Abstract

The Rosellinia sanctae-cruciana extract was subjected to detailed liquid chromatography tandem mass spectrometry studies. A total of 38 peaks were annotated to m/z 508.26, m/z 510.28, m/z 524.26, m/z 526.28, m/z 540.26, m/z 542.27, and m/z 584.28 [M + H]+. The accurate mass, mutually supported UV/vis spectra, and database search identified these compounds as cytochalasins. Systematic dereplication helped identify a peak at m/z 540.26 [M + H]+ as the new compound. Further, the identified compound was purified by high-performance liquid chromatography and characterized by 2D NMR to be 19,20-epoxycytochalasin N1, a new optical isomer of 19,20-epoxycytochalasin-N. It exhibited substantial cytotoxicity with IC50 values ranging from 1.34 to 19.02 μM. This study shows a fast approach for dereplicating and identifying novel cytochalasin metabolites in crude extracts.
1. Introduction
Cytochalasins are fungal metabolites with a polycyclic core and a polyketide-derived C-16 to C-18 carbocyclic ring that can take the form of a lactone, a carbocyclic ring, or a complex ring system fused with an isoindolone moiety. It is differentially substituted at the C-10 position and classified accordingly, namely, 10-phenyl (cytochalasins), 10-indole (chaetoglobosins), 10-hydroxy 10-p-methoxybenzyl group (pyrichalasins), and 2-methyl propyl group (aspochalasins).1−6 More than 100 cytochalasins and derivatives have been reported from the genera Penicillium, Aspergillus, Zygosporium, Phoma, Metarhizum, Chaetomium, Rosellinia, Ascochyta, Hypoxylon, Xylaria, and Phomopsis. In recent years, liquid chromatography–mass spectrometry (LC–MS) has received a lot of attention in terms of reforming metabolic fingerprinting and metabolomics. MS has recently gained credibility in the identification and dereplication of metabolites in situ, regardless of their low concentration. MS/MS, in conjunction with mutually supportive data such as retention time (tR) and/or UV/vis spectra, aids in the identification of metabolites.7−10
Typically, a database is needed for dereplication investigations in order to search for various molecular characteristics, including substructure search, UV spectroscopy, m/z, and high-resolution MS (HRMS). The database contains information as search filters. The Dictionary of Natural Products (DNP) is a comprehensive collection of the physical characteristics of 250,000 naturally occurring chemicals. To detect metabolites in extracts and biological samples, mass-based databases such as the WEIZMASS spectrum library (HRMS-based data of 3500 substances) and Metlin database are also helpful tools.11−16 HRMS allows for the prediction of the molecular formula, based on the isotopic distribution pattern of the parent ions and their daughter ions.17,18 This method also suggests a possible daughter ion chemical formula, which helps with the diagnosis of fragmentation loss in parent to daughter ions.
Using direct infusion MS (DI–MS), LC electrospray ionization MS (LC–ESI–MS), and LC–ESI–MS/MS analyses, we created a flexible dereplication approach in this study to characterize cytochalasin metabolites in crude microbial extracts. The study was initially aimed to isolate and characterize the novel cytotoxic compounds from a potent cytotoxic extract of Rosellinia sanctae-cruciana.
2. Results and Discussion
We previously described the isolation of cytochalasins from the fungus R. sanctae-cruciana.19,20 The endophytic fungus R. sanctae-cruciana belongs to the Xylariaceae family, which produces the majority of cytochalasins.21
The current study demonstrated the use of strategic dereplication for the rapid identification of potent cytotoxic compounds from the endophyte R. sanctae-cruciana. The crude extract was used to develop chromatographic methods, and the same method was used to record the UV and MS spectra by HPLC (PDA) and LC–MS/MS, respectively. The LC–MS/MS 2D plot of m/z versus retention time (tR) revealed multiple peaks in the range of m/z 450 to 700, eluted from 15 to 40 min of retention time (Figure S1 and Table 1). The UV spectra of all these peaks were similar (190 to 210 nm), suggesting that the corresponding compounds are from the same class.
Table 1. Molecular Formula, Retention Time (tR), Daughter Ions, Parts Per Million Accuracy, and DNP Database Report of Cytochalasinsa.
| molecular formula/calcd [M + H]+ | tR (min) | Δ (ppm) | ionic form | daughter ions | total chromatographic peaks | remark |
|---|---|---|---|---|---|---|
| C30H37NO8 (540.2592) | 20.6, 20.9, 21.1, 23.6, and 24.0 | –7.0 to −0.8 | +H, +NH4, and +Na | 91.05, 105.07, 120.08, 237.12, 402.20, 420.21, 444.21, 480.23, and 502.22 | 5 | two reported27 and three new derivatives of cytochalasin N |
| C30H37NO7 (524.2643) | 22.9, 24.0, 24.3, 24.8, 25.1, 25.9, 26.8, and 27.1 | –6.5 to 0.30 | +H, +NH4, and +Na | 91.05, 105.07, 120.08, 239.14, 386.21, 404.22, 428.22, 446.23, and 486.22 | 8 | all reported cytochalasins28 |
| C30H37NO6 (508.2694) | 24.5, 26.0, 27.5, 27.7,28.3, 28.7, 29.4, 29.9, 30.4, and 31.4 | –5.0 to −0.7 | +H, +NH4, and +Na | 91.055, 105.07, 120.08, 237.16, 265.16, 374.24, 402.24, 430.23, and 470.23 | 10 | eight reported28 and at least two new derivatives of cytochalasin C |
| C30H39NO7 (526.2799) | 21.6, 22.6, 23.1, and 23.1 | –4.5 to −0.8 | +H, +NH4, and +Na | 91.05, 105.07, 120.08, 239.14, 412.22, 430.23, 448.25, 470.23, and 488.24 | 4 | two reported28 and two new derivatives of cytochalasin P |
| C32H41NO9 (584.2854) | 21.8, 25.6, and 32.2 | –2.5 to 2.0 | +H, +NH4, and +Na | 91.05, 105.07, 120.08, 239.14, 386.21, 404.22, 428.23, 446.23, and 546.24 | 3 | three new acetylated derivatives of epoxycytochalasin C |
| C30H39NO8 (542.2748) | 17.5, 19.4, 19.8, and 20.3 | –6.0 to −0.8 | +H, +NH4, and +Na | 91.05, 105.07, 120.08, 239.13, 386.21, 404.22, 422.23, 446.23, 464.24, and 504.23 | 4 | one reported28 with three new derivatives of cytochalasin P1 |
| C30H39NO6 (510.2850) | 25.1, 26.5, 27.1, and 28.2 | –4.5 to −0.5 | +H, +NH4, and +Na | 91.05, 105.07, 120.08, 237.16, 384.23, 402.24, 430.23, and 470.22 | 4 | one reported28 with three new derivatives of cytochalasin Z11 |
| total | 38 | A total of 22 reported and 16 identified as new cytochalasins |
Abbreviations: Δ (ppm) = change in parts per million from the exact mass, m/z = mass-to-charge ratio, calcd = calculated, and tR = retention time. For the EIC (extracted ion chromatogram), mass accuracy, and tandem MS spectra, see Figures S2–S26 for MS/MS spectra.
In the visual examination of the total ion chromatogram, the major ionized components are m/z 508.26, m/z 510.28, m/z 524.26, m/z 526.28, m/z 540.26, m/z 542.27, and m/z 584.28. These peaks appeared frequently during chromatographic elution (tR = 15 to 40 min) with an acceptable mass accuracy of ±5 ppm. All these peaks were subjected to the tandem MS experiment.
During collision-induced fragmentation of major parent ions m/z 508.26, m/z 510.28, m/z 524.26, m/z 526.28, m/z 540.26, m/z 542.27, and m/z 584.28, we observed the most common daughter ions tropylium m/z = 91, (C7H7)+ and benzyl m/z 120 (C8H10N)+. The typical MS fragmentation of major peaks was also consistent with the previously reported data.20,24,25
It is worth noting that the peak at m/z 508.26 is 2 Da lower than the peak at m/z 510.26, indicating a difference of 2H, which is equivalent to one double bond. A 16 Da difference was found between the peaks at m/z 524.26 and 508.24, indicating a difference of one oxygen atom in their structure.26 A 16 Da difference was also noticed among the peaks at m/z 540.24 and 524.24. Furthermore, a difference equivalent to the acetyl group (60 Da) was observed between the peaks at m/z 584.28 and 524.24 (Table 1).
2.1. Strategic Dereplication
The recorded data were entered into the database DNP as an input value, and it was then processed in the way that is represented in the screenshots, as given in the Supporting Information Figures S2–S21. A precise mass range, a molecular formula along with elements and the substructure, and a benzyl unit were among the inputs. The outcome of the search was the list of recognized cytochalasins. It is important to note that the majority of the same m/z appeared at various retention times, which suggests the existence of various isomers. In several instances, only the major compounds are published, and the remaining isomers can be thought of as unique. For instance, the peak m/z 540.26 [M + H]+ eluted at tR: 20.6, 20.9, 21.2, 23.7, and 24.1 min (Figures S14–S16); however only two cytochalasins, 19,20-epoycytochalasin N and cytochalasin Q, are reported (Figures S14–S16). As a result, the remaining three peaks are thought to be new. When the results of the 38 cytochalasin peaks were compared, 22 were found to be previously reported and 16 were discovered to be novel cytochalasins (Table 1).22,23,27,28 Apparently, without isolation we cannot say which compound is new among the identified peaks of cytochalasins. Furthermore, to characterize the novel compound, purification followed by spectroscopic characterization needs to be performed.
LC–MS/MS can only help with the annotation of new compounds in this case, and additional spectroscopic data, such as NMR spectra, are necessary to assign the novelty. Because the newly annotated compounds are isomeric, the LC–MS/MS fragmentation pattern cannot identify the position of any structural isomer’s functional group. However, due to the unique nature of the elution, an HPLC method for the isolation of cytochalasin from the crude extract was developed.
As previously stated, m/z 540.26 [M + H]+ appears at five different retentions, and only two cytochalasins, 19,20-epoycytochalasin N and cytochalasin Q, are reported. A minor peak at tR = 19.4 min recorded m/z 540.2588 [M + H]+ , and its molecular formula was deduced to be C30H37NO8 (calculated for 540.2592 with Δ ppm = 0.74). It was subjected to HPLC purification for isolation, and colorless needles were obtained in 2.5 mg, which were subjected to full structural characterization.
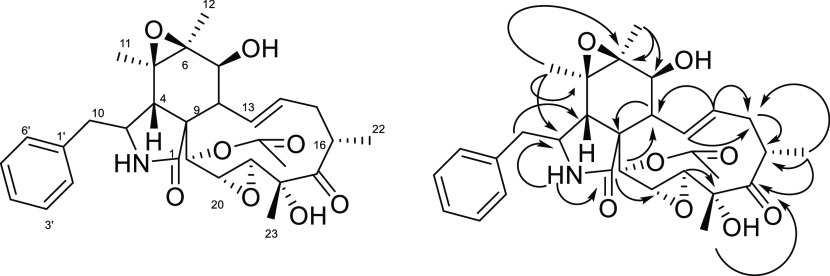
The mass spectra of the isolated compound m/z 540.2588 [M + H]+ corresponds to C30H37NO8, indicative of 13 degrees of unsaturation. In its 1H NMR spectrum, it displayed five methyl group proton signals at δH 1.55 (s, H-11), 1.16 (s, H-12), 1.04 (d, 6.7 Hz, H-22), 1.64 (s, H-23), and 2.18 (s, acetyl), two olefinic proton signals at δH 6.77 (dd, J = 15.55, 10.7 Hz, H-13) and 5.83 (ddd, 15.6, 10.3, 5.4 Hz, H-14), two pairs of ortho-coupled aromatic proton signals at δH 7.13 (2H, m, H-2, 6) and 7.29 (2H, m, H-3′, 5′), and several aliphatic proton signals in the range δH 1.04–4.12 (Table 2). Analysis of the 13C NMR and heteronuclear single quantum coherence (HSQC) data for 19,20-epoxycytochalasin N revealed three carbonyls (C-1, C-17, and acetyl), four quaternary carbons, including two oxygen-bearing quaternary carbons (C-5, C-6, C-9, and C-18), 16 methines, including four oxymethines (C-5, C-6, C-19, and C-20) and seven aromatic methines (C-2, C-3, C-4, C-5, C-6, C-13, and C-14), two methylene units (C-10 and C-15), and five methyl units (C-11, C-12, C-22, C-23, and acetyl). The 1H and 13C NMR data of compound m/z 540.2588 [M + H]+ were similar to those of the already reported compound 5,6,19,20-diepoxycytochalasin N, except the additional signals of the double bond at δH 6.67 (1H, dd, J = 15.5, 10.7 Hz, H-13) and 5.83 (1H, ddd, J = 15.6, 10.3, 5.4 Hz, H-14) and at δC 131.87 (C-13) and 132.67 (C-14). The two epoxide signals corresponding to δC 63.37 (C-5) and 65.57 (C-6) and another epoxide signal at δH 3.78 (1H, d, J = 1.9 Hz, H-19)/δC 60.49 (C-19) and 4.16 (1H, s, H-20)/δC 53.84 (C-20) suggested that the double bond at C-19/C-20 is replaced by an epoxide bond (Figure 1). Further, the heteronuclear multiple bond correlations (HMBCs) of H-21/C-19, C-20, and H-23/C-19 confirmed the above conclusion (Figures S25–S32).
Table 2. 1H and 13C NMR Data for 1 (at 500/125 MHz in Pyridine-d5), δ in Parts Per Million, and J in Hertz.
| 19,20 epoxycytochalasin N1 |
|||
|---|---|---|---|
| position | δC | δH | 1H–13C HMBC |
| 1 | 174.6 qC | ||
| 2 | NH | 9.59 (s) | H1, C3, C4 |
| 3 | 56.82 CH | 4.04 (t, 6.9) | H3, C4, C5 |
| 4 | 49.28 CH | 2.95 (d, 2) | H4, C1, C5, C9, C10, C21 |
| 5 | 63.37 qC | ||
| 6 | 65.57 qC | ||
| 7 | 69.85 CH | 4.12 (s) | |
| 8 | 43.72 CH | 3.59 (t, 10.4) | H8, C1, C9, C14 |
| 9 | 54.56 qC | ||
| 10 | 45.02 CH2 | 3.02(m) | H10, C4, C3 |
| 11 | 14.40 CH3 | 1.55(s) | H11, C4, C7 |
| 12 | 19.61 CH3 | 1.16 (s) | H12, C4, C5 |
| 13 | 131.87 CH | 6.77 (dd, 15.5, 10.7) | H13, C15 |
| 14 | 132.67 CH | 5.83 (ddd, 15.6, 10.3,5.4) | H14, C8, C15 |
| 15 | 38.47 CH2 | 2.71 (dd, 23,12), 2.01 (m) | H15, C13, C16 |
| 16 | 41.80 CH | 3.22 (m) | |
| 17 | 216.18 qC | ||
| 18 | 76.95 qC | ||
| 19 | 60.49 CH | 3.28 (d, 2.9) | |
| 20 | 53.84 CH | 4.16 (s) | |
| 21 | 73.96 CH | 6.07 (s) | |
| 22 | 18.61 CH3 | 1.04 (d, 6.7) | H22, C15, C17 |
| 23 | 22.14 CH3 | 1.64 (s) | H23, C15, C16, C17 |
| 1′ | 138.03 qC | ||
| 2′,6′ | 129.95 | 7.26 (m) | |
| 3′,5′ | 128.85 | 7.26 (m) | |
| 4′ | 126.90 | 7.26 (m) | |
| CH3CO– | 20.30 CH3 | 2.18 (s) | |
| CH3CO– | 170.75 qC | ||
2D NMR (COSY, HSQC, HMBC, and NOESY) (Figures S25–S32).
Figure 1.
Structure and key HMBCs (H → C) of 19,20-epoxycytochalasin N1.
The relative configuration of the compound was established by a combination of the coupling constant and nuclear Overhauser effect spectroscopy (NOESY) experiment. In the 1H NMR spectrum, the coupling constant J = 10.4 Hz for H-7 and H-8 suggested the cis orientation of the proton pair, and the trans geometry of the Δ13 double bond was deduced from the coupling constant J = 15.6 Hz for H-13 and H-14. The NOE correlation of H4–H3 and H3-12, H-4/H8 indicated that H-4, H3-12, H-8, and H3-11 were in the opposite orientation (β). The NOE correlation of H-7/H-13, H-14/H-8 and H-15b, H-15a/H-22, H-16/H-23 indicated that H-13 and H-22 were the same as α-oriented, and H-16 and H-23 were β-oriented. The NOE correlation of H-20/H-23, H-4/H-21 indicated that H-19, H-20, and H-21 were β-oriented. The optical rotation [α]D25 +6, (1 mg/mL in MeOH), UVmax (MeOH): 210 nm, while the optical rotation of reported 19,20-epoxycytochalasin N was [α]D −4.27 Thus, compound m/z 540.2588 [M + H]+ was established to be a new stereoisomer of cytochalasin N and was named cytochalasin N1. We recorded and compared the experimental circular dichroism spectra of six cytochalasins, which also substantiated the above conclusion (Figures S34 and S35).
2.2. Biological Activity
Using the sulforhodamine B (SRB) method and paclitaxel and 5-fluorouracil (5-FU) as the positive control, 19,20-epoxycytochalasin N1 was biologically evaluated for in vitro cytotoxicity against a panel of human tumor cell lines [lung (A549), prostate (PC-3), HCT-116 (colon), HT-29 (colon), SW-620 (colon), and breast (MCF-7)]. Table 2 shows the IC50 values for 19,20-epoxycytochalasin N1 against cell lines, which range from 1.34 to 19.02 μg/mL. However, 19,20-epoxycytochalasin N1 exhibited cytotoxic activity against a breast epithelial cell line (FR-2) at concentrations greater than 30 μg/mL (Table 3). These preliminary results confirm the potent cytotoxic action against colon cancer cell lines.
Table 3. Cytotoxicity Profiling of Isolated Cytochalasin (1).
3. Conclusions
The limitation of this LC–MS-based dereplication is that it always needs a comprehensive mass-based database, and another potential limitation is that the identification of the isomeric compound is not possible if not resolved chromatographically. However, the current study overcomes the above limitations because of the good resolution in the chromatographic profile. The use of HR-ESI–MS/MS enabled the identification of cytochalasin in fungal crude extracts in a timely manner (Figures S1–S24). We also provide tandem MS data for all 38 peaks, which greatly aids in the rapid identification of cytochalasin compounds in the crude extract. With IC50 values of 1.34 ± 0.46 μM, 19.20-eopxycytochalasin N1 demonstrated significant cytotoxicity against a panel of colon tumor cell lines, SW-620. As a result, the information presented here could be useful in identifying cytochalasin in fungal crude extracts.
4. Experimental Section
4.1. General Experimental Procedures
All the chemicals were obtained from Sigma-Aldrich Company and used as received. The 1H and 13C NMR spectra were recorded on Bruker-AVANCE III HD 500 and 125 MHz NMR instruments, respectively. Chemical shifts are reported in parts per million (ppm) downfield from tetramethylsilane and are referenced to the residual proton/carbon in the NMR solvent [CDCl3, 7.26/77.1 ppm; MeOD, 3.31/49.00 ppm; DMSO-d6, 2.50/39.5 ppm, and pyridin-d5, 7.19, 7.55, 8.71/123.5 (triplet)]. Potato dextrose agar (PDA) (Himedia, India), methanol, acetonitrile, and formic acid of LC–MS grade were purchased from Sigma-Aldrich, and LC–MS grade water (Merck, India) was used throughout the study.
4.2. Fungal Strain and Culture Conditions
The fungal endophyte R. sanctae-cruciana was obtained from our institutional microbial repository [Col Sir R. N. Chopra Repository, Council of Scientific and Industrial Research, Indian Institute of Integrative Medicine (CSIR-IIIM, Jammu, India)], and the identification details are described in previous studies.19,20 The PDA medium was used for fungal culture, and the primary fungal culture in the Petri dish was maintained at 28 °C for 7 days. For the seed culture, 1000 mL Erlenmeyer flasks containing 400 mL of autoclaved potato dextrose broth (PDB) medium was inoculated with agar plugs. A total of 30 Erlenmeyer flasks (each containing 400 mL of autoclaved PDB and 40 mL of seed culture) were kept in a shaking incubator (28 °C, 150 rpm) for 10 days.
4.3. Cell Lines and Cytotoxic Evaluation of 19,20-Epoxycytochalasin N1
NCI-60: National Cancer Institute provided human cancer cell lines for lung (A-549), prostate (PC-3), colon (HCT-116, HT-29, and SW-620), and breast (MCF-7) research. Human normal breast epithelial FR2 cells was purchased from American Type Culture Collection (ATCC), Manassas, USA. The cancer-screening panel’s cell lines were cultured in RPMI-1640 media with 10% fetal calf serum, penicillin (100 units/mL), and streptomycin (100 μg/mL).
The cell cultures were cultivated at 37 °C in a CO2 incubator with a humidity of 98% and a CO2 concentration of 5%.
Depending on the doubling time of the cells, 100 μL of the cell suspension (density = 7000 to 12,000 cells per well) was seeded in 96-well flat-bottom plates (NUNC). The plates were incubated for 24 h under the conditions 37 °C, 5% CO2, 95% air, and 100% relative humidity. After 24 h, the cells were exposed to various concentrations of cytochalasins. Paclitaxel and 5-FU were used as a positive control for 48 h under identical conditions. Cells were fixed in situ with cold TCA for 60 min at 4 °C after 48 h. Plates were then cleaned three times with water and dried in the air. Each well was filled with the SRB solution (0.4 % SRB in 1 percent acetic acid) and incubated for 30 min at room temperature. The plates were then rinsed three times with 1 % acetic acid and air-dried after staining. The protein-bound dye was solubilized in a 10 mM Tris base buffer solution. On a microplate reader, the absorbance was measured at 540 nm (Thermo Scientific). GraphPad Prism, version 5, was used to plot the optical density versus concentration to determine growth inhibition (GIC50).19,20
4.4. Extraction and Sample Preparation
After 10 days of culture, 15 L of fermentation broth was obtained, and 1500 mL of methanol was added to the broth before homogenization. The supernatant was extracted twice with an equal volume of ethyl acetate after centrifugation (10,000 rpm, 5 min, room temperature). The organic layer was concentrated under vacuum using a rotary evaporator, and 250 g of the brown oil extract was obtained. 10 mg of the extract was dissolved in 1 mL of HPLC-grade methanol and filtered using a 0.2 μm syringe filter before being subjected to LC–MS/MS analysis. Semipreparative HPLC-based compound isolation from the crude extract was performed, yielding 3 mg of a white color compound. Bruker NMR and pyridine-d5 NMR spectra were recorded at 500 MHz (Figures S25 and S32).
4.5. Semi-Preparative Isolation of Cytochalasin
The Shimadzu HPLC system was equipped with an Agilent RP-18 column (RP-18, 9.4 × 250 mm, 5 μm). The separation was carried out at a flow rate of 2.0 mL min–1 using a gradient program with eluent A and eluent B composed of 0.1 percent v/v formic acid in water and acetonitrile, respectively. The gradient system is as follows: 20 to 60% B in 20 min, 60 to 90% B from 20 to 30 min, and 90 to 10% B from 30 to 32 min, and the total run time was 35 min. 100 mg of the extract was dissolved in 1 mL of methanol and filtered through a 0.2 μm syringe filter, and a 100 μL injection volume was used for each HPLC run. The peak eluted at tR = 19.7 min was collected and subjected for mass spectrometric and NMR studies (Figure S36).
4.6. Profiling of the Extract by HPLC and LC–MS by Tandem MS Analysis
A suitable LC method was developed to profile the extract. The chromatographic separation was carried out on a Merck Chromolith column RP18e (4.6 × 100 mm) at a flow rate of 0.5 mL min–1 using a gradient program with eluent A and eluent B composed of 0.1% v/v formic acid in water and acetonitrile, respectively. The gradient system was used in the following manner: 5–60% of B in 0–30 min, 60–80% of B in 30–40 min, 80 to 5% of B in 40–42 min, and hold for 3 min; the total run time was 45 min. The LC method is used for the HPLC- and LC–MS-based profiling on a Shimadzu HPLC system equipped with a PDA detector and an Agilent 1290 HPLC system equipped with an Agilent UHD Q-TOF 6540 and an Agilent 6410 B triple quadrupole mass spectrometer, respectively.
In current study, an Agilent UHD Q-TOF 6540 and an Agilent 6410 B triple quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA, USA) were used for HRMS data, profiling, and precursor ion scan, respectively.
The LC–ESI–HRMS data were collected in the ESI positive mode, with the capillary voltage set to 3000 V; the gas temperature was set to 300 °C; the drying gas flow was set to 12 L min–1; and the nebulizer was set to 35 psi. For analysis, the scan source parameters of a skimmer voltage of 65 V, a fragmentor voltage of175 V, and an octapole RF peak at 700 V were used. For the MS scan, the mass scan range of 100–2000 Da was used in both the positive and negative ESI modes, and for MS/MS acquisition, the fix collision energy of 30 eV was used in the positive mode for target compounds.
Acknowledgments
This work was supported by the Council of Scientific and Industrial Research (CSIR) 12th FYP project (BSC-0108), MLP-1009, and the Department of Science and Technology (DST) grant no. ECR/2017/001381. The authors gratefully acknowledge Rajneesh Anand for analytical support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c03037.
Spectroscopic data of compounds (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Luo Y.-F.; Zhang M.; Dai J.-G.; Pedpradab P.; Wang W.-J.; Wu J. Cytochalasins from mangrove endophytic fungi Phomopsis spp. xy21 and xy22. Phytochem. Lett. 2016, 17, 162–166. 10.1016/j.phytol.2016.07.027. [DOI] [Google Scholar]
- Aldridge D. C.; Armstrong J. J.; Speake R. N.; Turner W. B. The cytochalasins, a new class of biologically active mould metabolites. Chem. Commun. 1967, 26–27. 10.1039/c19670000026. [DOI] [Google Scholar]
- Andersen B.; Smedsgaard J.; Frisvad J. C. Penicillium expansum: consistent production of patulin, chaetoglobosins, and other secondary metabolites in culture and their natural occurrence in fruit products. J. Agric. Food Chem. 2004, 52, 2421–2428. 10.1021/jf035406k. [DOI] [PubMed] [Google Scholar]
- Sekita S.; Yoshihira K.; Natori S.; Kuwano H. Chaetoglobosins Cytotoxic 10-(Indo-3-yl)-[13] cytochalasans from Chaetomium spp. II. Structures of Chaetoglobosins A, B, and D. Chem. Pharm. Bull. 1982, 30, 1618–1628. 10.1248/cpb.30.1618. [DOI] [PubMed] [Google Scholar]
- Scherlach K.; Boettger D.; Remme N.; Hertweck C. The chemistry and biology of cytochalasans. Nat. Prod. Rep. 2010, 27, 869–886. 10.1039/b903913a. [DOI] [PubMed] [Google Scholar]
- Yahara I.; Harada F.; Sekita S.; Yoshihira K.; Natori S. Correlation between effects of 24 different cytochalasins on cellular structures and cellular events and those on actin in vitro. J. Cell Biol. 1982, 92, 69–78. 10.1083/jcb.92.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G.; Mayhudin N. A.; Mitova M. I.; Sun L.; van der Sar S.; Blunt J. W.; Cole A. L. J.; Ellis G.; Laatsch H.; Munro M. H. G. Evolving trends in the dereplication of natural product extracts: New methodology for rapid, small-scale investigation of natural product extracts. J. Nat. Prod. 2008, 71, 1595–1599. 10.1021/np8002222. [DOI] [PubMed] [Google Scholar]
- El-Elimat T.; Figueroa M.; Ehrmann B. M.; Cech N. B.; Pearce C. J.; Oberlies N. H. High-resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products. J. Nat. Prod. 2013, 76, 1709–1716. 10.1021/np4004307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitgaard A.; Iversen A.; Andersen M. R.; Larsen T. O.; Frisvad J. C.; Nielsen K. F. Aggressive dereplication using UHPLC-DAD-QTOF: screening extracts for up to 3000 fungal secondary metabolites. Anal. Bioanal. Chem. 2014, 406, 1933–1943. 10.1007/s00216-013-7582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A. F.; Moraes L. A. Liquid chromatography-tandem mass spectrometry characterization of five new leucinostatins produced by Paecilomyces lilacinus CG-189. J. Antibiot. 2015, 68, 178–184. 10.1038/ja.2014.120. [DOI] [PubMed] [Google Scholar]
- Hou Y.; Braun D. R.; Michel C. R.; Klassen J. L.; Adnani N.; Wyche T. P.; Bugni T. S. Microbial strain prioritization using metabolomics tools for the discovery of natural products. Anal. Chem. 2012, 84, 4277–4283. 10.1021/ac202623g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.; Tianero M. D. B.; Kwan J. C.; Wyche T. P.; Michel C. R.; Ellis G. A.; Vazquez-Rivera E.; Braun D. R.; Rose W. E.; Schmidt E. W.; Bugni T. S. Structure and biosynthesis of the antibiotic Bottromycin D. Org. Lett. 2012, 14, 5050–5053. 10.1021/ol3022758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge P. J.; Challis G. L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- Shahaf N.; Rogachev I.; Heinig U.; Meir S.; Malitsky S.; Battat M.; Wyner H.; Zheng S.; Wehrens R.; Aharoni A. The WEIZMASS spectral library for high-confidence metabolite identification. Nat. Commun. 2016, 7, 12423. 10.1038/ncomms12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Carver J. J.; Phelan V. V.; Sanchez L. M.; Garg N.; Peng Y.; Nguyen D. D.; Watrous J.; Kapono C. A.; Luzzatto-Knaan T.; Porto C.; Bouslimani A.; Melnik A. V.; Meehan M. J.; Liu W.-T.; Crüsemann M.; Boudreau P. D.; Esquenazi E.; Sandoval-Calderón M.; Kersten R. D.; Pace L. A.; Quinn R. A.; Duncan K. R.; Hsu C.-C.; Floros D. J.; Gavilan R. G.; Kleigrewe K.; Northen T.; Dutton R. J.; Parrot D.; Carlson E. E.; Aigle B.; Michelsen C. F.; Jelsbak L.; Sohlenkamp C.; Pevzner P.; Edlund A.; McLean J.; Piel J.; Murphy B. T.; Gerwick L.; Liaw C.-C.; Yang Y.-L.; Humpf H.-U.; Maansson M.; Keyzers R. A.; Sims A. C.; Johnson A. R.; Sidebottom A. M.; Sedio B. E.; Klitgaard A.; Larson C. B.; Boya P C. A.; Torres-Mendoza D.; Gonzalez D. J.; Silva D. B.; Marques L. M.; Demarque D. P.; Pociute E.; O’Neill E. C.; Briand E.; Helfrich E. J. N.; Granatosky E. A.; Glukhov E.; Ryffel F.; Houson H.; Mohimani H.; Kharbush J. J.; Zeng Y.; Vorholt J. A.; Kurita K. L.; Charusanti P.; McPhail K. L.; Nielsen K. F.; Vuong L.; Elfeki M.; Traxler M. F.; Engene N.; Koyama N.; Vining O. B.; Baric R.; Silva R. R.; Mascuch S. J.; Tomasi S.; Jenkins S.; Macherla V.; Hoffman T.; Agarwal V.; Williams P. G.; Dai J.; Neupane R.; Gurr J.; Rodríguez A. M. C.; Lamsa A.; Zhang C.; Dorrestein K.; Duggan B. M.; Almaliti J.; Allard P.-M.; Phapale P.; Nothias L.-F.; Alexandrov T.; Litaudon M.; Wolfender J.-L.; Kyle J. E.; Metz T. O.; Peryea T.; Nguyen D.-T.; VanLeer D.; Shinn P.; Jadhav A.; Müller R.; Waters K. M.; Shi W.; Liu X.; Zhang L.; Knight R.; Jensen P. R.; Palsson B. O.; Pogliano K.; Linington R. G.; Gutiérrez M.; Lopes N. P.; Gerwick W. H.; Moore B. S.; Dorrestein P. C.; Bandeira N. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 2016, 34, 828–837. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Y.; Sanchez L. M.; Rath C. M.; Liu X.; Boudreau P. D.; Bruns N.; Glukhov E.; Wodtke A.; de Felicio R.; Fenner A.; Wong W. R.; Linington R. G.; Zhang L.; Debonsi H. M.; Gerwick W. H.; Dorrestein P. C. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco G.; Buchicchio A.; Lelario F.; Cataldi T. R. Molecular formula analysis of fragment ions by isotope-selective collision-induced dissociation tandem mass spectrometry of pharmacologically active compounds. J. Mass Spectrom. 2014, 49, 1322–1329. 10.1002/jms.3468. [DOI] [PubMed] [Google Scholar]
- Singh S. B.; Zhang C.; Zink D. L.; Herath K.; Ondeyka J.; Masurekar P.; Jayasuriya H.; Goetz M. A.; Tormo J. R.; Vicente F.; Martín J.; González I.; Genilloud O. Occurrence, distribution, dereplication and efficient discovery of thiazolyl peptides by sensitive-resistant pair screening. J. Antibiot. 2013, 66, 599–607. 10.1038/ja.2013.54. [DOI] [PubMed] [Google Scholar]
- Sharma N.; Kushwaha M.; Arora D.; Jain S.; Singamaneni V.; Sharma S.; Shankar R.; Bhushan S.; Gupta P.; Jaglan S. New cytochalasin from Rosellinia sanctae-cruciana, an endophytic fungus of Albizia lebbeck. J. Appl. Microbiol. 2018, 125, 111–120. 10.1111/jam.13764. [DOI] [PubMed] [Google Scholar]
- Kushwaha M.; Qayum A.; Jain S. K.; Singh J.; Srivastava A. K.; Srivastava S.; Sharma N.; Abrol V.; Malik R.; Singh S. K.; Vishwakarma R. A.; Jaglan S. Tandem MS-Based Metabolite Profiling of 19,20-Epoxycytochalasin C Reveals the Importance of a Hydroxy Group at the C7 Position for Biological Activity. ACS Omega 2021, 6, 3717–3726. 10.1021/acsomega.0c05307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley A. J. S.; Edwards R. L. The Xylariaceae: A case study in biological and chemical diversity. Pure Appl. Chem. 1998, 70, 2123. [Google Scholar]
- Chen Z.; Chen Y.; Huang H.; Yang H.; Zhang W.; Sun Y.; Wen J. Cytochalasin P1, a new cytochalasin from the marine-derived fungus Xylaria sp. SOF11. Z. Naturforsch., C: J. Biosci. 2017, 72, 129–132. 10.1515/znc-2016-0122. [DOI] [PubMed] [Google Scholar]
- Yan B. C.; Wang W. G.; Kong L. M.; Tang J. W.; Du X.; Li Y.; Puno P. T. Cytochalasans from the Endophytic Fungus Phomopsis sp. shj2 and Their Antimigratory Activities. J. Fungi 2022, 8, 543. 10.3390/jof8050543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral L. S.; Fill T. P.; Santos L. F. A.; Rodrigues-Filho E. Biosynthesis and mass spectral fragmentation pathways of (13)C and (15)N labeled cytochalasin D produced by Xylaria arbuscula. J. Mass Spectrom. 2017, 52, 239–247. 10.1002/jms.3922. [DOI] [PubMed] [Google Scholar]
- Prasain J. K.; Ueki M.; Stefanowicz P.; Osada H. Rapid screening and identification of cytochalasins by electrospray tandem mass spectrometry. J. Mass Spectrom. 2002, 37, 283–291. 10.1002/jms.282. [DOI] [PubMed] [Google Scholar]
- Jain S. K.; Pathania A. S.; Parshad R.; Raina C.; Ali A.; Gupta A. P.; Kushwaha M.; Aravinda S.; Bhushan S.; Bharate S. B.; Vishwakarma R. A. Chrysomycins A-C, antileukemic naphthocoumarins from Streptomyces sporoverrucosus. RSC Adv. 2013, 3, 21046–21053. 10.1039/c3ra42884b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R. L.; Maitland D. J.; Whalley A. J. Metabolites of the higher fungi. Part 24. Cytochalasin N, O, P, Q, and R. New cytochalasins from the fungus Hypoxylon terricola Mill. J. Chem. Soc., Perkin Trans. 1 1989, 1, 57–65. 10.1039/p19890000057. [DOI] [Google Scholar]
- https://dnp.chemnetbase.com (Accessed on 20 March 2021)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.