Abstract
Animals have evolved sophisticated temperature-sensing systems and mechanisms to detect and respond to ambient temperature changes. As a relict species endemic to the Qinghai-Tibet Plateau, hot-spring snake (Thermophis baileyi) survived the dramatic changes in climate that occurred during plateau uplift and ice ages, providing an excellent opportunity to explore the evolution of temperature sensation in ectotherms. Based on distributional information and behavioral experiments, we found that T. baileyi prefer hot-spring habitats and respond more quickly to warmth than other two snakes, suggesting that T. baileyi may evolve an efficient thermal-sensing system. Using high-quality chromosome-level assembly and comparative genomic analysis, we identified cold acclimation genes experiencing convergent acceleration in high-altitude lineages. We also discovered significant evolutionary changes in thermosensation- and thermoregulation-related genes, including the transient receptor potential (TRP) channels. Among these genes, TRPA1 exhibited three species-specific amino acid replacements, which differed from those found in infrared imaging snakes, implying different temperature-sensing molecular strategies. Based on laser-heating experiments, the T. baileyi-specific mutations in TRPA1 resulted in an increase in heat-induced opening probability and thermal sensitivity of the ion channels under the same degree of temperature stimulation, which may help the organism respond to temperature changes more quickly. These results provide insight into the genetic mechanisms underpinning the evolution of temperature-sensing strategies in ectotherms as well as genetic evidence of temperature acclimation in this group.
Keywords: hot-spring snake, comparative genomics, TRP channel, thermosensation, thermoregulation
Graphical abstract

Public summary
-
•
Hot-spring snakes prefer hot-spring habitats on the Qinghai-Tibet Plateau
-
•
Genetic variation in the snakes contribute to the temperature acclimation
-
•
Unique mutations in TRPA1 increase thermal sensitivity of the ion channel
-
•
Different temperature-sensing strategies existed across snakes
Introduction
Ambient temperature is one of the most critical environmental factors for organisms affecting many fundamental biological processes such as growth, survival, and reproduction. To adapt to the diverse range of thermal environments in the biosphere, animals have evolved a variety of behavioral, physiological, and molecular strategies for thermoregulation, resulting in a set of physiological and ecological traits appropriate to their habitat.1 These thermoregulatory processes require sensitive thermal sensors, which are essential for detecting the spatiotemporal variation in environmental temperatures.1, 2, 3 Therefore, evolutionary changes in thermosensory systems may improve thermal perception and responses, thereby facilitating animal survival and adaptation to the corresponding thermal niches.
The Qinghai-Tibet Plateau (QTP), also called the “Third Pole,”4 contains unique biodiversity due to its remarkable orogenic history and significant glacial climate fluctuations.5,6 In addition to strong ultraviolet (UV) radiation and relatively low oxygen partial pressure, organisms living on the plateau are exposed to low temperatures and extreme temperature fluctuations. Plateau endotherms, such as wild yaks (Bos grunniens), Tibetan plateau pika (Ochotona curzoniae), and ground tit (Parus humilis) cope with cold by producing sufficient energy via lipid metabolism and by retaining heat through compact coverings.7, 8, 9 However, unfavorable conditions on the plateau pose great challenges for ectotherms given their dependence on environmental heat sources. Temperature sensing is a prerequisite for behavioral thermoregulation in ectotherms, which is critical for maintaining behavioral and physiological functions.10 The ectothermic vertebrates have evolved the ability to exploit the spatiotemporal distribution of environmental temperatures to maximize energy utilization and survival.11
The ectothermic hot-spring snake (Thermophis baileyi), which is endemic to the QTP, shows a strong preference for habitats with hot springs as an adaptive strategy to climate change.5,12, 13, 14 These snakes have the highest altitudinal distribution among reptiles, ranging from 3600 to 4900 m above sea level.12 During the QTP uplift and ice age, ancestral populations of these snakes are thought to have found glacial refuges near geothermal resources and hot springs and may have evolved unique adaptive strategies to cope with the major geological and climatic events.12 As such, these snakes provide an ideal opportunity to explore the genetic adaptations of ectotherms to extreme environments. We previously reported a draft genome of the hot-spring snake and revealed the molecular basis underlying UV and low-temperature adaptation.15 However, the genetic basis of temperature acclimation and the adaptive changes in thermosensory systems in hot-spring snakes remain unknown. Notably, the relatively fragmented draft genomes reported thus far may introduce certain biases and miss key signals.16
Here, by integrating distributional information and behavioral comparisons, we revealed the preference of T. baileyi for hot-spring habitats and their rapid response to warm stimuli in cold environments, suggesting their thermosensory systems may have evolved to cope with temperature fluctuations. We also established a high-quality chromosome-level genome assembly of the hot-spring snake to gain insight into thermosensory and thermoregulatory adaptation. Comparative genomic analysis was performed to explore the evolutionary changes in coding genes and conserved non-coding elements, with a focus on the molecular basis of thermal perception. Genomic and experimental evidence showed significant evolutionary changes in temperature response, thermoregulation, and transient receptor potential (TRP) channel-related genes. Upon heating, T. baileyi-specific functional mutations in TRPA1 increased the heat-induced opening probability and thermal sensitivity of the ion channels, which may help in discovering suitable habitats more rapidly.
Results
Hot-spring snakes prefer hot springs
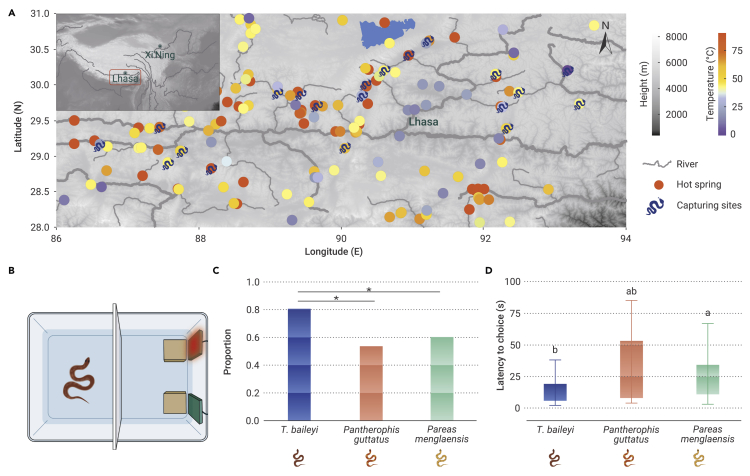
We evaluated the preference of hot-spring snakes for hot-spring environments by integrating the distributional coordinates of both (supplemental Tables 1 and 2). As seen in the distribution map, the hot-spring snakes showed a preference for habitats with hot springs, with roughly half occurring in 40°C–60°C thermal springs (Figure 1A; supplemental Table 1).
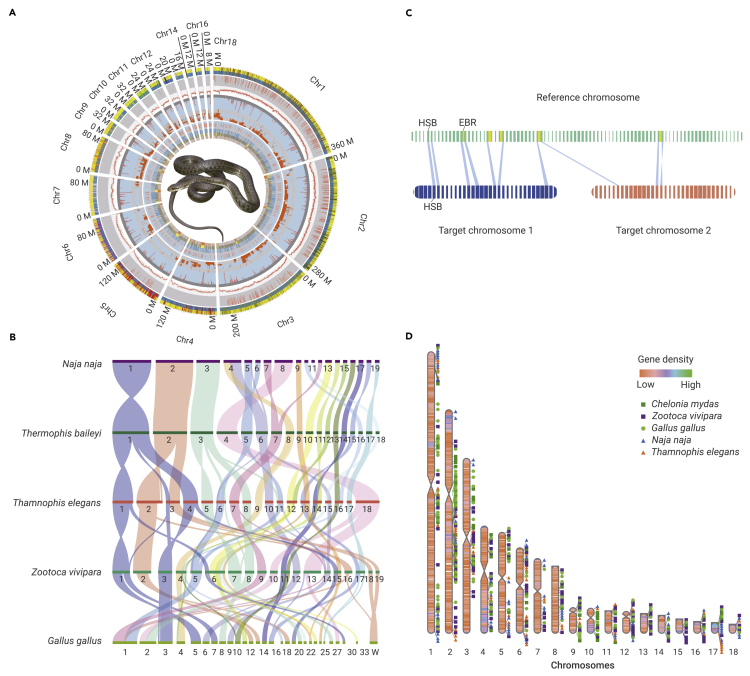
Figure 2.
Chromosome evolution of T. baileyi genome
(A) Circos plot of 18 chromosomes of T. baileyi. Protein-coding genes, highly divergent regions, GC-content, non-coding element regions, positively selected genes, and EBRs relative to Thamnophis elegans, Zootoca vivipara, and Gallus gallus are plotted on different levels of the circus from outer to inner.
(B) Synteny tracker of genomes. Synteny regions are colored according to chromosome of T. baileyi.
(C) Schematic of strategy to identify EBRs. An EBR was defined as an interval (in sulfur yellow) that is demarcated by the rearrangement of homology synteny blocks.
(D) EBR distribution in T. baileyi genome. The color bands on the chromosome marks the gene density in T. baileyi genome.
Figure 1.
Distribution of T. baileyi and thermotropic behavior experiments
(A) Geographical distribution of T. baileyi and hot springs on the QTP. Hot-spring temperatures are marked according to their highest records (supplemental Table 1). Distribution sites of T. baileyi are depicted with a snake.
(B) The schematic diagrams of the setup for the behavioral test (top view). We put the snake in a plastic box (L × W × H: 66 × 47 × 40 cm), which is divided into two halves (adaptation area and selection area) by a dummy plate (white). Two identical stones (brown) in the selection area serve as resting places for the snakes. Two heating pads are on the two stones, against the wall.
(C) The proportion of three kinds of snakes selecting high-temperature zone. ∗P < 0.05, exact binomial test.
(D) The latency of three kinds of snakes selecting high-temperature zone. Kruskal-Wallis one-way analysis of variance on ranks, different superscript letters indicate significant differences among different treatments (P < 0.05). T. baileyi, n = 36; Pantherophis guttatus, n = 15; Pareas menglaensis, n = 30.
It has been hypothesized that T. baileyi retreated to the micro-refugia created by hot springs and geothermal resources to survive changing climates, eg, during plateau uplift 20–60 million years ago.12 As such, they may have evolved efficient temperature-perception capabilities and unique thermoregulation mechanisms to detect suitable habitats and cope with low temperatures and temperature fluctuations. Thus, we assessed and compared the ability of hot-spring snakes to detect ambient temperature by recording their choices of warm environments in response to cold stimulation (Figure 1B). Compared with other two snakes, hot-spring snakes demonstrated a strong preference for the high-temperature zone (T. baileyi, 80.56%, Pantherophis guttatus, 53.33%, Pareas menglaensis, 60.00%; Figure 1C). Furthermore, hot-spring snakes located the warm environments more quickly (T. baileyi, 14.56 ± 10.86 s, Pantherophis guttatus, 30.53 ± 29.03 s, Pareas menglaensis, 24.6 ± 17.43 s; Figure 1D; supplemental Videos 1, 2, and 3). These findings suggest that T. baileyi may have evolved an efficient thermosensory system.
Genome assembly and chromosome evolution
In the current study, we sequenced the genome of a female hot-spring snake to explore the genetic changes in temperature-sensing systems using paired-end sequencing, single-molecule real-time sequencing, and Hi-C data (supplemental Table 3). A hybrid strategy (combined Illumina reads and PacBio long reads) was used to generate initial contigs, resulting in an assembly with a contig N50 of 4.02 Mb (supplemental Table 4) after polishing. The contigs were anchored onto chromosomes with Hi-C data, yielding a final chromosome-level assembly of 1.85 Gb with 91.47% of bases anchored onto 18 chromosomes (2n = 36) (Figure 2A and supplemental Figure 1). Based on Benchmarking Universal Single-Copy Ortholog (BUSCO) assessment of single-copy orthologous genes, genome completeness was 97.10% (supplemental Figure 2; supplemental Table 4). A total of 0.94 Gb of repetitive sequences were identified, accounting for 51.18% of the whole genome (supplemental Figure 3). We also annotated 22 292 coding genes. Compared with previously published snake genomes, the genome assembly was of relatively high quality (supplemental Figure 2; supplemental Tables 4 and 5).
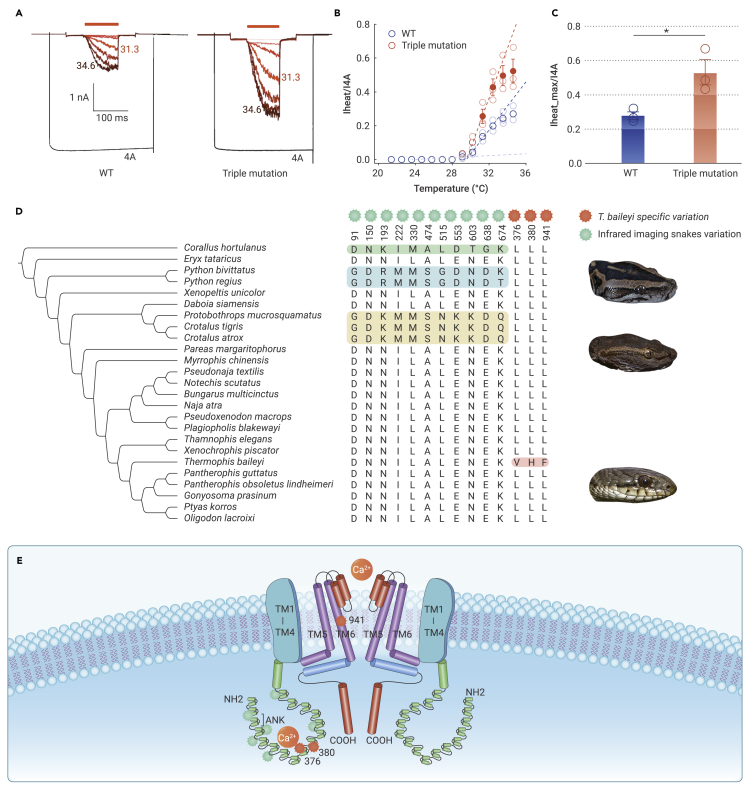
Figure 5.
Laser-heating experiment and functional comparison of T. baileyi TRPA1 gene
(A) Laser-induced heat activation of two types of TRPA1. 4-aminodiphe-nylamine is agonist of TRPA1.
(B) Thermal activation curve of wild-type TRPA1 and mutant TRPA1. Channel activity was initially small and steadily increased with subsequent stimulations. Channel activity of two types of TRPA1 showed different growth rates.
(C) Comparison of peak activity of two types of TRPA1. ∗P < 0.01.
(D) Variation in TRPA1 among snakes. The alignment in the left block shows specific variation in infrared imaging snakes, and the right block shows specific variation in T. baileyi.
(E) Schematic of TRPA1 transmembrane structure. TM1–TM6, transmembrane regions 1 to 6; ANK, Ankyrin repeats.
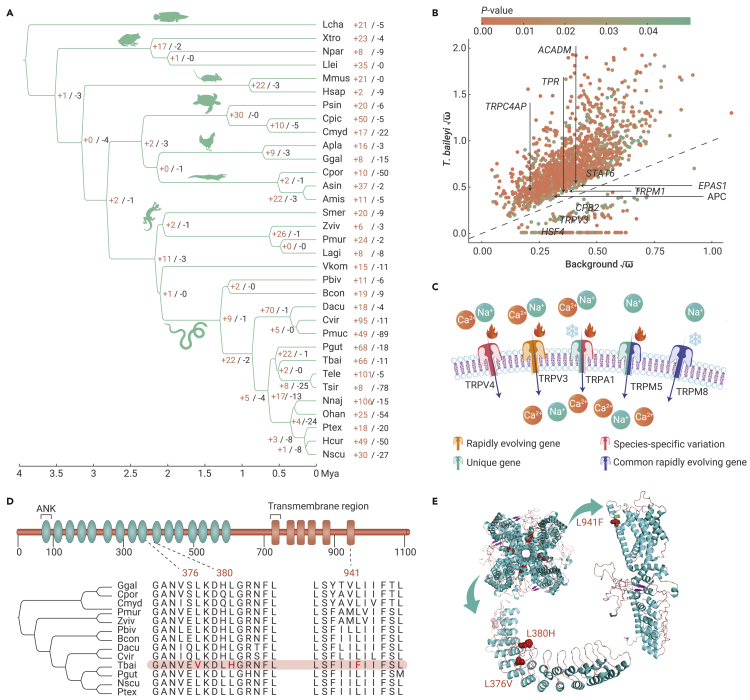
Chromosome evolution is a fundamental driving force of evolution and is related to genome size, gene family evolution, and speciation.17 Collinearity analysis found a high degree of collinearity within snakes but significant variation from birds and lizards (Figure 2B and supplemental Figure 4). Genes located within evolutionary breakpoint regions (EBRs) are associated with lineage-specific biology,18, 19, 20, 21 and 610 EBR genes were identified in T. baileyi (Figures 2C and 2D). Based on Gene Ontology (GO) enrichment analysis, we found that the EBR genes were significantly enriched in immune- and metabolism-related terms (supplemental Table 6). Furthermore, several genes were involved in DNA repair (eg, PAXX, PARK7, and TAOK1), hypoxia response (eg, ENDOG, PSD10, and PSMD5), and temperature acclimation (eg, TRPC5, EHMT1, MFAP2, and WNT10) (supplemental Table 7), suggesting that chromosome evolution in hot-spring snakes may be related to adaptation to high-altitude environments. However, the effects of chromosome evolution on the function of these genes and adaptive mechanisms of chromosome evolution in hot-spring snakes require further research.
Genome-wide convergent features within high-altitude lineages
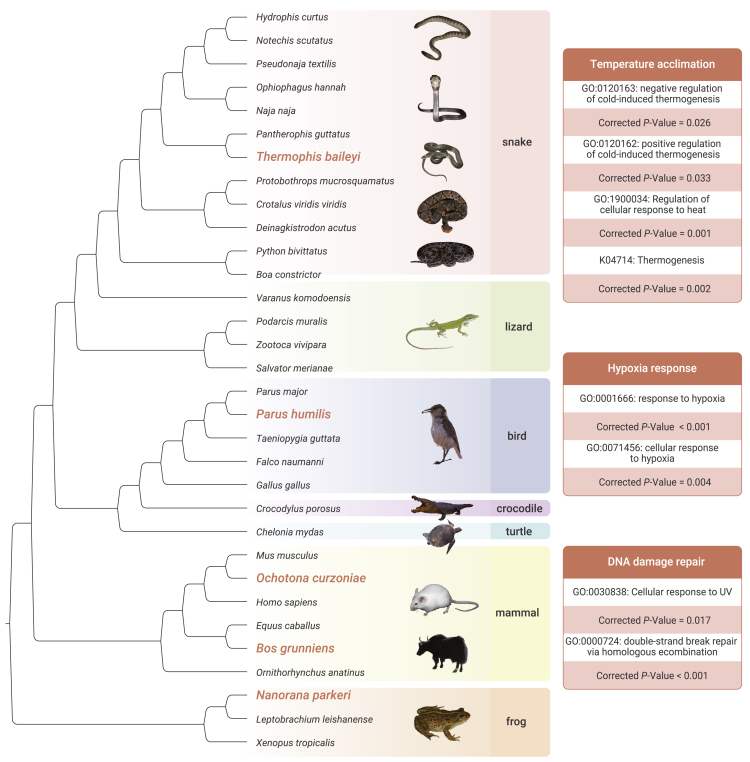
Expansion to the QTP occurred independently in vertebrates. Here, we explored convergent molecular signals associated with high-altitude adaptation between high-altitude endotherms (eg, yaks, Tibetan plateau pika, and ground tit) and ectotherms (T. baileyi and Nanorana parkeri). Little molecular convergence was identified at the sequence level (convergent amino acid site), yet there was convergent acceleration in gene-wide rates of molecular evolution in these species. After setting high-altitude species as foreground branches, a total of 2109 genes were identified as common rapidly evolving genes. Among them, 72 were assigned in temperature-acclimation-related pathways or GO terms, including thermogenesis (k04714, P adjusted = 0.002), regulation of cellular response to heat (GO: 1900034, P adjusted = 0.001), negative regulation of cold-induced thermogenesis (GO: 0120163, P adjusted = 0.026), and positive regulation of cold-induced thermogenesis (GO: 0120162, P adjusted = 0.033) (Figure 3; supplemental Tables 8 and 9). Within these terms, TRPM8 gene encodes a prototypical cold sensor in vertebrates, whose maximum cold activation is positively correlated with habitat temperature and is mainly determined by side-chain hydrophobicity and solvent accessibility in the pore domain.22 Furthermore, many identified genes have thermogenic function, such as Lpin1, Per2, and Lama4, which are involved in cold response by regulating brown adipose tissue.23, 24, 25 Brown adipose tissue is a unique thermogenic tissue in mammals,26 thus the function of these thermogenesis-related genes in birds and ectotherms needs further investigation. In addition, several genes involved in hypoxia response and DNA repair were also identified (Figure 3; supplemental Tables 8 and 9). For instance, Hus1−/− cells show heightened sensitivity to UV light, implicating its function in the maintenance of genomic stability and UV response;27 AMAD12 and AMAD17 are involved in hypoxia-induced impairment of neural vascular barrier function and are also considered as positively selected genes (PSGs) in yaks.28,29 These genes are critical for high-altitude adaptation, allowing animals to cope with extreme environmental stresses, such as low temperatures, strong UV radiation, and hypoxia.
Figure 3.
Gene-wide convergent features of high-altitude species
Five high-altitude species (orange) were set as foreground branches to detect rapidly evolving genes. Tree topology is supported by bootstrap supports of 100 for all nodes. The right panel is the Gene Ontology enrichment results.
Evolutionary changes associated with thermosensation in T. baileyi
Ectothermic vertebrates can exploit the distribution of ambient heat to regulate their own temperature and survive,11 which requires a sensitive thermosensory system. We performed comparative genomic analysis to explore the evolutionary changes in thermosensation- and thermoregulation-related genes. Here, we identified a set of 763 genes from 66 significantly expanded gene families (Figure 4A) involving basic biological processes, such as cold acclimation, cell differentiation, and development (supplemental Table 10). We then evaluated adaptive divergence in coding regions between hot-spring snakes and other Squamata species. A total of 1939 rapidly evolving genes (REGs) and 963 PSGs were detected in the hot-spring snake lineages using branch and branch-site models implemented in PAML.30 In addition to several genes related to DNA repair and hypoxia adaptation, which support life at high elevation, a set of temperature-acclimation-related genes were also identified (supplemental Table 7).
Figure 4.
Molecular adaptation of T. baileyi to hot-spring habitats
(A) Phylogenetic status of T. baileyi relative to other vertebrates. The number of significant expanded (orange) and contracted (black) gene families is designated on each node. The detailed species names are shown in the supplemental information.
(B) Non-synonymous/synonymous substitution ratio (dn/ds) of T. baileyi temperature-sensing genes compared with background species. P-values of chi-square from likelihood ratio tests were marked in different colors.
(C) Evolutionary changes in temperature-related TRP channel genes in hot-spring snake.
(D) Protein alignment and variations of TRPA1. The distributions of three amino acid replacements are marked.
(E) Three-dimensional structure of TRPA1 channel consists of four subunits. Red dots represent T. baileyi-specific replacements. The L941F is located on the TM6 domain, which is in proximity to the pore region of the homotetramer.
Among the REGs, 28 were associated with temperature sensing and response (Figure 4B; supplemental Table 7). For example, TRPV3 is activated at warm temperature (33°C) and is responsible for thermotaxis, with this ability reported to be impaired in TRPV3−/− mice.31,32 CPB2 (HSP47) is involved in heat-stress responses,33 and HSF4 is required for induction of certain non-classic heat-shock genes.34 Among the PSGs, nine were associated with temperature response (supplemental Table 7), eg, CAMK2 is involved in the transmission of the signaling response to cold stress.35,36 Taken together, the genes involved in thermosensation, thermotaxis, and heat/cold-stress response appear to have undergone adaptive changes, which may have contributed to species adaptation to the extreme plateau climate.
Evolution of temperature-sensing TRP channel genes
As TRP channel genes are considered important signal transducers for thermoreceptors,37 we explored the features of all TRP channel genes in our analysis results. A total of 21 TRP channel genes were annotated in the genome, 12 of which exhibited T. baileyi-specific features (supplemental Table 11). In addition to the TRP channel genes mentioned above, TRPC4AP, TRPC7, and TRPM1 showed rapid evolution; TRPA1, TRPC7, TRPM5, and TRPM7 were identified as T. baileyi unique genes; and TRPV4AP also showed positive selection. Furthermore, T. baileyi-specific replacements in TRPV4 and TRPA1 were identified. TRPM7 is located in the 20 kb region up- and downstream of rapidly evolving non-coding elements, which were likely serve as proximal cis-regulatory elements.
Among these TRP channel genes, TRPA1, TRPV4, TRPV3, TRPC5, TRPM5, and TRPM8 are temperature-sensing-related genes (Figure 4C, supplemental Table 11).37 TRPC5 and TRPM8 are involved in cold sensation.38,39 The TRP vanilloid (TRPV) channel family is activated by temperature,40 and both TRPV3 and TRPV4 are activated under warm conditions (26°C–39°C).37 TRPM5 is thought to be activated by warm temperature.37 TRPA1 is also a key molecular sensor involved in temperature detection.37,41,42 As ion channels, TRPs can be gated directly and cause action potentials by sensory stimuli.32 The extensive evolutionary changes in hot-spring snake TRP channel genes suggest adaptive changes in their perception systems, including thermosensation.
We next performed functional prediction to evaluate the role of species-specific replacements in TRPA1. Three T. baileyi-specific replacement sites were identified in the TRPA1 protein, which were found to be highly conserved in snakes (Figure 4D and supplemental Figure 5). Two replacements (L376V and L380H) were located on the intramembranous region of the transmembrane protein, ie, within the ninth and tenth ankyrin repeats domain (ANK). Another replacement (L941F) was located on the sixth membrane-spanning α-helical domain (transmembrane region 6 [TM6]) (Figures 4D and 4E). Two pore helices linked the fifth and sixth TM regions at the extracellular surface, which regulates cation influx.43 Three-dimensional structure prediction indicated that the structures between Pantherophis guttatus and T. baileyi were conserved (template modeling score >0.5) and the replacement sites may change the spatial conformation of TM5 and TM6, as well as the ANK “tail” (Figure S6). Functional prediction was performed using PROVEAN v.1.1.5 (Protein Variation Effect Analyzer),44 which classified two replacements (L380H and L941F) as intolerant, suggesting that these replacements would likely result in functional changes in TRPA1 (Table S12). A T. baileyi-specific replacement (T500K) located on the ion transport domain of TRPV4 was also discovered (Figure S7). These results suggest that changes in thermotaxis- and temperature-response-related genes have occurred in hot-spring snakes, potentially providing powerful temperature detection and response capabilities to cope with extreme environments. Thus, we performed functional experiments to test how these changes affect the thermosensory system.
Laser-heating experiments and functional comparison of TRPA1 gene
TRPA1 is a heat-sensitive ion channel and contributes to thermal detection and thermotaxis behavior in reptiles.10,37,45 We performed electrophysiological and laser irradiation experiments to examine divergence between wild-type and mutant TRPA1 (ie, with three T. baileyi-specific replacements) in response to heat. Figure 5A shows the temperature responses of the two types of TRPA1 after repeated stimulation with different temperatures produced by laser irradiation.46 The TRPA1 agonist (4-aminodiphe-nylamine) is potent against reptile TRPA1 orthologs,10 activating the channel with a high probability of opening in the absence of heat (Figure 5A). Whole-cell recordings indicated that heat activation of TRPA1 was normalized to agonist-induced currents (Figure 5A). We found that the temperature threshold (∼29°C–30°C) was comparable between the wild-type and mutated TRPA1 (Figure 5B). Subsequently, the peak currents of the wild-type and mutant channels significantly increased upon heating (Figure 5B). We then compared the current amplitudes at the end of each stimulus. Results showed that heat sensitivity of the TRPA1 mutant was higher than that of the wild type (Figure 5C). Functional analysis showed that the three T. baileyi-specific replacements in the TRPA1 did not alter the temperature-response threshold values but increased heat sensitivity to some extent.
The molecular mechanism of TRPA1 in thermal detection in hot-spring snakes may differ from that of snakes with pit organs. In infrared imaging snakes, TRPA1 shows an extremely robust response to infrared heat sources, mainly by lowering the thermal activity threshold.45 Interestingly, the positions of the replacement sites in hot-spring snake differed from the specific amino acid replacements in infrared imaging snakes (Figure 5D). Most infrared imaging snake-specific sites are distributed in the N-terminal domain (ANK1 to TM1) (Figure 5E). Structural prediction suggests that their structures are conserved (template modeling score >0.5) but with spatial conformation differences within their ANK tail (supplemental Figure 8), suggesting a potential region responsible for infrared detection capacity.47 The T. baileyi-specific amino acid replacements on TM6 may alter the conformation of the ion channel constructed by TM5 and TM6 and, in turn, alter the openness under the same temperature stimulus (Figures 5D and 5E; supplemental Figure 6). The other two replacements located within the ANK region may provide a cytoplasmic surface for interactions with ligands and conformational changes, thereby opening the channel.43
Genomic adaptation in other temperature-acclimation genes
Neural circuit mechanisms are critical for thermoregulation, acting as a signal carrier in thermosensation.48 Here, we identified 2437 unique genes that overlapped with T. baileyi-unique regions, with several significantly enriched terms related to neurons and synapses (eg, neuron projection, nervous system development, synapse, and neuron projection morphogenesis) (supplemental Table 13). We also found that several genes regulated by divergent non-coding elements were associated with neurogenesis (eg, synapse assembly, nervous system development, and neuron development) (supplemental Table 14). These results indicate that the evolution of both coding and non-coding elements may have contributed to the physiological basis of temperature perception. Although hot springs provide microhabitats and environments suitable for Thermophis species, those with excessively high temperatures may be a risk. We identified several genes involved in the transmission of thermal pain-evoked signals, such as PRDM12 (EBR gene) and OPRK1 (PSG) (supplemental Table 7). Previous studies have shown that individuals with PRDM12 mutations exhibit impaired noxious heat or cold pain sensitivity49 and that OPRK1 knockout mice exhibit enhanced thermal hyperalgesia.50, 51, 52 Thus, hot-spring snakes may have evolved sensitive sensory systems to avoid harmful temperature stimuli.
Discussion
The QTP is considered a “natural laboratory” for research on adaptations to extreme environments.4,6 In this study, we demonstrated the thermal sensitivity of hot-spring snakes through behavioral experiments and discovered extensive evolutionary changes in genes associated with thermal adaptation based on a high-quality genome assembly. These findings were not detected in our earlier draft genome,15 which was primarily due to the incompleteness of the assembly and annotation (eg, TRPA1 and TRPM8). The improved high-quality genome provides an important genetic resource for studying the biological characteristics of this rare species.
Similar environmental stress may drive species to acquire convergent phenotypes or physiology. In this study, little convergent amino acid replacement was identified at sequence level, but convergent signatures of acceleration in gene-wide rates of molecular evolution were detected across high-altitude endotherms and ectotherms, including hypoxia response, cold response, and UV-induced DNA-damage repair, which are necessary to cope with extreme altitude environment. A study on the high-altitude domestic mammals found similar patterns: little convergent evolution at the DNA sequence level occurring among different high-altitude domestic mammals, but convergent signature of positive selection in genes associated with hypoxia response existed.53 This suggests that although endotherms and ectotherms have very different physiological characteristics, they tend to change identical genes in order to respond to similar environmental stresses. Temperature-induced adaptation is often synergistic with physiological regulation,11 which is conducive to cold acclimation. In this study, we identified several REGs, PSGs, and common REGs involved in thermogenesis. Some of these genes, such as TBC1D7, UTP15, and GCLC were also identified as PSGs in opah, a warm-blooded fish,54 suggesting their role in thermal adaptations in vertebrates. However, the function of these genes has been elucidated only in endotherms, remaining unknown in ectothermic reptiles, especially the effects of cold-induced thermogenesis-related genes, and thus require further investigation.
Cold stress requires organisms to evolve sensitive thermal sensors and regulatory mechanisms, which is important for ectotherms that rely on environmental heat sources. TRP channels are an important class of molecules that contribute to thermosensation in vertebrates.32,55 Here, extensive evolutionary changes in TRP channels were identified in hot-spring snakes, implying the evolution of their temperature-sensing system. TRPA1 mediates thermal detection and thermotaxis behavior.10,37,45,56 Our results demonstrated that minimal changes in protein sequence (three amino acid replacements) are sufficient to generate a wide diversity of heat-induced opening probabilities and thermal sensitivities in TRPA1.57 This finding is consistent with the behavioral experiments, which showed strong and sensitive thermotaxis in the hot-spring snakes. Furthermore, TRPA1 also mediates the perception of the rate of temperature change,55 which may help organisms respond to temperature changes more quickly.
The differences in TRPA1 mutations and function between T. baileyi and infrared imaging snakes further suggest the adoption of two distinct molecular-sensing strategies, one of which modulates the temperature-sensing range by reducing the minimum activation threshold (eg, infrared imaging snakes),45 and the other of which adjusts heat-induced opening probability and thermal sensitivity of TRPA1, thus responding more quickly and sensitively to temperature changes. Structural variations of TRPA1 ion channel determine its diverse function and performance. We found that features between the two molecular-sensing strategies were mainly characterized by spatial conformation changes in ANK tail and TM5–TM6 domain. ANK tail is responsible for the diverse functions of proteins, such as ion transporters and signal transducers, and are important for the function of thermal nociception in Drosophila.58,59 ANK can communicate with the pore and be responsible for thermal or chemical responsiveness.60 The pore of TRPA1 is formed by the pore-forming helices in TM6 of the tetrameric channel, and point mutations in TM6 would reduce heat sensitivity.61,62 Given that changes at the TRPA1 sequence level may mediate different response mechanisms, it is important to determine the functional consequences of TRPA1 mutations and associated physiological processes in future studies. Overall, evolutionary changes in the thermosensory system of hot-spring snakes may have contributed to their survival in hot-spring refuges under changing geological and climatic conditions, as well as their adaptation to high-altitude plateau environments under temperature fluctuation.
Methods
Data and code availability
The genome assembly and raw sequencing data have been deposited in National Genomics Data Center (NGDC) (https://ngdc.cncb.ac.cn) under NGDC: PRJCA007342 (GWH: GWHBJWY00000000, GSA: CRA006131, GSA: CRA006150).
Experimental model and subject details
This study was approved by the Animal Ethics Committee of the CIB, and animal experiments were carried out in line with the institution’s guidelines.
Acknowledgments
We thank Hussam Zaher (University of São Paulo) for his advices on languages. We thank Chang-Jun Peng (Chengdu Institute of Biology, Chinese Academy of Sciences) for assistance with data analysis. We thank Jin-Long Ren (Chengdu Institute of Biology, Chinese Academy of Sciences) for his help in fieldwork. This work was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (CAS) (XDB31000000); the National Natural Science Foundation of China (32100396); the Second Tibetan Plateau Scientific Expedition and Research Program (STEP) (2019QZKK0501); Biodiversity Survey, Monitoring and Assessment Project of Ministry of Ecology and Environment, The People’s Republic of China, China (2019HB2096001006); Key Research Program of Frontier Sciences, CAS (QYZDB-SSW-SMC058); the International Partnership Program of Chinese Academy of Sciences (151751KYSB20190024); and the Sichuan Science and Technology Program (2021JDJQ0002; 2021YJ0088).
Author contributions
J.-T.L. initiated and conceived the current work. C.Y., W.W., and Y.L., completed omics data analysis. W.D., and S.Y., performed the electrophysiology and laser irradiation experiments. B.Z. performed the behavioral experiments. J.C. provided important suggestions. W.W., C.Y., B.Z., and J.-T.L. wrote the manuscript, with input from all authors.
Declaration of interests
The authors declare no competing interests.
Published Online: August 1, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xinn.2022.100295.
Lead contact website
http://sourcedb.cib.cas.cn/zw/rck/201303/t20130318_3795275.html.
Supplemental information
References
- 1.Saito S., Tominaga M. Evolutionary tuning of TRPA1 and TRPV1 thermal and chemical sensitivity in vertebrates. Temperature. 2017;4:141–152. doi: 10.1080/23328940.2017.1315478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saito S., Ohkita M., Saito C.T., et al. Evolution of heat sensors drove shifts in thermosensation between Xenopus species adapted to different thermal niches. J. Biol. Chem. 2016;291:11446–11459. doi: 10.1074/jbc.M115.702498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laursen W.J., Schneider E.R., Merriman D.K., et al. Low-cost functional plasticity of TRPV1 supports heat tolerance in squirrels and camels. Proc. Natl. Acad. Sci. USA. 2016;113:11342–11347. doi: 10.1073/pnas.1604269113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wingfield J.C., Patrick Kelley J., Angelier F., et al. Organism-environment interactions in a changing world: a mechanistic approach. J. Ornithol. 2011;152:S279–S288. [Google Scholar]
- 5.Hofmann S. Population genetic structure and geographic differentiation in the hot spring snake Thermophis baileyi (Serpentes, Colubridae): indications for glacial refuges in southern-central Tibet. Mol. Phylogenet. Evol. 2012;63:396–406. doi: 10.1016/j.ympev.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Deng T., Wu F., Zhou Z., Su T. Tibetan Plateau: an evolutionary junction for the history of modern biodiversity. Sci. China Earth Sci. 2020;63:172–187. [Google Scholar]
- 7.Li J., Yang Q., Bai Z., et al. Chronic cold exposure results in subcutaneous adipose tissue browning and altered global metabolism in Qinghai-Tibetan plateau pika (Ochotona curzoniae) Biochem. Biophys. Res. Commun. 2018;500:117–123. doi: 10.1016/j.bbrc.2018.03.147. [DOI] [PubMed] [Google Scholar]
- 8.Jia C., Wang H., Li C., et al. Genome-wide detection of copy number variations in polled yak using the Illumina BovineHD BeadChip. BMC Genom. 2019;20:376. doi: 10.1186/s12864-019-5759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu Y., Zhao H., Han N., et al. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat. Commun. 2013;4:2071. doi: 10.1038/ncomms3071. [DOI] [PubMed] [Google Scholar]
- 10.Ye Y.Z., Zhang H., Li J., et al. Molecular sensors for temperature detection during behavioral thermoregulation in turtle embryos. Curr. Biol. 2021;31:2995–3003.e4. doi: 10.1016/j.cub.2021.04.054. [DOI] [PubMed] [Google Scholar]
- 11.Hutchison V.H., Maness J.D. The role of behavior in temperature acclimation and tolerance in ectotherms. Am. Zool. 1979;19:367–384. [Google Scholar]
- 12.Cloudsley-Thompson J. The ecological specialist, Thermophis baileyi (Wall, 1907) – new records, distribution and biogeographic conclusions. Herpetol. Bull. 2007;101:8–12. [Google Scholar]
- 13.Huang S., Liu S.y., Guo P., et al. What are the closest relatives of the hot-spring snakes (Colubridae, Thermophis), the relict species endemic to the Tibetan Plateau? Mol. Phylogenet. Evol. 2009;51:438–446. doi: 10.1016/j.ympev.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Ren J.-L., Yan C., Peng Z.-L., Li J.-T. Sichuan hot-spring snakes imperiled: reason, situation, and protection. Zool. Res. 2022;43:95–97. doi: 10.24272/j.issn.2095-8137.2021.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J.-T., Gao Y.D., Xie L., et al. Comparative genomic investigation of high-elevation adaptation in ectothermic snakes. Proc. Natl. Acad. Sci. USA. 2018;115:8406–8411. doi: 10.1073/pnas.1805348115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallick S., Gnerre S., Muller P., Reich D. The difficulty of avoiding false positives in genome scans for natural selection. Genome Res. 2009;19:922–933. doi: 10.1101/gr.086512.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eichler E.E., Sankoff D. Structural dynamics of eukaryotic chromosome evolution. Science. 2003;301:793–797. doi: 10.1126/science.1086132. [DOI] [PubMed] [Google Scholar]
- 18.Farré M., Kim J., Proskuryakova A.A., et al. Evolution of gene regulation in ruminants differs between evolutionary breakpoint regions and homologous synteny blocks. Genome Res. 2019;29:576–589. doi: 10.1101/gr.239863.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groenen M.A.M., Archibald A.L., Uenishi H., et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491:393–398. doi: 10.1038/nature11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ullastres A., Farré M., Capilla L., Ruiz-Herrera A. Unraveling the effect of genomic structural changes in the rhesus macaque - implications for the adaptive role of inversions. BMC Genom. 2014;15:530. doi: 10.1186/1471-2164-15-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farré M., Narayan J., Slavov G.T., et al. Novel insights into chromosome evolution in birds, archosaurs, and reptiles. Genome Biol. Evol. 2016;8:2442–2451. doi: 10.1093/gbe/evw166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang S., Lu X., Wang Y., et al. A paradigm of thermal adaptation in penguins and elephants by tuning cold activation in TRPM8. Proc. Natl. Acad. Sci. USA. 2020;117:8633–8638. doi: 10.1073/pnas.1922714117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadra K., Médard J.J., Mul J.D., et al. Cell autonomous Lipin 1 function is essential for development and maintenance of white and Brown adipose tissue. Mol. Cell Biol. 2012;32:4794–4810. doi: 10.1128/MCB.00512-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chappuis S., Ripperger J.A., Schnell A., et al. Role of the circadian clock gene Per2 in adaptation to cold temperature. Mol. Metab. 2013;2:184–193. doi: 10.1016/j.molmet.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaicik M.K., Blagajcevic A., Ye H., et al. The absence of laminin a4 in male mice results in enhanced energy expenditure and increased beige subcutaneous adipose tissue. Endocrinology. 2018;159:356–367. doi: 10.1210/en.2017-00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ballinger M.A., Andrews M.T. Nature’s fat-burning machine: Brown adipose tissue in a hibernating mammal. J. Exp. Biol. 2018;121:1–10. doi: 10.1242/jeb.162586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss R.S., Enoch T., Leder P. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 2000;14:1886–1898. [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu Q., Zhang G., Ma T., et al. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012;44:946–949. doi: 10.1038/ng.2343. [DOI] [PubMed] [Google Scholar]
- 29.Cui D., Arima M., Takubo K., et al. ADAM12 and ADAM17 are essential molecules for hypoxia-induced impairment of neural vascular barrier function. Sci. Rep. 2015;5:12796. doi: 10.1038/srep12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 31.Moqrich A., Hwang S.W., Earley T.J., et al. Impaired thermosensation in mice lacking TRPV3, a heat and camphor sensor in the skin. Science. 2005;307:1468–1472. doi: 10.1126/science.1108609. [DOI] [PubMed] [Google Scholar]
- 32.Dhaka A., Viswanath V., Patapoutian A. TRP ion channels and temperature sensation. Annu. Rev. Neurosci. 2006;29:135–161. doi: 10.1146/annurev.neuro.29.051605.112958. [DOI] [PubMed] [Google Scholar]
- 33.Abdelnasir A., Sun J.R., Cheng Y.F., et al. Evaluation of Hsp47 expression in heat-stressed rat myocardial cells in vitro and in vivo. Genet. Mol. Res. 2014;13:10787–10802. doi: 10.4238/2014.December.18.20. [DOI] [PubMed] [Google Scholar]
- 34.Fujimoto M., Oshima K., Shinkawa T., et al. Analysis of HSF4 binding regions reveals its necessity for gene regulation during development and heat shock response in mouse lenses. J. Biol. Chem. 2008;283:29961–29970. doi: 10.1074/jbc.M804629200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu D., You Q., Chi C., et al. Transcriptional response to low temperature in the yellow drum (Nibea albiflora) and identification of genes related to cold stress. Comp. Biochem. Physiol. Part D: Genomics Proteomics. 2018;28:80–89. doi: 10.1016/j.cbd.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Ju Z., Dunham R.A., Liu Z. Differential gene expression in the brain of channel catfish (Ictalurus punctatus) in response to cold acclimation. Mol. Genet. Genomics. 2002;268:87–95. doi: 10.1007/s00438-002-0727-9. [DOI] [PubMed] [Google Scholar]
- 37.Xiao R., Xu X.Z.S. Temperature sensation: from molecular thermosensors to neural circuits and coding principles. Annu. Rev. Physiol. 2021;83:205–230. doi: 10.1146/annurev-physiol-031220-095215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brauchi S., Orio P., Latorre R. Clues to understanding cold sensation: thermodynamics and electrophysiological analysis of the cold receptor TRPM8. Proc. Natl. Acad. Sci. USA. 2004;101:15494–15499. doi: 10.1073/pnas.0406773101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zimmermann K., Lennerz J.K., Hein A., et al. Transient receptor potential cation channel, subfamily C, member 5 (TRPC5) is a cold-transducer in the peripheral nervous system. Proc. Natl. Acad. Sci. USA. 2011;108:18114–18119. doi: 10.1073/pnas.1115387108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu B., Yao J., Zhu M.X., Qin F. Hysteresis of gating underlines sensitization of TRPV3 channels. J. Gen. Physiol. 2011;138:509–520. doi: 10.1085/jgp.201110689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caspani O., Heppenstall P.A. TRPA1 and cold transduction: an unresolved issue? J. Gen. Physiol. 2009;133:245–249. doi: 10.1085/jgp.200810136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cortright D.N., Krause J.E., Broom D.C. TRP channels and pain. Biochim. Biophys. Acta. 2007;1772:978–988. doi: 10.1016/j.bbadis.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Paulsen C.E., Armache J.P., Gao Y., et al. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature. 2015;520:511–517. doi: 10.1038/nature14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi Y., Sims G.E., Murphy S., et al. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gracheva E.O., Ingolia N.T., Kelly Y.M., et al. Molecular basis of infrared detection by snakes. Nature. 2010;464:1006–1011. doi: 10.1038/nature08943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao J., Liu B., Qin F. Kinetic and energetic analysis of thermally activated TRPV1 channels. Biophys. J. 2010;99:1743–1753. doi: 10.1016/j.bpj.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geng J., Liang D., Jiang K., Zhang P. Molecular evolution of the infrared sensory gene TRPA1 in snakes and implications for functional studies. PLoS One. 2011;6:e28644. doi: 10.1371/journal.pone.0028644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liedtke W.B. Deconstructing mammalian thermoregulation. Proc. Natl. Acad. Sci. USA. 2017;114:1765–1767. doi: 10.1073/pnas.1620579114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y.C., Auer-Grumbach M., Matsukawa S., et al. Transcriptional regulator PRDM12 is essential for human pain perception. Nat. Genet. 2015;47:803–808. doi: 10.1038/ng.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmisano M., Caputi F.F., Mercatelli D., et al. Dynorphinergic system alterations in the corticostriatal circuitry of neuropathic mice support its role in the negative affective component of pain. Genes Brain Behav. 2019;18:e12467. doi: 10.1111/gbb.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu M., Petraschka M., McLaughlin J.P., et al. Neuropathic pain activates the endogenous κ opioid system in mouse spinal cord and induces opioid receptor tolerance. J. Neurosci. 2004;24:4576–4584. doi: 10.1523/JNEUROSCI.5552-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aita M., Byers M.R., Chavkin C., Xu M. Trigeminal injury causes kappa opioid-dependent allodynic, glial and immune cell responses in mice. Mol. Pain. 2010;6:8. doi: 10.1186/1744-8069-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu D.D., Yang C.P., Wang M.S., et al. Convergent genomic signatures of high-Altitude adaptation among domestic mammals. Natl. Sci. Rev. 2020;7:952–963. doi: 10.1093/nsr/nwz213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X., Qu M., Liu Y., et al. Genomic basis of evolutionary adaptation in a warm-blooded fish. Innovation. 2022;3:100185. doi: 10.1016/j.xinn.2021.100185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo J., Shen W.L., Montell C. TRPA1 mediates sensation of the rate of temperature change in Drosophila larvae. Nat. Neurosci. 2017;20:34–41. doi: 10.1038/nn.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Corfas R.A., Vosshall L.B. The cation channel TRPA1 tunes mosquito thermotaxis to host temperatures. Elife. 2015;4:e11750. doi: 10.7554/eLife.11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jabba S., Goyal R., Sosa-Pagán J.O., et al. Directionality of temperature activation in mouse TRPA1 ion channel can be inverted by single-point mutations in ankyrin repeat six. Neuron. 2014;82:1017–1031. doi: 10.1016/j.neuron.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hwang R.Y., Stearns N.A., Tracey W.D. The ankyrin repeat domain of the TRPA protein painless is important for thermal nociception but not mechanical nociception. PLoS One. 2012;7:e30090. doi: 10.1371/journal.pone.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bork P. Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins. 1993;17:363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- 60.Cordero-Morales J.F., Gracheva E.O., Julius D. Cytoplasmic ankyrin repeats of transient receptor potential A1 (TRPA1) dictate sensitivity to thermal and chemical stimuli. Proc. Natl. Acad. Sci. USA. 2011;108:E1184–E1191. doi: 10.1073/pnas.1114124108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meents J.E., Ciotu C.I., Fischer M.J.M. Trpa1: a molecular view. J. Neurophysiol. 2019;121:427–443. doi: 10.1152/jn.00524.2018. [DOI] [PubMed] [Google Scholar]
- 62.Wang H., Schupp M., Zurborg S., Heppenstall P.A. Residues in the pore region of Drosophila transient receptor potential A1 dictate sensitivity to thermal stimuli. J. Physiol. 2013;591:185–201. doi: 10.1113/jphysiol.2012.242842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome assembly and raw sequencing data have been deposited in National Genomics Data Center (NGDC) (https://ngdc.cncb.ac.cn) under NGDC: PRJCA007342 (GWH: GWHBJWY00000000, GSA: CRA006131, GSA: CRA006150).