1:1 (v/v) DCM-DMF mixture, and palmitoleic acid was coupled to the side-chain amine of Dapa209 using 91.2 mg O-(7-Azabenzotriazol-1-yl)-N,N,N’,N’-tetramethyluronium hexafluorophosphate (HATU, P3 Biosystems), 63.5 mg palmitoleic acid and 4.2 μL N,N-Diisopropylethylamine (Sigma-Aldrich) in 1 mL 1:1 DCM-DMF mixture at RT for 1 hour. The N-terminal Fmoc group was then deprotected using 4 mL 20% piperidine in DMF (2 × 10 minutes at RT), and the peptide was then cleaved and deprotected using the same TFA cocktail and condition to WNT3Ap. Cocktail containing peptide was filtered using a fritted syringe into 45mL ice-cold diethyl ether. Precipitated peptide was collected, thoroughly washed (3×10 mL) with diethyl ether, dissolved in 10mL HPLC solvent (A: B = 50: 50), and lyophilized.
To form the disulfide bond between Cys205 and Cys212, the peptide was dissolved at 0.1 mM in a buffer containing 100 mM Tris-HCl 8.0, 6 M Gu-HCl and 10% (v/v) DMSO and stirred overnight under N2 protection. The reaction was then acidified using TFA to pH 2, diluted with equal volume of a solution containing 2 M Gu-HCl, 20% acetonitrile and 0.1% TFA. The solution was filtered, and filtrate was applied to a Semi-prep column (DiscoveryBIO wide pore C5, 10×250 mm, 5 μm). Bound peptide was fractionated using a gradient in which percentage of solvent B increases from 30% to 60% in 60 mL.
Iodine treatment procedures to form the disulfide between Cys203 and Cys217 were identical to those that gave rise to WNT3Ap. The residual following rotary evaporation was taken up in 1 mL 6 M GuHCl containing 1% TFA and applied to a semi-prep column (DiscoveryBIO wide pore C5, 10×250 mm, 5 μm). Bound peptide was fractionated using a gradient in which percentage of solvent B increases from 40% to 60% in 90 mL. Fractions containing pure pamWNT3Ap was lyophilized. ESI-MS: m/z calculated for [M+3H]3+ (C124H196N33O28S5) 918.451, found 918.446.
Construction of model
The PORCN protein structure was modeled as follows. The atomic coordinates were constructed from the 7URA. Hydrogen atoms were added using H-build from CHARMM 56. A disulfide bond was constructed between Cys17 and Cys209. The amino acid sidechains coordinated to the zinc ion were modeled as follows: His382 was protonated at Nε and Cys370, Cys376, and Cys380 were modeled with anionic sidechains. An initial protonation pattern was obtained by calculating the pKa values of all titratable residues. For this, electrostatic energy computations were carried out with PROPKA 57. Based on these results, a model was built to represent the protonation pattern at pH 7. This model included a protonated amino acid sidechain of Asp413.
Next, the protein structure was modeled in the lipid bilayer of the ER. The transmembrane domain (TMD) helices were inserted into the lipid bilayer using the CHARMM-GUI 58 and OPM database 59. The lipid bilayer consists of an upper leaflet (direction of cytosol) and a lower leaflet (direction of extracellular space). Both upper and lower leaflets contain a 10: 4: 2: 1: 1 mixture of didecanoyl-D-glycero-3-Phosphatidylcholine (DDPC): dilauroyl-D-glycero-1-Phosphatidylethanolamine (DLPE): di-myristoyl-inositol (DMPI): dilauroyl-D-glycero-1-Phosphatidylserine (DLPS): cholesterol, respectively. The protein-membrane system was next solvated in water with 0.15 M KCl concentration, resulting in K+ (50) and Cl− (36) ions to neutralize charge, and 12,861 explicit TIP3 water molecules 60. The total system consists of 59,914 atoms and was simulated in a rectangular box of dimension 79.4 Å × 79.4 Å × 106.7 Å.
The initial coordinates of the palmitoleoyl-CoA molecule in “curled-up” and “curled-down” configurations were taken from 7URA. Avogadro (http://avogadro.cc/) was used to build hydrogen atoms on palmitoleoyl-CoA, and the simulation parameters for CoA were obtained using the CGenFF program 61. A simulation model was constructed for the PORCN-WNT complex; the atomic coordinates were predicted from AlphaFold. Hydrogen atoms were added using H-build from CHARMM 56. The following twelve disulfide bonds were constructed: Cys42-Cys56, Cys77-Cys88, Cys128-Cys136, Cys138-Cys155, Cys203-Cys217, Cys205-Cys212, Cys281-Cys312, Cys297-Cys307, Cys311-Cys351, Cys327-Cys342, Cys329-Cys339, Cys334-Cys335. The composition of the lipid bilayer, as well as the position of PORCN’s TMD in the bilayer, is identical to that in the model without WNT. The protein-membrane system was next solvated in water with 0.15 M KCl concentration, resulting in K+ (211) and Cl− (153) ions to neutralize charge, and 53,357 explicit TIP3 water molecules 60. The total system consists of 216,448 atoms and was simulated in a rectangular box of dimension 126.7 Å × 126.7 Å × 148.7 Å.
Geometry optimizations and molecular dynamics
The initial geometry of each solvated protein-membrane complex was optimized with 10,000 steps of conjugate gradient energy minimization to remove any close contacts. All energy minimizations used the all-atom CHARMM36 parameter set for the protein, lipid, and ion molecules 62 and the TIP3P model for water molecules 60. Next, the solvated protein-membrane complex was simulated with molecular dynamics (MD) at 310 K according to the following protocol with NAMD 63: 1) equilibration MD with Langevin dynamics (time step of 1 fs) for 375 ps followed by Langevin dynamics (time step 2 fs) for 1.5 ns; 2) production MD with Langevin dynamics (time step 2 fs) for 100 ns. To simulate a continuous system, periodic boundary conditions were applied. Electrostatic interactions were summed with the Particle Mesh Ewald method 64 (grid spacing ~0.88 Å; for PORCN fftx 90, ffty 90, fftz 120 and for PORCN-WNT fftx 144, ffty 144, fftz 160). A nonbonded cutoff of 16.0 Å was used, and Heuristic testing was performed at each energy call to evaluate whether the non-bonded pair list should be updated.
Interaction energy calculations
The interaction energy (electrostatic and van der Waal energies) between the palmitoleoyl-CoA and the surrounding amino acids in PORCN was calculated for the “curled-up” and “curled-down” CoA conformations at regular intervals of each 100 ns simulation and averaged. The energies were calculated using the INTER module in CHARMM with the CHARMM36 parameter set 62. The following amino acids of PORCN were included in the selection: Phe246, His247, Val294, Val296, Val297, Thr298, Trp300, Asn301, Met304, Ser305, Leu308, Asn309, Phe313, Thr321, Val325, Thr328, Tyr329, Ser332, His336, Leu342, Val345, Leu346, Leu349, Thr353, Tyr354, His357, Lys361, Lys374, Leu405, Leu408, and Phe412.
Data Availability
Sequences of the anti-PORCN antibody candidates were analyzed with the IMGT database (http://www.imgt.org/). The OPM database (https://opm.phar.umich.edu/) was used to insert PORCN into the lipid bilayer for MD simulation. The 3D cryo-EM maps have been deposited in the Electron Microscopy Data Bank under the accession numbers 26707, 726708, 26709, 26710 and 26711. Atomic coordinates for the atomic model have been deposited in the Protein Data Bank under the accession numbers 7URA, 7URC, 7URD, 7URE and 7URF. Additional data supporting the findings in this study are provided as source data and supplementary information to this manuscript.
Extended Data
Extended Data Fig. 1. LC-MS analysis of PORCN-mediated WNT3Ap acylation and MS analysis of pamWNT3Ap.
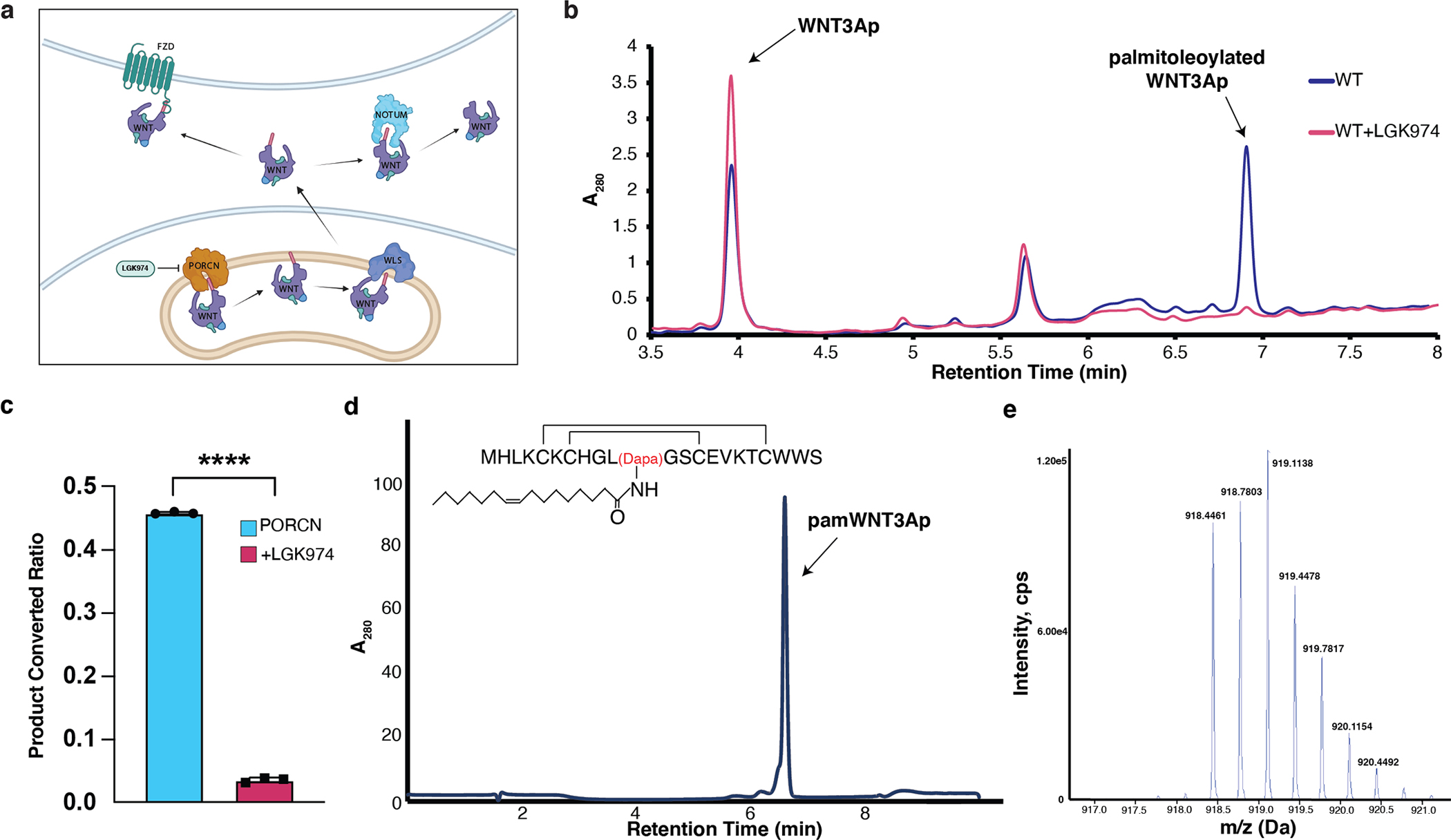
a, The secretion pathway of WNT ligand. In the endoplasmic reticulum (yellow), WNT (purple) is modified by PORCN (orange). Transport of the modified WNT is mediated by WLS (blue). WNTs are secreted to extracellular space via vesicles. After binding to FZD (green), WNTs trigger the signal transduction. Notum (light blue) acts as a deacylase, removing the lipid of WNT to abolish the signal. The structure of palmitoleated WNT3A hairpin 2 is shown. b, Representative LC chromatograms of PORCN-mediated WNT3Ap acylation with (red) and without (blue) LGK974. ESI-MS: m/z calculated for [M+3H]3+ 840.041, found 840.039, and m/z calculated for [palmitoleated WNT3Ap+3H]3+ 918.779, found 918.778. c, Quantitative analysis of the product ratio after the reaction. Data are mean ± s.d. (n=3 biologically independent experiments). ****P ≤ 0.0001, two-tailed unpaired t-test using GraphPad Prism 8. d, LC analysis of pamWNT3Ap. The samples (panels b and d) were applied to a HPLC column (DiscoveryBIO wide pore C5, 4.6×150mm, 5 μm) using a gradient in which percentage of solvent B increases from 20% to 80% in 6 minutes at 1.5 mL/min. e, Mass spectrometry analysis of [pamWNT3Ap+3H]3+. ESI-MS: m/z calculated for [M+3H]3+ (C124H196N33O28S5) 918.451, found 918.446.
Extended Data Fig. 2. Protein Purification and data processing of palmitoleoyl-CoA-bound and LGK974-bound PORCN.
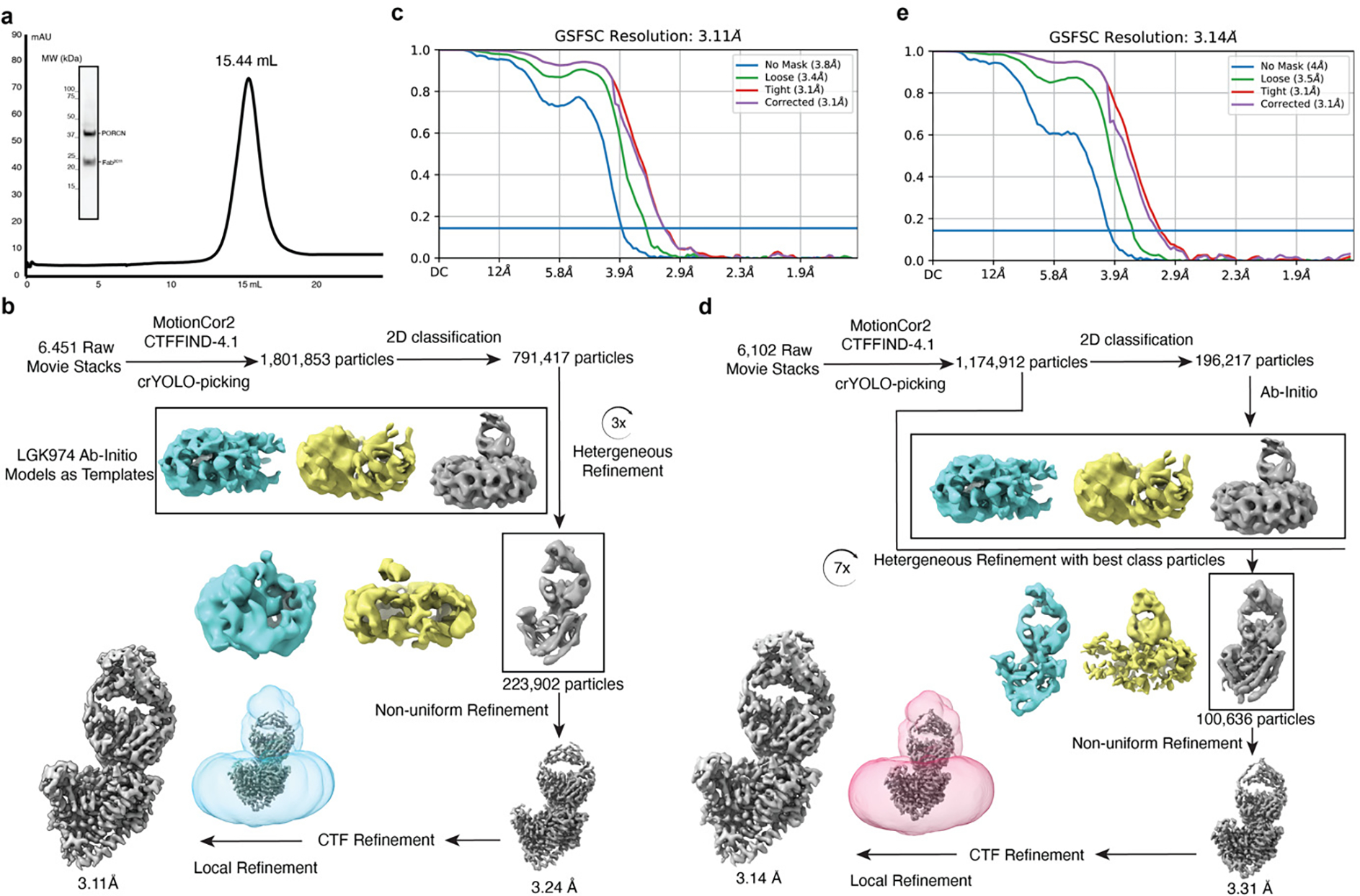
a, Representative Superose 6 increase 10/300 GL gel-filtration chromatogram of PORCN complex with Fab2C11. The peak fraction is shown on SDS-PAGE with molecular markers. Each protein is indicated. For gel source data, see Supplementary Fig. 1a. b, The data processing of palmitoleoyl-CoA-bound PORCN. The cryo-EM 3D classes as well as the mask used for the refinement are shown. The final cryo-EM map after cryoSPARC refinement was sharpened with a B-factor value of −170 Å2. c, Fourier shell correlation (FSC) curve of palmitoleoyl-CoA-bound PORCN map as a function of resolution using cryoSPARC output. d, The data processing of LGK974-bound PORCN. The cryo-EM 3D classes as well as the mask used for the refinement are shown. The final cryo-EM map after cryoSPARC refinement was sharpened with a B-factor value of −130 Å2. e, Fourier shell correlation (FSC) curve of LGK974-bound PORCN map as a function of resolution using cryoSPARC output.
Extended Data Fig. 3. cryo-EM map of structural elements of palmitoleoyl-CoA-bound and LGK974-bound PORCN.
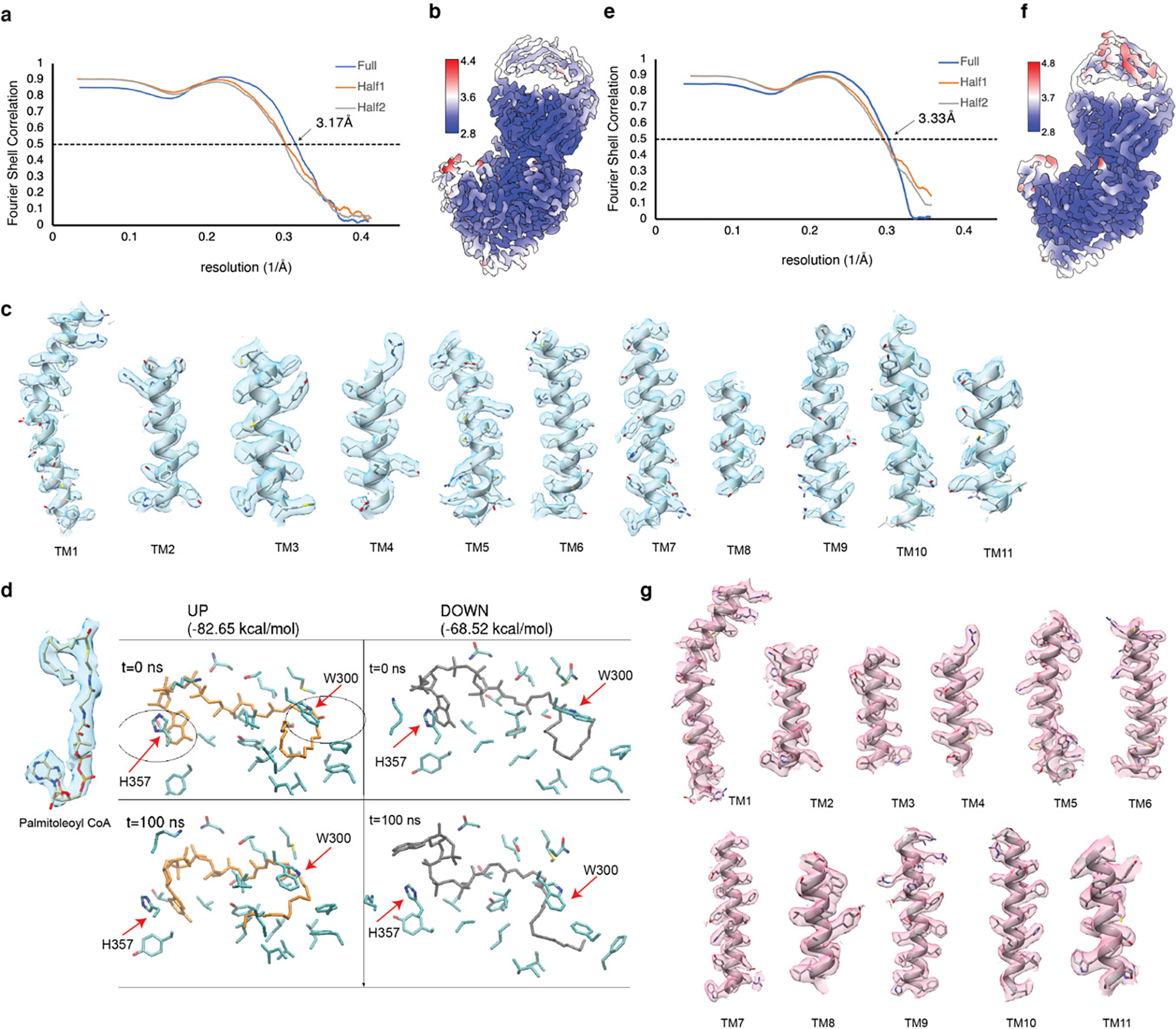
a, The Fourier shell correlation (FSC) curves calculated between the refined structure model and the half map used for refinement (yellow), the other half map (gray) and the full map (blue) of palmitoleoyl-CoA-bound PORCN. b, Density map colored by local resolution estimation using cryoSPARC. c, The major helices of PORCN. d, MD simulation suggests that palmitoleoyl-CoA binds to PORCN in the curled-up conformation. After 100 ns simulations, the interaction between residues W300, H357 and the curled-down palmitoleoyl-CoA disrupts (the right bottom panel). The palmitoleoyl-CoA in either conformation with the cryo-EM map is shown. e, The Fourier shell correlation (FSC) curves calculated between the refined structure model and the half map used for refinement (yellow), the other half map (gray) and the full map (blue) of palmitoleoylated WNT3A-bound PORCN. f, Density map colored by local resolution estimation using cryoSPARC. g, The major helices of PORCN.
Extended Data Fig. 4. Data processing of WNT3Ap/LGK974-bound PORCN and pamWNT3Ap-bound PORCN.
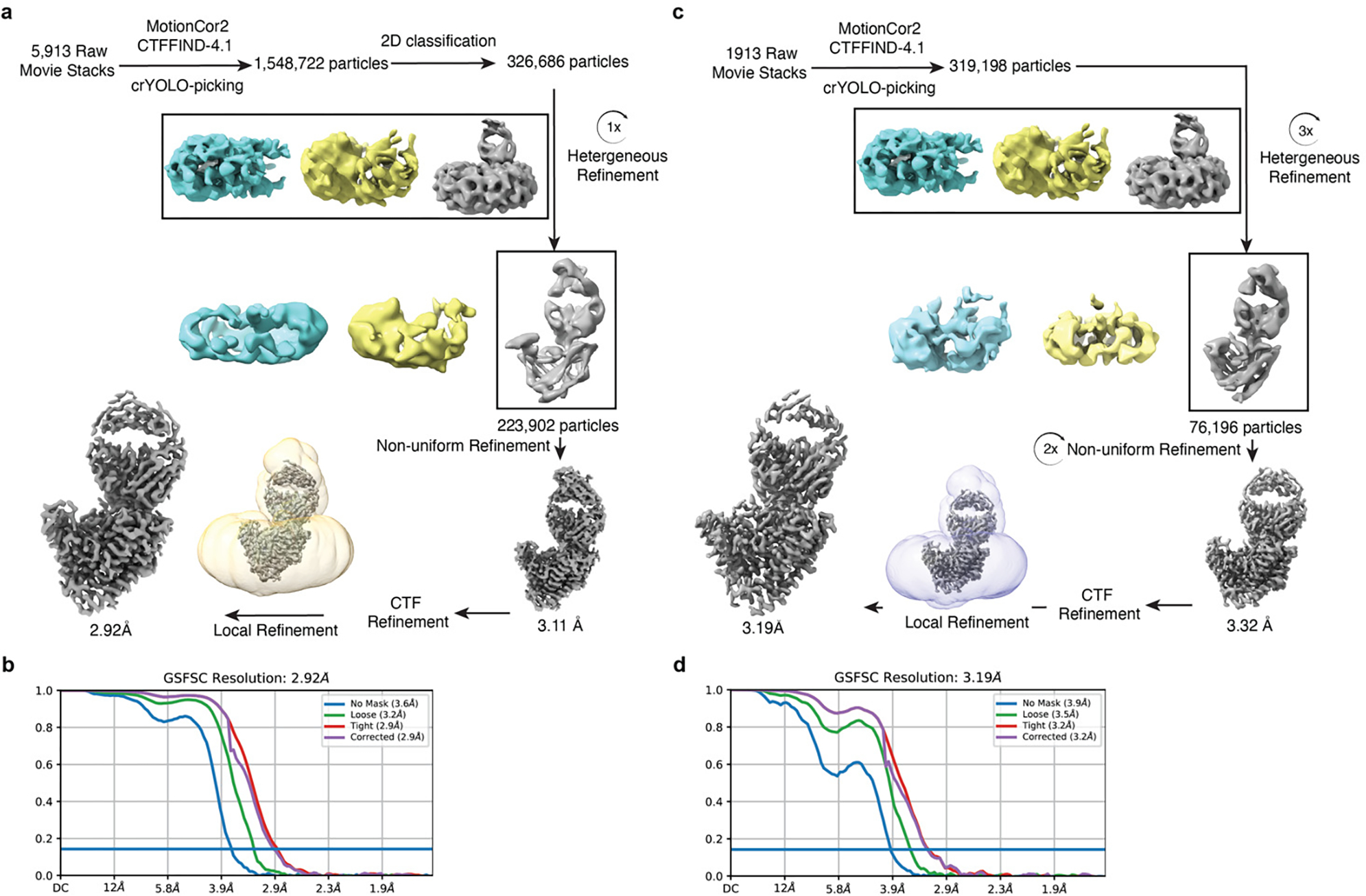
a, The data processing of WNT3Ap/LGK974-bound PORCN. The cryo-EM 3D classes as well as the mask used for the refinement are shown. The final cryo-EM map after cryoSPARC refinement was sharpened with a B-factor value of −147 Å2. b, Fourier shell correlation (FSC) curve of WNT3Ap/LGK974-bound PORCN map as a function of resolution using cryoSPARC output. c, The data processing of pamWNT3Ap-bound PORCN. The cryo-EM 3D classes as well as the mask used for the refinement are shown. The final cryo-EM map after cryoSPARC refinement was sharpened with a B-factor value of −140 Å2. d, Fourier shell correlation (FSC) curve of pamWNT3Ap-bound PORCN map as a function of resolution using cryoSPARC output.
Extended Data Fig. 5. cryo-EM map of structural elements of WNT3Ap/LGK974-bound PORCN and pamWNT3Ap-bound PORCN.
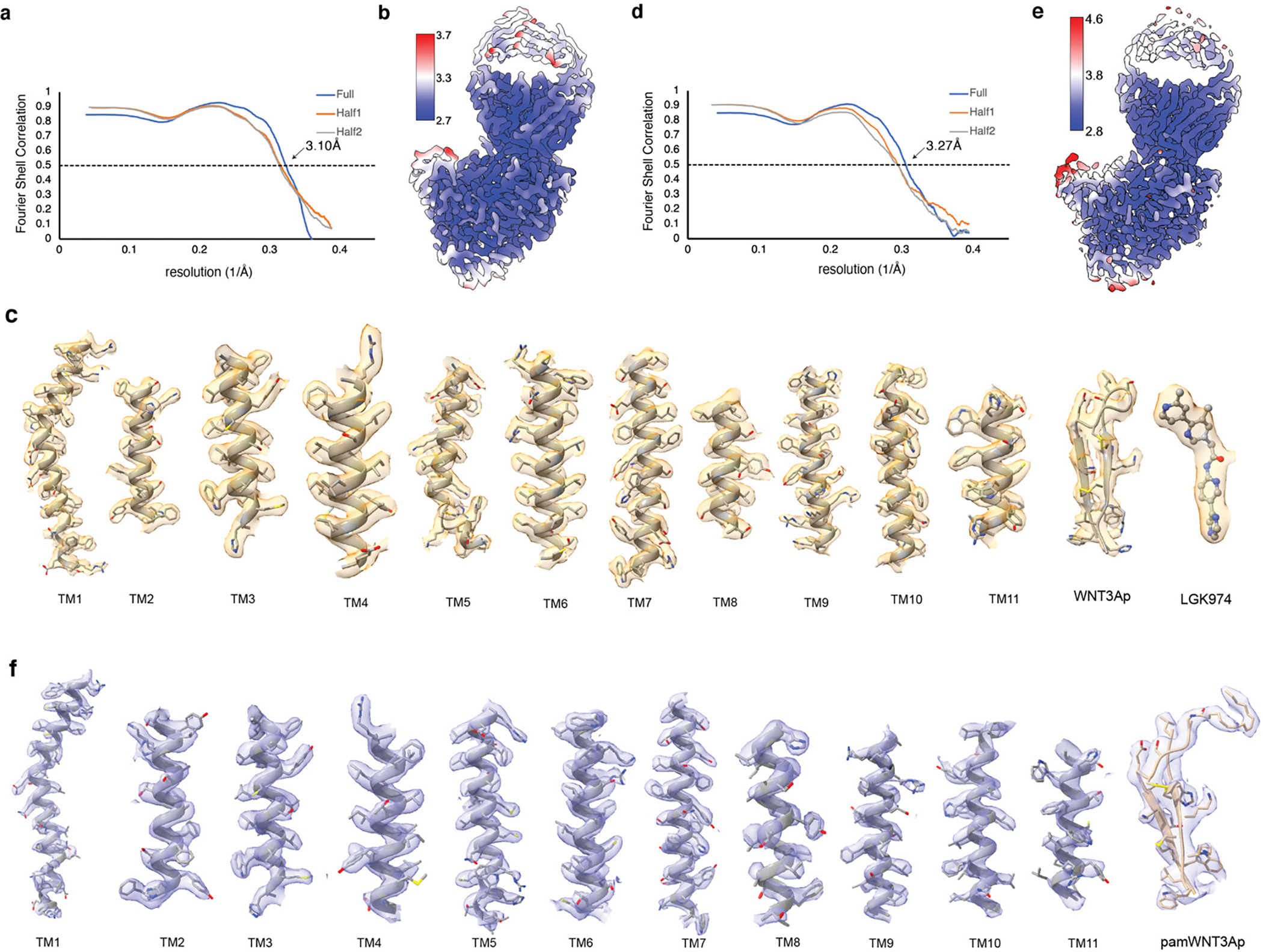
a, The Fourier shell correlation (FSC) curves calculated between the refined structure model and the half map used for refinement (yellow), the other half map (gray) and the full map (blue) of WNT3Ap/LGK974-bound PORCN. b, Density map colored by local resolution estimation using cryoSPARC. c, The major helices of PORCN, WNT3Ap and LGK974. d, The Fourier shell correlation (FSC) curves calculated between the refined structure model and the half map used for refinement (yellow), the other half map (gray) and the full map (blue) of pamWNT3Ap-bound PORCN. e, Density map colored by local resolution estimation using cryoSPARC. f, The major helices of PORCN and pamWNT3Ap.
Extended Data Fig. 6. The structure of DHHC20 and the structural comparison of PORCN (gray) with the other MBOAT proteins.
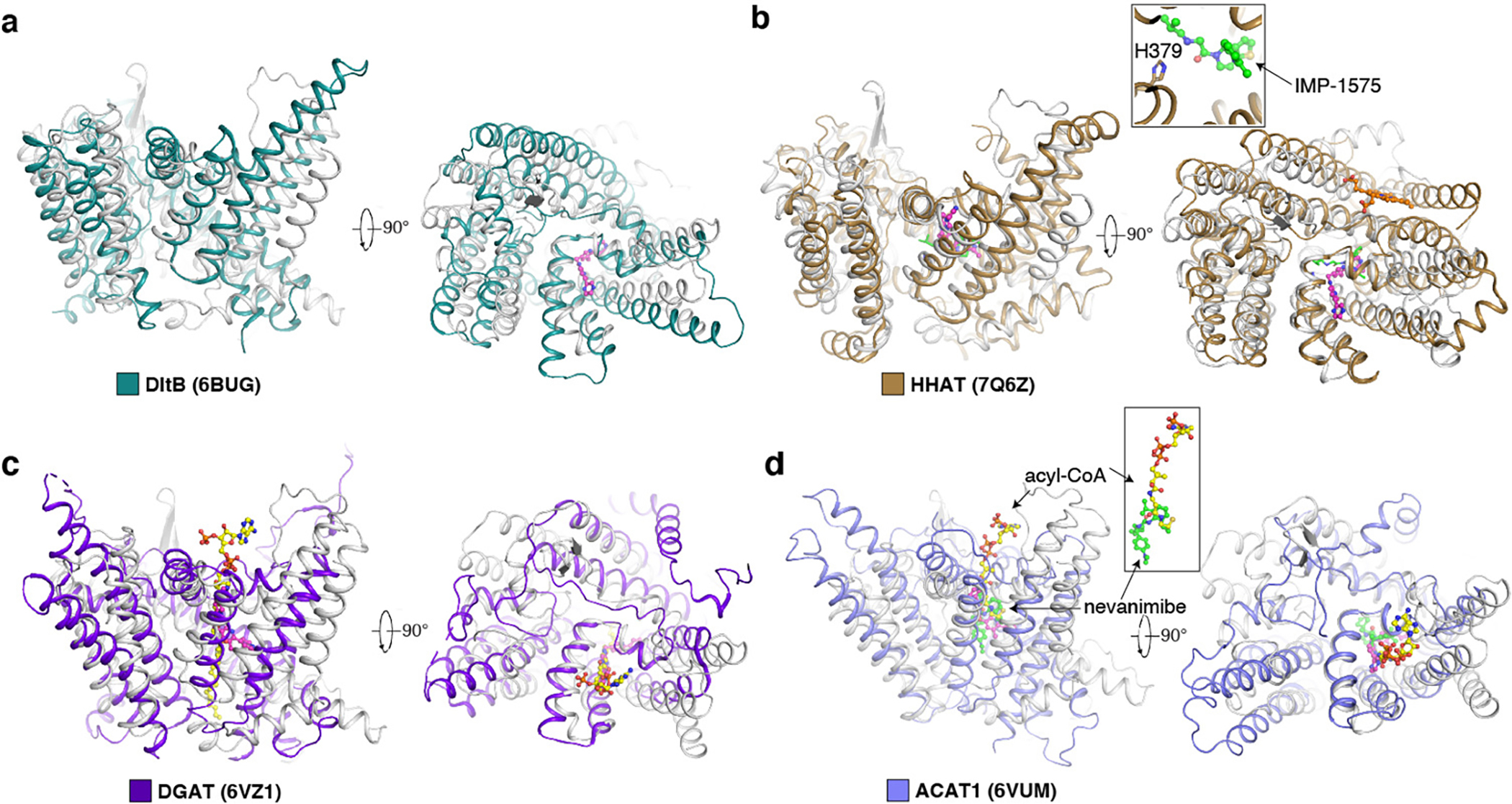
a, The structure of DHHC20. The zinc ions are shown as gray balls. b, The structural comparison with DltB. c, The structural comparison with HHAT. IMP-1575, an HHAT inhibitor, is shown as green sticks. The catalytic H379 of HHAT is shown as sticks. d, The structural comparison with DGAT1. e, The structural comparison with ACAT1. The LGK974 is shown as magenta sticks. Nevanimibe, an ACAT1 inhibitor, is shown as green sticks. The acyl-CoA in the DGAT and ACAT1 is shown as yellow sticks.
Extended Data Fig. 7. Purification of human PORCN variants for activity assays.
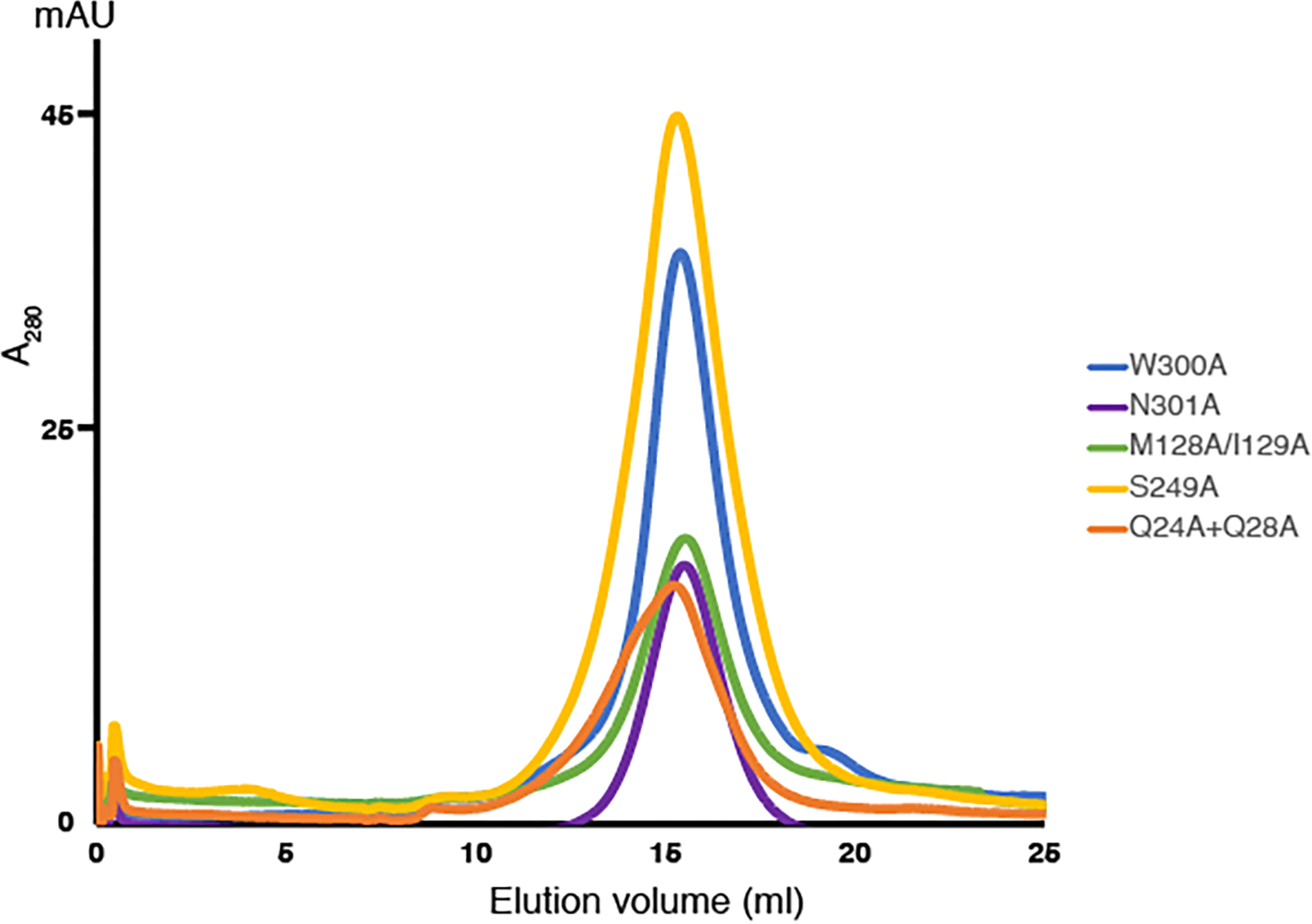
Representative Superose 6 increase 10/300 GL gel-filtration chromatogram of PORCN variants in buffer containing 20 mM HEPES, pH 7.5, 150 mM NaCl, and 0.06% Digitonin.
Extended Data Fig. 8. Superimposing the WNT8A-FZD and WNT8A-WLS complexes into the structure of pamWNT3Ap-bound PORCN and the model of WNT3A-bound PORCN.
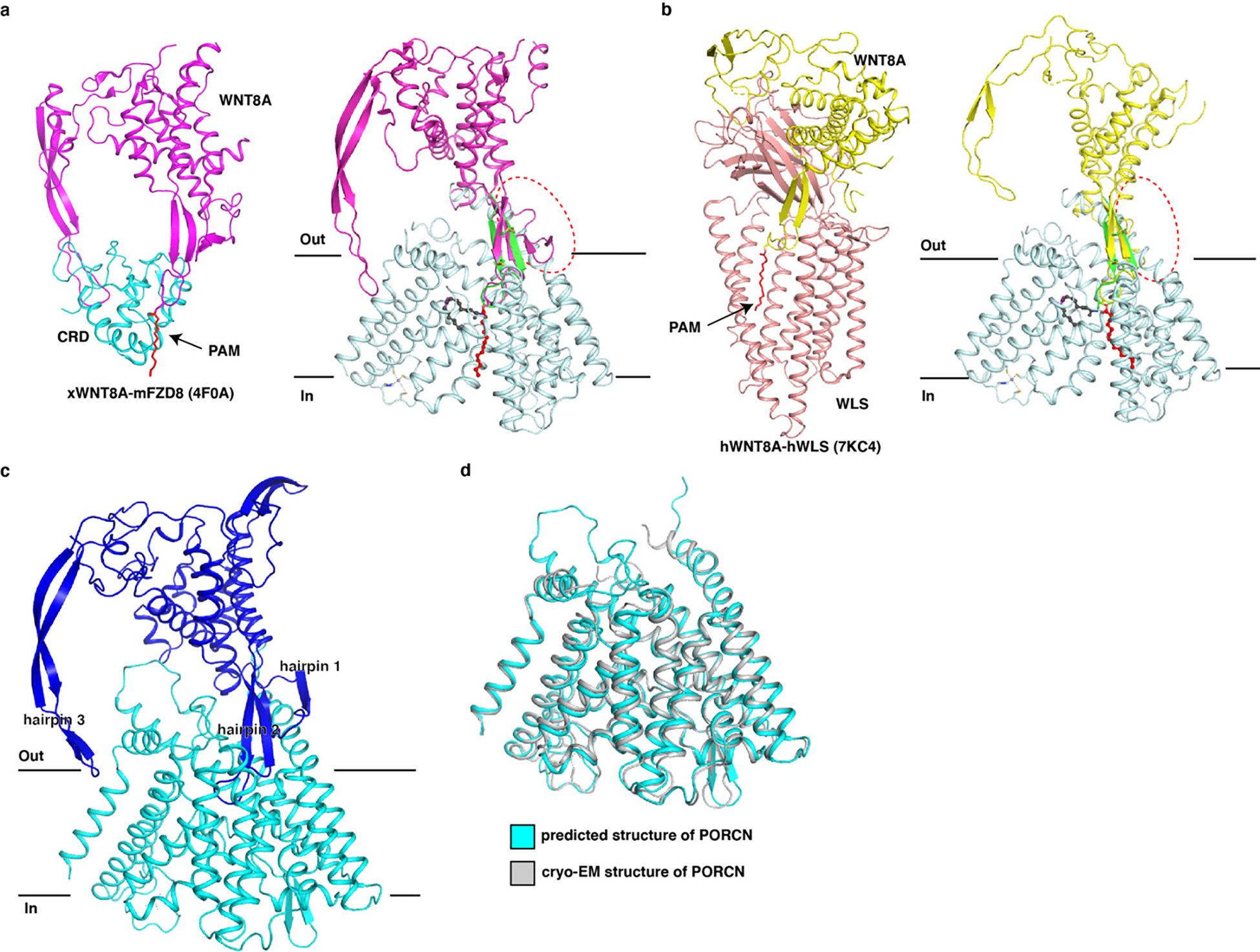
a, Superimposing the hairpin 2 of the structure of WNT8A-FZD complex into PORCN-pamWNT3Ap complex. The clash between WNT8A ligand and PORCN is indicated by a dashed circle. b, Superimposing the hairpin 2 of the structure of WNT8A-WLS complex into PORCN-pamWNT3Ap complex. The clash between WNT8A ligand and PORCN is indicated by a dashed circle. PAM, palmitoleic acid (red sticks). c, The predicted structure of WNT3A-bound PORCN. Three hairpins of WNT3A have been indicated. d, Structural comparison of the predicted PORCN structure to the cryo-EM structure (PDB:7URA).
Extended Data Fig. 9. The structure of HHAT-SHH-N complex.
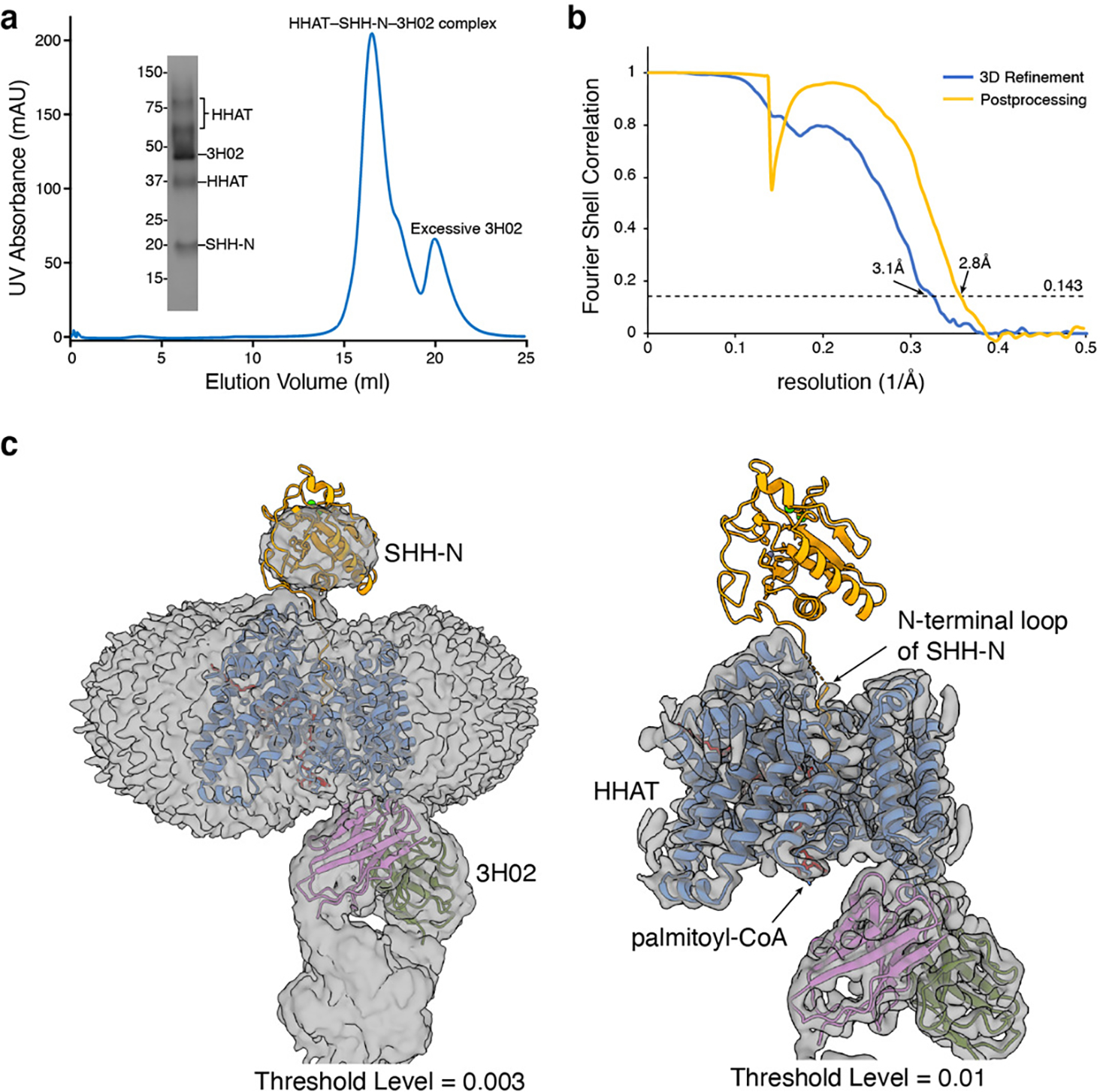
a, Representative Superose 6 increase 10/300 GL gel-filtration chromatogram of HHAT-SHH-N complex with Fab3H02. The peak fraction is shown on SDS-PAGE with molecular markers. Each protein is indicated. For gel source data, see Supplementary Fig. 1b. b, Fourier shell correlation (FSC) curve as a function of resolution using RELION-3 output. c, Cryo-EM map of the complex after 3D refinement reveals the mean body of SHH-N protein at the threshold level of 0.003 but not 0.01.
Extended Data Table. 1.
Cryo-EM data collection, refinement and validation statistics
| Palmitoleoyl-CoA-PORCN (EMDB-26707) (PDB-7URA) |
LGK974-PORCN (EMDB-26708) (PDB-7URC) |
LGK974- WNT3Ap- PORCN (EMDB-26709) (PDB-7URD) |
pamWNT3Ap- PORCN (EMDB-26710) (PDB-7URE) |
SHH-N-HHAT (EMDB-26711) (PDB-7URF) |
|
|---|---|---|---|---|---|
|
| |||||
| Data collection and processing | 80 | 79 | 78 | 73 | 74 |
| Magnification | 60024 | 60024 | 60024 | 60024 | 59382 |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 |
| Electron exposure (e-/Å2 | 60 | 60 | 60 | 60 | 60 |
| Defocus range (μm) | −1.0 to −2.0 | −1.0 to −2.0 | −1.0 to −2.0 | −0.7 to −2.0 | −0.8 to −2.0 |
| Pixel size (Å) | 0.83 | 0.83 | 0.83 | 0.83 | 0.842 |
| Symmetry imposed | C1 | C1 | C1 | C1 | C1 |
| Initial particle images (no.) | 1,801,853 | 1,174,912 | 1,548,772 | 475,594 | 1,524,337 |
| Final particle images (no.) | 223,902 | 100,636 | 397,217 | 113,566 | 274,370 |
| Map resolution (Å) | 3.11 | 3.14 | 2.92 | 3.19 | 2.8 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
|
| |||||
| Refinement | |||||
| Initial model used (PDB code) | AlphaFold2 | 7URA | 7URA | 7URA | 7MHZ, 6E1H |
| Model resolution (Å) | 3.17 | 3.33 | 3.10 | 3.27 | 2.84 |
| FSC threshold | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Map sharpening B factor (Å2) | −170 | −130 | −146.7 | −140 | −93.6 |
| Non-hydrogen atoms | 5400 | 5561 | 5534 | 5450 | 6133 |
| Protein residues | 658 | 658 | 678 | 678 | 722 |
| Ligands | 6 | 12 | 6 | 3 | 8 |
| B factors (Å2) | |||||
| Protein | 56.27 | 88.66 | 73.03 | 58.93 | 54.66 |
| Ligand | 78.36 | 105.44 | 83.52 | 72.64 | 58.13 |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.005 | 0.004 | 0.005 | 0.004 | 0.003 |
| Bond angles (°) | 0.737 | 0.680 | 0.794 | 0.667 | 0.641 |
| Validation | |||||
| MolProbity score | 1.91 | 1.80 | 1.92 | 1.91 | 1.48 |
| Clashscore | 13.09 | 11.35 | 11.46 | 9.91 | 5.50 |
| Poor rotamers (%) | 0 | 0 | 0 | 0 | 0 |
| Ramachandran plot | |||||
| Favored (%) | 95.83 | 96.45 | 95.05 | 94.27 | 96.91 |
| Allowed (%) | 4.17 | 3.55 | 4.95 | 5.73 | 3.09 |
| Disallowed (%) | 0 | 0 | 0 | 0 | 0 |
Supplementary Material
The PORCN is colored in cyan and the WNT3A is colored in blue. The disulfide bonds are shown in sticks.
The PORCN-TM11 is colored in red.
Acknowledgements
Cryo-EM Data were collected at the UT Southwestern Medical Center Cryo-EM Facility (funded in part by the CPRIT Award RP170644). We thank D. Stoddard, L. Friedberg, L. Esparza and Y. Qin for technical support, A. Lemoff at the UTSW Proteomics Core for mass spectrometry analysis, and C. Chattergee and Z. Chen for discussion, E. Debler and P. Schmiege for editing the manuscript. This work was supported by NIH P01 HL020948, R01 GM135343 and Welch Foundation (I-1957) (to X.L.). N.E.-M. was funded in part by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—EXC 2008-390540038-UniSysCat. X.Q. is a recipient of DDBrown Fellowship of Life Sciences Research Foundation. B.W. is a Southwestern Medical Foundation Scholar in Biomedical Research. X.L. is a Damon Runyon-Rachleff Innovator supported by the Damon Runyon Cancer Research Foundation (DRR-53S-19) and a Rita C. and William P. Clements Jr. Scholar in Biomedical Research at UT Southwestern Medical Center.
Footnotes
Competing interests: The authors declare no competing interests.
References:
- 1.Nusse R & Clevers H Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 169, 985–999, doi: 10.1016/j.cell.2017.05.016 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Taipale J & Beachy PA The Hedgehog and Wnt signalling pathways in cancer. Nature 411, 349–354, doi: 10.1038/35077219 (2001). [DOI] [PubMed] [Google Scholar]
- 3.MacDonald BT, Tamai K & He X Wnt/beta-catenin signaling: components, mechanisms, and diseases. Developmental cell 17, 9–26, doi: 10.1016/j.devcel.2009.06.016 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhan T, Rindtorff N & Boutros M Wnt signaling in cancer. Oncogene 36, 1461–1473, doi: 10.1038/onc.2016.304 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willert K et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423, 448–452, doi: 10.1038/nature01611 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Zhai L, Chaturvedi D & Cumberledge S Drosophila wnt-1 undergoes a hydrophobic modification and is targeted to lipid rafts, a process that requires porcupine. The Journal of biological chemistry 279, 33220–33227, doi: 10.1074/jbc.M403407200 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Hofmann K A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends in biochemical sciences 25, 111–112, doi: 10.1016/s0968-0004(99)01539-x (2000). [DOI] [PubMed] [Google Scholar]
- 8.Janda CY, Waghray D, Levin AM, Thomas C & Garcia KC Structural basis of Wnt recognition by Frizzled. Science 337, 59–64, doi: 10.1126/science.1222879 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodon J et al. Phase 1 study of single-agent WNT974, a first-in-class Porcupine inhibitor, in patients with advanced solid tumours. Br J Cancer 125, 28–37, doi: 10.1038/s41416-021-01389-8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nusse R Wnts and Hedgehogs: lipid-modified proteins and similarities in signaling mechanisms at the cell surface. Development 130, 5297–5305, doi: 10.1242/dev.00821 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Qi X & Li X Mechanistic Insights into the Generation and Transduction of Hedgehog Signaling. Trends in biochemical sciences 45, 397–410, doi: 10.1016/j.tibs.2020.01.006 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi X, Schmiege P, Coutavas E, Wang J & Li X Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 560, 128–132, doi: 10.1038/s41586-018-0308-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi X, Schmiege P, Coutavas E & Li X Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science, doi: 10.1126/science.aas8843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Proffitt KD & Virshup DM Precise regulation of porcupine activity is required for physiological Wnt signaling. The Journal of biological chemistry 287, 34167–34178, doi: 10.1074/jbc.M112.381970 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Routledge D & Scholpp S Mechanisms of intercellular Wnt transport. Development 146, doi: 10.1242/dev.176073 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Kadowaki T, Wilder E, Klingensmith J, Zachary K & Perrimon N The segment polarity gene porcupine encodes a putative multitransmembrane protein involved in Wingless processing. Genes & development 10, 3116–3128, doi: 10.1101/gad.10.24.3116 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Resh MD Targeting protein lipidation in disease. Trends Mol Med 18, 206–214, doi: 10.1016/j.molmed.2012.01.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian H et al. Structural basis for catalysis and substrate specificity of human ACAT1. Nature 581, 333–338, doi: 10.1038/s41586-020-2290-0 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Long T, Sun Y, Hassan A, Qi X & Li X Structure of nevanimibe-bound tetrameric human ACAT1. Nature 581, 339–343, doi: 10.1038/s41586-020-2295-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L et al. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature 581, 329–332, doi: 10.1038/s41586-020-2280-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sui X et al. Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature 581, 323–328, doi: 10.1038/s41586-020-2289-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan C et al. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nature communications 11, 2478, doi: 10.1038/s41467-020-16288-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long T, Liu Y & Li X Molecular structures of human ACAT2 disclose mechanism for selective inhibition. Structure, doi: 10.1016/j.str.2021.07.009 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y, Benz TL & Long SB Substrate and product complexes reveal mechanisms of Hedgehog acylation by HHAT. Science 372, 1215–1219, doi: 10.1126/science.abg4998 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee CJ, Rana MS, Bae C, Li Y & Banerjee A In vitro reconstitution of Wnt acylation reveals structural determinants of substrate recognition by the acyltransferase human Porcupine. The Journal of biological chemistry 294, 231–245, doi: 10.1074/jbc.RA118.005746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grzeschik KH et al. Deficiency of PORCN, a regulator of Wnt signaling, is associated with focal dermal hypoplasia. Nature genetics 39, 833–835, doi: 10.1038/ng2052 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Wang X et al. Mutations in X-linked PORCN, a putative regulator of Wnt signaling, cause focal dermal hypoplasia. Nature genetics 39, 836–838, doi: 10.1038/ng2057 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Liu J et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proceedings of the National Academy of Sciences of the United States of America 110, 20224–20229, doi: 10.1073/pnas.1314239110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang X et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America 110, 12649–12654, doi: 10.1073/pnas.1307218110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madan B et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene 35, 2197–2207, doi: 10.1038/onc.2015.280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asciolla JJ, Miele MM, Hendrickson RC & Resh MD An in vitro fatty acylation assay reveals a mechanism for Wnt recognition by the acyltransferase Porcupine. The Journal of biological chemistry 292, 13507–13513, doi: 10.1074/jbc.C117.800136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rana MS et al. Fatty acyl recognition and transfer by an integral membrane S-acyltransferase. Science 359, doi: 10.1126/science.aao6326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coupland CE et al. Structure, mechanism, and inhibition of Hedgehog acyltransferase. Molecular cell 81, 5025–5038 e5010, doi: 10.1016/j.molcel.2021.11.018 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holm L & Rosenstrom P Dali server: conservation mapping in 3D. Nucleic acids research 38, W545–549, doi: 10.1093/nar/gkq366 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma D et al. Crystal structure of a membrane-bound O-acyltransferase. Nature 562, 286–290, doi: 10.1038/s41586-018-0568-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rios-Esteves J & Resh MD Stearoyl CoA desaturase is required to produce active, lipid-modified Wnt proteins. Cell reports 4, 1072–1081, doi: 10.1016/j.celrep.2013.08.027 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu J et al. Structural model of human PORCN illuminates disease-associated variants and drug-binding sites. Journal of cell science 134, doi: 10.1242/jcs.259383 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Kakugawa S et al. Notum deacylates Wnt proteins to suppress signalling activity. Nature 519, 187–192, doi: 10.1038/nature14259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rios-Esteves J, Haugen B & Resh MD Identification of key residues and regions important for porcupine-mediated Wnt acylation. The Journal of biological chemistry 289, 17009–17019, doi: 10.1074/jbc.M114.561209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nygaard R et al. Structural Basis of WLS/Evi-Mediated Wnt Transport and Secretion. Cell 184, 194–206 e114, doi: 10.1016/j.cell.2020.11.038 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jumper J et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589, doi: 10.1038/s41586-021-03819-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuladhar R et al. Stereoselective fatty acylation is essential for the release of lipidated WNT proteins from the acyltransferase Porcupine (PORCN). The Journal of biological chemistry 294, 6273–6282, doi: 10.1074/jbc.RA118.007268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Y et al. Molecular basis of cholesterol efflux via ABCG subfamily transporters. Proceedings of the National Academy of Sciences of the United States of America 118, doi: 10.1073/pnas.2110483118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q et al. A Combination of Human Broadly Neutralizing Antibodies against Hepatitis B Virus HBsAg with Distinct Epitopes Suppresses Escape Mutations. Cell Host Microbe 28, 335–349 e336, doi: 10.1016/j.chom.2020.05.010 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51, doi: 10.1016/j.jsb.2005.07.007 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Li X et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, doi: 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner T et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221, doi: 10.1107/S0907444909052925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Pettersen EF et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82, doi: 10.1002/pro.3943 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brooks BR et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. Journal of computational chemistry 4, 30 (1983). [Google Scholar]
- 57.Li H, Robertson AD & Jensen JH Very fast empirical prediction and rationalization of protein pKa values. Proteins 61, 704–721, doi: 10.1002/prot.20660 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Jo S, Kim T, Iyer VG & Im W CHARMM-GUI: a web-based graphical user interface for CHARMM. Journal of computational chemistry 29, 1859–1865, doi: 10.1002/jcc.20945 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Lomize MA, Lomize AL, Pogozheva ID & Mosberg HI OPM: orientations of proteins in membranes database. Bioinformatics 22, 623–625, doi: 10.1093/bioinformatics/btk023 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Jorgensen W, Chandrasekhar J, Madura J, Impey R & Klein M Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics 79, 9 (1983). [Google Scholar]
- 61.Vanommeslaeghe K et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. Journal of computational chemistry 31, 671–690, doi: 10.1002/jcc.21367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacKerell AD et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102, 3586–3616, doi: 10.1021/jp973084f (1998). [DOI] [PubMed] [Google Scholar]
- 63.Phillips JC et al. Scalable molecular dynamics with NAMD. Journal of computational chemistry 26, 1781–1802, doi: 10.1002/jcc.20289 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Essmann U et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 16 (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The PORCN is colored in cyan and the WNT3A is colored in blue. The disulfide bonds are shown in sticks.
The PORCN-TM11 is colored in red.
Data Availability Statement
Sequences of the anti-PORCN antibody candidates were analyzed with the IMGT database (http://www.imgt.org/). The OPM database (https://opm.phar.umich.edu/) was used to insert PORCN into the lipid bilayer for MD simulation. The 3D cryo-EM maps have been deposited in the Electron Microscopy Data Bank under the accession numbers 26707, 726708, 26709, 26710 and 26711. Atomic coordinates for the atomic model have been deposited in the Protein Data Bank under the accession numbers 7URA, 7URC, 7URD, 7URE and 7URF. Additional data supporting the findings in this study are provided as source data and supplementary information to this manuscript.


