Abstract

Ethyl 4-(4-fluorophenylamino)-2,6-bis(4-(trifluoromethyl)phenyl)-1-(4-fluoro-phenyl)-1,2,5,6-tetrahydropyridine-3-carboxylate (FTEAA) has been synthesized efficiently in an iodine-catalyzed five-component reaction of 4-fluoroaniline, 4-trifluoromethyl benzaldehyde, and ethyl acetoacetate in methanol at 55 °C for 12 h. Various spectro-analytical techniques such as 1H and 13C NMR and Fourier-transform infrared spectroscopy have validated the structure of FTEAA. Further confirmation of the structure of FTEAA has been established on the basis of single-crystal X-ray diffraction analysis. The supramolecular assembly of FTEAA in terms of strong and comparatively weak noncovalent interactions is fully investigated by Hirshfeld surface analysis, the interaction energy between pairs of molecules, and energy frameworks. The void analysis is conducted to explore the strength and stability of the crystal structure. Furthermore, molecular docking analysis was computationally performed to see the potential intermolecular interactions between the selected proteins and FTEAA. The binding interaction energies are found to be −8.8 and −9.6 kcal/mol for the proteins MAO-B (PDB ID: 2V5Z) and MAO-A (PDB ID: 2Z5X), respectively. These reasonably good binding energies (more negative values) indicate the efficient associations between the FTEAA and target proteins. The proteins and FTEAA were also analyzed for intermolecular interactions. FTEAA and proteins interact in a variety of ways, like conventional hydrogen bonds, carbon–hydrogen bonds, alkyl, π-alkyl, and halide interactions.
1. Introduction
The piperidine scaffold, the most important structural feature of many pharmaceuticals and found in the top-selling small molecule pharmaceutical, is ubiquitous in many natural products and medicinal products. Moreover, the piperidine moiety behaves as the main chromophore in several commercial drugs, including Aricept, alogliptin, desloratadine, raloxifene, and Ritalin (Figure 1).1,2
Figure 1.
Commercial drugs bearing the piperidine moiety.
In addition, piperidines have widespread pharmacological applications as antibacterial, antifungal, anticancer, anti-tuberculosis, elastase inhibitors, antiobesity, antianalgesic, acetylcholine serine protease inhibitor, antihypertensive, antidepressant, neuroprotective, and antipsychotics.3,4 Due to the significant relevance of piperidine derivatives belonging to the pharmacologically active scaffold in different therapeutic drugs, their synthesis has been extensively studied in the literature.5,6
Dysthymia and nervous ailments such as Parkinson’s and Alzheimer’s diseases arise because of an over-release of monoamine oxidases, which diminishes the levels of monoaminergic transmitters and, hence, the communication in the central nervous system. The communication and intracellular levels of monoamine oxidases can be maintained by administering monoamine oxidase inhibitors.7 Among monoamine oxidases, monoamine oxidase A has the potential to catalyze the deamination of adrenaline and noradrenaline as well as serotonin; however, monoamine oxidase B catalyzes the disintegration of phenyl ethyl amine (a central nervous stimulant) and phenyl methyl amine.8 Additionally, monoamine oxidases A and B can break down dopamine, tryptamine, and tyramine.9 Monoamine oxidases are administrated by the use of cordyline,10 moclobemide (MAO-A),11 and selegiline (MOA-B).12
Owing to their tendency to break down monoamine neurotransmitters, monoamine oxidases are effective protein targets for drug development. The molecules disclosing inhibition potential in opposition to MAO-A and B could serve as potential drugs to cure depression and Parkinson’s and Alzheimer’s diseases.13
Recently, cis- and trans-isomers of 1-propargyl-4-styrylpiperidine have been reported as potent inhibitors of MAO-A and MAO-B having IC50 values of 0.7261 + 0.0269 and 0.3422 + 0.0224 μM, respectively14 (Figure 2).
Figure 2.
(a) Piperidine-based geometric isomers selectively inhibit monoamine oxidase A and B. (b) Tetrahydropyridine-based inhibitors of monoamine oxidase A and B.
Tetrahydropyridines, structures with one double bond in the piperidine nucleus, possess good monoamine oxidase substrate properties. 1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and its analogues are inhibitors of monoamine oxidase B.15 Installation of various substituents on the tetrahydropyridine has shown an increase in activity toward monoamine oxidase B inhibition (Figure 2b).16 The introduction of fluorine and trifluoromethyl groups onto the core structure of MPTP provided better enzyme substrates for monoamine oxidase A and B17 as the fluorine and trifluoromethyl groups are known to impart enhanced metabolic stability, lipophilicity, and dipole moment to drug-like molecules.18,19
The current study involved the synthesis and characterization of a densely substituted tetrahydropyridine-based derivative, ethyl 4-(4-fluorophenylamino)-2,6-bis(4-(trifluoromethyl)phenyl)-1-(4-fluoro-phenyl)-1,2,5,6-tetrahydropyridine-3-carboxylate (FTEAA). Furthermore, the synthesized derivative is evaluated for potential dual inhibition of MAO-A and B proteins. Several important structural aspects are determined through the X-ray diffraction (XRD) technique for the single crystal structure of FTEAA. As intermolecular interactions play a vital role in crystal formation, the supramolecular assembly of FTEAA in terms of strong as well as, comparatively, weak noncovalent interactions is fully investigated by Hirshfeld surface (HS) analysis. Furthermore, docking studies to examine the binding interactions of FTEAA within the active pockets of MAO-A and B proteins supplement the current experimental findings regarding inhibition activity.
2. Results and Discussion
2.1. Single Crystal Analysis
Single crystal XRD is utilized for crystal structure determination. The Cambridge structure database search ratifies the originality of the crystal structure of FTEAA. The experimental details are listed in Table 1, while selected bond lengths and bond angles are given in Table 2.
Table 1. Single-crystal Analysis of FTEAA.
| crystal data | FTEAA |
|---|---|
| CCDC | 2173544 |
| chemical formula | C34H26F8N2O2 |
| Mr | 646.57 |
| crystal system, space group | triclinic, P1̅ |
| temperature (K) | 150 (2) |
| a, b, c (Å) | 9.9381 (6), 11.7975 (7), 14.0204 (9) |
| α, β, γ (deg) | 106.603 (5), 105.040 (6), 99.032 (5) |
| V (Å3) | 1473.11 (17) |
| Z | 2 |
| density (calculated)g/cm–3 | 1.458 |
| F(000) | 664 |
| radiation type | Cu Kα |
| wavelength (λ) | 1.54184 |
| μ (mm–1) | 1.088 |
| crystal size (mm) | 0.28 × 0.24 × 0.17 |
| data collection | |
| diffractometer | Bruker APEXII CCD |
| absorption correction | multiscan (SADABS; Bruker, 2007) |
| no. of measured, independent, and observed [I > 2σ(I)] reflections | 9288, 4663, 3772 |
| Rint | 0.034 |
| θ range for data collection (deg) | 3.474–62.769 |
| index ranges | –11 ≤ h ≤ 11, –13 ≤ k ≤ 12, –15 ≤ l ≤ 16 |
| data refinement | |
| R[F2 > 2σ(F2)], wR(F2), S | 0.042, 0.116, 1.04 |
| no. of reflections | 4663 |
| no. of parameters | 426 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å–3) | 0.32, −0.27 |
Table 2. Selected Bond Lengths (Å) and Bond Angles (deg) of FTEAA.
| selected bond lengths | selected bond angles | ||
|---|---|---|---|
| N1–C1 | 1.470 (2) | C1–N1–C29 | 121.33 (15) |
| N1–C5 | 1.453 (2) | C5–N1–C29 | 119.98 (14) |
| N1–C29 | 1.394 (2) | C1–N1–C5 | 118.59 (14) |
| N2–C3 | 1.354 (3) | C3–N2–C16 | 125.50 (14) |
| N2–C16 | 1.431 (3) | C2–C13–O1 | 124.40 (18) |
| O1–C13 | 1.231 (2) | C2–C13–O2 | 113.77 (15) |
| O2–C13 | 1.343 (2) | O1–C13–O2 | 121.81 (16) |
| O2–C14 | 1.451 (2) | C13–O2–C14 | 116.17 (14) |
| F4–C19 | 1.354 (3) | O2–C14–C15 | 107.23 (16) |
| F8–C32 | 1.365 (2) | C18–C19–F4 | 118.9 (2) |
In FTEAA (Figure 3a and Table 1), the 1,2,3,6-tetrahydropyridine group A (C1–C5/N1) is puckered with a total puckering amplitude of 0.681 (2) Å, θ = 84.69 (17)° and φ = 248.83(17) Å. The C-atom (C4) deviation from the mean plane of group A is greater than that of other atoms in the mean plane, with a value of 0.4187 (1) Å. The conformation of the tetrahydropyridine ring is “a flatted boat” conformation (Figure 3b), which is substantiated by the puckering of C1 and C4 atoms to the extent of 0.5010 (3) and 0.6573 (3) Å, respectively, from the least square plane defined by (C2/C3/C5/N1) atoms. The flattened boat conformation is also confirmed by the torsional angles associated with the tetrahydropyridine ring that are C(5)–N(1)–C(1)–C(2) = 37.1 (2)°, N(1)–C(1)–C(2)–C(3) = −46.8 (2)°, C(1)–C(2)–C(3)–C(4) = 2.7 (2)°, C(2)–C(3)–C(4)–C(5) = 49.0 (2)°, C(1)–N(1)–C(5)–C(4) = 12.6 (2)°, and C(3)–C(4)–C(5)–N(1) = −55.94 (19)°. The phenyl ring B (C6–C11), ethyl formate group C (C13–C15/O1/O2), 4-fluoroaniline group D (C16–C21/N2/F4), phenyl ring E (C22–C27), and fluorophenyl ring F (C29–C34/F8) are planar with root mean square deviations of 0.0087, 0.0126, 0.0171, 0.0039, and 0.0076 Å, respectively. The mean plane of group A makes the dihedral angles of 82.32 (6), 32.79 (9), 72.39 (6), 82.42 (6), and 28.04 (9)° with ring B and groups D, E, and F, respectively. The dihedral angles infer that group A is nearly perpendicular to ring B and group E. One of the trifluoro groups (F1A–F3A) is disordered over two sets of locations with an occupancy ratio of 0.508(6): 0.492(6). The mean plane of the major part (F1A–F3A) is orientated at a dihedral angle of 11.89 (3)°, relative to the mean plane of the minor part (F1B–F3B). The anisotropic displacement parameter (ADP) of the major part of each disordered atom is made equal to the ADP of the minor part. The DFIX and DANG restraints are used for stabilizing disordered parts in refinement. The molecular configuration is stabilized by intramolecular N–H···O bonding to form an S (6) loop. The molecules are connected and arranged in the shape of dimers via C–H···F bonding (Figure 4). As a result of dimerization, an R22(8) loop is formed. The dimers are then interconnected via C–H···O and N–H···F bonding. An R2(14) loop is generated by C–H···O bonding. The crystal packing is further stabilized by intra and as well as intermolecular C–H···π interactions (Table 2). Intermolecular C–H···π interactions connect the molecular units in the form of a dimer. One such dimer is shown in Figure 5.
Figure 3.

(a) ORTEP diagram of FTEAA drawn at a probability level of 50%. Hydrogen atoms are visible in the form of small circles of arbitrary radii. Only the main portion of the disordered trifluoro moiety is displayed for simplicity. (b) Conformation of the tetrahydropyridine ring in FTEAA with all atoms other than the tetrahydropyridine ring was omitted for clarity.
Figure 4.
Packing diagram of FTEAA. Only selected hydrogen atoms and the main part of the disordered trifluoro group are shown for simplicity.
Figure 5.

Graphical illustration of C–H···π interactions in FTEAA.
The Cambridge structural database (CSD 9) provides nine crystal structures that are closely related to the crystal structure of FTEAA. The reference codes of these nine structures are COJCIP,20 GATCUB,21 KAGSAQ,22 KAGSEU,17 LETBET,23 MEJTAZ,24 MEWTOZ,25 MONVAO,26 and WIHCOH.27 The S (6) intramolecular H-bonding loop was formed through intramolecular N–H···O bonding between the amino group and carbonyl O-atom in all the nine crystal structures of the CSD. The same H-bonded loop is formed in FTEAA by the intramolecular H-bonding between the amino group and carbonyl O-atom. The tetrahydropyridine ring adopted a boat conformation, with some deviation in all the nine crystal structures of the CSD except in the crystal structures with reference code COJCIP and MEJTAZ. The authors of the crystal structure with reference code MEJTAZ did not describe the conformation of the tetrahydropyridine ring as well as intra- and intermolecular H-bonding. The tetrahydropyridine ring adopted a twisted chair conformation in the crystal structure with reference code COJCIP. Just like in FTEAA, the intermolecular C–H···O and C–H···F H-bonding interactions are present in the crystal structures with reference codes COJCIP, LETBET, MEWTOZ, and WIHCOH, whereas C–H···F bonding is absent in KAGSEU and C–H···O bonding is absent in KAGSAQ. Figure S1 shows the simulated powder pattern of FTEAA. The sharp peaks indicated high crystallinity, and the most intense peaks in the simulated powder XRD pattern indicated the preferred growth direction.
2.2. HS Exploration
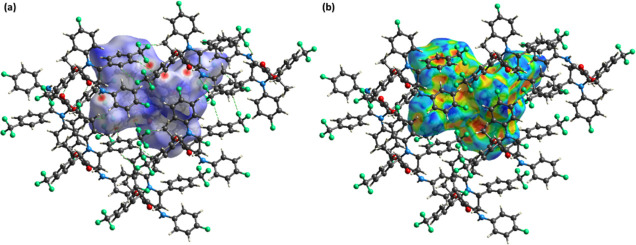
HS analysis is achieved on Crystal Explorer version 21.5.28 It is used for the exploration of the noncovalent interactions in the crystal packing of single crystals.29 The properties of the single crystal depend on the packing of molecules and noncovalent interactions that interlinked the molecules with each other. The HS concept was developed when scientists were trying to divide the electronic density of crystals into molecular fragments for integration purposes.30,31 HS can be plotted by using various properties like dnorm, shape index, curvedness, and so forth. The hydrogen bonding information can be extracted by drawing a graph of the HS over dnorm, where dnorm is the normalized distance.32,33 This surface contained three colors namely red, blue, and white. The red and blue regions show the short interatomic contacts and long interatomic contacts, respectively.34,35 The white regions stand for the contacts having an interatomic distance equal to the sum of van der Waals radii. The short contacts or the contacts that are suitable for the hydrogen bonding interactions are depicted by the red spots on the HS (Figure 6a). The green dotted lines in Figure 6a represent H-bonding interactions. π···π stacking interactions can be visualized by HS that is plotted over the shape index.36,37 The existence of consecutive red and blue triangle areas on the HS around aromatic rings indicates their involvement in π···π stacking interactions.38 The presence of such regions on the HS indicates the existence of the π···π stacking interactions in FTEAA (Figure 6b). However, these π···π stacking interactions are very weak as the intercentroid distance for this interaction ranges from 4.5501 (13) to 5.9959 (12) Å.
Figure 6.
HS plotted over (a) dnorm in the range of −0.2348 to 1.5584 au; (b) shape index in the range of −1 to 1 au.
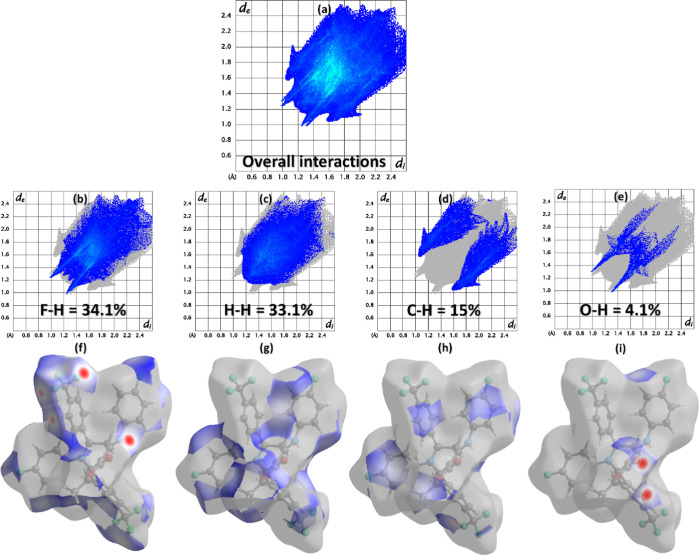
2D fingerprint plots express the involvement of the interatomic contacts in the packing of the crystal.39,40 di and de represents the distances from the HS to the nearest atom both inside and outside of it, respectively. Figure 7a is the 2D plot for all kinds of probable interactions in FTEAA. The fluorine atoms play an important role in the packing of the crystal, as the most significant contact for FTEAA is H–F or F–H (34.1%), as shown in Figure 7b. Figure 7f represents HS associated with H–F or F–H contact, which clearly shows the involvement of the fluorine atoms in the crystal packing interactions. The interacting region is shown by the red and blue regions of the surface. The other important contact is H–H, whose contribution to crystal packing is very close to that of H–F contacts (Figure 7c). The blue regions around H-atoms in Figure 7g show short H–H contacts. As expected, while looking at the hydrogen bond geometry table (Table 3), C–H contacts must have a noteworthy involvement (15%) in the packing of the crystal. The most important interatomic contact that involves O-atoms is O–H, with a percentage contribution of 4.1% (Figure 7e). The contacts with the relatively smaller contribution to the packing of the crystal are manifested in Figure S2.
Figure 7.
(a) 2D plots for overall interaction. (b–i) 2D plots of important contacts and their HS.
Table 3. Hydrogen-bond Geometry (Å, deg)a.
| D–H···A | D–H | H···A | D···A | ∠(D–H···A)° |
|---|---|---|---|---|
| N2–H2···O1 | 0.88 | 2.11 | 2.683 (2) | 122 |
| N2–H2···F5i | 0.88 | 2.34 | 3.067 (2) | 140 |
| C11–H11···O1ii | 0.95 | 2.44 | 3.345 (2) | 159 |
| C24–H24···F7iii | 0.95 | 2.51 | 3.356 (2) | 148 |
| C15–H15C···F7iv | 0.98 | 2.62 | 3.570 (3) | 164 |
| C31–H31···F8v | 0.95 | 2.48 | 3.412 (2) | 168 |
| C–H···π | C–H | H···π | C··· π | ∠(C–H··· π)° |
| C21–H21···Cg1 | 0.95 | 2.95 | 3.508 (2) | 119 |
| C15–H15B···Cg2vi | 0.98 | 2.73 | 3.624 (2) | 152 |
Symmetry codes: (i) −x + 2, −y + 2, −z + 1; (ii) −x + 2, −y + 1, −z + 1; (iii) −x + 1, −y + 2, −z + 1; (iv) x, y–1, z; (v) −x, −y + 1, −z; (vi) 1 – x, 1 – y, 1 – z. Cg1 and Cg2 are the centroid of phenyl rings (C22–C27) and (C29–C34), respectively.
The enrichment ratio for the pair of chemical moieties in the crystal provides the tendency of that pair of chemical species to make the crystal packing interactions. The enrichment ratio is obtained by dividing the proportion of the actual contact in the crystal packing with the proportion of random contact calculated theoretically. Table S1 shows the enrichment ratio of the pair of chemical moieties in FTEAA. The O–H contact has the maximum tendency to generate crystal packing interactions in FTEAA, with an enrichment ratio of 1.43. The other contacts of higher propensity are C–H, F–H, and F–C with enrichment ratios of 1.19, 1.05, and 1.04, respectively.
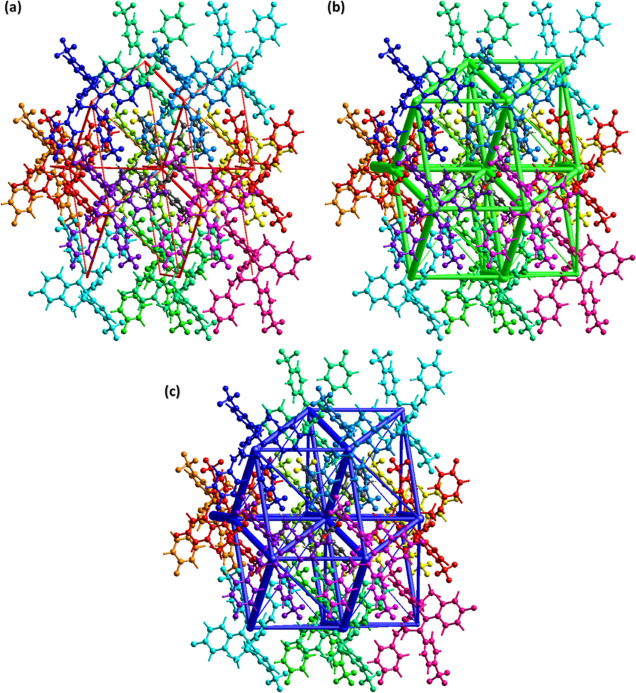
To explore the interactions between pairs of molecules, interaction energy is calculated by creating a bunch of molecules of size 3.8 Å around the reference molecule (Figure 8a). The B3LYP/6-31G(d,p) level electron density model is used for the calculations.41 Fifteen symmetry-related molecules are present around the reference molecule, and the coordinates of atoms that are involved in the interaction energy calculation are given in Table S2. The total energy between the molecular pair is the sum of four energies known as electrostatic, polarization, dispersion, and repulsion. The negative energy value designates that the energy is attractive, whereas the positive energy value indicates repulsive energy. The total energy is greater for the molecule pairs for which the distance between the molecular center is small. Figure 8b shows that for most of the molecular pairs, the dispersion energy term dominates over other energy terms. Electrostatic energy is mostly attractive but, in some cases, it may be repulsive. For example, in the present case, it is repulsive for the molecular pairs, with the distance between the molecular centers being 10.04, 13.21, and 16.36 Å.
Figure 8.
(a) Chemical diagram of the color-coded interaction mapping inside 3.8 Å of the reference molecule; (b) interaction energy results.
Energy frameworks for a single crystal are important to realize the topology of the single crystal.42 Interaction energies are used as input to form energy frameworks. The size or width of the cylinders is directly proportional to the strength of the interaction energy type. The width of the cylinders is smaller in the case of electrostatic energy (Figure 9a) as compared to the width of the cylinders in the case of dispersive energy (Figure 9b), which indicates that the contribution of dispersive energy is greater in defining the total energy as compared to the contribution of electrostatic energy. Figure 9c is the energy frame for the total energy, which is obtained by adding up the contributions of electrostatic energy and dispersive energy.
Figure 9.
Energy framework for (a) coulomb energy, (b) dispersion energy, and (c) total energy.
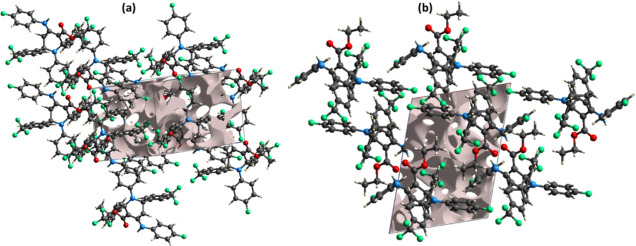
A void analysis is important to understand the response of the single crystals to the applied stress. It is related to the mechanical properties of single crystals. The voids in the crystal packing are calculated by using the Hartree–Fock theory.43,44 The electron density of all the atoms present in the molecule is added up while assuming that the atoms are spherically symmetrical (Figure 10). The volume of the void is determined to be 216.02 Å3. As voids occupy a very small amount of space (14.6%) in the crystal packing, this suggests that the molecules are tightly packed and that the crystal packing has no big cavities.
Figure 10.
Graphical views of voids in the crystal packing of FTEAA along (a) c-axis and (b) b-axis.
2.3. 1H NMR and 13C NMR Spectroscopy
In the 1H NMR spectrum, three protons of the methyl group were detected as a triplet at δ = 1.47 ppm. Two methylene protons (OCH2) were observed as a multiplet at δ = 4.51–4.32 ppm. Two methylene protons present in the piperidine ring were observed as two doublets of doublets representing one proton each at δ = 2.65 and 2.81 ppm. A multiplet for N–CH–CH2 of the ring was found at δ = 5.09–5.19 ppm. Another N–CH– was found as a singlet at δ = 6.39 ppm. Protons present in the trifluoro methylated aromatic ring were observed downfield as compared to the fluoro-containing aromatic ring. All the sixteen aromatic protons were observed between δ = 7.60–6.72 ppm. The proton present in the −NH was found as a broad singlet at δ = 10.19 ppm.
Similarly, the 13C NMR spectrum of FTEAA has shown carbon peaks in total agreement with the carbon atoms present in FTEAA. The carbon atom of the methyl group was found at δ = 14.43 ppm, methylene attached to methyl was found at δ = 33.26 ppm, the carbonyl carbon was observed at δ = 167.54 ppm, and the alkenic carbon attached to carbonyl was found at δ = 97.11 ppm, whereas the second alkenic carbon attached to nitrogen atom was observed at δ = 137.08 ppm. All the aromatic carbon atoms were observed in the range of δ = 155.39–112.51 ppm. Trifluoro methylated carbon atom was found as a quartet at 125.01 ppm.
2.4. Monoamine Oxidase Inhibition
FTEAA was evaluated for its inhibitory potential against MAO-A and MAO-B enzymes. For this purpose, clorgyline and deprenyl were utilized as the standard drugs for MAO-A and MAO-B, respectively. FTEAA showed excellent inhibition of both MAOs in the lower micromolar range. The inhibitory concentration (IC50) values of the tested compound and standard inhibitors are shown in Table 4.
Table 4. Monoamine Oxidase A and B Inhibition.
| IC50 ± SEM (μM)/%age inhibition |
||
|---|---|---|
| code | MAO-A | MAO-B |
| FTEAA | 0.52 ± 0.03 | 1.02 ± 0.11 |
| clorgyline | 0.0045 ± 0.0003 | 61.35 ± 1.13 |
| deprenyl | 67.25 ± 1.02 | 0.0196 ± 0.001 |
3. Computational Procedure
3.1. Binding Energy and Intermolecular Analysis
The FTEAA was analyzed after the docking that was carried out with two of the selected proteins, 2V5Z and 2Z5X. The docking studies gave rise to nine different conformational modes in which FTEAA could fit into the binding pocket of proteins. However, for the analysis of resulting interactions of FTEAA, its binding mode with the lowest root mean square deviation value was preferred over others. The binding energies were −8.8 and −9.6 kcal/mol for proteins 2V5Z and 2Z5X, respectively (see Table 5). These reasonably good binding energies (more negative values) indicate the effective docking process and efficient associations between the FTEAA and proteins. The protein and FTEAA were then analyzed for intermolecular interactions. There are diverse interactions between FTEAA and proteins, including conventional hydrogen bonds; carbon–hydrogen bonds; and alkyl, π-alkyl, and halide interactions. Visualizing these interactions is essential and tells us the extent of association between the macromolecules (proteins) and FTEAA. The docking interaction visualization results of the FTEAA with the 2V5Z protein have been verified and shown in Figure 11. It is obvious that favorable hydrogen bonding has been developed with ARG36 and ARG38 residues at 2.24 and 2.2 Å distances, respectively. Furthermore, the halide interactions can be seen with GLU390 and ASP37 residues. The alkyl and π-alkyl interactions can be observed with PRO234, ARG36, and ARG38 amino acid residues.
Table 5. Binding Energies (kcal/mol) between FTEAA and Selected Proteins, the Number of H-bonds, and Hydrophobic and Electrostatic Interactions for Docked Proteins.
Figure 11.
Interactions of the FTEAA with the 2V5Z protein; (a) overview of FTEAA adjustment in the binding pocket of protein with interacting residues, where green lines show hydrogen bond interactions between FTEAA and amino acid residues of 2V5Z; (b) 3-D diagram of the focused view having FTEAA and protein residues around it; (c) clear view having various residues and FTEAA interactions in a 2-dimensional illustration.
The docking interaction visualization results of the FTEAA with the 2Z5X protein have been demonstrated in Figure 12. The docking developed desirable hydrogen bonding between the FTEAA and ARG76 with a 2.3 Å interaction distance. Another hydrogen bond emerged with GLN474 at the 2.73 Å interaction length. Other considerable associations include halide (fluorine) interactions with several residues, including GLU474, LYS199, VAL473, and GLU450. The π-alkyl associations were observed with LYS440 and TRP472. Carbon–hydrogen bonding was also observed with certain residues.
Figure 12.
Interactions of FTEAA with the 2Z5X protein; (a) overview of FTEAA adjustment in the binding pocket of the protein with interacting residues, where green lines show hydrogen bond interactions between FTEAA and amino acid residues of 2Z5X; (b) 3-D diagram of the focused view having FTEAA and protein residues around it and interaction distances; (c) 2-D diagram of the clear view has various residues and FTEAA interactions.
3.2. ADMET and Drug Likeness
Usually, the in silico computationally designed drugs undergo analysis for their pharmacokinetic properties before being experimented on in vivo or practicing them clinically. These drugs should follow four out of five of Lipinski’s rules, which include that the molecular weight of the drug must not exceed 500 g/mol, hydrogen bond donors must be under five, hydrogen bond acceptors must essentially be less than ten, its lipophilicity (log P) should not cross the digit five, and the number of rotatable bonds must be less than 10. Table 6 represents the potential of FTEAA to be used as an inhibitor drug. It may be seen that FTEAA truly follows the rule of five, and its properties come under the prescribed range. This indicates the physiochemical favorability of this drug to inhibit the proteins efficiently and control the disease spread.
Table 6. Predicted Pharmacokinetic Analysis of FTEAA.
| compound | intestinal absorption (%) | total clearance (log/mL/min/kg) | max. tolerable dose (log mg/kg/day) | RO5 rule |
|---|---|---|---|---|
| FTEAA | 95.664 | 0.812 | 0.493 | yes |
The toxicological traits and pharmacokinetics of the FTEAA are determined by evaluating ADMET analysis. This analysis is done immediately after the docking process so that inhibitors with poor excretion and absorption properties and hepatotoxicity may be rejected early. In the FTEAA, it may be seen that intestinal absorption is high, at about 95.66% (see Table 6). The total clearance is 0.446 log/mL/min/kg for the entitled compound. The tolerable dose is prescribed to be 0.513 log mg/kg/day in the case of both the proteins.
4. Conclusions
Efficient synthesis of highly functionalized piperidine FTEAA was accomplished by following a single-pot five-component procedure. The structure of FTEAA was validated with NMR (1H and 13C), Fourier-transform infrared (FT-IR) spectroscopy, and single-crystal XRD analysis. The supramolecular environment is elaborated by HS analysis, from which it is inferred that the H–F/F–H contact is a noteworthy donor to the crystal packing, and the O–H contact has a higher propensity to make the crystal packing interactions relative to other contacts. The void analysis confirmed the strength of the crystal packing. The supramolecular environment is further explored by the interaction energy between the molecular pairs and by the construction of the energy frameworks. FTEAA is found to be an effective inhibitor of both monoamine oxidase A and B with IC50 values of 0.52 ± 0.03 and 1.02 ± 0.11 μM. Additionally, the in silico molecular docking analysis also indicated that there is a reasonably good association between the FTEAA ligand and selected MAO proteins. The binding interaction energies were calculated through molecular docking methods. The entitled ligand shows binding interaction energies of −8.8 and −9.6 kcal/mol for proteins 2V5Z and 2Z5X, respectively. The negative binding energies were further supported by the study and visualization of intermolecular interactions among entitled ligand and proteins. The ADMET and drug-likeness studies indicated the potential of FTEAA to be used as a monoamine oxidase inhibitor.
5. Experimental Section
5.1. Material and Techniques
All the chemicals, reagents, and solvents were acquired from Sigma-Aldrich and utilized without further purification. The melting point of FTEAA was measured by using the Gallenkamp melting point apparatus (MP-D). NMR spectra (1H and 13C) were measured on a Bruker AV-400 spectrometer (400 MHz). The FT-IR spectrum was recorded in the ATR (attenuated total reflectance) mode by using a Shimadzu FT-IR spectrophotometer.
5.1.1. Preparation of Ethyl 4-(4-fluorophenylamino)-2,6-bis(4-(trifluoromethyl)phenyl)-1-(4-fluorophenyl)-1,2,5,6-tetrahydropyridine-3-carboxylate
To prepare compound FTEAA, a 5 mL methanolic solution of 4-fluoroaniline (2 mmol, 186 mg), ethyl acetoacetate (1 mmol, 130 mg), and molecular iodine (10 mol %, 25 mg) was stirred at 55 °C for 25 min and 4-trifluoromethyl benzaldehyde (2 mmol, 348 mg) was added. After 12 h, the contents were cooled down to room temperature and subsequent precipitates were filtered, washed with absolute ethanol, dried, and recrystallized from 10% ethyl acetate/n-hexane to obtain FTEAA.
Colorless solid; yield 73%; mp 218–220 °C; solubility: acetone, chloroform, dichloromethane, ethyl acetate and ethanol; 1H NMR (400 MHz, CDCl3): δ ppm 1.47 (t, J = 7.15 Hz, 3H), 2.65 (dd, J = 15.31, 2.76 Hz, 1H), 2.81 (dd, J = 15.31, 5.77 Hz, 1H), 4.28–4.41 (m, 1H), 4.41–4.57 (m, 1H), 5.09–5.19 (m, 1H), 6.26–6.32 (m, 2H), 6.33–6.38 (m, 2H) 6.39 (s, 1H), 6.72–6.87 (m, 4 H), 7.20–7.31 (m, 2H), 7.43 (d, J = 8.28 Hz, 2H), 7.52–7.60 (m, 4H) 10.19 (br s, 1H) (Figure S3). 13C NMR (101 MHz, CDCl3): δ ppm 14.43, 33.26, 54.65, 57.35, 59.66, 76.36, 76.68, 97.11, 112.51, 116.67, 122.44, 122.56, 124.93, 124.97, 125.01, 125.28, 125.31, 125.40, 125.82, 126.36, 126.54, 128.28, 128.66, 128.84, 129.44, 137.08, 145.92, 146.27, 146.28, 147.73, 155.39, 167.54 (Figure S4). FT-IR ν(cm–1): 3242 (N–H), 3080 (ArCH), 2981 (CH2), 1649 (C=O), 1591 (C=C), 1115 (C–O), 764 (C=C).
The one-step five-component synthetic route was implemented to access the desired piperidine FTEAA as shown in Scheme 1. The reported procedure was followed to prepare the desired compound.45,46 The purity was established based on thin-layer chromatography. The structure of FTEAA was confirmed on the basis of 1H and 13C NMR, FT-IR, and single-crystal XRD analysis.
Scheme 1. Synthetic Route to Access FTEAA.
Conditions: 4-fluoroaniline (2.0 mmol), ethyl acetoacetate (1.0 mmol), molecular iodine (10 mol %), 4-trifluoromethyl benzaldehyde (2.0 mmol), methanol, 12 h, 55 °C.
5.2. Single Crystal Analysis Details
For the single crystal analysis of FTEAA by X-ray, a Bruker Kappa Apex-II CCD diffractometer is used along with APEX-II software for the collection of data. The diffractometer contains an X-ray source that produces Cu Kα radiations, and data is collected at a low temperature of 150 K for the best possible results. SHELXT-201447 and SHELXL 2019/248 are used for the solution and refinement of the crystal structure, respectively. All the nonhydrogen atoms are assigned ADP in refinement, whereas all the H-atoms are assigned isotropic displacement parameters and refined with the help of the riding model. Initially, one of the trifluoro groups has unusual thermal ellipsoids. The problem is identified and resolved by solving the disorder in the trifluoro group by using DFIX, DANG, and EADP restraints. ORTEP-3,49 PLATON,50 and Mercury51 were used for the graphical illustration of the single-crystal XRD data.
5.3. Monoamine Oxidase Activity
The assay was conducted in white 96-well plates as previously described.52 Before use, the enzymes MAO-A and MAO-B were irreversibly blocked at 25 °C for 15–20 min with clorgyline and deprenyl, respectively. The assay volume for MAO-A was 200 μL, containing 145 μL buffer, 10 μL of test compound (100 μM end concentration), and 20 μL (26 μg end concentration) of the freshly prepared enzyme. The assay volume for MAO-B was 100 μL, containing 45 μL of buffer, 10 μL of test compound (100 μM end concentration), and 20 μL (5 μg end concentration) of the freshly prepared enzyme. The reaction mixture was incubated at 37 °C for 10 min separately. After 15 min, 10 μL of the substrate (0.3 mM final concentration) was added, and at the end, 10 μL of Amplex red (fluorogenic substrate) was added to each well. The change in fluorescence was observed by using a FLUOstar Omega fluorescence plate reader (BMG Labtech GmbH, Ortenberg, Germany). The species with more than 50% inhibition of either MAO-A or MAO-B were further investigated and IC50 values were calculated with the help of the PRISM 5.0 (GraphPad, San Diego, California, USA) program.
6. Computational Methodology
6.1. Protein Preparation
The 3-dimensional structures of MAO-B (PDB ID: 2V5Z, resolution 1.60 Å) and MAO-A (PDB ID: 2Z5X) with a resolution of 2.20 Å were attained from the Protein Bank database.53 The MGL Tools were used to prepare the proteins for molecular docking analysis.54 Co-crystalized ligands and water molecules were removed from macromolecules and polar hydrogens, and Kollman Charges were added.55
6.2. Ligand Preparation
The experimental structure of the FTEAA was fully optimized by using density functional theory methods with B3LYP, along with a 6-311G** basis set.56 The FTEAA was prepared in the PDBQT format in the MGL tool, and torsions were set by converting rotatable bonds into nonrotatable ones so that the FTEAA could not undergo structural variation or bond modification during the docking process. Some further details for molecular docking methodology can be seen from our previous work with similar types of calculations.57,58
6.3. Molecular Docking
The prepared proteins and FTEAA were subjected to molecular docking using Autodock Vina 4.2. A configuration file was prepared, and the grid centers and grid dimensions were set according to the prereported literature. However, the grid dimensions were also confirmed by locating the residues interacting with co-crystallized inhibitors.59 After docking, the 3-D and 2-D interactions of FTEAA with amino acid residues were visualized by the Biovia Discovery Studio visualizer (2020).
6.4. ADMET Analysis
The physicochemical properties of the FTEAA were estimated through ADMET analysis, which includes adsorption, desorption, metabolism, excretion, and toxicity. This analysis was performed using a reliable online tool called pkCSM.60 ADMET properties indicate the drug-like properties of the FTEAA.
Acknowledgments
B.A.K. is thankful to HEC Pakistan for the financial support (NRPU-6455). The authors from King Khalid University extend their appreciation to the Deanship of Scientific Research at King Khalid University, Saudi Arabia for funding this work through Small Groups RGP.1/318/43. For computer time, this research used the resources of the Supercomputing Laboratory at King Abdullah University of Science & Technology (KAUST) in Thuwal, Saudi Arabia.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c03909.
Crystallographic data for FTEAA (CIF)
Stimulated powder pattern of FTEAA; remaining 2D plots (a-e) of FTEAA; 1H NMR and 13C NMR of compound FTEAA; enrichment ratio of the pair of the chemical species in FTEAA; and Cartesian coordinates of all atoms [16 molecules], 1152 atoms, that are involved in the interaction energy calculation (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- a Yoneda Y.; Kawajiri S.; Hasegawa A.; Kito F.; Katano S.; Takano E.; Mimura T. Synthesis of polyamine derivatives having non-hypotensive Ca2+-permeable AMPA receptor antagonist activity. Bioorg. Med. Chem. Lett. 2001, 11, 1261–1264. 10.1016/s0960-894x(01)00170-6. [DOI] [PubMed] [Google Scholar]; b Zheng Y.-Y.; Guo L.; Zhen X.-C.; Li J.-Q. Synthesis and antidepressant activity of arylalkanol-piperidine derivatives as triple reuptake inhibitors. Eur. J. Med. Chem. 2012, 54, 123–136. 10.1016/j.ejmech.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Funakoshi T.; Chaki S.; Kawashima N.; Suzuki Y.; Yoshikawa R.; Kumagai T.; Nakazato A.; Kameo K.; Goto M.; Okuyama S. In vitro and in vivo pharmacological profile of 5-{2-[4-(6-fluoro-1H-indole-3-yl)piperidin-1-yl]ethyl}-4-(4-fluorophenyl)thiazole-2-carboxylic acid amide (NRA0562), a novel and putative atypical antipsychotic. Life Sci. 2002, 71, 1371–1384. 10.1016/s0024-3205(02)01863-5. [DOI] [PubMed] [Google Scholar]
- a Pati H. N.; Das U.; Quail J. W.; Kawase M.; Sakagami H.; Dimmock J. R. Cytotoxic 3,5-bis(benzylidene)piperidin-4-ones and N-acyl analogs displaying selective toxicity for malignant cells. Eur. J. Med. Chem. 2008, 43, 1–7. 10.1016/j.ejmech.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wardell J. L.; De Souza M. V.; Wardell S. M.; Lourenço M. C. Mefloquine derivatives: Crystal structures and anti-tubercular activities of diphenyl [((R*, S*)-2, 8-bis (trifluoromethyl) quinolin-4-yl)-piperidin-2-yl-methanolato-O, N] boron and (±)-erythro-mefloquinium tetraphenylborate solvates. J. Mol. Struct. 2011, 990, 67–74. 10.1016/j.molstruc.2011.01.019. [DOI] [Google Scholar]
- Trapella C.; Fischetti C.; Pela’ I.; Lazzari R.; Guerrini A.; Calo’ V.; Rizzi D. G.; Camarda J.; Lambert D.; McDonald S.; Regoli D.; Salvadori S. Structure-activity studies on the nociceptin/orphanin FQ receptor antagonist 1-benzyl-N-{3-[spiroisobenzofuran-1(3H),4′-piperidin-1-yl]propyl} pyrrolidine-2-carboxamide. Bioorg. Med. Chem. 2009, 17, 5080–5095. 10.1016/j.bmc.2009.05.068. [DOI] [PubMed] [Google Scholar]
- Santana L.; González-Díaz H.; Quezada E.; Uriarte E.; Yáñez M.; Viña D.; Orallo F. Quantitative Structure–Activity Relationship and Complex Network Approach to Monoamine Oxidase A and B Inhibitors. J. Med. Chem. 2008, 51, 6740–6751. 10.1021/jm800656v. [DOI] [PubMed] [Google Scholar]
- Chhabria M. T.; Patel S.; Modi P.; Brahmkshatriya P. S. Thiazole: A review on chemistry, synthesis and therapeutic importance of its derivatives. Curr. Top. Med. Chem. 2016, 16, 2841–2862. 10.2174/1568026616666160506130731. [DOI] [PubMed] [Google Scholar]
- Pisani L.; Catto M.; Leonetti F.; Nicolotti O.; Stefanachi A.; Campagna F.; Carotti A. Targeting Monoamine Oxidases with Multipotent Ligands: An Emerging Strategy in the Search of New Drugs Against Neurodegenerative Diseases. Curr. Med. Chem. 2011, 18, 4568–4587. 10.2174/092986711797379302. [DOI] [PubMed] [Google Scholar]
- Lewinsohn R.; Glover V.; Sandler M. β-Phenylethylamine and benzylamine as substrates for human monoamine oxidase A: A source of some anomalies?. Biochem. Pharmacol. 1980, 29, 777–781. 10.1016/0006-2952(80)90556-0. [DOI] [PubMed] [Google Scholar]
- Youdim M. B.; Edmondson D.; Tipton K. F. The therapeutic potential of monoamine oxidase inhibitors. Nat. Rev. Neurosci. 2006, 7, 295–309. 10.1038/nrn1883. [DOI] [PubMed] [Google Scholar]
- Fowler C. J.; Mantle T. J.; Tipton K. F. The nature of the inhibition of rat liver monoamine oxidase types A and B by the acetylenic inhibitors clorgyline, l-deprenyl and pargyline. Biochem. Pharmacol. 1982, 31, 3555–3561. 10.1016/0006-2952(82)90575-5. [DOI] [PubMed] [Google Scholar]
- Esfahani A. N.; Mirzaei M. Flavonoid Derivatives for Monoamine Oxidase–A Inhibition. Adv. J. Chem., Sect. B 2019, 1, 17–22. 10.33945/sami/ajcb.2019.1.4. [DOI] [Google Scholar]
- Clarke A.; Brewer F.; Johnson E. S.; Mallard N.; Hartig F.; Taylor S.; Corn T. H. A new formulation of selegiline: improved bioavailability and selectivity for MAO-B inhibition. J. Neural. Transm. 2003, 110, 1241–1255. 10.1007/s00702-003-0036-4. [DOI] [PubMed] [Google Scholar]
- Finberg J. P.; Rabey J. M. Inhibitors of MAO-A and MAO-B in Psychiatry and Neurology. Front. Pharmacol. 2016, 7, 340. 10.3389/fphar.2016.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knez D.; Colettis N.; Iacovino L. G.; Sova M.; Pišlar A.; Konc J.; Lešnik S.; Higgs J.; Kamecki F.; Mangialavori I.; Dolšak A.; Žakelj S.; Trontelj J.; Kos J.; Binda C.; Marder M.; Gobec S. Stereoselective activity of 1-propargyl-4-styrylpiperidine-like analogues that can discriminate between monoamine oxidase isoforms A and B. J. Med. Chem. 2020, 63, 1361–1387. 10.1021/acs.jmedchem.9b01886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L.; Murray S.; Castagnoli K.; Castagnoli N. Studies on 1,2,3,6-tetrahydropyridine derivatives as potential monoamine oxidase inactivators. Chem. Res. Toxicol. 1992, 5, 625–633. 10.1021/tx00029a006. [DOI] [PubMed] [Google Scholar]
- a Kalgutkar A. S.; Castagnoli N. Synthesis of novel MPTP analogs as potential monoamine oxidase B (MAO-B) inhibitors. J. Med. Chem. 1992, 35, 4165–4174. 10.1021/jm00100a023. [DOI] [PubMed] [Google Scholar]; b Youngster S. K.; McKeown K. A.; Jin Y. Z.; Ramsay R. R.; Heikkila R. E.; Singer T. P. Oxidation of Analogs of l-Methyl-4-Phenyl-1,2,3,6-Tetrahydropyridine by Monoamine Oxidases A and B and the Inhibition of Monoamine Oxidases by the Oxidation Products. J. Neurochem. 1989, 53, 1837–1842. 10.1111/j.1471-4159.1989.tb09250.x. [DOI] [PubMed] [Google Scholar]
- Riachi N. J.; Arora P. K.; Sayre L. M.; Harik S. I. Potent neurotoxic fluorinated 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine analogs as potential probes in models of Parkinson disease. J. Neurochem. 1988, 50, 1319–1321. 10.1111/j.1471-4159.1988.tb10610.x. [DOI] [PubMed] [Google Scholar]
- a Böhm H.-J.; Banner D.; Bendels S.; Kansy M.; Kuhn B.; Müller K.; Obst-Sander U.; Stahl M. Fluorine in Medicinal Chemistry. Chembiochem 2004, 5, 637. 10.1002/cbic.200301023. [DOI] [PubMed] [Google Scholar]; b Kirk K. L. Fluorine in medicinal chemistry: Recent therapeutic applications of fluorinated small molecules. J. Fluorine Chem. 2006, 127, 1013–1029. 10.1016/j.jfluchem.2006.06.007. [DOI] [Google Scholar]; c Purser S.; Moore P. R.; Swallow S.; Gouverneur V. Fluorine in medicinal chemistry. Chem. Soc. Rev. 2008, 37, 320–330. 10.1039/b610213c. [DOI] [PubMed] [Google Scholar]
- a Jeschke P. The Unique Role of Fluorine in the Design of Active Ingredients for Modern Crop Protection. ChemBioChem 2004, 5, 570–589. 10.1002/cbic.200300833. [DOI] [PubMed] [Google Scholar]; b Kirsch P.; Bremer M. Nematic Liquid Crystals for Active Matrix Displays: Molecular Design and Synthesis. Angew. Chem., Int. Ed. 2000, 39, 4216–4235. . [DOI] [PubMed] [Google Scholar]; c Kirk K. L. Fluorination in Medicinal Chemistry: Methods, Strategies, and Recent Developments. Org. Process Res. Dev. 2008, 12, 305–321. 10.1021/op700134j. [DOI] [Google Scholar]
- Chinnaraja D.; Rajalakshmi R. Atom and step economic multicomponent synthesis of highly functionalized novel N-alkyl piperidines: structural elucidation through spectral studies and single crystallographic analysis. RSC Adv. 2014, 4, 41314–41322. 10.1039/c4ra06505k. [DOI] [Google Scholar]
- Ramachandran R.; Jayanthi S.; Jeong Y. T. One-pot synthesis of highly diversified tetrahydropyridines by tandem condensation of aldehydes, amines, and β-ketoesters. Tetrahedron 2012, 68, 363–369. 10.1016/j.tet.2011.09.105. [DOI] [Google Scholar]
- Anand P. S.; Sethukumar A.; Kumar C. U.; Krishnasamy K.; Senthan S.; Manikandan G.; Prakasam B. A. Synthesis, stereochemical, single crystal X-ray structural and antimicrobial studies of some isobutyl-1,2,6-triaryl-4-(arylamino)-1,2,5,6-tetrahydropyridine-3-carboxylates: Exploring RAHB with S(6) graph set. J. Mol. Struct. 2021, 1227, 129563. 10.1016/j.molstruc.2020.129563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthal S.; Brahmachari G.; Das S.; Kant R.; Gupta V. K. Ethyl 4-anilino-2,6-bis(4-fluorophenyl)-1-phenyl-1,2,5,6-tetrahydropyridine-3-carboxylate. Acta Crystallogr., Sect. E: Struct. Rep. Online 2013, 69, o299–o300. 10.1107/s1600536813002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh R.; Maheswari S.; Arivazhagan M.; Malecki J. G.; Lalitha A. Cyanuric chloride catalyzed metal-free mild protocol for the synthesis of highly functionalized tetrahydropyridines. Tetrahedron Lett. 2017, 58, 3905–3909. 10.1016/j.tetlet.2017.08.074. [DOI] [Google Scholar]
- Anthal S.; Brahmachari G.; Das S.; Kant R.; Gupta V. K. Methyl 4-(4-fluoroanilino)-1,2,6-tris(4-fluorophenyl)-1,2,5,6-tetrahydropyridine-3-carboxylate. Acta Crystallogr., Sect. E: Struct. Rep. Online 2013, 69, o373–o374. 10.1107/s160053681300370x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajadikhah S. S.; Hazeri N.; Maghsoodlou M. T.; Habibi-Khorassani S. M.; Willis A. C. Trityl chloride as an efficient organic catalyst for one-pot, five-component and diastereoselective synthesis of highly substituted piperidines. Res. Chem. Intermed. 2014, 40, 723–736. 10.1007/s11164-012-0997-8. [DOI] [Google Scholar]
- Anthal S.; Brahmachari G.; Das S.; Kant R.; Gupta V. K. Ethyl 2,6-bis(4-chlorophenyl)-4-(4-fluoroanilino)-1-(4-fluorophenyl)-1,2,5,6-tetrahydropyridine-3-carboxylate. Acta Crystallogr., Sect. E: Struct. Rep. Online 2013, 69, o506–o507. 10.1107/s1600536813006090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman P. R.; Turner M. J.; McKinnon J. J.; Wolff S. K.; Grimwood D. J.; Jayatilaka D.; Spackman M. A. CrystalExplorer: a program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54, 1006. 10.1107/S1600576721002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargar H.; Fallah-Mehrjardi M.; Behjatmanesh-Ardakani R.; Torabi V.; Munawar K. S.; Ashfaq M.; Tahir M. N. Sonication-assisted synthesis of new Schiff bases derived from 3-ethoxysalicylaldehyde: Crystal structure determination, Hirshfeld surface analysis, theoretical calculations and spectroscopic studies. J. Mol. Struct. 2021, 1243, 130782. 10.1016/j.molstruc.2021.130782. [DOI] [Google Scholar]
- a Ashfaq M.; Munawar K. S.; Tahir M. N.; Dege N.; Yaman M.; Muhammad S.; Alarfaji S. S.; Kargar H.; Arshad M. U. Synthesis, Crystal Structure, Hirshfeld Surface Analysis, and Computational Study of a Novel Organic Salt Obtained from Benzylamine and an Acidic Component. ACS Omega 2021, 6, 22357–22366. 10.1021/acsomega.1c03078. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Madni M.; Ahmed M. N.; Hafeez M.; Ashfaq M.; Tahir M. N.; Gil D. M.; Galmés B.; Hameed S.; Frontera A. Recurrent π-π stacking motifs in three new 4,5-dihydropyrazolyl-thiazole-coumarin hybrids: X-ray characterization, Hirshfeld surface analysis and DFT calculations. New J. Chem. 2020, 44, 14592–14603. 10.1039/d0nj02931a. [DOI] [Google Scholar]
- Taia A.; Ibrahimi B.; Benhiba F.; Ashfaq M.; Tahir M. N.; Essaber M.; Aatif A.; Hökelek T.; Mague J. T.; Sebbar N. K.; Essassi E. M. Syntheses, single crystal X-ray structure, Hirshfeld surface analyses, DFT computations and Monte Carlo simulations of New Eugenol derivatives bearing 1,2,3-triazole moiety. J. Mol. Struct. 2021, 1234, 130189. 10.1016/j.molstruc.2021.130189. [DOI] [Google Scholar]
- a Spackman M. A.; Jayatilaka D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. 10.1039/b818330a. [DOI] [Google Scholar]; b Tahir M. N.; Ashfaq M.; de la Torre F. D. L. T.; Caballero J.; Hernández-Rodríguez E. W.; Ali A. Rationalizing the stability and interactions of 2,4-diamino-5-(4-chlorophenyl)-6-ethylpyrimidin-1-ium 2-hydroxy-3,5-dinitrobenzoate salt. J. Mol. Struct. 2019, 1193, 185–194. 10.1016/j.molstruc.2019.05.003. [DOI] [Google Scholar]; c Ashfaq M.; Tahir M. N.; Kuznetsov A.; Mirza S. H.; Khalid M.; Ali A. DFT and single crystal analysis of the pyrimethamine-based novel co-crystal salt: 2,4-diamino-5-(4-chloro-phenyl)-6-ethylpyrimidin-1-ium:4-hydroxybenzoate:methanol:hydrate (1:1:1:1) (DEHMH). J. Mol. Struct. 2020, 1199, 127041. 10.1016/j.molstruc.2019.127041. [DOI] [Google Scholar]
- a Ali A.; Khalid M.; Rehman M. F. U.; Haq S.; Ali A.; Tahir M. N.; Ashfaq M.; Rasool F.; Braga A. A. C. Efficient synthesis, SC-XRD, and theoretical studies of O-benzenesulfonylated pyrimidines: role of noncovalent interaction influence in their supramolecular network. ACS Omega 2020, 5, 15115–15128. 10.1021/acsomega.0c00975. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ali A.; Khalid M.; Tahir M. N.; Imran M.; Ashfaq M.; Hussain R.; Assiri M. A.; Khan I. Synthesis of Diaminopyrimidine Sulfonate Derivatives and Exploration of Their Structural and Quantum Chemical Insights via SC-XRD and the DFT Approach. ACS Omega 2021, 6, 7047. 10.1021/acsomega.0c06323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kargar H.; Forootan P.; Fallah-Mehrjardi M.; Behjatmanesh-Ardakani R.; Amiri Rudbari H. A.; Shahzad Munawar K. S.; Ashfaq M.; Nawaz Tahir M. N. Novel oxovanadium and dioxomolybdenum complexes of tridentate ONO-donor Schiff base ligand: Synthesis, characterization, crystal structures, Hirshfeld surface analysis, DFT computational studies and catalytic activity for the selective oxidation of benzylic alcohols. Inorg. Chim. Acta 2021, 523, 120414. 10.1016/j.ica.2021.120414. [DOI] [Google Scholar]; b Kargar H.; Fallah-Mehrjardi M.; Behjatmanesh-Ardakani R.; Munawar K. S.; Ashfaq M.; Tahir M. N. Titanium(IV) complex containing ONO-tridentate Schiff base ligand: Synthesis, crystal structure determination, Hirshfeld surface analysis, spectral characterization, theoretical and computational studies. J. Mol. Struct. 2021, 1241, 130653. 10.1016/j.molstruc.2021.130653. [DOI] [Google Scholar]
- a Raza H.; Yildiz I.; Yasmeen F.; Munawar K. S.; Ashfaq M.; Abbas M.; Ahmed M.; Younus H. A.; Zhang S.; Ahmad N. Synthesis of a 2D copper(II)-carboxylate framework having ultrafast adsorption of organic dyes. J. Colloid Interface Sci. 2021, 602, 43–54. 10.1016/j.jcis.2021.05.169. [DOI] [PubMed] [Google Scholar]; b Kargar H.; Fallah-Mehrjardi M.; Behjatmanesh-Ardakani R.; Tahir M. N.; Ashfaq M.; Munawar K. S. Synthesis, crystal structure determination, Hirshfeld surface analysis, spectral characterization, theoretical and computational studies of titanium (IV) Schiff base complex. J. Coord. Chem. 2021, 74, 2682. 10.1080/00958972.2021.1972984. [DOI] [Google Scholar]; c Ali A.; Kuznetsov A.; Khan M. U.; Tahir M. N.; Ashfaq M.; Raza A. R.; Muhammad S.; Muhammad S. 2-Amino-6-methylpyridine based co-crystal salt formation using succinic acid: Single-crystal analysis and computational exploration. J. Mol. Struct. 2021, 1230, 129893. 10.1016/j.molstruc.2021.129893. [DOI] [Google Scholar]
- Ashfaq M.; Munawar K. S.; Bogdanov G.; Ali A.; Tahir M. N.; Ahmed G.; Ramalingam A.; Alam M. M.; Imran M.; Sambandam S.; Munir B. Single crystal inspection, Hirshfeld surface investigation and DFT study of a novel derivative of 4-fluoroaniline: 4-((4-fluorophenyl) amino)-4-oxobutanoic acid (BFAOB). J. Iran. Chem. Soc. 2021, 19, 1953. 10.1007/s13738-021-02432-4. [DOI] [Google Scholar]
- Kargar H.; Fallah-Mehrjardi M.; Ashfaq M.; Munawar K. S.; Tahir M. N.; Behjatmanesh-Ardakani R.; Amiri Rudbari H.; Adabi Ardakani A.; Sedighi-Khavidak S. Zn (II) complexes containing O, N, N, O-donor Schiff base ligands: synthesis, crystal structures, spectral investigations, biological activities, theoretical calculations and substitution effect on structures. J. Coord. Chem. 2021, 74, 2720. 10.1080/00958972.2021.1990271. [DOI] [Google Scholar]
- Malik A. N.; Kuznetsov A.; Ali A.; Ashfaq M.; Tahir M. N.; Siddique A. Imine-based Zwitterion: Synthesis, single-crystal characterization, and computational investigation. J. Mol. Struct. 2022, 1253, 132237. 10.1016/j.molstruc.2021.132237. [DOI] [Google Scholar]
- a McKinnon J. J.; Jayatilaka D.; Spackman M. A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Commun. Chem. 2007, 3814–3816. 10.1039/b704980c. [DOI] [PubMed] [Google Scholar]; b Ashfaq M.; Bogdanov G.; Glebov V.; Ali A.; Tahir M. N.; Abdullah S. Single crystal investigation, Hirshfeld surface analysis and DFT exploration of the pyrimethamine-based novel organic salt: 2, 4-diamino-5-(4-chlorophenyl)-6-ethylpyrimidin-1-ium 3-carboxybenzoate hydrate (1: 1: 1). J. Mol. Struct. 2021, 1224, 129309. 10.1016/j.molstruc.2020.129309. [DOI] [Google Scholar]; c Ashfaq M.; Ali A.; Kuznetsov A.; Tahir M. N.; Khalid M. DFT and single-crystal investigation of the pyrimethamine-based novel co-crystal salt: 2,4-diamino-5-(4-chlorophenyl)-6-ethylpyrimidin-1-ium-4-methylbenzoate hydrate (1:1:1) (DEMH). J. Mol. Struct. 2021, 1228, 129445. 10.1016/j.molstruc.2020.129445. [DOI] [Google Scholar]
- a Kargar H.; Behjatmanesh-Ardakani R.; Fallah-Mehrjardi M.; Torabi V.; Munawar K. S.; Ashfaq M.; Tahir M. N. Ultrasound-based synthesis, SC-XRD, NMR, DFT, HSA of new Schiff bases derived from 2-aminopyridine: Experimental and theoretical studies. J. Mol. Struct. 2021, 1233, 130105. 10.1016/j.molstruc.2021.130105. [DOI] [Google Scholar]; b Ashfaq M.; Bogdanov G.; Ali A.; Tahir M. N.; Abdullah S. Pyrimethamine-Based Novel Co-Crystal Salt: Synthesis, Single-Crystal Investigation, Hirshfeld surface analysis and DFT inspection of the 2,4-diamino-5-(4-chlorophenyl)-6-ethylpyrimidin-1-ium 2,4-dichlorobenzoate (1:1) (DECB). J. Mol. Struct. 2021, 1235, 130215. 10.1016/j.molstruc.2021.130215. [DOI] [Google Scholar]; c Kargar H.; Bazrafshan M.; Fallah-Mehrjardi M.; Behjatmanesh-Ardakani R.; Rudbari H. A.; Munawar K. S.; Ashfaq M.; Tahir M. N. Synthesis, characterization, crystal structures, Hirshfeld surface analysis, DFT computational studies and catalytic activity of novel oxovanadium and dioxomolybdenum complexes with ONO tridentate Schiff base ligand. Polyhedron 2021, 202, 115194. 10.1016/j.poly.2021.115194. [DOI] [Google Scholar]
- Turner M. J.; Grabowsky S.; Jayatilaka D.; Spackman M. A. Accurate and efficient model energies for exploring intermolecular interactions in molecular crystals. J. Phys. Chem. Lett. 2014, 5, 4249–4255. 10.1021/jz502271c. [DOI] [PubMed] [Google Scholar]
- Mackenzie C. F.; Spackman P. R.; Jayatilaka D.; Spackman M. A. CrystalExplorermodel energies and energy frameworks: extension to metal coordination compounds, organic salts, solvates and open-shell systems. IUCrJ 2017, 4, 575–587. 10.1107/s205225251700848x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Turner M. J.; McKinnon J. J.; Jayatilaka D.; Spackman M. A. Visualisation and characterisation of voids in crystalline materials. CrystEngComm 2011, 13, 1804–1813. 10.1039/c0ce00683a. [DOI] [Google Scholar]; b Ashfaq M.; Tahir M. N.; Muhammad S.; Munawar K. S.; Ali A.; Bogdanov G.; Alarfaji S. S. Single-Crystal Investigation, Hirshfeld Surface Analysis, and DFT Study of Third-Order NLO Properties of Unsymmetrical Acyl Thiourea Derivatives. ACS Omega 2021, 6, 31211–31225. 10.1021/acsomega.1c04884. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Ashfaq M.; Munawar K. S.; Tahir M. N.; Dege N.; Yaman M.; Muhammad S.; Alarfaji S. S.; Kargar H.; Arshad M. U. Synthesis, crystal structure, Hirshfeld surface analysis, and computational study of a novel organic salt obtained from benzylamine and an acidic component. ACS Omega 2021, 6, 22357–22366. 10.1021/acsomega.1c03078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Haroon M.; Akhtar T.; Yousuf M.; Tahir M. N.; Rasheed L.; Zahra S. S.; Haq M.; Ashfaq M. Synthesis, crystal structure, Hirshfeld surface investigation and comparative DFT studies of ethyl 2-[2-(2-nitrobenzylidene)hydrazinyl]thiazole-4-carboxylate. BMC Chem. 2022, 16, 18. 10.1186/s13065-022-00805-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kargar H.; Ashfaq M.; Fallah-Mehrjardi M.; Behjatmanesh-Ardakani R.; Munawar K. S.; Tahir M. N. Unsymmetrical Ni (II) Schiff base complex: Synthesis, spectral characterization, crystal structure analysis, Hirshfeld surface investigation, theoretical studies, and antibacterial activity. J. Mol. Struct. 2022, 1265, 133381. 10.1016/j.molstruc.2022.133381. [DOI] [Google Scholar]
- Khan B. A.; Rehman O. u.; Alsfouk A. A.; Ejaz S. A.; Channar P. A.; Saeed A.; Ghafoor A.; Ujan R.; Mughal E. U.; Kumar R.; Yousuf S.; Hökelek T. Substituted piperidine as a novel lead molecule for the treatment of Parkinson’s disease: Synthesis, crystal structure, hirshfeld surface analysis, and molecular modeling. J. Mol. Struct. 2022, 1265, 133350. 10.1016/j.molstruc.2022.133350. [DOI] [Google Scholar]
- Hamdani S. S.; Khan B. A.; Saeed A.; Larik F. A.; Hameed S.; Channar P. A.; Ahmad K.; Mughal E. U.; Abbas Q.; Amin N. U.; Ghumro S. A.; Maitlo H.; Hassan M.; Raza H.; Seo S.-Y. Densely substituted piperidines as a new class of elastase inhibitors: Synthesis and molecular modeling studies. Arch. Pharm. 2019, 352, 1900061. 10.1002/ardp.201900061. [DOI] [PubMed] [Google Scholar]
- SHELXT S. G. Integrating space group determination and structure solution. Acta Crystallogr., Sect. A: Found. Adv. 2014, 70, C1437. 10.1107/s2053273314085623. [DOI] [Google Scholar]
- Sheldrick G. M. Crystal structure refinement withSHELXL. Acta Crystallogr., Sect. C: Struct. Chem. 2015, 71, 3–8. 10.1107/s2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrugia L. J. WinGXandORTEP for Windows: an update. J. Appl. Crystallogr. 2012, 45, 849–854. 10.1107/s0021889812029111. [DOI] [Google Scholar]
- Spek A. L. Structure validation in chemical crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2009, 65, 148–155. 10.1107/s090744490804362x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae C. F.; Sovago I.; Cottrell S. J.; Galek P. T.; McCabe P.; Pidcock E.; Platings M.; Shields G. P.; Stevens J. S.; Towler M.; Wood P. A. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Crystallogr. 2020, 53, 226–235. 10.1107/s1600576719014092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan B. A.; Hamdani S. S.; Ahmed M. N.; Rashid U.; Hameed S.; Ibrahim M. A. A.; Iqbal J.; Granados C. C.; Macías M. A.; Macedas M. A. Design, synthesis, crystal structures, computational studies, in vitro and in silico monoamine oxidase-A&B inhibitory activity of two novel S-benzyl dithiocarbamates. J. Mol. Struct. 2022, 1265, 133317. 10.1016/j.molstruc.2022.133317. [DOI] [Google Scholar]
- Jin Z.; Du X.; Xu Y.; Deng Y.; Liu M.; Zhao Y.; Zhang B.; Li X.; Zhang L.; Peng C.; Duan Y.; Yu J.; Wang L.; Yang K.; Liu F.; Jiang R.; Yang X.; You T.; Liu X.; Yang X.; Bai F.; Liu H.; Liu X.; Guddat L. W.; Xu W.; Xiao G.; Qin C.; Shi Z.; Jiang H.; Rao Z.; Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Dallakyan S.MGLTools, Reference Source, 2010.
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obot I.; Macdonald D.; Gasem Z. Density functional theory (DFT) as a powerful tool for designing new organic corrosion inhibitors. Part 1: an overview. Corros. Sci. 2015, 99, 1–30. 10.1016/j.corsci.2015.01.037. [DOI] [Google Scholar]
- Muhammad S.; Hassan S. H.; Al-Sehemi A. G.; Shakir H. A.; Khan M.; Irfan M.; Iqbal J. Exploring the new potential antiviral constituents of Moringa oliefera for SARS-COV-2 pathogenesis: An in silico molecular docking and dynamic studies. Chem. Phys. Lett. 2021, 767, 138379. 10.1016/j.cplett.2021.138379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Zia M.; Muhammad S.; Shafiq-urRehman S.; Bibi S. W.; Abbasi A. G.; Al-Sehemi A. R.; Chaudhary F. Q.; Bai F. Q. Exploring the potential of novel phenolic compounds as potential therapeutic candidates against SARS-CoV-2, using quantum chemistry, molecular docking and dynamic studies. Bioorg. Med. Chem. Lett. 2021, 43, 128079. 10.1016/j.bmcl.2021.128079. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Muhammad S.; Saba A.; Khera R. A.; Al-Sehemi A. G.; Algarni H.; Iqbal J.; Alshahrani M. Y.; Chaudhry A. R. Virtual screening of potential inhibitor against breast cancer-causing estrogen receptor alpha (ERα): molecular docking and dynamic simulations. Mol. Simul. 2022, 48, 1163. 10.1080/08927022.2022.2072840. [DOI] [Google Scholar]
- Selvaraj K.; Daoud A.; Alarifi S.; Idhayadhulla A. Tel-Cu-NPs Catalyst: Synthesis of Naphtho[2,3-g]phthalazine Derivatives as Potential Inhibiters of Tyrosinase Enzymes and Their Investigation in Kinetic, Molecular Docking, and Cytotoxicity Studies. Catalysts 2020, 10, 1442. 10.3390/catal10121442. [DOI] [Google Scholar]
- Pires D. E.; Blundell T. L.; Ascher D. B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.