Abstract
Excision and formation of a covalently closed circular transposon molecule are required for conjugative transposition of Tn916 but are not the only factors that limit the frequency of conjugative transposition from one host to another. We found that in gram-positive bacteria, an increase in the frequency of excision and circularization of Tn916 caused by expression of integrase (Int) and excisionase (Xis) from a xylose-inducible promoter does not lead to an increase in the frequency of conjugative transposition. We also found that the concentration of Int and Xis in the recipient cell does not limit the frequency of conjugative transposition and that increased excision does not result in increased expression of transfer functions required to mobilize a plasmid containing the Tn916 origin of transfer. We conclude that in gram-positive hosts in which the Tn916 functions Int and Xis are overexpressed, the frequency of conjugative transposition is limited by the availability of transfer functions.
Tn916 was first isolated from Enterococcus faecalis (8) and is the prototype of a family of conjugative transposons that includes the closely related element Tn1545. Members of this family are found in a wide variety of gram-positive and a few gram-negative bacterial species. They mediate their own transfer between different bacterial species and do not seem to be subject to restriction barriers. Most conjugative transposons encode antibiotic resistance determinants, including tet(M), and for this reason they are important in the spread of drug resistance to and among gram-positive bacterial pathogens (reference 9; for recent reviews, see references 6 and 21).
The first step in conjugative transposition is excision of Tn916 from the donor DNA molecule. Excision requires two transposon-encoded proteins: integrase (Int) and excisionase (Xis). Int is a member of the λ-Int family of site-specific recombinases (18), and Xis is a small basic protein, similar to the λ family of Xis proteins, that binds to both ends of Tn916 (19). Cleavage by Int results in 5′ single-stranded overhangs consisting of the 6 bp that flank the transposon, called coupling sequences (14, 24). Ligation of the excised transposon ends produces a covalently closed circular (CCC) Tn916 molecule with a 6-bp heteroduplex at the circle joint (22). The CCC form of Tn916 is unable to replicate and is the substrate for conjugative transfer.
Conjugative transfer of Tn916 is similar to that of conjugative plasmids. Like conjugative plasmids, Tn916 contains an origin of transfer (11) and a single strand of the excised CCC Tn916 molecule is transferred to the recipient cell (20), where the complementary strand is synthesized. This results in a double-stranded circular form of Tn916 which inserts into the target DNA molecule, usually at a region that includes several adenines followed by several thymines and often contains a static bend (13).
Excision of Tn916 from the donor DNA molecule is the first step in conjugative transposition in gram-positive hosts and is thought to be the rate-limiting step for several reasons. First, excision is required for conjugative transposition since Tn916 mutants defective in excision are unable to undergo this process (23). Second, the excised CCC form of Tn916 does not accumulate in gram-positive hosts in which conjugative transposition is observed, presumably because it is rapidly transferred to a new cell and inserted into a target site. In contrast, in Escherichia coli, where conjugative transposition is difficult to detect, the CCC form of Tn916 accumulates (22). Finally, in E. faecalis there is a report of a correlation between the number of CCC Tn916 molecules detected by PCR and the donor potential of the strain in matings (15). An alternative explanation for all these observations might be that excision and conjugation are coregulated.
The frequency of Tn916 excision is limited by the concentrations of Int and Xis in the host cell. Overexpression of Int or Xis alone does not affect the excision frequency of Tn916, but when they are overexpressed together, there is an increase in the frequency of excision of at least 1,000-fold in Bacillus subtilis and in E. faecalis (16). An increase in the concentration of both products of the excision reaction, the repaired donor DNA molecule and the excised CCC form of Tn916, can be detected when Int and Xis are overexpressed (16). To test the hypothesis that Tn916 excision limits the frequency of conjugative transposition, we determined the effect of increased excision on conjugative transposition.
MATERIALS AND METHODS
Media and growth conditions.
B. subtilis strains were grown in Luria-Bertani (LB) medium, and streptomycin (Sm) was used at a concentration of 1,200 μg/ml when appropriate. Expression from P-xylA in B. subtilis was induced by including 2% xylose in the growth medium. E. faecalis strains were grown in Todd-Hewitt medium supplemented with 0.2% yeast extract, and antibiotics were used at the following concentrations: erythromycin, 10 μg/ml; fusidic acid, 25 μg/ml; Sm, 1,000 μg/ml. Spectomycin (Spec) and tetracycline (Tc) were used at concentrations of 100 μg/ml and 10 μg/ml, respectively, for both species.
Matings.
Matings were performed as described previously (1), except that E. faecalis matings were performed by using Todd-Hewitt medium plus 0.2% yeast extract, and 2% xylose was added to the mating plates to induce expression of Int and/or Xis. All matings were repeated at least three times.
Construction of the Tn916-oriT-containing plasmid pEU358.
The Tn916-oriT-containing plasmid pAM5160 (11) was cut with EcoRI and ligated to the 2.2-kb EcoRI fragment of pUC4ΩKan (17). The resulting plasmid was named pEU358 and contains the Tn916-oriT, the ΩKan resistance cassette, the p15A origin of replication that functions in E. coli, and the pIP501 origin of replication that functions in gram-positive bacteria.
Determination of the relative abundance of the CCC Tn916 molecules in high- and low-frequency E. faecalis donors.
Total cell DNA was extracted from E. faecalis and used as a template in PCR. The CCC Tn916 joint-specific primers OTL3 (CTCGAAAGCACATAGAATAAGGC) and OTR1R (GGATAAATCGTCGTATCAAAGC) along with the tet(M)-specific primers CM8 (GCGGATCACTATCTGAGATTTCC) and CM9 (CGAATCTGAACAATGGGATACGG) were used with Taq polymerase to amplify the template DNA. [α-32P]dATP was added to each reaction mixture, and the reactions were cycled 20 times in a thermocycler under the following conditions: 95°C for 1 min, 52°C for 1 min, and 72°C for 1 min. The resulting products were separated on a 5% polyacrylamide gel. The intensity of the tet(M)-specific band was compared to the intensity of the joint-specific band for each sample by using a Molecular Dynamics PhosphorImager and Imagequant software.
Detection of excised CCC transposon DNA by Southern blotting.
B. subtilis CKS102/pEU327 and CKS102/pEU354 were grown in LB medium at 37°C to an optical density at 600 nm of 0.5. Xylose was added at 2%, and incubation was continued for 2 h. Cells were pelleted by centrifugation and suspended in 50 mM Tris HCl (pH 8.0)–50 mM EDTA–25% sucrose. Lysozyme was added to a final concentration of 4 mg/ml, and the suspension was incubated at 37°C for 15 min. The cells were lysed by the addition of an equal volume of 10 mM Tris-HCl (pH 8.0), 5 mM EDTA, and 1% sodium dodecyl sulfate. After lysis, 1/10 volume of proteinase K (4 mg/ml) and RNaseA (10 mg/ml) were added and the lysate was incubated for 4 h at 37°C. The lysates were extracted twice with phenol and once with chloroform, and the DNA was precipitated first with isopropanol and then with ethanol. DNA preparations were digested with restriction enzymes, subjected to electrophoresis on 0.4% agarose gels, and transferred by blotting to a nylon membrane. Tn916 DNA was detected by hybridization with a fragment internal to the tet(M) gene by using the Amersham ECL direct nucleic acid labeling and detection system. The tet(M) fragment was made by PCR amplification using the primers CM8 (GCGGATCAGTATCTGAGATTTCC) and CM9 (CGAATCTGAACAATGGGATACGG).
RESULTS
Effect of Int and Xis overexpression on conjugative transposition.
To test the hypothesis that the frequency of excision and circularization of Tn916 limits the frequency of conjugative transposition, we compared the conjugative transposition frequency in overnight matings of several strains containing a single copy of Tn916 under conditions that produce different frequencies of Tn916 excision. The plasmid pEU354 contains int and xis downstream of the P-xylA promoter and overexpresses both proteins in gram-positive bacteria. Because an increase of at least 1,000-fold in the frequency of Tn916 excision is produced by overexpression of Int and Xis from pEU354 in B. subtilis and E. faecalis (16), transposition was measured when pEU354 was present or absent from the donor strain. The conjugative transposition frequency was defined as the number of transconjugants resistant to tetracycline (TcR; the marker on Tn916) divided by the total number of donors after overnight coincubation of the strains on nonselective plates.
In E. faecalis DMM103 (20), overexpression of Int and Xis from pEU354 causes a 1,000-fold increase in the frequency of Tn916 excision (16). In contrast, the presence of pEU354 had no effect on the conjugative transposition frequency of DMM103 (2.9 × 10−6 ± 2.8 × 10−6 transconjugants per donor without pEU354 compared to 1.8 × 10−6 ± 1.1 × 10−6 transconjugants per donor with pEU354 present). Similarly, the frequency of conjugative transposition of the B. subtilis CKS101 (3) and CKS102 (22), each of which contains a single copy of Tn916 in the chromosome, was unaffected by overexpression of Int and Xis from pEU354 (1.8 × 10−6 ± 1.1 × 10−6 transconjugants per donor for CKS101 compared to 4.2 × 10−6 ± 1.9 × 10−6 for CKS101 containing pEU354 and 7.5 × 10−6 ± 3.5 × 10−6 for CKS102 compared to 4.6 × 10−6 ± 2.4 × 10−6 for CKS102 containing pEU354), although excision and CCC formation is increased at least 6,000-fold when Int and Xis are overexpressed in these strains (16). Therefore, increased excision of Tn916 in the donor does not result in increased conjugative transposition for either of the two gram-positive species tested. Since similar results were observed in E. faecalis and B. subtilis, subsequent experiments were performed in CKS102 because transcription of the conjugative transfer functions of Tn916 has been studied in this strain (4).
The circular form of Tn916 is supercoiled in strains overexpressing Int and Xis.
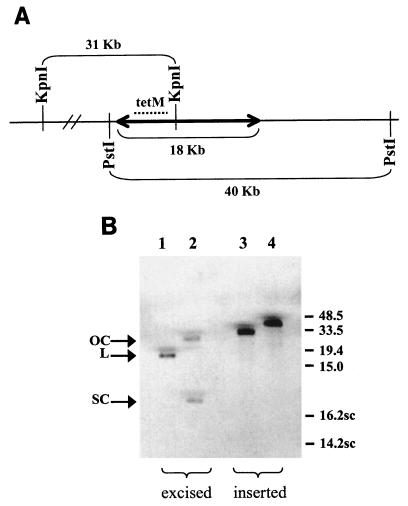
Previous assays to detect the circular form of Tn916 in strains containing pEU354 did not differentiate between a supercoiled form and an open circular form (16). To find out if the circular Tn916 molecule produced in B. subtilis strains overexpressing Int and Xis was supercoiled, we determined its mobility on an agarose gel. DNA from CKS102/pEU327 (vector alone) and from xylose-induced CKS102/pEU354 (overexpressing Int and Xis) was digested with KpnI, which cuts once within Tn916, or with PstI, which does not cut Tn916, separated on a 0.4% agarose gel, and hybridized to a tet(M) DNA probe. Following digestion with each enzyme, the DNA from CKS102/pEU327 contained a single tet(M)-complementary fragment of the size expected from the sequence of this chromosomal region (Fig. 1A and 1B, lanes 3 and 4). Digestion of DNA from the strain overexpressing Int and Xis, CKS102/pEU354, with KpnI yielded a single tet(M)-reactive band of the size expected for a linear 18-kb fragment, which is presumably the linearized circular transposon molecule (Fig. 1B, lane 1). The 31.4-kb band was no longer visible, consistent with the previous conclusion that all detectable copies of Tn916 had excised (16). When PstI, which does not cut within Tn916, was used to digest the DNA from CKS102/pEU354, two major species hybridized with the probe (Fig. 1B, lane 2). Comparison with the mobilities of CCC and nicked plasmid of a similar size in control experiments indicated that the faster migrating species in lane 2 corresponds to a supercoiled molecule of 18 kb and that the slower migrating species is probably nicked DNA, which might have been produced during experimental manipulations. Therefore, these results support the conclusion that when Int and Xis are overexpressed, at least half of the excised Tn916 molecules are supercoiled circles.
FIG. 1.
(A) Map showing KpnI and PstI restriction enzyme cleavage sites surrounding the insertion of Tn916 (double-headed arrow) in B. subtilis CKS102. The sizes of the fragments (not to scale) expected from cleavage with either KpnI or PstI are indicated. tet(M) (dotted line) indicates the position of the tet(M) gene, to which the probe used in Fig. 1B hybridizes. (B) Southern blot analysis of excised Tn916 DNA following induction of Int and Xis expression. The Southern blot has been hybridized with a probe specific for the tet(M) gene of Tn916. DNA from CKS102/pEU354 digested with KpnI (lane 1) and PstI (lane 2) and DNA from CKS102/pEU327 digested with KpnI (lane 3) and PstI (lane 4) are shown. OC, open circular; L, linear; SC, supercoiled. The numbers to the right of the blot show the mobilities of linear and supercoiled (16.2sc and 14.2sc) DNA markers in kilobases. Excised and inserted refer to the state of Tn916.
Overexpression of Int and/or Xis in the recipient does not inhibit insertion of Tn916.
The surprising result that the presence of 1,000 times more excised Tn916 CCC molecules does not lead to an increase in conjugative transposition suggests the possibility that integration of the transposon in the recipient cell is limiting in these conditions. Because overexpression of λ-Xis in E. coli inhibits insertion of the phage into the chromosomal target site (12), it seemed possible that overexpression of Tn916 Xis and/or Int might inhibit insertion of the transposon. Conjugative transposition of Tn916 requires the expression of Int only in the donor and not in the recipient cell, indicating that the Int protein can be transferred into the recipient during mating (2). It is possible, especially under conditions of overexpression, that Xis might also be transferred into the recipient during mating. To test the hypothesis that overexpression of Xis and/or Int in the recipient inhibits insertion of Tn916, matings were performed, using CKS102 as a donor, with a recipient B. subtilis strain (W168Sm) carrying pEU354 overexpressing Int and Xis, pEU329 overexpressing Int, and pEU357 overexpressing Xis. The conjugation frequency observed using W168Sm lacking pEU354 as recipient was 2.5 × 10−6 ± 2.3 × 10−6 transconjugants per donor. Overexpression of Int alone (6.0 × 10−7 ± 0.6 × 10−7 transconjugants per donor), Xis alone (2.6 × 10−6 ± 0.6 × 10−6 transconjugants per donor), or Int and Xis together (4.1 × 10−7 ± 2.1 × 10−7 transconjugants per donor) in the recipient had no significant effect on the frequency of Tn916 conjugative transposition. Similarly, no significant effect was seen when both donor and recipient contained pEU354 (3.1 × 10−6 ± 3.0 × 10−6 transconjugants per donor).
Since overexpression of Int and Xis from pEU354 in B. subtilis results in cells that no longer contain the plasmid (16), we were concerned that the transconjugants containing Tn916 represented the plasmid-free subclass. Therefore, we screened the W168Sm transconjugants for spectinomycin resistance (SpecR), the marker on the expression plasmid, to be sure they contained the plasmid. The number of SpecR W168Sm/pEU354 transconjugant cells after a mating with a Tn916-containing donor (16 of 252) was similar to the number of SpecR W168Sm/pEU354 cells in a culture that had not been exposed to a Tn916-containing donor (14 of 200). Likewise, the fractions of SpecR W168Sm/pEU329 (66 of 150) and W168Sm/pEU357 (65 of 150) transconjugant cells were similar to the fractions of SpecR W168Sm/pEU329 (17 of 50) and W168Sm/pEU357 (20 of 50) cells not exposed to a Tn916-containing donor strain. This indicates that the presence of the expression plasmid had no effect on the ability of the cell to serve as a recipient.
Mobilization of a plasmid containing Tn916-oriT by CKS102.
To determine if the tra genes were expressed at a level adequate for conjugation when Int and Xis were overexpressed from pEU354 in CKS102, we used a plasmid containing the Tn916 origin of transfer (oriT) that can be mobilized by the conjugation functions of Tn916 (11). The plasmid pEU358 includes the Tn916-oriT and the aphA3 gene (17) that confers kanamycin resistance (KmR). In E. coli, pEU358 replicates by using the p15A origin (5) and in gram-positive bacteria it replicates by using a derivative of the pIP501 origin (7). There was no significant difference in the frequency at which CKS102 and CKS102/pEU354 mobilized pEU358 (4.1 × 10−9 ± 2.1 × 10−9 and 1.0 × 10−8 ± 1.2 × 10−8 pEU358-containing transconjugants per donor, respectively). This indicates that the conjugation functions of Tn916 are active even when Int and Xis are overexpressed.
Excision frequency is not correlated with conjugative transposition frequency.
Tn916 insertions into identical target sites in E. faecalis have markedly different conjugative transposition frequencies, ranging from 4.1 × 10−4 to <10−8. The insertions differ only in the 6-bp coupling sequences that flank the inserted transposon (10). Therefore, it appears that the bases of the coupling sequences affect conjugative transposition frequency, and it was proposed that this correlates with the frequency of excision (10). Because we did not find that an increase in excision leads to an increase in conjugation in the system we studied, we compared the amount of excised CCC form of Tn916 from a high-frequency E. faecalis donor, OG1RF/pAM5100, to the amount in a low-frequency donor, OG1RF/pAM5106. In these strains Tn916 is present on a plasmid. We used quantitative PCR with labeled dATP (see Materials and Methods) to determine the relative number of Tn916 molecules in the CCC form compared to the total number of Tn916 molecules present in each strain. Primers within tet(M) were used to determine the total amount of Tn916, and this was compared to the total amount of the CCC form of Tn916 determined by using primers designed to amplify the circle joint. No difference was detected in the number of excised CCC Tn916 molecules between the high-frequency and the low-frequency donor strains (the ratio between high-frequency and low-frequency donors was 1.3 in total cellular DNA). Therefore, the composition of the coupling sequences did not affect the excision frequency in these strains.
Supercoiling of the CCC form is not different in high- and low-frequency donor strains.
A difference in the amount of CCC Tn916 molecules in the plasmid DNA fraction was reported between the high- and low-frequency donor E. faecalis strains (10). Since we saw no difference in the amount of CCC Tn916 molecules between these strains in the unfractionated DNA, it seemed possible that this difference was in the amount of the CCC form that was supercoiled. To address this question, we repeated the assay used to detect the CCC form of Tn916 following the DNA fractionation procedure used by Jaworski and Clewell (10). However, even in the fractionated DNA, no difference in the amount of the CCC Tn916 molecule was observed between the high- and low-frequency donor strains (the ratio between high-frequency and low-frequency donors was 1.4 in plasmid DNA). This result is further evidence that the frequency of excision of Tn916 does not limit conjugative transposition.
DISCUSSION
The conjugative transposition frequency of gram-positive strains containing Tn916 ranges from 10−4 to <10−9 per donor cell, with each strain having a characteristic frequency (6). Excision of Tn916 is the first step in conjugative transposition and results in a circular transposon molecule, which does not accumulate in gram-positive hosts. Since it had been reported earlier that (i) excision is required for conjugative transposition, (ii) the excised circular form of Tn916 does not accumulate, and (iii) the number of excised circles correlates with conjugation frequency, the hypothesis that excision is the rate-limiting step in conjugative transposition was proposed. We tested this hypothesis and have shown that the frequency of excision can be increased with no effect on the frequency of conjugative transposition. Therefore, we conclude that excision does not limit the rate at which Tn916 transfers to a new gram-positive host cell. We also found that the excised transposon form appears to be physically normal and should be an effective substrate for conjugative transposition.
Excised Tn916 molecules can accumulate in gram-positive hosts, and the number of CCC Tn916 molecules is not always correlated with the frequency of conjugative transposition.
There have been two reports of situations in which the number of excised CCC Tn916 molecules correlates with conjugative transposition frequency. In one, Manganelli et al. (15) reported a correlation between the frequency of conjugative transposition in E. faecalis and the number of CCC Tn916 molecules per chromosome determined by nested PCR, limited dilution, and use of the Poisson equation. It seems possible that in the strain with the largest difference, which contained two copies of Tn916, such a correlation exists, although the error in the measurement of conjugative transposition frequency was not reported. If the frequency of conjugative transposition were limited by the frequency of transposon excision, as these authors propose, it is difficult to understand why there is an apparent accumulation of a 600-fold excess of CCC transposon molecules in this strain.
The other case of a correlation between transposition and excision frequency was reported by Jaworski and Clewell (10) for strains they identified as having high (∼10−4 per donor) and low (∼10−7 per donor) donor potentials that differed only in the composition of the 6-bp coupling sequences flanking the inserted Tn916. Excised CCC Tn916 molecules were detected only in the high-donor-potential strain (with the “good” coupling sequence). We confirmed the high and low conjugation frequencies of the strains (data not shown) and repeated the PCR experiment used to detect excised CCC Tn916 molecules. However, using an internal control to correct for differences in PCR amplification, we found no significant difference in the amount of excised CCC Tn916 between these two strains. Since the good coupling sequence led to increased conjugational transfer and insertion into the replicating chromosome of the recipient while the circular intermediate form did not accumulate, it appears that the composition of the good coupling sequence increased both the frequency of excision of Tn916 and its transfer.
What limits the frequency of conjugative transposition?
The current model for Tn916 conjugative transposition includes excision and circularization of the transposon, transfer of a single DNA strand of the excised circular molecule into the recipient cell, synthesis of the complement of the transferred strand to form a closed circle again, and insertion into a target DNA molecule. The composition of the coupling sequence could influence the frequency of conjugative transposition by affecting any or all of these steps. Because only one strand of the CCC transposon is transferred from the donor to the recipient cell (20), the coupling sequence on one end of the Tn916 insertion is not transferred to the recipient. Since the coupling sequence on either end of Tn916 in the donor can affect the frequency of conjugative transposition (10), it is unlikely that the primary effect of the coupling sequence occurs in the recipient. In agreement with this, we found that overexpression of Int and/or Xis in the recipient had no effect, regardless of whether or not Int and Xis were overexpressed in the donor. In addition to demonstrating that this is not the limitation for conjugative transposition in our experiments, it shows that Tn916 differs from phage λ, in which insertion is inhibited by Xis overexpression (12).
Therefore, in B. subtilis strains overexpressing Int and Xis, conjugative transposition appears to be limited by the frequency of conjugal transfer. Because a transcript that encodes several of the proposed Tn916 tra genes was detected only if Tn916 is excised and circularized (4), it was proposed that excision to form the CCC Tn916 molecule was required to stimulate tra gene transcription. Following excision, transcription beginning in the left end of Tn916 proceeds though the circle joint, which contains the coupling sequence, into the right end of the transposon where the tra genes are located. Thus, because they lie between the tra genes and a promoter that drives expression of these genes, the nucleotide content of the coupling sequences could affect transcription of the tra genes.
The concentration of Tn916-Tra proteins required for conjugal transfer should determine the frequency of mobilization of a nonconjugative plasmid containing Tn916-oriT. When Int and Xis overexpression resulted in 1,000 times more CCC transposon in B. subtilis, we found no increase in the frequency of mobilization of the oriT plasmid, indicating that increased excision alone does not result in increased expression of the Tn916 conjugation functions. Furthermore, whether pEU354 (overexpressing Int and Xis) was present or absent from B. subtilis GVA61, which has one copy of Tn916 in its chromosome (16), made no difference in the amount of tra gene transcript for orf23 and orf16 (data not shown). The tra gene orf23 is located on the left side of Tn916-oriT and has homology to mbeA, a relaxase required for mobilization of ColE1 (7a). Orf16 is located on the right side of Tn916-oriT. Thus, when extra copies of the CCC form of Tn916 are produced following abnormal overproduction of Int and Xis, it appears that some regulatory function(s) remains to restrict overproduction of the transfer functions. Although excision is required for conjugative transposition and may also be necessary for transcription of tra genes, it is clearly not sufficient for either. We suggest, therefore, that excision and conjugal transfer of Tn916 are coregulated.
Consistent with this suggestion, when Int and Xis are removed from normal regulatory mechanisms and overexpressed, the resulting CCC transposons accumulate in B. subtilis (16). These Tn916 molecules, which appear to be the same physically as the CCC form that serves as an intermediate for conjugative transposition, are then lost from the population. This is presumably because the transfer functions remain under normal regulation since nothing has been done to overexpress them. The proposed coregulation of excision and transfer functions would have the advantage to the transposon that it would tend to prevent loss caused by excision in the absence of transfer to a new host and insertion into a replicating DNA molecule. Further studies of control of expression of the tra genes of conjugative transposons will be needed to determine the accuracy of this proposal.
ACKNOWLEDGMENTS
We thank Jennifer G. Smith for able technical assistance.
This work was supported in part by grant GM50376 from NIH, and D.M. was supported in part by NIH Training Grant T32 A107470.
REFERENCES
- 1.Bringel F, Van Alstine G L, Scott J R. A host factor absent from Lactococcus lactis subspecies lactis MG1363 is required for conjugative transposition. Mol Microbiol. 1991;5:2983–2993. doi: 10.1111/j.1365-2958.1991.tb01858.x. [DOI] [PubMed] [Google Scholar]
- 2.Bringel F G, Van Alstine G L, Scott J R. Conjugative transposition of Tn916: the transposon int gene is required only in the donor. J Bacteriol. 1992;174:4036–4041. doi: 10.1128/jb.174.12.4036-4041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caparon M G, Scott J R. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell. 1989;59:1027–1034. doi: 10.1016/0092-8674(89)90759-9. [DOI] [PubMed] [Google Scholar]
- 4.Celli J, Trieu-Cuot P. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol Microbiol. 1998;28:103–117. doi: 10.1046/j.1365-2958.1998.00778.x. [DOI] [PubMed] [Google Scholar]
- 5.Chang A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clewell D B, Flannagan S E, Jaworski D D. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 1995;3:229–236. doi: 10.1016/s0966-842x(00)88930-1. [DOI] [PubMed] [Google Scholar]
- 7.Evans R P J, Macrina F L. Streptococcal R plasmid pIP501: endonuclease site map, resistance determinant location, and construction of novel derivatives. J Bacteriol. 1983;154:1347–1355. doi: 10.1128/jb.154.3.1347-1355.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Flannagan S E, Zitzow L A, Su Y A, Clewell D B. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid. 1994;32:350–354. doi: 10.1006/plas.1994.1077. [DOI] [PubMed] [Google Scholar]
- 8.Franke A E, Clewell D B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of conjugative transfer in the absence of a conjugative plasmid. J Bacteriol. 1981;145:494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horaud T, de Cespedes G, Clermont D, David F, Delbos F. Variability of chromosomal genetic elements in streptococci. In: Dunny G M, Cleary P P, McKay L L, editors. Genetics and molecular biology of streptococci, lactococci, and enterococci. Washington, D.C: American Society for Microbiology; 1991. pp. 16–20. [Google Scholar]
- 10.Jaworski D D, Clewell D B. Evidence that coupling sequences play a frequency-determining role in conjugative transposition of Tn916 in Enterococcus faecalis. J Bacteriol. 1994;176:3328–3335. doi: 10.1128/jb.176.11.3328-3335.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaworski D D, Clewell D B. A functional origin of transfer (oriT) on the conjugative transposon Tn916. J Bacteriol. 1995;177:6644–6651. doi: 10.1128/jb.177.22.6644-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leffers G G, Gottesman S. Lambda Xis degradation in vivo by Lon and FtsH. J Bacteriol. 1998;180:1573–1577. doi: 10.1128/jb.180.6.1573-1577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu F, Churchward G. Tn916 target DNA sequences bind the C-terminal domain of integrase protein with different affinities that correlate with transposon insertion frequency. J Bacteriol. 1995;177:1938–1946. doi: 10.1128/jb.177.8.1938-1946.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manganelli R, Ricci S, Pozzi G. Conjugative transposon Tn916: evidence for excision with formation of 5′-protruding termini. J Bacteriol. 1996;178:5813–5816. doi: 10.1128/jb.178.19.5813-5816.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manganelli R, Romano L, Ricci S, Zazzi M, Pozzi G. Dosage of Tn916 circular intermediates in Enterococcus faecalis. J Bacteriol. 1995;34:48–57. doi: 10.1006/plas.1995.1032. [DOI] [PubMed] [Google Scholar]
- 16.Marra D, Scott J R. Regulation of excision of the conjugative transposon Tn916. Mol Microbiol. 1999;31:609–622. doi: 10.1046/j.1365-2958.1999.01201.x. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Casal J, Caparon M G, Scott J R. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poyart-Salmeron C, Trieu-Cuot P, Carlier C, Courvalin P. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 1989;8:2425–2433. doi: 10.1002/j.1460-2075.1989.tb08373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudy C K, Scott J R, Churchward G. DNA binding by the Xis protein of the conjugative transposon Tn916. J Bacteriol. 1997;179:2567–2572. doi: 10.1128/jb.179.8.2567-2572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott J R, Bringel F, Marra D, Van Alstine G, Rudy C K. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol Microbiol. 1994;11:1099–1108. doi: 10.1111/j.1365-2958.1994.tb00386.x. [DOI] [PubMed] [Google Scholar]
- 21.Scott J R, Churchward G G. Conjugative transposition. Annu Rev Microbiol. 1995;49:367–397. doi: 10.1146/annurev.mi.49.100195.002055. [DOI] [PubMed] [Google Scholar]
- 22.Scott J R, Kirchman P A, Caparon M G. An intermediate in the transposition of the conjugative transposon Tn916. Proc Natl Acad Sci USA. 1988;85:4809–4813. doi: 10.1073/pnas.85.13.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Storrs M J, Poyart-Salmeron C, Trieu-Cuot P, Courvalin P. Conjugative transposition of Tn916 requires the excisive and integrative activities of the transposon-encoded integrase. J Bacteriol. 1991;173:4347–4352. doi: 10.1128/jb.173.14.4347-4352.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor K L, Churchward G G. Specific DNA cleavage mediated by the integrase of conjugative transposon Tn916. J Bacteriol. 1997;179:1117–1125. doi: 10.1128/jb.179.4.1117-1125.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]