Abstract
Pseudomonas aeruginosa can utilize arginine and other amino acids as both carbon and nitrogen sources. Earlier studies have shown that the specific porin OprD facilitates the diffusion of basic amino acids as well as the structurally analogous beta-lactam antibiotic imipenem. The studies reported here showed that the expression of OprD was strongly induced when arginine, histidine, glutamate, or alanine served as the sole source of carbon. The addition of succinate exerted a negative effect on induction of oprD, likely due to catabolite repression. The arginine-mediated induction was dependent on the regulatory protein ArgR, and binding of purified ArgR to its operator upstream of the oprD gene was demonstrated by gel mobility shift and DNase assays. The expression of OprD induced by glutamate as the carbon source, however, was independent of ArgR, indicating the presence of more than a single activation mechanism. In addition, it was observed that the levels of OprD responded strongly to glutamate and alanine as the sole sources of nitrogen. Thus, that the expression of oprD is linked to both carbon and nitrogen metabolism of Pseudomonas aeruginosa.
Pseudomonas aeruginosa, an opportunistic pathogen of clinical relevance, is difficult to treat because of its high intrinsic resistance to many antibiotics. This resistance is partly due to its low outer membrane permeability relative to other gram-negative bacteria such as Escherichia coli (1, 8, 27). To overcome restrictions in nutrient uptake, gram-negative bacteria have channel-forming proteins which can be divided into three classes: general porins, specific porins, and highly substrate-specific gated channels which translocate their ligands in an energy-dependent process (8, 15). General porins form aqueous channels allowing the nonspecific diffusion of small hydrophilic molecules. Specific porins are differentiated from general porins by possessing a substrate-specific binding site that facilitates the diffusion of bound molecules at a much higher rate than other molecules of comparable size (15). While P. aeruginosa demonstrates weakly functional general porins (8), several specific porins have been described. Examples of the latter type include the phosphate-specific porin OprP from P. aeruginosa, which is induced by phosphate starvation (7), and the glucose-inducible porin OprB, which facilitates the diffusion of carbohydrates such as glucose, mannitol, and glycerol (6, 24, 25). Similarly, OprD has been shown to possess a binding site for basic amino acids and facilitates the diffusion of these and small peptides containing basic amino acids (23). OprD also serves as the channel for the antibiotic imipenem, which structurally resembles basic amino acids (23). Although OprD is classified as a specific porin, the channel can be utilized by the structurally unrelated compound gluconate under growth-limiting conditions in a binding-site-independent manner (9). Unlike the situation for OprB, substrate inducibility of OprD expression has not been previously demonstrated.
P. aeruginosa can utilize amino acids, including the basic amino acid arginine, as sole sources of nitrogen and carbon. When arginine is utilized as a carbon source under aerobic conditions, it is converted to glutamate by the enzymes of the arginine-succinyl transferase (AST) pathway (5). The enzymes of the AST pathway are strongly induced by exogenous arginine (5). Recently, the arginine-responsive regulatory protein ArgR was characterized (18). ArgR belongs to the AraC family of transcriptional regulators and activates the expression of the aru gene cluster, encoding enzymes of the AST pathway, and the aotJQMOP operon, encoding an arginine transport system (16, 18). ArgR also represses synthesis of enzymes involved in the biosynthesis of arginine (18). Inactivation of argR abolishes growth with arginine and ornithine as sources of carbon but not as sources of nitrogen (18).
We investigated the production of OprD in response to its putative substrates, basic amino acids, and found that the amounts of OprD were strongly increased when P. aeruginosa was grown with certain amino acids as the sole sources of carbon and that OprD was also regulated in response to various nitrogen sources. Furthermore, ArgR activated oprD expression in response to arginine, and purified ArgR protected a DNA region upstream of the oprD start codon from DNase I digestion.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. aeruginosa wild-type strain PAO1 (4) and strain PAO501 (argR::Gmr) (18) were grown in minimal medium containing 62 mM potassium phosphate buffer (pH 7), 0.5 mM MgSO4, 20 μM FeSO4, and carbon and nitrogen sources at final concentrations of 20 mM unless otherwise indicated. Gentamicin (15 μg/ml) was added to cultures of strain PAO501. The oprD-negative strain H729 (oprD::Kmr) (9) was grown in minimal medium containing succinate, ammonium sulfate, and kanamycin (150 μg/ml).
Materials.
Unless otherwise mentioned, chemicals were obtained from Sigma Chemical (St. Louis, Mo.); acrylamide and alkaline phosphatase-conjugated goat anti-mouse antibodies were obtained from Bio-Rad (Richmond, Calif.); molecular weight standards for protein gels were obtained from Bio-Rad or Pharmacia Biotec Inc. (Baie d’Urfe, Quebec, Canada). Enzymes were obtained from Gibco BRL (Burlington, Ontario, Canada).
Characterization of outer membranes.
P. aeruginosa was grown to mid-log phase (optical density at 600 nm of 0.45 to 0.55), and outer membrane proteins were isolated as the N-lauryl sarcosinate-insoluble fraction (20). Equal amounts of protein were loaded on each gel and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on gels containing 12.5% acrylamide (6). OprD was routinely identified by Western immunoblotting in each outer membrane protein preparation. Immunoblotting was performed as described previously (14). The mouse anti-OprD monoclonal antibody used in this study was a kind gift of Naomasa Gotoh (Kyoto Pharmaceutical University, Kyoto, Japan).
DNA footprinting and gel retardation experiments.
The experiments were performed essentially as previously described (19). Briefly, a DNA fragment carrying the regulatory region of the oprD gene (nucleotides 436 to 754 in Fig. 2) was amplified by PCR using two oligonucleotides designed to generate BamHI restriction sites: oligo-1 (5′-ATAGGATCCGTACGGAACATGACAT-3′) and oligo-2 (5′-ATAGGATCCACTTCATCACTTTCATT-3′). The amplified fragment was digested with BamHI and ligated into the BamHI site of pUC19 (26). The orientation and nucleotide sequence of the insert of one of the resulting plasmids, pOPD100, were confirmed by nucleotide sequencing. For gel retardation assays, the EcoRI/HindIII fragment of pOPD100 containing the oprD regulatory region was purified from 1% agarose gels and labeled with both [α-32P]dATP and [α-32P]dGTP, using Klenow polymerase. The radioactively labeled DNA probe (10−12 M) was allowed to interact with different concentrations of purified ArgR (19) in 20 μl of 50 mM Tris-HCl (pH 7.5)–50 mM KCl–1 mM EDTA–5% (vol/vol) glycerol–50 μg of bovine serum albumin per ml. The mixtures were allowed to equilibrate for 20 min at 25°C, reactions were terminated by the addition of an excess of unlabeled DNA probe (10−10 M), and then the reactants were applied to a 5% polyacrylamide gel while the gel was running. For DNase I footprinting experiments, the radioactively labeled probe described above was further digested with either SmaI or PstI to identify the footprinting pattern of the antisense or sense strands, respectively. The reaction mixture (200 μl) contained 10−10 M oprD operator DNA, 0.62 to 10 nM ArgR protein, 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 1 μg of sheared salmon sperm DNA, and 10 μg of bovine serum albumin. After incubation for 30 min at 25°C, pancreatic DNase I (0.2 μg; Boehringer Mannheim) was added. The digestion was allowed to proceed for 2 min and then terminated by the addition of 20 μl of 3 M sodium acetate, 10 μg of yeast tRNA, and 600 μl of ethanol. After precipitation with ethanol at −70°C, the pellet was dissolved in 20 μl of formamide dye mixture and the reaction products were analyzed on a 6% denaturing polyacrylamide sequencing gel against a guanine sequencing ladder (12).
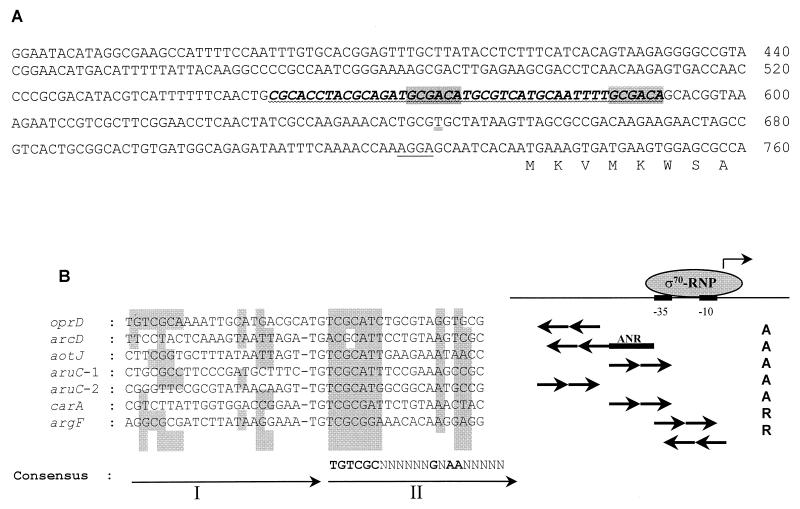
FIG. 2.
(A) Nucleotide sequence of the P. aeruginosa oprD promoter. The Shine-Dalgarno sequence (single line) is indicated. The DNA segment with high similarity to the consensus sequence for ArgR binding is underlined (wavy line) and the position of the conserved TGTCGC motifs present on the antisense strand (GCGACA on the sense strand shown here) are shaded. The DNA region protected by ArgR from DNase I digestion is shown in bold italic letters. The transcription start site of oprD is underlined (double line). (B) Sequence alignment of ArgR binding sites. The sequences were obtained from the results of DNase I footprintings (11, 16, 19) and aligned with the Clustal W program (22). The first and second repeats of the binding sites (I and II) are depicted by arrows. The consensus sequence was deduced from the second repeats, which are more conserved. Nucleotides identical to those of the consensus site are shaded. ς70-RNP, RNA polymerase holoenzyme; −35 and −10, promoter regions recognized by ς70. A, activation; R, repression. The ANR binding site of the arc promoter (11) is also indicated.
Primer extension experiments.
Total RNA (RNeasy Mini Kit; Qiagen, Mississauga, Ontario, Canada) was isolated from strain PAO1 after growth in minimal medium with succinate and glutamate or ammonium sulfate as carbon and nitrogen sources. A primer which could hybridize to bp 794 to 774 (5′-AACTGAGTGCTACCTGCGGA-3′) of the oprD gene was end labeled with [γ32P]ATP (Amersham Canada Ltd., Oakville, Ontario, Canada) and annealed to approximately 5 μg of total RNA in 250 mM Tris-Cl (pH 8.3)–100 mM KCl in a final volume of 10 μl. For extending the primer, the following reagents were added to the annealing mixture: 100 U of reverse transcriptase (Superscript; Gibco BRL), 1.5 μl of 1 M Tris-Cl (pH 8), 1.35 μl of 1 M KCl, 1.8 μl of 0.1 M MgCl2, 0.6 μl of 100 mM deoxynucleoside triphosphates, 0.3 μl of RNase inhibitor (Gibco BRL), and 14 μl of water. The reaction mixture was incubated at 45°C for 60 min, and then the reaction was stopped by addition of 1 μl of 0.5 M EDTA (pH 8.0). The samples were then treated with DNase-free RNase (Boehringer Mannheim) for 15 min at 37°C followed by ethanol precipitation of the cDNA. The dried samples were resuspended in 3 to 4 μl of loading buffer, denatured, and loaded on a 6% DNA sequencing gel. Dideoxy-sequencing reactions (Fmol DNA cycle sequencing system; Promega, Madison, Wis.) were generated from the oprD carrying plasmid pXH2 (9) with the same primer as used for the primer extension reactions. The following controls were performed: tRNA was used instead of total RNA, and the total RNA was treated with RNase prior to cDNA synthesis.
Quantification of oprD transcript by dot blot analysis.
Total RNA was isolated from strains PAO1 and PAO501, and from strain H729 as a negative control, by using an RNeasy Mini Kit (Qiagen). RNA concentrations were determined spectrometrically, and 2 μg of RNA was denatured and spotted onto positively charged nylon membranes (Boehringer Mannheim). An oprD-specific probe was generated by amplification of a 828-bp internal fragment of the oprD gene, and a rpoB-specific probe was generated by amplification of a 324-bp internal fragment of the rpoB gene. Both probes were labeled with [α-32P]dCTP (Amersham Canada Ltd.) by using a Rediprime II random prime labeling kit (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). Hybridization at 42°C overnight was followed by two low-stringency washes, two moderate-stringency washes, and two high-stringency washes essentially as described elsewhere (21). The amounts of bound radioactive probe were detected by using a PhosphorImager SI system and ImageQuant version 1.1 software (Molecular Dynamics Inc., (Sunnyvale, Calif.). The amounts of rpoB transcript were used as a reference to normalize the amounts of oprD transcript. Dot blot experiments were performed with three individual RNA preparations from each strain grown under the appropriate conditions.
Nucleotide sequence accession number.
The oprD nucleotide sequence is filed under GenBank accession no. Z14065.
RESULTS
Induction of OprD by amino acids provided as sole sources of carbon.
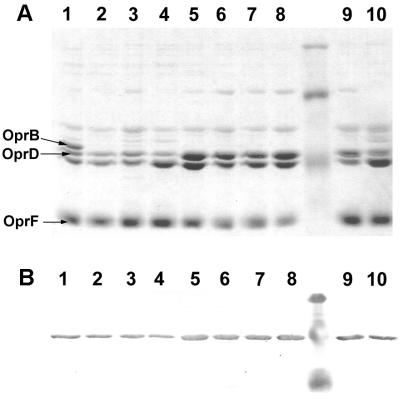
Previous studies indicated that OprD was involved in the uptake of basic amino acids and small peptides containing those amino acids (23). To determine whether the synthesis of OprD could be influenced by its substrates, P. aeruginosa PAO1 was grown in minimal medium with various amino acids as the sole sources of carbon and with ammonium sulfate as a nitrogen source. P. aeruginosa preferentially utilizes succinate or other tricarboxylic (TCA) cycle intermediates as carbon sources over glucose or other carbonhydrates (2). Consequently, succinate, which represses the utilization of other carbon sources, was used as a negative control. All cultures were harvested in mid-log phase, and outer membrane proteins were characterized by SDS-PAGE. The identification of OprD was confirmed by Western immunoblotting using an OprD-specific monoclonal antibody. After growth with succinate, OprD was present only at moderate levels (Table 1; Fig. 1, lane 4). In contrast, the amounts of OprD in outer membranes isolated from cultures grown with different amino acids (arginine, histidine, alanine, or glutamate) were 3.5- to 5.6-fold higher (Table 1; Fig. 1, lanes 5 to 8). These results suggested that amino acids, regardless of their charge or size, induced the synthesis of OprD to similar extents. This observation raised two questions: first, was the response to amino acids repressible by succinate?, and second, would carbon sources other than amino acids or TCA cycle intermediates lead to increased levels of OprD? To address the first question, outer membrane proteins were isolated from PAO1 grown with succinate and either glutamate or alanine as carbon sources. The presence of succinate suppressed the production of high levels of OprD mediated by glutamate completely and the induction mediated by alanine partially (Table 1; Fig. 5, lanes 1, 4, and 7). To assess whether the induction of OprD was an amino acid-specific response, outer membrane proteins were isolated from cultures grown with gluconate, glucose, or glycerol. Neither glucose nor glycerol induced OprD synthesis, and the amounts of OprD in outer membrane preparations were only slightly higher than those from succinate-grown cultures (Table 1; Fig. 1, lanes 1, 3, and 4). In contrast, the utilization of glucose strongly induced synthesis of OprB as expected (6). Although OprB also facilitates the passage of glycerol, induction of OprB mediated by glycerol seems to occur only after many generations of growth (24). Therefore, our findings indicated that amino acids when provided as the sole sources of carbon selectively induced the synthesis of OprD. The carbon source gluconate also failed to induce high level production of OprD when provided at a concentration of 20 mM. However, when PAO1 was grown with growth-limiting concentrations of 4 mM gluconate or 4 mM succinate and harvested 1 h after growth had ceased, the amounts of OprD were each 2.1-fold higher than those seen in cultures grown with 20 mM succinate (Table 1; Fig. 1 [compare lanes 2 and 9 and lanes 4 and 10]).
TABLE 1.
Influence of carbon sources on the production of OprD in P. aeruginosa PAO1
| Carbon source(s)a | Relative amt of OprDb |
|---|---|
| Succinate | 1.0 |
| Low succinate (4 mM) | 2.1 |
| Glucose (0.4%) | 1.2 |
| Gluconate | 1.0 |
| Low gluconate (4 mM) | 2.1 |
| Glycerol (1%) | 1.8 |
| Arginine | 4.6 |
| Histidine | 3.5 |
| Glutamate | 4.3 |
| Glutamate + succinate | 0.9 |
| Alanine | 5.6 |
| Alanine + succinate | 2.4 |
| Glucose (0.4%) + arginine | 1.9 |
| Glycerol (1%) + arginine | 3.0 |
All carbon sources were provided at 20 mM unless indicated otherwise. All cultures contained 7 mM (NH4)2SO4 as the nitrogen source.
The amount of OprD protein was estimated by densitometric analysis of SDS-polyacrylamide gels of individual outer membrane preparations by using ImageQuant version 1.1 (Molecular Dynamics). Values were normalized against the amount of the constitutively expressed outer membrane protein OprF present in each preparation. The amount of OprD observed after growth with succinate and ammonium sulfate was defined as 1.0, and the amounts observed with other carbon sources were calculated relative to this value.
FIG. 1.
Effects of different carbon sources on expression of OprD. (A) SDS-PAGE analysis of outer membrane proteins from P. aeruginosa PAO1; (B) Western immunoblot of outer membrane proteins probed with an OprD-specific monoclonal antibody. Molecular weight standards: (A) 94,000, 67,000, and 43,000; (B) 81,000, 47,700, and 34,600. The positions of OprD, OprB, and OprF are indicated by arrows on the left. Lane 1, 0.4% glucose; lane 2, 20 mM gluconate; lane 3, 1% glycerol; lane 4, 20 mM succinate; lane 5, 20 mM arginine; lane 6, 20 mM glutamate; lane 7, 20 mM histidine; lane 8, 20 mM alanine; lane 9, 4 mM gluconate; lane 10, 4 mM succinate.
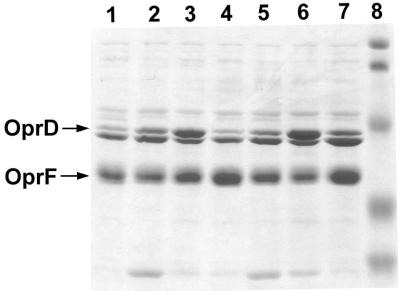
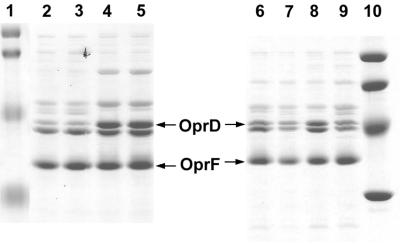
FIG. 5.
Regulation of OprD expression in response to nitrogen sources, determined by SDS-PAGE analysis of outer membrane proteins isolated from strain PAO1 grown with 7 mM ammonium sulfate (lane 1), 20 mM arginine (lane 2), 20 mM glutamate (lane 3), 20 mM glutamate and 7 mM ammonium sulfate (lane 4), 20 mM histidine (lane 5), 20 mM alanine (lane 6), or 20 mM alanine and 7 mM ammonium sulfate (lane 7) as sources of nitrogen. All cultures contained succinate as the carbon source. Molecular weight standards in lane 8: 104,000, 81,000, 47,700, 34,600, and 28,300. The positions of OprD and OprF are marked by arrows.
Binding of ArgR to the oprD operator.
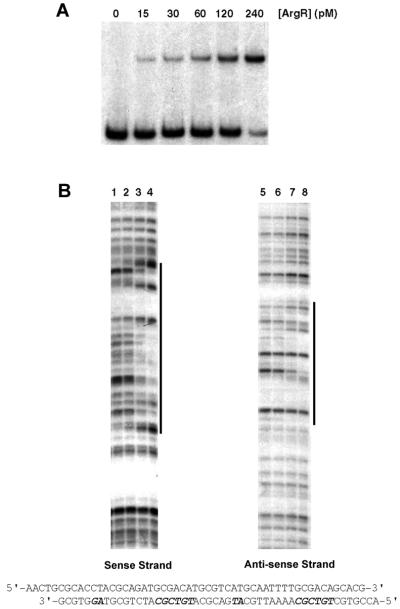
Since OprD production was activated when strain PAO1 was grown with arginine as the carbon source, we investigated whether the arginine-responsive regulator ArgR was involved in the control of oprD expression. Recently, ArgR was shown to be absolutely required for the utilization of arginine as a carbon source by activating the synthesis of enzymes of the catabolic AST pathway (18). Binding of purified ArgR to the operators of the carA, argF, aotJ, and aruC genes led to the identification of a consensus binding sequence, TGTCGCN8AAN5 (19). This motif appears twice in a direct repeat arrangement in each binding site (19). A search of the DNA sequence upstream of the start codon of oprD for possible ArgR binding sites identified a putative site present on the antisense strand, and the TGTCGC sequence was oriented in the opposite direction relative to the oprD gene. The two repeats were highly conserved and separated by a single nucleotide (Fig. 2). To determine whether ArgR binds to the oprD promoter, gel retardation assays were performed (Fig. 3). Increasing concentrations of purified ArgR were incubated with a DNA fragment containing the regulatory region of oprD. At ArgR concentrations from 15 to 240 pM, a single retarded band of progressively increasing intensity was observed (Fig. 3A). When ArgR was used at a higher concentration, however, a second retarded band appeared (data not shown). To experimentally define the target site for ArgR binding, DNase I footprinting analysis was performed (Fig. 3B). ArgR protected a 47-bp region from nuclease digestion. This region included the entire predicted binding site. An alignment of this sequence with the ArgR binding sites within the argF, car, aru, aot, and arc promoters (Fig. 2B) showed that the consensus TGTCGC sequence is present not only in the usually more conserved second repeat of the ArgR binding sites but also in the first repeat. The ArgR operator of oprD is the only one identified so far that possesses a second TGTCGC sequence in the more degenerate first repeat. However, it is also the only site that has an extra nucleotide between the two repeats and an AG sequence instead of a conserved AA sequence in the second repeat. Apparently, these nonconserved features found in the oprD regulatory region did not prevent ArgR from binding, and whether this unique feature of having two TGTCGC sequences accounted for the second retarded band observed in the gel retardation experiments but not in the DNase I protection experiments remains to be elucidated.
FIG. 3.
Analyses of ArgR binding to the oprD operator. (A) Gel retardation experiments. The radioactive 32P-labeled oprD operator DNA (1 pM) was incubated with increasing concentrations of purified ArgR as indicated. (B) DNase I footprintings. The DNA fragment (100 pM) was specifically labeled at the 3′ end of each strand as described in Materials and Methods. Lanes 1 and 5, DNase I digestion in the absence of ArgR; lanes 2 and 6, 2.5 nM ArgR; lanes 3 and 7, 5 nM ArgR; lanes 4 and 8, 10 nM ArgR. The nucleotide sequence of the ArgR-protected region is indicated.
Determination of the transcription start site.
Primer extension experiments were performed to determine the transcription start site of oprD in cells grown in either succinate and glutamate, succinate, glutamate, and ammonium sulfate, or succinate and ammonium sulfate. A strong signal corresponding to nucleotide T-647 in the promoter region of oprD was observed. There were also two weaker signals corresponding to nucleotides C-660 and C-663, which likely represented degradation products of the larger mRNA or were caused by premature termination of the cDNA synthesis. Alternatively, these signals could represent secondary transcription start sites.
Activation of OprD expression by ArgR.
Outer membranes were isolated from P. aeruginosa PAO1 and its argR-deficient mutant PAO501 (argR::Gmr) after growth in minimal medium with ammonium sulfate as the nitrogen source and glycerol or glucose as the carbon source in the presence or absence of arginine. As indicated above, neither glycerol nor glucose induced the synthesis of OprD in either strain (Table 2; Fig. 4, lanes 7 and 9). The addition of arginine to glycerol or glucose-containing cultures, however, resulted in an increase of OprD in strain PAO1 but not in PAO501 (argR::Gmr) (Table 2; Fig. 4, lanes 6 and 8). This observation was confirmed by determination of the amounts of oprD transcript (Table 2). The doubling times of PAO1 and PAO501 when grown with glycerol were the same (2.8 h). The addition of arginine reduced the doubling time of PAO1 to 1.65 h but had no effect on the growth of strain PAO501. This result indicated that PAO1 utilized arginine and that ArgR activated the expression of oprD in response to exogenous arginine. Since growth with amino acids other than arginine also strongly increased the amounts of OprD in strain PAO1, levels of OprD in strains PAO1 and PAO501 were compared after growth with succinate or glutamate. As expected, after growth with succinate, OprD was present at moderate levels, while glutamate induced the production of OprD in PAO1 and its argR mutant derivative PAO501 (Table 2; Fig. 4, lanes 2 to 5), indicating that induction by glutamate was independent of ArgR.
TABLE 2.
Influence of carbon sources on the production of OprD and oprD mRNA in P. aeruginosa PAO1 and its argR derivative PAO501
| Carbon source(s)a | Relative OprD level
|
|||
|---|---|---|---|---|
| PAO1
|
PAO501
|
|||
| Proteinb | mRNAc | Protein level | mRNA | |
| Succinate | 1.0 | 1.0 | 1.0 | 1.0 |
| Glucose (0.4%) | 1.2 | NDd | 1.4 | ND |
| Glycerol (1%) | 1.8 | 1.3 | 1.9 | 1.0 |
| Glutamate | 4.3 | 2.4e | 4.2 | 2.5 |
| Glucose (0.4%) + arginine | 1.9 | ND | 1.2 | ND |
| Glycerol (1%) + arginine | 3.0 | 2.0 | 2.1 | 1.0 |
All carbon sources were provided at 20 mM unless indicated otherwise. All cultures contained 7 mM (NH4)2SO4 as the nitrogen source.
The amount of OprD protein was estimated by densitometric analysis of SDS-polyacrylamide gels of individual outer membrane preparations by using ImageQuant version 1.1 (Molecular Dynamics). Values were normalized against the amount of the constitutively expressed outer membrane protein OprF present in each preparation. The amount of OprD observed after growth with succinate and ammonium sulfate was defined as 1.0, and the amounts observed with other carbon sources were calculated relative to this value.
The amount of oprD-specific mRNA was determined from three individual RNA preparations and is expressed as an average value relative to that determined after growth with succinate and ammonium sulfate which was defined as 1.0.
ND, not determined.
Represents the mean of two determinations only.
FIG. 4.
ArgR-dependent induction of OprD, determined by SDS-PAGE analysis of outer membrane proteins isolated from strains PAO1 and PAO501 (argR deficient). All cultures contained ammonium sulfate as the nitrogen source and carbon sources as indicated. Lane 2, PAO1 with succinate (20 mM); lane 3, PAO501 with succinate (20 mM); lane 4, PAO1 with glutamate (20 mM); lane 5, PAO501 with glutamate (20 mM); lane 6, PAO501 with 1% glycerol and arginine (20 mM); lane 7, PAO501 with 1% glycerol; lane 8, PAO1 with 1% glycerol and arginine (20 mM); lane 9, PAO1 with 1% glycerol. Molecular standards are, from top to bottom, 104,000, 81,000, 47,700, and 34,600 in lane 1 and 94,000, 67,000, 43,000, and 30,000 in lane 10. The positions of OprD and OprF are indicated by arrows.
Regulation of OprD in response to nitrogen sources.
The above studies concentrated on amino acids presented as carbon sources. To determine whether the expression of OprD would be also regulated in response to nitrogen sources, strain PAO1 was grown in minimal medium with succinate as the carbon source and various nitrogen sources (Table 3; Fig. 5). Glutamate or alanine induced OprD (lanes 3 and 6) 4.0-fold compared to ammonium sulfate (lane 1). The induction by glutamate was suppressed completely in the presence of ammonium sulfate (lanes 4), while induction mediated by alanine was partially suppressed (lane 7). Growth with the basic amino acids histidine or arginine also increased OprD synthesis (lanes 2 and 5), although to a lesser extent (2.8- to 3.2-fold) than glutamate or alanine. Since the amino acids used have different numbers of nitrogen atoms, experiments were also carried out with equimolar (20 mM) amounts of nitrogen. Again, the strongest increase of OprD was observed with glutamate or alanine. The utilization of arginine or potassium nitrate as a nitrogen source resulted in 2.8- to 3.0-fold-higher levels of OprD compared to ammonium sulfate, while asparagine or urea as a nitrogen source did not induce OprD (Table 3). Conditions of severe nitrogen starvation were also tested. PAO1 was grown with ammonium sulfate at 0.7 mM, i.e., 1/10 of the concentration routinely used, and harvested 1 h after cessation of growth. The levels of OprD observed under these conditions did not differ from those found with excess ammonium sulfate (Table 3).
TABLE 3.
Influence of nitrogen sources on OprD production in P. aeruginosa PAO1
| Nitrogen source(s)a | Relative amt of OprDb |
|---|---|
| 7 mM (NH4)2SO4 | 1.0 |
| 20 mM glutamate | 4.0 |
| 20 mM glutamate + 7 mM (NH4)2SO4 | 0.9 |
| 20 mM alanine | 4.0 |
| 20 mM alanine + 7 mM (NH4)2SO4 | 2.4 |
| 20 mM arginine | 3.2 |
| 5 mM arginine | 2.8 |
| 20 mM histidine | 2.8 |
| 20 mM KNO3 | 3.0 |
| 10 mM asparagine | 0.8 |
| 10 mM urea | 1.4 |
| 0.7 mM (NH4)2SO4 | 1.2 |
All cultures were grown with 20 mM succinate as the carbon source.
The amount of OprD protein was estimated by densitometric analysis of SDS-polyacrylamide gels of individual outer membrane preparations by using ImageQuant version 1.1 (Molecular Dynamics). Values were normalized against the amount of the constitutively expressed outer membrane protein OprF present in each preparation. The amount of OprD observed after growth with succinate and ammonium sulfate was defined as 1.0, and the amounts observed with other carbon sources were calculated relative to this value.
DISCUSSION
The results presented here demonstrate that OprD was induced by amino acids when provided as the sole sources of carbon and that succinate suppressed this amino acid-mediated induction. Although catabolite repression control in P. aeruginosa is poorly understood, it is well established that intermediates of the TCA cycle such as succinate repress the catabolic pathways of other carbon sources (2). Carbon starvation after growth with succinate or growth with the relatively poor carbon source glycerol led to a slight increase in OprD levels. These results indicated that carbon starvation enhanced the synthesis of OprD, possibly due to relief of catabolite repression. It is therefore proposed that the expression of oprD was subject to catabolite repression by succinate. However, high expression of oprD required inducing conditions such as the utilization of amino acids.
The activation of oprD expression by arginine was dependent on the arginine-responsive regulator ArgR. The response to glutamate, however, was not altered in an argR-negative mutant, demonstrating that the induction of oprD expression mediated by glutamate was independent of ArgR. Clearly, different mechanisms exist for activating the synthesis of OprD in response to different amino acids when these are used as carbon sources.
The expression of OprD was also regulated in response to various nitrogen sources. Glutamate or alanine led to a strong increase in OprD levels compared to the amounts observed with ammonium sulfate, and ammonium sulfate reduced the glutamate- or alanine-mediated induction. Nitrogen starvation after growth with ammonium sulfate, however, did not lead to enhanced amounts of OprD. This finding indicated that growth with certain specific nitrogen sources, rather than nitrogen deprivation, stimulated induction of OprD. Thus, the expression of OprD seems to be regulated in response to both the nitrogen and carbon sources available.
To date, ArgR is the only regulator for which a direct involvement in the regulation of oprD expression has been shown. Sequence analysis of the oprD operator revealed no conserved −10 and −35 sequences upstream of the experimentally determined transcription start site (Fig. 2), which is not unusual for promoters regulated by members of the AraC family (3), to which ArgR belongs. ArgR responds to exogenous arginine and activates the expression of enzymes of the catabolic AST pathway, the aotJQMOP arginine transport operon, and the arcDABC operon, encoding the enzymes of the arginine deiminase pathway. It also represses the expression of arginine biosynthetic enzymes (16, 18, 11). A putative ArgR binding site was present in the oprD operator, and results of gel retardation and DNase I footprinting experiments were consistent with the hypothesis that purified ArgR bound to the oprD operator. The ArgR binding sequence identified here is on the antisense strand of the oprD regulatory region, centered at 77 bp upstream of the transcription start point (Fig. 2). This arrangement was similar to that found with the arc promoter (11) but differed from that found for the ArgR-dependent aot and aru operons, where ArgR binding sites were on the sense strand and overlapped with the −35 region. Interestingly, expression from the arc promoter is activated not only by ArgR but also by the ANR protein, equivalent to the FNR protein of E. coli (28), and binding of ArgR immediately upstream of ANR increases the ANR-dependent induction. The similarity in positions of the ArgR binding site in the operators of oprD and arc could reflect the possible binding of other regulators to the oprD operator as indeed occurs in the arc promoter (11).
Other factors than those addressed in this study, have been indicated to influence the production of OprD. The multidrug resistant nfxC phenotype is characterized by overproduction of the MexEFOprN efflux system and simultaneous repression of OprD, which causes imipenem resistance (10, 13). The physiological conditions under which mexEFoprN is expressed are not known. Overexpression of a putative LysR-type regulator, mexT, reduced oprD transcription, imipenem susceptibility, and caused the multidrug resistance phenotype (10, 17). Under the growth conditions used in this study, we did not observe expression of the outer membrane protein OprN, as tested by probing Western immunoblots with an OprN-specific monoclonal antibody, under conditions leading at the same time to low levels of OprD (data not shown). Therefore, MexT was likely not involved in the expression of oprD under any of the growth conditions used here.
Trias and Nikaido (23) identified the basic amino acid specific binding site in OprD by competition studies with the antibiotic imipenem. The uptake of imipenem also competed with small peptides containing basic amino acids, and it was concluded that these peptides and basic amino acids are likely to diffuse efficiently through OprD. However, imipenem has a much higher affinity for the binding site than l-amino acids, and therefore basic amino acids might not be the optimal substrates for the OprD channel (23). Although an oprD-negative mutant could grow with arginine as the sole source of carbon (data not shown), indicating that OprD is not required for growth with arginine, the coregulation of oprD expression with enzymes involved in the catabolism of arginine suggests that higher amounts of OprD could increase the influx of arginine. Since ArgR responds to exogenous arginine, OprD seems specifically induced when arginine is used as the carbon source. A similar regulatory mechanism might also occur specifically with histidine. One can easily understand why OprD should be induced by its specific basic substrates, but what about the strongly inducing acidic and hydrophobic amino acids glutamate and alanine? To make use of the specific binding site in OprD, certain criteria must be fulfilled, namely, an α-amino group and a positive charge at a certain distance from the carboxyl group (23). Neither glutamate nor alanine meets these criteria, and alanine has been shown not to compete with imipenem (23). At this time, it can only be speculated that induction of OprD by alanine or glutamate might occur, because they are degradation products of peptide substrates of OprD. Only further studies can address this question.
ACKNOWLEDGMENTS
This work was supported by a grant from the Medical Research Council of Canada to R.E.W.H. and NIH research grant GM47926 to A.A. R.E.W.H. was an MRC Distinguished Scientist and M.M.O. was a Postdoctoral Fellow of the Deutsche Forschungsgemeinschaft and the Canadian Cystic Fibrosis Foundation.
M.M.O. thanks Margaret K. Pope for enjoyable discussions and advice on primer extension experiments.
REFERENCES
- 1.Angus B L, Carey A M, Caron D A, Kropinski A M B, Hancock R E W. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild type with an antibiotic-supersusceptible mutant. Antimicrob Agents Chemother. 1982;21:299–309. doi: 10.1128/aac.21.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collier D N, Hager P W, Phibbs P V., Jr Catabolite repression control in the Pseudomonads. Res Microbiol. 1996;147:551–561. doi: 10.1016/0923-2508(96)84011-3. [DOI] [PubMed] [Google Scholar]
- 3.Gallegos M-T, Schleif R, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haas D, Holloway B W, Schambock A, Leisinger T. The genetic organization of arginine biosynthesis in Pseudomonas aeruginosa. Mol Gen Genet. 1977;154:7–22. doi: 10.1007/BF00265571. [DOI] [PubMed] [Google Scholar]
- 5.Haas D, Galimands M, Gamper M, Zimmermann A. Arginine network of Pseudomonas aeruginosa: specific and global controls. In: Silver S, Chakrabarty A M, Iglewski B, Kaplan S, editors. Pseudomonas: biotransformations, pathogenesis, and evolving biotechnology. Washington, D.C: American Society for Microbiology; 1990. pp. 303–316. [Google Scholar]
- 6.Hancock R E W, Carey A M. Protein D1—a glucose inducible, pore-forming protein from the outer membrane of Pseudomonas aeruginosa. FEMS Microbiol Lett. 1980;8:105–109. [Google Scholar]
- 7.Hancock R E W, Benz R. Demonstration and chemical modification of a specific phosphate binding site in the phosphate-starvation-inducible outer membrane porin protein P of Pseudomonas aeruginosa. Biochim Biophys Acta. 1986;860:699–707. doi: 10.1016/0005-2736(86)90569-9. [DOI] [PubMed] [Google Scholar]
- 8.Hancock R E W. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997;5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 9.Huang H, Hancock R E W. Genetic definition of the substrate selectivity of outer membrane porin protein OprD of Pseudomonas aeruginosa. J Bacteriol. 1993;175:7793–7800. doi: 10.1128/jb.175.24.7793-7800.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohler, T., M. Michea-Hamzehpour, U. Henze, N. Gotoh, L. K. Curty, and J.-C. Pechere. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345–354. [DOI] [PubMed]
- 11.Lu C-D, Winteler H, Abdelal A, Haas D. ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J Bacteriol. 1999;181:2459–2464. doi: 10.1128/jb.181.8.2459-2464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 13.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mutharia L M, Hancock R E W. Surface localization of Pseudomonas aeruginosa outer membrane protein F by using monoclonal antibodies. Infect Immun. 1983;42:1027–1033. doi: 10.1128/iai.42.3.1027-1033.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikaido H. Porins and specific diffusion channels in bacterial outer membranes. J Biol Chem. 1994;269:3905–3908. [PubMed] [Google Scholar]
- 16.Nishijyo T, Park S-M, Lu C-D, Itoh Y, Abdelal A T. Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J Bacteriol. 1998;180:5559–5566. doi: 10.1128/jb.180.21.5559-5566.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ochs M, Mc Cusker M P, Bains M, Hancock R E W. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob Agents Chemother. 1999;43:1085–1090. doi: 10.1128/aac.43.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park S-M, Lu C-D, Abdelal A T. Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J Bacteriol. 1997;179:5300–5308. doi: 10.1128/jb.179.17.5300-5308.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park S-M, Lu C-D, Abdelal A T. Purification and characterization of an arginine regulatory protein, ArgR, from Pseudomonas aeruginosa and its interactions with the control regions for the car, argF, and aru operons. J Bacteriol. 1997;179:5309–5317. doi: 10.1128/jb.179.17.5309-5317.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poxton I R, Bell G T, Barclay G R. The association on SDS-polyacrylamide gels of lipopolysaccharide and outer membrane proteins of Pseudomonas aeruginosa as revealed by monoclonal antibodies and Western blotting. FEMS Microbiol Lett. 1985;27:247–251. [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Thompson J D, Higgins D G, Gilson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4674–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trias J, Nikaido H. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J Biol Chem. 1990;265:15680–15684. [PubMed] [Google Scholar]
- 24.Williams S G, Greenwood J A, Jones C W. The effect of nutrient limitation on glycerol uptake and metabolism in continuous cultures of Pseudomonas aeruginosa. Microbiology. 1994;140:2961–2969. doi: 10.1099/13500872-140-11-2961. [DOI] [PubMed] [Google Scholar]
- 25.Wylie J L, Worobec E A. The OprB porin plays a central role in carbohydrate uptake in Pseudomonas aeruginosa. J Bacteriol. 1995;177:3021–3026. doi: 10.1128/jb.177.11.3021-3026.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 27.Yoshimura F, Nikaido H. Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. J Bacteriol. 1982;152:636–642. doi: 10.1128/jb.152.2.636-642.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmermann A, Reimmann C, Galim M, Haas D. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol Microbiol. 1991;5:1483–1490. doi: 10.1111/j.1365-2958.1991.tb00794.x. [DOI] [PubMed] [Google Scholar]