Abstract
Intervertebral disc degeneration (IVDD) is a prevalent cause of low back pain. IVDD is characterized by abnormal expression of extracellular matrix components such as collagen and aggrecan. In addition, it results in dysfunctional growth, senescence, and death of intervertebral cells. The biological pathways involved in the development and progression of IVDD are not fully understood. Therefore, a better understanding of the molecular mechanisms underlying IVDD could aid in the development of strategies for prevention and treatment. Autophagy is a cellular process that removes damaged proteins and dysfunctional organelles, and its dysfunction is linked to a variety of diseases, including IVDD and osteoarthritis. In this review, we describe recent research findings on the role of autophagy in IVDD pathogenesis and highlight autophagy-targeting molecules which can be exploited to treat IVDD. Many studies exhibit that autophagy protects against and postpones disc degeneration. Further research is needed to determine whether autophagy is required for cell integrity in intervertebral discs and to establish autophagy as a viable therapeutic target for IVDD.
Keywords: intervertebral disc degeneration, autophagy, low back pain, autophagy-targeting compounds, ECM degradation
1. Introduction
Low back pain is an extremely common musculoskeletal condition that is experienced by nearly everyone at some point in their lives [1,2]. As low back pain is one of the leading sources of years lived with disability worldwide, more attention is urgently needed to ease its impact on health care systems [3,4,5].
Intervertebral disc degeneration (IVDD) is a prevalent musculoskeletal disorder defined as the degeneration of one or more intervertebral discs (IVDs) that separate the vertebrae, which produces pain in the back, neck, or extremities [6]. People of all ages are affected by IVDD, which decreases their quality of life and increases clinical and economic expenditures [7,8,9]. Over 40% of cases of low back pain result from IVDD progression, which can be caused by excessive stress, aging, injury, spine deformity, and hereditary predisposition [10,11,12,13]. Mutations in IVD proteins, such as collagens, proteoglycans, cytokines, and proteolytic enzymes, and the vitamin D receptor, are linked to IVDD [14,15]. Although much research has been conducted to investigate the molecular mechanisms of IVDD, the pathogenic process underlying the disc degeneration is still not fully understood.
Autophagy is a cellular process that removes damaged proteins and malfunctioning organelles and, thereby, acts as an organismal homeostasis mechanism [16,17]. Growing evidence implicates autophagy failure in a variety of diseases, including IVDD and osteoarthritis [18,19,20,21,22]. Thus, a synthesis of the literature on the role of autophagy in IVDD is needed to chart paths toward more effective therapeutic options. In this review, we describe recent studies linking autophagy to IVDD and highlight potential treatment options based on known underlying mechanisms of the disease.
2. Genetics of IVDD
2.1. IVD Composition
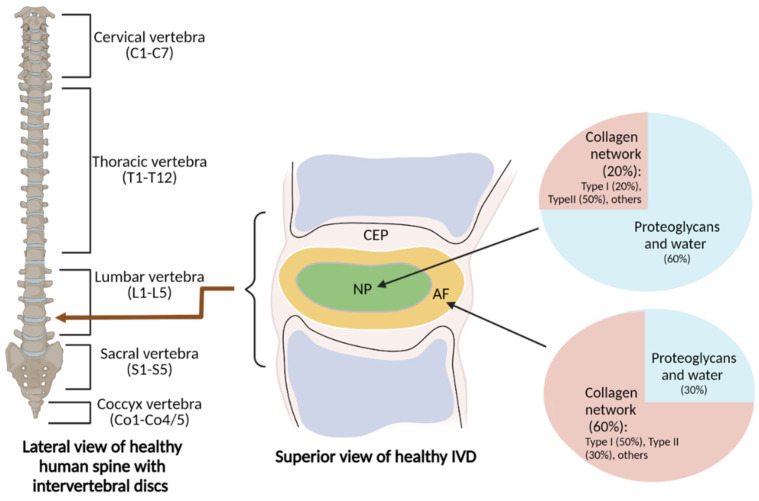
IVDs support the spinal column and act as shock absorbers against the human body’s natural axial loading. The spinal column, which starts at the skull base and terminates at the coccyx, is made up of vertebrae, and IVDs consist of five regions: cervical (C1-C7), thoracic (T1-T12), lumbar (L1-L5), sacral (S1-S5), and coccygeal (Co1-Co4/5) [23]. IVDs are divided into two parts: the central nucleus pulposus (NP) and the outer annulus fibrosus (AF) area [24,25] (Figure 1). The NP is mostly made up of proteoglycans and water. It functions as a hydrogel that helps maintain hydrostatic pressure by trapping water [26]. The NP also contains collagen network components including collagen type I, collagen type II, and other materials (e.g., chondrocyte-like cells) [27]. The composition of the NP allows it to remain elastic, compressible, and flexible against stress forces. The AF mostly consists of a highly structured collagen fiber network containing collagen type I, collagen type II, and other materials (e.g., fibroblast-like cells) that efficiently restrict the NP in place [28]. Sharpey’s fibres connect the AF to the vertebral body, forming a fibrous framework around the NP [29]. The IVD also contains interdependent and structurally connected cartilaginous endplates (CEPs).
Figure 1.
The major components of intervertebral discs. Intervertebral discs (IVDs) are located across the cervical, thoracic, lumbar, sacral, and coccygeal regions of the spinal column. IVDs consist of a central NP (nucleus pulposus), outer AF (annulus fibrosus) area, and interdependent and structurally connected CEP (cartilaginous endplate). The region-specific ECM structures, which include the proteoglycan-rich NP and collagen-rich AF, are maintained by separate cell populations.
2.2. IVDD
IVDD most commonly affects IVDs in the lower (i.e., lumbar) region of the spine but can affect any spinal region. IVDD can produce periodic or chronic discomfort in the back or neck depending on the location of the afflicted IVDs. Pain is typically exacerbated upon sitting, bending, twisting, or lifting objects. The spine can be vulnerable to stress and aberrant gene expression due to certain biological and environmental variables. As a result, IVDs may experience increased catabolic activity and decreased anabolic activity, resulting in their degeneration. Indeed, research shows that IVDD has a complicated pathophysiology related to factors such as aging, spine abnormalities and disorders, spine injuries, and genetic factors [26,30]. However, the molecular mechanisms underlying the disease are not yet fully understood.
2.3. Genes Associated with IVDD Progression
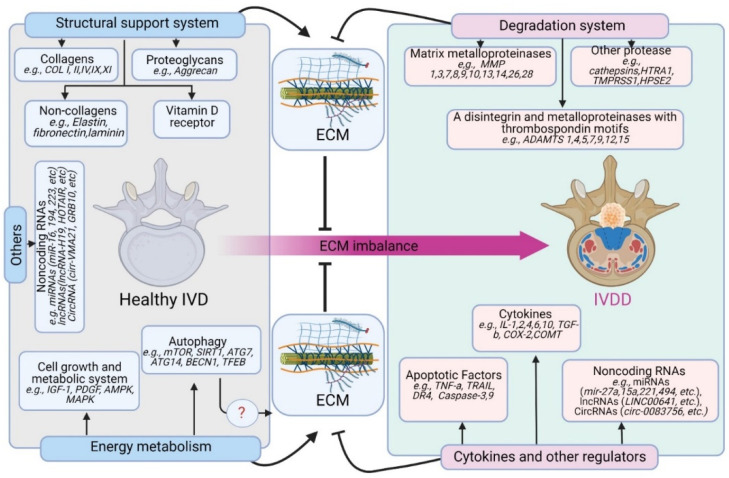
In vitro and in vivo studies of IVDD reveal several genes that impact IVD extracellular matrix (ECM) anabolism and catabolism [31,32,33,34,35]. Although IVDD is a heterogeneous disorder influenced by both genetic and environmental factors, emerging data indicate that the genetic influence greatly outweighs the environmental factors. Recent research reports a six times higher risk of developing IVDD among individuals with a genetic predisposition compared with the general population [36]. Thus, IVDD has been classified as a common, oligogenic, multifactorial genetic disorder [14]. Mutations in IVD-related proteins, such as collagens, proteoglycans, cytokines, and proteolytic enzymes, and the vitamin D receptor appear to play a role in IVDD pathogenesis [15]. The genes associated with IVDD progression are depicted in Figure 2.
Figure 2.
IVDD-associated genes. Mutations in IVD genes, such as structural proteins (e.g., collagens, proteoglycans, and others) and degradation enzymes (e.g., MMPs, ADAMTS, and others) are involved in the pathophysiology of IVDD associating with extracellular matrix (ECM) homeostasis. Many other gene products such as cytokines, apoptotic factors, as well as noncoding RNAs are associated with IVDD progression. Alterations in autophagy and energy metabolism genes facilitate the progression of IVDD, although their underlying mechanisms are not clear.
2.3.1. Gene Products Related to the Synthesis of ECM
The ECM is a non-cellular three-dimensional macromolecular structure made up of roughly 300 proteins that are found in the extracellular environment of all tissues and participate in various cellular activities [37,38,39]. Collagens, proteoglycans, and non-collagenous proteins are the primary components of the ECM in IVDs [40,41]. Proper physiological functioning of IVDs depends on balanced ECM dynamics [7,9,42,43]. IVD cells are dispersed across a complex network of interconnected sugar molecules and collagen that communicates reciprocally with the neighboring ECM [40,44].
Collagens
Collagens (collagens I, II, III, IX, and XI) are crucial structural elements of the ECM. Among the twenty-eight types of collagen discovered in different parts of the IVD matrix, the AF and NP have the highest quantities of collagen I, II, III, IV, V, IV, IX, X, and XI [45]. Although some studies have investigated potential links between polymorphisms in collagen genes and risk of IVDD, they have produced inconsistent findings due to variations in participant ethnicity, age, and site of single nucleotide polymorphism genetic predisposition [15,46,47]. Table 1 lists the ECM synthesis-related collagen genes linked to IVDD pathophysiology based on the latest studies.
Table 1.
ECM synthesis-related collagen genes associated with IVDD.
| Protein | Encoded Genes | Remarks | Ref. |
|---|---|---|---|
| Collagen I α-1 | COL1A1 | The rs1800012 polymorphism in COL1A1 is linked to IVDD susceptibility. | [48,49] |
| Collagen I α-2 | COL1A2 | COL1A2 regulates collagen activity during development and mature tissue regeneration. | [50,51] |
| Collagen II α-1 | COL2A1 | COL2A1 polymorphisms are linked to IVDD risk and clinical pathological characteristics in a Chinese Han population. | [52] |
| Collagen IX α-1 | COL9A1 | COL9A1 is strongly cross-linked to type II collagen and other type IX collagen molecules in NP cells. | [53] |
| Collagen IX α-2 | COL9A2 | IVDD is linked to collagen IX α chains encoded by different alleles. | [54,55,56] |
| Collagen IX α-3 | COL9A3 | IL-1β gene mutation alters COL9A3 gene polymorphism, which modifies the effect of obesity on IVDD. | [57,58] |
| Collagen XI α-1 | COL11A1 | A COL11A1 genetic variation is functionally linked to IVDD in a Chinese population. | [59] |
| Collagen XI α-2 | COL11A2 | COL11A2 polymorphism is linked to IVDD in a Chinese Han population. | [60] |
IVDD: intravertebral disc degeneration; ECM: extracellular matrix; NP: nucleus pulposus; IL-1β: interleukin 1 beta.
Proteoglycans
Aggrecan is a major and functionally important type of proteoglycan in the IVD extracellular matrix. Various numbers of tandem repeat (VNTR) polymorphisms within the aggrecan CS1 domain are unique to humans [61]. As a result, several recent studies have investigated the link between aggrecan VNTR polymorphism and the risk of IVDD, with conflicting findings [62,63,64,65,66,67,68,69,70,71].
Non-Collagenous Proteins
The IVD matrix contains a variety of non-collagenous proteins that govern pathologic pathways and induce disc dysfunction and degeneration [72,73,74,75].
2.3.2. Proteolytic Proteins Responsible for ECM Degradation
The ECM is characterized by a dynamic architectural framework that is continually modified in both normal and pathological conditions, driven by many matrix-degrading proteolytic enzymes such as matrix metalloproteinases (MMPs), a disintegrin, and metalloprotease with thrombospondin motifs (ADAMTS), and other proteases [37].
MMPs
MMPs are a group of proteolytic enzymes capable of degrading key critical elements (i.e., collagens and proteoglycans) of IVDs [76]. MMP expression is low in normal IVD tissue but is increased in degenerative IVD tissue. Higher MMP expression was observed as IVDD worsens [77,78,79,80]. The elevated expression of MMP-1 and -3 in inflammatory cells in degenerating discs implies that these proteinases are linked to IVDD [81,82,83]. Table 2 lists the MMP proteolytic genes linked to the pathophysiology of IVDD based on current research findings.
Table 2.
Proteolytic MMP genes associated with ECM degradation in IVDD.
| Protein | Alternative Name | Encoded Genes | Remarks | Ref |
|---|---|---|---|---|
| MMP-1 | Interstitial collagenase, fibroblast collagenase | MMP1 | MMP-1 IHC score predicts the degree of disc degeneration, and inhibition of the matrix-degrading function of MMP-1 attenuates early-stage IVDD. | [84,85] |
| MMP-2 | Gelatinase A | MMP2 | IVD AF cells use MMP-2 to facilitate local collagen breakdown and remodeling. | [86] |
| MMP-3 | Stromelysin-1 | MMP3 | Controlling the TNF-dependent production of MMP-3 helps halt IVDD and ECM catabolism. | [87] |
| MMP-7 | Matrilysin | MMP7 | Larger numbers of MMP-7 immuno-positive NP cells are indicative of intermediate and severe degrees of IVDD. | [88] |
| MMP-8 | Neutrophil collagenase, PMNL collagenase | MMP8 | MMP-8 is moderately upregulated in IVDD. | [80] |
| MMP-9 | Gelatinase B, progelatinase B | MMP9 | MMP-9 is released in the early stages of secondary damage during IVDD, and its presence in the CSF is indicative of serious IVDD and poor prognosis. | [89,90] |
| MMP-10 | Transin-2 | MMP10 | Increased MMP-10, nerve growth factor, and substance P expression are associated with painful IVDD. | [91] |
| MMP-13 | Collagenase 3 | MMP13 | IL-17 may activate NF-kB signaling and lead to upregulated MMP-13 expression and ECM degradation in IVDD. | [92] |
| MMP-14 | Membrane-type MMP | MMP14 | MMP-2 activity is linked to MMP-14 levels in IVDD. | [93] |
| MMP-26 | Matrilysin-2, endometase | MMP26 | MMP-26 is constitutively expressed in human IVDs in vitro and in vivo. | [94] |
| MMP-28 | Epilysin | MMP28 | MMP-28 exists in the ECM of more deteriorated discs and is constitutively produced in human IVD tissue. | [95] |
IVD: intravertebral disc; IVDD: intravertebral disc degeneration; ECM: extracellular matrix; IHC: immunohistochemistry, AF: annulus fibrosus; NP: nucleus pulposus; CSF: cerebrospinal fluid; IL-17: interleukin 17.
ADAMTS
Aggrecans are cleaved at a specific “aggrecanase” site, which involves several members of the ADAMTS family [96,97,98,99,100,101,102]. Although the role of MMPs in degenerative disorders has received much attention, the most prevalent proteoglycan degradation product in IVDD contains a terminal amino acid sequence structure suggestive of aggrecanase activity [98]. Therefore, aggrecanase activity could be a key player in the progression of IVDD [103,104,105,106,107,108].
Other Proteases
Cathepsins are cysteine proteases of the peptidases family that break down proteins in endosomes and lysosomes; however, they are also produced in the extracellular space and are linked to ECM degradation [109,110,111,112]. The presence of cathepsins D, G, L, and K at the site of degeneration implies that these proteinases are linked to CEP separation and AF disorganization in degenerative diseases [113,114,115,116]. Other proteases linked to anabolic imbalance and pathogenesis during IVDD include high-temperature requirement A1 (HTRA1), transmembrane serine protease 1 (TMPRSS1), and heparanase isoforms (HPSE1 and HPSE2) [117,118,119,120,121].
2.3.3. Other Gene Products Related to IVDD Pathogenesis
Inflammatory Cytokines
As inflammation contributes to IVDD, genomic alterations in inflammatory and anti-inflammatory markers may have a particular impact on IVDD. The balance of pro-inflammatory (e.g., IL-1, IL-6, and TNF-α) and anti-inflammatory (e.g., IL-10, TGF-β, and IL4) cytokines is associated with the onset and severity of IVDD [122,123]. As a result, the amount of cell death in IVDD is determined by the interplay between inflammatory and anti-inflammatory cytokines as well as other components of the IVD ECM. For example, TNF-α, IL-1, IL-2, IL-4, and IL-10 single nucleotide polymorphisms are strongly associated with IVDD in the Iranian population, suggesting that genetic modifications in anti-inflammatory cytokines contribute to IVD homeostasis imbalance and degenerative changes [123,124,125,126,127]. The expression levels of inflammatory pain mediators, prostaglandin E2 (PGE2), and cyclooxygenase 2 (COX-2) were found to be greater in degraded IVD cells [128]. In addition, human disc degeneration is linked to a new catechol-O-methyltransferase (COMT) variation [129].
Vitamin D Receptor
Vitamin D receptor mutations may contribute to pathologies such as osteoporosis, osteoarthritis, and IVDD because they affect bone mineralization and remodeling [130,131,132]. The vitamin D receptor gene, which is found on human chromosome 12 (12q12–q14) and has eight protein-coding and six untranslated exons, is one of the most highly investigated genes linked to IVDD [133,134,135,136,137,138,139]. In particular, the t allele of VDR Taq I is associated with a higher risk of developing IVDD and disc herniation, especially in people under the age of 40 [140,141].
Apoptotic and Growth Factors
Degenerated discs exhibit considerably higher apoptosis, according to studies on the molecular underpinnings of IVDD [142,143,144]. Even though the underlying mechanism for IVD cell death is still under investigation, mutation in some death genes has been linked to an elevated risk of IVDD, such as tumor necrosis factor alpha (TNF-α), caspase-3, capase-9, TNF-related apoptosis-inducing ligand (TRAIL), and death receptor-4 (DR-4) [145,146,147,148,149,150].
Back pain is also related to the ingrowth of blood vessels and nerves into the IVD during degeneration. Therefore, some growth factors are considered as a possible therapeutic to improve IVD tissue regeneration. Indeed, platelet-derived growth factor (PDGF), insulin-like growth factor 1 (IGF-1), growth differentiation factor 5 (GDF-5), bone morphogenetic protein 2 (BMP-2), and bone morphogenetic protein 7 (BMP-7) have been explored as viable treatment alternatives for IVD regeneration [151,152,153,154,155,156,157,158,159,160,161,162,163]. Furthermore, VEGF is highly expressed in the injured IVD, and mutational variations in the VEGF gene are associated with lumbar disc degeneration in a young Korean population [164].
Non-Coding RNAs (ncRNAs)
Several non-coding RNAs (ncRNAs) are dysregulated in IVDD, suggesting a role in the development of the disease [165,166,167]. Some ncRNAs’ expression profiles in IVDD patients differ from those in healthy subjects [165]. ncRNAs control apoptosis, proliferation, ECM deterioration, and inflammation during the course of the disease [167,168,169,170,171]. Other ncRNAs inhibit apoptosis and ECM degradation in normal IVD tissues through their target genes or pathways [172,173,174]. In this section, we catalog recent findings on the role of small and circular RNA in IVDD.
One study has shown that twenty-nine microRNAs (miRs) were differentially expressed in IVDD: six upregulated and twenty-three downregulated [175]. Another study has found twenty-five elevated and twenty-six miRNAs in IVDD patients [176]. miR-27a [177], miR-494 [178,179], miR-30d [180], miR-222-3p [181], miR-15a [182], miR-143 [183], miR-532 [184], miR-138-5p [185], miR-106a-5p [186], miR-34a [187], and miR-221 [188] have been suggested as potentially pro-apoptotic in IVDD. On the other hand, miR-155 [175], miR-21 [189], miR-499a-5p [190], miR-486-5p [191], miR-125a [192], miR-145 [193], and miR-573 [194] were significantly downregulated in IVDD tissues. This subsequently suppressed apoptosis through the interaction with mRNA of PTEN, SOX4, FOXO1, TP53INP1, ADAM17, and Bax, respectively. Moreover, miR-16 [195], miR-194 [196], and miR-223 [197] exhibit a protective effect against LPS-induced inflammation and IVDD.
Studies have demonstrated the role of circular RNAs (circRNAs) in IVDD. HDC-Circ 0083756 promotes IVDD by inhibiting the miR-558/TREM1 axis, representing a therapeutic target for IVDD [198]. An abnormally high amount of miR-200c promotes apoptosis of NP cells and ECM degradation by suppressing XIAP. CircVMA21 attenuates the activity of miR-200c, but it is highly suppressed in IVDD tissues [199]. Similarly, Circ-GRB10 is downregulated in degenerative NP cells. Overexpressing GRB10 reduces apoptosis of NP cells via miR-328-5p and activation of the ErbB pathway [200]. In addition, Circ-4099 induces collagen II and aggrecan production and inversely reduces the production of pro-inflammatory molecules, including IL-1, TNF-, and PGE2 via miR-616-5p inhibition [201].
In summary, individual genes and external factors may interact in the etiology of IVDD, producing gene-to-gene, gene-to-environment, and gene-to-age interactions [47]. More research, including linkage analysis and whole-genome scans of people of diverse ethnicities across the lifespan, is needed to improve our understanding of the genetic influence on IVDD and to uncover additional genes involved in IVDD pathogenesis.
3. Current Treatment Approaches for IVDD
There is an increase in the prevalence of intervertebral disc diseases in both young and older populations [202,203,204]. Effective therapeutic approaches are needed. There is a substantial body of research on the treatment of IVDD, which we can divide into three major categories: (a) conservative or non-surgical management, (b) surgical treatment, and (c) molecular and biological therapy [204,205,206].
-
(a)
Conservative or non-surgical management
Some IVDD patients benefit from conservative care, which includes physiotherapy, oral analgesics, vitamins, exercise, heat, cold, corsetry, acupuncture, radio frequency/shock waves, and varying intensities of massage [204,205,207]. However, the condition worsens for some patients or frequently recurs [208]. This approach is generally safe but with modest outcomes.
-
(b)
Surgical treatment
Surgery may include decompression to relieve neurological symptoms [209,210], fusion to stop motion at a functional spinal unit [211,212], motion preservation/modifying surgery using disc replacement/dynamic fixation devices [213,214], and deformity surgery to correct biomechanics across several functional spinal units [215,216]. Patients who undergo surgery have poorer baseline conditions but recover and return to normal quicker, even though the long-term outcomes may vary [210]. Nevertheless, many people have post-spinal surgery syndrome (PSSS), which is chronic or recurring LBP following surgery [217,218,219]. In addition, the effectiveness of these surgical procedures is not always adequate and may result in unfavorable side effects, including a recurrence of nucleus pulposus (NP) herniation [220,221,222], neighboring segment infection [223], or surgical site infection [224]. Therefore, there is a need for more direct and efficient therapies.
-
(c)
Molecular and biological therapies
Molecular therapies hold great promise for the treatment of IVDD. Compared to the conservative approach, they do not focus on pain relief or invasive surgeries that replace or fuse the affected disc. Several experimental and clinical trials have been devised to regenerate and repair the damaged disc. These include manipulating different growth factors with or without carriers [158,225,226,227,228], cells with or without scaffolds [32,229,230,231,232], and gene therapy [233,234]. Effective gene transfer to target cells inside IVD in animal models offers promising new opportunities for IVDD therapy, for example, gene transfer to disc cells through targeting RNA interference (RNAi), clustered regularly interspaced short palindromic repeats (CRISPR), and mammalian target of rapamycin (mTOR) signaling [234,235]. Currently, a limited number of efficient biological agents for IVD regeneration have only been tested in animal models [236,237,238,239]. The role of autophagy in the development of IVDD has been rigorously studied. Autophagy may be a double-edged sword in developing IVDD [240,241,242,243,244,245]. Modulating autophagy represents a potential target for IVDD therapy.
4. Autophagy as a Potential Therapeutic Target for IVDD
4.1. Autophagy and Its Implications for IVDD
Autophagy is a self-digesting system that allows cells to sequester internal and external substrates in double-membrane structures known as autophagosomes. Autophagosomes merge with lysosomes to degrade their contents. Autophagy maintains cellular homeostasis and contributes biosynthesis and energy generation in the face of nutritional and metabolic stresses [246]. Several proteins are essential for the localization of autophagy-related gene (ATG) proteins to the autophagosome formation site, called the pre-autophagosomal structure [247]. A complex containing unc-51-like kinase 1 (ULK1) and members of the ATG family, which includes ATG13, focal adhesion kinase family interacting protein of 200 kDa (FIP200), and ATG101, initiates the autophagic process [248,249]. The ULK1 complex recruits the vacuolar protein sorting 34 (VPS34) complex, which contains VPS34, BECN1, VPS15, and ATG14L at autophagy initiation sites [248,250]. VPS34 complex proteins help form the phagophore that is then processed into an autophagosome through lipidation of LC3 with phosphatidylethanolamine (PE) by another complex that includes ATG16, ATG5, and ATG12 [251]. Autophagy makes an essential contribution to the development of IVDD, with most recent studies indicating that abnormal autophagy levels and aberrant nutrition in IVDs are important factors leading to IVDD [252,253,254,255]. However, the role of autophagy in the IVDD process is still debated. Although most recent studies show that autophagy protects against and delays IVDD, some studies show that autophagy can accelerate IVDD.
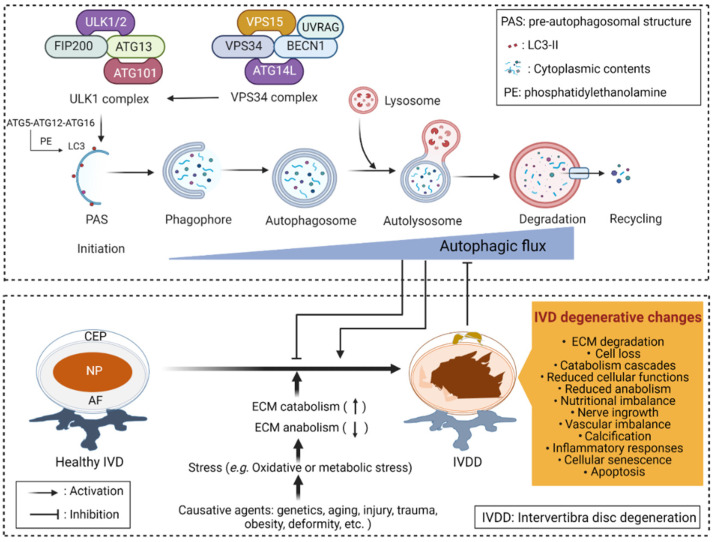
Several studies show that autophagic flux, which encompasses the complete dynamic process of autophagy, is linked to the development of IVDD (Figure 3). For example, restoring the autophagic flux protects against IVDD by reversing oxidative damage and mitochondrial dysfunction [256]. Thus, activating autophagy can be a potential target for treating IVDD.
Figure 3.
Autophagy and IVDD. Autophagic flux refers to the entire process of autophagy, including autophagosome formation, maturation, fusion with lysosomes, subsequent breakdown, and macromolecule release back into the cytosol. Excessive stress, aging, injury, spine deformity, genetic predisposition, and other pathogenic factors contribute to IVDD progression. IVDD is associated with structural and morphologic abnormalities, nutritional imbalances, nerve and vascular in-growth, decreased cellular anabolic function, activating catabolic cascades, elevated chondrocyte calcification, increased cell senescence and apoptosis, and altered expression of ECM and inflammatory markers. Autophagic flux may either delay or accelerate the onset of IVDD.
4.2. Autophagy-Targeting Therapeutic Approaches for IVDD
Autophagy is a regulatory mechanism whereby autophagosomes containing organelles or cytosolic proteins are ultimately destroyed by lysosomes to preserve normal physiological equilibrium and stability [257,258]. Autophagy and apoptosis of NP cells in response to oxidative stress are linked to IVDD [241,259]. Despite the existence of many studies investigating autophagy and IVDD, therapeutic approaches that target autophagy are still limited. In this section, we discuss molecules and compounds that have been used to target autophagy for the treatment of IVDD.
4.2.1. Autophagy-Targeting ncRNAs in IVDD
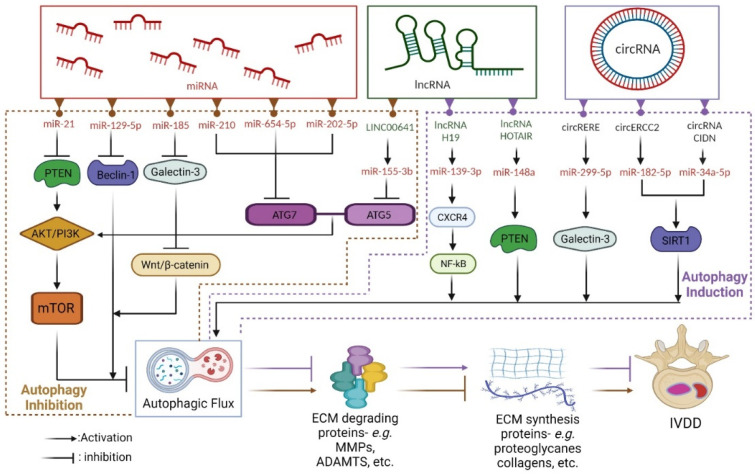
ncRNA is a type of RNA that is transcribed from DNA but does not have the ability to be translated into proteins or peptides. ncRNA includes short hairpin RNA (shRNA), small interfering RNA (siRNA), antisense RNA, microRNA (miRNA), long non-coding RNA (lncRNA), circular RNA (circRNA), and extracellular RNA [260,261,262]. Although evidence suggests that miRNA, lncRNA, and circRNA contribute to the progression of IVDD by affecting the processes of apoptosis, cell proliferation, ECM degradation, and inflammation [263,264,265,266], the role of ncRNA in autophagy is unclear. Figure 4 depicts autophagy-targeting ncRNAs that can be exploited to treat IVDD.
Figure 4.
Autophagy-targeting ncRNAs for the treatment of IVDD. ncRNAs including miRNA, lncRNA, and circRNA could play important roles in IVDD treatment. These molecules can either activate (purple dotted box) or inhibit (brown dotted box) the autophagic flux, resulting in less degradation of ECM synthesis genes in IVDD.
miRNA
miRNAs, which consist of a unique family of 19–25-nucleotide microscopic ncRNAs, are hypothesized to govern several biological activities including cell development and death, cellular senescence, and inflammatory cytokine release [267,268,269,270,271]. Growing evidence indicates that miRNAs play a major role in the genesis and progression of malignancies by acting as tumor suppressor genes or oncogenes [272,273,274,275,276]. Recent research also reports that miRNAs are related to IVDD. For example, one study reports that miR-184 expression is elevated in degenerative NP cells and is positively associated with IVDD severity [277]. By contrast, another study shows that miR-573 expression is downregulated in degenerative NP cells, along with reduced Bcl-2 expression and elevated Bax, cleaved caspase-9, and cleaved caspase-3 expression [194]. The results of several recent studies suggest that miRNA-regulated autophagy can be a potential therapeutic target in IVDD. In addition, the increased expression of miR-21 suppresses autophagy in many cell types including chondrocytes [278,279,280]. In human degenerated NP cells, miR-21 specifically induces ECM breakdown, which markedly downregulates PTEN expression and consequently results in Akt activation [174]. By targeting SIRT1 via PTEN/PI3K/Akt signaling, miR-138-5p enhances TNF-induced apoptosis in human IVDD [185]. miR-10b targets HOXD10 and stimulates NP cell proliferation via the RhoC-Akt pathway in IVDD [281]. Furthermore, activating the Akt/mTOR pathway inhibits autophagy, which enhances MMP-3 and -9 production and facilitates Col II and aggrecan breakdown [282]. Notably, miR-129-5p inhibits autophagy by targeting beclin-1 in NP cells. Indeed, the autophagy activity decreases in human NP cells transfected with a miR-129-5P mimic. By contrast, its activity increases upon treatment of a miR-129-5P inhibitor via modulation of Beclin-1 expression [283]. Moreover, overexpression of miR-185 exhibits a decrease in the autophagy activity by suppressing the Wnt/β-catenin signaling pathway via galectin-3 [284]. Similarly, miR-210 suppresses autophagy in human degenerated NP cells by directly targeting ATG7 and upregulating MMP-3 and -13 expression, leading to enhanced degradation of Col II and aggrecan [285]. Another functional assay suggests that miR-654-5p inhibits autophagy and facilitates ECM degradation by boosting MMP-3, -9, and -13 expression and lowering collagen I, collagen II, SOX9, and aggrecan expression via increased levels of phosphorylated (p)-PI3K, p-AKT, and p-mTOR [286]. Furthermore, expression of MMP-14 is induced by miR-193a-3p downregulation, which accelerates the loss of type II collagen and, hence, promotes IVDD [287]. Notably, overexpression of miR-15a promotes NP cell proliferation and triggers apoptosis by downregulating MAP3K9, upregulating Bax and caspase-3, and downregulating Bcl-2 in degenerative NP tissue and cells [182]. In addition, miR-96 stimulates the growth of human degenerated NP cells by targeting ARID2 via the Akt pathway, suggesting that it could serve as a therapeutic target for IVDD [288].
lncRNA
lncRNA has a limited ability to code for proteins and is involved in a variety of biological processes including transcription, protein activity, and aging-related degenerative musculoskeletal disorders such as IVDD [289]. In IVDD, dysregulated lncRNA plays a key role in altering NP cell activity [290]. Increasing evidence suggests that lncRNA interacts with miRNA, inducing cell autophagy and death through a lncRNA–miRNA–mRNA competitive endogenous RNA (ceRNA) network [167,291]. The lncRNA-FAM83H-AS1 pathway preserves IVD tissue homeostasis and reduces inflammation-related discomfort by inhibiting miR-22-3p, which promotes NP cell proliferation [292]. lncRNA-H19 increases autophagy and death in NP cells, exacerbating IVDD via the miR-139/CXCR4/NF-B axis [293]. In addition, the lncRNA HOTAIR induces apoptosis, senescence, and ECM catabolism in NP cells by upregulating autophagy [294]. HOTAIR is highly abundant in degenerative NP cells, and si-HOTAIR reduces apoptosis and autophagy in degenerative NP cells by boosting PTEN expression as a ceRNA of miR-148a [295]. HOTAIR regulates degenerative changes in IVDs via the Wnt/β-catenin pathway [296] and impacts cell proliferation in IVDD via the miR-130b/PTEN/AKT axis [297]. Furthermore, under nutrition deficiency stress, long intergenic non-protein coding RNA 641 (LINC00641) regulates autophagy and the development of IVDD by serving as a ceRNA of miR-153-3p [298].
circRNA
circRNA regulates gene transcription and translation as well as inflammatory cytokine release, ECM metabolism, and cell proliferation and death [266], suggesting that it could be therapeutically targeted in IVDD. However, our understanding of the mechanism by which circRNA impacts IVDD is still limited. circRNAs were recently reported to influence pathogenic processes of IVDD including inflammation, ECM metabolism, NP cell proliferation, autophagy, and apoptosis by acting as ceRNAs [266,299,300,301]. Thus, these newly discovered ceRNA interaction sites could be important targets for IVDD treatment [167,291]. A growing body of evidence suggests that circRNA and miRNA control cell death and autophagy in IVDD. For example, through the miR-299-5p/Galectin-3 axis, circRNA-RERE promotes oxidative stress-induced apoptosis and autophagy in NP cells [302]. Similarly, circRNA-ERCC2 alleviates IVDD by regulating autophagy and apoptosis via the miR-182-5p/SIRT1 axis [303]. Moreover, circRNA-CIDN binds to miR-34a-5p and prevents compression loading-induced NP cell injury by targeting SIRT1, suggesting that it could be therapeutically targeted to treat IVDD [304].
4.2.2. Autophagy-Targeting Compounds in IVDD
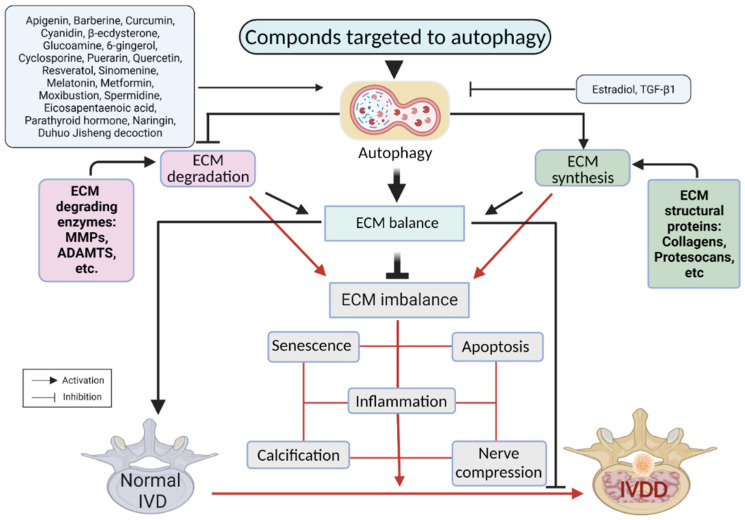
Although autophagy may have a dual role in IVDD, most recent investigations show that it protects against IVDD. Autophagy reduces NP cell apoptosis, ECM degradation, senescence, and CEP inflammation and calcification, thereby balancing levels of IVD ECM components and preventing IVDD [22,254,285,305,306,307,308,309]. A growing number of studies suggest that targeting autophagy could be a promising therapeutic strategy for treating IVDD. A variety of compounds have been suggested to slow IVDD progression by regulating autophagy activity in vivo and in vitro through different signaling pathways and functional proteins (Figure 5).
Figure 5.
Autophagy-targeting compounds for the treatment of IVDD. Activating compounds (i.e., apigenin) targeting autophagy could reduce IVD cell apoptosis, ECM breakdown, senescence, and CEP inflammation and calcification, thereby balancing levels of IVD ECM components and preventing IVDD. Autophagy inhibiting compounds (i.e., estradiol) reverse this reaction. Black and red lines represent reactions at the balanced or imbalanced ECM condition, respectively.
Alleviating Apoptosis, Cell Senescence, and ECM Degradation in IVDD
Inhibiting apoptosis is a promising strategy for treating IVDD. The exogenous death receptor pathway, endogenous mitochondrial pathway, and endoplasmic reticulum stress pathway are among the apoptotic pathways linked to IVDD [310,311]. These pathways may be effective therapeutic targets because they cause IVDD by inducing IVD cell death, which is the most prevalent apoptotic component in IVDD [312]. For example, puerarin, an 8-C-glucoside of daidzein extracted from Pueraria plants, prevents apoptosis and cell death in in vitro human NP mesenchymal stem cells (NPMSCs) and in a rat compression model by maintaining mitochondrial membrane potential and reducing reactive oxygen species generation by activating the PI3K/Akt pathway [313]. Additionally, cyclosporine reduces apoptosis in NPMSCs by reducing mitochondrial malfunction and oxidative stress [314]. In rat NPMSCs, naringin, a bio-flavonoid extracted from tomatoes, grapefruit, and related citrus fruits, inhibits hydrogen peroxide (H2O2)-induced apoptosis via the PI3K/Akt pathway. Autophagy induced by oxidative stress induces apoptosis of NP cells, and autophagy mediated by mechanical stress induces apoptosis of AF cells [241]. On the other hand, autophagy triggered by hypoxia or metformin protects NP cells from apoptosis [242,244].
As an intricate interplay between autophagy and apoptosis maintains healthy IVDs, apoptosis signaling pathways or proteins targeting autophagy could be exploited to treat IVDD. For example, upregulation of transcription factor EB (TFEB) increases autophagy of NP cells, reverses TBHP-induced autophagic flux degradation, and decreases cleaved caspase-3 expression [22]. Similarly, downregulation of STING reduces apoptosis, senescence, and ECM degradation [315]. Berberine inhibits apoptosis and ECM breakdown in NP cells in a mouse model of disc degeneration [316]. Cyanidin inhibits rat NP cell apoptosis and IVDD via modulation of autophagy and the JAK2/STAT3 signaling pathway in vitro and in vivo [317]. Sinomenine, an alkaloid monomer extracted from the Sinomenium acutum, reduces IVDD by inhibiting apoptosis and autophagy in vitro and in vivo [318]. Ecdysterone protects NP cells from apoptosis and attenuates IVDD by stimulating autophagy [319]. Furthermore, moxibustion, a traditional Chinese medical intervention, enhances autophagy and reduces the apoptosis of NP cells via the HIF-1α/VEGF pathway [320].
In addition to increased apoptosis that diminishes IVD cells and ECM, IVDD is also associated with increased cellular senescence [321,322,323]. Cellular senescence in IVDD is linked to decreased cell proliferation, hampered self-repair, an increased inflammatory response, and increased catabolic metabolism [324,325,326]. Autophagy, a well-known and widely conserved intracellular degradation mechanism, protects cells by removing senescent organelles and misfolded proteins [327]. Several studies show that autophagic flux is linked to cell senescence and apoptosis in IVDD. For example, by restoring autophagic flux, TFEB protects NP cells from apoptosis and senescence [22]. An in vitro and in vivo study suggests that BRD4 inhibition suppresses the senescence and apoptosis of NP cells by inducing autophagy in IVDD [328]. SIRT3 helps prevent IVDD by delaying oxidative stress-induced NP cell senescence [329]. Apigenin is a flavonoid that inhibits tert-butyl hydroperoxide (TBHP)-induced apoptosis, senescence, and ECM degradation by restoring autophagic flux in vitro and slows the progression of IVDD in rats in vivo [330]. The mTOR pathway activated by parathyroid hormone 1–34 reduces senescence in rat NP cells by inducing autophagy [331]. Moreover, metformin protects NP cells from apoptosis and senescence by stimulating autophagy and alleviating IVDD in vivo [242].
Damage and limited ability to replenish the ECM in the NP negatively impact the biomechanical properties of IVDs, which compromises spinal stability during disc degeneration [7,9,332,333]. Thus, therapeutic targets for maintaining ECM equilibrium may be crucial for preventing disc degeneration. For example, hydroxysafflor yellow A inhibits TBHP-induced oxidative stress in an NP cell line and modulates ECM equilibrium through carbonic anhydrase 12 (CA XII) [334]. Under oxidative stress, resveratrol promotes ECM production in NP cells by activating autophagy via the PI3K/Akt pathway [335]. ECM degradation and apoptosis are suppressed by eicosapentaenoic acid-induced autophagy, which slows the development of IVDD [336]. Melatonin inhibits the development of IVDD in vivo and in vitro by promoting autophagy via the NF-B signaling pathway, which prevents ECM degradation in IVDs [337]. Berberine inhibits the NF-kB pathway, which protects human NP cells from IL1-induced ECM breakdown and death [338]. Simvastatin inhibits IL-1-induced apoptosis and ECM degradation in NP cells by suppressing the NF-kB and MAPK pathways [339]. Through modulation of p38 MAPK-mediated autophagy, quercetin improves ECM integrity in IVDD patients by raising collagen II and aggrecan levels and reducing MMP13 levels [340]. In a rat model of IVDD, Duhuo Jisheng decoction inhibits ECM degradation and death in human NP cells and alleviates disc degeneration [341]. Spermidine controls the expression of anabolic and catabolic proteins involved in ECM synthesis and breakdown [342]. In NP cells, metformin boosts the expression of anabolic genes such as Col2a1 and Acan and reduces the expression of catabolic genes such as Mmp3 and Adamts5 [242]. The antioxidant and anti-inflammatory agent 6-gingerol reduces reactive oxygen species levels, inhibits NPMSC apoptosis mediated by H2O2, and protects the ECM from degradation [343].
Preventing Inflammation and Endplate Chondrocyte Calcification
During the degeneration of IVDs, secreted inflammatory mediators such as TNF-α, IL-1, IL-6, IL-8, IL-2, IL-17, IL-10, IL-4, IFN-α, and PGE2 promote ECM degradation and IVD cell autophagy, senescence, and apoptosis, ultimately resulting in low back pain [143,308,344,345]. Therefore, these mediators could be exploited to treat IVDD. Acacetin reduces inflammation and ECM degradation in NP cells and attenuates IVDD in vitro [346]. Ligustilide protects NP cells from IL-1-induced inflammation, apoptosis, and ECM degradation and slows IVDD in vivo [347]. In addition, glucosamine reduces IL-1β-induced inflammation in IVDD by triggering autophagy via the mTOR-dependent pathway [348].
IVDs obtain nutrients mainly through diffusion via the calcification of endplate chondrocytes, which inhibits nutrient utilization and metabolite exchange and promotes IVDD [349,350,351]. Therefore, preventing the calcification of endplate chondrocytes is critical. CEP stem cells (CESCs) may increase NP cell proliferation via a paracrine mechanism mediated in part by the SDF-1/CXCR4 axis via the ERK1/2 signaling pathway [352]. Chlorogenic acid, through inhibiting NF-κB signaling, slows CEP degeneration and alleviates IVDD [353]. Furthermore, autophagy acts as a protective response against oxidative damage to endplate chondrocytes [354]. For example, autophagy protects endplate chondrocytes against calcification caused by cyclic mechanical stress [309]. Exosomes released by CESCs to NP cells stimulate Akt/autophagy, which inhibits IVDD [355]. In addition, curcumin promotes autophagy, which reduces the deterioration of CEP caused by stress [356].
4.2.3. Suppressing Autophagy as a Therapeutic Option for IVDD
Several studies have linked autophagy with the initiation of disk degeneration and the development of IVDD [240,245,252,357,358]. The NP and AF cells isolated from non-degenerative adult rats exhibit a basal level of autophagy activity under normal physiological conditions. Therefore, autophagy is required for preserving normal disk cell integrity and survival [359,360]. However, other studies of degenerative rat NP and AF cells have shown heightened autophagy activity and upregulated autophagy-related genes, such as Beclin-1, LC3, Atg12, presenilin 1, and cathepsin B, in comparison to healthy AF tissues [252]. Likewise, IVDD progresses with age, possibly through activation of macroautophagy and chaperone-mediated autophagy (CMA) [357,358]. These findings show that the interaction between autophagy and apoptosis or ROS-dependent endoplasmic reticulum stress could cause IVDD [245].
Blocking autophagy can lessen IVDD. For example, autophagy induced by oxidative or mechanical stress triggers apoptosis in NP and AF cells [41,245]. Indeed, in a rat model of menopause, estradiol reduces IVDD by modulating antioxidant enzymes and inhibiting autophagy [361]. In addition, TGF-b1 rescues AF cells from starvation-induced apoptosis by suppressing excessive autophagy through PI3K-AKT-mTOR and MAPK-ERK1/2 [362]. Furthermore, TGF-b1 inhibits autophagy and apoptosis evoked by exogenous H2O2 in AF cells by downregulating ERK expression through the upregulation of GPx-1 [363].
5. Challenges and Future Applications of Autophagy-Based Therapies for IVDD
Autophagy appears to play a critical role in degenerative IVD cells, although the direction of this role varies by condition. Therefore, future research should focus on determining the specific functional aspects of autophagy in disk degeneration. Furthermore, autophagy-targeted natural molecules and/or gene therapy might be an alternative to conventional IVDD therapy. We suggest that pharmacologically targeting autophagy or gene therapy for autophagy-related genes such as mTOR, LC3, Beclin-1, p62, or other ATG genes are potential therapeutic options for degenerative disc diseases. Since autophagy is required for homeostasis under normal conditions and adaptive pathological responses, the best autophagy-targeted treatment approach for IVDD should attempt to normalize the levels of autophagy inside the IVD.
Although RNAi-mediated mTOR signal inhibition protects human disc NP cells from degeneration, pharmacological modulators are preferable to gene-silencing treatment in clinical settings [364,365]. Indeed, the mTORC1 inhibitor rapamycin and its derivatives, such as everolimus and temsirolimus, increase mammalian lifespan and protect chondrocytes [366,367]. Recently, we reported that delphinidin protects chondrocytes against age-related oxidative stress through autophagy, with potential application in treating osteoarthritis [368]. Thus, we envision extending the use of natural compounds such as delphinidin as autophagy-based therapy for IVDD. In order to properly define the efficacy and safety of autophagy-targeted treatment, preclinical animal and clinical studies are urgently needed.
6. Conclusions
Accumulating evidence indicates that autophagy plays a critical role in the progression of IVDD and, thus, could be used as a novel therapeutic target. This review of recent studies on autophagy-regulated IVD degeneration lays a foundation for further research aimed at elucidating the underlying mechanisms of IVDD and understanding the contribution of autophagy, the ultimate goal of which is to develop novel approaches to IVDD treatment and prevention.
Acknowledgments
We created the figures with BioRender.com (accessed on 1 October 2020).
Abbreviations
ADAMTS: A disintegrin and metalloprotease with thrombospondin motifs; AF: annulus fibrosus; ATGs: autophagy-related genes; ceRNA: competitive endogenous RNA; CEP: cartilaginous endplate; circRNA: circular RNA; ECM: extracellular matrix; EPC: endplate chondrocytes; ERK1/2: Extracellular signal-regulated protein kinase 1//2; IL: interleukin; IVDD: intervertebral disc degeneration; IVDs: intervertebral discs; JNK: Jun N-terminal kinase family; lncRNA: long non-coding RNA; MAPK: mitogen-activated protein kinase; miRNA: microRNA; MMPs: matrix metalloproteinases; mTOR: mammalian target of rapamycin; ncRNAs: noncoding RNAs; NP: nucleus pulposus; PI3K: phosphatidylinositol 3 kinase; shRNA: short hairpin RNA; siRNA: small interfering RNA; SNP: single nucleotide polymorphism; TGF-β: tumor growth factor beta; TNF-α: tumor necrosis factor-alpha; ULK1: unc-51-like kinase 1; VDR: vitamin D receptor; VNTR: various numbers of tandem repeat.
Author Contributions
M.E.B. and D.R.K. designed and conceived this review. M.E.B. and D.R.K. drafted the manuscript and prepared the figures. J.S.H., M.A., T.H.L., T.M.P., O.E., K.-M.A., J.Y. and D.-H.K. edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Education, Science, and Technology (2021R1F1A1059969) and the Ministry of Science and ICT (MSIT) of the Korean government (2015R1A5A2008833 and 2020R1A2C2011416).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Deyo R.A., Mirza S.K. Herniated Lumbar Intervertebral Disk. N. Engl. J. Med. 2016;374:1763–1772. doi: 10.1056/NEJMcp1512658. [DOI] [PubMed] [Google Scholar]
- 2.Slomkowski Z., Golebiowski J., Dreczka J., Sarna R., Michaluk A., Erdenberger M. Clinical picture of medial prolapse of a lumbar intervertebral disk. Neurol. Neurochir. Pol. 1997;31:915–926. [PubMed] [Google Scholar]
- 3.Hoy D., March L., Brooks P., Blyth F., Woolf A., Bain C., Williams G., Smith E., Vos T., Barendregt J., et al. The global burden of low back pain: Estimates from the Global Burden of Disease 2010 study. Ann. Rheum. Dis. 2014;73:968–974. doi: 10.1136/annrheumdis-2013-204428. [DOI] [PubMed] [Google Scholar]
- 4.Hoy D., Brooks P., Blyth F., Buchbinder R. The Epidemiology of low back pain. Best Pract. Res. Clin. Rheumatol. 2010;24:769–781. doi: 10.1016/j.berh.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Wu A., March L., Zheng X., Huang J., Wang X., Zhao J., Blyth F.M., Smith E., Buchbinder R., Hoy D. Global low back pain prevalence and years lived with disability from 1990 to 2017: Estimates from the Global Burden of Disease Study 2017. Ann. Transl. Med. 2020;8:299. doi: 10.21037/atm.2020.02.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dagenais S., Caro J., Haldeman S. A systematic review of low back pain cost of illness studies in the United States and internationally. Spine J. 2008;8:8–20. doi: 10.1016/j.spinee.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Setton L.A., Chen J. Mechanobiology of the intervertebral disc and relevance to disc degeneration. J. Bone Jt. Surg. Am. 2006;88((Suppl. 2)):52–57. doi: 10.2106/JBJS.F.00001. [DOI] [PubMed] [Google Scholar]
- 8.Daly C., Ghosh P., Jenkin G., Oehme D., Goldschlager T. A review of animal models of intervertebral disc degeneration: Pathophysiology, regeneration, and translation to the clinic. Biomed. Res. Int. 2016;2016:5952165. doi: 10.1155/2016/5952165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vergroesen P.P., Kingma I., Emanuel K.S., Hoogendoorn R.J., Welting T.J., van Royen B.J., van Dieen J.H., Smit T.H. Mechanics and biology in intervertebral disc degeneration: A vicious circle. Osteoarthr. Cartil. 2015;23:1057–1070. doi: 10.1016/j.joca.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 10.Clark S., Horton R. Low back pain: A major global challenge. Lancet. 2018;391:2302. doi: 10.1016/S0140-6736(18)30725-6. [DOI] [PubMed] [Google Scholar]
- 11.Foster N.E., Anema J.R., Cherkin D., Chou R., Cohen S.P., Gross D.P., Ferreira P.H., Fritz J.M., Koes B.W., Peul W., et al. Prevention and treatment of low back pain: Evidence, challenges, and promising directions. Lancet. 2018;391:2368–2383. doi: 10.1016/S0140-6736(18)30489-6. [DOI] [PubMed] [Google Scholar]
- 12.Ikegawa S. The genetics of common degenerative skeletal disorders: Osteoarthritis and degenerative disc disease. Annu. Rev. Genom. Hum. Genet. 2013;14:245–256. doi: 10.1146/annurev-genom-091212-153427. [DOI] [PubMed] [Google Scholar]
- 13.Kanayama M., Togawa D., Takahashi C., Terai T., Hashimoto T. Cross-sectional magnetic resonance imaging study of lumbar disc degeneration in 200 healthy individuals. J. Neurosurg. Spine. 2009;11:501–507. doi: 10.3171/2009.5.SPINE08675. [DOI] [PubMed] [Google Scholar]
- 14.Feng Y., Egan B., Wang J. Genetic factors in intervertebral disc degeneration. Genes Dis. 2016;3:178–185. doi: 10.1016/j.gendis.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanaei S., Abdollahzade S., Khoshnevisan A., Kepler C.K., Rezaei N. Genetic aspects of intervertebral disc degeneration. Rev. Neurosci. 2015;26:581–606. doi: 10.1515/revneuro-2014-0077. [DOI] [PubMed] [Google Scholar]
- 16.Pietrocola F., Izzo V., Niso-Santano M., Vacchelli E., Galluzzi L., Maiuri M.C., Kroemer G. Regulation of autophagy by stress-responsive transcription factors. Semin. Cancer Biol. 2013;23:310–322. doi: 10.1016/j.semcancer.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Galluzzi L., Pietrocola F., Levine B., Kroemer G. Metabolic control of autophagy. Cell. 2014;159:1263–1276. doi: 10.1016/j.cell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y., Lin J., Chen J., Huang C., Zhang Z., Wang J., Wang K., Wang X. Mfn2 is involved in intervertebral disc degeneration through autophagy modulation. Osteoarthr. Cartil. 2020;28:363–374. doi: 10.1016/j.joca.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Lin J., Zheng X., Zhang Z., Zhuge J., Shao Z., Huang C., Jin J., Chen X., Chen Y., Wu Y., et al. Inhibition of LRRK2 restores parkin-mediated mitophagy and attenuates intervertebral disc degeneration. Osteoarthr. Cartil. 2021;29:579–591. doi: 10.1016/j.joca.2021.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Xu L., Wu Z., He Y., Chen Z., Xu K., Yu W., Fang W., Ma C., Moqbel S.A.A., Ran J., et al. MFN2 contributes to metabolic disorders and inflammation in the aging of rat chondrocytes and osteoarthritis. Osteoarthr. Cartil. 2020;28:1079–1091. doi: 10.1016/j.joca.2019.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Hu S., Chen L., Al Mamun A., Ni L., Gao W., Lin Y., Jin H., Zhang X., Wang X. The therapeutic effect of TBK1 in intervertebral disc degeneration via coordinating selective autophagy and autophagic functions. J. Adv. Res. 2021;30:1–13. doi: 10.1016/j.jare.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng G., Pan Z., Zhan Y., Tang Q., Zheng F., Zhou Y., Wu Y., Zhou Y., Chen D., Chen J., et al. TFEB protects nucleus pulposus cells against apoptosis and senescence via restoring autophagic flux. Osteoarthr. Cartil. 2019;27:347–357. doi: 10.1016/j.joca.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Schnake K.J., Hoffmann C.H., Kandziora F. Cervical disc herniation. Z. Orthop. Unf. 2012;150:657–673. doi: 10.1055/s-0032-1328067. [DOI] [PubMed] [Google Scholar]
- 24.Amin R.M., Andrade N.S., Neuman B.J. Lumbar disc herniation. Curr. Rev. Musculoskelet. Med. 2017;10:507–516. doi: 10.1007/s12178-017-9441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clouet J., Vinatier C., Merceron C., Pot-Vaucel M., Hamel O., Weiss P., Grimandi G., Guicheux J. The intervertebral disc: From pathophysiology to tissue engineering. Jt. Bone Spine. 2009;76:614–618. doi: 10.1016/j.jbspin.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Zielinska N., Podgorski M., Haladaj R., Polguj M., Olewnik L. Risk factors of intervertebral disc pathology—A point of view formerly and today—A review. J. Clin. Med. 2021;10:409. doi: 10.3390/jcm10030409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sive J.I., Baird P., Jeziorsk M., Watkins A., Hoyland J.A., Freemont A.J. Expression of chondrocyte markers by cells of normal and degenerate intervertebral discs. Mol. Pathol. 2002;55:91–97. doi: 10.1136/mp.55.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eyre D.R., Muir H. Types I and II collagens in intervertebral disc. Interchanging radial distributions in annulus fibrosus. Biochem. J. 1976;157:267–270. doi: 10.1042/bj1570267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buckwalter J.A., Cooper R.R., Maynard J.A. Elastic fibers in human intervertebral discs. J. Bone Jt. Surg. Am. 1976;58:73–76. doi: 10.2106/00004623-197658010-00013. [DOI] [PubMed] [Google Scholar]
- 30.Hadjipavlou A.G., Tzermiadianos M.N., Bogduk N., Zindrick M.R. The pathophysiology of disc degeneration: A critical review. J. Bone Jt. Surg. Br. 2008;90:1261–1270. doi: 10.1302/0301-620X.90B10.20910. [DOI] [PubMed] [Google Scholar]
- 31.Mern D.S., Thome C. Identification and characterization of human nucleus pulposus cell specific serotypes of adeno-associated virus for gene therapeutic approaches of intervertebral disc disorders. BMC Musculoskelet. Disord. 2015;16:341. doi: 10.1186/s12891-015-0799-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kandel R., Roberts S., Urban J.P. Tissue engineering and the intervertebral disc: The challenges. Eur. Spine J. 2008;17((Suppl. 4)):480–491. doi: 10.1007/s00586-008-0746-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mern D.S., Beierfuss A., Thome C., Hegewald A.A. Enhancing human nucleus pulposus cells for biological treatment approaches of degenerative intervertebral disc diseases: A systematic review. J. Tissue Eng. Regen. Med. 2014;8:925–936. doi: 10.1002/term.1583. [DOI] [PubMed] [Google Scholar]
- 34.Mern D.S., Beierfubeta A., Fontana J., Thome C., Hegewald A.A. Imbalanced protein expression patterns of anabolic, catabolic, anti-catabolic and inflammatory cytokines in degenerative cervical disc cells: New indications for gene therapeutic treatments of cervical disc diseases. PLoS ONE. 2014;9:e96870. doi: 10.1371/journal.pone.0096870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hegewald A.A., Ringe J., Sittinger M., Thome C. Regenerative treatment strategies in spinal surgery. Front. Biosci. 2008;13:1507–1525. doi: 10.2741/2777. [DOI] [PubMed] [Google Scholar]
- 36.Chan D., Song Y., Sham P., Cheung K.M. Genetics of disc degeneration. Eur. Spine J. 2006;15((Suppl. 3)):S317–S325. doi: 10.1007/s00586-006-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theocharis A.D., Skandalis S.S., Gialeli C., Karamanos N.K. Extracellular matrix structure. Adv. Drug Deliv. Rev. 2016;97:4–27. doi: 10.1016/j.addr.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Karamanos N.K., Theocharis A.D., Piperigkou Z., Manou D., Passi A., Skandalis S.S., Vynios D.H., Orian-Rousseau V., Ricard-Blum S., Schmelzer C.E.H., et al. A guide to the composition and functions of the extracellular matrix. FEBS J. 2021;288:6850–6912. doi: 10.1111/febs.15776. [DOI] [PubMed] [Google Scholar]
- 39.Karamanos N.K., Piperigkou Z., Passi A., Gotte M., Rousselle P., Vlodavsky I. Extracellular matrix-based cancer targeting. Trends Mol. Med. 2021;27:1000–1013. doi: 10.1016/j.molmed.2021.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Roughley P.J., Melching L.I., Heathfield T.F., Pearce R.H., Mort J.S. The structure and degradation of aggrecan in human intervertebral disc. Eur. Spine J. 2006;15((Suppl. 3)):S326–S332. doi: 10.1007/s00586-006-0127-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKee T.J., Perlman G., Morris M., Komarova S.V. Extracellular matrix composition of connective tissues: A systematic review and meta-analysis. Sci. Rep. 2019;9:10542. doi: 10.1038/s41598-019-46896-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sivan S.S., Wachtel E., Roughley P. Structure, function, aging and turnover of aggrecan in the intervertebral disc. Biochim. Biophys. Acta. 2014;1840:3181–3189. doi: 10.1016/j.bbagen.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 43.Fearing B.V., Hernandez P.A., Setton L.A., Chahine N.O. Mechanotransduction and cell biomechanics of the intervertebral disc. JOR Spine. 2018;1:e1026. doi: 10.1002/jsp2.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manou D., Caon I., Bouris P., Triantaphyllidou I.E., Giaroni C., Passi A., Karamanos N.K., Vigetti D., Theocharis A.D. The complex interplay between extracellular matrix and cells in tissues. Methods Mol. Biol. 2019;1952:1–20. doi: 10.1007/978-1-4939-9133-4_1. [DOI] [PubMed] [Google Scholar]
- 45.Kalb S., Martirosyan N.L., Kalani M.Y., Broc G.G., Theodore N. Genetics of the degenerated intervertebral disc. World Neurosurg. 2012;77:491–501. doi: 10.1016/j.wneu.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 46.Kelempisioti A., Eskola P.J., Okuloff A., Karjalainen U., Takatalo J., Daavittila I., Niinimaki J., Sequeiros R.B., Tervonen O., Solovieva S., et al. Genetic susceptibility of intervertebral disc degeneration among young Finnish adults. BMC Med. Genet. 2011;12:153. doi: 10.1186/1471-2350-12-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalichman L., Hunter D.J. The genetics of intervertebral disc degeneration. Associated genes. Jt. Bone Spine. 2008;75:388–396. doi: 10.1016/j.jbspin.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 48.Pluijm S.M., van Essen H.W., Bravenboer N., Uitterlinden A.G., Smit J.H., Pols H.A., Lips P. Collagen type I alpha1 Sp1 polymorphism, osteoporosis, and intervertebral disc degeneration in older men and women. Ann. Rheum. Dis. 2004;63:71–77. doi: 10.1136/ard.2002.002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong B., Huang D., Ma K., Deng X., Shi D., Wu F., Shao Z. Association of COL1A1 rs1800012 polymorphism with musculoskeletal degenerative diseases: A meta-analysis. Oncotarget. 2017;8:75488–75499. doi: 10.18632/oncotarget.20797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bedore J., Quesnel K., Quinonez D., Seguin C.A., Leask A. Targeting the annulus fibrosus of the intervertebral disc: Col1a2-Cre(ER)T mice show specific activity of Cre recombinase in the outer annulus fibrosus. J. Cell Commun. Signal. 2016;10:137–142. doi: 10.1007/s12079-016-0329-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ponticos M., Abraham D., Alexakis C., Lu Q.L., Black C., Partridge T., Bou-Gharios G. Col1a2 enhancer regulates collagen activity during development and in adult tissue repair. Matrix Biol. 2004;22:619–628. doi: 10.1016/j.matbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Deng Y., Tan X.T., Wu Q., Wang X. Correlations between COL2A and aggrecan genetic polymorphisms and the risk and clinicopathological features of intervertebral disc degeneration in a Chinese han population: A case-control study. Genet. Test. Mol. Biomark. 2017;21:108–115. doi: 10.1089/gtmb.2016.0256. [DOI] [PubMed] [Google Scholar]
- 53.Wu J.J., Eyre D.R. Intervertebral disc collagen. Usage of the short form of the alpha1(IX) chain in bovine nucleus pulposus. J. Biol. Chem. 2003;278:24521–24525. doi: 10.1074/jbc.M302431200. [DOI] [PubMed] [Google Scholar]
- 54.Annunen S., Paassilta P., Lohiniva J., Perala M., Pihlajamaa T., Karppinen J., Tervonen O., Kroger H., Lahde S., Vanharanta H., et al. An allele of COL9A2 associated with intervertebral disc disease. Science. 1999;285:409–412. doi: 10.1126/science.285.5426.409. [DOI] [PubMed] [Google Scholar]
- 55.Karppinen J., Paakko E., Raina S., Tervonen O., Kurunlahti M., Nieminen P., Ala-Kokko L., Malmivaara A., Vanharanta H. Magnetic resonance imaging findings in relation to the COL9A2 tryptophan allele among patients with sciatica. Spine. 2002;27:78–83. doi: 10.1097/00007632-200201010-00018. [DOI] [PubMed] [Google Scholar]
- 56.Wrocklage C., Wassmann H., Paulus W. COL9A2 allelotypes in intervertebral disc disease. Biochem. Biophys. Res. Commun. 2000;279:398–400. doi: 10.1006/bbrc.2000.3967. [DOI] [PubMed] [Google Scholar]
- 57.Solovieva S., Lohiniva J., Leino-Arjas P., Raininko R., Luoma K., Ala-Kokko L., Riihimaki H. Intervertebral disc degeneration in relation to the COL9A3 and the IL-1ss gene polymorphisms. Eur. Spine J. 2006;15:613–619. doi: 10.1007/s00586-005-0988-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Solovieva S., Lohiniva J., Leino-Arjas P., Raininko R., Luoma K., Ala-Kokko L., Riihimaki H. COL9A3 gene polymorphism and obesity in intervertebral disc degeneration of the lumbar spine: Evidence of gene-environment interaction. Spine. 2002;27:2691–2696. doi: 10.1097/00007632-200212010-00008. [DOI] [PubMed] [Google Scholar]
- 59.Liu W., Sun G., Guo L., Wang L., Fan W., Lang M., Chen D., Yi X. A genetic variant in COL11A1 is functionally associated with lumbar disc herniation in Chinese population. J. Genet. 2017;96:867–872. doi: 10.1007/s12041-017-0874-8. [DOI] [PubMed] [Google Scholar]
- 60.Yang X., Jia H., Xing W., Li F., Li M., Sun K., Zhu Y. Genetic variants in COL11A2 of lumbar disk degeneration among Chinese Han population. Mol. Genet. Genom. Med. 2019;7:e00524. doi: 10.1002/mgg3.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doege K.J., Coulter S.N., Meek L.M., Maslen K., Wood J.G. A human-specific polymorphism in the coding region of the aggrecan gene. Variable number of tandem repeats produce a range of core protein sizes in the general population. J. Biol. Chem. 1997;272:13974–13979. doi: 10.1074/jbc.272.21.13974. [DOI] [PubMed] [Google Scholar]
- 62.Casa N.L., Junior A.J.C., Melo A.V., Teodoro L.S., Nascimento G.R., Sousa A.F., Flausino T.C., Brito D., Bergamini R., Minasi L.B., et al. CASE-REPORT Association between an ACAN gene variable number tandem repeat polymorphism and lumbar disc herniation: A case control study. Genet. Mol. Res. 2016;15:gmr15048867. doi: 10.4238/gmr15048867. [DOI] [PubMed] [Google Scholar]
- 63.Cong L., Zhu Y., Tu G. Association between the expression of Aggrecan and the distribution of Aggrecan gene variable number of tandem repeats with symptomatic lumbar disc herniation. Zhonghua Wai Ke Za Zhi. 2015;53:116–120. [PubMed] [Google Scholar]
- 64.Cong L., Zhu Y., Pang H., Guanjun T.U. The interaction between aggrecan gene VNTR polymorphism and obesity in predicting incident symptomatic lumbar disc herniation. Connect. Tissue Res. 2014;55:384–390. doi: 10.3109/03008207.2014.959117. [DOI] [PubMed] [Google Scholar]
- 65.Eser O., Eser B., Cosar M., Erdogan M.O., Aslan A., Yildiz H., Solak M., Haktanir A. Short aggrecan gene repetitive alleles associated with lumbar degenerative disc disease in Turkish patients. Genet. Mol. Res. 2011;10:1923–1930. doi: 10.4238/vol10-3gmr1222. [DOI] [PubMed] [Google Scholar]
- 66.Kim N.K., Shin D.A., Han I.B., Yoo E.H., Kim S.H., Chung S.S. The association of aggrecan gene polymorphism with the risk of intervertebral disc degeneration. Acta Neurochir. 2011;153:129–133. doi: 10.1007/s00701-010-0831-2. [DOI] [PubMed] [Google Scholar]
- 67.Cong L., Pang H., Xuan D., Tu G.J. Association between the expression of aggrecan and the distribution of aggrecan gene variable number of tandem repeats with symptomatic lumbar disc herniation in Chinese Han of Northern China. Spine. 2010;35:1371–1376. doi: 10.1097/BRS.0b013e3181c4e022. [DOI] [PubMed] [Google Scholar]
- 68.Mashayekhi F., Shafiee G., Kazemi M., Dolati P. Lumbar disk degeneration disease and aggrecan gene polymorphism in northern Iran. Biochem. Genet. 2010;48:684–689. doi: 10.1007/s10528-010-9350-3. [DOI] [PubMed] [Google Scholar]
- 69.Kawaguchi Y., Osada R., Kanamori M., Ishihara H., Ohmori K., Matsui H., Kimura T. Association between an aggrecan gene polymorphism and lumbar disc degeneration. Spine. 1999;24:2456–2460. doi: 10.1097/00007632-199912010-00006. [DOI] [PubMed] [Google Scholar]
- 70.Roughley P., Martens D., Rantakokko J., Alini M., Mwale F., Antoniou J. The involvement of aggrecan polymorphism in degeneration of human intervertebral disc and articular cartilage. Eur. Cells Mater. 2006;11:1–7. doi: 10.22203/eCM.v011a01. [DOI] [PubMed] [Google Scholar]
- 71.Noponen-Hietala N., Kyllonen E., Mannikko M., Ilkko E., Karppinen J., Ott J., Ala-Kokko L. Sequence variations in the collagen IX and XI genes are associated with degenerative lumbar spinal stenosis. Ann. Rheum. Dis. 2003;62:1208–1214. doi: 10.1136/ard.2003.008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anderson D.G., Markova D., Adams S.L., Pacifici M., An H.S., Zhang Y. Fibronectin splicing variants in human intervertebral disc and association with disc degeneration. Spine. 2010;35:1581–1588. doi: 10.1097/BRS.0b013e3181c6ef1a. [DOI] [PubMed] [Google Scholar]
- 73.Anderson D.G., Li X., Tannoury T., Beck G., Balian G. A fibronectin fragment stimulates intervertebral disc degeneration in vivo. Spine. 2003;28:2338–2345. doi: 10.1097/01.BRS.0000096943.27853.BC. [DOI] [PubMed] [Google Scholar]
- 74.Speer J., Barcellona M., Jing L., Liu B., Lu M., Kelly M., Buchowski J., Zebala L., Luhmann S., Gupta M., et al. Integrin-mediated interactions with a laminin-presenting substrate modulate biosynthesis and phenotypic expression for cells of the human nucleus pulposus. Eur. Cells Mater. 2021;41:793–810. doi: 10.22203/eCM.v041a50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bridgen D.T., Gilchrist C.L., Richardson W.J., Isaacs R.E., Brown C.R., Yang K.L., Chen J., Setton L.A. Integrin-mediated interactions with extracellular matrix proteins for nucleus pulposus cells of the human intervertebral disc. J. Orthop. Res. 2013;31:1661–1667. doi: 10.1002/jor.22395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goupille P., Jayson M.I., Valat J.P., Freemont A.J. Matrix metalloproteinases: The clue to intervertebral disc degeneration? Spine. 1998;23:1612–1626. doi: 10.1097/00007632-199807150-00021. [DOI] [PubMed] [Google Scholar]
- 77.Wang W.J., Yu X.H., Wang C., Yang W., He W.S., Zhang S.J., Yan Y.G., Zhang J. MMPs and ADAMTSs in intervertebral disc degeneration. Clin. Chim. Acta. 2015;448:238–246. doi: 10.1016/j.cca.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 78.Xu H., Mei Q., He J., Liu G., Zhao J., Xu B. Correlation of matrix metalloproteinases-1 and tissue inhibitor of metalloproteinases-1 with patient age and grade of lumbar disk herniation. Cell Biochem. Biophys. 2014;69:439–444. doi: 10.1007/s12013-014-9815-9. [DOI] [PubMed] [Google Scholar]
- 79.Xu H., Mei Q., Xu B., Liu G., Zhao J. Expression of matrix metalloproteinases is positively related to the severity of disc degeneration and growing age in the East Asian lumbar disc herniation patients. Cell Biochem. Biophys. 2014;70:1219–1225. doi: 10.1007/s12013-014-0045-y. [DOI] [PubMed] [Google Scholar]
- 80.Bachmeier B.E., Nerlich A., Mittermaier N., Weiler C., Lumenta C., Wuertz K., Boos N. Matrix metalloproteinase expression levels suggest distinct enzyme roles during lumbar disc herniation and degeneration. Eur. Spine J. 2009;18:1573–1586. doi: 10.1007/s00586-009-1031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matsui Y., Maeda M., Nakagami W., Iwata H. The involvement of matrix metalloproteinases and inflammation in lumbar disc herniation. Spine. 1998;23:863–868. doi: 10.1097/00007632-199804150-00005. [DOI] [PubMed] [Google Scholar]
- 82.Eser B., Eser O., Yuksel Y., Aksit H., Karavelioglu E., Tosun M., Sekerci Z. Effects of MMP-1 and MMP-3 gene polymorphisms on gene expression and protein level in lumbar disc herniation. Genet. Mol. Res. 2016;15:gmr.15038669. doi: 10.4238/gmr.15038669. [DOI] [PubMed] [Google Scholar]
- 83.Zigouris A., Batistatou A., Alexiou G.A., Pachatouridis D., Mihos E., Drosos D., Fotakopoulos G., Doukas M., Voulgaris S., Kyritsis A.P. Correlation of matrix metalloproteinases-1 and -3 with patient age and grade of lumbar disc herniation. J. Neurosurg. Spine. 2011;14:268–272. doi: 10.3171/2010.9.SPINE09935. [DOI] [PubMed] [Google Scholar]
- 84.Hsu H.T., Yue C.T., Teng M.S., Tzeng I.S., Li T.C., Tai P.A., Huang K.F., Chen C.Y., Ko Y.L. Immuohistochemical score of matrix metalloproteinase-1 may indicate the severity of symptomatic cervical and lumbar disc degeneration. Spine J. 2020;20:124–137. doi: 10.1016/j.spinee.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 85.Hingert D., Nawilaijaroen P., Ekstrom K., Baranto A., Brisby H. Human levels of MMP-1 in degenerated disks can be mitigated by signaling peptides from mesenchymal stem cells. Cells Tissues Organs. 2020;209:144–154. doi: 10.1159/000509146. [DOI] [PubMed] [Google Scholar]
- 86.Rastogi A., Kim H., Twomey J.D., Hsieh A.H. MMP-2 mediates local degradation and remodeling of collagen by annulus fibrosus cells of the intervertebral disc. Arthritis Res. Ther. 2013;15:R57. doi: 10.1186/ar4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang X., Wang H., Yang H., Li J., Cai Q., Shapiro I.M., Risbud M.V. Tumor necrosis factor-alpha-and interleukin-1beta-dependent matrix metalloproteinase-3 expression in nucleus pulposus cells requires cooperative signaling via syndecan 4 and mitogen-activated protein kinase-NF-kappaB axis: Implications in inflammatory disc disease. Am. J. Pathol. 2014;184:2560–2572. doi: 10.1016/j.ajpath.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Le Maitre C.L., Freemont A.J., Hoyland J.A. Human disc degeneration is associated with increased MMP 7 expression. Biotech. Histochem. 2006;81:125–131. doi: 10.1080/10520290601005298. [DOI] [PubMed] [Google Scholar]
- 89.Levine J.M., Ruaux C.G., Bergman R.L., Coates J.R., Steiner J.M., Williams D.A. Matrix metalloproteinase-9 activity in the cerebrospinal fluid and serum of dogs with acute spinal cord trauma from intervertebral disk disease. Am. J. Vet. Res. 2006;67:283–287. doi: 10.2460/ajvr.67.2.283. [DOI] [PubMed] [Google Scholar]
- 90.Nagano S., Kim S.H., Tokunaga S., Arai K., Fujiki M., Misumi K. Matrix metalloprotease-9 activity in the cerebrospinal fluid and spinal injury severity in dogs with intervertebral disc herniation. Res. Vet. Sci. 2011;91:482–485. doi: 10.1016/j.rvsc.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 91.Richardson S.M., Doyle P., Minogue B.M., Gnanalingham K., Hoyland J.A. Increased expression of matrix metalloproteinase-10, nerve growth factor and substance P in the painful degenerate intervertebral disc. Arthritis Res. Ther. 2009;11:R126. doi: 10.1186/ar2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yao Z., Nie L., Zhao Y., Zhang Y., Liu Y., Li J., Cheng L. Salubrinal suppresses IL-17-induced upregulation of MMP-13 and extracellular matrix degradation through the NF-kB pathway in human nucleus pulposus cells. Inflammation. 2016;39:1997–2007. doi: 10.1007/s10753-016-0435-y. [DOI] [PubMed] [Google Scholar]
- 93.Rutges J.P., Kummer J.A., Oner F.C., Verbout A.J., Castelein R.J., Roestenburg H.J., Dhert W.J., Creemers L.B. Increased MMP-2 activity during intervertebral disc degeneration is correlated to MMP-14 levels. J. Pathol. 2008;214:523–530. doi: 10.1002/path.2317. [DOI] [PubMed] [Google Scholar]
- 94.Gruber H.E., Hoelscher G.L., Ingram J.A., Hanley E.N., Jr. Matrix metalloproteinase-26, a novel MMP, is constitutively expressed in the human intervertebral disc in vivo and in vitro. Exp. Mol. Pathol. 2012;92:59–63. doi: 10.1016/j.yexmp.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 95.Gruber H.E., Ingram J.A., Hoelscher G.L., Zinchenko N., Norton H.J., Hanley E.N., Jr. Matrix metalloproteinase 28, a novel matrix metalloproteinase, is constitutively expressed in human intervertebral disc tissue and is present in matrix of more degenerated discs. Arthritis Res. Ther. 2009;11:R184. doi: 10.1186/ar2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Seki S., Asanuma-Abe Y., Masuda K., Kawaguchi Y., Asanuma K., Muehleman C., Iwai A., Kimura T. Effect of small interference RNA (siRNA) for ADAMTS5 on intervertebral disc degeneration in the rabbit anular needle-puncture model. Arthritis Res. Ther. 2009;11:R166. doi: 10.1186/ar2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sandy J.D., Flannery C.R., Neame P.J., Lohmander L.S. The structure of aggrecan fragments in human synovial fluid. Evidence for the involvement in osteoarthritis of a novel proteinase which cleaves the Glu 373-Ala 374 bond of the interglobular domain. J. Clin. Investig. 1992;89:1512–1516. doi: 10.1172/JCI115742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lohmander L.S., Neame P.J., Sandy J.D. The structure of aggrecan fragments in human synovial fluid. Evidence that aggrecanase mediates cartilage degradation in inflammatory joint disease, joint injury, and osteoarthritis. Arthritis. 1993;36:1214–1222. doi: 10.1002/art.1780360906. [DOI] [PubMed] [Google Scholar]
- 99.Kuno K., Okada Y., Kawashima H., Nakamura H., Miyasaka M., Ohno H., Matsushima K. ADAMTS-1 cleaves a cartilage proteoglycan, aggrecan. FEBS Lett. 2000;478:241–245. doi: 10.1016/S0014-5793(00)01854-8. [DOI] [PubMed] [Google Scholar]
- 100.Rodriguez-Manzaneque J.C., Westling J., Thai S.N., Luque A., Knauper V., Murphy G., Sandy J.D., Iruela-Arispe M.L. ADAMTS1 cleaves aggrecan at multiple sites and is differentially inhibited by metalloproteinase inhibitors. Biochem. Biophys. Res. Commun. 2002;293:501–508. doi: 10.1016/S0006-291X(02)00254-1. [DOI] [PubMed] [Google Scholar]
- 101.Somerville R.P., Longpre J.M., Jungers K.A., Engle J.M., Ross M., Evanko S., Wight T.N., Leduc R., Apte S.S. Characterization of ADAMTS-9 and ADAMTS-20 as a distinct ADAMTS subfamily related to Caenorhabditis elegans GON-1. J. Biol. Chem. 2003;278:9503–9513. doi: 10.1074/jbc.M211009200. [DOI] [PubMed] [Google Scholar]
- 102.Collins-Racie L.A., Flannery C.R., Zeng W., Corcoran C., Annis-Freeman B., Agostino M.J., Arai M., DiBlasio-Smith E., Dorner A.J., Georgiadis K.E., et al. ADAMTS-8 exhibits aggrecanase activity and is expressed in human articular cartilage. Matrix Biol. 2004;23:219–230. doi: 10.1016/j.matbio.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 103.Huo J.Z., Ji X.H., Su Z.Y., Shang P., Gao F. Association of ADAMTS4 and ADAMTS5 polymorphisms with musculoskeletal degenerative diseases: A systematic review and meta-analysis. Biosci. Rep. 2018;38:BSR20181619. doi: 10.1042/BSR20181619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hatano E., Fujita T., Ueda Y., Okuda T., Katsuda S., Okada Y., Matsumoto T. Expression of ADAMTS-4 (aggrecanase-1) and possible involvement in regression of lumbar disc herniation. Spine. 2006;31:1426–1432. doi: 10.1097/01.brs.0000219954.67368.be. [DOI] [PubMed] [Google Scholar]
- 105.Liu S., Wu N., Liu J., Liu H., Su X., Liu Z., Zuo Y., Chen W., Liu G., Chen Y., et al. Association between ADAMTS-4 gene polymorphism and lumbar disc degeneration in Chinese Han population. J. Orthop. Res. 2016;34:860–864. doi: 10.1002/jor.23081. [DOI] [PubMed] [Google Scholar]
- 106.Wu N., Chen J., Liu H., Zhao L., Liu S., Liu J., Su X., Wu W., Cong J., Qiu G., et al. The involvement of ADAMTS-5 genetic polymorphisms in predisposition and diffusion tensor imaging alterations of lumbar disc degeneration. J. Orthop. Res. 2014;32:686–694. doi: 10.1002/jor.22582. [DOI] [PubMed] [Google Scholar]
- 107.Pockert A.J., Richardson S.M., Le Maitre C.L., Lyon M., Deakin J.A., Buttle D.J., Freemont A.J., Hoyland J.A. Modified expression of the ADAMTS enzymes and tissue inhibitor of metalloproteinases 3 during human intervertebral disc degeneration. Arthritis Rheum. 2009;60:482–491. doi: 10.1002/art.24291. [DOI] [PubMed] [Google Scholar]
- 108.Yu H., Zhu Y. Expression of ADAMTS-7 and ADAMTS-12 in the nucleus pulposus during degeneration of rat caudal intervetebral disc. J. Vet. Med. Sci. 2012;74:9–15. doi: 10.1292/jvms.10-0556. [DOI] [PubMed] [Google Scholar]
- 109.Lutgens S.P., Cleutjens K.B., Daemen M.J., Heeneman S. Cathepsin cysteine proteases in cardiovascular disease. FASEB J. 2007;21:3029–3041. doi: 10.1096/fj.06-7924com. [DOI] [PubMed] [Google Scholar]
- 110.Wu H., Du Q., Dai Q., Ge J., Cheng X. Cysteine protease cathepsins in atherosclerotic cardiovascular diseases. J. Atheroscler. Thromb. 2018;25:111–123. doi: 10.5551/jat.RV17016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cheng X.W., Huang Z., Kuzuya M., Okumura K., Murohara T. Cysteine protease cathepsins in atherosclerosis-based vascular disease and its complications. Hypertension. 2011;58:978–986. doi: 10.1161/HYPERTENSIONAHA.111.180935. [DOI] [PubMed] [Google Scholar]
- 112.Menou A., Duitman J., Crestani B. The impaired proteases and anti-proteases balance in Idiopathic Pulmonary Fibrosis. Matrix Biol. 2018;68–69:382–403. doi: 10.1016/j.matbio.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 113.Ariga K., Yonenobu K., Nakase T., Kaneko M., Okuda S., Uchiyama Y., Yoshikawa H. Localization of cathepsins D, K, and L in degenerated human intervertebral discs. Spine. 2001;26:2666–2672. doi: 10.1097/00007632-200112150-00007. [DOI] [PubMed] [Google Scholar]
- 114.Konttinen Y.T., Kaapa E., Hukkanen M., Gu X.H., Takagi M., Santavirta S., Alaranta H., Li T.F., Suda A. Cathepsin G in degenerating and healthy discal tissue. Clin. Exp. Rheumatol. 1999;17:197–204. [PubMed] [Google Scholar]
- 115.Gruber H.E., Ingram J.A., Hoelscher G.L., Zinchenko N., Norton H.J., Hanley E.N., Jr. Constitutive expression of cathepsin K in the human intervertebral disc: New insight into disc extracellular matrix remodeling via cathepsin K and receptor activator of nuclear factor-kappaB ligand. Arthritis Res. Ther. 2011;13:R140. doi: 10.1186/ar3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kague E., Turci F., Newman E., Yang Y., Brown K.R., Aglan M.S., Otaify G.A., Temtamy S.A., Ruiz-Perez V.L., Cross S., et al. 3D assessment of intervertebral disc degeneration in zebrafish identifies changes in bone density that prime disc disease. Bone Res. 2021;9:39. doi: 10.1038/s41413-021-00156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Akhatib B., Onnerfjord P., Gawri R., Ouellet J., Jarzem P., Heinegard D., Mort J., Roughley P., Haglund L. Chondroadherin fragmentation mediated by the protease HTRA1 distinguishes human intervertebral disc degeneration from normal aging. J. Biol. Chem. 2013;288:19280–19287. doi: 10.1074/jbc.M112.443010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tiaden A.N., Klawitter M., Lux V., Mirsaidi A., Bahrenberg G., Glanz S., Quero L., Liebscher T., Wuertz K., Ehrmann M., et al. Detrimental role for human high temperature requirement serine protease A1 (HTRA1) in the pathogenesis of intervertebral disc (IVD) degeneration. J. Biol. Chem. 2012;287:21335–21345. doi: 10.1074/jbc.M112.341032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang X., Li D., Wu H., Liu F., Liu F., Zhang Q., Li J. LncRNA TRPC7-AS1 regulates nucleus pulposus cellular senescence and ECM synthesis via competing with HPN for miR-4769-5p binding. Mech. Ageing Dev. 2020;190:111293. doi: 10.1016/j.mad.2020.111293. [DOI] [PubMed] [Google Scholar]
- 120.Rodrigues L.M., Oliveira L.Z., Pinhal M.A. Expression of heparanase isoforms in intervertebral discs classified according to Pfirrmann grading system for disc degeneration. Spine. 2013;38:1112–1118. doi: 10.1097/BRS.0b013e3182894cf4. [DOI] [PubMed] [Google Scholar]
- 121.Rodrigues L.M., Theodoro T.R., Matos L.L., Mader A.M., Milani C., Pinhal M.A. Heparanase isoform expression and extracellular matrix remodeling in intervertebral disc degenerative disease. Clinics. 2011;66:903–909. doi: 10.1590/S1807-59322011000500030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Valdes A.M., Hassett G., Hart D.J., Spector T.D. Radiographic progression of lumbar spine disc degeneration is influenced by variation at inflammatory genes: A candidate SNP association study in the Chingford cohort. Spine. 2005;30:2445–2451. doi: 10.1097/01.brs.0000184369.79744.a5. [DOI] [PubMed] [Google Scholar]
- 123.Hanaei S., Abdollahzade S., Sadr M., Mirbolouk M.H., Fattahi E., Khoshnevisan A., Rezaei N. The role of interleukin 4 and IL-4RA in intervertebral disc degeneration: Investigation of single nucleotide polymorphisms in genes and a systematic review & meta-analysis of IL-4 expression level. Br. J. Neurosurg. 2020;34:66–71. doi: 10.1080/02688697.2019.1698010. [DOI] [PubMed] [Google Scholar]
- 124.Abdollahzade S., Hanaei S., Sadr M., Mirbolouk M.H., Fattahi E., Rezaei N., Khoshnevisan A. Significant association of TNF-alpha, but not other pro-inflammatory cytokines, single nucleotide polymorphisms with intervertebral disc degeneration in Iranian population. Clin. Neurol. Neurosurg. 2018;173:77–83. doi: 10.1016/j.clineuro.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 125.Solovieva S., Kouhia S., Leino-Arjas P., Ala-Kokko L., Luoma K., Raininko R., Saarela J., Riihimaki H. Interleukin 1 polymorphisms and intervertebral disc degeneration. Epidemiology. 2004;15:626–633. doi: 10.1097/01.ede.0000135179.04563.35. [DOI] [PubMed] [Google Scholar]
- 126.Hanaei S., Abdollahzade S., Sadr M., Mirbolouk M.H., Fattahi E., Khoshnevisan A., Rezaei N. Association of interleukin 2, interleukin 12, and interferon-gamma with intervertebral disc degeneration in Iranian population. BMC Med. Genet. 2020;21:143. doi: 10.1186/s12881-020-01081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hanaei S., Abdollahzade S., Sadr M., Mirbolouk M.H., Khoshnevisan A., Rezaei N. Association of IL10 and TGFB single nucleotide polymorphisms with intervertebral disc degeneration in Iranian population: A case control study. BMC Med. Genet. 2018;19:59. doi: 10.1186/s12881-018-0572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Willems N., Tellegen A.R., Bergknut N., Creemers L.B., Wolfswinkel J., Freudigmann C., Benz K., Grinwis G.C., Tryfonidou M.A., Meij B.P. Inflammatory profiles in canine intervertebral disc degeneration. BMC Vet. Res. 2016;12:10. doi: 10.1186/s12917-016-0635-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gruber H.E., Sha W., Brouwer C.R., Steuerwald N., Hoelscher G.L., Hanley E.N., Jr. A novel catechol-O-methyltransferase variant associated with human disc degeneration. Int. J. Med. Sci. 2014;11:748–753. doi: 10.7150/ijms.8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Darwish H., DeLuca H.F. Vitamin D-regulated gene expression. Crit. Rev. Eukaryot. Gene Expr. 1993;3:89–116. [PubMed] [Google Scholar]
- 131.Christakos S., Barletta F., Huening M., Dhawan P., Liu Y., Porta A., Peng X. Vitamin D target proteins: Function and regulation. J. Cell. Biochem. 2003;88:238–244. doi: 10.1002/jcb.10349. [DOI] [PubMed] [Google Scholar]
- 132.Uitterlinden A.G., Fang Y., Bergink A.P., van Meurs J.B., van Leeuwen H.P., Pols H.A. The role of vitamin D receptor gene polymorphisms in bone biology. Mol. Cell. Endocrinol. 2002;197:15–21. doi: 10.1016/S0303-7207(02)00274-5. [DOI] [PubMed] [Google Scholar]
- 133.Xue J., Song Y., Liu H., Liu L., Li T., Gong Q. Vitamin D receptor gene polymorphisms and risk of intervertebral disc degeneration: An updated meta-analysis based on 23 studies. Medicine. 2021;100:e25922. doi: 10.1097/MD.0000000000025922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vieira L.A., Dos Santos A.A., Peluso C., Barbosa C.P., Bianco B., Rodrigues L.M.R. Influence of lifestyle characteristics and VDR polymorphisms as risk factors for intervertebral disc degeneration: A case-control study. Eur. J. Med. Res. 2018;23:11. doi: 10.1186/s40001-018-0309-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vieira L.A., De Marchi P.L., dos Santos A.A., Christofolini D.M., Barbosa C.P., Fonseca F.L., Bianco B., Rodrigues L.M. Analysis of FokI polymorphism of vitamin D receptor gene in intervertebral disc degeneration. Genet. Test. Mol. Biomark. 2014;18:625–629. doi: 10.1089/gtmb.2014.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Colombini A., Brayda-Bruno M., Lombardi G., Croiset S.J., Vrech V., Maione V., Banfi G., Cauci S. FokI polymorphism in the vitamin D receptor gene (VDR) and its association with lumbar spine pathologies in the Italian population: A case-control study. PLoS ONE. 2014;9:e97027. doi: 10.1371/journal.pone.0097027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Uitterlinden A.G., Fang Y., Van Meurs J.B., Pols H.A., Van Leeuwen J.P. Genetics and biology of vitamin D receptor polymorphisms. Gene. 2004;338:143–156. doi: 10.1016/j.gene.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 138.Videman T., Leppavuori J., Kaprio J., Battie M.C., Gibbons L.E., Peltonen L., Koskenvuo M. Intragenic polymorphisms of the vitamin D receptor gene associated with intervertebral disc degeneration. Spine. 1998;23:2477–2485. doi: 10.1097/00007632-199812010-00002. [DOI] [PubMed] [Google Scholar]
- 139.Miyamoto K., Kesterson R.A., Yamamoto H., Taketani Y., Nishiwaki E., Tatsumi S., Inoue Y., Morita K., Takeda E., Pike J.W. Structural organization of the human vitamin D receptor chromosomal gene and its promoter. Mol. Endocrinol. 1997;11:1165–1179. doi: 10.1210/mend.11.8.9951. [DOI] [PubMed] [Google Scholar]
- 140.Cheung K.M., Chan D., Karppinen J., Chen Y., Jim J.J., Yip S.P., Ott J., Wong K.K., Sham P., Luk K.D., et al. Association of the Taq I allele in vitamin D receptor with degenerative disc disease and disc bulge in a Chinese population. Spine. 2006;31:1143–1148. doi: 10.1097/01.brs.0000216530.41838.d3. [DOI] [PubMed] [Google Scholar]
- 141.Kawaguchi Y., Kanamori M., Ishihara H., Ohmori K., Matsui H., Kimura T. The association of lumbar disc disease with vitamin-D receptor gene polymorphism. J. Bone Jt. Surg. Am. 2002;84:2022–2028. doi: 10.2106/00004623-200211000-00018. [DOI] [PubMed] [Google Scholar]
- 142.Ding F., Shao Z.W., Xiong L.M. Cell death in intervertebral disc degeneration. Apoptosis. 2013;18:777–785. doi: 10.1007/s10495-013-0839-1. [DOI] [PubMed] [Google Scholar]
- 143.Kadow T., Sowa G., Vo N., Kang J.D. Molecular basis of intervertebral disc degeneration and herniations: What are the important translational questions? Clin. Orthop. Relat. Res. 2015;473:1903–1912. doi: 10.1007/s11999-014-3774-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kepler C.K., Ponnappan R.K., Tannoury C.A., Risbud M.V., Anderson D.G. The molecular basis of intervertebral disc degeneration. Spine J. 2013;13:318–330. doi: 10.1016/j.spinee.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 145.Purmessur D., Walter B.A., Roughley P.J., Laudier D.M., Hecht A.C., Iatridis J. A role for TNFalpha in intervertebral disc degeneration: A non-recoverable catabolic shift. Biochem. Biophys. Res. Commun. 2013;433:151–156. doi: 10.1016/j.bbrc.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ohnishi T., Yamada K., Iwasaki K., Tsujimoto T., Higashi H., Kimura T., Iwasaki N., Sudo H. Caspase-3 knockout inhibits intervertebral disc degeneration related to injury but accelerates degeneration related to aging. Sci. Rep. 2019;9:19324. doi: 10.1038/s41598-019-55709-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mu J., Ge W., Zuo X., Chen Y., Huang C. Analysis of association between IL-1beta, CASP-9, and GDF5 variants and low-back pain in Chinese male soldier: Clinical article. J. Neurosurg. Spine. 2013;19:243–247. doi: 10.3171/2013.4.SPINE12782. [DOI] [PubMed] [Google Scholar]
- 148.Martirosyan N.L., Patel A.A., Carotenuto A., Kalani M.Y., Belykh E., Walker C.T., Preul M.C., Theodore N. Genetic Alterations in Intervertebral Disc Disease. Front. Surg. 2016;3:59. doi: 10.3389/fsurg.2016.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xu S., Liang T., Li S. Correlation between polymorphism of TRAIL gene and condition of intervertebral disc degeneration. Med. Sci. Monit. 2015;21:2282–2287. doi: 10.12659/MSM.894157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tan H., Zhao J., Jiang J., Ren Y. Association of the polymorphism of DR4 with the risk and severity of lumbar disc degeneration in the Chinese Han population. Scand. J. Clin. Lab. Investig. 2012;72:576–579. doi: 10.3109/00365513.2012.713176. [DOI] [PubMed] [Google Scholar]
- 151.Paglia D.N., Singh H., Karukonda T., Drissi H., Moss I.L. PDGF-BB delays degeneration of the intervertebral discs in a rabbit preclinical model. Spine. 2016;41:E449–E458. doi: 10.1097/BRS.0000000000001336. [DOI] [PubMed] [Google Scholar]
- 152.Zhang W., Gong Y., Zheng X., Qiu J., Jiang T., Chen L., Lu F., Wu X., Cheng F., Hong Z. Platelet-derived growth factor-BB inhibits intervertebral disc degeneration via suppressing pyroptosis and activating the MAPK signaling pathway. Front. Pharmacol. 2021;12:799130. doi: 10.3389/fphar.2021.799130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Presciutti S.M., Paglia D.N., Karukonda T., Soung Do Y., Guzzo R., Drissi H., Moss I.L. PDGF-BB inhibits intervertebral disc cell apoptosis in vitro. J. Orthop. Res. 2014;32:1181–1188. doi: 10.1002/jor.22638. [DOI] [PubMed] [Google Scholar]
- 154.Takayama B., Sekiguchi M., Yabuki S., Kikuchi S., Konno S. Localization and function of insulin-like growth factor 1 in dorsal root ganglia in a rat disc herniation model. Spine. 2011;36:E75–E79. doi: 10.1097/BRS.0b013e3181d56208. [DOI] [PubMed] [Google Scholar]
- 155.Gruber H.E., Hoelscher G.L., Ingram J.A., Bethea S., Hanley E.N. IGF-1 rescues human intervertebral annulus cells from in vitro stress-induced premature senescence. Growth Factors. 2008;26:220–225. doi: 10.1080/08977190802273814. [DOI] [PubMed] [Google Scholar]
- 156.Luo X.W., Liu K., Chen Z., Zhao M., Han X.W., Bai Y.G., Feng G. Adenovirus-mediated GDF-5 promotes the extracellular matrix expression in degenerative nucleus pulposus cells. J. Zhejiang Univ. Sci. B. 2016;17:30–42. doi: 10.1631/jzus.B1500182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Guo S., Cui L., Xiao C., Wang C., Zhu B., Liu X., Li Y., Liu X., Wang D., Li S. The mechanisms and functions of GDF-5 in intervertebral disc degeneration. Orthop. Surg. 2021;13:734–741. doi: 10.1111/os.12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Feng C., Liu H., Yang Y., Huang B., Zhou Y. Growth and differentiation factor-5 contributes to the structural and functional maintenance of the intervertebral disc. Cell. Physiol. Biochem. 2015;35:1–16. doi: 10.1159/000369670. [DOI] [PubMed] [Google Scholar]
- 159.Yoon S.T., Kim K.S., Li J., Park J.S., Akamaru T., Elmer W.A., Hutton W.C. The effect of bone morphogenetic protein-2 on rat intervertebral disc cells in vitro. Spine. 2003;28:1773–1780. doi: 10.1097/01.BRS.0000083204.44190.34. [DOI] [PubMed] [Google Scholar]
- 160.Kim H., Lee J.U., Moon S.H., Kim H.C., Kwon U.H., Seol N.H., Kim H.J., Park J.O., Chun H.J., Kwon I.K., et al. Zonal responsiveness of the human intervertebral disc to bone morphogenetic protein-2. Spine. 2009;34:1834–1838. doi: 10.1097/BRS.0b013e3181ae18ba. [DOI] [PubMed] [Google Scholar]
- 161.Kim D.J., Moon S.H., Kim H., Kwon U.H., Park M.S., Han K.J., Hahn S.B., Lee H.M. Bone morphogenetic protein-2 facilitates expression of chondrogenic, not osteogenic, phenotype of human intervertebral disc cells. Spine. 2003;28:2679–2684. doi: 10.1097/01.BRS.0000101445.46487.16. [DOI] [PubMed] [Google Scholar]
- 162.Xu J., E X.-Q., Wang N.X., Wang M.N., Xie H.X., Cao Y.H., Sun L.H., Tian J., Chen H.J., Yan J.L. BMP7 enhances the effect of BMSCs on extracellular matrix remodeling in a rabbit model of intervertebral disc degeneration. FEBS J. 2016;283:1689–1700. doi: 10.1111/febs.13695. [DOI] [PubMed] [Google Scholar]
- 163.Wang Z., Fu C., Chen Y., Xu F., Wang Z., Qu Z., Liu Y. FoxC2 enhances BMP7-mediated anabolism in nucleus pulposus cells of the intervertebral disc. PLoS ONE. 2016;11:e0147764. doi: 10.1371/journal.pone.0147764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Han I.B., Ropper A.E., Teng Y.D., Shin D.A., Jeon Y.J., Park H.M., Shin D.E., Park Y.S., Kim K.N., Kim N.K. Association between VEGF and eNOS gene polymorphisms and lumbar disc degeneration in a young Korean population. Genet. Mol. Res. 2013;12:2294–2305. doi: 10.4238/2013.July.8.10. [DOI] [PubMed] [Google Scholar]
- 165.Guo H.Y., Guo M.K., Wan Z.Y., Song F., Wang H.Q. Emerging evidence on noncoding-RNA regulatory machinery in intervertebral disc degeneration: A narrative review. Arthritis Res. Ther. 2020;22:270. doi: 10.1186/s13075-020-02353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Adams M.A., Roughley P.J. What is intervertebral disc degeneration, and what causes it? Spine. 2006;31:2151–2161. doi: 10.1097/01.brs.0000231761.73859.2c. [DOI] [PubMed] [Google Scholar]
- 167.Zhu J., Zhang X., Gao W., Hu H., Wang X., Hao D. lncRNA/circRNAmiRNAmRNA ceRNA network in lumbar intervertebral disc degeneration. Mol. Med. Rep. 2019;20:3160–3174. doi: 10.3892/mmr.2019.10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wan Z.Y., Song F., Sun Z., Chen Y.F., Zhang W.L., Samartzis D., Ma C.J., Che L., Liu X., Ali M.A., et al. Aberrantly expressed long noncoding RNAs in human intervertebral disc degeneration: A microarray related study. Arthritis Res. Ther. 2014;16:465. doi: 10.1186/s13075-014-0465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chen Y., Ni H., Zhao Y., Chen K., Li M., Li C., Zhu X., Fu Q. Potential Role of lncRNAs in Contributing to Pathogenesis of Intervertebral Disc Degeneration Based on Microarray Data. Med. Sci. Monit. 2015;21:3449–3458. doi: 10.12659/MSM.894638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang C., Wang W.J., Yan Y.G., Xiang Y.X., Zhang J., Tang Z.H., Jiang Z.S. MicroRNAs: New players in intervertebral disc degeneration. Clin. Chim. Acta. 2015;450:333–341. doi: 10.1016/j.cca.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 171.Wang S., Sun J., Yang H., Zou W., Zheng B., Chen Y., Guo Y., Shi J. Profiling and bioinformatics analysis of differentially expressed circular RNAs in human intervertebral disc degeneration. Acta Biochim. Biophys. Sin. 2019;51:571–579. doi: 10.1093/abbs/gmz036. [DOI] [PubMed] [Google Scholar]
- 172.Ma X., Lin Y., Yang K., Yue B., Xiang H., Chen B. Effect of lentivirus-mediated survivin transfection on the morphology and apoptosis of nucleus pulposus cells derived from degenerative human disc in vitro. Int. J. Mol. Med. 2015;36:186–194. doi: 10.3892/ijmm.2015.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Le Maitre C.L., Pockert A., Buttle D.J., Freemont A.J., Hoyland J.A. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem. Soc. Trans. 2007;35:652–655. doi: 10.1042/BST0350652. [DOI] [PubMed] [Google Scholar]
- 174.Liu H., Huang X., Liu X., Xiao S., Zhang Y., Xiang T., Shen X., Wang G., Sheng B. miR-21 promotes human nucleus pulposus cell proliferation through PTEN/AKT signaling. Int. J. Mol. Sci. 2014;15:4007–4018. doi: 10.3390/ijms15034007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang H.Q., Yu X.D., Liu Z.H., Cheng X., Samartzis D., Jia L.T., Wu S.X., Huang J., Chen J., Luo Z.J. Deregulated miR-155 promotes Fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J. Pathol. 2011;225:232–242. doi: 10.1002/path.2931. [DOI] [PubMed] [Google Scholar]
- 176.Zhao B., Yu Q., Li H., Guo X., He X. Characterization of microRNA expression profiles in patients with intervertebral disc degeneration. Int. J. Mol. Med. 2014;33:43–50. doi: 10.3892/ijmm.2013.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Liu G., Cao P., Chen H., Yuan W., Wang J., Tang X. MiR-27a regulates apoptosis in nucleus pulposus cells by targeting PI3K. PLoS ONE. 2013;8:e75251. doi: 10.1371/journal.pone.0075251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kang L., Yang C., Song Y., Zhao K., Liu W., Hua W., Wang K., Tu J., Li S., Yin H., et al. MicroRNA-494 promotes apoptosis and extracellular matrix degradation in degenerative human nucleus pulposus cells. Oncotarget. 2017;8:27868–27881. doi: 10.18632/oncotarget.15838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang T., Li P., Ma X., Tian P., Han C., Zang J., Kong J., Yan H. MicroRNA-494 inhibition protects nucleus pulposus cells from TNF-alpha-induced apoptosis by targeting JunD. Biochimie. 2015;115:1–7. doi: 10.1016/j.biochi.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 180.Lv J., Li S., Wan T., Yang Y., Cheng Y., Xue R. Inhibition of microRNA-30d attenuates the apoptosis and extracellular matrix degradation of degenerative human nucleus pulposus cells by up-regulating SOX9. Chem. Biol. Interact. 2018;296:89–97. doi: 10.1016/j.cbi.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 181.Liu J., Yu J., Jiang W., He M., Zhao J. Targeting of CDKN1B by miR-222-3p may contribute to the development of intervertebral disc degeneration. FEBS Open Bio. 2019;9:728–735. doi: 10.1002/2211-5463.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cai P., Yang T., Jiang X., Zheng M., Xu G., Xia J. Role of miR-15a in intervertebral disc degeneration through targeting MAP3K9. Biomed. Pharmacother. 2017;87:568–574. doi: 10.1016/j.biopha.2016.12.128. [DOI] [PubMed] [Google Scholar]
- 183.Zhao K., Zhang Y., Kang L., Song Y., Wang K., Li S., Wu X., Hua W., Shao Z., Yang S., et al. Epigenetic silencing of miRNA-143 regulates apoptosis by targeting BCL2 in human intervertebral disc degeneration. Gene. 2017;628:259–266. doi: 10.1016/j.gene.2017.07.043. [DOI] [PubMed] [Google Scholar]
- 184.Sun Z., Jian Y., Fu H., Li B. MiR-532 downregulation of the Wnt/beta-catenin signaling via targeting Bcl-9 and induced human intervertebral disc nucleus pulposus cells apoptosis. J. Pharmacol. Sci. 2018;138:263–270. doi: 10.1016/j.jphs.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 185.Wang B., Wang D., Yan T., Yuan H. MiR-138-5p promotes TNF-alpha-induced apoptosis in human intervertebral disc degeneration by targeting SIRT1 through PTEN/PI3K/Akt signaling. Exp. Cell Res. 2016;345:199–205. doi: 10.1016/j.yexcr.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 186.Hai B., Ma Y., Pan X., Yong L., Liang C., He G., Yang C., Zhu B., Liu X. Melatonin benefits to the growth of human annulus fibrosus cells through inhibiting miR-106a-5p/ATG7 signaling pathway. Clin. Interv. Aging. 2019;14:621–630. doi: 10.2147/CIA.S193765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chen H., Wang J., Hu B., Wu X., Chen Y., Li R., Yuan W. MiR-34a promotes Fas-mediated cartilage endplate chondrocyte apoptosis by targeting Bcl-2. Mol. Cell. Biochem. 2015;406:21–30. doi: 10.1007/s11010-015-2420-4. [DOI] [PubMed] [Google Scholar]
- 188.Sheng B., Yuan Y., Liu X., Zhang Y., Liu H., Shen X., Liu B., Chang L. Protective effect of estrogen against intervertebral disc degeneration is attenuated by miR-221 through targeting estrogen receptor alpha. Acta Biochim. Biophys. Sin. 2018;50:345–354. doi: 10.1093/abbs/gmy017. [DOI] [PubMed] [Google Scholar]
- 189.Cheng X., Zhang G., Zhang L., Hu Y., Zhang K., Sun X., Zhao C., Li H., Li Y.M., Zhao J. Mesenchymal stem cells deliver exogenous miR-21 via exosomes to inhibit nucleus pulposus cell apoptosis and reduce intervertebral disc degeneration. J. Cell. Mol. Med. 2018;22:261–276. doi: 10.1111/jcmm.13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sun J.C., Zheng B., Sun R.X., Meng Y.K., Wang S.M., Yang H.S., Chen Y., Shi J.G., Guo Y.F. MiR-499a-5p suppresses apoptosis of human nucleus pulposus cells and degradation of their extracellular matrix by targeting SOX4. Biomed. Pharmacother. 2019;113:108652. doi: 10.1016/j.biopha.2019.108652. [DOI] [PubMed] [Google Scholar]
- 191.Chai X., Si H., Song J., Chong Y., Wang J., Zhao G. miR-486-5p inhibits inflammatory response, matrix degradation and apoptosis of nucleus pulposus cells through directly targeting FOXO1 in intervertebral disc degeneration. Cell. Physiol. Biochem. 2019;52:109–118. doi: 10.33594/000000008. [DOI] [PubMed] [Google Scholar]
- 192.Ma J.F., Zang L.N., Xi Y.M., Yang W.J., Zou D. MiR-125a Rs12976445 polymorphism is associated with the apoptosis status of nucleus pulposus cells and the risk of intervertebral disc degeneration. Cell. Physiol. Biochem. 2016;38:295–305. doi: 10.1159/000438630. [DOI] [PubMed] [Google Scholar]
- 193.Zhou J., Sun J., Markova D.Z., Li S., Kepler C.K., Hong J., Huang Y., Chen W., Xu K., Wei F., et al. MicroRNA-145 overexpression attenuates apoptosis and increases matrix synthesis in nucleus pulposus cells. Life Sci. 2019;221:274–283. doi: 10.1016/j.lfs.2019.02.041. [DOI] [PubMed] [Google Scholar]
- 194.Wang R., Wen B., Sun D. miR-573 regulates cell proliferation and apoptosis by targeting Bax in nucleus pulposus cells. Cell. Mol. Biol. Lett. 2019;24:2. doi: 10.1186/s11658-018-0132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Du X., Chen S., Cui H., Huang Y., Wang J., Liu H., Li Z., Liang C., Zheng Z., Wang H. Circular RNA hsa_circ_0083756 promotes intervertebral disc degeneration by sponging miR-558 and regulating TREM1 expression. Cell Prolif. 2022;55:e13205. doi: 10.1111/cpr.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Du K., He X., Deng J. MicroRNA-16 inhibits the lipopolysaccharide-induced inflammatory response in nucleus pulposus cells of the intervertebral disc by targeting TAB3. Arch. Med. Sci. 2021;17:500–513. doi: 10.5114/aoms.2018.74950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kong L., Sun M., Jiang Z., Li L., Lu B. MicroRNA-194 Inhibits Lipopolysaccharide-Induced Inflammatory Response in Nucleus Pulposus Cells of the Intervertebral Disc by Targeting TNF Receptor-Associated Factor 6 (TRAF6) Med. Sci. Monit. 2018;24:3056–3067. doi: 10.12659/MSM.907280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang H., Hao P., Zhang H., Xu C., Zhao J. MicroRNA-223 inhibits lipopolysaccharide-induced inflammatory response by directly targeting Irak1 in the nucleus pulposus cells of intervertebral disc. IUBMB Life. 2018;70:479–490. doi: 10.1002/iub.1747. [DOI] [PubMed] [Google Scholar]
- 199.Cheng X., Zhang L., Zhang K., Zhang G., Hu Y., Sun X., Zhao C., Li H., Li Y.M., Zhao J. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann. Rheum. Dis. 2018;77:770–779. doi: 10.1136/annrheumdis-2017-212056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Guo W., Zhang B., Mu K., Feng S.Q., Dong Z.Y., Ning G.Z., Li H.R., Liu S., Zhao L., Li Y., et al. Circular RNA GRB10 as a competitive endogenous RNA regulating nucleus pulposus cells death in degenerative intervertebral disk. Cell Death Dis. 2018;9:319. doi: 10.1038/s41419-017-0232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang H., He P., Pan H., Long J., Wang J., Li Z., Liu H., Jiang W., Zheng Z. Circular RNA circ-4099 is induced by TNF-alpha and regulates ECM synthesis by blocking miR-616-5p inhibition of Sox9 in intervertebral disc degeneration. Exp. Mol. Med. 2018;50:1–14. doi: 10.1038/s12276-018-0056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sharma A., Sargar K., Salter A. Temporal Evolution of Disc in Young Patients with Low Back Pain and Stress Reaction in Lumbar Vertebrae. AJNR Am. J. Neuroradiol. 2017;38:1647–1652. doi: 10.3174/ajnr.A5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gerhardt J., Bette S., Janssen I., Gempt J., Meyer B., Ryang Y.M. Is eighty the new sixty? Outcomes and complications after lumbar decompression surgery in elderly patients over 80 years of age. World Neurosurg. 2018;112:e555–e560. doi: 10.1016/j.wneu.2018.01.082. [DOI] [PubMed] [Google Scholar]
- 204.Wu P.H., Kim H.S., Jang I.T. Intervertebral disc diseases Part 2: A review of the current diagnostic and treatment strategies for intervertebral disc disease. Int. J. Mol. Sci. 2020;21:2135. doi: 10.3390/ijms21062135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Eisenstein S.M., Balain B., Roberts S. Current treatment options for intervertebral disc pathologies. Cartilage. 2020;11:143–151. doi: 10.1177/1947603520907665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Knezevic N.N., Mandalia S., Raasch J., Knezevic I., Candido K.D. Treatment of chronic low back pain–new approaches on the horizon. J. Pain Res. 2017;10:1111–1123. doi: 10.2147/JPR.S132769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ohnishi T., Iwasaki N., Sudo H. Causes of and molecular targets for the treatment of intervertebral disc degeneration: A review. Cells. 2022;11:394. doi: 10.3390/cells11030394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hestbaek L., Leboeuf-Yde C., Manniche C. Low back pain: What is the long-term course? A review of studies of general patient populations. Eur. Spine J. 2003;12:149–165. doi: 10.1007/s00586-002-0508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ruetten S., Komp M., Merk H., Godolias G. Full-endoscopic interlaminar and transforaminal lumbar discectomy versus conventional microsurgical technique: A prospective, randomized, controlled study. Spine. 2008;33:931–939. doi: 10.1097/BRS.0b013e31816c8af7. [DOI] [PubMed] [Google Scholar]
- 210.Atlas S.J., Keller R.B., Wu Y.A., Deyo R.A., Singer D.E. Long-term outcomes of surgical and nonsurgical management of lumbar spinal stenosis: 8 to 10 year results from the maine lumbar spine study. Spine. 2005;30:936–943. doi: 10.1097/01.brs.0000158953.57966.c0. [DOI] [PubMed] [Google Scholar]
- 211.Lee C.S., Hwang C.J., Lee D.H., Kim Y.T., Lee H.S. Fusion rates of instrumented lumbar spinal arthrodesis according to surgical approach: A systematic review of randomized trials. Clin. Orthop. Surg. 2011;3:39–47. doi: 10.4055/cios.2011.3.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Boden S.D., Kang J., Sandhu H., Heller J.G. Use of recombinant human bone morphogenetic protein-2 to achieve posterolateral lumbar spine fusion in humans: A prospective, randomized clinical pilot trial: 2002 Volvo Award in clinical studies. Spine. 2002;27:2662–2673. doi: 10.1097/00007632-200212010-00005. [DOI] [PubMed] [Google Scholar]
- 213.Zigler J.E., Delamarter R.B. Five-year results of the prospective, randomized, multicenter, Food and Drug Administration investigational device exemption study of the ProDisc-L total disc replacement versus circumferential arthrodesis for the treatment of single-level degenerative disc disease. J. Neurosurg. Spine. 2012;17:493–501. doi: 10.3171/2012.9.SPINE11498. [DOI] [PubMed] [Google Scholar]
- 214.Kong L., Ma Q., Meng F., Cao J., Yu K., Shen Y. The prevalence of heterotopic ossification among patients after cervical artificial disc replacement: A systematic review and meta-analysis. Medicine. 2017;96:e7163. doi: 10.1097/MD.0000000000007163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Roussouly P., Nnadi C. Sagittal plane deformity: An overview of interpretation and management. Eur. Spine J. 2010;19:1824–1836. doi: 10.1007/s00586-010-1476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Worley N., Marascalchi B., Jalai C.M., Yang S., Diebo B., Vira S., Boniello A., Lafage V., Passias P.G. Predictors of inpatient morbidity and mortality in adult spinal deformity surgery. Eur. Spine J. 2016;25:819–827. doi: 10.1007/s00586-015-4104-x. [DOI] [PubMed] [Google Scholar]
- 217.Han I., Ropper A.E., Konya D., Kabatas S., Toktas Z., Aljuboori Z., Zeng X., Chi J.H., Zafonte R., Teng Y.D. Biological approaches to treating intervertebral disk degeneration: Devising stem cell therapies. Cell Transplant. 2015;24:2197–2208. doi: 10.3727/096368915X688650. [DOI] [PubMed] [Google Scholar]
- 218.Dowdell J., Erwin M., Choma T., Vaccaro A., Iatridis J., Cho S.K. Intervertebral disk degeneration and repair. Neurosurgery. 2017;80:S46–S54. doi: 10.1093/neuros/nyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Li W., Liu T., Wu L., Chen C., Jia Z., Bai X., Ruan D. Blocking the function of inflammatory cytokines and mediators by using IL-10 and TGF-beta: A potential biological immunotherapy for intervertebral disc degeneration in a beagle model. Int. J. Mol. Sci. 2014;15:17270–17283. doi: 10.3390/ijms151017270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Tsujimoto T., Sudo H., Todoh M., Yamada K., Iwasaki K., Ohnishi T., Hirohama N., Nonoyama T., Ukeba D., Ura K., et al. An acellular bioresorbable ultra-purified alginate gel promotes intervertebral disc repair: A preclinical proof-of-concept study. EBioMedicine. 2018;37:521–534. doi: 10.1016/j.ebiom.2018.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ukeba D., Sudo H., Tsujimoto T., Ura K., Yamada K., Iwasaki N. Bone marrow mesenchymal stem cells combined with ultra-purified alginate gel as a regenerative therapeutic strategy after discectomy for degenerated intervertebral discs. EBioMedicine. 2020;53:102698. doi: 10.1016/j.ebiom.2020.102698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Shin E.H., Cho K.J., Kim Y.T., Park M.H. Risk factors for recurrent lumbar disc herniation after discectomy. Int. Orthop. 2019;43:963–967. doi: 10.1007/s00264-018-4201-7. [DOI] [PubMed] [Google Scholar]
- 223.Phan K., Nazareth A., Hussain A.K., Dmytriw A.A., Nambiar M., Nguyen D., Kerferd J., Phan S., Sutterlin C., 3rd, Cho S.K., et al. Relationship between sagittal balance and adjacent segment disease in surgical treatment of degenerative lumbar spine disease: Meta-analysis and implications for choice of fusion technique. Eur. Spine J. 2018;27:1981–1991. doi: 10.1007/s00586-018-5629-6. [DOI] [PubMed] [Google Scholar]
- 224.Takahashi J., Shono Y., Hirabayashi H., Kamimura M., Nakagawa H., Ebara S., Kato H. Usefulness of white blood cell differential for early diagnosis of surgical wound infection following spinal instrumentation surgery. Spine. 2006;31:1020–1025. doi: 10.1097/01.brs.0000214895.67956.60. [DOI] [PubMed] [Google Scholar]
- 225.Kennon J.C., Awad M.E., Chutkan N., DeVine J., Fulzele S. Current insights on use of growth factors as therapy for Intervertebral Disc Degeneration. Biomol. Concepts. 2018;9:43–52. doi: 10.1515/bmc-2018-0003. [DOI] [PubMed] [Google Scholar]
- 226.Zhu J., Xia K., Yu W., Wang Y., Hua J., Liu B., Gong Z., Wang J., Xu A., You Z., et al. Sustained release of GDF5 from a designed coacervate attenuates disc degeneration in a rat model. Acta Biomater. 2019;86:300–311. doi: 10.1016/j.actbio.2019.01.028. [DOI] [PubMed] [Google Scholar]
- 227.Cho H., Lee S., Park S.H., Huang J., Hasty K.A., Kim S.J. Synergistic effect of combined growth factors in porcine intervertebral disc degeneration. Connect. Tissue Res. 2013;54:181–186. doi: 10.3109/03008207.2013.775258. [DOI] [PubMed] [Google Scholar]
- 228.Masuda K. Biological repair of the degenerated intervertebral disc by the injection of growth factors. Eur. Spine J. 2008;17((Suppl. 4)):441–451. doi: 10.1007/s00586-008-0749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Benneker L.M., Andersson G., Iatridis J.C., Sakai D., Hartl R., Ito K., Grad S. Cell therapy for intervertebral disc repair: Advancing cell therapy from bench to clinics. Eur. Cells Mater. 2014;27:5–11. doi: 10.22203/eCM.v027sa02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Jandial R., Aryan H.E., Park J., Taylor W.T., Snyder E.Y. Stem cell-mediated regeneration of the intervertebral disc: Cellular and molecular challenge. Neurosurg. Focus. 2008;24:E21. doi: 10.3171/FOC/2008/24/3-4/E20. [DOI] [PubMed] [Google Scholar]
- 231.Choi Y., Park M.H., Lee K. Tissue engineering strategies for intervertebral disc treatment using functional polymers. Polymers. 2019;11:872. doi: 10.3390/polym11050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Moriguchi Y., Mojica-Santiago J., Grunert P., Pennicooke B., Berlin C., Khair T., Navarro-Ramirez R., Ricart Arbona R.J., Nguyen J., Hartl R., et al. Total disc replacement using tissue-engineered intervertebral discs in the canine cervical spine. PLoS ONE. 2017;12:e0185716. doi: 10.1371/journal.pone.0185716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sobajima S., Kim J.S., Gilbertson L.G., Kang J.D. Gene therapy for degenerative disc disease. Gene Ther. 2004;11:390–401. doi: 10.1038/sj.gt.3302200. [DOI] [PubMed] [Google Scholar]
- 234.Krupkova O., Cambria E., Besse L., Besse A., Bowles R., Wuertz-Kozak K. The potential of CRISPR/Cas9 genome editing for the study and treatment of intervertebral disc pathologies. JOR Spine. 2018;1:e1003. doi: 10.1002/jsp2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Yurube T., Ito M., Kakiuchi Y., Kuroda R., Kakutani K. Autophagy and mTOR signaling during intervertebral disc aging and degeneration. JOR Spine. 2020;3:e1082. doi: 10.1002/jsp2.1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ju D.G., Kanim L.E., Bae H.W. Intervertebral disc repair: Current concepts. Glob. Spine J. 2020;10:130S–136S. doi: 10.1177/2192568219872460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yamada K., Iwasaki N., Sudo H. Biomaterials and cell-based regenerative therapies for intervertebral disc degeneration with a focus on biological and biomechanical functional repair: Targeting treatments for disc herniation. Cells. 2022;11:602. doi: 10.3390/cells11040602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhao L., Manchikanti L., Kaye A.D., Abd-Elsayed A. Treatment of Discogenic Low Back Pain: Current Treatment Strategies and Future Options—A Literature Review. Curr. Pain Headache Rep. 2019;23:86. doi: 10.1007/s11916-019-0821-x. [DOI] [PubMed] [Google Scholar]
- 239.Li X., Dou Q., Kong Q. Repair and regenerative therapies of the annulus fibrosus of the intervertebral disc. J. Coll. Physicians Surg. Pak. 2016;26:138–144. [PubMed] [Google Scholar]
- 240.Zhang S.J., Yang W., Wang C., He W.S., Deng H.Y., Yan Y.G., Zhang J., Xiang Y.X., Wang W.J. Autophagy: A double-edged sword in intervertebral disk degeneration. Clin. Chim. Acta. 2016;457:27–35. doi: 10.1016/j.cca.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 241.Chen J.W., Ni B.B., Li B., Yang Y.H., Jiang S.D., Jiang L.S. The responses of autophagy and apoptosis to oxidative stress in nucleus pulposus cells: Implications for disc degeneration. Cell. Physiol. Biochem. 2014;34:1175–1189. doi: 10.1159/000366330. [DOI] [PubMed] [Google Scholar]
- 242.Chen D., Xia D., Pan Z., Xu D., Zhou Y., Wu Y., Cai N., Tang Q., Wang C., Yan M., et al. Metformin protects against apoptosis and senescence in nucleus pulposus cells and ameliorates disc degeneration in vivo. Cell Death Dis. 2016;7:e2441. doi: 10.1038/cddis.2016.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ouyang Z.H., Wang W.J., Yan Y.G., Wang B., Lv G.H. The PI3K/Akt pathway: A critical player in intervertebral disc degeneration. Oncotarget. 2017;8:57870–57881. doi: 10.18632/oncotarget.18628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.He R., Wang Z., Cui M., Liu S., Wu W., Chen M., Wu Y., Qu Y., Lin H., Chen S., et al. HIF1A Alleviates compression-induced apoptosis of nucleus pulposus derived stem cells via upregulating autophagy. Autophagy. 2021;17:3338–3360. doi: 10.1080/15548627.2021.1872227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Chen J., Lin Z., Deng K., Shao B., Yang D. Tension induces intervertebral disc degeneration via endoplasmic reticulum stress-mediated autophagy. Biosci. Rep. 2019;39:BSR20190578. doi: 10.1042/BSR20190578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Bahar E., Han S.Y., Kim J.Y., Yoon H. Chemotherapy resistance: Role of mitochondrial and autophagic components. Cancers. 2022;14:1462. doi: 10.3390/cancers14061462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kawamata T., Kamada Y., Kabeya Y., Sekito T., Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell. 2008;19:2039–2050. doi: 10.1091/mbc.e07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Zachari M., Ganley I.G. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017;61:585–596. doi: 10.1042/EBC20170021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.e08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Pavel M., Rubinsztein D.C. Mammalian autophagy and the plasma membrane. FEBS J. 2017;284:672–679. doi: 10.1111/febs.13931. [DOI] [PubMed] [Google Scholar]
- 251.Hanada T., Noda N.N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 252.Gruber H.E., Hoelscher G.L., Ingram J.A., Bethea S., Hanley E.N., Jr. Autophagy in the degenerating human intervertebral disc: In vivo molecular and morphological evidence, and induction of autophagy in cultured annulus cells exposed to proinflammatory cytokines-implications for disc degeneration. Spine. 2015;40:773–782. doi: 10.1097/BRS.0000000000000865. [DOI] [PubMed] [Google Scholar]
- 253.Zhang T.W., Li Z.F., Dong J., Jiang L.B. The circadian rhythm in intervertebral disc degeneration: An autophagy connection. Exp. Mol. Med. 2020;52:31–40. doi: 10.1038/s12276-019-0372-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Gong C.Y., Zhang H.H. Autophagy as a potential therapeutic target in intervertebral disc degeneration. Life Sci. 2021;273:119266. doi: 10.1016/j.lfs.2021.119266. [DOI] [PubMed] [Google Scholar]
- 255.Zhang F., Zhao X., Shen H., Zhang C. Molecular mechanisms of cell death in intervertebral disc degeneration (Review) Int. J. Mol. Med. 2016;37:1439–1448. doi: 10.3892/ijmm.2016.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Kang L., Xiang Q., Zhan S., Song Y., Wang K., Zhao K., Li S., Shao Z., Yang C., Zhang Y. Restoration of Autophagic Flux Rescues Oxidative Damage and Mitochondrial Dysfunction to Protect against Intervertebral Disc Degeneration. Oxid. Med. Cell. Longev. 2019;2019:7810320. doi: 10.1155/2019/7810320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Yang Z., Klionsky D.J. Eaten alive: A history of macroautophagy. Nat. Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018;20:521–527. doi: 10.1038/s41556-018-0092-5. [DOI] [PubMed] [Google Scholar]
- 259.Jiang W., Zhang X., Hao J., Shen J., Fang J., Dong W., Wang D., Zhang X., Shui W., Luo Y., et al. SIRT1 protects against apoptosis by promoting autophagy in degenerative human disc nucleus pulposus cells. Sci. Rep. 2014;4:7456. doi: 10.1038/srep07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Matsui M., Corey D.R. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 2017;16:167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ning B., Yu D., Yu A.M. Advances and challenges in studying noncoding RNA regulation of drug metabolism and development of RNA therapeutics. Biochem. Pharmacol. 2019;169:113638. doi: 10.1016/j.bcp.2019.113638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Hasvik E., Schjolberg T., Jacobsen D.P., Haugen A.J., Grovle L., Schistad E.I., Gjerstad J. Up-regulation of circulating microRNA-17 is associated with lumbar radicular pain following disc herniation. Arthritis Res. Ther. 2019;21:186. doi: 10.1186/s13075-019-1967-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Moen A., Jacobsen D., Phuyal S., Legfeldt A., Haugen F., Roe C., Gjerstad J. MicroRNA-223 demonstrated experimentally in exosome-like vesicles is associated with decreased risk of persistent pain after lumbar disc herniation. J. Transl. Med. 2017;15:89. doi: 10.1186/s12967-017-1194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Huang H., Xing D., Zhang Q., Li H., Lin J., He Z., Lin J. LncRNAs as a new regulator of chronic musculoskeletal disorder. Cell Prolif. 2021;54:e13113. doi: 10.1111/cpr.13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Li H., Tian L., Li J., Li Y., Du L., Huo Z., Xu B. The Roles of circRNAs in Intervertebral Disc Degeneration: Inflammation, Extracellular Matrix Metabolism, and Apoptosis. Anal. Cell. Pathol. 2022;2022:9550499. doi: 10.1155/2022/9550499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Farazi T.A., Spitzer J.I., Morozov P., Tuschl T. miRNAs in human cancer. J. Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Mo Y.Y. MicroRNA regulatory networks and human disease. Cell Mol. Life Sci. 2012;69:3529–3531. doi: 10.1007/s00018-012-1123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Duchaine T.F., Fabian M.R. Mechanistic insights into MicroRNA-mediated gene silencing. Cold Spring Harb. Perspect. Biol. 2019;11:a032771. doi: 10.1101/cshperspect.a032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ferland-McCollough D., Ozanne S.E., Siddle K., Willis A.E., Bushell M. The involvement of microRNAs in Type 2 diabetes. Biochem. Soc. Trans. 2010;38:1565–1570. doi: 10.1042/BST0381565. [DOI] [PubMed] [Google Scholar]
- 271.Yang W.L., Wang J.P., Cui Q.H., Yang J.C. Noncoding RNA in the Regulation of Hepatic Glucose and Lipid Metabolism. Sheng Li Ke Xue Jin Zhan. 2016;47:194–202. [PubMed] [Google Scholar]
- 272.Jiao X., Zhao L., Ma M., Bai X., He M., Yan Y., Wang Y., Chen Q., Zhao X., Zhou M., et al. MiR-181a enhances drug sensitivity in mitoxantone-resistant breast cancer cells by targeting breast cancer resistance protein (BCRP/ABCG2) Breast Cancer Res. Treat. 2013;139:717–730. doi: 10.1007/s10549-013-2607-x. [DOI] [PubMed] [Google Scholar]
- 273.Wang Z., Wang N., Liu P., Chen Q., Situ H., Xie T., Zhang J., Peng C., Lin Y., Chen J. MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget. 2014;5:7013–7026. doi: 10.18632/oncotarget.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Huang J., Zhang S.Y., Gao Y.M., Liu Y.F., Liu Y.B., Zhao Z.G., Yang K. MicroRNAs as oncogenes or tumour suppressors in oesophageal cancer: Potential biomarkers and therapeutic targets. Cell Prolif. 2014;47:277–286. doi: 10.1111/cpr.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kestens C., Siersema P.D., van Baal J.W. Current understanding of the functional roles of aberrantly expressed microRNAs in esophageal cancer. World J. Gastroenterol. 2016;22:1–7. doi: 10.3748/wjg.v22.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Li M., Yu M., Liu C., Zhu H., He X., Peng S., Hua J. miR-34c works downstream of p53 leading to dairy goat male germline stem-cell (mGSCs) apoptosis. Cell Prolif. 2013;46:223–231. doi: 10.1111/cpr.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Li W., Wang P., Zhang Z., Wang W., Liu Y., Qi Q. MiR-184 Regulates Proliferation in Nucleus Pulposus Cells by Targeting GAS1. World Neurosurg. 2017;97:710–715.e711. doi: 10.1016/j.wneu.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 278.Huang Z., Wu S., Kong F., Cai X., Ye B., Shan P., Huang W. MicroRNA-21 protects against cardiac hypoxia/reoxygenation injury by inhibiting excessive autophagy in H9c2 cells via the Akt/mTOR pathway. J. Cell. Mol. Med. 2017;21:467–474. doi: 10.1111/jcmm.12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.He C., Dong X., Zhai B., Jiang X., Dong D., Li B., Jiang H., Xu S., Sun X. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget. 2015;6:28867–28881. doi: 10.18632/oncotarget.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zhu H., Yan X., Zhang M., Ji F., Wang S. miR-21-5p protects IL-1beta-induced human chondrocytes from degradation. J. Orthop. Surg. Res. 2019;14:118. doi: 10.1186/s13018-019-1160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Yu X., Li Z., Shen J., Wu W.K., Liang J., Weng X., Qiu G. MicroRNA-10b promotes nucleus pulposus cell proliferation through RhoC-Akt pathway by targeting HOXD10 in intervetebral disc degeneration. PLoS ONE. 2013;8:e83080. doi: 10.1371/journal.pone.0083080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 282.Wang W.J., Yang W., Ouyang Z.H., Xue J.B., Li X.L., Zhang J., He W.S., Chen W.K., Yan Y.G., Wang C. MiR-21 promotes ECM degradation through inhibiting autophagy via the PTEN/akt/mTOR signaling pathway in human degenerated NP cells. Biomed. Pharmacother. 2018;99:725–734. doi: 10.1016/j.biopha.2018.01.154. [DOI] [PubMed] [Google Scholar]
- 283.Zhao K., Zhang Y., Kang L., Song Y., Wang K., Li S., Wu X., Hua W., Shao Z., Yang S., et al. Methylation of microRNA-129-5P modulates nucleus pulposus cell autophagy by targeting Beclin-1 in intervertebral disc degeneration. Oncotarget. 2017;8:86264–86276. doi: 10.18632/oncotarget.21137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Yun Z., Wang Y., Feng W., Zang J., Zhang D., Gao Y. Overexpression of microRNA-185 alleviates intervertebral disc degeneration through inactivation of the Wnt/beta-catenin signaling pathway and downregulation of Galectin-3. Mol. Pain. 2020;16:1744806920902559. doi: 10.1177/1744806920902559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Wang C., Zhang Z.Z., Yang W., Ouyang Z.H., Xue J.B., Li X.L., Zhang J., Chen W.K., Yan Y.G., Wang W.J. MiR-210 facilitates ECM degradation by suppressing autophagy via silencing of ATG7 in human degenerated NP cells. Biomed. Pharmacother. 2017;93:470–479. doi: 10.1016/j.biopha.2017.06.048. [DOI] [PubMed] [Google Scholar]
- 286.Wang S., Guo Y., Zhang X., Wang C. miR6545p inhibits autophagy by targeting ATG7 via mTOR signaling in intervertebral disc degeneration. Mol. Med. Rep. 2021;23:444. doi: 10.3892/mmr.2021.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Ji M.L., Zhang X.J., Shi P.L., Lu J., Wang S.Z., Chang Q., Chen H., Wang C. Downregulation of microRNA-193a-3p is involved in invertebral disc degeneration by targeting MMP14. J. Mol. Med. 2016;94:457–468. doi: 10.1007/s00109-015-1371-2. [DOI] [PubMed] [Google Scholar]
- 288.Tao B., Yi J., Huang C., Xu W., Qin C., Chen L., Chen J., Gao Y., Wang R. microRNA96 regulates the proliferation of nucleus pulposus cells by targeting ARID2/AKT signaling. Mol. Med. Rep. 2017;16:7553–7560. doi: 10.3892/mmr.2017.7560. [DOI] [PubMed] [Google Scholar]
- 289.Chen W.K., Yu X.H., Yang W., Wang C., He W.S., Yan Y.G., Zhang J., Wang W.J. lncRNAs: Novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50:e12313. doi: 10.1111/cpr.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Li Z., Li X., Chen C., Li S., Shen J., Tse G., Chan M.T.V., Wu W.K.K. Long non-coding RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif. 2018;51:e12483. doi: 10.1111/cpr.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Zhang Y.H., Song J., Shen L., Shao J. Systematic identification of lncRNAs and circRNAs-associated ceRNA networks in human lumbar disc degeneration. Biotech. Histochem. 2019;94:606–616. doi: 10.1080/10520295.2019.1622782. [DOI] [PubMed] [Google Scholar]
- 292.Jiang X., Chen D. LncRNA FAM83H-AS1 maintains intervertebral disc tissue homeostasis and attenuates inflammation-related pain via promoting nucleus pulposus cell growth through miR-22-3p inhibition. Ann. Transl. Med. 2020;8:1518. doi: 10.21037/atm-20-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Sun Z., Tang X., Wang H., Sun H., Chu P., Sun L., Tian J. LncRNA H19 aggravates intervertebral disc degeneration by promoting the autophagy and apoptosis of nucleus pulposus cells through the miR-139/CXCR4/NF-kappaB Axis. Stem Cells Dev. 2021;30:736–748. doi: 10.1089/scd.2021.0009. [DOI] [PubMed] [Google Scholar]
- 294.Zhan S., Wang K., Xiang Q., Song Y., Li S., Liang H., Luo R., Wang B., Liao Z., Zhang Y., et al. lncRNA HOTAIR upregulates autophagy to promote apoptosis and senescence of nucleus pulposus cells. J. Cell. Physiol. 2020;235:2195–2208. doi: 10.1002/jcp.29129. [DOI] [PubMed] [Google Scholar]
- 295.Zhang S., Song S., Cui W., Liu X., Sun Z. Mechanism of long noncoding RNA HOTAIR in nucleus pulposus cell autophagy and apoptosis in intervertebral disc degeneration. Evid. Based Complement. Altern. Med. 2022;2022:8504601. doi: 10.1155/2022/8504601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Zhan S., Wang K., Song Y., Li S., Yin H., Luo R., Liao Z., Wu X., Zhang Y., Yang C. Long non-coding RNA HOTAIR modulates intervertebral disc degenerative changes via Wnt/beta-catenin pathway. Arthritis Res. Ther. 2019;21:201. doi: 10.1186/s13075-019-1986-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Chen W.K., Zhang H.J., Zou M.X., Wang C., Yan Y.G., Zhan X.L., Li X.L., Wang W.J. LncRNA HOTAIR influences cell proliferation via miR-130b/PTEN/AKT axis in IDD. Cell Cycle. 2022;21:323–339. doi: 10.1080/15384101.2021.2020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Wang X.B., Wang H., Long H.Q., Li D.Y., Zheng X. LINC00641 regulates autophagy and intervertebral disc degeneration by acting as a competitive endogenous RNA of miR-153-3p under nutrition deprivation stress. J. Cell. Physiol. 2019;234:7115–7127. doi: 10.1002/jcp.27466. [DOI] [PubMed] [Google Scholar]
- 299.Li Z., Chen X., Xu D., Li S., Chan M.T.V., Wu W.K.K. Circular RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif. 2019;52:e12704. doi: 10.1111/cpr.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Zang J., Lu D., Xu A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J. Neurosci. Res. 2020;98:87–97. doi: 10.1002/jnr.24356. [DOI] [PubMed] [Google Scholar]
- 301.Huo Z., Li H., Tian L., Li J., Zhang K., Li Z., Li G., Du L., Xu H., Xu B. Construction of a potentially functional circRNA-miRNA-mRNA network in intervertebral disc degeneration by bioinformatics analysis. Biomed. Res. Int. 2021;2021:8352683. doi: 10.1155/2021/8352683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Wang R., Zhou X., Luo G., Zhang J., Yang M., Song C. CircRNA RERE promotes the oxidative stress-induced apoptosis and autophagy of nucleus pulposus cells through the miR-299-5p/Galectin-3 Axis. J. Healthc. Eng. 2021;2021:2771712. doi: 10.1155/2021/2771712. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 303.Xie L., Huang W., Fang Z., Ding F., Zou F., Ma X., Tao J., Guo J., Xia X., Wang H., et al. CircERCC2 ameliorated intervertebral disc degeneration by regulating mitophagy and apoptosis through miR-182-5p/SIRT1 axis. Cell Death Dis. 2019;10:751. doi: 10.1038/s41419-019-1978-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Xiang Q., Kang L., Wang J., Liao Z., Song Y., Zhao K., Wang K., Yang C., Zhang Y. CircRNA-CIDN mitigated compression loading-induced damage in human nucleus pulposus cells via miR-34a-5p/SIRT1 axis. EBioMedicine. 2020;53:102679. doi: 10.1016/j.ebiom.2020.102679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Ma K., Chen S., Li Z., Deng X., Huang D., Xiong L., Shao Z. Mechanisms of endogenous repair failure during intervertebral disc degeneration. Osteoarthr. Cartil. 2019;27:41–48. doi: 10.1016/j.joca.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 306.van Deursen J.M. The role of senescent cells in ageing. Nature. 2014;509:439–446. doi: 10.1038/nature13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Chen J., Xie J.J., Jin M.Y., Gu Y.T., Wu C.C., Guo W.J., Yan Y.Z., Zhang Z.J., Wang J.L., Zhang X.L., et al. Sirt6 overexpression suppresses senescence and apoptosis of nucleus pulposus cells by inducing autophagy in a model of intervertebral disc degeneration. Cell Death Dis. 2018;9:56. doi: 10.1038/s41419-017-0085-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Ruiz-Fernandez C., Francisco V., Pino J., Mera A., Gonzalez-Gay M.A., Gomez R., Lago F., Gualillo O. Molecular relationships among obesity, inflammation and intervertebral disc degeneration: Are adipokines the common link? Int. J. Mol. Sci. 2019;20:2030. doi: 10.3390/ijms20082030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Xu H.G., Yu Y.F., Zheng Q., Zhang W., Wang C.D., Zhao X.Y., Tong W.X., Wang H., Liu P., Zhang X.L. Autophagy protects end plate chondrocytes from intermittent cyclic mechanical tension induced calcification. Bone. 2014;66:232–239. doi: 10.1016/j.bone.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 310.Wang H., Liu H., Zheng Z.M., Zhang K.B., Wang T.P., Sribastav S.S., Liu W.S., Liu T. Role of death receptor, mitochondrial and endoplasmic reticulum pathways in different stages of degenerative human lumbar disc. Apoptosis. 2011;16:990–1003. doi: 10.1007/s10495-011-0644-7. [DOI] [PubMed] [Google Scholar]
- 311.Zhao C.Q., Zhang Y.H., Jiang S.D., Jiang L.S., Dai L.Y. Both endoplasmic reticulum and mitochondria are involved in disc cell apoptosis and intervertebral disc degeneration in rats. Age. 2010;32:161–177. doi: 10.1007/s11357-009-9121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Taylor J.L., Mayer R.T., Himel C.M. Conformers of acetylcholinesterase: A mechanism of allosteric control. Mol. Pharmacol. 1994;45:74–83. [PubMed] [Google Scholar]
- 313.Huang D., Peng Y., Ma K., Qing X., Deng X., Li Z., Shao Z. Puerarin relieved compression-induced apoptosis and mitochondrial dysfunction in human nucleus pulposus mesenchymal stem cells via the PI3K/Akt Pathway. Stem Cells Int. 2020;2020:7126914. doi: 10.1155/2020/7126914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Li Z., Chen S., Ma K., Lv X., Lin H., Hu B., He R., Shao Z. CsA attenuates compression-induced nucleus pulposus mesenchymal stem cells apoptosis via alleviating mitochondrial dysfunction and oxidative stress. Life Sci. 2018;205:26–37. doi: 10.1016/j.lfs.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 315.Guo Q., Zhu D., Wang Y., Miao Z., Chen Z., Lin Z., Lin J., Huang C., Pan L., Wang L., et al. Targeting STING attenuates ROS induced intervertebral disc degeneration. Osteoarthr. Cartil. 2021;29:1213–1224. doi: 10.1016/j.joca.2021.04.017. [DOI] [PubMed] [Google Scholar]
- 316.Chen Y., Zheng Z., Wang J., Tang C., Khor S., Chen J., Chen X., Zhang Z., Tang Q., Wang C., et al. Berberine suppresses apoptosis and extracellular matrix (ECM) degradation in nucleus pulposus cells and ameliorates disc degeneration in a rodent model. Int. J. Biol. Sci. 2018;14:682–692. doi: 10.7150/ijbs.24081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Bai X., Jiang M., Wang J., Yang S., Liu Z., Zhang H., Zhu X. Cyanidin attenuates the apoptosis of rat nucleus pulposus cells and the degeneration of intervertebral disc via the JAK2/STAT3 signal pathway in vitro and in vivo. Pharm. Biol. 2022;60:427–436. doi: 10.1080/13880209.2022.2035773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Gao Z., Lin Y., Zhang P., Cheng Q., Ye L., Wu F., Chen Y., Fu M., Cheng C., Gao Y. Sinomenine ameliorates intervertebral disc degeneration via inhibition of apoptosis and autophagy in vitro and in vivo. Am. J. Transl. Res. 2019;11:5956–5966. [PMC free article] [PubMed] [Google Scholar]
- 319.Wen F., Yu J., He C.J., Zhang Z.W., Yang A.F. betaecdysterone protects against apoptosis by promoting autophagy in nucleus pulposus cells and ameliorates disc degeneration. Mol. Med. Rep. 2019;19:2440–2448. doi: 10.3892/mmr.2019.9861. [DOI] [PubMed] [Google Scholar]
- 320.Zhang B., Zhao Q., Li Y., Zhang J. Moxibustion alleviates intervertebral disc degeneration via activation of the HIF-1alpha/VEGF pathway in a rat model. Am. J. Transl. Res. 2019;11:6221–6231. [PMC free article] [PubMed] [Google Scholar]
- 321.Kim K.W., Ha K.Y., Lee J.S., Na K.H., Kim Y.Y., Woo Y.K. Senescence of nucleus pulposus chondrocytes in human intervertebral discs. Asian Spine J. 2008;2:1–8. doi: 10.4184/asj.2008.2.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Kim K.W., Chung H.N., Ha K.Y., Lee J.S., Kim Y.Y. Senescence mechanisms of nucleus pulposus chondrocytes in human intervertebral discs. Spine J. 2009;9:658–666. doi: 10.1016/j.spinee.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 323.Roberts S., Evans E.H., Kletsas D., Jaffray D.C., Eisenstein S.M. Senescence in human intervertebral discs. Eur. Spine J. 2006;15((Suppl. 3)):S312–S316. doi: 10.1007/s00586-006-0126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Feng C., Liu H., Yang M., Zhang Y., Huang B., Zhou Y. Disc cell senescence in intervertebral disc degeneration: Causes and molecular pathways. Cell Cycle. 2016;15:1674–1684. doi: 10.1080/15384101.2016.1152433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Wang F., Cai F., Shi R., Wang X.H., Wu X.T. Aging and age related stresses: A senescence mechanism of intervertebral disc degeneration. Osteoarthr. Cartil. 2016;24:398–408. doi: 10.1016/j.joca.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 326.Chen Y., Tang L. Stem cell senescence: The obstacle of the treatment of degenerative disk disease. Curr. Stem Cell Res. Ther. 2019;14:654–668. doi: 10.2174/1574888X14666190906163253. [DOI] [PubMed] [Google Scholar]
- 327.Deretic V. Autophagy in inflammation, infection, and immunometabolism. Immunity. 2021;54:437–453. doi: 10.1016/j.immuni.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Zhang G.Z., Chen H.W., Deng Y.J., Liu M.Q., Wu Z.L., Ma Z.J., He X.G., Gao Y.C., Kang X.W. BRD4 inhibition suppresses senescence and apoptosis of nucleus pulposus cells by inducing autophagy during intervertebral disc degeneration: An in vitro and in vivo study. Oxid. Med. Cell. Longev. 2022;2022:9181412. doi: 10.1155/2022/9181412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Lin J., Du J., Wu X., Xu C., Liu J., Jiang L., Cheng X., Ge G., Chen L., Pang Q., et al. SIRT3 mitigates intervertebral disc degeneration by delaying oxidative stress-induced senescence of nucleus pulposus cells. J. Cell. Physiol. 2021;236:6441–6456. doi: 10.1002/jcp.30319. [DOI] [PubMed] [Google Scholar]
- 330.Xie C., Shi Y., Chen Z., Zhou X., Luo P., Hong C., Tian N., Wu Y., Zhou Y., Lin Y., et al. Apigenin Alleviates Intervertebral Disc Degeneration via Restoring Autophagy Flux in Nucleus Pulposus Cells. Front. Cell Dev. Biol. 2021;9:787278. doi: 10.3389/fcell.2021.787278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Wang X.Y., Jiao L.Y., He J.L., Fu Z.A., Guo R.J. Parathyroid hormone 134 inhibits senescence in rat nucleus pulposus cells by activating autophagy via the mTOR pathway. Mol. Med. Rep. 2018;18:2681–2688. doi: 10.3892/mmr.2018.9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Boos N., Weissbach S., Rohrbach H., Weiler C., Spratt K.F., Nerlich A.G. Classification of age-related changes in lumbar intervertebral discs: 2002 Volvo Award in basic science. Spine. 2002;27:2631–2644. doi: 10.1097/00007632-200212010-00002. [DOI] [PubMed] [Google Scholar]
- 333.Weiler C., Schietzsch M., Kirchner T., Nerlich A.G., Boos N., Wuertz K. Age-related changes in human cervical, thoracal and lumbar intervertebral disc exhibit a strong intra-individual correlation. Eur. Spine J. 2012;21((Suppl. 6)):S810–S818. doi: 10.1007/s00586-011-1922-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Yang S., Liao W. Hydroxysafflor yellow A attenuates oxidative stress injury-induced apoptosis in the nucleus pulposus cell line and regulates extracellular matrix balance via CA XII. Exp. Ther. Med. 2022;23:182. doi: 10.3892/etm.2021.11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Gao J., Zhang Q., Song L. Resveratrol enhances matrix biosynthesis of nucleus pulposus cells through activating autophagy via the PI3K/Akt pathway under oxidative damage. Biosci. Rep. 2018;38:BSR20180544. doi: 10.1042/BSR20180544. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 336.Lin Z., Ni L., Teng C., Zhang Z., Wu L., Jin Y., Lu X., Lin Z. Eicosapentaenoic acid-induced autophagy attenuates intervertebral disc degeneration by suppressing endoplasmic reticulum stress, extracellular matrix degradation, and apoptosis. Front. Cell Dev. Biol. 2021;9:745621. doi: 10.3389/fcell.2021.745621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Chen F., Liu H., Wang X., Li Z., Zhang J., Pei Y., Zheng Z., Wang J. Melatonin activates autophagy via the NF-kappaB signaling pathway to prevent extracellular matrix degeneration in intervertebral disc. Osteoarthr. Cartil. 2020;28:1121–1132. doi: 10.1016/j.joca.2020.05.011. [DOI] [PubMed] [Google Scholar]
- 338.Lu L., Hu J., Wu Q., An Y., Cui W., Wang J., Ye Z. Berberine prevents human nucleus pulposus cells from IL1betainduced extracellular matrix degradation and apoptosis by inhibiting the NFkappaB pathway. Int. J. Mol. Med. 2019;43:1679–1686. doi: 10.3892/ijmm.2019.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Tu J., Li W., Zhang Y., Wu X., Song Y., Kang L., Liu W., Wang K., Li S., Hua W., et al. Simvastatin Inhibits IL-1beta-Induced apoptosis and extracellular matrix degradation by suppressing the NF-kB and MAPK pathways in nucleus pulposus cells. Inflammation. 2017;40:725–734. doi: 10.1007/s10753-017-0516-6. [DOI] [PubMed] [Google Scholar]
- 340.Zhang S., Liang W., Abulizi Y., Xu T., Cao R., Xun C., Zhang J., Sheng W. Quercetin alleviates intervertebral disc degeneration by modulating p38 MAPK-Mediated autophagy. Biomed. Res. Int. 2021;2021:6631562. doi: 10.1155/2021/6631562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Liu W., Jin S., Huang M., Li Y., Wang Z., Wang P., Zhao X., Xia P., Feng J. Duhuo jisheng decoction suppresses matrix degradation and apoptosis in human nucleus pulposus cells and ameliorates disc degeneration in a rat model. J. Ethnopharmacol. 2020;250:112494. doi: 10.1016/j.jep.2019.112494. [DOI] [PubMed] [Google Scholar]
- 342.Zheng Z., Wang Z.G., Chen Y., Chen J., Khor S., Li J., He Z., Wang Q., Zhang H., Xu K., et al. Spermidine promotes nucleus pulposus autophagy as a protective mechanism against apoptosis and ameliorates disc degeneration. J. Cell. Mol. Med. 2018;22:3086–3096. doi: 10.1111/jcmm.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Nan L.P., Wang F., Liu Y., Wu Z., Feng X.M., Liu J.J., Zhang L. 6-gingerol protects nucleus pulposus-derived mesenchymal stem cells from oxidative injury by activating autophagy. World J. Stem Cells. 2020;12:1603–1622. doi: 10.4252/wjsc.v12.i12.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Risbud M.V., Shapiro I.M. Role of cytokines in intervertebral disc degeneration: Pain and disc content. Nat. Rev. Rheumatol. 2014;10:44–56. doi: 10.1038/nrrheum.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Sampara P., Banala R.R., Vemuri S.K., Av G.R., Gpv S. Understanding the molecular biology of intervertebral disc degeneration and potential gene therapy strategies for regeneration: A review. Gene Ther. 2018;25:67–82. doi: 10.1038/s41434-018-0004-0. [DOI] [PubMed] [Google Scholar]
- 346.Wang H., Jiang Z., Pang Z., Zhou T., Gu Y. Acacetin Alleviates Inflammation and Matrix Degradation in Nucleus Pulposus Cells and Ameliorates Intervertebral Disc Degeneration in vivo. Drug Des. Dev. Ther. 2020;14:4801–4813. doi: 10.2147/DDDT.S274812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Wang K., Chen T., Ying X., Zhang Z., Shao Z., Lin J., Xu T., Chen Y., Wang X., Chen J., et al. Ligustilide alleviated IL-1beta induced apoptosis and extracellular matrix degradation of nucleus pulposus cells and attenuates intervertebral disc degeneration in vivo. Int. Immunopharmacol. 2019;69:398–407. doi: 10.1016/j.intimp.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 348.Jiang L., Jin Y., Wang H., Jiang Y., Dong J. Glucosamine protects nucleus pulposus cells and induces autophagy via the mTOR-dependent pathway. J. Orthop. Res. 2014;32:1532–1542. doi: 10.1002/jor.22699. [DOI] [PubMed] [Google Scholar]
- 349.Moore R.J. The vertebral endplate: Disc degeneration, disc regeneration. Eur. Spine J. 2006;15((Suppl. 3)):S333–S337. doi: 10.1007/s00586-006-0170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Yuan W., Che W., Jiang Y.Q., Yuan F.L., Wang H.R., Zheng G.L., Li X.L., Dong J. Establishment of intervertebral disc degeneration model induced by ischemic sub-endplate in rat tail. Spine J. 2015;15:1050–1059. doi: 10.1016/j.spinee.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 351.Urban J.P., Smith S., Fairbank J.C. Nutrition of the intervertebral disc. Spine. 2004;29:2700–2709. doi: 10.1097/01.brs.0000146499.97948.52. [DOI] [PubMed] [Google Scholar]
- 352.He Z., Jia M., Yu Y., Yuan C., Wang J. Roles of SDF-1/CXCR4 axis in cartilage endplate stem cells mediated promotion of nucleus pulposus cells proliferation. Biochem. Biophys. Res. Commun. 2018;506:94–101. doi: 10.1016/j.bbrc.2018.10.069. [DOI] [PubMed] [Google Scholar]
- 353.Ge Q., Ying J., Shi Z., Mao Q., Jin H., Wang P.E., Chen J., Yuan W., Tong P., Li J. Chlorogenic Acid retards cartilaginous endplate degeneration and ameliorates intervertebral disc degeneration via suppressing NF-kappaB signaling. Life Sci. 2021;274:119324. doi: 10.1016/j.lfs.2021.119324. [DOI] [PubMed] [Google Scholar]
- 354.Chen K., Lv X., Li W., Yu F., Lin J., Ma J., Xiao D. Autophagy is a protective response to the oxidative damage to endplate chondrocytes in intervertebral disc: Implications for the treatment of degenerative lumbar disc. Oxid. Med. Cell. Longev. 2017;2017:4041768. doi: 10.1155/2017/4041768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Luo L., Jian X., Sun H., Qin J., Wang Y., Zhang J., Shen Z., Yang D., Li C., Zhao P., et al. Cartilage endplate stem cells inhibit intervertebral disc degeneration by releasing exosomes to nucleus pulposus cells to activate Akt/autophagy. Stem Cells. 2021;39:467–481. doi: 10.1002/stem.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Xiao L., Ding B., Gao J., Yang B., Wang J., Xu H. Curcumin prevents tension-induced endplate cartilage degeneration by enhancing autophagy. Life Sci. 2020;258:118213. doi: 10.1016/j.lfs.2020.118213. [DOI] [PubMed] [Google Scholar]
- 357.Ye W., Zhu W., Xu K., Liang A., Peng Y., Huang D., Li C. Increased macroautophagy in the pathological process of intervertebral disc degeneration in rats. Connect. Tissue Res. 2013;54:22–28. doi: 10.3109/03008207.2012.715702. [DOI] [PubMed] [Google Scholar]
- 358.Ye W., Xu K., Huang D., Liang A., Peng Y., Zhu W., Li C. Age-related increases of macroautophagy and chaperone-mediated autophagy in rat nucleus pulposus. Connect. Tissue Res. 2011;52:472–478. doi: 10.3109/03008207.2011.564336. [DOI] [PubMed] [Google Scholar]
- 359.Kong C.G., Park J.B., Kim M.S., Park E.Y. High glucose accelerates autophagy in adult rat intervertebral disc cells. Asian Spine J. 2014;8:543–548. doi: 10.4184/asj.2014.8.5.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Jiang L., Zhang X., Zheng X., Ru A., Ni X., Wu Y., Tian N., Huang Y., Xue E., Wang X., et al. Apoptosis, senescence, and autophagy in rat nucleus pulposus cells: Implications for diabetic intervertebral disc degeneration. J. Orthop. Res. 2013;31:692–702. doi: 10.1002/jor.22289. [DOI] [PubMed] [Google Scholar]
- 361.Jin L.Y., Lv Z.D., Wang K., Qian L., Song X.X., Li X.F., Shen H.X. Estradiol Alleviates intervertebral disc degeneration through modulating the antioxidant enzymes and inhibiting autophagy in the model of menopause rats. Oxid. Med. Cell. Longev. 2018;2018:7890291. doi: 10.1155/2018/7890291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Ni B.B., Li B., Yang Y.H., Chen J.W., Chen K., Jiang S.D., Jiang L.S. The effect of transforming growth factor beta1 on the crosstalk between autophagy and apoptosis in the annulus fibrosus cells under serum deprivation. Cytokine. 2014;70:87–96. doi: 10.1016/j.cyto.2014.07.249. [DOI] [PubMed] [Google Scholar]
- 363.Ni B., Shen H., Wang W., Lu H., Jiang L. TGF-beta1 reduces the oxidative stress-induced autophagy and apoptosis in rat annulus fibrosus cells through the ERK signaling pathway. J. Orthop. Surg. Res. 2019;14:241. doi: 10.1186/s13018-019-1260-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Kakiuchi Y., Yurube T., Kakutani K., Takada T., Ito M., Takeoka Y., Kanda Y., Miyazaki S., Kuroda R., Nishida K. Pharmacological inhibition of mTORC1 but not mTORC2 protects against human disc cellular apoptosis, senescence, and extracellular matrix catabolism through Akt and autophagy induction. Osteoarthr. Cartil. 2019;27:965–976. doi: 10.1016/j.joca.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 365.Ito M., Yurube T., Kakutani K., Maeno K., Takada T., Terashima Y., Kakiuchi Y., Takeoka Y., Miyazaki S., Kuroda R., et al. Selective interference of mTORC1/RAPTOR protects against human disc cellular apoptosis, senescence, and extracellular matrix catabolism with Akt and autophagy induction. Osteoarthr. Cartil. 2017;25:2134–2146. doi: 10.1016/j.joca.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 366.Zaytseva Y.Y., Valentino J.D., Gulhati P., Evers B.M. mTOR inhibitors in cancer therapy. Cancer Lett. 2012;319:1–7. doi: 10.1016/j.canlet.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 367.Carames B., Taniguchi N., Otsuki S., Blanco F.J., Lotz M. Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthritis Rheum. 2010;62:791–801. doi: 10.1002/art.27305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Lee D.Y., Park Y.J., Song M.G., Kim D.R., Zada S., Kim D.H. Cytoprotective effects of delphinidin for human chondrocytes against oxidative stress through activation of autophagy. Antioxidants. 2020;9:83. doi: 10.3390/antiox9010083. [DOI] [PMC free article] [PubMed] [Google Scholar]