Abstract
Simple Summary
A growing number of studies have focused their attention on the potential role of microRNAs (miRNA) as biomarkers for several diseases. However, very few evaluated the role of miRNAs in the aetiogenesis of frailty, a multidimensional geriatric syndrome, characterized by an individual and dynamic state of impairments in one or more domains, such as physical, cognitive, psychological, and social. In this review, we first provided an overview on the different frailty domains, current assessment tools and plasma/blood biomarkers. Then, we collected the evidence linking changes of miRNAs expression to impairment of frailty in physical and cognitive domains, with the ultimate aim of finding those that are common. In silico analyses prioritized ten top-ranked miRNAs and their targets, the three most significant regulating processes involved in inflammation and energy homeostasis pathway. Such miRNAs, through the integration with existing markers, may be useful for an early and accurate diagnosis of frailty in the elderly population.
Abstract
The past years have seen an increasing concern about frailty, owing to the growing number of elderly people and the major impact of this syndrome on health and social care. The identification of frail people passes through the use of different tests and biomarkers, whose concerted analysis helps to stratify the populations of patients according to their risk profile. However, their efficiency in prognosis and their capability to reflect the multisystemic impairment of frailty is discussed. Recent works propose the use of miRNAs as biological hallmarks of physiological impairment in different organismal districts. Changes in miRNAs expression have been described in biological processes associated with phenotypic outcomes of frailty, opening intriguing possibilities for their use as biomarkers of fragility. Here, with the aim of finding reliable biomarkers of frailty, while considering its complex nature, we revised the current literature on the field, for uncovering miRNAs shared across physical and cognitive frailty domains. By applying in silico analyses, we retrieved the top-ranked shared miRNAs and their targets, finally prioritizing the most significant ones. From this analysis, ten miRNAs emerged which converge into two main biological processes: inflammation and energy homeostasis. Such markers, if validated, may offer promising capabilities for early diagnosis of frailty in the elderly population.
Keywords: miRNA, frailty, physical domain, cognitive domain, biomarkers, multisystemic impairment
1. Introduction
1.1. The Frailty Domains
Frailty is a common clinical syndrome in older adults characterized by a multisystem impairment which ranges from musculoskeletal, to pulmonary, cardiovascular and neurological systems and by a marked vulnerability to adverse health outcomes, including an increased risk of disability, admission to long-term care and increased mortality [1,2]. Aging, which causes a progressive decrease in the physiological reserves and an overall loss of homeostasis, is the main risk factor for frailty. The pace of this progressive decay is accelerated by genetic factors, epigenetic events and environmental stressors [3].
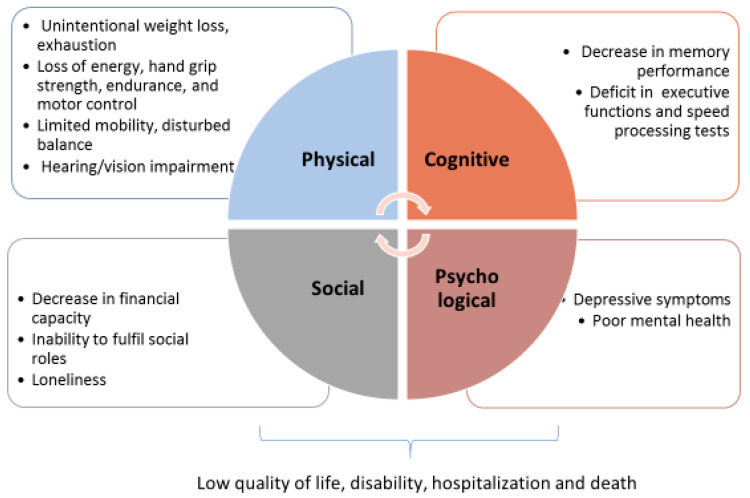
Literature still lacks a consensus for a comprehensive definition of frailty, although the classification of frailty which most of all embraces its complex nature is that of a multi-domain phenotype, firstly proposed by Abellan Van Kan et al. [4] and further developed by many authors [5,6,7,8,9]. As outlined in Figure 1, the multi-domain model includes physical, cognitive, psychological and social domains of frailty, while Box 1 describes the criteria generically used for the classification of physical, cognitive and psychological/social declines, applied by many frailty tools such as that of Fried et al. [10], Rockwood and Mitnitski [11], Gobbens et al. [12], Peters et al. [13], Morley et al. [14] and Wieland and Hirth [15]. The tools comprehend tests which can be used as endophenotypes (defined as quantitative biological traits which reflect the function of a discrete biological system), closely related to the specifical domain respect to the broad phenotype [16,17,18].
Figure 1.
The multi-domain model of frailty.
Box 1. Criteria used for the classification of impairments in the physical, cognitive and psychological/social domains of frailty, applied by many frailty tools as described in the text.
The physical domain
Physical frailty was reported by Maxwell and Wang [8] as “characterized by gradual loss of energy, strength, endurance, and motor control”. It is defined basically considering ≥4 of 8 criteria, reflecting the screening of muscle health and functional status: unintentional weight loss, exhaustion, strength, perceived health, walking, balance, hearing and vision impairments [19]. A series of socio-demographic, lifestyle, and health-related factors have been shown to be associated with physical frailty, such as age, female sex, cardiovascular diseases, multimorbidity, BMI, and smoking.
The cognitive domain
Cognitive impairment in absence of dementia is considered a relevant domain of frailty. Cognitive impairment is defined as <10th percentile on global cognitive functioning, detected with cognitive tests, such as the MMSE (Mini Mental State Examination) score [20] and the cognitive abilities screening instrument (CASI) [21].
The psychological and social domains
Psychological frailty is defined on the base of two criteria such as depressive symptoms and mental health, and is measured by Geriatric Depression Scale and Mental Health Inventory 5 (MHI-5) [22]. A higher psychological frailty risk is associated with the female sex, low educational level, smoking, a short sleep duration and multi-morbidity, while being married, a long sleep duration and being physically active are normally associated with a lower risk of being psychologically frail.
Social frailty is measured as ≥2 of 3 criteria among loneliness, social support and social participation.
Common to all the frailty assessment tools is the physical domain, while the cognitive domain is included in only 50% of them. This because in the past there was still some uncertainty about the relationship between frailty, cognitive impairment and dementia, so that some authors exclude people severely cognitively compromised in the analyzed cohorts. Recently, cognitive impairment was recognized as a significant determinant of frailty and consequently a novel target for the prevention [23].
Growing evidences suggest that the different frailty domains are not independent entities, but share subcellular pathophysiological mechanisms, not only increasing the vulnerability and frailty prevalence [24] but also forming a substrate for the development of chronic age-related diseases, such as Alzheimer diseases (AD), where physical frailty was proposed as a non-cognitive manifestation that precedes the onset of dementia [25]. In addition, social and physical frailty were considered as a risk factor for the development of the other, considering that a decline in social roles precedes the activity of daily living disabilities among community-dwelling independent older adults, with an impact on the mortality risk [26,27].
Although at a population level the overlap among impairments in different frailty domains can be limited to a small proportion of the oldest population, with around 17% of people found frail at two or more domains [19], evidences were reported that multi-domain interventions are able to improve health status in elderly people at-risk from the general population [28]. The co-occurrence of physical, cognitive and emotional decline was named triad of impairment (TOI) and it was used in the past as a surrogate marker of frailty in some studies, such as the Cardiovascular Health study [10] and Aberdeen Birth Cohort study [29].
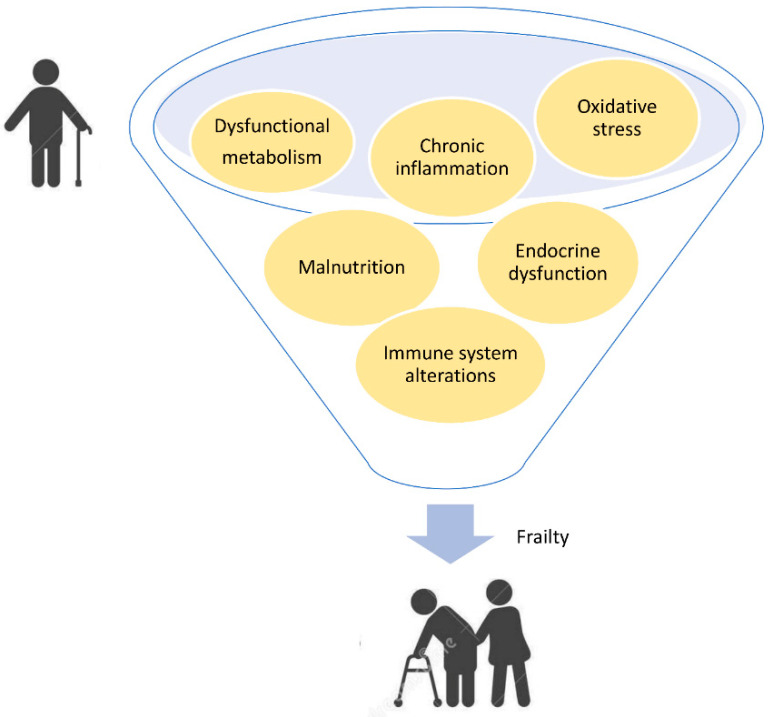
The overlap among different phenotypic domains of frailty account for pathogenic pathways shared among multiple districts, which provide a background prone to dysfunction, driving the accumulation of deficits which concur with frailty development. By filtering the extensive literature [30,31,32] (and references therein), seven most-cited and replicated biological mechanisms dysregulated in frailty can be retrieved, which are common to different domains, as schematized in Figure 2.
Figure 2.
Impaired biological mechanisms shared among different frailty domains.
Such deficiencies, such as chronic inflammation, immune and endocrine impairment, dysregulated metabolism, oxidative stress and malnutrition, are common and interconnected denominators observed in both physical and cognitive decline status, occurring in a very early step of frailty onset and contributing to its progression.
1.2. Current Biomarkers of Frailty
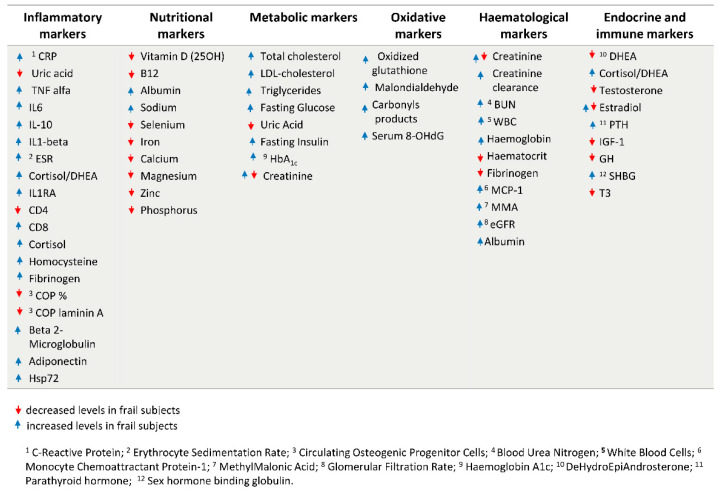
The identification of the above biological hallmarks has driven the research on the identification of circulating biomarkers whose screening, in association with a complete geriatric assessment, including cognitive and physical tests, can be helpful for a primary frailty diagnosis of the patient. Figure 3 [33,34,35,36,37] resumes the most common identified blood and biochemical markers of frailty covering different sets of physiological parameters, which includes inflammatory markers [38] oxidative markers [39,40], nutritional and metabolic markers, hematological markers, endocrine and immune markers [36].
Figure 3.
Circulating biomarkers of frailty divided in six sets of physiological parameters. The red and blue arrows indicate if increased or decreased levels were found in frail subjects, according to literature evidence, as reported in the text.
However, while for some of them the association with frailty is clear, in other cases their efficiency as prognostic markers of frailty is still under discussion [41,42]. This is the case for creatinine, whose levels can reflect both kidney insufficiency and sarcopenia: thus, by using creatinine as a biomarker of frailty alone, a misidentification of frail subjects may occur [43].
Some biomarkers, such as endocrine ones, are age- and sex-specific, with different and sometimes opposite trends of association observed in males and females. For example, some studies reported that in men aged 65–90 years higher estradiol levels were associated with a decreased frailty [44], while in females higher estradiol levels were associated with increased frailty up to the age of 79, but not thereafter (Carcaillon et al. [45] for the Spanish Toledo Study for Healthy Aging). Moreover, an interaction between hormones and inflammation was found in women with CRP (C-Reactive Protein) levels. Thus, even though it is difficult to compare different studies, due to the application of different frailty measures, these evidences suggest that the intertwining among different physiological systems can drive the associations with frailty phenotype, further confirming the importance of considering the whole set of markers in an integrated approach [46,47].
2. Search for Novel Biomarkers of Frailty: The Role of miRNAs
Novel biomarkers of frailty can be reasonably chosen among factors acting in pathophysiological mechanism common to the physical and cognitive domains, and should be highly sensitive to minimum changes in physiological conditions. Such markers should be integrated with those previously discussed, to complete the picture of changes in the physiological status consequent to the occurrence of frailty. In this context, the evaluation of circulating microRNAs (miRNAs) was proposed as non-invasive diagnostic biomarkers of frailty, potentially regulating very large sets of genes (even hundreds of putative gene targets are known) and targeting different pathways at the same time [48].
MiRNAs are small non-coding single-stranded RNAs approximately 21–25 nucleotides long, regulating gene expression by binding to complementary messenger RNAs (mRNAs) and preventing the production of the corresponding protein [49]. MiRNA biogenesis is a step-wise process which starts with pri-miRNAs, long primary transcripts produced by RNA polymerase II and then processed into a pre-miRNA (~70- to 120-nucleotide-long) by a multiprotein complex containing the nuclear RNase III enzyme, known as Drosha. Then, this pre-miRNA is exported into the cytoplasm, to be processed into a mature duplex (~18- to 23-nucleotide-long) by the RNase III Dicer-1. A ribonuclear-protein complex, called RNA-induced silencing complex (RISC), composed by the guide strand of the duplex along with Argonaute proteins, directs to target mRNA and through sequence complementarity causes its translational repression [50]. A “seed region” (nucleotides 2 to 8) at the 5′ end of the mature miRNA can mediate the recognition of the mRNAs target site [51], usually by binding the target sequences at the 3′ UTR but sometimes also in 5′ UTR and open reading frame [52]. Each miRNA targets hundreds of transcripts, regulating fundamental cell processes such as proliferation, apoptosis, differentiation, migration, metabolism and stress response [53]. In addition to intracellular miRNAs, circulating extracellular miRNAs have been detected in different biofluids including blood, plasma, serum, saliva, urine and pleural effusions [54]. They can circulate as free molecules or be bound to carriers such as low-density lipoproteins (LDL), high-density lipoproteins (HDL), ribonucleoproteins and extracellular vesicles (EVs) [54]. In relation to different pathophysiological conditions, biofluids can have specific circulating miRNAs [55].
Several authors analyzed miRNA levels in pathological conditions [56,57,58] and in relation to aging [59,60,61,62,63], further supporting the use of miRNA panels as potential diagnostic biomarkers. A critical role of these small molecules has been documented relative to the frailty-associated phenotypes, i.e., in muscle development and homeostasis [64], but also in neuronal processes such as brain development, synaptic plasticity, and learning and memory functions [65]. A study of network biology, by analyzing the interactome of frailty-related genes, prioritized 10 miRNA markers indirectly associated with frailty through the association of their targets [66]. To date, only three studies directly evaluated changes in blood plasma miRNAs in frail/non frail subjects [67,68,69]. Rusanova measured levels of the inflammation-related miRNAs, miR-21, miR-146a, miR-223, and those of miR-483, associated with the control of melatonin synthesis, reporting an association between the expression of miR-21 with the presence of frailty [67]. Ipson and co-workers, instead, examined the levels of plasma-derived exosome miRNA and identified eight miRNAs significantly enriched in frailty subjects: miR-10a-3p, miR-92a-3p, miR-185-3p, miR-194-5p, miR-326, miR-532-5p, miR-576-5p, and miR-760 [68]. Interestingly, many of these markers modulate on/off switching of crucial cellular mechanism involved in frailty, such as miR-194-5p, which is both associated with cellular senescence and ROS production [70] and was reported to regulate muscle cell homeostasis [71]. Very recently, Carini and co-workers [69] carried out a larger study by profiling a total of 41 frail/non frail subjects for a miRNA set of 2654 markers. They found two miRNAs downregulated in the frailty group, namely miR-101-3p and miR-142-5p, both previously associated with oxidative stress-induced apoptosis and immune-inflammatory responses. As a whole, these researches indicate miRNAs mechanistically involved in the aetiogenesis of frailty, pinpointing the main pathways (as inflammation or ROS production) in common among different frailty domains, thus making them good candidates as frailty biomarkers.
In the following sections we reviewed the knowledge on miRNAs associated with physical and cognitive decline. In particular, we considered the literature that found associations between miRNAs and phenotypes related to physical frailty, such as muscle loss, sarcopenia, cachexia, or participating in the proliferation and differentiation of myogenic progenitors and myotubes, applying keywords such as “sarcopenia”, “physical impairment at old age”, “physical frailty”, “muscle loss” and “skeletal muscle decline”. In the case of the cognitive domain, we used keywords such as “cognitive decline”, “cognitive impairment” and “neurodegeneration”. Furthermore, for the retrieval of the list of relevant miRNAs, we excluded studies carried out in model organisms, focusing on human studies (cells, tissues or individuals) but excluding those related to severe pathological age-related conditions. In the case of cognitive domain of frailty, because our interest was to identify early markers of neurodegeneration, we considered early stages of Mild Cognitive Impairment (MCI).
3. MiRNAs as Biomarkers of the Physical Domain of Frailty
By reviewing the literature on phenotypes related to physical frailty, we identified a total of 57 miRNAs associated with physical phenotypes, resumed in Table S1 [67,68,69,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111]. An inspection of their characteristics revealed that they belong to three main groups: muscle-related miRNAs, inflamm-miRNAs and mitochondrial miRNAs (mitomiRNA).
Among muscle-related miRNAs a sub-group, classified as myomiRs and comprising miR-1, miR-206, miR-208a, miR-208b, miR-133a, miR-133b, miR-486 and miR-499 [112] has been extensively investigated. These miRNAs regulate myoblast proliferation, differentiation and regeneration i.e., increasing the expression of targets such as the myogenic factors MYOG (myogenin), MYF5 (myogenic factor 5), MYOD1 (myogenic differentiation 1) and PAX7 (paired box protein 7), in order to induce muscle regeneration and to prevent fibrosis [113]. Moreover, beyond acting at muscle level, some of them can exert additional functions such as cell fate regulation, chromatin remodeling and oxidative stress control [57]. They can regulate, or be regulated by, factors involved in the IGF-1/Akt/mTOR signaling pathway, known to control skeletal muscle protein synthesis and muscle protein breakdown, processes controlled by anabolic stimuli, such as physical activity and nutritional status [88,94,97,114,115].
Many miRNAs related to physical frailty are involved in inflammation, which is recognized as the underlying pathway in sarcopenia and muscle loss. The best known are miR-21 and miR-146a, proposed as inflamm-miRNA owing to their ability to master (NF-κB)-driven inflammatory pathways [116]. MiR-21 was proposed by Rusanova et al. [67] as a biomarker of human muscle frailty. Its levels were found to correlate with AOPP (Advanced oxidation protein products) levels, mediators of the proinflammatory effect of oxidative stress [117].
MiR-146a is one of the most prevalent miRNAs in the literature in instances of chronic inflammatory disorders and oxidative stress, both in the muscle [118,119] and in the brain [120] and is a regulator of osteogenesis and angiogenesis [121].
Other families of miRNAs related to both physical frailty and inflammatory status are miR-19 and miR-181 [122,123].
Finally, some miRNAs listed also regulate mitochondrial functions and are known as mitomiR [124]. Most of them are nuclear-encoded; some mitomiRs modulate mitochondrial function by binding mRNA in the cytoplasm (examples can be found among miR-27a, miR-34, mir-155 or miR-181a), while others are imported into mitochondria as part of RISC or pre-RISC and target mitochondrial-encoded mRNA (i.e., mir-151a and miR-181c).
4. MiRNAs as Biomarkers of the Cognitive Domain of Frailty
Table S2 [68,69,120,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154] summarizes the 43 miRNAs reported in association with cognitive impairment/decline in older adults.
Actually, it is known that the brain expresses about 70% of experimentally detectable miRNAs [155].
The critical role of miRNAs in the central nervous system was demonstrated in model organisms, where the disruption of the miRNA biogenesis machinery [156,157,158] or the knock-out of specific miRNAs produced worsened long-term memory, enhanced Aβ burden, and increased tau pathology, revealing a possible influence of miRNAs on many genes of the tau subnetwork [159,160,161]. As for humans, in recent years two papers emerged with evidence for the discrimination power of miRNAs in respect to cognitive decline in old age. First, Kondo and collaborators (2019) found a positive correlation between low serum levels of miR-20a, miR-27a, and miR-103a and MMSE scores in Japanese individuals with early-stage cognitive decline [126]. After, Gullet et al. (2020) by an in-silico analysis proposed three miRNAs (miR-140-5p, miR-197-3p, miR-501-3p) as blood-based biomarkers of cognitive aging, and top-ranked predictors of multiple cognitive outcomes in healthy older adults [146].
Pathways targeted by multiple miRNAs include Aβ genesis, regulation of AMPAR subunits, autophagy homeostasis, apoptosis, microglial activation, NF-κB signaling, blood-brain barrier maintenance, synaptic plasticity and neurogenesis [120].
Interestingly, many miRNAs related to cognition also have roles in inflammation, confirming the inflammatory pathway as a major component of neurodegenerative diseases and a plausible mechanism at the crossroad between several frailty domains. Among them, some were addressed by some authors as NeurimmiRs (the best known are miR-9, miR-21, miR-124, miR-132, miR-135, miR-146a, miR-155, miR-186, miR-223, in addition to the miR-29 family) affecting both immune and neuroinflammatory processes [162]. The evidence documenting their role in different pathological conditions characterized by cognitive decline demonstrates the major role of miRNAs in the neuroimmune interface, acting as ‘negotiators’ between these two interacting compartments, through direct or indirect alterations of neuron-glia and/or brain-to-body signaling [162]. Considering the importance of the network nervous–endocrine–immune system in maintaining the overall homeostasis [163], these miRNAs may be at the crossroad between the cognitive domain and the other domains of frailty.
5. In Silico Analysis of Shared miRNAs between Cognitive and Physical Domains
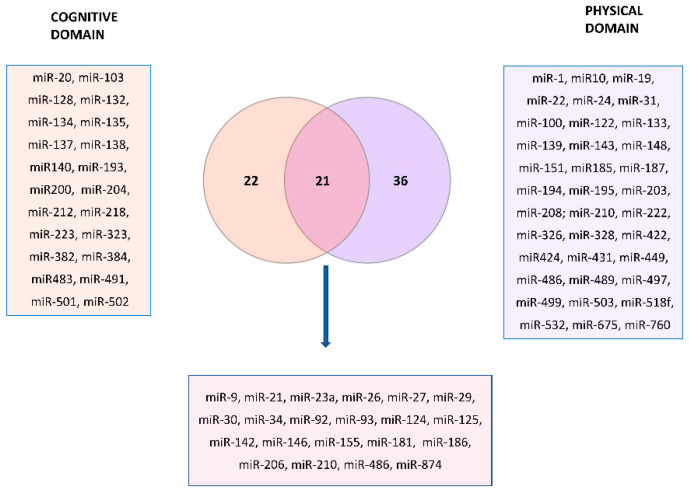
Figure 4 represents the Venn diagram describing the overlapping miRNAs between cognitive and physical domains, indicated in bold in the Tables S1 and S2.
Figure 4.
Venn diagram highlighting the (21) miRNAs markers shared between the cognitive and physical domains of frailty (listed in the violet square). The list of markers belonging to the cognitive domain (22) and physical domain (36) is also reported in the pink and indigo squares.
There are 22 miRNAs related exclusively to the cognitive domain of frailty and 36 only to the physical domain, while 21 were common. We performed miRNA-target enrichment of the 21 miRNAs identified by a target prediction tool, MIENTURNET (http://userver.bio.uniroma1.it/apps/mienturnet/, accessed on 11 July 2022) [164] by using experimentally validated (miRTarBase) miRNA-target interactions for discover the targets of the candidate miRNA list. With an FDR cut-off <0.05, and a minimum number of two interactions of gene-miRNAs, we retrieved 55 genes significantly associated with those miRNA markers (Table 1).
Table 1.
Gene targets significantly associated with the 21 miRNA markers candidates for the in silico analysis.
| miRNA-Target Enrichment Results | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | p-Value | FDR | O Rt |
Interacting miRs |
miR 1 | miR 2 | mioR 3 | miR 4 | miR 5 | miR 6 | miR 7 | miR 8 |
| PTEN | 2.75 × 10−6 | 0.011 | 0.126 | 8 | miR-26a | miR-29b | miR-23a | miR-92a | miR-155 | miR-34a | miR-486 | miR-21 |
| ATP1A1 | 3.02 × 10−5 | 0.016 | 0.048 | 4 | miR-93 | miR-92a | miR-26a | miR-155 | ||||
| CIT | 4.20 × 10−5 | 0.016 | 0.052 | 4 | miR-93 | miR-29b | miR-92a | miR-486 | ||||
| GATA3 | 1.59 × 10−5 | 0.016 | 0.018 | 3 | miR-92a | miR-29b | miR-27a | |||||
| GIT2 | 3.70 × 10−5 | 0.016 | 0.024 | 3 | miR-26a | miR-210 | miR-92a | |||||
| MYC | 2.84 × 10−5 | 0.016 | 0.140 | 7 | miR-26a | miR-30a | miR-92a | miR-23a | miR-125a | miR-34a | miR-29b | |
| PPP1R2 | 4.20 × 10−5 | 0.016 | 0.052 | 4 | miR-210 | miR-142 | miR-30a | miR-34a | ||||
| RGS5 | 3.13 × 10−5 | 0.016 | 0.079 | 5 | miR-142 | miR-92a | miR-124 | miR-9 | miR-23a | |||
| STAT3 | 2.64 × 10−5 | 0.016 | 0.107 | 6 | miR-92a | miR-874 | miR-21 | miR-23a | miR-124 | miR-29b | ||
| TKT | 3.70 × 10−5 | 0.016 | 0.024 | 3 | miR-92a | miR-26a | miR-206 | |||||
| CAMKV | 6.57 × 10−5 | 0.020 | 0.058 | 4 | miR-92a | miR-26a | miR-23a | miR-874 | ||||
| CCL8 | 6.22 × 10−5 | 0.020 | 0.008 | 2 | miR-23a | miR-92a | ||||||
| PDS5B | 6.22 × 10−5 | 0.020 | 0.008 | 2 | miR-27a | miR-92a | ||||||
| CTC1 | 7.08 × 10−5 | 0.020 | 0.127 | 6 | miR-93 | miR-92a | miR-26a | miR-29b | miR-181a | miR-874 | ||
| COL4A2 | 9.55 × 10−5 | 0.024 | 0.032 | 3 | miR-29b | miR-92a | miR-210 | |||||
| HES1 | 9.55 × 10−5 | 0.024 | 0.032 | 3 | miR-23a | miR-92a | miR-9 | |||||
| CPEB4 | 1.07 × 10−4 | 0.025 | 0.101 | 5 | miR-26a | miR-34a | miR-92a | miR-874 | miR-27a | |||
| KIAA1671 | 1.25 × 10−4 | 0.026 | 0.068 | 4 | miR-93 | miR-29b | miR-92a | miR-30a | ||||
| LDHB | 1.23 × 10−4 | 0.026 | 0.0350 | 3 | miR-186 | miR-23a | miR-210 | |||||
| CNOT1 | 1.94 × 10−4 | 0.029 | 0.040 | 3 | miR-93 | miR-92a | miR-23a | |||||
| CSAG1 | 1.85 × 10−4 | 0.029 | 0.012 | 2 | miR-186 | miR-93 | ||||||
| ESR1 | 2.04 × 10−4 | 0.029 | 0.116 | 5 | miR-206 | miR-29b | miR-26a | miR-142 | miR-874 | |||
| FUK | 1.85 × 10−4 | 0.029 | 0.012 | 2 | miR-93 | miR-92a | ||||||
| MCM3 | 1.56 × 10−4 | 0.029 | 0.037 | 3 | miR-93 | miR-92a | miR-210 | |||||
| P2RX7 | 2.15 × 10−4 | 0.029 | 0.078 | 4 | miR-9 | miR-146a | miR-186 | miR-125a | ||||
| PFDN2 | 1.56 × 10−4 | 0.029 | 0.037 | 3 | miR-93 | miR-210 | miR-92a | |||||
| PIK3CG | 2.15 × 10−4 | 0.029 | 0.078 | 4 | miR-29b | miR-27a | miR-142 | miR-26a | ||||
| PSMC3 | 1.85 × 10−4 | 0.029 | 0.01 | 2 | miR-92a | miR-23a | ||||||
| WDR77 | 1.94 × 10−4 | 0.029 | 0.076 | 4 | miR-27a | miR-93 | miR-186 | miR-125a | ||||
| PPARD | 2.38 × 10−4 | 0.031 | 0.043 | 3 | miR-92a | miR-29b | miR-30a | |||||
| KCTD5 | 2.89 × 10−4 | 0.036 | 0.084 | 4 | miR-92a | miR-26a | miR-125a | miR-34a | ||||
| TET2 | 2.87 × 10−4 | 0.036 | 0.045 | 3 | miR-92a | miR-29b | miR-26a | |||||
| APC | 3.43 × 10−4 | 0.037 | 0.048 | 3 | miR-210 | miR-27a | miR-142 | |||||
| CPEB2 | 3.47 × 10−4 | 0.037 | 0.088 | 4 | miR-210 | miR-26a | miR-92a | miR-142 | ||||
| FAU | 3.69 × 10−4 | 0.037 | 0.016 | 2 | miR-92a | miR-23a | ||||||
| GPD1L | 3.69 × 10−4 | 0.037 | 0.016 | 2 | miR-210 | miR-142 | ||||||
| IRAK1 | 3.43 × 10−4 | 0.037 | 0.048 | 3 | miR-93 | miR-92a | miR-142 | |||||
| PHB | 3.69 × 10−4 | 0.037 | 0.016 | 2 | miR-27a | miR-26a | ||||||
| SCAF8 | 3.69 × 10−4 | 0.037 | 0.016 | 2 | miR-29b | miR-92a | ||||||
| VMAC | 3.43 × 10−4 | 0.037 | 0.048 | 3 | miR-146a | miR-186 | miR-125a | |||||
| HMGCR | 4.05 × 10−4 | 0.039 | 0.051 | 3 | miR-92a | miR-29b | miR-27a | |||||
| HECTD1 | 4.74 × 10−4 | 0.043 | 0.053 | 3 | miR-210 | miR-142 | miR-92a | |||||
| TBC1D16 | 4.74 × 10−4 | 0.043 | 0.053 | 3 | miR-26a | miR-186 | miR-210 | |||||
| TUT1 | 4.74 × 10−4 | 0.043 | 0.053 | 3 | miR-93 | miR-92a | miR-26a | |||||
| ABCB9 | 6.12 × 10−4 | 0.046 | 0.020 | 2 | miR-210 | miR-26a | ||||||
| CDH1 | 6.34 × 10−4 | 0.046 | 0.059 | 3 | miR-92a | miR-23a | miR-9 | |||||
| DUSP5 | 5.51 × 10−4 | 0.046 | 0.056 | 3 | miR-27a | miR-92a | miR-26a | |||||
| IL6 | 6.34 × 10−4 | 0.046 | 0.059 | 3 | miR-142 | miR-26a | miR-125a | |||||
| INPP5A | 6.12 × 10−4 | 0.046 | 0.020 | 2 | miR-210 | miR-142 | ||||||
| KIF20A | 6.12 × 10−4 | 0.046 | 0.020 | 2 | miR-92a | miR-23a | ||||||
| NEK6 | 6.12 × 10−4 | 0.046 | 0.020 | 2 | miR-92a | miR-23a | ||||||
| PDE4B | 6.12 × 10−4 | 0.046 | 0.020 | 2 | miR-26a | miR-34a | ||||||
| SOCS6 | 6.34 × 10−4 | 0.046 | 0.059 | 3 | miR-23a | miR-27a | miR-142 | |||||
| UBE2R2 | 6.12 × 10−4 | 0.046 | 0.020 | 2 | miR-93 | miR-92a | ||||||
| ZNF618 | 6.34 × 10−4 | 0.046 | 0.059 | 3 | miR-21 | miR-27a | miR-210 | |||||
Significance was considered under a level of FDR < 0.05, and a minimum number of 2 interactions gene-miRNAs. In italics are indicated the 10 top-ranked targets (p < 4.5 × 10−5). FDR: False Discovery Rate; OR: Odd ratio.
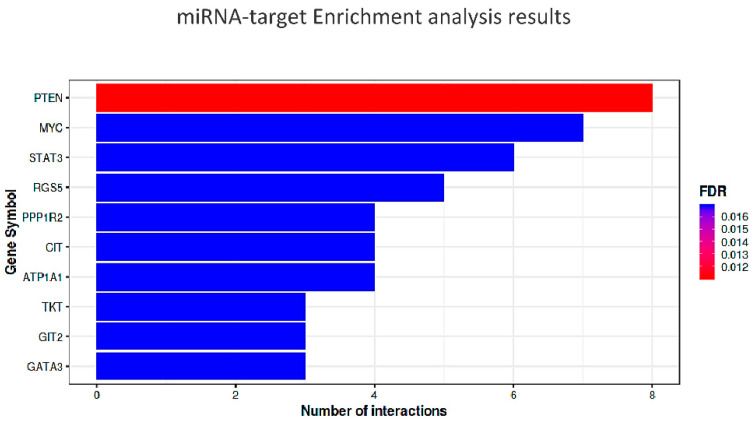
The top ten most significant targets (minimum of three gene-miRNA interactions and p < 4.5 × 10−5) are indicated in italics and represented in the diagram of Figure 5.
Figure 5.
Bar plot of the results of the enrichment analysis, obtained by using miTarBASE as reference dataset. In the Y-axis are reported the top ten target genes of the submitted miRNAs, while the X-axis represents the number of miRNAs targeting them. The color code reflects the adjusted p-values (FDR) increasing from red to blue. The threshold for the minimum number of miRNA-target interactions was set up to 2.
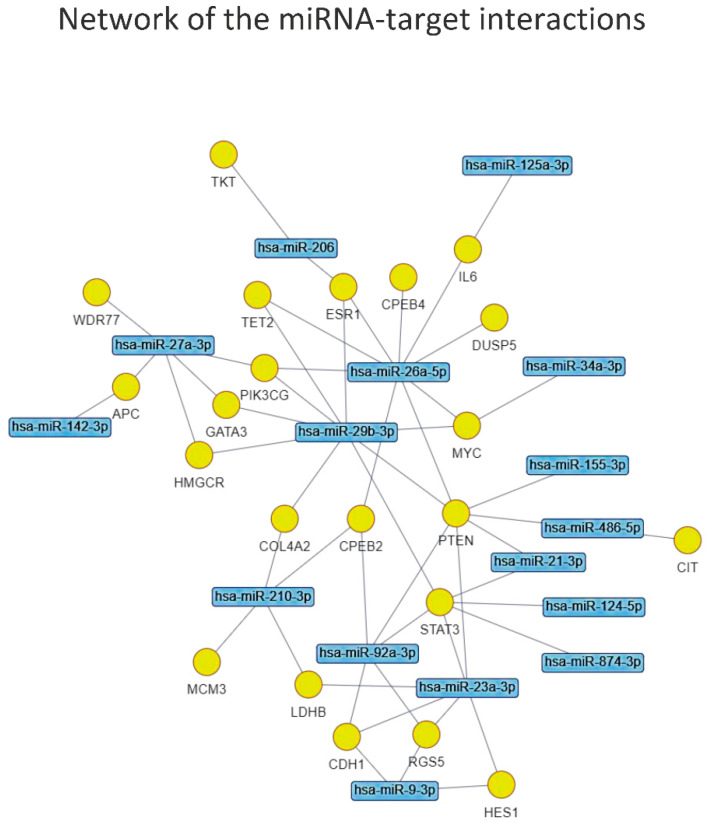
The network of miRNA-target interactions identified by the enrichment analysis (Figure 6) obtained by applying the same FDR < 0.05 allowed us to prioritize the most interesting miRNAs to submit for the functional enrichment analysis, which were: miR-26a-5p, miR-29b-3p, miR-23a-3p, miR-27a-3p, miR-92a-3p, miR-210-3p, miR-9-3p, miR-206,-miR-21-3p and miR-486-5p.
Figure 6.
Graphical representation of the network of miRNA-target interactions identified by the enrichment analysis.
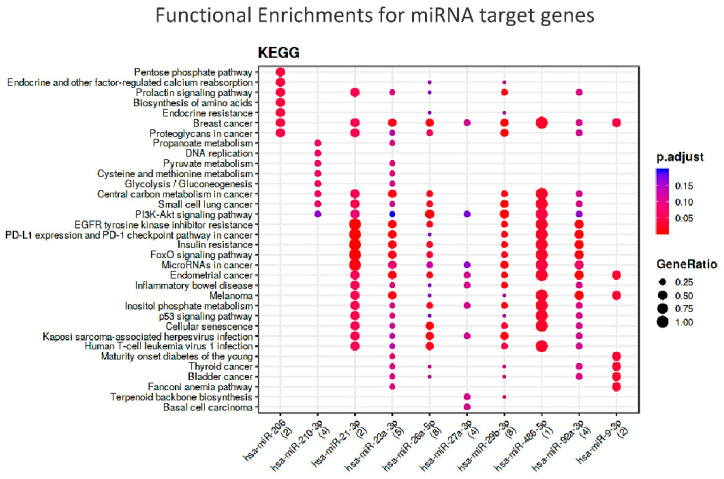
To understand which could be the common molecular pathways linking the two frailty domains, we performed a functional enrichment analysis of target genes of these 10 selected miRNAs. Significant enrichment was found for miR-21-3p, miR-206 and mir486a-5p and the relevant pathways, as shown in Figure 7.
Figure 7.
Functional Enrichments for miRNA target genes according to KEGG db. N.B.: The colors of the dots represent the adjusted p-values (FDR), whereas the size of the dots represents gene ratio (i.e., the number of miRNA targets found annotated in each category over the total number of recognized targets indicated in round brackets).
These results deserve a brief discussion. PTEN (Phosphatase and Tensin Homolog), which is the master gene tagged by eight of the candidate miRNAs, encodes for a multi-functional phosphatase belonging to the PI3K/AKT/mTOR pathway. This evolutionarily conserved component, most prominently known for its function in tumorigenesis, is increasingly seen as having a metabolic role as a negative regulator of the IIS (Insulin Signaling pathway) [165].
In humans, recent results indicate a role of PTEN in the control of adipose tissue growth with aging in cultured cells, associated with a higher insulin sensitivity [166] and insulin resistance in the pathogenesis of T2D [167]. Interestingly, the KEGG analysis showed an enrichment of molecular pathways related to nutrient regulation, such as insulin resistance, FOXO signaling, biosynthesis of amino acids and penthose phosphate pathway. Among the miRNAs prioritized by the KEGG analysis, miR-21, a PTEN inhibitor and a downstream effector of AKT, is reported to be able to reverse high glucose- and high insulin-induced Insulin Resistance in 3T3-L1 adipocytes, through modulating the PTEN-AKT pathway [168].
Moreover, as previously stated, miR-21 is, an inflamm-miRNA particularly linked to sarcopenia [67] but with a relevance in human age-related diseases [116].
The importance of inflammation as a crucial pathway in frailty comes from the interrogation of the STRING database (https://string-db.org/ accessed on 11 July 2022) showing that PTEN is functionally interrelated with two further significant targets of the miRNA-target enrichment: the nuclear phosphoprotein MYC (MYC Proto-Oncogene, BHLH Transcription Factor) and the transcription activator STAT3 (Signal Transducer and Activator Of Transcription 3). This trio is part of an inflammatory regulatory network, particularly linked to the downstream program of IL6, which exerts both pro- and anti-inflammatory activities, eliciting different biological responses on different cell types [169,170,171]. Overall, these evidences further confirm the relevance of the energetic metabolism and inflammation pathway in human frailty. This observation is further confirmed by the other two miRNAs merged from the KEGG analysis, miR-206 and miR-486-5p. Known as myomiRNA, miR-206 suppresses IGF1 [172] and regulates also inflammation [173]. In turn, miR-486-5p inhibits inflammatory response [174] and is known to up-regulate the expression of silent information regulator 1 (SIRT1), a major regulator of lifespan and metabolic disorders [175], whose levels have been associated with frailty in older adults [176]. Thus, it appears very probable that these miRs may represent the link between nutrition and inflammation as master regulators of frailty.
6. Conclusions and Final Remarks
The study of miRNAs represents an emerging area of interest in the aging research. For their possibility to target several biological processes, they may represent efficient diagnostic and prognostic biomarkers for frailty, which is characterized by impairment in multiple domains, likely sharing common molecular pathways. Here we investigated this issue by reviewing the literature on miRNAs associated with phenotypes of physical and cognitive dysfunctions, with the aim of identifying regulators common to both frailty domains. A target enrichment and functional enrichment approach provided us with a panel of 10 miRNAs, which target energy metabolism and inflammatory pathways, further confirming the relevance of these mechanisms in the failure of homeostasis related to frailty, and which could be tested as signatures of impairment in both frailty areas.
Although the study of differentially expressed miRNAs in frailty is at its infancy and further studies are necessary, we are very confident that the combination of miRNA-panels and traditional biomarkers may have a clinical value, representing a promising tool for the screening of the population at risk of frailty, to reach the aim of modeling health trajectories toward positive outcomes.
Acknowledgments
The work has been made possible by the collaboration with Gruppo Baffa (Sadel Spa, Sadel San Teodoro srl, Sadel CS srl, Casa di Cura Madonna dello Scoglio, AGI srl, Casa di Cura Villa del Rosario srl, Savelli Hospital srl, Casa di Cura Villa Ermelinda). The authors would like to thank Valerio Licursi for the support in using MIENTURNET tool.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology11081151/s1, Table S1: A summary of microRNAs (miRNAs) associated with pathways related to physical frailty in human; Table S2: A summary of microRNAs (miRNAs) associated with pathways related to cognitive frailty in humans.
Author Contributions
Conceptualization, S.D. and G.R.; formal analysis and data curation S.D.; writing—original draft preparation, F.I., P.C. and S.D.; writing—review and editing, P.C., G.R. and G.P.; funding acquisition, G.P. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This research was supported by “SI.F.I.PA.CRO.DE. –Sviluppo e industrializzazione farmaci innovativi per terapia molecolare personalizzata PA.CRO.DE.” PON ARS01_00568 granted by MIUR (Ministry of Education, University and Research) Italy to G.P.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abate M., Di Iorio A., Di Renzo D., Paganelli R., Saggini R., Abate G. Frailty in the elderly: The physical dimension. Eura Medicophys. 2007;43:407–415. [PubMed] [Google Scholar]
- 2.Gordon A.L., Masud T., Gladman J.R. Now that we have a definition for physical frailty, what shape should frailty medicine take? Age Ageing. 2014;43:8–9. doi: 10.1093/ageing/aft161. [DOI] [PubMed] [Google Scholar]
- 3.Sathyan S., Verghese J. Genetics of frailty: A longevity perspective. Trans. Res. 2020;221:83–96. doi: 10.1016/j.trsl.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abellan van Kan G., Rolland Y.M., Morley J.E., Vellas B. Frailty: Toward a clinical definition. J. Am. Med. Dir. Assoc. 2008;9:71–72. doi: 10.1016/j.jamda.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Ernsth Bravell M., Westerlind B., Midlöv P., Östgren C.J., Borgquist L., Lannering C., Mölstad S. How to assess frailty and the need for care? Report from the Study of Health and Drugs in the Elderly (SHADES) in community dwellings in Sweden. Arch. Gerontol. Geriatr. 2011;53:40–45. doi: 10.1016/j.archger.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Mohandas A., Reifsnyder J., Jacobs M., Fox T. Current and future directions in frailty research. Popul. Health Manag. 2011;14:277–283. doi: 10.1089/pop.2010.0066. [DOI] [PubMed] [Google Scholar]
- 7.Manthorpe J., Iliffe S. The many meanings of frailty: Is there a shared understanding? Nurs. Resid. Care. 2015;17:575–576. doi: 10.12968/nrec.2015.17.10.575. [DOI] [Google Scholar]
- 8.Maxwell C.A., Wang J. Understanding Frailty: A Nurse’s Guide. Nurs. Clin. N. Am. 2017;52:349–361. doi: 10.1016/j.cnur.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Panza F., Lozupone M., Solfrizzi V., Sardone R., Dibello V., Di Lena L., D’Urso F., Stallone R., Petruzzi M., Giannelli G., et al. Different Cognitive Frailty Models and Health- and Cognitive-related Outcomes in Older Age: From Epidemiology to Prevention. J. Alzheimers Dis. 2018;62:993–1012. doi: 10.3233/JAD-170963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fried L.P., Tangen C.M., Walston J., Newman A.B., Hirsch C., Gottdiener J., Seeman T., Tracy R., Kop W.J., Burke G., et al. Cardiovascular Health Study Collaborative Research Group. Frailty in older adults: Evidence for a phenotype. J. Gerontol. A Biol. Sci. Med. Sci. 2001;56:M146–M156. doi: 10.1093/gerona/56.3.M146. [DOI] [PubMed] [Google Scholar]
- 11.Rockwood K., Mitnitski A. Frailty defined by deficit accumulation and geriatric medicine defined by frailty. Clin. Geriatr. Med. 2011;27:17–26. doi: 10.1016/j.cger.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Gobbens R.J.J., Luijkx K.G., van Assen M.A.L.M. Explaining quality of life of older people in the Netherlands using a multidimensional assessment of frailty. Qual. Life Res. 2013;22:2051–2061. doi: 10.1007/s11136-012-0341-1. [DOI] [PubMed] [Google Scholar]
- 13.Peters L.L., Boter H., Buskens E., Slaets J.P. Measurement properties of the Groningen Frailty Indicator in home-dwelling and institutionalized elderly people. J. Am. Med. Dir. Assoc. 2012;13:546–551. doi: 10.1016/j.jamda.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Morley J.E., Malmstrom T.K., Miller D.K. A simple frailty questionnaire (FRAIL) predicts outcomes in middle aged African Americans. J. Nutr. Health Aging. 2012;16:601–608. doi: 10.1007/s12603-012-0084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wieland D., Hirth V. Comprehensive geriatric assessment. Cancer Control. 2003;10:454–462. doi: 10.1177/107327480301000603. [DOI] [PubMed] [Google Scholar]
- 16.Gottesman I.I., Gould T.D. The endophenotype concept in psychiatry: Etymology and strategic intentions. Am. J. Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 17.Cannon T.D., Keller M.C. Endophenotypes in the genetic analyses of mental disorders. Annu Rev. Clin. Psychol. 2006;2:267–290. doi: 10.1146/annurev.clinpsy.2.022305.095232. [DOI] [PubMed] [Google Scholar]
- 18.Meyer-Lindenberg A., Weinberger D.R. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat. Rev. Neurosci. 2006;7:818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- 19.van Oostrom S.H., van der A.D.L., Rietman M.L., Picavet H.S.J., Lette M., Verschuren W.M.M., de Bruin S.R., Spijkerman A.M.W. A four-domain approach of frailty explored in the Doetinchem Cohort Study. BMC Geriatr. 2017;17:196. doi: 10.1186/s12877-017-0595-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Folstein M.F., Folstein S.E., McHugh P.R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 21.Teng E.L., Hasegawa K., Homma A., Imai Y., Larson E., Graves A., Sugimoto K., Yamaguchi T., Sasaki H., Chiu D. The Cognitive Abilities Screening Instrument (CASI): A practical test for cross-cultural epidemiological studies of dementia. Int. Psychogeriatr. 1994;6:45–62. doi: 10.1017/S1041610294001602. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell A.J., Coyne J.C. Screening for Depression in Clinical Practice: An Evidence-Based Guide. Oxford University Press; Oxford, UK: 2009. [Google Scholar]
- 23.Ruan Q., Yu Z., Chen M., Bao Z., Li J., He W. Cognitive frailty, a novel target for the prevention of elderly dependency. Ageing Res. Rev. 2015;20:1–10. doi: 10.1016/j.arr.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Grande G., Haaksma M.L., Rizzuto D., Melis R.J.F., Marengoni A., Onder G., Welmer A.K., Fratiglioni L., Vetrano D.L. Co-occurrence of cognitive impairment and physical frailty, and incidence of dementia: Systematic review and meta-analysis. Neurosci. Biobehav. Rev. 2019;107:96–103. doi: 10.1016/j.neubiorev.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Buchman A.S., Schneider J.A., Leurgans S., Bennett D.A. Physical frailty in older persons is associated with Alzheimer disease pathology. Neurology. 2008;71:499–504. doi: 10.1212/01.wnl.0000324864.81179.6a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujiwara Y., Shinkai S., Kumagai S., Amano H., Yoshida Y., Yoshida H., Kim H., Suzuki T., Ishizaki T., Haga H., et al. Longitudinal changes in higher-level functional capacity of an older population living in a Japanese urban community. Arch. Gerontol. Geriatr. 2003;36:141–153. doi: 10.1016/S0167-4943(02)00081-X. [DOI] [PubMed] [Google Scholar]
- 27.Makizako H., Shimada H., Doi T., Tsutsumimoto K., Hotta R., Nakakubo S., Makino K., Lee S. Social Frailty Leads to the Development of Physical Frailty among Physically Non-Frail Adults: A Four-Year Follow-Up Longitudinal Cohort Study. Int. J. Environ. Res. Public. Health. 2018;15:490. doi: 10.3390/ijerph15030490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee W.J., Peng L.N., Lin C.H., Chen R.C., Lin S.Z., Loh C.H., Kao S.L., Hung T.S., Chang C.Y., Huang C.F., et al. Effects of incorporating multidomain interventions into integrated primary care on quality of life: A randomised controlled trial. Lancet. 2021;2:e712–e723. doi: 10.1016/S2666-7568(21)00248-8. [DOI] [PubMed] [Google Scholar]
- 29.Whalley L.J., Murray A.D., Staff R.T., Starr J.M., Deary I.J., Fox H.C., Lemmon H., Duthie S.J., Collins A.R., Crawford J.R. How the 1932 and 1947 mental surveys of Aberdeen schoolchildren provide a framework to explore the childhood origins of late onset disease and disability. Maturitas. 2011;69:365–372. doi: 10.1016/j.maturitas.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Evans W.J., Paolisso G., Abbatecola A.M., Corsonello A., Bustacchini S., Strollo F., Lattanzio F. Frailty and muscle metabolism dysregulation in the elderly. Biogerontology. 2010;11:527–536. doi: 10.1007/s10522-010-9297-0. [DOI] [PubMed] [Google Scholar]
- 31.Fried L.P., Cohen A.A., Xue Q.L., Walston J., Bandeen-Roche K., Varadhan R. The physical frailty syndrome as a transition from homeostatic symphony to cacophony. Nat. Aging. 2021;1:36–46. doi: 10.1038/s43587-020-00017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwak D., Thompson L.V. Frailty: Past, present, and future? Sports Med. Health Sci. 2021;3:1–10. doi: 10.1016/j.smhs.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J.K., Choi S.R., Choi M.J., Kim S.G., Lee Y.K., Noh J.W., Kim H.J., Song Y.R. Prevalence of and factors associated with sarcopenia in elderly patients with end-stage renal disease. Clin. Nutr. 2014;33:64–68. doi: 10.1016/j.clnu.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Ramakrishnan P., Alyousefi N., Abdul-Rahman P.S., Kamaruzzaman S.B., Chin A.V., Tan M.P. A systematic review of studies comparing potential biochemical biomarkers of frailty with frailty assessments? Eur. Geriatr. Med. 2017;8:397–407. doi: 10.1016/j.eurger.2017.07.010. [DOI] [Google Scholar]
- 35.Sargent L., Nalls M., Starkweather A., Hobgood S., Thompson H., Amella E.J., Singleton A. Shared biological pathways for frailty and cognitive impairment: A systematic review. Ageing Res. Rev. 2018;47:149–158. doi: 10.1016/j.arr.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saedi A.A., Feehan J., Phu S., Duque G. Current and emerging biomarkers of frailty in the elderly. Clin. Interv. Aging. 2019;14:389–398. doi: 10.2147/CIA.S168687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Supriya R., Singh K.P., Gao Y., Li F., Dutheil F., Baker J.S. A Multifactorial Approach for Sarcopenia Assessment: A Literature Review. Biology. 2021;10:1354. doi: 10.3390/biology10121354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viña J., Borras C., Gomez-Cabrera M.C. A free radical theory of frailty. Free Radic. Biol. Med. 2018;124:358–363. doi: 10.1016/j.freeradbiomed.2018.06.028. [DOI] [PubMed] [Google Scholar]
- 39.Serviddio G., Romano A.D., Greco A., Rollo T., Bellanti F., Altomare E., Vendemiale G. Frailty syndrome is associated with altered circulating redox balance and increased markers of oxidative stress. Int. J. Immunopathol. Pharmacol. 2009;22:819–827. doi: 10.1177/039463200902200328. [DOI] [PubMed] [Google Scholar]
- 40.Wu B., Fukuo K., Suzuki K., Yoshino G., Kazumi T. Relationships of systemic oxidative stress to body fat distribution, adipokines and inflammatory markers in healthy middle-aged women. Endocr. J. 2009;56:773–782. doi: 10.1507/endocrj.K08E-332. [DOI] [PubMed] [Google Scholar]
- 41.Ju S.Y., Lee J.Y., Kim D.H. Low 25-hydroxyvitamin D levels and the risk of frailty syndrome: A systematic review and dose-response meta-analysis. BMC Geriatr. 2018;18:206. doi: 10.1186/s12877-018-0904-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J., Liu B., Liang C., Li Y., Song Y.H. Cytokine Signaling in Skeletal Muscle Wasting. Trends Endocrinol. Metab. 2016;27:335–347. doi: 10.1016/j.tem.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Kameda M., Teruya T., Yanagida M., Kondoh H. Reduced uremic metabolites are prominent feature of sarcopenia, distinct from antioxidative markers for frailty. Aging. 2021;13:20915–20934. doi: 10.18632/aging.203498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swiecicka A., Eendebak R.J.A.H., Lunt M., O’Neill T.W., Bartfai G., Casanueva F.F., Forti G., Giwercman A., Han T.S., Slowikowska-Hilczer J., et al. European Male Ageing Study Group. Reproductive Hormone Levels Predict Changes in Frailty Status in Community-Dwelling Older Men: European Male Ageing Study Prospective Data. J. Clin. Endocrinol. Metab. 2018;103:701–709. doi: 10.1210/jc.2017-01172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carcaillon L., García-García F.J., Tresguerres J.A., Gutiérrez Avila G., Kireev R., Rodríguez-Mañas L. Higher levels of endogenous estradiol are associated with frailty in postmenopausal women from the toledo study for healthy aging. J. Clin. Endocrinol. Metab. 2012;97:2898–2906. doi: 10.1210/jc.2012-1271. [DOI] [PubMed] [Google Scholar]
- 46.Picca A., Coelho-Junior H.J., Cesari M., Marini F., Miccheli A., Gervasoni J., Bossola M., Landi F., Bernabei R., Marzetti E., et al. The metabolomics side of frailty: Toward personalized medicine for the aged. Exp. Gerontol. 2019;126:110692. doi: 10.1016/j.exger.2019.110692. [DOI] [PubMed] [Google Scholar]
- 47.Picca A., Calvani R. Biomarkers of frailty: Moving the field forward. Exp. Gerontol. 2020;133:110868. doi: 10.1016/j.exger.2020.110868. [DOI] [PubMed] [Google Scholar]
- 48.Zafari S., Backes C., Leidinger P., Meese E., Keller A. Regulatory microRNA networks: Complex patterns of target pathways for disease-related and housekeeping microRNAs. Genom. Proteom. Bioinform. 2015;13:159–168. doi: 10.1016/j.gpb.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hammond S.M. An overview of microRNAs. Adv. Drug. Deliv. Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheng P., Fields C., Aadland K., Wei T., Kolaczkowski O., Gu T., Kolaczkowski B., Xie M. Dicer cleaves 5’-extended microRNA precursors originating from RNA polymerase II transcription start sites. Nucleic Acids Res. 2018;46:5737–5752. doi: 10.1093/nar/gky306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang F., Wang D. The Pattern of microRNA Binding Site Distribution. Genes. 2017;8:296. doi: 10.3390/genes8110296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moretti F., Thermann R., Hentze M.W. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA. 2010;16:2493–2502. doi: 10.1261/rna.2384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Denzler R., McGeary S.E., Title A.C., Agarwal V., Bartel D.P., Stoffel M. Impact of MicroRNA Levels, Target-Site Complementarity, and Cooperativity on Competing Endogenous RNA-Regulated Gene Expression. Mol. Cell. 2016;64:565–579. doi: 10.1016/j.molcel.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sohel M.H. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achiev. Life Sci. 2016;10:175–186. doi: 10.1016/j.als.2016.11.007. [DOI] [Google Scholar]
- 55.Bayraktar R., Van Roosbroeck K., Calin G.A. Cell-to-cell communication: microRNAs as hormones. Mol. Oncol. 2017;11:1673–1686. doi: 10.1002/1878-0261.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jung H.J., Suh Y. Circulating miRNAs in Ageing and Ageing-Related Diseases. J. Genet. Genom. 2014;41:465–472. doi: 10.1016/j.jgg.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Horak M., Novak J., Bienertova-Vasku J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016;410:1–13. doi: 10.1016/j.ydbio.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 58.Wang H., Peng R., Wang J., Qin Z., Xue L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenetics. 2018;10:59. doi: 10.1186/s13148-018-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.ElSharawy A., Keller A., Flachsbart F., Wendschlag A., Jacobs G., Kefer N., Brefort T., Leidinger P., Backes C., Meese E., et al. Genome-wide miRNA signatures of human longevity. Aging Cell. 2012;11:607–616. doi: 10.1111/j.1474-9726.2012.00824.x. [DOI] [PubMed] [Google Scholar]
- 60.Olivieri F., Capri M., Bonafè M., Morsiani C., Jung H.J., Spazzafumo L., Viña J., Suh Y. Circulating miRNAs and miRNA shuttles as biomarkers: Perspective trajectories of healthy and unhealthy aging. Mech. Ageing Dev. 2017;165:162–170. doi: 10.1016/j.mad.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serna E., Gambini J., Borras C., Abdelaziz K.M., Belenguer A., Sanchis P., Avellana J.A., Rodriguez-Mañas L., Viña J. Centenarians, but not octogenarians, up-regulate the expression of microRNAs. Sci. Rep. 2012;2:961. doi: 10.1038/srep00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noren Hooten N., Fitzpatrick M., Wood W.H., 3rd, De S., Ejiogu N., Zhang Y., Mattison J.A., Becker K.G., Zonderman A.B., Evans M.K. Age-related changes in microRNA levels in serum. Aging. 2013;5:725–740. doi: 10.18632/aging.100603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith-Vikos T., Slack F.J. MicroRNAs and their roles in aging. J. Cell Sci. 2012;125:7–17. doi: 10.1242/jcs.099200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jung H.J., Lee K.P., Kwon K.S., Suh Y. MicroRNAs in Skeletal Muscle Aging: Current Issues and Perspectives. J. Gerontol. A Biol. Sci. Med. Sci. 2019;74:1008–1014. doi: 10.1093/gerona/gly207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woldemichael B.T., Mansuy I.M. Micro-RNAs in cognition and cognitive disorders: Potential for novel biomarkers and therapeutics. Biochem. Pharmacol. 2016;104:1–7. doi: 10.1016/j.bcp.2015.11.021. [DOI] [PubMed] [Google Scholar]
- 66.Gomez-Verjan J.C., Ramírez-Aldana R., Pérez-Zepeda M.U., Quiroz-Baez R., Luna-López A., Gutierrez Robledo L.M. Systems biology and network pharmacology of frailty reveal novel epigenetic targets and mechanisms. Sci. Rep. 2019;9:10593. doi: 10.1038/s41598-019-47087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rusanova I., Diaz-Casado M.E., Fernández-Ortiz M., Aranda-Martínez P., Guerra-Librero A., García-García F.J., Escames G., Mañas L., Acuña-Castroviejo D. Analysis of Plasma MicroRNAs as Predictors and Biomarkers of Aging and Frailty in Humans. Oxid. Med. Cell. Longev. 2018;2018:7671850. doi: 10.1155/2018/7671850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ipson B.R., Fletcher M.B., Espinoza S.E., Fisher A.L. Identifying Exosome-Derived MicroRNAs as Candidate Biomarkers of Frailty. J. Frailty Aging. 2018;7:100–103. doi: 10.14283/jfa.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carini G., Musazzi L., Bolzetta F., Cester A., Fiorentini C., Ieraci A., Maggi S., Popoli M., Veronese N., Barbon A. The Potential Role of miRNAs in Cognitive Frailty. Front. Aging Neurosci. 2021;13:763110. doi: 10.3389/fnagi.2021.763110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee Y.H., Kim S.Y., Bae Y.S. Upregulation of miR-760 and miR-186 is associated with replicative senescence in human lung fibroblast cells. Mol. Cells. 2014;37:620–627. doi: 10.14348/molcells.2014.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morton S.U., Sefton C.R., Zhang H., Dai M., Turner D.L., Uhler M.D., Agrawal P.B. microRNA-mRNA profile of skeletal muscle differentiation and relevance to congenital myotonic dystrophy. Int. J. Mol. Sci. 2021;22:2692. doi: 10.3390/ijms22052692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X.H. MicroRNA in myogenesis and muscle atrophy. Curr. Opinion Clin. Nutr. Metab. Care. 2013;16:258–266. doi: 10.1097/MCO.0b013e32835f81b9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Russell A.P., Lamon S., Boon H., Wada S., Güller I., Brown E.L., Chibalin A.V., Zierath J.R., Snow R.J., Stepto N., et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J. Physiol. 2013;591:4637–4653. doi: 10.1113/jphysiol.2013.255695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Coolen M., Katz S., Bally-Cuif L. miR-9: A versatile regulator of neurogenesis. Front. Cell Neurosci. 2013;7:220. doi: 10.3389/fncel.2013.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J., Gu Z., Ni P., Qiao Y., Chen C., Liu X., Lin J., Chen N., Fan Q. NF-kappaB P50/P65 hetero-dimer mediates differential regulation of CD166/ALCAM expression via interaction with micoRNA-9 after serum deprivation, providing evidence for a novel negative auto-regulatory loop. Nucleic Acids Res. 2011;39:6440–6455. doi: 10.1093/nar/gkr302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weilner S., Skalicky S., Salzer B., Keider V., Wagner M., Hildner F., Gabriel C., Dovjak P., Pietschmann P., Grillari-Voglauer R., et al. Differentially circulating miRNAs after recent osteoporotic fractures can influence osteogenic differentiation. Bone. 2015;79:43–51. doi: 10.1016/j.bone.2015.05.027. [DOI] [PubMed] [Google Scholar]
- 77.Yanai K., Kaneko S., Ishii H., Aomatsu A., Ito K., Hirai K., Ookawara S., Ishibashi K., Morishita Y. MicroRNAs in Sarcopenia: A Systematic Review. Front. Med. 2020;7:180. doi: 10.3389/fmed.2020.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang G., He M., Wu P., Zhang X., Zhou K., Li T., Zhang T., Xie K., Dai G., Wang J. MicroRNA-27b-3p Targets the Myostatin Gene to Regulate Myoblast Proliferation and Is Involved in Myoblast Differentiation. Cells. 2021;10:423. doi: 10.3390/cells10020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Borja-Gonzalez M., Casas-Martinez J.C., McDonagh B., Goljanek-Whysall K. Inflamma-miR-21 Negatively Regulates Myogenesis during Ageing. Antioxidants. 2020;9:345. doi: 10.3390/antiox9040345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li T., Li H., Li T., Fan J., Zhao R.C., Weng X. MicroRNA expression profile of dexamethasone-induced human bone marrow-derived mesenchymal stem cells during osteogenic differentiation. J. Cell Biochem. 2014;115:1683–1691. doi: 10.1002/jcb.24831. [DOI] [PubMed] [Google Scholar]
- 81.Seeliger C., Karpinski K., Haug A.T., Vester H., Schmitt A., Bauer J.S., van Griensven M. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J. Bone Miner. Res. 2014;29:1718–1728. doi: 10.1002/jbmr.2175. [DOI] [PubMed] [Google Scholar]
- 82.Panach L., Mifsut D., Tarín J.J., Cano A., García-Pérez M.A. Serum Circulating MicroRNAs as Biomarkers of Osteoporotic Fracture. Calcif. Tissue Int. 2015;97:495–505. doi: 10.1007/s00223-015-0036-z. [DOI] [PubMed] [Google Scholar]
- 83.Dey B.K., Gagan J., Yan Z., Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012;26:2180–2191. doi: 10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y., Pan Y., Xie C., Zhang Y. miR-34a exerts as a key regulator in the dedifferentiation of osteosarcoma via PAI-1-Sox2 axis. Cell Death Dis. 2018;9:777. doi: 10.1038/s41419-018-0778-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li J., Chan M.C., Yu Y., Bei Y., Chen P., Zhou Q., Cheng L., Chen L., Ziegler O., Rowe G.C., et al. miR-29b contributes to multiple types of muscle atrophy. Nat. Commun. 2017;8:15201. doi: 10.1038/ncomms15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Pelt D.W., Vechetti I.J., Jr., Lawrence M.M., Van Pelt K.L., Patel P., Miller B.F., Butterfield T.A., Dupont-Versteegden E.E. Serum extracellular vesicle miR-203a-3p content is associated with skeletal muscle mass and protein turnover during disuse atrophy and regrowth. Am. J. Physiol. Cell Physiol. 2020;319:C419–C431. doi: 10.1152/ajpcell.00223.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Z.C., Wang Z.Z., Ma H.J., Wang C.C., Wang H.T. Attenuation of the hypoxia-induced miR-34a protects cardiomyocytes through maintenance of glucose metabolism. Biochem. Biophys. Res. Commun. 2018;498:375–381. doi: 10.1016/j.bbrc.2017.06.030. [DOI] [PubMed] [Google Scholar]
- 88.Iannone F., Montesanto A., Cione E., Crocco P., Caroleo M.C., Dato S., Rose G., Passarino G. Expression Patterns of Muscle-Specific miR-133b and miR-206 Correlate with Nutritional Status and Sarcopenia. Nutrients. 2020;12:297. doi: 10.3390/nu12020297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He N., Zhang Y.L., Zhang Y., Feng B., Zheng Z., Wang D., Zhang S., Guo Q., Ye H. Circulating MicroRNAs in Plasma Decrease in Response to Sarcopenia in the Elderly. Front. Genet. 2020;11:167. doi: 10.3389/fgene.2020.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Y., Li L., Moore B.T., Peng X.H., Fang X., Lappe J.M., Recker R.R., Xiao P. MiR-133a in human circulating monocytes: A potential biomarker associated with postmenopausal osteoporosis. PLoS ONE. 2012;7:e34641. doi: 10.1371/journal.pone.0034641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dobrowolny G., Martone J., Lepore E., Casola I., Petrucci A., Inghilleri M., Morlando M., Colantoni A., Scicchitano B.M., Calvo A., et al. A longitudinal study defined circulating microRNAs as reliable biomarkers for disease prognosis and progression in ALS human patients. Cell Death Discov. 2021;7:4. doi: 10.1038/s41420-020-00397-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Feng Y., Wan P., Yin L., Lou X. The Inhibition of MicroRNA-139-5p Promoted Osteoporosis of Bone Marrow-Derived Mesenchymal Stem Cells by Targeting Wnt/Beta-Catenin Signaling Pathway by NOTCH1. J. Microbiol. Biotechnol. 2020;30:448–458. doi: 10.4014/jmb.1908.08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu W., Ye Y., Zhang W., Wang J., Chen A., Guo F. miR 142 3p promotes osteoblast differentiation by modulating Wnt signaling. Mol. Med. Rep. 2013;7:689–693. doi: 10.3892/mmr.2012.1207. [DOI] [PubMed] [Google Scholar]
- 94.Margolis L.M., Lessard S.J., Ezzyat Y., Fielding R.A., Rivas D.A. Circulating MicroRNA Are Predictive of Aging and Acute Adaptive Response to Resistance Exercise in Men. J. Geront. A Biol Sci Med Sci. 2017;72:1319–1326. doi: 10.1093/gerona/glw243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Q., Deng J., Qiu Y., Gao J., Li J., Guan L., Lee H., Zhou Q., Xiao J. Non-coding RNA basis of muscle atrophy. Mol. Ther. Nucleic Acids. 2021;26:1066–1078. doi: 10.1016/j.omtn.2021.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Onodera Y., Teramura T., Takehara T., Itokazu M., Mori T., Fukuda K. Inflammation-associated miR-155 activates differentiation of muscular satellite cells. PLoS ONE. 2018;13:e0204860. doi: 10.1371/journal.pone.0204860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Drummond M.J., McCarthy J.J., Sinha M., Spratt H.M., Volpi E., Esser K.A., Rasmussen B.B. Aging and microRNA expression in human skeletal muscle: A microarray and bioinformatics analysis. Physiol. Genom. 2011;43:595–603. doi: 10.1152/physiolgenomics.00148.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Antoniou A., Mastroyiannopoulos N.P., Uney J.B., Phylactou L.A. miR-186 inhibits muscle cell differentiation through myogenin regulation. J. Biol. Chem. 2014;289:3923–3935. doi: 10.1074/jbc.M113.507343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garmilla-Ezquerra P., Sañudo C., Delgado-Calle J., Pérez-Nuñez M.I., Sumillera M., Riancho J.A. Analysis of the bone microRNome in osteoporotic fractures. Calcif. Tissue Int. 2015;96:30–37. doi: 10.1007/s00223-014-9935-7. [DOI] [PubMed] [Google Scholar]
- 100.Meng J., Zhang D., Pan N., Sun N., Wang Q., Fan J., Zhou P., Zhu W., Jiang L. Identification of miR-194-5p as a potential biomarker for postmenopausal osteoporosis. Peer J. 2015;3:e971. doi: 10.7717/peerj.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sato T., Yamamoto T., Sehara-Fujisawa A. miR-195/497 induce postnatal quiescence of skeletal muscle stem cells. Nat. Commun. 2014;5:4597. doi: 10.1038/ncomms5597. [DOI] [PubMed] [Google Scholar]
- 102.Okugawa Y., Toiyama Y., Hur K., Yamamoto A., Yin C., Ide S., Kitajima T., Fujikawa H., Yasuda H., Koike Y., et al. Circulating miR-203 derived from metastatic tissues promotes myopenia in colorectal cancer patients. J. Cachexia Sarcopenia Muscle. 2019;10:536–548. doi: 10.1002/jcsm.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cui S., Sun B., Yin X., Guo X., Chao D., Zhang C., Zhang C.Y., Chen X., Ma J. Time-course responses of circulating microRNAs to three resistance training protocols in healthy young men. Sci. Rep. 2017;7:2203. doi: 10.1038/s41598-017-02294-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cao Z., Moore B.T., Wang Y., Peng X.H., Lappe J.M., Recker R.R., Xiao P. MiR-422a as a potential cellular microRNA biomarker for postmenopausal osteoporosis. PLoS ONE. 2014;9:e97098. doi: 10.1371/journal.pone.0097098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Connolly M., Paul R., Farre-Garros R., Natanek S.A., Bloch S., Lee J., Lorenzo J.P., Patel H., Cooper C., Sayer A.A., et al. miR-424-5p reduces ribosomal RNA and protein synthesis in muscle wasting. J. Cachexia Sarcopenia Muscle. 2018;9:400–416. doi: 10.1002/jcsm.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee K.P., Shin Y.J., Panda A.C., Abdelmohsen K., Kim J.Y., Lee S.M., Bahn Y.J., Choi J.Y., Kwon E.S., Baek S.J., et al. miR-431 promotes differentiation and regeneration of old skeletal muscle by targeting Smad4. Genes Dev. 2015;29:1605–1617. doi: 10.1101/gad.263574.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cheung T.H., Quach N.L., Charville G.W., Liu L., Park L., Edalati A., Yoo B., Hoang P., Rando T.A. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–528. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sarkar S., Dey B.K., Dutta A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol. Biol. Cell. 2010;21:2138–2149. doi: 10.1091/mbc.e10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guo X., Wei S., Xu F., Cai X., Wang H., Ding R. MicroRNA-532-5p is implicated in the regulation of osteoporosis by forkhead box O1 and osteoblast differentiation. BMC Musculoskelet Disord. 2020;21:296. doi: 10.1186/s12891-020-03317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lewis A., Lee J.Y., Donaldson A.V., Natanek S.A., Vaidyanathan S., Man W.D., Hopkinson N.S., Sayer A.A., Patel H.P., Cooper C., et al. Increased expression of H19/miR-675 is associated with a low fat-free mass index in patients with COPD. J. Cachexia Sarcopenia Muscle. 2016;7:330–344. doi: 10.1002/jcsm.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mei L., Li M., Zhang T. MicroRNA miR-874-3p inhibits osteoporosis by targeting leptin (LEP) Bioengineered. 2021;12:11756–11767. doi: 10.1080/21655979.2021.2009618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brown D.M., Goljanek-Whysall K. microRNAs: Modulators of the underlying pathophysiology of sarcopenia? Ageing Res. Rev. 2015;24:263–273. doi: 10.1016/j.arr.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 113.Nakasa T., Ishikawa M., Shi M., Shibuya H., Adachi N., Ochi M. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J. Cell. Mol. Med. 2010;14:2495–2505. doi: 10.1111/j.1582-4934.2009.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pasiakos S.M., Cao J.J., Margolis L.M., Sauter E.R., Whigham L.D., McClung J.P., Rood J.C., Carbone J.W., Combs G.F., Jr., Young A.J. Effects of high-protein diets on fat-free mass and muscle protein synthesis following weight loss: A randomized controlled trial. FASEB J. 2013;27:3837–3847. doi: 10.1096/fj.13-230227. [DOI] [PubMed] [Google Scholar]
- 115.Camera D.M., Ong J.N., Coffey V.G., Hawley J.A. Selective Modulation of MicroRNA Expression with Protein Ingestion Following Concurrent Resistance and Endurance Exercise in Human Skeletal Muscle. Front. Physiol. 2016;7:87. doi: 10.3389/fphys.2016.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Olivieri F., Prattichizzo F., Giuliani A., Matacchione G., Rippo M.R., Sabbatinelli J., Bonafè M. miR-21 and miR-146a: The microRNAs of inflammaging and age-related diseases. Ageing Res. Rev. 2021;70:101374. doi: 10.1016/j.arr.2021.101374. [DOI] [PubMed] [Google Scholar]
- 117.Dimassi S., Karkeni E., Laurant P., Tabka Z., Landrier J.F., Riva C. Microparticle miRNAs as Biomarkers of Vascular Function and Inflammation Response to Aerobic Exercise in Obesity? Obesity. 2018;26:1584–1593. doi: 10.1002/oby.22298. [DOI] [PubMed] [Google Scholar]
- 118.Fiorillo A.A., Heier C.R., Novak J.S., Tully C.B., Brown K.J., Uaesoontrachoon K., Vila M.C., Ngheim P.P., Bello L., Kornegay J.N., et al. TNF-α-Induced microRNAs Control Dystrophin Expression in Becker Muscular Dystrophy. Cell. Rep. 2015;12:1678–1690. doi: 10.1016/j.celrep.2015.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fiorillo A.A., Tully C.B., Damsker J.M., Nagaraju K., Hoffman E.P., Heier C.R. Muscle miRNAome shows suppression of chronic inflammatory miRNAs with both prednisone and vamorolone. Physiol. Genom. 2018;50:735–745. doi: 10.1152/physiolgenomics.00134.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Juźwik C.A., S Drake S., Zhang Y., Paradis-Isler N., Sylvester A., Amar-Zifkin A., Douglas C., Morquette B., Moore C.S., Fournier A.E. microRNA dysregulation in neurodegenerative diseases: A systematic review. Prog. Neurobiol. 2019;182:101664. doi: 10.1016/j.pneurobio.2019.101664. [DOI] [PubMed] [Google Scholar]
- 121.Liu N., Bezprozvannaya S., Shelton J.M., Frisard M.I., Hulver M.W., McMillan R.P., Wu Y., Voelker K.A., Grange R.W., Richardson J.A., et al. Mice lacking microRNA 133a develop dynamin 2–dependent centronuclear myopathy. J. Clin. Investig. 2011;121:3258–3268. doi: 10.1172/JCI46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Morsiani C., Terlecki-Zaniewicz L., Skalicky S., Bacalini M.G., Collura S., Conte M., Sevini F., Garagnani P., Salvioli S., Hackl M., et al. Circulating miR-19a-3p and miR-19b-3p characterize the human aging process and their isomiRs associate with healthy status at extreme ages. Aging Cell. 2021;20:e13409. doi: 10.1111/acel.13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tsukamoto H., Kouwaki T., Oshiumi H. Aging-Associated Extracellular Vesicles Contain Immune Regulatory microRNAs Alleviating Hyperinflammatory State and Immune Dysfunction in the Elderly. Iscience. 2020;23:101520. doi: 10.1016/j.isci.2020.101520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Purohit P.K., Saini N. Mitochondrial microRNA (MitomiRs) in cancer and complex mitochondrial diseases: Current status and future perspectives. Cell. Mol. Life Sci. 2021;78:1405–1421. doi: 10.1007/s00018-020-03670-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jung H.J., Coffinier C., Choe Y., Beigneux A.P., Davies B.S., Yang S.H., Barnes R.H., 2nd, Hong J., Sun T., Pleasure S.J., et al. Regulation of prelamin A but not lamin C by miR-9, a brain-specific microRNA. Proc. Natl. Acad. Sci. USA. 2012;109:E423–E431. doi: 10.1073/pnas.1111780109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kondo M., Yamada H., Munetsuna E., Yamazaki M., Hatta T., Iwahara A., Ohashi K., Ishikawa H., Tsuboi Y., Inoue T., et al. Associations of serum microRNA-20a, -27a, and -103a with cognitive function in a Japanese population: The Yakumo study. Arch. Gerontol. Geriatr. 2019;82:155–160. doi: 10.1016/j.archger.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 127.Bai X., Bian Z. MicroRNA-21 Is a Versatile Regulator and Potential Treatment Target in Central Nervous System Disorders. Front. Mol. Neurosci. 2022;15:842288. doi: 10.3389/fnmol.2022.842288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Weinberg R.B., Mufson E.J., Counts S.E. Evidence for a neuroprotective microRNA pathway in amnestic mild cognitive impairment. Front. Neurosci. 2015;9:430. doi: 10.3389/fnins.2015.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Absalon S., Kochanek D.M., Raghavan V., Krichevsky A.M. MiR-26b, upregulated in Alzheimer’s disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J. Neurosci. 2013;33:14645–14659. doi: 10.1523/JNEUROSCI.1327-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Marttila S., Rovio S., Mishra P.P., Seppälä I., Lyytikäinen L.P., Juonala M., Waldenberger M., Oksala N., Ala-Korpela M., Harville E., et al. Adulthood blood levels of hsa-miR-29b-3p associate with preterm birth and adult metabolic and cognitive health. Sci. Rep. 2021;11:9203. doi: 10.1038/s41598-021-88465-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nagaraj S., Laskowska-Kaszub K., Dębski K.J., Wojsiat J., Dąbrowski M., Gabryelewicz T., Kuźnicki J., Wojda U. Profile of 6 microRNA in blood plasma distinguish early stage Alzheimer’s disease patients from non-demented subjects. Oncotarget. 2017;8:16122–16143. doi: 10.18632/oncotarget.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhao Y., Jaber V.R., LeBeauf A., Sharfman N.M., Lukiw W.J. microRNA-34a (miRNA-34a) Mediated Down-Regulation of the Post-synaptic Cytoskeletal Element SHANK3 in Sporadic Alzheimer’s Disease (AD) Front. Neurol. 2019;10:28. doi: 10.3389/fneur.2019.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.van den Berg M.M.J., Krauskopf J., Ramaekers J.G., Kleinjans J.C.S., Prickaerts J., Briedé J.J. Circulating microRNAs as potential biomarkers for psychiatric and neurodegenerative disorders. Prog. Neurobiol. 2020;185:101732. doi: 10.1016/j.pneurobio.2019.101732. [DOI] [PubMed] [Google Scholar]
- 134.Zhang J., Sun P., Zhou C., Zhang X., Ma F., Xu Y., Hamblin M.H., Yin K.J. Regulatory microRNAs and vascular cognitive impairment and dementia. CNS Neurosci. Ther. 2020;26:1207–1218. doi: 10.1111/cns.13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Han D., Dong X., Zheng D., Nao J. MiR-124 and the Underlying Therapeutic Promise of Neurodegenerative Disorders. Front. Pharmacol. 2020;10:1555. doi: 10.3389/fphar.2019.01555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Banzhaf-Strathmann J., Benito E., May S., Arzberger T., Tahirovic S., Kretzschmar H., Fischer A., Edbauer D. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 2014;33:1667–16680. doi: 10.15252/embj.201387576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sheinerman K.S., Tsivinsky V.G., Crawford F., Mullan M.J., Abdullah L., Umansky S.R. Plasma microRNA biomarkers for detection of mild cognitive impairment. Aging. 2012;4:590–605. doi: 10.18632/aging.100486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hadar A., Milanesi E., Walczak M., Puzianowska-Kuźnicka M., Kuźnicki J., Squassina A., Niola P., Chillotti C., Attems J., Gozes I., et al. SIRT1, miR-132 and miR-212 link human longevity to Alzheimer’s Disease. Sci. Rep. 2018;8:8465. doi: 10.1038/s41598-018-26547-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xie B., Zhou H., Zhang R., Song M., Yu L., Wang L., Liu Z., Zhang Q., Cui D., Wang X., et al. Serum miR-206 and miR-132 as Potential Circulating Biomarkers for Mild Cognitive Impairment. J. Alzheimers Dis. 2015;45:721–731. doi: 10.3233/JAD-142847. [DOI] [PubMed] [Google Scholar]
- 140.Cha D.J., Mengel D., Mustapic M., Liu W., Selkoe D.J., Kapogiannis D., Galasko D., Rissman R.A., Bennett D.A., Walsh D.M. miR-212 and miR-132 Are Downregulated in Neurally Derived Plasma Exosomes of Alzheimer’s Patients. Front. Neurosci. 2019;13:1208. doi: 10.3389/fnins.2019.01208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang X.M., Jia R.H., Wei D., Cui W.Y., Jiang W. MiR-134 blockade prevents status epilepticus like-activity and is neuroprotective in cultured hippocampal neurons. Neurosci. Lett. 2014;572:20–25. doi: 10.1016/j.neulet.2014.04.049. [DOI] [PubMed] [Google Scholar]
- 142.Konovalova J., Gerasymchuk D., Arroyo S.N., Kluske S., Mastroianni F., Pereyra A.V., Domanskyi A. Human-Specific Regulation of Neurotrophic Factors MANF and CDNF by microRNAs. Int. J. Mol. Sci. 2021;22:9691. doi: 10.3390/ijms22189691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang T.T., Liu C.G., Gao S.C., Zhang Y., Wang P.C. The Serum Exosome Derived MicroRNA-135a, -193b, and -384 Were Potential Alzheimer’s Disease Biomarkers. Biomed. Environ. Sci. 2018;31:87–96. doi: 10.3967/bes2018.011. [DOI] [PubMed] [Google Scholar]
- 144.Siegert S., Seo J., Kwon E.J., Rudenko A., Cho S., Wang W., Flood Z., Martorell A.J., Ericsson M., Mungenast A.E., et al. The schizophrenia risk gene product miR-137 alters presynaptic plasticity. Nat. Neurosci. 2015;18:1008–1016. doi: 10.1038/nn.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schröder J., Ansaloni S., Schilling M., Liu T., Radke J., Jaedicke M., Schjeide B.M., Mashychev A., Tegeler C., Radbruch H., et al. MicroRNA-138 is a potential regulator of memory performance in humans. Front. Hum. Neurosci. 2014;8:501. doi: 10.3389/fnhum.2014.00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gullett J.M., Chen Z., O’Shea A., Akbar M., Bian J., Rani A., Porges E.C., Foster T.C., Woods A.J., Modave F., et al. MicroRNA predicts cognitive performance in healthy older adults. Neurobiol. Aging. 2020;95:186–194. doi: 10.1016/j.neurobiolaging.2020.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wei W., Wang Z.Y., Ma L.N., Zhang T.T., Cao Y., Li H. MicroRNAs in Alzheimer’s Disease: Function and Potential Applications as Diagnostic Biomarkers. Front. Mol. Neurosci. 2020;13:160. doi: 10.3389/fnmol.2020.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ansari A., Maffioletti E., Milanesi E., Marizzoni M., Frisoni G.B., Blin O., Richardson J.C., Bordet R., Forloni G., Gennarelli M., et al. miR-146a and miR-181a are involved in the progression of mild cognitive impairment to Alzheimer’s disease. Neurobiol. Aging. 2019;82:102–109. doi: 10.1016/j.neurobiolaging.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 149.Satoh J., Kino Y., Niida S. MicroRNA-Seq Data Analysis Pipeline to Identify Blood Biomarkers for Alzheimer’s Disease from Public Data. Biomark. Insights. 2015;10:21–31. doi: 10.4137/BMI.S25132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu C.G., Song J., Zhang Y.Q., Wang P.C. MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer’s disease. Mol. Med. Rep. 2014;10:2395–2400. doi: 10.3892/mmr.2014.2484. [DOI] [PubMed] [Google Scholar]
- 151.Kaalund S.S., Venø M.T., Bak M., Møller R.S., Laursen H., Madsen F., Broholm H., Quistorff B., Uldall P., Tommerup N., et al. Aberrant expression of miR-218 and miR-204 in human mesial temporal lobe epilepsy and hippocampal sclerosis-convergence on axonal guidance. Epilepsia. 2014;55:2017–2027. doi: 10.1111/epi.12839. [DOI] [PubMed] [Google Scholar]
- 152.Chiu C.C., Yeh T.H., Chen R.S., Chen H.C., Huang Y.Z., Weng Y.H., Cheng Y.C., Liu Y.C., Cheng A.J., Lu Y.C., et al. Upregulated Expression of MicroRNA-204-5p Leads to the Death of Dopaminergic Cells by Targeting DYRK1A-Mediated Apoptotic Signaling Cascade. Front. Cell Neurosci. 2019;13:399. doi: 10.3389/fncel.2019.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhu Y., Li C., Sun A., Wang Y., Zhou S. Quantification of microRNA-210 in the cerebrospinal fluid and serum: Implications for Alzheimer’s disease. Exp. Ther. Med. 2015;9:1013–1017. doi: 10.3892/etm.2015.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hara N., Kikuchi M., Miyashita A., Hatsuta H., Saito Y., Kasuga K., Murayama S., Ikeuchi T., Kuwano R. Serum microRNA miR-501-3p as a potential biomarker related to the progression of Alzheimer’s disease. Acta Neuropathol. Commun. 2017;5:10. doi: 10.1186/s40478-017-0414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Abuelezz N.Z., Nasr F.E., AbdulKader M.A., Bassiouny A.R., Zaky A. MicroRNAs as Potential Orchestrators of Alzheimer’s Disease-Related Pathologies: Insights on Current Status and Future Possibilities. Front. Aging Neurosci. 2021;13:743573. doi: 10.3389/fnagi.2021.743573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Akhtar S., Patnaik S.R., Kotapati Raghupathy R., Al-Mubrad T.M., Craft J.A., Shu X. Histological Characterization of the Dicer1 Mutant Zebrafish Retina. J. Ophthalmol. 2015;2015:309510. doi: 10.1155/2015/309510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fiorenza A., Lopez-Atalaya J.P., Rovira V., Scandaglia M., Geijo-Barrientos E., Barco A. Blocking miRNA Biogenesis in Adult Forebrain Neurons Enhances Seizure Susceptibility, Fear Memory, and Food Intake by Increasing Neuronal Responsiveness. Cereb. Cortex. 2016;26:1619–1633. doi: 10.1093/cercor/bhu332. [DOI] [PubMed] [Google Scholar]
- 158.Sundermeier T.R., Sakami S., Sahu B., Howell S.J., Gao S., Dong Z., Golczak M., Maeda A., Palczewski K. MicroRNA-processing Enzymes Are Essential for Survival and Function of Mature Retinal Pigmented Epithelial Cells in Mice. J. Biol. Chem. 2017;292:3366–3378. doi: 10.1074/jbc.M116.770024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Smith P.Y., Hernandez-Rapp J., Jolivette F., Lecours C., Bisht K., Goupil C., Dorval V., Parsi S., Morin F., Planel E., et al. miR-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum. Mol. Genet. 2015;24:6721–6735. doi: 10.1093/hmg/ddv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Salta E., Sierksma A., Vanden Eynden E., De Strooper B. miR-132 loss de-represses ITPKB and aggravates amyloid and TAU pathology in Alzheimer’s brain. EMBO Mol. Med. 2016;8:1005–1018. doi: 10.15252/emmm.201606520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lau P., Bossers K., Janky R., Salta E., Frigerio C.S., Barbash S., Rothman R., Sierksma A.S., Thathiah A., Greenberg D., et al. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol. Med. 2013;5:1613–1634. doi: 10.1002/emmm.201201974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Soreq H., Wolf Y. NeurimmiRs: microRNAs in the neuroimmune interface. Trends Mol. Med. 2011;17:548–555. doi: 10.1016/j.molmed.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 163.Muthusami S., Vidya B., Shankar E.M., Vadivelu J., Ramachandran I., Stanley J.A., Selvamurugan N. The Functional Significance of Endocrine-immune Interactions in Health and Disease. Curr. Protein. Pept. Sci. 2020;21:52–65. doi: 10.2174/1389203720666191106113435. [DOI] [PubMed] [Google Scholar]
- 164.Licursi V., Conte F., Fiscon G., Paci P. MIENTURNET: An interactive web tool for microRNA-target enrichment and networkbased analysis. BMC Bioinform. 2019;20:1–10. doi: 10.1186/s12859-019-3105-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Park H.H., Hwang W., Ham S., Kim E., Altintas O., Park S., Son H.G., Lee Y., Lee D., Heo W.D., et al. A PTEN variant uncouples longevity from impaired fitness in Caenorhabditis elegans with reduced insulin/IGF-1 signaling. Nat. Commun. 2021;12:5631. doi: 10.1038/s41467-021-25920-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kirstein A.S., Kehr S., Nebe M., Hanschkow M., Barth L., Lorenz J., Penke M., Breitfeld J., Le Duc D., Landgraf K., et al. PTEN regulates adipose progenitor cell growth, differentiation, and replicative aging. J. Biol. Chem. 2021;297:100968. doi: 10.1016/j.jbc.2021.100968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li Y.Z., Di Cristofano A., Woo M. Metabolic Role of PTEN in Insulin Signaling and Resistance. Cold Spring Harb. Perspect. Med. 2020;10:a036137. doi: 10.1101/cshperspect.a036137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ling H.Y., Hu B., Hu X.B., Zhong J., Feng S.D., Qin L., Liu G., Wen G.B., Liao D.F. MiRNA-21 reverses high glucose and high insulin induced insulin resistance in 3T3-L1 adipocytes through targeting phosphatase and tensin homologue. Exp. Clin. Endocrinol. Diabetes. 2012;120:553–559. doi: 10.1055/s-0032-1311644. [DOI] [PubMed] [Google Scholar]
- 169.Yuan R., Zhang S., Yu J., Huang Y., Lu D., Cheng R., Huang S., Ao P., Zheng S., Hood L., et al. Beyond cancer genes: Colorectal cancer as robust intrinsic states formed by molecular interactions. Open Biol. 2017;7:170169. doi: 10.1098/rsob.170169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Iliopoulos D., Jaeger S.A., Hirsch H.A., Bulyk M.L., Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nowak D.G., Cho H., Herzka T., Watrud K., DeMarco D.V., Wang V.M., Senturk S., Fellmann C., Ding D., Beinortas T., et al. MYC Drives Pten/Trp53-Deficient Proliferation and Metastasis due to IL6 Secretion and AKT Suppression via PHLPP2. Cancer Discov. 2015;5:636–651. doi: 10.1158/2159-8290.CD-14-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yan B., Zhu C.D., Guo J.T., Zhao L.H., Zhao J.L. miR-206 regulates the growth of the teleost tilapia (Oreochromis niloticus) through the modulation of IGF-1 gene expression. J. Exp. Biol. 2013;216:1265–1269. doi: 10.1242/jeb.079590. [DOI] [PubMed] [Google Scholar]
- 173.Huang X., Gong F., Lu Z., Zhu J., Yu Q. Downregulated MiR-206 expression promotes the proliferation and migration of macrophages by regulating IL-17A/REG3A pathway. Eur. J. Inflamm. 2020;18:2058739220917490. doi: 10.1177/2058739220917490. [DOI] [Google Scholar]
- 174.Chai X., Si H., Song J., Chong Y., Wang J., Zhao G. miR-486-5p Inhibits Inflammatory Response, Matrix Degradation and Apoptosis of Nucleus Pulposus Cells through Directly Targeting FOXO1 in Intervertebral Disc Degeneration. Cell Physiol. Biochem. 2019;52:109–118. doi: 10.33594/000000008. [DOI] [PubMed] [Google Scholar]
- 175.Kim Y.J., Hwang S.H., Lee S.Y., Shin K.K., Cho H.H., Bae Y.C., Jung J.S. miR-486-5p induces replicative senescence of human adipose tissue-derived mesenchymal stem cells and its expression is controlled by high glucose. Stem Cells Dev. 2012;21:1749–1760. doi: 10.1089/scd.2011.0429. [DOI] [PubMed] [Google Scholar]
- 176.Ma L., Niu H., Sha G., Zhang Y., Liu P., Li Y. Serum SIRT1 Is Associated with Frailty and Adipokines in Older Adults. J. Nutr. Health Aging. 2019;23:246–250. doi: 10.1007/s12603-018-1149-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.