Abstract
Genes encoding a branched-chain α-keto acid dehydrogenase from Enterococcus faecalis 10C1, E1α (bkdA), E1β (bkdB), E2 (bkdC), and E3 (bkdD), were found to reside in the gene cluster ptb-buk-bkdDABC. The predicted products of ptb and buk exhibited significant homology to the phosphotransbutyrylase and butyrate kinase, respectively, from Clostridium acetobutylicum. Activity and redox properties of the purified recombinant enzyme encoded by bkdD indicate that E. faecalis has a lipoamide dehydrogenase that is distinct from the lipoamide dehydrogenase associated with the pyruvate dehydrogenase complex. Specific activity of the ptb gene product expressed in Escherichia coli was highest with the substrates valeryl-coenzyme A (CoA), isovaleryl-CoA, and isobutyryl-CoA. In cultures, a stoichiometric conversion of α-ketoisocaproate to isovalerate was observed, with a concomitant increase in biomass. We propose that α-ketoisocaproate is converted via the BKDH complex to isovaleryl-CoA and subsequently converted into isovalerate via the combined actions of the ptb and buk gene products with the concomitant phosphorylation of ADP. In contrast, an E. faecalis bkd mutant constructed by disruption of the bkdA gene did not benefit from having α-ketoisocaproate in the growth medium, and conversion to isovalerate was less than 2% of the wild-type conversion. It is concluded that the bkd gene cluster encodes the enzymes that constitute a catabolic pathway for branched-chain α-keto acids that was previously unidentified in E. faecalis.
The gram-positive, fermentative bacterium Enterococcus faecalis is a member of the lactic acid bacteria (LAB) which generally degrade glucose via the Embden-Meyerhof-Parnas (EMP) pathway. Anaerobically, pyruvate is mostly reduced to lactate, with 2 ATP formed per glucose consumed. Aerobically (or under glucose-limiting conditions) the homolactic fermentation is abandoned in favor of a fermentation with higher acetate production rates, yielding maximally 4 ATP per glucose. The conversion of pyruvate to acetate yields additional ATP via substrate level phosphorylation by the combined actions of the phosphotransacetylase and acetate kinase. The higher acetate production rates under aerobic conditions stem from a sparing effect of the NADH oxidase, relieving the stoichiometrical limits of the system by uncoupling NADH oxidation from carbon metabolism (7). Despite their relatively simple fermentative metabolism, LAB can utilize a much larger spectrum of Gibbs free energy sources, including gluconate, glycerol, pyruvate, malate, pentitols, and some amino acids, such as arginine (29).
The LAB are unable to synthesize heme and therefore lack respiratory cytochromes, catalase, and other heme proteins (12). These deficiencies account for the absence of both a functional electron transport chain and oxidative phosphorylation. The primary mechanism of ATP generation is via substrate-level phosphorylation. Although they also lack other typical aerobic enzymes, such as most of the enzymes of the tricarboxylic acid cycle, including the α-ketoglutarate dehydrogenase (OGDH) complex, the LAB do have a pyruvate dehydrogenase (PDH) complex (7). In E. faecalis, the PDH complex is active under anaerobic conditions, leading to acetate formation from pyruvate via the consecutive action of the PDH complex, phosphotransacetylase, and acetate kinase (53). The PDH complex has been purified from anaerobically grown E. faecalis, and four components corresponding to the E1α and E1β (PDH), E2 (dihydrolipoyl transacetylase), and E3 (lipoamide dehydrogenase) subunits were identified (54). In addition, the gene encoding the dihydrolipoyl transacetylase component, pdhC, has been cloned from E. faecalis (1). Of interest was the presence of two lipoyl domains in the E2 (pyruvate [E2p]) component, previously seen only in mammalian systems. Analysis of the E. faecalis pdhC gene and the flanking sequence revealed the E1β and E3 components, suggesting that the PDH complex is encoded in a gene cluster similar to that seen in Bacillus subtilis (24) and Bacillus stearothermophilus (3).
A second α-keto acid dehydrogenase complex, distinct from the PDH, has been identified in E. faecalis—the branched-chain α-keto acid dehydrogenase (BKDH) complex. This was purified from E. faecalis and found to be active with the branched-chain α-keto acids α-ketoisocaproic, α-ketoisovaleric, and α-keto-β-methylvaleric acid (49). The BKDH complex catalyzes the oxidative decarboxylation of the branched-chain α-keto acids usually derived from the transamination of the branched-chain amino acids valine, leucine, and isoleucine, generating the corresponding branched-chain acyl-coenzyme A’s (CoA’s). The BKDH complex has been purified from a number of other sources, including Pseudomonas putida (55), Pseudomonas aeruginosa (35), B. subtilis (30), rabbit liver (42), and rat and bovine kidneys (38, 45). The purified complexes are all composed of four polypeptides, E1α, E1β, E2, and E3. Cloning of the prokaryotic BKDH genes has been reported for P. putida (57), B. subtilis (61), Streptomyces avermitilis (10, 52), and Myxococcus xanthus (59). These complexes were all found in gene clusters with the expected gene order E1α-E1β-E2. The BKDH complexes serve a number of metabolic functions, including ATP generation in P. putida, production of branched-chain fatty acids for membrane biosynthesis in B. subtilis, cell-cell signaling in M. xanthus, and avermectin biosynthesis in S. avermitilis.
In this article we report on the cloning, characterization, and physiological role of the bkd gene cluster from E. faecalis, which contains the genetic determinants encoding the BKDH complex, an acylphosphotransferase, and an acyl kinase. Mutational inactivation of the gene cluster in E. faecalis abolishes the ability to utilize the branched-chain α-keto acids, demonstrating an essential role for the pathway in the catabolism of these compounds. This gene cluster has significant homology with one that was recently described in B. subtilis (8); however, significant differences—both at the genetic and the metabolic level—distinguish the two clusters, the most important of these being their respective roles. The B. subtilis system is involved in carbon assimilation, in contrast to the E. faecalis system, whose main function is most likely energy generation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains, plasmids, and DNA fragments used in this study are listed in Table 1. Enterococcus faecalis was grown aerobically at 37°C in M17 (58) or brain heart infusion (BHI) broth (Difco, Detroit, Mich.). Escherichia coli was grown aerobically at 37°C in TYP broth (16 g of Bacto tryptone, 16 g of yeast extract, 5 g of NaCl, and 2.5 g of K2HPO4 per liter) or in TY broth (10 g of Bacto tryptone, 5 g of yeast extract, and 5 g of NaCl [pH = 7.4]). When the λgt11 and λFIXII E. faecalis genomic libraries in E. coli Y1090 were grown and screened, maltose was included at a final concentration of 0.2% (wt/vol). Concentrations of antibiotics used in selective media were as follows: for chloramphenicol, ampicillin, and carbenicillin, 50 μg/ml; for tetracycline, 10 μg/ml; and for kanamycin, 25 μg/ml for E. coli and 1,000 μg/ml for E. faecalis.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Bacterial strains | ||
| E. faecalis | ||
| 10C1 | ATCC 11700 | |
| OG1RF | 13 | |
| OG1RF:bkdA | bkdA::mγδ-200; Kanr | This work |
| OG1RF:bkdC | bkdC::mγδ-200; Kanr | This work |
| E. coli | ||
| Y1090 | Δ(lac)U169 Δ(lon)? araD139 strA supF mcrA trpC22::Tn10(Tetr) (pMC9 Ampr Tetr) | |
| XL1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| TG-2 | Δ(lac-pro) thi recA supE hsdR hsdM F′(traD36 proA+ B+ lacIqlacZΔM15) | |
| JM109(DE3) | el4 (McrA+) recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1Δ(lac-proAB) 1(DE3) | Promega |
| DPWC | Gold Biotechnology | |
| BW26 | Gold Biotechnology | |
| CBK884 | 16 | |
| LW49 | 16 | |
| Plasmids | ||
| pBluescript | Cloning vector; Apr | Stratagene |
| pUC118/9 | Cloning vector; Apr | 68 |
| pMOB | Cloning vector; Apr | Gold Biotechnology |
| pOXO7 | T7 expression vector; Cmr | This work |
| pOXO4 | T7 expression vector; Cmr | 47 |
| pWKS30 | Cloning vector; Apr | 62 |
| pLD03 | pBluescript containing a 4.2-kb EcoRI insert from λlpd3; Apr | This work |
| pLD09 | pOXO4 containing a 2.4-kb PstI/BamHI insert from λlpd9; Cmr | This work |
| pBKD06 | pWKS30 containing a 5.0-kb BamHI/SalI insert from λbkd3; Apr | This work |
| pBKD09 | pBluescript containing a 2.3-kb KpnI/XbaI insert from pBKD06 containing bkdA and bkdB; Apr | This work |
| pBKD14 | pUC118 containing a 3.0-kb XbaI/KpnI insert from pBKD06 containing bkdC; Apr | This work |
| pBKD15 | pUC118 containing a 2.4-kb PstI/BamHI insert from pLD09 containing bkdD; Apr | This work |
| pPTB01 | pOXO7 containing a 2.3-kb KpnI/PstI insert from pLD03 containing bkdT; Cmr | This work |
| pBUK01 | pOXO7 containing a 1.2-kb XbaI/HindIII insert from pLD03 containing bkdK; Cmr | This work |
DNA isolation, manipulation, and amplification.
Recombinant DNA procedures, including plasmid and total DNA isolations, DNA-DNA hybridizations, and molecular cloning, were performed essentially as described previously (47, 48). DNA modification enzymes, including restriction endonucleases and T4 DNA ligase, were obtained from Promega (Madison, Wis.). DNA restrictions were performed in a “cuts all” reaction buffer (10× = 200 mM Tris-HCl [pH = 7.5], 70 mM MgCl2, 1 M KCl, 20 mM 2-mercaptoethanol) rather than those provided with the enzymes. In many cases, ligations were performed in Sea Plaque GTG agarose (FMC Bioproducts, Rockland, Maine) by using the conditions suggested by the manufacturer. For all ligations, phage T4 DNA ligase and 10× reaction buffer were obtained from Promega (Madison, Wis.). PCR amplification of genomic DNA was carried out as essentially described by Ross and Claiborne (48). In the case of the amplified bkdD gene, a second PCR with 10 μl of the initial reaction was necessary to obtain sufficient amounts for cloning and sequencing purposes.
The λgt11 and λFIXII libraries were constructed as previously described (48). The λgt11 library was screened by using the bkdD gene PCR product as a probe. The bkdD product was purified from 0.8% low-melting agarose gels by organic extraction as recommended by Maniatis et al. (33). The purified PCR product was then labelled with either 32P, by using a random primer kit obtained from Promega, or digoxigenin, by using the Boehringer Mannheim (Indianapolis, Ind.) Genius system. Hybridizations were performed on DNA transferred to a Boehringer Mannheim nylon membrane. Standard high-stringency hybridization conditions were generally employed according to specified conditions as recommended by Maniatis et al. (33). Alternatively, for medium- to low-stringency conditions, the hybridizations were incubated overnight at 37°C and washed for 15 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) three times at room temperature and twice at 37°C. The λFIXII library was screened by direct PCR screening (18). Synthetic oligonucleotides designed to the bkdD and bkdA genes were used in the PCR screening.
Cloning, sequencing, and identification of the ptb, buk, and bkdDABC genes.
We chose initially to amplify enterococcal DNA encoding lipoamide dehydrogenase (LipDH) by using primers designed to two highly conserved regions of the protein. The first of these, which includes the redox-active disulfide, was used to design a 40-mer, 5′-AAGGATCCGGIGGIACITG(T/C)(C/T)TIAA(C/T)GTIGGITG(C/T)ATICC. Since these primers were intended for use with DNA isolated from various sources, we decided to use inosine in many of the “wobble” positions in an effort to make them universal. Restriction enzyme sites were included to facilitate subsequent cloning of the PCR products. Another of these primers, LipDH3, was based on the C-terminal region containing the highly conserved histidine with the oligonucleotide designed in the antisense direction as follows: 5′-AAGAATTCGCTTCIIIIA(G/A)IGT(A/T)GG(A/G)TGIGCITG. The LipDH1 and LipDH3 primers were then used in a series of PCRs with genomic DNA from E. faecalis 10C1 and OG1X, a group A strain D420, a group C strain 090R, and Lactococcus lactis MG1363. All but the lactococcal strain yielded a product of 1.2 kb, which correlated well with the expected size, based on the sequence alignments (data not shown). Sequence analysis of the cloned PCR products exhibited 30 to 45% identity to LipDHs from various sources.
A λgt11 library of E. faecalis 10C1 was then screened by using the labelled 10C1 PCR product as a probe. Two of the positives that resulted were plaque purified, and their DNA was isolated. The entire 4.2-kb EcoRI insert from one (λbkdD3) was cloned into pBluescript to form pLD03 (Fig. 1). Initial sequencing and restriction analyses, however, immediately demonstrated that the clone was lacking the 3′ end of the gene. For this reason, DNA encoding the entire bkdD gene was cloned on a 2.0-kb PstI/BamHI fragment into pOXO4, with DNA isolated from the other λ clone (λbkdD9). The inserts in pLD03 and pLD09 overlap by 1.7 kb. Sequence analysis of the 3′ end of the pLD09 clone revealed a closely linked open reading frame (ORF) (bkdA) that was shown by TFASTA and BLAST analysis to have 51% identity to the E1α component of the BKDH complex from B. subtilis.
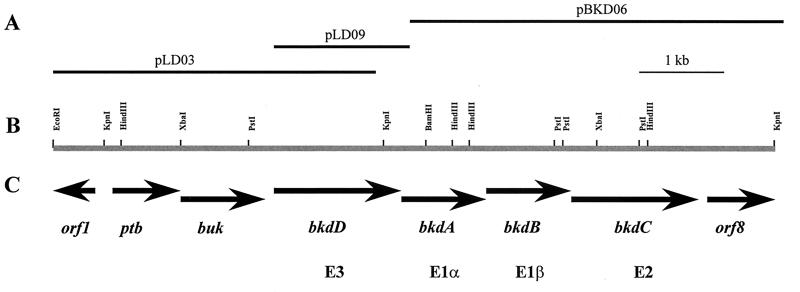
FIG. 1.
Molecular organization of the E. faecalis bkd gene cluster. (A) Clones used for sequencing and subsequent subcloning. (B) Restriction map of the region analyzed. (C) Structural genes of the bkd gene cluster.
For further cloning experiments, we chose to use a λFIXII library of E. faecalis 10C1, given the increase in insert size over λgt11. Based on the sequence obtained from the pLD09 clone, primers were designed corresponding to bkdD and bkdA. The first of these, LipDH10, was based on nucleotides 4000 to 4019, while the second (LipDH19) was based on nucleotides 4421 to 4442 and in the antisense direction. The LipDH10 and LipDH19 primers produced the expected 440-bp PCR product when either E. faecalis 10C1 genomic DNA or the λFIXII library was used as a template. The λFIXII library was then screened by using the direct PCR screening protocol (18). One of the positives that resulted (λbkd3) was plaque purified, and the DNA was isolated. PCR screening and restriction analyses of the 20-kb insert demonstrated that the desired downstream sequence was located on a 5-kb BamHI/KpnI fragment. This fragment was cloned into pWKS30 to form pBKD06 (Fig. 1). To be certain that only a single BamHI site existed in the region between the bkdD and bkdA genes, we PCR amplified the region from the genome of E. faecalis 10C1 and sequenced directly. Sequence analysis confirmed the presence of only a single BamHI site. The nucleotide sequence was determined on both strands from the inserts of pLD03, pLD09, and pBKD06 (Fig. 2).
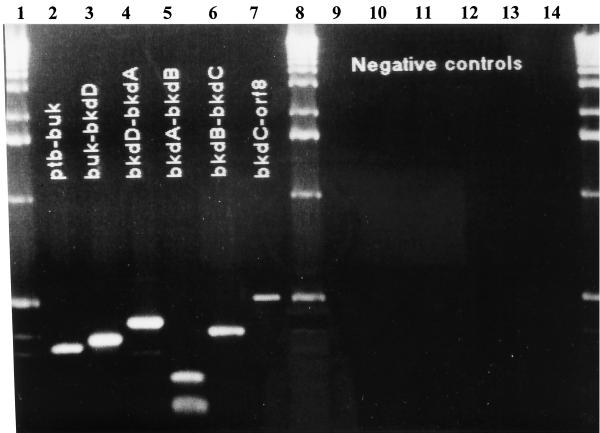
FIG. 2.
RT-PCR analysis of the bkd gene cluster. Molecular weight markers are from Bio-Rad. The following oligonucleotide pairs were used: KD24 and KD23 (lanes 2 and 9), KD26 and KD27 (lanes 3 and 10), LipDH19 and LipDH10 (lanes 4 and 11), KD31 and KD32 (lanes 5 and 12), KD33 and KD34 (lanes 6 and 13), and KD03 and KD14 (lanes 7 and 14). For the negative controls, Moloney murine leukemia virus reverse transcriptase was omitted from the reaction (lanes 9 to 14).
DNA sequencing and analysis.
DNA sequencing was carried out by the dideoxy-chain termination method of Sanger et al. (51) by using the Sequenase version 2.0 kit (U.S. Biochemical Corp., Cleveland, Ohio). Sequencing templates were either M13 (68) or double-stranded denatured plasmid DNA. When possible, sequencing information was first obtained from DNA subcloned into pUC119 and/or pBluescript with appropriate restriction endonucleases. In addition, the rapid accumulation of sequence data was facilitated by using the TN1000 system (Gold Biotechnology). Finally, gaps in the sequence on a particular strand were filled in by using synthetic oligonucleotides. Compressions were resolved by using dITP (U.S. Biochemical Corp.) in the sequencing reactions. Nucleic acid and amino acid sequence data were analyzed by using the Wisconsin Genetics Computer Group (GCG) software package (11) and the National Center for Biotechnology Information Blast network service (2).
Heterologous expression of ptb, buk, and bkdD in E. coli.
Expression of the ptb, buk, and bkdD genes was performed by modification of the method described for T7 expression (56) of the npr gene (47). This was achieved by first cloning the genes into the T7 vector pOXO4 or pOXO7. The plasmid pOXO7 is a derivative of pOXO4 in that the pUC19 origin of replication is replaced with the pACYC184 origin of replication. For ptb, this was accomplished by cloning the 2.3-kb KpnI/PstI fragment from pLD03, containing the intact ptb gene, into pOXO7 that was similarly cut and designated pPTB01. The buk gene was also cloned into pOXO7 by cutting pLD03 with XbaI/HindIII. The 1.2-kb fragment containing the intact buk gene was then cloned into similarly cut pOXO7 and designated pBUK01. The bkdD gene was cloned as a 2.4-kb PstI/BamHI insert from λbkdD9 into pOXO4. In all cases, the intact genes were cloned such that they were under control of the T7 promoter. The resultant plasmids were introduced into E. coli JM109(DE3), and the resultant transformed cells were then used to inoculate TYP medium supplemented with 30 mM glucose and chloramphenicol. The cultures were aerated vigorously until the optical density at 600 nm (OD600) reached approximately 1.0, at which point isopropyl-β-d-thiogalactopyranoside (IPTG) was added to 0.4 mM (final concentration) to induce expression of the T7 RNA polymerase. Overexpression of the genes was then monitored in lysates of these induced cells by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Induced proteins could later be visualized in the lysates after Coomassie blue staining of the gel. Low-molecular-mass standard proteins for molecular mass determination were obtained from Bio-Rad (Richmond, Calif.).
Enzyme assays.
For the purpose of enzyme assays, 50 ml of induced cells was harvested at the appropriate time point and washed once in an equal volume of 50 mM phosphate buffer (pH = 8.0). Following resuspension in 1 ml of the phosphate buffer, cell extracts were prepared by using the mini-Bead-Beater. The phosphotransferase was assayed by measuring the formation of CoASH upon its reaction with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) at 412 nm by using the conditions described by Wiesenborn et al. (65). Lipoamide dehydrogenase activity was assayed by monitoring the oxidation of NADH at 340 nm with lipoamide as a substrate. These assays were performed at 25°C in a total volume of 3 ml containing 0.16 mM NADH and 0.4 mM lipoamide in 50 mM phosphate buffer (pH = 8.0). Following the addition of enzyme, the decrease in absorbance was measured in a Gilford model 260 recording spectrophotometer. One unit of activity is defined as the amount of protein which catalyzes the conversion of 1 μmol of NADH to NAD+ per min at 25°C.
Purification of the recombinant LipDH.
Recombinant LipDH was purified to homogeneity as follows: 8 liters of E. coli JM109(DE3), harboring pLD09, was harvested after induction as described above, and the pellet was washed before being stored at −20°C. Cells were subsequently thawed, resuspended in 80 ml of 50 mM potassium phosphate (pH 7.0)–0.6 mM EDTA, and broken with a bead beater (BioSpec Products); the crude extract was centrifuged at 14,000 × g for 20 min. All procedures were carried out at 4°C, and all the buffers used in the purification contained 0.6 mM EDTA, unless otherwise indicated. Streptomycin sulfate (Sigma) was then added to a final concentration of 2.5%, and the suspension was stirred for 30 min before being centrifuged for 30 min at 14,000 × g. The supernatant was brought to 40% saturation with solid ammonium sulfate and stirred for 30 min. Precipitated proteins were removed by centrifugation, and the supernatant was applied to a 20-ml phenyl Sepharose 6 fast flow (Pharmacia Biotech) column equilibrated with 40% saturated ammonium sulfate in 50 mM potassium phosphate (pH 7.0). Column chromatography was carried out by using a Pharmacia Biotech fast-protein liquid chromatography system. The column was washed with the equilibration buffer until the A280 of the eluate was less than 0.1. A 250-ml gradient from 40 to 0% saturated ammonium sulfate was used to elute the LipDH. Peak fractions (4 ml each) with A450 greater than 0.04 were pooled and dialyzed three times against 4 liters of 50 mM potassium phosphate (pH 7.0). The dialyzed protein was loaded onto a 50-ml Q Sepharose fast flow column equilibrated with the same buffer as that used for dialysis. After an extensive washing, two yellow protein peaks were eluted from the column by applying a 250-ml gradient from 0 to 1 M NaCl in 50 mM potassium phosphate (pH 7.0). Fractions containing LipDH activity and having an A280/A450 ratio of <18 were pooled and dialyzed three times against 4 liters of 10 mM potassium phosphate (pH 7.0) without EDTA. The dialyzed protein was applied to a 20-ml Bio-Gel HTP (Bio-Rad) column equilibrated in the 10 mM phosphate buffer. After being washed, the LipDH was eluted with a gradient from 0.01 to 0.2 M potassium phosphate; fractions with an A280/A450 ratio of <10.5 were pooled and concentrated to a volume of 4 to 5 ml by ultrafiltration. The sample was then loaded onto a column of Sephacryl S-200 equilibrated in 50 mM potassium phosphate (pH 7.0). Fractions with an A280/A450 ratio of <5.6 were pooled; the purified recombinant LipDH has a specific activity of 270 U/mg by the standard assay and is 95% homogeneous as analyzed by SDS-PAGE (data not shown).
RNA isolation and RT-PCR.
RNA was isolated from E. faecalis as described by Platteeuw et al. (46). To show that the bkd gene cluster is transcribed as a single transcriptional unit, reverse transcriptase PCR (RT-PCR) was used. In brief, 1 μg of total RNA was treated with DNase I (Worthington Enzymes, Freehold, N.J.) by using the supplied buffer at 37°C for 30 min. The RNA was then ethanol precipitated, and the pellet was resuspended in a hybridization/extension buffer (125 mM Tris-HCl [pH = 8.3], 185 mM KCl, and 7.5 mM MgCl2) in a reaction volume of 10 μl containing 1 pmol of the appropriate oligonucleotide. Oligonucleotides were designed so that, after extension and subsequent amplification, the PCR product would cross the intergenic region between the two genes of interest. The following oligonucleotides were used in the primer extension reactions: for ptb-buk, KD24 (5′-GATGTTATGTGTTTCTAAAA-3′); buk-bkdD, KD26 (5′-GCGGATCCGCCTTTGTGCAAACAAGTGCC-3′); bkdD-bkdA, LipDH19 (5′-CAAAAGAACCCATTAAAATATC-3′); bkdA-bkdB, KD31 (5′-CCTTTGTCCCCGCCGACATCT-3′); bkdB-bkdC, KD33 (5′-ATCTCCTGGTTTAACTAACCA-3′); and bkdC-orf8, KD03 (5′-GCCTTTCACTTCTGAATTAAA-3′).
The hybridization was carried out by heating the samples at 70°C for 10 min before allowing them to cool to room temperature. The reaction was started by the addition of deoxynucleoside triphosphates (dNTPs) (final concentration, 100 μM each), dithiothreitol (DTT) (final concentration, 10 mM), 3.3 U of RNasin (Promega), and 300 U of Superscript II reverse transcriptase (Gibco BRL, Grand Island, N.Y.) in a final volume of 20 μl. The reaction was incubated at 42°C for 30 min. Following the extension reactions, the samples were boiled for 5 min, 20 U of RNase cocktail was added, and incubations were continued at 42°C for 20 min. The 20-μl extension reaction mixtures were added directly to the PCRs (final volume, 100 μl) with the following modifications: only 8 μl of the 10× Taq buffer was added instead of the usual 10 μl, and only 125 μM dNTPs were added. The oligonucleotide from the extension reaction served as one primer, and a second was then added for the PCR as follows: for ptb-buk, KD23 (5′-CTATTGATGTTGGAAATTGTT-3′); for buk-bkdD, KD27 (5′-GCGAATTCTATTGGCGAGATGGCCGTAG-3′); for bkdD-bkdA, LipDH10 (5′-GTCACGGATTTAATTGCCG-3′); for bkdA-bkdB, KD32 (5′-TAGAAGAGGGCTATTTAACAG-3′); for bkdB-bkdC, KD34 (5′-AATTGATGCCGAAATCGTCGA-3′); and for bkdC-orf8, KD14 (5′-AAGTTCTTGCGAGATGTTAAA-3′). The PCR was then carried out as previously described, and the products were separated on a 1.5% agarose gel.
Generation of chromosomal bkd mutations.
To generate transposon mutants for the construction of subsequent chromosomal mutations of the enterococcal bkd gene cluster, the transposon mγδ was chosen to mutagenize pBKD09 and pBKD14. The mγδ system has been previously used successfully to generate auxotrophic mutants in E. faecalis (28). Transposon mutagenesis of pBKD09 and pBKD14 was carried out as described by Li et al. (28). The positions of the mγδ insertions were determined by PCR by using an oligonucleotide specific to the transposon, GDIR (5′-TTTCGTTCCATTGGCCCTCAAACCCC-3′), which is complementary to either end of the transposon, and an oligonucleotide specific to the insert of pBKD09 or pBKD14. Constructs with mγδ insertions within the bkdA or bkdC gene loci were transformed into OG1RF by using the following protocol. For the preparation of electrocompetent cells, OG1RF was grown overnight in GM17 (M17 plus 50 mM glucose). The overnight culture was diluted 100-fold into fresh GM17 and grown at 37°C to an OD600 of 0.5. Cells were then harvested by centrifugation at 5,000 × g for 10 min and washed twice with 1/10 of the original volume of ice-cold 0.5 M sucrose. The washed cells were resuspended in 1/100 of the original volume of 0.5 M sucrose. The cells could then be used immediately or stored at −70°C for later use. For electroporation 2 to 5 μg of DNA was ethanol precipitated, washed with 70% ethanol, and dried. The DNA was resuspended in 50 μl of competent OG1RF and transferred immediately to a chilled 0.1-cm cuvette and electroporated with a Bio-Rad gene pulser apparatus at a capacitance of 25 μF, a resistance of 200 Ω, and a peak voltage of 2.5 kV. Cells were then incubated in 1 ml of BHI broth for 90 to 120 min at 37°C and plated on BHI agar plates containing kanamycin (1,000 μg/ml). After electroporation, colonies growing on selective plates after 24 h were streaked onto fresh BHI agar plates containing kanamycin. To determine the physical structure expected from homologous recombination, kanamycin-resistant colonies were analyzed by PCR by using the GeneAmp XL PCR kit (Perkin-Elmer, Branchburg, N.J.).
Determination of product formation in E. faecalis 10C1.
E. faecalis 10C1 was grown overnight in Evans medium (14) supplemented with 0.5% yeast extract and phosphate buffered (50 mM) in the presence of the appropriate carbon source. Cells were then diluted 100-fold with identical media and grown at 37°C. At appropriate time points, 1-ml samples were taken and the cells were removed by centrifugation, at which point 100 μl of 35% perchloric acid was added to the supernatant. This was incubated on ice for 10 min, followed by the addition of 50 μl of 7 N KOH. The resulting precipitate was removed by centrifugation, and the supernatant was analyzed for product formation by using the high-performance liquid chromatography conditions described by Snoep et al. (53).
Nucleotide sequence accession number.
The DNA sequence of the E. faecalis bkd gene cluster is available from GenBank under accession no. AF149712.
RESULTS
We set out to clone the gene encoding lipoamide dehydrogenase (LipDH) from the genome of E. faecalis, using primers to conserved sequence regions. This approach was successful in locating a gene encoding LipDH which was sequenced along with a number of ORFs to which it is linked (Fig. 1). We started a study on the characterization of this gene cluster and report here on the gene products, the transcriptional analysis, and on the metabolic route that the enzymes form.
Sequence analysis of 8,401 bp revealed eight ORFs, six of which are complete (accession no. AF149712). The six complete ORFs encoded by ptb-buk-bkdDABC are very tightly linked, with the largest intergenic region, being 23 bp, found between bkdB and bkdC and the smallest intergenic spacing, of 5 bp, found between buk and bkdD. The start codons (ATG) are all preceded by typical Shine-Dalgarno sequences which are complementary to the sequences near the 3′ end of the enterococcal 16S rRNA (31). Sequences comprising putative promoter −35 (TTGTAT) and −10 sites (TAGATT) were identified 33 bp upstream of the start codon of ptb. In addition to these sites, a sequence (TGTATGCGCTTACA, nucleotides 752 to 765) that is closely related to the cis-acting catabolite responsive element (CRE) of B. subtilis was identified (50). This sequence is identical to the 14-bp CRE consensus (TGWNANCGNTNWCA), with the exception of one base, and overlaps the putative −35 promoter region. As expected, the entire sequence exhibited a low G+C content of 39%, which is typical for E. faecalis (37). ORF1 was found 280 bp upstream of the second ORF, ptb, and is transcribed divergently. ORF8 was identified 123 bp downstream of bkdC and is transcribed divergently. ORF8 was identified 123 bp downstream of bkdC and is transcribed in the same orientation. Neither ORF1 nor ORF8 was homologous to any sequence in the database.
The ptb and buk gene products are homologous to clostridial phosphotransbutyrylase and butyrate kinase.
A homology search of the translated nucleotide and protein sequence databases by using the deduced amino acid sequence of ptb and buk revealed a moderate level of identity to phosphotransbutyrylase and butyrate kinase from Clostridium acetobutylicum and a lower level of identity to various phosphotransacetylases and acetate kinases as shown in Table 2. The predicted molecular mass of 29.5 and 39.3 kDa for the ptb and buk gene products, respectively, is also in good agreement with the value of 31 and 39 kDa determined for the purified clostridial phosphotransbutyrylase and butyrate kinase (20, 65). The consensus ATP-binding site reported by Flaherty et al. (15) [(I/L/V)X(I/L/V/C)DXG(T/S/G)(T/S/G)XX(R/K/C)] was also identified in buk. Interestingly, there is a substitution of the Asp in this sequence by an Asn which is found in acetate and butyrate kinases also.
TABLE 2.
Amino acid identity between proteins encoded by the E. faecalis bkd gene cluster and the corresponding proteins from various sources
| E. faecalis ORF (predicted protein), no. of codons, and predicted mass (kDa) | Homologous protein | Source | % Identity | Reference |
|---|---|---|---|---|
| ptb, 274, 29.5 | YqiS | Bacillus subtilis | 37.6 | 27 |
| Ptb | Clostridium acetobutylicum | 34.0 | 39, 60 | |
| Pta | Clostridium acetobutylicum | 23.8 | 4 | |
| buk, 361, 39.3 | YqiU | Bacillus subtilis | 47.3 | 27 |
| Buk | Clostridium acetobutylicum | 45.8 | 39, 60 | |
| Ack | Clostridium acetobutylicum | 16.6 | 4 | |
| Ack | Bacillus subtilis | 22.4 | 19 | |
| bkdD (E3), 470, 50.4 | E3 BKDH | Bacillus subtilis | 51.6 | 27 |
| E3 PDH | Enterococcus faecalis | 34.0 | 9 | |
| E3 PDH | Bacillus subtilis | 37.3 | 24 | |
| Lpd-Val | Pseudomonas putida | 31.8 | 6 | |
| Lpd-Glc | Pseudomonas putida | 38.2 | 40 | |
| bkdA (E1α), 331, 36.7 | E1α BKDH | Bacillus subtilis | 52.0 | 61 |
| E1α BKDH | Streptomyces avermitilis | 34.3 | 52 | |
| E1α BKDH | Streptomyces avermitilis | 31.7 | 10 | |
| E1α BKDH | Pseudomonas putida | 29.4 | 5 | |
| E1α BKDH | Myxococcus xanthus | 28.5 | 59 | |
| bkdB (E1β), 329, 35.7 | E1β BKDH | Bacillus subtilis | 60.4 | 61 |
| E1β BKDH | Pseudomonas putida | 44.8 | 5 | |
| E1β BKDH | Streptomyces avermitilis | 43.9 | 52 | |
| E1β BKDH | Myxococcus xanthus | 40.9 | 59 | |
| bkdC (E2), 433, 47.3 | E2 BKDH | Bacillus subtilis | 51.0 | 61 |
| E2 PDH | Enterococcus faecalis | 37.2 | 1 | |
| E2 BKDH | Pseudomonas putida | 28.2 | 57 |
The bkdDABC genes have homology to branched-chain α-keto acid dehydrogenase complexes.
The ptb and buk genes are followed by four closely linked genes which, together, most likely constitute an α-keto acid dehydrogenase complex. Homology searches of the databases with the bkdA, bkdB, and bkdC products revealed 51, 60, and 51% identity to the E1α, E1β, and E2 subunits, respectively, of the B. subtilis branched-chain α-keto acid dehydrogenase complex (61). Strong similarities were also found to the E1α, E1β, and E2 subunits of the other BKDH complexes from various sources as shown in Table 2. For this reason, these ORFs were designated bkdA, bkdB, and bkdC and most likely represent the E1α, E1β, and E2 subunits of the branched-chain α-keto acid dehydrogenase complex.
Analysis of the amino acid sequence of the E. faecalis E1α (BkdA) revealed the structural motifs common to all E1α subunits, including the TPP-binding motif (21), a putative subunit interaction site (64), and the phosphorylation sites (I and II) of the E1α components of the mammalian BKDH and PDH complexes (64). Alignment of the bkdB gene product with E1β’s from various sources revealed four regions of extensive similarity common to this subunit (64). Attempts to express the E1α or E1β, either separately or in conjunction, resulted in proteins found in the insoluble fraction, most likely as inclusion bodies.
Analysis of the E2b primary structure revealed the three-domain architecture common to all known E2 components, the lipoyl domain, the E1/E3 binding domain, and the catalytic domain (44). All three domains are separated by regions rich in alanine, proline, and charged residues typical of the linker regions (43). A single lipoyl domain was identified consistent with the BKDH complexes of P. putida (5), B. subtilis (61) and bovine (17) sources. The appearance of a single lipoyl domain in the enterococcal E2b does, however, clearly distinguish it from the enterococcal E2p, of the PDH complex, which contains two lipoyl domains (1). In the catalytic domain, both residues involved in the active site, His404 and Asp408, are conserved (25, 34). Of particular interest in the E2 polypeptide is the E1/E3 binding domain. It has been shown in P. putida that BkdD-Val is unable to complement the loss of BkdD-Glc, and this may be due to the inability of the BkdD-Val to bind to the E2p component of the PDH complex (40). Since E. faecalis also has distinct LipDH’s for both the PDH and BKDH complexes, it is likely that the E3p and E3b components are specific for their respective complexes, as in P. putida. Furthermore, while conserved in the E. faecalis PDH E2 and E3 components, the amino acid residues known to be involved in the E2/E3 binding of the Bacillus stearothermophilus PDH complex (32) are not in the E2 and E3 components of the BKDH complex.
The bkdD gene encodes the E3 subunit or lipoamide dehydrogenase. All four domains, the redox-active cysteines, and the active site His-Glu dyad were easily identified in the E. faecalis E3b. The presence of a gene encoding LipDH in the bkd gene cluster indicates that E. faecalis has at least two genes encoding LipDH. The other is found within the cluster of genes encoding the PDH complex. Multiple LipDH’s have previously been observed in P. putida (41) and B. subtilis (8), both of which have three, and Alcaligenes eutrophus, which contains two (22, 23). Similar to both P. putida and A. eutrophus, the amino acid identity between the E. faecalis PDH E3 (E3p) and BKDH E3 (E3b) is only moderate, with only 37% of the residues being identical.
Transcriptional analysis of the bkd gene cluster.
Initial attempts to detect bkd mRNA by Northern analysis by using probes to ptb, buk, bkdD, and bkdA proved unsuccessful. However, we were able to easily detect both the 1.5- and 6-kb transcripts of the E. faecalis NADH oxidase and pyruvate dehydrogenase operon under all the growth conditions tested (data not shown). Based on this result, it is feasible that the bkd transcript is either unstable or expressed at very low levels. Consequently, RT-PCR was used to determine whether all six complete ORFs (ptb-buk-bkdDABC) are expressed. Total RNA was isolated from E. faecalis grown in M17 containing 20 mM (each) branched-chain amino acid. RT-PCR gave rise to a PCR product of the correct size from each reaction as shown in Fig. 2. Interestingly, the predicted 500-bp product from the bkdC-orf8 reaction was also present, indicating that orf8 is also transcribed as part of the bkd gene cluster. Based on the RT-PCR results, all of the genes are, in fact, transcribed. However, it is not possible to confirm the presence of a single bkd transcript of at least 7.6 kb including all of the genes.
Overexpression of the ptb and buk gene products in E. coli.
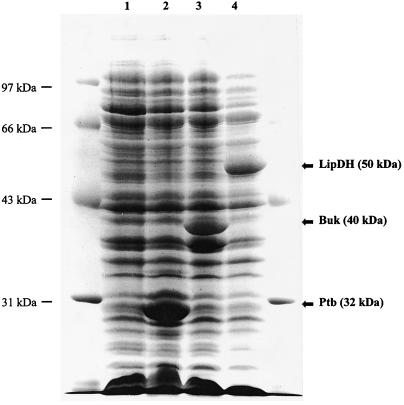
In an effort to further characterize the bkd gene cluster, the products of ptb, buk, and bkdD were overexpressed and characterized in E. coli. The ptb and buk genes were cloned into the T7 expression plasmid pOXO7 and designated pPTB01 and pBUK01, respectively (as described in Materials and Methods), and were overexpressed in E. coli. The ptb and buk gene products have molecular masses of 32 and 40 kDa, respectively, based on SDS-PAGE, in good agreement with that predicted from the translated nucleotide sequence as shown in Fig. 3. To determine the substrate specificity of the ptb gene product, crude extracts of E. coli expressing Ptb were assayed by using various acyl-CoA’s as substrates. Since E. coli does contain a phosphotransacetylase (Pta), E. coli JM109(DE3), harboring pOXO7, was assayed in parallel. The only background activity was observed with the substrate acetyl-CoA (5 U/mg), which was subtracted from the activity found with the extract harboring the ptb gene product (11 U/mg). The ptb gene product exhibited a broad substrate specificity in the acyl-phosphate-forming direction as shown in Table 3. It could utilize C2 to C8 straight-chain acyl-CoA’s, with valeryl-CoA having the highest activity at 108 U/mg. Branched-chain acyl-CoA’s such as isobutyryl-CoA and isovaleryl-CoA, which are the acyl-CoA derivatives of valine and leucine, also exhibited high activity. In addition, the buk gene product was shown to have kinase activity in the ATP-forming direction by using acetyl-phosphate and butyryl-phosphate as substrates, with specific activities of 4 and 8 U/mg, respectively. The lack of other commercially available acyl-phosphates limited further study in the ATP-forming direction.
FIG. 3.
SDS-PAGE analysis of the overexpression of the ptb, buk, and bkdD gene products from extracts of E. coli JM109(DE3). Lane 1, extracts from the JM109(DE3) host containing the T7 expression vector pOXO7; lane 2, extracts from the JM109(DE3) host containing pPTB01; lane 3, extracts from the JM109(DE3) host containing pBUK01; lane 4, extracts from the JM109(DE3) host containing pLD09.
TABLE 3.
Substrate specificity of the ptb gene product with respect to various acyl-CoA compoundsa
| Substrate | Relative rate (%)b |
|---|---|
| Acetyl-CoA | 5 |
| Propionyl-CoA | 32 |
| Butyryl-CoA | 58 |
| Isobutyryl-CoA | 78 |
| Valeryl-CoA | 100 |
| Isovaleryl-CoA | 80 |
| Hexanoyl-CoA | 15 |
| Octanoyl-CoA | 1 |
| Succinyl-CoA | 1 |
The phosphotransferase was assayed by measuring the formation of CoASH upon its reaction with DTNB at 412 nm (65).
One hundred percent activity is 108 U/mg.
Expression and purification of recombinant LipDH (BkdD).
Crude extracts of E. coli JM109(DE3), harboring pLD09, were assayed for lipoamide dehydrogenase activity following induction with IPTG. To take into account the background E. coli lipoamide dehydrogenase activity, E. coli JM109(DE3), harboring pOXO4, was also assayed. After this background activity (approximately 0.1 to 0.2 U/mg) was corrected for, a lipoamide dehydrogenase-specific activity of 3 to 4 U/mg was determined, attributable to bkdD expression. Unfortunately, almost 90% of the expressed protein was found in an inactive and insoluble form, probably inclusion bodies. Attempts to resolubilize the protein in an active form proved unsuccessful. Despite this, approximately 5 mg of pure enzyme was obtained from 8 liters of E. coli. The purified E. faecalis LipDH had a specific activity of 215 to 225 U/mg. The N-terminal amino acid sequence determined for the recombinant LipDH was AEQTDLLILGGGTGGYVAAIRAAQKA, which is identical to that predicted for the bkdD gene product, with the exception that the N-terminal methionine was absent, suggesting that it is modified in E. coli.
Redox properties of BkdD.
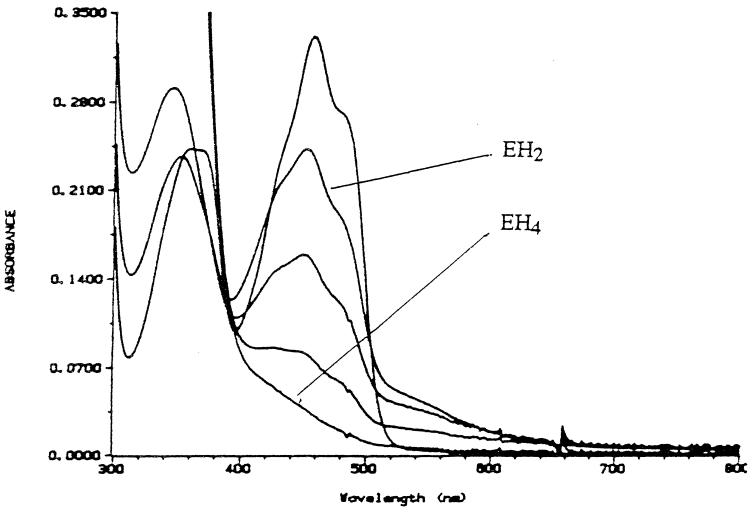
The recombinant LipDH exhibits the visible and UV absorbance characteristics of a typical flavoprotein, and the oxidized form closely resembles the oxidized spectra of both the E. coli and pig LipDH, as shown in Fig. 4. Anaerobic titration with NADH displayed the expected spectral changes. Upon the addition of 1 equivalent of NADH, the electrons are transferred via the flavin to the active site disulfide, resulting in the two-electron reduced EH2 species. This can be observed by the decrease in A450 and an increase in A540, due to the formation of a charge-transfer complex between the thiolate anion and the flavin adenine dinucleotide (FAD). Subsequent additions of 2.0, 5.0, and 6.5 equivalents of NADH resulted in a decrease in both A450 and A540. After the addition of 6.5 equivalents of NADH, the enzyme was fully reduced (EH4), i.e., both the disulfide and the FAD were reduced. Of interest was the observation that, unlike the E3p component from E. faecalis, the E3b component could be fully reduced to the EH4 redox state with NADH (although this required the addition of 6.5 equivalents of NADH per FAD). While the E3b enzyme is not as resistant to overreduction by NADH as the E. faecalis E3p, it is significantly more resistant than the E. coli enzyme, which is fully reduced by the addition of only 2 equivalents of NADH per FAD (66). Furthermore, it has been shown that the interaction of the Azotobacter vinelandii E3 component with the E2 core results in a stabilization of the EH2 redox state (63). It is conceivable that the association of the E3b with the E2 core would result in an even higher level of insensitivity to inhibition by NADH, thereby allowing the complex to be active in vivo anaerobically as well as aerobically.
FIG. 4.
Anaerobic titration of recombinant lipoamide dehydrogenase with NADH. The enzyme (29 mM in 50 mM phosphate [pH = 7.0], 0.6 mM EDTA) was made anaerobic in the presence of an oxygen-scrubbing system consisting of 0.02 U of protocatechuate dioxygenase and 40 mmol of protocatechuic acid. Aliquots of an anaerobic 2.5 mM NADH solution were added from the titrating syringe. Spectra shown correspond to oxidized enzyme and to enzyme after the addition of 1 (EH2), 2, 5, and 6.5 (EH4) equivalents of NADH/FAD.
Growth stimulation of E. faecalis OG1RF by the branched-chain α-keto acids.
In an effort to better understand the role of the bkd gene cluster in the metabolism of E. faecalis, gene disruptions were made within the cluster on the chromosome of E. faecalis OG1RF. This initially involved insertion of mγδ-200 into inserts carried on pBKD09 and pBKD14. The exact locations of the insertions were first mapped by PCR. Constructs that were shown to have insertions within the cloned fragment based on the PCR analysis were then sequenced by using the transposon-specific oligonucleotide mγδ-R. Using this protocol, we were able to introduce transposon insertions within the bkdA and bkdC genes. Plasmid DNA was then introduced into E. faecalis OG1RF, and transformants were selected for kanamycin resistance. Gene disruption occurred by homologous recombination between the OG1RF chromosome and the cloned DNA flanking the mγδ insertion. PCR mapping with oligonucleotides specific to the transposon and sequences specific to the OG1RF chromosome confirmed that the gene disruption had occurred by a single crossover event into the bkdA and bkdC genes.
The consequences of disrupting the gene cluster were then investigated by comparing the growth of the mutants to the wild type in the presence of α-ketoisocaproic acid, the α-keto acid derivative of leucine. E. faecalis OG1RF was grown aerobically in batch cultures (pH 7.0) with pyruvate (20 mM) as the energy source on minimal medium, phosphate buffered, supplemented with 0.5% yeast extract in the absence or presence of α-ketoisocaproate (KIC) (20 mM). In the absence of KIC, a final A600 of 0.91 was reached after the cells were depleted of pyruvate, while in the presence of KIC a final density of 1.35 was reached (Table 4). The KIC was converted stoichiometrically to isovalerate as expected (Table 4). In the cultures with the strains mutated in the bkdA or bkdC genes, no increase in final OD was observed upon the addition of KIC to the medium, and only traces of isovalerate were detected. A similar result was observed with the branched-chain α-keto acids α-ketoisovalerate (KIV) and α-keto-β-methylvalerate (KMV). The presence of both resulted in a higher final OD, and they were converted to their corresponding branched-chain carboxylic acids, isobutyrate and methylbutyrate, with rates comparable to that observed with KIC (data not shown). Similar to KIC, the mutants were unable to utilize either KIV or KMV. These results demonstrate that the bkd gene cluster is required for the catabolism of the branched-chain α-keto acids. Interestingly, there was no benefit to growth from the presence of the branched-chain amino acid leucine, valine, or isoleucine in the media, and there was no production of isovalerate, isobutyrate, or methylbutyrate (data not shown). These results suggest that E. faecalis is able to utilize only the α-keto acid forms as an energy source and not the amino acids.
TABLE 4.
Biomass formation and conversion of α-ketoisocaproate to isovalerate in E. faecalis OG1RF and in the bkdA mutant strain
| Strain | α-Ketoisocaproate addition | OD600a | Concn (mM) ofb:
|
|
|---|---|---|---|---|
| α-Ketoisocaproate | Isovalerate | |||
| OG1RF | − | 0.90 | 0 | 0 |
| + | 1.34 | 17.7 | 16.6 | |
| OG1RF::bkdA | − | 0.94 | 0 | 0 |
| + | 0.88 | 17.5 | 0.5 | |
OD600 is given for stationary-phase cultures after 18 h of growth.
Extracellular concentrations of α-ketoisocaproate and isovalerate are given before inoculation and after stationary phase was reached, respectively.
DISCUSSION
In this article we describe the elucidation of a previously undescribed pathway in E. faecalis for the utilization of the branched-chain α-keto acids KIC, KIV, and KMV as energy sources. These compounds are reductively decarboxylated by the BKDH complex generating the corresponding acyl-CoA’s, which are then converted to the free acids isovalerate, isobutyrate, and methylbutyrate through the combined actions of the ptb and buk gene products. The presence of these α-keto acids in the medium during growth of E. faecalis resulted in a higher final OD, which is most likely due to the generation of ATP via substrate-level phosphorylation by the combined actions of the ptb and buk gene products.
The observation that the substrates of the ptb gene product included a broad range of acyl-CoA’s offers important clues about its biological function. An increase in activity of the ptb gene product was observed with increasing chain length, the highest activity being observed with the five-carbon straight-chain substrate valeryl-CoA and the branched-chain substrates isobutyryl-CoA and isovaleryl-CoA. An increase in chain length to six or eight carbons resulted in a dramatic loss of activity. A similar situation was observed with the C. acetobutylicum phosphotransbutyrylase in that it also exhibited a relatively broad substrate specificity, and an increase in chain length resulted in higher activity until the chain length exceeded that of the natural substrate butyryl-CoA (65).
Based on these results and on the tight linkage of the ptb and buk genes to the structural genes encoding the branched-chain α-keto acid dehydrogenase complex, we propose that the natural substrates of the ptb and buk gene products are the branched-chain acyl-CoA’s and branched-chain acyl-phosphates. We will refer to the E. faecalis ptb and buk gene products as a branched-chain phosphotransacylase (Bct) and a branched-chain acyl kinase (Bck). The enterococcal ptb and buk genes are distinct from all known Pta-Ack and Ptb-Buk systems in that they are located on the chromosome adjacent to and are coexpressed with other catabolic genes which function in the same metabolic pathway. To our knowledge, this work represents the first cloning and identification of an acyltransferase and kinase which utilize branched-chain substrates.
During database searches, a gene cluster similar to the E. faecalis bkd gene cluster was identified in B. subtilis, which contained the previously published BKDH complex (61). In addition, it has recently been described in further detail by Debarbouille et al. (8) and they used the nomenclature based on the E. faecalis system described here. The bkd gene clusters in E. faecalis and B. subtilis have in common a number of unusual features. These include the tight linkage of the ptb and buk (and bcd in B. subtilis) genes to the genes encoding the BKDH complex. However, there are also a number of distinct differences between the E. faecalis and B. subtilis gene clusters, both from a genetic and a metabolic standpoint. These differences involve the inclusion of two additional genes in the B. subtilis gene cluster. The first of these, bcd, is found between the B. subtilis ptb and buk genes and is thought to encode leucine dehydrogenase (8). A gene disruption in the bcd gene resulted in the inability to utilize the branched-chain amino acids as the sole nitrogen source. The absence of the bcd loci in the E. faecalis bkd gene cluster may explain the organism’s inability to convert the branched-chain amino acids to the corresponding free acids. It has been shown that the branched-chain amino acids are required for normal growth of E. faecalis in a minimal defined medium (36). Thus, E. faecalis can uptake the individual amino acids, suggesting that the inability to utilize the branched-chain amino acids is due to a lack of an appropriate transaminase activity.
The second gene that is present in the B. subtilis system but absent in E. faecalis is bkdR. This gene has been shown to encode a positive transcriptional regulator of the B. subtilis bkd operon. In addition, it was found that expression of the operon was from a −12, −24 promoter and dependent on the sigma factor SigL, a member of the sigma 54 family (8). The presence of a putative CRE sequence found within the promoter region of the E. faecalis bkd gene cluster suggests a role of catabolite repression in the regulation. Sequence analysis of the B. subtilis bkd operon did not reveal the presence of a CRE-like sequence, and in fact the operon is expressed at high levels in the presence of glucose (8). The presence of the CRE within the E. faecalis promoter region is consistent with the locations of those CREs found upstream of the amyE gene and in the xyl, gnt, hut, and bgl operons in B. subtilis (50). In all of these cases, the CRE was found either within the promoter region or downstream of the transcriptional start; these systems are all negatively regulated by CcpA. This mode of regulation in B. subtilis and in many other gram-positive organisms involves the cis-acting CRE sequence, the catabolite control protein A (CcpA), and the HPr of the phosphotransferase system (50). Furthermore, the role of catabolite repression in the regulation of the bkd gene cluster would be consistent with its catabolic role in the utilization of the branched-chain α-keto acids as an alternate energy source. Regulatory studies are currently under way to address the role of the CRE and catabolite repression in the regulation of the bkd gene cluster.
Despite the fact that 21% of the membrane of the enterococci is composed of branched-chain fatty acids (26), there is no evidence linking the enterococcal bkd gene cluster to the biosynthesis of branched-chain fatty acids, as found in B. subtilis. This is also supported by the observation that the branched-chain α-keto acids KIC, KIV, and KMV are converted stoichiometrically to the corresponding free acids isovalerate, isobutyrate, and methylbutyrate. If the branched-chain acyl-CoA’s were being utilized for membrane biosynthesis, the ratio of the consumption of the α-keto acid to the production of the corresponding free acid would not be 1:1. Furthermore, if the bkd gene cluster was involved in membrane biosynthesis, the genes would be expected to be expressed constitutively (67). However, the presence of a CRE in the promoter region suggests a role for catabolite repression in the regulation of the complex. This type of regulation should be expected to occur only with systems involved in the catabolism of alternative carbon sources and not with genes that are constitutively expressed. Therefore, the E. faecalis bkd gene cluster is distinct from the B. subtilis system, both at the genetic and the metabolic levels, and these gene clusters appear to be transcriptionally regulated via very different mechanisms.
In conclusion, it is our belief that the α-keto acids KIC, KIV, and KMV serve as an alternative energy source to enterococci, thereby increasing their metabolic diversity and flexibility. Based on their inability to utilize the amino acids, these α-keto acids are not obtained by protein degradation but, rather, by degradation of membrane lipids or through the environment, as they apparently have a transport system for the branched-chain α-keto acids. We propose that the BKDH complex and Bct and Bck act in a concerted manner to convert the branched-chain α-keto acids KIC, KIV, and KMV to the corresponding free acids isovalerate, isobutyrate, and methylbutyrate and to synthesize ATP in a system analogous to the PDH complex and phosphotransacetylase and acetate kinase.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant GM-35394 and by National Science Foundation grant INT-9400123.
REFERENCES
- 1.Allen A G, Perham R N. Two lipoyl domains in the dihydrolipoamide acetyltransferase chain of the pyruvate dehydrogenase multienzyme complex of Streptococcus faecalis. FEBS Lett. 1991;287:206–210. doi: 10.1016/0014-5793(91)80052-5. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Borges A, Hawkins C F, Packman L C, Perham R N. Cloning and sequence analysis of the genes encoding the dihydrolipoamide acetyltransferase and dihydrolipoamide dehydrogenase components of the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus. Eur J Biochem. 1990;194:95–102. doi: 10.1111/j.1432-1033.1990.tb19432.x. [DOI] [PubMed] [Google Scholar]
- 4.Boyton Z L, Bennett G N, Rudolph F B. Cloning, sequence, and expression of genes encoding phosphotransacetylase and acetate kinase from Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1996;62:2758–2766. doi: 10.1128/aem.62.8.2758-2766.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns G, Brown T, Hatter K, Idriss J M, Sokatch J R. Similarity of the E1 subunits of branched-chain-oxoacid dehydrogenase from Pseudomonas putida to the corresponding subunits of mammalian branched-chain-oxoacid and pyruvate dehydrogenases. Eur J Biochem. 1988;176:311–317. doi: 10.1111/j.1432-1033.1988.tb14283.x. [DOI] [PubMed] [Google Scholar]
- 6.Burns G, Brown T, Hatter K, Sokatch J R. Sequence analysis of the lpdV gene for lipoamide dehydrogenase of branched-chain-oxoacid dehydrogenase of Pseudomonas putida. Eur J Biochem. 1989;179:61–69. doi: 10.1111/j.1432-1033.1989.tb14521.x. [DOI] [PubMed] [Google Scholar]
- 7.Condon S. Responses of lactic acid bacteria to oxygen. FEMS Microbiol Rev. 1987;46:269–280. [Google Scholar]
- 8.Debarbouille M, Rozenn G, Arnaud M, Rapoport G. Role of BkdR, a transcriptional activator of the SigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J Bacteriol. 1999;181:2059–2066. doi: 10.1128/jb.181.7.2059-2066.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Kok, A. 1996. Personal communication.
- 10.Denoya C D, Fedechko R W, Hafner E W, McArthur H A I, Morgenstern M R, Skinner D D, Stutzman-Engwall K, Wax R G, Wernau W C. A second branched-chain α-keto acid gene cluster (bkdFGH) from Streptomyces avermitilis: its relationship to avermectin biosynthesis and the construction of a bkdF mutant suitable for the production of novel antiparasitic avermectins. J Bacteriol. 1995;177:3504–3511. doi: 10.1128/jb.177.12.3504-3511.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolin M I. Cytochrome-independent electron transport enzymes of bacteria. In: Gunsalus I C, Stainer R Y, editors. The bacteria. II. New York, N.Y: Academic Press; 1961. pp. 425–460. [Google Scholar]
- 13.Dunny G M, Brown B L, Clewall D B. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci USA. 1978;75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans C G T, Herbert D, Tempest D W. The continuous culture of microorganisms. 2. Construction of a chemostat. In: Norris J R, Ribbons D W, editors. Methods in microbiology. Vol. 2. London, England: Academic Press; 1970. pp. 277–327. [Google Scholar]
- 15.Flaherty K M, McKay D B, Kabsch W, Holmes K C. Similarity of the three-dimensional structures of actin and the ATPase fragment of a 70-kDa heat shock cognate protein. Proc Natl Acad Sci USA. 1991;88:5041–5045. doi: 10.1073/pnas.88.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fogg G C, Gibson C M, Caparon M G. The identification of rofA, a positive-acting regulatory component of prtF expression: use of an mγδ-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol Microbiol. 1994;11:671–684. doi: 10.1111/j.1365-2958.1994.tb00345.x. [DOI] [PubMed] [Google Scholar]
- 17.Griffen T A, Lau K S, Chuang D T. Characterization of the inner E2 core domain structure of branched-chain α-keto acid dehydrogenase from bovine liver. J Biol Chem. 1988;263:14008–14014. [PubMed] [Google Scholar]
- 18.Griffin H G, l’Anson K J, Gasson M J. Rapid isolation of genes from bacterial lambda libraries by direct polymerase chain reaction screening. FEMS Microbiol Lett. 1993;112:49–54. doi: 10.1111/j.1574-6968.1993.tb06422.x. [DOI] [PubMed] [Google Scholar]
- 19.Grundy F J, Waters D A, George Allen S H, Henkin T M. Regulation of the Bacillus subtilis acetate kinase gene by CcpA. J Bacteriol. 1993;175:7348–7355. doi: 10.1128/jb.175.22.7348-7355.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartmanis M G N. Butyrate kinase from Clostridium acetobutylicum. J Biol Chem. 1987;262:617–621. [PubMed] [Google Scholar]
- 21.Hawkins C F, Borges A, Perham R N. A common structural motif in thiamin pyrophosphate-binding enzymes. FEBS Lett. 1989;255:77–82. doi: 10.1016/0014-5793(89)81064-6. [DOI] [PubMed] [Google Scholar]
- 22.Hein S, Steinbuchel A. Biochemical and molecular characterization of the Alcaligenes eutrophus pyruvate dehydrogenase complex and identification of a new type of dihydrolipoamide dehydrogenase. J Bacteriol. 1994;176:4394–4408. doi: 10.1128/jb.176.14.4394-4408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hein S, Steinbuchel A. Cloning and characterization of the Alcaligenes eutrophus 2-oxoglutarate dehydrogenase complex. FEMS Microbiol Lett. 1996;136:231–238. doi: 10.1111/j.1574-6968.1996.tb08054.x. [DOI] [PubMed] [Google Scholar]
- 24.Hemila H, Palva A, Paulin I, Arvidson S, Palva I. Secretory S complex of Bacillus subtilis: sequence analysis and identity to pyruvate dehydrogenase. J Bacteriol. 1990;172:5052–5063. doi: 10.1128/jb.172.9.5052-5063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendle J, Mattevi A, Westphal A H, Spee J, de Kok A, Teplyakov A, Hol W G J. Crystallographic and enzymatic investigations on the role of Ser558, His610, and Asn614 in the catalytic mechanism of Azotobacter vinelandii dihydrolipoamide acetyltransferase (E2p) Biochemistry. 1995;34:4287–4298. doi: 10.1021/bi00013a018. [DOI] [PubMed] [Google Scholar]
- 26.Kaneda T. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol Rev. 1991;55:288–302. doi: 10.1128/mr.55.2.288-302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi, Y., M. Mizuno, S. Masuda, K. Takemaru, S. Hosono, T. Sato, and M. Takeuchi. 1996. Systematic sequencing of the 283 kb region of the Bacillus subtilis genome containing the skin element. Unpublished accession no. D84432. [DOI] [PubMed]
- 28.Li X, Weinstock G M, Murray B E. Generation of auxotrophic mutants of Enterococcus faecalis. J Bacteriol. 1995;177:6866–6873. doi: 10.1128/jb.177.23.6866-6873.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.London J. Uncommon pathways of metabolism among lactic acid bacteria. FEMS Microbiol Rev. 1990;87:103–112. doi: 10.1111/j.1574-6968.1990.tb04882.x. [DOI] [PubMed] [Google Scholar]
- 30.Lowe P N, Hodgson J A, Perham R N. Dual role of a single multienzyme complex in the oxidative decarboxylation of pyruvate and branched chain 2-oxoacids in Bacillus subtilis. Biochem J. 1983;215:133–140. doi: 10.1042/bj2150133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludwig W, Seewaldt E, Kilper-Balz R, Schleifer K H, Magrum L, Woese C R, Fox G E, Stackebrandt E. The phylogenetic position of Streptococcus and Enterococcus. J Gen Microbiol. 1985;131:543–551. doi: 10.1099/00221287-131-3-543. [DOI] [PubMed] [Google Scholar]
- 32.Mande S S, Sarfaty S, Allen M D, Perham R N, Hol W G J. Protein-protein interactions in the pyruvate dehydrogenase multienzyme complex: dihydrolipoamide dehydrogenase with the binding domain of dihydrolipoamide acetyltransferase. Structure. 1996;4:277–286. doi: 10.1016/s0969-2126(96)00032-9. [DOI] [PubMed] [Google Scholar]
- 33.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 34.Mattevi A, Obmolova G, Kalk K H, Westphal A H, de Kok A, Hol W G. Refined crystal structure of the catalytic domain of dihydrolipoyl transacetylase from Azotobacter vinelandii at 2.6A resolution. J Mol Biol. 1993;230:1183–1199. doi: 10.1006/jmbi.1993.1235. [DOI] [PubMed] [Google Scholar]
- 35.McCully V, Burns G, Sokatch J R. Resolution of branched-chain oxo acid dehydrogenase complex of Pseudomonas aeruginosa. Biochem J. 1986;233:737–742. doi: 10.1042/bj2330737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray B E, Singh K V, Ross R P, Heath J D, Dunny G M, Weinstock G M. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J Bacteriol. 1993;175:5216–5223. doi: 10.1128/jb.175.16.5216-5223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura Y, Wada K, Wada Y, Doi H, Kanaya S, Gojobori T, Ikemura T. Codon usage tabulated from the international DNA sequence databases. Nucleic Acids Res. 1996;24:244–245. doi: 10.1093/nar/24.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Odessey R. Purification of rat kidney branched-chain oxo acid dehydrogenase complex with endogenous kinase activity. Biochem J. 1982;204:353–356. doi: 10.1042/bj2040353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oultram J D, Burr I D, Elmore M J, Minton N P. Cloning and sequence analysis of the genes encoding phosphotransbutyrylase and butyrate kinase from Clostridium acetobutylicum NCIMB 8052. Gene. 1993;131:107–112. doi: 10.1016/0378-1119(93)90677-u. [DOI] [PubMed] [Google Scholar]
- 40.Palmer J A, Hatter K, Sokatch J R. Cloning and sequence analysis of the LPD-glc structural gene of Pseudomonas putida. J Bacteriol. 1991;173:3109–3116. doi: 10.1128/jb.173.10.3109-3116.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer J A, Madhusudhan K T, Hatter K, Sokatch J R. Cloning, sequence and transcriptional analysis of the structural gene for LPD-3, the third lipoamide dehydrogenase of Pseudomonas putida. Eur J Biochem. 1991;202:231–240. doi: 10.1111/j.1432-1033.1991.tb16367.x. [DOI] [PubMed] [Google Scholar]
- 42.Paxton R, Harris R A. Isolation of rabbit liver branched-chain α-keto acid dehydrogenase and its regulation by phosphorylation. J Biol Chem. 1982;257:14433–14439. [PubMed] [Google Scholar]
- 43.Perham R N, Packman L C. 2-Oxo acid dehydrogenase multienzyme complexes: domains, dynamics, and design. Ann N Y Acad Sci. 1989;573:1–20. doi: 10.1111/j.1749-6632.1989.tb14983.x. [DOI] [PubMed] [Google Scholar]
- 44.Perham R N. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry. 1991;30:8501–8512. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- 45.Pettit F H, Yeaman S J, Reed L J. Purification and characterization of branched chain-ketoacid dehydrogenase complex of bovine kidney. Proc Natl Acad Sci USA. 1978;75:4881–4885. doi: 10.1073/pnas.75.10.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Platteeuw C, Simons G, de Vos W M. Use of the β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1994;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross R P, Claiborne A. Cloning, sequence, and overexpression of NADH peroxidase from Streptococcus faecalis 10C1. Structural relationship with the flavoprotein disulfide reductases. J Mol Biol. 1991;221:857–871. doi: 10.1016/0022-2836(91)80180-3. [DOI] [PubMed] [Google Scholar]
- 48.Ross R P, Claiborne A. Molecular cloning and analysis of the gene encoding the NADH oxidase from Streptococcus faecalis 10C1. Comparison with NADH peroxidase and the flavoprotein disulfide reductases. J Mol Biol. 1992;227:658–671. doi: 10.1016/0022-2836(92)90215-6. [DOI] [PubMed] [Google Scholar]
- 49.Rudiger H W, Langenbeck U, Goedde H W. Oxidation of branched chain α-keto acids in Streptococcus faecalis and its dependence on lipoic acid. Hoppe-Seyler’s Z Physiol Chem. 1972;353:875–882. doi: 10.1515/bchm2.1972.353.1.875. [DOI] [PubMed] [Google Scholar]
- 50.Saier M H, Jr, Chauvaux S, Cook G M, Deutscher J, Paulsen I T, Reizer J, Ye J J. Catabolite repression and inducer control in Gram-positive bacteria. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 51.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skinner D D, Morgenstern M R, Fedechko R W, Denoya C D. Cloning and sequencing of a cluster of genes encoding branched-chain α-keto acid dehydrogenase from Streptomyces avermitilis and the production of a functional E1[αβ] component in Escherichia coli. J Bacteriol. 1995;177:183–190. doi: 10.1128/jb.177.1.183-190.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snoep J L, Teixeira de Mattos M J, Postma P W, Neijssel O M. Involvement of pyruvate dehydrogenase in product formation in pyruvate-limited anaerobic chemostat cultures of Enterococcus faecalis NCTC 775. Arch Microbiol. 1990;154:50–55. doi: 10.1007/BF00249177. [DOI] [PubMed] [Google Scholar]
- 54.Snoep J L, Westphal A H, Benen J A E, Teixeira de Mattos M J, Neijssel O M, de Kok A. Isolation and characterization of the pyruvate dehydrogenase complex of anaerobically grown Enterococcus faecalis NCTC 775. Eur J Biochem. 1992;203:245–250. doi: 10.1111/j.1432-1033.1992.tb19853.x. [DOI] [PubMed] [Google Scholar]
- 55.Sokatch J R, McCully V, Roberts C M. Purification of a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981;148:647–652. doi: 10.1128/jb.148.2.647-652.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Studier F W, Moffat B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 57.Sykes P J, Burns G, Menard J, Hatter K, Sokatch J R. Molecular cloning of genes encoding branched-chain keto acid dehydrogenase of Pseudomonas putida. J Bacteriol. 1987;169:1619–1625. doi: 10.1128/jb.169.4.1619-1625.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Terzaghi B, Sandine W F. Improved medium for lactic streptococci and their bacteriophages. Arch Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toal D R, Clifton S W, Roe B A, Downard J. The esg locus Myxococcus xanthus encodes the E1α and E1β subunits of a branched-chain keto acid dehydrogenase. Mol Microbiol. 1995;16:177–189. doi: 10.1111/j.1365-2958.1995.tb02291.x. [DOI] [PubMed] [Google Scholar]
- 60.Walter K A, Nair R V, Cary J W, Bennett G N, Papoutsakis E T. Sequence and arrangement of the two genes of the butyrate-synthesis pathway of Clostridium acetobutylicum ATCC 824. Gene. 1993;134:107–111. doi: 10.1016/0378-1119(93)90182-3. [DOI] [PubMed] [Google Scholar]
- 61.Wang G, Kuriki T, Roy K L, Kaneda T. The primary structure of branched-chain α-oxo acid dehydrogenase from Bacillus subtilis and its similarity to other α-oxo acid dehydrogenases. Eur J Biochem. 1993;213:1091–1099. doi: 10.1111/j.1432-1033.1993.tb17858.x. [DOI] [PubMed] [Google Scholar]
- 62.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 63.Westphal A H, Fabisz-Kilowska A, Kester H, Obels P P, de Kok A. The interaction between lipoamide dehydrogenase and the peripheral-component-binding domain from the Azotobacter vinelandii pyruvate dehydrogenase complex. Eur J Biochem. 1995;234:861–870. doi: 10.1111/j.1432-1033.1995.861_a.x. [DOI] [PubMed] [Google Scholar]
- 64.Wexler I D, Hemalatha S G, Patel M S. Sequence conservation in the alpha and beta subunits of pyruvate dehydrogenase and its similarity to branched-chain alpha-keto acid dehydrogenase. FEBS Lett. 1991;282:209–213. doi: 10.1016/0014-5793(91)80479-m. [DOI] [PubMed] [Google Scholar]
- 65.Wiesenborn D P, Rudolph F B, Papoutsakis E T. Phosphotransbutyrylase from Clostridium acetobutylicum ATCC 824 and its role in acidogenesis. Appl Environ Microbiol. 1989;55:317–322. doi: 10.1128/aem.55.2.317-322.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilkinson K D, William C H., Jr NADH inhibition and NAD activation of Escherichia coli lipoamide dehydrogenase catalyzing the NADH-lipoamide reaction. J Biol Chem. 1981;256:2307–2314. [PubMed] [Google Scholar]
- 67.Willecke I D, Pardee A B. Fatty acid-requiring mutant of Bacillus subtilis defective in branched-chain α-keto acid dehydrogenase. J Biol Chem. 1971;246:5261–5272. [PubMed] [Google Scholar]
- 68.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]