Abstract
Over time, molecular biology and genomics techniques have been developed to speed up the early diagnosis and clinical management of cancer. These therapies are often most effective when administered to the subset of malignancies harboring the target identified by molecular testing. Important advances in applying molecular testing involve circulating-free DNA (cfDNA)- and cell-free RNA (cfRNA)-based liquid biopsies for the diagnosis, prognosis, prediction, and treatment of cancer. Both cfDNA and cfRNA are sensitive and specific biomarkers for cancer detection, which have been clinically proven through multiple randomized and prospective trials. These help in cancer management based on the noninvasive evaluation of size, quantity, and point mutations, as well as copy number alterations at the tumor site. Moreover, personalized detection of ctDNA helps in adjuvant therapeutics and predicts the chances of recurrence of cancer and resistance to cancer therapy. Despite the controversial diagnostic values of cfDNA and cfRNA, many clinical trials have been completed, and the Food and Drug Administration has approved many multigene assays to detect genetic alterations in the cfDNA of cancer patients. In this review, we underpin the recent advances in the physiological roles of cfDNA and cfRNA, as well as their roles in cancer detection by highlighting recent clinical trials and their roles as prognostic and predictive markers in cancer management.
Keywords: liquid biopsy, cancer, anticancer therapy, diagnosis, cfRNA, cfDNA, biomarkers
1. Introduction
Cancer is a complex and multifactorial disease with debilitating effects, orchestrated by a plethora of genetic and environmental factors and associated with serious comorbidities. As cancer is one of the leading causes of death after cardiovascular diseases [1], many researchers have spent their time, efforts, and resources on finding new cancer management solutions to reduce the mortality rate; ergo, the cancer death rate decreased by 32% between 1991 and 2019. Many efforts are focused on the early diagnosis and prediction of reoccurrence of disease; for that purpose, tissue biopsies and imaging-based technologies have been introduced. Liquid biopsies (mainly cell-free DNA and RNA) are more desirable and attractive options due to their non-invasiveness, easiness, and effectiveness.
Cell-free DNA (cfDNA) and cell-free RNA (cfRNA) are, respectively, fragmented DNA and RNA that move freely in body fluids (rather than being encapsulated inside the cells). cfDNA gained global recognition in the 1960s when it was hypothesized that it is somehow related to the metastasis of cancer; however, it was first reported in human blood by Mandel and Metais in 1948 [2]. In 1965, the relationship between cancer and cfDNA was hypothesized [3]; a year later, high levels of cfDNA were reported in systemic lupus erythematosus disease (SLE) [4]. The higher levels of cfDNA were reported in 1977 when the radioimmunoassay technique was used to compare the quantities of cfDNA of cancer patients and normal persons [5]. In 1989, the presence of cfDNA was detected in the plasma of cancer patients. After that, the presence of cfDNA was proven through various studies; scientists discovered that this DNA has unique mutations and epigenetic alternations. In the 1990s, the Human Genome Project demonstrated that cfDNA has tumor-specific mutations. Most of the cancer research was focused on cfDNA as compared to cfRNA because of its stability and detectability; however, numerous studies have speculated that cfRNA may have more potential compared to cfDNA [6,7,8].
Along with multiple types of cfDNA and cfRNA, various other entities are released by tumor cells, including circulating tumor cells (CTCs) and tissue DNA (tDNA). The existence of CTCs has been known since 1869 as these are commonly present in cancer patients. In 2007, CTCs were enlisted as biomarkers for cancer diagnoses; multiple kits are available on the market. However, CTCs are heterogenous seeds of cancer, and one in a million CTCs have a chance of developing cancer. CTCs are diagnosed by digital PCR, RNA sequencing, next generation sequencing (NGS), mass cytometry, and CellSearch; nonetheless, rare and low frequencies of tumor-causing CTCs make them difficult options for diagnoses [9]. Detection of tissue DNA (tDNA) was an attractive option a few years ago; however, its invasive nature, low frequency, tissue heterogeneity, and inaccuracy due to sampling locations resulted in scientists moving away from this option [10,11]; scientists have since developed noninvasive liquid biopsies, which detect cell-free DNA in blood and plasma [12]. There are three types of cell-free DNA, as explained below.
-
⯀
Circulating tumor DNA
-
⯀
Cell-Free fetal DNA
-
⯀
Cell-Free mitochondrial DNA
All types of cfDNA have their importance and applications which are explained in Table 1. The unique genetic makeup of cfDNA has given it multiple applications in diagnoses and therapeutics. For example, fetal cfDNA was discovered in 1997 in maternal circulation; since then, it has been used to monitor the health, gender, and genetic disorder of the fetus, especially Down’s syndrome [13]. Furthermore, mitochondrial cfDNA has application in diagnosing cardiovascular diseases [14], diabetes, acute myocardial infarction, cancer, and internal body trauma [15]. Finally, tumor cfDNA is an important diagnostic marker for cancer [16] and SLA diagnosis. This paper focuses on cell-free tumor DNA and cfRNA as diagnostic and prognostic markers for different types of cancer.
Table 1.
Comparison of various types of cell-free DNA.
| Sr number | Properties | Circulating Tumor DNA | Cell-Free Fetal DNA | Cell-Free Mitochondrial DNA |
|---|---|---|---|---|
| 1 | Strands | Single or double | Single or double | Double |
| 2 | Origin | Tumor cells | Trophoblastic cells [13] | Mitochondria or unknown [17] |
| 3 | Size | Less than 100 bp [18] | 200 bp with dominate peaks at 162 bp [19] | Shorter fragment = less than 1 kb; larger fragments = 21 kb [14] |
| 4 | Applications | Early cancer detection, mutation analysis, cancer prediction, noninvasive cancer detection | Prenatal testing, genetic disease detection in the fetus | Forensic sciences, detection of the geographical distribution of genes, gene flow identification, human remain recognition, cancer detection |
| 5 | Advantages | Sensitive than other cancer detection techniques, can detect mutations better than biopsy, detect heterogeneous tumor cells, predict cancer reoccurrence | Increased chances to detect chromosomal disorders, noninvasive, no side effects | Lacks genetic ambiguities, higher copy number, a diagnostic and prognostic marker for multiple diseases |
| 6 | Disadvantages | Cannot be detected by FISH or ICC techniques, expensive, lack standardization | Increased chances of false positives or false negatives | No heterogenicity, lower discrimination power |
In this review paper, we first discuss and summarize the secretion mechanisms and characteristics of cfDNA and cfRNA. Afterward, both are discussed as diagnostic and prognostic biomarkers for cancer. In addition, their applications in minimal residual disease detection are explained in detail. We believe that a thorough understanding of the size profiles, quantitative measurements, and analyses of genetic repeats in cfDNA and cfRNA could assure reliable results in clinical practice for cancer diagnoses and therapeutics. At the end of the paper, the roles of cfDNA and cfRNA in cancer therapeutics are discussed.
2. Secretion
The biological and molecular analyses of cancer cells reveal the release of intracell entities, such as proteins, vesicles, nucleic acids, and many others from cancer cells. Among these entities, cfDNA and cfRNA are the most notable due to their potential to improve cancer management. These are secreted by multiple methods using active and dead cells by using various pathways, which are explained here.
2.1. Secretion of cfDNA
Although cfDNA was discovered more than 80 years ago, its molecular origins and secretion mechanisms are poorly understood. Several pathways and sources have been reported in the literature to explain its origin. A few major sources of cfDNA in the blood include apoptosis, necrosis, NETosis, and extracellular vesicles. The details of the secretion of cfDNA are reviewed in this paper. Apoptosis was the first source of cfDNA that was reported by Wyllie et al. [20] and similarities were found between cfDNA and DNA extracted from apoptotic cells leading to the conclusion that there was indeed a connection between apoptosis and cfDNA. During apoptosis, the DNA molecules were usually cleaved into 50−300 kb size fragments, which were further degraded into 167 bp long nucleosome units. Furthermore, Stroun et al. reported that lymphocytes and cultured cell lines, including HL-60, spontaneously release a nucleoprotein complex within a homeostatic system and preferentially release newly-synthesized DNA [21].
Necrosis is another source of cfDNA in the blood and generates larger fragments of cfDNA. The size of cfDNA from the necrotic origin is typically ~10k bp [22]. Therefore, necrotic cfDNA is not seen as commonly as apoptotic cfDNA in the bloodstream [23]. The post-translational modifications are responsible for histone citrullination in NETosis, which causes chromatin de-condensation and cell death. Evidence of a relationship between cfDNA and NETosis has been found in many studies [24,25]. In one study, increased levels of cfDNA were found in septic patients and indicated a correlation of NETosis with cfDNA secretion [26]. Another important source of cfDNA is extracellular vehicles (EVs); several studies have demonstrated that the uptake of RNA and DNA is done specifically as genetic material, with specific sequences and properties only being found inside EVs. The apoptotic EVs are formed due to programmed cell death and membrane blebbing and are then released from the apoptotic bodies containing fragments of degraded DNA, including cfDNA [27]. The stability of DNA is increased by being packaged inside EVs, resulting in protection from the external environment and nucleases. It also prevents recognition by the immune system. Cancer cells generate tumor EVs, which carry mtDNA, tumor DNA, and other mutated genetic material [28]. These cells tell the status of genetic mutation and the level of amplification of oncogenes, i.e., c-Myc [29]. These EVs are responsible for the transfer of encapsulated DNA to the target fibroblasts. Each type of EV encapsulates DNA with a specific type of mutation; therefore, cfDNA secreted from EVs can be used as a potential biomarker for cancer diagnosis [30].
Lastly, pyroptosis (caspase 1-dependent cell death) is an important source of cfDNA, which occurs as a result of multiple stimuli, such as caspase activation, an immune-inflammatory reaction, or microbial stimulus [31]. Upon stimulation, pores that are 2.5 > micrometer size are formed, causing the inward flux of water and ions resulting in cell swelling and larger sizes. Proinflammatory substances, such as IL-1β and IL-18, are released into the bloodstream, triggering inflammatory and autoimmune responses. One study demonstrated that chronic inflammation activation helped prevent tumors and cancer [32]. The secretion mechanism of cfDNA is explained in Figure 1. The cfDNA was released along with other components of the cell after the membrane pore formation. Membrane lysis is not required in pyroptosis; instead, pores are enough to transport the material into the cell surroundings [33].
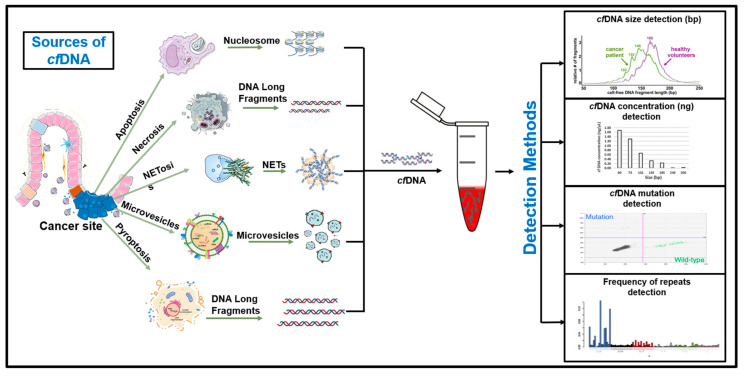
Figure 1.
Secretion of cfDNA from tumor cells. Cell death by apoptosis, pyroptosis, or necrosis is one of the most important sources of ctDNA in body fluids; however, even without cell death, cfDNA/ctDNA has been found in the medium. This means that live cells can actively release cfDNA. Due to autophagy and exosome activity, the active secretion of cfDNA through microvesicles has also been observed. Once the cfDNA is released into the body fluids, it is detected by various methods, based on its size, concentration, frequency of repeats, or presence of mutations.
2.2. Secretion of cfRNA
The presence of cfRNA has been confirmed in various body fluids, including blood, plasma, serum, and saliva. There are three main sources of cfRNA in the blood, as described in Figure 2.
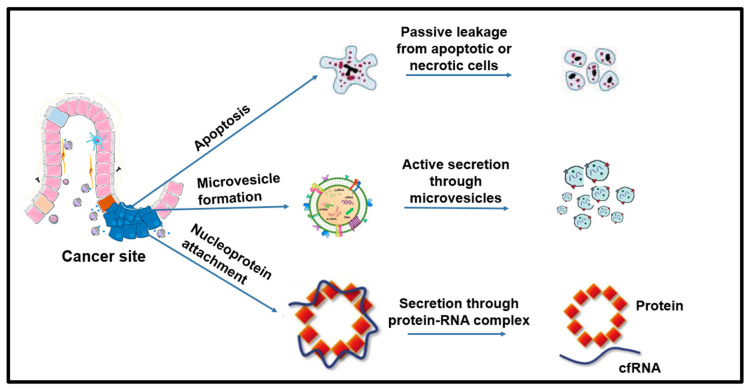
Figure 2.
Sources of cfRNA. Similar to ctDNA, it is secreted into the body fluids through cell death events or by attaching itself to the nuclear proteins. The presence of cfRNA plasma can reflect the phenotypic alterations of localized sites of cancer as well as a systemic host response.
-
⯀
Passive leakage from the tumor or apoptotic or necrotic cells;
-
⯀
Active secretion through microvesicles;
-
⯀
Secretion through nucleoproteins or protein-RNA complex.
The cfRNA is relatively unstable and easily degrades; therefore, it has to be packaged inside other molecular entities for their transport. Predominantly, extracellular membrane vesicles, exosomes, and microvesicles are used as transport vehicles; furthermore, nucleoproteins bind cfRNA and carry it outside the cell. The use of a vehicle depends on the origin of the cfRNA. For example, after programmed cell death, cfRNA is encapsulated inside extracellular membrane vesicles (EMVs) used as the delivery system [34]. The microvesicles are excellent cellular garbage disposals that transport bioactive cellular components from the cell. Their cargo also contains cfRNA along with other mRNAs, non-coding RNA, and other cellular entities [35]. Lastly, the attachment of cfRNA with nucleoproteins or high-density lipoproteins ensures the secretion into the bloodstream without any damage or degradation.
3. Discerning cfDNA and cfDRNA Molecular Alterations in Clinical Management
Different studies have proven that the genetic makeups of cfDNA and cfRNA of a healthy person and a cancer patient are different due to multiple mutations resulting in different genetic makeups, copy numbers, or repetitions, making them suitable candidates for diagnoses and therapeutics. Multiple techniques, including digital PCR, NGS, and genome-wide sequencing, are used to detect the molecular alterations in the cfDNA and cfRNA of cancer patients. These molecular alterations are mostly somatic, and several studies have proven the presence of mutations in RAS, Wnt, Hippo, Nrf2, TGFβ, PI3K, Notch, and P53 genes. C.H. Brian et al. studied 73 B-cell lymphoma and follicular lymphoma patients and observed lymphomagenesis. The study proved that both B-cell lymphoma and follicular lymphoma patients have different profiles of 5-hydroxymethylcytosines of cfDNA, which can be used to diagnose cancer. This study proposed using 5hmC-Seal as a detection tool, as it is an ultrasensitive method requiring only a 5 mL blood sample and nanograms of cfDNA [36]. M. Schwaederle et al. used the NGS method to detect the molecular alterations in the ctDNA of 670 patients. The results demonstrated that at least 63% of patients had one mutation. The most common mutation was present in TP53, which was found in 33% of patients. Other common mutations were found in EGFR, KRAS, and PIK3CA. The authors demonstrated the potential utility and feasibility of ctDNA in precision medicine for cancer treatment [37].
Through multiple studies, it has been established that ctDNA and cfRNA harbor multiple genomic alterations specific to the original tumor cells. Many commercially available kits detect these specific genomic alterations and mutations, i.e., the Cobas EGFR Mutation Test v2 detects mutations in EGFR. Cancer, a heterogeneous and diverse disease, tends to develop subclonal mutations, leading to tumor resistance development. A myriad of studies has found that mutations in KRAS, RAS, and EGFR T790M, and the amplification of the MET protooncogene, give rise to resistance against anti-EGFR therapy. Such mutations also alter the expressions of several oncogenic genes, such as KRAS, TP53, PKC epsilon, Akt, and several others [38,39]. These mutations are present in ctDNA and can be utilized to monitor the emergence of therapy resistance. In a clinical trial, Safe-SeqS was used to monitor the therapy resistance in the ctDNA of 42 patients after receiving the treatment and identified the emergence of KRAS mutations in 23 patients, which led to the conclusion that rather than immediately preceding surgery, neoadjuvant therapy strategies should be used as the first treatment step in cancer management.
In the case of cfRNA, the quantity is increased by 20Xin a cancer patient as compared to a healthy person. In patients with lung cancer, PD-L1 levels are used to monitor the response of therapy on the patients [40]. In addition, molecular expressions of RNA were used for the pathological staging and measurement of recurrence of colon cancer [41].
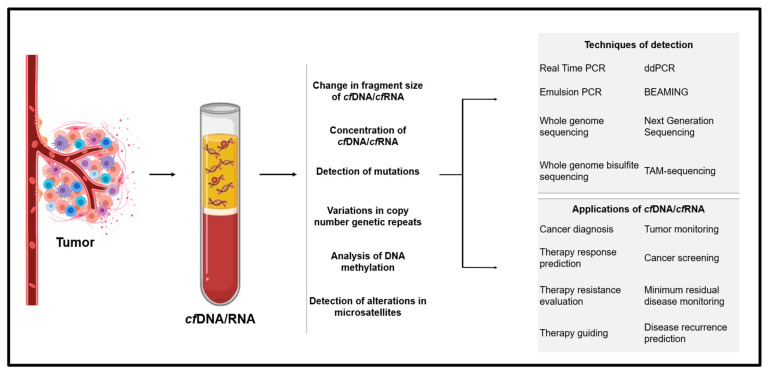
Overall, cfDNA and cfRNA have shown molecular alterations in their genetic makeups, which scientists can capture and utilize for diagnosis and treatment purposes. The details of molecular alterations, techniques that could detect them, and their applications are explained in Figure 3.
Figure 3.
For cell-free DNA or RNA screening, the blood sample is taken from the patient and analyzed through techniques, such as qPCR, NGS WGS, etc. The non-invasiveness of a liquid biopsy makes it suitable for a myriad of applications, including cancer diagnoses, tumor burden analyses, and therapeutic analyses.
4. Applications of cfDNA and cfRNA
Due to the lethal (and high) cancer mortality rates, scientists have been encouraged to discover new and efficient methods for cancer detection and therapeutics. Tissue biopsies were the preferred method for cancer detection; however, they involve invasive procedures and some cancerous sites are (sometimes) unable to be reached. Therefore, scientists have identified and verified cfDNA, cfRNA, and ctDNA as diagnostic markers for cancer. Furthermore, due to their molecular heterogeneities, they have various clinical applications as they are used as prognostic markers, measure residual disease, evaluate the treatment responses, and identify molecular alterations. The details of various applications of cfDNA and cfRNA are explained here.
4.1. Role of cfDNA and cfRNA in Cancer Diagnosis
Due to the strong responsiveness and extreme dynamic properties of cfDNA and cfRNA, they are considered excellent predictors and indicators of tumor cells and DNA damage; hence, both have the potential to be used in cancer diagnostics at the commercial level.
4.1.1. Role of cfDNA
cfDNA was discovered several decades ago; however, significant progress was observed after introducing NGS. In 1994, the presence of N-Ras mutations in the DNA of acute myelogenous leukemia patients [42] and K-ras sequences in pancreatic adenocarcinoma patients was reported [43]. These studies proved the elevated levels of mutations in DNA sequences of plasma, proving the presence of ctDNA. The concentration of ctDNA is negligible in an average person—it cannot be detected using typical sequencing methods; however, a cancer patient has a higher concentration, making it a potential noninvasive approach for cancer detection.
Not surprisingly, there is significant interest in the potential utility of ctDNA for the early noninvasive detection of cancer, with over USD 1 billion invested in companies developing such technologies in 2017 alone. Advanced PCR techniques, such as the amplification refractory mutation system (ARMS)-PCR, BEAMing (Beads, Emulsions, Amplification, and Magnetics), or droplet digital PCR (ddPCR) are utilized to identify ctDNA from the pool of normal cfDNA. The diagnostics of ctDNA are based on size, length, the presence of repeats, and mutations in a normal sequence. Advanced PCR techniques, such as an amplification refractory mutation system (ARMS)-PCR, BEAMing (Beads, Emulsions, Amplification, and Magnetics), or droplet digital PCR (ddPCR) are utilized to identify ctDNA from a pool of normal cfDNA. The diagnostics of ctDNA are based on size, length, the presence of repeats, and mutations in a normal sequence.
In the literature, the diagnoses of different types of cancer have been reported. In this regard, Panagopoulou et al. provided clinical evidence on the use of cfDNA as a diagnostic and prognostic marker for metastatic breast cancer [44]. In addition, Klein et al. and Rossi et al. corroborated the use of ctDNA as a potential prognostic and diagnostic marker for breast cancer [45,46,47,48]. Rossi et al. utilized the Guardant360 kit to analyze the cfDNA and Kaplan–Meier curves for the survival analysis of 90 patients. The results indicated that the average ctDNA fraction was 4.5% and commonly mutated genes were TP53, PIK3CA, and ERBB2. Additionally, in the baseline sample, the Fisher exact test found a strong association between the number of alterations and cfDNA percentage. In another study, Braicu et al. proposed ctDNA as a promising tool for the detection of ovarian cancer by providing clinical insights [49]. Moreover, the effectiveness and sensitivity of cfRNA for the detection of ovarian cancer were reported by Hannan et al. and Hulstaert et al. [50,51]. Powles et al. suggested the use of cfDNA with adjuvant atezolizumab to guide the immunotherapy in urothelial carcinoma patients [52]. Through various studies, clinical evidence of cfDNA as a potential diagnostic marker has been reported for lung cancer [53,54], pancreatic cancer [55,56], melanoma [57], B-cell lymphoma [58], and colorectal cancer [59,60,61]. Rizzo et al. reported the use of cfDNA for the diagnosis of biliary tract cancer [62]. Due to the presence of clinical evidence, cfDNA may be a potential biomarker for the detection of early–advanced-stage cancer. Different detection techniques are explained in Table 2.
Table 2.
Techniques for the detection of ctDNA and cfRNA.
| Sr number | Technique Name | Description | Sensitivity (Lower Limit of Detection) |
Specificity | Limitations | Cancer Type |
|---|---|---|---|---|---|---|
| 1 | Quantitative polymerase chain reaction | Amplifies the genes in real-time | 0.01–0.1% | 90% for breast cancer [70] | Needs standard, prone to errors, primer design depends on results | Non-small cell lung cancer, breast cancer |
| 2 | Droplet digital polymerase chain reaction | Water–oil emulsion droplet technology-based PCR | 0.01–0.1% | 100% [71] | Lower quantification, loss of linearity at a high concentration | Lung adenocarcinomas, squamous cell carcinoma, neck cancer, breast cancer, gastric cancer, and others |
| 3 | Beads, emulsion, amplification, and magnetics | Combination of emulsion PCR and flow cytometry ultrasensitive technique | <0.1% | ---- | Single mutation per test; lacks standard data | Blood cancer, colorectal cancer |
| 4 | Cancer personalized profiling by deep sequencing | NGS-based method for ctDNA detection | 0.01%–2.0% | 96% [72] | For selected alterations across targeted regions | Cervical squamous cancer, bladder cancer, esophageal, lung cancer |
| 5 | Whole-genome sequencing | Analysis of the whole genome | 2 × 10−3 [73], up to 10−5 shown by different studies [74]. | 98.4% for somatic SNV and indels [75] | Expensive, comparatively low sensitivity and specificity, large amounts of data | Gastric cancer, pancreatic cancer, breast cancer |
| 6 | TAm-Sequencing | Identify low-frequency mutations in ctDNA | 2% | 97% for ovarian cancer [76] | Less comprehensive | Breast cancer, hepatocellular carcinoma |
| 7 | Whole exome sequencing | A sequencing-based technique to study protein-coding regions in the genome | Generally 5% [77], some studies show 50% [78] | 99.9% [79] | Rare variants affect the sensitivity, restricted only to exon regions | Metastatic melanoma, multiple myeloma |
| 8 | Whole-genome bisulfite sequencing | NGS-based technique to find out the methylation status of cytosine | --- | 99% [80] | It is difficult to differentiate between substitutions and epiallele changes; single reference genomes are not enough to discriminate the changes. It is expensive and generates a large amount of data [81] | Breast cancer, prostate cancer |
4.1.2. Role of cfRNA
Elevated levels of cfRNA have been found in the blood of cancer patients; it can potentially be used as a non-invasive biomarker of cancer detection [63]. In 1999, Lo et al. reported on the elevated levels of cfRNA in nasopharyngeal carcinoma patients (~105 to 106 copies per cell) and suggested that it may be used as a diagnostic marker for nasopharyngeal carcinoma [64]. Cancer patients can have organ-specific transcripts of RNA, which could be detected to confirm the presence of cancer. RNA sequencing is another popular method that is used to study and research RNA. In a study, Larson et al. used transcriptome-wide characterization of cfRNA to identify breast cancer; they studied 57,820 genes and compared them with healthy humans. The results showed that 68% of genes present in cancer patients were different from healthy humans [65], which may be used for cancer diagnosis. Recently, Ring et al. used an epitope-independent approach for the isolation of whole transcriptome RNA-Seq of circulating tumor cells and proposed it to be used in the biopsies of macrometastases [47]. The genes associated with cancer were highly expressive in tumor cells indicating that the analysis of cfRNA can help in cancer discovery and understanding.
Various studies have advocated for the presence of mutations and different genetic information in the cfRNA of cancer patients when compared to healthy patients. Hieter et al. performed plasma cfRNA-sequencing to use cfRNA as a biomarker for hepatocellular carcinoma and multiple myeloma, showing that cfRNA has different expressions throughout the cancer progression stages; therefore, it is possible to identify the precancerous and cancerous conditions of patients through the cfRNA-based analysis [66]. The diagnosis of cancer through cfRNA can also be conducted by using a urine analysis; Kim et al. reported that the urine of bladder cancer patients has higher levels of cfRNA compared to healthy persons. Multiple studies have shown the effective diagnostic capabilities of cfRNA in different cancer types, including breast cancer [67], colorectal cancer [68], lung cancer [69], and many others.
4.1.3. Challenges in cfDNA and cfRNA Diagnoses
Due to the small quantities of both ctDNA and cfRNA, early cancer detection is filled with cautionary tales that highlight the challenges. For example, one study [82] proposed that the fundamental limitations for such a ctDNA-based early detection test, beyond the current state-of-the-art, require around 100x more sequencing bandwidth and improved variant interpretation. Subsequently, Phallen et al., in the most comprehensive study of early-stage cancers to date, reported the detection of somatic alterations in 50–75% of patients depending on histology [83].
A positive relationship between tumor burden and cfDNA has been found by Valpione et al. and Xu et al. [84,85]; however, a few studies suggested otherwise [86]. In the case of cfRNA, only a few nanograms are available for detection, which may not be enough for most detection techniques. cfDNA and cfRNA represent complex mechanisms of cancer biology and their relationship with tumor burden have yet to be fully explored, which is a challenge for cancer diagnosis.
False negatives and false positives are major challenges that arise during ctDNA detection. The major reasons for false negative results are the low signal-to-noise ratio, tumor destruction, and short half-life of ctDNA. A low signal-to-noise ratio is an important challenge that arises in ctDNA detection, especially in early cancer detection. When cancer is benign or just starting to metastasize, ctDNA represents a tiny percentage of cell-free DNA, perhaps less than 0.01% of a 5 mL sample of plasma. At such low rates, ctDNA detections will need to improve significantly and be specialized to reduce the chance of error. A similar situation is true for cfRNA because blood contains mRNA, miRNA, and RNA fragments, which act as noise and contribute to the false detection of RNA [69]. In addition, the shorter half-life of ctDNA and cfRNA is another challenge that causes a range of problems, from tracking tumor heterogeneity to precision treatment.
Clonal hematopoiesis is one of the factors that negatively affect ctDNA detection. In this process, somatic mutations in hematopoietic stem cells accumulate, which disguise themselves as tumor mutations and create biological noise in cancer detection. In this regard, Liu et al. and Razavi et al. provided clinical evidence of the prevalence of clonal hematopoiesis mutations in cfDNA, which can affect the detection of tumor mutations [87,88]. To avoid this noise, different models of high coverage depths and ultralow error rates for NGS have been presented; however, for now, these are not accessible due to their high costs and difficult approaches.
4.2. cfDNA and cfRNA Roles as Prognostic Markers
In recent years, anticancer therapies have explored tumor biology-driven therapeutics, mainly focusing on prognostic and predictive biomarkers of cancer to improve cancer therapeutics. The prognostic biomarkers objectively evaluate the future outcomes of the treatment plans for specific patients and the chances of cancer reoccurrence. This helps select the cancer patients that can benefit from specific treatment plans. Both cfDNA and cfRNA are considered prognostic markers for various cancer types.
4.2.1. cfDNA
Fernandez-Garcia et al. compared the conventional biomarkers for breast cancer with ctDNA and cfDNA for their potential as prognostic markers. They compared the levels of ctDNA and cfDNA of 194 patients with CA15-3 and alkaline phosphatase values. The results indicated that both ctDNA and cfDNA are good predictors of the overall response of survival; however, only cfDNA is the predictor of progression-free survival [89]. This study proved that both ctDNA and cfDNA are good prognostic markers of metastatic breast cancer. Liu et al. performed a meta-analysis to understand the prognostic relationship between prostate cancer and cfDNA. This study proved that the higher concentrations of cfDNA are related to progression-free survival and the overall survival of prostate cancer [90]. Apart from prostate cancer, cfDNA has been proved to be a potential prognostic marker for colorectal cancer. Basnet et al. reported the association between higher concentrations of cfDNA and recurrence-free survival and overall survival of colorectal cancer [91]. A few studies have explained the role of cfDNA in ovarian cancer, such as the one by No et al., who conducted the mutational analysis for the RAS oncogene family, beta-2-microglobulin, the ATP-binding cassette subfamily F member 2 and claudin 4, and levels of cfDNA. In this study, cfDNA levels from B2M and CA125 or CA19-9 found no relationship with cancer prognosis. However, the cfDNA of RAB25 had variations in the copy number, which could be used as a prognostic marker for ovarian cancer [92]. Bortolin et al. also demonstrated the relationship between cfDNA levels and lung cancer in 22 patients, proving that cfDNA could be used as a prognostic marker for high-risk NSCLC cancer patients [93]. These studies have proven that tracking ctDNA can help predict the effectiveness of therapy and the recurrence of cancer in patients.
4.2.2. cfRNA
Many studies have reported that cfRNA has organ-specific transcripts, which can undergo temporal changes due to the development of cancer and tumors [65,94,95]. cfRNA not only predicts cancer and tumors but also pregnancy deliveries and preterm births [96]. In addition, it can distinguish among cancer stages and types. In a recent study, Hieter et al. distinguished the cancers from their premalignant conditions using cfRNA profiles and predicted the occurrence of cancer [66]. Raez et al. analyzed and measured cfRNA expression to monitor the clinical responses of patients with non-small cell lung cancer and reported that cfRNA can be used as a predictive tool for cancer onset and progression. In another study, Pucciarelli et al. used cfRNA to monitor the tumor response of patients with rectal cancer while they were receiving chemotherapy; the results indicated that cfRNA along with telomere-specific reverse transcriptase mRNA can be used to predict and measure the effects of chemotherapy [97].
Although many studies mention that cfRNA may be an excellent tool for cancer prediction [61,98], it has yet to be fully explored. Moreover, other than cfRNA, other types of RNA, including miRNA, mRNA, and circular RNA, are found to be excellent predictors; several studies have proven their prognostic potential [99,100].
4.3. Role of cfDNA and cfRNA in Measuring Residual Disease
After cancer treatment, a small amount of cancer cells remains inside the body, resulting in a relapse of the disease, as these are capable of activation and proliferation after a few months or years. In general, in clinical practice, flow cytometry, PCR, and NGS are used for the detection. However, scientists have found that cfDNA and cfRNA can detect minimal residual disease (MRD). The clinical applications of liquid biopsy are described in Table 3.
Table 3.
Clinical applications of ctDNA.
| Cancer Types | Clinical Management | Conclusion | References |
|---|---|---|---|
| Breast Cancer | Assessment of early ctDNA dynamics in 149 patients | Assessment of early dynamic changes in ctDNA can predict the efficacy of targeted therapy | [106] |
| Prediction of recurrence of cancer | Predicted metastatic relapse with high accuracy | [103] | |
| Detection of PIK3CA mutations | CtDNA can be used for the detection of PIK3CA mutations | [107] | |
| Lung cancer | Cancer diagnosis and therapeutics | EGFR L858R and T790M mutations can be detected in ctDNA using NGS and ddPCR | [108] |
| Detection of cancer at IA, IB, and IIA stages | Found an association among tumor stage, subtype, and cfDNA concentration, and demonstrated the utility and feasibility of targeted sequencing for ctDNA monitoring | [109] | |
| Monitoring treatment response and drug resistance | Proved the decrease of mutant copies after treatment and monitored the emergence of secondary mutations | [110] | |
| Detection of copy number variations in tumor cells | Copy number changed in BAX, P53, CASP3, SOX2, GRB2, SOS1, MAPK1, and a few other genes of cancer patients | [111,112] | |
| Diagnostic marker | The presence of ctDNA in cancer patients was found to be eight times higher than in healthy persons | [113] | |
| Colon cancer | Adjuvant therapy | Provided evidence of the potential use of ctDNA with adjuvant novel drugs for cancer patients | [114] |
| Colorectal cancer | Marker for cancer recurrence | Postoperative ctDNA positivity may be associated with the recurrence of cancer | [115] |
| Predicting response to neoadjuvant chemoradiotherapy | Combination of MRI and ctDNA potentially improves the predictive performance of cancer | [116] | |
| Novel biomarker Optimizing chemotherapy |
Described that ctDNA can potentially be utilized for diagnosis and personalized medicine for colorectal cancer | [117] | |
| Melanoma | Monitoring of relapse in stage III | ctDNA identified the highest risk stage III melanoma patients and helped in adjuvant therapy decisions | [118] |
| Detection of ctDNA levels in patients treated with anti-PD1 therapy | The evaluation of ctDNA during anti-PD1 antibody therapy provided reliable information | [119] | |
| Leiomyosarcoma | Detection of genetic variants using Guardant360 | The identification of molecular alterations can help develop therapy, targeting TP53, cell cycle, and kinase signaling pathways | [120] |
4.3.1. ctDNA
A. A. Chaudhuri et al. analyzed the ctDNA samples of 40 patients (treated with curative-intent first-line therapies) and 54 healthy persons through PET-Scan and CAPP-seq techniques by targeting 128 genes. The dates pre- and post-treatment for lung cancer were compared. The results demonstrated that the patients in which ctDNA was detected had the recurrence of cancer. This study proved that ctDNA could potentially detect MRD and predict the relapse of the disease in patients [73]. C. Abbosh et al. provided a phylogenetic analysis of ctDNA, the proof of chemotherapy resistance, and the chance of lung cancer recurrence [101]. Leal et al. compared the alterations of ctDNA with the DNA of white blood cells of the same patients and proved that ctDNA could predict recurrence when analyzed within nine weeks after treatment [102]. Murillas et al. analyzed the potential of ctDNA to detect MRD and predict the relapse of disease after the treatment of breast cancer in 55 patients. The results demonstrated that ctDNA could accurately predict the relapse of the disease. They also used a mutation tracking analysis in this study, which increased the sensitivity and specificity of the detection [103].
4.3.2. cfRNA
cfRNA, similar to ctDNA, has the potential to measure the disease burden and MRD. Most of the studies report the capability of ctDNA to measure MRD; however, a few report that cfRNA can potentially measure MRD [104]. Rossi reported that cfRNA can detect MRD and the reoccurrence of cancer; its capacity and performance were compared via a flow cytometry-based analysis [105].
Detecting MRD requires highly sensitive approaches that can monitor MRD through patient-specific and mutation-specific analyses. The analysis of MRD may benefit humans as the major challenge in cancer treatment involves the recurrence of disease after completing painful treatments and surgeries. MRD is an issue that scientists have discussed and researched for decades. Currently, MRD has become a focus of research for the prediction of disease relapse. The detection of ctDNA after treatment can identify the patients with MRD who are at a higher risk of cancer recurrence. Multiple studies have demonstrated that ctDNA can be a potential diagnostic marker of MRD and predict the recurrence of the disease.
5. Clinical Trials
Scientists have shifted their interest to personalized medicines due to their high accuracy and ability to predict the recurrence of cancer. Oncologists have focused their research on ctDNA and cfDNA as biomarkers for tumor profiling. In addition, the noninvasive nature of a cfDNA-based analysis makes it a good treatment option for patients. As a result, several types of cfDNA and ctDNA-based cancer management systems have been developed and clinically tested.
5.1. ctDNA
The details of a few major clinical trials, which were multicentric, multinational, and used ctDNA as the detection method in larger population sizes, are given below.
5.1.1. TARGET Study
Tumor characterization to guide experimental targeted therapy, known as TARGET, was a phase I study conducted at Manchester Cancer Research Center on 550 patients to determine the feasibility of using ctDNA to diagnose cancer, utilizing mutational analyses of advanced-stage cancers. Results were compared to the tissue-based testing. The study was divided into two parts. The first part was conducted on 100 patients to assess the feasibility of the analytical workflow and data turnaround (of two to four weeks) for clinical decision-making. Then, the clinical utility of ctDNA was analyzed on the remaining 450 patients who were selected based on ctDNA and/or tumor genomic profiling. The results demonstrated that ctDNA could detect 54 out of 69 mutations, giving a 78.3% detection rate [121]. Foundation medicine testing was also conducted on 39 patients, where ctDNA results were compared with the FoundationOne panel. This trial proved that ctDNA is a feasible option for the guidance of selecting specific treatment regimens for cancer patients. The results of the TARGET study encourage routine implementation of ctDNA testing as an adjunct to tumor testing.
5.1.2. MONALEESA Study
The MONALEESA study was one of the largest clinical trials that studied the feasibility of using ctDNA as the biomarker for cancer diagnoses and therapeutics. In all three MONALEESA trials, a total of 1503 patients with advanced breast cancer were recruited for the analyses. The trials were conducted to observe the effects of the combination of ribociclib with endocrine therapy and analyses were conducted via a ctDNA evaluation using NGS. Due to the results, the researchers proposed the use of ribociclib with endocrine therapy as a first-line treatment for breast cancer patients. During the ctDNA analysis, genomic alterations in PIK3CA, TP53, ZNF703/FGFR1, and ESR1 were observed; however, irrespective of the genomic alterations, the treatment was found effective to reduce the disease progression [122].
5.1.3. I-PREDICT Study
I-PREDICT is the abbreviation for Investigation of Profile-Related Evidence Determining Individualized Cancer Therapy. It involved an open-label navigational investigation and a prospective study conducted at the University of California, San Diego Moores Cancer Center, and Avera Cancer Institute South Dakota on 149 patients. The objective of this study was to assess the success of individualized cancer therapy by analyzing the molecular profiles of the patients. Multiple techniques were utilized for molecular profiling, including ctDNA analysis, NGS, tumor mutational burden, programmed death-ligand 1 immunohistochemistry, and microsatellite instability status. A total of 49% of these stage IV patients were treated using personalized treatments consisting of customized drug regimens. The study proved that identifying molecular alterations for larger fractions of alterations is better than targeting fewer somatic alterations because it yields high matching scores, improving disease control rates and longer survival and progression-free rates. The success rate of this trial was higher than in other precision medicine trials. There were several factors responsible for the increased success rate, including the use of sophisticated diagnostic techniques, such as ctDNA, and combining the results with targeted drug delivery. In conclusion, this trial proposed that the paradigm for precision oncology can be improved by using multi-drug combinations targeting multiple identified molecular alterations [123].
In conclusion, the TARGET, I-PREDICT, and MONALEESA studies nicely demonstrated the feasibility of using some of these innovative approaches in cancer precision medicine, including ctDNA for identification of molecular alterations and guidance of patients to clinical trials, drug combinations to target as many molecular alterations in a patient as possible, and treatment algorithms based on the RNA expressions that necessitate normal adjacent tissue samples.
Apart from these clinical trials, to date, 20 different clinical trials have been conducted to prove the safety, efficacy, and efficiency of ctDNA, including PADA, Solar-1, BELLE 1, 2, 3, POSEIDON, MONALEESA I, II, III, and others.
As cfDNA-based diagnosis is an emerging field, several trials are ongoing. The clinical trial MERMAID-I is being conducted on lung cancer patients to study the adjuvant durvalumab along with chemotherapy and minimal residual disease, studied by analyzing the ctDNA of patients. Similar to MERMAID-I, MERMAID-II is a phase III clinical trial that is being conducted on patients who have completed their curative intent and are now in the surveillance mode for MRD and recurrence of disease through ctDNA analysis. Another ongoing trial is SUMMIT, which has a large population size (25,000), to validate the feasibility and accuracy of early cancer detection by using ctDNA. There are many other ongoing trials related to the use of ctDNA for cancer diagnoses, therapeutics, and MRD; only a few are described here.
5.2. cfRNA
Due to the potential of cfRNA, scientists have focused on ctDNA/cfDNA-based studies and several clinical trials have been conducted. In the case of cfRNA, not much data are available and fewer clinical trials were conducted (details are given below).
5.2.1. ICE-PAC
The clinical trial ICE-PAC was a phase II, multicentric, nonrandomized trial conducted in Australia between 2017 and 2019. The main objective of the trial was to analyze the effects of the PD-L1 checkpoint inhibitor avelumab in combination with stereotactic ablative body radiotherapy (SABR) in prostate cancer in 31 male patients. This trial also analyzed cfRNA as a biomarker and used it to measure pathogenicity. The results of this trial indicated that cfRNA has the potential to be used to measure the tumor response and the development/emergence of resistance [124].
5.2.2. NCT02853097
This was a research trial that was conducted to analyze the correlation of cfRNA with the progression of prostate cancer. For this purpose, the expressions of PD-L1, TIM-3, PCA3, AR, and AR-V7 were measured with the help of cfRNA, and the results were compared to healthy individuals. The results indicated that PCA3 was not detected in healthy humans; however, AR, AR-V, and TIM-3 were found in 41%, 7%, and 43% of cancer patients, respectively. The study concluded that cfRNA may be potentially used in the early diagnosis of cancer and reoccurrence of cancer after treatment [125].
Many clinical trials on cfDNA and ctDNA have proven their efficiency and utility; the FDA has even approved some of the kits. The literature has shown that weak and highly variable signal detections make cfRNA difficult to measure and analyze. Many research groups are attempting to update their protocols for the robust and reproducible detection of cfRNA, such as the nCounter platform [126]; we hope that we will have more results to publish and products to review in the future.
6. Food and Drug Administration Approved ctDNA and cfRNA Tests
For decades, scientists have attempted to develop noninvasive diagnostic and treatment methods for cancer patients. Due to the non-invasiveness, effectiveness, and sensitivity of ctDNA, many assays and detection kits have been developed. Many are still in the trial phase, and a few have been FDA-approved.
Cobas® EGFR Mutation Test v2 (Roche, Basel, Switzerland) was one of the earliest tests approved by the FDA (on 1 June 2016) to detect cancer, using blood. It is recommended for non-small cell lung cancer and identifies mutations in EGFR, including exon 19 deletions or exon 21 substitutions.
Signatera (Natera Inc., Austin, TX, USA) is a ctDNA-based cancer diagnostic assay that has been proven to be a suitable candidate for predicting the long-term outcomes of surgery and anticancer treatments.
Guardant360® (Guardant Health, Lansdale, PA, USA) is another FDA-approved liquid biopsy (7 August 2020) that has been tested on more than 150,000 patients and approved by 7000 oncologists. It is recommended for advanced cancer patients and is used as a companion diagnostic test for lung cancer patients who have EGFR mutations. In addition to Medicare, it is also covered by some private players. FDA approval of this test was an important milestone in the history of oncology. Currently, this test is recommended for colorectal, breast, lung, and prostate cancer.
FoundationOne CDx (Foundation Medicine, Cambridge, MA, USA) is one of the blood tests approved by the FDA (on 26 August 2020); it is used to detect tumors in cancer patients. It can detect the mutations of 300 genes in more than 30 cancer types. In addition to tumor profiling, it is used as a companion diagnostic test. It has been approved for solid cancers, including lung and prostate cancer. Apart from the mutation analysis, it also provides information about tumor burden and microsatellite instability.
Other than the assays mentioned above, several other kits are available commercially to detect ctDNA but are not FDA-approved. These kits include QIAamp Circulating NucleicAcid kit, the Maxwell® RSC ccfDNA Plasma kit, NucleoSpin Plasma XS, MagMAX™ Total Nucleic Acid Isolation kit, and the Chemagic Circulating NA kit. In the case of cfRNA, only one assay is approved, known as the PROGENSA PCA3 assay. It is an in-vitro test that amplifies nucleic acids and measures a specific marker of prostate cancer called PCA3 RNA. The result of this assay generates a score that correlates with the likelihood of a positive repeat biopsy. This test is suitable to analyze at-risk patient populations [127].
7. Other Techniques for Cancer Detection
The PET/CT technique is widely used for the detection and screening of cancer; however, it is associated with some challenges, i.e., inaccuracy, expensiveness, and radiation exposure. Currently, it is combined with other modalities to improve the issues related to accuracy. Sasamori et al. combined the PEC/CT scan with whole-body diffusion-weighted imaging and improved accuracy from 48% to 55% [128]. Moving a step forward, Lennon et al. combined cfDNA detection with PET/CT and reduced the chances of false positive results in cfDNA-based cancer diagnoses [129]. In addition, Kwee et al. and Woff et al. provided clinical evidence on the improvement of cancer detection by combining cfDNA with PET-based techniques [130,131]. Through these studies, it was found that cfDNA-based diagnosis is a promising method; however, it has yet to be fully developed, and by combining it with other techniques, the sensitivity and specificity can be improved. GeneCT is a deep learning, RNA-based classifier for the prediction of the status, stage, and origins of cancer. This system has shown good results, is expected to be further developed and may be used for commercial purposes [132].
8. Discussion
In clinical practice, tissue biopsy-based invasive methods are commonly used. Despite their accuracy, a few issues have been associated with them, shifting the focus of researchers toward non-invasive liquid biopsies. Although imaging-related techniques are available, their costs and false positive results make them nonpreferred techniques. Liquid biopsies have proven suitable for cancer management by providing early diagnoses, as well as MRD evaluation and tumor response analysis after treatment. They can improve cancer diagnosis and treatment by using minimally invasive or non-invasive means.
In addition to cfDNA and cfRNA, researchers are investigating circulating cells (e.g., circulating tumor cells, circulating hybrid cells, mRNA, miRNA, and tumor-associated macrophages). Currently, all of these methods are expensive, and their detections are challenging due to high variations in molecular genetics; nevertheless, it is expected that in the near future, other than cfDNA and cfRNA, there will be many options available for cancer management.
Although fewer studies are available for the analysis and exploration of cfRNA, it has great potential to be used as a biomarker for cancer diagnosis and treatment. It can not only detect cancer but also provide additional useful information, such as alterations in the molecular profiles, effects on the immune system or other systems of the body, and disease progression. Furthermore, it is stable in blood, making it easier for researchers to extract and analyze. In conclusion, cfDNA- and cfRNA-based liquid biopsies have shown promising results and will play important roles in cancer management and personalized medication, but there is still a need for further research.
9. Future Directions and Limitations
In the past decade, many studies have been conducted on the use of cfDNA as a potential biomarker for cancer diagnosis and therapeutics; however, there is still room for further research and improvements. Currently, ctDNA-based assays are potentially suitable for late-stage cancer diagnoses, prognoses, and disease recurrence predictions; however, early-stage cancer management is still uncertain. Secondly, ctDNA-based assays lack standardization; therefore, the standard should be developed to make this technique more appropriate and desirable. Furthermore, there is the issue of false negatives and false positives due to the presence of cfDNA in the normal cells of healthy humans; thus, the quantification techniques should be improved and updated to minimize the false predictions and diagnoses. Lastly, the cost is an essential factor that should be considered as the assays and kits available are not affordable for poorer populations.
One major challenge faced with a cfRNA analysis involves the absence of reference genes and unavailability of data; therefore, developing archives for reference data should be encouraged to solve it. Secondly, specific reference genes and tests are required for specific types of cancer; hence, the specificity of cfRNA-based tests should be increased. Non-specific detection of RNA has also been observed due to non-cancer-related factors, such as posttraumatic organ failure. Future research should focus on enhancing the specificities of these analyses and building standards/references for comparison purposes.
10. Conclusions
The analysis of cfDNA and cfRNA using body fluids is an attractive solution for cancer diagnosis, to solve, for example, the invasiveness, low frequency, and non-specificity of other techniques. They not only detect the tumor inside the body but can also analyze the effectiveness of anticancer therapy, evaluate the drug resistance capacity, and predict the chances of recurrence of cancer. There are limitations associated with these techniques, including relatively low sensitivity, lack of information about the origin, and low specificity; researchers are attempting to improve the sensitivities and specificities of these techniques. Multiple clinical trials and studies have been conducted, utilizing sophisticated sequencing and PCR-based techniques, resulting in a tremendous increase in the sensitivity and specificity of ctDNA and cfRNA. To the best of our knowledge, liquid biopsy-based assays and therapeutics are breakthroughs in the field of companion diagnostics and personalized medicine; however, concrete evidence is still required through clinical trials on a larger population.
Acknowledgments
The authors acknowledge Khushbakhat Khan for providing her support during the data collection.
Author Contributions
All authors contributed to the study conception and design. S.H. and A.S. reviewed the literature and drafted the manuscript. S.A.K., W.M. and S.K. assisted with the literature review and data collection. Y.-S.L. critically reviewed the manuscript and assisted with the literature review. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hassan S., Ali M.N., Ghafoor B. Evolutionary perspective of drug eluting stents: From thick polymer to polymer free approach. J. Cardiothorac. Surg. 2022;17:65. doi: 10.1186/s13019-022-01812-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandel P., Metais P. Nuclear Acids In Human Blood Plasma. Comptes Rendus Seances Soc. Biol. Fil. 1948;142:241–243. [PubMed] [Google Scholar]
- 3.Bendich A., Wilczok T., Borenfreund E. Circulating dna as a possible factor in oncogenesis. Science. 1965;148:374–376. doi: 10.1126/science.148.3668.374. [DOI] [PubMed] [Google Scholar]
- 4.Tan E.M., Schur P.H., Carr R.I., Kunkel H.G. Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J. Clin. Investig. 1966;45:1732–1740. doi: 10.1172/JCI105479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leon S.A., Shapiro B., Sklaroff D.M., Yaros M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 6.Pös O., Biró O., Szemes T., Nagy B. Circulating cell-free nucleic acids: Characteristics and applications. Eur. J. Hum. Genet. 2018;26:937–945. doi: 10.1038/s41431-018-0132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tosevska A., Morselli M., Basak S.K., Avila L., Mehta P., Wang M.B., Srivatsan E.S., Pellegrini M. Cell-Free RNA as a Novel Biomarker for Response to Therapy in Head & Neck Cancer. Front. Oncol. 2022;12:869108. doi: 10.3389/fonc.2022.869108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raez L.E., Danenberg K., Sumarriva D., Usher J., Sands J., Castrellon A., Ferraro P., Milillo A., Huang E., Soon-Shiong P., et al. Using cfRNA as a tool to evaluate clinical treatment outcomes in patients with metastatic lung cancers and other tumors. Cancer Drug Resist. 2021;4:1061–1071. doi: 10.20517/cdr.2021.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andree K.C., van Dalum G., Terstappen L.W. Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol. 2016;10:395–407. doi: 10.1016/j.molonc.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samsuwan J., Somboonchokepisal T., Akaraputtiporn T., Srimuang T., Phuengsukdaeng P., Suwannarat A., Mutirangura A., Kitkumthorn N. A method for extracting DNA from hard tissues for use in forensic identification. Biomed. Rep. 2018;9:433–438. doi: 10.3892/br.2018.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee K.J., Mann E., da Silva L.M., Scalici J., Gassman N.R. DNA damage measurements within tissue samples with Repair Assisted Damage Detection (RADD) Curr. Res. Biotechnol. 2019;1:78–86. doi: 10.1016/j.crbiot.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seton-Rogers S. Closing in on cfDNA-based detection and diagnosis. Nat. Rev. Cancer. 2020;20:481. doi: 10.1038/s41568-020-0293-7. [DOI] [PubMed] [Google Scholar]
- 13.Rafi I., Chitty L. Cell-free fetal DNA and non-invasive prenatal diagnosis. Br. J. Gen. Pract. J. R. Coll. Gen. Pract. 2009;59:e146–e148. doi: 10.3399/bjgp09X420572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berezin A.J.T.B. The cell-free mitochondrial DNA: A novel biomarker of cardiovascular risk. Transl. Biomed. 2016;7:68–71. doi: 10.21767/2172-0479.100068. [DOI] [Google Scholar]
- 15.Al Amir Dache Z., Otandault A., Tanos R., Pastor B., Meddeb R., Sanchez C., Arena G., Lasorsa L., Bennett A., Grange T., et al. Blood contains circulating cell-free respiratory competent mitochondria. FASEB J. 2020;34:3616–3630. doi: 10.1096/fj.201901917RR. [DOI] [PubMed] [Google Scholar]
- 16.Pessoa L.S., Heringer M., Ferrer V.P. ctDNA as a cancer biomarker: A broad overview. Crit. Rev. Oncol. Hematol. 2020;155:103109. doi: 10.1016/j.critrevonc.2020.103109. [DOI] [PubMed] [Google Scholar]
- 17.An Q., Hu Y., Li Q., Chen X., Huang J., Pellegrini M., Zhou X.J., Rettig M., Fan G. The size of cell-free mitochondrial DNA in blood is inversely correlated with tumor burden in cancer patients. Precis. Clin. Med. 2019;2:131–139. doi: 10.1093/pcmedi/pbz014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Underhill H.R., Kitzman J.O., Hellwig S., Welker N.C., Daza R., Baker D.N., Gligorich K.M., Rostomily R.C., Bronner M.P., Shendure J. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12:e1006162. doi: 10.1371/journal.pgen.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan H.C., Blumenfeld Y.J., Chitkara U., Hudgins L., Quake S.R. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin. Chem. 2010;56:1279–1286. doi: 10.1373/clinchem.2010.144188. [DOI] [PubMed] [Google Scholar]
- 20.Wyllie A., Morris R., Smith A., Dunlop D. Chromatin cleavage in apoptosis: Association with condensed chromatin morphology and dependence on macromolecular synthesis. J. Pathol. 1984;142:67–77. doi: 10.1002/path.1711420112. [DOI] [PubMed] [Google Scholar]
- 21.Stroun M., Lyautey J., Lederrey C., Olson-Sand A., Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin. Chim. Acta. 2001;313:139–142. doi: 10.1016/S0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 22.Aucamp J., Bronkhorst A.J., Badenhorst C.P.S., Pretorius P.J. The diverse origins of circulating cell-free DNA in the human body: A critical re-evaluation of the literature. Biol. Rev. 2018;93:1649–1683. doi: 10.1111/brv.12413. [DOI] [PubMed] [Google Scholar]
- 23.Heitzer E., Auinger L., Speicher M.R. Cell-Free DNA and Apoptosis: How Dead Cells Inform About the Living. Trends Mol. Med. 2020;26:519–528. doi: 10.1016/j.molmed.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Jackson Chornenki N.L., Coke R., Kwong A.C., Dwivedi D.J., Xu M.K., McDonald E., Marshall J.C., Fox-Robichaud A.E., Charbonney E., Liaw P.C. Comparison of the source and prognostic utility of cfDNA in trauma and sepsis. Intensive Care Med. Exp. 2019;7:29. doi: 10.1186/s40635-019-0251-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goggs R., Jeffery U., LeVine D.N., Li R.H.L. Neutrophil-Extracellular Traps, Cell-Free DNA, and Immunothrombosis in Companion Animals: A Review. Vet. Pathol. 2020;57:6–23. doi: 10.1177/0300985819861721. [DOI] [PubMed] [Google Scholar]
- 26.Li R.H.L., Tablin F. A Comparative Review of Neutrophil Extracellular Traps in Sepsis. Front. Vet. Sci. 2018;5:291. doi: 10.3389/fvets.2018.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elzanowska J., Semira C., Costa-Silva B. DNA in extracellular vesicles: Biological and clinical aspects. Mol. Oncol. 2021;15:1701–1714. doi: 10.1002/1878-0261.12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salehi B., Mishra A.P., Nigam M., Kobarfard F., Javed Z., Rajabi S., Khan K., Ashfaq H.A., Ahmad T., Pezzani R. Multivesicular liposome (Depofoam) in human diseases. Iran. J. Pharm. Res. IJPR. 2020;19:9. doi: 10.22037/ijpr.2020.112291.13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balaj L., Lessard R., Dai L., Cho Y.J., Pomeroy S.L., Breakefield X.O., Skog J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thakur B.K., Zhang H., Becker A., Matei I., Huang Y., Costa-Silva B., Zheng Y., Hoshino A., Brazier H., Xiang J., et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014;24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abe J.-I., Morrell C. Pyroptosis as a Regulated Form of Necrosis: PI+/Annexin V-/High Caspase 1/Low Caspase 9 Activity in Cells = Pyroptosis? Circul. Res. 2016;118:1457–1460. doi: 10.1161/CIRCRESAHA.116.308699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou J., Zhao R., Xia W., Chang C.W., You Y., Hsu J.M., Nie L., Chen Y., Wang Y.C., Liu C., et al. PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis. Nat. Cell Biol. 2020;22:1264–1275. doi: 10.1038/s41556-020-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang Y., Tian S., Pan Y., Li W., Wang Q., Tang Y., Yu T., Wu X., Shi Y., Ma P., et al. Pyroptosis: A new frontier in cancer. Biomed. Pharmacother. 2020;121:109595. doi: 10.1016/j.biopha.2019.109595. [DOI] [PubMed] [Google Scholar]
- 34.Alqurashi H., Ortega Asencio I., Lambert D.W. The Emerging Potential of Extracellular Vesicles in Cell-Free Tissue Engineering and Regenerative Medicine. Tissue Eng. Part B Rev. 2021;27:530–538. doi: 10.1089/ten.teb.2020.0222. [DOI] [PubMed] [Google Scholar]
- 35.Guo Y., Wang H., Huang L., Ou L., Zhu J., Liu S., Xu X. Small extracellular vesicles-based cell-free strategies for therapy. MedComm. 2021;2:17–26. doi: 10.1002/mco2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiu B.C.H., Chen C., You Q., Chiu R., Venkataraman G., Zeng C., Zhang Z., Cui X., Smith S.M., He C., et al. Alterations of 5-hydroxymethylation in circulating cell-free DNA reflect molecular distinctions of subtypes of non-Hodgkin lymphoma. NPJ Genom. Med. 2021;6:11. doi: 10.1038/s41525-021-00179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwaederle M., Chattopadhyay R., Kato S., Fanta P.T., Banks K.C., Choi I.S., Piccioni D.E., Ikeda S., Talasaz A., Lanman R.B., et al. Genomic Alterations in Circulating Tumor DNA from Diverse Cancer Patients Identified by Next-Generation Sequencing. Cancer Res. 2017;77:5419–5427. doi: 10.1158/0008-5472.CAN-17-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan K., Safi S., Abbas A., Badshah Y., Dilshad E., Rafiq M., Zahra K., Shabbir M. Unravelling Structure, Localization, and Genetic Crosstalk of KLF3 in Human Breast Cancer. Biomed. Res. Int. 2020;2020:1354381. doi: 10.1155/2020/1354381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zahra K., Shabbir M., Badshah Y., Trembley J.H., Badar Z., Khan K., Afsar T., Almajwal A., Alruwaili N.W., Razak S. Determining KLF14 tertiary structure and diagnostic significance in brain cancer progression. Sci. Rep. 2022;12:8039. doi: 10.1038/s41598-022-12072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raez L., Danenberg K., Huang E., Usher J., Danenberg P., Sumarriva D. P14. 04 cfRNA from Liquid Biopsies Is More Abundant Than cfDNA, Informs Treatment Outcome and Is Concordant with Tissue. J. Thorac. Oncol. 2021;16:S330. doi: 10.1016/j.jtho.2021.01.510. [DOI] [Google Scholar]
- 41.Marisa L., de Reyniès A., Duval A., Selves J., Gaub M.P., Vescovo L., Etienne-Grimaldi M.-C., Schiappa R., Guenot D., Ayadi M., et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasioukhin V., Anker P., Maurice P., Lyautey J., Lederrey C., Stroun M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br. J. Haematol. 1994;86:774–779. doi: 10.1111/j.1365-2141.1994.tb04828.x. [DOI] [PubMed] [Google Scholar]
- 43.Sorenson G.D., Pribish D.M., Valone F.H., Memoli V.A., Bzik D.J., Yao S.-L. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol. Prev. Biomark. 1994;3:67–71. [PubMed] [Google Scholar]
- 44.Panagopoulou M., Karaglani M., Balgkouranidou I., Biziota E., Koukaki T., Karamitrousis E., Nena E., Tsamardinos I., Kolios G., Lianidou E., et al. Circulating cell-free DNA in breast cancer: Size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene. 2019;38:3387–3401. doi: 10.1038/s41388-018-0660-y. [DOI] [PubMed] [Google Scholar]
- 45.Panagopoulou M., Esteller M., Chatzaki E. Circulating Cell-Free DNA in Breast Cancer: Searching for Hidden Information towards Precision Medicine. Cancers. 2021;13:728. doi: 10.3390/cancers13040728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossi G., Mu Z., Rademaker A.W., Austin L.K., Strickland K.S., Costa R.L.B., Nagy R.J., Zagonel V., Taxter T.J., Behdad A., et al. Cell-Free DNA and Circulating Tumor Cells: Comprehensive Liquid Biopsy Analysis in Advanced Breast Cancer. Clin. Cancer. Res. 2018;24:560. doi: 10.1158/1078-0432.CCR-17-2092. [DOI] [PubMed] [Google Scholar]
- 47.Ring A., Campo D., Porras T.B., Kaur P., Forte V.A., Tripathy D., Lu J., Kang I., Press M.F., Jeong Y.J., et al. Circulating Tumor Cell Transcriptomics as Biopsy Surrogates in Metastatic Breast Cancer. Ann. Surg. Oncol. 2022;29:2882–2894. doi: 10.1245/s10434-021-11135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klein E.A., Richards D., Cohn A., Tummala M., Lapham R., Cosgrove D., Chung G., Clement J., Gao J., Hunkapiller N., et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021;32:1167–1177. doi: 10.1016/j.annonc.2021.05.806. [DOI] [PubMed] [Google Scholar]
- 49.Braicu E.I., du Bois A., Sehouli J., Beck J., Prader S., Kulbe H., Eiben B., Harter P., Traut A., Pietzner K., et al. Cell-Free-DNA-Based Copy Number Index Score in Epithelial Ovarian Cancer—Impact for Diagnosis and Treatment Monitoring. Cancers. 2022;14:168. doi: 10.3390/cancers14010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hannan N.J., Cohen P.A., Beard S., Bilic S., Zhang B., Tong S., Whitehead C., Hui L. Transcriptomic analysis of patient plasma reveals circulating miR200c as a potential biomarker for high-grade serous ovarian cancer. Gynecol. Oncol. Rep. 2022;39:100894. doi: 10.1016/j.gore.2021.100894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hulstaert E., Levanon K., Morlion A., Van Aelst S., Christidis A.-A., Zamar R., Anckaert J., Verniers K., Bahar-Shany K., Sapoznik S., et al. RNA biomarkers from proximal liquid biopsy for diagnosis of ovarian cancer. Neoplasia. 2022;24:155–164. doi: 10.1016/j.neo.2021.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Powles T., Assaf Z.J., Davarpanah N., Banchereau R., Szabados B.E., Yuen K.C., Grivas P., Hussain M., Oudard S., Gschwend J.E., et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature. 2021;595:432–437. doi: 10.1038/s41586-021-03642-9. [DOI] [PubMed] [Google Scholar]
- 53.Garcia J., Gauthier A., Lescuyer G., Barthelemy D., Geiguer F., Balandier J., Edelstein D.L., Jones F.S., Holtrup F., Duruisseau M. Routine Molecular Screening of Patients with Advanced Non-SmallCell Lung Cancer in Circulating Cell-Free DNA at Diagnosis and During Progression Using OncoBEAM TM EGFR V2 and NGS Technologies. Mol. Diagn. Ther. 2021;25:239–250. doi: 10.1007/s40291-021-00515-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pellini B., Chaudhuri A.A. Circulating tumor DNA minimal residual disease detection of non–small-cell lung cancer treated with curative intent. J. Clin. Oncol. 2022;40:567–575. doi: 10.1200/JCO.21.01929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bunduc S., Gede N., Váncsa S., Lillik V., Kiss S., Dembrovszky F., Eross B., Szakács Z., Gheorghe C., Mikó A. Prognostic role of cell-free DNA biomarkers in pancreatic adenocarcinoma: A systematic review and meta–analysis. Crit. Rev. Oncol. 2022;169:103548. doi: 10.1016/j.critrevonc.2021.103548. [DOI] [PubMed] [Google Scholar]
- 56.Kim H., Heo C.M., Oh J., Chung H.H., Lee E.M., Park J., Lee S.-H., Lee K.H., Lee K.T., Lee J.K. Clinical significance of circulating tumor cells after chemotherapy in unresectable pancreatic ductal adenocarcinoma. Transl. Oncol. 2022;16:101321. doi: 10.1016/j.tranon.2021.101321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bustamante P., Tsering T., Coblentz J., Mastromonaco C., Abdouh M., Fonseca C., Proença R.P., Blanchard N., Dugé C.L., Andujar R.A.S. Circulating tumor DNA tracking through driver mutations as a liquid biopsy-based biomarker for uveal melanoma. J. Exp. Clin. Cancer Res. 2021;40:196. doi: 10.1186/s13046-021-01984-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bobillo S., Crespo M., Escudero L., Mayor R., Raheja P., Carpio C., Rubio-Perez C., Tazón-Vega B., Palacio C., Carabia J. Cell free circulating tumor DNA in cerebrospinal fluid detects and monitors central nervous system involvement of B-cell lymphomas. Haematologica. 2021;106:513. doi: 10.3324/haematol.2019.241208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salvianti F., Gelmini S., Mancini I., Pazzagli M., Pillozzi S., Giommoni E., Brugia M., Di Costanzo F., Galardi F., De Luca F. Circulating tumour cells and cell-free DNA as a prognostic factor in metastatic colorectal cancer: The OMITERC prospective study. Br. J. Cancer. 2021;125:94–100. doi: 10.1038/s41416-021-01399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye P., Cai P., Xie J., Wei Y. The diagnostic accuracy of digital PCR, ARMS and NGS for detecting KRAS mutation in cell-free DNA of patients with colorectal cancer: A systematic review and meta-analysis. PLoS ONE. 2021;16:e0248775. doi: 10.1371/journal.pone.0248775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morris V.K., Strickler J.H. Use of Circulating Cell-Free DNA to Guide Precision Medicine in Patients with Colorectal Cancer. Annu. Rev. Med. 2021;72:399–413. doi: 10.1146/annurev-med-070119-120448. [DOI] [PubMed] [Google Scholar]
- 62.Rizzo A., Ricci A.D., Tavolari S., Brandi G. Circulating Tumor DNA in Biliary Tract Cancer: Current Evidence and Future Perspectives. Cancer Genom. Proteom. 2020;17:441–452. doi: 10.21873/cgp.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fernando M.R., Norton S.E., Luna K.K., Lechner J.M., Qin J. Stabilization of cell-free RNA in blood samples using a new collection device. Clin. Biochem. 2012;45:1497–1502. doi: 10.1016/j.clinbiochem.2012.07.090. [DOI] [PubMed] [Google Scholar]
- 64.Lo K.-W., Lo Y.M.D., Leung S.-F., Tsang Y.-S., Chan L.Y.S., Johnson P.J., Hjelm N.M., Lee J.C.K., Huang D.P. Analysis of Cell-free Epstein-Barr Virus-associated RNA in the Plasma of Patients with Nasopharyngeal Carcinoma. Clin. Chem. 1999;45:1292–1294. doi: 10.1093/clinchem/45.8.1292. [DOI] [PubMed] [Google Scholar]
- 65.Larson M.H., Pan W., Kim H.J., Mauntz R.E., Stuart S.M., Pimentel M., Zhou Y., Knudsgaard P., Demas V., Aravanis A.M., et al. A comprehensive characterization of the cell-free transcriptome reveals tissue- and subtype-specific biomarkers for cancer detection. Nat. Commun. 2021;12:2357. doi: 10.1038/s41467-021-22444-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roskams-Hieter B., Kim H.J., Anur P., Wagner J.T., Callahan R., Spiliotopoulos E., Kirschbaum C.W., Civitci F., Spellman P.T., Thompson R.F., et al. Plasma cell-free RNA profiling distinguishes cancers from pre-malignant conditions in solid and hematologic malignancies. NPJ Precis. Oncol. 2022;6:28. doi: 10.1038/s41698-022-00270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwarzenbach H. Clinical relevance of circulating, cell-free and exosomal microRNAs in plasma and serum of breast cancer patients. Oncol. Res. Treat. 2017;40:423–429. doi: 10.1159/000478019. [DOI] [PubMed] [Google Scholar]
- 68.Molnár B., Galamb O., Kalmár A., Barták B.K., Nagy Z.B., Tóth K., Tulassay Z., Igaz P., Dank M. Circulating cell-free nucleic acids as biomarkers in colorectal cancer screening and diagnosis-an update. Expert Rev. Mol. Diagn. 2019;19:477–498. doi: 10.1080/14737159.2019.1613891. [DOI] [PubMed] [Google Scholar]
- 69.Hasegawa N., Kohsaka S., Kurokawa K., Shinno Y., Nakamura I.T., Ueno T., Kojima S., Kawazu M., Suehara Y., Ishijima M. Highly sensitive fusion detection using plasma cell-free RNA in non-small-cell lung cancers. Cancer Sci. 2021;112:4393. doi: 10.1111/cas.15084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Savino M., Parrella P., Copetti M., Barbano R., Murgo R., Fazio V.M., Valori V.M., Carella M., Garrubba M., Santini S.A. Comparison between real-time quantitative PCR detection of HER2 mRNA copy number in peripheral blood and ELISA of serum HER2 protein for determining HER2 status in breast cancer patients. Cell. Oncol. Off. J. Int. Soc. Cell. Oncol. 2009;31:203–211. doi: 10.1155/2009/206068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang R., Chen B., Tong X., Wang Y., Wang C., Jin J., Tian P., Li W. Diagnostic accuracy of droplet digital PCR for detection of EGFR T790M mutation in circulating tumor DNA. Cancer Manag. Res. 2018;10:1209–1218. doi: 10.2147/CMAR.S161382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Newman A.M., Bratman S.V., To J., Wynne J.F., Eclov N.C., Modlin L.A., Liu C.L., Neal J.W., Wakelee H.A., Merritt R.E., et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chaudhuri A.A., Chabon J.J., Lovejoy A.F., Newman A.M., Stehr H., Azad T.D., Khodadoust M.S., Esfahani M.S., Liu C.L., Zhou L., et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017;7:1394–1403. doi: 10.1158/2159-8290.CD-17-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zviran A., Schulman R.C., Shah M., Hill S.T.K., Deochand S., Khamnei C.C., Maloney D., Patel K., Liao W., Widman A.J., et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat. Med. 2020;26:1114–1124. doi: 10.1038/s41591-020-0915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roepman P., de Bruijn E., van Lieshout S., Schoenmaker L., Boelens M.C., Dubbink H.J., Geurts-Giele W.R., Groenendijk F.H., Huibers M.M., Kranendonk M.E., et al. Clinical validation of Whole Genome Sequencing for routine cancer diagnostics. J. Mol. Diagn. 2020;23:816–833. doi: 10.1016/j.jmoldx.2021.04.011. [DOI] [PubMed] [Google Scholar]
- 76.Forshew T., Murtaza M., Parkinson C., Gale D., Tsui D.W., Kaper F., Dawson S.-J., Piskorz A.M., Jimenez-Linan M., Bentley D. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 77.Bohers E., Viailly P.-J., Jardin F. cfDNA Sequencing: Technological Approaches and Bioinformatic Issues. Pharmaceuticals. 2021;14:596. doi: 10.3390/ph14060596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bos M.K., Angus L., Nasserinejad K., Jager A., Jansen M.P.H.M., Martens J.W.M., Sleijfer S. Whole exome sequencing of cell-free DNA—A systematic review and Bayesian individual patient data meta-analysis. Cancer Treat. Rev. 2020;83:101951. doi: 10.1016/j.ctrv.2019.101951. [DOI] [PubMed] [Google Scholar]
- 79.Rennert H., Eng K., Zhang T., Tan A., Xiang J., Romanel A., Kim R., Tam W., Liu Y.-C., Bhinder B., et al. Development and validation of a whole-exome sequencing test for simultaneous detection of point mutations, indels and copy-number alterations for precision cancer care. NPJ Genom. Med. 2016;1:16019. doi: 10.1038/npjgenmed.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oxnard G.R., Klein E.A., Seiden M., Hubbell E., Venn O., Jamshidi A., Zhang N., Beausang J.F., Gross S., Kurtzman K.N., et al. Simultaneous multi-cancer detection and tissue of origin (TOO) localization using targeted bisulfite sequencing of plasma cell-free DNA (cfDNA) J. Glob. Oncol. 2019;5:44. doi: 10.1200/JGO.2019.5.suppl.44. [DOI] [Google Scholar]
- 81.Paun O., Verhoeven K.J.F., Richards C.L. Opportunities and limitations of reduced representation bisulfite sequencing in plant ecological epigenomics. New Phytol. 2019;221:738–742. doi: 10.1111/nph.15388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aravanis A.M., Lee M., Klausner R.D. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell. 2017;168:571–574. doi: 10.1016/j.cell.2017.01.030. [DOI] [PubMed] [Google Scholar]
- 83.Phallen J., Sausen M., Adleff V., Leal A., Hruban C., White J., Anagnostou V., Fiksel J., Cristiano S., Papp E., et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017;9:eaan2415. doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu X., Yu Y., Shen M., Liu M., Wu S., Liang L., Huang F., Zhang C., Guo W., Liu T. Role of circulating free DNA in evaluating clinical tumor burden and predicting survival in Chinese metastatic colorectal cancer patients. BMC Cancer. 2020;20:1006. doi: 10.1186/s12885-020-07516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valpione S., Gremel G., Mundra P., Middlehurst P., Galvani E., Girotti M.R., Lee R.J., Garner G., Dhomen N., Lorigan P.C., et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur. J. Cancer. 2018;88:1–9. doi: 10.1016/j.ejca.2017.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nygaard A.D., Holdgaard P.C., Spindler K.L.G., Pallisgaard N., Jakobsen A. The correlation between cell-free DNA and tumour burden was estimated by PET/CT in patients with advanced NSCLC. Br. J. Cancer. 2014;110:363–368. doi: 10.1038/bjc.2013.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu J., Chen X., Wang J., Zhou S., Wang C.L., Ye M.Z., Wang X.Y., Song Y., Wang Y.Q., Zhang L.T., et al. Biological background of the genomic variations of cf-DNA in healthy individuals. Ann. Oncol. 2019;30:464–470. doi: 10.1093/annonc/mdy513. [DOI] [PubMed] [Google Scholar]
- 88.Razavi P., Li B.T., Brown D.N., Jung B., Hubbell E., Shen R., Abida W., Juluru K., De Bruijn I., Hou C., et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 2019;25:1928–1937. doi: 10.1038/s41591-019-0652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fernandez-Garcia D., Hills A., Page K., Hastings R.K., Toghill B., Goddard K.S., Ion C., Ogle O., Boydell A.R., Gleason K., et al. Plasma cell-free DNA (cfDNA) as a predictive and prognostic marker in patients with metastatic breast cancer. Breast Cancer Res. 2019;21:149. doi: 10.1186/s13058-019-1235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu H., Gao Y., Vafaei S., Gu X., Zhong X. The Prognostic Value of Plasma Cell-Free DNA Concentration in the Prostate Cancer: A Systematic Review and Meta-Analysis. Front. Oncol. 2021;11:599602. doi: 10.3389/fonc.2021.599602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Basnet S., Zhang Z.-Y., Liao W.-Q., Li S.-H., Li P.-S., Ge H.-Y. The Prognostic Value of Circulating Cell-Free DNA in Colorectal Cancer: A Meta-Analysis. J. Cancer. 2016;7:1105–1113. doi: 10.7150/jca.14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.No J.H., Kim K., Park K.H., Kim Y.-B. Cell-free DNA Level as a Prognostic Biomarker for Epithelial Ovarian Cancer. Anticancer Res. 2012;32:3467. [PubMed] [Google Scholar]
- 93.Bortolin M.T., Tedeschi R., Bidoli E., Furlan C., Basaglia G., Minatel E., Gobitti C., Franchin G., Trovò M., De Paoli P. Cell-free DNA as a prognostic marker in stage I non-small-cell lung cancer patients undergoing stereotactic body radiotherapy. Biomarkers. 2015;20:422–428. doi: 10.3109/1354750X.2015.1094139. [DOI] [PubMed] [Google Scholar]
- 94.Ibarra A., Zhuang J., Zhao Y., Salathia N., Huang V., Acosta A., Aballi J., Toden S., Karns A., Purnajo I., et al. Non-invasive characterization of human bone marrow stimulation and reconstitution by cell-free messenger RNA sequencing. Nat. Commun. 2020;11:400. doi: 10.1038/s41467-019-14253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guan S., Lu Y. Dissecting organ-specific transcriptomes through RNA-sequencing. Plant Methods. 2013;9:42. doi: 10.1186/1746-4811-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Camunas-Soler J., Gee E.P.S., Reddy M., Mi J.D., Thao M., Brundage T., Siddiqui F., Hezelgrave N.L., Shennan A.H., Namsaraev E., et al. Predictive RNA profiles for early and very early spontaneous preterm birth. Am. J. Obstet. Gynecol. 2022;227:72.e1–72.e16. doi: 10.1016/j.ajog.2022.04.002. [DOI] [PubMed] [Google Scholar]
- 97.Pucciarelli S., Rampazzo E., Briarava M., Maretto I., Agostini M., Digito M., Keppel S., Friso M.L., Lonardi S., De Paoli A., et al. Telomere-Specific Reverse Transcriptase (hTERT) and Cell-free RNA in Plasma as Predictors of Pathologic Tumor Response in Rectal Cancer Patients Receiving Neoadjuvant Chemoradiotherapy. Ann. Surg. Oncol. 2012;19:3089–3096. doi: 10.1245/s10434-012-2272-z. [DOI] [PubMed] [Google Scholar]
- 98.Zaporozhchenko I.A., Ponomaryova A.A., Rykova E.Y., Laktionov P.P. The potential of circulating cell-free RNA as a cancer biomarker: Challenges and opportunities. Expert Rev. Mol. Diagn. 2018;18:133–145. doi: 10.1080/14737159.2018.1425143. [DOI] [PubMed] [Google Scholar]
- 99.Wang W., Lou W., Ding B., Yang B., Lu H., Kong Q., Fan W. A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging. 2019;11:2610. doi: 10.18632/aging.101933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.López-Urrutia E., Bustamante Montes L.P., Ladrón de Guevara Cervantes D., Pérez-Plasencia C., Campos-Parra A.D. Crosstalk between long non-coding RNAs, micro-RNAs and mRNAs: Deciphering molecular mechanisms of master regulators in cancer. Front. Oncol. 2019;9:669. doi: 10.3389/fonc.2019.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Abbosh C., Birkbak N.J., Wilson G.A., Jamal-Hanjani M., Constantin T., Salari R., Le Quesne J., Moore D.A., Veeriah S., Rosenthal R., et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Leal A., van Grieken N.C.T., Palsgrove D.N., Phallen J., Medina J.E., Hruban C., Broeckaert M.A.M., Anagnostou V., Adleff V., Bruhm D.C., et al. White blood cell and cell-free DNA analyses for detection of residual disease in gastric cancer. Nat. Commun. 2020;11:525. doi: 10.1038/s41467-020-14310-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Garcia-Murillas I., Schiavon G., Weigelt B., Ng C., Hrebien S., Cutts R.J., Cheang M., Osin P., Nerurkar A., Kozarewa I., et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 2015;7:302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 104.Del Giudice I., Raponi S., Della Starza I., De Propris M.S., Cavalli M., De Novi L.A., Cappelli L.V., Ilari C., Cafforio L., Guarini A., et al. Minimal Residual Disease in Chronic Lymphocytic Leukemia: A New Goal? Front. Oncol. 2019;9:689. doi: 10.3389/fonc.2019.00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rossi G., Minervini M.M., Carella A.M., de Waure C., di Nardo F., Melillo L., D’Arena G., Zini G., Cascavilla N. Comparison between multiparameter flow cytometry and WT1-RNA quantification in monitoring minimal residual disease in acute myeloid leukemia without specific molecular targets. Leuk. Res. 2012;36:401–406. doi: 10.1016/j.leukres.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 106.Pascual J., Cutts R.J., Kingston B., Hrebien S., Kilburn L.S., Kernaghan S., Moretti L., Wilkinson K., Wardley A.M., Macpherson I.R., et al. Assessment of early ctDNA dynamics to predict efficacy of targeted therapies in metastatic breast cancer: Results from plasmaMATCH trial. Cancer Res. 2021;81:PS5-02. doi: 10.1158/1538-7445.SABCS20-PS5-02. [DOI] [Google Scholar]
- 107.Higgins M.J., Jelovac D., Barnathan E., Blair B., Slater S., Powers P., Zorzi J., Jeter S.C., Oliver G.R., Fetting J., et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin. Cancer. Res. 2012;18:3462–3469. doi: 10.1158/1078-0432.CCR-11-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li Y., Lv J., Wan S., Xin J., Xie T., Li T., Zhu W., Zhang G., Wang Y., Tang Y., et al. High Sensitive and Non-invasive ctDNAs Sequencing Facilitate Clinical Diagnosis And Clinical Guidance of Non-small Cell Lung Cancer Patient: A Time Course Study. Front. Oncol. 2018;8:491. doi: 10.3389/fonc.2018.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen K.Z., Lou F., Yang F., Zhang J.B., Ye H., Chen W., Guan T., Zhao M.Y., Su X.X., Shi R., et al. Circulating Tumor DNA Detection in Early-Stage Non-Small Cell Lung Cancer Patients by Targeted Sequencing. Sci. Rep. 2016;6:31985. doi: 10.1038/srep31985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee J.Y., Qing X., Xiumin W., Yali B., Chi S., Bak S.H., Lee H.Y., Sun J.M., Lee S.H., Ahn J.S., et al. Longitudinal monitoring of EGFR mutations in plasma predicts outcomes of NSCLC patients treated with EGFR TKIs: Korean Lung Cancer Consortium (KLCC-12-02) Oncotarget. 2016;7:6984–6993. doi: 10.18632/oncotarget.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kutilin D.S., Airapetova T.G., Anistratov P.A., Pyltsin S.P., Leiman I.A., Karnaukhov N.S., Kit O.I. Copy Number Variation in Tumor Cells and Extracellular DNA in Patients with Lung Adenocarcinoma. Bull. Exp. Biol. Med. 2019;167:771–778. doi: 10.1007/s10517-019-04620-y. [DOI] [PubMed] [Google Scholar]
- 112.Peng H., Lu L., Zhou Z., Liu J., Zhang D., Nan K., Zhao X., Li F., Tian L., Dong H., et al. CNV Detection from Circulating Tumor DNA in Late Stage Non-Small Cell Lung Cancer Patients. Genes. 2019;10:926. doi: 10.3390/genes10110926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sozzi G., Conte D., Leon M., Cirincione R., Roz L., Ratcliffe C., Roz E., Cirenei N., Bellomi M., Pelosi G. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J. Clin. Oncol. 2003;21:3902–3908. doi: 10.1200/JCO.2003.02.006. [DOI] [PubMed] [Google Scholar]
- 114.Naidoo M., Gibbs P., Tie J. ctDNA and Adjuvant Therapy for Colorectal Cancer: Time to Re-Invent Our Treatment Paradigm. Cancers. 2021;13:346. doi: 10.3390/cancers13020346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen G., Peng J., Xiao Q., Wu H.-X., Wu X., Wang F., Li L., Ding P., Zhao Q., Li Y., et al. Postoperative circulating tumor DNA as markers of recurrence risk in stages II to III colorectal cancer. J. Hematol. Oncol. 2021;14:80. doi: 10.1186/s13045-021-01089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang Y., Yang L., Bao H., Fan X., Xia F., Wan J., Shen L., Guan Y., Bao H., Wu X. Utility of ctDNA in predicting response to neoadjuvant chemoradiotherapy and prognosis assessment in locally advanced rectal cancer: A prospective cohort study. PLoS Med. 2021;18:e1003741. doi: 10.1371/journal.pmed.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Osumi H., Shinozaki E., Yamaguchi K. Circulating tumor DNA as a novel biomarker optimizing chemotherapy for colorectal cancer. Cancers. 2020;12:1566. doi: 10.3390/cancers12061566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tan L., Sandhu S., Lee R.J., Li J., Callahan J., Ftouni S., Dhomen N., Middlehurst P., Wallace A., Raleigh J., et al. Prediction and monitoring of relapse in stage III melanoma using circulating tumor DNA. Ann. Oncol. 2019;30:804–814. doi: 10.1093/annonc/mdz048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Seremet T., Jansen Y., Planken S., Njimi H., Delaunoy M., El Housni H., Awada G., Schwarze J.K., Keyaerts M., Everaert H., et al. Undetectable circulating tumor DNA (ctDNA) levels correlate with favorable outcome in metastatic melanoma patients treated with anti-PD1 therapy. J. Transl. Med. 2019;17:303. doi: 10.1186/s12967-019-2051-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Arshad J., Barreto-Coelho P., Jonczak E., Espejo A., D’Amato G., Trent J.C. Identification of Genetic Alterations by Circulating Tumor DNA in Leiomyosarcoma: A Molecular Analysis of 73 Patients. J. Immunother. Precis. Oncol. 2020;3:64–68. doi: 10.36401/JIPO-20-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Krebs M., Dive C., Dean E.J., Rothwell D.G., Brognard J., Wallace A., Miller C., Cook N., Rafii S., Ayub M., et al. TARGET trial: Molecular profiling of circulating tumour DNA to stratify patients to early phase clinical trials. J. Clin. Oncol. 2016;34:TPS11614. doi: 10.1200/JCO.2016.34.15_suppl.TPS11614. [DOI] [Google Scholar]
- 122.Yardley D.A. MONALEESA clinical program: A review of ribociclib use in different clinical settings. Future Oncol. 2019;15:2673–2686. doi: 10.2217/fon-2019-0130. [DOI] [PubMed] [Google Scholar]
- 123.Sicklick J.K., Kato S., Okamura R., Schwaederle M., Hahn M.E., Williams C.B., De P., Krie A., Piccioni D.E., Miller V.A., et al. Molecular profiling of cancer patients enables personalized combination therapy: The I-PREDICT study. Nat. Med. 2019;25:744–750. doi: 10.1038/s41591-019-0407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kwan E.M., Spain L., Anton A., Gan C.L., Garrett L., Chang D., Liow E., Bennett C., Zheng T., Yu J., et al. Avelumab Combined with Stereotactic Ablative Body Radiotherapy in Metastatic Castration-resistant Prostate Cancer: The Phase 2 ICE-PAC Clinical Trial. Eur. Urol. 2022;81:253–262. doi: 10.1016/j.eururo.2021.08.011. [DOI] [PubMed] [Google Scholar]
- 125.Grussie E., Danenberg G., Kollengode K.A., Danenberg K., Rabizadeh S., Huang E., Usher J.L., Danenberg P.V., Pinski J.K. Gene expression differences in cell-free RNA between prostate cancer patients and healthy individuals. J. Clin. Oncol. 2018;36:e17077. doi: 10.1200/JCO.2018.36.15_suppl.e17077. [DOI] [Google Scholar]
- 126.Huang C.-Y.A., Warren S., Reeves J., Wurden M., Riordan T., Boykin R., Liu M., Henderson D., Hanson D., Gerlach J. Profiling plasma cell-free RNA (cfRNA) with the NanoString low input nCounter assay; Proceedings of the AACR Annual Meeting; Atlanta, GA, USA. 29 March–3 April 2019; p. 406. [Google Scholar]
- 127.Durand X., Moutereau S., Xylinas E., de la Taille A. Progensa™ PCA3 test for prostate cancer. Expert Rev. Mol. Diagn. 2011;11:137–144. doi: 10.1586/erm.10.122. [DOI] [PubMed] [Google Scholar]
- 128.Sasamori H., Uno K., Wu J. Usefulness of both PET/CT with F18-FDG and whole-body diffusion-weighted imaging in cancer screening: A preliminary report. Ann. Nucl. Med. 2019;33:78–85. doi: 10.1007/s12149-018-1307-3. [DOI] [PubMed] [Google Scholar]
- 129.Lennon A.M., Buchanan A.H., Kinde I., Warren A., Honushefsky A., Cohain A.T., Ledbetter D.H., Sanfilippo F., Sheridan K., Rosica D., et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science. 2020;369:eabb9601. doi: 10.1126/science.abb9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kwee S., Song M.-A., Cheng I., Loo L., Tiirikainen M. Measurement of Circulating Cell-Free DNA in Relation to 18F-Fluorocholine PET/CT Imaging in Chemotherapy-Treated Advanced Prostate Cancer. Clin. Transl. Sci. 2012;5:65–70. doi: 10.1111/j.1752-8062.2011.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Woff E., Kehagias P., Vandeputte C., Ameye L., Guiot T., Paesmans M., Hendlisz A., Flamen P. Combining 18F-FDG PET/CT—Based Metabolically Active Tumor Volume and Circulating Cell-Free DNA Significantly Improves Outcome Prediction in Chemorefractory Metastatic Colorectal Cancer. J. Nucl. Med. 2019;60:1366–1372. doi: 10.2967/jnumed.118.222919. [DOI] [PubMed] [Google Scholar]
- 132.Sun K., Wang J., Wang H., Sun H. GeneCT: A generalizable cancerous status and tissue origin classifier for pan-cancer biopsies. Bioinformatics. 2018;34:4129–4130. doi: 10.1093/bioinformatics/bty524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.