Abstract
The Pseudomonas aeruginosa fabI structural gene, encoding enoyl-acyl carrier protein (ACP) reductase, was cloned and sequenced. Nucleotide sequence analysis revealed that fabI is probably the last gene in a transcriptional unit that includes a gene encoding an ATP-binding protein of an ABC transporter of unknown function. The FabI protein was similar in size and primary sequence to other bacterial enoyl-ACP reductases, and it contained signature motifs for the FAD-dependent pyridine nucleotide reductase and glucose/ribitol dehydrogenase families, respectively. The chromosomal fabI gene was disrupted, and the resulting mutant was viable but possessed only 62% of the total enoyl-ACP reductase activity found in wild-type cell extracts. The fabI-encoded enoyl-ACP reductase activity was NADH dependent and inhibited by triclosan; the residual activity in the fabI mutant was also NADH dependent but not inhibited by triclosan. An polyhistidine-tagged FabI protein was purified and characterized. Purified FabI (i) could use NADH but not NADPH as a cofactor; (ii) used both crotonyl-coenzyme A and crotonyl-ACP as substrates, although it was sixfold more active with crotonyl-ACP; and (iii) was efficiently inhibited by low concentrations of triclosan. A FabI Gly95-to-Val active-site amino acid substitution was generated by site-directed mutagenesis, and the mutant protein was purified. The mutant FabI protein retained normal enoyl-ACP reductase activity but was highly triclosan resistant. When coupled to FabI, purified P. aeruginosa N-butyryl-l-homoserine lactone (C4-HSL) synthase, RhlI, could synthesize C4-HSL from crotonyl-ACP and S-adenosylmethionine. This reaction was NADH dependent and inhibited by triclosan. The levels of C4-HSL and N-(3-oxo)-dodecanoyl-l-homoserine lactones were reduced 50% in a fabI mutant, corroborating the role of FabI in acylated homoserine lactone synthesis in vivo.
The important opportunistic pathogen Pseudomonas aeruginosa contains a type II or dissociated fatty acid synthetase system, in which the individual reactions are catalyzed by separate proteins (22, 24). Although the overall organization of the P. aeruginosa fab genes is similar to that in Escherichia coli (for reviews see references 9 and 28), some potentially significant differences exist. First, the P. aeruginosa fabA and fabB genes, encoding β-hydroxyacyl-acyl carrier protein (ACP) dehydratase and β-ketoacyl-ACP synthase I, form an operon (22), whereas in E. coli these genes map to separate genetic loci. Second, unlike in E. coli and several other gram-negative bacteria, the fabH gene, encoding β-ketoacyl-synthase III, is absent from the fabD-fabG-acpP-fabF gene cluster, encoding malonyl-coenzyme A (CoA):ACP transacylase, β-ketoacyl-ACP reductase, ACP, and β-ketoacyl-ACP synthase II (25). Searches of the P. aeruginosa genome database revealed several potential fabH homologs located elsewhere on the genome.
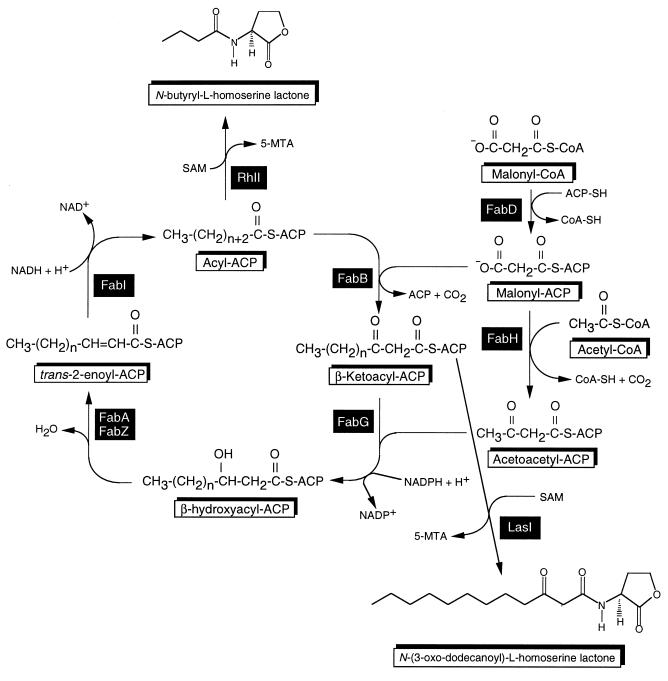
Our current model for fatty acid biosynthesis in P. aeruginosa is shown in Fig. 1. Three β-ketoacyl-ACP synthases, KAS I (FabB), KAS II (FabF), and KAS III (FabH), play pivotal roles in fatty acid synthesis. Initiation requires malonyl-CoA and malonyl-ACP. Malonyl-CoA is synthesized from acetyl-CoA via the acetyl carboxylase reaction (5). Malonyl-ACP is derived from malonyl-CoA and ACP by malonyl-CoA:ACP transacylase (FabD) (24, 25). Although a P. aeruginosa fabH homolog has only tentatively been identified, the first cycle of elongation is probably initiated by KAS III (FabH), which condenses malonyl-ACP with acetyl-CoA. Subsequent cycles are then initiated by condensation of malonyl-ACP with acyl-ACP, catalyzed by KAS I (FabB) for saturated fatty acid substrates and KAS II (FabF) for unsaturated fatty acid substrates. In the second step, the resulting β-ketoester is reduced to a β-hydroxyacyl-ACP by a NADPH-dependent β-ketoacyl-ACP reductase (FabG). It has recently been shown that P. aeruginosa contains a FabG homolog, RhlG, which presumably functions as a NADPH-dependent β-ketoacyl-ACP reductase specific for rhamnolipid synthesis (6). The third step in the cycle is catalyzed by either the fabA- or fabZ-encoded β-hydroxyacyl-ACP dehydratase. The final step in each cycle involves conversion of trans-2-enoyl-ACP to acyl-ACP, a reaction catalyzed by NADH-dependent enoyl-ACP reductase (FabI). Physiologically, FabI is an important enzyme because (i) reduction of enoyl-ACP derivatives is thought to regulate the ratio of saturated to unsaturated fatty acids and to coordinate fatty acid and phospholipid syntheses (16, 23); and (ii) FabI plays a determinant role in completing cycles of fatty acid elongation (15). FabI belongs to the short-chain alcohol dehydrogenase family and in E. coli is the target of a group of antibacterial compounds, the diazoborines (44) and triclosan (19, 33). In Mycobacterium tuberculosis, the FabI homolog InhA is one of the targets for the clinically used antimycobacterial isoniazid (2), and genetic evidence was obtained that M. smegmatis InhA is a triclosan target (32). More recently, it has been shown that E. coli mutants resistant to the antiseptic triclosan contain fabI mutations (33), and a subsequent study confirmed that triclosan and other related compounds indeed target FabI (19). Although P. aeruginosa PAO1 is resistant to triclosan due to constitutive expression of the mexAB-oprM-encoded efflux pump, Δ(mexAB-oprM) mutants are susceptible to triclosan (41). The E. coli FabI protein has been cocrystallized with NADH and thienodiazoborine (18), or NAD+ (26), and these studies allowed definition of the enzyme active site and amino acid residues important in substrate and drug interactions.
FIG. 1.
Current model for fatty acid biosynthesis in P. aeruginosa and proposed role of butyryl-ACP as acyl donor in N-butyryl-l-homoserine lactone synthesis. The genes for all enzymes except one (fabH) have been identified and characterized; for fabH, only a tentative identification has been made among several paralogs. Abbreviations: ACP, acyl carrier protein; ACP-SH, ACP with 4′-phosphopantetheine co-factor; CoA-SH; coenzyme A; FabA (and FabZ), β-hydroxyacyl-ACP dehydratase; FabB, β-ketoacyl synthase I; FabD; malonyl-CoA:ACP transacylase; FabF, β-ketoacyl synthase II; FabG, β-ketoacyl-ACP reductase; FabH, β-ketoacyl synthase III; FabI; enoyl-ACP reductase; 5-MTA, 5-methylthioadenosine; SAM, S-adenosylmethionine; LasI, N-(3-oxo)-dodecanoyl homoserine lactone synthase; RhlI, N-butyryl homoserine lactone synthase.
Besides providing fatty acid intermediates for a multitude of cellular constituents (phospholipids [9], lipid A [37], etc.), the Fab pathway has been implicated in providing the acyl groups for the acylated homoserine lactones (HSLs) that are the signalling molecules in quorum sensing (14, 35, 39, 45). In P. aeruginosa, quorum sensing is a mechanism that regulates virulence factor gene expression (46), biofilm formation in vitro (10) and in natural settings (31), twitching motility (13), and other important cellular processes. According to the current model, quorum sensing in P. aeruginosa involves two separate systems, LasR-LasI and RhlR-RhlI. These proteins are encoded by two separate, tandemly arranged transcriptional units. In these two systems, the LasI and RhlI proteins are HSL synthases that direct the synthesis of N-(3-oxo)-dodecanoyl-l-homoserine lactone (3-oxo-C12-HSL) and N-butyryl-l-homoserine lactone (C4-HSL), respectively. According to the current model, the synthases react with acyl-ACPs and S-adenosylmethionine (SAM) to form the cognate HSLs, with concomitant release of 5-methylthioadenosine (35, 39). For C4-HSL synthesis, the rhlI-encoded P. aeruginosa HSL synthase RhlI catalyzes the formation of C4-HSL in a reaction utilizing butyryl-ACP as the acyl donor and SAM as the nucleophile in a lactonization reaction (35, 39).
In this report, we describe the characterization of the P. aeruginosa fabI gene and its product, show evidence for the presence of at least one other enoyl-ACP reductase in this bacterium, provide biochemical evidence that FabI is a triclosan target, and demonstrate its role in C4-HSL synthesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and media.
The relevant strains, plasmids, and primers are described in Table 1. LB (Luria-Bertani) medium (34) was routinely used as the rich medium for all bacterial strains. For growth of the fabI(Ts) mutant JP1111, the NaCl concentration of LB medium was reduced to 0.05%. For some experiments, RB medium (20) was used instead of LB. The minimal medium used for growth of P. aeruginosa was VBMM (40). The antibiotics used in selection media were as follows: for E. coli, ampicillin (100 μg/ml) and gentamicin (15 μg/ml); for P. aeruginosa, carbenicillin (500 μg/ml) and gentamicin (200 μg/ml).
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description or sequencea | Reference or source |
|---|---|---|
| E. coli | ||
| DH5αF′ | [F+ φ80lacZΔM15] Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 [rK− mK+] supE44 thi-1 gyrA relA1 | 27 |
| JP1111 | Hfr galE45 fabI392(Ts) relA1 spoT1 | 44 |
| BL21(DE3) | E. coli B; F−ompT rB− mB− (λDE3) | 43 |
| P. aeruginosa | ||
| PAO1 | PAO1 | B. H. Holloway |
| PAO200 | PAO1 with Δ(mexAB-oprM) | 41 |
| PAO234 | PAO1 with fabI::Gmr-FRT | This study |
| PAO235 | PAO1 with fabI::FRT | This study |
| Plasmids | ||
| pET-15b | Apr; hexahistidine fusion and expression vector | Novagen |
| pWSK29/30 | Apr; low-copy-number cloning and T7 expression vectors | 47 |
| pFLP2 | Apr; source of Flp recombinase | 21 |
| pPS856 | Apr Gmr; source of Gmr-FRT cassette | 21 |
| pUCP21T | Apr; broad-host-range cloning vector | 42 |
| pEX18Tc | Tcr; sacB-based gene replacement vector | 21 |
| pPS922 | AprfabI+ (ligation of a 880-bp BamHI-HindIII PCR fragment between the same sites of pWSK30; fabI-transcription driven by Plac) | This study |
| pPS925 | AprfabI::Gmr-FRT (ligation of a 1,053-bp blunt-ended SacI fragment from pPS856 into the SmaI site of pPS922) | This study |
| pPS933 | AprsacB+ fabI::Gmr-FRT (ligation of a 1.95-kb BamHI-HindIII fragment between the same sites of pEX18Tc) | This study |
| pPS935 | Apr; H6-FabI expression vector (PCR-amplified 0.85-kb NdeI-BamHI fragment cloned between same sites of pET-15b) | This study |
| pPS967 | AprfabI+ (pUCP21T with BamHI-HindIII fragment from pPS922; fabI transcription driven by Plac) | This study |
| pPS1098 | Apr; H6-FabI expression vector (PCR-amplified 0.85-kb NdeI-BamHI fragment cloned between same sites of pET-15b); expresses H6-FabI mutant protein | This study |
| Primers | ||
| FabIU | HindIII-GCAAGCttGCGGAAAACTGAGTAGCAGA | |
| FabID | BamHI-ATGGatCCGCTTGAAGCGGCATTTTTC | |
| FabI-Nde | NdeI-GGcatATGGGATTTCTCACAGGAAAAC | |
| FabI G>V | CCACTCCGTCGtATTCGCTCCAG |
Plac, E. coli lac operon promoter. Primer sequences are printed 5′ to 3′; lowercase letters indicate nonmatching oligonucleotides used to either form the indicated motif as underlined or introduce other nonmotif changes indicated in lowercase letters.
General DNA procedures.
Routine DNA were performed as previously described (21). The details underlying construction of pPS921 were as follows. For PCR amplification of a fragment from chromosomal DNA, two mutagenic oligonucleotides were synthesized. The FabIU and FabID primers were designed to introduce a HindIII site upstream of fabI and a BamHI site downstream of fabI, respectively (Table 1). These oligonucleotides were used to prime synthesis from PAO1 chromosomal DNA. PCRs were performed on a PTC-100 PCR system thermocycler (MJ Research, Watertown, Mass.) as previously described (21, 22). The ca. 870-bp PCR fragment was ligated to BamHI-HindIII-digested pWSK30 (47) DNA to form pPS921. Nucleotide sequences were determined and analyzed as previously described (22). Motif searches were performed by using the on-line E-Motif program provided by Stanford University, Palo Alto, Calif.
A two-step method was used for site-directed mutagenesis for fabI. First, PCRs were set up as described above but with pPS935 DNA as the template, and DNA synthesis was primed with the T7 terminator primer (Novagen, Madison, Wis.) and a mutagenic primer, FabI G>V (Table 1), that was designed to (i) introduce a single G-to-T change which leads to a Gly95-to-Val amino acid change and (ii) yield a PCR product that would properly prime to pPS935 DNA in a second round of PCR even when T-tailed by Taq DNA polymerase. The single 1,046-bp PCR product was digested with BamHI plus NdeI, and the resulting 840-bp BamHI-NdeI fragment was gel purified and ligated to BamHI-NdeI-digested pET-15b DNA. The presence of the single G-to-T change was verified by nucleotide sequence analysis.
Disruption of the fabI gene.
The fabI strain PAO235 was isolated in several steps. A plasmid-borne fabI mutation was constructed by insertion of a blunt-ended 1,053-bp gentamicin resistance (Gmr)-FRT cassette from pPS856 (21) into the SmaI site located within the fabI coding sequence. The resulting mutation was returned to the P. aeruginosa chromosome after subcloning into the sacB-based suicide vector pEX18Tc (21) to derive the fabI::Gmr-FRT mutant PAO234. From PAO234, the unmarked fabI::FRT mutant PAO235 was then derived by in vivo excision of the Gmr-FRT cassette with Flp recombinase (21). The mutants were confirmed by performing colony PCR (21) utilizing the FabIU and FabID primers.
Purification of an polyhistidine-tagged FabI fusion protein.
A hexahistidine-FabI (H6-FabI) expression vector was constructed by PCR amplifying the fabI coding sequence from PAO1 genomic DNA with primers FabI-Nde, creating a NdeI site at the fabI ATG initiation codon, and FabID, which creates a BamHI site immediately downstream of fabI. The PCR fragment was digested with NdeI and BamHI, and the resulting 840-bp BamHI-NdeI fragment was ligated between the same restriction sites of pET-15b to form pPS935, which was then transformed into E. coli BL21(DE3). Expression of H6-FabI, cell lysis, and purification of the soluble fusion protein on a Ni2+-agarose column (Qiagen) were performed as previously described (17) except that the cells were grown in LB medium containing ampicillin (LB-Ap medium). The same procedure was used for purification of a mutant FabI protein.
Protein concentrations were determined by using the Bradford dye binding assay (Bio-Rad Laboratories, Hercules, Calif.) and bovine serum albumin as the standard. Proteins were analyzed by electrophoresis on 0.1% sodium dodecyl sulfate–10% polyacrylamide gels (SDS-PAGE) as previously described (29). The proteins were visualized by quick staining with Coomassie brilliant blue R-250 (7).
Enoyl-ACP reductase assays.
P. aeruginosa cell extracts were prepared from exponentially growing cells (A540 of ∼0.8 to 1.0). Cells grown in LB medium at 37°C were harvested by centrifugation and then suspended and washed in 0.1 M sodium phosphate buffer (pH 6.5). Cell lysates were prepared by passing the cell suspensions three times through a French pressure cell (19,000 lb/in2). Cell debris was removed by ultracentrifugation for 1 h at 260,000 × g, and the supernatants were saved as cell extracts.
Enoyl-ACP reductase activity of purified H6-FabI, as well as in cell extracts, was determined by using crotonyl-ACP as the substrate. This was done by measuring spectrophotometrically the decrease in absorbance at 340 nm at room temperature due to consumption of NADH (or NADPH) during reduction of crotonyl-ACP (3). In 500 μl, a typical assay mixture contained 0.1 M sodium phosphate (pH 7.5), 0.1 mM NADH, and 1 μg of purified H6-FabI. The reactions were initiated by addition of 20 μM crotonyl-ACP. In some instances, 0.1 mM NADPH was used as the reductant. For some experiments, enzyme and substrate concentrations were varied as indicated in the figure legends. A stock solution of triclosan was prepared in 95% ethanol, and appropriate amounts of ethanol were added to control experiments to assess possible inhibitory effects of the solvent on enzyme activities.
Crotonyl-ACP was prepared by using a previously published procedure (48), with slight modifications. Briefly, 20 mg of purified P. aeruginosa ACP was precipitated as previously described (8) except that the hydroxylamine step was omitted. After precipitation of the protein with 10% trichloroacetic acid for 30 min on ice, the precipitate was pelleted in a microcentrifuge and then washed once with 0.2 M citrate buffer (pH 4.0) and once with water. The protein was resuspended in 0.1 M sodium phosphate buffer (pH 8.0) containing 2 μmol of dithiothreitol and reacted with a 10-fold molar excess of crotonic anhydride (Aldrich, Milwaukee, Wis.) for 30 min at 4°C with constant stirring. The reaction mixture was loaded on a PD10 column (Pharmacia Biotech, Uppsala, Sweden) that had been equilibrated with 0.1 M sodium phosphate buffer (pH 7.5), and the crotonyl-ACP was eluted with 3.5 ml of the same buffer. The crotonyl-ACP concentration was determined by measuring the difference in the number of free thiol groups of an untreated versus a hydroxylamine-treated sample by the Ellmann technique (12). The treatment of crotonyl-ACP (20 μl of the column eluate) was performed for 5 min at room temperature in a 1 M hydroxylamine solution in 0.1 M sodium phosphate buffer (pH 7.5). For thiol group determinations, hydroxylamine-treated or untreated crotonyl-ACP was reacted for 30 min at room temperature with 0.15 mM 5,5′-dithiobis (2-nitrobenzoic acid) (Aldrich) in a final volume of 0.8 ml of 0.1 M phosphate buffer (pH 7.5), and the absorption was measured at 400 nm. According to this determination, 80% of the ACP was acylated.
Determination of acyl HSL concentrations in culture fluid.
Acyl HSLs were detected in culture fluids of LB-grown P. aeruginosa strains as previously described (36), using E. coli XL-1 Blue/pECP61.5 for detection of C4-HSL and E. coli MG4/pKDT17 for detection of 3-oxo-C12-HSL. Standard curves were prepared by using synthetic HSLs to ensure that bioassays were always performed in the linear range.
C4-HSL assays.
The in vitro C4-HSL synthesis reactions contained (in 500 μl) Moré buffer (10 mM Tris-HCl [pH 7.4], 330 mM NaCl, 15% glycerol, 0.7 mM dithiothreitol, 2 mM EDTA, 25 mM MgSO4, 0.1 mM FeSO4) (35), 0.1 mM SAM, 0.2 mM NADH, 1 μg of purified H6-RhlI (details to be published elsewhere), and various amounts of H6-FabI. The reaction mixtures were incubated at 37°C for 1 h and then extracted three times with 250 μl of ethyl acetate. The ethyl acetate extracts were vacuum dried, and the residue was suspended in 20 μl of high-pressure liquid chromatography-grade acetonitrile (Fisher Scientific, Fair Lawn, N.J.). For detection of C4-HSL, the Chromobacterium violaceum bioassay was used (30). Aliquots (5 μl) of the extracted reactions were spotted onto a Whatman (Clifton, N.J.) C18 reverse-phase thin-layer chromatography (TLC) plate and allowed to air dry. The solvent used for chromatography was 60% methanol–40% water (vol/vol). For detection, 2.5 ml of an overnight culture of strain CV026 grown at room temperature in LB medium was diluted into 25 ml of 0.3% LB agar kept at 45°C, and the agar mixture was used to overlay the dried TLC plate. After solidification of the overlay, the TLC plate was incubated overnight at room temperature in a wet box. The presence of C4-HSL in the extracted reactions was scored by appearance of a violet spot on the TLC plates.
Nucleotide sequence accession number.
The DNA sequence of the fabI region has been deposited in GenBank under accession no. AF104262.
RESULTS
Cloning of the fabI gene of P. aeruginosa.
At the onset of this project, an examination of the incomplete Pseudomonas genome database release revealed two contigs which contained sequences that in BLAST searches showed extensive similarities to E. coli fabI. The amino and carboxy termini were located on two different contigs. The putative fabI coding sequence was successfully amplified from P. aeruginosa chromosomal DNA by using two oligonucleotide primers, FabIU and FabID (Table 1). These two primers were designed to introduce a unique HindIII site upstream of fabI and its putative ribosome binding site and to introduce a unique BamHI site downstream of fabI. PCR amplification was very specific and yielded a 880-bp product that was digested with BamHI plus HindIII and ligated to similarly digested pWSK30 DNA to form pPS922. This procedure placed fabI under transcriptional control of the lac promoter (Plac) since the cloned fabI gene presumably lacks its own promoter. The low-copy-number (five to eight copies per cell) cloning vector pWSK30 (47) was used to avoid potential toxicity problems that we had observed during the cloning of other fab genes (25). The fabI(Ts) E. coli strain JP1111 does not grow at 42°C and has a low-salt tolerance at the nonpermissive temperature; e.g., it does not grow on regular LB medium containing 0.5% NaCl. This strain was transformed with pPS922 DNA, and Apr transformants were selected at 30°C on LB-Ap medium. The transformants were then transferred to LB-Ap medium with and without 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubated at 42°C. Within 24 h, only the colonies plated on LB-Ap medium with IPTG grew under these conditions, indicating that the cloned fabI (i) complemented the temperature-sensitive (Ts) and low-salt-tolerance phenotype of the E. coli fabI(Ts) mutant and (ii) does not contain its own promoter since transcription from Plac requires IPTG induction in the lac repressor producing strain JP1111. When JP1111 was transformed with pPS967, a multicopy plasmid expressing fabI from Plac (Table 1), complementation was observed in the absence of IPTG since the amounts of chromosomally encoded lac repressor were insufficient to fully repress fabI transcription.
Sequence analysis of the fabI gene.
Since the fabI gene sequence in the incomplete genome database was contained on two separate contigs, we sequenced the cloned 874-bp HindIII-BamHI fragment which contained a single open reading frame (ORF), and BLAST searches revealed significant identity of the protein to other FabI proteins. Alignments showed the greatest similarities with FabI proteins from E. coli and Salmonella typhimurium (69% identity; 81% similarity) and Haemophilus influenzae (63% identity; 76% similarity). This similarity also extended to FabIs from gram-positive bacteria, including Bacillus subtilis (49% identity; 66% similarity).
Analysis of the deduced FabI amino acid sequence using the E-Motif program revealed a glucose/ribitol dehydrogenase family signature motif (P..[KR].....[DE][FILVY].....[FWY].[AST]), as well as a signature motif for the FAD-dependent pyridine nucleotide reductase family (D..[FILMV]..[IV]G..P...L]. The latter sequence includes Gly95, which is equivalent to a glycine found in position 93 of E. coli FabI. In E. coli, Gly93-to-Ser and Gly93-to-Ala changes, which lead to resistance to diazoborine (4) and triclosan (33), respectively, map close to the nucleotide binding site.
Examination of the latest P. aeruginosa genome database revealed a complete fabI gene sequence. Our sequence was identical to the database sequence with the exception of an additional G residue. This discrepancy arose from a mistake in the previous contig sequence after which our primer FabID was modeled. The fabI gene is followed by a sequence that could assume a stem-loop structure (ΔG = −48.2 kcal/mol) indicative of Rho-independent transcriptional terminators (38). Analysis of the fabI upstream sequence did not reveal any obvious promoter/regulatory sequences. Instead, the fabI gene is separated by 21 nucleotides from another ORF whose gene product exhibits significant similarity to many bacterial ATP-binding proteins that are components of ABC transporters; this ORF is 58% identical and 74% similar to the hypothetical E. coli ABC transporter ATP-binding protein YejF (GenBank accession no. U00008).
Disruption of the chromosomal fabI gene.
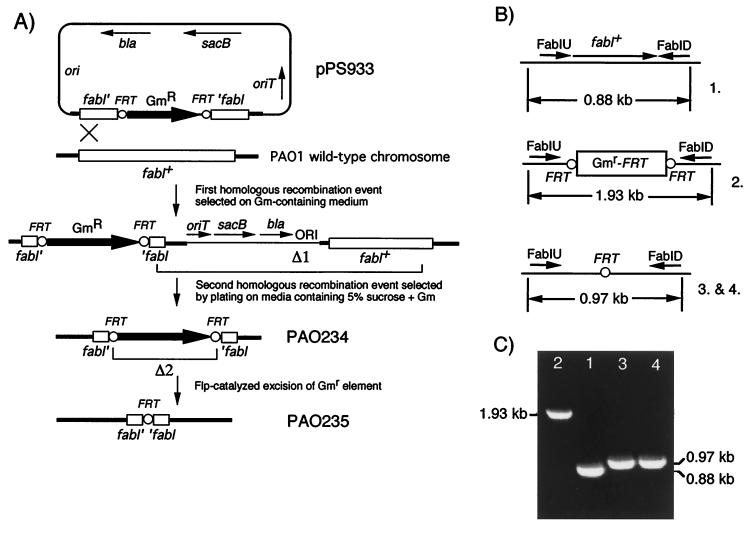
A defined pPS933-borne fabI::Gmr insertion was constructed as described in Table 1, and the deletion was returned to the P. aeruginosa chromosome as illustrated in Fig. 2A. After conjugal transfer of the nonreplicative pPS933 from E. coli SM10 into PAO1, merodiploids were obtained by selecting for Gmr. From these, colonies having undergone Δ1 were selected as sucrose resistant, Gmr, and carbenicillin resistant. The unmarked fabI::FRT mutant PAO235 was then derived from PAO234 by Flp-catalyzed excision of the Gmr marker. During its expression in the recipient, Flp recombinase acted at the FRT sites to catalyze excision of the Gmr element (labeled Δ2 in Fig. 2A), leaving behind a short FRT-containing sequence.
FIG. 2.
(A) Strategy for isolation of an unmarked chromosomal fabI mutation via sacB-counterselected gene replacement (Δ1) and Flp-mediated excision of the Gmr marker (Δ2). (B) Genetic organization of the fabI region in wild-type PAO1 (1.), the fabI::Gmr-FRT insertion mutant (2.), and the unmarked fabI mutant after Flp-mediated excision of the Gmr marker (3. and 4.). (C) PCR analysis of genomic DNA from the strains depicted in panel B. The PCRs were primed with the primers FabIU and FabID (B). Abbreviations: bla, β-lactamase-encoding gene; FRT, Flp recombinase target sites; Gm, Gmr marker; ori, pMB1 origin of replication; oriT, origin of transfer; sacB, levansucrase-encoding gene.
Successful execution of the steps labeled Δ1 and Δ2 in Fig. 2A was monitored by colony PCR analysis using the FabIU and FabID primers. As expected, the primers amplified a 880-bp fabI-containing fragment from wild-type PAO1 DNA (Fig. 2C, lane 1). In PAO234 (lane 2), this fragment was increased to 1.93 kb by insertion of the 1,053-bp Gmr-FRT cassette. After Flp-mediated excision of the Gmr marker, the PCR fragment was reduced to ∼970 bp (880 bp and one 86-bp FRT-containing sequence from pPS856) (lanes 3 and 4). These events were further verified by genomic Southern analyses utilizing Gmr and fabI-specific DNA segments as the probes (data not shown).
fabI mutants contain reduced levels of enoyl-ACP reductase activity.
The growth rates of wild-type PAO1 and the fabI mutant PAO235 were superimposable when grown in RB medium and other media (data not shown). Since PAO235 contains only a 86-bp nonpolar insertion at a SmaI site that is located late in the fabI gene, we considered the possibility that this strain could still express a functional FabI protein. To rule out this possibility, we PCR amplified the fabI region from PAO235 by using the primers FabIU and FabID and subcloned the resulting BamHI-HindIII fragment into pWSK30. Under inducing conditions, this DNA segment no longer complemented the Ts and low-salt-tolerance phenotypes of E. coli fabI(Ts) strain JP1111.
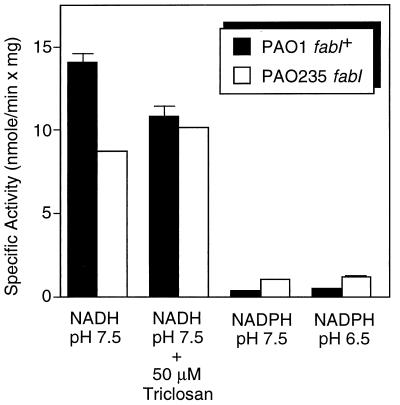
Determination of enzyme activities in cell extracts of wild-type and mutant PAO235 confirmed the existence of residual NADH-dependent activity in the mutant that amounted to 62% of the total activity observed in PAO1 extracts (Fig. 3). Whereas total NADH-dependent enoyl-ACP reductase in PAO1 was inhibited by the addition of 50 μM triclosan, this inhibitor had no effect on the residual activity found in PAO235, even when its concentration was increased to 100 μM (data not shown). Cell extracts from both strains showed very little NADPH-dependent enoyl-ACP reductase activity when assayed at pH 7.5 or 6.5.
FIG. 3.
Enoyl-ACP reductase activities in cell extracts of wild-type PAO1 and the fabI mutant PAO235. The 500-μl reactions contained in phosphate buffer with the indicated pH, 0.1 mM NADH or NADPH, and 50 to 100 μg of PAO1 extract or 300 μg of PAO235 extract for determination of NADH- or NADPH-dependent enoyl-ACP reductase activities, respectively. The reactions were initiated by addition of 13 μM crotonyl-ACP, and the decrease in absorbance at 340 nm was recorded. The results shown represent the means plus standard deviations of three experiments.
Purification and properties of purified FabI.
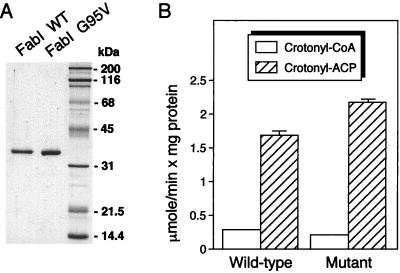
An expression vector was constructed, and FabI was purified as an H6-FabI protein. When transformed into E. coli JP1111, the expression vector could complement the fabI(Ts) mutation of this strain, indicating expression of a functional H6-FabI protein. The purified FabI protein (Fig. 4A) could utilize crotonyl-CoA and crotonyl-ACP as substrates (Fig. 4B), although it was 5.9-fold more active with crotonyl-ACP as the substrate. The enzyme was unable to utilize NADPH as cofactor (not shown), which may be a consequence of its purification at alkaline pH, conditions that may inactivate its NADPH-dependent activity, as has been observed with the E. coli enzyme (3).
FIG. 4.
(A) SDS-PAGE of purified wild-type (WT) FabI and FabI G95V mutant proteins; (B) substrate preference and specific activities of wild-type and mutant FabI proteins. (A) The proteins were expressed as H6-tagged proteins and purified by Ni2+ affinity chromatography. One microgram of each protein was analyzed by SDS-PAGE, and proteins were detected by Coomassie blue staining. The protein standards (Bio-Rad Laboratories) in the rightmost lane were (top to bottom) myosin, β-galactosidase, bovine serum albumin, ovalbumin, carbonic anhydrase, trypsin inhibitor, and lysozyme; molecular masses are shown on the right. (B) Enoyl-ACP reductase activities were measured with 20 μM crotonyl-CoA or 13 μM crotonyl-ACP as substrate and 0.1 mM NADH as cofactor. The reaction mixture contained either 1 μg of wild-type FabI or 1 μg of mutant FabI protein. The results shown represent the means plus standard deviations of three experiments.
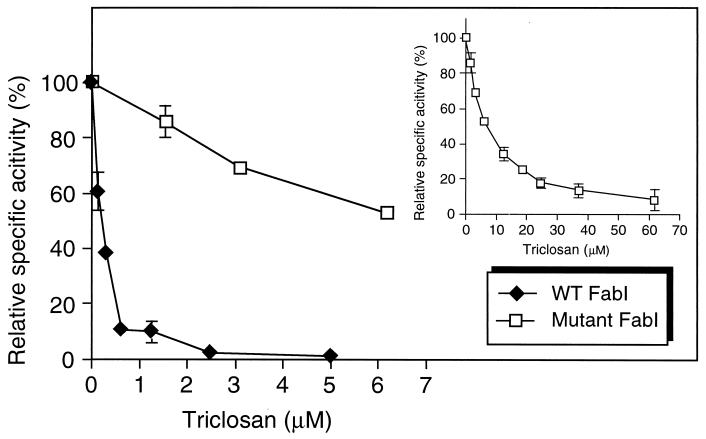
The enoyl-ACP reductase activity was inhibited by very low concentrations of triclosan (Fig. 5). Triclosan-resistant E. coli strains contain changes in amino acid residues that are thought to line the enzyme active site (1, 33), and the most resistant strains contained Gly93-to-Val substitutions (33). We therefore changed the corresponding Gly95 in the P. aeruginosa enzyme to a Val and purified the resulting H6-tagged mutant protein. The mutant protein (Fig. 4A) was slightly less active than wild-type FabI with crotonyl-CoA as the substrate and slightly more active with crotonyl-ACP as the substrate (Fig. 4B). The mutant enzyme was significantly more resistant to triclosan (Fig. 5).
FIG. 5.
Inhibition of FabI activity by triclosan. The reaction mixtures (500 μl) in phosphate buffer (pH 7.5) contained either 1 μg of purified wild-type (WT) FabI or 1 μg of mutant FabI protein, 0.1 mM NADH, 13 μM crotonyl-ACP, and increasing concentrations of triclosan. The insert shows the inhibition curve for the mutant FabI protein over a wider triclosan range; the first four values are identical to the ones plotted in the larger graph. Each value shown represents the mean plus standard deviation of three independent enzymatic reactions.
Role of FabI in HSL synthesis in vitro and in vivo.
Reduction of crotonyl-ACP by FabI to form butyryl-ACP is the last step in the first cycle of fatty acid elongation following condensation of malonyl-ACP with acetyl-CoA (Fig. 1). Theoretically, butyryl-ACP generated by FabI could serve as a substrate for transacylation of RhlI, followed by a lactonization reaction utilizing SAM as the substrate to form C4-HSL, and inhibition of FabI or mutational inactivation should therefore lead to reduced levels of HSL production.
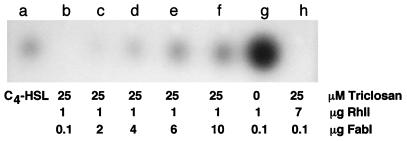
To test this hypothesis, we first set up an in vitro C4-HSL synthesis system with purified H6-FabI and H6-RhlI. In this system, RhlI synthesized C4-HSL from crotonyl-ACP in the presence of FabI, NADH, and SAM (Fig. 6, lane g). Under the same conditions, C4-HSL synthesis was completely inhibited by 25 μM triclosan (lane b), and this inhibition was partially relieved by increasing concentrations of FabI (lane c to f). Inhibition of C4-HSL synthesis by triclosan could not be relieved by increasing the RhlI concentration sevenfold (lane h).
FIG. 6.
In vitro synthesis of C4-HSL and inhibition by triclosan. In vitro synthesis reaction mixtures containing the indicated components plus 20 μM crotonyl-ACP, 0.1 mM SAM, and 0.2 mM NADH were extracted with ethyl acetate, and portions of the concentrated residues were separated by TLC. C4-HSL in the samples was detected by overlaying the TLC plate with the C. violaceum indicator strain CV026. Lane a contained 50 pmol of synthetic C4-HSL.
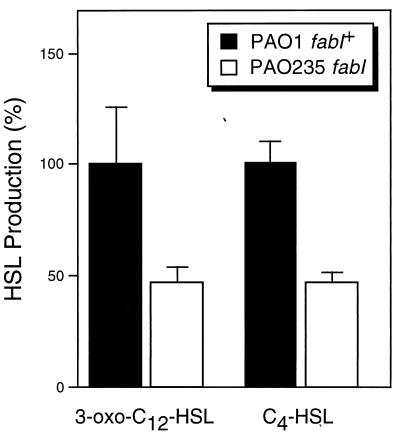
To confirm the in vivo role of fabI in HSL synthesis, HSL levels were measured in culture fluids of wild-type PAO1 and fabI mutant PAO235. The fabI mutant produced only 50% of the 3-oxo-C12-HSL and C4-HSL levels of wild-type PAO1 (Fig. 7), paralleling the reduction of FabI enoyl-ACP reductase activity observed in this mutant (Fig. 3).
FIG. 7.
HSL levels in wild-type and a fabI P. aeruginosa mutant. HSLs were extracted from overnight culture supernatants, and relative levels were determined by using E. coli-based C4-HSL and 3-oxo-C12-HSL bioassays. Values were determined in triplicate, and the values shown represent the means plus standard deviations.
DISCUSSION
The fabI gene encodes an enoyl-ACP reductase.
We have cloned and characterized a gene from P. aeruginosa, encoding an enoyl-ACP reductase, by PCR amplification from genomic DNA and complementation of a defined E. coli fabI(Ts) mutant. Sequence analysis revealed that this gene may be transcriptionally linked to an upstream ORF encoding an ATP-binding protein of an ABC transporter of unknown function, and the presence of strong downstream Rho-independent transcriptional terminator indicated that it is probably the last gene in a transcriptional unit.
Although our experiments showed that P. aeruginosa contains several enoyl-ACP reductases, several lines of evidence strongly support the notion that the cloned gene is fabI. First, the deduced FabI sequence is 69% identical to FabI sequences from E. coli and S. typhimurium and contains the signature motif (D..[FILMV]..[IV]G..P...L]) for the FAD-dependent pyridine nucleotide reductase family. Second, this motif contains the amino acid residues forming the enzyme active site that when mutationally altered confer resistance to specific inhibitors of FabI enoyl-ACP reductase activity. This included Gly95, which is equivalent to Gly93 found in E. coli FabI. In E. coli, Gly93-to-Ser and Gly93-to-Ala changes led to resistance to diazoborine (4) and triclosan (33), respectively. In M. tuberculosis, a Ser94-to-Ala mutation, which leads to isoniazid resistance, lies in the same region of the mycobacterial FabI homolog, InhA (11).
In E. coli, fabI is an essential gene and fabI knockout mutants are not viable. Mutants containing single-amino-acid substitutions can be isolated, but they grow at reduced rates (33) or contain Ts alleles (4) that grow only at the permissive temperature. We were able to disrupt the chromosomal fabI gene and showed that the disrupted gene no longer complemented an E. coli fabI(Ts) allele. This was expected since the insertion at a SmaI site disrupted amino acid residues that form an integral part of the enzyme active site (18, 26). Since we could isolate a fabI insertion mutant and since this mutant was not altered in its growth rate, we concluded that unlike E. coli, P. aeruginosa must contain more than one enoyl-ACP reductase activity. This was corroborated by the presence of significant levels of NADH-dependent enoyl-ACP reductase activities in cell extracts of the fabI mutant PAO235 (Fig. 3). Since the activity found in the mutant amounted to 62% of the total activity found in the wild type, we concluded that FabI contributes a substantial part of the total enoyl-ACP reductase activity found in P. aeruginosa. This was corroborated by triclosan inhibition studies which indicated that ∼23% of the total enoyl-ACP reductase activity in the wild type was inhibited by this drug, whereas the residual activity in PAO235 was triclosan insensitive. Searches of the P. aeruginosa genome database revealed at least 18 possible paralogs exhibiting on average 25% identity and 40% similarity to FabI, but none of them contained the signature motifs found in FabI proteins.
In cell extracts prepared at pH 6.5, E. coli FabI accepted both NADH and NADPH as cofactors, but when cell extracts were prepared at 7.5, only the NADH-dependent enoyl-ACP reductase activity remained (3). However, the NADPH-dependent FabI activity amounted to only 14 to 22% of the NADH-dependent enzyme. Although we isolated our cell extracts at pH 6.5, the NADPH-dependent FabI activity in wild-type or mutant cell extracts was very low, even at pH 6.5, and required three to six times more enzyme for detection (Fig. 3). Under these conditions, the NADPH-dependent activities were 3.5 or 10% of the NADH-dependent activities observed with wild-type and mutant extracts, respectively. We do not yet know what proportion, if any, of this activity is attributable to FabI, since our purified FabI protein does not exhibit NADPH-dependent enoyl-ACP reductase activity (see below).
To assess the substrate specificity of FabI, we expressed and purified FabI as an H6-tagged protein. As its E. coli counterpart, the purified FabI protein could use both crotonyl-CoA and crotonyl-ACP as substrates, although it was much more active with crotonyl-ACP (Fig. 4B). The purified enzyme could not use NADPH as cofactor (data not shown). We do not yet know whether this reflects an intrinsic property of the P. aeruginosa FabI enzyme or whether the NADPH-dependent activity was lost during the purification steps that involve buffers with alkaline pH. We were unable to differentiate between these two possibilities since H6 fusion proteins cannot be purified at pH 6.5.
FabI is a triclosan target.
It has recently been shown that in E. coli triclosan targets lipid synthesis, and since triclosan-resistant mutants harbor mutations in fabI, it was concluded that the most likely target is enoyl-ACP reductase (33); this was subsequently confirmed in assays using purified FabI (19). To examine whether P. aeruginosa FabI is a triclosan target, we used the purified H6-FabI in triclosan inhibition studies (Fig. 5). The purified protein was efficiently inhibited by very low concentrations of triclosan, showing 50% inhibition (50% inhibitory concentration [IC50]) with 0.2 μM triclosan. McMurry et al. (33) and Heath et al. (18, 19) showed that their most triclosan-resistant mutants contained Gly93-to-Val substitutions. To assess whether changing a glycine residue at the equivalent position 95 in P. aeruginosa FabI would lead to synthesis of a triclosan-resistant FabI enzyme, we introduced a Gly95-to-Val change by site-directed mutagenesis and then expressed and purified the mutant H6-FabI protein. Although the mutant protein exhibited a slightly increased enoyl-ACP reductase activity (Fig. 4B), it showed an IC50 of 7 μM for triclosan and was not yet completely inhibited by the highest concentration (62 μM) tested in our experiments (Fig. 5, insert). While our studies neared completion, Heath et al. (18, 19) arrived at similar results after comparing the purified E. coli FabI with Gly93-to-Ser and Gly93-to-Val mutant enzymes; the triclosan IC50s were 2 μM for FabI, 8 μM for the Gly93-to-Ser mutant enzyme (19), and 10 μM for the Gly93-to-Val mutant enzyme (18). Although both proteins were purified by using the same expression vector and same NH2-terminal H6 extension, P. aeruginosa FabI was ∼10-fold more sensitive than the E. coli enzyme to triclosan. Although we do not know the reason(s) for this observation, the differences may be partially due to the different enoyl-ACP reductase assays used for the inhibition studies. We used the natural substrate crotonyl-ACP, whereas Heath et al. (18, 19) used the synthetic substrate trans-2-octadecenoyl-N-cysteamine, and the latter assay required substantially more FabI (12 μg, versus 1 μg in our assays). The recent determination of the X-ray structure of the E. coli FabI-NAD+-triclosan complex confirmed that triclosan interacts with both the NAD+ and the protein via hydrogen and hydrophobic bonds, forming a stable ternary complex (18, 26). Mutations in the FabI active site interfere with the formation of a stable FabI-NAD+-triclosan ternary complex and thus confer resistance to the drug.
We have previously shown that P. aeruginosa PAO1 is resistant to triclosan due to efflux by the tripartite MexAB-OprM multidrug efflux system, but Δ(mexAB-oprM) mutants are susceptible to triclosan (41). The same efflux system seems also partially responsible for diazoborine resistance. Both wild-type PAO1 and fabI mutant PAO235 showed high levels of resistance to diazoborine (the MIC in both strains was >160 μg/ml), and the MIC for diazoborine was significantly lower (80 μg/ml) in the Δ(mexAB-opM) mutant PAO200.
FabI participates in homoserine lactone synthesis.
According to the current model, butyryl-ACP serves as the most likely acyl donor in C4-HSL synthesis. To confirm that FabI can indeed provide butyryl-ACP for C4-HSL synthesis by RhlI, we set up an in vitro synthesis system containing purified FabI, RhlI, the cofactor NADH, and the substrates crotonyl-ACP and SAM. This reconstituted system allowed synthesis of biologically active C4-HSL that comigrated with synthetic C4-HSL (Fig. 6). Synthesis of C4-HSL was efficiently inhibited by triclosan. This inhibition was relieved by increasing concentrations of FabI but not RhlI, indicating that FabI and not RhlI is the triclosan target. Corroborating evidence that FabI plays a central role in HSL synthesis in vivo was provided by the observation that a fabI mutant produced only 50% of the HSLs found in wild-type cells. This is the first documented evidence that an enzyme of the fatty acid biosynthetic pathway plays a crucial role in HSL synthesis, and the establishment of an in vitro HSL synthesis system will facilitate the search for novel antimicrobials aimed at interfering with the pathways involved in HSL biosynthesis. The design and development of new FabI inhibitors will form an integral part of such strategies.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant GM56685 from the National Institutes of Health. Additional funding to H.P.S. was provided by a grant from the Colorado State University Research Council of the College of Veterinary Medicine and Biomedical Sciences.
We thank Barbara Iglewski and Anatol Eberhard for the generous gifts of synthetic HSLs, and we thank Matt Parsek for providing the HSL bioassay strains and protocols. Triclosan (Irgasan) was provided as a gift from Ciba Corporation, and diazoborine was provided by Friederike Turnowsky. We also thank Charles O. Rock for providing information on the mechanism of triclosan inhibition of FabI prior to publication.
REFERENCES
- 1.Baldock C, Rafferty J B, Sedelnikova S E, Baker P J, Stuitje A R, Slabas A R, Hawkes T R, Rice D W. A mechanism of drug action revealed by structural studies of enoyl reductase. Science. 1996;274:2107–2110. doi: 10.1126/science.274.5295.2107. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um K S, Wilson T, Collins D, de Lisle G, Jacobs W R. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 3.Bergler H, Fuchsbichler S, Hoegenauer G, Turnowsky F. The enoyl-[acyl-carrier-protein] reductase (FabI) of Escherichia coli, which catalyzes a key regulatory step in fatty acid biosynthesis, accepts NADH and NADPH as cofactors and is inhibited by palmitoyl-CoA. Eur J Biochem. 1996;242:689–694. doi: 10.1111/j.1432-1033.1996.0689r.x. [DOI] [PubMed] [Google Scholar]
- 4.Bergler H, Hoegenauer G, Turnowski F. Sequences of the envM gene and of two mutated alleles in Escherichia coli. J Gen Microbiol. 1992;138:2093–2100. doi: 10.1099/00221287-138-10-2093. [DOI] [PubMed] [Google Scholar]
- 5.Best E, Knauf V C. Organization and nucleotide sequence of the genes encoding the biotin carboxyl carrier protein and biotin carboxylase protein of Pseudomonas aeruginosa acetyl coenzyme A carboxylase. J Bacteriol. 1993;175:6881–6889. doi: 10.1128/jb.175.21.6881-6889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campos-Garcia J, Caro A D, Najera R, Miller-Maier R M, Al-Tahhan R A, Soberon-Chavez G. The Pseudomonas aeruginosa rhlG gene encodes an NADPH-dependent β-ketoacyl reductase which is specifically involved in rhamnolipid synthesis. J Bacteriol. 1998;180:4442–4451. doi: 10.1128/jb.180.17.4442-4451.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Cheng H, Bjerkness M. One-step Coomassie Brilliant Blue R-250 staining of proteins in polycarylamide gel. Anal Biochem. 1993;212:295–296. doi: 10.1006/abio.1993.1330. [DOI] [PubMed] [Google Scholar]
- 8.Cronan J E, Klages A L. Chemical synthesis of acyl thioesters of acyl carrier protein with native structure. Proc Natl Acad Sci USA. 1981;78:5440–5444. doi: 10.1073/pnas.78.9.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cronan J E, Rock C O. Biosynthesis of membrane lipids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 612–636. [Google Scholar]
- 10.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 11.Dessen A, Quemard A, Blanchard J S, Jacobs W R, Sacchettini J C. Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science. 1995;267:1638–1641. doi: 10.1126/science.7886450. [DOI] [PubMed] [Google Scholar]
- 12.Ellmann G L. Tissue sulfhydryl groups. Anal Biochem. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 13.Glessner A, Smith R S, Iglewski B H, Robinson J B. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of twitching motility. J Bacteriol. 1999;181:1623–1629. doi: 10.1128/jb.181.5.1623-1629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenberg E P. Quorum sensing in Gram-negative bacteria. ASM News. 1997;63:371–377. [Google Scholar]
- 15.Heath R J, Rock C O. Enoyl-acyl carrier protein reductase (fabI) plays a determinant role in completing cycles of fatty acid elongation in Escherichia coli. J Biol Chem. 1995;270:26538–26542. doi: 10.1074/jbc.270.44.26538. [DOI] [PubMed] [Google Scholar]
- 16.Heath R J, Rock C O. Regulation of fatty acid elongation and initiation by acyl-acyl carrier protein in Escherichia coli. J Biol Chem. 1996;271:1833–1836. doi: 10.1074/jbc.271.4.1833. [DOI] [PubMed] [Google Scholar]
- 17.Heath R J, Rock C O. Roles of the FabA and FabZ β-hydroxyacyl-acyl carrier protein dehydratase in Escherichia coli fatty acid biosynthesis. J Biol Chem. 1996;271:27795–27801. doi: 10.1074/jbc.271.44.27795. [DOI] [PubMed] [Google Scholar]
- 18.Heath R J, Rubin J R, Holland D R, Zhang E, Snow M E, Rock C O. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J Biol Chem. 1999;274:11110–11114. doi: 10.1074/jbc.274.16.11110. [DOI] [PubMed] [Google Scholar]
- 19.Heath R J, Yu Y-T, Shapiro M A, Olson E, Rock C O. Broad spectrum antimicrobial biocides target the FabI component of fatty acid biosynthesis. J Biol Chem. 1998;273:30316–30320. doi: 10.1074/jbc.273.46.30316. [DOI] [PubMed] [Google Scholar]
- 20.Henry M F, Cronan J E. A new mechanism of transcriptional regulation: release of an activator triggered by small molecule binding. Cell. 1992;70:671–679. doi: 10.1016/0092-8674(92)90435-f. [DOI] [PubMed] [Google Scholar]
- 21.Hoang T T, Karkhoff-Schweizer R R, Kutchma A J, Schweizer H P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 22.Hoang T T, Schweizer H P. Fatty acid biosynthesis in Pseudomonas aeruginosa: cloning and characterization of the fabAB operon encoding β-hydroxyacyl-acyl carrier protein dehydratase (fabA) and β-ketoacyl-acyl carrier protein synthase I (fabB) J Bacteriol. 1997;179:5326–5332. doi: 10.1128/jb.179.17.5326-5332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kater M M, Konigstein G M, Nijkamp H J J, Stuitje A R. The use of a hybrid genetic system to study the functional relationship between prokaryotic and plant multi-enzyme fatty acid synthase complexes. Plant Mol Biol. 1994;25:771–790. doi: 10.1007/BF00028873. [DOI] [PubMed] [Google Scholar]
- 24.Kutchma A J. Characterization of a gene cluster containing the Pseudomonas aeruginosa fabD, fabG, acpP and fabF genes involved in fatty acid biosynthesis. M.S. thesis. Fort Collins, Colo: Colorado State University; 1997. [Google Scholar]
- 25.Kutchma A J, Hoang T T, Schweizer H P. Characterization of a Pseudomonas aeruginosa fatty acid biosynthetic gene cluster: purification of acyl carrier protein (ACP) and malonyl-coenzyme A:ACP transacylase (FabD) J Bacteriol. 1999;181:5498–5504. doi: 10.1128/jb.181.17.5498-5504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy C W, Roujeinikova A, Sedelnikova S, Baker P J, Stuitje A R, Slabas A R, Rice D W, Rafferty J B. Molecular basis of triclosan activity. Nature. 1999;398:383–384. doi: 10.1038/18803. [DOI] [PubMed] [Google Scholar]
- 27.Liss L. New M13 host: DH5αF′ competent cells. Focus. 1987;9:13. [Google Scholar]
- 28.Magnuson K, Jackowski S, Rock C O, Cronan J E. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makowski G S, Ramsby M L. pH modification to enhance the molecular sieving properties of sodium dodecyl sulfate-10% polyacrylamide gel. Anal Biochem. 1993;212:283–285. doi: 10.1006/abio.1993.1324. [DOI] [PubMed] [Google Scholar]
- 30.McClean K H, Winson M K, Fish L, Taylor A, Chhabra S R, Camara M, Daykin M, Lamb J H, Swift S, Bycroft B W, Stewart G S A B, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production for the detection of N-acylhomoserine lactones. Microbiology. 1998;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 31.McLean R J C, Whiteley M, Stickler D J, Fuqua W C. Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol Lett. 1997;154:259–263. doi: 10.1111/j.1574-6968.1997.tb12653.x. [DOI] [PubMed] [Google Scholar]
- 32.McMurry L M, McDermott P F, Levy S B. Genetic evidence that InhA of Mycobacterium smegmatis is a target for triclosan. Antimicrob Agents Chemother. 1999;43:711–713. doi: 10.1128/aac.43.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMurry L M, Oethinger M, Levy S B. Triclosan targets lipid synthesis. Nature. 1998;394:531–532. doi: 10.1038/28970. [DOI] [PubMed] [Google Scholar]
- 34.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 35.Moré M I, Finger D, Stryker J L, Fuqua C, Eberhard A, Winans S C. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 36.Parsek M R, Schaefer A L, Greenberg E P. Analysis of random and site-directed mutations in rhlI, a Pseudomonas aeruginosa gene encoding an acylhomoserine lactone synthase. Mol Microbiol. 1997;26:301–310. doi: 10.1046/j.1365-2958.1997.5741935.x. [DOI] [PubMed] [Google Scholar]
- 37.Raetz C H R. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1035–1063. [Google Scholar]
- 38.Rosenberg M C, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 39.Schaefer A L, Val D L, Hanzelka B L, Cronan J E, Greenberg E P. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci USA. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schweizer H P. The agmR gene, an environmentally responsive gene, complements defective glpR, which encodes the putative activator for glycerol metabolism in Pseudomonas aeruginosa. J Bacteriol. 1991;173:6798–6806. doi: 10.1128/jb.173.21.6798-6806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweizer H P. Intrinsic resistance to inhibitors of fatty acid biosynthesis in Pseudomonas aeruginosa is due to efflux: application of a novel technique for generation of unmarked chromosomal mutations for the study of efflux systems. Antimicrob Agents Chemother. 1998;42:394–398. doi: 10.1128/aac.42.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schweizer H P, Klassen T R, Hoang T. Improved methods for gene analysis and expression in Pseudomonas. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: American Society for Microbiology; 1996. pp. 229–237. [Google Scholar]
- 43.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 44.Turnowsky F, Fuchs K, Jeschek C, Hoegenauer G. envM genes of Salmonella typhimurium and Escherichia coli. J Bacteriol. 1989;171:6555–6565. doi: 10.1128/jb.171.12.6555-6565.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Val D L, Cronan J E. In vivo evidence that S-adenosylmethionine and fatty acid intermediates are the substrates for the LuxI family of autoinducer synthases. J Bacteriol. 1998;180:2644–2651. doi: 10.1128/jb.180.10.2644-2651.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Delden C, Iglewski B H. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 48.Weeks G, Wakil S J. Studies on the mechanisms of fatty acid synthesis. J Biol Chem. 1968;243:1180–1189. [PubMed] [Google Scholar]