Abstract
Endometrial cancer (EC) is the most common gynecologic malignancy, and it arises in the inner part of the uterus. Identification of serum biomarkers is essential for diagnosing the disease at an early stage. In this study, we selected 44 healthy controls and 44 type I EC at tumor stage 1, and we used the Immuno-oncology panel and the Target 96 Oncology III panel to simultaneously detect the levels of 92 cancer-related proteins in serum, using a proximity extension assay. By applying this methodology, we identified 20 proteins, associated with the outcome at binary logistic regression, with a p-value below 0.01 for the first panel and 24 proteins with a p-value below 0.02 for the second one. The final multivariate logistic regression model, combining proteins from the two panels, generated a model with a sensitivity of 97.67% and a specificity of 83.72%. These results support the use of the proposed algorithm after a validation phase.
Keywords: biomarkers, proteins, endometrial cancer, targeted proteomics, PEA
1. Introduction
Endometrial cancer (EC) arises in the inner part of the uterus and represents the fourth most common female malignancy in Europe [1]. Unlike other cancers, the incidence and mortality of EC are rapidly increasing worldwide, especially in North America and Western Europe (incidence 12.9–20.2 per 100,000 women and mortality 2.0–3.7 per 100,000 women) [2]. Although genetic predisposition and racial background might promote EC development, the most important EC-predisposing factors seem to be associated with health and lifestyle conditions (e.g., obesity, metabolic syndrome, diabetes, polycystic ovary syndrome, high estrogen levels) [3,4,5,6].
EC is classified into two subtypes with distinct clinical, pathological and molecular features. Commonly, type I ECs display a low grade (I or II) endometrioid morphology and are estrogen-dependent; thus, they are associated with a good prognosis. Type II ECs include non-endometrioid adenocarcinomas, serous clear cell, undifferentiated carcinomas and carcinosarcomas, usually hormone-receptor negative high-grade tumors, with poor prognosis [7].
Type I EC comprises the large majority of endometrial cancers, ~90%, while type II EC comprises ~10%. In type I EC, stage 1 is the most frequent [8]. In EC, 80% of patients are in the early stages, while 20% are in more advanced phases [9]. At stage I, the five-year survival rate is 95%, and the survival decreases dramatically to 14% for stage IV [10]. The identification of biomarkers at an early stage would lead to a prompt diagnosis, reduce inappropriate and invasive examinations and improve patient care and prognosis [11]. Patients in more advanced stages require more specific care, such as radio therapy or chemotherapy. For this purpose, identification of biomarkers at an early stage is important. Harsh treatment can be avoided for many patients when EC is discovered at an early stage [12].
Growing pieces of evidence showed that, by releasing soluble mediators and extracellular vesicles, EC cells are able to recruit stromal cell, immune system cells and macrophages [13,14], which, in turn, favor tumor growth.
Interestingly, inflammation can exert a double role in cancer development. On one hand, triggers of inflammation activate immune cells that, in turn, produce suppressive tumor growth cytokines [15]. Thus, inflammation can induce the response of the immune system against cancer. On the other hand, inflammation is involved in the promotion of tumorigenesis [16], for example, several cytokines such as IL-6 [17], TNFα [18], TGF-β [19], and IFNs [20] are involved in cancer development and progression.
Cytokines’ profiles are promising in the diagnosis and prognosis of pancreatic [21], colorectal [22] and ovarian cancers [23]. Multi-omics approaches allowed us to understand several tumor mechanisms and, of note, the identification of diagnostic and prognostic markers [24]. The proteomic approach allowed for the identification of several candidate biomarkers in the serum of EC patients, such as CLU, C1R, and SERPINC1 [25], SBSN [26], and PAK1 [27].
In this study, we used proximity extension assays (PEAs) for target biomarker analysis in serum from EC patients. Such technology uses high-multiplex matched pairs of antibodies, labelled with unique DNA oligonucleotides, which bind to their specific/relative proteins in the samples [28]. Several studies used PEA for targeted biomarker analysis in different types of cancers, such as esophageal squamous cell carcinomas [29], cervical cancer [30] and ovarian cancer [31]. The Olink PEA platform is a high-multiplex protein biomarker platform with a high-throughput capacity with high specificity and sensitivity in combination with minimal sample volume [32]. This powerful technology has been applied for the screening of proteins in blood [33,34], and on other matrices, e.g., cells [35], urine [36], peritoneal fluid [37], and exosomes [38]. Moreover, recently it has been applied to study immunological mechanisms in the multisystem inflammatory syndrome in children with COVID-19 [39].
Our study aims to apply the PEA technology to screen candidate serum protein markers of early-stage of type I EC.
2. Materials and Methods
2.1. Patients
For this study, a total of 88 women (44 suffering from EC and 44 non-EC controls) were recruited at the Institute for Maternal and Child Health—IRCCS “Burlo Garofolo” (Trieste, Italy) from 2019 to 2021. All EC patients had type I endometrioid adenocarcinomas at tumor stage 1. Type I EC comprises the large majority of endometrial cancers, ~90%, while the type II EC comprises ~10%. In type I EC, stage 1 is the most frequent. For this reason, in this study we focused on type I EC at stage 1 patients to identify candidate serum protein biomarkers.
All procedures complied with the Declaration of Helsinki and were approved by the Institute’s Technical and Scientific Committee. All patients signed informed consent forms. In Supplementary Table S1, we describe the clinical and pathological characteristics of the patients. The median age of patients was 67 years (Inter quartile range 55–71), with a minimum of 44 and a maximum of 81, while the median age of controls was 35 years (IQR 27–51), with a minimum of 22 and a maximum of 77 years. Controls were chosen excluding oncologic patients, human immunodeficiency virus (HIV), hepatitis B virus (HBV), hepatitis C virus (HCV) seropositive subjects, and patients with leiomyomas or adenomyosis. For EC cases, we ruled out women with other oncologic pathologies, human immunodeficiency virus (HIV), hepatitis B virus (HBV), hepatitis C virus (HCV) seropositive patients, and patients with leiomyomas or adenomyosis. We excluded control patients with benign tumors (myoma), chronic inflammatory disease (adenomyosis) or viral infections because these pathologies may influence the abundance of several proteins in serum and, consequently, affect the proteomic analysis.
2.2. Serum Sample Collection and the PEA
To obtain serum, blood was centrifuged at 5000 rcf × 5 min. Once obtained, the serum was preserved at −80 °C. Sera were shipped to Olink® Proteomic (Dag Hammarskjölds väg 52B, SE-752 37 Uppsala, Sweden). In total, 40 µL of serum was used for PEA analysis in the Immuno-oncology panel and the Target 96 Oncology III panel with 96-wells, in which 92 oligonucleotide-labeled antibody probe pairs bind to their specific targeted proteins. The protein names, gene names, and abbreviations for the 92 proteins of the Immuno-oncology panel and Target 96 Oncology III panel are reported in Tables S2 and S3. The PEA technology includes three core steps. It starts with an overnight incubation of 16–22 h. The 92 antibody pairs, labelled with DNA oligonucleotides, bind to their respective protein in the samples. During the second step, we have 2 h of extension and amplification. Oligonucleotides that are brought into proximity hybridize, and are extended using a DNA polymerase. This newly created piece of DNA barcode is amplified by PCR. In the last step, we have 4.5 h of detection. The amount of each DNA barcode is quantified by microfluidic qPCR.
Negative Control for Olink Explore is also included in triplicate on each plate and consists of buffer run as a normal sample. These are used to monitor any background noise generated when DNA-tags come in close proximity without prior binding to the appropriate protein. The negative controls set the background levels for each protein assay and are used to calculate the limit of detection (LOD) and to assess the potential contamination of the assays. The Plate Control was another control included in triplicate on each plate. The median of the Plate Control triplicates is used to normalize each assay and compensate for the potential variation between runs and plates. Once the data were obtained from the plate reading, they were analyzed, including normalization and linearization, by protocols of the manufacturer. The protein level is expressed as NPX, Normalized Protein eXpression, an arbitrary unit in Log2 scale. It is calculated from Ct values, and data pre-processing (normalization) is performed to minimize both intra- and inter-assay variation. NPX data allow users to identify changes in individual protein levels across their sample set, and then use these data to establish protein signatures.
Olink Target 96 Immuno-oncology panels include proteins associated with biological functions linked to immune response and immuno-oncology diseases. The biomarkers in this panel include proteins involved in processes such as promotion and inhibition of tumor immunity, chemotaxis, vascular and tissue remodeling, apoptosis and cell killing and metabolism and autophagy. The Olink Target 96 oncology III panel comprises 92 human proteins that participate in biological mechanisms that are central to the initiation and progression of cancer, e.g., angiogenesis, cell communication, cellular metabolic processes, apoptosis, cell proliferation/differentiation, etc. In Supplement Tables S2 and S3, all the proteins that make up the two panels are reported. These panels do not focus on specific malignancies. The categorization of the proteins included in the panel was carried out via referral to widely used public-access bioinformatic databases, including Uniprot, Human Protein Atlas, Gene Ontology (GO) and DisGeNET.
2.3. Bioinformatic Analysis
Proteins used for statistical analysis for both the panels were analyzed by gProfiler [40] (https://biit.cs.ut.ee/gprofiler/gost) accessed on 4 July 2022 classification systems and categorized according to their: molecular function involvement, biological processes, and protein class. Pathway analysis was done by REACTOME tool [41] (https://reactome.org/PathwayBrowser/#TOOL=AT) accessed on 4 July 2022.
2.4. Statistical Analyses
Each of the two panels comprised 92 proteins. We first excluded all proteins with more than 25% values below the limit of detection (LOD). Olink suggests excluding assays in the range of less than 25–50% of samples above LOD (https://www.olink.com/faq/how-is-the-limit-of-detection-lod-estimated-and-handled/) accessed on 4 July 2022, but we adopted a more restrictive approach and chose to have at least 75% of samples above the LOD. After excluding these, for each of the remaining proteins, we calculated the median value, and the interquartile range. We carried out binary logistic regressions to study the association with the outcome. Before proceeding to the multivariate logistic regression, for each panel, we selected the proteins more strongly associated with the dependent variable, on the basis of the binary logistic regression p-value result. Our first approach was to carry out a first selection of proteins with a least absolute shrinkage and selection operator (LASSO) multivariate logistic regression approach, but the results were unfortunately not comparable with a traditional binary logistic regression approach. For each panel separately, the selected proteins were simultaneously considered in a multivariate logistic regression model. A downward selection was applied to exclude, one at a time, the proteins with the highest p-value, if p ≥ 0.05. We thus obtained two final predictive models which included only proteins significantly and simultaneously associated with the outcome. For each model, we reported the Pseudo-R-squared value, the Area under the Receiver Operating Characteristic (ROC) Curve (AUC), sensitivity and specificity. Finally, we decided to consider the two final models together in a multivariate logistic regression model. we hypothesize that improving the predictive models we might obtain a group of proteins that could be included in an ad hoc panel. Again, we adopted a stepdown procedure and obtained a third model.
3. Results
In the first panel—the Immuno-oncology panel—ten proteins had more than 25% values below the LOD (IL_1_alpha, FGF2, IL2, IL33, CD28, IL5, PTN, CXCL12, IL4, IL13) and were excluded from further analyses. Of the remaining proteins, median values and interquartile ranges are reported for cases and controls, as well as the odds ratios, 95% confidence intervals and p-values of the binary logistic regression (Table 1).
Table 1.
Immuno-oncology panel: result of descriptive analysis, and binary logistic regression against the outcome.
| Immuno-Oncology | Cases | Controls | Binary Logistic Regression |
|---|---|---|---|
| Proteins | Median (IQR) | Median (IQR) | OR (95% CI); p-Value |
| IL8 | 6.044 (5.629–6.517) | 5.66 (5.336–6.106) | 1.291 (0.817–2.039); 0.274 |
| TNFRSF9 | 6.485 (6.147–6.626) | 6.043 (5.905–6.246) | 17.67 (3.931–79.432); 0.000 |
| TIE2 | 7.485 (7.269–7.708) | 7.487 (7.229–7.572) | 1.45 (0.302–6.968); 0.643 |
| MCP-3 | 2.057 (1.847–2.303) | 1.762 (1.402–2.24) | 2.264 (1.031–4.975); 0.042 |
| CD40-L | 8.018 (7.329–8.390) | 7.889 (6.814–8.276) | 1.195 (0.808–1.766); 0.372 |
| CD244 | 6.385 (6.192–6.586) | 6.299 (6.128–6.476) | 1.542 (0.395–6.020); 0.533 |
| EGF | 9.054 (7.886–9.7600) | 9.146 (7.360–9.413) | 1.164 (0.829–1.634); 0.381 |
| ANGPT1 | 9.769 (9.487–9.968) | 9.811 (9.624–9.877) | 0.588 (0.129–2.691); 0.494 |
| IL7 | 5.952 (5.601–6.553) | 6.095 (5.696–6.534) | 0.889 (0.450–1.756); 0.735 |
| PGF | 8.295 (8.186–8.506) | 8.051 (7.945–8.245) | 16.516 (3.089–88.316); 0.001 |
| IL6 | 3.539 (3.022–4.38) | 2.462 (1.888–3.612) | 1.850 (1.263–2.708); 0.002 |
| ADGRG1 | 1.796 (1.570–2.764) | 1.627 (1.364–1.878) | 3.126 (1.410–6.930); 0.005 |
| MCP-1 | 11.488 (11.293–11.832) | 11.147 (10.71–11.44) | 4.318 (1.689–11.038); 0.002 |
| CRTAM | 5.762 (5.335–5.985) | 5.473 (5.131–5.894) | 2.086 (0.928–4.692); 0.075 |
| CXCL11 | 7.766 (6.873–8.155) | 7.388 (6.871–7.799) | 1.568 (0.864–2.845); 0.139 |
| MCP-4 | 11.037 (10.626–11.529) | 10.638 (10.213–10.873) | 2.116 (1.073–4.176); 0.031 |
| TRAIL | 8.094 (7.763–8.288) | 7.870 (7.551–8.133) | 3.787 (1.134–12.65); 0.030 |
| CXCL9 | 7.049 (6.571–7.811) | 6.054 (5.777–6.732) | 3.690 (1.932–7.049); 0.000 |
| CD8A | 9.223 (8.884–9.765) | 9.497 (9.191–9.789) | 0.642 (0.321–1.286); 0.211 |
| CAIX | 4.500 (4.237–4.932) | 4.241 (3.967–4.693) | 3.354 (1.274–8.826); 0.014 |
| MUC-16 | 2.053 (1.785–2.757) | 2.034 (1.758–2.585) | 1.118 (0.626–1.994); 0.707 |
| ADA | 5.288 (4.916–5.892) | 5.109 (4.856–5.507) | 1.755 (0.903–3.408); 0.097 |
| CD4 | 3.475 (3.261–3.727) | 3.279 (3.094–3.446) | 8.973 (1.963–41.013); 0.005 |
| NOS3 | 2.06 (1.689–2.355) | 1.782 (1.580–1.988) | 3.899 (1.392–10.92); 0.010 |
| Gal-9 | 7.903 (7.443–8.121) | 7.358 (7.162–7.523) | 15.109 (4.003–57.025); 0.000 |
| VEGFR-2 | 8.229 (8.034–8.477) | 8.24 (8.119–8.532) | 0.542 (0.137–2.143); 0.383 |
| CD40 | 9.506 (9.302–9.752) | 9.302 (9.059–9.508) | 4.179 (1.224–14.262); 0.022 |
| IL18 | 8.915 (8.372–9.222) | 8.539 (8.198–8.891) | 3.243 (1.311–8.024); 0.011 |
| GZMH | 3.875 (3.185–4.346) | 3.207 (2.837–4.018) | 1.885 (1.094–3.249); 0.022 |
| KIR3DL1 | 1.930 (0.995–2.674) | 1.789 (0.567–2.509) | 1.098 (0.753–1.601); 0.628 |
| LAP TGF-beta-1 | 9.665 (9.136–9.978) | 9.599 (9.333–9.889) | 1.080 (0.429–2.716); 0.871 |
| CXCL1 | 9.135 (8.801–9.48) | 8.988 (8.716–9.405) | 1.239 (0.532–2.888); 0.619 |
| TNFSF14 | 6.359 (5.559–6.991) | 6.250 (5.312–7.117) | 1.112 (0.740–1.672); 0.609 |
| TWEAK | 8.485 (8.272–8.749) | 8.600 (8.291–8.790) | 0.505 (0.147–1.741); 0.279 |
| PDGF subunit B | 10.638 (10.589–10.686) | 10.636 (10.595–10.674) | 1.665 (0.009–302.191); 0.848 |
| PDCD1 | 4.551 (4.369–4.836) | 4.682 (4.410–4.853) | 0.640 (0.188–2.175); 0.475 |
| FASLG | 6.765 (6.439–6.969) | 7.175 (6.813–7.377) | 0.171 (0.058–0.505); 0.001 |
| CCL19 | 10.521 (9.986–11.038) | 10.327 (9.858–10.934) | 1.305 (0.857–1.988); 0.215 |
| MCP-2 | 8.183 (7.845–8.681) | 8.031 (7.578–8.473) | 1.880 (0.904–3.909); 0.091 |
| CCL4 | 6.813 (6.568–7.372) | 6.626 (6.289–6.890) | 3.144 (1.268–7.794); 0.013 |
| IL15 | 5.212 (4.91–5.463) | 5.029 (4.797–5.233) | 4.885 (1.306–18.273); 0.018 |
| Gal-1 | 6.801 (6.594–6.966) | 6.455 (6.234–6.597) | 353.644 (27.173–4602.582); 0.000 |
| PD-L1 | 5.647 (5.414–5.886) | 5.497 (5.374–5.740) | 4.928 (0.994–24.444); 0.051 |
| CD27 | 8.181 (7.874–8.406) | 7.902 (7.697–8.119) | 6.518 (1.706–24.899); 0.006 |
| CXCL5 | 12.024 (11.577–12.602) | 11.966 (11.315–12.334) | 1.246 (0.687–2.260); 0.469 |
| HGF | 9.226 (8.754–9.662) | 9.115 (8.600–9.356) | 2.108 (0.977–4.548); 0.057 |
| GZMA | 7.127 (6.771–7.371) | 6.933 (6.514–7.267) | 2.623 (0.952–7.226); 0.062 |
| HO-1 | 12.008 (11.764–12.304) | 11.886 (11.619–12.107) | 2.405 (0.865–6.693); 0.093 |
| CX3CL1 | 4.005 (3.683–4.300) | 3.842 (3.615–4.053) | 3.479 (0.971–12.459); 0.055 |
| CXCL10 | 9.027 (8.522–9.557) | 8.389 (7.868–8.740) | 2.746 (1.454–5.186); 0.002 |
| CD70 | 3.865 (3.46–4.199) | 3.511 (3.26–3.760) | 3.926 (1.433–10.756); 0.008 |
| IL10 | 3.752 (3.312–4.120) | 3.383 (3.105–3.858) | 1.877 (0.994–3.543); 0.052 |
| TNFRSF12A | 4.851 (4.528–5.126) | 4.456 (4.168–4.656) | 11.654 (3.29–41.277); 0.000 |
| CCL23 | 10.191 (9.773–10.574) | 10.088 (9.936–10.469) | 0.775 (0.302–1.991); 0.597 |
| CD5 | 6.029 (5.763–6.276) | 5.816 (5.612–6.044) | 2.649 (0.892–7.865); 0.079 |
| CCL3 | 6.250 (5.928–6.685) | 5.864 (5.431–6.112) | 4.251 (1.715–10.538); 0.002 |
| MMP7 | 12.696 (12.569–12.849) | 12.256 (12.061–12.455) | 481.625 (36.262–6396.808); 0.000 |
| ARG1 | 5.561 (5.000–6.640) | 5.198 (4.880–6.030) | 1.634 (1.002–2.666); 0.049 |
| NCR1 | 3.714 (3.431–3.922) | 3.509 (3.253–3.769) | 2.378 (0.725–7.804); 0.153 |
| DCN | 4.535 (4.37–4.725) | 4.315 (4.206–4.437) | 111.093 (8.317–1483.879); 0.000 |
| TNFRSF21 | 7.544 (7.398–7.662) | 7.522 (7.373–7.628) | 2.68 (0.361–19.901); 0.335 |
| TNFRSF4 | 6.205 (5.943–6.493) | 5.912 (5.759–6.194) | 6.216 (1.657–23.316); 0.007 |
| MIC-A/B | 4.905 (4.395–5.533) | 4.978 (4.537–5.412) | 0.842 (0.590–1.200); 0.340 |
| CCL17 | 10.277 (9.562–10.915) | 10.105 (9.632–10.604) | 1.377 (0.812–2.335); 0.235 |
| ANGPT2 | 5.975 (5.62–6.302) | 5.885 (5.634–6.270) | 2.031 (0.675–6.116); 0.208 |
| IFN-gamma | 6.359 (5.913–6.877) | 5.964 (5.427–6.330) | 1.471 (0.931–2.324); 0.098 |
| LAMP3 | 4.745 (4.284–5.43) | 4.358 (4.113–4.677) | 2.183 (1.12–4.255); 0.022 |
| CASP-8 | 5.047 (4.103–6.314) | 3.851 (3.396–4.414) | 2.019 (1.338–3.048); 0.001 |
| ICOSLG | 5.940 (5.659–6.089) | 5.983 (5.875–6.218) | 0.608 (0.175–2.111); 0.433 |
| MMP12 | 7.046 (6.772–7.771) | 6.780 (6.33–7.309) | 2.181 (1.171–4.063); 0.014 |
| CXCL13 | 8.299 (8.071–8.524) | 8.269 (7.877–8.605) | 0.698 (0.307–1.588); 0.392 |
| PD-L2 | 3.200 (2.792–3.464) | 3.000 (2.794–3.283) | 1.953 (0.613–6.22); 0.258 |
| VEGFA | 9.720 (9.345–10.084) | 9.558 (9.332–9.887) | 1.768 (0.723–4.326); 0.212 |
| LAG3 | 4.523 (4.258–4.77) | 4.462 (4.112–4.672) | 4.164 (1.31–13.237); 0.016 |
| IL12RB1 | 1.888 (1.711–2.114) | 1.725 (1.565–1.934) | 9.144 (1.67–50.067); 0.011 |
| CCL20 | 5.710 (5.477–6.405) | 5.499 (5.049–5.740) | 2.036 (1.119–3.705); 0.020 |
| TNF | 3.914 (3.620–4.217) | 3.831 (3.571–4.143) | 1.515 (0.623–3.679); 0.359 |
| KLRD1 | 6.55 (6.182–6.934) | 5.953 (5.63–6.409) | 5.074 (2.083–12.36); 0 |
| GZMB | 2.507 (2.139–2.804) | 2.410 (2.135–2.804) | 1.462 (0.623–3.429); 0.383 |
| CD83 | 2.814 (2.665–3.039) | 2.671 (2.523–2.903) | 5.185 (1.065–25.239); 0.042 |
| IL12 | 7.326 (6.955–7.723) | 7.135 (6.782–7.510) | 2.032 (0.928–4.447); 0.076 |
| CSF-1 | 10.421 (10.227–10.561) | 10.270 (10.083–10.440) | 4.818 (0.927–25.049); 0.062 |
Note: IQR = Interquartile Range (25th and 75th centiles); OR = Odds Ratio; CI = Confidence Interval.
The binary logistic regression analyses allowed us to identify the proteins more strongly associated with the outcome. For the first panel, we selected 20 proteins with a binary logistic regression p-value below 0.01. These 20 proteins were considered together in a multivariate logistic regression model (Table 2). After applying a downward selection as described in the Methods section, four proteins remain, significantly and simultaneously associated with the outcome (Table 3).
Table 2.
Multivariate logistic regression model including all variables from the immune-oncology panel with p < 0.01 at binary logistic regression.
| Protein | OR (95% CI) | p-Value |
|---|---|---|
| TNFRSF9 | 13.785 (0.014–1.38 × 104) | 0.457 |
| CXCL9 | 2.164 (0.198–23.641) | 0.527 |
| Gal-9 | 5.09 × 10−5 (6.36 × 10−10–4.073) | 0.086 |
| Gal-1 | 2.82 × 108 (93.752–8.50 × 1014) | 0.011 |
| TNFRSF12A | 0.245 (0.003–20.627) | 0.534 |
| MMP7 | 7348.889 (3.198–1.69 × 107) | 0.024 |
| DCN | 0.472 (2.88 × 10−5–7731.066) | 0.880 |
| KLRD1 | 0.644 (0.015–27.749) | 0.819 |
| PGF | 0.001 (1.75 × 10−8–20.145) | 0.163 |
| IL6 | 2.910 (0.509–16.642) | 0.230 |
| ADGRG1 | 2.162 (0.156–29.909) | 0.565 |
| MCP-1 | 0.082 (0.001–7.585) | 0.279 |
| CD4 | 638.942 (0.106–3.84 × 106) | 0.146 |
| FASLG | 2.32 × 10−4 (8.23 × 10−8–0.655) | 0.039 |
| CD27 | 1.740 (0.011–268.23) | 0.829 |
| CXCL10 | 1.066 (0.073–15.616) | 0.963 |
| CD70 | 5.843 (0.194–175.917) | 0.310 |
| CCL3 | 2.238 (0.128–39.183) | 0.581 |
| TNFRSF4 | 1.107 (0–2636.098) | 0.979 |
| CASP-8 | 0.791 (0.184–3.402) | 0.753 |
Note: OR = Odds Ratio; CI = Confidence Interval.
Table 3.
Immuno-oncology panel: result of the stepdown procedure applied to the saturated multivariate logistic regression model.
| p-Value | OR (95% CI) | Coefficient (95% CI) | |
|---|---|---|---|
| Gal-9 | 0.025 | 0.035 (0.002–0.652) | −3.352554 (−6.277541–−0.4275675) |
| Gal-1 | 0.001 | 18657.14 (60.882–5717447) | 9.833984 (4.108936–15.55903) |
| MMP7 | 0.001 | 243.809 (9.623–6177.328) | 5.496387 (2.264132–8.728641) |
| FASLG | 0.003 | 0.047 (0.006–0.358) | −3.052633 (−5.07917–−1.026096) |
| Constant | 0.000 | 1.5 × 10−38 (8.12 × 10−59–2.79 × 10−18) | −87.09041 (−133.7588–−40.42205) |
Note: OR = Odds Ratio; CI = Confidence Interval; Coefficient = logistic regression model coefficient.
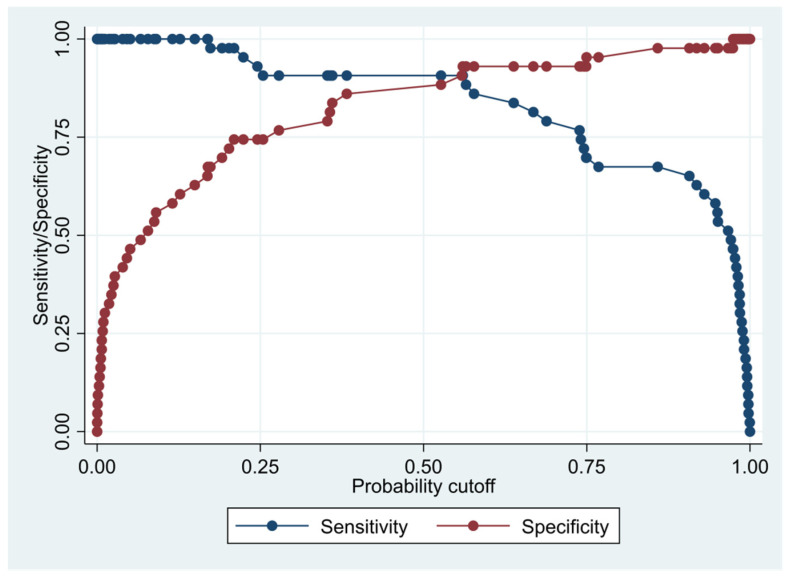
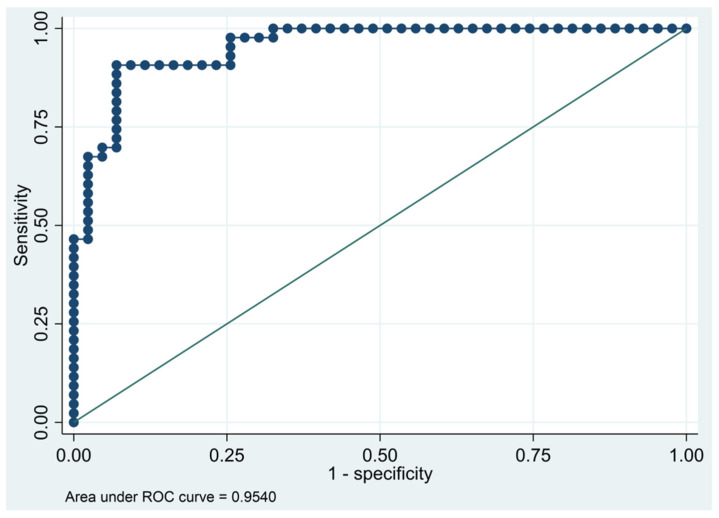
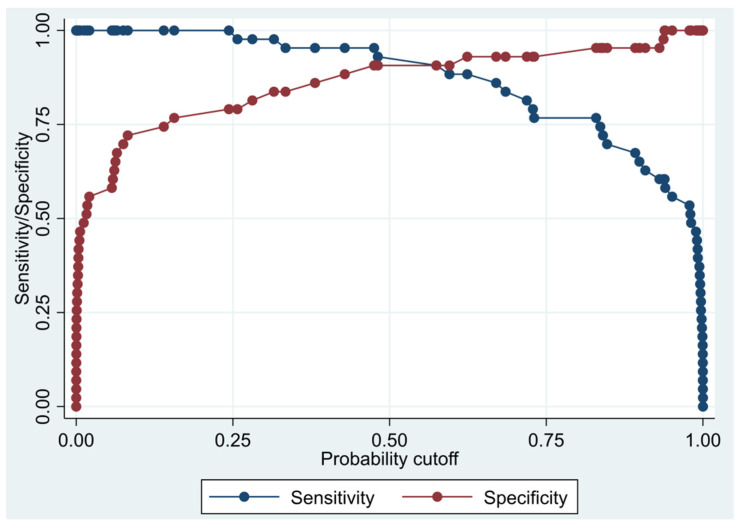
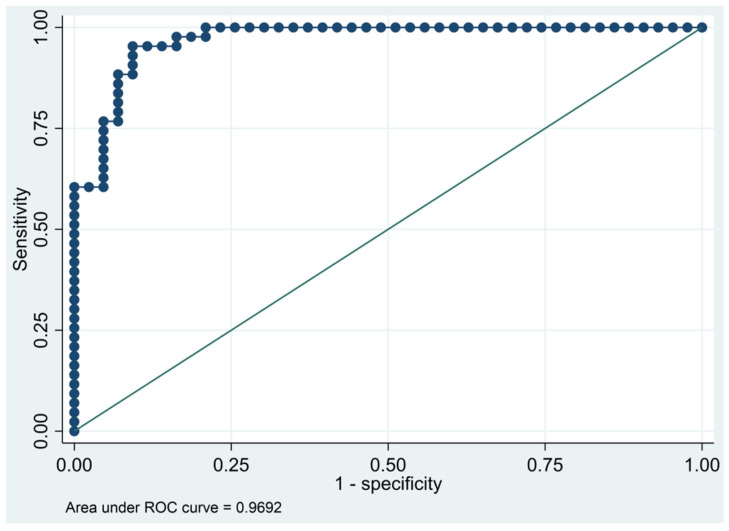
This model has a Pseudo R-squared = 0.605, an AUC = 95.4% (95% CI 91.5–99.3%), reaching a sensitivity of 97.67% with a specificity of 74.42% (Table 4 and Figure 1 and Figure 2). For regression coefficients reported in Table 3 and predicted probability cut points reported in Table 4, the following model will identify cases and controls with the specified sensitivity and specificity:
Table 4.
Immuno-oncology panel: sensitivity/specificity plot for the resulting multivariate logistic regression model, and for sensitivity levels higher than specificity levels.
| Predicted Probability Cut Point | Sensitivity | Specificity |
|---|---|---|
| ≥0.1696234 | 100.00% | 67.44% |
| ≥0.2097338 | 97.67% | 74.42% |
Figure 1.
Sensitivity and specificity plot of the final multivariate logistic regression model based on cytokines from the immune-oncology panel.
Figure 2.
Receiver operating characteristics curve of the final multivariate logistic regression model based on cytokines from the immune-oncology panel.
Predicted probability = 1/(1 + exp(–(–87.09041 − 3.352554 × Gal-9 + 9.833984 × Gal-1 + 5.496387 × MMP7 − 3.052633 × FASLG))).
In the second panel—the Target 96 Oncology III panel—there were 19 proteins with more than 25% values below the limit of detection (TBL1X, IL17F, TPMT, KLK4, NT5C3A, GAMT, HEXA, TNFAIP8, AIF1, CNPY2, SEPT9, CDC27, CXCL14, LAP3, SPINK4, YTHDF3, ACTN4, GGA1, TPT1) so they were excluded from further analyses. Of the remaining proteins, in Table 5 we report median values and interquartile ranges for cases and controls, as well as the odds ratios, 95% confidence intervals and p-values of the binary logistic regression.
Table 5.
Oncology panel: result of descriptive analysis, and binary logistic regression against the outcome.
| Oncology | Cases | Controls | Binary Logistic Regression |
|---|---|---|---|
| Proteins | Median (IQR) | Median (IQR) | OR (95% CI): p-Value |
| CD22 | 5.633 (5.371–5.891) | 5.945 (5.565–6.088) | 0.355 (0.126–1.003); 0.051 |
| INPP1 | 1.444 (1.124–1.907) | 1.144 (0.934–1.456) | 1.607 (0.834–3.095); 0.156 |
| ALPP | 6.802 (5.775–7.713) | 5.836 (4.828–6.321) | 1.637 (1.172–2.287); 0.004 |
| CGB3 | 5.052 (4.473–5.432) | 3.732 (2.946–4.752) | 2.642 (1.627–4.291); 0.000 |
| NAMPT | 2.006 (1.525–3.161) | 1.557 (1.194–2.358) | 1.334 (0.957–1.859); 0.089 |
| VMO1 | 2.266 (1.916–2.404) | 1.919 (1.522–2.236) | 2.874 (1.156–7.142); 0.023 |
| IFNGR2 | 4.891 (4.646–5.139) | 4.775 (4.582–5.196) | 1.478 (0.679–3.217); 0.325 |
| ERP44 | 5.901 (5.621–6.108) | 5.612 (5.318–5.899) | 3.633 (1.156–11.418); 0.027 |
| CBLN4 | 3.444 (3.298–3.680) | 3.698 (3.314–3.977) | 0.343 (0.120–0.985); 0.047 |
| ACAA1 | 2.427 (1.879–3.125) | 1.923 (1.734–2.484) | 1.704 (1.033–2.808); 0.037 |
| S100A16 | 2.739 (2.354–3.118) | 2.937 (2.509–3.202) | 0.732 (0.405–1.326); 0.304 |
| PSPN | 2.927 (2.576–3.874) | 3.335 (2.624–3.773) | 0.885 (0.523–1.498); 0.649 |
| DCTPP1 | 4.963 (4.706–5.173) | 4.683 (4.342–4.924) | 6.91 (2.082–22.935); 0.002 |
| MANSC1 | 5.962 (5.765–6.178) | 6.081 (5.803–6.264) | 0.843 (0.226–3.149); 0.800 |
| GFER | 3.559 (3.172–4.142) | 3.329 (3.056–3.578) | 2.775 (1.291–5.967); 0.009 |
| RP2 | 1.799 (1.344–2.267) | 1.534 (1.200–1.905) | 1.886 (0.988–3.603); 0.055 |
| JCHAIN | 3.420 (3.081–3.917) | 3.483 (3.202–3.705) | 1.173 (0.474–2.903); 0.730 |
| RAB6A | 4.359 (3.629–5.391) | 4.064 (3.434–4.728) | 1.211 (0.879–1.669); 0.242 |
| C1QA | 6.135 (6.007–6.325) | 6.205 (5.946–6.351) | 1.273 (0.259–6.243); 0.766 |
| AKR1B1 | 3.743 (3.193–4.232) | 3.273 (2.876–4.084) | 1.204 (0.763–1.902); 0.425 |
| SCG2 | 3.605 (3.291–3.910) | 3.677 (3.424–3.875) | 0.805 (0.286–2.268); 0.681 |
| RFNG | 2.5100 (2.276–2.715) | 2.280 (2.130–2.463) | 9.739 (2.099–45.182); 0.004 |
| MLN | 3.789 (2.327–4.461) | 2.639 (1.850–3.436) | 1.771 (1.210–2.593); 0.003 |
| ARHGAP25 | 1.381 (0.660–2.393) | 1.111 (0.614–1.910) | 1.283 (0.848–1.942); 0.238 |
| IL1B | 1.013 (0.548–1.575) | 0.638 (0.309–1.319) | 1.207 (0.844–1.726); 0.304 |
| CCT5 | 0.806 (0.558–1.347) | 0.492 (0.387–0.782) | 3.433 (1.389–8.483); 0.008 |
| CASP2 | 1.619 (1.216–2.204) | 1.465 (0.942–2.340) | 0.987 (0.686–1.420); 0.943 |
| ELOA | 1.234 (0.766–2.042) | 0.905 (0.584–1.308) | 1.384 (0.933–2.052); 0.106 |
| NCS1 | 8.279 (8.000–8.556) | 7.923 (7.704–8.144) | 9.272 (2.593–33.153); 0.001 |
| LSP1 | 4.291 (3.626–4.770) | 4.224 (3.446–4.731) | 1.198 (0.676–2.126); 0.536 |
| AFP | 5.767 (5.330–6.316) | 5.373 (4.979–5.866) | 2.166 (1.133–4.141); 0.019 |
| GOPC | 1.306 (0.968–1.783) | 1.042 (0.833–1.247) | 2.382 (1.071–5.302); 0.033 |
| USO1 | 2.507 (1.866–3.311) | 2.011 (1.725–2.592) | 1.705 (1.067–2.723); 0.026 |
| AIMP1 | 2.507 (1.830–3.010) | 1.935 (1.625–2.538) | 1.513 (0.925–2.476); 0.099 |
| SCGN | 2.439 (2.033–2.646) | 2.173 (1.948–2.525) | 1.858 (0.801–4.311); 0.149 |
| TXNDC15 | 6.925 (6.815–7.071) | 6.932 (6.830–7.047) | 1.132 (0.142–9.049); 0.907 |
| ICAM5 | 5.189 (4.957–5.424) | 5.321 (4.973–5.628) | 0.731 (0.254–2.110); 0.563 |
| FUS | 3.247 (2.068–4.311) | 2.673 (1.860–3.261) | 1.328 (0.943–1.871); 0.104 |
| PTP4A1 | 1.073 (0.871–1.364) | 1.078 (0.838–1.420) | 1.105 (0.457–2.670); 0.825 |
| FOXO3 | 1.495 (1.120–1.875) | 1.126 (0.885–1.530) | 1.709 (0.928–3.150); 0.086 |
| VWA1 | 5.145 (4.880–5.544) | 4.985 (4.497–5.479) | 1.605 (0.748–3.445); 0.225 |
| FLT3 | 2.666 (2.469–2.914) | 2.890 (2.611–3.112) | 0.198 (0.053–0.741); 0.016 |
| COL9A1 | 1.159 (0.880–1.520) | 1.595 (1.043–2.031) | 0.398 (0.184–0.861); 0.019 |
| BRK1 | 1.457 (1.167–1.692) | 1.324 (1.118–1.458) | 4.147 (1.076–15.988); 0.039 |
| NELL1 | 8.846 (8.385–9.075) | 8.883 (8.633–9.226) | 0.492 (0.188–1.29); 0.149 |
| SFTPA1 | 1.152 (0.889–1.426) | 1.079 (0.835–1.282) | 1.01 (0.539–1.895); 0.974 |
| VPS37A | 0.873 (0.615–1.420) | 0.555 (0.395–0.919) | 2.768 (1.249–6.135); 0.012 |
| DRG2 | 1.151 (0.601–1.674) | 0.721 (0.434–1.236) | 1.878 (1.037–3.400); 0.037 |
| HMBS | 3.947 (3.284–4.464) | 3.489 (2.393–4.040) | 1.840 (1.121–3.017); 0.016 |
| CLIP2 | 2.185 (1.866–2.682) | 2.286 (1.661–2.622) | 0.966 (0.584–1.597); 0.891 |
| HSPB6 | 6.899 (6.393–7.351) | 6.128 (5.733–6.771) | 5.084 (2.308–11.198); 0.000 |
| ATP6V1D | 1.075 (0.881–1.581) | 0.950 (0.771–1.149) | 3.140 (1.198–8.230); 0.020 |
| LACTB2 | 4.843 (4.193–5.294) | 4.115 (3.522–4.703) | 1.955 (1.205–3.171); 0.007 |
| HBQ1 | 4.388 (3.830–4.951) | 3.911 (2.598–5.099) | 1.440 (1.029–2.016); 0.033 |
| SCLY | 6.124 (5.682–6.764) | 5.778 (5.538–6.111) | 3.588 (1.504–8.56); 0.004 |
| MYO9B | 1.528 (1.094–2.021) | 1.251 (0.923–1.775) | 1.401 (0.815–2.408); 0.223 |
| CD300E | 4.070 (3.832–4.544) | 3.779 (3.583–4.164) | 4.269 (1.611–11.311); 0.004 |
| CDHR2 | 2.589 (1.804–3.275) | 1.132 (0.693–2.067) | 2.820 (1.712–4.644); 0.000 |
| CPVL | 4.479 (4.015–4.876) | 4.811 (4.555–4.915) | 0.295 (0.110–0.791); 0.015 |
| ICAM4 | 5.253 (4.979–5.448) | 5.229 (4.960–5.545) | 1.229 (0.536–2.818); 0.626 |
| PSMD9 | 3.598 (2.964–4.455) | 3.048 (2.400–3.848) | 1.693 (1.121–2.557); 0.012 |
| VPS53 | 0.483 (0.167–1.176) | 0.221 (−0.016–0.712) | 1.542 (0.917–2.592); 0.102 |
| CALCOCO1 | 6.179 (5.618–6.844) | 5.753 (4.989–6.350) | 1.285 (0.892–1.850); 0.178 |
| UBAC1 | 3.990 (3.465–4.495) | 3.645 (3.041–4.149) | 2.162 (1.154–4.053); 0.016 |
| PTPRM | 4.227 (3.989–4.393) | 4.210 (4.138–4.367) | 0.562 (0.117–2.706); 0.473 |
| GALNT7 | 4.344 (4.166–4.576) | 4.505 (4.259–4.645) | 0.497 (0.129–1.915); 0.310 |
| FLT1 | 4.235 (4.075–4.415) | 4.157 (4.064–4.266) | 3.489 (0.56–21.742); 0.181 |
| VAT1 | 4.014 (3.678–4.483) | 3.801 (3.528–4.051) | 3.569 (1.323–9.627); 0.012 |
| L1CAM | 6.812 (6.638–7.042) | 6.801 (6.658–6.970) | 3.636 (0.589–22.451); 0.165 |
| GPA33 | 6.351 (5.596–7.801) | 6.893 (5.512–8.086) | 0.952 (0.710–1.276); 0.743 |
| HLA-E | 0.831 (0.757–0.955) | 0.775 (0.611–0.863) | 13.044 (1.56–109.096); 0.018 |
| PCDH1 | 5.926 (5.777–6.047) | 5.837 (5.742–5.953) | 18.508 (1.399–244.885); 0.027 |
| NPY | 3.913 (3.519–4.458) | 4.042 (3.690–4.556) | 0.758 (0.416–1.381); 0.365 |
Note: IQR = Interquartile Range (25th and 75th centiles); OR = Odds Ratio; CI = Confidence Interval.
With the results of the binary logistic regression, for the second panel, we selected 24 proteins with a binary logistic regression p-value below 0.02, as we only had 11 proteins with a p-value below 0.01 (Table 6). These 24 proteins were considered together in the multivariate logistic regression model. After applying a downward selection, we were left with five proteins, significantly and simultaneously associated with the outcome (Table 7).
Table 6.
Multivariate logistic regression model including all variables from the oncology panel with p < 0.02 at binary logistic regression.
| Protein | OR (95% CI) | p-Value |
|---|---|---|
| CGB3 | 1.938 (0.586–6.415) | 0.586 |
| HSPB6 | 0.626 (0.062–6.360) | 0.062 |
| CDHR2 | 2.752 (0.937–8.085) | 0.937 |
| NCS1 | 41.559 (0.371–4649.940) | 0.371 |
| DCTPP1 | 1.713 (0.016–178.523) | 0.016 |
| LMLN | 3.302 (0.987–11.044) | 0.987 |
| ALPP | 0.739 (0.262–2.078) | 0.262 |
| SCLY | 41.021 (0.694–2425.919) | 0.694 |
| CD300E | 1.863 (0.125–27.774) | 0.125 |
| rfng | 91.252 (0.288–28,947.58) | 0.288 |
| LACTB2 | 0.478 (0.022–10.402) | 0.022 |
| CCT5 | 0.003 (4.95 × 10−6–1.415) | 0.000 |
| GFER | 0.156 (0.010–2.423) | 0.010 |
| VPS37A | 0.352 (0.033–3.791) | 0.033 |
| VAT1 | 3.957 (0.053–296.511) | 0.053 |
| PSMD9 | 67.170 (1.007–4479.84) | 1.007 |
| KLK4 | 3.246 (0.433–24.334) | 0.433 |
| CPVL | 0.103 (0.007–1.423) | 0.007 |
| FLT3 | 0.002 (9.21 × 10−6–0.398) | 0.000 |
| HMBS | 0.500 (0.012–21.041) | 0.012 |
| UBAC1 | 0.998 (0.013–74.526) | 0.013 |
| HLA-E | 0.252 (0.001–62.466) | 0.001 |
| AFP | 0.771 (0.138–4.319) | 0.138 |
| COL9A1 | 0.054 (0.004–0.774) | 0.004 |
Note: OR = Odds Ratio; CI = Confidence Interval.
Table 7.
Oncology panel: result of the stepdown procedure applied to the saturated multivariate logistic regression model.
| p-Value | OR (95% CI) | Coefficient (95% CI) | |
|---|---|---|---|
| CDHR2 | 0.002 | 2.368 (1.372–4.085) | 0.8618858 (0.3165523–1.407219) |
| NCS1 | 0.001 | 30.429 (4.28–216.339) | 3.415408 (1.45397–5.376846) |
| MLN | 0.026 | 1.914 (1.082–3.384) | 0.6490679 (0.0790204–1.219115) |
| FLT3 | 0.011 | 0.054 (0.006–0.507) | −2.915442 (−5.151296–−0.6795876) |
| COL9A1 | 0.034 | 0.285 (0.089–0.909) | −1.256405 (−2.417216–−0.0955934) |
| Constant | 0.002 | 3.52 × 10−10 (4.36 × 10−16–2.84 × 10−4) | −21.76806 (−35.36822–−8.167891) |
Note: OR = Odds Ratio; CI = Confidence Interval; Coefficient = logistic regression model coefficient.
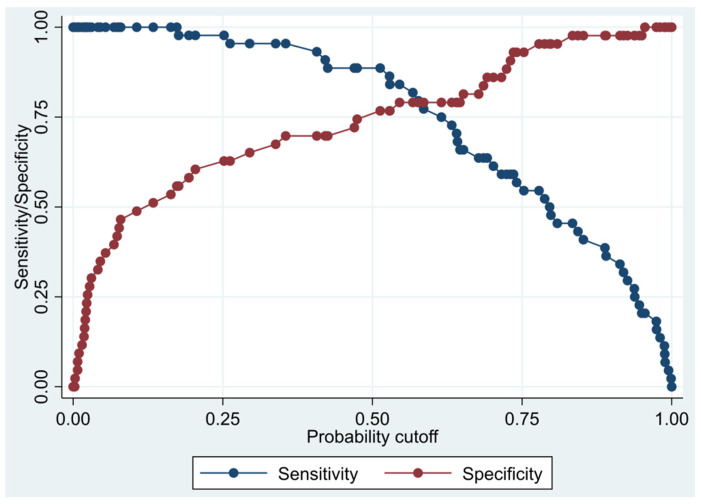
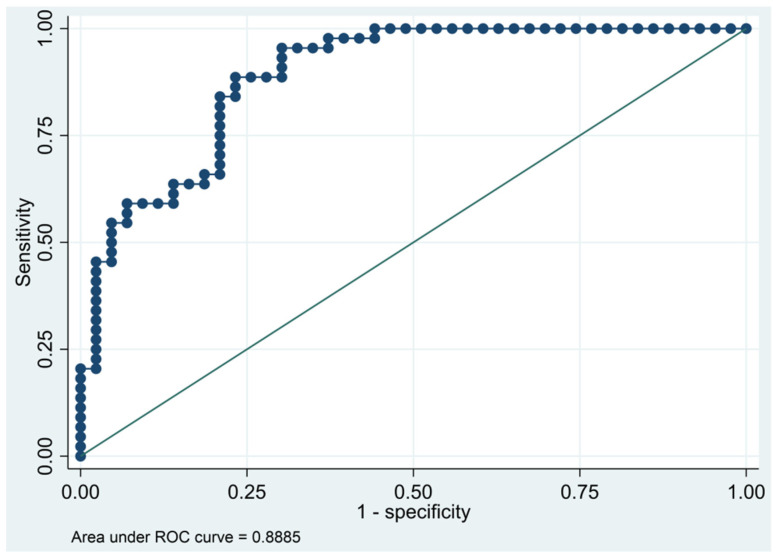
This model has a Pseudo R-squared = 0.436, an AUC = 88.9% (82.1–95.6%), reaching a sensitivity of 95.45% with a specificity of 69.77% (Table 8 and Figure 3 and Figure 4). For the regression coefficients reported in Table 7 and predicted probability cut points reported in Table 8, the following model identified cases and controls with the specified sensitivity and specificity:
Table 8.
Oncology panel: sensitivity/specificity plot for the resulting multivariate logistic regression model, and for sensitivity levels higher than specificity levels.
| Predicted Probability Cut Point | Sensitivity | Specificity |
|---|---|---|
| ≥0.1731692 | 100.00% | 55.81% |
| ≥0.2520203 | 97.73% | 62.79% |
| ≥0.3550157 | 95.45% | 69.77% |
| ≥0.5128943 | 88.64% | 76.74% |
| ≥0.5453203 | 84.09% | 79.07% |
Figure 3.
Sensitivity and specificity plot of the final multivariate logistic regression model based on cytokines from the oncology panel.
Figure 4.
Receiver operating characteristics curve of the final multivariate logistic regression model based on cytokines from the oncology panel.
Predicted probability = 1/(1 + exp(−(−21.76806 + 0.8618858 × CDHR2 + 3.415408 × NCS1 + 0.6490679 × MLN − 2.915442 × FLT3 − 1.256405 × COL9A1))).
The third model was generated by considering in a multivariate logistic regression all proteins included in the two final models, i.e., Gal-1, Gal-9, MMP7. FASLG, CDHR2, NCS1, MLN, FLT3 and COL9A1 (Table 9). After the stepdown procedure, the final model included all variables previously included in the immune-oncology final model, plus COL9A1 (Table 10). This model has a Pseudo R-squared = 0.691, an AUC = 96.9% (93.9–99.9%), reaching a sensitivity of 97.67% with a specificity of 83.72% (Table 11, Figure 5 and Figure 6). Regression coefficients are reported in Table 10. Predicted probability cut points for specificity higher than sensitivity are reported in Table 11. The predicted probability can be calculated from the following model:
Table 9.
Multivariate logistic regression model including all variables from the final model of the immuno-oncology and oncology panels.
| Protein | OR (95% CI) | p-Value |
|---|---|---|
| Gal-1 | 99.147 × 104 (65.740–1.50 × 1010) | 0.005 |
| MMP7 | 40.640 × 104 (6.623–2.49 × 1010) | 0.022 |
| Gal-9 | 4.82 × 10−4 (8.37 × 10−7–0.277) | 0.018 |
| FASLG | 1.665 × 10−2 (2.505 × 10−4–1.107) | 0.056 |
| CDHR2 | 1.738 (0.426–7.083) | 0.441 |
| NCS1 | 5.054 (0.252–101.451) | 0.290 |
| MLN | 2.360 (0.771–7.227) | 0.132 |
| FLT3 | 6.315 × 10−3 (4.39 × 10−5–0.909) | 0.046 |
| COL9A1 | 3.174 × 10−2 (1.50 × 10−3–0.673) | 0.027 |
Note: OR = Odds Ratio; CI = Confidence Interval.
Table 10.
Joint panel: result of the stepdown procedure applied to the saturated multivariate logistic regression model.
| p-Value | OR (95% CI) | Coefficient (95% CI) | |
|---|---|---|---|
| Gal-1 | 0.001 | 7.30 × 104 (7.38 × 101–7.21 × 107) | 11.198 (4.302–18.094) |
| MMP7 | 0.001 | 1.04 × 104 (4.71 × 101–2.29 × 106) | 9.249 (3.852–14.646) |
| Gal-9 | 0.010 | 8.98 × 10−3 (2.47 × 10−4–3.27 × 10−1) | −4.713 (−8.307–−1.119) |
| FASLG | 0.003 | 1.56 × 10−2 (9.58 × 10−4–2.53 × 10−1) | −4.163 (−6.951–−1.375) |
| COL9A1 | 0.008 | 6.80 × 10−2 (9.34 × 10−3–4.96 × 10−1) | −2.688 (−4.673–−0.702) |
| Constant | 0.001 | 1.72 × 10−53 (2.49 × 10−83–1.18 × 10−23) | −121.497 (−190.203–−52.791) |
Note: OR = Odds Ratio; CI = Confidence Interval; Coefficient = logistic regression model coefficient.
Table 11.
Joint panel: sensitivity/specificity plot for the resulting multivariate logistic regression model, and for sensitivity levels higher than specificity levels.
| Predicted Probability Cut Point | Sensitivity | Specificity |
|---|---|---|
| ≥0.2436392 | 100.00% | 79.07% |
| ≥0.3154508 | 97.67% | 83.72% |
| ≥0.4752594 | 95.35% | 90.70% |
Figure 5.
Sensitivity and specificity plot of the final multivariate logistic regression model based on cytokines resulting from the two final models based on both the immune-oncology and the oncology panel.
Figure 6.
Receiver operating characteristics curve of the final multivariate logistic regression model based on cytokines resulting from the two final models based on both the immune-oncology and the oncology panel.
Predicted probability = 1/(1 + exp(−(−121.4969 − 4.713017 × Gal-9 + 11.1979 × Gal-1 + 9.248928 × MMP7 − 4.163016 × FASLG − 2.687621 × COL9A1))).
Bioinformatic Analysis
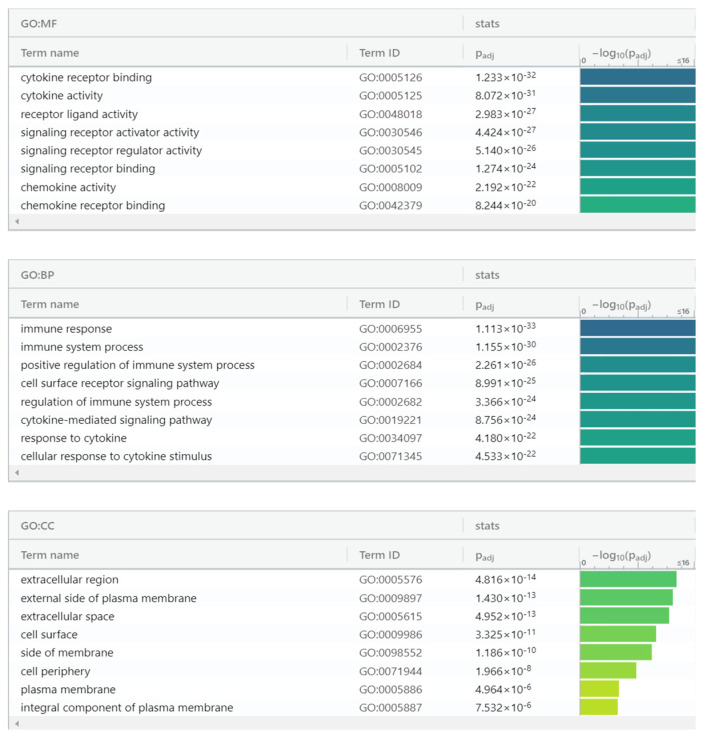
We used gProfiler as the classification tool for proteomic enrichment data analysis. For the Immuno-oncology panel, proteins (Figure 7) are classified into groups according to their molecular function, biological processes, and protein classes. Regarding molecular function, proteins were categorized into: cytokine receptor binding, cytokine activity, receptor-ligand activity, signaling receptor activator activity, signaling receptor regulator activity, signaling receptor binding, chemokine activity, and chemokine receptor binding. For biological processes, proteins were categorized into: immune response, immune system process, positive regulation of immune system process, cell surface receptor signaling pathway, regulation of immune system process, cytokine-mediated signaling pathway, response to cytokine, and cellular response to cytokine stimulus. Contrastingly, for protein class, proteins were categorized into: extracellular region, external side of plasma membrane, extracellular space, cell surface, side of membrane, cell periphery, plasma membrane, and integral component of plasma membrane. Reactome tool grouped these proteins into eight pathways: chemokine receptors bind chemokines, interleukin-10 signaling, cytokine signaling in immune system, immune system, signaling by interleukins, peptide ligand-binding receptors, TNFR2 non-canonical NF-kB pathway, class A/1 (Rhodopsin-like receptors).
Figure 7.
gProfiler classification of Immuno-oncologic panel proteins in the EC serum according to their molecular function, biological processes, cellular component.
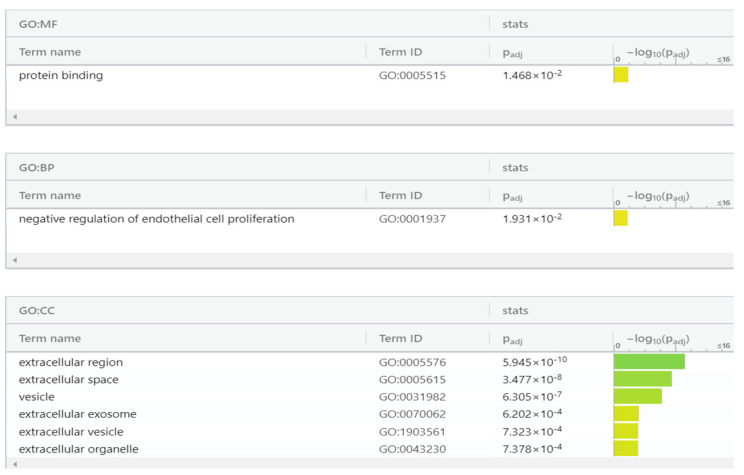
The same analysis was performed for the Target 96 Oncology III panel (Figure 8). Proteins were classified into groups according to their molecular function, biological processes, and protein class. The molecular function categories were: protein binding. The biological processes were: negative regulation of endothelial cell proliferation. The protein class categories were: extracellular region, extracellular space, vesicle, extracellular exosome, extracellular vesicle, extracellular organelle, and extracellular membrane-bounded organelle. Reactome tool analysis classified these proteins into four pathways: CLEC7A/inflammasome pathway, Defective CSF2RA causes SMDP4, and Defective CSF2RB causes SMDP5, Innate Immune System.
Figure 8.
gProfiler classification of Oncology III panel proteins in the EC serum according to their molecular function, biological processes, cellular component.
4. Discussion
Biomarkers play a key role in oncological applications, including diagnosis of the disease, prognosis and determination of personalized treatment [42]. High throughput technology of TMT LC\MS-MS allowed for the identification and quantification of several candidate biomarkers in a large cohort of patients [43]. In recent years, there has been a great effort to identify and validate biomarkers in EC, but, until now, no candidate biomarker has reached the clinical stage [43,44].
To identify early candidate biomarkers in EC, we exploited for the first time PEA technology in targeted proteomics, which had already been used successfully for identification of biomarkers in several pathologies. From the 92 proteins of the Immuno-oncologic panel, only 20 were selected with a binary logistic regression p-value below 0.01. A multivariate logistic regression analysis based on four proteins (Gal-9, Gal-1, MMP7, FASLG) allowed us to separate cases from controls with an AUC = 95.4%. From the Target 96 Oncology III, only 24 proteins were selected with a binary logistic regression p-value below 0.02. A multivariate logistic regression analysis based on five proteins (CDHR2, NCS1, MLN, FLT3, COL9A1) allowed us to separate cases from controls with an AUC = 88.9%. According to these results, the performance of the Immuno-oncologic panel was better than the Target 96 Oncology III panel. To further improve the model, we performed a multivariate logistic regression, including all proteins from the first (Gal-1, Gal-9, MMP7, FASLG) and one from the second model (COL9A1) obtaining an AUC = 96.9%.
Thanks to PEA technology, several candidate biomarkers were identified in different pathologies, such as FABP2, FGF5, LPL, and LTA in coronary artery disease (CAD) pathogenesis [45]; ITGAV, EpCAM, IL18, SLAMF7 and IL8 were identified as biomarkers in inflammatory bowel disease (IBD) [46]; and ten candidate biomarkers, including CHIT1, SMOC2, MMP-10, LDLR, CD200, EIF4EBP1, ALCAM, RGMB, tPA and STAMBP, were identified in early Alzheimer’s disease [47]. In endometriosis, PEA technology identified seven proteins up-regulated (IL-6, IL-8, CCL 19, SCF, VEGF-D, IL-6RA, MIA9) and ten proteins down-regulated (ICOSLG, EGFR, SELE, ErbB2/HER2, IL-6RA, VEGFR-2, Flt3L, CXCL10, HE4, FR-alpha). Moreover, in endometriosis patients, dysregulation of these proteins has been reported to induce immune response modulation, angiogenesis, cell proliferation, cell adhesion and inhibition of apoptosis [38].
In this work, we identified five proteins, namely, Gal-1, Gal-9, MMP7, FASLG, and COL9A1, that based on their known function in endometrial and other cancers might represent useful early-stage EC biomarkers, upon a validation phase.
Indeed, Extracellular Gal-9 induces apoptosis of effector T-cells through mucin domain-containing molecule 3 (Tim-3) [48]. This protein induces the differentiation and suppressive activity of regulatory T-cells [49]. In their work, Chuan-xia Zhang et al. highlighted a novel mechanism involving Gal-9 in creating an immune-suppressive microenvironment, which favors tumor progression [50].
Gal-1 is a small lectin that binds beta-galactoside and a wide array of complex carbohydrates. This protein acts as an immunosuppressive molecule and is expressed by different types of cancer cells [51]. Once secreted, Gal-1 binds to the glycosylated receptor of immune cells, leading to their inhibition and consequently to the immune escape of cancer cells [52]. In EC, Mylonas and colleagues performed an immunohistochemistry study of Gal-1 and Gal-9, finding a correlation between these proteins and EC clinicopathological features. [53] Indeed, high expression of Gal-1 is associated with poor prognosis, while high expression of Gal-9 is associated with early pathological changes. In addition, Gal-1 also correlates with lymphangiosis, a poor prognostic marker in EC [54].
MMP7 is a small enzyme that degrades several types of galectins, casein and fibronectin [55]. MMP7 promotes tumor cell invasion and migration, digesting the extracellular matrix (ECM) and components of cell surface proteins [56]. This protein is a promising diagnostic and prognostic biomarker of pancreatic cancer [57] and bladder cancer [58]. Its biochemical characteristics make MMP7 a potential target in several types of cancers [56]. Downregulation of MMP7 leads to a reduced proliferation and migration of tumor cells in gallbladder cancer [59] and reduces the cisplatin resistance in NCSLC cells [60]. In EC, high expression of MMP7 correlates with higher lymph node invasion [61] and increased risk of metastasis [62]. These data are supported also by in vitro assays (Misugi et al.), confirming that increased expression of MMP-7 in high-grade ECs may be correlated with tumor invasion and the protein may be a prognostic marker in EC [63].
COL9A1 is a structural component of hyaline cartilage and vitreous of the eye; the COL9A1 gene localizes in chromosome 16q13 [64]. Several studies correlates COL9A1 with breast cancer [65] and oral squamous cell carcinoma [66]. In EC, computational analysis of RNAseq data deposited in the TCGA database show that COL9A1 expression is increased both in primary and metastatic tumor (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7173926/) accessed on 4 July 2022.
Lastly, FASLG binds to TNFRSF6/FAS receptor inducing apoptosis [67]. This protein is important to maintain the immune homeostasis and for the elimination of cancer cells [68]. Epithelial mesenchymal transition (EMT) and stiffness make cancer cells more aggressive by excessive production and secretion of FASLG [69]. McGlorthan et al. described a possible mechanism explaining how progesterone and calcitriol induce apoptosis in EC, relying on the induction of FasL, Fas, and FADD expression, which, in turn, activates the caspase-8 pathway [70]. Accordingly, a genetic study showed that the homozygous CC variant of FASL −844 T>C polymorphism confers protection against EC [71].
The Reactome analysis of our five candidate proteins also identified several pathways related with the immunity system. Of them, chemokines and chemokine receptors not only take part in immune regulation but also play a key role in tumor development. Moreover, chemokines and chemokine receptors are related with angiogenesis, metastasis, drug resistance, and immunity of breast cancer [72]. For example, IL-10 plays a key role on the regulation of several genes in gastric cancer cells involved in cell proliferation and migration [73]. Interestingly, cytokines are small proteins that play an important role in cellular functions, such as proliferation, differentiation, and survival, as well as the response to pathogens. These proteins induce the activation of the JAK-STAT pathway, which is fundamental in the regulation of immune system and tumor surveillance [74].
Altogether, in our study, we found a panel of proteins whose secretion in blood correlates with early EC and can be exploited as a diagnostic biomarker upon further validation, although we know that there are some limitations. First, since it was very difficult for us to find controls with the same age as the EC patients, the controls are slightly younger than the EC patients. We acknowledge this is a limitation and that levels of some proteins might change with age. The next step will be the validation of these results with age-matched cases and controls.
Another limit of our study is the small number of patients, which does not allow us to generalize to the overall population. Even if the results are satisfactory, more studies are certainly required to confirm and consolidate these findings.
5. Conclusions
In conclusion, by combining proteins from the Immuno-oncology panel and the Target 96 Oncology III panel, we were able to generate an algorithm that was able to discriminate early EC type I patients from controls with high specificity and sensitivity thanks to the analysis of Gal-1, Gal-9, MMP7, COL9A1, and FASLG serum levels. Although Gal-1, Gal-9, MMP7, COL9A1, and FASLG are overexpressed in different kinds of cancers, the analysis of their serum levels allows one to discriminate between healthy controls and woman affected by type I EC, and combining this analysis with a typical clinical manifestation of EC, such us bleeding and pelvic pain, might help early EC diagnosis, avoiding invasive diagnostic techniques.
Acknowledgments
The authors thank Martina Bradaschia for the English revision of the manuscript.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines10081857/s1, Table S1: The clinical and pathological characteristics of the patients. Table S2: List of proteins of 92 human proteins of Olink Target 96 oncology III. Table S3: List of proteins of 92 human proteins of Olink Target 96 Immuno-Oncology Panels.
Author Contributions
Conceptualization, B.U. and L.M.; methodology, M.A.; software, L.M.; formal analysis, V.C.; investigation, G.D.L. and F.R.; resources, G.R.; data curation, L.M.; writing—original draft preparation, B.U., L.M. and V.C.; writing—review and editing, L.M., V.C. and M.A.; visualization, G.R.; supervision, L.M.; project administration, G.D.L. and F.R.; funding acquisition, G.R. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of IRCCS Burlo Garofolo and Regional Ethics Committee (protocol code RC18/19 approved in 2019 and CEUR-2020-Os-030).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethical reasons.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Italian Health Ministry, grant number RC 18/19.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ferlay J., Colombet M., Soerjomataram I., Parkin D.M., Piñeros M., Znaor A., Bray F. Cancer statistics for the year 2020: An overview. Int. J. Cancer. 2021;149:778–789. doi: 10.1002/ijc.33588. [DOI] [PubMed] [Google Scholar]
- 2.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Barry J.A., Azizia M.M., Hardiman P.J. Risk of endometrial, ovarian and breast cancer in women with polycystic ovary syndrome: A systematic review and meta-analysis. Hum. Reprod. Update. 2014;20:748–758. doi: 10.1093/humupd/dmu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calle E.E., Rodriguez C., Walker-Thurmond K., Thun M.J. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N. Engl. J. Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 5.Colombo N., Creutzberg C., Amant F., Bosse T., González-Martín A., Ledermann J., Marth C., Nout R., Querleu D., Mirza M.R., et al. ESMO-ESGO-ESTRO Consensus Conference on Endometrial Cancer: Diagnosis, treatment and follow-up. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2016;27:16–41. doi: 10.1093/annonc/mdv484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saed L., Varse F., Baradaran H.R., Moradi Y., Khateri S., Friberg E., Khazaei Z., Gharahjeh S., Tehrani S., Sioofy-Khojine A.-B., et al. The effect of diabetes on the risk of endometrial Cancer: An updated a systematic review and meta-analysis. BMC Cancer. 2019;19:527. doi: 10.1186/s12885-019-5748-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu K.H., Broaddus R.R. Endometrial Cancer. N. Engl. J. Med. 2020;383:2053–2064. doi: 10.1056/NEJMra1514010. [DOI] [PubMed] [Google Scholar]
- 8.Brennan D.J., Hackethal A., Mann K.P., Mutz-Dehbalaie I., Fiegl H., Marth C., Obermair A. Serum HE4 detects recurrent endometrial cancer in patients undergoing routine clinical surveillance. BMC Cancer. 2015;15:33. doi: 10.1186/s12885-015-1028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lax S.F. Pathology of endometrial carcinoma. Adv. Exp. Med. Biol. 2017;943:75–96. doi: 10.1007/978-3-319-43139-0_3. [DOI] [PubMed] [Google Scholar]
- 10.Office for National Statistics Cancer Survival in England. [(accessed on 5 July 2022)]; Available online: https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/bulletins/cancersurvivalinengland/adultstageatdiagnosisandchildhoodpatientsfollowedupto2016/previous/v1#cancer-survival-in-england-patients-diagnosed-between-2011-and-2015-and-followed-up-to-2016-national-statistics.
- 11.O’Flynn H., Ryan N.A.J., Narine N., Shelton D., Rana D., Crosbie E.J. Diagnostic accuracy of cytology for the detection of endometrial cancer in urine and vaginal samples. Nat. Commun. 2021;12:952. doi: 10.1038/s41467-021-21257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baskar R., Lee K.A., Yeo R., Yeoh K.W. Cancer and radiation therapy: Current advances and future directions. Int. J. Med. Sci. 2019;9:193–199. doi: 10.7150/ijms.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Nola R., Menga A., Castegna A., Loizzi V., Ranieri G., Cicinelli E., Cormio G. The Crowded Crosstalk between Cancer Cells and Stromal Microenvironment in Gynecological Malignancies: Biological Pathways and Therapeutic Implication. Int. J. Mol. Sci. 2019;20:2401. doi: 10.3390/ijms20102401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanderstraeten A., Tuyaerts S., Amant F. The immune system in the normal endometrium and implications for endometrial cancer development. J. Reprod. Immunol. 2015;109:7–16. doi: 10.1016/j.jri.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Lan T., Chen L., Wei X. Inflammatory cytokines in cancer: Comprehensive understanding and clinical Progress in Gene Therapy. Cells. 2021;10:100. doi: 10.3390/cells10010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greten F.R., Grivennikov S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity. 2019;51:27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumari N., Dwarakanath B.S., Das A., Bhatt A.N. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol. 2016;37:11553–11572. doi: 10.1007/s13277-016-5098-7. [DOI] [PubMed] [Google Scholar]
- 18.van Horssen R., Ten Hagen T.L., Eggermont A.M. TNF-alpha in cancer treatment: Molecular insights, antitumor effects, and clinical utility. Oncologist. 2006;11:397–408. doi: 10.1634/theoncologist.11-4-397. [DOI] [PubMed] [Google Scholar]
- 19.Marson A., Levine S.S., Cole M.F., Frampton G.M., Brambrink T., Johnstone S., Guenther M.G., Johnston W.K., Wernig M., Newman J., et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNab F., Mayer-Barber K., Sher A., Wack A., O’Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choa Tan H.N., Velasco R.N., San Juan M.D., Cornelio G.H. Cytokines as a prognostic biomarker of overall survival in pancreatic cancer. J. Clin. Oncol. 2020;38:e16753. doi: 10.1200/JCO.2020.38.15_suppl.e16753. [DOI] [Google Scholar]
- 22.Burgdorf S.K., Claesson M.H., Nielsen H.J., Rosenberg J. Changes in cytokine and biomarker blood levels in patients with colorectal cancer during dendritic cell-based vaccination. Acta Oncol. 2009;48:1157–1164. doi: 10.3109/02841860903099964. [DOI] [PubMed] [Google Scholar]
- 23.Dalal V., Kumar R., Kumar S., Sharma A., Kumar L., Sharma J.B., Roy K.K., Singh N., Vanamail P. Biomarker potential of IL-6 and VEGF-A in ascitic fluid of epithelial ovarian cancer patients. Clin. Chim. Acta. 2018;482:27–32. doi: 10.1016/j.cca.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 24.Chakraborty S., Hosen I., Ahmed M., Shekhar H.U. Onco-Multi-OMICS Approach: A New Frontier in Cancer Research. Biomed Res. Int. 2018;2018:9836256. doi: 10.1155/2018/9836256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ura B., Biffi S., Monasta L., Arrigoni G., Battisti I., Lorenzo G.D., Romano F., Aloisio M., Celsi F., Addobbati R. Two Dimensional-Difference in Gel Electrophoresis (2D-DIGE) Proteomic Approach for the Identification of Biomarkers in Endometrial Cancer Serum. Cancers. 2021;13:3639. doi: 10.3390/cancers13143639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Celsi F., Monasta L., Arrigoni G., Battisti I., Licastro D., Aloisio M., Lorenzo G.D., Romano F., Ricci Blendi Ura B. Gel-based proteomic identification of suprabasin as a potential new candidate biomarker in endometrial cancer. Int. J. Mol. Sci. 2021;23:2076. doi: 10.3390/ijms23042076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uyar D.S., Huang Y.W., Chesnik M.A., Doan N.B., Mirza S.P. Comprehensive serum proteomic analysis in early endometrial cancer. J. Proteomics. 2021;234:104099. doi: 10.1016/j.jprot.2020.104099. [DOI] [PubMed] [Google Scholar]
- 28.Assarsson E., Lundberg M., Holmquist G., Björkesten J., Thorsen S.B., Ekman D., Eriksson A., Dickens E.R., Ohlsson S., Edfeldt G. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE. 2014;9:e95192. doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X., Suo C., Zhang T., Yin X., Man J., Yuan Z., Yu J., Jin L., Chen X., Lu M., et al. Targeted proteomics-derived biomarker profile develops a multi-protein classifier in liquid biopsies for early detection of esophageal squamous cell carcinoma from a population-based case-control study. Biomark Res. 2021;9:12. doi: 10.1186/s40364-021-00266-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berggrund M., Enroth S., Lundberg M., Assarsson E., Stålberg K., Lindquist D., Hallmans G., Grankvist K., Olovsson M., Gyllensten U. Identification of candidate Plasma Protein Biomarkers for Cervical Cancer Using the Multiplex Proximity Extension Assay. Mol. Cell Proteom. 2019;18:735–743. doi: 10.1074/mcp.RA118.001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graumann J., Finkernagel F., Reinartz S., Stief T., Brödje D., Renz H., Jansen J.M., Wagner U., Worzfeld T., von Strandmann E.P., et al. Multi-platform Affinity Proteomics Identify Proteins Linked to Metastasis and Immune Suppression in Ovarian Cancer Plasma. Front. Oncol. 2019;9:1150. doi: 10.3389/fonc.2019.01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wik L., Nordberg N., Broberg J., Björkesten J., Assarsson E., Henriksson S., Grundberg I., Pettersson E., Westerberg C., Liljeroth E., et al. Proximity extension assay in combination with next-generation sequencing for high-throughput proteome-wide analysis. Mol. Cell Proteomics. 2021;20:100168. doi: 10.1016/j.mcpro.2021.100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraser D.D., Cepinskas G., Patterson E.K., Slessarev M., Martin C., Daley M., Patel M.A., Miller M.R., O’Gorman D.B., Gill S.E., et al. Novel outcome biomarkers identified with targeted proteomic analyses of plasma from critically Ill coronavirus disease 2019 patients. Crit. Care Explor. 2020;2:e0189. doi: 10.1097/CCE.0000000000000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bom M.J., Levin E., Driessen R.S., Danad I., Van Kuijk C.C., van Rossum A.C., Narula J.N., Min J.K., Leipsic J.A., Belo Pereira J.P., et al. Predictive value of targeted proteomics for coronary plaque morphology in patients with suspected coronary artery disease. EBioMedicine. 2019;39:109–117. doi: 10.1016/j.ebiom.2018.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Consiglio C.R., Cotugno N., Sardh F., Pou C., Amodio D., Rodriguez L., Tan Z., Zicari S., Ruggiero A., Pascucci G.R., et al. The immunology of multisystem inflammatory syndrome in children with COVID-19. Cell. 2020;183:968–981. doi: 10.1016/j.cell.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansson P., Krona C., Kundu S., Doroszko M., Baskaran S., Schmidt L., Vinel C., Almstedt E., Elgendy R., Elfineh L., et al. A patient-derived cell atlas informs precision targeting of glioblastoma. Cell Rep. 2020;32:107897. doi: 10.1016/j.celrep.2020.107897. [DOI] [PubMed] [Google Scholar]
- 37.Fellström B., Helmersson-Karlqvist J., Lind L., Soveri I., Wu P.H., Thulin M., Ärnlöv J., Larsson A. Associations between apolipoprotein A1, high-density lipoprotein cholesterol, and urinary cytokine levels in elderly males and females. J. Interferon Cytokine Res. 2020;40:71–74. doi: 10.1089/jir.2019.0074. [DOI] [PubMed] [Google Scholar]
- 38.Perricos A., Wenzl R., Husslein H., Eiwegger T.H., Gstoettner M., Weinhaeusel A., Beikircher G., Kuessel L. Does the use of the “Proseek® Multiplex Oncology I Panel” on peritoneal fluid allow a better insight in the pathophysiology of endometriosis, and in particular deep-infiltrating endometriosis? J. Clin. Med. 2020;9:2009. doi: 10.3390/jcm9062009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larssen P., Wik L., Czarnewski P., Eldh M., Löf L., Ronquist K.G., Dubois L., Freyhult E., Gallant C.J., Oelrich J., et al. Tracing cellular origin of human exosomes using multiplex proximity extension assays. Mol. Cell Proteom. 2017;16:502–511. doi: 10.1074/mcp.M116.064725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reimand J., Kull M., Peterson H., Hansen J., Vilo J. g:Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic. Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vastrik I., D’Eustachio P., Schmidt E., Gopinath G., Croft D., de Bono B., Gillespie M., Jassal B., Lewis S., Matthews L., et al. Reactome: A knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kucuk O. Cancer biomarkers. Mol. Aspects Med. 2015;45:1–2. doi: 10.1016/j.mam.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez H., Pennington S.R. Revolutionizing Precision Oncology through Collaborative Proteogenomics and Data Sharing. Cell. 2018;173:535–539. doi: 10.1016/j.cell.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogelstein B., Kinzler K.W. Cancer genes and the pathways they control. Nat. Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 45.Bretherick A.D., Canela-Xandri O., Joshi P.K., Clark D.W., Rawlik K., Boutin T.H.S., Zeng Y., Amador C., Navarro P., Rudan I., et al. Linking protein to phenotype with Mendelian Randomization detects 38 proteins with causal roles in human diseases and traits. PLoS Genet. 2020;16:e1008785. doi: 10.1371/journal.pgen.1008785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalla R., Adams A.T., Bergemalm D., Vatn S., Kennedy N.A., Ricanek P., Lindstrom J., Ocklind A., Hjelm F., Ventham N.T., et al. Serum proteomic profiling at diagnosis predicts clinical course, and need for intensification of treatment in inflammatory bowel disease. J. Crohn’s Colitis. 2021;15:699–708. doi: 10.1093/ecco-jcc/jjaa230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whelan C.D., Mattsson N., Nagle M.W., Vijayaraghavan S., Hyde C., Janelidze S.H., Stomrud E., Lee J., Fitz L., Tarek ASamad T.A., et al. Multiplex proteomics identifies novel CSF and plasma biomarkers of early Alzheimer’s disease. Acta Neuropathol. Commun. 2019;7:169. doi: 10.1186/s40478-019-0795-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gleason K.M., Lenvik T.R., McCullar V., Felices M., O’Brien M.S.H., Cooley S.A., Verneris M.R., Cichocki F., Holman C.J., Panoskaltsis-Mortari A. Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to galectin-9. Blood. 2012;119:3064–3072. doi: 10.1182/blood-2011-06-360321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.John S., Mishra R. Galectin-9: From cell biology to complex disease dynamics. J. Biosci. 2016;41:507–534. doi: 10.1007/s12038-016-9616-y. [DOI] [PubMed] [Google Scholar]
- 50.Zhang C.X., Huang D.J., Baloche V., Zhang L., Xu J.X., Li B.W., Zhao X.R., He J., Mai H.Q., Chen Q.Y. Galectin-9 promotes a suppressive microenvironment in human cancer by enhancing STING degradation. Oncogenesis. 2020;9:65. doi: 10.1038/s41389-020-00248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Postow M.A., Callahan M.K., Wolchok J.D. Immune Checkpoint Blockade in Cancer Therapy. J. Clin. Oncol. 2015;33:1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y., Wang H.C., Zhao J., Wu M.H., Shih T.C. Immunosuppressive Roles of Galectin-1 in the Tumor Microenvironment. Biomolecules. 2021;11:1398. doi: 10.3390/biom11101398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fei Sun X., Yan Dai S.H. The Significance of Galectin-1 and Galectin-9 Expression in Endometrial Carcinoma. Gynecol Obstet Invest. 2020;85:34–40. doi: 10.1159/000502787. [DOI] [PubMed] [Google Scholar]
- 54.Mylonas I., Mayr D., Walzel H., Shabani N., Dian D., Kuhn C., Kunze S., Jeschke U., Friese K. Mucin 1, Thomsen-Friedenreich expression and galectin-1 binding in endometrioid adenocarcinoma: An immunohistochemical analysis. Anticancer Res. 2007;27:1975–1980. [PubMed] [Google Scholar]
- 55.Quantin B., Murphy G., Breathnach R. Pump-1 cDNA codes for a protein with characteristics similar to those of classical collagenase family members. Biochemistry. 1989;28:5327–5334. doi: 10.1021/bi00439a004. [DOI] [PubMed] [Google Scholar]
- 56.Liao H.Y., Da C.M., Liao B., Zhang H.H. Roles of matrix metalloproteinase-7 (MMP-7) in cancer. Clin. Biochem. 2021;92:9–18. doi: 10.1016/j.clinbiochem.2021.03.003. [DOI] [PubMed] [Google Scholar]
- 57.Wang S.C., Parekh J.R., Porembka M.R., Nathan H., D’Angelica M.I., DeMatteo R.P., Fong Y., Kingham T.P., Jarnagin W.R., Allen P.J. A pilot study evaluating serum MMP7 as a preoperative prognostic marker for pancreatic ductal adenocarcinoma patients. J. Gastrointest. Surg. 2016;20:899–904. doi: 10.1007/s11605-015-3057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szarvas T., Jäger T., Becker M., Tschirdewahn S., Niedworok C., Kovalszky I., Rübben H., Ergün S., vom Dorp F. Validation of circulating MMP-7 level as an independent prognostic marker of poor survival in urinary bladder cancer. Pathol. Oncol. Res. 2011;17:325–332. doi: 10.1007/s12253-010-9320-4. [DOI] [PubMed] [Google Scholar]
- 59.Sharma K.L., Misra S., Kumar A., Mittal B. Higher risk of matrix metalloproteinase (MMP-2, 7, 9) and tissue inhibitor of metalloproteinase (TIMP-2) genetic variants to gallbladder cancer. Liver Int. 2012;32:1278–1286. doi: 10.1111/j.1478-3231.2012.02822.x. [DOI] [PubMed] [Google Scholar]
- 60.Sanli M., Akar E., Pehlivan S., Bakır K., Tuncozgur B., Isik A.F., Pehlivan M., Elbeyli L. The relationship of metalloproteinase gene polymorphisms and lung cancer. J. Surg. Res. 2013;183:517–523. doi: 10.1016/j.jss.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 61.Graesslin O., Cortez A., Fauvet R., Lorenzato M., Birembaut P., Daraï E. Metalloproteinase-2, -7 and -9 and tissue inhibitor of metalloproteinase-1 and -2 expression in normal, hyperplastic and neoplastic endometrium: A clinical-pathological correlation study. Ann. Oncol. 2006;17:637–645. doi: 10.1093/annonc/mdj129. [DOI] [PubMed] [Google Scholar]
- 62.Ueno H., Yamashita K., Azumano I., Inoue M., Okada Y. Enhanced production and activation of matrix metalloproteinase-7 (matrilysin) in human endometrial carcinomas. Int. J. Cancer. 1999;84:470–477. doi: 10.1002/(SICI)1097-0215(19991022)84:5<470::AID-IJC4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 63.Misugi F., Sumi T., Okamoto E., Nobeyama H., Hattori K., Yoshida H., Matsumoto Y., Yasui T., Honda K.I., Ishiko O. Expression of matrix metalloproteinases and tissue inhibitors of metalloproteinase in uterine endometrial carcinoma and a correlation between expression of matrix metalloproteinase-7 and prognosis. Int. J. Mol. Med. 2005;16:541–546. [PubMed] [Google Scholar]
- 64.McCormick D., van der Rest M., Goodship J., Lozano G., Ninomiya Y., Olsen B.R. Structure of the glycosaminoglycan domain in the type IX collagen-proteoglycan. Proc. Natl. Acad. Sci. USA. 1987;84:4044–4048. doi: 10.1073/pnas.84.12.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Piotrowski A., Benetkiewicz M., Menzel U., de Ståhl T.D., Mantripragada K., Grigelionis G., Buckley P.G., Jankowski M., Hoffman J., Bała D., et al. Microarray-based survey of CpG islands identifies concurrent hyper- and hypomethylation patterns in tissues derived from patients with breast cancer. Genes Chromosomes Cancer. 2006;45:656–667. doi: 10.1002/gcc.20331. [DOI] [PubMed] [Google Scholar]
- 66.Roi A., Andreescu N.I., Roi C.I., Negrut M.L., Sinescu C., Rivis M., Boruga M.V., Rusu L.C. Comparative Analysis of COL9A1 Genotyping in Oral Squamous Cell Carcinoma Diagnosis: A Pilot Study. Appl. Sci. 2021;11:11102. doi: 10.3390/app112311102. [DOI] [Google Scholar]
- 67.Ruiz-García R., Mora S., Sánchez G.L., Lostao L.M., Artal E.P., Contreras J.R., Anel A., González-Granado L.I., Pérez D.M., Allende L.M. Decreased activation-induced cell death by EBV-transformed B-cells from a patient with autoimmune lymphoproliferative syndrome caused by a novel FASLG mutation. Pediatr. Res. 2015;78:603–608. doi: 10.1038/pr.2015.170. [DOI] [PubMed] [Google Scholar]
- 68.Peter M.E., Hadji A., Murmann A.E., Brockway S., Putzbach W., Pattanayak A., Ceppi P. The role of CD95 and CD95 ligand in cancer. Cell Death Differ. 2015;22:549–559. doi: 10.1038/cdd.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hadji A., Ceppi P., Murmann A.E., Brockway S., Pattanayak A., Bhinder B., Hau A., De Chant S., Parimi V., Kolesza P., et al. Death induced by CD95 or CD95 ligand elimination. Cell Rep. 2014;7:208–222. doi: 10.1016/j.celrep.2014.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McGlorthan L., Paucarmayta A., Casablanca Y., Maxwell G.L., Syed V. Progesterone induces apoptosis by activation of caspase-8 and calcitriol via activation of caspase-9 pathways in ovarian and endometrial cancer cells in vitro. Apoptosis. 2021;26:184–194. doi: 10.1007/s10495-021-01657-1. [DOI] [PubMed] [Google Scholar]
- 71.Nallapalle S.R., Daripally S., Vidudala Prasad V.T.S. Promoter polymorphism of FASL confers protection against female-specific cancers and those of FAS impact the cancers divergently. Tumour. Biol. 2015;36:2709–2724. doi: 10.1007/s13277-014-2896-7. [DOI] [PubMed] [Google Scholar]
- 72.Liu H., Yang Z., Lu W., Chen Z., Chen L., Han S., Wu X., Cai T., Cai Y. Chemokines and chemokine receptors: A new strategy for breast cancer therapy. Cancer Med. 2020;9:3786–3799. doi: 10.1002/cam4.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen L., Shi Y., Zhu X., Guo W., Zhang M., Che Y., Tang L., Yang X., You Q., Liu Z. IL-10 secreted by cancer-associated macrophages regulates proliferation and invasion in gastric cancer cells via c-Met/STAT3 signaling. Oncol. Rep. 2019;42:595–604. doi: 10.3892/or.2019.7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gutiérrez-Hoya A., Soto-Cruz I. Role of the JAK/STAT Pathway in Cervical Cancer: Its Relationship with HPV E6/E7. Oncoproteins Cells. 2020;9:2297. doi: 10.3390/cells9102297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethical reasons.