Abstract
The recombination properties of Escherichia coli strains expressing the red genes of bacteriophage λ and lacking recBCD function either by mutation or by expression of λ gam were examined. The substrates for recombination were nonreplicating λ chromosomes, introduced by infection; Red-mediated recombination was initiated by a double-strand break created by the action of a restriction endonuclease in the infected cell. In one type of experiment, two phages marked with restriction site polymorphisms were crossed. Efficient formation of recombinant DNA molecules was observed in ruvC+ recG+, ruvC recG+, ruvC+ recG, and ruvC recG hosts. In a second type of experiment, a 1-kb nonhomology was inserted between the double-strand break and the donor chromosome’s restriction site marker. In this case, recombinant formation was found to be partially dependent upon ruvC function, especially in a recG mutant background. In a third type of experiment, the recombining partners were the host cell chromosome and a 4-kb linear DNA fragment containing the cat gene, with flanking lac sequences, released from the infecting phage chromosome by restriction enzyme cleavage in the cell; the formation of chloramphenicol-resistant bacterial progeny was measured. Dependence on RuvC varied considerably among the three types of cross. However, in all cases, the frequency of Red-mediated recombination was higher in recG than in recG+. These observations favor models in which RecG tends to push invading 3′-ended strands back out of recombination intermediates.
As many as 15 to 20 proteins are thought to be directly involved in homologous recombination in Escherichia coli (for reviews, see references 19 and 23). The biochemical activities of most of these proteins have been extensively characterized. Even so, it is difficult to specify the precise roles of most of the recombination proteins. One main reason for the difficulty is that only one of the known proteins, RecA, is strictly required for recombination to take place. Certain recombination events, involving strand annealing rather than strand invasion, can take place in a recA null mutant, but these require bacteriophage-encoded recombination functions (36). The nearly complete deficiency of recA mutants, blocked at an early step in recombination (5), combined with extensive biochemical characterization of RecA protein have yielded a relatively clear description of RecA’s role in recombination: synapsis and strand invasion (for a review, see reference 32.
Several of the recombination proteins of E. coli are helicases. Recombinant formation following conjugation or generalized transduction depends upon one of the helicases, the RecBCD protein, which is also a complex nuclease. In the absence of RecBCD, efficient recombination can take place if the sbcB gene, as well as the sbcC or sbcD gene, encoding other nucleases, is mutated. In this setting, recombination depends upon several helicases, including RecG, RecQ, and RuvAB (19). Another helicase, PriA, is important for recombination in otherwise-wild-type cells (18, 33).
One recombination protein, RuvC, is a junction-specific endonuclease that has the properties expected of a Holliday junction resolvase (8). Recombination in wild-type E. coli is not highly dependent upon RuvC, though, suggesting either that there is another resolvase or that cutting a Holliday junction is only one of two or more pathways to the production of recombinants. The latter explanation appears to be the case: Lloyd (21) found that recombination in a ruvC mutant is dependent on RecG, and biochemical studies of RecG protein have uncovered helicase but not endonuclease activity (24). Rather, Whitby and coworkers (38, 39) have proposed that RecG helicase can process three-stranded junctions in such a way as to generate recombinants without endonucleolytic resolution of a Holliday junction.
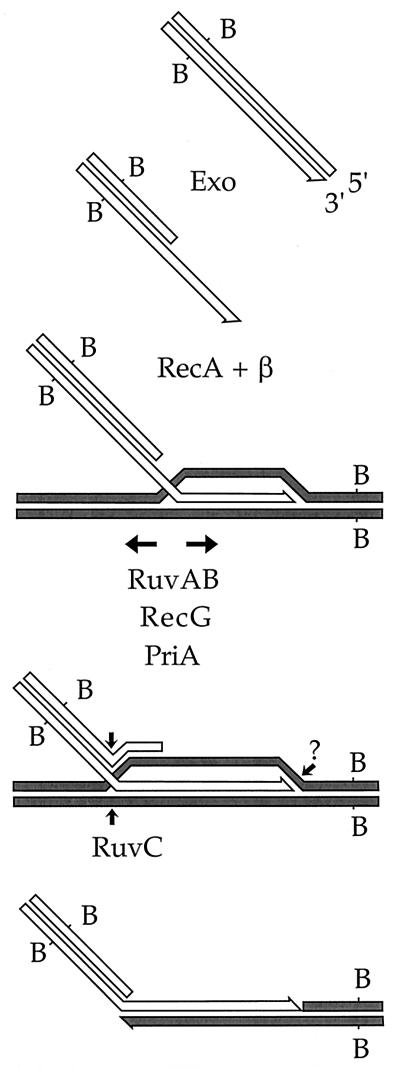
The bacteriophage λ homologous recombination system, known as Red, normally promotes highly efficient double-strand break repair and recombination transiently in λ-infected E. coli. The Red-induced “hyper-rec” state of E. coli can be made permanent if the λ recombination proteins are expressed in the cell in the absence of infecting phage (29, 31). In addition to operating at higher efficiency than other homologous recombination pathways in E. coli, Red-mediated recombination is possibly simpler in its early steps (diagrammed in Fig. 1). The λ Gam protein shuts down all the enzymatic activities of RecBCD (16, 28). Double-stranded ends initiate Red-mediated recombination (35, 37). The λ exonuclease (Redα) loads onto double-stranded ends and processively degrades the 5′-ended strand, leaving a 3′ single-stranded tail (20). RecA and λ Redβ protein then cooperate in promoting invasion of the 3′-ended strand into an unbroken homologous duplex (17, 27, 31). (Redβ can promote recombination by strand annealing in the absence of RecA protein, but only if a partner with a double-stranded end at a nonallelic site is provided [36].) In the absence of RecBCD, the only λ proteins required for efficient double-strand break repair and recombination are Redα and Redβ; host proteins carry out all other steps.
FIG. 1.
Possible mechanism of Red-mediated recombination involving strand invasion. The steps, and the roles of the recombination proteins, are described in the text.
An E. coli strain containing λ recombination proteins provides an experimental setting in which recombination between nonreplicating λ phages occurs at high frequency at the site of a double-strand break (37). Formation of recombinants between physically marked chromosomes can be monitored directly by extraction and analysis of DNA from infected cells. It depends upon (i) double-strand breaks, (ii) RecA, (iii) Red proteins, and (iv) inactivation of RecBCD, either by mutation or by λ Gam protein (31).
In this study, we examined the properties of Red-expressing E. coli bearing mutations in ruvC and recG, singly and in combination. We found that the dependence of Red-mediated recombination on RuvC was variable among the different types of crosses tested. Red-mediated recombination is more efficient in a recG mutant than in a recG+ strain, in contrast to recombination via other pathways (RecBCD, RecF, or RecE), which is reduced in recG mutant (22). However, as in wild-type E. coli, this Red-mediated recombination was, in most cases, highly dependent upon RuvC in the recG mutant. These observations distinguish between models that have been proposed for the roles of RuvC and RecG in recombination (38, 39).
MATERIALS AND METHODS
Bacteria.
Except as noted, all bacterial strains used in this study were derivatives of E. coli AB1157 (argE3 his-4 leuB6 proA2 thr-1 ara-14 galK2 lacY1 mtl-1 xyl-5 thi-1 rpsL31 tsx-33 supE44); it and JC15329 [Δ(srlR-recA)306::Tn10] were obtained from A. J. Clark. Strains CS85 (eda-51::Tn10 ruvC53) (34) and N2731 (recG258::kan) (22) were obtained from F. W. Stahl. Strain CS85 was reconstructed by transduction of AB1157 with a P1 lysate grown on strain CS85, with selection for tetracycline resistance. A tetracycline-sensitive derivative, TP438, was subsequently selected on the basis of fusaric acid resistance (26). TP440 (ruvC53 recG258::kan) was constructed by transduction of TP438 with a P1 lysate grown on N2731, with selection for kanamycin resistance. Phage recombination functions and the EcoRI restriction-modification system were introduced into these strains by simultaneous transformation with plasmids pTP223 and pMB4, as described previously (31).
Strain KM22, Δ(recC ptr recB recD)::Plac-bet-exo kan, has been described previously (29). KM32, Δ(recC ptr recB recD)::Ptac-gam-bet-exo cat, was constructed similarly.
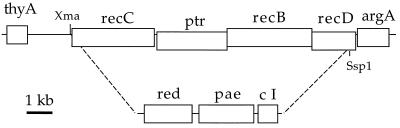
Strain TP507, in which the recC, ptr, recB, and recD genes are replaced by a cassette consisting of Ptac-gam-bet-exo, the PaeR7 restriction-modification system, and Plac-cI, was constructed by crossing KM22 with a linear DNA fragment as described previously (29). Recombinants were selected on the basis of their immunity to λ infection. The linear fragment was generated by digestion with NdeI and BamHI of plasmid pTP822, which was constructed from a series of intermediates as follows. (i) Plasmid pTP800 was made by ligation of a polycloning site, consisting of the partially complementary oligonucleotides AATTGGGCCCAGATCTCCATGGCCGCGGTCTAGAGCTC and AATTGAGCTCTAGACCGCGGCCATGGAGATCTGGGCCC, into the EcoRI site of pKM125 (29). One isolate, in which the orientation of the inserted sequence was thyA-ApaI-BglII-NcoI-SacII-XbaI-SacI-argA (see Fig. 4), was selected. (ii) The source of the Ptac-gam-bet-exo sequence was plasmid pTP234 (Ptac-gam-bet-exo operon fusion inserted into the EcoRI site of pBR322, containing λ sequences from the SalI site immediately upstream from gam to the AccI site immediately downstream from exo), which served as a template for PCR with oligonucleotides GACATAAGATCTCCGACATCATAACGGTTCTGGCAA and GACATAAGATCTTTGCGCCTACCCGGATATTATCGT. The resulting 2.1-kb fragment was digested with BglII and ligated into the BglII site of pTP800. One isolate, in which the orientation of the inserted sequence is such that the λ red genes are transcribed in the direction thyA-argA, was selected and designated pTP806. (iii) The source of the PaeR7 restriction-modification system (pae) was plasmid pAORM3.8 (12), which served as a template for PCR with oligonucleotides ACAATTCCATGGGCCGATGATTTAGTGAGGTCGTCA and ACAATTTCTAGACCTCCAAGCCCGAATGATCGAGAA. The resulting 2.5-kb fragment was digested with NcoI and XbaI and ligated between the corresponding sites in pTP806, generating plasmid pTP808. (iv) The source of Plac-cI was plasmid pKB280 (4), which served as a template for PCR with oligonucleotides AGTTGCTCTAGAACTCATTAGGCACCCCAGGCTTTA and AGTTGCTCTAGATTATCAGCTATGCGCCGACCAGAA. The resulting 0.9-kb fragment was digested with XbaI and ligated into the XbaI site of pTP808. One isolate, in which the orientation of the inserted sequence is such that the λ cI gene is transcribed in the direction thyA-argA, was selected and designated pTP810. (v) An ApaI-SacI fragment from pTP810 containing Ptac-gam-bet-exo pae Plac-cI was inserted between the ApaI and SacI sites in pKM145-6 to generate pTP822. pKM145-6 is similar in structure to pKM125 but has no antibiotic resistance determinant between the recBCD-flanking sequences (29a).
FIG. 4.
Map of the Δ(recC-recD)::red-pae-cI substitution in strain TP507 and derivatives. Sequences between the XmaI site upstream from recC and the SspI site near the C-terminus-encoding end of recD are replaced. The block of sequence designated “red” includes (from left to right) the promoter Ptac and λ genes gam, redβ, and redα. pae includes the restriction endonuclease and methylase genes of Pseudomonas aeruginosa R7, transcribed rightward. The λ cI gene is fused to the promoter PlacUV5 and is also transcribed rightward.
The chromosomal Ptac-gam-bet-exo pae Plac-cI operon of strain TP507 represses expression of lacZ from the PL promoter of an infecting λ phage at least 1,000-fold and restricts the efficiency of plaque formation by PaeR7-unmodified λ imm P22 h80 approximately 104- to 105-fold (data not shown). Although the λ recombination and repressor genes in the substitution are nominally under the control of the wild-type lacI gene of this strain, effective expression is seen even in the absence of inducer. Derivatives of TP507 bearing rec and ruv mutations were constructed by P1 transduction, with selection for antibiotic resistance. Donor strains were as listed above.
Phages.
λ wild type and λ nin5 were obtained from F. W. Stahl. λ sR1° RFLP381 sR3° sR4° sR5° and sR1° RFLP382 sR3° sR4° sR5°, referred to below as λ RFLP381 and λ RFLP382, have been described previously (31). Additional RFLP substitutions were crossed into the λ chromosome as described previously (31); recombinants were selected as described or on the basis of plaque formation on a phage P2 lysogen (Spi− phenotype) and were given the numerical designations of their plasmid parents (e.g., λ RFLP835 was made by crossing λ wild type with plasmid pTP835). Phages were propagated in a Δ(recC ptr recB recD)::Ptac-bet-exo cat derivative of W3110 strA594 lac gal, which was constructed by P1 transduction with KM32 as the donor and selection for chloramphenicol resistance.
New substitution sequences were assembled by making a variety of derivatives of plasmid pTP368 (31). This plasmid contains λ sequences from 21,226 to 23,130 and 33,498 to 34,499; sequences cloned between the λ segments, on recombination with phage, replace λ sequences normally between them. The intermediate plasmids were made as follows. For pTP812, the cat gene from pCDK3 (9) was amplified by PCR with oligonucleotides ATCATCGCTAGCATGAGAACGTTGATCGGCACGTAAG and ATCATCGGTACCGGGCCCGACCGGGTCGAATTTGCTTTCGAA. The resulting 0.9-kb fragment was digested with KpnI and NheI and ligated between the KpnI and NheI sites in pTP368. pTP813 was constructed by conversion of the XhoI site of pTP368 to an ApaI site by insertion of an oligonucleotide, TCGATCGGGCCCGA. For pTP815, the XhoI site in pTP812 was eliminated by digestion with XhoI, filling in the ends with DNA polymerase and deoxynucleoside triphosphates, and ligating. pTP816 was constructed by conversion of the AflII site in pTP815 to a PaeR7 (XhoI) site by substitution of a corresponding segment, bounded by NheI and SacI sites, from pTP817. pTP817 was constructed by conversion of the AflII site in pTP813 to a PaeR7 (XhoI) site by insertion of an oligonucleotide, TTAACGCCTCGAGGCG. pTP835 and pTP836 were constructed by conversion of the ApaI sites of pTP816 and pTP817 to BglII sites by insertion of an oligonucleotide, GCAGATCTGCGGCC. For pTP863 and pTP864, the PaeR7 (XhoI) sites of pTP835 and pTP836, respectively, were eliminated by digestion with XhoI, filling in the ends with DNA polymerase and deoxynucleoside triphosphates, and ligating with a HindIII linker (CAAGCTTG).
Derivatives of λ designed to monitor recombination events with the cell chromosome were constructed as described above. Two plasmid intermediates were involved. pTP818 was constructed by conversion of the AflII site in pTP368 to a PaeR7 (XhoI) site by insertion of an oligonucleotide, TTAACGCCTCGAGGCG. For pTP819, sequences between the two XhoI sites of pTP818 were replaced by a segment in which the cat gene is flanked by lac sequences, assembled from three PCR-generated parts: (i) lacZ sequences amplified from E. coli AB1157 DNA with primers GACGCACTCGAGGCGTTAACCGTCACGA and CTCCAGTGGCCGGCGATGATGCAGCGGCGTCAGCAGTTGTT; (ii) cat sequences amplified from DNA from an E. coli strain bearing transposon Tn9 with primers ATCATCGCCGGCCACTGGAGCACCTCAAAAACACCA and TCATCAGGGCCCGACCGGGTCGAATTTGCTTTCGAA; and (iii) lacZ and lacY sequences amplified from E. coli AB1157 DNA with primers ACCCGGTCGGGCCCTGATGACCCGTGCACCGCTGGATAACG and GACGCACTCGAGGCACACAGCGCCCAG. Parts i and ii and parts ii and iii were joined in pairs by PCR, making use of the overlaps of their primer sequences, and subcloned in intermediate plasmids via their XhoI and NgoM1 and their ApaI and XhoI ends, respectively. The two subassemblies were recombined in vitro by cutting and joining at the NcoI site in cat. This construction makes use of two sites in the E. coli lac operon that differ by 1 bp from PaeR7 sites, in such a way that when the lac::cat819 segment is cut from the phage that bears it, the ends of the resulting DNA fragment match the cell chromosome exactly.
Crosses.
Bacterial host strains were cultured and infected with parent phages at a multiplicity of seven each as described previously (31). When plasmid-bearing strains were used as hosts, retention of the plasmid pMB4 bearing the EcoRI restriction-modification system was measured as described previously (31); in cases in which less than 85% of the cells retained pMB4, the experiment was discontinued.
Phage DNA was extracted from 10-ml samples of λ RFLP-infected cell cultures by the use of lysozyme and a phenol-chloroform-isoamyl alcohol mixture as described previously (31); in later experiments, phenol was substituted for the phenol-chloroform-isoamyl alcohol mixture. DNA samples (10% of the total) were digested overnight at 37°C with 15 U of BglII and 2.5 U of RNase A and then subjected to electrophoresis in 0.7% agarose gels and transferred by capillary flow to Zeta-Probe (Bio-Rad) membranes. In experiments with λ RFLP381 and λ RFLP382, blots were probed with 32P-labeled RNA as described previously (31). In experiments with other λ RFLP phages, the prehybridization mixture was composed of 1 mM EDTA, 0.5 M sodium phosphate buffer [pH 7.2], 7% sodium dodecyl sulfate, and 200 μg of yeast RNA (baker’s yeast RNA, type III; Sigma). Following 30 min of prehybridization at 60°C, 32P-labeled cat gene probe (107 cpm) was added. Following incubation at 60°C overnight, the filters were washed and processed as described previously (31).
The cat gene probe was generated as follows. Plasmid pTP801 (made by cloning the cat gene, in the form of a DNA fragment generated by PCR with oligonucleotide primers CGGGATCCCGTGAGACGTTGATCGGCACGTAAGA and CGGGATCCCGGACCGGGTCGAATTTGCTTTCGAA, into the BamHI site of pUC19) was cut with BamHI to generate a 0.9-kb fragment, which was purified by electrophoresis in a 0.7% agarose gel. The 0.9-kb band was removed, and the DNA was extracted by centrifugation of the gel slice through an Easy Clean agarose gel DNA extraction filter (Primm Labs). Approximately 25 ng of the purified DNA was denatured by boiling it for 10 min and then placed on ice and added to a random priming reaction mixture containing 3 μl of a mixture of 0.167 mM (each) dATP, dGTP, and TTP (50 μCi of [α32P]dCTP; 3,000 Ci/mmol), 5 U of Klenow polymerase, and 2 μl of Boehringer Mannheim hexanucleotide mix (10× concentration) in a total volume of 20 μl. After incubation at 37°C for 30 min, the reaction mixture was extracted twice with phenol and then once with ether, and DNA was precipitated twice with ethanol and then redissolved in 100 μl of water and boiled for 10 min before being added to the membranes.
Autoradiographs were scanned in a Molecular Dynamics Personal Densitometer SI. Quantitation of bands was done by the use of ImageQuant software on an Apple computer.
In crosses between λ lac::cat819 and bacterial hosts, the bacteria were cultured and infected with phage as described previously (31). The multiplicity of infection was 5, except as noted. After 1 h at 37°C, the infected cells were titered on Luria-Bertani agar plates and on Luria-Bertani agar plates supplemented with chloramphenicol (20 μg/ml), isopropylthiogalactopyranoside (250 μg/ml), and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (133 μg/ml) for detection of recombinants.
RESULTS
Red-mediated recombination independent of RuvC and RecG.
Genetic and biochemical studies of recombination in E. coli, summarized in the introduction, suggest that a recG ruvC double mutant, which is highly defective in recombination, is specifically blocked at the resolution step. A prediction that follows from this idea is that unresolved recombination intermediates might accumulate in replication-blocked crosses between λ phages in a recG ruvC double mutant. To test this, we constructed single- and double-mutant strains bearing plasmids that express red genes, λ cI repressor, and the EcoRI restriction-modification system. The somewhat surprising result of phage crosses in these strains (Fig. 2) was that the frequency of recombination relative to that in the wild type was increased by mutation of recG and only slightly affected by mutation of ruvC, either alone or in combination with recG. In seeking to understand this lack of dependence on either RuvC or RecG, we considered and tested a number of possible explanations.
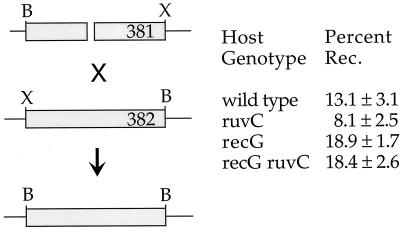
FIG. 2.
Recombination (Rec.) between nonreplicating λ chromosomes, one of which has a double-strand break. The phages, λ RFLP381 (381) and λ RFLP382 (382), bear a substitution that replaces λ genes between orf194 and ssb with unrelated sequences; the ends of the substitution are marked with different restriction sites: B, BglII, and X, XbaI. The host cells bear two plasmids: pTP223, which expresses λ genes cI, gam, redβ, and redα, and pMB4, which encodes the EcoRI restriction-modification system. The phages each have a single EcoRI restriction site in the substitution sequence; in the case of λ RFLP382, it was made uncuttable by prior passage through an EcoRI-modifying host. DNA was extracted from infected cells 60 min after infection, digested with BglII, and analyzed on Southern gels. Parental and recombinant bands from three experiments were quantitated; the means and standard errors are indicated.
One explanation for the recombination proficiency of the ruvC recG double mutant might be that the strain had acquired a suppressing mutation. The rusA mutation described by Mahdi et al. (25), in particular, activates a RuvC-like resolvase encoded by a cryptic prophage. However, tests (data not shown) of the ruvC recG strain showed that it is as recombination-deficient in conjugative crosses and transduction, and as UV sensitive, as the original strain described by Lloyd (21).
Another explanation for the high frequency of Red-mediated recombination might be that the phages themselves express a resolving function. The most likely candidate for such a function would be the rap gene in the λ nin region (15), which Mahdi et al. (25) have proposed to be a Holliday junction resolvase. This explanation is unlikely for two reasons: (i) the crosses are carried out in host cells that overexpress λ repressor from a plasmid, so few λ genes (none from the nin region) should be transcribed, and (ii) in redesigning the tester phages (see below), we have deleted the nin region and still detected efficient recombination in the ruvC recG mutant host.
The explanation we consider most likely is that the process observed in the λ crosses does not necessarily involve simple break-join recombination but rather can proceed via branch migration and heteroduplex formation, as diagrammed in Fig. 3a. A control experiment showed that mismatch repair would necessarily be involved in this alternative pathway. We constructed mismatched heteroduplexes with the structure that would be produced in an event of the type pictured above and found that they could not be digested with the restriction enzyme (BglII) used to characterize phage chromosomes extracted from the infected cells (data not shown); therefore, branch migration alone cannot account for the apparent recombination. We then attempted to test the role of mismatch repair genetically. A ruvC recG mutS triple mutant was constructed and transformed with Red, cI, and EcoRI-expressing plasmids. However, the resulting strain grew poorly, particularly in liquid culture, and the phage crosses could not be carried out.
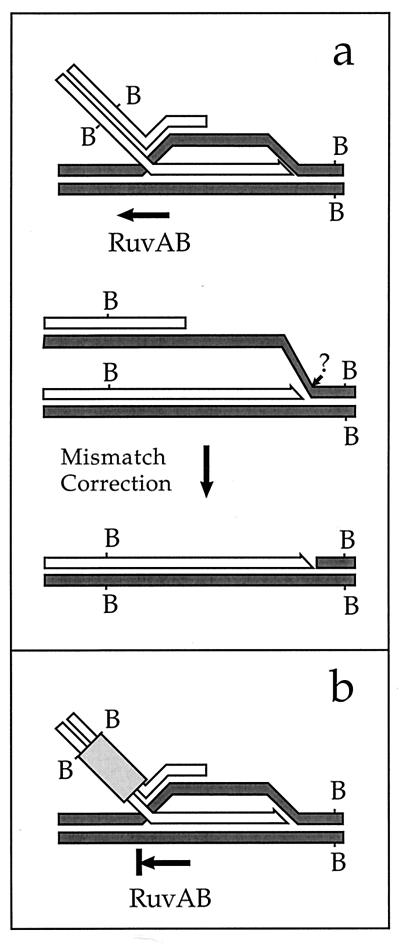
FIG. 3.
(a) Possible mechanism of recombinant formation via the Red pathway, independent of RuvC and RecG. The first steps (not shown) would be the same as in Fig. 1. Branch migration, possibly directionally biased due to the absence of RecG, leads to heteroduplex formation at the site of marker B. Mismatch repair then converts the heteroduplex to a homoduplex, bypassing the need for endonucleolytic processing of a Holliday junction. On the right side, endonucleolytic cleavage and trimming of the displaced strand or, possibly, extension of the invading 3′ end would produce a structure that would be counted as a recombinant following extraction, digestion with BglII, and analysis on a Southern gel. (b) A heterologous sequence inserted between the invading end and the marker B is predicted to block the pathway shown in panel a, making recombination more dependent upon RuvC and/or RecG.
The alternative, branch migration-heteroduplex formation-mismatch repair pathway to recombinant formation could in principle be blocked by insertion of a heterologous sequence between the double-strand break and the marker used to score recombination (a restriction site, in this case), as pictured in Fig. 3b. Recombination might then be channeled through a resolvase-requiring pathway.
Redesign of phages and hosts for detection of resolvase-dependent recombination events.
For reasons described above, we constructed new variants of the λ RFLP phages (see Fig. 5 [top]). The λ sequences replaced by the new RFLP substitutions are the same as in the previous versions, but three main changes were made. (i) In the new phages, the leftmost 1 kb of sequence in the substitution is either the same as before or has been replaced by a heterologous sequence, the cat gene of transposon Tn9. (ii) A unique PaeR7 restriction site has been introduced near the middle of the 5-kb substitution sequence (near the EcoRI site of the previous versions). This change was introduced because other studies had demonstrated the greater efficiency of the PaeR7 restriction enzyme in promoting recombination events in vivo, relative to the EcoRI system employed previously (36). PaeR7-uncuttable partners were constructed by insertion of HindIII linkers into the PaeR7 sites. (iii) The nin region was eliminated by crossing in the deletion nin5.
FIG. 5.
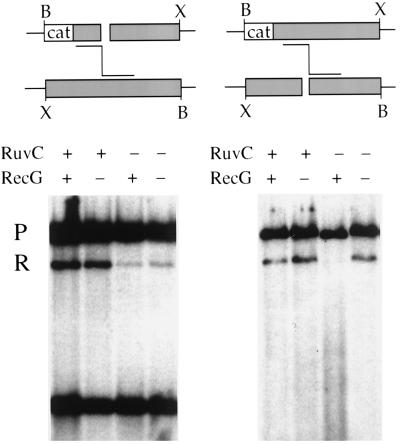
(Top) Recombination between λ RFLP phages bearing a heterologous sequence. As indicated, production of a recombinant follows different potential pathways, depending upon which phage is cut. On the left, the invading partner has homology (1 kb) at the end but has a heterologous sequence (the cat gene; 1 kb) between the end and the restriction site marker B (BglII). Formation of recombinants by the mechanism diagrammed in Fig. 3 is blocked. On the right, the invading strand encounters no substantial heterology, only the restriction site polymorphism. Formation of recombinants by the mechanism diagrammed in Fig. 3 is permitted. The cross on the left involves phages λ RFLP835 nin5 (above) and λ RFLP864 nin5 (below); that on the right involves λ RFLP863 nin5 (above) and λ RFLP836 nin5 (below). (Bottom) Autoradiographs of Southern gels of BglII-digested DNA extracted from host cells 60 min after infection and probed with cat sequences. The positions of parental (P) and recombinant (R) bands are indicated. The lowest band results from cutting with PaeR7 in vivo and with BglII in vitro. The bacterial strains were all Δ(recC-recD)::red-pae-cI. +, present; −, absent.
The extraordinary recombination proficiency of E. coli strains bearing λ red genes in their chromosomes in place of recBCD (29) led us to place the other functions needed for λ RFLP crosses—cI repressor and the PaeR7 restriction-modification system—in the chromosome as well (Fig. 4). This approach eliminates the need for plasmids in the host strains, a significant advantage. Some bacterial strains lacking recombination functions grow relatively poorly when they bear plasmids. The plasmids’ replication, moreover, is affected by mutations of the host recombination genes, as well as expression of phage recombination functions. For example, in bacteria in which RecBCD is inactivated, much plasmid DNA is found in the form of linear multimers, apparently as the result of rolling-circle replication (7).
As detailed in Materials and Methods, we constructed a cassette bearing the λ genes gam, bet, and exo, the PaeR7 restriction-modification system, and λ cI repressor and recombined it into the chromosome of E. coli AB1157 in place of recBCD (Fig. 4). The resulting strain, TP507, is recombination proficient in general and hyper-rec with respect to recombination with short linear DNA fragments, as described previously (29). Mutant alleles of recG and ruvC were introduced into the TP507 background by P1 transduction.
Resolvase-dependent Red-mediated recombination.
Crosses between the new λ RFLP phages were predicted to have different outcomes, depending upon which partner was cut, as diagrammed in Fig. 5 (top). Cutting the non-cat-bearing phage should result in the production of recombinants independent of resolution functions, as seen previously. However, according to the scheme outlined in Fig. 3, a double-strand break in the cat-bearing phage should lead to the production of a recombination intermediate that can be processed into a recombinant only by the action of a resolvase.
The involvement of resolution functions in recombination in the new system was tested in the experiment shown in Fig. 5 (bottom). In this experiment, DNA was extracted from cells infected with RFLP phages, digested with BglII endonuclease, run in a Southern gel, and hybridized with a cat-specific probe. In crosses in which the cat-bearing phage was cut in vivo, three bands were seen: parental, recombinant, and a 1,990-bp fragment generated by PaeR7 cutting in vivo and BglII cutting in vitro. The last band was not present in DNA extracted from crosses in which the non-cat-bearing phage was cut. The three bands correspond to ones seen previously (31).
The fraction of label in recombinant bands in Fig. 5 (bottom) and similar autoradiograms was quantitated; the results are shown in Table 1. As seen previously, the two phages recombine efficiently in the red-expressing host that lacks RecBCD but is otherwise wild type for recombination functions. In contrast to previous results, however, recombination was reduced more than 15-fold by mutation of ruvC. The RuvC dependence of recombination was seen regardless of which parent was cut. Recombination was increased by mutation of recG; this effect, too, was independent of which parent was cut. In a recG mutant background, however, loss of ruvC function resulted in a 16-fold decrease in recombination when the cat-bearing chromosome was cut but only a slight (possibly insignificant) decrease when the non-cat-bearing chromosome was cut.
TABLE 1.
Effects of ruvC and recG mutations on λ Red-mediated recombination between phage chromosomes
| Bacterial strainb | Percent recombinanta
|
|
|---|---|---|
| cat-bearing phage cut | Non-cat-bearing phage cut | |
| Wild type | 8.4 ± 5.4 | 13 ± 4.0 |
| ruvC | 0.6 ± 0.4 | 0.3 ± 0.3 |
| recG | 21 ± 9.3 | 28 ± 1.3 |
| ruvC recG | 1.3 ± 0.2 | 23 ± 2.6 |
The percent of total cat gene sequences found in recombinant DNA molecules in the crosses shown in Fig. 5 and two similar experiments were measured as described in the text. The means and standard errors are shown.
All strains bear the Δ(recC-recD)::red-pae-cI substitution.
Biological-recombinant formation.
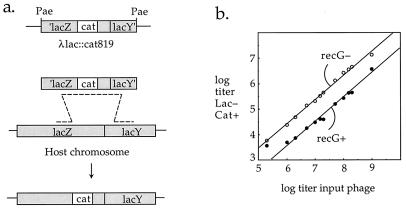
We sought to address the question of whether the formation of recombinant DNA molecules in λ RFLP crosses correlates with the production of biological recombinants. We constructed a variant of the RFLP phages, λ lac::cat819, which, upon infecting an immune host with the PaeR7 restriction-modification system, is cut by the PaeR7 nuclease, releasing a 4-kb fragment with lacZ and lacY sequences at its ends and the cat gene in the middle (Fig. 6a). Red-mediated recombination results in replacement of the chromosomal lacZ gene with a mutant allele that is defective for lacZ function but confers chloramphenicol resistance on the recombinant. According to the rationale diagrammed in Fig. 3, production of chloramphenicol-resistant recombinants would be expected to require the same functions as production of recombinant DNA molecules in λ RFLP crosses in which the cat-bearing chromosome is cut (Fig. 5, top left).
FIG. 6.
(a) Recombination between λ lac::cat819 nin5 and the bacterial chromosome. Phage repressor in the infected cell prevents transcription of phage genes. PaeR7 restriction endonuclease releases a DNA fragment that recombines with the chromosome, generating a chloramphenicol-resistant, Lac− recombinant. (b) Production of recombinants following infection with λ lac::cat819 nin5. Log-phase cells (approximately 108/ml) were infected at various multiplicities and, after 1 h of further aeration, were plated for total and recombinant (cat+ lac) titers. Open circles, Δ(recC-recD)::red-pae-cI recG258; solid circles, Δ(recC-recD)::red-pae-cI. Total cell counts increased approximately fourfold during the experiment.
Infection of a ΔrecBCD::red-pae-cI recG host with λ lac::cat819 nin5 at low multiplicity produced Lac− chloramphenicol-resistant recombinants at the rate of approximately 0.02 per infecting phage. This frequency was independent of multiplicity over a range of 0.001 to 1, indicating that infection with a single phage is sufficient to produce a recombinant (Fig. 6b). When this strain was infected with λ lac::cat819 nin5 at high multiplicity, as many as 18% of the infected cells gave rise to chloramphenicol-resistant recombinants (data not shown).
The abilities of (ΔrecBCD::red-pae-cI) ruvC, recG, and recG ruvC mutants to recombine with λ lac::cat819 and λ lac::cat819 nin5 are compared in Table 2. The two phages gave comparable frequencies of recombination in all hosts, indicating that λ genes in the nin region did not significantly affect recombinant formation. Loss of ruvC function reduced the recombination frequency two- to threefold, while loss of recG function increased it five- to eightfold. The effect of the ruvC mutation was much greater in a recG mutant background, resulting in a 20- to 40-fold decrease in recombination. A recA mutation, by comparison, reduced recombination 90-fold.
TABLE 2.
Effects of ruvC and recG mutations on gene replacement frequency
| Bacterial straina | Relative frequency of Lac− Cmr recombinantsb
|
|
|---|---|---|
| λ lac::cat819 | λ lac::cat819 nin5 | |
| Wild type | 1.0 | 1.0 |
| ruvC | 0.32 ± 0.11 | 0.49 ± 0.07 |
| recG | 4.8 ± 1.7 | 7.8 ± 4.1 |
| ruvC recG | 0.20 ± 0.12 | 0.20 ± 0.13 |
| recA | NT | 0.011 ± 0.004 |
All strains bear the Δ(recC-recD)::red-pae-cI substitution.
Wild-type values ranged from 0.19 to 0.57%. The means and standard errors from two to four experiments are given. NT, not tested.
An interesting and as yet unexplained aspect of recombination between λ lac::cat819 and E. coli strains bearing the ΔrecBCG::red-pae-cI substitution is that up to one-third of the chloramphenicol-resistant recombinants in some crosses were LacZ+. Production of these LacZ+ recombinants appears to depend on the same functions as the LacZ− recombinants—in particular, recA and, in a recG background, ruvC (data not shown). Their retention of lacZ function implies that the recombination event that generated them was not simple gene replacement. The LacZ+ recombinants may contain duplications of the lac region. This idea is favored by the observation that many of them gave rise to spontaneous LacZ− colonies on restreaking (unpublished observations).
DISCUSSION
The results of experiments examining Red-mediated recombination between linear and circular partners suggest two main interpretations concerning the roles of RecG and RuvC. (i) the main pathway to recombinant formation involves RuvC and (ii) RecG impedes this main recombination pathway. However, RecG may also stimulate recombination via a RuvC-independent pathway.
In the presence of RecG function, the degree of dependence of Red-mediated recombination on RuvC varied substantially among the three types of crosses described in the preceding section. The dependence was greatest in crosses between nonreplicating phage chromosomes in non-plasmid-bearing cells (Fig. 5). In the other cases, recombination appeared to be nearly independent of RuvC.
The contribution of RecG to Red-mediated recombination was less variable among experiments: in all cases, elimination of RecG function increased the frequency of recombination. In both types of crosses that were carried out in non-plasmid-bearing cells—between nonreplicating λ chromosomes and between short linear DNA fragments and the bacterial chromosome—the resulting higher-frequency recombination was also more dependent on RuvC (except when recombinant formation could in theory proceed via branch migration, heteroduplex formation, and mismatch repair, as pictured in Fig. 3). The simplest interpretation of these observations is that RecG opposes the main pathway of Red-mediated recombination, which is RuvC dependent. The data in Table 2 suggest that RecG may also promote an alternative, RuvC-independent recombination pathway: recombination frequencies in the ruvC mutant were slightly higher than in the ruvC recG double mutant.
An alternative interpretation of the effects on Red-mediated recombination of the recG258 allele is that they do not result from inactivation of recG but rather from a function expressed by the mini-Tn10 used to generate the allele (22). This interpretation is ruled out by the observation that a simple deletion allele of recG exhibits the same stimulation of Red-mediated recombination as recG258 (unpublished observations).
Our description of the roles of RecG and RuvC is similar to a model proposed by Whitby and coworkers (39), in which RecG actually aborted genetic exchanges resulting from RecA-mediated strand invasion but then allowed RecBCD to catalyze exchanges at the ends of the incoming DNA by an unspecified mechanism. Although these researchers found biochemical evidence for RecG’s ability to act in this way, the fact that a recG mutant is recombination deficient relative to wild type apparently led them to favor other interpretations of RecG’s activity in recombination (1, 38).
The generation of late-arising revertants of lacZ mutants under conditions of selection is another cellular activity that apparently is impeded by RecG (10, 13). This activity, sometimes called “adaptive mutability,” is dependent upon recombination functions (6, 13). Its dependence upon recombination functions may be a consequence of the involvement of gene amplification in the process (3). Harris et al. (13) proposed a model for recombination associated with adaptive mutation, in which RecG aborts 3′-end invasion and promotes 5′-end invasion.
The initial steps of Red-mediated recombination are perhaps less complex than those of RecBCD-mediated recombination or of RecBCD-dependent adaptive mutation. The λ exonuclease specifically and processively degrades the 5′-ended strand of double-stranded DNA (20), leaving exclusively 3′-ended single strands for synapsis and strand invasion. Production of these 3′-ended single-strand tails in λ-infected cells has been observed directly (14). The observation that RecG inhibits Red-mediated recombination that is constrained to proceed via strand invasion favors models in which RecG tends to push out invading 3′ ends.
The model diagrammed in Fig. 1 and 3 accounts reasonably well for recombination between linear and circular partners in a red-expressing cell lacking recBCD and recG functions: double-stranded ends are channeled nearly exclusively through a pathway that involves the creation of 3′-ended single-strand tails, which invade an unbroken homologous duplex. If and only if branch migration is impeded by a significant nonhomology, recombinant formation is dependent upon nucleolytic resolution of the resulting Holliday junction by RuvC. In the absence of RuvC, presumably, a recombination intermediate something like the structure diagrammed in Fig. 3b accumulates. We have not observed such an intermediate, but the methods employed in the extraction and restriction enzyme digestion of DNA from λ-infected cells might be expected to favor its dissociation by spontaneous branch migration.
The model diagrammed in Fig. 1 and 3 does not account so well, by itself, for what happens in a recG+ cell. A more complete model would account for three questions raised by the data in Fig. 2 and Tables 1 and 2. (i) Why was recombination between nonreplicating λ chromosomes, with no substantial nonhomologies, dependent upon RuvC in one set of crosses (Table 1, non-cat-bearing phage cut) and independent in another (Fig. 2)? One possible explanation is that the interacting DNA sequences in question were not identical in the two experiments and branch migration could proceed all the way past the restriction site polymorphism in the face of opposition by RecG in one case but not in the other. (ii) The crosses diagrammed in Fig. 5 (left side) and Fig. 6 both involve recombination of a linear cat-bearing chromosome with a circular chromosome. Why was recombinant formation dependent upon RuvC in the former case and independent in the latter? A key difference between the two crosses is that replication of both partners is blocked in the λ cross (Fig. 5) whereas replication of only the linear partner is blocked in the linear-by-host chromosome cross (Fig. 6). The passage of a replication fork may potentiate alternative pathways for the resolution of recombination intermediates. (iii) How does RecG promote, rather than impede, RecBCD-mediated recombination? One possibility is that RecBCD-mediated recombination proceeds primarily via 5′-ended strand invasion, but extensive studies of the activities of RecBCD, both in vitro (2) and in vivo (11), favor the view that RecBCD generates invading 3′-ended strands. Together, these observations are consistent with the idea that RecBCD may function in the processing or resolution of recombination intermediates that it participates in generating (5, 30). Perhaps the normal role of RecG is to facilitate this function of RecBCD.
ACKNOWLEDGMENTS
We thank Rik Myers and Susan Rosenberg for helpful discussions and Michael Volkert for critical reading of the manuscript.
This work was supported by Public Health Service grant GM51609 from the National Institutes of Health.
REFERENCES
- 1.Al-Deib A A, Mahdi A A, Lloyd R G. Modulation of recombination and DNA repair by the RecG and PriA helicases of Escherichia coli K-12. J Bacteriol. 1996;178:6782–6789. doi: 10.1128/jb.178.23.6782-6789.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson D G, Kowalczykowski S C. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- 3.Anderson D I, Slechta E S, Roth J R. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science. 1998;282:1133–1135. doi: 10.1126/science.282.5391.1133. [DOI] [PubMed] [Google Scholar]
- 4.Backman K, Ptashne M. Maximizing gene expression on a plasmid using recombination in vitro. Cell. 1978;13:65–71. doi: 10.1016/0092-8674(78)90138-1. [DOI] [PubMed] [Google Scholar]
- 5.Birge E A, Low K B. Detection of transcribable recombination products following conjugation in Rec+, RecB−, and RecC− strains of Escherichia coli K12. J Mol Biol. 1974;83:447–457. doi: 10.1016/0022-2836(74)90506-3. [DOI] [PubMed] [Google Scholar]
- 6.Cairns J, Foster P L. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen A, Clark A J. Synthesis of linear plasmid multimers in Escherichia coli K-12. J Bacteriol. 1986;167:327–335. doi: 10.1128/jb.167.1.327-335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunderdale H J, Benson F E, Parsons C A, Sharples G J, Lloyd R G, West S C. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature. 1991;354:506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- 9.Dykstra C C, Prasher D, Kushner S R. Physical and biochemical analysis of the cloned recB and recC genes of Escherichia coli K-12. J Bacteriol. 1984;157:21–27. doi: 10.1128/jb.157.1.21-27.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster P L, Trimarchi J M, Maurer R A. Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation in Escherichia coli. Genetics. 1996;142:25–37. doi: 10.1093/genetics/142.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman-Ohana R, Cohen A. Heteroduplex joint formation in Escherichia coli recombination is initiated by pairing of a 3′-ending strand. Proc Natl Acad Sci USA. 1998;95:6909–6914. doi: 10.1073/pnas.95.12.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gingeras T R, Brooks J E. Cloned restriction/modification system from Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1983;80:402–406. doi: 10.1073/pnas.80.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris R S, Ross K J, Rosenberg S M. Opposing roles of the Holliday junction processing systems of Escherichia coli in recombination-dependent adaptive mutation. Genetics. 1996;142:681–691. doi: 10.1093/genetics/142.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill S A, Stahl M M, Stahl F W. Single-strand DNA intermediates in phage λ’s Red recombination pathway. Proc Natl Acad Sci USA. 1997;94:2951–2956. doi: 10.1073/pnas.94.7.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollifield W, Kaplan E, Huang H. Efficient RecABC-dependent, homologous recombination between coliphage lambda and plasmids requires a phage ninR region gene. Mol Gen Genet. 1987;210:248–255. doi: 10.1007/BF00325690. [DOI] [PubMed] [Google Scholar]
- 16.Karu A E, Sakaki Y, Echols H, Linn S. The γ protein specified by bacteriophage λ. J Biol Chem. 1975;250:7377–7387. [PubMed] [Google Scholar]
- 17.Kmiec E, Holloman W K. β protein of bacteriophage λ promotes renaturation of DNA. J Biol Chem. 1981;256:12636–12639. [PubMed] [Google Scholar]
- 18.Kogoma T, Cadwell G W, Barnard K G, Asai T. The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J Bacteriol. 1996;178:1258–1264. doi: 10.1128/jb.178.5.1258-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Little J W. An exonuclease induced by bacteriophage λ. II. Nature of the enzymic reaction. J Biol Chem. 1967;242:679–686. [PubMed] [Google Scholar]
- 21.Lloyd R G. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J Bacteriol. 1991;173:5414–5418. doi: 10.1128/jb.173.17.5414-5418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lloyd R G, Buckman C. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J Bacteriol. 1991;173:1004–1011. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lloyd R G, Low K B. Homologous recombination. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 2236–2255. [Google Scholar]
- 24.Lloyd R G, Sharples G J. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 1993;12:17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahdi A A, Sharples G J, Mandal T N, Lloyd R G. Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J Mol Biol. 1996;257:561–573. doi: 10.1006/jmbi.1996.0185. [DOI] [PubMed] [Google Scholar]
- 26.Maloy S R, Nunn W D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981;145:1110–1112. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muniyappa K, Radding C M. The homologous recombination system of phage λ: pairing activities of β protein. J Biol Chem. 1986;261:7472–7478. [PubMed] [Google Scholar]
- 28.Murphy K C. λ gam protein inhibits the helicase and χ-stimulated recombination activities of Escherichia coli RecBCD enzyme. J Bacteriol. 1991;173:5808–5821. doi: 10.1128/jb.173.18.5808-5821.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy K C. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Murphy, K. C. Unpublished data.
- 30.Myers R S, Stahl F W. Chi and the RecBCD enzyme of Escherichia coli. Annu Rev Genet. 1994;28:49–70. doi: 10.1146/annurev.ge.28.120194.000405. [DOI] [PubMed] [Google Scholar]
- 31.Poteete A R, Fenton A C. Efficient double-strand break-stimulated recombination promoted by the general recombination systems of phages λ and P22. Genetics. 1993;134:1013–1021. doi: 10.1093/genetics/134.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roca A I, Cox M M. RecA protein: structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 33.Sandler S J, Samra H S, Clark A J. Differential suppression of priA2::kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA, and dnaC. Genetics. 1996;143:5–13. doi: 10.1093/genetics/143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharples G J, Benson F E, Illing G T, Lloyd R G. Molecular and functional analysis of the ruv region of Escherichia coli K-12 reveals three genes involved in DNA repair and recombination. Mol Gen Genet. 1990;221:219–226. doi: 10.1007/BF00261724. [DOI] [PubMed] [Google Scholar]
- 35.Stahl F W, Kobayashi I, Stahl M M. In phage λ, cos is a recombinator in the Red pathway. J Mol Biol. 1985;181:199–209. doi: 10.1016/0022-2836(85)90085-3. [DOI] [PubMed] [Google Scholar]
- 36.Stahl M M, Thomason L, Poteete A R, Tarkowski T, Kuzminov A, Stahl F W. Annealing vs. invasion in phage λ recombination. Genetics. 1997;147:961–977. doi: 10.1093/genetics/147.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thaler D S, Stahl M M, Stahl F W. Double-chain-cut sites are recombination hotspots in the Red pathway of phage λ. J Mol Biol. 1987;195:75–87. doi: 10.1016/0022-2836(87)90328-7. [DOI] [PubMed] [Google Scholar]
- 38.Whitby M C, Lloyd R G. Branch migration of three-strand recombination intermediates by RecG, a possible pathway for securing exchanges initiated by 3′-tailed duplex DNA. EMBO J. 1995;14:3302–3310. doi: 10.1002/j.1460-2075.1995.tb07337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitby M C, Ryder L, Lloyd R G. Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell. 1993;75:341–350. doi: 10.1016/0092-8674(93)80075-p. [DOI] [PubMed] [Google Scholar]