Abstract
During sporulation, the Bacillus subtilis transcription factor ςK is activated by regulated proteolytic processing. I have used a system that facilitates the analysis of the contributions of a modified form of the processing enzyme, SpoIVFB-GFP, and the regulatory proteins BofA and SpoIVFA to the conversion of pro-ςK to ςK. The results show that in the presence of BofA, SpoIVFA levels increase by greater than 20-fold, SpoIVFA is substantially stabilized, and pro-ςK processing is inhibited. In addition, enhanced accumulation of the SpoIVFA protein in the absence of BofA (achieved through the use of an ftsH null mutation) substantially inhibits pro-ςK processing. These results suggest that during growth, increased accumulation of the SpoIVFA protein inhibits the activity of SpoIVFB-GFP and regulates the activation of ςK.
During the process of sporulation in the bacterium Bacillus subtilis, asymmetric positioning of a septum partitions the developing cell into two compartments. Subsequently, the smaller compartment (the forespore) is engulfed by the larger compartment (the mother cell), to create a protoplast within a cell. In the developing sporangium, transcription is controlled by a cascade of four sporulation-specific transcription factors (ς factors), whose activities are compartment specific and regulated (6, 8, 15, 25). The mother cell transcription factors ςE and ςK are activated by proteolytic removal of their NH2 termini, 27 amino acids in the case of ςE (19) and 20 amino acids in the case of ςK (3, 14, 24). These regulated proteolytic events are governed by signal transduction pathways that originate in the forespore (11, 25). While the components and regulation of the two pathways differ, both delay gene expression in the mother cell until a signal has been received from the developing forespore and both are thought to be important for coordinating the two-compartment programs of gene expression (25).
Following engulfment, processing of pro-ςK to ςK takes place in the mother cell and requires the protein SpoIVFB (3, 4, 16, 21, 22). SpoIVFB is the processing enzyme or a required regulator of a presently unidentified processing complex (16, 22) and, consistent with its proposed function, contains a motif associated with Zn2+-dependent endopeptidases (14a, 16). SpoIVFB is inferred to be held inactive by the regulators BofA and SpoIVFA (3, 4, 10, 22, 23). All three proteins are produced in the mother cell (4, 10, 23) and are integral membrane proteins (21, 26). SpoIVFA and SpoIVFB localize to the boundary between the mother cell and the forespore (21) and are encoded in the two-cistron spoIVF operon (4). The localization pattern of BofA is unknown. Activation of SpoIVFB requires the signal protein SpoIVB, which is produced in and presumably secreted from the forespore (2, 7). In the absence of the signal protein, processing of pro-ςK and ςK-directed gene expression can be activated by mutations in bofA or specialized mutations in spoIVFA (bofB mutations); in these cases, processing occurs about 1 h earlier than in wild-type cells (3, 4, 23).
Vegetative B. subtilis cells engineered to induce two proteins normally made only during sporulation, the proprotein pro-ςK and a modified form of SpoIVFB (SpoIVFB-GFP; see Materials and Methods), convert the inactive transcription factor pro-ςK to the active transcription factor ςK (21, 22). The additional induction of SpoIVFA stimulates the processing reaction and is correlated with an increase in the level of SpoIVFB-GFP (22). (The increase in the level of SpoIVFB-GFP and the stimulation of the processing reaction are seen only when the spoIVF operon with the spoIVFB-gfp gene fusion substituted for wild-type spoIVFB is expressed, not when spoIVFA and spoIVFB-gfp are expressed independently during growth [22a].) The additional induction of BofA blocks processing of pro-ςK, in a SpoIVFA-dependent manner, without affecting the accumulation of SpoIVFB-GFP (22). Therefore, BofA and SpoIVFA are the only sporulation proteins needed to inhibit SpoIVFB-GFP-mediated processing, and inhibition is exerted at the level of the function of SpoIVFB (22).
How does BofA inhibit processing of pro-ςK? Using the vegetative processing system, I show that (i) in cells engineered to produce BofA, the level of the SpoIVFA protein is increased by greater than 20-fold; (ii) enhanced accumulation of the SpoIVFA protein does not require SpoIVFB-GFP; (iii) stabilization of SpoIVFA by BofA accounts for the accumulation of SpoIVFA; and, finally, (iv) a null mutation in the ftsH gene (an ATP- and Zn2+-dependent protease) facilitates the accumulation of the SpoIVFA protein in the absence of BofA and substantially inhibits pro-ςK processing without affecting the accumulation of SpoIVFB-GFP. Together, these results suggest that BofA-mediated inhibition of pro-ςK processing is exerted by altering the level of the SpoIVFA protein.
MATERIALS AND METHODS
Strains, growth, and media.
The B. subtilis strains used in this study are as follows: OR9 (PY79, wild-type B. subtilis), OR910 [trpC2 sigK::pOR286 Pxyl-sigK (spc) spoIIEΔ::kn], OR918 [trpC2 sigK::pOR286 Pxyl-sigK (spc) spoIIEΔ::kn amyE::Pxyl-spoIVFA, spoIVFB-gfp (cm)], OR920 [trpC2 sigK::pOR286 Pxyl-sigK (spc), spoIIEΔ::kn amyE::Pxyl-spoIVFA (cm spc)], OR948 [thrC::Pxyl-bofA (erm phleo)], and OR956 [trpC2 sigK::pOR286 Pxyl-sigK (spc), spoIIEΔ::kn thrC::Pxyl-bofA (erm phleo), amyE::Pxyl-spoIVFA, spoIVFB-gfp (cm)] (22). The in-frame spoIVFB-gfp gene fusion used in the above-described strains, and those described below, was made by fusing the coding sequence for green fluorescent protein (GFP) from Aquorea victoria to the 3′ terminus of spoIVFB (21, 22). This construction created a protein fusion at the fifth amino acid from the COOH terminus of SpoIVFB to GFP (21, 22). The Escherichia coli strains used in this study were DH5α (Life Technologies, Inc.), BL21(DE3)pLysS (Novagen), and OR692 (SAE97) (1). OR692 is identical to strain SAE92 [RL1196, BL21(DE3)pSA32] (21), except that it also contains the plasmid pLysS(cm).
For the routine growth of B. subtilis and E. coli at 37°C, Luria-Bertani (LB) medium was used (18). To induce promoters under xylose control during growth, d-xylose was added to a final concentration of 20 mM when the cultures were at an optical density at 600 nm of 0.25 to 0.30.
For sporulation assays, B. subtilis strains were grown for 23 h at 37°C in DS medium (9) supplemented with 20 mM d-xylose and 50 μg of threonine per ml. The cells were serially diluted, and 0.1 ml was plated on LB agar (viable cells), while the remainder was incubated at 80°C for 20 min and then 0.1 ml was plated on LB agar (heat-resistant cells). Colonies were counted following an overnight incubation at 37°C.
For growth under conditions of osmotic stress, B. subtilis strains were grown at 37°C to mid-log phase (optical density at 600 nm of about 0.5) in LB medium supplemented with 20 mM d-xylose. The cultures were split, and NaCl was added to a final concentration of 1.2 M to one portion and an equal volume of H2O was added to the second portion. The growth of all the cultures was monitored at 37°C for 9 h.
Antibiotics were used at the following concentrations for selection in B. subtilis: chloramphenicol at 5 μg/ml; erythromycin and lincomycin (MLS) at 1 and 25 μg/ml, respectively; spectinomycin at 100 μg/ml; kanamycin at 10 μg/ml; and tetracycline at 10 μg/ml. For selection in E. coli, chloramphenicol was used at 25 μg/ml and ampicillin was used at 100 μg/ml. To inhibit protein synthesis in B. subtilis (see Fig. 3) chloramphenicol was used at 200 μg/ml and was added 60 min after induction by xylose (see above).
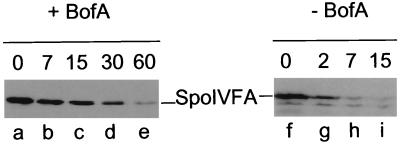
FIG. 3.
BofA protects SpoIVFA from degradation. Turnover of SpoIVFA in the presence or absence of BofA. A Western blot analysis of the disappearance of SpoIVFA in strains OR918 and OR956 following blocking of protein synthesis with chloramphenicol is shown. Cells were induced, chloramphenicol was added after 60 min to inhibit protein synthesis, and samples were taken at the times (in minutes) indicated above the blot. Lanes correspond to strains: a to e, OR956; f to i, OR918.
Amylase activity was examined by growing cells overnight on 1% starch plates (Sigma 9765) and then staining the agar with Gram’s iodine solution (Sigma HT90-2-32).
Chromosomal DNA from B. subtilis was prepared as described previously (9).
B. subtilis strains were made competent as described previously (9); for auxotrophic strains, the necessary amino acid supplements were added to 50 μg/ml.
Strain construction.
Strain OR958 was made by transforming chromosomal DNA from strain OR948 [PY79 × pOR350, thrC::Pxyl-bofA (erm phleo)] (22) into competent cells of strain OR920, selecting for MLS resistance.
Strain OR1017 was made by transforming strain OR918 with chromosomal DNA from strain OR866(PY79, ftsH::tet). [Strain OR866 was made by transforming OR9(PY79) with chromosomal DNA from strain ED04 (ftsH::tet) (5), selecting for tetracycline resistance.] Since the ftsH locus is physically close to the spoIIE locus on the B. subtilis chromosome, the majority of tetracycline resistant transformants had crossed out the spoIIE mutation. Strain OR1017 was selected because it retained resistance to kanamycin, indicating that it retained the spoIIE mutation.
Western blot analysis.
Samples of 2 ml were collected at the indicated intervals during growth. Whole-cells extracts were prepared as described previously (22). The protein concentrations of cell extracts were determined by using the Bio-Rad protein assay reagent. Portions of 11 μg of protein from whole-cell extracts (except in the experiment in Fig. 3, lanes f to i, where 44 μg of protein from whole-cell extracts was used) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12.5% polyacrylamide) and electroblotted to an Immobilon-P membrane (Millipore). The blots were processed for immunodetection as described previously (22), except that 5% dry milk was used for blocking. Two different methods were used to detect the binding of the primary antibody: a chemiluminescence detection system (Amersham) and a colorimetric detection system (ProtoBlot Western blot AP system; Promega); unless noted, the chemiluminescence system was used.
To estimate the fold difference in the accumulation of SpoIVFA in extracts from strains OR918 and OR956, Western blot analysis was performed with extracts from strains OR956 and OR918, 60 min after induction with xylose. The cell extract from OR956 was diluted to produce a signal of similar intensity to the extract from OR918 (data not shown).
Antibodies used were rabbit polyclonal antisera against SpoIVFA (affinity purified), ςK (polyclonal antiserum [21]), or GFP (polyclonal; Clontech no. 8361-2). The antibodies were used at 1/250, 1/10,000, and 1/2,000 dilutions, respectively.
Polyclonal antiserum against SpoIVFA was affinity purified, following heat treatment at 50°C for 30 min, with His6-SpoIVFA from strain OR692 as described previously (21). A detailed protocol for the affinity purification of the serum is available upon request. The histidine-tagged fusion protein, which contains the COOH-terminal 116 amino acids of SpoIVFA, was purified on Ni-nitrilotriacetic acid spin columns (Qiagen) by a denaturing method.
Image quantification.
For half-life determinations, images were quantified with NIH Image V1.61. The program was developed at the National Institutes of Health and is available on the World Wide Web (19a).
Image processing.
Images were copied on an Epson 636 scanner, processed with Adobe Photoshop and Deneba Canvas on a Macintosh PowerPC, and printed on a FUJIX3000 printer.
RESULTS
BofA-mediated inhibition of pro-ςK processing during growth is correlated with an enhanced accumulation of SpoIVFA.
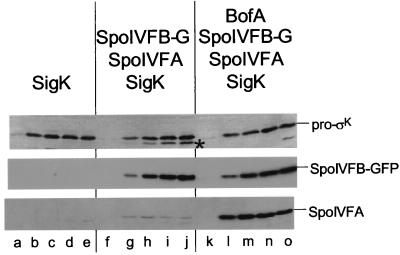
It was previously shown that vegetative B. subtilis cells engineered to produce pro-ςK do not process pro-ςK to ςK (Fig. 1, top, lanes a to e). However, cells engineered to produce pro-ςK, SpoIVFA, and SpoIVFB-GFP process pro-ςK to ςK efficiently (lanes f to j). The additional induction of the gene encoding BofA substantially inhibits the processing reaction (lanes k to o) (22). (The faster-migrating protein evident in lane o is an antibody-reactive species unrelated to ςK.) BofA was previously inferred to inhibit the activity of SpoIVFB-GFP because there is no obvious difference in the level of SpoIVFB-GFP in cells that process pro-ςK to ςK compared to cells in which this reaction is inhibited (Fig. 1, middle, lanes f to j versus lanes k to o) (22). Here I extend those results and show that, surprisingly, there is a greater than 20-fold difference in the accumulation of SpoIVFA in extracts from cells that process pro-ςK to ςK compared to cells in which the processing reaction is inhibited (Fig. 1, bottom, lanes f to j versus lanes k to o) (see Materials and Methods for how the difference in the level of SpoIVFA in cell extracts from the two strains was estimated). (The faster-migrating protein seen in lanes a to o is an antibody-reactive species unrelated to SpoIVFA.) These results show that induction of the gene encoding BofA is correlated with enhanced accumulation of SpoIVFA and inhibition of pro-ςK processing.
FIG. 1.
BofA-mediated inhibition of pro-ςK processing during growth is correlated with an enhanced accumulation of the SpoIVFA protein. Western blots of cell extracts from B. subtilis cells are shown. The proteins and protein fusion shown above the blots were induced during growth, and cell extracts were analyzed with antibodies that recognize pro-ςK and ςK (top panel, colorimetric detection), GFP (middle panel), or SpoIVFA (bottom panel). SigK refers to the pro-ςK protein, and SpoIVFB-G refers to the SpoIVFB-GFP protein fusion. Lowercase letters below the blots refer to individual lanes. ςK is identified by a black star. The first lane in each set of five corresponds to the zero time point, the second lane corresponds to 30 min after induction, and each subsequent lane corresponds to an additional 30 min of induction. For specific details of the induction protocol, see Materials and Methods. Lanes correspond to strains: a to e, OR910; f to j, OR918; k to o, OR956.
BofA is sufficient to enhance the accumulation of SpoIVFA during growth.
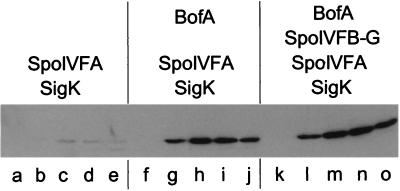
Figure 2 (lanes f to j) shows that the enhanced accumulation of SpoIVFA evident in extracts from cells engineered to produce pro-ςK, SpoIVFA, and BofA is not seen in extracts from cells engineered to produce pro-ςK and SpoIVFA alone (lanes a to e). Therefore, during growth, overexpression of BofA is sufficient to substantially enhance the accumulation of SpoIVFA. A further very modest enhancement is seen in extracts from cells which additionally produce SpoIVFB-GFP (compare lanes f to j with lanes k to o). Finally, the low accumulation of SpoIVFA seen in Fig. 1 (bottom, lanes f to j) and 2 (lanes a to e) is not detectably affected by the presence or absence of SpoIVFB-GFP. Therefore, SpoIVFB-GFP (a putative Zn2+-dependent endopeptidase [14a, 16]) is not involved in maintaining the low level of SpoIVFA.
FIG. 2.
Induction of the gene encoding BofA is sufficient to enhance the accumulation of SpoIVFA during growth. A Western blot of cell extracts from B. subtilis cells is shown. The proteins and protein fusion shown above the blot were induced during growth (details were given in the legend to Fig. 1), and cell extracts were analyzed with an antibody that recognizes SpoIVFA. Lanes correspond to strains: a to e, OR920; f to j, OR958; k to o, OR956.
In these cells, spoIVFA is not under the control of its natural sporulation-specific promoter but is transcribed by PxylA. A simple explanation for the enhanced accumulation of SpoIVFA in cells engineered to produce BofA is that BofA increases the expression of the promoter PxylA. β-Galactosidase assays showed that expression of a transcriptional fusion of E. coli lacZ to PxylA and a 5′ portion of the spoIVFA gene was similar in the presence and absence of BofA (data not shown). Therefore, the observed difference in the level of the SpoIVFA protein (Fig. 1 and 2) is not due to transcriptional regulation of PxylA and must be due to a postinitiation event.
SpoIVFA accumulation during growth is regulated at the level of protein stability.
To test whether the difference in SpoIVFA accumulation in the presence and absence of BofA is regulated by proteolysis, pro-ςK, SpoIVFA and SpoIVFB-GFP were induced in cells in which BofA was additionally induced (strain OR956) or not induced (strain OR918), and the turnover of SpoIVFA was compared in the two strains by blocking protein synthesis and monitoring the disappearance of the protein by Western blot analysis. As seen in Fig. 3 (lanes f to i), SpoIVFA was degraded rapidly in the absence of BofA with a half-life of <1.5 min. In contrast, in the presence of BofA, the SpoIVFA protein was stabilized with a half-life of 10 min (lanes a to e). These results show that SpoIVFA was unstable and that proteolysis of SpoIVFA was substantially inhibited by the presence or expression of BofA.
A null mutation in a gene encoding a protease is sufficient to enhance the accumulation of SpoIVFA and inhibit pro-ςK processing during growth.
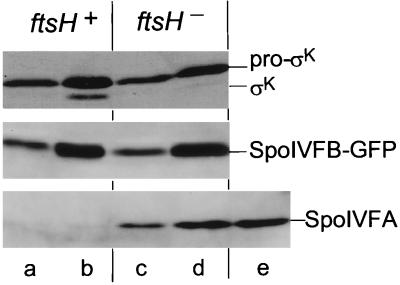
SpoIVFA is an integral membrane protein (21). In E. coli, the essential ftsH gene (20) encodes a membrane-bound ATP- and Zn2+-dependent protease that degrades several membrane substrates (SecY, YccA, and subunit a of the F0 part of the H+-ATPase [12]). In B. subtilis, while the ftsH gene is essential to sporulation, it is dispensable for growth (5, 17). Therefore, I tested whether a null mutation of the ftsH gene affects the accumulation of SpoIVFA during growth. Pro-ςK, SpoIVFA, and SpoIVFB-GFP were induced in cells which contain a wild-type copy (ftsH+, strain OR918) or a null mutation (ftsH, strain OR1017) of the ftsH gene, and the accumulation of SpoIVFA was compared in extracts from the two strains by Western blot analysis. Figure 4 shows that the level of SpoIVFA was markedly increased in extracts from cells that contained a null mutation of the ftsH gene (Fig. 4, bottom, lanes c and d) compared to extracts from cells that contained the ftsH+ gene (bottom, lanes a and b). As a control, the accumulation of another integral membrane protein, SpoIVFB-GFP (21), was not affected by a null mutation of the ftsH gene (middle, compare lanes a and b to lanes c and d).
FIG. 4.
A null mutation in the gene encoding FtsH facilitates the accumulation of the SpoIVFA protein and inhibits pro-ςK processing. Western blots of cell extracts were analyzed with an antibody that recognizes pro-ςK and ςK (top panel, colorimetric detection), GFP (middle panel), or SpoIVFA (bottom panel). ftsH+ refers to the wild-type ftsH gene, ftsH− refers to a null mutation of the ftsH gene. Lanes: a and c, (30 min postinduction); b, d, and e, 120 min postinduction. Lanes correspond to strains: a and b, OR918; c and d, OR1017; e, OR956.
The level of SpoIVFA seen in the ftsH mutant strain at 120 min postinduction (Fig. 4, bottom, lane d) was comparable to that seen in extracts from cells that contain a wild-type copy of the ftsH gene but, additionally, induced BofA (lane e). As described above, in these cells an enhanced level of SpoIVFA is correlated with an inhibition of pro-ςK processing (22) (Fig. 1, lanes k to o). Therefore, I investigated whether a null mutation in the ftsH gene, in the absence of BofA, would similarly affect pro-ςK processing during growth. Western blot analysis with anti-ςK antibodies showed that pro-ςK processing was substantially reduced in extracts from cells that contained a null mutation of the ftsH gene (Fig. 4, top, lanes c and d) compared to extracts from cells that contained the ftsH+ gene (lanes a and b). SpoIVFB-GFP levels were similar in the presence and absence of FtsH (middle, lanes a to d). Consistent with the Western blot analysis, plate tests showed that expression of lacZ fused to a sporulation gene (gerE) under the control of ςK was also substantially reduced in cells that contained a null mutation of the ftsH gene compared to cells that contained the ftsH+ gene (data not shown). Therefore, during growth, expression of SpoIVFA could inhibit pro-ςK processing and is inferred to do so by regulating the activity of SpoIVFB-GFP. BofA-mediated inhibition of pro-ςK processing may be more efficient than a null mutation of the ftsH gene since no ςK was visible up to 4 h after induction of the gene encoding BofA (22), while in extracts from cells that contain a null mutation of the ftsH gene, a small amount of ςK was seen 120 min postinduction (Fig. 4, top, lane d).
Induction of BofA during growth does not inhibit sporulation and does not result in a salt-sensitive growth phenotype.
The finding that the level of SpoIVFA was markedly increased in extracts from cells that contained a null mutation of the ftsH gene suggested that BofA might act by inhibiting the activity of FtsH. FtsH is required for both spore formation and growth under conditions of osmotic stress (5, 17). Therefore I tested whether induction of BofA during growth inhibited spore formation or affected growth under conditions of osmotic stress. As can be seen in Table 1, induction of BofA during growth (strain OR948) did not affect the viability of the culture or spore formation as compared to wild-type cells (PY79). In contrast, a null mutation of the ftsH gene (strain OR866) affected the viability of the culture and the remaining cells did not sporulate.
TABLE 1.
Induction of BofA during growth does not inhibit sporulationa
| Strain | Genotype | No. of cells/mlb
|
|
|---|---|---|---|
| Viable | Heat resistant | ||
| PY79 | Wild type | 4.7 × 108 | 1.3 × 108 |
| OR948 | thrC::PxylA-bofA | 8.4 × 108 | 2.8 × 108 |
| OR866 | ftsH::tet | 1.2 × 106 | <100 |
Cells were sporulated in DS medium containing 20 mM d-xylose and 50 μg of threonine per ml. For details, see Materials and Methods.
The numbers represent the averages from two experiments.
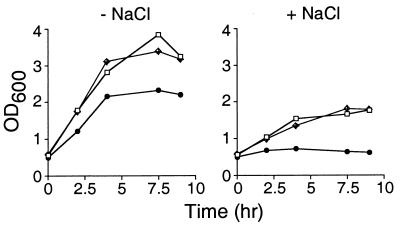
To test whether the induction of BofA during growth resulted in a salt-sensitive phenotype, exponentially growing cells were challenged with 1.2 M NaCl and their growth was monitored in the presence or absence of salt. As can be seen in Fig. 5 (left), the induction of BofA (strain OR948) did not affect the growth of the culture compared to that of wild-type cells (PY79). In contrast, and consistent with previous reports (5, 17), cells containing an ftsH null mutation (strain OR866) grew at the same rate as wild-type cells but did not reach the same density. Wild-type cells and cells in which BofA was induced responded to osmotic shock by resuming growth (Fig. 5, right), while cells containing an ftsH null mutation did not resume growth effectively. These results, and those described above, are consistent with the conclusion that the induction of BofA during growth does not result in an ftsH phenotype.
FIG. 5.
Induction of BofA during growth does not result in a salt-sensitive growth phenotype. Growth of wild-type cells (PY79) or cells containing PxylA-bofA (OR948) or ftsH::tet (OR866) in the presence or absence of NaCl is shown. For details, see Materials and Methods. NaCl was added at the zero time point depicted on these graphs. Symbols: open squares, PY79; crossed diamonds, OR948; solid circles, OR866. OD600, optical density at 600 nm.
DISCUSSION
I have investigated BofA-mediated inhibition of pro-ςK processing in the absence of sporulation. The most important finding of the present study is that BofA overexpression is associated with a greater than 20-fold increase in the accumulation of the SpoIVFA protein. This finding is consistent with the previous demonstration that BofA-mediated inhibition of processing requires SpoIVFA (22). Herein I have also shown that in the presence of BofA, the increased level of SpoIVFA is correlated with a substantial inhibition of the otherwise rapid degradation of SpoIVFA. In addition, I have shown that enhanced accumulation of the SpoIVFA protein, achieved through the use of an ftsH null mutation in the absence of BofA, is concurrent with inhibition of pro-ςK processing. The level of SpoIVFB-GFP is not affected, suggesting that SpoIVFA inhibits the activity of SpoIVFB-GFP.
A simple interpretation that encompasses the results presented in this study is that BofA protects SpoIVFA from proteolysis by promoting the assembly and maintenance of a membrane complex composed of BofA, SpoIVFA, and SpoIVFB-GFP. Since all three proteins are integral membrane proteins, such a complex would be membrane bound (3, 21, 23, 26). In this view, uncomplexed SpoIVFA would be subject to proteolysis. This would be reminiscent of the degradation of uncomplexed SecY by FtsH in E. coli (13). The finding that a null mutation in the gene encoding FtsH enhances the level of SpoIVFA in the absence of BofA suggests that FtsH, either directly or indirectly, regulates SpoIVFA degradation during growth. BofA could increase the level of SpoIVFA by inhibiting the activity of FtsH and/or another protease or by interacting with SpoIVFA, thereby protecting it from proteolysis. Since the induction of BofA does not result in an ftsH phenotype, it seems unlikely that BofA inhibits the activity of FtsH.
The pro-ςK processing complex in sporulating cells is believed to be held inactive by the regulators BofA and SpoIVFA for about 1 h (3, 4, 10, 22, 23). While it is possible that BofA protects SpoIVFA from proteolysis only during vegetative growth (the conditions used in this study), the genetics of the phenotypes of bofA mutants are consistent with such a role for BofA during sporulation also. Extracts from sporulating cells that contain null mutations in bofA have lower levels of the SpoIVFA protein than do extracts from wild-type cells (21, 22b). Also, extracts from sporulating cells that contain specialized mutations in spoIVFA (bofB5 and bofB8) (3, 4) have lower levels of the SpoIVFA protein than do extracts from wild-type cells (21, 22b), suggesting that these mutant proteins might not interact effectively with BofA and/or SpoIVFB. Consistent with this suggestion, pro-ςK processing occurs about 1 h earlier in the above-described strains than in wild-type sporulating cells (3, 4).
Does FtsH regulate the level of SpoIVFA in sporulating cells? It has recently been shown that FtsH does persist during sporulation (27), but there is presently no evidence that FtsH is directly involved in the ςK pathway during sporulation. FtsH is required for at least two sporulation-specific events that occur prior to the synthesis of SpoIVFA (1a, 5, 17); therefore, this hypothesis has not been tested directly in sporulating cells. Principal challenges for the future will include elucidating the molecular mechanism(s) of action of BofA and SpoIVFA.
ACKNOWLEDGMENTS
Strains OR958, OR866, and SAE97 were made in the laboratory of R. Losick (Harvard University). I thank P. Stragier for pDG795; W. Schumann and E. Deuerling for chromosomal DNA from strain ED04; S. Alper for strain SAE97; members of the Weisberg laboratory and the vegetable data club at the National Institutes of Health and D. Garsin, R. Losick, and D. Rudner for many helpful comments and suggestions; R. Weisberg and the National Institute of Child Health and Human Development for hospitality; and A. Decatur, L. Duncan, S. Gottesman, J. Nodwell, P. Stragier, and R. Weisberg for critical reading of the manuscript.
REFERENCES
- 1.Alper S. Ph.D. thesis. Cambridge, Mass: Harvard University; 1996. [Google Scholar]
- 1a.Cutting S, Anderson M, Lysenko E, Page A, Tomoyasu T, Tatematsu K, Tatsuta T, Kroos L, Ogura T. SpoVM, a small protein essential to development in Bacillus subtilis interacts with the ATP-dependent protease FtsH. J Bacteriol. 1997;179:5534–5542. doi: 10.1128/jb.179.17.5534-5542.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-ςK processing in Bacillus subtilis. Genes Dev. 1991;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- 3.Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. A forespore checkpoint for mother cell gene expression during development in B. subtilis. Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- 4.Cutting S, Roels S, Losick R. Sporulation operon spoIVF and the characterization of mutations that uncouple mother cell from forespore gene expression in Bacillus subtilis. J Mol Biol. 1991;221:1237–1256. doi: 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- 5.Deuerling E, Mogk A, Richter C, Purucker M, Schumann W. The ftsH gene of Bacillus subtilis is involved in major cellular processes such as sporulation, stress adaptation and secretion. Mol Microbiol. 1997;23:921–933. doi: 10.1046/j.1365-2958.1997.2721636.x. [DOI] [PubMed] [Google Scholar]
- 6.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez M, Cutting S, Stragier P. Transcription of spoIVB is the only role of ςG that is essential for pro-ςK processing during spore formation in Bacillus subtilis. J Bacteriol. 1995;177:4825–4827. doi: 10.1128/jb.177.16.4825-4827.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harwood C R, Cutting S M. Molecular biological methods for Bacillus. New York, N.Y: John Wiley & Sons, Inc.; 1990. [Google Scholar]
- 10.Ireton K, Grossman A D. Interactions among mutations that cause altered timing of expression during sporulation in Bacillus subtilis. J Bacteriol. 1992;174:3185–3195. doi: 10.1128/jb.174.10.3185-3195.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaiser D, Losick R. How and why bacteria talk to each other. Cell. 1993;73:873–885. doi: 10.1016/0092-8674(93)90268-u. [DOI] [PubMed] [Google Scholar]
- 12.Kihara A, Akiyama Y, Ito K. Different pathways for protein degradation by the FtsH/HflKC membrane-embedded protease complex: and implication from the interference by a mutant form of a new substrate protein, YccA. J Mol Biol. 1998;279:175–188. doi: 10.1006/jmbi.1998.1781. [DOI] [PubMed] [Google Scholar]
- 13.Kihara A, Akiyama Y, Ito K. FtsH is required for proteolytic elimination of uncomplexes forms of SecY, an essential protein translocase subunit. Proc Natl Acad Sci USA. 1995;92:4532–4536. doi: 10.1073/pnas.92.10.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kroos L, Kunkel B, Losick R. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science. 1989;243:526–529. doi: 10.1126/science.2492118. [DOI] [PubMed] [Google Scholar]
- 14a.Lewis A P, Thomas P J. A novel clan of zinc metallopeptidases with possible intramembrane cleavage properties. Prot Sci. 1999;8:439–442. doi: 10.1110/ps.8.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Losick R, Stragier P. Crisscross regulation of cell-type specific gene expression during development in B. subtilis. Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- 16.Lu S, Cutting S, Kroos L. Sporulation protein SpoIVFB from Bacillus subtilis enhances processing of the sigma factor precursor pro-ςK in the absence of other sporulation gene products. J Bacteriol. 1995;177:1082–1085. doi: 10.1128/jb.177.4.1082-1085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lysenko E, Ogura T, Cutting S M. Characterization of the ftsH gene of Bacillus subtilis. Microbiology. 1997;143:971–978. doi: 10.1099/00221287-143-3-971. [DOI] [PubMed] [Google Scholar]
- 18.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 19.Miyao A, Theeragool G, Takeuchi M, Kobayashi Y. Bacillus subtilis spoVE gene is transcribed by ςE-associated RNA polymerase. J Bacteriol. 1993;175:4081–4086. doi: 10.1128/jb.175.13.4081-4086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.National Institutes of Health. 20 December 1996, release date. NIH Image software. [Online]. http://rsb.info.nih.gov/nih-image/. [10 June 1999, last date accessed.]
- 20.Ogura T, Tomoyasu T, Yuki T, Morimura K, Begg J, Donachie W D, Mori H, Niki H, Hiraga S. Structure and function of the ftsH gene in Escherichia coli. Res Microbiol. 1991;142:279–282. doi: 10.1016/0923-2508(91)90041-8. [DOI] [PubMed] [Google Scholar]
- 21.Resnekov O, Alper S, Losick R. Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Genes Cells. 1996;1:529–542. doi: 10.1046/j.1365-2443.1996.d01-262.x. [DOI] [PubMed] [Google Scholar]
- 22.Resnekov O, Losick R. Negative regulation of the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Proc Natl Acad Sci USA. 1998;95:3162–3167. doi: 10.1073/pnas.95.6.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Resnekov, O. Unpublished data.
- 22b.Resnekov, O., and R. Losick. Unpublished data.
- 23.Ricca E, Cutting S, Losick R. Characterization of bofA, a gene involved in intercompartmental regulation of pro-ςK processing during sporulation in Bacillus subtilis. J Bacteriol. 1992;174:3177–3184. doi: 10.1128/jb.174.10.3177-3184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stragier P, Kunkel B, Kroos L, Losick R. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science. 1989;243:507–512. doi: 10.1126/science.2536191. [DOI] [PubMed] [Google Scholar]
- 25.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 26.Varcamonti M, Marasco, De Felice R, Sacco M. Membrane topology analysis of the Bacillus subtilis BofA protein involved in pro-ςK processing. Microbiology. 1997;143:1053–1058. doi: 10.1099/00221287-143-4-1053. [DOI] [PubMed] [Google Scholar]
- 27.Zhang B, Hofmeister A, Kroos L. The prosequence of pro-ςK promotes membrane association and inhibits RNA polymerase core binding. J Bacteriol. 1998;180:2434–2441. doi: 10.1128/jb.180.9.2434-2441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]