Abstract
Colorimetric loop-mediated DNA isothermal amplification-based assays have gained momentum in the diagnosis of COVID-19 owing to their unmatched feasibility in low-resource settings. However, the vast majority of them are restricted to proprietary pH-sensitive dyes that limit downstream assay optimization or hinder efficient result interpretation. To address this problem, we developed a novel dual colorimetric RT-LAMP assay using in-house pH-dependent indicators to maximize the visual detection and assay simplicity, and further integrated it with the artificial intelligence (AI) operated tool (RT-LAMP-DETR) to enable a more precise and rapid result analysis in large scale testing. The dual assay leverages xylenol orange (XO) and a newly formulated lavender green (LG) dye for distinctive colorimetric readouts, which enhance the test accuracy when performed and analyzed simultaneously. Our RT-LAMP assay has a detection limit of 50 viral copies/reaction with the cycle threshold (Ct) value ≤ 39.7 ± 0.4 determined by the WHO-approved RT-qPCR assay. RT-LAMP-DETR exhibited a complete concordance with the results from naked-eye observation and RT-qPCR, achieving 100% sensitivity, specificity, and accuracy that altogether render it suitable for ultrasensitive point-of-care COVID-19 screening efforts. From the perspective of pandemic preparedness, our method offers a simpler, faster, and cheaper (∼$8/test) approach for COVID-19 testing and other emerging pathogens with respect to RT-qPCR.
Keywords: Colorimetric RT-LAMP, SARS-CoV-2, Machine learning, LAMP-DETR
Graphical abstract
1. Introduction
The spread of novel coronavirus (COVID-19) disease, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has claimed a tally of nearly 4.3 million lives worldwide with accumulated infections rapidly soaring beyond 200 million cases in August 2021 [1]. Although mass vaccination programs have already been rolled out in many countries, it is still unclear when herd immunity will be achieved due to the existing challenges associated with vaccine efficacy, development, distribution, and hesitation [2,3]. Hence, broad access to testing is still essential to keep the COVID-19 pandemic under control.
The World Health Organization (WHO) has published guidelines that favor the use of real-time reverse transcription polymerase chain reaction (RT-qPCR). Albeit robust, qPCR critically lacks the accessibility required for mass screening, especially in the landscape of public health infrastructure of developing nations. To address the existing challenge, various testing modalities that can be operated de-centrally, such as antigen test [4,5], antibody test [6,7], and isothermal detection, including CRISPR-Cas-based assays [8,9], were leveraged for rapid SARS-CoV-2 diagnosis in resource-limited settings.
Among the nucleic amplification technologies (NAAT), loop-mediated isothermal amplification (LAMP) presents the competitive advantage that lends itself exploitable for COVID-19 detection. LAMP mechanism inherently permits its further integration with colorimetric or visual readouts by which results can easily be observed with the naked eye [10]. Mechanistically, as LAMP propagates, pyrophosphates and protons (H+) are generated as by-products by the strand-displacement activity of Bst polymerase [11]. In a weak buffering environment, the excess of protons causes a dramatic pH drop that ensues a spontaneous change in the optical property of a pH-sensitive dye whose conversion point matches the operational pH range of the Bst DNA polymerase.
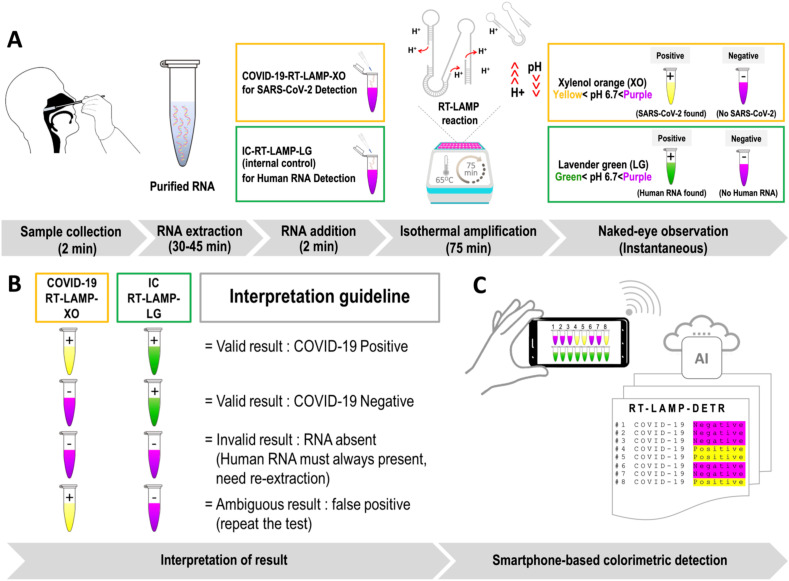
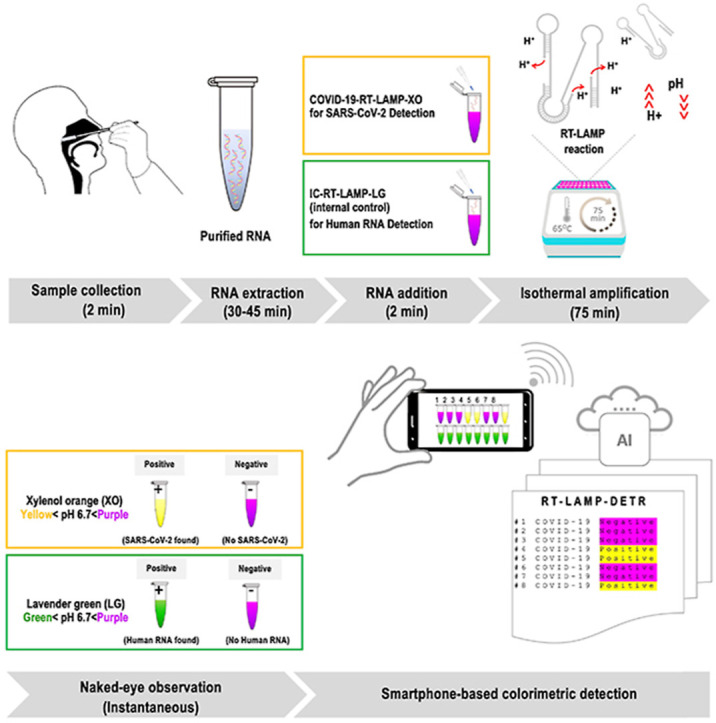
Both RT-LAMP assays presented herein share a common utilization of XO (yellow < pH 6.7 < purple) that is traditionally used as an indicator for industrial titration of various metal ions that offers a higher contrast between the positive (yellow) and negative (purple) test outcomes relative to other dyes, e.g. phenol red [12], leuco crystal violet [13], hydroxy naphthol blue (HNB) [14], and calcein [15]. The development of COVID-19-RT-LAMP-XO has ushered in a co-development of another composite colorimetric dye system that incorporates the existing XO [16,17] with malachite green (MG) [18]. This new colorimetric system, which shall be addressed as lavender green (LG), is exploited for the internal control detection of human 18 S rRNA (IC-RT-LAMP-LG) performed as an alternative approach to conventional spectrophotometric analysis after RNA extraction (Fig. 1 A and B).
Fig. 1.
Diagnostic principle of the combined colorimetric COVID-19 RT-LAMP-XO and IC-RT-LAMP-LG assay with AI-assisted automated result analysis (RT-LAMP-DETR). A RNA extracted from commercial RNA extraction kits can be eluted in water, and directly supplied to the COVID-19-RT-LAMP-XO and IC-RT-LAMP-LG reactions. The two tests were designed to complement each other to ensure the accuracy of test results. During the incubation at 65 °C, in the presence of target RNAs (Nsp 9 for COVID-19-RT-LAMP-XO, and human 18 S rRNA for IC-RT-LAMP-LG), the buildup of excess protons (H+) in the reactions will cause a dramatic pH drop, and change the hues of pH-sensitive dyes that can be further analyzed with the naked-eye or an AI model for automated, high-volume testing. With the exception of RNA extraction, the entire operation takes less than 2 hours to finish with very limited hands-on time. B The colorimetric results of the combined COVID-19-RT-LAMP-XO and IC-RT-LAMP-LG assay could be cross-compared according to the graphical guideline to accurately interpret the test results. For the COVID-19-RT-LAMP-XO assay, the presence of SARS-CoV-2 in the test samples will trigger the change of reaction hue from purple to yellow. On the contrary, for the IC-RT-LAMP-LG assay, the presence of human 18 S rRNA in the test samples will trigger the change of reaction hue from purple to green. In both tests, a lack of detection targets will preserve the original purple hue of the reaction. In the context of point-of-care testing, the outcomes of IC-RT-LAMP-LG assay provide a qualitative assessment of the RNA extraction process, and can be used as an alternative method to the standard spectrophotometry for nucleic acid quality determination. C Images of multiple reaction tubes can be taken via a smartphone camera for AI-assisted automated result analysis by the RT-LAMP-DETR mode. Details of the RT-LAMP-DETR analysis pipeline are in Fig. 2. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
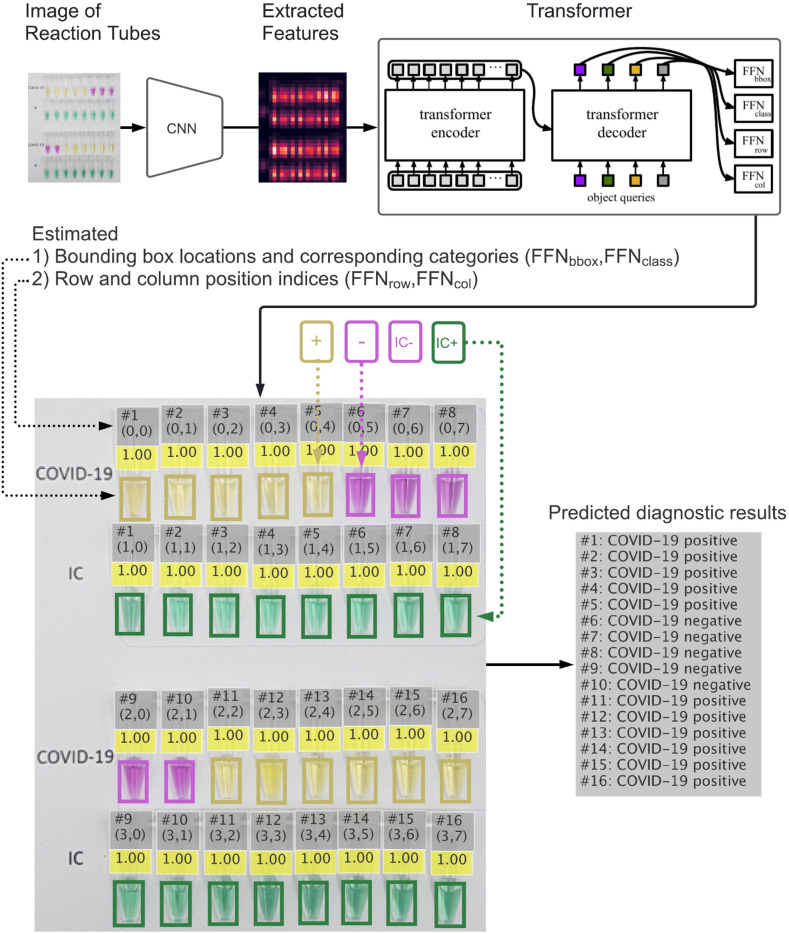
We utilized deep learning to enable high-throughput colorimetric analysis based on images of multiple reaction tubes (Fig. 1C). The analysis can be done on a mobile phone by taking a picture with our mobile application, and the result will be displayed as an overlay on the original image as shown in Fig. 2 . We adapt the detection transformer (DETR) model [19] to efficiently solve the task. While most object detection methods require multiple hand-designed components, such as region proposals [20] and anchor generations [21], DETR combines the transformer model architecture [22] and a set-based objective function to eliminate these requirements and streamline the object detection pipeline. In our proposed deep learning model, entitled RT-LAMP-DETR, we added additional prediction pathways that estimate the row and column position indices to simultaneously analyze the colorimetric results of both COVID-19-RT-LAMP-XO and IC-RT-LAMP-LG assays. Therefore, the novelty of this research lies in the 1) the integration of Xylenol Orange (XO) and newly formulated Lavender Green (LG) in colorimetric RT-LAMP for COVID-19 diagnosis. For the first time, we demonstrated LG for its ability to clearly discriminate the positive from negative reactions. 2) Aside from providing easy-to-read colorimetric results, we addressed the critical need for high-throughput screening of our RT-LAMP assay in the future by developing an AI-based analysis tool that could help determine the colorimetric results more accurately and rapidly. An overview of our RT-LAMP-DETR analysis pipeline is illustrated in Fig. 2.
Fig. 2.
Analysis pipeline of RT-LAMP-DETR. Image features are extracted from an image of reaction tubes that has been taken by a smartphone camera using the pretrained ResNet50 model, and then used as inputs to the proposed encoder-decoder model which outputs the estimated 1) reaction tubes' locations with their corresponding categories and 2) row and column position indices of the tubes. Each tube's location is represented by a bounding box with its color indicating the predicted category. The number above each bounding box represents the model's confidence in the estimated category (1.0 = highest, and 0.0 = lowest). The row and column position indices of the tubes are used to identify which RNA sample (e.g. from #1 to #16) each tube belongs to. Once the sample is identified, we combine the two estimated reaction tubes' categories with the same identifier to form the final predicted result: COVID-19 positive, COVID-19 negative, RNA absent, and False positive (inconclusive result due to a combination of positive RT-LAMP-XO and negative RT-LAMP-LG). If a sample cannot be identified, its predicted result is marked as Void. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2. Materials and methods
2.1. Primer designs and optimization
We explored a fragment of 216 bp in the Nsp9 of the ORF1ab gene (GenBank accession number: NC_045512.2) as an alternative single target for SARS-CoV-2 detection to avoid the previously reported targets of N, E, and ORF1ab genes [12,23,24]. In addition to the 6 common LAMP primers, we designed 4 additional primers (loop forward and backward 2; LF2 and LB2, and forward inner and backward inner 2; FIP2 and BIP2) as shown in Fig. S1 to further improve the reaction kinetics [25]. Primers were examined for possible cross dimerization by basic local alignment search tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Those for the IC-RT-LAMP-LG (internal control) assay targeting the human 18 S RNA were from our previous study [26]. Primers are listed in (Table S1). The optimal condition of the COVID-19-RT-LAMP-XO was determined empirically through the variation of incubation temperature in the 63–70 °C range and time (30–75 min).
2.2. Development of LG colorimetric system for IC-RT-LAMP-LG
Optimization of the LG system was performed by varying the working concentration of XO (0–0.5 mM) in fixed 0.02 mM MG in the IC assay. The optimal balance of XO and MG in the novel LG system was investigated through the limit of detection (LoD) of human 18 S rRNA (total RNA) target and the clarity of contrast between negative and positive RT-LAMP reactions.
2.3. Isothermal amplification of COVID-19-RT-LAMP-XO and IC-RT-LAMP-LG
Unless otherwise stated, all reagents were purchased from New England Biolabs (MA, USA). To prepare a 25-μL premix solution, the individual components were combined according to the specified volumes and concentrations as follows: 1.4 mM dNTP mix, 0.4 M Betaine (Merck Millipore, MA, USA), 6 mM MgSO4, 0.12% Triton X-100 (Merck Millipore, MA, USA), 8 UμL−1 Bst 2.0 WarmStart™ DNA polymerase, 15 UμL−1 WarmStart™ RTx Reverse Transcriptase and 1 × isothermal buffer. The primer set of COVID-19-RT-LAMP-XO comprises 0.2 μM each of the outer primers (F3 and B3), 1 μM each of the inner primers (FIP-BIP and FIP2-BIP2) and 1 μM each of the loop primer (LF-LB and LF2-LB2). The primer set of IC-RT-LAMP-LG comprises 0.2 μM each of the outer primers (F3 and B3), 2 μM each of the inner primers (FIP-BIP) and 2 μM each of the loop primer (LF-LB). Additionally, the COVID-19-RT-LAMP-XO requires 0.12 mM XO from the 5 mM stock, while IC-RT-LAMP-LG requires a total of 1 × LG solution (see Reagent setup in Supplementary Materials). The reaction volume was adjusted to 25 μL with DNase-, RNase-free water prior to template addition.
The reaction mixture was aliquoted into individual PCR tubes to which negative (blank) controls were supplied with 25 μL water. In a space designated for template addition, 25 μL of RNA template (see in vitro RNA preparation in Supplementary Materials) were added into the PCR tubes assigned for testing. The reactions were incubated at 65 °C for 75 min. Once the run is finished, the reaction tubes were set at room temperature for an additional of 2 min to allow the color to fully develop.
2.4. Specificity and sensitivity of COVID-19-RT-LAMP-XO
The specificity of COVID-19-RT-LAMP-XO assay was examined using a panel of human respiratory viruses and other disease agents that include SARS-CoV-2, MERS-CoV, RSV, Influenza A virus subtype H1N1, Influenza A virus subtype H3N2, Influenza B virus (Yamagata lineage), Influenza B virus (Victoria lineage), Influenza B virus (B/Lee/40), Mycobacterium tuberculosis, Klebsiella pneumoniae strain ATCC 700603, Acinetobacter baumannii strain ATCC19606, Pseudomonas aeruginosa strain ATCC 27853, Bacillus cereus strain BCC 6386, Streptococcus pneumonia, Listeria monocytogenes strain ATCC 19115 and Porcine epidemic diarrhea virus strain AVCT12.
The analytical sensitivity of COVID-19-RT-LAMP-XO assay was investigated first by using in vitro RNA transcripts prepared in serial dilutions ranging from 1000 to 0 copies/reaction (N = 8 per dilution). Once the analytical sensitivity was determined by in vitro RNA transcript, we repeated the sensitivity analysis with COVID-19 infected patient-derived total RNA templates that were serially diluted by a factor of 1,000, 2,000, 10,000, 50,000 and 100,000 (N = 12 per dilution), followed by comparing the results to that of the WHO reported RT-qPCR (references assay) [27]. For both types of the template, the number of positive reactions based on colorimetric results was used to calculate the positive rate of detection for each dilution. The last dilution whose positive rate was still at 100% was regarded as the LoD of the method.
2.5. Data collection and augmentation for image analysis
To train our RT-LAMP-DETR model, 60 images of a set of 25-μL and 50-μL reaction tubes were captured using a smartphone camera (Samsung Galaxy S7): 29 and 31 images taken under controlled and uncontrolled lighting conditions, respectively. Each image contains an even number of tubes, ranging from 8 to 32 tubes which correspond to a minimum and maximum of 4 and 16 RNA samples, respectively. For each reaction tube in the images, we drew a rectangular bounding box that enclosed the solution region of the tube and categorized it based on colors: yellow (COVID-19-RT-LAMP-XO positive), green (IC-RT-LAMP-LG positive) and purple (COVID-19-RT-LAMP-XO negative or IC-RT-LAMP-LG negative). Each bounding box is represented by four numbers (x,y,h,w) where x and y are the (x,y)-coordinates of the upper-left corner of the box; w and h are the width and height of the box. We also assigned the row and column position indices to each bounding box to identify the RNA sample that a reaction tube belongs to. For example, the tubes with indices (0,0) and (1,0) belong to RNA sample #1, and the tubes with indices (2,7) and (3,7) belong to RNA sample #16, as shown in Fig. 2. We split the annotated images into two groups: 54 images as training data and 6 images as validation data. The training data were used to optimize the proposed RT-LAMP-DETR model. The validation data were used to monitor the training process, select the hyperparameters of the model, and prevent model overfitting.
To assess the performance of the optimized model on unseen test data, we used the captured images of de-identified 213 RNA samples as a test set for model validation. The numbers of images and RNA samples were summarized in Table S2. For each image in the training set, we applied a sequence of image transformations consisting of brightness-contrast-saturation perturbations, image rotations, image translations, and shear mapping to increase the size of the training set (example images shown in Fig. S2).
2.6. The proposed RT-LAMP-DETR model and training
We extended the standard DETR architecture [19] with ResNet50 [28] as the backbone to include additional prediction heads to predict the row and column position indices of the reaction tubes for automatic association of tubes to its RNA samples as shown in Fig. 2. The proposed model uses six encoding layers and six decoding layers with hidden dimensions of 256. The number of attention heads and the number of queries used are 8 and 64, respectively. Each feedforward layer has 2,048 nodes. Our proposed neural network model takes an image as its input and outputs the predicted row position indices, column position indices, category, and bounding box locations. Mathematically, we have
where , , , are the predicted row position indices, column position indices, category, and bounding box locations, respectively, is the input image, and is a function describing our neural network model with 36,799,318 parameters in total. Since different values of the parameters give rise to different functional behaviors, we optimize the parameters by solving the following optimization problem:
where is in the universe of all possible neural network models, and
, , , are the target (i.e., true) row position indices, column position indices, category, and bounding box locations, respectively. is the cross-entropy loss function. is the L1 loss function (also known as mean absolute error). is the generalized intersection over union (GIoU) loss function [29]. , , , , and are the regularization parameters that can be modified to assign a different contribution to each term in the objective function .
To accelerate the model training, we attached the additional prediction heads with He initialization [30] to the pretrained DETR model [19] and then optimized the model parameters by minimizing our proposed loss function for 400 epochs using AdamW [31] with the learning rates of 10−5 and 10−4 for the CNN backbone and the rest of the model, respectively. We dropped these learning rates to 10−6 and 10−5 at epoch 200 and used the batch size of 3, weight decay of 10−4, and dropout of 0.3. The auxiliary loss [32] was also used to assist with the training. We set , , and . The entire training process took approximately 3.6 h on a Tesla P100 12 GB GPU. As the number of epochs increases, the losses decrease and saturate at around 300 epochs (Fig. S3). The model that achieved the lowest validation loss was considered the best model and then used to evaluate the test set.
2.7. Clinical validation of RT-LAMP assays by visual detection and the RT-LAMP-DETR model
The total of blinded, de-identified 213 RNA samples were extracted from patient's derived nasopharyngeal swabs collected at the Tropical Medicine Hospital, Mahidol University, Thailand, under the approval of the Ethics Committee (EC) of Mahidol University's Institutional Review Board (IRB) with the Ethics Committee document No. MUTM 2021-004-01. They were stored initially in the viral transport medium (VTM; BioTrend, Germany). Total RNA was extracted from 150 μL of the original VTM stock by using QIAamp Viral RNA Mini kit (Qiagen, Germany) according to the manufacturer's instruction. RNA was reconstituted in 50 μL DNase-, RNase-free water, and then used to validate the COVID-19-RT-LAMP-XO and IC-RT-LAMP-LG assays that were performed in parallel. For each assay, 25 μL of RNA samples were added into individual reaction tubes prior to incubation at 65 °C for 75 min after which the colorimetric results were assessed according to the guideline shown in Fig. 1 B. Briefly, a test result of an individual sample is valid only when its IC-RT-LAMP-LG result is positive (green). Test results of the COVID-19-RT-LAMP-XO assay were compared with those of RT-qPCR targeting ORF1ab and N genes (Da An Gene Co., Ltd. of Sun Yat-Sen University, China). Test outcomes of both assays were analyzed based on the naked-eye colorimetric interpretations prior to taking images of reaction tubes with a smartphone camera for validation by RT-LAMP-DETR.
3. Results
3.1. Optimization, molecular specificity and sensitivity of colorimetric COVID-19-RT-LAMP-XO
During assay optimization, using the incubation temperature of 65 °C enabled the detection of template down to 100 copies (Fig. S4 A and B). We next varied the time of amplification that resulted in the clearest colorimetric observation (Fig. S4 C). Although results could be observed after 45 min, 75 min was selected as a standard RT-LAMP assay time to allow the final reaction color to fully develop without compromising the turnaround time. The optimal condition (65 °C for 75 min) established here was then used in all following RT-LAMP reactions.
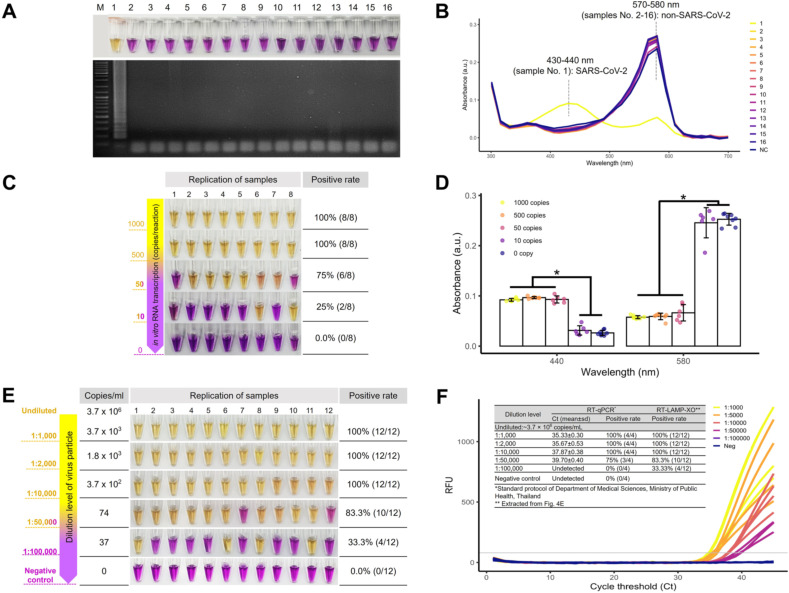
Our COVID-19-RT-LAMP-XO exhibited its exclusive specificity toward SARS-CoV-2 when tested against a panel of respiratory disease agents (Fig. 3 A). The visual difference between positive (yellow) and negative (purple) reactions also translated to their discrete spectrophotometric fingerprints based on the UV–Vis absorptions at 430–440 and 570–580 nm, respectively, (Fig. 3 B). Regarding the analytical sensitivity, our assay could detect down to 500 and 50 copies of in vitro RNA transcripts of the target Nsp9 segment with the positive rate of 100% (N = 8) and 75% (N = 8), respectively (Fig. 3C). The UV–Vis analysis at 430–440 and 570–580 nm of these in vitro RNA transcripts also revealed highly distinguishable absorption spectra between positive and negative test results (Fig. 3 D).
Fig. 3.
Specificity and analytical sensitivity of COVID-19-RT-LAMP-XO for in vitro RNA transcripts and viral RNA isolate. A The molecular specificity of COVID-19-RT-LAMP-XO by the naked-eye observation (top) and AGE (bottom) for (1) SARS-CoV-2 with respect to the following pathogens: (2) Middle East Respiratory Syndrome Coronavirus, (3) Respiratory Syncytial Virus, (4) Influenza A virus subtype H1N1, (5) Influenza A virus subtype H3N2, (6) Influenza B virus (Yamagata lineage), (7) Influenza B virus (Victoria lineage), (8) Influenza B virus (B/Lee/40), (9) Porcine epidemic diarrhea virus strain AVCT12, (10) Klebsiella pneumoniae strain ATCC 700603, (11) Acinetobacter baumannii strain ATCC 19606, (12) Pseudomonas aeruginosa strain ATCC 27853, (13) Bacillus cereus BCC 6386, (14) Streptococcus pneumoniae, (15) Listeria monocytogenes strain ATCC 19115 and (16) Mycobacterium tuberculosis. B UV–Vis absorption spectra of samples (1)–(16) as shown in A. SARS-CoV-2 positive sample (yellow line) exhibits the peak at 430–440 nm while the remainder of SARS-CoV-2 negative samples exhibits the peak at 570–580 mm that corresponds to the purple hue of the reaction mixture. C The sensitivity of COVID-19-RT-LAMP-XO for in vitro RNA transcripts of the Nsp9 target where the limit of detection was shown to be 500 copies and 50 copies/reaction with a positive rate of 100% and 75%, respectively (N = 8). D UV–Vis absorption measurements at 430–440 nm and 570–580 nm of the colorimetric results shown in C, demonstrating a statistical difference between the positive (POS) and negative outcomes (NEG). E The sensitivity of COVID-19-RT-LAMP-XO for total RNA of the SARS-CoV-2 isolate that was serially diluted down to a factor of 100,000. The limit of detection was shown at 1:10,000 dilution (3.7 × 102 copies/mL-equivalent), which was the last dilution level to achieve a 100% positive rate (N = 12). F Real-time RT-PCR [27] fluorogram of the total RNA isolates shown in E, with summarized sensitivity and cross-comparison with the COVID-19-RT-LAMP-XO results at different dilution levels (inset Table). Both methods showed a comparable detection limit with a 100% positive rate in the Ct ≤ 37.87 ± 0.38 (3.7 × 102 copies/mL-equivalent). M: the DNA marker/ladder. (*, p < 0.00001 Two-Sample t-test). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In the context of whole viral particles, our assay demonstrated a 100% (N = 12) positive rate of detection when total RNA was diluted by 10,000 folds (370 viral particles/mL by theoretical estimation) (Fig. 3 E). It is worth mentioning that the assay was still able to detect 1:50,000 diluted total RNA (∼74 copies/mL) with a lower positive rate of 83% (N = 12) that generally still aligns with that of the WHO reported RT-qPCR method [27] at this dilution level (Fig. 3 F) with the Ct of 39.7 ± 0.4 (Fig. 3 F, inset table). Based on this preliminary demonstration, the last reproducibly detectable dilution of total RNA at 1:10,000 has a Ct value of 37.87 ± 0.4 (Fig. 3 F), highlighting the ability of our COVID-19-RT-LAMP-XO assay that is on par with RT-qPCR for its ability to detect late-Ct samples (Ct > 35).
3.2. Development of the LG colorimetric indicator for IC-RT-LAMP-LG assay
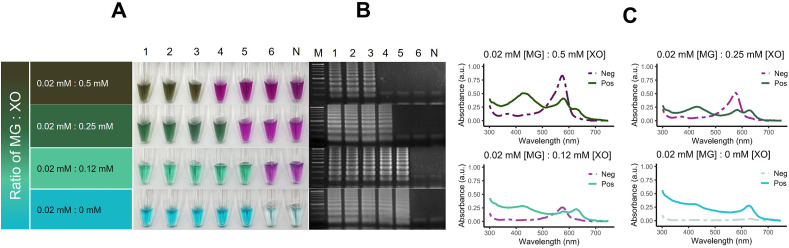
In all variations of XO concentration, the contrast between positive and negative results was observable most clearly at 0.12 mM XO where the final reaction hue of the positive reaction is green (Fig. 4 A). The LoD of human 18 S rRNA was improved as the concentration of XO in the reaction dropped from 0.5 mM to 0.12 mM where the LoD lies at 10 pg as shown in Fig. 4 A and B. The UV–Vis analysis revealed the absorption spectra of LG that combined the characteristics of XO and MG where peaks at 440 nm, 580 nm, and 620 nm were observed (Fig. 4C).
Fig. 4.
Development of the composite lavender green (LG) pH-sensitive indicator for IC-RT-LAMP-LG. A The lavender green indicator is a combination of two pH-sensitive indicators, XO (XO) and malachite green (MG). The determination of the optimal balance between the two dye components was performed through the iteration of xylenol orange concentration while the working concentration of MG was fixed at 0.02 mM in the RT-LAMP reaction that targets human 18 S rRNA. The stoichiometric ratio of dyes at 0.02 mM MG: 0.12 mM XO offers the clearest visual contrast between positive and negative results and the highest limit of detection (LoD) at 10 pg total RNA by which the incorporation of XO did not affect the assay's intrinsic LoD. B Corresponding AGE confirmation of the colorimetric results. C UV–Vis analysis of the positive and negative IC-RT-LAMP-LG reactions across stoichiometric variations of MG and XO where the positive reaction is characterized by the presence of the absorption peak at 620 nm that corresponds to the blue shade of MG in sub-neutral pH environment. Each line represents the average absorption of samples within the positive and negative groups. Lanes 1, 2, 3, 4, 5, 6, M and N: 100 ng, 10 ng, 1 ng, 100 pg, 10 pg, 1 pg, molecular marker and negative control (template-free reaction), respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Clinical validation by the visual and AI-integrated analyses with respect to RT-qPCR
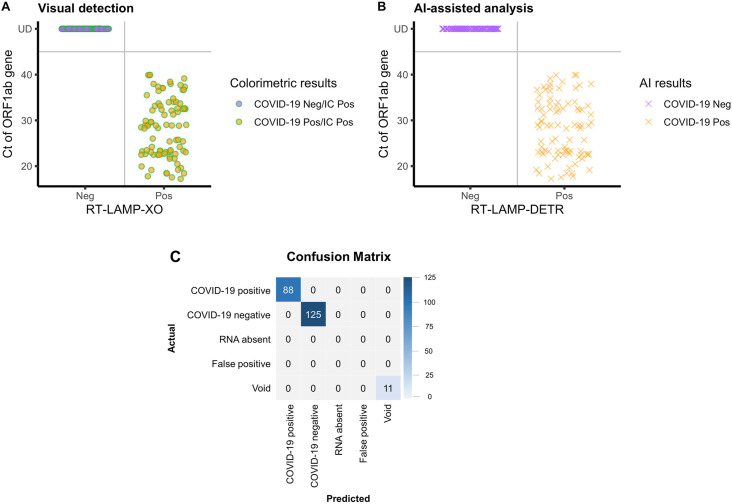
The diagnostic results of visual analysis and RT-LAMP-DETR were classified according to the breakdowns of Ct values of samples (Table 1 ). Attributes of diagnostic performance, such as sensitivity, specificity and accuracy, were calculated according to equations provided in Table S3. Based on the RT-qPCR results of the ORF1ab gene, the COVID-19-RT-LAMP-XO and IC-RT-LAMP-LG could correctly identify all samples across the breakdowns of Ct values (Fig. 5 A). Note that RT-qPCR was performed simultaneously on both the ORF1ab and N gene targets, but the ORF1ab results were chosen as representative because the Ct values of both genes are highly correlated (Fig. S5).
Table 1.
Clinical validation of the COVID-19-RT-LAMP-XO by visual observation and RT-LAMP-DETR against RT-qPCR on ORF1ab gene.
| Result | Ct value (ORF1ab gene) | RT-LAMP-XO by visual observation/RT-LAMP-DETR |
|||
|---|---|---|---|---|---|
| Positive | Negative | Sum (N) | |||
| RT-qPCR | Positive | 0–25.0 | 34/34 | 0/0 | 34 |
| 25.1–30.0 | 19/19 | 0/0 | 19 | ||
| 30.1–35.0 | 19/19 | 0/0 | 19 | ||
| 35.1–40.0 | 16/16 | 0/0 | 16 | ||
| Negative | >40/Undetectable | 0/0 | 125/125 | 125 | |
| Total | 88/88 | 125/125 | 213 | ||
| % Concordance with respect to RT-qPCR result | 100 | 100 | |||
N denotes sample size.
Fig. 5.
Clinical validation of the colorimetric COVID-19 assays by visual detection and RT-LAMP-DETR. The total of 213 total RNA extracted from samples collected by nasopharyngeal swab were subject to COVID-19-RT-LAMP-XO and IC-RT-LAMP-LG assays. The colorimetric results were determined by two modes of analysis: (A) naked-eye observation and (B) automated RT-LAMP-DETR. A Distribution of colorimetric results of the combined COVID-19-RT-LAMP-XO and IC-RT-LAMP-LG assays with respect to the Ct values of RT-qPCR performed on the ORF1ab gene and N gene. All test samples were determined positive by the IC-RT-LAMP-LG assay. B Distribution of automated predictions of the RT-LAMP-DETR model with respect to the Ct values. C Confusion matrix of automated results generated by RT-LAMP-DETR, which simultaneously analyzes the colorimetric outcomes of both COVID-19-RT-LAMP-XO and IC-RT-LAMP-LG assays, and categorizes the test results according to the detected colors of reaction tubes of individual samples. Diagnostic outcomes were compared with RT-qPCR results (Ct value), and the concordance of test results is summarized in Table 1. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
RT-LAMP-DETR conforms with visual analysis in all test samples (Fig. 5 B). Taking only 4.73 ms per RNA sample, it correctly generated the bounding boxes and categories, row and column position indices of the tubes, and colorimetric diagnostic results (Fig. 2). The quality of the bounding boxes was evaluated using the mean Average Precision (mAP), a well-established metric to compare the performance of object detectors. RT-LAMP-DETR achieved an mAP of 0.99 out of 1 on the test samples. The diagnostic results of the whole test set were summarized into a 5 × 5 confusion matrix (Fig. 5C). Non-zero values only appeared along the diagonal of the matrix, suggesting no misdiagnoses. RT-LAMP-DETR was able to identify all the 11 missing tubes in the test set by outputting the Void category. All predictions made by RT-LAMP-DETR were in perfect agreement with the “manual” visual detection (Table S3 and Fig. 5). This includes 16 late-Ct samples categorized in the 35.1–40.0 Ct bracket of the ORF1ab detection results (Table 1) which linearly correlate with those of the N gene (Table S4). Both approaches achieved 100% sensitivity, specificity and accuracy (Table S3).
4. Discussion
Our dual one-step colorimetric RT-LAMP assays offer the versatility of analysis that accommodates testing at scale in the context of mass population screening. The two-color systems employed in this research help users quickly identify the assays, offering a simple solution to prevent clerical errors during result analysis. This instrument-free colorimetric analysis lends our technique readily exploitable for decentralized applications. The recommended use of IC-RT-LAMP-LG as a control assay for all test samples has never been reported elsewhere, and offers a critical means for sample quality assurance to rule out the possibility of false negatives owing to unqualified samples, which underlie a major pitfall of many colorimetric assays [33].
Despite being developed around a lesser-known molecular target of a highly conserved Nsp9 segment of SARS-CoV-2 polyprotein, the COVID-19-RT-LAMP assay is highly sensitive and able to achieve a detection limit that is on par with the standard RT-qPCR assay for late-Ct samples that could be challenging to detect via rapid NAAT without producing the nonspecific outcomes. Based on the available genomic data of existing variants of concern (VOCs), such as B.1.1.7 (Alpha) [34] or B.1.617 (Delta) [35], our COVID-19-RT-LAMP assay should also be able to detect these emerging strains since none of the known mutations are in the proximity of the target region. To highlight their robustness, our RT-LAMP assays were shown to retain activity up after 5 freeze-thaw cycles in an accelerated stability test (Fig. S6). The COVID-19-RT-LAMP-XO was proven to be active after 6 months of storage at a regular freezer temperature (−20 °C) (Fig. S7).
The development of the composite lavender green (LG) pH-sensitive indicator demonstrated in IC-RT-LAMP-LG has laid a vital foundation for improving the repertoire of novel colorimetric indicators compatible with isothermal amplification in emerging applications. The establishment of the LG system can trace its root back to the proof-of-concept LAMP-XO [16] that provides an unequivocal contrast between positive and negative results relative to other common dyes, e.g. phenol red and HNB. Since then, the assay has served as a springboard for various other assays that address the diagnostic needs in a wide range of emerging point-of-need applications, predominantly those in aquaculture [17,36] where demand for simple, robust one-step test kits is high.
Compared to other existing techniques, our assay is superior to other recently reported COVID-19 colorimetric platforms that leveraged lateral flow chromatographic CRISPR [9,37] and DNA-functionalized gold nanoparticles-dependent RT-LAMP [38] with respect to the ease of use as our technique abrogates the need for post-amplification workflow associated with hybridization and readout development. While the uniqueness of our method is attributed to colorimetry that offers high contrast between positive and negative outcomes, we further improved its efficiency by integrating it with a powerful AI model (RT-LAMP-DETR) for high-throughput analysis without requiring complicated data acquisition setups. This feature, together with the advent of LG dye, has set our platform unique from the current research that goes beyond its own colorimetric RT-LAMP modality, such as the CRISPR-Cas technology and rapid antigen/antibody testing (Table S5). The prospect of having a fully integrated automated system to effortlessly sweep across the test outcomes with high accuracy will also minimize interpersonal bias in result interpretation that involves a large volume of samples. Thus, our study also serves to showcase the utility of AI-based object detection that has recently gained traction in clinical diagnosis, and needless to say, the implication of our platform in providing an accessible mass screening in the future pandemics.
Specifically, our work features an implementation of a deep learning model, which has been shown to revolutionize how images are analyzed [28,[39], [40], [41], [42]]. While using convolution as the main building block is doubtlessly effective and still the norm, the self-attention mechanism has attracted more interest in recent years for its ability to provide data representation based on the global information in the data [22,[43], [44], [45], [46]]. In this work, we use the self-attention mechanism to complement convolution, equipping the proposed RT-LAMP-DETR model with the ability to effectively capture both local and global dependencies between the pixels in the intermediate image features. The dependencies captured by the model can be investigated through the visualization of the encoder and decoder attention maps. As shown in Fig. S8, the decoder attention maps show sharper localization than the encoder attention maps as expected. With the self-attention mechanism, the model learns to pay attention to each of the reaction tubes and provide its corresponding category based on the pixels within the same tube. We have demonstrated the potential of RT-LAMP-DETR in the high-throughput colorimetric analysis that needs to associate two reaction tubes from the same RNA sample to form the final prediction. In future applications, RT-LAMP-DETR can be easily extended to provide high-throughput analysis of any colorimetric assays that may require multiple reaction tubes to be associated by the model to form the prediction in an end-to-end manner.
Regarding the limitation of our assay, fundamentally, the introduction of LAMP-XO has served as a basis for the advent of the LG indicator that offers a different end result color from its parent dyes. Under a certain condition, though, the color development of the lavender green system is adversely affected by the presence of mineral oil often added into the LAMP reaction to prevent its content from evaporating when the choice of heat source is not itemized with a heated lid control [[47], [48], [49], [50], [51]]. Thus, the chemical nature of this composite indicator still needs to be investigated in order to utilize it to the fullest potential. We speculate that MG can be reduced to its more lipophilic analog, leucomalachite green [52], in an excess of protons in the positive LAMP reaction, and eventually loses its desired optical property after becoming soluble in oil. Until a better solution has been proposed, the total exclusion of mineral oil is highly suggested for any LAMP reactions that contain LG. RNA samples must also be eluted in DNase-, RNase-free water only. Any other non-low buffer solution, e.g. Tris-EDTA (TE), will interfere with the color development of the RT-LAMP assay [10,53]. We acknowledge that the current protocol still employs a standard RNA extraction technique to prepare the test samples. Nevertheless, rapid nucleic acid extractions have been shown to be compatible with colorimetric isothermal amplification reactions [17,54,55]. We highly recommend readers who are interested in implementing this protocol at their own laboratories to further evaluate a suitable rapid sample preparation technique that coherently yields reasonable amounts of RNA.
5. Conclusions
Our test has been validated with 213 patient samples, offering a 100% accuracy, and additional ease of analysis that makes high volume testing feasible in low-resource settings. With the cost per assay of approximately $8 for each reaction and the turnaround time of 75 min with a convenient result readout, the effectiveness of COVID-19 screening in the population can be considerably improved as testing can be done more frequently and widespread. We hope that the test platform established in this research will serve as a toolkit that will expedite the developmental pipeline of new diagnostics for emerging pathogens.
Author contributions
Wansadaj Jaroenram designed the study, investigated, conducted experiments, analyzed results, drafted, edited, and reviewed the manuscript. Jantana Kampeera, Sukanya Pengpanich, Rapheephat Suvannakad conducted experiments, analyzed results, and reviewed the manuscript. Pakapreud Khumwan conducted experiments, drafted, edited and reviewed the manuscript. Sarawut Sirithammajak, Benyatip Tondee, Narong Arunrut and Sirintip Dangtip conducted experiments. Pornsawan Leaungwutiwong, Sirasate Bantuchai, Wang Nguitragool and Jetsumon Sattabongkot provided clinical samples and conducted experiments. Wansika Kiatpathomchai conceptualized, investigated, reviewed the manuscript and acquired funding. Itthi Chatnuntawech, Surat Teerapittayanon developed and implemented AI models, conceptualized, designed and conducted AI experiments, analyzed AI results, drafted, edited and reviewed the manuscript. Suchawit Wongwaroran conducted a preliminary AI experiment. Paisan Khanchaitit conceptualized AI experiments, acquired funding for computational resources, and reviewed the manuscript. All authors have given approval of the final version of the manuscript.
Data and materials availability
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank colleagues who kindly provided viral samples for assay development and optimization (obtained from Virology and Cell Technology Research Team (AVCT), National Center for Genetic Engineering and Biotechnology (BIOTEC), the Faculty of Tropical Medicine, Mahidol University, Thailand, and the Department of Medical Science, Ministry of Public Health, Thailand). We would also like to acknowledge the Department of Medical Science, Ministry of Public Health, Thailand, and Dr. Anan Jongkaewwattana for providing samples for specificity testing, Dr. Noppadol Prasertsincharoen for Malachite Green, and Prof. Timothy Flegel for reviewing and editing the manuscript. Funding: This research is financially supported by National Center for Genetic Engineering and Biotechnology(BIOTEC) (Project number P2050651).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2022.123375.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.World Health Organization, WHO Coronavirus Disease (COVID-19) Dashboard, (n.d.). https://covid19.who.int/(accessed 2 January, 2021).
- 2.Sharma O., Sultan A.A., Ding H., Triggle C.R. A Review of the progress and challenges of developing a vaccine for COVID-19. Front. Immunol. 2020;11:585354. doi: 10.3389/fimmu.2020.585354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dror A.A., Eisenbach N., Taiber S., Morozov N.G., Mizrachi M., Zigron A., et al. Vaccine hesitancy: the next challenge in the fight against COVID-19. Eur. J. Epidemiol. 2020;35:775–779. doi: 10.1007/s10654-020-00671-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert E., Torres I., Bueno F., Huntley D., Molla E., Fernández-Fuentes M.Á., et al. Field evaluation of a rapid antigen test (PanbioTM COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centres. Clin. Microbiol. Infect. 2021;27 doi: 10.1016/j.cmi.2020.11.004. 472.e7-472.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamayoshi S., Sakai-Tagawa Y., Koga M., Akasaka O., Nakachi I., Koh H., et al. Comparison of rapid antigen tests for COVID-19. Viruses. 2020;12 doi: 10.3390/v12121420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J.-L., Tseng W.-P., Lin C.-H., Lee T.-F., Chung M.-Y., Huang C.-H., et al. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J. Infect. 2020;81:435–442. doi: 10.1016/j.jinf.2020.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spicuzza L., Montineri A., Manuele R., Crimi C., Pistorio M.P., Campisi R., et al. Reliability and usefulness of a rapid IgM-IgG antibody test for the diagnosis of SARS-CoV-2 infection: a preliminary report. J. Infect. 2020;81 doi: 10.1016/j.jinf.2020.04.022. e53–e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshikawa R., Abe H., Igasaki Y., Negishi S., Goto H., Yasuda J. Development and evaluation of a rapid and simple diagnostic assay for COVID-19 based on loop-mediated isothermal amplification. PLoS Neglected Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanner N.A., Zhang Y., Evans T.C. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques. 2015;58:59–68. doi: 10.2144/000114253. [DOI] [PubMed] [Google Scholar]
- 11.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., et al. 2020. Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP. MedRxiv. [DOI] [Google Scholar]
- 13.El-Tholoth M., Bau H.H., Song J. A single and two-stage, closed-tube, molecular test for the 2019 Novel Coronavirus (COVID-19) at home, clinic, and points of entry. ChemRxiv. 2020 [Google Scholar]
- 14.Goto M., Honda E., Ogura A., Nomoto A., Hanaki K.-I. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques. 2009;46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- 15.Tomita N., Mori Y., Kanda H., Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 16.Jaroenram W., Cecere P., Pompa P.P. Xylenol orange-based loop-mediated DNA isothermal amplification for sensitive naked-eye detection of Escherichia coli. J. Microbiol. Methods. 2019;156:9–14. doi: 10.1016/j.mimet.2018.11.020. [DOI] [PubMed] [Google Scholar]
- 17.Dangtip S., Kampeera J., Suvannakad R., Khumwan P., Jaroenram W., Sonthi M., et al. Colorimetric detection of scale drop disease virus in Asian sea bass using loop-mediated isothermal amplification with xylenol orange. Aquaculture. 2019;510:386–391. doi: 10.1016/j.aquaculture.2019.05.071. [DOI] [Google Scholar]
- 18.Nzelu C.O., Cáceres A.G., Guerrero-Quincho S., Tineo-Villafuerte E., Rodriquez-Delfin L., Mimori T., et al. A rapid molecular diagnosis of cutaneous leishmaniasis by colorimetric malachite green-loop-mediated isothermal amplification (LAMP) combined with an FTA card as a direct sampling tool. Acta Trop. 2016;153:116–119. doi: 10.1016/j.actatropica.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 19.Carion N., Massa F., Synnaeve G., Usunier N., Kirillov A., Zagoruyko S. In: Computer Vision – ECCV 2020: 16th European Conference, Glasgow, UK, August 23–28, 2020, Proceedings, Part I. Vedaldi A., Bischof H., Brox T., Frahm J.-M., editors. Springer International Publishing; Cham: 2020. End-to-End object detection with transformers; pp. 213–229. [DOI] [Google Scholar]
- 20.Ren S., He K., Girshick R., Sun J., Faster R.-C.N.N. Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39:1137–1149. doi: 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- 21.Lin T.-Y., Goyal P., Girshick R., He K., Dollar P. 2017 IEEE International Conference on Computer Vision (ICCV) IEEE; 2017. RetinaNet - focal loss for dense object detection; pp. 2999–3007. [DOI] [Google Scholar]
- 22.Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., et al. 2017. Attention is all you need. ArXiv Preprint ArXiv:1706.03762. [Google Scholar]
- 23.Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., et al. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin. Chem. 2020;66:975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park G.-S., Ku K., Baek S.-H., Kim S.-J., Kim S.I., Kim B.-T., et al. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 26.Arunrut N., Kampeera J., Sappat A., Tuantranont A., Avihingsanon Y., Benjachat T., et al. 2021. Primers and A Screening and Quantification Method for Urine IP-10 mRNA Biomarker of Lupus Nephritis, 2003000051 (Application Number) [Google Scholar]
- 27.Department of Medical Sciences, Ministry of Public Health, Thailand . WHO; 2020. Diagnostic Detection of Novel Coronavirus 2019 by Real-Time RT-PCR. [Google Scholar]
- 28.He K., Zhang X., Ren S., Sun J. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) IEEE; 2016. Deep residual learning for image recognition; pp. 770–778. [DOI] [Google Scholar]
- 29.Rezatofighi H., Tsoi N., Gwak J., Sadeghian A., Reid I., Savarese S. 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) IEEE; 2019. Generalized intersection over union: a metric and a loss for bounding box regression; pp. 658–666. [DOI] [Google Scholar]
- 30.He K., Zhang X., Ren S., Sun J. 2015 IEEE International Conference on Computer Vision (ICCV) IEEE; 2015. Delving deep into rectifiers: surpassing human-level performance on ImageNet classification; pp. 1026–1034. [DOI] [Google Scholar]
- 31.Loshchilov I., Hutter F. 2017. Decoupled weight decay regularization. ArXiv Preprint ArXiv:1711.05101. [Google Scholar]
- 32.Al-Rfou R., Choe D., Constant N., Guo M., Jones L. Character-level language modeling with deeper self-attention. AAAI. 2019;33:3159–3166. doi: 10.1609/aaai.v33i01.33013159. [DOI] [Google Scholar]
- 33.de Oliveira Coelho B., Sanchuki H.B.S., Zanette D.L., Nardin J.M., Morales H.M.P., Fornazari B., et al. Essential properties and pitfalls of colorimetric Reverse Transcription Loop-mediated Isothermal Amplification as a point-of-care test for SARS-CoV-2 diagnosis. Mol. Med. 2021;27:30. doi: 10.1186/s10020-021-00289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramírez J.D., Muñoz M., Patiño L.H., Ballesteros N., Paniz-Mondolfi A. Will the emergent SARS-CoV2 B.1.1.7 lineage affect molecular diagnosis of COVID-19? J. Med. Virol. 2021;93:2566–2568. doi: 10.1002/jmv.26823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pango lineages . 2021. B.1.617.2, Grinch | Global Report Investigating Novel Coronavirus Haplotypes.https://cov-lineages.org/global_report_B.1.617.2.html [Google Scholar]
- 36.Tatulli G., Cecere P., Maggioni D., Galimberti A., Pompa P.P. A rapid colorimetric assay for on-site authentication of cephalopod species. Biosensors. 2020;10 doi: 10.3390/bios10120190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020;4:1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- 38.Ventura B.D., Cennamo M., Minopoli A., Campanile R., Censi S.B., Terracciano D., et al. Colorimetric test for fast detection of SARS-CoV-2 in nasal and throat swabs. ACS Sens. 2020;5:3043–3048. doi: 10.1021/acssensors.0c01742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krizhevsky A., Sutskever I., Hinton G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM. 2012;60:84–90. doi: 10.1145/3065386. [DOI] [Google Scholar]
- 40.Simonyan K., Zisserman A. Very deep convolutional networks for large-scale image recognition. ArXiv Preprint ArXiv. 2014:1409–1556. [Google Scholar]
- 41.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., et al. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115:211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 42.Hu J., Shen L., Sun G. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. IEEE; 2018. Squeeze-and-Excitation networks; pp. 7132–7141. [DOI] [Google Scholar]
- 43.Cheng J., Dong L., Lapata M. Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics; Stroudsburg, PA, USA: 2016. Long short-term memory-networks for machine reading; pp. 551–561. [DOI] [Google Scholar]
- 44.Parikh A., Täckström O., Das D., Uszkoreit J. Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics; Stroudsburg, PA, USA: 2016. A decomposable attention model for natural language inference; pp. 2249–2255. [DOI] [Google Scholar]
- 45.Lin Z., Feng M., Santos C.N., Yu M., Xiang B., Zhou B., et al. 2017. A structured self-attentive sentence embedding. ArXiv Preprint ArXiv:1703.03130. [Google Scholar]
- 46.Paulus R., Xiong C., Socher R. 2017. A deep reinforced model for abstractive summarization. ArXiv Preprint ArXiv:1705.04304. [Google Scholar]
- 47.Fischbach J., Xander N.C., Frohme M., Glökler J.F. Shining a light on LAMP assays--a comparison of LAMP visualization methods including the novel use of berberine. Biotechniques. 2015;58:189–194. doi: 10.2144/000114275. [DOI] [PubMed] [Google Scholar]
- 48.Cammilleri G., Ferrantelli V., Pulvirenti A., Drago C., Stampone G., Del Rocio Quintero Macias G., et al. Validation of a commercial loop-mediated isothermal amplification (LAMP) assay for the rapid detection of anisakis spp. DNA in processed fish products. Foods. 2020:9. doi: 10.3390/foods9010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar Y., Bansal S., Jaiswal P. Loop-mediated isothermal amplification (LAMP): a rapid and sensitive tool for quality assessment of meat products. Comp. Rev. Food Sci. Food Safety. 2017;16:1359–1378. doi: 10.1111/1541-4337.12309. [DOI] [PubMed] [Google Scholar]
- 50.Tian X., Feng J., Wang Y. Direct loop-mediated isothermal amplification assay for on-site detection of Staphylococcus aureus. FEMS Microbiol. Lett. 2018;365 doi: 10.1093/femsle/fny092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foo P.C., Nurul Najian A.B., Muhamad N.A., Ahamad M., Mohamed M., Yean Yean C., et al. Loop-mediated isothermal amplification (LAMP) reaction as viable PCR substitute for diagnostic applications: a comparative analysis study of LAMP, conventional PCR, nested PCR (nPCR) and real-time PCR (qPCR) based on Entamoeba histolytica DNA derived from faecal sample. BMC Biotechnol. 2020;20:34. doi: 10.1186/s12896-020-00629-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitrowska K., Posyniak A., Zmudzki J. Determination of malachite green and leucomalachite green in carp muscle by liquid chromatography with visible and fluorescence detection. J. Chromatogr., A. 2005;1089:187–192. doi: 10.1016/j.chroma.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 53.Huang W.E., Lim B., Hsu C.-C., Xiong D., Wu W., Yu Y., et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez N.M., Linnes J.C., Fan A., Ellenson C.K., Pollock N.R., Klapperich C.M. Paper-based RNA extraction, in situ isothermal amplification, and lateral flow detection for low-cost, rapid diagnosis of Influenza A (H1N1) from clinical specimens. Anal. Chem. 2015;87:7872–7879. doi: 10.1021/acs.analchem.5b01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klein S., Müller T.G., Khalid D., Sonntag-Buck V., Heuser A.-M., Glass B., et al. SARS-CoV-2 RNA extraction using magnetic beads for rapid large-scale testing by RT-qPCR and RT-LAMP. Viruses. 2020;12 doi: 10.3390/v12080863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.