Abstract
Sporulation mutants of Streptomyces coelicolor appear white because they are defective in the synthesis of the grey polyketide spore pigment, and such white (whi) mutants had been used to define eight sporulation loci, whiA, whiB, whiD, whiE, whiG, whiH, whiI, and whiJ (K. F. Chater, J. Gen. Microbiol. 72:9–28, 1972; N. J. Ryding, Ph.D. thesis, University of East Anglia, 1995). In an attempt to identify new whi loci, we mutagenized S. coelicolor M145 spores with nitrosoguanidine and identified 770 mutants with colonies ranging from white to medium grey. After excluding unstable strains, we examined the isolates by phase-contrast microscopy and chose 115 whi mutants with clear morphological phenotypes for further study. To exclude mutants representing cloned whi genes, self-transmissible SCP2*-derived plasmids carrying whiA, whiB, whiG, whiH, or whiJ (but not whiD, whiE, or whiI) were introduced into each mutant by conjugation, and strains in which the wild-type phenotype was restored either partially or completely by any of these plasmids were excluded from further analysis. In an attempt to complement some of the remaining 31 whi mutants, an SCP2* library of wild-type S. coelicolor chromosomal DNA was introduced into 19 of the mutants by conjugation. Clones restoring the wild-type phenotype to 12 of the 19 strains were isolated and found to represent five distinct loci, designated whiK, whiL, whiM, whiN, and whiO. Each of the five loci was located on the ordered cosmid library: whiL, whiM, whiN, and whiO occupied positions distinct from previously cloned whi genes; whiK was located on the same cosmid overlap as whiD, but the two loci were shown by complementation to be distinct. The phenotypes resulting from mutations at each of these new loci are described.
In the filamentous, gram-positive bacterium Streptomyces coelicolor, dispersal is achieved through a simple differentiation process that results in the release of exospores (7). During this process, multigenomic aerial hyphae divide into unigenomic prespore compartments by synchronized multiple septation at regular intervals along their length. These cylindrical prespore compartments subsequently mature to give rise to chains of 50 to 100 ovoid, thick-walled spores. During this maturation phase, colonies develop a grey color, due to the synthesis of a polyketide spore pigment.
This pigmentation has been exploited to isolate developmental mutants; all of the original sporulation-deficient mutants were identified by virtue of their inability to synthesize wild-type levels of the spore pigment, resulting in colonies that remained white, even on prolonged incubation (18). Fifty of these white (whi) mutants were analyzed genetically, and current information suggests that they represent eight separate loci, whiA, whiB, whiD, whiE, whiG, whiH, whiI, and whiJ (6, 9, 31). Of these, whiE is a complex locus which encodes the enzymes that synthesize the spore pigment itself, and whiE mutants do not appear to be morphologically defective (6, 12, 25). whiA, whiB, and whiG mutants are completely blocked in sporulation septation and are white in appearance. whiH and whiI mutants produce some sporulation septa; while whiI mutants are completely white, whiH mutants are pale grey and make low but detectable levels of the whiE transcripts that specify the spore pigment (6, 7, 9, 22, 25, 31, 32). whiJ mutants produce low numbers of apparently normal spore chains, are pale grey, and again make low but detectable levels of the whiE transcripts (22, 31). Mutation in the remaining locus, whiD, causes the formation of spores of highly irregular size and shape which are defective in wall thickening and lyse extensively (6, 25, 28). Two other loci, whiC and whiF, are no longer included: the only known whiC mutant has been lost, and whiF99 was shown to be an unusual allele of whiG (5, 31). The eight known whi loci have all been cloned and sequenced (1, 11–13, 27, 31, 32).
Chater (6) suggested that the genetic map of whi loci might not be saturated on the grounds that several loci are poorly represented in the original collection of whi mutants analyzed; examples are whiD (one allele), whiB (two alleles), whiE (two alleles) and whiJ (two alleles). The subsequent discovery by reverse genetics of the involvement of the sigF locus in sporulation supported this viewpoint (21, 22, 29). As a consequence, we have attempted to identify novel whi loci through the isolation of new whi mutants and their complementation, taking advantage of previously cloned whi genes to exclude known loci from the screen. We report the identification, phenotypic characterization, mapping, and cloning of five new whi loci.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, protoplast transformation, and chemical mutagenesis.
S. coelicolor A3(2) strains used were the wild-type strain 1147 (prototrophic, SCP1+ SCP2+ Pgl+ [19]), M145 (prototrophic, SCP1− SCP2− Pgl+ [19]), J1501 (hisA1 uraA1 strA1 SCP1− SCP2− Pgl− [10]), 1258 (proA1 hisC9 argA1 cysD18 uraA1 strA1 SCP1NF [SCP2 status uncertain] Pgl+ [19]), and J243 (uraA1 strA1 whiD16 SCP1+ [SCP2 status uncertain] Pgl+ [8]). S. coelicolor strains were cultured on minimal medium MM (19) containing 0.5% (wt/vol) mannitol as the carbon source or on MS (mannitol plus soya flour) agar (17). Protoplasts were made and transformed as described by Hopwood et al. (19). S. coelicolor M145 spores were mutagenized with nitrosoguanidine (NTG) as described previously (14, 18). Plasmids used were pIJ698 (23) and pSET152 (4).
Construction of a genomic library and complementation of mutants.
Total chromosomal DNA from wild-type S. coelicolor was partially digested with Sau3AI and size fractionated on a sucrose gradient, and fragments in the size range 15 to 22 kb were treated with calf intestinal alkaline phosphatase and ligated with the self-transmissible, single-copy SCP2*-derived plasmid, pIJ698, cut with BglII. The ligation mix was introduced into the histidine and uracil auxotroph S. coelicolor J1501 by protoplast transformation, and 4,000 of the resulting transformants were arrayed on master plates (MM containing histidine, uracil, and 50 μg of hygromycin per ml), with 100 patches per plate.
For preparation of an inoculum for mating, developmental mutants were grown on cellophane discs on MM agar, scraped into 3 ml of 10.3% (wt/vol) sucrose, and homogenized in a manual glass homogenizer. This suspension was diluted with 10.3% (wt/vol) sucrose to 9 ml of final volume, and 200 μl was spread and dried on each mating plate (MM containing histidine and uracil). Genomic library master plates were replicated onto the mating plates, and after incubation to allow mating and growth, exconjugants of the developmental mutant carrying the plasmid were selected by replica plating to MM lacking auxotrophic supplements but containing 50 μg of hygromycin per ml.
Complementation of mutants with known whi genes.
Self-transmissible SCP2*-derived plasmids carrying whiA (pIJ6204 [31]), whiB (pIJ2157 [13]), whiG (pIJ597 [31]), whiH (pIJ6201 [32]), or whiJ (pIJ6205 [31]) were transferred by conjugation from S. coelicolor J1501 into new whi mutants by replica plating and subsequent counterselection of the auxotrophic donor strain as described for the genomic library.
Conjugation from Escherichia coli into Streptomyces developmental mutants.
Because many whi mutants do not sporulate, the method of Flett et al. (16) was adapted to promote conjugal transfer from E. coli into Streptomyces by using mycelial fragments, rather than spores, as an inoculum; high frequencies of exconjugants were obtained without difficulty. pSET152 and its derivatives were introduced by transformation or electroporation into the dam dcm hsdS E. coli strain ET12567 containing the RK2 derivative pUZ8002 (35). pUZ8002 supplies transfer functions to oriT-carrying plasmids, such as pSET152, but is not efficiently transferred itself because of a mutation in its own oriT. E. coli containing pSET152 or its derivatives was grown in L broth to an A600 of 0.4, washed twice with an equal volume of fresh medium, and resuspended in 1/10 the volume of L broth. Four-day-old lawns of the whi mutants, grown on MS agar, were harvested by pipetting 3 to 4 ml of 20% (vol/vol) glycerol onto the surface and gently dislodging the aerial mycelium with a sterile loop. The resulting suspension of mycelial fragments was vortexed for 1 min, and 0.5 ml was mixed with 0.5 ml of washed E. coli cells. After harvesting by centrifugation, the pellet, containing Streptomyces mycelial fragments and E. coli cells, was resuspended in the residual medium and plated on MS agar containing 10 mM MgCl2. Following incubation for 16 to 20 h at 30°C, each plate was overlaid with 1 ml of water containing 0.5 mg of nalidixic acid (to kill E. coli) and 1 mg of apramycin (to select Streptomyces exconjugants).
Physical mapping of cloned DNA.
Inserts from complementing clones were gel isolated, 32P radiolabelled by random priming, and used to probe Qiabrane membranes on which the entire minimal, ordered cosmid library of Redenbach et al. (30) had been arrayed.
Genetic mapping of whiL mutants.
A suspension of mycelial fragments was prepared from each of the mutants R214, R349, and R491 in the same way as the inoculum for the library matings. Each suspension was mixed with spores of S. coelicolor 1258 and plated on appropriately supplemented MM. After incubation to allow mating and growth, serial dilutions of harvested mycelium and spores were plated on MM containing uracil, proline, arginine, cystine, and 10 μg of streptomycin per ml, selecting for His+ streptomycin-resistant recombinants. One hundred recombinants from each cross were arrayed on master plates of the same medium, incubated for several days, and then replicated to MM lacking one of the auxotrophic supplements (uracil, proline, arginine, or cystine). Recombinants were scored for the whiL phenotype by phase-contrast microscopy and for growth on each medium.
Scanning electron microscopy.
For scanning electron microscopy, colonies were mounted on the surface of an aluminum stub with O.C.T. compound (BDH Laboratory Supplies, Poole, United Kingdom), plunged into liquid nitrogen slush at approximately −210°C to cryopreserve the material, and transferred to the cryostage of a CT1500HF cryotransfer system (Oxford Instruments, Oxford, United Kingdom) attached to a Philips XL30 FEG scanning electron microscope (Philips Electron Optics, FEI UK Ltd., Cambridge, United Kingdom). Surface frost was sublimated at −95°C for 3 min before sputter coating the sample with platinum for 2 min at 10 mA, at below −110°C. Finally, the sample was moved onto the cryostage in the main chamber of the microscope, held at approximately −140°C, and viewed at 1.2 to 5.0kV. Photographs were taken with Ilford FP4 120 roll film in a Linhof camera.
RESULTS
Isolation of new whi mutants.
S. coelicolor M145 spores were mutagenized with NTG and plated directly on MM at a density of approximately 200 colonies per plate; 1,500 mutant colonies with aerial surface color varying from white to medium grey were picked from a total population of approximately 30,000. From the 1,500 patches, 770 strains were streaked for single colonies, and those that showed unstable phenotypes (approximately one-third of the total) were discarded. A large majority of the unstable mutants exhibited a hypervariable phenotype similar to that described for strains of Streptomyces ambofaciens that have undergone large chromosomal rearrangements (24, 34). Coverslip impressions of the remaining strains were examined, and 431 isolates with stable phenotypes distinguishable from that of the wild type were identified. These strains could be divided into three groups: 71 that failed to produce abundant aerial hyphae, 144 that produced abundant aerial hyphae but showed clear defects in sporulation; and 216 mutants that sporulated abundantly but produced either spores with aberrant size or shape or spores of normal appearance but with reduced pigmentation. Primary interest was in the second group (144 strains).
Complementation with known whi genes.
In an attempt to exclude mutants that represented whi genes that had already been cloned, self-transmissible SCP2*-derived plasmids carrying whiA, whiB, whiG, whiH, or whiJ (but not whiD, whiE, or whiI) were introduced into 115 of the 144 whi mutants by mating. The morphological defects shown by the mutants made it unlikely that they were caused by mutations in the whiE spore pigment biosynthesis cluster, and self-transmissible clones were not available for whiD or whiI. Complementation was judged initially by looking for the restoration of grey pigmentation to the aerial surface of exconjugants and subsequently by inspection of coverslip impressions in a phase-contrast microscope. Of the 115 strains, 6 were fully complemented by whiA, 15 were fully complemented by whiB, 19 were fully complemented by whiG, 16 were fully complemented by whiH, and 3 were fully complemented by whiJ. A striking observation was that in two of the strains complemented by whiB, five of the strains complemented by whiG, and seven of the strains complemented by whiH, one or more of the other whi genes also increased the level of grey pigmentation. In some cases, there was also some degree of morphological correction as well. In a further 25 strains, no fully complementing whi gene was identified, but one or more of the five whi genes increased the level of grey pigmentation.
All strains in which the wild-type phenotype was restored, either partially or completely, by whiA, whiB, whiG, whiH, or whiJ were excluded from further analysis. This left 31 whi mutants.
Complementation and phenotypes of novel whi mutants.
Attempts were made to complement 19 of the remaining 31 whi mutants. An SCP2* library of chromosomal DNA isolated from wild-type S. coelicolor was constructed in the auxotrophic strain J1501. Four thousand clones from this library were arrayed on master plates and mated with each mutant in turn by replica plating, and transconjugants were isolated by a second round of replica plating to a selective medium as described in Materials and Methods. Potentially complementing clones were identified initially by looking for grey patches and subsequently by inspection of coverslip impressions in a phase-contrast microscope. In each case, we confirmed plasmid linkage of the phenotype by reintroduction of the cloned DNA into the corresponding whi mutant(s), either by protoplast transformation using the primary SCP2* clone itself or by mating from E. coli, having first subcloned the insert from SCP2* into the intergeneric, conjugative vector pSET152, which integrates site specifically into the S. coelicolor chromosome at the phage φC31 attB site (4). In all, complementing clones were found for 12 of the 19 strains, and the mutations were allocated to five loci, according to their complementation groups. A summary of information on these 12 strains is given in Table 1.
TABLE 1.
Characteristics of the novel S. coelicolor whi mutants
| Locus | Cosmid | Allele | Aerial surface color | Phenotype |
|---|---|---|---|---|
| whiK | D63 | R273 | White | Long tight coils but with septation |
| R318 | White | Long tight coils but with septation | ||
| R655 | White | Undifferentiated aerial hyphae with very rare chains of curved spores | ||
| whiL | 4A7 | R139 | Pale grey | Long curved and straight spores |
| R214 | Very pale grey | Long curved and straight spores | ||
| R349 | Medium grey | Short coils and long and short fragments | ||
| R491 | Pale grey | Long curved and straight spores | ||
| R514 | Pale grey | Long curved and straight spores | ||
| whiM | I51 | R432 | White | Undifferentiated hyphae with occasional spore chains of normal appearance; aerial hyphae lyse rapidly |
| whiN | E68 | R112 | White | Undifferentiated hyphae with rare spore chains sometimes showing irregular septum placement |
| R650 | Medium grey | Slightly oligosporogenous; spores longer than those of the wild type | ||
| whiO | 6G4 | R589 | Medium grey | Undifferentiated hyphae with frequent spore chains of normal appearance |
whiK.
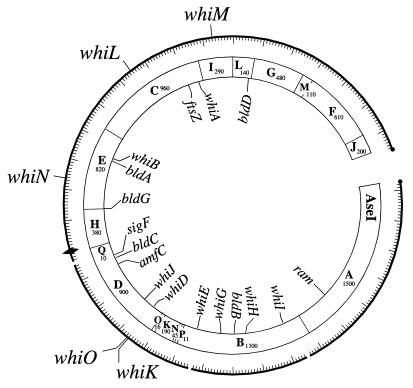
One clone, pIJ6700, complemented three of the new whi mutants, R273, R318, and R655. Microscopic examination showed that the plasmid restored wild-type levels of sporulation to all three strains. Hybridization, using the insert from pIJ6700 as a probe, localized the complementing DNA to the overlap between cosmids 6G4 and D63, in the 7 o’clock region of the chromosome (Fig. 1). whiD also maps to this overlap (27, 30), but further analysis showed that whiD did not complement R273, R318, or R655 and that whiK did not complement the whiD strain J243 (26). Accordingly, the new locus was designated whiK.
FIG. 1.
Locations of developmental genes on the combined physical and genetic map of S. coelicolor. Positions of the five new whi loci are shown on the outside of the circle, and positions of previously identified developmental genes are shown on the inside (adapted from reference 30). Sizes of the AseI fragments are in kilobases. ⧫, oriC; ●, telomeres.
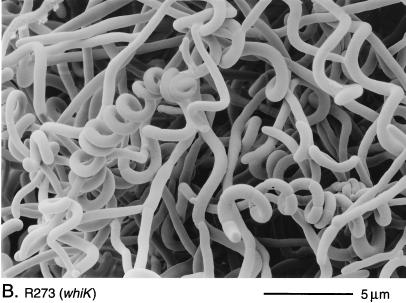
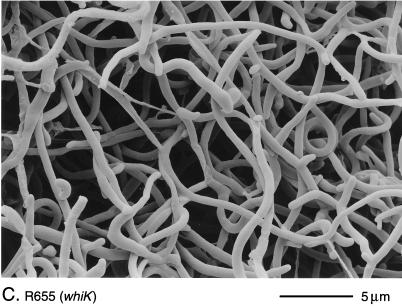
The phenotypes of R273 and R318 were indistinguishable. Both mutants produced white colonies with long, tightly coiled aerial hyphae with frequent septation (Fig. 2B). In contrast, the third whiK mutant, R655, showed a more severe phenotype; although very occasional short chains of spores were detected, R655 produced, almost entirely, straight, undifferentiated aerial hyphae (Fig. 2C).
FIG. 2.
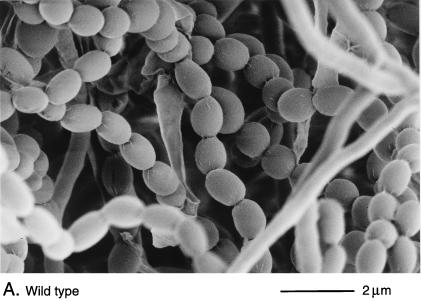
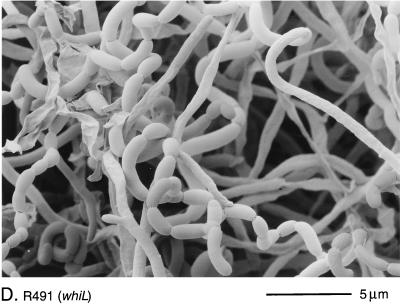
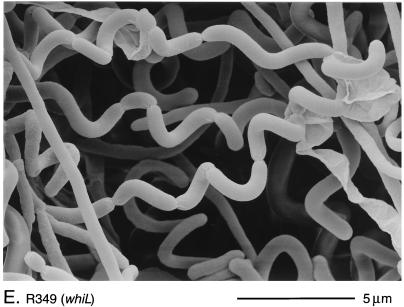
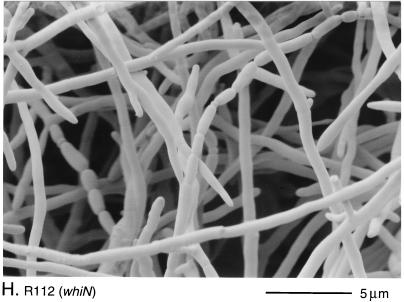
Scanning electron micrographs of the aerial hyphae of the morphologically wild-type strain M145 (A), the whiK mutant R273 (B), the whiK mutant R655 (C), the whiL mutant R491 (D), the whiL mutant R349 (E), the whiL mutant R349 carrying pIJ6710 (a derivative of the vector pSET152 carrying the whole whiL insert derived from the original SCP2* clone pIJ6702 [3]) (F), the whiM mutant R432 (G), the whiN mutant R112 (H), and the whiO mutant R589 (I). All strains were grown on MM containing 0.5% (wt/vol) mannitol. Scale bars are shown in each panel.
whiL.
Three clones, pIJ6701, pIJ6702, and pIJ6703, complemented five of the new whi mutants, R139, R214, R349, R491, and R514. Southern hybridization showed that these three clones overlapped and that they mapped to the unique region of cosmid 4A7, in the 10 o’clock region of the chromosome (Fig. 1). This locus was designated whiL.
There was significant phenotypic variation among the whiL mutants. R139, R491, and R514 had pale grey colonies that produced long, curved and straight spores (Fig. 2D). R214 was morphologically indistinguishable from these three strains but produced less spore pigment. The most striking phenotype among the whiL mutants was that of R349. R349 produced medium grey colonies with tightly coiled aerial hyphae that had septa at long intervals, giving rise to corkscrew-like fragments of irregular length, often several times longer than a wild-type prespore compartment (Fig. 2E). Although the cloned DNA restored the wild-type phenotype to four of the whiL strains, it did not completely complement R349; introduction of the cloned DNA restored wild-type levels of colony pigmentation and regular septation, but the spores produced were still noticeably longer than those of the wild type (Fig. 2F). One possible explanation for this observation is that R349 carries a second mutation that contributes to its morphological phenotype, in addition to the mutation in whiL.
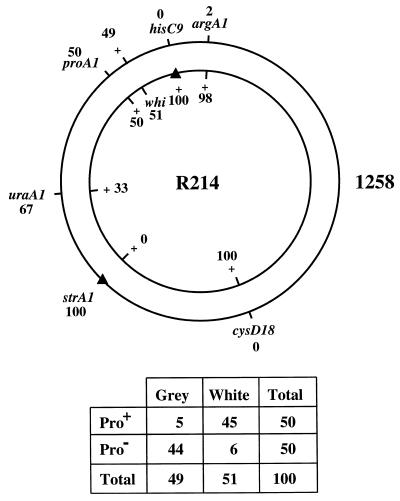
The variation in the phenotypes of the strains apparently complemented by whiL, and the fact that whiL clones did not restore a full wild-type phenotype to R349 raised the possibility that not all of these strains carried allelic mutations and that some of the effects observed might represent suppression. In an attempt to address this question, we determined approximate genetic map locations for the whi mutations carried by R214, R349, and R491, three strains with distinguishable phenotypes, to see if the genetic map locations of these mutations were compatible with the physical map location of the complementing DNA. In all three cases, the mutation mapped between proA and hisC but closer to proA (e.g., Fig. 3). proA has been mapped physically to the unique region of cosmid C123 (30). Cosmid 4A7, which carries whiL, lies between proA and hisC, 10 cosmids distant from proA and 18 cosmids distant from hisC. The approximate genetic map locations of the mutations in R214, R349, and R491 are therefore consistent with the physical map location of the DNA that complements these strains.
FIG. 3.
Genetic mapping of the whiL mutant R214. R214 (inner circle) was crossed with strain 1258 (outer circle), and hisC+ and strA1 (indicated by solid triangles) were selected. Numbers in the diagram are the frequencies of alleles among the recombinant progeny. The segregation of the whi mutation with respect to proA1 is tabulated below. Of the three whiL mutants analyzed, R214 showed the largest number of Pro+ grey (multiple-crossover class) recombinants. The crosses with R349 and R491 gave 3 and 0 such recombinants, respectively.
whiM.
One clone, pIJ6704, complemented R432, and hybridization showed that the cloned DNA mapped to the unique region of cosmid I51, in the 12 o’clock region of the chromosome (Fig. 1). This locus was designated whiM.
R432 colonies were white and produced undifferentiated aerial hyphae with occasional spore chains of normal appearance (Fig. 2G), a phenotype similar to that of whiJ mutants. However, unlike those of whiJ mutants, the aerial hyphae of R432 lysed very rapidly (data not shown), and presumably as a consequence, the strain showed very poor viability on prolonged incubation on plates in comparison to the other whi mutants examined.
whiN.
Two clones, pIJ6705 and pIJ6706, complemented R112 and R650. Both clones restored wild-type levels of sporulation. Southern hybridization showed that these clones overlapped and that they mapped to the unique region of cosmid E68, in the 9 o’clock region of the chromosome (Fig. 1). This locus was designated whiN.
The morphological phenotypes of the two whiN mutants were different. On MM, colonies of R650 were pale to medium grey and produced frequent spores that were longer than those of the wild type. In contrast, colonies of R112 were white and produced long, straight, undifferentiated hyphae, although occasional spore chains, sometimes showing highly irregular septum placement, were observed (Fig. 2H). The R112 mutation also showed clear signs of pleiotropic effects; in addition to the defects in sporulation, on some media, such as MM, R112 produced significantly less aerial mycelium than the parental strain M145. In this respect, R112 does not fit the classical definition of a whi mutant, which should be solely defective in sporulation (6). All aspects of the pleiotropic phenotype of R112 were fully complemented by both clones.
whiO.
One clone, pIJ6707, complemented R589. Southern hybridization showed that the cloned DNA mapped to the unique region of cosmid 6G4 in the 7 o’clock region of the chromosome (Fig. 1). whiD and whiK map very close by, in the overlap between cosmids 6G4 and D63, but further Southern analysis confirmed that the insert from pIJ6707 did not overlap either of these genes. Therefore, this locus was designated whiO.
R589 colonies were medium grey in color and had an oligosporogenous phenotype; by scanning electron microscopy, some undifferentiated aerial hyphae, but with fairly frequent spore chains of normal appearance, were observed (Fig. 2I).
DISCUSSION
The poor representation of several loci among the original collection of 50 whi mutants analyzed suggested that the genetic map of whi loci was not saturated (6, 31), a conclusion borne out by the results presented here. The primary criterion for the selection of sporulation mutants in the new screen remained the loss of production of the grey spore pigment, because, in contrast to the endospores of Bacillus and Clostridium, the exospores of S. coelicolor are not particularly resistant to high temperatures or chemical attack, and screens for sensitivity to these kinds of stresses therefore could not be used. However, we recognized that some developmental mutants could have severe morphological defects but be only partially blocked in production of spore pigment (as is true for whiH and whiJ mutants), and so during the selection process we attempted to include all mutants in which spore pigmentation was reduced to a noticeable extent. Two important variables between the first and second screens whose influence cannot readily be assessed are that (i) in the first screen (18) a rich medium was used, whereas in this screen MM was used throughout for the assessment of phenotype; and (ii) some of the mutations in the earlier screen were induced by UV light (18), whereas all the mutations in this screen were induced by NTG. Here, genetic mapping was not relied on heavily as a means of determining the positions of mutations, because the library-based complementation required less time and effort and ultimately led to the physical mapping of the new loci. Like the old whi loci, the new loci are located in the 6-Mb central region of the linear S. coelicolor chromosome where virtually all genes identified by classical genetics have mapped (Fig. 1), in contrast to the two ∼1-Mb regions at the chromosome ends that are almost devoid of classical markers (30).
From the 770 mutants that were streaked for single colonies, 431 were stable and could be distinguished from the wild type by spore pigmentation, and the vast majority of these also showed morphological defects. As in the first whi mutant analysis (6), emphasis was placed on mutants showing clear morphological defects that could easily be distinguished from the wild type by phase-contrast microscopy, thereby making the scoring of complementation relatively straightforward. However, this left as subjects for further study a large number of more subtle mutants that showed variation in spore size or shape. This group contained strains that showed phenotypes similar to those of sigF (29) and whiD (6) and could be used to expand the number of genes known to be involved in the later stages of spore development. Both whiL (Fig. 2D and E) and whiN (Fig. 2H) mutants showed unusual intervals between sporulation septa. The control of septal placement in bacteria is mostly studied in rod-shaped unicellular bacteria, but further studies of these Streptomyces mutants may provide distinctive and novel insights into this process.
Of the 19 whi strains that received the library, 12 were complemented. The 12 mutants were grouped by complementation into five loci, named whiK to whiO. The failure to complement the other seven strains, and the small numbers of complementing clones isolated for each of the new loci (whiK, one; whiL, three; whiM, one; whiN, two; and whiO, one) indicated that the library was not fully representative. An attractive alternative strategy to complement the remaining strains would be to derive an approximate genetic map position for each mutation and then to use the minimal, ordered cosmid library of Redenbach et al. (30) to “walk” across the corresponding interval of the combined physical and genetic map. Although these cosmids cannot replicate autonomously in S. coelicolor, selection for kanamycin resistance after protoplast transformation results in the recovery of isolates in which the cosmid has integrated into the chromosome via insert-directed homologous recombination. The cosmids can therefore be used to clone genes by complementation (30), an approach that has been used to isolate whiD (27, 28), whiI (1), and bldC (20).
The new screen showed a wide variation in the number of mutations isolated at each whi locus (whiA, 6; whiB, 15; whiG, 19; whiH, 16; whiJ, 3; whiK, 3; whiL, 5; whiM, 1; whiN, 2; whiO, 1), as was found in the previous screen (6). Part of this disparity can be explained by the fact that some of the mutants (particularly whiA, whiB, and whiG) yield colonies that are snowy white and have a raised aerial mycelium, but others (especially some of those representing the new loci described here) produce some spore pigment and so were less likely to be picked out from a field of colonies.
Of the 115 mutants that were checked for complementation by clones of whiA, whiB, whiG, whiH, and whiJ carried on self-transmissible plasmids, 59 showed full complementation by one of the clones, 25 showed partial complementation by one or more of the clones, and 31 strains were unaffected, although many of the strains that were fully complemented by one of the existing clones were also partially complemented by another. The 25 mutants that were only partially complemented by one of the five clones could, in principle, represent partially dominant alleles that do not allow full restoration of the wild-type phenotype, but it seems much more likely that they contain mutations in genes other than whiA, whiB, whiG, whiH, and whiJ which are partially suppressed by the introduction of the cloned gene on the SCP2* low-copy-number plasmid. In cases where more than one of these five genes were able partially to restore the wild-type phenotype, there can be no doubt that suppression is involved. Suppression effects have not previously been reported in developmental work in Streptomyces, although the ability of an additional copy of whiG partially to suppress the spore pigment defect of whiH mutants without affecting their morphological phenotype has recently been noted (15). Suppression effects caused by additional copies of genes have been both informative and problematic in the analysis of the regulation of antibiotic biosynthesis in Streptomyces (2).
In the first genetic screen for whi loci, most of the mutants assigned to a given locus were similar in phenotype (6), as might be expected if most of the alleles were null or close to null. The one notable exception was a mild allele of whiG that caused the formation of long spores, rather than the more characteristic block in sporulation septum formation, and indeed this mutant was originally designated whiF because of this wide phenotypic difference (5, 6, 31). In contrast, in the work described here, there was wide phenotypic variation within each of the three loci represented by multiple alleles, whiK, whiL, and whiN. This again raised the possibility that some of these effects might represent suppression rather than true complementation (although the three clones complementing whiL mutants overlapped, as did the two clones complementing whiN mutants). However, in genetic mapping experiments using three whiL strains each having a distinct phenotype (R214, R349, and R491), the genetic map locations of the three mutations were consistent with the physical map location of the complementing DNA.
The approach taken here has enabled us simultaneously to identify, map, and clone five new whi loci and should facilitate further rapid progress. Our prospects in these endeavors will undoubtedly be enhanced by the current S. coelicolor genome sequencing project (33).
ACKNOWLEDGMENTS
We thank Gabriella Kelemen, David Hopwood, and Mervyn Bibb for helpful comments on the manuscript, Sue Bunnewell for hand-printing the scanning electron micrographs, and Tobias Kieser for help in preparing Fig. 1.
This work was funded by BBSRC grants CAD 04380 (to K.F.C.) and 83/P07658 (to M. J. Buttner), by a Lister Institute research fellowship (to M. J. Buttner), by a John Innes Foundation studentship (to V.M.), and by a grant-in-aid to the John Innes Centre from the BBSRC.
REFERENCES
- 1.Ainsa, J., H. D. Parry, and K. F. Chater. Unpublished data.
- 2.Bibb M J. The regulation of antibiotic production in Streptomyces coelicolor A3(2) Microbiology. 1996;142:1335–1344. doi: 10.1099/13500872-142-6-1335. [DOI] [PubMed] [Google Scholar]
- 3.Bibb, M. J., and M. J. Buttner. Unpublished data.
- 4.Bierman M, Logan R, O’Brien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 5.Bruton, C. J. Personal communication.
- 6.Chater K F. A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J Gen Microbiol. 1972;72:9–28. doi: 10.1099/00221287-72-1-9. [DOI] [PubMed] [Google Scholar]
- 7.Chater K F. Taking a genetic scalpel to the Streptomyces colony. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- 8.Chater, K. F. Unpublished data.
- 9.Chater K F, Merrick M J. Approaches to the study of differentiation in Streptomyces coelicolor A3(2) In: MacDonald K D, editor. Second International Symposium on the Genetics of Industrial Micro-Organisms. London, England: Academic Press; 1976. pp. 583–593. [Google Scholar]
- 10.Chater K F, Bruton C J, King A A, Suarez J E. The expression of Streptomyces and Escherichia coli drug resistance determinants cloned into the Streptomyces phage φC31. Gene. 1982;19:21–32. doi: 10.1016/0378-1119(82)90185-8. [DOI] [PubMed] [Google Scholar]
- 11.Chater K F, Bruton C J, Plaskitt K A, Buttner M J, Méndez C, Helmann J D. The developmental fate of Streptomyces coelicolor hyphae depends upon a gene product homologous with the motility sigma factor of Bacillus subtilis. Cell. 1989;59:133–143. doi: 10.1016/0092-8674(89)90876-3. [DOI] [PubMed] [Google Scholar]
- 12.Davis N K, Chater K F. Spore colour in Streptomyces coelicolor A3(2) involves the developmentally regulated synthesis of a compound biosynthetically related to polyketide antibiotics. Mol Microbiol. 1990;4:1679–1691. doi: 10.1111/j.1365-2958.1990.tb00545.x. [DOI] [PubMed] [Google Scholar]
- 13.Davis N K, Chater K F. The Streptomyces coelicolor whiB gene encodes a small transcription factor-like protein dispensable for growth but essential for sporulation. Mol Gen Genet. 1992;232:351–358. doi: 10.1007/BF00266237. [DOI] [PubMed] [Google Scholar]
- 14.Delić V, Hopwood D A, Friend E J. Mutagenesis by N-methyl-N′-nitro-N-nitrosoguanidine (NTG) in Streptomyces coelicolor. Mutat Res. 1970;9:167–182. doi: 10.1016/0027-5107(70)90055-2. [DOI] [PubMed] [Google Scholar]
- 15.Flärdh, K. Personal communication.
- 16.Flett F, Mersinias V, Smith C P. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett. 1997;155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 17.Hobbs G, Frazer C, Gardner D C J, Cullum J, Oliver S G. Dispersed growth of Streptomyces in liquid culture. Appl Microbiol Biotechnol. 1989;31:272–277. [Google Scholar]
- 18.Hopwood D A, Wildermuth H, Palmer H M. Mutants of Streptomyces coelicolor defective in sporulation. J Gen Microbiol. 1970;61:397–408. doi: 10.1099/00221287-61-3-397. [DOI] [PubMed] [Google Scholar]
- 19.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, England: The John Innes Foundation; 1985. [Google Scholar]
- 20.Kelemen, G. H. Personal communication.
- 21.Kelemen G H, Brown G L, Kormanec J, Potúc̆ková L, Chater K F, Buttner M J. The positions of the sigma factor genes, whiG and sigF, in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2) Mol Microbiol. 1996;21:593–603. doi: 10.1111/j.1365-2958.1996.tb02567.x. [DOI] [PubMed] [Google Scholar]
- 22.Kelemen G H, Brian P, Flärdh K, Chamberlin L, Chater K F, Buttner M J. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2) J Bacteriol. 1998;180:2515–2521. doi: 10.1128/jb.180.9.2515-2521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kieser T, Melton R E. Plasmid pIJ699, a multi-copy positive-selection vector for Streptomyces. Gene. 1988;65:83–91. doi: 10.1016/0378-1119(88)90419-2. [DOI] [PubMed] [Google Scholar]
- 24.Leblond P, Decaris B. New insights into the genetic instability of Streptomyces. FEMS Microbiol Lett. 1994;123:225–232. doi: 10.1111/j.1574-6968.1994.tb07229.x. [DOI] [PubMed] [Google Scholar]
- 25.McVittie A. Ultrastructural studies on sporulation in wild-type and white colony mutants of Streptomyces coelicolor. J Gen Microbiol. 1974;81:291–302. doi: 10.1099/00221287-81-2-291. [DOI] [PubMed] [Google Scholar]
- 26.Molle, V., and M. J. Buttner. Unpublished data.
- 27.Molle, V., W. J. Palframan, and M. J. Buttner. Unpublished data.
- 28.Palframan W J. The whiD locus of Streptomyces coelicolor A3(2). Ph.D. thesis. Norwich, England: University of East Anglia; 1998. [Google Scholar]
- 29.Potúc̆ková L, Kelemen G H, Findlay K C, Lonetto M A, Buttner M J, Kormanec J. A new RNA polymerase sigma factor, ςF, is required for the late stages of morphological differentiation in Streptomyces sp. Mol Microbiol. 1995;17:37–48. doi: 10.1111/j.1365-2958.1995.mmi_17010037.x. [DOI] [PubMed] [Google Scholar]
- 30.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 31.Ryding N J. Analysis of sporulation genes in Streptomyces coelicolor A3(2). Ph.D. thesis. Norwich, England: University of East Anglia; 1995. [Google Scholar]
- 32.Ryding N J, Kelemen G H, Whatling C A, Flärdh K, Buttner M J, Chater K F. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2) Mol Microbiol. 1998;29:343–357. doi: 10.1046/j.1365-2958.1998.00939.x. [DOI] [PubMed] [Google Scholar]
- 33.Streptomyces coelicolor Genome Project Web Site. [Online.] http://www.sanger.ac.uk/Projects/S_coelicolor/. Sanger Centre, Wellcome Trust Genome Campus, Hinxton, Cambridge, United Kingdom.
- 34.Volff J-N, Altenbuchner J. Genetic instability of the Streptomyces chromosome. Mol Microbiol. 1998;27:239–246. doi: 10.1046/j.1365-2958.1998.00652.x. [DOI] [PubMed] [Google Scholar]
- 35.Wilson, J., and D. H. Figurski. Personal communication.