Highlights
-
•
circ_0038138 is highly expressed in GAC tissues and plasma-Exos.
-
•
Exosomal circ_0038138 induces glycolysis to boost the growth of GC cells.
-
•
Loss of circ_0038138 inhibits the expression of EZH2, a miR-198 target gene.
-
•
Exosomal circ_0038138 enhances gastric tumorigenesis via miR-198/EZH2.
-
•
This study may provide a theocratic target for Exo-based therapy to treat GC.
Keywords: Gastric adenocarcinoma, Exosomes, circ_0038138, microRNA-198, EZH2
Abstract
This study aims to decipher the impact and downstream mechanisms of the bioinformatically identified circ_0038138 delivered by cancer-derived exosomes in gastric adenocarcinoma (GAC). Expression of circ_0038138 in clinical GAC tissues and exosomes (Exos) from clinical plasma samples (plasma-Exos) was predicted by bioinformatics analysis and validated by RT-qPCR. The binding affinity between circ_0038138, miR-198 and EZH2 was identified using luciferase activity, RIP, and RNA pull-down assays. GAC cells (AGS) were co-cultured with Exos isolated from GAC cell supernatant (GC9811-P). After co-culture, the behaviors of GAC cells including proliferation and glycolysis were assessed to identify the biological effect of exosomal circ_0038138. Also, in vivo effects of exosomal circ_0038138 on the tumorigenesis and lung metastasis of GAC cells were evaluated by developing nude mouse xenograft and metastatic models. circ_0038138 upregulation was detected in GAC tissues and plasma-Exos. Exos delivered circ_0038138 to GAC cells and potentiated the proliferative, migratory, invasive, and glycolytic potentials of GAC cells. Mechanistically, circ_0038138 competitively bound to miR-198, which in turn targeted EZH2 by binding to its 3′-UTR. Silencing of EZH2 promoted CXXC4 expression and inhibited Wnt/β-catenin pathway activation, thus repressing the malignancy and glycolysis of GAC cells. In vivo assay confirmed that exosomal circ_0038138 induced tumorigenesis and lung metastasis by regulating the miR-198/EZH2 axis. Collectively, our work suggests that the Exo-mediated transfer of circ_0038138 potentially facilitates the glycolysis, growth and metastasis of GAC cells via miR-198/EZH2 axis, which offers a potential prognostic marker and a therapeutic target for GAC.
Introduction
Gastric cancer (GC) is one of gastrointestinal cancers that are more prevalent in Asian countries than in the other regions of the world [1]. Among the GCs, adenocarcinomas are the most common histological type and phenotypically and genotypically heterogeneous [2]. Risk factors for gastric adenocarcinoma (GAC) includes Helicobacter pylori infection, Epstein-Barr virus (EBV) infection, family history of GC, hereditary diffuse GC, pernicious anaemia, and other environmental and genetic factors [3,4]. GAC is accompanied by high mortality rates and resistance to treatment modalities owing to its heterogeneity [5]. Discovering the molecular targets for GAC is of the utmost importance.
Exosomes (Exos) are single-membrane vesicles that have the same topology as the cell, a diameter of 30–200 nm, which are enriched in plenty of lipids, proteins, nucleic acids and glycoconjugates [6]. Circular RNAs (circRNAs) are known as a class of single-stranded closed RNA molecules formed by precursor mRNA back-splicing or skipping events of genes in eukaryotes [7,8]. Studies have demonstrated enrichment and stability of circRNAs in Exos and potential applications of exosomal circRNAs as disease biomarkers and therapeutic targets [9,10]. The RNA22 database predicted the presence of binding sites between a novel circRNA circ_0038138 and microRNA-198 (miR-198). miRNAs, a class of noncoding RNA molecules, exert crucial roles in the cancer progression, serving either as oncomiRs or as anti-oncomiRs through diverse molecular mechanisms [11,12]. Specifically, miR-198 can suppress the proliferative and clonogenic properties of GC cells via targeting FGFR1 [13]. Interestingly, reduced miR-198 expression links to larger tumor size, advanced tumor-node-metastasis (TNM) stage, the presence of lymph node metastasis (LNM), and poor prognosis in patients with GC [14].
The prediction results from the RNA22 database revealed miR-198 binding sites in the 3′-untranslated region (3′-UTR) of enhancer of zeste homolog 2 (EZH2) mRNA. EZH2 represents an essential component of the polycomb repressive complex 2, which facilitates carcinogenesis by restricting tumor suppressor genes [15]. EZH2 is upregulated in GC tissues while knockdown of its expression impairs cellular proliferative and invasive capacities [16]. EZH2 has been highlighted to be intensively associated with the tumor glycolysis [17,18], which has been considered to be vital for uninterrupted cancer growth, metabolic reprogramming, and immune evasion [19]. We therefore proposed a hypothesis that exosomal circ_0038138 may participate in the glycolysis, growth and metastasis of GAC via the mechanism involving the binding of miR-198 to EZH2.
To address this hypothesis, we isolated Exos from GAC patient plasma samples (plasma-Exos) and cancer cell supernatant and conducted co-culture experiments to examine the effect of plasma-Exos on GAC cells. Mechanistic investigations were performed based on in silico analysis and testified in the settings of in vitro and in vivo.
Materials and methods
Ethics statement
The current study was performed with the approval of the Ethics Committee of the Affiliated Hospital of Jiangnan University. All participants or their guardians signed informed consent documentation before enrollment. Animal experiments were approved by the Animal Ethics Committee of the Affiliated Hospital of Jiangnan University with procedures in line with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
In silico prediction
The significantly highly expressed circRNAs in the plasma-Exos of GC were determined based on the sequencing data from the existing literature, the name of which was identified through retrieval on the circBase database. R language “limma” package was used for differential analysis on the GC-related circRNA expression dataset GSE78092 (6 samples) retrieved from the GEO database with p < 0.05 as the threshold. These significantly highly expressed circRNAs were then intersected to identify the candidate circRNAs. circRNA sequences were obtained from the circBank database, miRNA mature body sequences from the mirBase database, and gene sequences from the NCBI database. RNA22 database was adopted to predict the binding sites between circRNA and miRNA, and between miRNA and mRNA. Gene expression in GC was determined through analysis of the STAD data in TCGA database using the GEPIA tool.
Sample collection
Totally, 66 patients with GAC underwent radical surgery at Jiangdu People's Hospital Affiliated to Medical College of Yangzhou University from 2017 to 2019 were enrolled. GAC tissues and corresponding adjacent non-neoplastic tissues and 5 mL of venous blood samples were collected from patients with GAC. Among them, 24 cases were younger than 60 years old, 42 cases were older than 60 years old. Histological grades (41 cases with metastasis, and 25 cases without metastasis) were determined through morphological observation and diagnosis by two or more associate chief physicians at the Department of Pathology in accordance with the World Health Organization (WHO) classification criteria [20]. Tumor staging (28 cases at stage I + II and 38 cases at stage III) was based on the sixth edition of the American Joint Committee on Cancer tumor node metastasis (AJCC/TNM) system [21]. None of the enrolled patients underwent chemotherapy or radiotherapy before surgery. Patients meeting the following criteria were excluded: (a) unknown grade, (b) unknown tumor size, (c) positive/unknown number of checkpoints. In addition, fresh normal plasma samples were obtained from 66 gender- and age-matched healthy volunteers. The basic information of patients was collected from the medical record room, and all patients were followed up to document the patients’ condition and clinical outcome after treatment, and perfect the clinical data of the patients.
Immunohistochemistry
Paraffin-embedded GAC tissues were cut into 5 μm-thick serial sections. Following dehydration, the sections were treated with 3% hydrogen peroxide for 10 min and blocked with normal goat serum for 10 min. Next, immunostaining was performed with primary antibodies: rabbit anti-EZH2 (1:50, #5246, Cell Signaling Technologies [CST], Beverly, MA), rabbit anti-CXXC4 (1:200, ab105400, Abcam Inc., Cambridge, UK), mouse anti-β-catenin (1:1000, 51067-2-AP, Proteintech, Wuhan, Hubei, China) overnight at 4 °C. Biotin-labeled goat anti-mouse or goat anti-rabbit (1:200, BA1001/BA1003, Boster, Wuhan, Hubei, China) served as secondary antibody. Thereafter, the sections were treated with SP solution, developed with DAB, and counterstained with hematoxylin before observation under a microscope. The primary antibody was substituted with IgG as negative control (NC).
Cell culture
Human GAC cell line AGS and human embryonic kidney cell HEK-293T cell line were purchased from the American Type Culture Collection (ATCC; Manassas, VA). Human GAC cell line GC9811-P was purchased from Shanghai Zishi Biotechnology Co., Ltd. (Shanghai, China) and XGC-9811 cells were isolated and established from a human malignant ascites specimen with multi-organ metastasis ability [22]. Human normal gastric mucosal cell line GES1 was purchased from Shanghai EK-Bioscience Biotechnology Co., Ltd. (Shanghai, China). GC9811-P and AGS cells were cultured in RPMI 1640 medium (Gibco BRL, Grand Island, NY) containing 10% fetal bovine serum (FBS; Gibco) and 1% double antibody (100 U/mL penicillin and 100 μg/mL streptomycin) in a 5% CO2 incubator at 37 °C. HEK-293T, XGC-9811 and GES1 cells were cultured in DMEM containing 10% fetal bovine serum (FBS) and 1% double antibody in a 5% CO2 incubator at 37 °C. The culture medium was changed every 24 h, and cells were digested with 0.25% trypsin (HyClone Laboratories, Logan, UT) every 72 h and passaged.
Isolation and identification of Exos
GC9811-P and GES1 cells were cultured in Exo-depleted medium containing 10% FBS (Exos were removed by ultracentrifugation at 100,000 × g and 4 °C overnight), and the cell culture supernatant was collected after 48 h. Next, ultracentrifugation was performed to separate the Exos from the cell supernatant and plasma [23]. Following filtration through a 0.22 μm filter (Merck Millipore, Billerica, MA), the supernatant was collected and ultracentrifuged at 140,000 g and 4 °C for 3 h. The pellet was washed with 10 mL of PBS and centrifuged at 140,000 × g and 4 °C for 2 h. The obtained pellet was then resuspended in PBS to obtain Exos, the number of which was quantified using the EXOCET Exosome Quantification Kit (System Biosciences Inc.) according to the instructions. The Exos were further treated for RNA or protein extraction. The total exosomal RNA was harvested with the use of TRIzol reagent (Thermo Fisher Scientific). Epoch (BioTek Instruments, Inc., Winooski, VT) was utilized to measure the protein quality of the harvested Exos.
The morphology of the isolated Exos was then observed under a JEM-2000EX transmission electron microscope (TEM; JEOL, Tokyo, Japan). Nanoparticle tracking analyzer (NS300, Malvern Instruments, Malvern, UK) was applied to measure the size distribution of Exos. The expression of Exo surface markers (CD63, CD81, Alix) and the non-Exo marker Calnexin was detected using western blot analysis, with the used antibodies including CD63 (1:1000, 25682-1-AP, Proteintech ProteinTech Group, Chicago, IL), CD81 (1:1000, 66866-1-Ig, Proteintech), Alix (1:1000, 12422-1-AP, Proteintech), and Calnexin (1:1000, ab133615, Abcam) [24].
Uptake of Exos by GAC cells
The purified Exos labeled by PKH67 (Sigma-Aldrich, St Louis, MO) were centrifuged at 120,000 g for 90 min. The labeled Exos by PKH67 were resuspended in basal medium, and co-cultured with the target cells for 12 h at 37 °C. Thereafter, the cells were stained with DAPI (Sigma-Aldrich) for 10 min and observed under an IX53 fluorescence microscope (Olympus, Tokyo, Japan) [25].
Dual-luciferase reporter assay
DNA fragments binding to the miR-198 sequence were cloned into the pGL3-basic vector (Promega) at the XbaI restriction site to generate the firefly luciferase reporter gene vector pGL3-basic-circ_0038138-wild type (WT) (circ_0038138-WT). At the same time, the pGL3-basic-circ_0038138-mutant type (MUT) was constructed. In the same way, luciferase reporter gene vectors containing EZH2-WT sequence and EZH2-MUT sequence were constructed, namely EZH2-3′-UTR-WT and EZH2-3′-UTR-MUT. These reporter plasmids were then introduced together with NC mimic/miR-198 mimic into HEK-293T cells using the Lipofectamine 2000 reagent. After 24 h, luciferase activity was measured using the Dual-Luciferase® Reporter Assay System (E1910, Promega).
RNA pull-down assay
Cells were transfected with biotinylated circ_0038138 (WT), circ_0038138 (MUT) and NC probes synthesized by Guangzhou RiboBio Co., Ltd. (Guangdong, China). The cell lysates were then incubated with M-280 streptavidin magnetic beads (Invitrogen Inc., Carlsbad, CA). Magnetic RNA extraction kit (A27828, Thermo Fisher Scientific) was applied to perform RNA extraction and analysis according to the supplier's instructions. Cells were transfected with a biotinylated probe, and the cell lysates were incubated with the M-280 streptavidin magnetic beads (Invitrogen) [26]. The expression of miR-198 was measured by reverse transcription quantitative polymerase chain reaction (RT-qPCR) (Supplementary Table 1).
Chromatin immunoprecipitation (ChIP)
Cells were lysed with cell lysis buffer (20 mM Tris-HCL, pH 8.0, 85 mM KCL, 0.5% NP40 and protease inhibitor) and SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCL, pH 8.1 and protease inhibitor) and subjected to ultrasonic treatment. Next, the lysate was incubated with antibodies against EZH2 (1:100, #5246, CST) and IgG (1:100, ab185913, Abcam, serving as NC) at 4 °C overnight. Pierce protein A/G beads (88803, Thermo Fisher Scientific) were used to immunoprecipitate the lysate. The DNA-protein complex was incubated at 65 °C overnight to reverse the cross-linking following centrifugation. The recovered and purified DNA fragments were used as amplification templates. The immunoprecipitated CXXC4 was detected by RT-qPCR using iQ SYBR Green Supermix (BioRad) and sequence (Forward: 5′-TGCCTCCTGTTAGTCATTAGA-3′ and Reverse: 5′-CAATATTCTGCACACCTGTTG-3′) [27].
Lentivirus transduction
sh-circ_0038138 recombinant lentivirus and sh-NC lentivirus (GenePharma, Shanghai, China) were prepared and titrated to 109 TU/mL. GC9811-P cells were transduced with lentivirus carrying sh-circ_0038138 and sh-NC. After 72 h, the medium containing 4 μg/mL puromycin was used for at least 14 days of culture. Puromycin-resistant cells were expanded in a medium containing 2 μg/mL puromycin for 9 days, and then cultured with a puromycin-free medium to obtain stable circ_0038138-silencing GC9811-P cells. Exos were extracted for subsequent animal experiments [28].
Cell transfection
Based on the sequences of β-catenin, EZH2, CXXC4, and miR-198 in NCBI, Sangon Biotechnology (Shanghai, China) was commissioned to synthesize small interfering RNA (si)-NC, si-β-catenin (S: 5′-GAUGGUGUCUGCUAUUGUACG-3′; AS: 5′-UACAAUAGCAGACACCAUCUG-3′), si-EZH2 (S: 5′-CCAUGUUUACAACUAUCAATT-3′; AS: 5′-UUGAUAGUUGUAAACAUGGTT-3′), si-CXXC4 (S: 5′-GGGAAUGCAUGAACAAGCUTT-3′; AS: 5′-AGCUUGUUCAUGCAUUCCCTT-3′), oe-NC, oe-EZH2, mimic NC, inhibitor NC, miR-198 mimic, and miR-198 inhibitor. GC9811-P cells were subjected to transfection with the above-mentioned siRNAs/plasmids using Lipofectamine 2000 reagent (11668-019, Invitrogen). AGS cells were treated with PBS, Exos isolated from GC9811-P cells (GC9811-P-Exo), Exos isolated from sh-NC-treated GC9811-P cells (Exo-sh-NC), or Exos isolated from sh-circ_0038138-treated GC9811-P cells (Exo-sh-circ_0038138). The used concentration of each plasmid referred to the reagent instructions. After 6–8 h, the medium was renewed with complete medium, and 48 h later, RNA and protein were extracted for the following experiments [29,30].
Lactate production detection
The lactic acid dehydrogenase (LDH) level was detected with LDH detection kit II (MAK065, Sigma-Aldrich). The cell sample or tissue sample was mixed with 4 times the volume of lactic acid analysis buffer, and centrifuged at 13,000 g for 10 min, with the pellet discarded. Ultracentrifugation was conducted in a 10 KDa ultracentrifuge tube to remove LDH, and the supernatant (50 μL) was added to a 96-well plate. Each well was supplemented with lactic acid analysis buffer (50 μL), mechanically dissociated, and then incubated for 30 min. The optical density (OD) value was finally measured at 450 nm [31].
Glucose detection
A high-sensitivity glucose detection kit (G3293, Sigma-Aldrich) was employed to detect glucose levels. Cell (1 × 106) or tissue samples were mixed with 106 μL of glucose and assay buffer, centrifuged at 12,000 g for 5 min. The supernatant was collected and a spin filter with a molecular weight of 10 KDa was applied to remove protein from the sample. Next, each well in a 96-well plate was added with 25 μL of samples and then with glucose assay buffer to a final volume of 50 μL. Afterwards, the samples were dispersed and incubated at 37 °C in the dark for 30 min. Finally, the OD value was measured at 340 nm [31].
Adenosine triphosphate (ATP) detection
Cells were seeded in a black 96-well plate (Corning, Toledo, OH; 1 × 106 cells/well), and a control well containing a cell-free medium was set to obtain the background luminescence value. ATP detection lysis buffer was utilized to lyse cells or tissue samples. ATP measurement was performed in the obtained lysate with the application of ATP detection kit (MAK190, Sigma-Aldrich). OD value at 570 nm was measured using a microplate reader. The average ATP level of each cell was calculated based on the number of cells in each well [32].
Cell counting kit-8 (CCK-8) assay
GAC cells in a 96-well plate (8 × 103 cells/well) were incubated for 48 h before assay. Next, 10 µL of CCK-8 solution (Sigma-Aldrich) was loaded and incubated at 37 °C for 1 h. The OD value at 450 nm was measured using an Epoch Mini Flat Spectrophotometer (Bio-Tek, Winooski, VT) [33].
Transwell assays
Transwell chamber was coated with 50 μL Matrigel (354234, BD Biosciences, Franklin Lakes, NJ). Thereafter, 100 μL of cell suspension (2.5 × 104 cells/mL) was added to each well in the upper chamber, and 500 μL of culture medium containing 10% FBS was supplemented to the lower chamber. After 24 h, the chamber was taken out whereupon the cells in the upper chamber were removed by cotton swabs. The cells were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and observed under an inverted microscope (IXplore Pro, Olympus, Japan). The number of invaded cells in five randomly selected areas was counted. The procedures of cell migration assay were the same as above except for the addition of Matrigel [34].
RNA isolation and quantitation
Total RNA was extracted from GAC tissues and cells with TRIzol reagent (15596026, Invitrogen). For mRNA detection, the extracted RNA was reverse-transcribed into complementary DNA (cDNA) using the PrimeScript RT reagent Kit (RR047A, Takara, Japan). With β-actin regarded as an internal reference, RT-qPCR was performed with SYBR Premix Ex Taq II (Takara) to evaluate the expression of mRNA. Besides, PP1A was selected as the reference gene for circRNA detection. For the detection of miRNA, total miRNA was extracted from tissues and cells using miRNeasy Mini kit (217004, Qiagen company, Hilden, Germany), and reverse transcribed according to the instructions of TaqMan MiRNA Reverse Transcription Kit (Applied Biosystems). miRNA expression, as normalized to U6, was quantified using TaqMan miRNA assays (Applied Biosystems). The fold changes were calculated using relative quantification (the 2−ΔΔCt method) [35]. The primer sequences are shown in Supplementary Table 1.
Western blot analysis
Total protein extraction from GAC tissues and cells was conducted with radioimmunoprecipitation assay lysis buffer (Beyotime, Shanghai, China) supplemented with 1% phenylmethylsulphonyl fluoride. After concentration determination by a bicinchoninic acid (BCA) kit (Pierce Biotechnology Inc., Rockford, IL), 50 μg protein was separated by electrophoresis and then transferred onto polyvinylidene fluoride membranes. The membrane was blocked using 5% skimmed milk powder for 1 h, followed by overnight incubation at 4 °C with primary antibodies: rabbit anti-EZH2 (1:1000, #5246, CST), rabbit anti-CXXC4 (1:500, ab105400, Abcam), mouse anti-β-catenin (1:5000, 51067-2-AP, Proteintech), rabbit anti-GSK-3β (1:1000, #12456, CST), rabbit anti-phosphorylated (p-)GSK-3β (Ser9, 1:1000, #5558, CST), mouse anti-β-actin (1:10000, AC004, ABclonal, China), with β-actin serving as the loading control. The next day, the membrane was incubated with horseradish peroxidase-labeled goat anti-mouse/rabbit IgG (1:10000, BA1056, Boster, Wuhan, Hubei, China) for 1 h. The immunocomplexes were visualized using enhanced chemiluminescence reagent (Thermo Fisher Scientific) and band intensities were quantified using Image J software [36]. The detailed experimental reagents and instruments are listed in Supplementary Table 2.
Orthotopic xenograft and lung metastasis models in nude mice
Twenty 4-to-5-week-old male BALB/c nude mice (weighing 18–22 g; Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) were housed in a specific pathogen-free animal room at 25–27 °C and 45–50% humidity with a 12 h light/dark cycle for 1 week to allow acclimatization. The mice were fasted for 12 h before administration, and they were given ad libitum access to food and water at other times. Totally, 20 tumor-bearing mice were subjected to treatment with PBS and GC9811-P-Exo (n = 10). In detail, 1 × 107 AGS cells were injected subcutaneously into the back of mice and the tumor was excised on the 15th day and cut into small pieces. The small pieces were implanted into the gastric subserosa hematoma [37]. One week later, 100 μg Exos or 100 μL PBS were injected to each mouse via tail vein, twice a week. About 7 weeks after the injection, the nude mice were euthanized by intravenous injection with overdose (3 times the concentration) sodium pentobarbital (3%; P3761, Sigma-Aldrich) and tissue samples were gained and stored at −80 °C for subsequent experiments. The lung tissues of mice were fixed with 4% paraformaldehyde, and the lung metastasis of GAC cells was examined by hematoxylin-eosin (HE) staining.
HE staining
The lung tissues were fixed with 4% paraformaldehyde, paraffin-embedded and cut into sections at the thickness of 5 μm. Paraffin-embedded sections were dewaxed and hydrated, followed by staining with hematoxylin (Solarbio, Beijing, China). The sections were immersed in 1% hydrochloric acid-ethanol and counterstained with eosin. The tissue morphological changes were observed under an optical microscope (XP-330, Bingyu Optical Instrument Co., Ltd., Shanghai, China) and the number of metastatic nodes in the lung tissues was counted [38].
Statistical analysis
All data were analyzed using SPSS 22.0 statistical software (IBM Corp. Armonk, NY) and GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA). The measurement data were described as mean ± standard deviation. Chi-square test was utilized for determining the correlation between gene expression and clinical characteristics of GAC patients. Normally-distributed paired data with homogeneity of variance between GAC tissues and adjacent non-neoplastic tissues were compared using paired t-test and those normally-distributed unpaired data with homogeneity of variance between two groups were compared by unpaired t-test. Differences among multiple groups were statistically analyzed employing one-way analysis of variance (ANOVA). The value of p < 0.05, p < 0.01 and p < 0.001 was statistically significant.
Results
circ_0038138 is highly expressed in GAC tissues and plasma-Exos and closely related to the metastasis of GAC
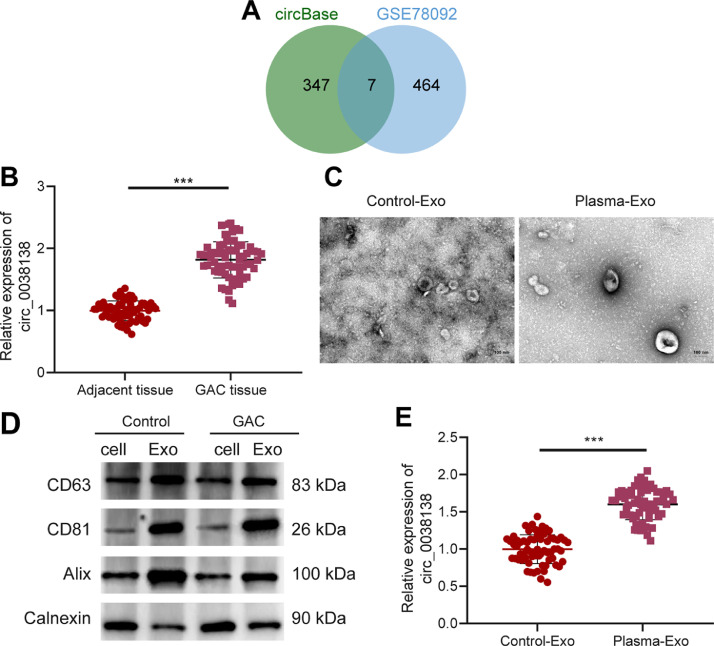
Analysis on the sequencing data of the existing literature revealed 620 significantly highly expressed circRNAs in the plasma-Exos and further retrieval on the circBase database suggested 354 circRNAs. Differential analysis of the GC-related circRNA expression dataset GSE78092 yielded 471 significantly upregulated circRNAs in GC samples. Followingly, Venn diagram analysis of these circRNAs further exhibited seven key circRNAs in the intersection, including circ_0038138, circ_0009004, circ_0003770, circ_0005957, circ_0002818, circ_0001542, and circ_0084678 (Fig. 1A). Among them, circ_0038138 was upregulated at largest fold change in Exos (p = 5.10E-06) (Supplementary Fig. 1A–F) and, therefore, selected as the target for subsequent experiments.
Fig. 1.
Prominent expression of circ_0038138 in GAC tissues and plasma-Exos. A, Venn diagram of the significantly upregulated circRNAs in the plasma-Exos from literature sequencing and from the GSE78092 dataset. B, Expression of circ_0038138 in GC tissues (n = 66) and adjacent non-neoplastic tissues (n = 66) determined by RT-qPCR. C, Morphological characterization of the plasma-Exos from GAC patients and healthy volunteers observed using a TEM. D, Western blot analysis of Exo surface makers CD63, CD81, Alix and non-Exo marker Calnexin in plasma-Exos from GAC patients and healthy volunteers. E, Expression of circ_0038138 in plasma-Exos from GAC patients and healthy volunteers determined by RT-qPCR. *** p < 0.001.
As illustrated in Fig. 1B, compared with adjacent non-neoplastic tissues, circ_0038138 expression was considerably higher in GAC tissues. Higher expression of circ_0038138 in GAC tissues corresponded to the higher tumor stage, moreLNM and higher histological type of GAC patients, while its was not correlated with the tumor depth, tumor position, and EBV-positive of GAC patients (Supplementary Table 3).
Under a TEM, the size of Exos isolated from plasma samples of GAC patients and healthy volunteers, the size of which ranged between 50 and 150 nm (Fig. 1C). The results of western blot analysis showed an abundance of Exo markers CD63, CD81 and Alix in the isolated Exos and the absence of non-Exo marker Calnexin (Fig. 1D). Moreover, RT-qPCR results displayed the augmented expression of circ_0038138 in plasma-Exos of GAC patients (Fig. 1E).
These results suggest that circ_0038138 is robustly induced in GAC tissues and plasma-Exos, and positively correlates with the growth and metastasis of GAC.
GAC cell-Exos deliver circ_0038138 to promote glycolysis and malignancy of GAC cells
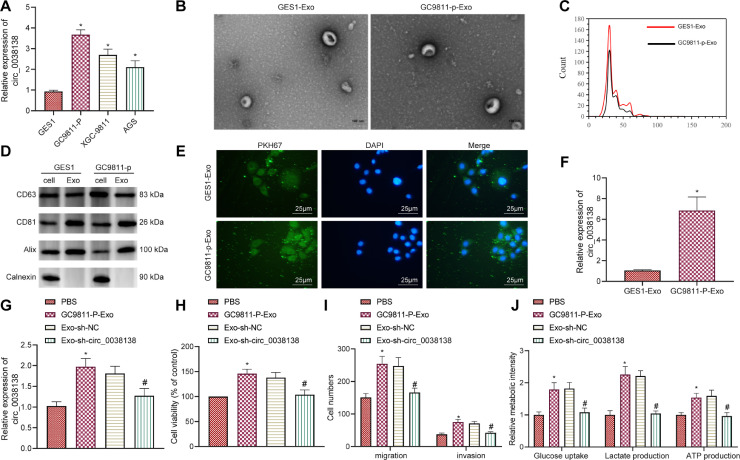
Next, we conducted cell experiments to explore the effect of circ_0038138 delivered by GAC cell-Exos on the biological functions of GAC cells. RT-qPCR characterized higher expression of circ_0038138 in human GAC cell lines GC9811-P, XGC9811 and AGS than in normal human gastric mucosal cells GES1 (Fig. 2A). GC9811-P cells exhibited the highest circ_0038138 expression and therefore were selected for subsequent experiments.
Fig. 2.
GAC cell-Exos deliver circ_0038138 to stimulate glycolysis and malignant phenotypes of GAC cells. A, Expression of circ_0038138 determined by RT-qPCR in GAC cells (GC9811-P, XGC-9811, AGS) and normal gastric mucosal cells (GES1). * p < 0.05, compared with GES1 cells. B, Morphological characterization of GC9811-P-Exos and GES1-Exos observed using a TEM as well as the size distribution. C, Quantitative analysis of size distribution analyzed by nanoparticle tracking analyzer. D, Western blot analysis of Exo makers CD63, CD81, and Alix and non-Exo marker Calnexin in the GC9811-P-Exos and GES1-Exos. E, Uptake of GC9811-P-Exos and GES1-Exos by GAC cells. F, Expression of circ_0038138 determined by RT-qPCR in GC9811-P-Exos and GES1-Exos. * p < 0.05, compared with GES1-Exos. G, Expression of circ_0038138 determined by RT-qPCR in AGS cells co-cultured with GC9811-P-Exos or Exo-sh-circ_0038138. H, Viability of AGS cells co-cultured with GC9811-P-Exos or Exo-sh-circ_0038138 measured by CCK-8 assay. I, The number of migrated and invaded AGS cells co-cultured with GC9811-P-Exos or Exo-sh-circ_0038138 measured by Transwell assay. J, Glucose uptake, and production of lactate and ATP in AGS cells co-cultured with GC9811-P-Exos or Exo-sh-circ_0038138. In panel G-J, * p < 0.05, compared with AGS cells treated with PBS. # p < 0.05, compared with AGS cells co-cultured with Exo-sh-NC. The cell experiment was conducted three times independently.
The Exos isolated from GC9811-P and GES1 cells were observed to be cup-shaped or elliptical-shaped, with a typical morphological characteristic of Exos, and their size ranged 30–150 nm. The Exo markers CD63, CD81, and Alix were all highly expressed in Exos, while non-Exo marker Calnexin was hardly expressed (Fig. 2B–D). In addition, PKH67-labeled Exos could be internalized by the recipient cells (Fig. 2E). As shown in Fig. 2F, circ_0038138 expression was much higher in GC9811-P-Exos than that in GES1 cell-Exos.
Additionally, RT-qPCR detection results demonstrated an increase in the circ_0038138 expression in the AGS cells co-cultured with GC9811-P-Exos while a decline was noted in AGS cells co-cultured with Exo-sh-circ_0038138 (Fig. 2G). Treatment with GC9811-P-Exo led to enhanced proliferative, migratory and invasive potentials of AGS cells, which was negated by Exo-sh-circ_0038138 (Fig. 2H,I). Furthermore, the glucose uptake, and production of lactate and ATP were increased in AGS cells co-cultured with GC9811-P-Exo, while treatment with Exo-sh-circ_0038138 reversed the increases (Fig. 2J).
The above-mentioned data indicate that Exos can deliver circ_0038138 to GAC cells and facilitate glycolysis of GAC cells and malignant progression.
circ_0038138 competitively binds to miR-198 to downregulate its expression
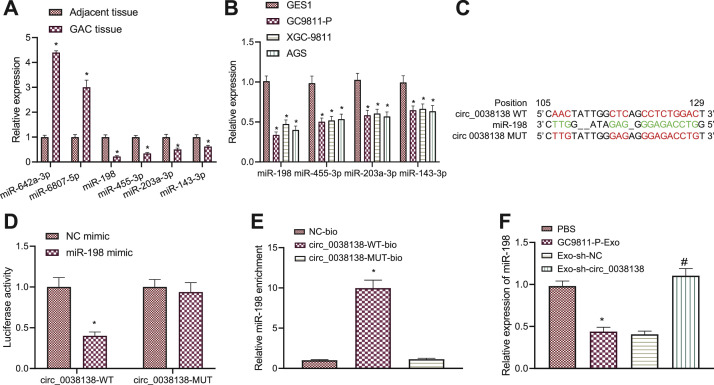
The mechanism by which circ_0038138 promoted the malignant phenotype of GAC cells was our next focus. The circBank database predicted 23 miRNAs bound by circ_0038138, of which, miR-642a-3p, miR-6807-5p, miR-198, miR-455-3p, miR-203a-3p, and miR-143-3p shared an association with GAC. Compared with adjacent non-neoplastic tissues, miR-642a-3p and miR-6807-5p were upregulated in GAC tissues, while miR-198, miR-455-3p, miR-203a-3p, and miR-143-3p showed low expression, of which miR-198 expression had the lowest expression (Fig. 3A). Meanwhile, consistent results were obtained in the GC9811-P, XGC-9811 and AGS cells in contrast to GES1 cells (Fig. 3B). Therefore, miR-198 was selected as the target for follow-up research.
Fig. 3.
circ_0038138 competitively binds to miR-198 and suppresses its expression. A, Expression of miR-642a-3p, miR-6807-5p, miR-198, miR-455-3p, miR-203a-3p, and miR-143-3p in GAC tissues and adjacent non-neoplastic tissues determined by RT-qPCR. * p < 0.05, compared with adjacent non-neoplastic tissues. B, Expression of miR-642a-3p, miR-6807-5p, miR-198, miR-455-3p, miR-203a-3p, and miR-143-3p in GAC cells (GC9811-P, XGC-9811, AGS), and normal gastric mucosal cells (GES1) determined by RT-qPCR. * p < 0.05, compared with GES1 cells. C, The predicted binding sites or mutation sites between circ_0038138 and miR-198 from the RNA22 database. Red indicates binding sites or mutation sites. D, Binding of miR-198 to circ_0038138 confirmed by dual-luciferase reporter assay in HEK-293T cells. * p < 0.05, compared with HEK-293T cells transfected with NC mimic. E, Relative miR-198 enrichment in cells transfected with circ_0038138-WT-bio or circ_0038138-MUT-bio assessed by RNA pull-down assay. * p < 0.05, compared with cells transfected with NC-bio or circ_0038138-MUT-bio. F, Expression of miR-198 in AGS cells co-cultured with GC9811-P-Exos or Exo-sh-circ_0038138 determined by RT-qPCR. * p < 0.05, compared with AGS cells treated with PBS. # p < 0.05, compared with AGS cells co-cultured with Exo-sh-NC. The cell experiment was conducted three times independently.
The binding sites between circ_0038138 and miR-198 were predicted by the RNA22 database (Fig. 3C). Dual-luciferase reporter assay results further presented that overexpression of miR-198 decreased the luciferase activity of circ_0038138-WT without altering that of circ_0038138-MUT (Fig. 3D), indicating that circ_0038138 can bind to miR-198. RNA pull-down assay data confirmed that circ_0038138 could enrich miR-198 (Fig. 3E). In addition, the results of RT-qPCR revealed a reduction of miR-198 expression in AGS cells treated with GC9811-P-Exos while an increase was observed after treatment with Exo-sh-circ_0038138 (Fig. 3F).
These findings demonstrate that circ_0038138 could competitively bind to miR-198 and reduce its expression.
circ_0038138 elevates the expression of EZH2 by reducing miR-198, thus promoting the glycolysis and malignant phenotype of GAC cells
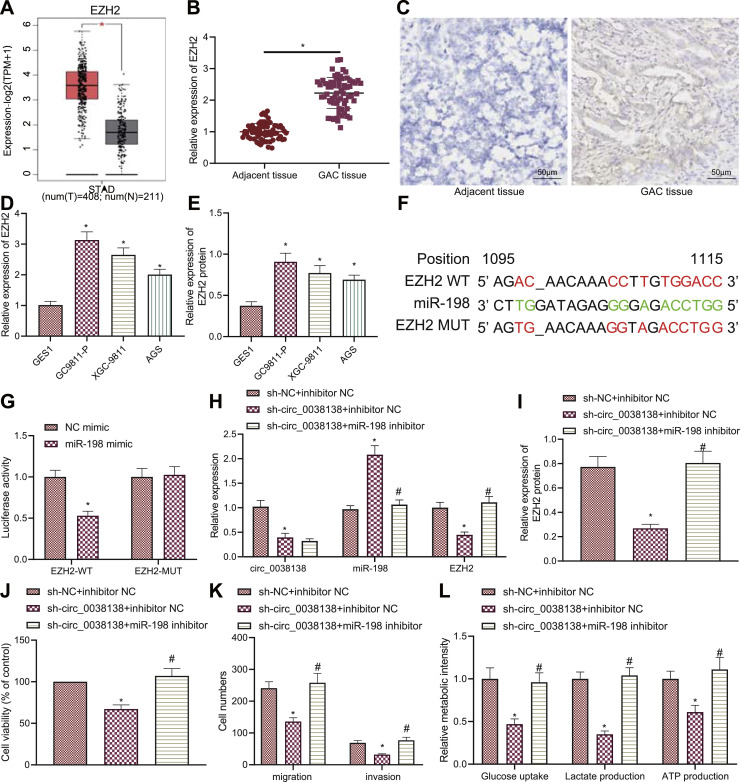
Studies have shown that EZH2, highly expressed in GC tissues and cells, is related to the occurrence and development of GC [39,40]. GEPIA analysis of STAD data from the TCGA database suggested an upregulation of EZH2 in GC samples (Fig. 4A). The results of RT-qPCR, immunohistochemistry, and Western blot assay validated high expression of EZH2 in GAC tissues (Fig. 4B,C) and GAC cell lines (Fig. 4D, E).
Fig. 4.
circ_0038138 increases the expression of EZH2 by binding to the miR-198, thus stimulating GAC cell malignancy and glycolysis. A, A box plot of EZH2 expression in TCGA-STAD data by the GEPIA analysis. The red box on the left indicates EZH2 expression in GC samples, and the gray box on the right indicates EZH2 expression in normal samples. * p < 0.05. B, mRNA expression of EZH2 in GAC tissues and adjacent non-neoplastic tissues determined by RT-qPCR. * p < 0.05, compared with adjacent non-neoplastic tissues. C, Immunohistochemical analysis of EZH2 protein expression in GAC tissues and adjacent non-neoplastic tissues. D, mRNA expression of EZH2 in GAC cells (GC9811-P, XGC-9811, AGS), and normal gastric mucosal cells (GES1) determined by RT-qPCR. * p < 0.05, compared with GES1 cells. E, Western blot analysis of EZH2 protein in GAC cells (GC9811-P, XGC-9811, AGS), and normal gastric mucosal cells (GES1). * p < 0.05, compared with GES1 cells. F, Predicted binding sites or mutation sites between miR-198 and EZH2 by the RNA22 database. Red indicates binding sites or mutation sites. G, Binding of miR-198 to EZH2 confirmed by dual-luciferase reporter assay in HEK-293T cells. * p < 0.05, compared with HEK-293T cells transfected with NC mimic. H, Expression of circ_0038138, EZH2 and miR-198 in GC9811-P cells treated with sh-circ_0038138 or combined with miR-198 inhibitor determined by RT-qPCR. I, Western blot analysis of EZH2 protein in GC9811-P cells treated with sh-circ_0038138 or combined with miR-198 inhibitor. J, Viability of GC9811-P cells treated with sh-circ_0038138 or combined with miR-198 inhibitor measured by CCK-8 assay. K, The number of migrated and invaded GC9811-P cells treated with sh-circ_0038138 or combined with miR-198 inhibitor assessed by Transwell assay. L, Glucose uptake, and production of lactate and ATP in GC9811-P cells treated with sh-circ_0038138 or combined with miR-198 inhibitor. In panel H-L, * p < 0.05, compared with GC9811-P cells treated with sh-NC + inhibitor NC. # p < 0.05, compared with GC9811-P cells treated with sh-circ_0038138 + inhibitor NC. The cell experiment was conducted three times independently.
Prediction results of the RNA22 database revealed the presence of binding sites between miR-198 and EZH2 (Fig. 4F). Analysis using the dual-luciferase reporter assay further presented a reduction in the luciferase activity of EZH2-WT following miR-198 mimic transfection while no alterations were found in the luciferase activity of EZH2-MUT (Fig. 4G). These results indicate that miR-198 can bind to EZH2.
Elevated miR-198 expression was detected in GC9811-P cells after circ_0038138 silencing. Conversely, miR-198 inhibitor decreased miR-198 expression and elevated EZH2 expression in the circ_0038138-silencing GC9811-P cells (Fig. 4H,I). Additionally, circ_0038138 silencing caused inhibition in the proliferating, migrating and invading properties of GC9811-P cells, which was abrogated by further inhibition of miR-198 (Fig. 4J,K). Also, circ_0038138 silencing decreased the glucose uptake, and production of lactate and ATP in GC9811-P cells while miR-198 inhibitor abolished the effect of sh-circ_0038138 (Fig. 4L).
Overall, these findings support that circ_0038138 binds to miR-198 and upregulates the expression of EZH2, thereby inducing the glycolysis and malignant phenotype of GAC cells.
miR-198 targets EZH2 to upregulate CXXC4 and block Wnt/β-catenin pathway
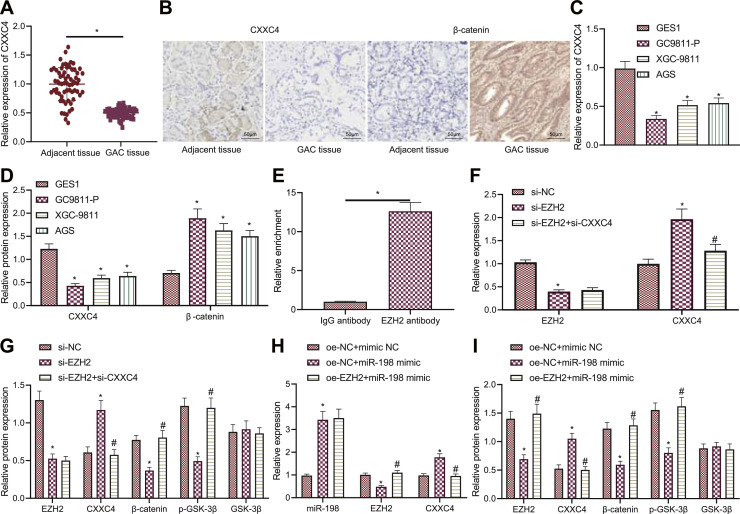
Lower CXXC4 mRNA and protein and higher β-catenin protein expression were detected in GAC tissues than adjacent non-neoplastic tissues (Fig. 5A,B). In addition, consistent downregulation of CXXC4 and upregulation of β-catenin were detected in GAC cell lines (Fig. 5C,D).
Fig. 5.
miR-198 increases CXXC4 expression and inactivates the Wnt/β-catenin pathway by targeting EZH2. A, mRNA expression of CXXC4 in GAC tissues and adjacent non-neoplastic tissues determined by RT-qPCR. * p < 0.05, compared with adjacent non-neoplastic tissues. B, Immunohistochemical analysis of CXXC4 protein in GAC tissues and adjacent non-neoplastic tissues. C, mRNA expression of CXXC4 in GAC cells (GC9811-P, XGC-9811, AGS), and normal gastric mucosal cells (GES1) determined by RT-qPCR. * p < 0.05, compared with GES1 cells. D, Western blot analysis of CXXC4 and β-catenin proteins in GAC cells (GC9811-P, XGC-9811, AGS), and normal gastric mucosal cells (GES1). * p < 0.05, compared with GES1 cells. E, Enrichment of EZH2 in the promoter region of CXXC4 gene detected by ChIP. * p < 0.05, compared with IgG antibody. F, mRNA expression of EZH2 and CXXC4 in GC9811-P cells treated with si-EZH2 or combined with si-CXXC4 determined by RT-qPCR. * p < 0.05, compared with si-NC-treated GC9811-P cells. # p < 0.05, compared with si-EZH2-treated GC9811-P cells. G, Western blot analysis of expression of EZH2, CXXC4 and β-catenin proteins along with p-GSK-3β extent in GC9811-P cells treated with si-EZH2 or combined with si-CXXC4. * p < 0.05, compared with si-NC-treated GC9811-P cells. # p < 0.05, compared with si-EZH2-treated GC9811-P cells. H, miR-198 expression and mRNA expression of EZH2 and CXXC4 in GC9811-P cells treated with miR-198 mimic or combined with oe-EZH2 determined by RT-qPCR. * p < 0.05, compared with oe-NC + mimic NC-treated GC9811-P cells. * p < 0.05, compared with oe-NC + mimic NC-treated GC9811-P cells. # p < 0.05, compared with oe-NC + miR-198 mimic-treated GC9811-P cells. I, Western blot analysis of expression of EZH2, CXXC4 and β-catenin proteins along with p-GSK-3β extent in GC9811-P cells treated with miR-198 mimic or combined with oe-EZH2. * p < 0.05, compared with GC9811-P cells treated with oe-NC + mimic NC. # p < 0.05, compared with GC9811-P cells treated with oe-NC + miR-198 mimic. The cell experiment was conducted three times independently.
ChIP data presented significant enrichment of EZH2 in the promoter region of CXXC4 gene (Fig. 5E). Furthermore, si-EZH2 transduction in GC9811-P cells reduced the expression of EZH2 and β-catenin as well as p-GSK-3β extent while enhancing CXXC4 expression. Meanwhile, further treatment ofsi-CXXC4 caused no influence in EZH2 expression, but an enhancement in β-catenin expression and p-GSK-3β extent as well as a decline in CXXC4 expression in the EZH2-silencing GC9811-P cells (Fig. 5F,G). The aforementioned results indicate that EZH2 can bind to the CXXC4 gene promoter and that silencing of EZH2 can promote the expression of CXXC4, thereby inhibiting activation of the Wnt/β-catenin pathway.
As shown in Fig. 5H,I, the expression of CXXC4 was augmented but that of EZH2 and β-catenin along with p-GSK-3β extent was decreased in GC9811-P cells overexpressing miR-198. In contrast, restoration of EZH2 counteracted the changes induced by miR-198 mimic.
Collectively, miR-198 can target EZH2 to upregulate CXXC4 expression and inactivate the Wnt/β-catenin pathway.
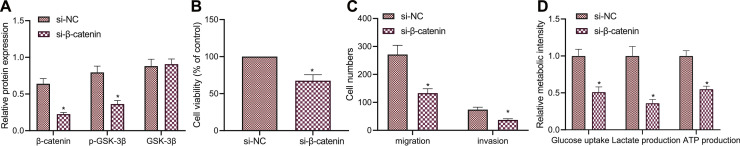
Inhibition of the Wnt/β-catenin pathway represses the glycolysis and malignancy of GAC cells
The aforementioned results allowed us to extensively validate the mechanism involving the Wnt/β-catenin pathway in GAC. Western blot analysis results showed a reduction in p-GSK-3β extent when in β-catenin was knocked down in GC9811-P cells (Fig. 6A). As a consequence of β-catenin silencing, proliferative, migratory and invasive abilities of GC9811-P cells were impeded (Fig. 6B,C). In addition, glucose uptake, and production of lactate and ATP were reduced by β-catenin knockdown (Fig. 6D).
Fig. 6.
Inactivated Wnt/β-catenin pathway arrests the malignant phenotypes and glycolysis of GAC cells. A, Western blot analysis of the Wnt/β-catenin pathway-related proteins in GC9811-P cells upon transduction of si-β-catenin. B, Viability of GC9811-P cells upon silencing of β-catenin measured by CCK-8 assay. C, The number of migrated and invaded GC9811-P cells upon silencing of β-catenin measured by Transwell assay. D, Glucose uptake, and production of lactate and ATP in GC9811-P cells upon silencing of β-catenin. * p < 0.05, compared with si-NC-treated GC9811-P cells. The cell experiment was conducted three times independently.
Taken together, these lines of evidence indicate that inhibition of the Wnt/β-catenin pathway can restrict the malignant phenotypes and glycolysis in vitro.
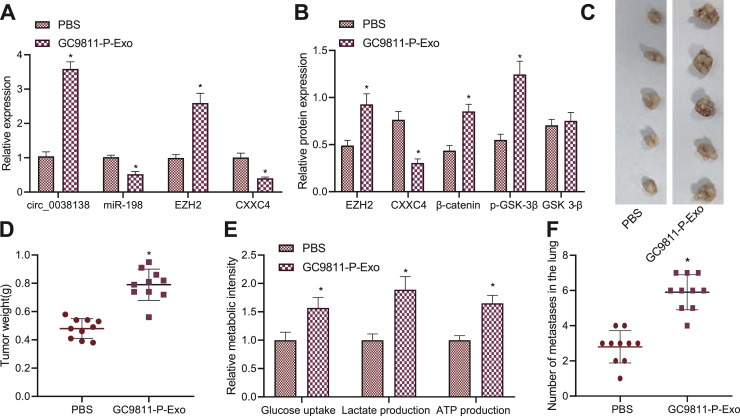
Exosomal circ_0038138 facilitates glycolysis, tumorigenesis and lung metastasis in vivo
Finally, we sought to characterize the effect of circ_0038138 delivered by GAC cell-Exos on orthotopic xenograft and lung metastasis in vivo. RT-qPCR results showed increased expression of circ_0038138 and EZH2 and decreased expression of miR-198 and CXXC4 in tumor tissues of GC9811-P-Exo-treated mice (Fig. 7A). Meanwhile, western blot analysis results exhibited downregulation of CXXC4 expression and elevations in EZH2 and β-catenin protein levels along with p-GSK-3β extent in tumor tissues of GC9811-P-Exo-treated mice (Fig. 7B). Treatment with GC9811-P-Exos increased the tumor weight of mice (Fig. 7C,D) as well as glucose uptake, and production of lactate and ATP (Fig. 7E). Analysis of the tumor tissues using HE staining suggested that the lung metastasis was accelerated following treatment with GC9811-P-Exos (Fig. 7F).
Fig. 7.
Exosomal circ_0038138 boosts glycolysis, tumorigenesis and lung metastasis of GAC cells by regulating the miR-198/EZH2 axis in vivo. A, miR-198 expression and mRNA expression of CXXC4, circ_0038138 and EZH2 in tumor tissues of GC9811-P-Exo-treated mice. B, Western blot analysis of expression of EZH2, CXXC4 and β-catenin proteins along with p-GSK-3β extent in tumor tissues of GC9811-P-Exo-treated mice. C, Representative images showing xenografts in GC9811-P-Exo-treated mice at 7 week. D, Tumor weight of GC9811-P-Exo-treated mice at 7 week. E, Glucose uptake, and production of lactate and ATP in tumor tissues of GC9811-P-Exo-treated mice. F, The number of metastatic nodes in the tumor tissues of GC9811-P-Exo-treated mice. * p < 0.05, compared with mice treated with PBS. n = 10 for mice following each treatment.
Cumulatively, these results provided evidence suggesting that exosomal circ_0038138 can regulate the miR-198/EZH2 axis to promote glycolysis, tumorigenesis and lung metastasis in vivo.
Discussion
Exosomal circRNAs have emerged as promising prognostic markers and therapeutic targets for human cancers [41]. The findings gained from this study supported the promoting effect of exosomal circ_0038138 on the glycolysis, growth, and metastasis of GAC via regulation of the miR-198/EZH2/CXXC4/Wnt/β-catenin pathway, providing mechanistic insights into the role of exosomal circ_0038138 in GAC progression.
Cancer-derived Exos have been shown to carry tumor-promotive circRNAs. For instance, circ_IARS expression is upregulated in pancreatic cancer tissues and plasma-Exos of patients with pancreatic cancer; the exosomal circ-IARS promotes tumor invasion and metastasis [23]. In addition, circ_PACRGL shows amplified expression in colorectal cancer cells and tumor-derived Exos and the exosomal circ_PACRGL acts as an oncogene to stimulate cancer proliferative and metastatic potential [42]. Another circRNA, circ_RanGAP1, has been identified to be upregulated in both GC tissues and plasma-Exo from GC patients, and this upregulation is strongly related to advanced TNM stage and LNM [43]. The current study first revealed that circ_0038138 was highly expressed in GAC tissues and plasma-Exos, which was closely associated with the growth and metastasis of GAC. In addition, our data suggested tumor-supporting properties of exosomal circ_0038138 via boosting glycolysis and malignant progression in GAC.
The subsequent mechanistic findings in the current study demonstrated that circ_0038138 competitively could bind to miR-198 to attenuate its effect on the target gene EZH2. Indeed, studies have revealed that circRNAs can act as miRNA sponges, and thus reduce their regulatory effects on the target mRNAs. CircRNA LPAR3 function as a ceRNA of miR-198 to upregulate MET whereby contributing to tumor metastasis [44]. Similarly, circAKT3 (hsa_circ_0000199) promotes cisplatin resistance by sponging miR-198 and impairing its effect on PIK3R1 [45]. A previous study has confirmed the decreased expression of miR-198 in GC tissues compared to that in corresponding noncancerous tissues which correlates with the cancer progression [14]. Consistent with our findings, miR-198 overexpression can markedly impair the malignant capabilities of GC cells [46]. Moreover, EZH2 expression shows a remarkable increase in GC cell lines and tissues and its hyper-activation can negate the suppressive effect of miR-625-3p on the malignant progression of GC cells [47]. Additionally, amounting evidence has highlighted that EZH2 overexpression leads to a significant induction in tumor glycolysis [18,48]. Also, the present results elaborated that exosomal circ_0038138 could facilitate glycolysis, tumorigenesis and lung metastasis by regulating the miR-198/EZH2 axis in vivo, which was consistent with the in vitro results. These results suggested that the circ_0038138, miR-198, and EZH2 may be promising prognostic markers and therapeutic targets for GAC.
Further analysis exhibited that miR-198 targeted EZH2 to elevate CXXC4 expression and inhibit Wnt/β-catenin pathway activation. Consistently, previous literature has identified CXXC4 as a target of EZH2 and that EZH2 can trigger the activation of the Wnt pathway by downregulating CXXC4, thus accelerating the growth of GC cells [27]. CXXC4 has been validated to express poorly in clinical samples of GC patients and act as a tumor suppressor in GC [49,50]. Strikingly, CXXC4 can disrupt the Wnt/β-catenin pathway activation in breast cancer cells by regulating the phosphorylation of GSK-3β, thus affecting the integrity of the β-catenin degradation complex [51]. Inhibiting the Wnt/β-catenin pathway results in delayed malignant progression of GC [52]. Furthermore, inactivation of the Wnt/β-catenin pathway contributes to inhibition of glycolysis in cancer cells [31,53], which was further evidenced in this study that inhibition of the Wnt/β-catenin pathway repressed the malignancy and glycolysis of GAC cells.
Overall, our study indicates that Exo-mediated transfer of circ_0038138 contributes to the glycolysis, growth, and metastasis of GAC by regulating the miR-198/EZH2/CXXC4/Wnt/β-catenin axis (Fig. 8). These findings provide better understanding of molecular mechanisms involving gastric carcinogenesis, which further aids in the development of early detection molecular markers and targets to improve the prognosis of GAC. Meanwhile, due to the limited sample size, the exact mechanism of circ_0038138 is not fully elucidated, and therefore, further large-scale studies are required to illustrate the underlying mechanism. In addition, GES1 cells are SV40-transformed epithelial gastric cells, which maintain a normal cytoskeleton, are positive in the PAS reaction, and are not tumorigenic in nude mice [54]. However, the transformed GES1 cells may have some characteristics that normal (primary) human gastric mucosal cells do not possess, such as the expression of transcription factors (snail, slug and twist) [55]. Besides, only β-actin was used as an internal reference, which may lead to the bias in qPCR results. Therefore, additional studies are required to validate the current findings.
Fig. 8.
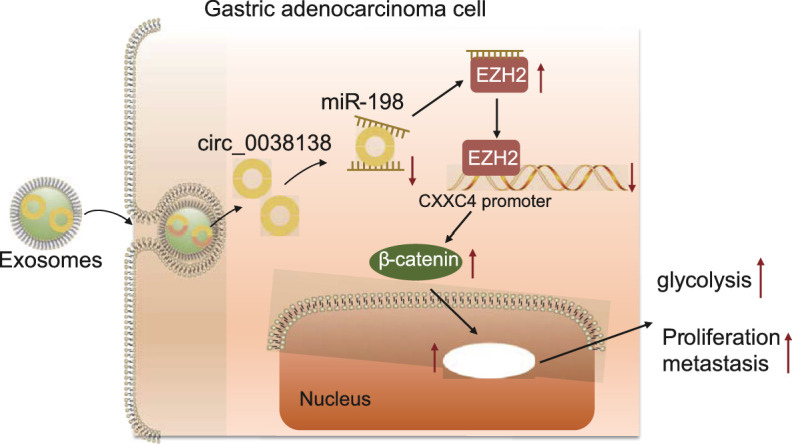
Schematic diagram of the mechanism of cancer-derived exosomal circ_0038138 in GAC. Exosomal delivery of circ_0038138 binds to miR-198, thereby upregulating EZH2, inhibiting the expression of CXXC4, and activating the Wnt/β-catenin pathway, thereby accelerating glycolysis, proliferation, and metastasis of GAC.
Funding
The study was supported by the Scientific research project of Wuxi Municipal Health Commission (No. M202051)
Supplementary Fig. 1 Expression of circ_0009004 (A), circ_0003770 (B), circ_0005957 (C), circ_0002818 (D), circ_0001542 (E) and circ_0084678 (F) in the GAC tissues and adjacent non-neoplastic tissues determined by RT-qPCR. * p < 0.05, ** p < 0.01, compared with adjacent non-neoplastic tissues.
CRediT authorship contribution statement
Yuanyuan Zheng: Writing – review & editing. Ping Li: Conceptualization, Resources, Writing – original draft. Jianghui Ma: Data curation, Formal analysis, Supervision. Chengxi Yang: Validation, Investigation, Project administration. Saimin Dai: Writing – review & editing. Changyong Zhao: Writing – review & editing.
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101479.
Appendix. Supplementary materials
References
- 1.Arnold M., Abnet C.C., Neale R.E., Vignat J., Giovannucci E.L., McGlynn K.A., et al. Global burden of 5 major types of gastrointestinal cancer. Gastroenterology. 2020;159(1):335–349e315. doi: 10.1053/j.gastro.2020.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajani J.A., Lee J., Sano T., Janjigian Y.Y., Fan D., Song S. Gastric adenocarcinoma. Nat. Rev. Dis. Prim. 2017;3:17036. doi: 10.1038/nrdp.2017.36. [DOI] [PubMed] [Google Scholar]
- 3.Banks M., Graham D., Jansen M., Gotoda T., Coda S., di Pietro M., et al. British society of gastroenterology guidelines on the diagnosis and management of patients at risk of gastric adenocarcinoma. Gut. 2019;68(9):1545–1575. doi: 10.1136/gutjnl-2018-318126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okabe A., Huang K.K., Matsusaka K., Fukuyo M., Xing M., Ong X., et al. Cross-species chromatin interactions drive transcriptional rewiring in Epstein-Barr virus-positive gastric adenocarcinoma. Nat. Genet. 2020;52(9):919–930. doi: 10.1038/s41588-020-0665-7. [DOI] [PubMed] [Google Scholar]
- 5.Ramezankhani R., Solhi R., Es H.A., Vosough M., Hassan M. Novel molecular targets in gastric adenocarcinoma. Pharmacol. Ther. 2021;220 doi: 10.1016/j.pharmthera.2020.107714. [DOI] [PubMed] [Google Scholar]
- 6.Pegtel D.M., Gould S.J. Exosomes. Annu. Rev. Biochem. 2019;88:487–514. doi: 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- 7.Bach D.H., Lee S.K., Sood A.K. Circular RNAs in cancer. Mol. Ther. Nucleic Acids. 2019;16:118–129. doi: 10.1016/j.omtn.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 3.Smyth E.C., Nilsson M., Grabsch H.I., van Grieken N.C., Lordick F. Gastric cancer. Lancet. 2020;396(10251):635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 10.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markopoulos G.S., Roupakia E., Tokamani M., Chavdoula E., Hatziapostolou M., Polytarchou C., et al. A step-by-step microRNA guide to cancer development and metastasis. Cell. Oncol. 2017;40(4):303–339. doi: 10.1007/s13402-017-0341-9. (Dordr) [DOI] [PubMed] [Google Scholar]
- 12.Treiber T., Treiber N., Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019;20(1):5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 13.Gu J., Li X., Li H., Jin Z., Jin J. microRNA-198 inhibits proliferation and induces apoptosis by directly suppressing FGFR1 in gastric cancer. Biosci. Rep. 2019;39(6) doi: 10.1042/BSR20181258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui Z., Zheng X., Kong D. Decreased miR-198 expression and its prognostic significance in human gastric cancer. World J. Surg. Oncol. 2016;14:33. doi: 10.1186/s12957-016-0784-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eich M.L., Athar M., Ferguson J.E., Varambally S. EZH2-targeted therapies in cancer: hype or a reality. Cancer Res. 2020;80(24):5449–5458. doi: 10.1158/0008-5472.CAN-20-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J., Wang Z., Lu W., Jiang H., Lu J., Qiu J., et al. EZH2 promotes gastric cancer cells proliferation by repressing p21 expression. Pathol. Res. Pract. 2019;215(6) doi: 10.1016/j.prp.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Wang M., Wei W., Han D., Chen X., Hu Q., et al. Disruption of the EZH2/miRNA/beta-catenin signaling suppresses aerobic glycolysis in glioma. Oncotarget. 2016;7(31):49450–49458. doi: 10.18632/oncotarget.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng M., Cao M.X., Luo X.J., Li L., Wang K., Wang S.S., et al. EZH2 promotes invasion and tumour glycolysis by regulating STAT3 and FoxO1 signalling in human OSCC cells. J. Cell. Mol. Med. 2019;23(10):6942–6954. doi: 10.1111/jcmm.14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganapathy-Kanniappan S. Linking tumor glycolysis and immune evasion in cancer: emerging concepts and therapeutic opportunities. Biochim. Biophys. Acta Rev. Cancer. 2017;1868(1):212–220. doi: 10.1016/j.bbcan.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Lebeau A., Kriegsmann M., Burandt E., Sinn H.P. Invasive breast cancer: the current WHO classification. Pathologe. 2014;35(1):7–17. doi: 10.1007/s00292-013-1841-7. [DOI] [PubMed] [Google Scholar]
- 21.Kwan M.L., Haque R., Lee V.S., Joanie Chung W.L., Avila C.C., Clancy H.A., et al. Validation of AJCC TNM staging for breast tumors diagnosed before 2004 in cancer registries. Cancer Causes Control. 2012;23(9):1587–1591. doi: 10.1007/s10552-012-0026-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu S., Guo X., Xie H., Du Y., Pan Y., Shi Y., et al. Phage display selection of peptides that inhibit metastasis ability of gastric cancer cells with high liver-metastatic potential. Biochem. Biophys. Res. Commun. 2006;341(4):964–972. doi: 10.1016/j.bbrc.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 23.Li J., Li Z., Jiang P., Peng M., Zhang X., Chen K., et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J. Exp. Clin. Cancer Res. 2018;37(1):177. doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen F., Xu B., Li J., Yang X., Gu J., Yao X., et al. Hypoxic tumour cell-derived exosomal miR-340-5p promotes radioresistance of oesophageal squamous cell carcinoma via KLF10. J. Exp. Clin. Cancer Res. 2021;40(1):38. doi: 10.1186/s13046-021-01834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei S., Peng L., Yang J., Sang H., Jin D., Li X., et al. Exosomal transfer of miR-15b-3p enhances tumorigenesis and malignant transformation through the DYNLT1/Caspase-3/Caspase-9 signaling pathway in gastric cancer. J. Exp. Clin. Cancer Res. 2020;39(1):32. doi: 10.1186/s13046-019-1511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng Z., Zhao G., Zhu H., Nie L., He L., Liu J., et al. LncRNA FOXD3-AS1 promoted chemo-resistance of NSCLC cells via directly acting on miR-127-3p/MDM2 axis. Cancer Cell Int. 2020;20:350. doi: 10.1186/s12935-020-01402-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu H., Sun J., Wang F., Feng L., Ma Y., Shen Q., et al. Enhancer of zeste homolog 2 activates wnt signaling through downregulating CXXC finger protein 4. Cell Death Dis. 2013;4:e776. doi: 10.1038/cddis.2013.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao K., Lin Y., Gao W., Xiao Z., Medina R., Dmitriev P., et al. Blocking lncRNA MALAT1/miR-199a/ZHX1 axis inhibits glioblastoma proliferation and progression. Mol. Ther. Nucleic Acids. 2019;18:388–399. doi: 10.1016/j.omtn.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang R., Huang H., Cui S., Zhou Y., Zhang T., Zhou Y. IFN-gamma promoted exosomes from mesenchymal stem cells to attenuate colitis via miR-125a and miR-125b. Cell Death Dis. 2020;11(7):603. doi: 10.1038/s41419-020-02788-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye M., Dong S., Hou H., Zhang T., Shen M. Oncogenic role of long noncoding RNAMALAT1 in thyroid cancer progression through regulation of the miR-204/IGF2BP2/m6A-MYC signaling. Mol. Ther. Nucleic Acids. 2021;23:1–12. doi: 10.1016/j.omtn.2020.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan Q., Yang L., Zhang X., Ma Y., Li Y., Dong L., et al. Autophagy promotes metastasis and glycolysis by upregulating MCT1 expression and Wnt/beta-catenin signaling pathway activation in hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. 2018;37(1):9. doi: 10.1186/s13046-018-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Zhang H., Yang H., Bai M., Ning T., Deng T., et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 2020;14(3):539–555. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S., Cheng M., Zheng X., Zheng L., Liu H., Lu J., et al. Interactions between lncRNA TUG1 and miR-9-5p modulate the resistance of breast cancer cells to doxorubicin by regulating eIF5A2. OncoTargets Ther. 2020;13:13159–13170. doi: 10.2147/OTT.S255113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui H., Wang Q., Lei Z., Feng M., Zhao Z., Wang Y., et al. DTL promotes cancer progression by PDCD4 ubiquitin-dependent degradation. J. Exp. Clin. Cancer Res. 2019;38(1):350. doi: 10.1186/s13046-019-1358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Shu M., Zheng X., Wu S., Lu H., Leng T., Zhu W., et al. Targeting oncogenic miR-335 inhibits growth and invasion of malignant astrocytoma cells. Mol. Cancer. 2011;10:59. doi: 10.1186/1476-4598-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S., Tang D., Wang W., Yang Y., Wu X., Wang L., et al. circLMTK2 acts as a sponge of miR-150-5p and promotes proliferation and metastasis in gastric cancer. Mol. Cancer. 2019;18(1):162. doi: 10.1186/s12943-019-1081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen G., Ran X., Li B., Li Y., He D., Huang B., et al. Sodium butyrate inhibits inflammation and maintains epithelium barrier integrity in a TNBS-induced inflammatory bowel disease mice model. EBioMedicine. 2018;30:317–325. doi: 10.1016/j.ebiom.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan Y.M., Wang C.G., Zhu M., Xing R., Cui J.T., Li W.M., et al. STAT3 signaling drives EZH2 transcriptional activation and mediates poor prognosis in gastric cancer. Mol. Cancer. 2016;15(1):79. doi: 10.1186/s12943-016-0561-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang B., Mu P., Yu Y., Zhu W., Jiang T., Deng R., et al. Inhibition of EZH2 and activation of ERRgamma synergistically suppresses gastric cancer by inhibiting FOXM1 signaling pathway. Gastric Cancer. 2021;24(1):72–84. doi: 10.1007/s10120-020-01097-x. [DOI] [PubMed] [Google Scholar]
- 41.Dai J., Su Y., Zhong S., Cong L., Liu B., Yang J., et al. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020;5(1):145. doi: 10.1038/s41392-020-00261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shang A., Gu C., Wang W., Wang X., Sun J., Zeng B., et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-beta1 axis. Mol. Cancer. 2020;19(1):117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu J., Wang Y.H., Yoon C., Huang X.Y., Xu Y., Xie J.W., et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. doi: 10.1016/j.canlet.2019.11.038. [DOI] [PubMed] [Google Scholar]
- 44.Shi Y., Fang N., Li Y., Guo Z., Jiang W., He Y., et al. Circular RNA LPAR3 sponges microRNA-198 to facilitate esophageal cancer migration, invasion, and metastasis. Cancer Sci. 2020;111(8):2824–2836. doi: 10.1111/cas.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang X., Li Z., Zhang Q., Wang W., Li B., Wang L., et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol. Cancer. 2019;18(1):71. doi: 10.1186/s12943-019-0969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quan X.Q., Xie Z.L., Ding Y., Feng R., Zhu X.Y., Zhang Q.X. miR-198 regulated the tumorigenesis of gastric cancer by targeting toll-like receptor 4 (TLR4) Eur. Rev. Med. Pharmacol. Sci. 2018;22(8):2287–2296. doi: 10.26355/eurrev_201804_14817. [DOI] [PubMed] [Google Scholar]
- 47.Li Y., Zhou H.C., Zhang Y., Huang H. microRNA-625-3p inhibits gastric cancer metastasis through modulating EZH2. Eur. Rev. Med. Pharmacol. Sci. 2020;24(3):1177–1185. doi: 10.26355/eurrev_202002_20169. [DOI] [PubMed] [Google Scholar]
- 48.Zhou X., Gao W., Hua H., Ji Z. LncRNA-BLACAT1 facilitates proliferation, migration and aerobic glycolysis of pancreatic cancer cells by repressing CDKN1C via EZH2-induced H3K27me3. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.539805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li P., Luo X., Xie Y., Li P., Hu F., Chu J., et al. GC-derived EVs enriched with microRNA-675-3p contribute to the MAPK/PD-L1-mediated tumor immune escape by targeting CXXC4. Mol. Ther. Nucleic Acids. 2020;22:615–626. doi: 10.1016/j.omtn.2020.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Byun H.J., Yoon J.H., Lee S.K. LUCAT1 epigenetically downregulates the tumor suppressor genes CXXC4 and SFRP2 in gastric cancer. Yonsei Med. J. 2020;61(11):923–934. doi: 10.3349/ymj.2020.61.11.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu Y., Wang Z., Luo C., Wang Y., Wang Y., Zhong X., et al. Downregulation of CXXC finger protein 4 leads to a tamoxifen-resistant phenotype in breast cancer cells through activation of the Wnt/beta-catenin pathway. Transl. Oncol. 2020;13(2):423–440. doi: 10.1016/j.tranon.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li L., Meng Q., Li G., Zhao L. BASP1 suppresses cell growth and metastasis through inhibiting Wnt/beta-catenin pathway in gastric cancer. Biomed. Res. Int. 2020;2020 doi: 10.1155/2020/8628695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li R., Li P., Wang J., Liu J. STIP1 down-regulation inhibits glycolysis by suppressing PKM2 and LDHA and inactivating the Wnt/beta-catenin pathway in cervical carcinoma cells. Life Sci. 2020;258 doi: 10.1016/j.lfs.2020.118190. [DOI] [PubMed] [Google Scholar]
- 54.Ke Y., Ning T., Wang B. [Establishment and characterization of a SV40 transformed human fetal gastric epithelial cell line-GES-1] Zhonghua Zhong Liu Za Zhi. 1994;16(1):7–10. [PubMed] [Google Scholar]
- 55.Wang B.J., Zhang Z.Q., Ke Y. Conversion of cadherin isoforms in cultured human gastric carcinoma cells. World J. Gastroenterol. 2006;12(6):966–970. doi: 10.3748/wjg.v12.i6.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.