Summary
The methods for the culture and cardiomyocyte differentiation of human embryonic stem cells, and later human induced pluripotent stem cells (hiPSC), have moved from a complex and uncontrolled systems to simplified and relatively robust protocols, using the knowledge and cues gathered at each step. HiPSC-derived cardiomyocytes have proven to be a useful tool in human disease modelling, drug discovery, developmental biology, and regenerative medicine. In this protocol review, we will highlight the evolution of protocols associated with hPSC culture, cardiomyocyte differentiation, sub-type specification, and cardiomyocyte maturation. We also discuss protocols for somatic cell direct reprogramming to cardiomyocyte-like cells.
Subject areas: Cell culture, Stem Cells, Cell Differentiation
The methods for the culture and cardiomyocyte differentiation of human embryonic stem cells, and later human induced pluripotent stem cells (hiPSC), have moved from a complex and uncontrolled systems to simplified and relatively robust protocols, using the knowledge and cues gathered at each step. HiPSC-derived cardiomyocytes have proven to be a useful tool in human disease modelling, drug discovery, developmental biology, and regenerative medicine. In this protocol review, we will highlight the evolution of protocols associated with hPSC culture, cardiomyocyte differentiation, sub-type specification, and cardiomyocyte maturation. We also discuss protocols for somatic cell direct reprogramming to cardiomyocyte-like cells.
Introduction
Cardiovascular disease remains the leading cause of death in the United States and worldwide (Virani et al., 2021). A major restriction in progressing our understanding of cardiovascular disease and developing therapies is that human cardiomyocytes cannot be isolated easily or non-invasively from patients, requiring alternative approaches to generate de novo cardiomyocytes. For the last 20 years, the culture and cardiomyocyte differentiation of human embryonic stem cells (hESC), and later human induced pluripotent stem cells (hiPSC), have provided a valuable source of these cells. More recently, direct reprogramming of human somatic cells to cardiomyocytes has presented a potential secondary source of these cells. Since then, human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs) have proven to be a useful tool in human disease modelling, drug discovery, developmental biology, and regenerative medicine. Furthermore, one of the most powerful applications of hPSC-CMs has been demonstrated to be modeling an individual patient’s disease, validating its genomic nature, and potentially designing genome-informed therapies.
For all the applications of hPSC-CMs, it is essential to have simple and reproducible tools that enable both the appropriate pluripotent culture and differentiation of cardiomyocytes that accurately recapitulate the infantile and adult cardiomyocyte phenotypes. Knowledge gleaned over more than 2 decades of work have provided us with an understanding of mechanisms associated with maintenance of human pluripotency and pathways associated with differentiation. This knowledge is now used to develop more robust, reproducible, and cost-effective protocols that enable the generation of cardiomyocytes and cardiac tissues in larger scales, with specific subtypes, and with a more mature phenotype. However, further improvements can still be made, including increased robustness and scalability, decreased time and costs, and improved direct reprogramming that skips the pluripotency phase entirely.
In this protocol review, we will highlight the development of protocols associated with hPSC culture, cardiomyocyte differentiation, sub-type specification, cardiomyocyte maturation (Figure 1), and somatic cell direct reprogramming (Figure 2).
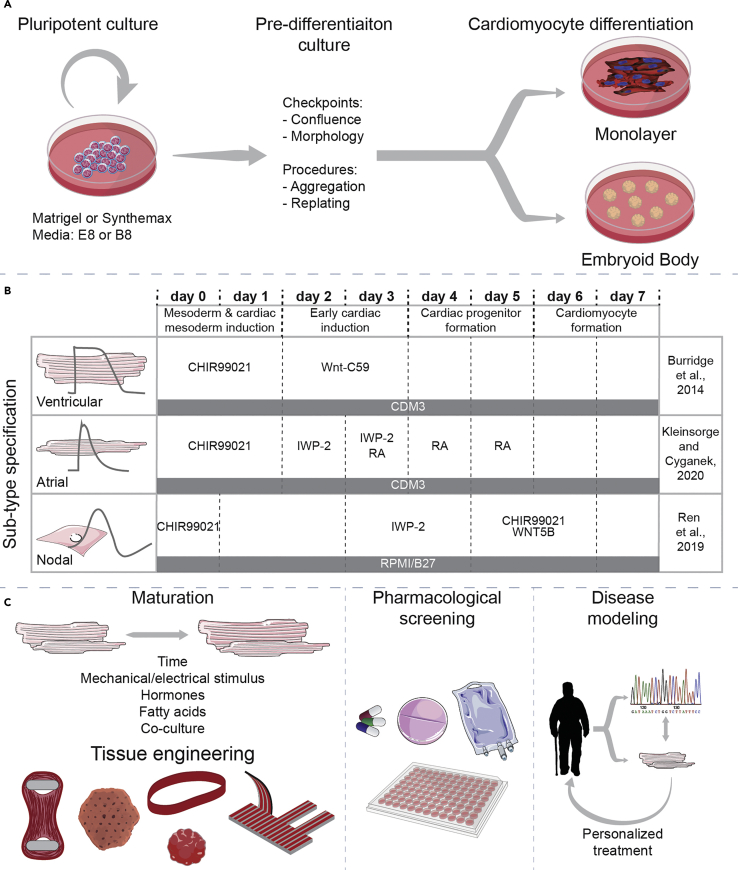
Figure 1.
hPSC culture, differentiation, and applications
(A) hiPSC are cultured on Matrigel or Synthemax using chemically defined media until differentiation, with continuous monitoring of morphology and confluence levels. Before differentiation, hiPSC can also be replated or aggregated depending on the differentiation protocol.
(B) Examples of differentiation protocols to obtain ventricular (also Table 1), atrial, and nodal (Table 2) hiPSC-CM. (C) Further culture approaches and applications of hiPSC-CM using protocols described in this paper.
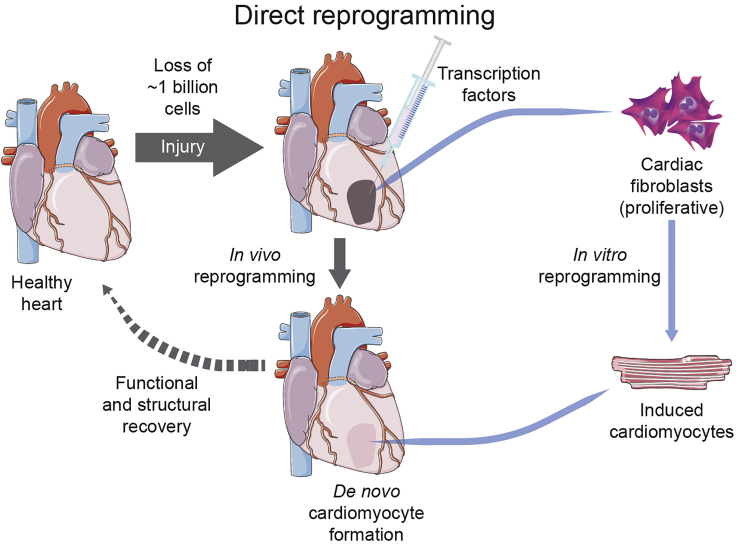
Figure 2.
Direct reprogramming of CFs into induced cardiomyocytes
In vivo reprogramming is performed via injection of transcription factors into the injured area of the heart that will lead to de novo cardiomyocyte formation. In vitro reprogramming is performed via treatment of (cardiac) fibroblasts with different cocktails to generate iCMs. These cells can later be re-transplanted into the heart if needed to recover injured areas.
Methods for hiPSC culture
Since the first derivation of hESC in 1998, methods for the culture of hPSC have progressed rapidly, with the ambition to develop conditions that are as simple to use and as defined as possible. Initially, hESC were cultured in the basal media DMEM with 20% fetal bovine serum (FBS) on a feeder layer of mouse embryonic fibroblasts (MEF) (Thomson et al., 1998). FBS was quickly eliminated, settling on a formula consisting of DMEM/F12 with 10%–20% knockout serum replacement (KSR) and low levels of FGF2 (4 ng/mL) commonly called “hESC media” (Schulz et al., 2004). Modifications were then made to allow the elimination of the MEF feeder layer, replacing it with Matrigel, and the use of high levels of FGF2 (100 ng/mL) (Xu et al., 2005) and addition of TGFB1 (Amit et al., 2004) or activin A (Beattie et al., 2005). KSR, a propriety FBS replacement (Garcia-Gonzalo and Izpisua Belmonte, 2008), was eventually replaced with bovine or human albumin in the defined formulae of TeSR1 (Ludwig et al., 2006), DC-HAIF (Wang et al., 2007), hESF9T (Furue et al., 2008; Yamasaki et al., 2014), FTDA (Frank et al., 2012), CDM-BSA (Hannan et al., 2013; Vallier et al., 2005), and i-DEAL (Marinho et al., 2015). The first chemically defined (except for bovine transferrin) hPSC culture media, E8, eliminated the need for albumin (Chen et al., 2011). We subsequently developed a further optimized, fully chemically defined variant B8 which was specifically optimized for cost-efficiency (Kuo et al., 2020).
Many commercial versions of the above media exist for the culture of hPSC, typically costing around $500 per liter. These commercial media are made in a controlled manner, which reduces the need for quality control to be performed in lab. Nevertheless, we would not recommend using any commercial media that do not have a published formula as this will prevent any further development and customization of protocols. Both E8 and B8 have been commercialized as “Essential-8” and “HiDef B8” respectively. We strongly recommend starting with the commercial versions of either of these media. Once your usage increases enough to justify the additional effort, you can generate and validate mediate in house.
Although albumin-containing media are thought to be more robust when culturing hPSC as low-density colonies, these media also appear to be more prone to supporting partially reprogrammed cells and allowing spontaneous differentiation. The highest growth rates have been demonstrated in chemically defined media such as E8 or B8 with near-monolayer culture (Kuo et al., 2020) using EDTA-passaging and a rigid 3- or 4-day passage schedule. A protocol for making your own B8 is available (Lyra-Leite et al., 2021) allowing for production at around $10 per liter. Ideally, the future of hiPSC culture will not use recombinant proteins like insulin, transferrin, FGF2, and TGFB1, as these are the most expensive components. Potential replacements of these recombinant proteins include peptides and small molecules. The AKIT formula (Yasuda et al., 2018) is a promising step in this direction, although we did not find this successful for our cell lines.
hPSC were traditionally passaged with collagenase or dispase when cultured on MEF or with trypsin or TrypLE when cultured on Matrigel. The use of these enzymes required the use of a centrifugation step, which can become laborious with numerous cell lines in culture. This has subsequently been replaced with an incubation in 0.5 mM EDTA (in DPBS−/−) for approximately 6 min (Beers et al., 2012); the EDTA can then be aspirated, and the cells suspended in E8/B8, without centrifugation. Alternatives such as the use of sodium citrate also exist (Nie et al., 2014). After passage, especially in the case of 2D culture, a small molecule Rho kinase inhibitor is typically added for 24 h to enhance cell survival (Chen et al., 2010). 10 μM Y27632 is most commonly used, but 2 μM thiazovivin is just as effective (Kuo et al., 2020; Xu et al., 2010). Thiazovivin also provides the best combination of availability in bulk and potency. Alternatives to ROCK inhibitors include commercial products such as RevitaCell and CloneR, and the recently published CEPT cocktail (Chen et al., 2021).
hiPSC are typically grown on growth-factor reduced Matrigel, and we have found that a high dilution ratio (1:800) is just as effective for growth, and that alternatives such as Geltrex or Cultrex at 1:800 are just as suitable (Kuo et al., 2020). We have not found any advantage of using “hESC-qualified” versions of these matrices. Users may choose to batch test their Matrigel, although we do not find it necessary if the concentrations specified in the certificates of analysis are within a range (10–12 mg/mL). Additionally, numerous alternative chemically defined matrices have been found to support the growth of hPSC (Burridge et al., 2014), yet only Synthmax II-SC, a tethered synthetic vitronectin peptide (at 1:320), offers a suitable combination of cost and ease of use (although this still costs around three times the cost of 1:800 Matrigel). All other recombinant proteins (e.g., vitronectin or laminin-521) cost many times more on a per-well basis and are thus not suitable. Ideally, a synthetic product at a similar cost to 1:800 Matrigel, could be developed and even made in house (Lambshead et al., 2018), but this may require processes such as UV-ozone cleaning and/or UV crosslinking not needed for Synthemax II-SC.
Methods for cardiac differentiation
A similar process of evolution seen in hPSC culture also happened in the realm of cardiac differentiation from hPSC, with an additional component: dimensionality (Table 1). In this case, architectures ranging from monolayers to uncontrolled or controlled 3D aggregates have been historically used and combined with other culture conditions to generate a wide range of cardiac cells. Initial differentiation protocols relied on spontaneous differentiation of stem cells aggregated into embryoid bodies in KO-DMEM and 20% FBS (Kehat et al., 2001), followed by dissociation and selection the desired cell types. In the Kehat protocol, 8% of embryoid bodies presented spontaneous contraction. This was followed by a co-culture system that also provided a platform for early generation of cardiomyocytes from hESC without need for further control of parameters, but with low yield and purity of tissues (Mummery et al., 2003). In the mid-2000s, new protocols arose, informed by developmental biology cues, utilizing growth factors such as activin A, FGF2, and BMP4. These protocols showed improved differentiation efficiency versus spontaneous conditions both in embryoid body (Burridge et al., 2007) and monolayers (Laflamme et al., 2007), the latter being one of the earliest applications of the RPMI/B27 combination that would become almost standard in the following decade. This was followed by the first application of small molecules to improve differentiation, in this case the p38 MAPK inhibitor SB203580 (Graichen et al., 2008), and the first chemically-defined medium for the differentiation of cardiomyocytes, indicating that insulin potentially presents a negative effect on differentiation (Xu et al., 2008). Further developments led to increased efficiency in the generation of beating embryoid bodies either via Wnt inhibition (Yang et al., 2008), optimizing concentrations of growth factors mixed with FBS (Takei et al., 2009), identifying developmental stage-specific signaling requirements (Kattman et al., 2011), or improving chemically-defined conditions (Burridge et al., 2011).
Table 1.
Evolution of differentiation approaches to generate hPSC-CMs
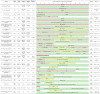 |
Pink cells indicate positive inducers (growth factors, sera, small molecules); yellow cells indicate inhibitors; green cells indicate media and additional supplements used. Acronyms: 2ME, ß-mercaptoethanol; 1TG, monothioglycerol; BIO, 6-bromoindirubin-30-oxime; BMP4, bone morphogenetic protein 4; DKK1, Dickkopf-related protein-1; EB, embryoid body; EDTA, Ethylenediaminetetraacetic acid; END-2, endoderm-like cell line; FBS, fetal bovine serum; FGF2, basic fibroblast growth factor; HF, human fibroblasts; HSA, human serum albumin; ITS, insulin, transferrin, selenium supplement; IWR-1, inhibitor of Wnt response-1; IWP-2, inhibitor of Wnt production 2; IWP-4, inhibitor of Wnt production 4; KO-DMEM, KnockOut DMEM; KSR, KnockOut Serum Replacement; MEF, mouse embryonic fibroblasts; MEF-CM, MEF conditioned medium; NEAA, nonessential amino acids; N/S, not stated; PFHM, Protein-Free Hybridoma mixture; PI3K, phosphoinositide 3-kinases; PVA, polyvinyl alcohol; SCF, stem cell factor; VEGFA, vascular endothelial growth factor.
The knowledge acquired with spontaneous differentiation and developmental biology cues enabled complex, yet inefficient protocols. These protocols were further optimized, but they still relied on the use of sera, proteins, and/or growth factors that are expensive and lead to more variability. After the cross-talk between the BMP4/activin A and Wnt/β-catenin pathways was identified as the essential element for cardiac differentiation (Paige et al., 2010), further protocols were developed exploring these elements. The next step was the simplification and replacement of expensive components with others that are easier to use and more widely available, hence we saw the exploration of small molecules to either enhance or replace growth factors. An initial modification was the replacement of DKK1 with small molecule-based Wnt inhibition with the use of IWR-1 either combined with sera (Ren et al., 2011) or in a serum-free system (Willems et al., 2011). Another development was the adaptation of protocols for use on both monolayer and embryoid body cultures (Elliott et al., 2011). Finally, in 2011, the first protocol using only small molecules for a stage-specific modulation of the Wnt/β-catenin was developed (Gonzalez et al., 2011).
In 2012, a similar approach was shown to work with cells grown in a defined pluripotent media; it explored the RPMI/B27 combination first utilized in 2007, now modified to remove insulin during the first 5 days of differentiation, and became known as the GiWi small molecule differentiation protocol (Lian et al., 2012, 2013). In addition to small molecules, the use of protein transduction was also shown effective in inducing cardiomyocyte differentiation in embryonic stem cells (Fonoudi et al., 2013). The time-dependent inhibition of BMP by a small molecule (DMH1) in a complex system using DMEM/F12 supplemented with B27 was also shown to promote cardiomyocyte differentiation (Aguilar et al., 2015). In 2014, we reported the first chemically defined, small molecule-based protocol for the differentiation of hiPSCs, called CDM3. We identified the components of the B27 supplement that were essential for cardiac differentiation when utilizing RPMI, namely ascorbic acid and albumin, while applying CHIR99021 and Wnt-C59 to modulate the Wnt/β-catenin pathway (Burridge et al., 2014). In addition, we showed that it was possible to remove albumin, but with a significant reduction in cardiomyocyte yield. To overcome this limitation, Lian and colleagues suggested the addition of other components from the B27 supplement in a stage-specific manner, namely putrescine, progesterone, and sodium selenite, combined with the reduction of the CHIR and IWP-2 concentrations (Lian et al., 2015). In a more complex system, utilizing KO-DMEM as basal medium, Zhang and colleagues were also able to differentiate cardiomyocytes in an albumin-free condition (Zhang et al., 2015). Another protocol proposed replacing albumin with heparin in a time-dependent treatment, also combined with lipids, in an E8-based system (Lin et al., 2017). However, even though the system was able to generate a significant number of TNNT2-positive cells, contraction was not as robust. Thus, supplementation with albumin was recommended in the STAR Protocols version (Lin and Zou, 2020). Tan and colleagues suggested supplementing RPMI with L-ascorbic acid and N-acetyl-L-cysteine, while using CHIR (during day 0 of differentiation) and IWR-1 (between days 3 and 8) to create an albumin-free differentiation system (Tan et al., 2018). This combination is also the basis of the S12 medium, which combines RPMI, ascorbic acid, N-acetyl-L-cysteine, insulin, transferrin, sodium selenite, ethanolamine, carnitine, linoleic and linolenic acids, methyl-β-cyclodextrin, Trolox, and sodium pyruvate to generate cardiomyocytes (Pei et al., 2017). Another albumin-free protocol uses DMEM/F12 as basal medium, supplemented with sodium selenite, ascorbic acid, transferrin, and lipids. It was used to identify how the IGF pathway regulates mesoderm differentiation and to show that PI3K inhibition, instead of Wnt inhibition, might be enough to induce hESC differentiation into cardiomyocytes (Yang et al., 2019b).
However, the proposed albumin-free protocols have not been optimized or explored in depth, and CDM3- and RPMI/B27-based protocols dominate the landscape of hiPSC-derived cardiomyocyte (hiPSC-CM) differentiation due to their reliability, yield, and cost. The majority of recent developments have focused on small improvements to these protocols, such as with the addition of transferrin to the CDM3 basal medium (Zhang et al., 2021a), and on increased scalability via the use of bioreactors either with CDM3 (Manstein et al., 2021a, 2021b) or RPMI/B27 (Laco et al., 2020) systems. Similarly, the re-modulation of the Wnt/β-catenin pathway at different time points has been proposed to increase the yield of cardiomyocytes in an RPMI/B27-based differentiation (Buikema et al., 2020; Maas et al., 2021; Sharma et al., 2018). Other developments based on CDM3 or the GiWi differentiation protocol focused on:
-
(1)
The development of controlled subtype specification differentiations (Cyganek et al., 2018; Funakoshi et al., 2021; Kleinsorge and Cyganek, 2020; Lee et al., 2017)
-
(2)
Strategies for hiPSC-CM maturation (Feyen et al., 2020; Giacomelli et al., 2020; Hu et al., 2018; Ribeiro et al., 2015b)
-
(3)
The application of hiPSC-CMs towards disease modeling either in simple 2D culture systems (Hanses et al., 2020; Magdy et al., 2021a, 2021b; McKeithan et al., 2020) or tissue engineered applications (Mills et al., 2017; Querdel et al., 2021) using functional measurements (Zhang et al., 2021b)
Finally, the role of other small molecules as enhancers of differentiation has also been investigated. For example, as an evolution of protocols proposed in the late 2000s and early 2010s (Kattman et al., 2011; Yang et al., 2008), the casein kinase 1 inhibitor CKI-7 has been combined with VEGFA and FGF2 in a time dependent manner using an embryoid body system to improve the recapitulation of drug-induced QT prolongation in vitro (Shinozawa et al., 2017). Rapamycin has also been tested as a potential enhancer of early mesoderm induction, specifically its addition in tandem with CHIR for 4 days was shown to increase the purity of hiPSC-CMs population compared to CHIR alone, also increasing the percentage of beating embryoid bodies (Qiu et al., 2017).
As a field, we have moved from a complex and uncontrolled system to simplified and relatively robust protocols, using the knowledge and cues gathered at each step; then we moved from a world with the use of complex sera and feeder layers to media that are chemically defined. We now have a family of small molecules that have been shown to be effective at modulating the Wnt/β-catenin pathways, especially CHIR99021, and the Wnt inhibitors: IWR-1, IWP-2 (Lin et al., 2017; Santoro et al., 2021), IWP-4, XAV939 (Wang et al., 2011), KY02111 (Minami et al., 2012; Qiu et al., 2017), TA01 (Laco et al., 2018), and Wnt-C59 (Table 1). We also understand better the role of common media supplements such as insulin (Freund et al., 2008; McDevitt et al., 2005; Tran et al., 2009) and ascorbic acid (Cao et al., 2012; Passier et al., 2005). However, even though we developed a vast understanding of the genetic pathways associated with differentiation, we still do not have a clear picture of the nutritional requirements, which is seen in a variety of basal media being suitable for differentiation. Additionally, protocols still rely on the use of proteins as differentiation enhancers (such as albumin and heparin) as a legacy feature, even though these proteins are not necessarily mechanistically relevant for differentiation. Hence, identifying minimum nutritional requirements and being able to fully remove proteins without an impact on tissue functionally should be the focus for the next generation of hiPSC-CM differentiation protocols.
Directed differentiation into defined cardiomyocyte subtypes
The human heart is composed of a highly diversified set of CM subtypes. These ventricular, atrial, and sinoatrial nodal CMs differ in their unique molecular, cellular, and physiological features, highly adapted to their defined localization and function within the heart (Litviňuková et al., 2020). Hence, the utilization of a highly homogeneous population of hPSC-CMs with a defined cardiac subtype is relevant for investigation of potentially subtype-related disease mechanisms, for the development of chamber-specific drugs, and for toxicity screening approaches. Although it is believed that the cardiac differentiation procedure yields a heterogeneous mixture of various CM subtypes, the most commonly applied differentiation protocols are exclusively based on Wnt signaling modulation (Burridge et al., 2014; Lian et al., 2013) and predominantly promote the differentiation of hPSC towards the ventricular lineage. Therefore, these protocols are typically used to obtain ventricular hPSC-CMs with cultures composed of around 90% of ventricular-like cells, with atrial- and nodal-like CMs being underrepresented. Significant efforts have been made to modify and optimize differentiation protocols by modulation of additional signaling pathways to direct the differentiation process towards enriched populations of atrial and nodal cells (Table 2). Once again, researchers used hints from developmental biology to determine what signaling pathways to target.
Table 2.
Protocols to generate subtype-defined hPSC-CMs
 |
Pink cells indicate positive inducers (growth factors, sera, small molecules); yellow cells indicate inhibitors; green cells indicate media and additional supplements used. Acronyms: 1TG monothioglycerol; BMP4, bone morphogenetic protein 4; BPEL, Bovine Serum Albumin (BSA) Polyvinylalchohol Essential Lipids medium containing IMDM/F12, lipids, Protein-Free Hybridoma mixture II, insulin, transferrin, sodium selenite, ascorbic acid, L-alanyl-l-glutamine, penicillin. streptomycin; DKK1, Dickkopf-related protein-1; EB, embryoid body; FGF2, basic fibroblast growth factor; HSA, human serum albumin; IWP-2, inhibitor of Wnt production 2; MEF-CM, MEF conditioned medium; VEGFA, vascular endothelial growth factor.
Retinoic acid (RA), the biologically active derivative of vitamin A, plays a pivotal role in multiple stages of heart development including defining the second heart field that further contributes to the formation of the atria (Stefanovic and Zaffran, 2017). Here, the cardiac subtype fate is believed to be determined early in development during mesodermal to cardiac progenitor stage. Consequently, activation of RA during early stages of in vitro differentiation has been proposed to facilitate cardiac subtype specification. Studies in murine and human ESC revealed that RA administration at initial stages of differentiation indeed increased the ratio of the atrial subtype population in cultures by suppressing the expression of ventricular genes (Gassanov et al., 2008; Zhang et al., 2011).
Devalla and colleagues determined that cardiac subtype specification occurs post-mesoderm formation and prior to the onset of cardiac progenitor stage (Devalla et al., 2015). By applying a cytokine cocktail in EB-based suspension culture of hESC to initiate CM differentiation, RA treatment (1 μM RA between day 4 and day 7) resulted in approximately 50% CMs predominantly of the atrial lineage, as verified by transcriptomic and electrophysiological analyses. In an extensive study, Lee and colleagues phenotyped the mesodermal population that is induced by cytokines during EB-based differentiation of hESC and hiPSC (Lee et al., 2017). The authors identified distinct mesoderm patterning based on the activin A and BMP4 concentrations applied: a CD235a+ population at higher cytokine concentrations mainly giving rise to ventricular CMs and a RA-responsive RALDH2+ population at lower cytokine concentrations predominantly inducing specification into the atrial subtype (0.5 μM RA or 2 μM retinol was applied from day 3 to day 5). CMs obtained via this differentiation process were validated at the molecular, electrophysiological, pharmacological, as well as on cardiac tissue level (Goldfracht et al., 2020; Lee et al., 2017).
In contrast to the EB-based suspension culture protocols, we adapted the efficient monolayer-based differentiation approach from Burridge and colleagues (Burridge et al., 2014), to establish a simple, robust and easy-to-apply protocol for the directed differentiation into the atrial lineage. By optimizing timing and concentration of RA application (1 μM RA between day 3 and day 6), a highly enriched population of atrial hiPSC-CMs could be obtained (Cyganek et al., 2018). The atrial fate of hiPSC-CMs derived via this protocol was verified on molecular, electrophysiological and 3D cardiac tissue level. By applying this protocol, efficiencies of approximately 90% of the intended atrial subtype within cultures could be achieved. The detailed protocol is available in STAR Protocols (Kleinsorge and Cyganek, 2020).
Recent studies also improved our understanding in directing hPSC towards pacemaker CMs possessing molecular profiles from the sinoatrial node. Protze and colleagues proposed that a stage-specific modulation of several signaling pathways simultaneously promote the directed differentiation of hPSC into nodal-like CMs (Protze et al., 2017). By using their established EB-based differentiation based on cytokines, the combined activation of BMP4, VEGFA, and RA (0.25 μM) as well as signaling inhibition of Wnt (by applying 0.5 μM IWP-2), TGF-β (by applying 5.4 μM SB431542), and FGF2 (by applying 0.5–1 nM PD173074) between day 3 and day 6 resulted in enriched populations of nodal CMs. Although the overall differentiation efficiency was reduced, cultures yielded more than 50% of NKX2.5- SHOX2+ TNNT2+ nodal CMs and only low percentages of the atrial or ventricular subtype. Molecular and electrophysiological analyses as well as transplantation experiments revealed that resulting CMs display the expected pacemaker activity in vitro and in vivo. A further study applied a slightly different cocktail of cytokines and inhibitors to hESC and hiPSC monolayer culture with the combined administration of BMP4, PD173074, and RA antagonist BMS189453 from day 5 to day 7 displaying the best efficiency (around 50%) for pacemaker enriched cultures (Liu et al., 2020).
Ren and colleagues showed that the pacemaker cardiogenesis depends on Wnt signaling at the cardiac precursor stage (Ren et al., 2019). In their approach, Wnt activation by 3 μM CHIR99021 treatment at day 5 for 48 h was sufficient to boost the hESC culture towards the nodal lineage. By applying this protocol, around 50% of CMs displayed molecular and functional properties of cardiac pacemaker CMs. Liang and colleagues hypothesized that lowering the Wnt signaling inhibition typically applied to initiate the mesoderm to cardiac mesoderm transition might suffice to favor pacemaker differentiation (Liang et al., 2020). Indeed, low concentrations of Wnt inhibitor Wnt-C59 (0.01 μM instead of 2 μM) enriched the pacemaker population within hESC and hiPSC monolayer cultures, but to a lower extent (20%–30% TNNT2+ CMs) than other protocols.
In summary, current protocols allow us to efficiently generate nearly homogenous populations of ventricular, atrial, or nodal CMs that highly resemble the major characteristics of the respective subtypes.
Maturation and tissue engineering
Despite the enormous progress in the development of differentiation protocols, the immaturity of hPSC-CMs with embryonic to fetal-like properties might remain a limiting factor to model genetic cardiac diseases, which generally appear in adulthood. In this section, we summarize the major differences between immature and mature CMs, which have been broadly discussed in a number of reviews (Ahmed et al., 2020; Guo and Pu, 2020; Karbassi et al., 2020). Subsequently, we highlight the different strategies proposed for enhancing the maturity of hPSC-CMs.
The degree of CM maturation can be determined by comparing the key features of morphology and structure, electrophysiology, calcium handling, metabolism, and transcriptional signatures. Unlike adult CM morphology, hPSC-CMs cultured in 2D monolayer are typically smaller in size and round or polygonal (with a typical aspect ratio of 2–3:1); they display shorter and less organized sarcomeres and a less developed sarcoplasmic reticulum, and they lack transverse tubules (T-tubules). Immature hPSC-CMs show an electrophysiological phenotype characterized by spontaneous beating and an action potential with decreased duration and a less negative resting membrane potential. In addition, hPSC-CMs show different calcium handling kinetics due to the reduced volume and calcium stores of the sarcoplasmic reticulum, the lack of T-tubules and the lower expression of calcium handling proteins (such as SERCA and RYR2), leading to an underdeveloped excitation-contraction coupling. At the metabolic level, mature CMs mainly rely on fatty acid β-oxidation rather than glycolysis as the major source of energy production. This switch to fatty acid consumption, which occurs during postnatal development, is accompanied by an increase in the number, area, and cristae density of mitochondria. Maturation of hPSC-CMs can be also assessed by measuring sarcomeric isoforms due to the transition of these myofilament proteins during development. For example, adult CMs express the β-MHC isoform (MYH7) instead of α-MHC (MYH6), the shorter titin isoform N2B instead of N2BA, and the cardiac troponin isoform cTnI (TNNI3) instead of the slow skeletal muscle isoform ssTnI (TNNI1) (Ahmed et al., 2020; Guo and Pu, 2020; Karbassi et al., 2020).
Recently, different approaches have been proposed to promote the maturation of hPSC-CM cultures to at least some extent (Figure 3). As one of the first strategies, Lundy and colleagues showed that long-term culture of hESC- and hiPSC-CMs for up to 120 days presented structural changes such as increased cell size and increased myofibrillar density and alignment. They also showed an improvement in contractile performance, calcium handling, and electrophysiological properties (Lundy et al., 2013). Interestingly, long-term culture resulted in myofibrils more tightly packed with the appearance of mature Z-, A-, H-, and I-bands in 180-day-old EBs and M-bands only after 1 year of EB culture (Kamakura et al., 2013). Although prolonged culture can promote maturation, it is not beneficial in terms of time and costs and other methods are preferable.
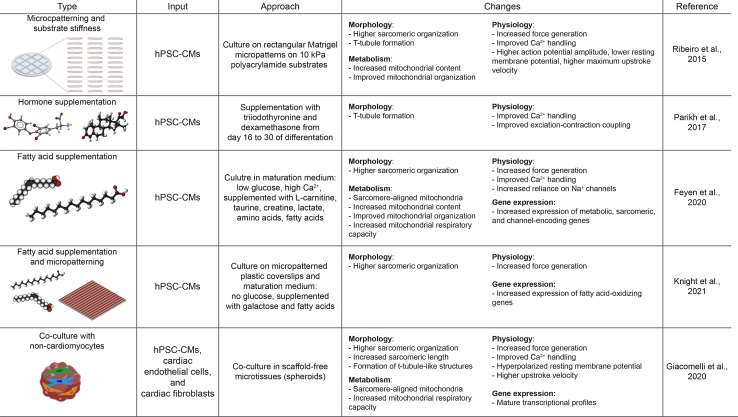
Figure 3.
Different maturation approaches for hPSC-CMs
Mechanical, chemical, and cellular-based cues can be used to induced maturation of hPSC-CMs. Each method has the potential to change to different degrees the morphology, metabolism, physiology, and gene expression of cardiomyocytes.
Alternative efforts to mimic the cardiac environment and promote CM maturation have been tested in vitro. Co-culture with cardiac fibroblasts (CFs) and/or endothelial cells could induce maturation through cell-cell interaction and paracrine signals (Dunn et al., 2019; Ieda et al., 2009; Lee et al., 2015). Recently, Giacomelli and collaborators showed enhanced maturation in micro-tissues generated by combining CMs, CFs, and cardiac endothelial cells, all derived from hiPSCs (Giacomelli et al., 2020). They proved that maturation was induced at least in part by increasing cAMP levels in CMs, which enhanced the assembly of connexin 43 gap junctions between CFs and CMs. In other studies, the cardiac stiffness, much smaller in the myocardium than in culture dishes (∼10 kPa vs. ∼1 GPa), has been mimicked by the use of soft matrices such as polydimethylsiloxane, hydrogels, or polyacrylamide, showing improved features of CM maturation (Feaster et al., 2015; Herron et al., 2016). Ribeiro and colleagues showed that hiPSC-CMs cultured on polyacrylamide with a physiological stiffness (10 kPa) generated more force than those on stiffer substrates (Ribeiro et al., 2015a). Interestingly, they also proved that CMs cultured on rectangular micropatterns with substrates with physiological stiffness and aspect ratio presenting higher alignment of sarcomeric myofibrils, improved calcium flow, higher degree of mitochondrial organization, matured electrophysiology, T-tubule formation, and higher contractile performance.
The use of biochemical cues like hormones can also promote maturation. Treatment of hiPSC-CM cultures for 1 week with the thyroid hormone triiodothyronine (T3), essential for heart development, enhanced the CM maturity state by increasing cell size and sarcomere length and by improving calcium handling kinetics, mitochondrial activity, and contractile force (Yang et al., 2014). Interestingly, the combination of T3 with glucocorticoid hormones (also important during fetal and postnatal heart development) within the culture medium could initiate the development of T-tubules and enhance the excitation-contraction coupling (Parikh et al., 2017). Additionally, the supplementation of fatty acids to the culture medium is gaining relevance to induce a more “adult-like” CM phenotype. Except during the short-term selection period, when lactate is the major energy source, most of the cardiac differentiation protocols are based on glucose-containing medium (Burridge et al., 2014; Lian et al., 2013). In contrast, most of the energy consumed by adult CMs is produced by oxidative phosphorylation using lipid sources (Lopaschuk and Jaswal, 2010). Hence, shifting from glucose-containing to fatty acid-containing medium can enhance maturation of hPSC-CMs by increasing cell size and increasing myofibril density and alignment, as well as by improving contractile force, calcium and action potential kinetics, and mitochondrial oxidative capacity (Correia et al., 2017; Yang et al., 2019a). Recently, Feyen and colleagues developed a defined metabolic maturation medium supplemented with a mixture of fatty acids and containing lower concentrations of glucose and calcium, compared to typically utilized culture media (Feyen et al., 2020). In addition to improved structural, electrophysiological, and functional properties of hiPSC-CMs cultured in maturation medium, the more mature cells also showed similarities to disease phenotypes of Long QT syndrome and dilated cardiomyopathy that could not be reliably modelled in high glucose medium. In parallel, the Song lab integrated the use of a maturation medium rich in fatty acids together with the use of micropatterned surfaces (plastic coverslips patterned with grooves via lapping) to produce structurally and functionally matured human iPSC-CMs (Knight et al., 2021b). Interestingly, by using this combinatorial approach, they were able to suppress baseline hypertrophy, typically associated to prolonged glucose-based culture, and thereby to better model the hypertrophic phenotype in Danon disease. Their detailed two-step protocol is available in STAR Protocols (Knight et al., 2021a).
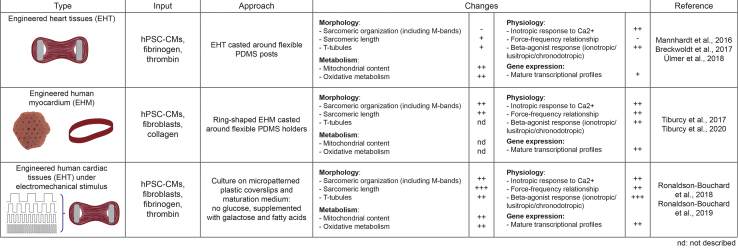
Finally, bioengineering approaches are considered the gold standard for to structural and functional maturation by more closely reflecting the native physiological environment of the human heart muscle (Figure 4). These approaches rely on the generation of 3D cardiac tissues using hPSC-derived CMs (and non-myocytes) embedded in scaffolds and extracellular matrix components. The Hansen lab developed engineered heart tissues by embedding hPSC-CMs into fibrin hydrogels with tissues attached to flexible silicone posts allowing performing contraction analysis under auxotonic stretch conditions (Breckwoldt et al., 2017; Mannhardt et al., 2016). Besides a structural and contractile maturation like native human heart tissue, these CMs had a higher number of mitochondria and an increase in oxidative phosphorylation rather than anaerobic glycolysis (Ulmer et al., 2018). The Zimmermann group constructed engineered heart muscles under defined serum-free conditions, consisting of a defined mixture of hPSC-CMs and (cardiac) fibroblasts or stromal cells cast in collagen hydrogels. They verified advanced morphological, molecular, and functional maturation (Tiburcy et al., 2017). Importantly, addition of the non-myocytes facilitated extracellular matrix remodeling and compaction of the tissues and significantly improved contractile performance with an optimal force performance at an input ratio of 70% CMs and 30% non-CMs. In their STAR Protocol (Tiburcy et al., 2020), the authors provide a step-by-step tutorial to generate engineered heart muscle loops in a 48-well plate format which allows to easily study contractile function under different physiological, pathological, or pharmacological conditions.
Figure 4.
Tissue engineered approaches for further culture of hPSC-CMs
Post-differentiation hPSC-CMs can be engineered into different types of constructs for disease modeling, pharmacological screenings, and developmental studies. Furthermore, each engineered construct has the potential to promote phenotypical changes in the hPSC-CMs, as listed here.
Naturally, these engineered tissues can be combined with further approaches that enhance maturity of CMs in vitro, such as fatty acid supplementation (Mills et al., 2017), mechanical stress, and electrical stimulation (Ruan et al., 2016). Electrical pacing has become in an attractive strategy for biomimicking electrical signals and enhancing CM maturation in vitro. Increasing stimulation constantly from 1 Hz to 6 Hz for one week in engineered cardiac micro-tissues (hPSC-CMs seeded with fibroblasts, endothelial cells and smooth muscle cells on a silicone template), improved major maturation features such as myofibril organization and electrophysiological parameters (Nunes et al., 2013; Zhao et al., 2019). Based on these studies, the Vunjak-Novakovic lab established a protocol to produce cardiac tissues (engineered from hPSC-CMs and fibroblasts in a fibrin hydrogel) with remarkable maturity features by early-stage conditioning with mechanical loading and ramped electrical stimulation (2 weeks at a frequency increasing from 2 Hz to 6 Hz, followed by 1 week at 2 Hz) (Ronaldson-Bouchard et al., 2019). These tissues demonstrated sarcomeric re-arrangement, T-tubule maturation, highly improved contractile performance, and enhanced metabolic features, closely resembling an adult-like phenotype (Ronaldson-Bouchard et al., 2018).
Despite these advances in CM maturation, cost-effective protocols with improved standardization and automation are still needed to facilitate cardiovascular disease modelling and drug screening.
Protocols to generate cardiomyocytes via direct cardiac reprogramming of fibroblasts
Direct cardiac reprogramming, i.e., converting fibroblasts into cardiomyocytes bypassing progenitor stages, is an established and valued strategy for the de novo generation of cardiomyocytes for heart repair (Srivastava and DeWitt, 2016). In general, this is achieved via viral delivery or transient transduction followed by an average 2- to 4-weeks of culture, whereby the forced expression of protein and/or microRNA combinations can convert fibroblasts into induced cardiomyocytes (iCMs) featured with aligned sarcomeres, spontaneous calcium flux, and contractility. In the past decade, different approaches using various cocktails and culture systems have been reported with diverse outcomes. Here, we reviewed the mainstream approaches and aimed to provide a clear guidance for their future applications.
In Tables 3 and 4, we comprehensively list the components of each direct reprogramming approach according to the original reports, which have been repeatedly used in published research. We have divided the reprogramming strategies into five major categories:
-
1.
MGT-core cocktail (Ieda et al., 2010; Mohamed et al., 2017; Muraoka et al., 2014, 2019; Wang et al., 2015a; Yamakawa et al., 2015)
-
2.
GHMT-based cocktail (Garry et al., 2021; Nam et al., 2014; Song et al., 2012; Zhang et al., 2019a, 2021c; Zhou et al., 2015)
-
3.
microRNA combo (Hu et al., 2021; Jayawardena et al., 2012)
-
4.
direct human cardiac reprogramming (Fu et al., 2013; Garbutt et al., 2020; Nam et al., 2013; Wada et al., 2013)
-
5.
chemical only approach (Cao et al., 2016; Fu et al., 2015)
Table 3.
Direct reprogramming approaches
| Features | Cell types |
Cocktails | Efficiency | Reference | |
|---|---|---|---|---|---|
| From | To | ||||
| GMT-based cocktail | CF, TTF | MYH6-GFP+ | GMT | ∼20% | leda et al., 2010 |
| microRNA | CF, MEF | Beating cells | GMT, miR-133 | 7 -old more | Muraoka et al. (2014) |
| HCF | TNNT2+ | GMT, MESP1, MYOCD, miR-133 | 23–27% | ||
| Chemical compound | CF | MYH6-GFP+ | GMT | ∼30% | Mohamed et al. (2017) |
| HCF | TNNT2-GCaMP5+ | GMT, MYOCD, ESRRG, MESP1, ZFPM2 | ∼12% | ||
| MEF, TTF | Beating cells | GMT or GHMT | 100-fold | Yamakawa et al. (2015) | |
| TTF | Beating cells | GMT or GHMT | 4-fold | Muraoka et al. (2019) | |
| Polycistronic puro selection | CF | Beating cells | polycistronic MGT | 10-fold | Wang et al. (2015a) |
| Optimized GHMT-based cocktail | CFs,TTFs | MYH6-GFP+/TNNT2+ | GHMT | 5%–20% | Song et al. (2012) |
| MEF | Beating cells | GHMT Akt1 | 50.0% | Zhou et al. (2015) | |
| CF, TTF | ∼0.8% | ||||
| MEF, CF, TTF | MYH6-GFP+/TNNT2+ | GHMT, Akt1, PHF7 | > 20% | Garry et al. (2021) | |
| Beating cells | ∼2-fold | ||||
| HCF | MYH6-GFP+/TNNT2+ | GHMT, Myocd, PHF7 | ∼ 3% | Garry et al. (2021) | |
| Chamber-specific subtype | MEF, TTF | Sarcomere+/HCN4-GFP+ | GHMT | 1% × ∼32% | Nam et al. (2014) |
| Sarcomere+/HCN4-GFP−/MYL2+ | 1% × ∼22% | ||||
| Sarcomere+/HCN4-GFP−/NPPA+ | 1% × ∼35% | ||||
| MEF | MYL7+/TNNI3+ | Polycistronic M-G-T-H | ∼26% | Zhang et al. (2019a)(2021c) | |
| MYL2+/TNNI3+ | ∼16% | ||||
| Sarcomere+, beating cells | ∼5–6-fold | ||||
| Human Fibroblasts | H9F | MYH6-mCherry+/TNNT2+ | GMT, ESRRG, MESP1, MYOCD, ZFPM2 (7F) | 13.0% ± 9.3% | Fu et al. (2013) |
| HCF, HDF | ACTN2+/TNNT2+ | 1–4% | |||
| HFF, AHCF, AHDF | TNNT2+ | GHT, Myocd, miR-1, miR-133 (6F) | ∼10–35% | Nam et al. (2013) | |
| HCF, HDF | ACTN2+ or TNNT2+ | GMT, Mesp1, Myocd (GMTMM) | ∼ 5% | Wada et al. (2013) | |
| H9F, HCF | TNNT2+ | Polycistronic hMGT + miR-133 (MGT133) | 40–60% | Garbutt et al. (2020) | |
| MicroRNA combo | MEF, CF | MYH6-CFP+ | miR-1, miR-133, miR-208, miR-499 (miRcombo) |
∼13–27% | Jayawardena et al. (2012) |
| Chemical only approach | MEF, TTF | ACTN2+ or MYH6+ | small-molecule combination CRFVPT |
∼10–15% | Fu et al. (2015) |
| HFF | TNNT2+ cells | 9 chemical compounds (9C) | 6.6 ± 0.4% | Cao et al. (2016) | |
| HLF | 5.5% | ||||
Acronyms: AHCF, adult human cardiac fibroblasts; AHDF, adult human dermal fibroblasts; CF, cardiac fibroblasts; H9F, H9-derived fibroblasts; HFF, neonatal human foreskin fibroblasts; HLF, human fetal lung fibroblasts; MEF, mouse embryonic fibroblasts; TTF, tail tip fibroblasts.
Table 4.
Direct reprogramming protocols
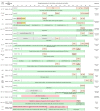 |
Pink cells indicate positive inducers (growth factors, sera, small molecules); yellow cells indicate inhibitors; green cells indicate media and additional supplements used; gray cells indicate selection steps; orange cells indicate measurements. Acronyms: AHCF, adult human CFs; AHDF, adult human dermal fibroblasts; CF, CFs; H9F, H9-derived fibroblasts; HFF, neonatal human foreskin fibroblasts; HLF, human fetal lung fibroblasts; MEF, mouse embryonic fibroblasts; TTF, TTFs; 2ME, ß-mercaptoethanol; FBS, fetal bovine serum; FGF2, basic fibroblast growth factor; FGF10, fibroblast growth factor 10; Glutamax, l-alanyl-l-glutamine; ITS, insulin, transferrin, selenium supplement; KSR, KnockOut Serum Replacement; LIF, leukemia inhibitory factor; NEAA, non-essential amino acids supplement; VEGFA, vascular endothelial growth factor.
In 2010, Ieda and colleagues reported that retroviral delivery of cardiac-enriched transcriptional factors (TFs) Gata4, Mef2c, and Tbx5 (GMT) is sufficient to convert ∼20% of mouse CFs into alpha myosin heavy chain (Myh7)-GFP reporter positive iCMs (Ieda et al., 2010). After 3–4 weeks of culture in iCM medium, iCMs also exhibited calcium flux and spontaneous beating. Optimization of this protocol has been achieved by addition of microRNAs (miR-1 and miR-133) into reprogramming cocktail (Muraoka et al., 2014), treatment of chemicals (Mohamed et al., 2017; Muraoka et al., 2019; Yamakawa et al., 2015), and stoichiometry control of TFs (Wang et al., 2015a, 2015b). The early stage of chemical treatment (SB431542 and XAV939) shortened the appearance of beating cells from 5 weeks to 10 days. Using FFV medium supplemented with FGF2, FGF10, and VEGFA starting from reprogramming day 14 promoted iCM maturation and gave rise to ∼100-fold more beating cells (Mohamed et al., 2017). Moreover, taking advantage of the polycistronic vector expressing factors in the order of Mef2c, Gata4, and Tbx5 (MGT), Li Wang and colleagues generated 10-fold more beating iCMs from CFs (Wang et al., 2015a). Puromycin selection was also performed 2 days after infection to enrich MGT-transduced cells and reduce heterogeneity during reprogramming. The GMT-based approaches have been widely used in the field to answer a variety of questions (Liu et al., 2016, 2017; Stone et al., 2019; Wang et al., 2020; Zhou et al., 2016, 2017, 2018).
Alternatively, independent laboratories established a platform using GMT plus Hand2 (GHMT) as a minimal cocktail to generate induced cardiomyocyte-like myocytes from different types of fibroblasts. Besides the change of cocktail, cardiac induction medium was also modified by including horse serum, additional nutrients and 10% conditioned medium obtained from neonatal rat or mouse cardiomyocytes (Song et al., 2012). Repeated viral infection was applied at the first 2 days. Furthermore, the same group found that addition of auxiliary factors such as AKT serine/threonine kinase 1 (Akt1) and histone reader PHD finger protein 7 (Phf7), led to more efficient generation of beating cells from relatively reluctant MEF (MEFs) and tail-tip fibroblast (TTFs) and adult CFs (Garry et al., 2021; Zhou et al., 2015). Of note, several studies determined the subtype cardiomyocytes generated with GHMT via combinational marker expression. Nam and colleagues adopted multi-immunostaining and assigned a unique identity for pacemaker CMs, ventricular, and atrial CMs (Nam et al., 2014; Zhang et al., 2021c). They found 1% of GHMT-transduced cells with well aligned sarcomeres, among which ∼32% Hcn4-GFP+ cells were pacemaker like cells; ∼ 22% were Hcn4-GFP−/Myl2+ ventricular myocytes; ∼35% were Hcn4-GFP−/Nppa+ atrial like myocytes. Recently, Hand2 was revealed as a key factor that regulates chromatin accessibility to induce pacemaker lineage conversion (Fernandez-Perez et al., 2019), highlighting the application of GHMT-based approach to study direct reprogramming of chamber or subtype specific cardiomyocytes.
Development of an efficient and reproducible platform for human direct cardiac reprogramming is critical for moving to clinical applications. Many efforts have been made to generate human iCMs, but current protocols are still limited. Due to an unknown molecular basis, adult human fibroblasts are unable to be converted to iCMs by the mouse core TF combination GMT. Through screening led by independent laboratories, human direct cardiac reprogramming could be induced with three different combinations of TFs/miRs: 7F (GMT, ESRRG, MESP1, MYOCD, ZFPM2) (Fu et al., 2013), 6F (GHT, MYOCD, miR-1, miR-133) (Nam et al., 2013), and GMTMM (MGT, MESP1, MYOCD) (Wada et al., 2013). Although additional factors were delivered via retroviruses, human fibroblasts were delayed in reprogramming into CM-like cells with much lower efficiency in calcium flux and rare spontaneous beating compared to mouse reprogramming. Similar stoichiometry control of MGT and puro selection simplified the cocktail components and improved the human cardiac reprogramming in terms of efficiency, yield, and reproducibility (Garbutt et al., 2020; Zhou et al., 2019). The field still struggles to obtain spontaneously beating human iCMs. Most studies showing contractility of human iCMs are in a long-term co-culture system with functionally beating CMs derived from rat/mouse or PSC differentiation. The success of reprogramming requires highly efficient viral infection and robust transgene expression (Garbutt et al., 2020). In addition, the use of human fibroblasts primarily derived from adult tissue and well-maintained proliferating fibroblasts without excessive passaging typically yields better outcomes.
Unlike viral delivery of TFs, a combination of microRNAs, including miR-1, miR-133, miR-208, and miR-499, termed as miRcombo, was transiently transfected in fibroblasts and induced CMs with treatment of JAK inhibitor I (Jayawardena et al., 2012). The miRcombo-induced CMs were characterized by cardiac marker expression, sarcomeric structure and CM-like calcium flux. Furthermore, the same miRcombo was transduced in adult human cardiac fibroblasts and led to ∼15% of cells expressing cardiac troponin T (TNNT2) (Paoletti et al., 2020). Although this approach seems convenient and integration-free, miRcombo-induced CMs have not been fully characterized in terms of electrophysiological function. Also, it remains unknown what are the similarities and differences between TF- and miRcombo-induced CMs. Additional transcriptomic or functional characterization of miRcombo-induced CMs or side-by-side comparison using TF- and miR-based reprogramming approaches would help us to further evaluate these two approaches.
Direct conversion of fibroblasts into CMs via a chemical-only approach is attractive and valuable as it has several advantages over genetic methods mentioned above. Delivery of small molecules is simple, efficient, and controllable. A protocol with two-stage chemical induction from mouse fibroblasts has been reported to efficiently generate chemical-induced cardiomyocytes (ciCMs) (Fu et al., 2015). Of note, through a lineage tracing assay, the authors found the generation of a mixed progenitor population after CRFVPT induction (Fu et al., 2015). Similarly, a 9-chemical cocktail initiated the two-stage cell fate conversion of human fibroblasts mimicking cardiogenetic program from cardiac progenitor cells to differentiated beating CMs (Cao et al., 2016). Considering the appearance of progenitor stage, the current chemical-induced approach is not the same as transgene-derived direct cardiac reprogramming. Unlike the transgene approaches mentioned above, chemical approaches have not been widely investigated and questions remain: are chemical approaches applicable for in vivo reprogramming?; how can chemical cocktails be specifically administrated into fibroblasts in the heart?
The approaches used to evaluate reprogramming efficiency is different and inconsistent between various studies. Flow cytometry is widely used to quantify the proportion of cells labeled by cardiac sarcomere proteins (Table 3). Results from flow cytometry are straightforward and intuitive to compare and analyze, but the expression of only one or two proteins is insufficient to indicate the quantity and quality of iCMs. For instance, flow cytometry shows no significant difference in TNNT2 and MYH6, while qRT-PCR and microarray show significant differences in global gene expression (Yamakawa et al., 2015). To address these limitations, we evaluated reprogramming with additional assessments, such as Ca2+ flux/ oscillations, beating clusters/cells, and epigenomic profiling. It should be noted that it is not reasonable to compare the number of beating cells provided by independent laboratories. Some groups showed beating iCM number in each high-power field (Miyamoto et al., 2018; Zhou et al., 2015), some used number of beating clusters per field (Fu et al., 2015; Testa et al., 2020), others counted the number of beating loci in each culture well (Wang et al., 2015a; Zhang et al., 2019b). A panel of relatively standardized measurements might be helpful for the field, and those should include the evaluation of morphological, genomic, epigenomic, and functional markers.
Virus packaging protocols
Since most approaches used lenti- or retro-viral delivery method to overexpress reprogramming cocktails, we also summarized the virus packaging protocols in Table 5. The differences in viral generation, including different constructs, host cell lines, and transfection reagents, are likely to be part of the reasons for the variations reported by different laboratories. The high expression levels of reprogramming factors are key for successful cardiac reprogramming (Liu et al., 2017). For clinical applications, alternative integration-free and efficient delivery methods have been tested during direct cardiac reprogramming. Adenoviral vectors (Ad) and chimeric adeno-associated viruses (AAV) expressing GMT showed equivalent reprogramming efficacy in CFs according to the immune florescence staining of sarcomere proteins (Mathison et al., 2017; Yoo et al., 2018). However, no detailed functional or transcriptome-wide characterization of iCMs generated by Ad or AAV has been reported. Additionally, Miyamoto and colleagues used sendai virus vectors expressing GMT to generate iCMs in vitro and in vivo (Miyamoto et al., 2018). They showed high transgene expression shortly after infection and more efficient generation of beating iCMs when compared to retroviral delivery. As a safe and non-immunogenic gene delivery method, modified mRNA (modRNA) was used to deliver a cocktail including GHMT, dominant-negative (DN)-TGF-β, DN-Wnt8a, and acid ceramidase into mouse fibroblasts (Kaur et al., 2021). Cardiac marker expression showed 57% reprogramming efficiency in vitro after repeated transfection of modRNA mixture during the first two weeks of reprogramming. The potential limitation for modRNA might be the relatively short transgene expression time and unknown cell type specificity for in vivo treatment. Nevertheless, these studies provide additional alternatives for future therapeutic studies and applications.
Table 5.
Viral packaging for direct reprogramming
| Cells | Overexpressed genes | Virus | Backbone | Transfection | Polybrene | Positive selection | Induction system | Reference |
|---|---|---|---|---|---|---|---|---|
| Platinum E cells | GMT | Retrovirus | pMXs | FuGene6 (Roche) | leda et al., 2010 | |||
| Platinum E cells | Myc-tagged GHMT | Retrovirus | pBabeX | FuGene6 (Roche) | 6 μg/mL | Song et al. (2012) | ||
| HEK293 FT | GMT, ESRRG, MESP1, MYOCD, ZFPM2 (7F) | Lentivirus | pLVX-tetO-cDNA | Lipofectamine 2000 | 4 μg/mL | 1 μg/mL Dox | Fu et al. (2013) | |
| pLVX-Tight-Puro | ||||||||
| Retrovirus | pMXs | FuGene6 (Promega) | ||||||
| Retro-X-Tet-ON | 5 μg/mL | puromycin & neomycin | ||||||
| pRetro-X-Tight-cDNA | 100 ng/mL Dox | |||||||
| Platinum A cells | GMT, Mesp1, Myocd | Retrovirus | pBabeX | FuGene6 (Promega) | 6 μg/mL | Nam et al. (2013) | ||
| PLAT-GP cells | GMT, MESP1, MYOCD (GMTMM) | Retrovirus | pMXs | TremeGene 9 (Roche) | 4 μg/mL | Wada et al. (2013) | ||
| 293FT cells | Lentivirus | pLVX-Tight-Puro | Lipofectamine 2000 | 4 μg/mL | 1 μg/mL Dox | |||
| pLVX-rtTA | ||||||||
| pLVX-tetO-cDNA | ||||||||
| Platinum A cells | Myc-tagged GHMT | Retrovirus | pBabeX | FuGene6 (Promega) | 6 μg/mL | Nam et al. (2014) | ||
| Platinum E cells | GMT | Retrovirus | pMXs | FuGene6 (Roche) | Muraoka et al. (2014) | |||
| HEK293 | human GMT, MESP1, MYOCD | Lentivirus | CSII-CMV-RfA | Lipofectamine 2000 | ||||
| Platinum E cells | Polycistronic M-G-T | Retrovirus | pMXs | Lipofectamine 2000 | 4 μg/mL | puromycin 2 μg/mL | Wang et al. (2015a) | |
| Platinum A cells | GHMT | Retrovirus | pBabeX | FuGene6 (Promega) | 6 μg/μL | Zhou et al. (2015) | ||
| Akt1 | pMxs-MyrAkt1 | |||||||
| Platinum E cells | GHMT, MESP1, MYOCD | Retrovirus | pMXs | FuGene6 (Promega) | Yamakawa et al. (2015) | |||
| Platinum E cells | GMT | Retrovirus | pMXs | FuGene HD (Roche) | 0.6 μg/mL | Mohamed et al. (2017) | ||
| Platinum E cells | GMT or GHMT | Retrovirus | pMXs | FuGene6 | 10 μM diclofenac | Muraoka et al. (2019) | ||
| Platinum E cells | polycistronic M-G-T-H | Retrovirus | pBabeX | FuGene6 (Promega) | 6 μg/mL | Zhang et al. (2019a)(2021c) | ||
| 293T cells | Polycistronic human M-G-T, miRNA-133 | Retrovirus | pMXs | Nanofect | 8 μg/mL | puromycin 1 μg/mL | Garbutt et al. (2020) | |
| pBabe | ||||||||
| Platinum E cells | GHMT, Myocd, PHF7 | Retrovirus | pMXs | FuGene6 (Promega) | 8 μg/mL | Garry et al. (2021) |
Acronyms: Dox, doxycycline.
Conclusion
In the past 20 years, we moved from using hESC to hiPSC providing significant gains in the ability to study patient-relevant models, understand genetic diseases, and perform effective drug screenings. The methods for culturing the cells also evolved, from the use of poorly defined sera to chemically defined media supplementations such as E8 and B8. The same can be said about differentiation strategies that progressed from uncontrolled and non-specific spontaneous differentiation protocols with embryoid bodies that relied on sera and expensive growth factors, to more chemically defined approaches such as CDM3 that works for both monolayers and 3D constructs. These approaches enabled the development of strategies to generate subtype-specific tissues, and to mature the cardiomyocytes with extracellular cues, avoiding long-term cultures, speeding up research, and broadening the knowledge about heart diseases and their causes. Additionally, we saw the evolution of direct reprogramming, with initial studies identifying the GMT cocktail, but more recent approaches focusing on the use of other TFs, in addition to replacing retroviruses with sendai or other reprogramming techniques. However, at present cardiomyocytes generated by directed reprogramming strategies are not comparable to those generated by hiPSC differentiation techniques and have not been demonstrated to be suitable for disease modeling or drug testing.
Some legacy elements of the methods to culture and differentiate hiPSC and complete directed reprogramming remain. The use of old basal media, like DMEM/F12, for the generation and maintenance of stem cells, or the reliance on albumin for consistent differentiation and potential maturation of cardiomyocytes should be addressed, as should the correct choice of TFs for reprogramming and their use in a potential non-integrative way. HiPSC have become the tool of choice for disease modelling, drug screening, personalized medicine, and potential regenerative medicine applications. To ensure further improvements, subsequent developments should concentrate on scalability, reproducibility, and lowering costs of both culture and differentiation. With an increased availability of cells at lower costs, we can use the combined knowledge around the world to improve the present applications and develop new systems that will make the next 20 years even more exciting.
Acknowledgments
This work was supported by NIH grants CA220002 and CA261898 (to P.W.B), R01 HL153220 (to Y.Z.); American Heart Association Postdoctoral Fellowship 874276 (to D.M.L-L.); the German Federal Ministry of Education and Research (BMBF)/German Center for Cardiovascular Research (DZHK) (to L.C.); the German Research Foundation (DFG) EXC 2067/1–390729940, CY 90/1-1 and SFB1002 S01 (to L.C.); and the Else-Kröner-Fersenius Foundation 2019_A75 (to L.C.). Figures were produced with the aid of SMART Servier Medical Art (www.smart.servier.com, Attribution 3.0 Unported (CC BY 3.0)), and Marcel Tisch via bioicons.com (CC0 1.0 Universal (CC0 1.0) Public Domain Dedication).
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101560.
Supplemental information
Document S4. Excel version of Table 2, “Protocols to generate subtype-defined hPSC-CMs”
References
- Aguilar J., Begum A., Alvarez J., Zhang X.B., Hong Y., Hao J. Directed cardiomyogenesis of human pluripotent stem cells by modulating Wnt/β-catenin and BMP signalling with small molecules. Biochem. J. 2015;469:235–241. doi: 10.1042/bj20150186. [DOI] [PubMed] [Google Scholar]
- Ahmed R.E., Anzai T., Chanthra N., Uosaki H. A Brief review of current maturation methods for human induced pluripotent stem cells-derived cardiomyocytes. Front. Cell Dev. Biol. 2020;8:178. doi: 10.3389/fcell.2020.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit M., Shariki C., Margulets V., Itskovitz-Eldor J. Feeder layer- and serum-free culture of human embryonic stem Cells1. Biol. Reprod. 2004;70:837–845. doi: 10.1095/biolreprod.103.021147. [DOI] [PubMed] [Google Scholar]
- Beattie G.M., Lopez A.D., Bucay N., Hinton A., Firpo M.T., King C.C., Hayek A. Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cell. 2005;23:489–495. doi: 10.1634/stemcells.2004-0279. [DOI] [PubMed] [Google Scholar]
- Beers J., Gulbranson D.R., George N., Siniscalchi L.I., Jones J., Thomson J.A., Chen G. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat. Protoc. 2012;7:2029–2040. doi: 10.1038/nprot.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckwoldt K., Letuffe-Breniere D., Mannhardt I., Schulze T., Ulmer B., Werner T., Benzin A., Klampe B., Reinsch M.C., Laufer S., et al. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc. 2017;12:1177–1197. doi: 10.1038/nprot.2017.033. [DOI] [PubMed] [Google Scholar]
- Buikema J.W., Lee S., Goodyer W.R., Maas R.G., Chirikian O., Li G., Miao Y., Paige S.L., Lee D., Wu H., et al. Wnt activation and reduced cell-cell Contact Synergistically induce Massive expansion of functional human iPSC-derived cardiomyocytes. Cell Stem Cell. 2020;27:50–63.e5. doi: 10.1016/j.stem.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P.W., Anderson D., Priddle H., Barbadillo Munoz M.D., Chamberlain S., Allegrucci C., Young L.E., Denning C. Improved human embryonic stem cell embryoid body homogeneity and cardiomyocyte differentiation from a novel V-96 plate aggregation system highlights interline variability. Stem Cell. 2007;25:929–938. doi: 10.1634/stemcells.2006-0598. [DOI] [PubMed] [Google Scholar]
- Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M., et al. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P.W., Thompson S., Millrod M.A., Weinberg S., Yuan X., Peters A., Mahairaki V., Koliatsos V.E., Tung L., Zambidis E.T. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao N., Huang Y., Zheng J., Spencer C.I., Zhang Y., Fu J.D., Nie B., Xie M., Zhang M., Wang H., et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science. 2016;352:1216–1220. doi: 10.1126/science.aaf1502. [DOI] [PubMed] [Google Scholar]
- Cao N., Liu Z., Chen Z., Wang J., Chen T., Zhao X., Ma Y., Qin L., Kang J., Wei B., et al. Ascorbic acid enhances the cardiac differentiation of induced pluripotent stem cells through promoting the proliferation of cardiac progenitor cells. Cell Res. 2012;22:219–236. doi: 10.1038/cr.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E., et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Hou Z., Gulbranson D.R., Thomson J.A. Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell. 2010;7:240–248. doi: 10.1016/j.stem.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Tristan C.A., Chen L., Jovanovic V.M., Malley C., Chu P.-H., Ryu S., Deng T., Ormanoglu P., Tao D., et al. A versatile polypharmacology platform promotes cytoprotection and viability of human pluripotent and differentiated cells. Nat. Methods. 2021;18:528–541. doi: 10.1038/s41592-021-01126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia C., Koshkin A., Duarte P., Hu D., Teixeira A., Domian I., Serra M., Alves P.M. Distinct carbon sources affect structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Sci. Rep. 2017;7:8590. doi: 10.1038/s41598-017-08713-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyganek L., Tiburcy M., Sekeres K., Gerstenberg K., Bohnenberger H., Lenz C., Henze S., Stauske M., Salinas G., Zimmermann W.H., et al. Deep phenotyping of human induced pluripotent stem cell-derived atrial and ventricular cardiomyocytes. JCI Insight. 2018;3:99941. doi: 10.1172/jci.insight.99941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devalla H.D., Schwach V., Ford J.W., Milnes J.T., El-Haou S., Jackson C., Gkatzis K., Elliott D.A., Chuva de Sousa Lopes S.M., Mummery C.L., et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 2015;7:394–410. doi: 10.15252/emmm.201404757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn K.K., Reichardt I.M., Simmons A.D., Jin G., Floy M.E., Hoon K.M., Palecek S.P. Coculture of endothelial cells with human pluripotent stem cell-derived cardiac progenitors reveals a differentiation stage-specific Enhancement of cardiomyocyte maturation. Biotechnol. J. 2019;14 doi: 10.1002/biot.201800725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D.A., Braam S.R., Koutsis K., Ng E.S., Jenny R., Lagerqvist E.L., Biben C., Hatzistavrou T., Hirst C.E., Yu Q.C., et al. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat. Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- Feaster T.K., Cadar A.G., Wang L., Williams C.H., Chun Y.W., Hempel J.E., Bloodworth N., Merryman W.D., Lim C.C., Wu J.C., et al. Matrigel Mattress: a method for the generation of single contracting human-induced pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2015;117:995–1000. doi: 10.1161/circresaha.115.307580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Perez A., Sathe A.A., Bhakta M., Leggett K., Xing C., Munshi N.V. Hand2 Selectively Reorganizes chromatin accessibility to induce pacemaker-like transcriptional reprogramming. Cell Rep. 2019;27:2354–2369.e7. doi: 10.1016/j.celrep.2019.04.077. e2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyen D.A., McKeithan W.L., Bruyneel A.A., Spiering S., Hörmann L., Ulmer B., Zhang H., Briganti F., Schweizer M., Hegyi B., et al. Metabolic maturation media improve physiological function of human iPSC-derived cardiomyocytes. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonoudi H., Yeganeh M., Fattahi F., Ghazizadeh Z., Rassouli H., Alikhani M., Mojarad B.A., Baharvand H., Salekdeh G.H., Aghdami N. ISL1 protein transduction promotes cardiomyocyte differentiation from human embryonic stem cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0055577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S., Zhang M., Scholer H.R., Greber B. Small molecule-assisted, line-independent maintenance of human pluripotent stem cells in defined conditions. PLoS One. 2012;7 doi: 10.1371/journal.pone.0041958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund C., Ward-van Oostwaard D., Monshouwer-Kloots J., van den Brink S., van Rooijen M., Xu X., Zweigerdt R., Mummery C., Passier R. Insulin redirects differentiation from cardiogenic mesoderm and endoderm to neuroectoderm in differentiating human embryonic stem cells. Stem Cell. 2008;26:724–733. doi: 10.1634/stemcells.2007-0617. [DOI] [PubMed] [Google Scholar]
- Fu J.D., Stone N., Liu L., Spencer C., Qian L., Hayashi Y., Delgado-Olguin P., Ding S., Bruneau B., Srivastava D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. 2013;1:235–247. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Huang C., Xu X., Gu H., Ye Y., Jiang C., Qiu Z., Xie X. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res. 2015;25:1013–1024. doi: 10.1038/cr.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi S., Fernandes I., Mastikhina O., Wilkinson D., Tran T., Dhahri W., Mazine A., Yang D., Burnett B., Lee J., et al. Generation of mature compact ventricular cardiomyocytes from human pluripotent stem cells. Nat. Commun. 2021;12:3155. doi: 10.1038/s41467-021-23329-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furue M.K., Na J., Jackson J.P., Okamoto T., Jones M., Baker D., Hata R.I., Moore H.D., Sato J.D., Andrews P.W. Heparin promotes the growth of human embryonic stem cells in a defined serum-free medium. Proc. Natl. Acad. Sci. USA. 2008;105:13409–13414. doi: 10.1073/pnas.0806136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbutt T.A., Zhou Y., Keepers B., Liu J., Qian L. An optimized protocol for human direct cardiac reprogramming. STAR Protoc. 2020;1 doi: 10.1016/j.xpro.2019.100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F.R., Izpisua Belmonte J.C. Albumin-associated lipids regulate human embryonic stem cell self-renewal. PLoS One. 2008;3:e1384. doi: 10.1371/journal.pone.0001384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garry G.A., Bezprozvannaya S., Chen K., Zhou H., Hashimoto H., Morales M.G., Liu N., Bassel-Duby R., Olson E.N. The histone reader PHF7 cooperates with the SWI/SNF complex at cardiac super enhancers to promote direct reprogramming. Nat. Cell Biol. 2021;23:467–475. doi: 10.1038/s41556-021-00668-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassanov N., Er F., Zagidullin N., Jankowski M., Gutkowska J., Hoppe U.C. Retinoid acid-induced effects on atrial and pacemaker cell differentiation and expression of cardiac ion channels. Differentiation. 2008;76:971–980. doi: 10.1111/j.1432-0436.2008.00283.x. [DOI] [PubMed] [Google Scholar]
- Giacomelli E., Meraviglia V., Campostrini G., Cochrane A., Cao X., van Helden R.W., Krotenberg Garcia A., Mircea M., Kostidis S., Davis R.P., et al. Human-iPSC-Derived cardiac stromal cells enhance maturation in 3D cardiac Microtissues and reveal non-cardiomyocyte Contributions to heart disease. Cell Stem Cell. 2020;26:862–879.e11. doi: 10.1016/j.stem.2020.05.004. e811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfracht I., Protze S., Shiti A., Setter N., Gruber A., Shaheen N., Nartiss Y., Keller G., Gepstein L. Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat. Commun. 2020;11:75. doi: 10.1038/s41467-019-13868-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Lee J.W., Schultz P.G. Stepwise chemically induced cardiomyocyte specification of human embryonic stem cells. Angew Chem. Int. Ed. Engl. 2011;123:11377–11381. doi: 10.1002/ange.201103909. [DOI] [PubMed] [Google Scholar]
- Graichen R., Xu X., Braam S.R., Balakrishnan T., Norfiza S., Sieh S., Soo S.Y., Tham S.C., Mummery C., Colman A., et al. Enhanced cardiomyogenesis of human embryonic stem cells by a small molecular inhibitor of p38 MAPK. Differentiation. 2008;76:357–370. doi: 10.1111/j.1432-0436.2007.00236.x. [DOI] [PubMed] [Google Scholar]
- Guo Y., Pu W.T. Cardiomyocyte maturation: new phase in development. Circ. Res. 2020;126:1086–1106. doi: 10.1161/circresaha.119.315862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan N.R.F., Segeritz C.P., Touboul T., Vallier L. Production of hepatocyte-like cells from human pluripotent stem cells. Nat. Protoc. 2013;8:430–437. doi: 10.1038/nprot.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanses U., Kleinsorge M., Roos L., Yigit G., Li Y., Barbarics B., El-Battrawy I., Lan H., Tiburcy M., Hindmarsh R., et al. Intronic CRISPR repair in a preclinical model of Noonan syndrome-associated cardiomyopathy. Circulation. 2020;142:1059–1076. doi: 10.1161/circulationaha.119.044794. [DOI] [PubMed] [Google Scholar]
- Herron T.J., Rocha A.M.D., Campbell K.F., Ponce-Balbuena D., Willis B.C., Guerrero-Serna G., Liu Q., Klos M., Musa H., Zarzoso M., et al. Extracellular matrix-mediated maturation of human pluripotent stem cell-derived cardiac monolayer structure and electrophysiological function. Circ Arrhythm Electrophysiol. 2016;9 doi: 10.1161/circep.113.003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D., Linders A., Yamak A., Correia C., Kijlstra J.D., Garakani A., Xiao L., Milan D.J., van der Meer P., Serra M., et al. Metabolic maturation of human pluripotent stem cell-derived cardiomyocytes by inhibition of HIF1α and LDHA. Circ. Res. 2018;123:1066–1079. doi: 10.1161/circresaha.118.313249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Hodgkinson C.P., Pratt R.E., Lee J., Sullenger B.A., Dzau V.J. Enhancing cardiac reprogramming via synthetic RNA oligonucleotides. Mol. Ther. Nucleic Acids. 2021;23:55–62. doi: 10.1016/j.omtn.2020.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M., Tsuchihashi T., Ivey K.N., Ross R.S., Hong T.T., Shaw R.M., Srivastava D. Cardiac fibroblasts regulate myocardial proliferation through β1 Integrin signaling. Dev. Cell. 2009;16:233–244. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena T.M., Egemnazarov B., Finch E.A., Zhang L., Payne J.A., Pandya K., Zhang Z., Rosenberg P., Mirotsou M., Dzau V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012;110:1465–1473. doi: 10.1161/circresaha.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakura T., Makiyama T., Sasaki K., Yoshida Y., Wuriyanghai Y., Chen J., Hattori T., Ohno S., Kita T., Horie M., et al. Ultrastructural maturation of human-induced pluripotent stem cell-derived cardiomyocytes in a long-term culture. Circ. J. 2013;77:1307–1314. doi: 10.1253/circj.cj-12-0987. [DOI] [PubMed] [Google Scholar]
- Karbassi E., Fenix A., Marchiano S., Muraoka N., Nakamura K., Yang X., Murry C.E. Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol. 2020;17:341–359. doi: 10.1038/s41569-019-0331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman S.J., Witty A.D., Gagliardi M., Dubois N.C., Niapour M., Hotta A., Ellis J., Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kaur K., Hadas Y., Kurian A.A., Zak M.M., Yoo J., Mahmood A., Girard H., Komargodski R., Io T., Santini M.P., et al. Direct reprogramming induces vascular regeneration post muscle ischemic injury. Mol. Ther. 2021;29:3042–3058. doi: 10.1016/j.ymthe.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehat I., Kenyagin-Karsenti D., Snir M., Segev H., Amit M., Gepstein A., Livne E., Binah O., Itskovitz-Eldor J., Gepstein L. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J. Clin. Invest. 2001;108:407–414. doi: 10.1172/jci200112131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinsorge M., Cyganek L. Subtype-directed differentiation of human iPSCs into atrial and ventricular cardiomyocytes. STAR Protoc. 2020;1 doi: 10.1016/j.xpro.2020.100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight W.E., Cao Y., Dillon P., Song K. A simple protocol to produce mature human-induced pluripotent stem cell-derived cardiomyocytes. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2021.100912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight W.E., Cao Y., Lin Y.H., Chi C., Bai B., Sparagna G.C., Zhao Y., Du Y., Londono P., Reisz J.A., et al. Maturation of pluripotent stem cell-derived cardiomyocytes enables modeling of human hypertrophic cardiomyopathy. Stem Cell Rep. 2021;16:519–533. doi: 10.1016/j.stemcr.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H.H., Gao X., DeKeyser J.M., Fetterman K.A., Pinheiro E.A., Weddle C.J., Fonoudi H., Orman M.V., Romero-Tejeda M., Jouni M., et al. Negligible-cost and weekend-free chemically defined human iPSC culture. Stem Cell Rep. 2020;14:256–270. doi: 10.1016/j.stemcr.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laco F., Lam A.T.L., Woo T.L., Tong G., Ho V., Soong P.L., Grishina E., Lin K.H., Reuveny S., Oh S.K.W. Selection of human induced pluripotent stem cells lines optimization of cardiomyocytes differentiation in an integrated suspension microcarrier bioreactor. Stem Cell Res. Ther. 2020;11:118. doi: 10.1186/s13287-020-01618-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laco F., Woo T.L., Zhong Q., Szmyd R., Ting S., Khan F.J., Chai C.L., Reuveny S., Chen A., Oh S. Unraveling the Inconsistencies of cardiac differentiation efficiency induced by the GSK3β inhibitor CHIR99021 in human pluripotent stem cells. Stem Cell Rep. 2018;10:1851–1866. doi: 10.1016/j.stemcr.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme M.A., Chen K.Y., Naumova A.V., Muskheli V., Fugate J.A., Dupras S.K., Reinecke H., Xu C., Hassanipour M., Police S., et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- Lambshead J.W., Meagher L., Goodwin J., Labonne T., Ng E., Elefanty A., Stanley E., O'Brien C.M., Laslett A.L. Long-term maintenance of human pluripotent stem cells on cRGDfK-presenting synthetic surfaces. Sci. Rep. 2018;8:701. doi: 10.1038/s41598-018-19209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Chen J.H., Lundy D., Liu C.H., Hwang S.M., Pabon L., Shieh R.C., Chen C.C., Wu S.N., Yan Y.T., et al. Defined MicroRNAs induce aspects of maturation in mouse and human embryonic-stem-cell-derived cardiomyocytes. Cell Rep. 2015;12:1960–1967. doi: 10.1016/j.celrep.2015.08.042. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Protze S.I., Laksman Z., Backx P.H., Keller G.M. Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell. 2017;21:179–194.e4. doi: 10.1016/j.stem.2017.07.003. e174. [DOI] [PubMed] [Google Scholar]
- Lian X., Bao X., Zilberter M., Westman M., Fisahn A., Hsiao C., Hazeltine L.B., Dunn K.K., Kamp T.J., Palecek S.P. Chemically defined, albumin-free human cardiomyocyte generation. Nat. Methods. 2015;12:595–596. doi: 10.1038/nmeth.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Zhang J., Azarin S.M., Zhu K., Hazeltine L.B., Bao X., Hsiao C., Kamp T.J., Palecek S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Han P., Kim E.H., Mak J., Zhang R., Torrente A.G., Goldhaber J.I., Marbán E., Cho H.C. Canonical Wnt signaling promotes pacemaker cell specification of cardiac mesodermal cells derived from mouse and human embryonic stem cells. STEM CELLS. 2020;38:352–368. doi: 10.1002/stem.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Linask K.L., Mallon B., Johnson K., Klein M., Beers J., Xie W., Du Y., Liu C., Lai Y., et al. Heparin promotes cardiac differentiation of human pluripotent stem cells in chemically defined albumin-free medium, enabling consistent Manufacture of cardiomyocytes. Stem Cells Transl Med. 2017;6:527–538. doi: 10.5966/sctm.2015-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Zou J. Differentiation of cardiomyocytes from human pluripotent stem cells in fully chemically defined conditions. STAR Protoc. 2020;1:100015. doi: 10.1016/j.xpro.2020.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litviňuková M., Talavera-López C., Maatz H., Reichart D., Worth C.L., Lindberg E.L., Kanda M., Polanski K., Heinig M., Lee M., et al. Cells of the adult human heart. Nature. 2020;588:466–472. doi: 10.1038/s41586-020-2797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Fang Y., Hou X., Yan Y., Xiao H., Zuo D., Wen J., Wang L., Zhou Z., Dang X., et al. Enrichment differentiation of human induced pluripotent stem cells into sinoatrial node-like cells by combined modulation of BMP, FGF, and RA signaling pathways. Stem Cell Res. Ther. 2020;11:284. doi: 10.1186/s13287-020-01794-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Chen O., Zheng M., Wang L., Zhou Y., Yin C., Liu J., Qian L. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res. 2016;16:507–518. doi: 10.1016/j.scr.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wang L., Welch J.D., Ma H., Zhou Y., Vaseghi H.R., Yu S., Wall J.B., Alimohamadi S., Zheng M., et al. Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature. 2017;551:100–104. doi: 10.1038/nature24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopaschuk G.D., Jaswal J.S. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J. Cardiovasc. Pharmacol. 2010;56:130–140. doi: 10.1097/fjc.0b013e3181e74a14. [DOI] [PubMed] [Google Scholar]
- Ludwig T.E., Levenstein M.E., Jones J.M., Berggren W.T., Mitchen E.R., Frane J.L., Crandall L.J., Daigh C.A., Conard K.R., Piekarczyk M.S., et al. Derivation of human embryonic stem cells in defined conditions. Nat. Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- Lundy S.D., Zhu W.Z., Regnier M., Laflamme M.A. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev. 2013;22:1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyra-Leite D.M., Fonoudi H., Gharib M., Burridge P.W. An updated protocol for the cost-effective and weekend-free culture of human induced pluripotent stem cells. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2020.100213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas R.G., Lee S., Harakalova M., Snijders Blok C.J., Goodyer W.R., Hjortnaes J., Doevendans P.A., Van Laake L.W., van der Velden J., Asselbergs F.W., et al. Massive expansion and cryopreservation of functional human induced pluripotent stem cell-derived cardiomyocytes. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2021.100334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdy T., Jiang Z., Jouni M., Fonoudi H., Lyra-Leite D., Jung G., Romero-Tejeda M., Kuo H.H., Fetterman K.A., Gharib M., et al. RARG variant predictive of doxorubicin-induced cardiotoxicity identifies a cardioprotective therapy. Cell Stem Cell. 2021;28:2076–2089.e7. doi: 10.1016/j.stem.2021.08.006.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdy T., Jouni M., Kuo H.H., Weddle C.J., Lyra-Leite D., Fonoudi H., Romero-Tejeda M., Gharib M., Javed H., Fajardo G., Ross C.J., Carleton B.C., Bernstein D., Burridge P.W. Identification of drug Transporter genomic variants and inhibitors that Protect against doxorubicin-induced cardiotoxicity. Circulation. 2022;145:279–294. doi: 10.1161/circulationaha.121.055801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannhardt I., Breckwoldt K., Letuffe-Brenière D., Schaaf S., Schulz H., Neuber C., Benzin A., Werner T., Eder A., Schulze T., et al. Human engineered heart tissue: analysis of contractile force. Stem Cell Rep. 2016;7:29–42. doi: 10.1016/j.stemcr.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manstein F., Ullmann K., Kropp C., Halloin C., Triebert W., Franke A., Farr C.M., Sahabian A., Haase A., Breitkreuz Y., et al. High density bioprocessing of human pluripotent stem cells by metabolic control and in silico modeling. Stem Cells Transl Med. 2021;10:1063–1080. doi: 10.1002/sctm.20-0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manstein F., Ullmann K., Triebert W., Zweigerdt R. Process control and in silico modeling strategies for enabling high density culture of human pluripotent stem cells in stirred tank bioreactors. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2021.100988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinho P.A., Chailangkarn T., Muotri A.R. Systematic optimization of human pluripotent stem cells media using Design of Experiments. Sci. Rep. 2015;5:9834. doi: 10.1038/srep09834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathison M., Singh V.P., Chiuchiolo M.J., Sanagasetti D., Mao Y., Patel V.B., Yang J., Kaminsky S.M., Crystal R.G., Rosengart T.K. In situ reprogramming to transdifferentiate fibroblasts into cardiomyocytes using adenoviral vectors: implications for clinical myocardial regeneration. J. Thorac. Cardiovasc. Surg. 2017;153:329–339.e3. doi: 10.1016/j.jtcvs.2016.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt T.C., Laflamme M.A., Murry C.E. Proliferation of cardiomyocytes derived from human embryonic stem cells is mediated via the IGF/PI 3-kinase/Akt signaling pathway. J. Mol. Cell. Cardiol. 2005;39:865–873. doi: 10.1016/j.yjmcc.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeithan W.L., Feyen D.A., Bruyneel A.A., Okolotowicz K.J., Ryan D.A., Sampson K.J., Potet F., Savchenko A., Gómez-Galeno J., Vu M., et al. Reengineering an Antiarrhythmic drug using patient hiPSC cardiomyocytes to improve therapeutic potential and reduce toxicity. Cell Stem Cell. 2020;27:813–821.e6. doi: 10.1016/j.stem.2020.08.003. e816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.J., Titmarsh D.M., Koenig X., Parker B.L., Ryall J.G., Quaife-Ryan G.A., Voges H.K., Hodson M.P., Ferguson C., Drowley L., et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. USA. 2017;114:E8372–E8381. doi: 10.1073/pnas.1707316114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami I., Yamada K., Otsuji T., Yamamoto T., Shen Y., Otsuka S., Kadota S., Morone N., Barve M., Asai Y., et al. A small molecule that promotes cardiac differentiation of human pluripotent stem cells under defined, cytokine- and xeno-free conditions. Cell Rep. 2012;2:1448–1460. doi: 10.1016/j.celrep.2012.09.015. [DOI] [PubMed] [Google Scholar]
- Miyamoto K., Akiyama M., Tamura F., Isomi M., Yamakawa H., Sadahiro T., Muraoka N., Kojima H., Haginiwa S., Kurotsu S., et al. Direct in vivo reprogramming with sendai virus vectors improves cardiac function after myocardial infarction. Cell Stem Cell. 2018;22:91–103.e5. doi: 10.1016/j.stem.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Mohamed T.M.A., Stone N.R., Berry E.C., Radzinsky E., Huang Y., Pratt K., Ang Y.S., Yu P., Wang H., Tang S., et al. Chemical Enhancement of in vitro and in vivo direct cardiac reprogramming. Circulation. 2017;135:978–995. doi: 10.1161/circulationaha.116.024692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummery C., Ward-van Oostwaard D., Doevendans P., Spijker R., van den Brink S., Hassink R., van der Heyden M., Opthof T., Pera M., de la Riviere A.B., et al. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–2740. doi: 10.1161/01.cir.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- Muraoka N., Nara K., Tamura F., Kojima H., Yamakawa H., Sadahiro T., Miyamoto K., Isomi M., Haginiwa S., Tani H., et al. Role of cyclooxygenase-2-mediated prostaglandin E2-prostaglandin E receptor 4 signaling in cardiac reprogramming. Nat. Commun. 2019;10:674. doi: 10.1038/s41467-019-08626-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka N., Yamakawa H., Miyamoto K., Sadahiro T., Umei T., Isomi M., Nakashima H., Akiyama M., Wada R., Inagawa K., et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. Embo j. 2014;33:1565–1581. doi: 10.15252/embj.201387605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y.J., Lubczyk C., Bhakta M., Zang T., Fernandez-Perez A., McAnally J., Bassel-Duby R., Olson E.N., Munshi N.V. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development. 2014;141:4267–4278. doi: 10.1242/dev.114025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y.J., Song K., Luo X., Daniel E., Lambeth K., West K., Hill J.A., DiMaio J.M., Baker L.A., Bassel-Duby R., Olson E.N. Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. USA. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y., Walsh P., Clarke D.L., Rowley J.A., Fellner T. Scalable passaging of adherent human pluripotent stem cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0088012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes S.S., Miklas J.W., Liu J., Aschar-Sobbi R., Xiao Y., Zhang B., Jiang J., Masse S., Gagliardi M., Hsieh A., et al. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat. Methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige S.L., Osugi T., Afanasiev O.K., Pabon L., Reinecke H., Murry C.E. Endogenous Wnt/β-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti C., Divieto C., Tarricone G., Di Meglio F., Nurzynska D., Chiono V. MicroRNA-Mediated direct reprogramming of human adult fibroblasts toward cardiac phenotype. Front. Bioeng. Biotechnol. 2020;8:529. doi: 10.3389/fbioe.2020.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S.S., Blackwell D.J., Gomez-Hurtado N., Frisk M., Wang L., Kim K., Dahl C.P., Fiane A., Tønnessen T., Kryshtal D.O., et al. Thyroid and glucocorticoid hormones promote functional T-tubule development in human-induced pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2017;121:1323–1330. doi: 10.1161/circresaha.117.311920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passier R., Oostwaard D.W., Snapper J., Kloots J., Hassink R.J., Kuijk E., Roelen B., de la Riviere A.B., Mummery C. Increased cardiomyocyte differentiation from human embryonic stem cells in serum-free cultures. Stem Cell. 2005;23:772–780. doi: 10.1634/stemcells.2004-0184. [DOI] [PubMed] [Google Scholar]
- Pei F., Jiang J., Bai S., Cao H., Tian L., Zhao Y., Yang C., Dong H., Ma Y. Chemical-defined and albumin-free generation of human atrial and ventricular myocytes from human pluripotent stem cells. Stem Cell Res. 2017;19:94–103. doi: 10.1016/j.scr.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Protze S.I., Liu J., Nussinovitch U., Ohana L., Backx P.H., Gepstein L., Keller G.M. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol. 2017;35:56–68. doi: 10.1038/nbt.3745. [DOI] [PubMed] [Google Scholar]
- Qiu X.X., Liu Y., Zhang Y.F., Guan Y.N., Jia Q.Q., Wang C., Liang H., Li Y.Q., Yang H.T., Qin Y.W., et al. Rapamycin and CHIR99021 Coordinate robust cardiomyocyte differentiation from human pluripotent stem cells via reducing p53-dependent Apoptosis. J. Am. Heart Assoc. 2017;6:e005295. doi: 10.1161/jaha.116.005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querdel E., Reinsch M., Castro L., Köse D., Bähr A., Reich S., Geertz B., Ulmer B., Schulze M., Lemoine M.D., et al. Human engineered heart tissue Patches Remuscularize the injured heart in a Dose-dependent manner. Circulation. 2021;143:1991–2006. doi: 10.1161/circulationaha.120.047904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Han P., Ma X., Farah E.N., Bloomekatz J., Zeng X.X.I., Zhang R., Swim M.M., Witty A.D., Knight H.G., et al. Canonical Wnt5b signaling directs Outlying Nkx2.5+ mesoderm into pacemaker cardiomyocytes. Dev. Cell. 2019;50:729–743.e5. doi: 10.1016/j.devcel.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Lee M.Y., Schliffke S., Paavola J., Amos P.J., Ge X., Ye M., Zhu S., Senyei G., Lum L., et al. Small molecule Wnt inhibitors enhance the efficiency of BMP-4-directed cardiac differentiation of human pluripotent stem cells. J. Mol. Cell. Cardiol. 2011;51:280–287. doi: 10.1016/j.yjmcc.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro A.J.S., Ang Y.S., Fu J.D., Rivas R.N., Mohamed T.M.A., Higgs G.C., Srivastava D., Pruitt B.L. Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proc. Natl. Acad. Sci. USA. 2015;112:12705–12710. doi: 10.1073/pnas.1508073112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro M.C., Tertoolen L.G., Guadix J.A., Bellin M., Kosmidis G., D'Aniello C., Monshouwer-Kloots J., Goumans M.J., Wang Y.L., Feinberg A.W., et al. Functional maturation of human pluripotent stem cell derived cardiomyocytes in vitro--correlation between contraction force and electrophysiology. Biomaterials. 2015;51:138–150. doi: 10.1016/j.biomaterials.2015.01.067. [DOI] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K., Ma S.P., Yeager K., Chen T., Song L., Sirabella D., Morikawa K., Teles D., Yazawa M., Vunjak-Novakovic G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018;556:239–243. doi: 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K., Yeager K., Teles D., Chen T., Ma S., Song L., Morikawa K., Wobma H.M., Vasciaveo A., Ruiz E.C., et al. Engineering of human cardiac muscle electromechanically matured to an adult-like phenotype. Nat. Protoc. 2019;14:2781–2817. doi: 10.1038/s41596-019-0189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J.L., Tulloch N.L., Razumova M.V., Saiget M., Muskheli V., Pabon L., Reinecke H., Regnier M., Murry C.E. Mechanical stress conditioning and electrical stimulation promote contractility and force maturation of induced pluripotent stem cell-derived human cardiac tissue. Circulation. 2016;134:1557–1567. doi: 10.1161/circulationaha.114.014998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro F., Chien K.R., Sahara M. Isolation of human ESC-derived cardiac derivatives and embryonic heart cells for population and single-cell RNA-seq analysis. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2021.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz T.C., Noggle S.A., Palmarini G.M., Weiler D.A., Lyons I.G., Pensa K.A., Meedeniya A.C., Davidson B.P., Lambert N.A., Condie B.G. Differentiation of human embryonic stem cells to dopaminergic neurons in serum-free suspension culture. Stem Cell. 2004;22:1218–1238. doi: 10.1634/stemcells.2004-0114. [DOI] [PubMed] [Google Scholar]
- Sharma A., Zhang Y., Buikema J.W., Serpooshan V., Chirikian O., Kosaric N., Churko J.M., Dzilic E., Shieh A., Burridge P.W., et al. Stage-specific effects of Bioactive lipids on human iPSC cardiac differentiation and cardiomyocyte proliferation. Sci. Rep. 2018;8:6618. doi: 10.1038/s41598-018-24954-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozawa T., Nakamura K., Shoji M., Morita M., Kimura M., Furukawa H., Ueda H., Shiramoto M., Matsuguma K., Kaji Y., et al. Recapitulation of clinical individual Susceptibility to drug-induced QT prolongation in Healthy Subjects using iPSC-derived cardiomyocytes. Stem Cell Rep. 2017;8:226–234. doi: 10.1016/j.stemcr.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Nam Y.J., Luo X., Qi X., Tan W., Huang G.N., Acharya A., Smith C.L., Tallquist M.D., Neilson E.G., et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D., DeWitt N. 2016. In-Vivo Cellular Reprogramming: The Next Generation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S., Zaffran S. Mechanisms of retinoic acid signaling during cardiogenesis. Mech. Dev. 2017;143:9–19. doi: 10.1016/j.mod.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Stone N.R., Gifford C.A., Thomas R., Pratt K.J., Samse-Knapp K., Mohamed T.M., Radzinsky E.M., Schricker A., Ye L., Yu P., et al. Context-specific transcription factor functions regulate epigenomic and transcriptional Dynamics during cardiac reprogramming. Cell Stem Cell. 2019;25:87–102.e9. doi: 10.1016/j.stem.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei S., Ichikawa H., Johkura K., Mogi A., No H., Yoshie S., Tomotsune D., Sasaki K. Bone morphogenetic protein-4 promotes induction of cardiomyocytes from human embryonic stem cells in serum-based embryoid body development. Am. J. Physiol. Heart Circ. Physiol. 2009;296:H1793–H1803. doi: 10.1152/ajpheart.01288.2008. [DOI] [PubMed] [Google Scholar]
- Tan Y., Han P., Gu Q., Chen G., Wang L., Ma R., Wu J., Feng C., Zhang Y., Wang L., et al. Generation of clinical-grade functional cardiomyocytes from human embryonic stem cells in chemically defined conditions. J Tissue Eng Regen Med. 2018;12:153–163. doi: 10.1002/term.2381. [DOI] [PubMed] [Google Scholar]
- Testa G., Russo M., Di Benedetto G., Barbato M., Parisi S., Pirozzi F., Tocchetti C.G., Abete P., Bonaduce D., Russo T., Passaro F. Bmi1 inhibitor PTC-209 promotes Chemically-induced Direct Cardiac Reprogramming of cardiac fibroblasts into cardiomyocytes. Sci. Rep. 2020;10:7129. doi: 10.1038/s41598-020-63992-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tiburcy M., Hudson J.E., Balfanz P., Schlick S., Meyer T., Chang Liao M.L., Levent E., Raad F., Zeidler S., Wingender E., et al. Defined engineered human myocardium with advanced maturation for applications in heart Failure modeling and repair. Circulation. 2017;135:1832–1847. doi: 10.1161/circulationaha.116.024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiburcy M., Meyer T., Liaw N.Y., Zimmermann W.H. Generation of engineered human myocardium in a multi-well format. STAR Protoc. 2020;1 doi: 10.1016/j.xpro.2020.100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T.H., Wang X., Browne C., Zhang Y., Schinke M., Izumo S., Burcin M. Wnt3a-induced mesoderm formation and cardiomyogenesis in human embryonic stem cells. Stem Cell. 2009;27:1869–1878. doi: 10.1002/stem.95. [DOI] [PubMed] [Google Scholar]
- Ulmer B.M., Stoehr A., Schulze M.L., Patel S., Gucek M., Mannhardt I., Funcke S., Murphy E., Eschenhagen T., Hansen A. Contractile work contributes to maturation of energy metabolism in hiPSC-derived cardiomyocytes. Stem Cell Rep. 2018;10:834–847. doi: 10.1016/j.stemcr.2018.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L., Alexander M., Pedersen R.A. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- Virani S.S., Alonso A., Aparicio H.J., Benjamin E.J., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Cheng S., Delling F.N., et al. American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee Heart disease and Stroke Statistics-2021 Update: a report from the American heart association. Circulation. 2021;143:e254–e743. doi: 10.1161/cir.0000000000000950. [DOI] [PubMed] [Google Scholar]
- Wada R., Muraoka N., Inagawa K., Yamakawa H., Miyamoto K., Sadahiro T., Umei T., Kaneda R., Suzuki T., Kamiya K., et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. USA. 2013;110:12667–12672. doi: 10.1073/pnas.1304053110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hao J., Hong C.C. Cardiac induction of embryonic stem cells by a small molecule inhibitor of Wnt/β-catenin signaling. ACS Chem. Biol. 2011;6:192–197. doi: 10.1021/cb100323z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu Z., Yin C., Asfour H., Chen O., Li Y., Bursac N., Liu J., Qian L. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ. Res. 2015;116:237–244. doi: 10.1161/circresaha.116.305547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu Z., Yin C., Zhou Y., Liu J., Qian L. Improved generation of induced cardiomyocytes using a polycistronic construct expressing optimal ratio of Gata4, Mef2c and Tbx5. JoVE. 2015 doi: 10.3791/53426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Ma H., Huang P., Xie Y., Near D., Wang H., Xu J., Yang Y., Xu Y., Garbutt T., et al. Down-regulation of Beclin1 promotes direct cardiac reprogramming. Sci. Transl. Med. 2020;12:eaay7856. doi: 10.1126/scitranslmed.aay7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Schulz T.C., Sherrer E.S., Dauphin D.S., Shin S., Nelson A.M., Ware C.B., Zhan M., Song C.Z., Chen X., et al. Self-renewal of human embryonic stem cells requires insulin-like growth factor-1 receptor and ERBB2 receptor signaling. Blood. 2007;110:4111–4119. doi: 10.1182/blood-2007-03-082586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems E., Spiering S., Davidovics H., Lanier M., Xia Z., Dawson M., Cashman J., Mercola M. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ. Res. 2011;109:360–364. doi: 10.1161/circresaha.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Rosler E., Jiang J., Lebkowski J.S., Gold J.D., O'Sullivan C., Delavan-Boorsma K., Mok M., Bronstein A., Carpenter M.K. Basic fibroblast growth factor supports undifferentiated human embryonic stem cell growth without conditioned medium. Stem Cell. 2005;23:315–323. doi: 10.1634/stemcells.2004-0211. [DOI] [PubMed] [Google Scholar]
- Xu X.Q., Graichen R., Soo S.Y., Balakrishnan T., Bte Rahmat S.N., Sieh S., Tham S.C., Freund C., Moore J., Mummery C., et al. Chemically defined medium supporting cardiomyocyte differentiation of human embryonic stem cells. Differentiation. 2008;76:958–970. doi: 10.1111/j.1432-0436.2008.00284.x. [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhu X., Hahm H.S., Wei W., Hao E., Hayek A., Ding S. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc. Natl. Acad. Sci. USA. 2010;107:8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H., Muraoka N., Miyamoto K., Sadahiro T., Isomi M., Haginiwa S., Kojima H., Umei T., Akiyama M., Kuishi Y., et al. Fibroblast growth factors and vascular endothelial growth factor promote cardiac reprogramming under defined conditions. Stem Cell Rep. 2015;5:1128–1142. doi: 10.1016/j.stemcr.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S., Taguchi Y., Shimamoto A., Mukasa H., Tahara H., Okamoto T. Generation of human induced pluripotent stem (iPS) cells in serum- and feeder-free defined culture and TGF-β1 regulation of pluripotency. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Soonpaa M.H., Adler E.D., Roepke T.K., Kattman S.J., Kennedy M., Henckaerts E., Bonham K., Abbott G.W., Linden R.M., et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Yang X., Rodriguez M., Pabon L., Fischer K.A., Reinecke H., Regnier M., Sniadecki N.J., Ruohola-Baker H., Murry C.E. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J. Mol. Cell. Cardiol. 2014;72:296–304. doi: 10.1016/j.yjmcc.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Rodriguez M.L., Leonard A., Sun L., Fischer K.A., Wang Y., Ritterhoff J., Zhao L., Kolwicz S.C., Pabon L., et al. Fatty acids enhance the maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cell Rep. 2019;13:657–668. doi: 10.1016/j.stemcr.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ren Z., Xu F., Meng Y., Zhang Y., Ai N., Long Y., Fok H.I., Deng C., Zhao X., et al. Endogenous IGF signaling directs heterogeneous mesoderm differentiation in human embryonic stem cells. Cell Rep. 2019;29:3374–3384.e5. doi: 10.1016/j.celrep.2019.11.047. e3375. [DOI] [PubMed] [Google Scholar]
- Yasuda S.Y., Ikeda T., Shahsavarani H., Yoshida N., Nayer B., Hino M., Vartak-Sharma N., Suemori H., Hasegawa K. Chemically defined and growth-factor-free culture system for the expansion and derivation of human pluripotent stem cells. Nat Biomed Eng. 2018;2:173–182. doi: 10.1038/s41551-018-0200-7. [DOI] [PubMed] [Google Scholar]
- Yoo S.Y., Jeong S.N., Kang J.I., Lee S.W. Chimeric adeno-associated virus-mediated cardiovascular reprogramming for ischemic heart disease. ACS Omega. 2018;3:5918–5925. doi: 10.1021/acsomega.8b00904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Qiu H., Dong X., Wang C., Na J., Zhou J., Wang C. Transferrin improved the generation of cardiomyocyte from human pluripotent stem cells for myocardial infarction repair. J. Mol. Histol. 2021;52:87–99. doi: 10.1007/s10735-020-09926-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Z., Zhao S.R., Tu C., Pang P., Zhang M., Wu J.C. Protocol to measure contraction, calcium, and action potential in human-induced pluripotent stem cell-derived cardiomyocytes. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2021.100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Schulte J.S., Heinick A., Piccini I., Rao J., Quaranta R., Zeuschner D., Malan D., Kim K.-P., Röpke A., et al. Universal cardiac induction of human pluripotent stem cells in two and three-Dimensional formats: implications for in vitro maturation. STEM CELLS. 2015;33:1456–1469. doi: 10.1002/stem.1964. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Jiang J., Han P., Yuan Q., Zhang J., Zhang X., Xu Y., Cao H., Meng Q., Chen L., et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011;21:579–587. doi: 10.1038/cr.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Villalpando J., Zhang W., Nam Y.J. Chamber-specific protein expression during direct cardiac reprogramming. Cells. 2021;10:1513. doi: 10.3390/cells10061513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang A.D., Kim L.J., Nam Y.J. Ensuring expression of four core cardiogenic transcription factors enhances cardiac reprogramming. Sci. Rep. 2019;9:6362. doi: 10.1038/s41598-019-42945-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang W., Nam Y.J. Stoichiometric optimization of Gata4, Hand2, Mef2c, and Tbx5 expression for contractile cardiomyocyte reprogramming. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-51536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Rafatian N., Feric N.T., Cox B.J., Aschar-Sobbi R., Wang E.Y., Aggarwal P., Zhang B., Conant G., Ronaldson-Bouchard K., et al. A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell. 2019;176:913–927.e18. doi: 10.1016/j.cell.2018.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Dickson M.E., Kim M.S., Bassel-Duby R., Olson E.N. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc. Natl. Acad. Sci. USA. 2015;112:11864–11869. doi: 10.1073/pnas.1516237112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Alimohamadi S., Wang L., Liu Z., Wall J.B., Yin C., Liu J., Qian L. A Loss of function screen of Epigenetic Modifiers and Splicing factors during early stage of cardiac reprogramming. Stem Cells Int. 2018;2018:1–14. doi: 10.1155/2018/3814747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Liu Z., Welch J.D., Gao X., Wang L., Garbutt T., Keepers B., Ma H., Prins J.F., Shen W., et al. Single-cell transcriptomic analyses of cell fate transitions during human cardiac reprogramming. Cell Stem Cell. 2019;25:149–164.e9. doi: 10.1016/j.stem.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Wang L., Liu Z., Alimohamadi S., Yin C., Liu J., Qian L. Comparative gene expression analyses reveal distinct molecular signatures between differentially reprogrammed cardiomyocytes. Cell Rep. 2017;20:3014–3024. doi: 10.1016/j.celrep.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Wang L., Vaseghi H., Liu Z., Lu R., Alimohamadi S., Yin C., Fu J.D., Wang G., Liu J., Qian L. Bmi1 is a key Epigenetic Barrier to direct cardiac reprogramming. Cell Stem Cell. 2016;18:382–395. doi: 10.1016/j.stem.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S4. Excel version of Table 2, “Protocols to generate subtype-defined hPSC-CMs”