Abstract
The brain–gut axis (BGA) is an important bidirectional communication pathway for the development, progress and interaction of many diseases between the brain and gut, but the mechanisms remain unclear, especially the post-transcriptional regulation of BGA after traumatic brain injury (TBI). RNA methylation is one of the most important modifications in post-transcriptional regulation. N6-methyladenosine (m6A), as the most abundant post-transcriptional modification of mRNA in eukaryotes, has recently been identified and characterized in both the brain and gut. The purpose of this review is to describe the pathophysiological changes in BGA after TBI, and then investigate the post-transcriptional bidirectional regulation mechanisms of TBI-induced BGA dysfunction. Here, we mainly focus on the characteristics of m6A RNA methylation in the post-TBI BGA, highlight the possible regulatory mechanisms of m6A modification in TBI-induced BGA dysfunction, and finally discuss the outcome of considering m6A as a therapeutic target to improve the recovery of the brain and gut dysfunction caused by TBI.
Keywords: m6A RNA modification, brain-gut axis, traumatic brain injury
1. Introduction
Traumatic brain injury (TBI), one of the major causes of death and disability worldwide [1], impacts more than 50 million people every year, of whom 10 million die and nearly another 30 million remain physically disabled. Survivors experience a huge influence of physical, mental and cognitive disabilities, bringing considerable costs to their families and society [2,3]. In China, over 1 million new cases of TBI are reported each year [4]. In recent years, although the number of deaths caused by TBI has decreased significantly, unfortunately, effective treatments to promote the restoration of brain functions are still lacking. Thus, there is currently a major shift in TBI research in neural recovery and neurorehabilitation [5]. Increasing evidence has confirmed that the brain–gut axis (BGA) plays a key role in regulating behaviours associated with various mental and neurological diseases [6,7,8], including TBI [9]. A meta-analysis revealed that downregulated levels of anti-inflammatory butyrate-producing bacteria and upregulated levels of pro-inflammatory genera were consistently shared between major depressive disorder, psychosis and schizophrenia, bipolar disorder, and anxiety [10]; while a study showed that rifaximin improved depressive-like behaviour induced by chronic unpredictable mild stress via regulating the abundance of faecal microbial metabolites and microglial functions [11]. A clinical study showed significant alterations in gut microbiota and metabolome in faecal samples from Parkinson’s disease(PD) patients [12], while a meta-analysis indicated that inflammatory bowel disease(IBD) may increase PD risk, especially in patients over 65 years of age [13]. A pilot study suggested that the diversity of the microbial ecosystem in brain tumour patients was less than the healthy controls [14], while caecal content transfer experiments in mice revealed that cancer-associated alteration in caecal metabolites was involved in late-stage glioma progression [15]. In addition, there is clear evidence that TBI can cause gut microbiota dysbiosis [16], while disruption of the gut barrier and depletion of the gut microbiota are associated with neurologic outcomes following TBI [17]. With a growing understanding of BGA, people have a strong interest in studying the influences of BGA in neural recovery and neurorehabilitation after TBI.
However, the mechanisms of BGA in regulating mental and nervous system diseases induced by TBI are still unclear, especially in posttranscriptional regulation. Methylation modification is one of the most essential modifications in posttranscriptional regulation, including N6-methyladenosine (m6A), N1-methyladenosine, inosine, 5-methylcytidine, 5-hydroxymethylcytidine, pseudouridine, N6,2′-O-dimethyladenosine, N4-acetylcytidine, N7-methylguanosine, 8-oxoguanosine and 2′-O-methyl [18,19,20]. Among them, m6A, one of the most abundant mRNA posttranscriptional modifications in eukaryotes, has recently been identified and characterised in both the brain and gut. M6A modification is prevalent in the central nervous system, regulates the activation of various nerve conduction pathways and plays an irreplaceable role in the development, differentiation and regeneration of neurons [21,22,23]. At the same time, it exhibits an important impact on the communication between the gut microbiome and the host [24,25,26,27]. Therefore, m6A modification may also play a significant role in BGA. The main aim of this paper is to summarise the roles of m6A RNA methylation in post-TBI BGA to further highlight the possible regulatory mechanisms of m6A modification in TBI-induced BGA dysfunction, and, finally, to discuss the outcome of considering m6A as a therapeutic target to improve the recovery of the brain and gut dysfunction caused by TBI.
2. BGA
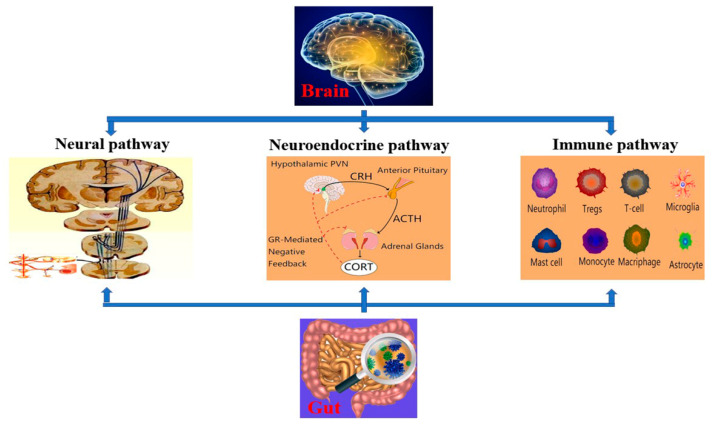
BGA, a bidirectional communication network connecting the central nervous system (CNS) and enteric nervous system (ENS) [28], is mainly composed of three pathways: the neural, neuroendocrine and immune pathways (Figure 1) [29]. (1). The neural pathway: the bidirectional exchange between the CNS and ENS, is the basis of BGA, and the autonomic nervous system (ANS), whose main branch is the ENS, is the centre of this interaction [9]. The afferent nerves of these systems are in key locations throughout the gut, including the mucosal surface. Both the sympathetic and parasympathetic systems are active, and the imbalance of input induces autonomic nerve disorders, which leads to gut dysfunction [30]. In addition, the gut microbiome can affect the production of neurotransmitters by “de novo synthesis” or by impacting neurotransmitter-related metabolism pathways [29]. (2). The neuroendocrine pathway: the hypothalamus-pituitary-adrenal (HPA) axis, is the key to neuroendocrine transmission and the stress response system [31,32]. Both the neural and nonneural components of the HPA axis have multiple regulatory controls to ensure that the response is suitable for both stressful and non-stressful stimuli, such as appetite, sexual experience, and even light exposure [33]. Meanwhile, the gut microbiome is indispensable in developing and functioning the HPA axis. The gut microbiome directly affects the generation of glucocorticoids and immune mediators, such as tumour necrosis factor-alpha (TNF-α) and interleukin-1 beta and 6 (IL-1β and IL-6), and then activates the HPA axis [34]. (3). Immune pathway: highly specialised macrophages in the CNS constitute microglia that release proinflammatory cytokines, such as TNF-α, IL-1 and IL-6, to maintain microglia-mediated inflammation and recruit other immune cells [35] in response to stress, thereby leading to gut maladaptation and inflammation. Furthermore, the development and integrity of the blood–brain barrier (BBB) and gut barrier depend on the gut microbiome. Changes in the gut microbiome downregulate the expression of tight junctions (TJs) [36], disclosing both organs to microorganisms and biomacromolecules and inducing neuroinflammatory processes [37].
Figure 1.
Schematic representation of the 3 pathways (neural, neuroendocrine and immune) that constitute the brain–gut axis. (1). Neural pathway: the brain regulates gut function mainly through ANS, while the gut microbiome exerts a feedback effect on the brain. (2). Neuroendocrine pathway: stimulation of the brain is transmitted to the gut via the HPA axis, while the gut microbiome influences the brain by affecting the production of immune mediators to activate the HPA axis. (3). Immune pathway: the brain causes gut dysfunction via releasing proinflammatory cytokines to recruit other immune cells, while the gut microbiome disrupts BBB integrity by downregulating TJs expression.
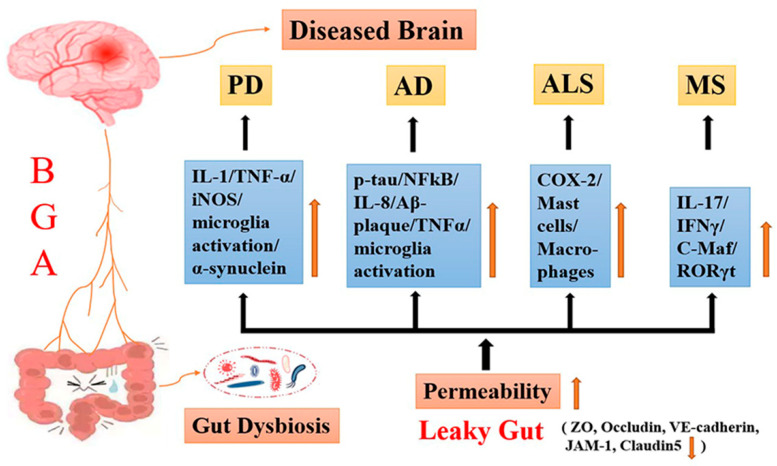
The bidirectional effects of BGA have been studied for an increasing number of neuropsychiatric diseases, including PD [38,39,40], Alzheimer’s disease (AD) [41,42], amyotrophic lateral sclerosis (ALS) [43,44,45] and multiple sclerosis (MS) [46,47,48]. (1). PD: compared to controls, the faecal aromatic amino acids and branched chain amino acids (BCAAs) concentrations are significantly reduced in PD patients [12]. Substantial lines of evidence have confirmed that inducible nitric oxide synthase (iNOS) is produced when lipopolysaccharides (LPS) are increased in substantia nigra [49]. In addition, a damaged gut barrier enhances levels of IL-1β, IL-6 and TNF-α in PD [50]. Gut dysbiosis induces microglial activation, which leads to the subsequent progression of PD [39,40]. (2). AD: a meta-analysis showed a significant reduction in gut microbiota diversity in patients with AD compared to healthy controls [51]. During ageing, the bacteria inhabiting in the gut releases mountains of LPS and amyloids when both the gastrointestinal tract and the BBB are more permeable [50]. Meanwhile, enhanced gut permeability upregulates the levels of proinflammatory cytokines, such as IL-1β, IL-6, IL-8, IL-12A, IL-18 and TNF-α, in the brain [52]. A study indicated that Bacteroides fragilis lipopolysaccharides (BFLPS) potentially activates the pro-inflammatory transcription factor nuclear factor-kappaB (NFκB), which leads to inflammation-mediated neurodegeneration [53]. (3). ALS: a clinical study revealed that compared to their spouses, the gut microbial communities of ALS patients are more diverse and are deficient in Prevotella spp. [54]. Additionally, due to augmented membrane permeability, increased invasion of macrophages and mast cells, and upregulated levels of cyclooxygenase-2 (COX2) result in inflammation in the brain [55]. (4). MS: a main pathological characteristic of MS is gut microbiota dysbiosis [47,48], while the alteration in the gut microbiome reduces C-C chemokine receptor type 9+ (CCR9+) cells, which express transcription factor c-Maf in high levels, leading to an increased expression of retinoic acid-related orphan receptor gamma t (RORγt) and proinflammatory cytokine IL-17/interferon-γ (IFNγ) in secondary progression MS (SPMS) patients [46] (Figure 2). Indeed, the impacts of BGA on the above diseases may be related to metabolic regulation and could trigger the release of many inflammatory transcription factors to activate the immune pathway, causing neurodegeneration [39,44,46,52,56]. However, the relationship between BGA and m6A RNA modification in these diseases is unknown.
Figure 2.
Diagrammatic representation of the gut dysbiosis leads to the development of neurodegenerative diseases and their potential mechanisms through activation of inflammatory pathways.
3. TBI and BGA
TBI-related pathophysiological influences directly associated with gut dysfunction have been increasingly studied. These influences significantly affect the host because BGA is considered to ensure the main bidirectional communication pathway between the brain and gut, including afferent and efferent signals, involving the crosstalk of neurons, hormones and immunity [57,58]. Such bidirectional interactions may cause sequelae, such as chronic fatigue of the gastrointestinal system, including its inability to function properly [59,60] (Figure 3). When TBI impacts this axis, disorders in digestion and absorption, afferent nerve signals, immunomodulation, gut barrier function, and the BBB may occur [5,61]. The effects of TBI on BGA may be realised via the influence of the neural pathway (autonomic nerve), the neuroendocrine pathway and the immune pathway. First, post-TBI autonomic dysfunction causes the altered production and release of key neurotransmitters involved in the sympathetic pathways [62]. When the integration of sympathetic and parasympathetic nerves is disrupted, the gut may also be influenced, involving symptoms of chronic pain and gut discomfort [61] and the dysbacteriosis of normal gut microbiomes. Second, overactivation of the post-TBI HPA axis affects neuroinflammation through the stress-induced initiation of microglia [63]. Dysfunction of the HPA axis occurs immediately after TBI, and the HPA axis is acutely activated by the stress of injury. Excessive glucocorticoids(GCs) caused by extreme activation of the HPA axis after TBI can induce microglial initiation in the brain and increase inflammatory cytokines in response to stress, leading to gut maladaptation and inflammation [63]. TBI activates systemic stress and motivates the HPA axis and the sympathetic branch of the ANS, which results in the release of GCs and catecholamines, respectively [64,65]. Excessive catecholamine release presumably causes gastrointestinal dysmotility involving gastroparesis and food intolerance [9]. Finally, at the cellular level, TBI pathophysiology is characterised by acute necrotic or delayed apoptotic neuronal death, production of cytokines and chemokines, peripheral immune cell infiltration, and the initiation of microglia and astrocytes [66], which likely affects BGA through the immune pathway, destroying gut mucosal integrity. A study showed that gut NFκB increased after TBI, inducing an acute inflammatory response, including increased expression of intracellular adhesion molecule-1 (ICAM-1) and the generation of TNF-α, IL-6 and other cytokines [67]. Inflammatory cytokines lead to gut mucosal damage in rats via two mechanisms. First, these signals probably induce the downregulation of tight junction proteins that cause increased gut permeability. Second, the inflammatory response (especially by the generation of TNF-α) likely induces intestinal epithelial cell (IEC) apoptosis [68,69,70]. Additionally, research in humans indicated that TBI-induced alterations in the gut microbiome [71]. However, whether TBI affects BGA via m6A RNA modification is unclear, and the potential mechanisms are currently areas of investigation for the development of underlying interventions.
Figure 3.
TBI caused dysbiosis through the BGA and its negative feedback mechanism.
4. m6A RNA Modification and BGA
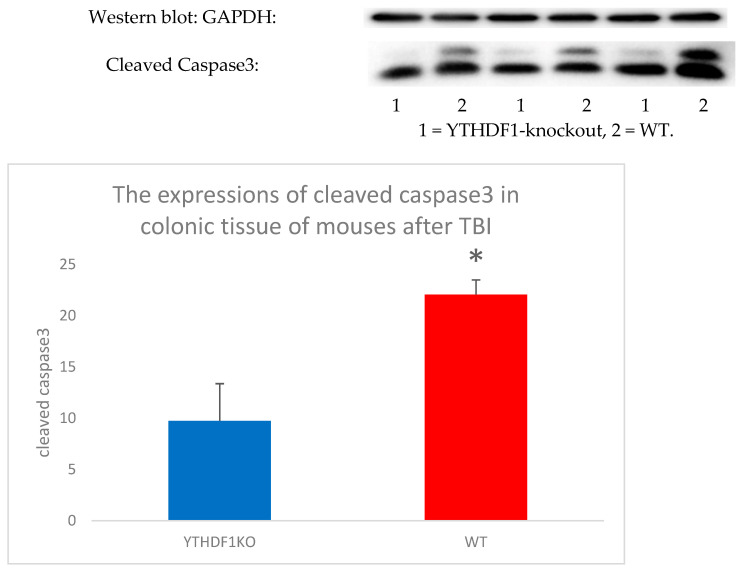
The accurate and controllable regulation of gene expression at the transcriptional and posttranscriptional levels is essential for the survival and function of eukaryotic cells. More than 100 RNA posttranscriptional modifications have been identified in eukaryotes. As one of the most common posttranscriptional RNA modifications in eukaryotes, m6A RNA modification can regulate gene expression [72,73]. Multiple lines of evidence from studies have shown that m6A modification influences almost all aspects of RNA metabolism, including RNA expression, splicing, nuclear output, translation, decay and RNA protein interactions [73,74,75,76]. Recent studies have revealed that m6A can delicately regulate a variety of spatial and temporal physiological processes, including gametogenesis, embryogenesis, sex determination, circadian rhythm, heat shock response, DNA damage response, cell fate determination, pluripotency, reprogramming and neuronal functions [75,77,78]. M6A plays a vital role in the development, differentiation, and regeneration of neurons [76,79,80]. It plays a crucial regulatory role in alterations in the gut microbiome [27], inflammatory gut diseases [81] and the development and progression of colon cancer [82]. Hui Wang et al. reported that the expression of m6A in the gut microbiome of patients with sepsis-associated encephalopathy (SAE) changed significantly [83]. In addition, our ongoing experiments revealed that the BGA dysfunction of YTHDF1-knockout mice following TBI showed significant differences compared to WT mice (Figure 4, preparing for publicity). Therefore, m6A exerts important effects on the bidirectional transmission of BGA.
Figure 4.
The expressions of cleaved caspase3 in colonic tissue of YTHDF1-knockout mice is significantly downregulated after TBI compared to WT mouses. * p < 0.05.
4.1. m6A Related Genes and the Regulatory Mechanism of m6A Modification
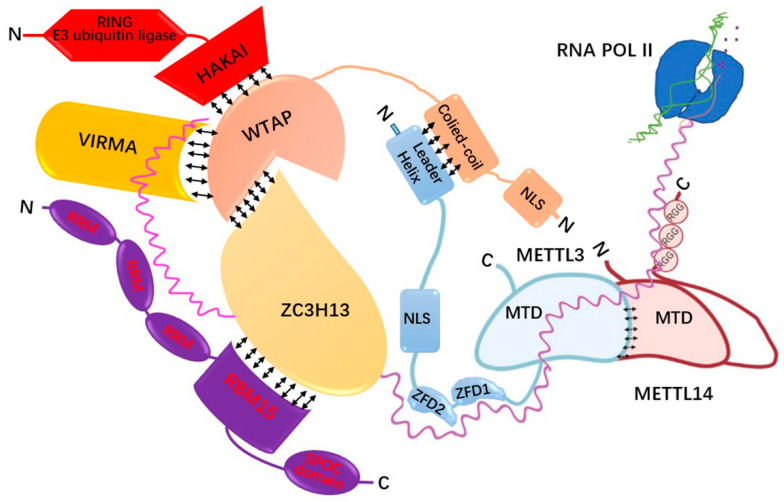
The regulatory mechanism of m6A modification involves a dynamic and reversible process of methylation and demethylation [84]. M6A modification requires multiple regulatory proteins encoded by writing genes (writers), erasing genes (erasers) and reading genes (readers) [80] (Table 1). M6A is catalysed by a writer complex, consisting of core proteins methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), and Wilms tumour 1-associated protein (WTAP) and several m6A-modified regulatory subunits. The first methyltransferase that binds to S-adenosylmethionine (SAM) in the methylase complex is METTL3 [85,86]. The binding of METTL3 and METTL14 is colocalised in nuclear spots to form a stable 1:1 heterodimer, which synergistically improves the methylation ability [87]. METT+L3/METTL14 heterodimers form in the cytoplasm and are recruited into the nucleus through nuclear localisation signals (NLSs) in METTL3 [88]. METTL3 can interact with the crimped part of WTAP via its lead helix regions and anchor it to RNA in the nucleus, which is rich in specific sites of premRNA processing factors [89]. WTAP interacts with zinc finger CCCH domain-containing protein 13 (ZC3H13) to keep the methyltransferase complex (MTC) in the nucleus, while ZC3H13 can interact with RNA binding motif protein 15 (RBM15) to recruit the MTC to U-rich sequences adjacent to the sequence RRACH (R = G/A; H = A/C/U) motif [90]. Another protein that interacts with WTAP is Virma, which provides sequence specificity and MTC recruitment to guide methylation in 3′UTR regions and near-stop codons [89]. Moreover, methyltransferase-like 16 (METTL16) [91], methyltransferase-like 5 (METTL5) and zinc finger CCHC domain containing 4 (ZCCHC4) act as linker proteins to guide MTC binding to its target mRNA [84]. To date, the last protein in MTC is Hakai, which plays a role in the sex-determining pathway and mediates sex-lethal splicing [18]. In addition, what is needed to maintain the integrity of m6A-METTL-Associated Complex (MACOM) is the ubiquitin domain of Hakai but not its activity (Figure 5). Hakai is essential to maintaining the functionality of m6A writers by maintaining the stability of MACOM components [92].
Table 1.
M6A related genes and their basic functions.
| Types | Regulators | Functions | References |
|---|---|---|---|
| m6A writers |
METTL3 | Catalyses the transfer of the methyl in single-stranded RNA (ssRNA) sequence motif DRACH (D = A, G or U; R = A or G; H = A, C or U) from S-Adenosyl methionine (SAM) to adenine. | [87,93] |
| METTL14 | Recognizes the RNA substrate that activates METTL3 and offers RNA binding sites as scaffolds to form a stable heterodimer with METTLL3. | [84,87,93] | |
| WTAP | The first one binds to METTL3-METTL14 heterodimer and recruits it to target RNA. The three proteins form together a conservative complex located in nuclear spot. | [84,94,95] | |
| METTL16 | Responsible for m6A modification of lncRNAs, U6 snRNA, and introns of pre-mRNAs. | [91,96,97,98] | |
| RBM15 | Binds m6A complex and recruits it to a special RNA site. | [84,99] | |
| VIRMA | Recruits m6A complex to a special RNA site and interacts with polyadenosine cleavage factors CPSF5 and CPSF6. |
[89,100] | |
| ZC3H13 | Bridges WTAP to mRNA binding factor Nito. | [84,101] | |
| METTL5 | Responsible for m6A modification of 18s rRNA. | [96,102,103] | |
| ZCCHC4 | Responsible for m6A modification of 28s rRNA. | [96,104,105,106] | |
| HAKAI | Exerts effects on gender determination and mediates lethal splicing, maintains the functions of m6A writers by ensuring the stability of MACOM components via the Hakai ubiquitin domain. | [92] | |
| m6A erasers |
FTO | Demethylates m6A, also harbours activity towards m6Am and m1A. | [96,107,108,109] |
| ALKBH5 | Mainly demethylates m6A. | [96,110,111] | |
| m6A readers |
YTHDF1/2/3 | Highly similar to m6A sites bound by YTHDF1, YTHDF2 or YTHDF3, and these three analogues together exerts effects on mediating mRNA degradation. | [93,112] |
| YTHDC1 | Promotes alternative splicing and RNA output. | [96,110,111] | |
| YTHDC2 | Boosts target RNA translation and reduces its abundance. | [96,113,114] | |
| HNRNPA2B1 | Mediates mRNA splicing and major microRNA processing. | [84,115] | |
| HNRNPC/ hn-RNPG |
Regulates mRNA structure and alternative splicing. | [96,116,117] | |
| EIF3 | Facilitates mRNA translation. | [84,118] | |
| IGF2BP1/2/3 | Enhances mRNA stability, storage capacity and translation. | [96,119] |
Figure 5.
Schematic diagram of the methyltransferase complex: The components and their interactions.
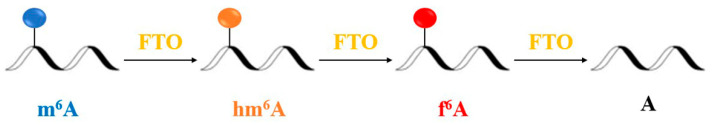
The demethylases (erasers) that reverse m6A methylation are FTO and ALKBH5. The FTO demethylation process requires ferrous ions and α-ketoglutarate to demethylate m6A via a sophisticated intermediate reaction [84]. First, FTO oxidises N6-methyladenosine to form N6-hydroxymethyladenosine (hm6A), which then catalyses the formation of N6-formyladenosine (f6A). Finally, f6A generates adenosine (A) (Figure 6), and the demethylation process is completed [120]. Intriguingly, a recent finding showed that FTO preferentially demethylated N6,2′-O-dimethyladenosine (m6Am) rather than m6A; by altering FTO levels, m6Am at the mRNA cap weakened the unwrapping effect of mRNA by decapping RNA2 (DCP2), and increasing m6Am levels through FTO knockout also improved the stability of mRNAs [121]. Additionally, FTO regulates reversible m6Am RNA methylation in small nuclear RNA (SnRNA) biogenesis [108,122]. These studies indicate that internal mRNA m6A sites may not be related to FTO substrates. However, Jiang Bo et al. found that FTO could effectively demethylate both m6A and cap m6Am from purified polyadenylated RNA with different substrate preferences in the nucleus and cytoplasm [107]. ALKBH5 is a kind of m6A demethylase that exhibits no activity against m6Am but does act strongly against m6A [121]. ALKBH5 mediates the stability of target mRNA [123] and reduces the output of mRNA to the cytoplasm [124]. These findings have begun to shed new light on multiple intricate m6A demethylation mechanisms.
Figure 6.
Schematic diagram of the process of FTO demethylates m6A.
M6A reader proteins modulate m6A modification biological functions to recognise m6A sites [125]. The proteins involving YTH domains are best characterised as m6A “readers” [126], and there are five YTH domain-involving proteins (YTHDF1-3, YTHDC1, and YTHDC2) in mammals [127]. YTHDF1 promotes mRNA translation in the classical model, YTHDF2 facilitates mRNA degradation, and YTHDF3 cooperates with YTHDF1 or YTHDF2 to promote either mRNA translation or degradation [19,126]. However, recent findings confirmed that the m6A sites binding with YTHDF1, YTHDF2, or YTHDF3 are highly similar, and that all three analogues act in concert to regulate the degradation of m6A-tagged mRNA [93,112]. Using dCasRx-M3 and dCasRx-A5 to regulate the methylation level of targeted sites at YTHDF paralogues, Zhen Xia et al. found that the m6A sites binding with YTHDF1, YTHDF2, and YTHDF3 all led to m6A-mediated mRNA degradation [93], which supported recent models [93,112] rather than the classical model [19,126]. YTHDC1 is the only YTH family protein located in the nucleus [128]. Previous studies have shown that YTHDC1 plays a crucial role in mediating mRNA splicing by recruiting serine/arginine-rich splicing Factor 3 (SRSF3), blocking serine/arginine-rich splicing Factor 10 (SRSF10) [110], and retaining m6A-modified exons during splicing [129]. Moreover, YTHDC1 also regulates the nuclear and cytoplasmic transport of methylated mRNAs [110]. Recently, however, Zhijian Qian’s team found that YTHDC1 KO did not influence the splicing of minichromosome maintenance deficient 4 (MCM4) in leukemic cells and did not affect the nuclear output and translation efficiency of MCM4 and MCM5 in acute myeloid leukaemia (AML) cells; in contrast, YTHDC1 mediated the expression of MCM2, MCM4 and MCM5 transcripts by controlling the stability of these transcripts in AML cells [128]. YTHDC2, the last member of the YTH protein family, which is chiefly enriched in the perinuclear region, can bind to ribosomes and function in small ribosomal subunits, decrease mRNA abundance and enhance hypoxia-inducible factor 1α (HIF-1α) mRNA translation efficiency by its helicase [114,130]. YTHDC2 harbours a unique domain that differs from other YTH family proteins. The YTHDC2 RNA binding domain, characterised by the R3H domain, binds YTHDC2 to intracellular RNA and plays an auxiliary role in the YTH domain [113,114]. In addition to YTH proteins, heterologous nuclear ribonucleoproteins A2B1 (HnRNPA2B1) and C (HNRNPC), two nuclear RNA-binding proteins, play a vital role in m6A-dependent nuclear RNA processing events [131]. HnRNPA2B1 directly binds to m6A sites of RNA and interacts with the microRNA microprocessor complex protein DiGeorge syndrome critical region gene 8 (Dgcr8) to promote METTL3-mediated microRNA processing [84,115]. HNRNPC interacts with m6A-modified mRNA to mediate structural transformation [117]. However, unlike HNRNPC, HNRNPG binds to a purine-rich motif of m6A-methylated RNAs, including the m6A site, via its low-complexity region at the C-terminus, which is self-assembled into large particles in vitro. Due to the overlap of the HNRNPG binding motif and the m6A site, HNRNPG binding may compete with the binding of direct m6A readers such as YTH domain proteins [116]. EIF3 is a large multiprotein complex composed of 13 subunits (a-m) [132]. By binding to 5′UTR m6A residues, EIF3 can directly recruit the 43S preinitiation complex to mRNA 5′UTRs to stimulate translation initiation [118]. Insulin-like growth Factor 2 mRNA binding protein 1, 2, and 3 (IGF2BP1, IGF2BP2 and IGF2BP3) also promotes target mRNAs’ stability and translation [119]. Intriguingly, Ras GTPase-activating protein-binding protein 1 (G3BP1) and G3BP2, two anti-readers of m6A, have been confirmed to date to act on the assembly of stress granules and are essential for embryonic development [133] but are expelled by the existence of m6A [134,135].
4.2. m6A Modification and Brain
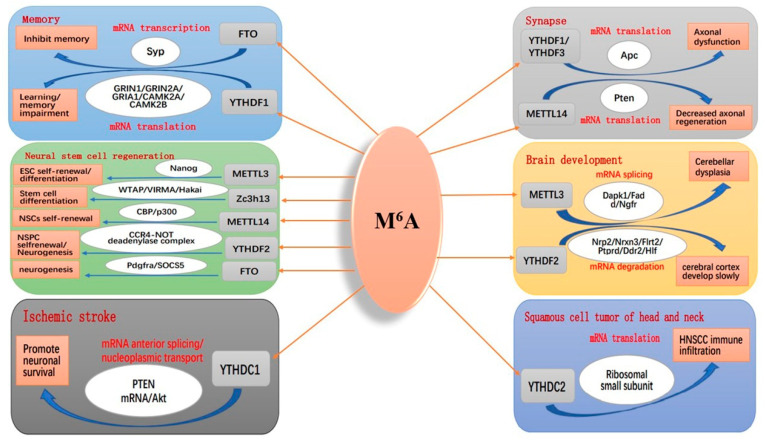
M6A modification is abundant in the brain, normalising the nervous system function by regulating mRNA metabolism. When the expression of key enzymes and/or binding proteins involved in m6A modification is abnormal, the level of m6A modification may fluctuate, leading to molecular dysfunctions such as mRNA metabolism disorder and transcription and translation disorders that cause physiological dysfunction of the nervous system [136,137,138]. Given its diversity and reversibility, m6A modification harbours more complicated physiological mechanisms in regulating CNS functions. For writers, METTL3 can influence the mRNA half-life of Dapk1, Fadd, and Ngfr. Indeed, the deletion of METTL3 can ectopically modify m6A, leading to cerebellar hypoplasia, long-term memory lesion, and effects on the self-renewal and differentiation of stem cells [139]. METTL14 can recognise and promote Pten protein translation, while METTL14 knockout can affect axonal regenerative ability [140]. ZC3h13 keeps the Zc3h13-WTAP-Virilizer-Hakai complex in the nucleus and enhances the level of mRNA m6A modification in a mouse embryonic stem cell (mESC). ZC3h13 knockout may cause the complex to be transferred from the nucleus to the cytoplasm, reducing the level of m6A, which destroys the self-renewal of mESCs and induces mESC differentiation [101]. For erasers, FTO can repress synaptophysin (Syp) gene mRNA transcription in synaptic ganglia and reduce Spy protein levels; downregulating the expression of FTO can also upregulate Syp protein levels and improve memory [141]. ALKBH5 decreases the m6A modification of C-X-C motif chemokine ligand 2 (CXCL2) and IFN-CD4 mRNA. It increases transcriptional stability and protein expression, which results in the enhanced response of CD4+ T cells and more neutrophils infiltrating the CNS in the process of neuroinflammation [142]. For readers, YTHDF1 recognises and binds the mRNAs of m6A-modified GRIN1, GRIN2A, GRIA1, CAMK2A, and CAMK2B genes to promote translation. Indeed, YTHDF1 deletion causes ectopic translation of related proteins, resulting in memory loss [79]. YTHDF2 reduces mRNA stability and facilitates its degradation by recruiting the CCR4-nondead enolase complex. Deleting YTHDF2 in the embryonic neocortex seriously influences the self-renewal of neural stem/progenitor cells (NSPCs). The spatiotemporal generation of neurons and other types of cells leads to death in the later stage of embryonic development [143]. YTHDF3 can specifically recognise m6A-modified Apc gene mRNA in the cytoplasm and regulate the translation process with YTHDF1. Deleting YTHDF1 and YTHDF3 can cause abnormal translation of Apc protein, which can induce ectopic neuronal development and attenuate synaptic transmission ability [144]. YTHDC1 plays a role in premRNA splicing and mediates the nuclear and cytoplasmic transport of methylated mRNAs, thus facilitating neuronal survival and reducing ischaemic stroke by destroying PTEN mRNA and promoting Akt phosphorylation [127]. YTHDC1 knockout downregulates the expression of the antiapoptotic protein Bcl2. It upregulates the expression of the proapoptotic protein cleavage caspase3, while the overexpression of YTHDC1 confirms its protective effect on neuronal death induced by oxygen-glucose deprivation (OGD) [127]. YTHDC2 can bind to ribosomes and act on small ribosomal subunits, decrease mRNA abundance and enhance HIF-1α mRNA translation efficiency by its helicase [114]. YTHDC2 can act as an independent gene as a prognostic marker of head and neck squamous cell carcinoma (HNSCC) [130]. Alteration of m6A modification exerts significant effects on brain tissue development, learning and memory ability, stem cell renewal and differentiation, synaptic regeneration, and other biological functions (Figure 7).
Figure 7.
The role of m6A modification in the CNS.
Therefore, the abnormal expression of m6A-related proteins may lead to the development of various neurological diseases, such as PD [145,146,147], AD [148,149,150,151,152], MS [153], tumours [154,155,156], epilepsy [157], and neuropsychiatric disorders [158]. TBI, as one of the major diseases of CNS, often causes memory problems in humans [159]. Several CNS traumas, both in vitro and in vivo experimental systems [147,160,161,162,163,164], have demonstrated that m6A modification also plays important roles in the aetiology and pathogenesis of brain injuries. Diao et al. reported that m6A levels in the total RNA of primary cultured rat hippocampal neurons increased markedly after hypoxia/reoxygenation (H/R) injury [161]. The expression of total RNA m6A modification also upregulated significantly in the cerebral cortex of rats and mice treated with middle cerebral arteryocclusion/reperfusion (MCAO/R) [160,162]. However, a decreased level of the total RNA m6A was observed with the elongation of reperfusion time in PC12 cells with oxygen-glucose deprivation/reoxygenation (OGD/R) treatment [162]. The total RNA m6A levels of the hippocampus of brain-injured mice decreased significantly at 6 h after injury compared with that in the sham operation group in a controlled cortical impact (CCI) mouse model [164], while the change in m6A methylation reduction only occurred in the striatum in a rat nerve injury model treated with 6-hydroxydop amine (6-OHDA) [147]. These studies revealed that the level of total RNA m6A modification differs between brain injury models, indicating that the cell type and brain region might affect the m6A modification level [165]. More importantly, the m6A-related enzymes are differentially expressed in CNS injuries and play significant roles in neuronal development, cognitive disorders, cell death, and apoptosis, suggesting that they may serve as a novel biomarker and therapeutic target for CNS injuries [166].
4.3. m6A Modification and Gut Microbiome
The gut microbiome influences host physiology, including host metabolism, gut morphology, immune system development, and behaviour [167,168]. Gut microbial metabolites and fermentation products, such as short-chain fatty acids (SCFAs), tryptophan metabolites, sphingolipids, and polyamines, have been known to partially regulate the effect of the gut microbiome on the host by modulating transcription and epigenetic modifications [169,170,171,172]. Sabrina Jabs et al. found that m6A modification spectra in both the mouse ceca and liver were affected by the presence of the gut microbiome by MERIP-Seq [27]. Xiaoyun Wang et al. carried out mass spectrometry and m6A sequencing on GF and SPF mice brain, gut, and liver tissues. The results revealed that the gut microbiome harbours a significant effect on the m6A mRNA modification of the host brain, gut, and liver, but especially in the brain [173]. In addition, dietary intake of non-starch polysaccharides (NSPs) may alter m6A RNA methylation to improve tumours by affecting the supply of methyl donors produced by the gut microbiome [174]. Multiple studies have shown that m6A modification is the key interaction mechanism between the gut microbiome and host, and m6A modification may also selectively modulate microbiome function by affecting gut inflammation. A recent study indicated that METTL3-knockout mice developed chronic gut inflammation at 3 months after birth, which resulted in gut microbiome disorders [175]. METTL14 deficiency interferes with the induction of immature T cells into inducted regulatory T cells (Tregs), which results in spontaneous colitis in mice [81]. Benyu Liu et al. found that CircZbtb20 promotes the interaction between Alkbh5 and Nr4a1 mRNA, resulting in the ablation of Nr4a1 mRNA m6A modification to enhance the stability of Nr4a1 mRNA. Nr4a1 initiates Notch2 signalling activation, which helps to maintain the dynamic balance of innate lymphoid cells (ILC3). The absence of Alkbh5 or Nr4a1 disrupts ILC3 homeostasis and causes serious gut diseases, such as inflammation [176]. YTHDF1 guides the translation of tumour necrosis factor receptor-associated factor 6 (TRAF6) mRNA, which encodes TRAF6, thus modulating the immune response by m6A modification near the transcript stop codon. Moreover, YTHDF1 recognises the target TRAF6 transcript to modulate the gut immune response to bacterial infection by the unique interaction mechanism between the P/Q/N-rich domain and host factor DDX60 death domain [177]. Ane Olazagoitia-Garmendia et al. demonstrated that a gluten-induced increase in YTHDF1-mediated XPO1 activates the NFκB pathway and then induces increased expression of IL8 in gut cells, resulting in the development and aggravation of celiac disease [178]. Dian Xu et al. used The Cancer Genome Atlas (TCGA) cohort data to analyse the expression patterns of m6A RNA methylation regulators in colonic adenocarcinoma (COAD) and their relationship with clinical features. The results suggested that YTHDF1 exhibited the highest diagnostic value for COAD, and survival analysis confirmed that highly expressed levels of YTHDF2 and YTHDF3 indicated a favourable prognosis. Furthermore, the expression levels of YTHDF3 were independent prognostic factors of 5-year total survival in COAD patients [82]. Tanabe et al. reported that YTHDC2 was upregulated in colon cancer and positively associated with tumour stages [179]. These findings confirm that host m6A modification also provides feedback to gut bacteria to sustain gut homeostasis.
5. The Role of m6A RNA Modification in TBI-Mediated BGA
5.1. m6A Modification and TBI
After TBI, excitatory amino acids such as glutamate release into the synapse which then overstimulates the N-methyl-D-aspartate (NMDA) receptor, which leads to Ca2+ overload [180,181]. Ca2+ overload and excitotoxicity induce reactive oxygen species (ROS) release [180], and excessive ROS causes oxidative stress and neuronal cell death [182,183]. Oxidative stress induces excessive free radicals, resulting in neuronal death, stimulating inflammation [184]. Oxidative stress occurs at the beginning and accompanies the whole process of TBI and is one of the significant mechanisms. A large number of studies have confirmed that m6A plays an important role in oxidative stress [151,185,186,187,188]. Anders et al. first found that the m6A signal accumulates in reaction to oxidative stress in stress granules and that YTHDF3 promotes target mRNA translocation into stress granules [189]. Ye Fu et al. observed that YTHDF1/3-knockout largely abolishes stress granule formation and mRNA localisation to stress granules. Reintroducing YTHDF proteins into knockout cells restores the formation of stress granules [190]. The regulation of phase separation by m6A weakens the stability and translation of mRNA, influencing cellular functions and biological processes [84]. Wang, J. et al. identified that METTL3 may have a protective effect against oxidative stress [191].
TBI-induced oxidative stress can alter genes expression and signal pathways, such as nucleotide-binding oligomerization domain-like receptor pyrin domain-containing-3 (NLRP3) [192], caveolin-1 [193], and p75-neurotrophin receptor (p75NTR) [194], and nuclear factor erythroid 2-related factor 2 (Nrf2) signalling [192,195], etc. These alterations may be correlated with posttranscriptional regulation, especially m6A. Considering the indispensable function of m6A modification in mediating protein translation and participating in various bioprocesses, it has been confirmed that m6A modification has also recently contributed to the aetiology and pathogenesis of TBI [163,164,165]. Yiqin Wang et al. investigated novel epigenetic alterations in TBI via genome-wide screening of m6A marker transcripts in the mouse hippocampus after TBI [164]. The results revealed that the expression of METTL3 and the total level of RNA m6A in the mouse hippocampus were distinctly decreased after TBI. The immunohistochemical results showed that METTL3 was mainly expressed in neurons, suggesting that cognitive dysfunction may be correlated with the downregulation of METTL3. Jiangtao Yu et al. found that the expression of METTL14 and FTO in the rat cortex was decreased after TBI and that there was a significant change in the m6A methylation levels of 1580 mRNAs. The study also revealed that FTO harboured a significant role in maintaining neurological functions in TBI rat that would be an intervention target for mitigating impairments caused by TBI [163]. However, it is not clear whether the change in TBI-induced m6A methylation is associated with BGA dysfunction.
5.2. The Role of m6A in TBI-Mediated BGA
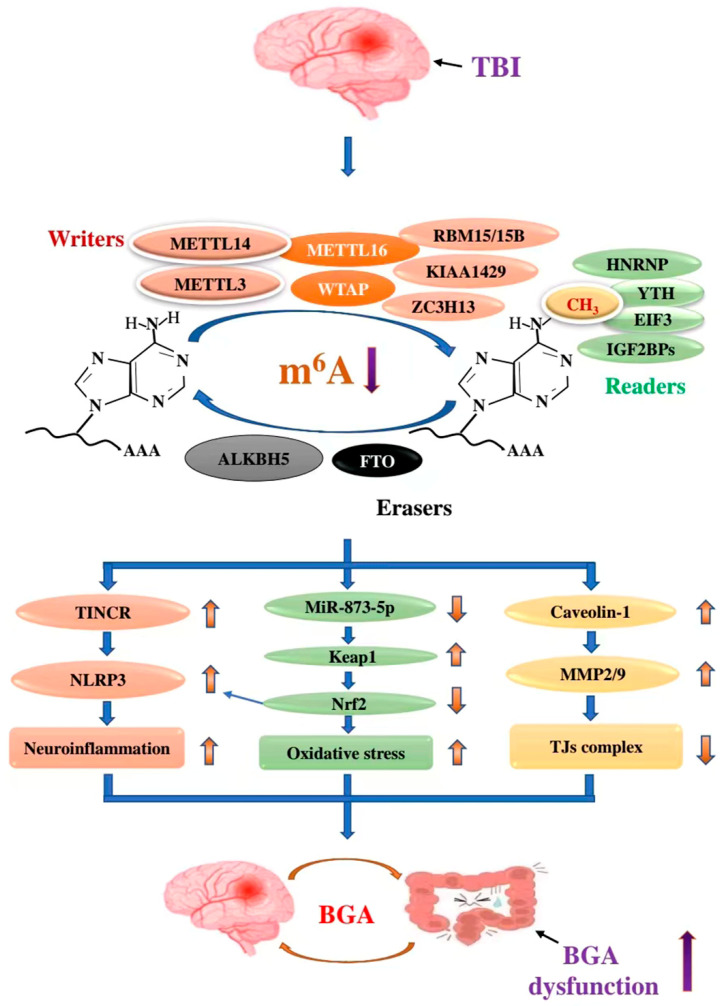
It is known that TBI can enhance the expression of NLRP3 [192,196] and caveolin-1 [193], and impede Nrf2 signalling [192,197], and these changes are also displayed in gut diseases [198,199,200]. Among them, NLRP3 and Nrf2 are the potential mediators in BGA dysfunction [198,201,202]. Moreover, there is clear evidence that m6A mediates gene expression and the signal pathway [191,203,204]. Hence, it is reasonable to speculate that m6A may take part in TBI-induced BGA dysfunction via regulating the expression of NLRP3, caveolin-1, and Nrf2 signalling (Figure 8).
Figure 8.
The possible pathways of m6A regulating the pathological process of BGA after TBI: TBI causes the downregulation of m6A, which is involved in TBI-induced BGA disfunction via METTL14/TINCR/NLRP3, METLL3/miR-873-5p/Keap1/Nrf2 signalling, and FTO/Caveolin-1/MMP2/9 pathways.
5.2.1. METTL14/TINCR/NLRP3 Axis
METTL14-mediated m6A modification induces inhibition of non-protein coding RNA (TINCR), and thus represses NLRP3 mRNA stability, preventing the development of pyroptosis [203]. In contrast, deletion of METTL14 causes upregulated NLRP3, cleaved caspase-1, and gasdermin D (GSDMD)-N [203]. NLRP3 inflammasome is a multiprotein complex and regulates inflammatory responses by activating caspase-1 for processing premature IL-1β and IL-18 [205]. NLRP3 inflammasome may be a potential target for treatments and biomarkers to improve TBI prognosis [206,207]. Multiple animal studies indicate an upregulation of the NLRP3 inflammasome at both gene and protein levels after a moderate TBI induced via CCI [208,209,210]. Furthermore, there is also clinical evidence of NLRP3 inflammasome potentiate in moderate and severe TBI [211,212]. Moreover, NLRP3 inflammasome is at the crossroads of BGA communications [213]. Taken together, TBI-induced METLL14 downregulation may lead to neuroinflammation through the METTL14/TINCR/NLRP3 axis, further causing BGA dysfunction via the immune pathway.
5.2.2. METLL3/miR-873-5p/Keap1/Nrf2 Signalling Pathway
METTL3-mediated m6A modification enhances DGCR8 recognition of pri-miR-873, promoting the generation of mature miR-873-5p. MiR-873-5p inhibits Kelch-like ECH-associated protein 1 (Keap1) and then activates the Nrf2//heme oxygenase-1 (HO-1) pathway [191]. TBI can impair autophagic flux and impede Nrf2 signalling, the major mediator in antioxidant response, thus leading to excessive oxidative stress [197]. Furthermore, the expression of NLRP3 inflammasome proteins can be induced by oxidative stress through increasing the expression of key proteins of the Nrf2/HO-1 pathway following TBI [192]. Additionally, Nrf2 activators have an influence on mitochondrial quality control and are correlated with a positive change in BGA [202]. Hence, TBI-induced METLL3 downregulation may induce oxidative stress and neuroinflammation via the METLL3/miR-873-5p/Keap1/Nrf2 signalling pathway, consequently leading to BGA dysfunction.
5.2.3. FTO/Caveolin-1/MMP2/9 Pathway
FTO directly targets the caveolin-1 mRNA and facilitates its degradation [204]. Thus, attenuated FTO in the rat cortex enhances caveolin-1 expression after TBI. Potentiated caveolin-1 expression and phosphorylation are associated with the decreased expression of occludin and claudin-5 [214,215]. Caveolin-1 activates matrix metalloproteinases (MMPs), especially MMP2 and MMP9, promoting BBB opening after injury [216,217,218]. MMPs are a group of Zn2+ dependent endonucleases that can cleave type IV collagen in the extracellular matrix of the BBB, inducing structure damage and leakage [219]. MMP2 and MMP9 activated by zinc accumulation in microvessels, especially, mainly cause the loss of TJs complex to destroy the BBB [220]. In addition, clinical datasets showed that disease-active patients displayed enhanced caveolin-1 level compared to normal controls or disease-inactive inflammatory gut disease groups [199]. Altogether, TBI-induced FTO downregulation may cause the loss of TJs complex via the FTO/caveolin-1/MMP2/9 pathway, ultimately leading to the destruction of the BBB and the gut barrier.
After TBI, a series of pathological reactions produced by brain tissue affect the integrity of the gut mucosa and the constituents and colonization of the gut microbiome via BGA signal transduction. Given that m6A plays an important role in regulating these pathological processes, it hints that m6A may take part in pathological processes of BGA after TBI via the above pathways. However, these studies are limited, and there may be other regulatory pathways that have not been found to date. The specific mechanisms behind m6A in BGA (especially after TBI) remain elusive. The important molecular information transmission processes m6A involved in post-TBI BGA require in-depth study.
6. Conclusions and Perspectives
M6A methylation plays an important role in the homeostasis regulation of tissues and organs, especially when injury occurs, such as TBI, oxidative stress, ischemic perfusion, etc. The role of m6A in the brain and gut and the microbiome has been demonstrated. Therefore, it is reasonable to speculate that m6A may take part in TBI-induced BGA dysfunction.
Although BGA posttranscriptional regulation studies have developed rapidly, many questions are still existing. First, although it has been confirmed that m6A plays a significant role in BGA, the specific role of each enzyme and protein in BGA and its detailed mechanism are not clear. Second, there is currently no report on the treatment of BGA dysfunction after TBI with specific agonists or inhibitors of m6A-related enzymes and proteins. It was reported that vitagenes play a key role in the pro-survival pathways involved in the programmed cell life, conferring protection against oxidative stress [221,222,223]. Vitagenes can promote the production of molecules (bilirubin, etc.) harbouring antioxidant and antiapoptotic activities [224]. To date, mountains of reports have shown that bilirubin is endowed with strong antioxidant activity, alleviating cellular damage induced by reactive oxygen species in vitro and in vivo models [225,226,227]. The process of vitagenes protect against oxidative stress causing cellular damage may be regulated by the Keap1/Nrf2/antioxidant response element (ARE) pathway [224]. The neuroprotective effects of the upregulation of the Keap1/Nrf2/ARE pathway have been confirmed in several in vitro and in vivo experimental systems [228,229]. Given that the Nrf2 signalling pathway plays an important role in neuroinflammation caused by oxidative stress after TBI [230,231], the upregulation of the Keap1/Nrf2/ARE pathway may block the occurrence of neuroinflammation caused by oxidative stress after TBI, thus producing neuroprotective effects. The polyphenolic compounds can attenuate oxidative stress-induced traumatic brain injury and reduce secondary injury via the Nrf2 signalling pathway [192,230,231]. Indeed, substantial lines of evidence have demonstrated that m6A plays an important role in oxidative stress by modulating the Nrf2 signalling pathway [187,191,232,233]. Meanwhile, as mentioned above, m6A may regulate TBI-induced BGA dysfunction via the METLL3/miR-873-5p/Keap1/Nrf2 signalling pathway. A member of our group, Xiang Zhong, reported that phytochemicals can interfere with the expression of m6A RNA methylation in preclinical experiments [234,235,236,237,238]. Thus, the polyphenolic compounds may act as potential drugs for the prevention and treatment of BGA dysfunction after TBI through regulating the m6A-related enzymes and proteins. Though there is still limited clinical evidence for these natural Nrf2 activators, we foresee that the combinational use of phytochemicals such as Nrf2 activators with gene and stem cell therapy such as agonists or inhibitors of specific RNA m6A-related enzymes and proteins will be a promising therapeutic strategy for TBI and TBI-induced BGA dysfunction in the future.
Acknowledgments
We acknowledge Meng Cheng for his English writing guidance.
Abbreviations
| m6A | N6-methyladenosine |
| TBI | Traumatic brain injury |
| BGA | Brain-gut axis |
| IBD | Inflammatory bowel disease |
| CNS | Central nervous system |
| ENS | Enteric nervous system |
| ANS | Autonomic nervous system |
| HPA | Hypothalamus-pituitary-adrenal |
| TNF-α | Tumour necrosis factor-alpha |
| IL-1β/6/1/8/17 | Interleukin-1 beta/6/1/8/17 |
| BBB | Blood–brain barrier |
| TJs | Tight junctions |
| PD | Parkinson’s disease |
| AD | Alzheimer’s disease |
| ALS | Amyotrophic lateral sclerosis |
| MS | Multiple sclerosis |
| BCAAs | Branched chain amino acids |
| LPS | Lipopolysaccharides |
| BFLPS | Bacteroides fragilis lipopolysaccharides |
| iNOS | Inducible nitric oxide synthase |
| p-tau | Phosphorylated tau |
| NFκB | Nuclear factor kappaB |
| COX-2 | Cyclooxygenase-2 |
| CCR9+ | C-C chemokine receptor type 9+ |
| IFNγ | Interferon-γ |
| c-Maf | C-musculoaponeurotic-fibrosarcoma |
| RORγt | Retinoic acid-related orphan receptor gamma t |
| SPMS | Secondary progression multiple sclerosis |
| ZO | Zonula occludens |
| VE-cadherin | Vascular endothelial–cadherin |
| JAM-1 | Junction adhesion molecule-1 |
| GCs | Glucocorticoids |
| ICAM-1 | Intracellular adhesion molecule-1 |
| IEC | Intestinal epithelial cell |
| SAE | Sepsis-associated encephalopathy |
| YTHDF1/2/3 | YTH domain family 1/2/3 |
| WT | Wild type |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| METTL3/14/16/5 | Methyltransferase-like 3/14/16/5 |
| WTAP | Wilms’tumor 1-associating protein |
| SAM | S-adenosylmethionine |
| NLSs | Nuclear localisation signals |
| ZC3H13 | Zinc finger CCCH-type containing 13 |
| MTC | Methyltransferase cpmplex |
| RBM15 | RNA binding motif protein 15 |
| ZCCHC4 | Zinc finger CCHC domain containing 4 |
| MACOM | m6A-METTL Associated Complex |
| RNA POL II | RNA polymerase II |
| ZFD1/2 | Zinc finger domain1/2 |
| RRM | RNA recognition motif |
| SPOC domain | SpenParalog and OrthologsC-terminal domain |
| ssRNA | Single-stranded RNA |
| snRNA | Small nuclear RNA |
| lncRNA | Long non-coding RNA, |
| CPSF5/6 | Cleavage and polyadenylation specific factor 5/6 |
| FTO | Fat mass and obesity-associated protein |
| ALKBH5 | AlkB homolog 5 |
| m6Am | N6, 2′-O-dimethyladenosine |
| M1A | N1-methyladenosine |
| YTHDC1/2 | YTH domain containing 1/2 |
| HNRNPA2B1 | Heterogenous nuclear ribonucleoprotein A2B1 |
| HNRNPC | Heterogenous nuclear ribonucleoprotein C |
| hn-RNPG | Heterogenous nuclear ribonucleoprotein G |
| EIF3 | Eukaryotic initiation factor 3 |
| IGF2BP1/2/3 | Insulin-like growth factor 2 mRNA-binding protein 1/2/3 |
| hm6A | N6-hydroxymethyladenosine |
| f6A | N6-formyladenosine |
| A | Adenosine |
| DCP2 | Decapping RNA2 |
| dCasRx-M3 | Dead RfxCas13d-Methyltransferase-like 3 |
| dCasRx-A5 | Dead RfxCas13d-AlkB homolog 5 |
| SRSF3/10 | Serine/arginine-rich splicing Factor 3/10 |
| MCM4/5/2 | Minichromosome maintenance deficient 4/5/2 |
| AML | Acute myeloid leukaemia |
| HIF-1α | Hypoxia-inducible factor 1α |
| Dgcr8 | DiGeorge syndrome critical region gene 8 |
| 3′/5′UTR | 3′/5′untranslated region |
| G3BP1/2 | Ras GTPase-activating protein-binding protein 1/2 |
| Dapk1 | Death Associated Protein Kinase 1 |
| Fadd | Fas-associated death domain |
| Ngfr | Nerve growth factor receptor |
| mESC | Mouse embryonic stem cell |
| SYP | Synaptophysin |
| CXCL2 | C-X-C motif chemokine ligand 2 |
| IFN-CD4 | Interferon-CD4 |
| GRIN1/2A | Glutamate receptor, ionotropic,N-methyl D-aspartate 1/2A |
| GRIA1 | Glutamate Receptor AMPA Type Subunit 1 |
| CAMK2A/2B | Calcium/Calmodulin Dependent Protein Kinase 2A/2B |
| NSPCs | Neural stem/progenitor cells |
| Apc | Adenomatous polyposis coli |
| PTEN | Phosphatase and tensin homolog deleted on chromosome ten |
| OGD | Oxygen-glucose deprivation |
| HNSCC | Head and neck squamous cell carcinoma |
| CBP | CREB binding proteins |
| SOCSS | Suppressors of cytokine signaling |
| H/R | Hypoxia/reoxygenation |
| MCAO/R | Middle cerebral arteryocclusion/reperfusion |
| OGD/R | Oxygen-glucose deprivation/reoxygenation |
| CCI | Controlled cortical impact |
| 6-OHDA | 6-hydroxydop amine |
| SCFAs | Short-chain fatty acids |
| MERIP-Seq | Methylated RNA immunoprecipitation sequencing |
| GF | Germ free |
| SPF | Specific pathogen free |
| NSPs | Nonstarch polysaccharides |
| ILC3 | Innate lymphoid cells |
| Nr4a1 | Nuclear receptor subfamily 4 group A member1 |
| TRAF6 | Tumour necrosis factor receptor-associated factor 6 |
| DDX60 | DEXD/H box helicase 60 |
| XPO1 | Exportin 1 |
| TCGA | The Cancer Genome Atlas |
| COAD | Colonic adenocarcinoma |
| NMDA | N-methyl-D-aspartate |
| ROS | Reactive oxygen stke |
| NLRP3 | Nucleotide-binding oligomerization domain-like receptor pyrin domain-containing-3 |
| p75NTR | P75-neurotrophin receptor |
| Nrf2 | Nuclear factor erythroid 2-related factor 2 |
| TINCR | Non-protein coding RNA |
| GSDMD | Gasdermin D |
| HO-1 | Heme oxygenase-1 |
| MMPs/2/9 | matrix metalloproteinases/2/9 |
| Keap1 | Kelch-like ECH-associated protein 1 |
| ARE | Antioxidant response element |
Author Contributions
All authors contributed to the study conception and design. The first draft of the manuscript was written by P.H. and all authors commented on previous versions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare that they have no conflict of interest. There are no competing interest of any nature to report. This research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (82172820 Jing Zhang, 82070541 Min Liu, 81771332 Chunlong Zhong, 81571184 Chunlong Zhong), the Natural Science Foundation of Shanghai (22ZR1451200 Chunlong Zhong), the Outstanding Leaders Training Program of Pudong Health Bureau of Shanghai (PWR12018-07 Chunlong Zhong), the Medical Discipline Construction Project of Pudong Health Committee of Shanghai (PWYgy 2021-07 Chunlong Zhong) and the Key Discipline Construction Project of Pudong Health Bureau of Shanghai (PWZxk 2017-23 Chunlong Zhong).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gu D., Ou S., Liu G. Traumatic Brain Injury and Risk of Dementia and Alzheimer’s Disease: A Systematic Review and Meta-Analysis. Neuroepidemiology. 2022;56:4–16. doi: 10.1159/000520966. [DOI] [PubMed] [Google Scholar]
- 2.Badhiwala J.H., Wilson J.R., Fehlings M.G. Global burden of traumatic brain and spinal cord injury. Lancet Neurol. 2019;18:24–25. doi: 10.1016/S1474-4422(18)30444-7. [DOI] [PubMed] [Google Scholar]
- 3.Singaravelu Jaganathan K., Sullivan K.A. Traumatic Brain Injury Rehabilitation: An Exercise Immunology Perspective. Exerc. Immunol. Rev. 2022;28:90–97. [PubMed] [Google Scholar]
- 4.Jiang J.Y., Gao G.Y., Feng J.F., Mao Q., Chen L.G., Yang X.F., Liu J.F., Wang Y.H., Qiu B.H., Huang X.J. Traumatic brain injury in China. Lancet Neurol. 2019;18:286–295. doi: 10.1016/S1474-4422(18)30469-1. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y., Wang Z., Peng J., Gerner S.T., Yin S., Jiang Y. Gut microbiota-brain interaction: An emerging immunotherapy for traumatic brain injury. Exp. Neurol. 2021;337:113585. doi: 10.1016/j.expneurol.2020.113585. [DOI] [PubMed] [Google Scholar]
- 6.Margolis K.G., Cryan J.F., Mayer E.A. The Microbiota-Gut-Brain Axis: From Motility to Mood. Gastroenterology. 2021;160:1486–1501. doi: 10.1053/j.gastro.2020.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Socala K., Doboszewska U., Szopa A., Serefko A., Włodarczyk M., Zielińska A., Poleszak E., Fichna J., Wlaź P. The role of microbiota-gut-brain axis in neuropsychiatric and neurological disorders. Pharmacol. Res. 2021;172:105840. doi: 10.1016/j.phrs.2021.105840. [DOI] [PubMed] [Google Scholar]
- 8.Agirman G., Hsiao E.Y. SnapShot: The microbiota-gut-brain axis. Cell. 2021;184:2524.e1. doi: 10.1016/j.cell.2021.03.022. [DOI] [PubMed] [Google Scholar]
- 9.Hanscom M., Loane D.J., Shea-Donohue T. Brain-gut axis dysfunction in the pathogenesis of traumatic brain injury. J. Clin. Investig. 2021;131:e143777. doi: 10.1172/JCI143777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikolova V.L., Hall M.R.B., Hall L.J., Cleare A.J., Stone J.M., Young A.H. Perturbations in Gut Microbiota Composition in Psychiatric Disorders: A Review and Meta-analysis. JAMA Psychiatry. 2021;78:1343–1354. doi: 10.1001/jamapsychiatry.2021.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., Xiang Y., Zhu Z., Wang W., Jiang Z., Zhao M., Cheng S., Pan F., Liu D., Ho R.C.M., et al. Rifaximin-mediated gut microbiota regulation modulates the function of microglia and protects against CUMS-induced depression-like behaviors in adolescent rat. J. Neuroinflamm. 2021;18:254. doi: 10.1186/s12974-021-02303-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan Z., Yang F., Cao J., Ding W., Yan S., Shi W., Wen S., Yao L. Alterations of gut microbiota and metabolome with Parkinson’s disease. Microb. Pathog. 2021;160:105187. doi: 10.1016/j.micpath.2021.105187. [DOI] [PubMed] [Google Scholar]
- 13.Xiang Z.-B., Xu R.-S., Zhu Y., Yuan M., Liu Y., Yang F., Chen W.-Z., Xu Z.-Z. Association between inflammatory bowel diseases and Parkinson’s disease: Systematic review and meta-analysis. Neural Regen. Res. 2022;17:344–353. doi: 10.4103/1673-5374.317981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang H., Zeng W., Zhang X., Pei Y., Zhang H., Li Y. The role of gut microbiota in patients with benign and malignant brain tumors: A pilot study. Bioengineered. 2022;13:7847–7859. doi: 10.1080/21655979.2022.2049959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herbreteau A., Aubert P., Croyal M., Naveilhan P., Billon-Crossouard S., Neunlist M., Delneste Y., Couez D., Aymeric L. Late-Stage Glioma Is Associated with Deleterious Alteration of Gut Bacterial Metabolites in Mice. Metabolites. 2022;12:290. doi: 10.3390/metabo12040290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weaver J.L., Eliceiri B., Costantini T.W. Fluoxetine reduces organ injury and improves motor function after traumatic brain injury in mice. J. Trauma Acute Care Surg. 2022;93:38–42. doi: 10.1097/TA.0000000000003646. [DOI] [PubMed] [Google Scholar]
- 17.Taraskina A., Ignatyeva O., Lisovaya D., Ivanov M., Ivanova L., Golovicheva V., Baydakova G., Silachev D., Popkov V., Ivanets T., et al. Effects of Traumatic Brain Injury on the Gut Microbiota Composition and Serum Amino Acid Profile in Rats. Cells. 2022;11:1409. doi: 10.3390/cells11091409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Zhang L., Ren H., Ma L., Guo J., Mao D., Lu Z., Lu L., Yan D. Role of Hakai in m6A modification pathway in Drosophila. Nat. Commun. 2021;12:2159. doi: 10.1038/s41467-021-22424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 20.Roundtree I.A., Evans M., Pan T., He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du B., Zhang Y., Liang M., Du Z., Li H., Fan C., Zhang H., Jiang Y., Bi X. N6-methyladenosine (m6A) modification and its clinical relevance in cognitive dysfunctions. Aging. 2021;13:20716–20737. doi: 10.18632/aging.203457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng Y., Zhu H., Xiao L., Liu C., Liu Y.-L., Gao W. Identification of the function and mechanism of m6A reader IGF2BP2 in Alzheimer’s disease. Aging. 2021;13:24086–24100. doi: 10.18632/aging.203652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang N., Ding C., Zuo Y., Peng Y., Zuo L. N6-methyladenosine and Neurological Diseases. Mol. Neurobiol. 2022;59:1925–1937. doi: 10.1007/s12035-022-02739-0. [DOI] [PubMed] [Google Scholar]
- 24.Zhuo R., Xu M., Wang X., Zhou B., Wu X., Leone V., Chang E.B., Zhong X. The regulatory role of N6-methyladenosine modification in the interaction between host and microbes. Wiley Interdiscip. Rev. RNA. 2022:e1725. doi: 10.1002/wrna.1725. [DOI] [PubMed] [Google Scholar]
- 25.Wu J., Zhao Y., Wang X., Kong L., Johnston L.J., Lu L., Ma X. Dietary nutrients shape gut microbes and intestinal mucosa via epigenetic modifications. Crit. Rev. Food Sci. Nutr. 2020;62:783–797. doi: 10.1080/10408398.2020.1828813. [DOI] [PubMed] [Google Scholar]
- 26.Sun L., Wan A., Zhou Z., Chen D., Liang H., Liu C., Yan S., Niu Y., Lin Z., Zhan S., et al. RNA-binding protein RALY reprogrammes mitochondrial metabolism via mediating miRNA processing in colorectal cancer. Gut. 2021;70:1698–1712. doi: 10.1136/gutjnl-2020-320652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jabs S., Biton A., Bécavin C., Nahori M.-A., Ghozlane A., Pagliuso A., Spanò G., Guérineau V., Touboul D., Gianetto Q.G., et al. Impact of the gut microbiota on the m6A epitranscriptome of mouse cecum and liver. Nat. Commun. 2020;11:1344. doi: 10.1038/s41467-020-15126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tait C., Sayuk G.S. The Brain-Gut-Microbiotal Axis: A framework for understanding functional GI illness and their therapeutic interventions. Eur. J. Intern. Med. 2021;84:1–9. doi: 10.1016/j.ejim.2020.12.023. [DOI] [PubMed] [Google Scholar]
- 29.Checa-Ros A., Jeréz-Calero A., Molina-Carballo A., Campoy C., Muñoz-Hoyos A. Current Evidence on the Role of the Gut Microbiome in ADHD Pathophysiology and Therapeutic Implications. Nutrients. 2021;13:249. doi: 10.3390/nu13010249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beopoulos A., Gea M., Fasano A., Iris F. Autonomic Nervous System Neuroanatomical Alterations Could Provoke and Maintain Gastrointestinal Dysbiosis in Autism Spectrum Disorder (ASD): A Novel Microbiome–Host Interaction Mechanistic Hypothesis. Nutrients. 2021;14:65. doi: 10.3390/nu14010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudo N. Microbiome, HPA Axis and Production of Endocrine Hormones in the Gut. Adv. Exp. Med. Biol. 2014;817:177–194. doi: 10.1007/978-1-4939-0897-4_8. [DOI] [PubMed] [Google Scholar]
- 32.Agirman G., Yu K.B., Hsiao E.Y. Signaling inflammation across the gut-brain axis. Science. 2021;374:1087–1092. doi: 10.1126/science.abi6087. [DOI] [PubMed] [Google Scholar]
- 33.Spencer R.L., Deak T. A users guide to HPA axis research. Physiol. Behav. 2017;178:43–65. doi: 10.1016/j.physbeh.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cussotto S., Sandhu K.V., Dinan T.G., Cryan J.F. The Neuroendocrinology of the Microbiota-Gut-Brain Axis: A Behavioural Perspective. Front. Neuroendocr. 2018;51:80–101. doi: 10.1016/j.yfrne.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Kumar A., Stoica B.A., Loane D.J., Yang M., Abulwerdi G., Khan N., Kumar A., Thom S.R., Faden A.I. Microglial-derived microparticles mediate neuroinflammation after traumatic brain injury. J. Neuroinflamm. 2017;14:47. doi: 10.1186/s12974-017-0819-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braniste V., Al-Asmakh M., Kowal C., Anuar F., Abbaspour A., Tóth M., Korecka A., Bakocevic N., Ng L.G., Kundu P., et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 2014;6:263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly J.R., Kennedy P.J., Cryan J.F., Dinan T.G., Clarke G., Hyland N.P. Breaking down the barriers: The gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front. Cell. Neurosci. 2015;9:392. doi: 10.3389/fncel.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renteria M.A., Gillett S.R., McClure L.A., Wadley V.G., Glasser S.P., Howard V.J., Kissela B.M., Unverzagt F.W., Jenny N.S., Manly J.J., et al. C-reactive protein and risk of cognitive decline: The REGARDS study. PLoS ONE. 2020;15:e0244612. doi: 10.1371/journal.pone.0244612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanscom M., Loane D.J., Aubretch T., Leser J., Molesworth K., Hedgekar N., Ritzel R.M., Abulwerdi G., Shea-Donohue T., Faden A.I. Acute colitis during chronic experimental traumatic brain injury in mice induces dysautonomia and persistent extraintestinal, systemic, and CNS inflammation with exacerbated neurological deficits. J. Neuroinflamm. 2021;18:24. doi: 10.1186/s12974-020-02067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baizabal-Carvallo J.F., Alonso-Juarez M. The Link between Gut Dysbiosis and Neuroinflammation in Parkinson’s Disease. Neuroscience. 2020;432:160–173. doi: 10.1016/j.neuroscience.2020.02.030. [DOI] [PubMed] [Google Scholar]
- 41.Cenit M.C., Sanz Y., Codoner-Franch P. Influence of gut microbiota on neuropsychiatric disorders. World J. Gastroenterol. 2017;23:5486–5498. doi: 10.3748/wjg.v23.i30.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ou Z., Deng L., Lu Z., Wu F., Liu W., Huang D., Peng Y. Protective effects of Akkermansia muciniphila on cognitive deficits and amyloid pathology in a mouse model of Alzheimer’s disease. Nutr. Diabetes. 2020;10:12. doi: 10.1038/s41387-020-0115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blacher E., Bashiardes S., Shapiro H., Rothschild D., Mor U., Dori-Bachash M., Kleimeyer C., Moresi C., Harnik Y., Zur M., et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature. 2019;572:474–480. doi: 10.1038/s41586-019-1443-5. [DOI] [PubMed] [Google Scholar]
- 44.Di Gioia D., Cionci N.B., Baffoni L., Amoruso A., Pane M., Mogna L., Gaggìa F., Lucenti M.A., Bersano E., Cantello R., et al. A prospective longitudinal study on the microbiota composition in amyotrophic lateral sclerosis. BMC Med. 2020;18:153. doi: 10.1186/s12916-020-01607-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright M.L., Fournier C., Houser M., Tansey M., Glass J., Hertzberg V.S. Potential Role of the Gut Microbiome in ALS: A Systematic Review. Biol. Res. Nurs. 2018;20:513–521. doi: 10.1177/1099800418784202. [DOI] [PubMed] [Google Scholar]
- 46.Kadowaki A., Saga R., Lin Y., Sato W., Yamamura T. Gut microbiota-dependent CCR9+CD4+ T cells are altered in secondary progressive multiple sclerosis. Brain. 2019;142:916–931. doi: 10.1093/brain/awz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boziki M.K., Kesidou E., Theotokis P., Mentis A.-F.A., Karafoulidou E., Melnikov M., Sviridova A., Rogovski V., Boyko A., Grigoriadis N. Microbiome in Multiple Sclerosis: Where Are We, What We Know and Do Not Know. Brain Sci. 2020;10:234. doi: 10.3390/brainsci10040234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reynders T., Devolder L., Valles-Colomer M., Van Remoortel A., Joossens M., De Keyser J., Nagels G., D’Hooghe M., Raes J. Gut microbiome variation is associated to Multiple Sclerosis phenotypic subtypes. Ann. Clin. Transl. Neurol. 2020;7:406–419. doi: 10.1002/acn3.51004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villaran R.F., Espinosa-Oliva A.M., Sarmiento M., De Pablos R.M., Argüelles S., Delgado-Cortés M.J., Sobrino V., Van Rooijen N., Venero J.L., Herrera A.J., et al. Ulcerative colitis exacerbates lipopolysaccharide-induced damage to the nigral dopaminergic system: Potential risk factor in Parkinson’s disease. J. Neurochem. 2010;114:1687–1700. doi: 10.1111/j.1471-4159.2010.06879.x. [DOI] [PubMed] [Google Scholar]
- 50.Banerjee A., Pradhan L.K., Sahoo P.K., Jena K.K., Chauhan N.R., Chauhan S., Das S.K. Unravelling the potential of gut microbiota in sustaining brain health and their current prospective towards development of neurotherapeutics. Arch. Microbiol. 2021;203:2895–2910. doi: 10.1007/s00203-021-02276-9. [DOI] [PubMed] [Google Scholar]
- 51.Hung C.C., Chang C.-C., Huang C.-W., Nouchi R., Cheng C.-H. Gut microbiota in patients with Alzheimer’s disease spectrum: A systematic review and meta-analysis. Aging. 2022;14:477–496. doi: 10.18632/aging.203826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rutsch A., Kantsjö J.B., Ronchi F. The Gut-Brain Axis: How Microbiota and Host Inflammasome Influence Brain Physiology and Pathology. Front. Immunol. 2020;11:604179. doi: 10.3389/fimmu.2020.604179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lukiw W.J. Bacteroides fragilis lipopolysaccharide and inflammatory signaling in Alzheimer’s Disease. Front. Microbiol. 2016;7:1544. doi: 10.3389/fmicb.2016.01544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hertzberg V.S., Singh H., Fournier C.N., Moustafa A., Polak M., Kuelbs C.A., Torralba M.G., Tansey M.G., Nelson K.E., Glass J.D. Gut microbiome differences between amyotrophic lateral sclerosis patients and spouse controls. Amyotroph. Lateral Scler. Front. Degener. 2022;23:91–99. doi: 10.1080/21678421.2021.1904994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garbuzova-Davis S., Hernandez-Ontiveros D.G., Rodrigues M.C., Haller E., Frisina-Deyo A., Mirtyl S., Sallot S., Saporta S., Borlongan C., Sanberg P.R. Impaired blood–brain/spinal cord barrier in ALS patients. Brain Res. 2012;1469:114–128. doi: 10.1016/j.brainres.2012.05.056. [DOI] [PubMed] [Google Scholar]
- 56.Paray B.A., Albeshr M.F., Jan A.T., Rather I.A. Leaky Gut and Autoimmunity: An Intricate Balance in Individuals Health and the Diseased State. Int. J. Mol. Sci. 2020;21:9770. doi: 10.3390/ijms21249770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dabrowski W., Siwicka-Gieroba D., Kotfis K., Zaid S., Terpilowska S., Robba C., Siwicki A.K. The Brain-gut Axis-where are we now and how can we Modulate these Connections? Curr. Neuropharmacol. 2021;19:1164–1177. doi: 10.2174/1570159X18666201119155535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma E.L., Smith A.D., Desai N., Cheung L., Hanscom M., Stoica B.A., Loane D.J., Shea-Donohue T., Faden A.I. Bidirectional brain-gut interactions and chronic pathological changes after traumatic brain injury in mice. Brain Behav. Immun. 2017;66:56–69. doi: 10.1016/j.bbi.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H., Sun J., Du J., Wang F., Fang R., Yu C., Xiong J., Chen W., Lu Z., Liu J. Clostridium butyricum exerts a neuroprotective effect in a mouse model of traumatic brain injury via the gut-brain axis. Neurogastroenterol. Motil. 2017;30:e13260. doi: 10.1111/nmo.13260. [DOI] [PubMed] [Google Scholar]
- 60.George A.K., Behera J., Homme R.P., Tyagi N., Tyagi S.C., Singh M. Rebuilding Microbiome for Mitigating Traumatic Brain Injury: Importance of Restructuring the Gut-Microbiome-Brain Axis. Mol. Neurobiol. 2021;58:3614–3627. doi: 10.1007/s12035-021-02357-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kharrazian D. Traumatic Brain Injury and the Effect on the Brain-Gut Axis. Altern. Ther. Health Med. 2015;21((Suppl. S3)):28–32. [PubMed] [Google Scholar]
- 62.Toklu H.Z., Sakarya Y., Tumer N. A Proteomic Evaluation of Sympathetic Activity Biomarkers of the Hypothalamus-Pituitary-Adrenal Axis by Western Blotting Technique Following Experimental Traumatic Brain Injury. Methods Mol. Biol. 2017;1598:313–325. doi: 10.1007/978-1-4939-6952-4_16. [DOI] [PubMed] [Google Scholar]
- 63.Tapp Z.M., Godbout J.P., Kokiko-Cochran O.N. A Tilted Axis: Maladaptive Inflammation and HPA Axis Dysfunction Contribute to Consequences of TBI. Front. Neurol. 2019;10:345. doi: 10.3389/fneur.2019.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McDonald S., Sharkey J.M., Sun M., Kaukas L.M., Shultz S.R., Turner R.J., Leonard A.V., Brady R.D., Corrigan F. Beyond the Brain: Peripheral Interactions after Traumatic Brain Injury. J. Neurotrauma. 2020;37:770–781. doi: 10.1089/neu.2019.6885. [DOI] [PubMed] [Google Scholar]
- 65.Taylor A.N., Rahman S.U., Sanders N.C., Tio D.L., Prolo P., Sutton R.L. Injury Severity Differentially Affects Short- and Long-Term Neuroendocrine Outcomes of Traumatic Brain Injury. J. Neurotrauma. 2008;25:311–323. doi: 10.1089/neu.2007.0486. [DOI] [PubMed] [Google Scholar]
- 66.Jassam Y.N., Izzy S., Whalen M., McGavern D.B., El Khoury J. Neuroimmunology of Traumatic Brain Injury: Time for a Paradigm Shift. Neuron. 2017;95:1246–1265. doi: 10.1016/j.neuron.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hang C.H., Shi J.-X., Li J.-S., Li W.-Q., Yin H.-X. Up-regulation of intestinal nuclear factor kappa B and intercellular adhesion molecule-1 following traumatic brain injury in rats. World J. Gastroenterol. 2005;11:1149–1154. doi: 10.3748/wjg.v11.i8.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen S., Liu H., Li Z., Tang J., Huang B., Zhi F., Zhao X. Epithelial PBLD attenuates intestinal inflammatory response and improves intestinal barrier function by inhibiting NF-kappaB signaling. Cell Death Dis. 2021;12:563. doi: 10.1038/s41419-021-03843-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hang C.H., Shi J.-X., Li J.-S., Li W.-Q., Wu W. Expressions of intestinal NF-κB, TNF-α, and IL-6 following traumatic brain injury in rats. J. Surg. Res. 2005;123:188–193. doi: 10.1016/j.jss.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 70.Bansal V., Costantini T., Kroll L., Peterson C., Loomis W., Eliceiri B., Baird A., Wolf P., Coimbra R. Traumatic Brain Injury and Intestinal Dysfunction: Uncovering the Neuro-Enteric AXIS. J. Neurotrauma. 2009;26:1353–1359. doi: 10.1089/neu.2008.0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Urban R.J., Pyles R.B., Stewart C.J., Ajami N., Randolph K.M., Durham W.J., Danesi C.P., Dillon E.L., Summons M.J.R., Singh C.K., et al. Altered Fecal Microbiome Years after Traumatic Brain Injury. J. Neurotrauma. 2020;37:1037–1051. doi: 10.1089/neu.2019.6688. [DOI] [PubMed] [Google Scholar]
- 72.Liu X., Liu L., Dong Z., Li J., Yu Y., Chen X., Ren F., Cui G., Sun R. Expression patterns and prognostic value of m6A-related genes in colorectal cancer. Am. J. Transl. Res. 2019;11:3972–3991. [PMC free article] [PubMed] [Google Scholar]
- 73.Coker H., Wei G., Brockdorff N. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim. Biophys. Acta Gene Regul. Mech. 2019;1862:310–318. doi: 10.1016/j.bbagrm.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 74.Roignant J.Y., Soller M. m6A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression. Trends Genet. 2017;33:380–390. doi: 10.1016/j.tig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 75.Sun T., Wu R., Ming L. The role of m6A RNA methylation in cancer. Biomed. Pharmacother. 2019;112:108613. doi: 10.1016/j.biopha.2019.108613. [DOI] [PubMed] [Google Scholar]
- 76.Zhang Y., Geng X., Li Q., Xu J., Tan Y., Xiao M., Song J., Liu F., Fang C., Wang H. m6A modification in RNA: Biogenesis, functions and roles in gliomas. J. Exp. Clin. Cancer Res. 2020;39:192. doi: 10.1186/s13046-020-01706-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pan Y., Ma P., Liu Y., Li W., Shu Y. Multiple functions of m6A RNA methylation in cancer. J. Hematol. Oncol. 2018;11:48. doi: 10.1186/s13045-018-0590-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma S., Chen C., Ji X., Liu J., Zhou Q., Wang G., Yuan W., Kan Q., Sun Z. The interplay between m6A RNA methylation and noncoding RNA in cancer. J. Hematol. Oncol. 2019;12:121. doi: 10.1186/s13045-019-0805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi H., Zhang X., Weng Y.-L., Lu Z., Liu Y., Lu Z., Li J., Hao P., Zhang Y., Zhang F., et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563:249–253. doi: 10.1038/s41586-018-0666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang J., Sha Y., Sun T. m6A Modifications Play Crucial Roles in Glial Cell Development and Brain Tumorigenesis. Front. Oncol. 2021;11:611660. doi: 10.3389/fonc.2021.611660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu T.X., Zheng Z., Zhang L., Sun H.-L., Bissonnette M., Huang H., He C. A New Model of Spontaneous Colitis in Mice Induced by Deletion of an RNA m6A Methyltransferase Component METTL14 in T Cells. Cell. Mol. Gastroenterol. Hepatol. 2020;10:747–761. doi: 10.1016/j.jcmgh.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu D., Shao J., Song H., Wang J. The YTH Domain Family of N6-Methyladenosine “Readers” in the Diagnosis and Prognosis of Colonic Adenocarcinoma. BioMed Res. Int. 2020;2020:9502560. doi: 10.1155/2020/9502560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang H., Wang Q., Chen J., Chen C. Association Among the Gut Microbiome, the Serum Metabolomic Profile and RNA m6A Methylation in Sepsis-Associated Encephalopathy. Front. Genet. 2022;13:859727. doi: 10.3389/fgene.2022.859727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu J., Frazier K., Zhang J., Gan Z., Wang T., Zhong X. Emerging role of m6A RNA methylation in nutritional physiology and metabolism. Obes. Rev. 2020;21:e12942. doi: 10.1111/obr.12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bedi R.K., Huang D., Eberle S.A., Wiedmer L., Śledź P., Caflisch A. Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer. ChemMedChem. 2020;15:744–748. doi: 10.1002/cmdc.202000011. [DOI] [PubMed] [Google Scholar]
- 86.Tuck M.T. Partial purification of a 6-methyladenine mRNA methyltransferase which modifies internal adenine residues. Pt 1Biochem. J. 1992;288:233–240. doi: 10.1042/bj2880233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X., et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scholler E., Weichmann F., Treiber T., Ringle S., Treiber N., Flatley A., Feederle R., Bruckmann A., Meister G. Interactions, localization, and phosphorylation of the m6A generating METTL3–METTL14–WTAP complex. RNA. 2018;24:499–512. doi: 10.1261/rna.064063.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morales D.G., Reyes J.L. A birds’-eye view of the activity and specificity of themRNA m6Amethyltransferase complex. Wiley Interdiscip. Rev. RNA. 2021;12:e1618. doi: 10.1002/wrna.1618. [DOI] [PubMed] [Google Scholar]
- 90.Balacco D.L., Soller M. The m6A Writer: Rise of a Machine for Growing Tasks. Biochemistry. 2019;58:363–378. doi: 10.1021/acs.biochem.8b01166. [DOI] [PubMed] [Google Scholar]
- 91.Warda A.S., Kretschmer J., Hackert P., Lenz C., Urlaub H., Höbartner C., Sloan K.E., Bohnsack M.T. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004–2014. doi: 10.15252/embr.201744940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bawankar P., Lence T., Paolantoni C., Haussmann I.U., Kazlauskiene M., Jacob D., Heidelberger J.B., Richter F.M., Nallasivan M.P., Morin V., et al. Hakai is required for stabilization of core components of the m6A mRNA methylation machinery. Nat. Commun. 2021;12:3778. doi: 10.1038/s41467-021-23892-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xia Z., Tang M., Ma J., Zhang H., Gimple R.C., Prager B.C., Tang H., Sun C., Liu F., Lin P., et al. Epitranscriptomic editing of the RNA N6-methyladenosine modification by dCasRx conjugated methyltransferase and demethylase. Nucleic Acids Res. 2021;49:7361–7374. doi: 10.1093/nar/gkab517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ping X.L., Sun B.-F., Wang L., Xiao W., Yang X., Wang W.-J., Adhikari S., Shi Y., Lv Y., Chen Y.-S., et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Y., Li Y., Toth J.I., Petroski M.D., Zhang Z., Zhao J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zeng C., Huang W., Li Y., Weng H. Roles of METTL3 in cancer: Mechanisms and therapeutic targeting. J. Hematol. Oncol. 2020;13:117. doi: 10.1186/s13045-020-00951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mendel M., Chen K.-M., Homolka D., Gos P., Pandey R.R., McCarthy A.A., Pillai R.S. Methylation of Structured RNA by the m6A Writer METTL16 Is Essential for Mouse Embryonic Development. Mol. Cell. 2018;71:986–1000.e11. doi: 10.1016/j.molcel.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pendleton K.E., Chen B., Liu K., Hunter O.V., Xie Y., Tu B.P., Conrad N.K. The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017;169:824–835.e14. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Patil D.P., Chen C.-K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., Cheng T., Gao M., Shu X., Ma H., et al. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wen J., Lv R., Ma H., Shen H., He C., Wang J., Jiao F., Liu H., Yang P., Tan L., et al. Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell. 2018;69:1028–1038.e6. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ignatova V.V., Stolz P., Kaiser S., Gustafsson T.H., Lastres P.R., Sanz-Moreno A., Cho Y.-L., Amarie O.V., Aguilar-Pimentel A., Klein-Rodewald T., et al. The rRNA m6A methyltransferase METTL5 is involved in pluripotency and developmental programs. Genes Dev. 2020;34:715–729. doi: 10.1101/gad.333369.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Van Tran N., Ernst F.G.M., Hawley B.R., Zorbas C., Ulryck N., Hackert P., Bohnsack K.E., Bohnsack M.T., Jaffrey S.R., Graille M., et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47:7719–7733. doi: 10.1093/nar/gkz619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pinto R., Vågbø C.B., Jakobsson M.E., Kim Y., Baltissen M.P., O’Donohue M.-F., Guzmán U.H., Małecki J.M., Wu J., Kirpekar F., et al. The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. 2020;48:830–846. doi: 10.1093/nar/gkz1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ren W., Lu J., Huang M., Gao L., Li D., Wang G.G., Song J. Structure and regulation of ZCCHC4 in m6A-methylation of 28S rRNA. Nat. Commun. 2019;10:5042. doi: 10.1038/s41467-019-12923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ma H., Wang X., Cai J., Dai Q., Natchiar S.K., Lv R., Chen K., Lu Z., Chen H., Shi Y.G., et al. N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 2019;15:88–94. doi: 10.1038/s41589-018-0184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wei J., Liu F., Lu Z., Fei Q., Ai Y., He P.C., Shi H., Cui X., Su R., Klungland A., et al. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell. 2018;71:973–985.e5. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mauer J., Sindelar M., Despic V., Guez T., Hawley B.R., Vasseur J.-J., Rentmeister A., Gross S.S., Pellizzoni L., Debart F., et al. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 2019;15:340–347. doi: 10.1038/s41589-019-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bartosovic M., Molares H.C., Gregorova P., Hrossova D., Kudla G., Vanacova S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 2017;45:11356–11370. doi: 10.1093/nar/gkx778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Roundtree I.A., Luo G.-Z., Zhang Z., Wang X., Zhou T., Cui Y., Sha J., Huang X., Guerrero L., Xie P., et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife. 2017;6:e31311. doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xiao W., Adhikari S., Dahal U., Chen Y.-S., Hao Y.-J., Sun B.-F., Sun H.-Y., Li A., Ping X.-L., Lai W.-Y., et al. Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 112.Zaccara S., Jaffrey S.R. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell. 2020;181:1582–1595.e18. doi: 10.1016/j.cell.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mao Y., Dong L., Liu X.-M., Guo J., Ma H., Shen B., Qian S.-B. m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 2019;10:5332. doi: 10.1038/s41467-019-13317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kretschmer J., Rao H., Hackert P., Sloan K.E., Höbartner C., Bohnsack M.T. The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′–3′ exoribonuclease XRN1. RNA. 2018;24:1339–1350. doi: 10.1261/rna.064238.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Alarcon C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F. HNRNPA2B1 Is a Mediator of m6A-Dependent Nuclear RNA Processing Events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu N., Zhou K.I., Parisien M., Dai Q., Diatchenko L., Pan T. N 6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45:6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.-B., Jaffrey S.R. 5′ UTR m6A Promotes Cap-Independent Translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., Zhao B.S., Mesquita A., Liu C., Yuan C.L., et al. Publisher Correction: Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2020;22:1288. doi: 10.1038/s41556-020-00580-y. [DOI] [PubMed] [Google Scholar]
- 120.Fu Y., Jia G., Pang X., Wang R.N., Wang X., Li C.J., Smemo S., Dai Q., Bailey K.A., Nobrega M.A., et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 2013;4:1798. doi: 10.1038/ncomms2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., Linder B., Pickering B.F., Vasseur J.-J., Chen Q., et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Garcia-Campos M.A., Edelheit S., Toth U., Safra M., Shachar R., Viukov S., Winkler R., Nir R., Lasman L., Brandis A., et al. Deciphering the “m6A Code” via Antibody-Independent Quantitative Profiling. Cell. 2019;178:731–747.e16. doi: 10.1016/j.cell.2019.06.013. [DOI] [PubMed] [Google Scholar]
- 123.Li X.-C., Jin F., Wang B.-Y., Yin X.-J., Hong W., Tian F.-J. The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of CYR61 mRNA. Theranostics. 2019;9:3853–3865. doi: 10.7150/thno.31868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.-M., Li C.J., Vågbø C.B., Shi Y., Wang W.-L., Song S.-H., et al. ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Allis C.D., Jenuwein T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 126.Zhang Y., Wang X., Zhang X., Wang J., Ma Y., Zhang L., Cao X. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc. Natl. Acad. Sci. USA. 2019;116:976–981. doi: 10.1073/pnas.1812536116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang Z., Wang Q., Zhao X., Shao L., Liu G., Zheng X., Xie L., Zhang Y., Sun C., Xu R. YTHDC1 mitigates ischemic stroke by promoting Akt phosphorylation through destabilizing PTEN mRNA. Cell Death Dis. 2020;11:977. doi: 10.1038/s41419-020-03186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sheng Y., Wei J., Yu F., Xu H., Yu C., Wu Q., Liu Y., Li L., Cui X.-L., Gu X., et al. A critical role of nuclear m6A reader YTHDC1 in leukemogenesis by regulating MCM complex–mediated DNA replication. Blood. 2021;138:2838–2852. doi: 10.1182/blood.2021011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roundtree I.A., He C. Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Trends Genet. 2016;32:320–321. doi: 10.1016/j.tig.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 130.Li Y., Zheng J.-N., Wang E.-H., Gong C.-J., Lan K.-F., Ding X. The m6A reader protein YTHDC2 is a potential biomarker and associated with immune infiltration in head and neck squamous cell carcinoma. PeerJ. 2020;8:e10385. doi: 10.7717/peerj.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Geuens T., Bouhy D., Timmerman V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016;135:851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Des Georges A., Dhote V., Kuhn L., Hellen C.U., Pestova T.V., Frank J., Hashem Y. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015;525:491–495. doi: 10.1038/nature14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Livneh I., Moshitch-Moshkovitz S., Amariglio N., Rechavi G., Dominissini D. The m6A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2019;21:36–51. doi: 10.1038/s41583-019-0244-z. [DOI] [PubMed] [Google Scholar]
- 134.Arguello A.E., DeLiberto A.N., Kleiner R.E. RNA Chemical Proteomics Reveals the N6-Methyladenosine (m6A)-Regulated Protein–RNA Interactome. J. Am. Chem. Soc. 2017;139:17249–17252. doi: 10.1021/jacs.7b09213. [DOI] [PubMed] [Google Scholar]
- 135.Edupuganti R.R., Geiger S., Lu Z., Wang S.-Y., Baltissen M.P.A., Jansen P.W.T.C., Rossa M., Müller M., Stunnenberg H.G., He C., et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 2017;24:870–878. doi: 10.1038/nsmb.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chang M., Wang Z., Gao J., Yang C., Feng M., Niu Y., Tong W.-M., Bao X., Wang R. METTL3-mediated RNA m6A Hypermethylation Promotes Tumorigenesis and GH Secretion of Pituitary Somatotroph Adenomas. J. Clin. Endocrinol. Metab. 2022;107:136–149. doi: 10.1210/clinem/dgab652. [DOI] [PubMed] [Google Scholar]
- 137.Lv Z., Xu T., Li R., Zheng D., Li Y., Li W., Yang Y., Hao Y. Downregulation of m6A Methyltransferase in the Hippocampus of Tyrobp–/– Mice and Implications for Learning and Memory Deficits. Front. Neurosci. 2022;16:739201. doi: 10.3389/fnins.2022.739201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lei C., Wang Q. The Progression of N6-methyladenosine Study and Its Role in Neuropsychiatric Disorders. Int. J. Mol. Sci. 2022;23:5922. doi: 10.3390/ijms23115922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang C.X., Cui G.-S., Liu X., Xu K., Wang M., Zhang X.-X., Jiang L.-Y., Li A., Yang Y., Lai W.-Y., et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018;16:e2004880. doi: 10.1371/journal.pbio.2004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Weng Y.L., Wang X., An R., Cassin J., Vissers C., Liu Y., Liu Y., Xu T., Wang X., Wong S.Z.H., et al. Epitranscriptomic m6A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron. 2018;97:313–325.e6. doi: 10.1016/j.neuron.2017.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Walters B.J., Mercaldo V., Gillon C., Yip M., Neve R.L., Boyce F.M., Frankland P.W., Josselyn S.A. The Role of The RNA Demethylase FTO (Fat Mass and Obesity-Associated) and mRNA Methylation in Hippocampal Memory Formation. Neuropsychopharmacology. 2017;42:1502–1510. doi: 10.1038/npp.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhou J., Zhang X., Hu J., Qu R., Yu Z., Xu H., Chen H., Yan L., Ding C., Zou Q., et al. m6A demethylase ALKBH5 controls CD4 + T cell pathogenicity and promotes autoimmunity. Sci. Adv. 2021;7:eabg0470. doi: 10.1126/sciadv.abg0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li M., Zhao X., Wang W., Shi H., Pan Q., Lu Z., Perez S.P., Suganthan R., He C., Bjørås M., et al. Ythdf2-mediated m6A mRNA clearance modulates neural development in mice. Genome Biol. 2018;19:69. doi: 10.1186/s13059-018-1436-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Merkurjev D., Hong W.-T., Iida K., Oomoto I., Goldie B.J., Yamaguti H., Ohara T., Kawaguchi S.-Y., Hirano T., Martin K.C., et al. Author Correction: Synaptic N6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 2018;21:1493. doi: 10.1038/s41593-018-0219-9. [DOI] [PubMed] [Google Scholar]
- 145.Quan W., Li J., Liu L., Zhang Q., Qin Y., Pei X., Chen J. Influence of N6-Methyladenosine Modification Gene HNRNPC on Cell Phenotype in Parkinson’s Disease. Park. Dis. 2021;2021:9919129. doi: 10.1155/2021/9919129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Qiu X., He H., Huang Y., Wang J., Xiao Y. Genome-wide identification of m6A-associated single-nucleotide polymorphisms in Parkinson’s disease. Neurosci. Lett. 2020;737:135315. doi: 10.1016/j.neulet.2020.135315. [DOI] [PubMed] [Google Scholar]
- 147.Chen X., Yu C., Guo M., Zheng X., Ali S., Huang H., Zhang L., Wang S., Huang Y., Qie S., et al. Down-Regulation of m6A mRNA Methylation Is Involved in Dopaminergic Neuronal Death. ACS Chem. Neurosci. 2019;10:2355–2363. doi: 10.1021/acschemneuro.8b00657. [DOI] [PubMed] [Google Scholar]
- 148.Xu C., Huang H., Zhang M., Zhang P., Li Z., Liu X., Fang M. Methyltransferase-Like 3 Rescues the Amyloid-beta protein-Induced Reduction of Activity-Regulated Cytoskeleton Associated Protein Expression via YTHDF1-Dependent N6-Methyladenosine Modification. Front. Aging Neurosci. 2022;14:890134. doi: 10.3389/fnagi.2022.890134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Feng Z., Zhou F., Tan M., Wang T., Chen Y., Xu W., Li B., Wang X., Deng X., He M.-L. Targeting m6A modification inhibits herpes virus 1 infection. Genes Dis. 2022;9:1114–1128. doi: 10.1016/j.gendis.2021.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cockova Z., Honc O., Telensky P., Olsen M.J., Novotny J. Streptozotocin-Induced Astrocyte Mitochondrial Dysfunction Is Ameliorated by FTO Inhibitor MO-I-500. ACS Chem. Neurosci. 2021;12:3818–3828. doi: 10.1021/acschemneuro.1c00063. [DOI] [PubMed] [Google Scholar]
- 151.Zhao F., Xu Y., Gao S., Qin L., Austria Q., Siedlak S.L., Pajdzik K., Dai Q., He C., Wang W., et al. METTL3-dependent RNA m6A dysregulation contributes to neurodegeneration in Alzheimer’s disease through aberrant cell cycle events. Mol. Neurodegener. 2021;16:70. doi: 10.1186/s13024-021-00484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shafik A.M., Zhang F., Guo Z., Dai Q., Pajdzik K., Li Y., Kang Y., Yao B., Wu H., He C., et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 2021;22:17. doi: 10.1186/s13059-020-02249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ye F., Wang T., Wu X., Liang J., Li J., Sheng W. N6-Methyladenosine RNA modification in cerebrospinal fluid as a novel potential diagnostic biomarker for progressive multiple sclerosis. J. Transl. Med. 2021;19:316. doi: 10.1186/s12967-021-02981-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li B., Wang F., Wang N., Hou K., Du J. Identification of Implications of Angiogenesis and m6A Modification on Immunosuppression and Therapeutic Sensitivity in Low-Grade Glioma by Network Computational Analysis of Subtypes and Signatures. Front. Immunol. 2022;13:871564. doi: 10.3389/fimmu.2022.871564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Miao Y.Q., Chen W., Zhou J., Shen Q., Sun Y., Li T., Wang S.-C. N(6)-adenosine-methyltransferase-14 promotes glioma tumorigenesis by repressing argininosuccinate synthase 1 expression in an m6A-dependent manner. Bioengineered. 2022;13:1858–1871. doi: 10.1080/21655979.2021.2018386. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 156.Zhang G., Zheng P., Lv Y., Shi Z., Shi F. m6A Regulatory Gene-Mediated Methylation Modification in Glioma Survival Prediction. Front. Genet. 2022;13:873764. doi: 10.3389/fgene.2022.873764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tan N.N., Tang H.-L., Lin G.-W., Chen Y.-H., Lu P., Li H.-J., Gao M.-M., Zhao Q.-H., Yi Y.-H., Liao W.-P., et al. Epigenetic Downregulation of Scn3a Expression by Valproate: A Possible Role in Its Anticonvulsant Activity. Mol. Neurobiol. 2017;54:2831–2842. doi: 10.1007/s12035-016-9871-9. [DOI] [PubMed] [Google Scholar]
- 158.Wang X.L., Wei X., Yuan J.-J., Mao Y.-Y., Wang Z.-Y., Xing N., Gu H.-W., Lin C.-H., Wang W.-T., Zhang W., et al. Downregulation of Fat Mass and Obesity-Related Protein in the Anterior Cingulate Cortex Participates in Anxiety- and Depression-Like Behaviors Induced by Neuropathic Pain. Front. Cell. Neurosci. 2022;16:884296. doi: 10.3389/fncel.2022.884296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Anderson K.S., Gosselin N., Sadikot A.F., Laguë-Beauvais M., Kang E.S.H., Fogarty A.E., Marcoux J., Dagher J., de Guise E. Pitch and Rhythm Perception and Verbal Short-Term Memory in Acute Traumatic Brain Injury. Brain Sci. 2021;11:1173. doi: 10.3390/brainsci11091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chokkalla A.K., Mehta S.L., Kim T., Chelluboina B., Kim J., Vemuganti R. Transient Focal Ischemia Significantly Alters the m6A Epitranscriptomic Tagging of RNAs in the Brain. Stroke. 2019;50:2912–2921. doi: 10.1161/STROKEAHA.119.026433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Diao M.Y., Zhu Y., Yang J., Xi S.-S., Wen X., Gu Q., Hu W. Hypothermia protects neurons against ischemia/reperfusion-induced pyroptosis via m6A-mediated activation of PTEN and the PI3K/Akt/GSK-3β signaling pathway. Brain Res. Bull. 2020;159:25–31. doi: 10.1016/j.brainresbull.2020.03.011. [DOI] [PubMed] [Google Scholar]
- 162.Si W., Li Y., Ye S., Li Z., Liu Y., Kuang W., Chen D., Zhu M. Methyltransferase 3 Mediated miRNA m6A Methylation Promotes Stress Granule Formation in the Early Stage of Acute Ischemic Stroke. Front. Mol. Neurosci. 2020;13:103. doi: 10.3389/fnmol.2020.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yu J., Zhang Y., Ma H., Zeng R., Liu R., Wang P., Jin X., Zhao Y. Epitranscriptomic profiling of N6-methyladenosine-related RNA methylation in rat cerebral cortex following traumatic brain injury. Mol. Brain. 2020;13:11. doi: 10.1186/s13041-020-0554-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang Y., Mao J., Wang X., Lin Y., Hou G., Zhu J., Xie B. Genome-wide screening of altered m6A-tagged transcript profiles in the hippocampus after traumatic brain injury in mice. Epigenomics. 2019;11:805–819. doi: 10.2217/epi-2019-0002. [DOI] [PubMed] [Google Scholar]
- 165.Wang Q., Liang Y., Luo X., Liu Y., Zhang X., Gao L. N6-methyladenosine RNA modification: A promising regulator in central nervous system injury. Exp. Neurol. 2021;345:113829. doi: 10.1016/j.expneurol.2021.113829. [DOI] [PubMed] [Google Scholar]
- 166.He P.C., He C. m6A RNA methylation: From mechanisms to therapeutic potential. EMBO J. 2021;40:e105977. doi: 10.15252/embj.2020105977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sommer F., Backhed F. Know your neighbor: Microbiota and host epithelial cells interact locally to control intestinal function and physiology. BioEssays. 2016;38:455–464. doi: 10.1002/bies.201500151. [DOI] [PubMed] [Google Scholar]
- 168.Wu J., Wang K., Wang X., Pang Y., Jiang C. The role of the gut microbiome and its metabolites in metabolic diseases. Protein Cell. 2021;12:360–373. doi: 10.1007/s13238-020-00814-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 170.Agus A., Planchais J., Sokol H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 171.Heaver S.L., Johnson E.L., Ley R.E. Sphingolipids in host–microbial interactions. Curr. Opin. Microbiol. 2018;43:92–99. doi: 10.1016/j.mib.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 172.Levy M., Thaiss C.A., Zeevi D., Dohnalová L., Zilberman-Schapira G., Mahdi J.A., David E., Savidor A., Korem T., Herzig Y., et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell. 2015;163:1428–1443. doi: 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang X., Li Y., Chen W., Shi H., Eren A.M., Morozov A., He C., Luo G.-Z., Pan T. Transcriptome-wide reprogramming of N6-methyladenosine modification by the mouse microbiome. Cell Res. 2019;29:167–170. doi: 10.1038/s41422-018-0127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Luo J., Yu J., Peng X. Could partial nonstarch polysaccharides ameliorate cancer by altering m6A RNA methylation in hosts through intestinal microbiota? Crit. Rev. Food Sci. Nutr. 2021:1–16. doi: 10.1080/10408398.2021.1927975. [DOI] [PubMed] [Google Scholar]
- 175.Tong J., Cao G., Zhang T., Sefik E., Vesely M.C.A., Broughton J.P., Zhu S., Li H.-B., Li B., Chen L., et al. m6A mRNA methylation sustains Treg suppressive functions. Cell Res. 2018;28:253–256. doi: 10.1038/cr.2018.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Liu B., Liu N., Zhu X., Yang L., Ye B., Li H., Zhu P., Lu T., Tian Y., Fan Z. Circular RNA circZbtb20 maintains ILC3 homeostasis and function via Alkbh5-dependent m6A demethylation of Nr4a1 mRNA. Cell. Mol. Immunol. 2021;18:1412–1424. doi: 10.1038/s41423-021-00680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zong X., Xiao X., Shen B., Jiang Q., Wang H., Lu Z., Wang F., Jin M., Min J., Wang F., et al. The N 6-methyladenosine RNA-binding protein YTHDF1 modulates the translation of TRAF6 to mediate the intestinal immune response. Nucleic Acids Res. 2021;49:5537–5552. doi: 10.1093/nar/gkab343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Olazagoitia-Garmendia A., Zhang L., Mera P., Godbout J.K., Sebastian-DelaCruz M., Garcia-Santisteban I., Mendoza L.M., Huerta A., Irastorza I., Bhagat G., et al. Gluten-induced RNA methylation changes regulate intestinal inflammation via allele-specific XPO1 translation in epithelial cells. Gut. 2022;71:68–76. doi: 10.1136/gutjnl-2020-322566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tanabe A., Tanikawa K., Tsunetomi M., Takai K., Ikeda H., Konno J., Torigoe T., Maeda H., Kutomi G., Okita K., et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett. 2016;376:34–42. doi: 10.1016/j.canlet.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 180.Ismail H., Shakkour Z., Tabet M., Abdelhady S., Kobaisi A., Abedi R., Nasrallah L., Pintus G., Al-Dhaheri Y., Mondello S., et al. Traumatic Brain Injury: Oxidative Stress and Novel Anti-Oxidants Such as Mitoquinone and Edaravone. Antioxidants. 2020;9:943. doi: 10.3390/antiox9100943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Barkhoudarian G., Hovda D.A., Giza C.C. The Molecular Pathophysiology of Concussive Brain Injury—An Update. Phys. Med. Rehabil. Clin. N. Am. 2016;27:373–393. doi: 10.1016/j.pmr.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 182.Tarudji A.W., Gee C.C., Romereim S.M., Convertine A.J., Kievit F.M. Antioxidant thioether core-crosslinked nanoparticles prevent the bilateral spread of secondary injury to protect spatial learning and memory in a controlled cortical impact mouse model of traumatic brain injury. Biomaterials. 2021;272:120766. doi: 10.1016/j.biomaterials.2021.120766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rizwana N., Agarwal V., Nune M. Antioxidant for Neurological Diseases and Neurotrauma and Bioengineering Approaches. Antioxidants. 2021;11:72. doi: 10.3390/antiox11010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Khatri N., Thakur M., Pareek V., Kumar S., Sharma S., Datusalia A.K. Oxidative Stress: Major Threat in Traumatic Brain Injury. CNS Neurol. Disord. Drug Targets. 2018;17:689–695. doi: 10.2174/1871527317666180627120501. [DOI] [PubMed] [Google Scholar]
- 185.Ding H., Li Z., Li X., Yang X., Zhao J., Guo J., Lu W., Liu H., Wang J. FTO Alleviates CdCl2-Induced Apoptosis and Oxidative Stress via the AKT/Nrf2 Pathway in Bovine Granulosa Cells. Int. J. Mol. Sci. 2022;23:4948. doi: 10.3390/ijms23094948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chen P.-B., Shi G.-X., Liu T., Li B., Jiang S.-D., Zheng X.-F., Jiang L.-S. Oxidative Stress Aggravates Apoptosis of Nucleus Pulposus Cells through m6A Modification of MAT2A Pre-mRNA by METTL16. Oxidative Med. Cell. Longev. 2022;2022:4036274. doi: 10.1155/2022/4036274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Xia C., Wang J., Wu Z., Miao Y., Chen C., Li R., Li J., Xing H. METTL3-mediated m6A methylation modification is involved in colistin-induced nephrotoxicity through apoptosis mediated by Keap1/Nrf2 signaling pathway. Toxicology. 2021;462:152961. doi: 10.1016/j.tox.2021.152961. [DOI] [PubMed] [Google Scholar]
- 188.Sun R., Tian X., Li Y., Zhao Y., Wang Z., Hu Y., Zhang L., Wang Y., Gao D., Zheng S., et al. The m6A reader YTHDF3-mediated PRDX3 translation alleviates liver fibrosis. Redox Biol. 2022;54:102378. doi: 10.1016/j.redox.2022.102378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Anders M., Chelysheva I., Goebel I., Trenkner T., Zhou J., Mao Y., Verzini S., Qian S.-B., Ignatova Z. Dynamic m6A methylation facilitates mRNA triaging to stress granules. Life Sci. Alliance. 2018;1:e201800113. doi: 10.26508/lsa.201800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fu Y., Zhuang X. m6A-binding YTHDF proteins promote stress granule formation. Nat. Chem. Biol. 2020;16:955–963. doi: 10.1038/s41589-020-0524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wang J., Ishfaq M., Xu L., Xia C., Chen C., Li J. METTL3/m6A/miRNA-873-5p Attenuated Oxidative Stress and Apoptosis in Colistin-Induced Kidney Injury by Modulating Keap1/Nrf2 Pathway. Front. Pharmacol. 2019;10:517. doi: 10.3389/fphar.2019.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhao M.X.J., Zhu M.H.-Y., Wang X.-L., Lu X.-W., Pan M.C.-L., Xu L., Liu M.X., Xu N., Zhang Z.-Y. Oridonin Ameliorates Traumatic Brain Injury-Induced Neurological Damage by Improving Mitochondrial Function and Antioxidant Capacity and Suppressing Neuroinflammation through the Nrf2 Pathway. J. Neurotrauma. 2022;39:530–543. doi: 10.1089/neu.2021.0466. [DOI] [PubMed] [Google Scholar]
- 193.Rui Q., Ni H., Lin X., Zhu X., Li D., Liu H., Chen G. Astrocyte-derived fatty acid-binding protein 7 protects blood-brain barrier integrity through a caveolin-1/MMP signaling pathway following traumatic brain injury. Exp. Neurol. 2019;322:113044. doi: 10.1016/j.expneurol.2019.113044. [DOI] [PubMed] [Google Scholar]
- 194.Hausburg M.A., Banton K.L., Roman P.E., Salgado F., Baek P., Waxman M.J., Tanner A., Yoder J., Bar-Or D. Effects of propofol on ischemia-reperfusion and traumatic brain injury. J. Crit. Care. 2020;56:281–287. doi: 10.1016/j.jcrc.2019.12.021. [DOI] [PubMed] [Google Scholar]
- 195.Xu M., Li L., Liu H., Lu W., Ling X., Gong M. Rutaecarpine Attenuates Oxidative Stress-Induced Traumatic Brain Injury and Reduces Secondary Injury via the PGK1/KEAP1/NRF2 Signaling Pathway. Front. Pharmacol. 2022;13:807125. doi: 10.3389/fphar.2022.807125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Tan S.W., Zhao Y., Li P., Ning Y.-L., Huang Z.-Z., Yang N., Liu D., Zhou Y.-G. HMGB1 mediates cognitive impairment caused by the NLRP3 inflammasome in the late stage of traumatic brain injury. J. Neuroinflamm. 2021;18:241. doi: 10.1186/s12974-021-02274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cui C., Wang C., Jin F., Yang M., Kong L., Han W., Jiang P. Calcitriol confers neuroprotective effects in traumatic brain injury by activating Nrf2 signaling through an autophagy-mediated mechanism. Mol. Med. 2021;27:118. doi: 10.1186/s10020-021-00377-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yi W., Cheng J., Wei Q., Pan R., Song S., He Y., Tang C., Liu X., Zhou Y., Su H. Effect of temperature stress on gut-brain axis in mice: Regulation of intestinal microbiome and central NLRP3 inflammasomes. Sci. Total Environ. 2021;772:144568. doi: 10.1016/j.scitotenv.2020.144568. [DOI] [PubMed] [Google Scholar]
- 199.Park S.H., Kim J., Moon Y. Caveolar communication with xenobiotic-stalled ribosomes compromises gut barrier integrity. Commun. Biol. 2020;3:270. doi: 10.1038/s42003-020-0994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Piotrowska M., Swierczynski M., Fichna J., Piechota-Polanczyk A. The Nrf2 in the pathophysiology of the intestine: Molecular mechanisms and therapeutic implications for inflammatory bowel diseases. Pharmacol. Res. 2021;163:105243. doi: 10.1016/j.phrs.2020.105243. [DOI] [PubMed] [Google Scholar]
- 201.Wang H., Yang F., Xin R., Cui D., He J., Zhang S., Sun Y. The gut microbiota attenuate neuroinflammation in manganese exposure by inhibiting cerebral NLRP3 inflammasome. Biomed. Pharmacother. 2020;129:110449. doi: 10.1016/j.biopha.2020.110449. [DOI] [PubMed] [Google Scholar]
- 202.Sadovnikova I., Gureev A., Ignatyeva D., Gryaznova M., Chernyshova E., Krutskikh E., Novikova A., Popov V. Nrf2/ARE Activators Improve Memory in Aged Mice via Maintaining of Mitochondrial Quality Control of Brain and the Modulation of Gut Microbiome. Pharmaceuticals. 2021;14:607. doi: 10.3390/ph14070607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Meng L., Lin H., Huang X., Weng J., Peng F., Wu S. METTL14 suppresses pyroptosis and diabetic cardiomyopathy by downregulating TINCR lncRNA. Cell Death Dis. 2022;13:38. doi: 10.1038/s41419-021-04484-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhou Y., Wang Q., Deng H., Xu B., Zhou Y., Liu J., Liu Y., Shi Y., Zheng X., Jiang J. N6-methyladenosine demethylase FTO promotes growth and metastasis of gastric cancer via m6A modification of caveolin-1 and metabolic regulation of mitochondrial dynamics. Cell Death Dis. 2022;13:72. doi: 10.1038/s41419-022-04503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Shi J., Guo J., Li Z., Xu B., Miyata M. Importance of NLRP3 Inflammasome in Abdominal Aortic Aneurysms. J. Atheroscler. Thromb. 2021;28:454–466. doi: 10.5551/jat.RV17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.O’Brien W.T., Pham L., Symons G.F., Monif M., Shultz S.R., McDonald S.J. The NLRP3 inflammasome in traumatic brain injury: Potential as a biomarker and therapeutic target. J. Neuroinflamm. 2020;17:104. doi: 10.1186/s12974-020-01778-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Irrera N., Russo M., Pallio G., Bitto A., Mannino F., Minutoli L., Altavilla D., Squadrito F. The Role of NLRP3 Inflammasome in the Pathogenesis of Traumatic Brain Injury. Int. J. Mol. Sci. 2020;21:6204. doi: 10.3390/ijms21176204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Liu H.D., Li W., Chen Z.-R., Hu Y.-C., Zhang D.-D., Shen W., Zhou M.-L., Zhu L., Hang C.-H. Expression of the NLRP3 Inflammasome in Cerebral Cortex After Traumatic Brain Injury in a Rat Model. Neurochem. Res. 2013;38:2072–2083. doi: 10.1007/s11064-013-1115-z. [DOI] [PubMed] [Google Scholar]
- 209.Ismael S., Nasoohi S., Ishrat T. MCC950, the Selective Inhibitor of Nucleotide Oligomerization Domain-Like Receptor Protein-3 Inflammasome, Protects Mice against Traumatic Brain Injury. J. Neurotrauma. 2018;35:1294–1303. doi: 10.1089/neu.2017.5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Xu X., Yin D., Ren H., Gao W., Li F., Sun D., Wu Y., Zhou S., Lyu L., Yang M., et al. Selective NLRP3 inflammasome inhibitor reduces neuroinflammation and improves long-term neurological outcomes in a murine model of traumatic brain injury. Neurobiol. Dis. 2018;117:15–27. doi: 10.1016/j.nbd.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 211.Lin C., Chao H., Li Z., Xu X., Liu Y., Bao Z., Hou L., Liu Y., Wang X., You Y., et al. Omega-3 fatty acids regulate NLRP3 inflammasome activation and prevent behavior deficits after traumatic brain injury. Exp. Neurol. 2017;290:115–122. doi: 10.1016/j.expneurol.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 212.Wallisch J., Simon D.W., Bayır H., Bell M.J., Kochanek P.M., Clark R.S.B. Cerebrospinal Fluid NLRP3 is Increased After Severe Traumatic Brain Injury in Infants and Children. Neurocrit. Care. 2017;27:44–50. doi: 10.1007/s12028-017-0378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Pellegrini C., Antonioli L., Calderone V., Colucci R., Fornai M., Blandizzi C. Microbiota-gut-brain axis in health and disease: Is NLRP3 inflammasome at the crossroads of microbiota-gut-brain communications? Prog. Neurobiol. 2020;191:101806. doi: 10.1016/j.pneurobio.2020.101806. [DOI] [PubMed] [Google Scholar]
- 214.Nag S., Venugopalan R., Stewart D.J. Increased caveolin-1 expression precedes decreased expression of occludin and claudin-5 during blood–brain barrier breakdown. Acta Neuropathol. 2007;114:459–469. doi: 10.1007/s00401-007-0274-x. [DOI] [PubMed] [Google Scholar]
- 215.Nag S., Manias J.L., Stewart D.J. Expression of endothelial phosphorylated caveolin-1 is increased in brain injury. Neuropathol. Appl. Neurobiol. 2009;35:417–426. doi: 10.1111/j.1365-2990.2008.01009.x. [DOI] [PubMed] [Google Scholar]
- 216.Gu Y., Zheng G.-Q., Xu M., Li Y., Chen X., Zhu W., Tong Y., Chung S.K., Liu K.J., Shen J. Caveolin-1 regulates nitric oxide-mediated matrix metalloproteinases activity and blood-brain barrier permeability in focal cerebral ischemia and reperfusion injury. J. Neurochem. 2012;120:147–156. doi: 10.1111/j.1471-4159.2011.07542.x. [DOI] [PubMed] [Google Scholar]
- 217.Gu Y., Dee C.M., Shen J. Interaction of free radicals, matrix metalloproteinases and caveolin-1 impacts blood-brain barrier permeability. Front. Biosci. 2011;3:1216–1231. doi: 10.2741/222. [DOI] [PubMed] [Google Scholar]
- 218.Blochet C., Buscemi L., Clément T., Gehri S., Badaut J., Hirt L. Involvement of caveolin-1 in neurovascular unit remodeling after stroke: Effects on neovascularization and astrogliosis. J. Cereb. Blood Flow Metab. 2020;40:163–176. doi: 10.1177/0271678X18806893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Klein T., Bischoff R. Physiology and pathophysiology of matrix metalloproteases. Amino Acids. 2011;41:271–290. doi: 10.1007/s00726-010-0689-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Qi Z., Liang J., Pan R., Dong W., Shen J., Yang Y., Zhao Y., Shi W., Luo Y., Ji X., et al. Zinc contributes to acute cerebral ischemia-induced blood–brain barrier disruption. Neurobiol. Dis. 2016;95:12–21. doi: 10.1016/j.nbd.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 221.Calabrese V., Cornelius C., Rizzarelli E., Owen J.B., Dinkova-Kostova A.T., Butterfield D.A. Nitric Oxide in Cell Survival: A Janus Molecule. Antioxid. Redox Signal. 2009;11:2717–2739. doi: 10.1089/ars.2009.2721. [DOI] [PubMed] [Google Scholar]
- 222.Calabrese V., Guagliano E., Sapienza M., Panebianco M., Calafato S., Puleo E., Pennisi G., Mancuso C., Butterfield D.A., Stella A.G. Redox Regulation of Cellular Stress Response in Aging and Neurodegenerative Disorders: Role of Vitagenes. Neurochem. Res. 2007;32:757–773. doi: 10.1007/s11064-006-9203-y. [DOI] [PubMed] [Google Scholar]
- 223.Mattson M.P. Activation of NF-kappaB protects hippocampal neurons against oxidative stress-induced apoptosis: Evidence for induction of manganese superoxide dismutase and suppression of peroxynitrite production and protein tyrosine nitration. J. Neurosci. Res. 1997;49:681–697. doi: 10.1002/(SICI)1097-4547(19970915)49:6<681::AID-JNR3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 224.Calabrese V., Cornelius C., Dinkova-Kostova A., Calabrese E.J., Mattson M.P. Cellular Stress Responses, The Hormesis Paradigm, and Vitagenes: Novel Targets for Therapeutic Intervention in Neurodegenerative Disorders. Antioxid. Redox Signal. 2010;13:1763–1811. doi: 10.1089/ars.2009.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Mancuso C., Pani G., Calabrese V. Bilirubin: An endogenous scavenger of nitric oxide and reactive nitrogen species. Redox Rep. 2006;11:207–213. doi: 10.1179/135100006X154978. [DOI] [PubMed] [Google Scholar]
- 226.Woronyczova J., Nováková M., Leníček M., Bátovský M., Bolek E., Cífková R., Vítek L. Serum Bilirubin Concentrations and the Prevalence of Gilbert Syndrome in Elite Athletes. Sports Med. Open. 2022;8:84. doi: 10.1186/s40798-022-00463-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Mancuso C., Capone C., Ranieri S.C., Fusco S., Calabrese V., Eboli M.L., Preziosi P., Galeotti T., Pani G. Bilirubin as an endogenous modulator of neurotrophin redox signaling. J. Neurosci. Res. 2008;86:2235–2249. doi: 10.1002/jnr.21665. [DOI] [PubMed] [Google Scholar]
- 228.Kraft A.D., Johnson D.A., Johnson J.A. Nuclear Factor E2-Related Factor 2-Dependent Antioxidant Response Element Activation by tert-Butylhydroquinone and Sulforaphane Occurring Preferentially in Astrocytes Conditions Neurons against Oxidative Insult. J. Neurosci. 2004;24:1101–1112. doi: 10.1523/JNEUROSCI.3817-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Calkins M.J., Jakel R.J., Johnson D.A., Chan K., Kan Y.W., Johnson J.A. Protection from mitochondrial complex II inhibition in vitro and in vivo by Nrf2-mediated transcription. Proc. Natl. Acad. Sci. USA. 2005;102:244–249. doi: 10.1073/pnas.0408487101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wu A.G., Yong Y.-Y., Pan Y.-R., Zhang L., Wu J.-M., Zhang Y., Tang Y., Wei J., Yu L., Law B.Y.-K., et al. Targeting Nrf2-Mediated Oxidative Stress Response in Traumatic Brain Injury: Therapeutic Perspectives of Phytochemicals. Oxidative Med. Cell. Longev. 2022;2022:1015791. doi: 10.1155/2022/1015791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Cordaro M., Salinaro A.T., Siracusa R., D’Amico R., Impellizzeri D., Scuto M., Ontario M., Crea R., Cuzzocrea S., Di Paola R., et al. Hidrox® Roles in Neuroprotection: Biochemical Links between Traumatic Brain Injury and Alzheimer’s Disease. Antioxidants. 2021;10:818. doi: 10.3390/antiox10050818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Arumugam T., Ghazi T., Chuturgoon A.A. Fumonisin B1 alters global m6A RNA methylation and epigenetically regulates Keap1-Nrf2 signaling in human hepatoma (HepG2) cells. Arch. Toxicol. 2021;95:1367–1378. doi: 10.1007/s00204-021-02986-5. [DOI] [PubMed] [Google Scholar]
- 233.Zhao T.X., Wang J.-K., Shen L.-J., Long C.-L., Liu B., Wei Y., Han L.-D., Wei Y.-X., Wu S.-D., Wei G.-H. Increased m6A RNA modification is related to the inhibition of the Nrf2-mediated antioxidant response in di-(2-ethylhexyl) phthalate-induced prepubertal testicular injury. Environ. Pollut. 2020;259:113911. doi: 10.1016/j.envpol.2020.113911. [DOI] [PubMed] [Google Scholar]
- 234.Wu J., Gan Z., Zhuo R., Zhang L., Wang T., Zhong X. Resveratrol Attenuates Aflatoxin B1-Induced ROS Formation and Increase of m6A RNA Methylation. Animals. 2020;10:677. doi: 10.3390/ani10040677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Yu J., Li Y., Wang T., Zhong X. Modification of N6-methyladenosine RNA methylation on heat shock protein expression. PLoS ONE. 2018;13:e0198604. doi: 10.1371/journal.pone.0198604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Wu J., Li Y., Yu J., Gan Z., Wei W., Wang C., Zhang L., Wang T., Zhong X. Resveratrol Attenuates High-Fat Diet Induced Hepatic Lipid Homeostasis Disorder and Decreases m6A RNA Methylation. Front. Pharmacol. 2020;11:568006. doi: 10.3389/fphar.2020.568006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Gan Z., Wei W., Wu J., Zhao Y., Zhang L., Wang T., Zhong X. Resveratrol and Curcumin Improve Intestinal Mucosal Integrity and Decrease m6A RNA Methylation in the Intestine of Weaning Piglets. ACS Omega. 2019;4:17438–17446. doi: 10.1021/acsomega.9b02236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lu N., Li X., Yu J., Li Y., Wang C., Zhang L., Wang T., Zhong X. Curcumin Attenuates Lipopolysaccharide-Induced Hepatic Lipid Metabolism Disorder by Modification of m6A RNA Methylation in Piglets. Lipids. 2018;53:53–63. doi: 10.1002/lipd.12023. [DOI] [PubMed] [Google Scholar]