Abstract
γ-Hexachlorocyclohexane (γ-HCH) is one of several highly chlorinated insecticides that cause serious environmental problems. The cellular proteins of a γ-HCH-degrading bacterium, Sphingomonas paucimobilis UT26, were fractionated into periplasmic, cytosolic, and membrane fractions after osmotic shock. Most of two different types of dehalogenase, LinA (γ-hexachlorocyclohexane dehydrochlorinase) and LinB (1,3,4,6-tetrachloro-1,4-cyclohexadiene halidohydrolase), that are involved in the early steps of γ-HCH degradation in UT26 was detected in the periplasmic fraction and had not undertaken molecular processing. Furthermore, immunoelectron microscopy clearly showed that LinA and LinB are periplasmic proteins. LinA and LinB both lack a typical signal sequence for export, so they may be secreted into the periplasmic space via a hitherto unknown mechanism.
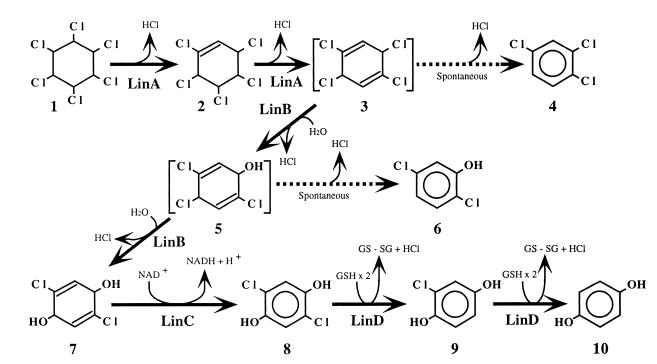
γ-Hexachlorocyclohexane (γ-HCH; also called γ-BHC and lindane) is a highly halogenated organic insecticide that has been used worldwide. Due to its toxicity and long persistence in soil, most countries have prohibited the use of γ-HCH. However, many contaminated sites remain throughout the world. Moreover, some countries are presently using γ-HCH, mainly for economic reasons, so new sites are continually being contaminated (3, 14). Sphingomonas (formerly Pseudomonas) paucimobilis UT26 is a unique microorganism that utilizes γ-HCH as its sole source of carbon and energy under aerobic conditions (11). We have cloned four genes (linA, linB, linC, and linD), the products of which are involved sequentially in the degradation of γ-HCH in UT26 (12, 15, 17, 18) (Fig. 1). Three of them encode different types of dehalogenases: dehydrochlorinase, halidohydrolase, and reductive dehalogenase. Among them, linA encodes an enzyme that catalyzes a unique dehydrochlorination reaction whose mechanism is still unknown. Dehalogenation is a key step in the degradation of halogenated compounds, so many enzymes that catalyze dehalogenation have been reported (7), and yet there is little or no information available about the distribution of dehalogenase in gram-negative bacteria.
FIG. 1.
Proposed degradation pathway of γ-HCH in S. paucimobilis UT26. Compounds: 1, γ-HCH (also called γ-BHC and lindane); 2, γ-pentachlorocyclohexene; 3, 1,3,4,6-tetrachloro-1,4-cyclohexadiene; 4, 1,2,4-trichlorobenzene; 5, 2,4,5-trichloro-2,5-cyclohexadiene-1-ol; 6, 2,5-dichlorophenol; 7, 2,5-dichloro-2,5-cyclohexadiene-1,4-diol; 8, 2,5-dichlorohydroquinone; 9, chlorohydroquinone; 10, hydroquinone.
Here we report that two different types of dehalogenase, LinA (γ-hexachlorocyclohexane dehydrochlorinase) (12, 16) and LinB (1,3,4,6-tetrachloro-1,4-cyclohexadiene halidohydrolase) (17, 20), that act on γ-HCH or its metabolite in S. paucimobilis are localized in the periplasmic space. As far as we know, this is the first report on the subcellular localization of dehalogenases that are involved in the degradation of halogenated xenobiotics in gram-negative bacteria.
MATERIALS AND METHODS
Subcellular fractionation of S. paucimobilis.
UT26 was cultured in 200 ml of 1/3 Luria broth (3.3 g of Bacto tryptone, 1.7 g of yeast extract, and 5 g of sodium chloride per liter). Cells were harvested in the exponential growth phase. The periplasmic fraction was prepared by osmotic shock (21). After osmotic shock, cells were disrupted by sonication (Sonifier 250; Branson, Danbury, Conn.). After centrifugation at 12,000 × g for 10 min, the pellet was discarded as debris. The supernatant was separated by centrifugation at 100,000 × g for 1 h into cytoplasmic and membrane fractions. The protein concentration was determined by using the protein assay kit (Bio-Rad Laboratories, Richmond, Calif.), with bovine serum albumin as a standard.
Enzyme assays.
Activities of glyceraldehyde-3-phosphate dehydrogenase, pyruvate dehydrogenase, and phosphatase were measured as described by Duggleby and Dennis (6), Friedemann and Haugen (8) and Hayakawa et al. (9), and Dassa et al. (5), respectively. The activities of LinA and LinB were measured spectrophotometrically at 460 nm with mercuric thiocyanate and ferric ammonium sulfate by the method of Iwasaki et al. (13) as described in previous studies (16, 20). One unit of enzyme activity of LinA and LinB was defined as the amount of enzyme required for the release of 1 μmol of chloride ion per min.
Western blot analysis.
Antibodies were raised against LinA and LinB by using purified enzymes produced in Escherichia coli (16, 20). Rabbits were injected subcutaneously four times at weekly intervals with 0.5 mg of purified LinA or LinB. The injections were given with 50% (vol/vol) Freund’s complete adjuvant. Sera were collected after four injections. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to the nitrocellulose membrane Hybond C (Amersham). The ECL (enhanced chemiluminescence) Western blotting system (Amersham) was used for detection.
Immunogold-labeling electron microscopy.
Cells were fixed in 2.5% glutaraldehyde in phosphate-buffered saline (PBS) for 3 h, dehydrated in graded ethanols and propylene oxide, and embedded in epoxy resins. Ultrathin sections were prepared by an ultramicrotome (LKB) and deposited on nickel grids. The grid was incubated in PBS containing 5% bovine serum albumin for 30 min and then in PBS containing the purified anti-LinA or anti-LinB immunoglobulin G for 30 min at room temperature. After pretreatment with 1% H2O2 for 3 min, the grid was incubated in PBS containing 3% goat anti-rabbit immunoglobulin G conjugated to 10-nm-diameter gold particles (Amersham) for 30 min at room temperature. The sections were stained with 4% uranyl acetate and 0.4% lead citrate in 0.1 M NaOH. The preparation was examined with electron microscopes (JEM-1200EX and HU-800).
Determination of N-terminal amino acid sequences.
The periplasmic proteins were fractionated by SDS-PAGE, transferred to a polyvinylidene difluoride-type membrane (Immobilon-PSQ; Millipore Corp.), and stained with Coomassie blue. The bands corresponding to LinA and LinB were excised and directly sequenced by automated Edman degradation with a model 477A protein sequencer equipped with a model 120A phenylthiohydantoin analyzer (Applied Biosystems).
RESULTS
Subcellular fractionation of S. paucimobilis UT26.
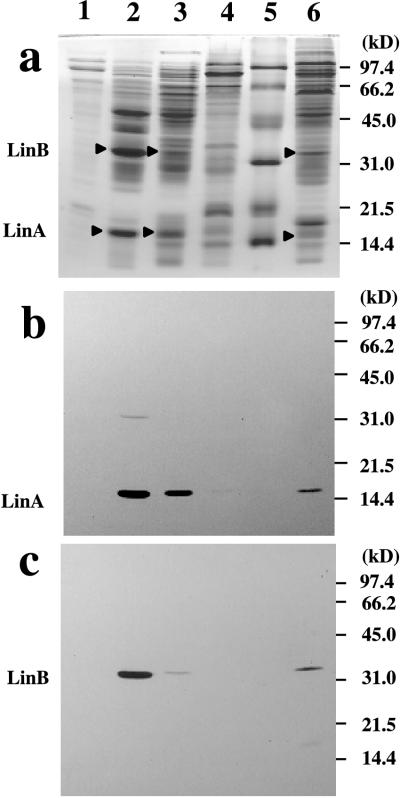
UT26, a nalidixic acid-resistant mutant of the first S. paucimobilis strain isolated, was cultured without an inducer, because the genes involved in the early steps of γ-HCH degradation are constitutively expressed in UT26 (19). Cells were harvested in the exponential growth phase, and the supernatant was saved and used as the extracellular fraction. The cellular proteins of UT26 were fractionated into periplasmic, cytosolic, and membrane fractions after osmotic shock. SDS-PAGE analysis showed a characteristic pattern of bands for each fraction (Fig. 2a). Table 1 shows a result typical of several independent experiments, because all of them showed the same tendency. The distributions of the cellular protein content across the periplasmic, cytoplasmic, and membrane fractions were 15.2, 67.8, and 17.0%, respectively (Table 1). Glyceraldehyde-3-phosphate dehydrogenase and pyruvate dehydrogenase, which are cytoplasmic enzymes, were mainly detected in the cytoplasmic fraction (98 and 82%, respectively) (Table 1), whereas only 34% of the phosphatase activity, which we expected to be periplasmic, was detected in the periplasmic fraction. Since 66% of the phosphatase activity was observed in the cytosolic fraction, the release of the periplasmic proteins was probably not complete. However, it is likely that little cytoplasmic proteins leaked into the periplasmic fraction during osmotic shock fractionation. These results support the validity of our subcellular fractionation technique.
FIG. 2.
SDS-PAGE of the fractionated cellular proteins in UT26. Cellular proteins in UT26 were fractionated into the periplasmic, cytoplasmic, and membrane fractions by using the osmotic shock method (21). Lanes: 1, extracellular fraction; 2, periplasmic fraction; 3, cytoplasmic fraction; 4, membrane fraction; 5, molecular mass markers (Bio-Rad); 6, total proteins in UT26. (a) Staining with Coomassie blue. (b) Western blot analysis for LinA protein. (c) Western blot analysis for LinB protein.
TABLE 1.
Cellular location of enzymic activities in S. paucimobilis UT26
| Cell fraction | Amt (%) of activity
|
|||||
|---|---|---|---|---|---|---|
| Protein (mg) | Phosphatase (pH 6.0) (103 U) | Glyceraldehyde-3-phosphate dehydrogenase (104 U) | Pyruvate dehydrogenase (104 U) | LinA (dehydrochlorinase) (U) | LinB (halidohydrolase) (U) | |
| Periplasmic | 9.1 (15.2) | 1.2 (34) | 0.040 (1.3) | 0.24 (15) | 3.4 (71) | 0.59 (83) |
| Cytoplasmic | 40.6 (67.8) | 2.3 (66) | 3.1 (98) | 1.3 (82) | 1.4 (29) | 0.12 (17) |
| Membrane | 10.2 (17) | NDa | 0.022 (0.7) | 0.043 (3) | ND | ND |
ND, not detectable.
Distribution of LinA and LinB in the subcellular fractions.
The LinA and LinB activities of each fraction were measured as described previously (16, 20); most of the LinA and LinB activities (71 and 83%, respectively) were detected in the periplasmic fraction. This result suggests that LinA and LinB are periplasmic proteins.
Next, antibodies were raised against LinA and LinB by using purified enzymes produced in E. coli (16, 20). Antibodies that specifically reacted with LinA and LinB were characterized (Fig. 2b and c, lanes 6). Western blot analysis of each cell fraction was performed. The amounts of proteins loaded were 4, 16, and 4 μg for the periplasmic, cytoplasmic, and membrane fractions, respectively. This ratio reflected approximately the ratio of total protein in each fraction (Table 1). It was revealed that most of the LinA and LinB protein was present in the periplasmic fraction (Fig. 2b and c, lanes 2, 3, and 4). LinA and LinB were not detected in the extracellular fraction (Fig. 2b and c, lanes 1), indicating that LinA and LinB are not secreted extracellularly.
Localization of LinA and LinB in the intact cell of S. paucimobilis UT26.
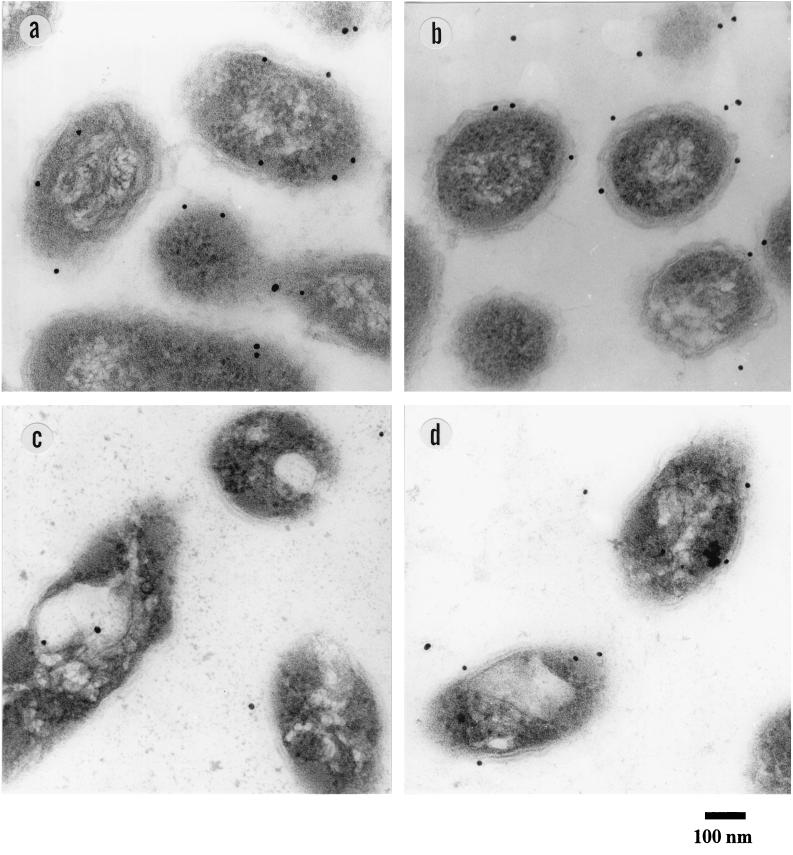
The periplasmic localization of LinA and LinB was confirmed by immunoelectron microscopy (Fig. 3 and Table 2 [by counting the number of gold particles in a field of vision]). In the wild-type strain, UT26, LinA and LinB were almost exclusively detected in the periphery of the cells (Fig. 3a and b and Table 2). On the other hand, in the spontaneous linA deletion mutant, YO5, only LinB was detected in the periphery of the cells (Fig. 3c and d and Table 2). This result excludes the possibility that the antibodies react nonspecifically with the periphery of the cells.
FIG. 3.
Immunogold-labeling electron microscopy. (a) Wild-type strain, UT26, with anti-LinA antibody. (b) UT26 with anti-LinB antibody. (c) The spontaneous linA deletion mutant, YO5, with anti-LinA antibody. (d) YO5 with anti-LinB antibody.
TABLE 2.
Distribution of gold particles in immunogold-labeling electron microscopy
| Strain | Antibody | No. (%) of particles distributed among cells
|
Area used for counting (μm2) | |||
|---|---|---|---|---|---|---|
| Inside | Periphery | Outside | Total | |||
| UT26 | Anti-LinA | 184 (24) | 441 (58) | 152 (20) | 777 (100) | 61 |
| UT26 | Anti-LinB | 111 (17) | 353 (53) | 203 (30) | 667 (100) | 54 |
| YO5 (ΔlinA) | Anti-LinA | 16 (29) | 5 (9) | 34 (62) | 55 (100) | 23 |
| YO5 (ΔlinA) | Anti-LinB | 26 (12) | 99 (46) | 88 (41) | 213 (100) | 32 |
Determination of N-terminal amino acid sequences of LinA and LinB in the periplasmic fraction.
In general, the signal peptide of periplasmic proteins is processed in the translocation process (22). However, the amino acid sequences of LinA and LinB, deduced from their nucleotide sequences, do not have a signal sequence that is typical of exported prokaryotic proteins; in fact, they remain the same size in both the cytoplasm and periplasm (Fig. 2b and c). To determine whether N-terminal regions of LinA and LinB are processed, the N termini of the proteins in the periplasmic fraction were sequenced. The sequences of the N-terminal residues of LinA and LinB in the periplasmic fraction were in perfect agreement with the deduced amino acid sequences (positions 2 to 10). These results indicate that LinA and LinB are not N-terminally processed during export, except for the removal of the N-terminal methionine residue. The N-terminal methionine is probably removed by the action of methionyl-aminopeptidase, which has a preference for proteins with a small side chain in the penultimate position (1, 10).
DISCUSSION
In this study, we determined that two different types of dehalogenase, dehydrochlorinase and halidohydrolase, which are involved in the early steps of γ-HCH degradation, accumulate in the periplasmic space without undergoing molecular processing. The periplasmic space lies between the inner and outer membranes of gram-negative bacteria and has many functions (2, 22, 23). For example, some proteins residing in the periplasmic space have important functions in the detection and processing of essential nutrients and their transport into the cell. Enzymes that detoxify antibiotics, such as β-lactamase, also exist in the periplasmic space (23). Our findings have added another function to the list of events that occur in periplasmic space: degradation of halogenated xenobiotics. Halogenated organic compounds constitute one of the largest groups of environmental pollutants as a result of both their widespread use as herbicides, insecticides, fungicides, solvents, etc., and their retention in the environment (7). Elimination of halogens from halogenated xenobiotic molecules is a key step in their degradation, because the carbon-halogen bond is relatively stable (7). These compounds may enter the periplasm through nonspecific porins in the outer membrane. Since dehalogenases degrade complex halogenated molecules into simpler ones for utilization and possibly for detoxification, the localization of dehalogenases in the periplasmic space seems reasonable.
We were surprised to discover that these dehalogenases are not subject to N-terminal processing during translocation to the periplasmic space. The present results show that dehalogenases of S. paucimobilis UT26 are exported by a secretion mechanism that differs from the signal peptide-based secretion mechanism that is common in prokaryotes. Generally, the translocation of bacterial proteins across the cytoplasmic membrane is directed by an N-terminal signal peptide that is removed during or shortly after the translocation step. Two mechanisms have been reported by which proteins that lack a typical N-terminal signal peptide and that are not processed during translocation are secreted (26). However, in both cases, the proteins cross both cell membranes without stopping in the periplasm, whereas the dehalogenases of S. paucimobilis UT26 are simply exported to the periplasm and are not secreted into the culture medium. A similar finding was made in a study of chitinase produced by Serratia marcescens (4). A novel unknown mechanism for protein accumulation in the periplasmic space may be one of several protein translocation pathways that operate in gram-negative bacteria. α-Enolase and glyceraldehyde-3-phosphate-dehydrogenase are known to be cell surface proteins in gram-positive bacteria, although they are synthesized without conserved signal peptides (24, 25).
ACKNOWLEDGMENTS
We thank S. Yamashita of The University of Tokyo, Tokyo, Japan, for technical help with immunoelectron microscopy. We thank K. Ohgi and her colleagues of the Hoshi College of Pharmacy, Tokyo, Japan, for determining the N-terminal amino acid sequences.
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Culture, and Sports of Japan. This work was performed with the facilities of the Biotechnology Research Center, The University of Tokyo.
REFERENCES
- 1.Ben-Bassat A, Bauer K, Chang S-Y, Myambo K, Boosman A, Chang S. Processing of the initiation methionine from proteins: properties of the Escherichia coli methionine aminopeptidase and its gene structure. J Bacteriol. 1987;169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beveridge T J. The periplasmic space and the periplasm in gram-positive and gram-negative bacteria. ASM News. 1995;61:125–130. [Google Scholar]
- 3.Blais J M, Schindler D W, Muir D C G, Kimpe L E, Donald D B, Rosenberg B. Accumulation of persistent organochlorine compounds in mountains of western Canada. Nature. 1998;395:585–588. [Google Scholar]
- 4.Brurberg M B, Eijsink V G H, Haandrikman A J, Venema G, Nes I F. Chitinase B from Serratia marcescens BJL200 is exported to the periplasm without processing. Microbiology. 1995;141:123–131. doi: 10.1099/00221287-141-1-123. [DOI] [PubMed] [Google Scholar]
- 5.Dassa E, Tesu C, Boquet P-L. Identification of the acid phosphatase (optimum pH 2.5) of Escherichia coli. FEBS Lett. 1980;113:275–278. doi: 10.1016/0014-5793(80)80608-9. [DOI] [PubMed] [Google Scholar]
- 6.Duggleby R G, Dennis D T. Nicotinamide adenine dinucleotide-specific glyceraldehyde 3-phosphate dehydrogenase from Pisum sativum. J Biol Chem. 1974;249:162–166. [PubMed] [Google Scholar]
- 7.Fetzner S, Lingens F. Bacterial dehalogenases: biochemistry, genetics, and biotechnological applications. Microbiol Rev. 1994;58:641–685. doi: 10.1128/mr.58.4.641-685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedemann T E, Haugen G E. Pyruvic acid 2. The determination of keto acids in blood and urine. J Biol Chem. 1943;47:415–442. [Google Scholar]
- 9.Hayakawa T, Hirashima M, Ide S, Hamada M, Okabe K, Koike M. Mammalian α-keto acid dehydrogenase complexes. J Biol Chem. 1966;241:4694–4699. [PubMed] [Google Scholar]
- 10.Hirel P, Schmitter J, Dessen P, Fayat G, Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc Natl Acad Sci USA. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai R, Nagata Y, Senoo K, Wada H, Fukuda M, Takagi M, Yano K. Dehydrochlorination of γ-hexachlorocyclohexane (γ-BHC) by γ-BHC-assimilating Pseudomonas paucimobilis. Agric Biol Chem. 1989;53:2015–2017. [Google Scholar]
- 12.Imai R, Nagata Y, Fukuda M, Takagi M, Yano K. Molecular cloning of a Pseudomonas paucimobilis gene encoding a 17-kilodalton polypeptide that eliminates HCl molecules from γ-hexachlorocyclohexane. J Bacteriol. 1991;173:6811–6819. doi: 10.1128/jb.173.21.6811-6819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwasaki I, Utsumi S, Ozawa T. New colorimetric determination of chloride using mercuric thiocyanate and ferric ion. Bull Chem Soc Jpn. 1952;25:226. [Google Scholar]
- 14.Iwata H, Tanabe S, Sakai N, Tatsukawa R. Distribution of persistent organochlorines in the oceanic air and surface seawater and the role of ocean on their global transport and fate. Environ Sci Technol. 1993;27:1080–1098. [Google Scholar]
- 15.Miyauchi K, Suh S-K, Nagata Y, Takagi M. Cloning and sequencing of a 2,5-dichlorohydroquinone reductive dehalogenase gene whose product is involved in degradation of γ-hexachlorocyclohexane by Sphingomonas paucimobilis. J Bacteriol. 1998;180:1354–1359. doi: 10.1128/jb.180.6.1354-1359.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagata Y, Hatta T, Imai R, Kimbara K, Fukuda M, Yano K, Takagi M. Purification and characterization of γ-hexachlorocyclohexane (γ-HCH) dehydrochlorinase (LinA) from Pseudomonas paucimobilis. Biosci Biotechnol Biochem. 1993;57:1582–1583. doi: 10.1271/bbb.57.703. [DOI] [PubMed] [Google Scholar]
- 17.Nagata Y, Nariya T, Ohtomo R, Fukuda M, Yano K, Takagi M. Cloning and sequencing of a dehalogenase gene encoding an enzyme with hydrolase activity involved in the degradation of γ-hexachlorocyclohexane in Pseudomonas paucimobilis. J Bacteriol. 1993;175:6403–6410. doi: 10.1128/jb.175.20.6403-6410.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagata Y, Ohtomo R, Miyauchi K, Fukuda M, Yano K, Takagi M. Cloning and sequencing of a 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase gene involved in the degradation of γ-hexachlorocyclohexane in Pseudomonas paucimobilis. J Bacteriol. 1994;176:3117–3125. doi: 10.1128/jb.176.11.3117-3125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagata Y, Fukuda M, Miyauchi K, Takagi M. Genes and enzymes for γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: American Society for Microbiology; 1996. pp. 58–70. [Google Scholar]
- 20.Nagata Y, Miyauchi K, Damborsky J, Manova K, Ansorgova A, Takagi M. Purification and characterization of haloalkane dehalogenase of a new substrate class from a γ-hexachlorocyclohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Appl Environ Microbiol. 1997;63:3707–3710. doi: 10.1128/aem.63.9.3707-3710.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nossal N, Heppel L. The release of enzymes by osmotic shock from Escherichia coli in exponential phase. J Biol Chem. 1966;241:3055–3062. [PubMed] [Google Scholar]
- 22.Oliver D B. Periplasm and protein secretion. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 1. Washington, D.C: American Society for Microbiology; 1987. pp. 56–69. [Google Scholar]
- 23.Oliver D B. Periplasm. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 88–103. [Google Scholar]
- 24.Pancholi V, Fischetti V A. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J Exp Med. 1992;176:415–426. doi: 10.1084/jem.176.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pancholi V, Fischetti V A. α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem. 1998;273:14503–14515. doi: 10.1074/jbc.273.23.14503. [DOI] [PubMed] [Google Scholar]
- 26.Salmond G P C, Reeves P J. Membrane traffic wardens and protein secretion in gram-negative bacteria. Trends Biochem Sci. 1993;18:7–12. doi: 10.1016/0968-0004(93)90080-7. [DOI] [PubMed] [Google Scholar]