Abstract
The biochemical properties of the d-glutamate-adding enzymes (MurD) from Escherichia coli, Haemophilus influenzae, Enterococcus faecalis, and Staphylococcus aureus were investigated to detect any differences in the activity of this enzyme between gram-positive and gram-negative bacteria. The genes (murD) that encode these enzymes were cloned into pMAL-c2 fusion vector and overexpressed as maltose-binding protein–MurD fusion proteins. Each fusion protein was purified to homogeneity by affinity to amylose resin. Proteolytic treatments of the fusion proteins with factor Xa regenerated the individual MurD proteins. It was found that these fusion proteins retain d-glutamate-adding activity and have Km and Vmax values similar to those of the regenerated MurDs, except for the H. influenzae enzyme. Substrate inhibition by UDP-N-acetylmuramyl-l-alanine, the acceptor substrate, was observed at concentrations greater than 15 and 30 μM for E. coli and H. influenzae MurD, respectively. Such substrate inhibition was not observed with the E. faecalis and S. aureus enzymes, up to a substrate concentration of 1 to 2 mM. In addition, the two MurDs of gram-negative origin were shown to require monocations such as NH4+ and/or K+, but not Na+, for optimal activity, while anions such as Cl− and SO42− had no effect on the enzyme activities. The activities of the two MurDs of gram-positive origin, on the other hand, were not affected by any of the ions tested. All four enzymes required Mg2+ for the ligase activity and exhibited optimal activities around pH 8. These differences observed between the gram-positive and gram-negative MurDs indicated that the two gram-negative bacteria may apply a more stringent regulation of cell wall biosynthesis at the early stage of peptidoglycan biosynthesis pathway than do the two gram-positive bacteria. Therefore, the MurD-catalyzed reaction may constitute a fine-tuning step necessary for the gram-negative bacteria to optimally maintain its relatively thin yet essential cell wall structure during all stages of growth.
Properly constructed peptidoglycan networks in the bacterial cell wall provide the rigidity, flexibility, and strength that are necessary for the bacterial cells to grow and divide while withstanding the extraordinarily high internal osmotic pressure and sometimes, harsh external environment. Most bacterial peptidoglycan networks share the same basic structural components that consist of repeating 2′-N-acetyl-disaccharide-tri (or tera)-peptide units (12, 24, 29). However, significant differences in cell wall structure exist between gram-positive and gram-negative bacteria (26). A better understanding on how bacteria coordinate and regulate the composition of these basic units may lead to development of more effective strategies to treat bacterial infections.
Earlier reports on the glutamate racemases (MurI or Dga) from several gram-positive bacteria and from Escherichia coli highlighted certain specific properties which distinguish enzymes from gram-positive and gram-negative bacteria (4, 8, 13, 17, 34). These earlier observations indicate that along the murein biosynthesis pathway, the reactions involved in d-glutamate formation and perhaps its incorporation into the cell wall network are potential fine-tuning points for total cell wall biosynthesis in certain bacterial species. We chose to examine the d-glutamate-adding enzymes (MurDs) from two gram-positive and two gram-negative organisms for differences that may provide insight into fine-tuning mechanisms important for cell wall synthesis. Furthermore, the enzymes that utilize amino acids with d configuration are especially attractive targets for broad-spectrum and selective antibacterial agents. The commonly applied beta-lactam (21) classes of antibiotics and the only effective agent for treating methicillin-resistant Staphylococcus aureus, vancomycin (22), are examples of antibacterial agents which take advantage of this unique feature of bacterial cell walls.
MurD (UDP-N-acetylmuramyl-l-alanine:d-glutamate ligase), the bacterial d-glutamate-adding enzyme, catalyzes the attachment of d-glutamate to a cytoplasmic peptidoglycan precursor, UDP-N-acetylmuramyl-l-alanine (5, 24, 25, 30). This reaction results in the formation of a peptide linkage between the amino function of d-glutamate and the carboxyl terminus of UDP-N-acetylmuramyl-l-alanine. A stoichiometric consumption of ATP supplies the energy needed for this peptide bond formation with concomitant generation of ADP and orthophosphate. In this report, we describe studies that were undertaken to investigate the basic biochemical properties of the d-glutamate-adding enzymes from two gram-positive bacteria (S. aureus and Enterococcus faecalis) and two gram-negative bacteria (E. coli and Haemophilus influenzae). The similarities and differences observed between these enzymes are discussed in detail.
MATERIALS AND METHODS
Preparation of pMAL::murD fusion constructs.
The pMAL-c2 vector (New England Biolabs) was used to express the various murD genes as fusions with the E. coli maltose-binding protein (MBP) gene (malE) according to the manufacturer’s recommendations. murD genes were isolated by PCR from the following strains by using the oligonucleotides listed in Table 1. PCR fragments were isolated and digested overnight with appropriate restriction enzymes (sites engineered within oligonucleotide primers). The digested fragments were ligated with pMAL-c2 which was similarly restriction digested and treated with alkaline phosphatase.
TABLE 1.
Oligonucleotides used to isolate murD genes by PCR
| Source of murD gene | GenBank accession no. | Sequences of oligonucleotides used for PCR |
|---|---|---|
| S. aureus RN4220 | U94706) | 5′-CATTGAATTCATGCTTAATTATACAGGGTTAGAA-3′ |
| 5′-CAATAAGCTTCATCCATCAATACTCACACCC-3′ | ||
| E. faecalis clinical isolate 24836 | U94707 | 5′-TTCGGAATTCATGAAAAAAATAACAACTTATCAAAAC-3′ |
| 5′-TAGCTCTAGATTACTAGTATCTTCATTTCCTCAC-3′ | ||
| E. coli K-12 | X17609 | 5′-TTCGGAATTCATGGCTGATTATCAGGG-3′ |
| 5′-TAGCTCTAGAGATAAACGCATCAACC-3′ | ||
| H. influenzae A9729 | U32793 | 5′-TTAAGAATTCATGAACGCCTATCAAAAC-3′ |
| 5′-TATAGAATTCGGTGTGATGCGTGTCCATTC-3′ |
Overexpression and purification of MurDs.
All E. coli/pMAL::murD clones were constructed by transforming E. coli JM109 with the individual pMAL::murD fusion plasmids. Each overnight culture was inoculated into 1 liter of SOC medium (Gibco BRL) containing 50 μg of ampicillin per ml, and the cultures were incubated at 37°C with aeration. Isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM) induction was initiated when the optical density of the culture at 600 nm reached 0.6. Two hours later, cells were harvested by centrifugation at 7,000 rpm for 30 min at 4°C. Cell pellets were resuspended in 25 ml of 50 mM Tris-HCl (pH 8.0) containing 1 mM EDTA, 1 mM β-mercaptoethanol, and protease inhibitors (Complete; Boehringer Mannheim). Cells were lysed by sonicating the cell suspension with cooling in an ethanol-ice bath. Cell debris was removed by centrifugation at 10,000 rpm for 30 min at 4°C. The MBP-MurD fusion proteins in the soluble fraction were absorbed onto amylose resin (New England Biolabs) via batch mixing at 4°C for 1 h and then eluted with buffer containing 10 mM maltose. These affinity-purified fusion proteins were further concentrated to a final concentration of approximately 10 mg/ml in an Amicon Centriprep 10 concentrator. After glycerol was added to a final concentration of 50%, these protein solutions were stored at −20°C. Regeneration of the individual MurD enzymes from their fusion proteins was carried out by subjecting the affinity-purified MBP-MurD fusion proteins to factor Xa treatment (based on the recommended conditions by New England Biolabs) for 24 to 72 h at 4°C. The proteolytic cleavage process was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of a small aliquot of the reaction mixture. Regenerated MurD was isolated from MBP by fast protein liquid chromatography (FPLC) using a Pharmacia MonoQ (5/5) anion-exchange column. Elution was carried out with a 0 to 1 M NaCl gradient in 50 mM Tris-HCl (pH 7.5) buffer containing 1 mM dithiothreitol and 10% glycerol. Fractions containing MurD were pooled and concentrated as described above.
Preparation of UDP-N-acetylmuramyl-l-alanine.
The UDP-linked substrate for MurD was prepared using stepwise enzymatic synthesis protocols with MBP-MurA, MBP-MurB, and MBP-MurC fusion proteins. The progress of product formation in each reaction mixture was monitored via high-pressure liquid chromatography (HPLC) analysis of small aliquots of reaction mixture over time. The individual fusion proteins used for substrate synthesis were prepared as reported previously (3, 6). The reaction mixture (640 ml) for UDP-N-acetylenolpyruvylglucosamine synthesis contained 20 mM Tris-HCl buffer (pH 7.5), 7 mM UDP-N-acetylglucosamine, 10 mM phosphoenolpyruvate, and 26 mg of MBP-MurA. After removal of enzyme, the product was purified by FPLC using a DEAE-cellulose column (2.5 by 30 cm) eluted with a 50 to 500 mM triethylammonium bicarbonate gradient at pH 8.0. For the synthesis of UDP-N-acetylmuramic acid, the reaction mixture (130 ml) contained 100 mM Tris-HCl buffer, (pH 7.5), 20 mM KCl, 22 mM UDP-N-acetylenolpyruvylglucosamine, 30 mM NADPH, 5 mM dithiothreitol, and 14 mg of MBP-MurB. The reaction product was purified by reverse-phase HPLC using a Bio-Rad Hi-Pore 318 column eluted with 50 mM NH4CO2H (pH 5.0). UDP-N-acetylmuramyl-l-alanine was synthesized in a reaction mixture (100 ml) containing 100 mM Tris-HCl (pH 7.5), 20 mM (NH4)2SO4, 1 mM β-mercaptoethanol, 5 mM MgCl2, 4 mM UDP-N-acetylmuramic acid, 22 mM l-alanine, 40 mM ATP, and 15 mg of MBP-MurC. The product was isolated by reverse-phase HPLC using the same Bio-Rad RP column eluted with 50 mM ammonium formate pH 3.5 to pH 5.0 gradient. At each purification step, small aliquots were analyzed by HPLC to identify those fractions containing the desired UDP-linked products. These fractions were then pooled and lyophilized to remove the excess salt contents. Identities of each UDP-linked product were further confirmed by HPLC, nuclear magnetic resonance, and mass spectrometry analyses.
Assay for MurD activities.
Three different protocols were used to monitor the d-glutamate-adding enzyme activities. All assays were carried out at 24°C and in duplicate. Initial detection of MurD activity was carried out in reaction mixtures containing 100 mM Tris-HCl (pH 8.0), 5 to 10 mM MgCl2, 1 mM β-mercaptoethanol, 2 mM ATP, 1 mM d-[C14]glutamate (16 nCi/nmol), 30 to 60 μM UDP-N-acetylmuramyl-l-alanine, and 20 mM (NH4)2SO4. d-[U-14C]glutamate was custom-prepared by Du-Pont NEN Research Products. An HPLC (Waters) assay with on-line UV (Waters 996 detector) and flow scintillation (Packard Radiomatic Flo/One scintillation counter) detectors was used to monitor the formation of UDP-N-acetylmuramyl-l-alanine-d-[14C]glutamate and ADP in each reaction mixture. The two other protocols were adapted to a 96-well microtiter plate format to allow parallel runs of multiple assays and eliminate the use of the radioactive tracer. One of the protocols follows the formation of orthophosphate generated during the reaction by adding malachite green (Sigma Chemical Co.)-molybdate (J.T. Baker Chemical Co.) reagent (15) to an aliquot of a reaction mixture at a designated time point. The absorbance at 660 nm was then measured with a Molecular Devices SpectraMax 250 plate reader. An accompanying series of orthophosphate standards was run for each plate. The third protocol takes advantage of the change of absorbance at 340 nm over time by coupling the MurD reaction with a significant excess of pyruvate kinase-lactate dehydrogenase (PK-LDH; Sigma), phosphoenolpyruvate, and NADH. This protocol monitors ADP formation in the MurD-catalyzed reaction. For kinetic parameter determinations, the concentrations of one of the three substrates were varied (as indicated in the footnotes to Table 2), while the other two were kept at saturating conditions. The coupled MurD-PK-LDH assay was used to determine the kinetic parameters of all MurDs except for that of E. coli, which was measured with the radioactive assays. The ions requirement was analyzed by HPLC and/or malachite green colorimetric assays. For the comparison of IC50s (concentrations that inhibit enzyme activity by 50%), concentrations of the substrates in the assay mixtures were kept near their Km values.
TABLE 2.
Relative efficiencies of MurDs from various bacterial species
| Origin | Form of MurD | Vmax (nmol/min · mg)a |
Km(app) (μM)a
|
Vmax/Km(app, UDPNacMur-l-Ala) | Relative efficiency | ||
|---|---|---|---|---|---|---|---|
| UDPNacMur-l-Alab | ATP | d-Glutamatec | |||||
| S. aureus | MBP-MurD fusion protein | 31,787 ± 3,668 | 245 ± 29 | 78 ± 9 | 660 ± 51 | 12,974 | 0.14 |
| Regenerated MurD | 32,118 ± 3,636 | 290 ± 34 | 84 ± 10 | 534 ± 25 | 11,075 | 0.12 | |
| E. faecalis | MBP-MurD fusion protein | 32,857 ± 3,647 | 38 ± 4 | 44 ± 5 | 121 ± 10 | 86,466 | 0.90 |
| Regenerated MurD | 29,823 ± 5,934 | 36 ± 7 | 47 ± 4 | 118 ± 14 | 82,842 | 0.87 | |
| E. coli | MBP-MurD fusion protein | 5,965 ± 297 | 5 ± 2 | 154 ± 23 | 27 ± 12 | 85,214 | 0.89 |
| Regenerated MurD | 4,783 ± 423 | 7 ± 3 | 135 ± 33 | 42 ± 11 | 95,660 | 1.00 | |
| H. influenzae | MBP-MurD fusion protein | NDd | ND | ND | ND | ND | ND |
| Regenerated MurD | 13,111 ± 2,199 | 8 ± 4 | 102 ± 6 | 169 ± 20 | 163,888 | 1.71 | |
Gram-positive MurD activity was determined with the coupled PK-LDH assay (up to 10 min).
Gram-negative MurD (1- to 2-min assay) was assayed by HPLC or coupled enzyme assay.
All assays were done in duplicate, and a minimal of three independent experiments were conducted for each kinetic determination.
Reaction rates within the linear region of product formation were used for parameter analysis.
0 to 2 mM UDP-N-acetylmuramyl-l-alanine (UDPNacMur-l-Ala) was used for gram-positive MurD assay; 0 to 200 μM was used for gram-negative MurD assay.
The ranges of ATP and d-glutamate used in gram-negative MurD assays were from 0 to 1 mM and from 0 to 2 mM, respectively.
The ranges of ATP and d-glutamate used in gram-positive MurD assays were from 0 to 1 mM and from 0 to 5 mM, respectively.
ND, not determined.
RESULTS
Overexpression and purification of MurD protein.
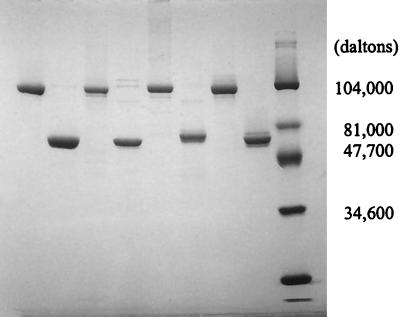
To obtain sufficient quantities of d-glutamate-adding enzymes from different bacterial hosts, the individual murD genes were PCR amplified, purified, and successfully subcloned into the pMAL-c2 vector as malE::murD fusions. Upon IPTG induction, those E. coli JM109 cells transformed with individual pMAL::murD plasmids significantly overproduced the corresponding MBP-MurD fusion protein. SDS-PAGE analysis of the crude lysates from all four cultures indicated that the individual fusion proteins accounted for 20 to 30% of the protein expressed in the host cells. The MBP-MurD fusion proteins recovered, after the amylose-resin affinity purification, were >90% pure by SDS-PAGE analysis. Between 30 and 40 mg of MBP-MurDs were obtained from 1-liter cultures of each of the four MurD-overexpressing clones. Regeneration of MurDs from MBP-MurD by factor Xa treatment was essentially quantitative (Fig. 1). All purified enzymes were stored in 50 mM Tris-HCl (pH 7.5)–1 mM β-mercaptoethanol–50% glycerol at −20°C. Under these conditions, enzymes are stable for several months with minimal loss of activity.
FIG. 1.
SDS-PAGE analysis of purified MBP-MurD fusion proteins and the corresponding MurDs regenerated by factor Xa treatment. Molecular masses for MBP and MurD are 42 and around 55 kDa, respectively. Regenerated MurD was purified from MBP by FPLC using a MonoQ anion-exchange column. Lanes: 1, MBP-MurD (E. coli); 2, regenerated MurD (E. coli); 3, MBP-MurD (H. influenzae); 4, regenerated MurD (H. influenzae); 5, MBP-MurD (S. aureus); 6, regenerated MurD (S. aureus); 7, MBP-MurD (E. faecalis); 8, regenerated MurD (E. faecalis); 9, Bio-Rad prestained low-molecular-weight range standards.
Characterization of MBP-MurDs and the regenerated MurDs.
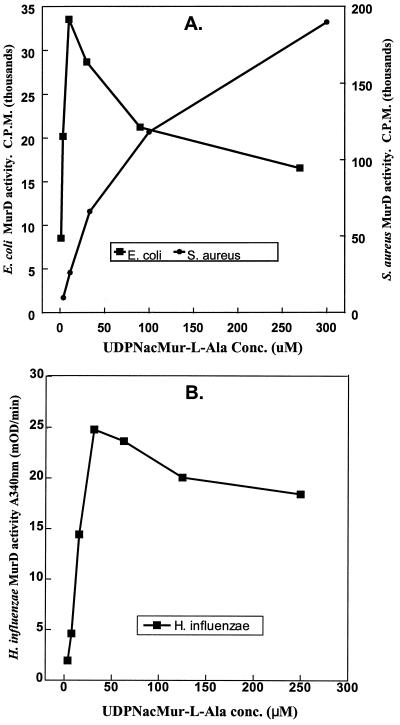
Both the MBP-MurD fusion proteins and the regenerated MurDs showed the d-glutamate-adding enzyme activities and the proper stoichiometry. The apparent Km [Km(app)] values for all three substrates of d-glutamate-adding enzymes are listed in Table 2. For the UDP-linked substrate, UDP-N-acetylmuramyl-l-alanine, the Km(app) values obtained from the two gram-positive bacterial MurDs were significantly higher than those determined with the two gram-negative bacterial MurDs. Furthermore, the activities of those enzymes from the two gram-negative bacteria were inhibited by this UDP-linked substrate at concentrations as low as 20 to 30 μM (Fig. 2). No substrate inhibition by the same UDP-linked substrate was detected in the gram-positive MurDs assays at concentrations as high as 2 mM. On the other hand, the binding affinities of the other two cosubstrates, ATP and d-glutamate, were fairly similar among three of the four MurDs. Except for the H. influenzae MurD, there were little differences in the Km(app) and Vmax values observed between the respective fusion proteins and the regenerated MurDs (Table 2). The fusion protein containing the MurD of the H. influenzae origin showed d-glutamate-adding activity significantly lower than that of the regenerated MurD.
FIG. 2.
Inhibition of gram-negative MurDs by UDP-N-acetylmuramyl-l-alanine. (A) E. coli and S. aureus MurDs. Determination of d-glutamate-adding enzyme activity was carried out with HPLC equipped with an on-line flow scintillation counter. The substrates, d-[14C]glutamate and ATP, were present at saturating concentration, while the concentration of UDP-N-acetylmuramyl-l-alanine was varied as shown. The amount of UDP-N-acetylmuramyl-l-alanyl-d-[14C]glutamate formed reflects the activity of MurD at various concentrations of UDP-N-acetylmuramyl-l-alanine. (B) H. influenzae MurD. Activity was monitored with the coupled PK-LDH assay.
Significant differences in the specific activities were observed among the four MurDs (Table 2). The specific activities of E. faecalis and S. aureus MurDs were two- to sixfold higher than those of the E. coli and H. influenzae MurDs, with E. coli MurD having the lowest activity of the four. However, when the affinity [Km(app)] of the UDP-linked substrate (UDP-N-acetylmuramyl-l-alanine) to each corresponding MurD was taken into consideration, MurDs from H. influenzae, E. coli, and E. faecalis all demonstrated similar efficiencies (Vmax/Km(app) [Table 2). The efficiency of S. aureus MurD was found to be 5- to 10-fold lower than those of the other three enzymes evaluated.
(i) Effects of bivalent cations.
As expected, all four MurDs investigated required Mg2+ ion for d-glutamate-adding enzyme activities. The highest concentration of Mg2+ ion evaluated was 25 mM. The optimal concentration of Mg2+ ion for the E. coli MurD activity was 5 mM. In the presence of higher concentrations of this bivalent cation, a decrease of E. coli MurD activity was observed. Activity at 25 mM Mg2+ was 12% lower than that at 5 mM. S. aureus and E. faecalis MurDs, on the other hand, demonstrated optimal activities at 20 to 25 and 10 to 25 mM Mg2+, respectively. Other divalent cations such as Mn2+ and Co2+ could be substituted for Mg2+; however, lower enzyme activities were observed.
(ii) Effect of monovalent cations.
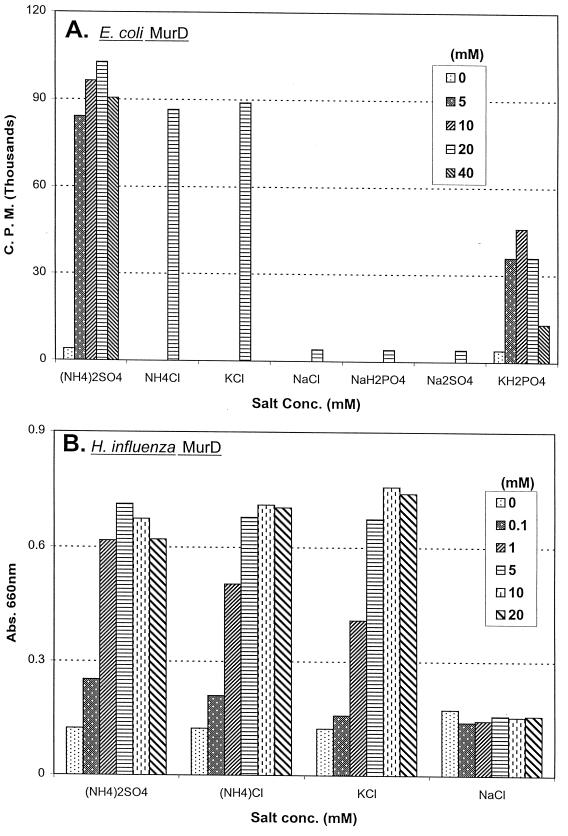
A stimulation of MurD activity by monovalent cations such as NH4+ and K+ was observed only with the two gram-negative MurDs. This stimulation of E. coli and H. influenzae MurD activities by either of these two monovalent cations is concentration dependent (Fig. 3). The most pronounced effect by these two monovalent cations was observed in the E. coli MurD assay. The E. coli d-glutamate-adding enzyme activity increased up to 20-fold in the presence of 20 mM (NH4)2SO4, NH4Cl, or KCl (Fig. 3A). The optimal E. coli MurD activity was observed when these monovalent cations reached concentrations of 20 mM or greater, except for (NH4)2SO4, which at 40 mM caused a 15% decrease of E. coli MurD activity. Potassium phosphate also stimulated the E. coli MurD activity, but to a lesser extent. Sodium salts, such as sodium phosphate, sodium sulfate, and sodium chloride, on the other hand, do not stimulate the E. coli enzyme activity. For the H. influenzae MurD, up to a 600% elevation of enzyme activity was observed in the presence of NH4+ or K+ salts (Fig. 3B). Optimal activities were also observed at 20 mM and greater concentrations of either of these two cations. Similar to the E. coli MurD, ammonium sulfate at 50 mM caused a 10 to 20% decrease of enzyme activity, compared with 10 to 20 mM ammonium sulfate. Also, like the E. coli MurD, sodium ion did not stimulate the H. influenzae MurD activity. In contrast, the activities of the MurD from either S. aureus or E. faecalis were not stimulated by the presence of any of the monovalent cations described above (data not shown). Similar patterns of ion requirement and ion-dependent stimulation of MurD activities were also observed with the corresponding MBP-MurD fusion proteins.
FIG. 3.
E. coli and H. influenzae MurDs required NH4+ or K+ for optimal d-glutamate-adding enzyme activities. (A) E. coli MurD activity was determined by monitoring the amount of UDP-N-acetylmuramyl-l-alanyl-d-[14C]glutamate formed with an HPLC equipped with on-line flow scintillation counter. (B) H. influenzae MurD activity was determined by using the malachite green reagent to determine the level of orthophosphate generated. The concentrations of selected salts present in each reaction mixture were as shown.
Inhibition of MurD activities by a transition state analog.
The differences exhibited between the MurDs from the two gram-positive and two gram-negative organisms were significant. To further evaluate the properties of these enzymes, the inhibition of the d-glutamate-adding enzyme activities by a transition state analog, [1(6-uridine diphospho)hexanamido](2,4-dicarboxybutyl)phosphinate, was evaluated. This compound was previously reported by Tanner et al. (28a) to inhibit the E. coli MurD activity by 50% at 0.7 μM (IC50). In our studies with the E. coli MurD, this compound had IC50s of 1.7 and 2.1 μM when assayed with the regenerated MurD and the MBP-MurD fusion, respectively. The IC50 determined with the H. influenzae MBP-MurD was 6.6 μM. Similar values were obtained for the MurDs of gram-positive origin. The IC50s obtained for E. faecalis MurD (regenerated) and MBP-MurD fusion were 5.7 and 7.0 μM, respectively. For the regenerated MurD and the MBP-MurD fusion from S. aureus, the IC50s were 10.1 and 8.6 μM, respectively. The results here show that this transition state analog inhibited all four MurDs to a similar extent.
DISCUSSION
In this report, we describe the successful cloning, overexpression, and preparation of the d-glutamate-adding-enzyme (MurD) from four bacterial species. These protocols provided the quantities of enzymes needed to further characterize the basic biochemical properties of these enzymes.
Although bacterial cell walls come in various sizes and shapes, most of them consist of the same basic type of disaccharide-tri (or tetra)-peptide subunits in their peptidoglycan network. The electron dense cell walls of gram-positive bacteria have thicknesses ranging between 20 and 50 nm (11, 27). In contrast, the gram-negative bacterial cell walls consist of a very thin peptidoglycan layer of about 1-nm thickness (2, 11). The demand for cytoplasmic peptidoglycan precursor building blocks in the gram-negative bacteria should therefore be significantly lower than that in the gram-positive bacteria. The complexity of coordinating and regulating peptidoglycan precursor biosynthesis with cell growth and division spans from the DNA level to the enzymatic level. Regulatory mechanisms at the enzymatic level allow the cells to instantaneously respond to their immediate environmental changes, without having to wait for changes occurring at the protein, RNA, or DNA level.
Several examples of cell wall precursor synthesis regulation at the enzymatic level have been reported. Feedback inhibitions of the enzymes in the earlier steps in the murein precursors biosynthesis pathway by the downstream precursors have been proposed as one of the ways that the early steps of cell wall synthesis are modulated (19, 35). The activity of E. coli glutamate racemase was previously shown to be uniquely regulated by UDP-N-acetylmuramyl-l-alanine (4, 13). This UDP-linked peptidoglycan precursor optimally stimulates E. coli glutamate racemase activity (50- to 80-fold increase) when the activator concentrations reach about 10 μM. Here we reported another regulatory action by this very molecule at the enzymatic level in E. coli. We showed that 10 μM UPD-N-acetylmuramyl-l-alanine is also the near optimal substrate level for the E. coli MurD activity (Fig. 2A). Furthermore, it has been reported that the cytoplasmic pool level of UDP-N-acetylmuramyl-l-alanine in E. coli cells growing at the log phase is also about 10 μM (18). Together, these results indicate that in E. coli cells growing in the logarithmic growth phase, both glutamate racemase and the d-glutamate-adding enzyme are functioning at their maximal level, which is influenced by the intracellular level of UDP-N-acetylmuramyl-l-alanine.
At concentrations greater than 15 μM, UDP-N-acetylmuramyl-l-alanine exhibits significant substrate inhibition to the E. coli d-glutamate-adding enzyme activity (Fig. 2A). Therefore, conditions that cause intracellular level of UDP-N-acetylmuramyl-l-alanine to elevate above 10 to 15 μM would ultimately decrease the cellular d-glutamate-adding enzyme activity and reduce the consumption of d-glutamate. Although under these conditions, E. coli glutamate racemase remains optimally active, this enzyme catalyzes the interconversion between d- and l-glutamate with equal efficiency in both directions (13). Unused d-glutamate can therefore be effectively converted to l-glutamate, without resulting in undesirable level of d-glutamate accumulation. It is also known that E. coli cells turn over up to 50% of its periplasmic peptidoglycan components in each cell cycle and recycle up to 90% of the material turned over (10, 23). The tripeptide, l-alanyl-d-glutamyl-meso-diaminopimelic acid (regenerated from the recycled muropeptides [10, 14]), is reattached to UDP-N-acetylmuramate and reintroduced into the UDP-N-acetylmuramyl-peptide biosynthesis pathway. The balance between the recycling pathway of the periplasmic peptidoglycan and the de novo biosynthesis of the cytoplasmic UDP-linked precursors would allow the most efficient usage of both the peptidoglycan precursors and enzymes involved in precursor synthesis such as MurD in E. coli. A similar pattern of substrate inhibition by UDP-N-acetylmuramyl-l-alanine to the d-glutamate-adding enzyme from H. influenzae, another gram-negative bacterium, was also demonstrated (Fig. 2B). Whether a similar regulatory mechanism to the glutamate racemase by UDP-N-acetylmuramyl-l-alanine also exists in H. influenzae has yet to be determined.
The stimulation of d-glutamate-adding enzyme activities by NH4+ or K+ ion is another characteristic shown to be shared only by the two gram-negative bacterial MurDs (Fig. 3). Stimulation of enzyme activity by these two monovalent cations has been well documented for a few bacterial (28, 32) and eukaryotic (9, 16, 33) enzymes. The suggested roles these cations play include inducing protein conformational change and stabilizing reaction intermediates. The mechanism(s) of E. coli and H. influenzae MurD stimulation by these cations remains unclear at this time. The observed lesser stimulation effect by potassium phosphate is likely due to the contribution of product inhibition by phosphate ion, which is also a product in the MurD-catalyzed reaction.
Contrary to what was observed with the two gram-negative MurDs, the d-glutamate-adding enzymes encoded by the two gram-positive (S. aureus and E. faecalis) murD genes were not subjected to substrate inhibition by UDP-N-acetylmuramyl-l-alanine at concentrations as high as 2 mM. Furthermore, there was no stimulation of the S. aureus and E. faecalis MurD activities by any of the ions tested. It is also true that the glutamate racemases from gram-positive bacteria such as Pediococcus pentosaceus and Lactobacillus fermenti were not stimulated by UDP-N-acetylmuramyl-l-alanine (7, 8, 13, 17). Although the intracellular levels of UDP-N-acetylmuramyl-l-alanine and other peptidoglycan precursors in these gram-positive organisms have yet to be determined, it has been shown the gram-positive bacteria do not recycle peptidoglycan components turned over during cell growth (1, 20, 31). In addition, the gram-positive bacterial cell walls contain a murein network 10- to 20-fold thicker than that of their gram-negative counterpart. A significantly higher demand for the de novo peptidoglycan precursors synthesis can therefore be expected. The enzymes involved in gram-positive UDP-N-acetylmuramylpeptide synthesis can probably be expected to operate at their maximal levels most if not all of the time. Fine-tuning cell wall precursors levels as closely as those observed in gram-negative bacteria, such as E. coli and H. influenzae, may therefore not be necessary. On the other hand, how the differences in the efficiency of each MurD shown here fit into the total picture of cell wall synthesis is more difficult to interpret. One would have to take into consideration the total number of active MurDs or the total activities present in each bacterial species and the respective peptidoglycan turnover rate (or recycle rate if present) to begin to interpret the true meanings of these data. It will also be interesting to see if these distinct properties influence shape determination of bacterial cells, i.e., rod versus sphere.
Despite the different characteristics present in the MurDs studied here, we found that the activities of these MurDs were inhibited to a similar extend by the transition state analog, [1(6-uridine diphospho)hexanamido](2,4-dicarboxybutyl)phosphinate. These results indicated that all four MurDs probably share a similar transition state conformation at the active site. Therefore, it should be possible to identify a broad spectrum MurD inhibitor in the future.
ACKNOWLEDGMENTS
This work was supported by Bristol Myers Squibb Pharmaceutical Research Institute, Infectious Diseases Department.
[1(6-Uridine diphospho)hexanamido](2,4-dicarboxybutyl)phosphinate was generously provided by Henry Wong at BMS-PRI. We are thankful to Stella Huang and Kevin Volk for their assistance in nuclear magnetic resonance and liquid chromatography-mass spectrometry analyses. Also, we are grateful for the helpful discussions and the critical reading of the manuscript by Thomas Dougherty and John Barrett.
REFERENCES
- 1.Blumel P, Uecker W, Giesbrecht P. Zero order kinetics of cell wall turnover in Staphylococcus aureus. Arch Microbiol. 1979;121:103–110. doi: 10.1007/BF00689972. [DOI] [PubMed] [Google Scholar]
- 2.De Petris S. Ultrastructure of the cell wall of Escherichia coli and chemical nature of its constituent layers. J Ultrastruct Res. 1967;19:45–83. doi: 10.1016/s0022-5320(67)80059-5. [DOI] [PubMed] [Google Scholar]
- 3.Dhalla A M, Yanchunas J, Jr, Ho H-T, Falk P J, Villafranca J J, Robertson J G. Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase. Biochemistry. 1995;34:5390–5402. doi: 10.1021/bi00016a010. [DOI] [PubMed] [Google Scholar]
- 4.Doublet P, van Heijenoort J, Mengin-Lecreulx D. The glutamate racemase activity from Escherichia coli is regulated by peptidoglycan precursor UDP-N-acetylmuramoyl-l-alanine. Biochemistry. 1994;33:5285–5290. doi: 10.1021/bi00183a035. [DOI] [PubMed] [Google Scholar]
- 5.El-Sherbeini M, Geissler W M, Pittman J, Yuan X, Wong K K, Pompliano D L. Cloning and expression of Staphylococcus aureus and Streptococcus pyogenes murD genes encoding uridine diphosphate-N-acetylmuramoyl-l-alanine:d-glutamate ligases. Gene. 1998;210:117–125. doi: 10.1016/s0378-1119(98)00059-6. [DOI] [PubMed] [Google Scholar]
- 6.Falk P J, Ervin K M, Volk K S, Ho H-T. Biochemical evidence for the formation of a covalent acyl-phosphate linkage between UDP-N-acetylmuramate and ATP in the Escherichia coli UDP-N-acetylmuramate:l-alanine ligase-catalyzed reaction. Biochemistry. 1996;35:1417–1422. doi: 10.1021/bi952078b. [DOI] [PubMed] [Google Scholar]
- 7.Gallo K A, Knowles J R. Purification, cloning, and cofactor independence of glutamate racemase from Lactobacillus. Biochemistry. 1993;32:3981–3990. doi: 10.1021/bi00066a019. [DOI] [PubMed] [Google Scholar]
- 8.Gallo K A, Knowles J R. Mechanism of the reaction catalyzed by glutamate racemase. Biochemistry. 1993;32:3991–3997. doi: 10.1021/bi00066a020. [DOI] [PubMed] [Google Scholar]
- 9.Gayathri J, Raghavendra A S. Ammonium ions stimulate in vitro the activity of phosphoenolpyruvate carboxylase from leaves of Amaranthus hypochondriacus, a C4 plant: evidence for allosteric activation. Biochem Mol Biol Int. 1994;33:337–343. [PubMed] [Google Scholar]
- 10.Goodell E W. Recycling of murein by Escherichia coli. J Bacteriol. 1985;163:305–310. doi: 10.1128/jb.163.1.305-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham L L, Beveridge T J. Effect of chemical fixatives on accurate preservation of Escherichia coli and Bacillus subtilis structure in cells prepared by freeze-substitution. J Bacteriol. 1990;172:2150–2159. doi: 10.1128/jb.172.4.2150-2159.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartman E, Konig H. Comparison of the biosynthesis of the methanobacterial pseudomurien and the eubacterial murien. Naturwissenschaften. 1990;77:472–475. doi: 10.1007/BF01135923. [DOI] [PubMed] [Google Scholar]
- 13.Ho H-T, Falk P J, Ervin K M, Krishnan B S, Discotto L F, Dougherty T J, Pucci M J. UDP-N-acetylmuramyl-l-alanine functions as an activator in the regulation of the Escherichia coli glutamate racemase activity. Biochemistry. 1995;34:2464–2470. doi: 10.1021/bi00008a009. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs C, Huang L J, Bartowsky E, Normark S, Park J T. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J. 1994;13:4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanzetta P A, Alvarez L J, Reinach P S, Candia O A. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 16.Laughlin L T, Reed G H. The monovalent cation requirement of rabbit muscle pyruvate kinase is eliminated by substitution of lysine for glutamate 117. Arch Biochem Biophys. 1997;348:262–267. doi: 10.1006/abbi.1997.0448. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Yoshimura T, Endo K, Esaki N, Soda K. Cloning and expression of the glutamate racemase gene of Bacillus pumilus. J Biochem (Tokyo) 1997;121:1155–1161. doi: 10.1093/oxfordjournals.jbchem.a021709. [DOI] [PubMed] [Google Scholar]
- 18.Mengin-Lecreulx D, Flouret B, van Heijenoort J. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J Bacteriol. 1982;151:1109–1117. doi: 10.1128/jb.151.3.1109-1117.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mengin-Lecreulx D, Flouret B, van Heijenoort J. Pool levels of UDP-N-acetylglucosamine and UDP-N-acetylglucosamine-enolpyruvate in Escherichia coli and correlation with peptidoglycan synthesis. J Bacteriol. 1983;154:1284–1290. doi: 10.1128/jb.154.3.1284-1290.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mobley H L T, Koch A L, Doyle R J, Streips U K. Insertion and fate of the cell wall in Bacillus subtilis. J Bacteriol. 1984;158:169–179. doi: 10.1128/jb.158.1.169-179.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neu H C. Antibacterial therapy: problems and promises, part I. Hosp Pract Off Ed. 1990;25:63–74. doi: 10.1080/21548331.1990.11703947. [DOI] [PubMed] [Google Scholar]
- 22.Nieto M, Perkins H R. Physical properties of vancomycin and iodovancomycin and the complexes with diacetyl-l-lysyl-d-alanyl-d-alanine. Biochem J. 1971;123:773–787. doi: 10.1042/bj1230773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J T. Turnover and recycling of the murein sacculus in oligopeptide permease-negative strains of Escherichia coli: indirect evidence for an alternative permease system and for a monolayered sacculus. J Bacteriol. 1993;175:7–11. doi: 10.1128/jb.175.1.7-11.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J T. The murein sacculus. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 48–57. [Google Scholar]
- 25.Pratviel-Sosa F, Mengin-Lecreulx D, van Heijenoort J. Over-production, purification and properties of the uridine diphosphate-N-acetylmuramoyl-l-alanine:d-glutamate ligase from Escherichia coli. Eur J Biochem. 1991;202:1169–1176. doi: 10.1111/j.1432-1033.1991.tb16486.x. [DOI] [PubMed] [Google Scholar]
- 26.Salton M R J. The bacterial cell envelope—a historical perspective. New Compr Biochem. 1994;27:12–16. [Google Scholar]
- 27.Suganuma A. Studies on the fine structure of Staphylococcus aureus. J Electron Microsc. 1966;15:257–261. [PubMed] [Google Scholar]
- 28.Takahashi M, Yamaguchi E, Uchida T J. Thermophilic DNA ligase. Purification and properties of the enzyme from Thermus thermophilus HB8. J Biol Chem. 1984;259:10041–10047. [PubMed] [Google Scholar]
- 28a.Tanner M E, Vaganay S, van Heijenoort J, Blanot D. Phosphinate inhibitors of the d-glutamic acid-adding enzyme of peptidoglycan biosynthesis. J Org Chem. 1996;61:1756–1760. doi: 10.1021/jo951780a. [DOI] [PubMed] [Google Scholar]
- 29.Togers H J, Perkins H R, Ward J B. Microbial cell walls and membranes. London, United Kingdom: Chapman & Hall, Ltd.; 1980. [Google Scholar]
- 30.Vaganay S, Tanner M E, van Heijenoort J, Blanot D. Study of the reaction mechanism of the d-glutamic acid-adding enzyme from Escherichia coli. Microb Drug Resist. 1996;2:51–54. doi: 10.1089/mdr.1996.2.51. [DOI] [PubMed] [Google Scholar]
- 31.Wong W, Young F E, Chatterjee A N. Regulation of bacterial cell walls: turnover of cell wall in Staphylococcus aureus. J Bacteriol. 1974;120:837–843. doi: 10.1128/jb.120.2.837-843.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wedler F C, Ley B W. Homoserine dehydrogenase-I (Escherichia coli): action of monovalent ions on catalysis and substrate association-dissociation. Arch Biochem Biophys. 1993;301:416–423. doi: 10.1006/abbi.1993.1165. [DOI] [PubMed] [Google Scholar]
- 33.Xiang B, Taylor J C, Markham G D. Monovalent cation activation and kinetic mechanism of inosine 5′-monophosphate dehydrogenase. J Biol Chem. 1996;271:1435–1440. doi: 10.1074/jbc.271.3.1435. [DOI] [PubMed] [Google Scholar]
- 34.Yagasaki M, Iwata K, Ashen S, Azuma M, Osaki A. Cloning, purification, and properties of a cofactor-independent glutamate racemase from Lactobacillus brevis ATCC 8287. Biosci Biotechnol Biochem. 1995;59:610–614. doi: 10.1271/bbb.59.610. [DOI] [PubMed] [Google Scholar]
- 35.Zemell R I, Anwar R A. Pyruvate-uridine diphospho-N-acetylglucosamine transferase. Purification to homogeneity and feedback inhibition. J Biol Chem. 1975;250:3185–3192. [PubMed] [Google Scholar]