Abstract
The positive control function of the bacterial enhancer-binding protein NtrC resides in its central domain, which is highly conserved among activators of ς54 holoenzyme. Previous studies of a small set of mutant forms specifically defective in transcriptional activation, called NtrC repressor [NtrC(Rep)] proteins, had enabled us to locate various functional determinants in the central domain. In this more comprehensive survey, the DNA encoding a major portion of the central domain was randomly mutagenized and mutated ntrC genes were introduced into the cell via multicopy expression plasmids. DNA sequencing of 95 isolates identified by a preliminary phenotypic screen revealed that the lesions in them caused 55 distinct single amino acid substitutions at 44 different positions. Assays of glnA transcription in vivo and in vitro yielded two conclusions. First, of the 41 mutant proteins that could be purified, 17 (1 known, 16 new) showed no detectable activity in either assay, thus qualifying them as true NtrC(Rep) proteins. These contained residue changes in six of the seven highly conserved regions in the central domain, including two never studied before. Second, some mutant proteins were inactive in vivo but were either marginally or fully active in vitro. Their surprising lack of activity in vivo may be accounted for by high levels of expression, which apparently decreased activation by these mutant proteins but not by wild-type NtrC (NtrCWT). Of particular interest were a subset of these proteins that exhibited greater transcriptional activation than NtrCWT at low concentrations. Their elevated activation capacities remain to be explained.
A prominent class of prokaryotic enhancer-binding proteins activates transcription by the ς54 holoenzyme form of RNA polymerase (23, 26, 30, 50). One such protein is nitrogen regulatory protein C (NtrC), which activates transcription in response to limitation of combined nitrogen in the medium. The NtrC protein from enteric bacteria has been well studied as an activator for the glnA gene; this gene encodes glutamine synthetase, an enzyme with a major role in assimilation of ammonia.
NtrC can function both as an activator and as a repressor of glnA transcription, depending on the nutritional status of the cell (41, 56). Positive and negative controls of expression are achieved at two different promoters: activation at a downstream ς54-dependent promoter and repression at an upstream ς70-dependent promoter (Fig. 1A). Under nitrogen-limiting conditions, NtrC is phosphorylated by the protein kinase NtrB (17, 32). Phosphorylated NtrC forms an unusual hexamer or octamer at the two sites that constitute the glnA enhancer (61). To activate transcription, this oligomer contacts ς54 holoenzyme at the glnA promoter by means of a DNA conformational change (55). It catalyzes isomerization of closed complexes between ς54 holoenzyme and the promoter to transcriptionally productive open complexes (29, 39, 46) in a manner that depends upon hydrolysis of ATP and an energy-coupling mechanism (37, 39, 55, 57). When NtrC is bound to the enhancer, it represses transcription from a secondary ς70-dependent promoter that lies in the enhancer region (41). Phosphorylation and oligomerization, which are essential for ATP hydrolysis and therefore for transcriptional activation (1, 40, 59, 60), are not required for repression of transcription (20).
FIG. 1.
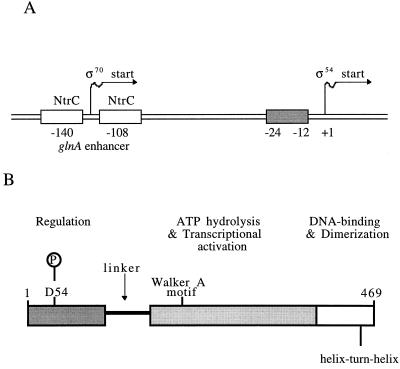
(A) Diagram of the glnA promoter-regulatory region from S. typhimurium (not to scale). The upstream binding sites for NtrC, which are centered at −140 and −108 with respect to the major ς54-dependent transcriptional start site at +1, function as a transcriptional enhancer (33, 42). Conserved promoter sequences recognized by the ς54 holoenzyme lie at −24 and −12, as indicated (23, 50). When phosphorylated, NtrC forms an oligomer at the enhancer (see text) and activates transcription by ς54 holoenzyme. Both phosphorylated and unphosphorylated NtrC can bind to the enhancer and repress transcription from the secondary ς70-dependent promoter that lies in the enhancer region (20, 41). (B) Domain structure of NtrC (not to scale) (reviewed by Kustu et al. [22], Weiss et al. [58], and Morett and Segovia [30]). An NtrC monomer (52,238 Da) contains 469 amino acid residues and is composed of three domains. The N-terminal receiver domain (∼120 residues) contains the site of phosphorylation, D54. Under nitrogen-limiting conditions, this aspartate residue receives a phosphate from the phosphorylated NtrB protein, a physiological signal that is necessary for NtrC-mediated transcriptional activation (1, 32, 57). NtrB and NtrC constitute a sensory kinase-response regulator pair in a “two-component” signal transduction system (21, 34). The N-terminal domain is linked by a glutamine-rich flexible linker (“Q-linker”) to the central domain (∼240 residues), which appears to be directly responsible for ATP hydrolysis and transcriptional activation by ς54 holoenzyme (see text). This domain is highly conserved among activators of ς54 holoenzyme (Fig. 2). The C-terminal domain (∼90 residues) contains a helix-turn-helix DNA-binding motif (40, 58) and the major dimerization determinants of the protein (19, 35).
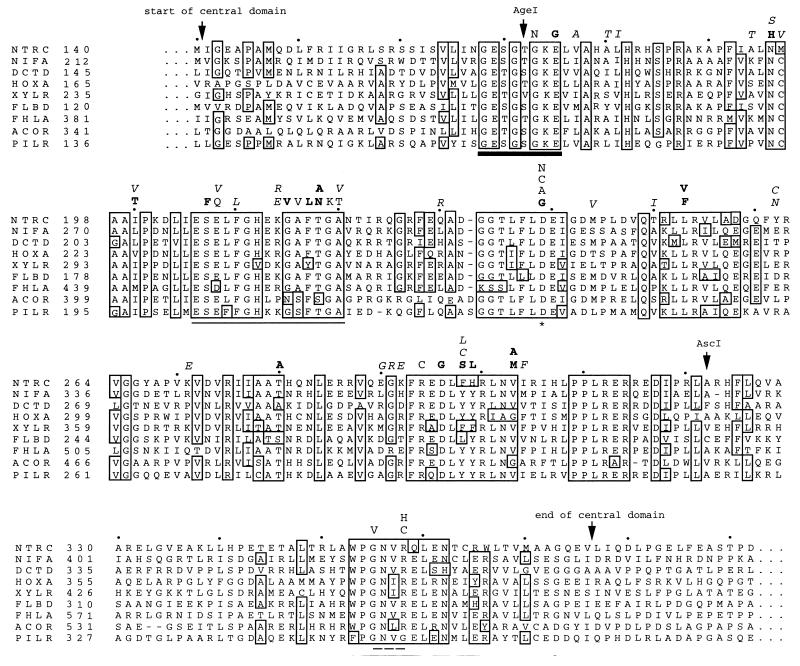
Each monomer of the dimeric NtrC protein is comprised of three domains (Fig. 1B). The N-terminal domain contains the site of phosphorylation, D54 (17, 32). The C-terminal domain is responsible for binding to the enhancer (7, 40). The central domain of NtrC, amino acid residues 141 to 376 (Fig. 2), is directly responsible for transcriptional activation (2, 10, 14, 15, 37), which appears to entail oligomerization, nucleotide binding and hydrolysis, and coupling of energy to a thermodynamically unfavorable change in the conformation of ς54 holoenzyme. The functional significance of the central domain of NtrC is underscored by the high degree of sequence similarity shared by all members of the ς54 activator family (4, 6, 44). Sequence comparisons among these homologous proteins have revealed seven highly conserved amino acid segments (designated C1 to C7) along the central domain (30). When such comparisons were combined with genetic and biochemical analyses and were expanded to include a broader range of purine nucleotide-binding proteins (38, 47), interesting parallels that provided insights into the distribution of the subfunctions of positive control within the central domain emerged. The C1 and C4 regions, respectively, were identified as likely Walker A and Walker B motifs, which are involved in ATP binding and hydrolysis (37, 38, 43, 44, 52, 57). The C3 region appears to be analogous to the “switch I” region of other purine nucleotide-binding proteins (37, 38), a region that lies between the Walker A and B motifs and plays an important role in coupling nucleotide hydrolysis to biological output (e.g., see references 3, 13, 18, and 48). The C7 region is required for nucleotide binding and is probably involved in binding the nucleotide base (38, 43). Finally, recent secondary structure predictions coupled with the use of recognition algorithms for protein folds indicated that the central domain of activators of ς54 holoenzyme adopts a mononucleotide-binding fold similar to those of the eukaryotic signaling protein p21ras and the G domain of the bacterial polypeptide elongation factor Tu (38).
FIG. 2.
Alignment of the central domains of nine activators of ς54 holoenzyme and the locations of amino acid substitutions in NtrC mutant proteins. The activators are chosen from members of the α, β, and δ subgroups of the purple bacteria. Respectively, the sequences belong to S. typhimurium NTRC, Klebsiella pneumoniae NIFA, R. leguminosarum DCTD, Alcaligenes eutrophus HOXA, Pseudomonas putida XYLR, Caulobacter crescentus FLBD, E. coli FHLA, A. eutrophus ACOR, and Pseudomonas aeruginosa PILR (original references given in reference 35). Residues that are identical in at least 6 activators are boxed. The common boundaries of their central domains are indicated by arrows, and their exact positions are according to Morett and Segovia (30). Another pair of arrows identify the locations of the AgeI and AscI restriction sites that correspond to the start and the end of the PCR mutagenesis target region (amino acids 173 to 321 in S. typhimurium NtrC). Four functional determinants are highlighted by symbols directly beneath their positions in the sequence alignment: the heavy black line denotes the glycine-rich segment that has been proposed to correspond to the Walker A motif of purine nucleotide-binding proteins (37, 38, 43, 44, 52, 57); the asterisk denotes the highly conserved aspartate at the end of the Walker B motif; the plain line denotes the putative switch I (effector) region (18, 37, 38); and the dashed line marks four residues believed to be involved in binding of the nucleotide base (38, 43). Letters above the sequence alignment designate all single amino acid substitutions for which in vitro measurements were made in this study and known substitutions resulting in NtrC(Rep) proteins. In cases where more than one such substitution was found for a given residue, the letters are arranged according to the severity of the effect, such that those resulting in the largest defects are placed closest to the alignment. Plain letters denote known NtrC(Rep) substitutions that had been described previously (37, 43, 56–58). Boldface letters indicate amino acid substitutions found in this study to yield NtrC(Rep) proteins. Like known NtrC(Rep) proteins, these new proteins were characterized by the absence of detectable transcriptional activities in vivo or in vitro. Italic letters represent other mutational substitutions generated in this study which resulted in little transcriptional activation and glutamine auxotrophy in vivo but readily detectable and different amounts of transcriptional activation in vitro (see Results).
Genetic studies of the NtrC protein of Salmonella typhimurium were facilitated by the isolation of a class of mutant proteins known as NtrC repressor [NtrC(Rep)] forms (37, 43, 56, 57). NtrC(Rep) proteins, which have been called “positive control” or “PC” forms in other systems, were defined as those that had lost the ability to activate transcription by ς54 holoenzyme but retained the ability to repress transcription by ς70 holoenzyme (37). Prior to this study, 13 NtrC(Rep) proteins carrying single residue changes in five of the seven conserved regions of the central domain (30) had been characterized. Their properties in vitro allowed us to distinguish between those regions involved specifically in Mg-ATP binding and those responsible for ATP hydrolysis per se (43) and to identify one region (C3 or switch I) required for transcriptional activation but not for nucleotide hydrolysis (37); by inference, the C3 region appears to be involved in interaction with ς54 holoenzyme, that is, in contact with the polymerase, and/or coupling of energy to a change in its conformation that allows it to form open complexes. Further advances in functional mapping were limited by the size of our mutant pool.
To characterize new functional determinants of NtrC, we performed random mutagenesis to scan most of the central domain of the S. typhimurium NtrC protein (residues 172 to 322) for new amino acid residues and regions important for positive control. We determined transcriptional activities of mutant proteins both in vivo and in vitro. As expected, these studies yielded additional NtrC(Rep) proteins. Unexpectedly, they also yielded a new and interesting class of mutant proteins that were more active than wild-type NtrC (NtrCWT) at low concentrations but had greatly decreased transcriptional activation capacities at high concentrations.
MATERIALS AND METHODS
Media.
Complex media were nutrient broth (NB), Luria broth (LB), and green indicator plates (5), which were supplemented with glutamine (2 mM) and/or ampicillin (100 μg/ml) when appropriate. The minimal medium, N−C− (12), was supplemented with glucose (0.4%) as the carbon source, glutamine (5 mM) as the nitrogen source, and l-histidine (0.2 mM) to satisfy an auxotrophic requirement. Ampicillin (50 μg/ml) was added for plasmid-carrying strains to prevent loss of plasmids.
Random mutagenesis with PCR.
Most of the DNA sequence encoding the central domain of NtrC (amino acids 141 to 376 or nucleotides 421 to 1128, where +1 designates the A of the ATG start codon for NtrC; Fig. 2) was subjected to random mutagenesis with PCR. The two oligonucleotide primers used to amplify this segment were 5′-ATTGGTCGGCTGTCGCGTTC-3′ (nucleotides 460 to 479; upstream primer) and 5′-ATTTGGCTTCCACGCCTAAT-3′ (nucleotides 1015 to 996; downstream primer). PCR conditions were chosen to exploit the inherent infidelity of DNA synthesis by Thermus aquaticus (Taq) DNA polymerase (24, 62). The template was pJES559 (Table 1), which carries malE-ntrC in an overexpression vector. The Mg2+ concentration was elevated to provide a large excess over the total deoxynucleoside triphosphates (dNTPs) present, a condition that further reduces the copying accuracy of the Taq polymerase (8). Reaction mixtures contained 5 ng of template, 100 ng of each primer, 50 μM (each) dNTP, 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 4 mM MgCl2 and 5 units of Taq DNA polymerase (AmpliTaq) in a total volume of 50 μl. PCRs were carried out on a Perkin-Elmer Cetus DNA Thermal Cycler with the following cycles programmed: 30 cycles of 1 min at 94°C, 1 min at 48°C, and 1 min at 70°C.
TABLE 1.
Bacterial strainsa and plasmids used in this study
| Strain or plasmid | Genotype and/or characteristic(s) | Construction,b reference, and/or source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Purchased as Subcloning Efficiency DH5α Competent Cells from Gibco BRL (cat. no. 18265-017) |
| NCM724 | HfrC(λ) ntrA6 argG::Tn10 | Weiss et al. (57) |
| NCM1099 | DH5α/pJES311 | Transformation; this work |
| NCM1237 | DH5α/pJES504 | Transformation; this work |
| NCM1946 | DH5α/pJES1065 | Transformation; this work |
| S. typhimurium strains | ||
| SK1489 | glnAp381 glnAp131 ntrA75 ΔhisF645 | McCarter et al. (27) |
| SK1490 | glnAp381 glnAp131 ΔhisF645 | McCarter et al. (27) |
| SK3044 | ntrC352::Tn10 putPA::[Kanr-Φ(glnA′-‵lacZ)] | Ikeda et al. (16) |
| SK3423 | glnAp381 glnAp131 ntrC352::Tn10 ΔhisF645 | SK1490, SK3044cd; this work |
| SK3424 | glnAp381 glnAp131 ntrC352::Tn10 ΔhisF645 putPA::[Kanr-Φ(glnA′-‵lacZ)] | SK3423, SK3044e; this work |
| SK3481 | glnAp381 glnAp131 ntrC352::Tn10 ntrA75 ΔhisF645 | SK1489, SK3424c; this work |
| SK3482 | glnAp381 glnAp131 ntrC352::Tn10 ntrA75 ΔhisF645 putPA::[Kanr-Φ(glnA′-‵lacZ)] | SK3481, SK3424e; this work |
| SK3489 | glnAp381 glnAp131 ntrA75 ΔhisF645 putPA::[Kanr-Φ(glnA′-‵lacZ)] | SK1489, SK3424e; this work |
| Plasmids | ||
| pJES311 | Overexpression vector for NtrCWT under control of a T7 promoterf | Weiss et al. (57) |
| pJES412 | Overexpression vector for NtrCS207F,g derived from pJES311 | Weiss et al. (57) |
| pJES504 | Overexpression vector for NtrCΔ412-469, derived from pJES311 | North and Kustu (36) |
| pJES534 | pJES436 derivative which contains a strong enhancer situated ∼460 bp from the glnA promoter | Porter et al. (40) |
| pJES559 | Overexpression vector for MBP-NtrC fusion protein under control of the tac promoter | Klose et al. (19) |
| pJES1065 | Overexpression vector for NtrCD239N,g derived from pJES311 | Rombel et al. (43) |
This table lists all strains used in this study except those listed in Table 2, which have the same chromosomal genotype as SK3424 (ntrC), SK3482 (ntrC ntrA), or NCM724 (E. coli).
For each phage P22-mediated transductional cross, we list the recipient and the donor.
Selected for tetracycline resistance.
Large colonies were selected to ensure that the transductants had not lost the promoter(Up) mutations (27).
Selected for kanamycin resistance.
φ10 promoter of phage T7 (dependent upon T7 RNA polymerase).
The PCR products were digested by a pair of restriction endonucleases, AgeI and AscI, which cut close to the ends of the fragments (Fig. 2). The resulting small fragments (nucleotides 514 to 963) were gel purified with Spinex columns (Amicon) and cloned into the NtrC overexpression plasmid pJES311, which had also been digested with AgeI and AscI. Ligation mixtures (with molar ratios of insert to vector of 3:1 and 4:1) were transformed into Gibco BRL DH5α competent cells according to the manufacturer’s specifications and were incubated overnight at 37°C on LB glutamine (Gln) ampicillin (Amp) selective plates.
Genetic screen.
A newly developed plate test was employed to discriminate between transformants carrying ntrC(Rep) alleles and those carrying the wild-type allele or ntrC null alleles. The screening medium was NB, which lacks sufficient glutamine to allow growth of glutamine auxotrophs. Because DH5α has an intact chromosomal ntrC gene, the introduction of an ntrC mutant allele on a multicopy expression plasmid (strain NCM1237 [Table 1]) did not affect glutamine-independent growth of the host strain. By contrast, when an ntrC(Rep) allele was introduced (strain NCM1946), there was no growth on NB because both glnA promoters on the host chromosome were inactivated. Interestingly, the control strain NCM1099, which expressed NtrCWT from pJES311, did not grow as well as the host strain on NB (phenotype intermediate between those of NCM1237 and NCM1946). This “semirepressed” phenotype is probably explained by the fact that there is only a single chromosomal copy of ntrB, and hence NtrC is not phosphorylated well enough to drive adequate expression from the downstream promoter.
To identify strains carrying NtrC(Rep) proteins, single colonies of 1,234 transformants were screened for growth on NB, as discussed above (all grew normally on NB Gln). One group was able to grow as well as NCM1237 (ntrC mutant allele). Another group grew noticeably worse, with a phenotype similar to NCM1099 (ntrC+ allele). The remainder of the transformants failed to grow on NB, a phenotype resembling that exhibited by NCM1946 (ntrC(Rep) allele). They were stored in glycerol at −80°C for further analysis.
DNA sequencing.
Strains were streaked out fresh from glycerol stocks, and plasmids were extracted with QIAGEN Miniprep kits. DNA sequencing of the entire mutagenized region was performed on an ABI373 automated sequencer using the upstream primer 5′-TATCGAGGTTAACGGCCCGA-3′ (nucleotides 390 to 409). For each sample, a second sequencing run was also performed to confirm the mutation from the reverse direction. The downstream primer was the same as that used previously for PCR mutagenesis.
Strain constructions.
Preparation of lysates of phage P22, transductional crosses, and the use of green indicator plates to purify transductants free of phage were as described previously (5).
Strain SK3424 {glnAp381 glnAp131 ntrC352::Tn10 ΔhisF645 putPA::[Kanr-Φ(glnA′-‵lacZ)]}, which allowed us to assess the effects of putative ntrC(Rep) alleles on glnA transcription in vivo, was constructed in two steps as described below. In step one, a phage lysate was prepared on strain SK3044, which carries the ntrC::Tn10 allele and was used to transduce SK1490 (glnAp381 glnAp131 ΔhisF645) to tetracycline resistance. Large transductant colonies were selected because their phenotype indicated that they had retained the promoter mutations (27). In step two, the same phage lysate was used to transduce the intermediate strain, SK3423, to kanamycin resistance to select for transfer of the Φ(glnA′-‵lacZ) fusion.
The glnA promoter(Up) mutations in SK3424 were introduced because a strain carrying the ntrC352::Tn10 allele alone grows poorly on glutamine as the sole nitrogen source (doubling time of ∼170 min versus ∼130 min for an ntrC+ strain) and such a strain transformed with plasmid pJES412 (an ntrC(Rep) allele) grows worse (doubling time, >220 min). The presence of the promoter(Up) mutations in SK3424 alleviated growth problems. Use of glutamine as the sole nitrogen source was chosen because it is a derepressing condition for glnA expression in an ntrC+ strain.
The construction of strain SK3482 {glnAp381 glnAp131 ntrC352::Tn10 ntrA75 hisF645 putPA::[Kanr-Φ(glnA′-‵lacZ)]}, which carries an ntrA null allele, was similarly accomplished in two cycles of P22-mediated transduction. In the first cycle, the ntrC352::Tn10 allele from strain SK3424 was introduced into strain SK1489, which carries a frameshift mutation in the ntrA gene (ntrA75) to yield strain SK3481 (Table 1). In the second cycle, the Φ(glnA′-‵lacZ) fusion at the put locus of strain SK3424 was transferred to strain SK3481 to yield strain SK3482. Strain SK3489 {glnAp381 glnAp131 ntrA75 hisF645 putPA::[Kanr-Φ(glnA′-‵lacZ)]}, which carries an ntrC+ allele in the same background as SK3482, was constructed by introducing the Φ(glnA′-‵lacZ) fusion from SK3424 into SK1489.
All plasmids were electroporated into the host strain SK3424. Selected plasmids were electroporated into strain SK3482.
Growth conditions and β-galactosidase assay.
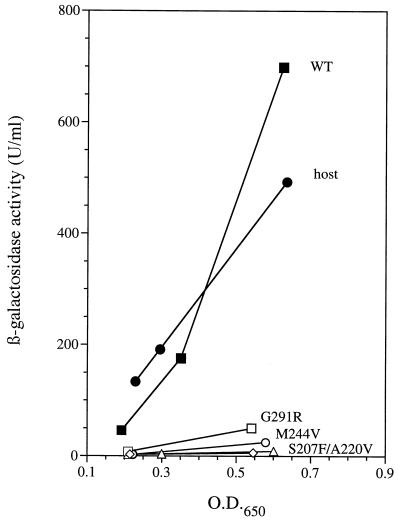
Cultures of strains carrying putative ntrC(Rep) alleles in the SK3424 background were adapted to growth on glutamine as the sole nitrogen source by overnight growth in minimal medium containing 5 mM glutamine and 2 mM NH4Cl. These precultures were subjected to centrifugation at room temperature. Cells were washed once with fresh medium containing 5 mM glutamine as the sole nitrogen source and were inoculated into 10 ml of this medium at an initial optical density at 650 nm (OD650) of 0.05. Cultures were incubated in baffled culture tubes on a New Brunswick model G76 water bath shaker at 37°C and 240 rpm. Samples in the exponential phase of growth were harvested in duplicate between OD650s of 0.2 and 0.6 and were immediately frozen on dry ice. Cultures of control strains SK3424 and SK3425 were grown along with each set of strains carrying putative ntrC(Rep) alleles in the overexpression vector. β-Galactosidase assays were carried out, and units of activity were calculated according to Miller (28) (see Fig. 3).
FIG. 3.
In vivo expression from the glnA promoter in three strains carrying mutant NtrC proteins. Expression of a Φ(glnA′-‵lacZ) fusion at the put locus of S. typhimurium was measured as a function of the OD650 for cultures grown under nitrogen-limiting conditions (see Materials and Methods). Curves labeled A220V (open diamonds), M244V (open circles), and G291R (open squares) give data for strains carrying plasmids that overexpress NtrC proteins with these three amino acid substitutions (strains SK3468, SK3458, and SK3447, respectively). In each case, two data points were used to determine the slope of the line, which reflects the differential rate of glnA′-‵lacZ expression. All three mutant NtrC proteins activated transcription better than NtrCWT at low concentrations in vitro (Fig. 4B through D). Curves labeled WT (closed squares) and S207F (open triangles) give data for control strains carrying plasmids that overexpress NtrCWT (strain SK3425) and NtrCS207F (strain SK3426), a known NtrC(Rep) protein. The curve labeled host (closed circles) gives data for the ntrC mutant host strain SK3424. For control strains, three data points were taken to allow reliable comparisons between experiments.
To assess the effect of an ntrA mutant allele on glnA′-‵lacZ expression for plasmids that yielded activity in the ntrA+ background, strains carrying such plasmids in strain SK3482 were grown as described above. β-Galactosidase assays were conducted as described above. Strain SK3483, which carries plasmid pJES311 (ntrC+ allele), and strain SK3489, which contains a single chromosomal ntrC+ gene, were used as controls.
Western blotting.
Cultures grown under the same conditions as those used for β-galactosidase assays (500 μl at an OD650 of 0.5) were subjected to centrifugation, and cell pellets were suspended in 20 μl of sodium dodecyl sulfate (SDS) gel-loading buffer (∼25-fold concentration). In each case, the entire sample was subjected to electrophoresis in an SDS–10% polyacrylamide gel and proteins were transferred to nitrocellulose membranes as described previously (45). Membranes were exposed to polyclonal mouse antiserum directed against the carboxy-terminal domain of the NtrC protein from S. typhimurium, and bands were detected with secondary antibodies directed against mouse immunoglobulin G and coupled to alkaline phosphatase (Bio-Rad). Purified NtrC and prestained markers (Broad Range; New England Biolabs) were used as standards.
To estimate the intracellular concentration of NtrC when the protein was expressed from a single chromosomal copy of the ntrC gene or from plasmid pJES311, we made the following assumptions: for S. typhimurium growing exponentially on minimal medium under nitrogen-limiting conditions, an OD650 of 1 corresponds to 0.37 mg (dry weight) per milliliter of culture (16) and cell volume is 2.0 μl/mg (dry weight) (16). Values of cell water content for enteric bacteria range from 1.9 to 2.5 μl/mg (dry weight) (25, 31, 53).
Overexpression and small-scale purification of NtrC proteins.
Plasmids for overproduction of NtrC proteins could not be maintained in Escherichia coli strains that contained T7 RNA polymerase (57); therefore, all of the 55 plasmids carrying putative ntrC(Rep) alleles were transformed into an Hfr strain of E. coli (NCM724), and induction was accomplished by infection with an M13 phage that carries T7 RNA polymerase under control of the lac promoter (M13mGP1-2). Cultures were grown in 50 ml of LB medium containing 2 mM glutamine and 100 μg of ampicillin per ml at 37°C with vigorous shaking. At an OD600 of ∼0.5, M13mGP1-2 was added to a multiplicity of infection of about 10 (49). IPTG (isopropyl-β-d-thiogalactoside) was also added, to 0.5 mM, to allow induction of T7 RNA polymerase. After 2 h, cultures were harvested by centrifugation. Cells were suspended in 5 ml of breakage buffer (50 mM Tris-acetate [pH 8.2], 200 mM KCl, 1 mM EDTA, 1 mM dithiothreitol [DTT]) and were lysed by two passages through a French pressure cell (SLM-Aminco) at 8,000 lb/in2. The resulting lysate was subjected to centrifugation at 27,000 × g for 30 min at 4°C, and ammonium sulfate (35% final concentration) was added to the supernatant at room temperature. The ammonium sulfate pellet was dissolved in 10 ml of B-50 buffer (10 mM Tris-acetate [pH 8.2], 50 mM KCl, 0.1 mM EDTA, 5% glycerol, 1 mM DTT) and loaded on DEAE Sephacel (Pharmacia Biotech) (1 ml of resin packed into Poly-Prep chromatography columns [Bio-Rad]) at room temperature. Columns were washed twice with 5 ml of B-50 buffer, and then the bottom of each column was closed and 2 ml of B-500 buffer (10 mM Tris-acetate [pH 8.2], 500 mM KCl, 0.1 mM EDTA, 5% glycerol, 1 mM DTT) was added. After 10 min at room temperature, the proteins were eluted by centrifugation at 2,500 × g for 5 min at 4°C. To each sample (2 ml), 18 ml of B-0 buffer (10 mM Tris-acetate [pH 8.2], 0.1 mM EDTA, 5% glycerol, 1 mM DTT) was added and the diluted samples were loaded on heparin Sepharose CL-6B (Pharmacia Biotech) (0.5 ml of resin packed in the columns described above) at room temperature. The columns were washed with 5 ml of B-50 buffer, and then the bottoms were closed and 2 ml of B-500 buffer was added. After 10 min of incubation at room temperature, the proteins were eluted and dialyzed into B-50 buffer overnight at 4°C. Proteins were assessed to be at least 90% pure by visual inspection of SDS–10% polyacrylamide gels stained with Coomassie blue (Sigma). NtrC concentrations were determined from absorption at 280 nm in the presence of 6 M guanidine hydrochloride (Pierce) by using an extinction coefficient of 44,907.8 M−1 cm−1 for NtrC (9). Because none of the amino acid substitutions in mutant proteins involved W residues, all mutant proteins had approximately the same extinction coefficient as NtrCWT.
The above procedure allowed us to purify all 41 mutant NtrC proteins that were soluble. SDS-polyacrylamide gels of various fractions indicated that purification was obtained by ammonium sulfate fractionation, because most proteins in crude cell supernatants remained in the ammonium sulfate supernatant, and upon binding to heparin Sepharose, because most of the remaining contaminating proteins did not bind (data not shown).
Assay for open complex formation.
The ability of NtrC proteins to catalyze the formation of open complexes by ς54 holoenzyme was assessed on a supercoiled plasmid template by using a single-cycle transcription assay, as described previously (10, 39, 55, 57). The template was plasmid pJES534 (1 nM) (40), which carries a “strong enhancer” situated ∼460 bp from the glnA promoter and directs synthesis of a 155-nucleotide transcript that contains no uracil (57). Final concentrations of reagents added to the buffer were as follows: core RNA polymerase, 30 nM; ς54, 50 nM; carbamyl phosphate, 10 mM; ATP, 4 mM; GTP, 400 μM; CTP, 100 μM containing 5 μCi of [α-32P]CTP. The NtrC concentration was varied. After allowing phosphorylation of NtrC by carbamyl phosphate for 10 min at 37°C, open complex formation was initiated by adding ATP to the reaction mixture. After 10 min, synthesis of transcripts was initiated by adding a mixture of heparin (100 μg/ml) and the remaining two nucleotides. After an additional 10 min, transcripts were precipitated, electrophoresed on 6% sequencing gels, and quantified with a Molecular Dynamics PhosphorImager. To test the transcriptional ability of the NtrC mutant proteins in the absence of phosphorylation, carbamyl phosphate was omitted from the reaction mixture. The detection limit with this assay was 0.05 fmol.
RESULTS
Random PCR mutagenesis yielded 55 different single amino acid substitutions in the central domain of NtrC.
To generate new mutant forms of NtrC defective in positive control, we performed PCR mutagenesis of the DNA encoding most of its central domain (see Materials and Methods). PCR-amplified DNA fragments were then substituted for the corresponding fragment of pJES311, which expresses wild-type NtrC from a strong T7 promoter and translational start. The resulting plasmids were transformed into E. coli DH5α to generate a random mutant library.
When subjected to the genetic screen described in Materials and Methods, 95 of 1,234 transformants in the mutant library (8%) were unable to grow on the selective medium NB in the absence of glutamine, indicating that the plasmids they carried encoded putative ntrC(Rep) alleles. Plasmid DNA was extracted from each of these transformants, and sequencing revealed that 75 plasmids carried base substitutions in the targeted region of ntrC that resulted in single amino acid substitutions in the NtrC protein (Table 2). Among these, 14 plasmids carried an additional silent mutation. The plasmids from these 75 transformants implicated 55 different amino acid substitutions (at 44 amino acid positions) in producing a “repressor-like” phenotype. Plasmids from the remaining 20 transformants were found to carry either two (19 plasmids) or three (1 plasmid) point mutations. They were not analyzed further. Tabulations of the frequency of each of the 12 possible base pair substitutions showed a clear predominance of A · T→G · C transition mutations (63% [data not shown]), a distinct feature of the base substitution specificity of Taq DNA polymerase (51).
TABLE 2.
Summary of DNA sequence changes and transcriptional activation at glnA in vivo and in vitro
| Amino acid substitutiona | Codon change |
S. typhimurium strainb
|
In vivo activityc (U/ml/OD650)
|
E. coli straind | In vitro activitye | Region of NtrC central domainf | ||
|---|---|---|---|---|---|---|---|---|
| ntrC | ntrC ntrA | ntrC | ntrC ntrA | |||||
| E175G | GAA→GGA | SK3431 | 9 | NCM3264 | − | C1 | ||
| V177A∗ | GTC→GCC | SK3430 | SK3496 | 130 | 7 | NCM3265 | + | C1-C2 |
| A180T | GCG→ACG | SK3427 | 13 | NCM3216 | +/− | C1-C2 | ||
| L181I | CTT→ATT | SK3432 | SK3485 | 86g | 73 | NCM3208 | + | C1-C2 |
| A190V | GCG→GTG | SK3472 | SK3527 | 105 | 64 | NCM3266 | ppt | C1-C2 |
| A194T∗ | GCA→ACA | SK3438 | 20 | NCM3078 | + | C2 | ||
| N196H | AAT→CAT | SK3473 | 6 | NCM3267 | − | C2 | ||
| N196S | AAT→AGT | SK3436 | 17 | NCM3048 | +/− | C2 | ||
| M197V | ATG→GTG | SK3439 | 21 | NCM3079 | +/− | C2 | ||
| I200T | ATC→ACC | SK3440 | 8 | NCM3080 | − | C2 | ||
| I200V | ATC→GTC | SK3441 | SK3497 | 196 | 44 | NCM3081 | + | C2 |
| S207F | TCC→TTC | SK3426 | 16 | NCM3268 | − | C3 | ||
| E208V | GAA→GTA | SK3460 | 37 | NCM3269 | +/− | C3 | ||
| F210L | TTT→TTG | SK3461 | 15 | NCM3217 | +/− | C3 | ||
| K214E | AAA→GAA | SK3463 | 23 | NCM3270 | +/− | C3 | ||
| K214R | AAA→AGA | SK3462 | SK3498 | 216 | 20 | NCM3271 | +/− | C3 |
| G215V∗ | GGC→GTC | SK3464 | 7 | NCM3218 | − | C3 | ||
| F217L | TTT→CTT | SK3466 | 10 | NCM3272 | − | C3 | ||
| T218A | ACC→GCC | SK3465 | 13 | NCM3219 | − | C3 | ||
| T218N | ACC→AAC | SK3467 | 7 | NCM3273 | − | C3 | ||
| A220V | GCG→GTG | SK3468 | 11 | NCM3274 | + | C3 | ||
| Q230R | CAG→CGG | SK3442 | 66 | NCM3204 | + | C3-C4 | ||
| F237S | TTT→TCT | SK3443 | 15 | NCM3220 | ppt | C4 | ||
| L238P | CTG→CCG | SK3475 | 32 | NCM3262 | ppt | C4 | ||
| D239G | GAC→GGC | SK3457 | 18 | NCM3261 | − | C4 | ||
| M244V | ATG→GTG | SK3458 | 61 | NCM3260 | + | C4-C5 | ||
| Q249R | CAG→CGG | SK3437 | 23 | NCM3123 | ppt | C4-C5 | ||
| T250I | ACT→ATT | SK3433 | SK3486 | 128 | 10 | NCM3121 | + | C4-C5 |
| L253F | TTA→TTT | SK3450 | 18 | NCM3259 | − | C5 | ||
| L253V | TTA→GTA | SK3434 | 18 | NCM3122 | − | C5 | ||
| L256P | CTG→CCG | SK3476 | SK3528 | 279 | 2 | NCM3206 | ppt | C5 |
| Y262C | TAC→TGC | SK3435 | 20 | NCM3253 | +/− | C5-C6 | ||
| Y262N∗ | TAC→AAC | SK3446 | 13 | NCM3252 | +/− | C5-C6 | ||
| K271E | AAA→GAA | SK3451 | SK3501 | 333 | 13 | NCM3221 | + | C5-C6 |
| V274E | GTG→GAG | SK3444 | SK3529 | 192 | 15 | NCM3245 | ppt | C6 |
| T280A | ACC→GCC | SK3469 | 7 | NCM3213 | − | C6 | ||
| L284H | CTC→CAC | SK3470 | 23 | NCM3209 | ppt | C6 | ||
| E290G∗ | GAA→GGA | SK3452 | SK3502 | 295 | 36 | NCM3246 | + | C6 |
| G291R | GGG→AGG | SK3447 | SK3503 | 127 | 7 | NCM3207 | + | C6 |
| K292E | AAA→GAA | SK3471 | SK3530 | 325 | 17 | NCM3205 | + | C6 |
| D296G | GAC→GGC | SK3474 | 6 | NCM3210 | − | C6 | ||
| L297P∗ | CTG→CCG | SK3477 | SK3488 | 467 | 40 | NCM3251 | ppt | C6 |
| F298C | TTC→TGC | SK3428 | 14 | NCM3254 | +/− | C6 | ||
| F298L | TTC→CTC | SK3453 | SK3531 | 268 | 3 | NCM3250 | + | C6 |
| F298S | TTC→TCC | SK3454 | 29 | NCM3255 | − | C6 | ||
| H299L | CAC→CTC | SK3448 | 17 | NCM3249 | − | C6 | ||
| L301P∗ | CTG→CCG | SK3478 | 37 | NCM3211 | ppt | C6 | ||
| V303A | GTG→GCG | SK3429 | 31 | NCM3215 | − | C6 | ||
| V303M | GTG→ATG | SK3459 | 21 | NCM3256 | − | C6 | ||
| I304F∗ | ATT→TTT | SK3455 | 21 | NCM3247 | +/− | C6 | ||
| I304N | ATT→AAT | SK3449 | 26 | NCM3257 | ppt | C6 | ||
| P309L∗ | CCG→CTG | SK3479 | 15 | NCM3214 | ppt | C6 | ||
| I318T | ATT→ACT | SK3445 | SK3505 | 98 | 6 | NCM3248 | ppt | C6-C7 |
| L321P | CTG→CCG | SK3480 | 38 | NCM3258 | ppt | C6-C7 | ||
| L321Q | CTG→CAG | SK3456 | 39 | NCM3212 | ppt | C6-C7 | ||
| None | SK3425 | SK3483 | 1,349 ± 343† | 30 ± 22‡ | NCM3120 | |||
| — | SK3424 | SK3482 | 838 ± 90‡ | 664 ± 206§ | NCM724 | |||
Eleven of the mutational substitutions in this column occurred multiple times (number in parentheses): E175G (3), N196S (6), M197V (2), I200T (2), S207F (5), F217L (2), L256P (2), Y262C (3), V274E (2), L301P (2), and I318T (2). In each case, one isolate was chosen for further analysis. ∗, in addition to the mutation causing the amino acid substitution indicated, the plasmid we used contained one “silent mutation” in the mutagenized region of ntrC; —, no plasmid in these strains.
Strains listed in this column were constructed by electroporating multicopy plasmids into SK3424 (ntrC mutant host strain) or SK3482 (ntrC ntrA mutant host strain). The plasmid carried in each strain was either pJES311 or a pJES311 derivative expressing mutant NtrC proteins with the specific amino acid substitution indicated in the first column.
Values presented in this column were calculated from the slopes of plots of β-galactosidase activities (U/ml) expressed from a glnA′-‵lacZ fusion versus the OD650 of the cultures. Where means ± standard deviations are given, values were calculated from 10 (†), 6 (‡), or 7 (§) independent experiments.
Strains listed in this column are NCM724 derivatives obtained by transforming this overexpression host strain with the plasmids described in footnote b.
The “+,” “+/−,” and “−” designations indicate that the mutant protein had, respectively, no detectable activity, less than the maximum activity of NtrCWT (observed at 100 nM dimer) at any concentration, or at least as much as the maximum activity of NtrCWT at some concentration; “ppt” means that these proteins were in the pellet after cells were broken with the French pressure cell and therefore could not be analyzed by in vitro transcription assays. Refer to Table 3 and Fig. 4 for more complete data.
Conserved regions of the NtrC central domain according to Morett and Segovia (30).
This strain gave two different values (447 and 697) when assayed in two other independent experiments.
Assays of glnA′-‵lacZ expression indicated that all 55 new mutant forms of NtrC were deficient in transcriptional activation in vivo.
We measured levels of expression of a Φ(glnA′-‵lacZ) fusion at the put locus of S. typhimurium for all 55 mutant NtrC proteins containing single residue changes to assess their residual ability to activate transcription (Table 2 and Fig. 3). The β-galactosidase activity of the host strain (SK3424 ntrC) was ∼850 U/ml/OD650 and is attributable to transcription from the upstream ς70-dependent promoter. When a multicopy expression vector encoding a known NtrC(Rep) protein, NtrCS207F (that is, NtrC with a change from serine to phenylalanine at position 207), was introduced into SK3424, the β-galactosidase activity dropped to 16 U/ml/OD650 because NtrCS207F represses transcription from the upstream promoter and is unable to activate transcription from the ς54-dependent promoter. By contrast, when the multicopy expression vector encoding the NtrCWT protein was introduced, expression increased to ∼1,350 U/ml/OD650 under the derepressing growth conditions employed (see Materials and Methods). This increase in glnA′-‵lacZ expression was attributed to NtrC-dependent transcriptional activation at the downstream ς54-dependent promoter because transcription from the upstream ς70-dependent promoter was presumably repressed. When similar experiments were done with plasmids encoding all 55 new mutant forms of NtrC, they allowed us to divide these forms into two classes. One class (40 of 55) yielded β-galactosidase values comparable to those of known NtrC(Rep) proteins, i.e., ≤70 U/ml/OD650. The remainder yielded values ranging from ∼90 to ∼500 U/ml/OD650, less than that of the ntrC mutant host strain.
Residual glnA′-‵lacZ expression in strains expressing mutant NtrC proteins is ς54 dependent.
For mutant NtrC proteins that yielded the highest residual glnA′-‵lacZ expression (90 to 500 U/ml/OD650), we wanted to determine whether this activity was due to residual transcriptional activation from the downstream ς54-dependent promoter or to loss of repression at the upstream ς70-dependent promoter. To do so we performed two sorts of experiments: first, we ascertained whether residual expression was dependent on function of the ntrA gene product, ς54; second, we assessed immunologically whether any of the mutant proteins were significantly degraded.
When plasmids encoding all of the mutant NtrC proteins that yielded 90 to 500 U/ml/OD650 of β-galactosidase activity (glnA′-‵lacZ expression) in an ntrA+ background were introduced into an ntrA mutant background (SK3482 host strain [see Materials and Methods and Table 1]), β-galactosidase activities dropped to values lower than 75 U/ml/OD650 in all cases (Table 2). Control strain SK3489, which contains a single chromosomal ntrC+ allele in the same background as SK3482, had activities ranging between 40 and 80 U/ml/OD650 in five independent experiments, and control strain SK3483, which carries plasmid pJES311 encoding NtrCWT, gave β-galactosidase values between 10 and 70 U/ml/OD650 in six independent experiments (Table 2).
To assess the integrity of all 55 mutant NtrC proteins, we performed Western blotting experiments with cells carrying plasmids encoding these proteins in the SK3424 background (ntrC glnA′-‵lacZ). Cells were grown under the same derepressing conditions used for β-galactosidase assays (Materials and Methods). Of these, 54 gave rise to only one prominent band which had the same mobility as NtrCWT (encoded by pJES311). No secondary bands or bands of higher mobility were observed, and intensities of staining appeared to be comparable in all cases (data not shown). The host strain SK3424 (ntrC) yielded no band. In only one case, NtrCL297P (SK3477), did we fail to detect any Western blot signal. Further analysis showed that strain SK3477 had lost its plasmid.
Taken together, the above results allowed us to conclude that the residual glnA′-‵lacZ expression in an ntrA+ background was, in most instances, due to residual transcriptional activation by mutant NtrC proteins: activity was dependent on ς54, the product of the ntrA gene (with the possible exceptions of proteins carrying the L181I [but see Table 2, footnote g] and A190V substitutions), and there was no sign of degradation of the mutant proteins. Moreover, experiments with control strains confirmed that expression of NtrC from a single chromosomal copy was sufficient for full repression (assessed in an ntrA mutant background), whereas expression of all proteins from plasmids was far in excess of this amount.
Forty-one of 55 mutant forms of NtrC were soluble and could be purified.
To characterize their transcriptional activation capacities in vitro, we attempted to purify all 55 NtrC mutant proteins by using the rapid protocol described in Materials and Methods. Fourteen of the proteins could not be purified by this protocol because they were in the pellet after cells were broken with the French pressure cell (Table 2). The 41 proteins that were purified yielded from 2.6 to 18 nmol of dimer per 50 ml of culture (0.85 to 6 μM), with the average yield around 6 nmol (2 μM). The variation in yield could be due to inefficient induction during overexpression or to differences in behavior during purification. All proteins were at least 90% pure based on visual inspection of SDS-polyacrylamide gels stained with Coomassie blue, and none showed evidence of degradation (data not shown).
Seventeen of 41 soluble mutant proteins failed to activate transcription by ς54 holoenzyme in vitro.
The 41 NtrC mutant proteins purified were tested for the ability to catalyze the formation of open complexes by ς54 holoenzyme at the glnA promoter (Table 2; see also Materials and Methods). Phosphorylated NtrCWT catalyzed the formation of open complexes at concentrations ranging from 10 nM dimer (the lowest concentration tested) up to 500 nM dimer (the highest concentration tested) (Fig. 4 and Table 3). In contrast, 17 of the 41 phosphorylated NtrC mutant proteins, which had amino acid substitutions in six of the seven highly conserved regions in the central domain, failed to catalyze open complex formation at 10, 20, 50, and 100 nM dimer (Table 2). Four of these 17 (N196H, F217L, L253V, and V303A) were also tested at 200 and 500 nM dimer and, in all cases, yielded <1% as many open complexes as the wild-type protein at the same concentrations (data not shown). The 17 mutant proteins that failed to activate transcription from the ς54-dependent glnA promoter in vitro also showed little glnA transcription in vivo (<70 U/ml/OD650 of β-galactosidase activity from the glnA′-‵lacZ fusion). Hence, they behaved like the NtrC(Rep) proteins characterized previously.
FIG. 4.
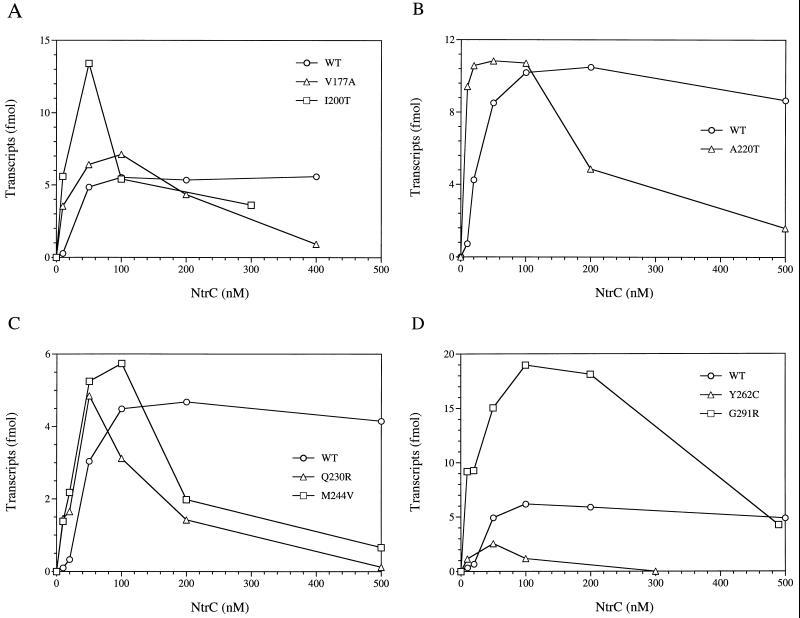
Transcriptional activation by selected mutant NtrC proteins in vitro. Formation of open complexes by ς54 holoenzyme was assessed by a single-round transcription assay as described in Materials and Methods. NtrC proteins were present at the concentrations indicated and were phosphorylated with carbamyl phosphate (10 mM). Effects of single amino acid substitutions were assessed by comparison to NtrCWT (open circles) in each panel. (A) V177A and I200V. (B) A220V. (C) Q230R and M244V. (D) Y262C and G291R. Proteins were chosen to illustrate their hyperactivity at low concentrations and the widespread locations of their amino acid substitutions within the central domain of NtrC: V177A between conserved regions 1 and 2 (C1-C2), I200V (C2), A220V (C3), Q230R (C3-C4), M244V (C4-C5), Y262C (C5), and G291R (C6). Data for NtrCWT is included in each plot to illustrate the variability between experiments.
TABLE 3.
Transcriptional activation by NtrC mutant proteins relative to the wild-type protein and by NtrCWT in different experiments
| Amino acid substitution or expt | Concna of NtrC (at indicated concn of dimer [nM])
|
||||||
|---|---|---|---|---|---|---|---|
| 10 | 20 | 50 | 100 | 200 | 300 | 500 | |
| Substitutionb | |||||||
| None | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| V177A (6) | 10.0 | 10.5 | 1.3 | 1.1 | 0.7 | nd | 0.2c |
| A180T (3) | 6.0 | 0.7 | 0.6 | 0.2 | 0.06 | nd | 0.03c |
| L181I (5) | 8.7 | 5.6 | 1.2 | 1.0 | 1.0 | nd | 0.7 |
| A194T (9) | 12.0 | nd | 1.7 | 0.5 | 0.07d | nd | nd |
| N196S (2) | 1.0 | 0.7 | 0.5 | 0.4 | 0.2 | nd | 0.05 |
| M197V (8) | 3.5 | nd | 0.6 | 0.6 | 0.4 | nd | nd |
| I200V (8) | 28.0 | nd | 2.8 | 1.1 | nd | 0.8 | nd |
| E208V (9) | 0.5 | nd | 0.1 | 0.01 | 0.0 | nd | nd |
| F210L (7) | 3.0 | nd | 0.4 | 0.2 | nd | 0.01 | nd |
| K214E (2) | 0.1 | 0.05 | 0.03 | 0.06 | 0.06 | nd | 0.02 |
| K214R (6) | 6.6 | 2.3 | 0.1 | 0.01 | 0.0 | nd | 0.0c |
| A220V (1) | 13.4 | 2.5 | 1.3 | 1.0 | 0.5 | nd | 0.2 |
| Q230R (4) | 15.0 | 5.7 | 1.6 | 0.7 | 0.3 | nd | 0.03 |
| M244V (4) | 14.0 | 7.3 | 1.7 | 1.3 | 0.4 | nd | 0.2 |
| T250I (9) | 28.5 | 10.2 | 2.2 | 1.5 | nd | nd | nd |
| Y262C (7) | 3.0 | nd | 0.5 | 0.2 | nd | 0.0 | nd |
| Y262N (3) | 5.0 | 0.5 | 0.2 | 0.2 | 0.03 | nd | 0.0c |
| K271E (8) | 1.0 | nd | 0.9 | 1.6 | nd | 1.3 | nd |
| E290G (7) | 17.0 | nd | 2.0 | 1.7 | nd | 1.0 | nd |
| G291R (5) | 23.0 | 13.3 | 3.0 | 3.0 | 3.0 | nd | 0.8 |
| K292E (7) | 25.0 | nd | 2.0 | 1.6 | nd | 0.8 | nd |
| F298C (9) | 1.0 | nd | 0.7 | 0.4 | 0.2 | nd | nd |
| F298L (8) | 13.0 | nd | 1.5 | 1.4 | nd | 1.4 | nd |
| I304F (1) | 4.0 | 1.5 | 0.8 | 0.4 | 0.06 | nd | 0.05 |
| Expt | |||||||
| 1 | 0.7 | 4.3 | 8.5 | 10.2 | 10.5 | nd | 8.7 |
| 2 | 1.2 | 4.2 | 9.7 | 10.7 | 10.2 | nd | 8.0 |
| 3 | 0.2 | 4.0 | 8.4 | 10.2 | 10.2 | nd | 8.6c |
| 4 | 0.1 | 0.3 | 3.0 | 4.5 | 4.7 | nd | 4.2 |
| 5 | 0.4 | 0.7 | 4.9 | 6.2 | 5.9 | nd | 4.9 |
| 6 | 0.4 | 0.6 | 5.0 | 6.2 | 6.4 | nd | 5.6c |
| 7 | 0.4 | nd | 4.8 | 5.8 | nd | 6.2 | nd |
| 8 | 0.2 | nd | 4.7 | 4.8 | 4.7 | 4.5 | nd |
| 9 | 0.2 | 0.6 | 3.0 | 4.2 | 4.0 | nd | nd |
| Avg | 0.4 ± 0.3 | 2.2 ± 1.9 | 5.3 ± 2.2 | 6.5 ± 2.4 | 7.2 ± 2.7 | 5.3 ± 1.0 | 6.5 ± 2.0 |
Data for individual amino acid substitutions are ratios of activities of mutant proteins to those of the wild-type protein. Data for the wild-type protein in individual experiments are presented in femtomoles. nd, not determined.
Numbers in parentheses refer to the corresponding experiment numbers (1 through 9 [below]), for which the data are wild-type values used for normalization.
Value at 400 nM dimer.
Value at 150 nM dimer.
The remaining 24 of 41 soluble mutant proteins did activate transcription by ς54 holoenzyme in vitro.
Unexpectedly, in vitro transcription assays revealed a large class of 24 mutant proteins that did catalyze formation of open complexes by ς54 holoenzyme in vitro. Among these proteins, 14 yielded essentially no glnA transcription in vivo (<70 U/ml/OD650 of β-galactosidase activity from the glnA′-‵lacZ fusion). Ten of these proteins (A180T, N196S, M197V, E208V, F210L, K214E, Y262C, Y262N, F298C, and I304F) failed to reach the maximum activity of phosphorylated NtrCWT in vitro (achieved at 100 nM dimer) at any concentration, and two of these (E208V and K214E) had very low levels of activity (Table 3 and Fig. 4D). The remaining four proteins of this subclass (A194T, A220V, Q230R, and M244V) did reach the maximum activity of NtrCWT (Table 3; Fig. 3, 4B, and 4C).
Ten of the 24 mutant proteins that activated transcription by ς54 holoenzyme in vitro also did so in vivo (between 90 and 500 U/ml/OD650 of β-galactosidase activity from the glnA′-‵lacZ fusion and dependent on an ntrA+ allele). Nine of these 10 proteins (V177A, L181I, I200V, T250I, K271E, E290G, G291R, K292E, and F298L) were fully active in vitro at a dimer concentration of 100 nM (Table 3; Fig. 3, 4A, and 4D), whereas 1 (K214R) yielded only 1% as many open complexes as NtrCWT at the same concentration and had very low levels of activity generally (Table 3).
Although it is reasonably common for mutant proteins to show less activity in vitro than in vivo, the reverse is unusual; therefore, we sought a specific explanation for why a large number of our mutant proteins showed activity in vitro but not in vivo. Because preliminary Western blot experiments had indicated that all proteins were greatly overexpressed even in the absence of T7 RNA polymerase, the 24 mutant proteins described in this section were tested for transcriptional activation at high concentrations (from 200 to 500 nM dimer). The activities of at least half dropped dramatically (Table 3; Fig. 4A through D). We infer that decreases in activity were due to abnormal oligomerization or aggregation because NtrCWT maintained maximal activity at 500 nM dimer (Table 3 and Fig. 4). For the 14 proteins that lacked activity in vivo, in vitro activities at 200 to 500 nM dimer dropped to values well below those for NtrCWT (Table 3; Fig. 4B through D). As shown below, the estimated in vivo concentrations of NtrC proteins from the overexpression plasmid were well above 500 nM, even in the absence of T7 RNA polymerase.
Nineteen of the 24 NtrC mutant proteins that activated transcription by ς54 holoenzyme in vitro showed hyperactivity at low concentrations.
Perhaps our most interesting result was the observation that 19 of the 24 mutant proteins that catalyze the formation of open complexes by ς54 holoenzyme in vitro (discussed above) did so better than NtrCWT at low concentrations. They yielded 3 to 20 times as many open complexes as NtrCWT at 10 nM dimer (Table 3; Fig. 4A through D). Figures 4A through D show some particularly striking examples of hyperactivity (V177A, I200V, A220V, Q230R, M244V, and G291R) and a single example (Y262C) in which hyperactivity at low concentrations is not accompanied by the ability to reach the maximum activity for NtrCWT at any concentration.
All 24 mutant proteins that are active in vitro must be phosphorylated.
Some NtrC mutant proteins, called NtrC constitutive [NtrC(Con)], can hydrolyze ATP and activate transcription in vitro without being phosphorylated. Amino acid substitutions in NtrC(Con) proteins have been localized to both the N-terminal regulatory domain and the central domain (10). To determine whether any of the 24 NtrC mutant proteins that were active in vitro were active without being phosphorylated, i.e., had the NtrC(Con) phenotype, all were tested for transcriptional activation in the absence of carbamyl phosphate, the in vitro phosphate donor. All 24 mutant proteins were tested at concentrations of 50 and 100 nM dimer, concentrations at which all of the phosphorylated proteins gave rise to clearly visible transcripts, as did the unphosphorylated NtrC(Con) protein NtrCS160F (10), which was used as a positive control. No visible transcripts were observed for the 24 new mutant proteins, showing that all must be phosphorylated to catalyze formation of open complexes by ς54 holoenzyme at the glnA promoter.
Levels of NtrC protein from the expression plasmid in the absence of T7 polymerase are hundreds of times higher than the chromosomal level.
The unexpected activities of many mutant NtrC proteins in vitro, together with the observation that their activities dropped considerably with an increase in protein concentration in many instances, led us to quantitate the overexpression of these proteins in vivo. Preliminary Western blot experiments performed with cells carrying plasmids that encoded either NtrCWT (pJES311) or the mutant proteins had already attracted our attention to the high levels of overexpression even in the absence of T7 RNA polymerase.
To estimate the degree of overexpression with respect to a single chromosomal ntrC+ allele, we lysed whole cells of strain SK3425, which carries pJES311, and strain SK1490, which contains a single chromosomal copy of ntrC, in SDS loading buffer and compared the intensity of the NtrC band in Western blots for dilutions of the lysate from SK3425 to those for undiluted lysate from SK1490. Comparable intensities were observed when the lysate of strain SK3425 was diluted on the order of 500-fold (Fig. 5A). To estimate the concentration of NtrC when it is overexpressed 500-fold, we next determined the absolute amount of NtrC in lysates of SK1490 by comparing the intensity of the NtrC band in Western blots for a lysate of this strain to those of different amounts of purified NtrC protein added to a lysate of strain SK3424 (ntrC) (Fig. 5B). Matched intensities were observed when 25 to 50 ng of purified NtrC protein was added to the lysate of SK3424. Assuming that the amount of NtrC expressed from a single chromosomal copy of ntrC is between 25 and 50 ng for the number of cells used (see below) and using the conversion factors given in Materials and Methods, we estimate that the concentration of NtrC is between 1.0 and 2.0 μM under nitrogen-limiting conditions (25 ng of NtrC dimer = 2.4 × 10−13 mol [the molecular mass of NtrC is 104,476 g/mol of dimer]; 0.5 ml of culture at an OD650 of 0.5 has 9.25 × 10−2 mg [dry weight]; 9.25 × 10−2 mg [dry weight] × 2 μl/mg [dry weight] = 0.19 μl ∴ 1.9 × 10−7 liter; [2.4 × 10−13 mol]/[1.9 × 10−7 liter] = 1.3 μM). Because the amount of NtrC protein expressed from plasmid pJES311 and its derivatives encoding mutant NtrC proteins (see above) appears to be on the order of 500-fold higher than that expressed from a single ntrC+ allele, the concentration of NtrC in strains carrying these plasmids is in the range of 0.5 to 1.0 mM, several hundredfold above the maximum concentration tested in vitro (0.5 μM). Thus, the apparent lack of activity of many NtrC mutant proteins in vivo appears to be accounted for by high levels of expression.
FIG. 5.
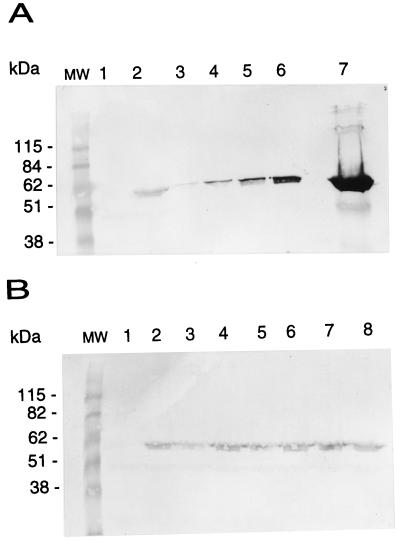
Amounts of NtrC protein expressed from a single chromosomal ntrC+ allele or from plasmid pJES311, which overexpresses NtrCWT. Cells were cultivated under nitrogen-limiting conditions as for assays of glnA′-‵lacZ expression (see Materials and Methods), and 0.5 ml of culture at an OD650 of 0.5 was concentrated and loaded on an SDS–10% polyacrylamide gel. Amounts of NtrC protein in crude lysates were determined by Western blotting with mouse polyclonal antiserum raised against the carboxy-terminal domain of NtrC. (A) Crude lysates of SK3424 (ntrC) (lane 1) and SK1490 (ntrC+) (lane 2) were compared with dilutions of lysates of SK3425(pJES311): lane 3, 1:1,250; lane 4, 1:625; lane 5, 1:125; lane 6, 1:25; lane 7, undiluted. (B) Crude lysates of strain SK1490 (lanes 2, 4, 6, and 8) were compared with different amounts of purified NtrCWT added to crude lysates of SK3424: lane 1, 0 ng; lane 3, 12.5 ng; lane 5, 25 ng; lane 7, 50 ng. MW, BenchMark molecular weight standards (Gibco BRL).
The above estimate for the concentration of NtrC in a cell carrying a single chromosomal ntrC+ allele is 10- to 20-fold higher than an estimate made previously (70 molecules/cell = ∼70 molecules/fl = ∼0.1 μM) (41). Nonetheless, if the amount of NtrC protein expressed from pJES311 and its derivatives is 500-fold higher than that expressed from a single ntrC+ allele, the concentration of NtrC in strains carrying these plasmids would be in the range of 50 μM, still 100-fold above the maximum concentration tested in vitro. Hence the conclusion stands.
DISCUSSION
New NtrC(Rep) proteins (positive control forms) have lesions in regions of the central domain that are not well studied.
In vitro assays of transcriptional activation at the ς54-dependent glnA promoter by 41 NtrC mutant proteins with decreased activity in vivo yielded 16 new forms that failed in transcriptional activation. Together with the 13 previously studied forms, lesions in these NtrC(Rep) proteins affect residues in or, in one case, directly adjacent to all seven of the conserved motifs in the central domain (30, 38). These include two regions—C2 and C5—in which such lesions had not been found previously and the unusually long conserved region C6, in which only a single lesion had been characterized (37, 43, 57). Lesions affecting C2 and C5 have not yet been characterized for other ς54-dependent activators, and few lesions in C6 have been characterized (54). In only 1 of 29 cases (T280A in the C6 region) was the amino acid residue in the inactive mutant form of NtrC found at the corresponding position of a homologous activator of ς54 holoenzyme (DctD of Rhizobium leguminosarum). Several of the residues that were altered in the inactive mutant forms of NtrC (N196, T218, D296, R358) had been predicted to be “functional” residues (38).
Unlike regions C1, C3, C4, and C7, which have obvious functional parallels in the large family of purine nucleotide-binding proteins with a mononucleotide fold (see the introduction), regions C2, C5, and C6 do not. Based on its location just downstream of the conserved aspartate of the Walker B motif, D239 in conserved region 4 of NtrC, it is possible that region C5 or the region between C4 and C5 corresponds to the switch II motif, critical in coupling of nucleotide hydrolysis to biological output. The one lesion in C6 that we have characterized previously, R294C, resulted in loss of ATP hydrolysis without effect on ATP binding or on oligomerization of NtrC (37, 43). It is possible that R294 is a catalytic residue. Oligomerization determinants of NtrC, which are thought to lie in its central domain (10), remain to be defined.
NtrC mutant forms that are hyperactive at low concentrations have lesions throughout the central domain.
Unexpectedly, 24 new mutant forms of NtrC that failed to activate transcription in vivo, or activated poorly, did activate in vitro. Because the activation by many of these forms decreased at high concentrations in vitro, their apparent failure to activate in vivo is probably accounted for by high levels of overexpression (∼500-fold [see Results]). Nineteen such forms were more active at low concentrations (10 to 50 nM) than NtrCWT, and many of these showed decreases in activity at higher concentrations (200 to 500 nM). These mutant forms have lesions that are widespread in the central domain of NtrC. They occur in the region between C1, the Walker A motif, and C2 (e.g., V177A and L181I); in or immediately adjacent to the C2 region (e.g., A194T and I200V); in the region between C4 and C5 (e.g., M244V and T250I); and in region C6, where they affect the three adjacent residues E290 to K292 and residue E298. There is one such lesion (A220V) in C3, the switch I region, and one (Q230R) between C3 and the Walker B motif, C4. The significance of the widespread occurrence of these forms is not understood, nor is the functional basis for their unexpected phenotype in vitro. Hyperactivity at low concentrations may be due to increased oligomerization and ATPase activity, increased biological output, or both, and the basis may be different for different lesions. Lesions to hyperactivity that occurred between, rather than within, conserved regions of NtrC tended to affect residues that were tree determining for the NtrC subfamily of ς54-dependent activators (e.g., V177, L181, Q230, and T250) (38). These residues were postulated to participate in unique aspects of oligomerization or of the response to signals from the regulatory domain.
Interestingly, amino acid covariation analysis indicated that region C6, a region in which several changes to hyperactivity occurred in NtrC, appears to make tertiary contacts with many of the other conserved regions in activators of ς54 holoenzyme. For several of the lesions in this region, the amino acid substitution that results in hyperactivity of the mutant form of NtrC at low concentrations is found naturally in the wild-type form of another activator. (This is also true for lesions to hyperactivity in other regions of NtrC; for a complete list, see the legend to Fig. 2.) A particularly interesting example is the G291R substitution, because this glycine residue is highly conserved among activators of ς54 holoenzyme and the substitution appears to be extreme. NtrCG291R has 20 times higher activity than NtrCWT at a concentration of 10 nM and continues to have 3-fold-more activity than NtrCWT when the wild-type protein has reached its maximal activity at 100 nM. An arginine is found at the corresponding position for the wild-type form of the E. coli FhlA protein (Fig. 2). Similar observations pertain regarding the extreme substitution at the adjacent position in C6, K292E, and for the F298L substitution (Fig. 2). Moreover, in the latter case, different amino acid substitutions at the same position result in either hyperactivity (F298L) or loss of activity (F298S). The only other positions at which both phenotypes were observed were position I200 in the C2 region (I200V resulted in hyperactivity, whereas I200T resulted in loss of activity) and position 220 in the C3, or switch I, region (see below).
Comments on new lesions in the Walker A (C1), Walker B (C4), and switch I (C3) regions.
The Walker A motif for activators of ς54 holoenzyme (C1) differs from that of other members of the purine nucleotide-binding protein family in ending with a conserved E or D, rather than the usual S or T, which is often involved in coordination of the divalent cation (38). NtrCE175G, in which the conserved glutamate is altered, was inactive in vivo and in vitro. The same was true when the corresponding glutamate in the homologous activator DctD was changed to either T or A (11). The mutant DctD proteins were shown to be greatly defective in ATP hydrolysis (11). Lesions in NtrC that altered the two hydrophobic residues preceding D239, the conserved aspartate of the Walker B motif, resulted in precipitation of the corresponding proteins (NtrCF237S and NtrCL238P). The hydrophobic residues normally present at these positions of purine nucleotide-binding proteins are part of a β-strand (38). Like previously characterized lesions at position 239 (D239A, N, and C [43]), each of which resulted in loss of ATPase activity but not ATP binding, an additional lesion at this position, D239G, resulted in loss of transcriptional activation. Interestingly, although NtrC has a glycine at position 242, as do other “DEXG proteins,” the E247G substitution at the corresponding position of Rhizobium meliloti DctD caused loss of transcriptional activation both in vivo and in vitro (54). This appears to be the only example in which changing a residue of a homologous activator to that found at the corresponding position of NtrC results in loss of activity.
The A220V substitution at the end of C3, the switch I region, results in hyperactivity at low concentrations, whereas the A220T substitution and an A-to-T substitution at the corresponding position of DctD (54) result in loss of transcriptional activation with little decrease in ATPase activity (37). Given that the switch I region does not appear to affect ATP hydrolysis per se but rather couples the energy made available to biological output, hyperactivity of NtrCA220V at low concentrations may be due to an improved efficiency of interaction with ς54 holoenzyme.
ACKNOWLEDGMENTS
We are indebted to O. Carmi for assistance in preparation of the manuscript.
This work was supported by a Brazilian Research Council (CNPq) postdoctoral fellowship to L.P. and by National Institutes of Health grant G738361 to S.K.
J.L. and L.P. contributed equally to this work and should both be considered the first author.
REFERENCES
- 1.Austin S, Dixon R. The prokaryotic enhancer binding protein NtrC has an ATPase activity which is phosphorylation and DNA dependent. EMBO J. 1992;11:2219–2228. doi: 10.1002/j.1460-2075.1992.tb05281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger D K, Narberhaus F, Kustu S. The isolated catalytic domain of NIFA, a bacterial enhancer-binding protein, activates transcription in vitro: activation is inhibited by NIFL. Proc Natl Acad Sci USA. 1994;91:103–107. doi: 10.1073/pnas.91.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature (London) 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 4.Buikema W J, Szeto W W, Lemley P V, Orme-Johnson W H, Ausubel F M. Nitrogen fixation specific regulatory genes of Klebsiella pneumoniae and Rhizobium meliloti share homology with the general nitrogen regulatory gene ntrC of K. pneumoniae. Nucleic Acids Res. 1985;13:4539–4555. doi: 10.1093/nar/13.12.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1980. [Google Scholar]
- 6.Drummond M, Whitty P, Wootton J. Sequence and domain relationships of ntrC and nifA from Klebsiella pneumoniae: homologies to other regulatory proteins. EMBO J. 1986;5:441–447. doi: 10.1002/j.1460-2075.1986.tb04230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drummond M H, Contreras A, Mitchenall L A. The function of isolated domains and chimaeric proteins constructed from the transcriptional activators NifA and NtrC of Klebsiella pneumoniae. Mol Microbiol. 1990;4:29–37. doi: 10.1111/j.1365-2958.1990.tb02012.x. [DOI] [PubMed] [Google Scholar]
- 8.Eckert K A, Kunkel T A. High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1990;18:3739–3744. doi: 10.1093/nar/18.13.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 10.Flashner Y, Weiss D S, Keener J, Kustu S. Constitutive forms of the enhancer-binding protein NtrC: evidence that essential oligomerization determinants lie in the central activation domain. J Mol Biol. 1995;249:700–713. doi: 10.1006/jmbi.1995.0330. [DOI] [PubMed] [Google Scholar]
- 11.Gao Y, Wang Y K, Hoover T R. Mutational analysis of the phosphate-binding loop of Rhizobium meliloti DctD, a ς54-dependent activator. J Bacteriol. 1998;180:2792–2795. doi: 10.1128/jb.180.10.2792-2795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutnick D, Calvo J M, Klopotowski T, Ames B N. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J Bacteriol. 1969;100:215–219. doi: 10.1128/jb.100.1.215-219.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilgenfeld R. Regulatory GTPases. Curr Opin Struct Biol. 1995;5:810–817. doi: 10.1016/0959-440x(95)80015-8. [DOI] [PubMed] [Google Scholar]
- 14.Huala E, Ausubel F M. The central domain of Rhizobium meliloti NifA is sufficient to activate transcription from the R. meliloti nifH promoter. J Bacteriol. 1989;171:3354–3365. doi: 10.1128/jb.171.6.3354-3365.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huala E, Stigter J, Ausubel F M. The central domain of Rhizobium leguminosarum DctD functions independently to activate transcription. J Bacteriol. 1992;174:1428–1431. doi: 10.1128/jb.174.4.1428-1431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda T P, Shauger A E, Kustu S. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J Mol Biol. 1996;259:589–607. doi: 10.1006/jmbi.1996.0342. [DOI] [PubMed] [Google Scholar]
- 17.Keener J, Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci USA. 1988;85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S-H, Privé G G, Milburn M V. Conformational switch and structural basis for oncogenic mutations of Ras proteins. In: Dickey B F, Birnbaumer L, editors. GTPases in biology I. Berlin, Germany: Springer-Verlag; 1993. pp. 177–194. [Google Scholar]
- 19.Klose K E, North A K, Stedman K M, Kustu S. The major dimerization determinants of the nitrogen regulatory protein NTRC from enteric bacteria lie in its carboxy-terminal domain. J Mol Biol. 1994;241:233–245. doi: 10.1006/jmbi.1994.1492. [DOI] [PubMed] [Google Scholar]
- 20.Klose K E, Weiss D S, Kustu S. Glutamate at the site of phosphorylation of nitrogen-regulatory protein NtrC mimics aspartyl-phosphate and activates the protein. J Mol Biol. 1993;232:67–78. doi: 10.1006/jmbi.1993.1370. [DOI] [PubMed] [Google Scholar]
- 21.Kofoid E C, Parkinson J S. Transmitter and receiver modules in bacterial signaling proteins. Proc Natl Acad Sci USA. 1988;85:4981–4985. doi: 10.1073/pnas.85.14.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kustu S, North A K, Weiss D S. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem Sci. 1991;16:397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- 23.Kustu S, Santero E, Keener J, Popham D, Weiss D. Expression of ς54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung D W, Chen E, Goeddel D V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 25.Lowry O H, Carter J, Ward J B, Glaser L. The effect of carbon and nitrogen sources on the level of metabolic intermediates in Escherichia coli. J Biol Chem. 1971;246:6511–6521. [PubMed] [Google Scholar]
- 26.Magasanik B. Reversible phosphorylation of an enhancer binding protein regulates the transcription of bacterial nitrogen utilization genes. Trends Biochem Sci. 1988;13:475–479. doi: 10.1016/0968-0004(88)90234-4. [DOI] [PubMed] [Google Scholar]
- 27.McCarter L, Krajewska-Grynkiewicz K, Trinh D, Wei G, Kustu S. Characterization of mutations that lie in the promoter-regulatory region for glnA, the structural gene encoding glutamine synthetase. Mol Gen Genet. 1984;197:150–160. doi: 10.1007/BF00327936. [DOI] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- 29.Morett E, Buck M. In vivo studies on the interaction of RNA polymerase-ς54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters. The role of NifA in the formation of an open promoter complex. J Mol Biol. 1989;210:65–77. doi: 10.1016/0022-2836(89)90291-x. [DOI] [PubMed] [Google Scholar]
- 30.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neidhardt F C, Umbarger H E. Chemical composition of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 13–16. [Google Scholar]
- 32.Ninfa A J, Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci USA. 1986;83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ninfa A J, Reitzer L J, Magasanik B. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell. 1987;50:1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- 34.Nixon B T, Ronson C W, Ausubel F M. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc Natl Acad Sci USA. 1986;83:7850–7854. doi: 10.1073/pnas.83.20.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.North A K, Klose K E, Stedman K M, Kustu S. Prokaryotic enhancer-binding proteins reflect eukaryote-like modularity: the puzzle of nitrogen regulatory protein C. J Bacteriol. 1993;175:4267–4273. doi: 10.1128/jb.175.14.4267-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.North A K, Kustu S. Mutant forms of the enhancer-binding protein NtrC can activate transcription from solution. J Mol Biol. 1997;267:17–36. doi: 10.1006/jmbi.1996.0838. [DOI] [PubMed] [Google Scholar]
- 37.North A K, Weiss D S, Suzuki H, Flashner Y, Kustu S. Repressor forms of the enhancer-binding protein NtrC: some fail in coupling ATP hydrolysis to open complex formation by ς54-holoenzyme. J Mol Biol. 1996;260:317–331. doi: 10.1006/jmbi.1996.0403. [DOI] [PubMed] [Google Scholar]
- 38.Osuna J, Soberón X, Morett E. A proposed architecture for the central domain of the bacterial enhancer-binding proteins based on secondary structure prediction and fold recognition. Protein Sci. 1997;6:543–555. doi: 10.1002/pro.5560060304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popham D L, Szeto D, Keener J, Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- 40.Porter S C, North A K, Wedel A B, Kustu S. Oligomerization of NtrC at the glnA enhancer is required for transcriptional activation. Genes Dev. 1993;7:2258–2273. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- 41.Reitzer L J, Magasanik B. Expression of glnA in Escherichia coli is regulated at tandem promoters. Proc Natl Acad Sci USA. 1985;82:1979–1983. doi: 10.1073/pnas.82.7.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reitzer L J, Magasanik B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell. 1986;45:785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- 43.Rombel I, Peters-Wendisch P, Mesecar A, Thorgeirsson T, Shin Y-K, Kustu S. MgATp binding and hydrolysis determinants of NtrC, a bacterial enhancer-binding protein. J Bacteriol. 1999;181:4628–4638. doi: 10.1128/jb.181.15.4628-4638.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ronson C W, Astwood P M, Nixon B T, Ausubel F M. Deduced products of C4-dicarboxylate transport regulatory genes of Rhizobium leguminosarum are homologous to nitrogen regulatory gene products. Nucleic Acids Res. 1987;15:7921–7934. doi: 10.1093/nar/15.19.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Sasse-Dwight S, Gralla J D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci USA. 1988;85:8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seefeldt L C, Dean D R. Role of nucleotides in nitrogenase catalysis. Accounts Chem Res. 1997;30:260–266. [Google Scholar]
- 48.Story R M, Steitz T A. Structure of the RecA protein-ADP complex. Nature (London) 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 49.Tabor S. Expression using the T7 RNA polymerase/promoter system. In: Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1990. pp. 16.2.1–16.2.11. [Google Scholar]
- 50.Thöny B, Hennecke H. The −24/−12 promoter comes of age. FEMS Microbiol Rev. 1989;5:341–357. doi: 10.1016/0168-6445(89)90028-4. [DOI] [PubMed] [Google Scholar]
- 51.Tindall K R, Kunkel T A. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988;27:6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- 52.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walsh K, Koshland D E., Jr Determination of flux through the branch point of two metabolic cycles. The tricarboxylic acid cycle and the glyoxylate shunt. J Biol Chem. 1984;259:9646–9654. [PubMed] [Google Scholar]
- 54.Wang Y K, Hoover T R. Alterations within the activation domain of the ς54-dependent activator DctD that prevent transcriptional activation. J Bacteriol. 1997;179:5812–5819. doi: 10.1128/jb.179.18.5812-5819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wedel A, Weiss D S, Popham D, Dröge P, Kustu S. A bacterial enhancer functions to tether a transcriptional activator near a promoter. Science. 1990;248:486–490. doi: 10.1126/science.1970441. [DOI] [PubMed] [Google Scholar]
- 56.Wei G R, Kustu S. Glutamine auxotrophs with mutations in a nitrogen regulatory gene, ntrC, that is near glnA. Mol Gen Genet. 1981;183:392–399. doi: 10.1007/BF00270646. [DOI] [PubMed] [Google Scholar]
- 57.Weiss D S, Batut J, Klose K E, Keener J, Kustu S. The phosphorylated form of the enhancer-binding protein NtrC has an ATPase activity that is essential for activation of transcription. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- 58.Weiss D S, Klose K E, Hoover T R, North A K, Porter S C, Wedel A B, Kustu S. Prokaryotic transcriptional enhancers. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 667–694. [Google Scholar]
- 59.Weiss V, Claverie-Martin F, Magasanik B. Phosphorylation of nitrogen regulator I of Escherichia coli induces strong cooperative binding to DNA essential for activation of transcription. Proc Natl Acad Sci USA. 1992;89:5088–5092. doi: 10.1073/pnas.89.11.5088. . (Erratum, 89:8856.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiss V, Magasanik B. Phosphorylation of nitrogen regulator I (NRI) of Escherichia coli. Proc Natl Acad Sci USA. 1988;85:8919–8923. doi: 10.1073/pnas.85.23.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wyman C, Rombel I, North A K, Bustamante C, Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 62.Zhou Y H, Zhang X P, Ebright R H. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 1991;19:6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]