Abstract
Tuberculosis (TB) is a contagious disease that predominantly affects the lungs, but can also spread to other organs via the bloodstream. TB affects about one-fourth population of the world. With age, the effectiveness of Bacillus Calmette-Guérin (BCG), the only authorized TB vaccine, decreases. In the quest for a prophylactic and immunotherapeutic vaccine, in this study, a hypothetical mRNA vaccine is delineated, named MT. P495, implementing in silico and immunoinformatics approaches to evaluate key aspects and immunogenic epitopes across the PstS1, a highly conserved periplasmic protein of Mycobacterium tuberculosis (Mtb). PstS1 elicited the potential to generate 99.9% population coverage worldwide. The presence of T- and B-cell epitopes across the PstS1 protein were validated using several computational prediction tools. Molecular docking and dynamics simulation confirmed stable epitope-allele interaction. Immune cell response to the antigen clearance rate was verified by the in silico analysis of immune simulation. Codon optimization confirmed the efficient translation of the mRNA in the host cell. With Toll-like receptors, the vaccine exhibited stable and strong interactions. Findings suggest that the MT. P495 vaccine probably will elicit specific immune responses against Mtb. This mRNA vaccine model is a ready source for further wet-lab validation to confirm the efficacy of this proposed vaccine candidate.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s11030-022-10515-4.
Keywords: Tuberculosis mRNA vaccine, Tuberculosis vaccine design, Immune cell activation/modulation, Epitope identification
Introduction
Tuberculosis (TB) is a transmissible disease that is characterized by the infection of the opportunistic bacterial species Mycobacterium tuberculosis (Mtb), primarily targeting the lung [1, 2]. TB is initiated when Mtb is deposited onto the surface of the lung alveoli from the airborne droplets containing the pathogen. Pathogen-containing droplets are mostly dispersed by people with active pulmonary or laryngeal TB. Inhalation of these droplets results in symptoms such as persistent cough, fever and night sweats [3, 4].
The most common cause of mortality caused by a single infectious agent is TB [5]. In 2020, 1.44 million individuals died, with 214,000 of them being HIV-positive [6]. Around 1.7 billion people were latently infected with Mtb in 2014, almost a quarter of the world population [7]. Generally, individuals with a healthy immune system can suppress the growth of Mtb, but the situation gets complicated for immunocompromised patients (e.g. HIV infected or patients with diabetes) who cannot generate enough immune response to suppress the progression of infection [1, 4, 5, 8].
Pulmonary macrophages play a critical role in the primary immune response against Mtb upon entry while a minimum of 12 days are required for the CD4+ T-cells after aerosol infection to respond [9–11]. During this period, Mtb increases its population number by > 20,000-fold [9]. Mtb is readily phagocytized by the macrophages present in alveoli. Most often the entering bacteria are killed by these macrophages. But some bacteria can escape from being phagocytized and they start to proliferate within the macrophage as an intracellular parasites [10].
To provide better protection against Mtb, vaccination is a must as the adaptive immune system requires a lot of time to be activated against this pathogen. Moreover, studies on Mtb infected patients have revealed the presence of various multidrug-resistant strains as well as extensively drug-resistant strains [12]. Currently, the only available approved vaccine against TB is Bacillus Calmette-Guérin, BCG [12, 13]. BCG is generally injected into newborn babies which protects them from Mtb till the age of 10 years [8]. For adults, the efficacy of the BCG vaccine varies greatly between 0 and 80% [12]. Several vaccines are in clinical trial, showing promising results. For instance, the M72/AS01E vaccine, a subunit vaccine, is showing very good results in its clinical trial [14]. Although there are several vaccines in the clinical trial phase, currently there is no mRNA-based vaccine developed for TB, neither in the clinical trial phase nor licensed.
mRNA vaccines are a rapidly developing area. A vast amount of preclinical evidence has been obtained recently, and several human clinical trials have begun [15]. Based on this evidence, mRNA is now considered a safe and effective alternative to (subunit) protein, chimeric virus, and even DNA-based therapies in the form of vaccination [16]. The transient expression and accumulation of selected antigens in the cytoplasm are induced by mRNA.
The proteasome in the cytoplasm of antigen-presenting cells (APCs) can breakdown the antigen into peptides. In the endoplasmic reticulum, the peptides with antigenic properties, are complexed with nascent major histocompatibility complex (MHC) class I molecules. The peptide–MHC complexes can then activate CD8+ cytotoxic T (Tc) cells when they are expressed on the surface of the cell membrane of APCs. CD4+ helper T (Th) cells are activated by MHC class II–peptide complexes expressed on the surface of the cell membrane of APCs. Antigens, secreted by, or released from dead cells that have uptaken and translated the exogenous mRNA, can bind with B-cells within the extracellular matrix, activating these cells. As a result, all adaptive immune effectors, including B lymphocytes, Tc-cells and Th-cells, will be activated by mRNA-based vaccines [16, 17].
In silico analysis of target proteins has simplified the identification of immunogenic B- and T-cell epitopes in the proteins, facilitating the detection of the antigenic epitopes specifically having the potential of eliciting an immune response in particular [18]. Because in silico predictions can minimize the number of experiments required, this strategy is both cost-effective and convenient [19, 20]. However, in silico vaccine design strategy seems to be quite effective, but it might not be efficient enough to catch pace with the advent of newer pathogens. All findings must be thoroughly and extensively analyzed in order to identify antigenic regions for designing an effective vaccine, which presents a substantial overhead and can be time-intensive [21]. This technique has been successfully implemented to develop a vaccine against various pathogens, for example, serogroup B Neisseria meningitides (MenB) [22].
In this study, an mRNA vaccine has been modeled, named MT. P495, using several bioinformatics tools targeting the phosphate-binding protein PstS1 of Mtb and also has been tested computationally for its ability to elicit immunogenic response and safety, predicted several types of T-cell and B-cell epitopes present within this antigen and their ability to generate an immune response within the host body. PstS1 protein is an immunodominant, TLR-2 agonist, inorganic phosphate up-taking lipoprotein found on the cell membrane surface of Mtb and also exhibits function as an adhesion molecule that facilitates binding with macrophage through mannose receptor (MR). This mRNA vaccine model thus serves as ready to test model in vivo by experimentalists and industries.
Materials and methods
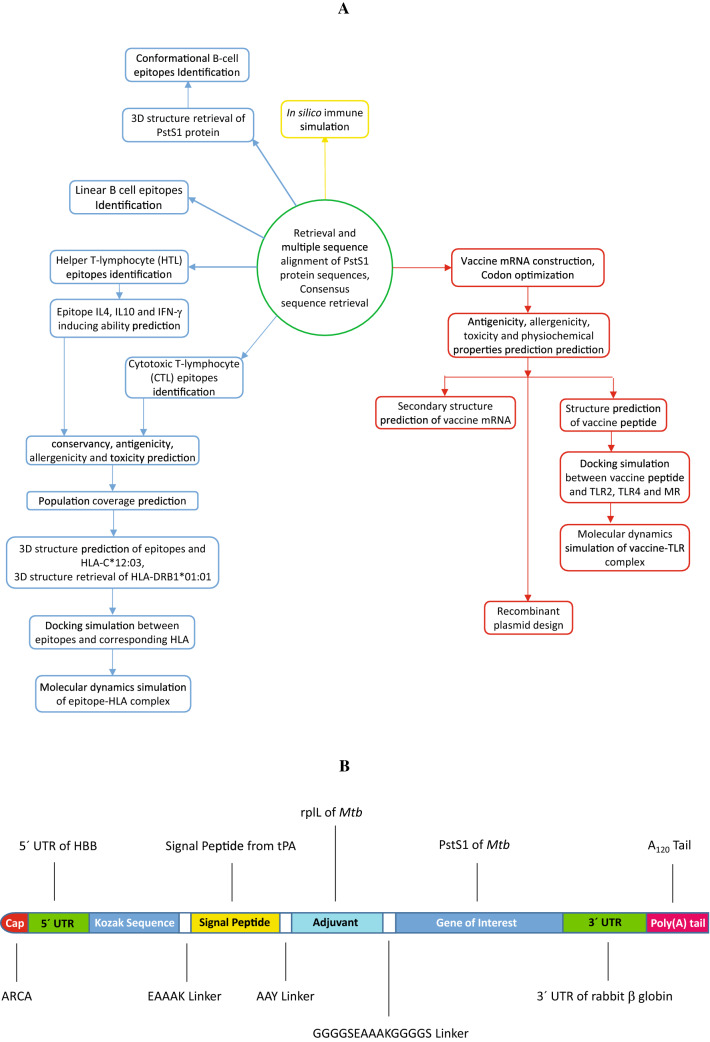
A graphical depiction of the workflow is represented in Fig. 1A.
Fig. 1.
Graphical presentation of A steps involved in modeling an mRNA-based vaccine and B mRNA of the MT. P495 vaccine
Retrieval of protein sequence and 3D structure
A total of 3470 sequences of phosphate-binding protein PstS1 of Mtb were retrieved from the NCBI [23] database. The retrieved protein sequences had a length of 374 amino acids. The first 23 amino acids were the signal peptide and the rest of them encoded the PstS1 protein. Multiple sequence alignment (MSA) between the retrieved sequences was performed using the Clustal Omega [24] and the result was analyzed using the JalView [25]. The consensus sequence was retrieved from JalView and was used for sequence-based epitope prediction. The consensus sequence was 100% identical to the reference sequence of PstS1 retrieved from UniProt [26] (UniProtKB accession number P9WGU1). The X-ray crystal structure Mtb H37Rv PstS1 was obtained from RCSB PDB [27] (PDB ID-1PC3) for the prediction of structure-based discontinuous B-cell epitope.
Identification of cytotoxic T-lymphocyte (CTL) epitopes
To predict CD8+ CTL epitopes, NetCTL 1.2 Server [28] was used. The NetCTL server predicts CTL epitopes in a given protein sequence using the stabilized matrix base method [29]. The threshold level for prediction of a CTL epitope was set at 0.5 with a specificity of 0.94 along with a sensitivity of 0.89. Further analysis was performed on predicted epitopes with a higher combined score.
SMMPMBEC prediction method [30] of the T-cell epitope prediction resource of the Immune Epitope Database (IEDB) [31] was utilized to predict both types of MHC-I-binding alleles, occurring frequently as well as non-frequently. The half-maximal inhibitory concentration (IC50) threshold was set at 250 nM.
Helper T-lymphocyte (HTL) epitopes identification
The T-cell epitope prediction resource of the IEDB server was used to predict promiscuous CD4+ HTL epitopes. The NN-align 2.3 (NetMHCII 2.3) method [32] was employed in this study to predict MHC-II-binding alleles. Peptide length was set to be 15. For further analysis, predicted peptides having an IC50 value less than 50 nM were considered.
Evaluation of the selected epitopes
For the analysis of antigenic, allergenic and toxic properties of all predicted epitopes, VaxiJen v2.0 server [33], AllerTOP v.2.0 server [34] and ToxinPred server [35] were employed respectively. Based on an alignment-independent manner, VaxiJen predicts the antigenic probability of a given peptide [33]. In this study, the target organism was set to bacteria and the threshold was set to 0.5. For the categorization of allergenic and nonallergenic peptides, AllerTOP v.2.0 utilizes the k-nearest neighbours (kNN) method [34]. Employing both the dipeptide-based SVM algorithm and the MEME/MAST algorithm, the ToxinPred can predict the toxicity of the given peptide [35]. Using the IEDB conservancy analysis tool, the conservancy of the epitopes was determined [36] using a 100% sequence identity threshold.
Cytokine inducing ability of the HTL epitopes was subject to further assessment. IL4pred server [37] was used to predict the interleukin-4 (IL4) inducing ability of the selected peptides while Interleukin-10 (IL10) inducing ability was assessed using the IL10pred server [38]. Another property that was assessed of the HTL peptides was the interferon-γ (IFN-γ) inducing probability. IFNepitope server [39] was used in this study.
Estimation of population coverage
The population coverage of the selected CTL and HTL epitopes was evaluated. Employing the population coverage tool [40] of the IEDB server, population coverage of the selected epitopes and their corresponding MHC HLA-binding alleles was predicted.
Prediction of three-dimensional (3D) structure of the epitopes and HLA proteins
After confirmation of epitope antigenicity, non-allergenicity, non-toxicity, conservancy and the availability of alleles, the 3D structure of the selected 8 CTL peptides, as well as 3 HTL peptides, were predicted using the PEP-FOLD server at the Ressource Parisienne en Bioinformatique Structurale (RPBS) Mobyle Portal [41]. The five most probable structures were predicted by the server for each peptide sequence. For further investigation, the best-predicted structure with the lowest energy model was chosen.
In this study, HLA-C*12:03 and HLA-DRB1*01:01 were used for the validation of the binding of selected epitope and HLA molecules. The 3D structure of HLA-DRB1*01:01 was retrieved from RCSB PDB (PDB ID 4I5B). 3D structure of HLA-C*12:03 being unavailable in PDB, SWISS-MODEL server [42] that performs homology-based modeling, was used for the prediction of the 3D structure of this molecule. The sequence of the HLA-C*12:03 was retrieved from IPD-IMGT/HLA Database [43] (IMGT/HLA Accession No. HLA00455). The predicted structure was validated using PROCHECK [44], MolProbity [45], ProSA-web [46] and QMEAN [47].
Molecular docking of the selected epitopes
To confirm the interaction between the selected alleles and their respective epitopes, at the DINC 2.0 web-server [48], molecular docking was carried out following the protocol described elsewhere [49]. The box centre was set at 13.7, − 60.0, and − 20.2 Å in the X, Y, and Z axes, respectively, to predict the binding energy of HLA-C*12:03 with an epitope. Similarly, to predict the binding affinity of HLA-DRB1*01:01 with an epitope, the box centre, in this case, was set at 39.9, 47.5 and 93.0 Å in the X, Y, and Z axes, respectively. In both cases, box size was also set to automatic (based on ligand). Results from this server were further analyzed using BIOVIA Discovery Studio [50] and UCSF Chimera [51].
Molecular dynamics simulation
Following a procedure described elsewhere [49], MD simulations were carried out employing GROMACS (v2021) [52]. For the simulation process, CHARMM36 all-atom additive force field [53] was used. The protein was cleaned using UCSF Chimera dock prep functionality, and then missing residues were added by using the Dunbrack rotamer Library, employing an in-house python script. GROMACS pdb2gmx tool was used in order to add hydrogens to the protein. The GROMACS program was used to generate the protein topology for HLA and epitope. The system was contained within a cubic box of a simple point charge extended (SPC/E) water model [53], with a minimum distance of 1.0 nm between the wall and any component of the protein. The system was neutralized by adding an aqueous solution of Na+ (sodium) and Cl− (chloride) to a 0.15 M ionic strength.
For 100 picoseconds (ps), the system was equilibrated utilizing the NVT and NPT ensemble. For the next 100 ps under an isothermal ensemble, soft coupling with the Berendsen thermostat (NVT) [54] was used to progressively heat the minimized system to target temperatures. Position restrictions to the ligand were imposed before performing NVT simulations to prevent the ligand from drifting away from the protein during equilibration. Employing the LINCS algorithm [55], all of the bonds were restricted. At a temperature of 300 K, the NPT ensemble (constant pressure, and constant temperature simulation criteria) was performed, employing periodic boundary conditions (PBC).
The system was then coupled to a Parrinello–Rahman barostat [56] for an equilibration period of 100 ps at 1 bar of pressure. The Particle Mesh Ewald (PME) method was employed to process the electrostatic interactions. With a cutoff of 1.0 nm, the short-range van der Waals cutoff (rvdW) interactions were computed. Simulations were run for 100 ns. GROMACS was used to compute the root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), solvent accessible surface area (SASA) (using van der Waals Volumes and Radii) [57] and hydrogen bond (H-bond). In-house Python script with Matplotlib [58] and NumPy [59] library, as well as [R] (version 3.6.3) [60] Peptides library [61] were used to generate trajectory plots and figures.
Conformational B-cell epitopes identification
The conformational B-cell epitopes were identified by using the ElliPro [62] tool of the IEDB. ElliPro can identify both the linear and conformational B-cell epitopes based on the 3D structure of the given protein. In this study, the minimum PI (protrusion index) score was set to 0.5 and the maximum distance was set to 6 Å for the prediction of conformational B-cell epitopes.
Linear B-cell (LBL) epitopes identification
To predict LBL epitope, linear B-cell epitope prediction tool from IEDB was used. Epitopes were predicted using Emini surface accessibility prediction [63], BepiPred Linear Epitope Prediction 2.0 [64], Kolaskar and Tongaonkar antigenicity [65] and Karplus and Schulz flexibility prediction method [66], present at the IEDB server. Epitopes having a length between 10 and 40 amino acids were selected. Linear epitopes predicted by ElliPro were also selected. Antigenicity, allergenicity, toxicity and conservancy of the selected epitopes were assessed as described earlier.
Immune simulation of the epitopes
At the C-ImmSim server [67], in silico immune simulation was carried out for the characterization of the immune response profile of the selected peptides. Two injections of the target antigen were administered 4 weeks apart at 1 and 84 time-steps (wherein, the first dose is administered at time = 0 and each time-step is corresponding to 8 h in real life) with a dose of 1000 antigen proteins each, containing no LPS. The simulation was conducted for 5000 simulation steps. Host HLAs were selected according to their occurring frequency. Frequently occurring HLA alleles were selected to perform the study. Other simulation parameters were kept default.
Designing the vaccine mRNA construct
The open reading frame (ORF) of a conventional mRNA-based vaccine consists of five fundamental parts. The target antigen, also known as the gene of interest (GOI) is linked with an adjuvant by a linker. This construct is flanked by 5′ and 3′ untranslated regions (UTRs) and a terminal poly(A) tail. The 5′ end is capped by Cap1 (m7GpppNm) Cap. In this study, PstS1 was used as the GOI which exhibited high immunogenic activity in several studies [68–70]. As an adjuvant, 50S ribosomal protein L7/L12 (UniProtKB accession number P9WHE3) from Mtb was used while the signal peptide from the tissue plasminogen activator (tPA, UniProtKB accession number P00750) of Homo sapiens was also used.
Two peptide linkers were used to join the polypeptide chains. GGGGSEAAAKGGGGS linker was used to link the GOI and the adjuvant. Another peptide linker, AAY was used between the signal peptide and the adjuvant. In this study, the 5′ UTR from human β-globin gene (NCBI accession number NM_000518.5) along with the 3′ UTR from rabbit β-globin gene (GenBank [71] accession number V00882.1) were used to flank the construct. A 120-nucleotides (nts) long poly(A) tail was added to complete the vaccine construct (Fig. 1B). The construct was named MT. P495.
Optimizing codons and predicting secondary structure of the vaccine mRNA
For the vaccine mRNA to be efficiently translated by the host cells, codon optimization is important. Therefore, the codons of the final vaccine construct were optimized for efficient expression in human cells using several codon optimization tools; JCat [72], GeneArt Instant Designer by Thermofisher, GenSmart™ Codon optimization by GenScript (GS), Codon Optimization Tool by Integrated DNA Technologies (IDT). The quality of the optimized codons was analyzed Using Rare Codon Analysis tools by GS. This tool can predict the efficiency of the translation of the mRNA expressed as the codon adaptation index (CAI) value. Also, the presence of any tandem unusual codons can be detected, shown as codon frequency distribution (CFD). Based on these parameters, the best-optimized sequence was chosen for further assessment.
The secondary structure of the mRNA construct was predicted using the RNAfold tool of ViennaRNA Package 2.0 [73]. Both the minimum free energy (MFE) structure and the centroid secondary structure of the mRNA were obtained from this tool along with their MFE.
Physicochemical properties assessment of the vaccine peptide
For the assessment of physiochemical properties, several bioinformatics tools were used. tPA signal sequence was excluded as this segment will be cleaved by the protease, only the adjuvant and the target antigen were assessed for various properties. To predict the antigenicity of this peptide segment, VaxiJen v2.0 [33] and ANTIGENpro [74] were used. Allergenicity of the peptide was predicted using AllerTOP v.2.0 [34]. The toxicity of the peptide was predicted using ToxinPred [35]. Various physiochemical properties [i.e., theoretical isoelectric point (pI), instability index (II), aliphatic index (AI), and grand average of hydropathicity (GRAVY)] were predicted using ProtParam [75]. Adhesin probability was checked using Vaxign [76].
Prediction of secondary and three-dimensional structure of vaccine peptide
To predict the secondary structure of the vaccine peptide the PSIPRED server was used [77]. The secondary structure of a particular protein sequence is predicted by this server employing position-specific scoring matrices [77, 78]. For the prediction of the 3D structure of the vaccine peptide, the Robetta server was utilized [79]. Robetta server predicts the 3D structure of a given protein sequence by combining the data from SPIDER3, PSIPRED, DeepConCNF, DISOPRED and TMHMM server. The predicted structures were validated using PROCHECK [44], MolProbity [45], ProSA-web [46] and QMEAN [47].
Molecular docking of the vaccine peptide
To confirm the interaction between the vaccine peptide and Toll-like receptors (TLRs) and human MR, the molecular docking analysis was performed at the ClusPro server [80]. Crystal structure of TLR-2 (PDB ID 6NIG), TLR-4 (PDB ID 4G8A) and CysR-CTLD3 fragment of human MR (PDB ID 5XTS) were retrieved from PDB and were used in the docking analysis as the receptor molecule. The vaccine peptide was used as the ligand molecule.
The binding energy of receptor-ligand interaction was predicted using the PROtein binDIng enerGY prediction (PRODIGY) tool of the HADDOCK server [81] from their bound 3D structure. The binding free energy (ΔG), as well as the dissociation constant (Kd), can be predicted by the PRODIGY server, based on intermolecular contacts and properties obtained from non-interface surfaces.
Molecular dynamics simulation of the vaccine–TLR4 complex
The molecular dynamics (MD) simulation was performed to check the stability of the vaccine–TLR4 complex employing the iMODS server [82]. The analysis of complex mobility (NMA B-factors), eigenvalues, deformability, covariance map and linking matrix based on the 3D structure of the given protein–protein complex were performed on this server.
In silico cloning
The manufacturing process of vaccine mRNA begins with the generation of a plasmid that contains a specific DNA-dependent RNA polymerase promoter along with the sequence of the mRNA construct and in vitro transcription (IVT) approach is widely used for large-scale production of mRNA [83]. To generate a recombinant plasmid, under SpeI and NheI restriction sites at the N and C terminals, the codon-optimized vaccine construct sequence was created as a cloning insert for the pJAZZ-OK® vector. The promoter and termination sequence specific to the T7 polymerase was introduced to the cloning insert. The recombinant plasmid was designed by inserting the adapted codon sequence of the vaccine construct into the pJAZZ-OK® cloning vector using the SnapGene software (from Insightful Science; available at snapgene.com). The pJAZZ-OK®, a linear plasmid was used as the cloning vector. For IVT mRNA production, linear plasmids can be used for the successful propagation of poly(A) tracts up to ≃500 bp in length [84].
Results
Multiple sequence alignment of PstS1 revealed high conservancy
A total of 3470 complete PstS1 protein sequences (Supplementary Table 1), available at NCBI, were retrieved and subject to MSA, which revealed that the PstS1 protein is highly conserved (Supplementary Fig. S1). Surprisingly, the consensus sequence was 100% identical with the reference sequence of that protein, retrieved from UniProt. The first 23 amino acids of this protein are annotated as a signal peptide. The rest of the amino acids were subject to analysis for vaccine formulation.
Several T-cell epitopes were identified
CTL epitopes were identified using the NetCTL 1.2 server. A set of 98 epitopes that interact with any of the MHC class I allele supertype families with a combined score above 0.50 were primarily selected. The IC50 value for the selected epitopes was predicted and all of them had an IC50 value less than 250 nM and were further analyzed.
The T-cell epitope prediction resource of the IEDB server was used for the identification of HTL epitopes. The NN-align 2.3 (NetMHCII 2.3) method was used for the identification. For further analysis, 162 epitopes with IC50 values less than 50 nM were chosen.
For the vaccine to be effective against the various strains of the targeted protein, epitope conservation is important. An epitope with a higher conservancy would be a better target for improved vaccine design. Conservation of epitopes across antigen of the primarily selected epitopes was checked. A total of 44 CTL epitopes and 149 HTL epitopes were identified with 100% conservancy and were analysed using VaxiJen to confirm their antigenicity.
The epitopes with antigenic potential were then analyzed employing AllerTOP v.2.0 as well as ToxinPred to ensure their safety as non-allergen and non-toxic, respectively. Finally, 8 CTL epitopes (Supplementary Table 2) and 17 HTL epitopes (Supplementary Table 3) were found to be antigenic, non-allergen and non-toxic. The cytokine production inducing potential of the 17 HTL epitopes was also assessed. Among the 17 HTL epitopes, 6 epitopes have IFN-γ inducing ability, 7 epitopes have IL4-inducing ability and 9 epitopes have IL10-inducing ability (Supplementary Table 3).
Population coverage estimation of the predicted epitopes
Around the world, HLA allele distribution varies by geographic region and ethnic group. As a result, when designing an effective, ethnically neutral vaccine, population coverage must be taken into account. The population coverage tool predicted 99.92% world population coverage (Supplementary Fig. S2) for the selected 25 epitopes (class I and II combined, 80.66% for class I and 99.61% for class II). The class combined population coverage was predicted to be highest in Europe, North America and West Africa (100%) and lowest in South Africa (67.77%). Highest population coverage for separated class I was predicted in Europe (86.07%) and for class II, in Europe and West Africa (99.99%). The lowest population coverage for separated class II was predicted in Central America (0.0%) and for class II, in South Africa (8.42%).
Prediction of three-dimensional structure of selected epitopes and HLA proteins
The potential of an epitope to interact with the MHC molecule and form a complex with it determines how effective a potential vaccine will be. As a response, corresponding alleles for the chosen epitopes were predicted and a docking analysis was conducted. Eight CTL epitopes and three HTL epitopes were selected for docking analysis. HLA-C*12:03 and HLA-DRB1*01:01 were used as receptor molecules. The 3D structure of the selected epitopes was predicted using the PEP-FOLD (Supplementary Fig. S3).
The 3D structure of HLA-DRB1*01:01 was obtained from PDB. Unwanted molecules bound with the crystal structure of the protein were removed prior to docking. The 3D structure of HLA-C*12:03 was predicted using the SWISS-MODEL server (Supplementary Fig. S4A). With 93.2% residues in most favoured regions, the Ramachandran plot indicated that the 3D structure of the allele is acceptable (Supplementary Fig. S4B). The Rama distribution Z-score (Supplementary Fig. S4C) from the MolProbity server was found to be 1.14 ± 0.49 and the QMEAN value (Supplementary Fig. S4D) from the SWISS-MODEL server was found to be 1.12. These findings confirmed the predicted 3D structure's high fidelity.
Molecular docking analysis confirmed binding of the selected epitopes with respective MHC molecule
The interaction of the epitope with MHC molecules was studied using the DINC 2.0 server. The side chains of the epitope protruded into pockets within the groove of the MHC molecules, according to docking analysis (Supplementary Figs. S5, S6) through several types of bonds. Each epitope has different postures in which it can attach to its specific MHC allele. The docked poses were analyzed using BIOVIA Discovery Studio; the pose with the highest binding energy (Table 1) and no unfavorable bond was selected as the best pose.
Table 1.
Predicted binding energy of selected epitopes with their corresponding HLA molecule
| MHC allele | Epitope | Binding energy (kcal/mol) |
|---|---|---|
| HLA-C*12:03 | AALNPGVNL | − 7.40 |
| FLFTQYLSK | − 8.10 | |
| GLMNIALAI | − 7.10 | |
| KLSDALIAT | − 6.70 | |
| NIGASDAYL | − 7.20 | |
| QIAALNPGV | − 7.00 | |
| SQRGLGEAQ | − 6.00 | |
| VNIGASDAY | − 7.50 | |
| HLA-DRB1*01:01 | AALNPGVNLPGTAVV | − 6.70 |
| NPGVNLPGTAVVPLH | − 5.70 | |
| GLMNIALAISAQQVN | − 6.60 |
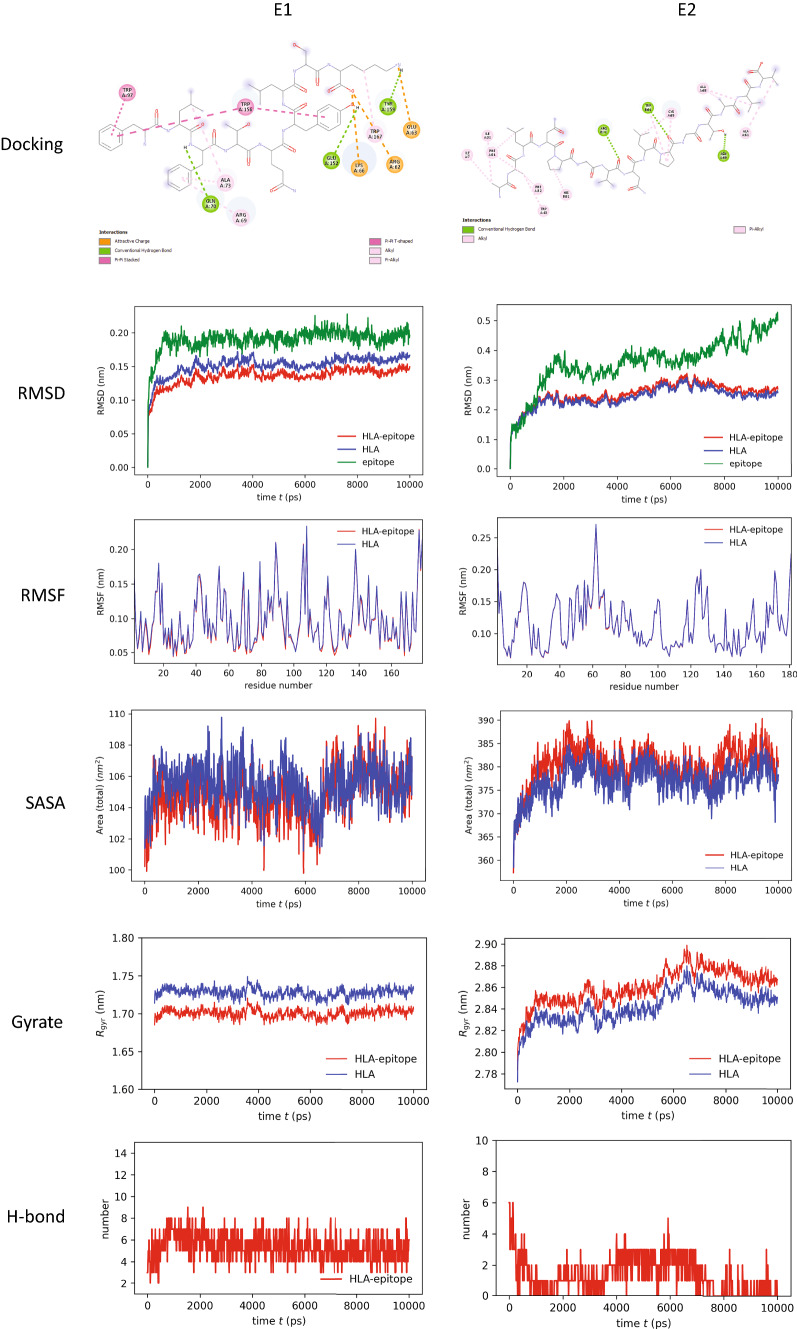
Docking analysis revealed that the epitope “FLFTQYLSK” binds with HLA-C*12:03 with the highest binding energy (− 8.1 kcal/mol) among the CTL epitopes. This indicates that HLA-C*12:03 and the epitope FLFTQYLSK have a strong affinity. The epitope was bound with three hydrogen bonds at GLN (A:70), GLU (A:152) and TYR (A:159) residues with the HLA molecule. Several other covalent bonds, in this case, attractive charge, Pi–Pi T-shaped, Pi–Pi stacked, alkyl and Pi-alkyl bonds, were observed in the ligand–receptor complex (Fig. 2, 1st row, 1st column).
Fig. 2.
Analysis of molecular docking and MD simulation trajectory plots: Bonds between HLA molecule and epitopes (1st row panel). Plot of root mean square deviation (RMSD, 2nd row panel), root mean square fluctuation (RMSF, 3rd row panel), solvent accessible surface area (SASA) with respect to time (ps, 4th row panel), radius of gyration (Rg, 5th row panel), and hydrogen bonds (6th row panel) during MD simulation of MHC-I/MHC-II in complex with respective selective epitope FLFTQYLSK (E1) and AALNPGVNLPGTAVV (E2). Red color indicates HLA–epitope complex, blue color for only HLA and green color for the epitope
Again, the epitope “AALNPGVNLPGTAVV” binds with HLA-DRB1*01:01 with the highest binding energy (− 6.7 kcal/mol) among the HTL epitopes. This epitope was bound with the MHC molecule by three hydrogen bonds at ARG (B:71), TRP (B:61) and ASN (A:69). Alkyl bonds and Pi-alkyl bonds were observed in the ligand–receptor complex (Fig. 2, 1st row, 2nd column).
Molecular dynamics (MD) simulation indicated stability of HLA–epitope complexes
MD simulation was performed to understand how stably identified epitopes interact with HLAs. The RMSD of the HLA–epitope complexes indicated that the systems were stabilized after 2 ns of simulation in both cases and tended to remain in the plateau phase (ranging from 0.15 to 0.25 nm) thereafter for the rest of the period (Fig. 2, 2nd row panel). The epitope had a similar RMSD pattern as well. A little bit of fluctuation in the RMSD plot of the MHC-II–epitope complex was observed after 80 ns which was again stabilized after 90 ns at a higher RMSD. This suggests that the systems have already attained a state of relative equilibrium. The RMSF represents the fluctuation of each atom in the entire simulation (Fig. 2, 3rd row panel). In both cases, the average RMSF value was 0.1 nm. The SASA of protein is a measurement of the surface area accessibility by the complex during the simulation (Fig. 2, 4th row panel). It can be observed that the complex's SASA is just slightly greater than HLAs alone. It indicates that the MHC is linked to the epitope. The minor rise in SASA values over time suggests a partial unfolding of the structure due to solvent exposure. Increased SASA values with epitope binding to MHC, on the other hand, indicate its accommodating tendency. As a measure of structural compactness, the radius of gyration (Rg) was calculated (Fig. 2, 5th row panel). If the protein is folded correctly, the Rg value will likely remain stable. The Rg of the MHC-I–epitope complex was about 1.7–1.75 nm and remained consistent over time; while the Rg of MHC-II, which has a big structure, was in the range of 2.8 to 2.9 nm. A slight fluctuation was observed in the case of the MHC-II–epitope complex after 60 ns which becomes stable after 80 ns. When the MHC protein is docked with the epitope, the total Rg value of the MHC protein increases somewhat, indicating that MHC opens up for the epitope. The trend of calculated hydrogen bonds over time (Fig. 2, 6th row panel) was similar to that of Rg. The steady interaction of complexes was confirmed by all studies from the MD simulation of selective HLA–epitopes.
Presence of B-cell epitopes was also observed
For conformational B-cell epitope prediction, the 3D structure of the protein is required. The X-ray crystal structure of the PstS1 was used for analysis using the ElliPro tool. This analysis revealed four conformational B-cell epitopes (Supplementary Fig. S7) and nine linear B-cell epitopes.
Linear B-cell epitope prediction is done based on the sequence of the protein. To predict the linear epitopes, various methods were used in this study (Supplementary Fig. S8). The BepiPred 2.0 method predicts the antigens based on a random forest algorithm. The threshold level was 0.5. For an effective binding of epitope and antibody, the epitopes need to be surface accessible. To predict surface accessibility, Emini surface accessibility prediction was done at a 1.0 threshold level. Again, the flexibility of the epitope also plays a role in antigenicity. Karplus and Schulz flexibility prediction was done at a threshold level of 1.001. The epitope is also influenced by the physicochemical properties of amino acid residues, as well as their frequency of occurrence. Kolaskar and Tongaonkar antigenicity prediction method was used for this purpose which can detect the epitopes based on a semi-empirical method from the given protein sequence. The threshold level was 1.025. Among 22 epitopes predicted, ranging from 10 to 40 residues in length, only 8 epitopes were highly conservancy, antigenic, non-allergen and non-toxic (Supplementary Table 4).
Selected B- and T-cell epitopes were able to elicit a strong immune response
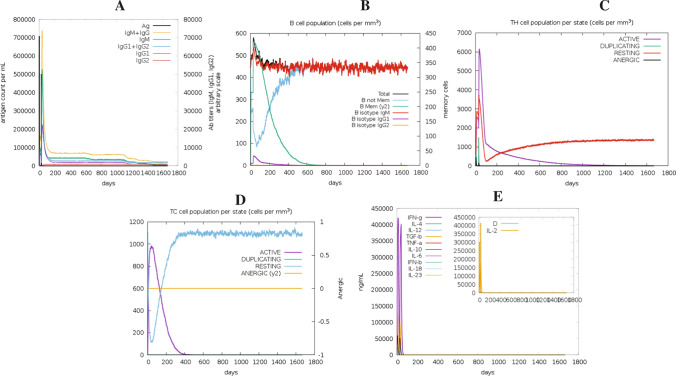
The target antigen contained several conserved B- and T-cell epitopes. The immune response of the antigen was studied using the C-ImmSim server. Results from this analysis depict that the antigen elicits strong primary and secondary immune responses, characterized by high levels of IgM. The first antibody isotype that the B-cell secretes in response to a foreign antigen is IgM. With a subsequent decrease in antigen concentration, levels of different antibody subclasses (i.e., IgG1 + IgG2, IgM, as well as IgG + IgM) and B-cell populations increased in the secondary and tertiary responses (Fig. 3A, B). This profile suggests the production of immune memory and, as a result, enhanced antigen clearance after subsequent exposures (Fig. 3C). The TH (helper) and TC (cytotoxic) cell populations showed a similarly high response, with corresponding memory production (Fig. 3C, D). Repeated administration with two injections (four weeks apart) resulted in increasing IgG1 levels and decreasing IgM levels, while IFN-γ concentration and TH-cell population remained high during the exposure period (Fig. 3E).
Fig. 3.
In silico simulation of the immunological response to the target antigen: A antigen and immunoglobulins, B B-cell population, C helper T-cell population, D cytotoxic T-cell population per state, and E cytokine and interleukin production with Simpson index (D) of the immune response
Optimizing codons and predicting secondary structure of the vaccine mRNA
An mRNA-based vaccination using the target antigen as the GOI was developed once the immunogenic potential of the target antigen had been established. Several tools were used to optimize the GOI codons for better expression in the human host, and the findings showed that the quality of the codons that were improved by merging the tools from IDT and GS was superior to the others (Supplementary Table 5). The mRNA sequence for the vaccine was 1856 nts long. The average GC content of the optimized sequence was 63%, suggesting that the vaccine candidate will be effectively expressed in humans. The range of 30 to 70% is the optimum percentage for GC content in human [85]. The potency of the optimized sequence was evaluated using the Rare Codon Analysis tools by GS (Supplementary Fig. S9A–C). The CAI value of the sequence was predicted to be 0.91, which is acceptable for the desired expression. The CFD was found to be 0%, indicating the absence of any tandem unusual codons, which might have a minimizing effect on translation efficiency or even disengage the translational machinery.
The RNAfold server was used to predict the secondary structures of the delineated mRNA and their corresponding free energy. The minimum free energy of the MFE structure was found to be − 680.10 kcal/mol and the MFE of the centroid secondary structure was found to be − 563.21 kcal/mol (Supplementary Fig. S9D, E). From the secondary structures, it could be concluded that the MT. P495 mRNA will be stable after being synthesized.
The vaccine peptide was predicted to be safe for human usage
To ensure the safety of the vaccine to be used for human usage, the antigenicity, allergenicity and toxicity of the antigen were analyzed and it was found that the antigen is antigenic, non-allergenic, and non-toxic and safe for the host body (Supplementary Table 6). The vaccine peptide contains 495 amino acids and has a molecular weight of 50,323.60 Da. According to the predicted theoretical pI of 4.71, the protein is acidic by nature. It was predicted that the peptide would have an II of 25.93, indicating that it would be stable in a test tube. The AI was determined to be 85.33. The predicted GRAVY for the GOI was 0.012. The positive GRAVY value indicates the hydrophobic nature of the protein. The half-life for the GOI was measured as 30 h for mammalian reticulocytes. The adhesin probability of the vaccine was 0.663.
Prediction and validation of the structure of the vaccine peptide
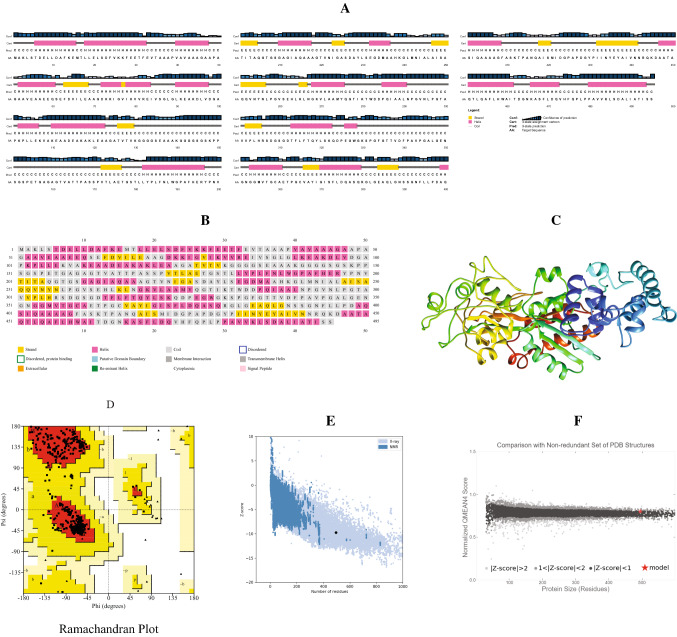
The secondary and 3D structure of the delineated vaccine peptide was predicted to perform the molecular docking analysis with the projected vaccine peptide. Using the PSIPRED server, the secondary structure was predicted. The structural features showed that the random coils are dominant in the structure with 263 amino acid residues in this region. 198 Residues were found in the alpha helix region while 61 residues were in the extended strands region (Fig. 4A, B).
Fig. 4.
Prediction and validation of structures of the vaccine peptide: A AnnotationGrid and B PSIPREDChart of the secondary structure. C 3D structure of the vaccine peptide, D Ramachandran plot analysis, Z-score analysis [E from Pro-SA web and F from QMEAN]
Employing the Robetta server, the 3D structure of the vaccine peptide was estimated with a confidence score of 0.67 (Fig. 4C). The Ramachandran plot (Fig. 4D) statistics showed 91.4% residues in most favoured regions and 8.6% in the additional allowed region. The MolProbity score was 0.92 (100th percentile), indicating that among structures with the comparable resolution, this is the highest. ProSA-web was used to review the protein 3D model for possible defects, and it projected a negative Z-score of 9.75 (Fig. 4E), suggesting that the model is of high consistency. For an environmental profile graph for the vaccine structure, another assessment method QMEAN gave various scores (Fig. 4F).
The vaccine peptide had a strong binding affinity with the receptors
For the validation of the interaction of the vaccine peptide with its receptors, molecular docking analysis was performed. For this purpose, two immune receptors, TLR2 (PDB ID 6NIG) and TLR4 (PDB ID 4G8A), as well as the CysR-CTLD3 fragment of human MR (PDB ID 5XTS) were selected. The molecular docking was carried out at the ClusPro server. TLRs and MR all were docked with the PstS1 protein as control (Supplementary Fig. S10). Using the PRODIGY server, the protein–protein complexes were further examined at 37 °C for binding affinities and dissociation constants (Table 2). The results illustrate that the delineated vaccine construct has a lower dissociation constant and a higher affinity for each of the studied receptors. Both of the ligand and peptide (vaccine and PstS1) showed a higher affinity and lower dissociation constant for TLR4 than the others.
Table 2.
Predicted binding affinity and dissociation constant of the vaccine–receptor complex
| Ligand | Receptor | Binding affinity, ΔG (kcal/mol) | Dissociation constant, Kd (M) |
|---|---|---|---|
| Vaccine peptide | 6NIG | − 13.7 | 2.4E−10 |
| 4G8A | − 23.3 | 3.9E−17 | |
| MR | − 10.1 | 8.1E−08 | |
| PstS1 | 6NIG | − 10.8 | 2.6E−08 |
| 4G8A | − 21.1 | 1.3E−15 | |
| MR | − 10.0 | 9.0E−08 |
The vaccine–TLR4 complex was stable during MD simulation
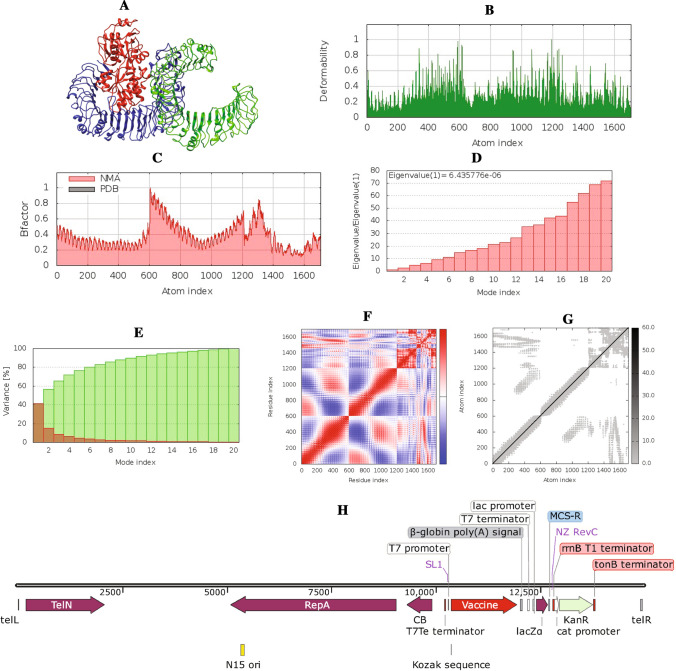
For the evaluation of the stability of the vaccine–TLR complex as well as the physical motions of the atoms of the vaccine peptide, MD simulation was performed during vaccine design. Normal mode analysis (NMA) of the vaccine–TLR4 docked complex (Fig. 5A) was performed. The deformable regions of the complex are displayed as peaks in the deformability graph (Fig. 5B). The deformability plot shows the residues formed a coiled structure that provides flexibility to the complex. A visualization of the relation between the NMA and the PDB region of the docked complex is represented in the B-factor graph (Fig. 5C). The eigenvalue of the TLR4 was 6.823114E−06 while the vaccine–TLR4 complex had an eigenvalue of 6.435776E−05 (Fig. 5D). These findings suggest that complex structures have higher rigidity and are more stable with a low deformation index. The covariance matrix (Fig. 5F) depicts the relationship between amino acid duplets, with correlated residues highlighted in red, anti-correlated residues highlighted in white, and non-correlated residues highlighted in blue, all interspersed in dynamical areas. The elastic network model (Fig. 5G), which can be visualized as a connecting matrix, categorizes which atom pairs are bound by springs. Each of the individual chains of the complex showed higher rigid regions.
Fig. 5.
NMA analysis and in silico cloning of the vaccine peptide: A vaccine–TLR4 complex; red color for the vaccine and blue and green color for TLR4, B deformability, C B-factor, D eigenvalues, E variance against mode index, F covariance matrix (correlated residues in red, uncorrelated in white or anticorrelated in blue motions), G elastic network model (darker gray regions indicate more stiffer regions) and H recombinant pJAZZOK® linear plasmid
In silico cloning of the vaccine construct
So far, the previous analyses indicate that the MT. P495 vaccine to be immunogenic and safe for human usage. For large-scale production and in silico cloning, a recombinant plasmid was designed by inserting the cloning insert (vaccine mRNA construct and accessory sequences) with SpeI restriction site at N terminal and NheI restriction site at C terminal into the pJAZZ-OK® expression vector. The cloning insert was 1935 nts long. The recombinant vector (Fig. 5H) that resulted was 14,977 bp long.
Discussion
TB is currently one of the major causes of death in the world [6]. Although this pathogen infects nearly a quarter of the world's population, it develops a latent chronic infection that reactivates when immune responses are compromised [1, 4, 5, 8]. BCG, the only approved Mtb vaccine, has been shown to have significant drawbacks, including failing to protect against TB in adults with pulmonary TB [86]. Live attenuated or inactivated vaccines (such as BCG) have also had a significant effect on public health. Other drawbacks include the chance of the pathogen reverting to its virulent state. Complicated cell-based manufacturing processes can make them ineffective against some strains [87, 88].
Vaccines based on individual subunit antigens are less effective, and adjuvants are required to improve immune responses. Furthermore, these vaccines fail to elicit CD8+ T-cell responses in humans, which are critical for infection clearance [87, 88]. Studies on the M72/AS01E subunit vaccine also showed the failure of CD8+ T-cell activation by the vaccine [89]. Activation of CD8+ T-cells within the body of an HIV patient is essential to fight against TB as HIV infection causes a progressive decrease of CD4+ T-cells [90]. These weaknesses have prompted researchers to look at alternative vaccine strategies. It is important to create a vaccine that is both safer and more immunogenic, as well as one that offers longer-lasting safety.
mRNA-based vaccines have the potential to become a more appealing choice than other types of vaccines. During the recent pandemic of SARS-CoV-2, mRNA vaccines against this deadly pathogen showed better protection than the conventional viral vector vaccine. The mRNA vaccines showed a higher efficacy (94.1% by mRNA-1273 vaccine [91] and 95% by BNT162b2 vaccine [92]) while the viral vectored vaccine showed a lower efficacy (70·4% by ChAdOx1 nCoV-19 vaccine [93]). BNT162b2 mRNA vaccine has shown evidence of stimulating CD4+ and CD8+ T-cells [94, 95] as well as B lymphocytes [94–96].
Phosphate-binding protein PstS1 of Mtb was used as the target antigen in this study to model a theoretical mRNA vaccine. Surprisingly, after alignment, it was found that the protein is highly conserved among reported strains (Supplementary Fig. S1). PstS1 is a well-known periplasmic protein that can induce adaptive immune effectors. Although it is a polymorphic protein, a relative immune reaction in human is not likely to be affected by polymorphism [97]. Conserved residues across the antigen were analyzed using several bioinformatics tools to validate their potential for immune effectors activation.
One of the key elements that contribute to the antigenicity of an antigen is the presence of T-cell epitopes. T-cell epitope immunogenicity is mostly determined by the intensity of T-cell epitopes' interactions with HLA molecules [98]. Peptides with IC50 value less than 50 nM are generally considered as high affinity while IC50 value less than 500 nM are considered intermediate affinity [99]. In this study, 98 CTL epitopes were identified with IC50 values less than 250 nM and 162 HTL epitopes were identified with IC50 values less than 50 nM. All of the identified T-cell epitopes were assessed using the IEDB conservancy analysis tool; 44 CTL epitopes, as well as 149 HTL epitopes, were observed to be 100% conserved all through the 3470 sequences. Antigenicity, allergenicity, and toxicity of the peptides were assessed in order to confirm the safety of these epitopes (Supplementary Tables 2, 3). The potential of using the PstS1 antigen as a candidate for vaccine development against Mtb is highlighted by the existence of safe and highly immunogenic epitopes.
The most important cells in adaptive immunity are helper T-cells. They participate in the activation of B-cells to release antibodies, cytotoxic T-cells to kill infected target cells, and macrophages to destroy ingested pathogens by secreting cytokines [100]. Therefore, the cytokine production inducing potential of the selected HTL epitopes was evaluated. Six of the selected 17 HTL epitopes demonstrated IFN-inducing potential, 7 exhibited IL4-inducing potential, and 9 showed IL10-inducing potential (Supplementary Table 3). According to a population coverage analysis, 99.92% of people worldwide are expected to elicit an immunological response to the T-cell epitopes reported in this study (Supplementary Fig. S2).
Several T-cell epitopes were previously identified by in vivo experiments on human as host. Interestingly it was found that, among the HTL epitopes that were finally selected in this study, several epitopes overlapped with previously identified epitopes [101–107] (Supplementary Table 3). These evidences prove that the epitopes identified in this study will be able to stimulate CD4+ T-cells to secrete cytokines. Proof of CD8+ T-cell stimulation by this antigen was also provided by several studies [108–110]. The epitopes, identified by those studies, were also identified at the primary stage of this study. But they were not finally selected as they were found to be allergenic, non-antigenic or not 100% conserved. Although the lowest percentage of protein sequence matches at identity at the 100% threshold was 98.10% (3404/3470) with a minimum identity of 77.78% (data not shown).
For the validation of the binding of the identified CTL and HTL epitopes with their corresponding HLA molecules, molecular docking was performed and this study revealed that epitopes could bind with their respective HLA molecule with higher binding energy (Table 1). To further confirm the stability of the bonds between the epitope and HLA, MD simulation was carried out (Fig. 2). Two epitope-HLA complexes were selected for this analysis: FLFTQYLSK-HLA-C*12:03 complex and AALNPGVNLPGTAVV-HLA-DRB1*01:01 complex. Interestingly both complexes showed stability throughout the time the simulation study was carried out. RMSD analysis of the complexes showed a similar pattern with their corresponding HLA allele. RMSF and SASA also indicate the stability of the epitope–HLA complex.
In silico immune simulation was carried out to confirm the immunogenic potential of the target antigen. The analysis revealed the activation of both B- as well as T-cells (Fig. 3). The activated effector cells persisted throughout the time the study was performed. Also, the level of antibodies was persistent, although a depletion was observed after 1100 days. The results also indicate a rise in the levels of IL-2 and IFN-γ, which are critical for immune system activation against any infection. These findings suggest that PstS1, an immunodominant protein from Mtb, has the potential to be used as a safe and effective antigen for developing a vaccine against Mtb.
Identification of the B-cell epitope is a vital step for vaccine design. Antibodies that are produced within the host body, bind to antigens at these sites. Conformational B-cell epitopes were identified based on the 3D structure of the antigen (Supplementary Fig. S7). Four epitopes were predicted; one of them consisted of 159 amino acid residues (Supplementary Fig. S8). A most recent study on human antibodies against Mtb revealed the presence of several PstS1-specific antibodies. These antibodies were able to reduce the Mtb burden at the lung by 50% [111]. All the interacting amino acids of PstS1 with those antibodies were also identified in this study as conformational B-cell epitopes. MSA showed that most of the interacting residues were 100% conserved in all the isolates of Mtb. Twenty-two linear epitopes were also identified which were further analyzed for their antigenic, allergenic and toxicity profile. After analysis, eight epitopes were found to be safe and immunogenic. Among them, several were found to overlap with previously identified epitopes [112, 113] (Supplementary Table 4). These evidences suggest that PstS1 would be able to efficiently stimulate the B-cell epitope.
In this study, a theoretical mRNA vaccine, MT. P495 was delineated against Mtb. To construct the MT. P495 mRNA, PstS1 was used as the GOI. To generate a stronger immune response, an adjuvant was added with the GOI. Adjuvant plays an important role in improving humoral and cellular immune response [114, 115]. 50S ribosomal protein L7/L12 protein from Mtb was used as an adjuvant. 50S ribosomal protein L7/L12 itself is a TLR-4 agonist and has the potential to induce dendritic cell maturation [116]. The translation efficiency is also influenced by the length of the poly(A) tail. mRNAs with an A120 tail showed increased, prolonged expression of the protein and a poly(A) tail longer than A120 did not show any significant effect on the expression of the target protein [117]. UTRs from two different genes were used in this vaccine construct: 5′ UTR of the human β-globin gene and 3′ UTR of the rabbit β-globin gene. These UTRs are important for the efficient translation of the mRNA. The UTRs of the human β-globin gene can increase the efficiency of mRNA translation [118]. The 3′ UTR of rabbit β-globin gene can influence seroconversion, antibody titer and cytokine profiles [117, 119]. To cap the 5′ end, Cap1 (m7GpppNm) was used. The half-life of mRNA is also increased as a result of a synergistic effect of 5′ end cap and 3′ end poly(A) tail [120]. Another peptide chain, the signal peptide was obtained from the tPA of H. sapiens which can improve the immunogenicity of the vaccine [121]. Two different linkers were used to join different components of the vaccine construct. Linker molecules can join two polypeptide chains, the protein moieties [122]. They are also used for increasing the stability of the final mRNA product, the protein [122, 123]. The linker GGGGSEAAAKGGGGS can increase both the thermal as well as the pH stability of the fused protein [124]. Another linker, AAY is an in vivo cleavable linker that acts as a cleavage site of proteasomes [125]. The final length of the mRNA construct was 1856 nts.
The codons of the delineated mRNA were optimized for efficient translation and better expression in the host (Supplementary Fig. S9; Supplementary Table 5). The properties of the optimized sequence suggested that host cells will efficiently express the mRNA. No tandem unusual codons were identified which is also an indicator of efficient translation. For optimization of the codons, we integrated two different codon optimization tools. In vivo experiments also suggest that the combined use of these two tools can increase protein expression efficiency [126]. The optimized codons have been used for the prediction of the secondary structure of the mRNA. Predicted free energy and the structures indicated that the mRNA will be stable.
The physiochemical properties of the vaccine peptide were assessed to ensure the safety of the vaccine (Supplementary Table 6). The peptide was predicted to be antigenic, non-allergenic, and non-toxic. So, it will be safe for human usage. Since the protein will have a longer half-life in mammalian reticulocytes, it is likely to remain stable inside the host cells and will be bioavailable for a long time. After the confirmation of safety in the host, the structure of the vaccine peptide (Fig. 4) was predicted to confirm the binding ability of the vaccine peptide with TLRs and MR by the molecular docking study. Both of the ligand peptides (adjuvant-linked GOI and GOI alone) could bind with the receptors with a high binding affinity (Table 2). The binding affinity also indicated that the vaccine peptide will be able to bind with the receptors with a higher binding affinity than the GOI alone. It was also found that our vaccine peptide could bind with the receptors 100 times more tightly than GOI. The stability of the receptor–ligand complex was checked (Fig. 5). To perform this analysis, the vaccine–TLR4 complex was selected. The deformability graph and covariance map indicated that the complex will be stable.
Instead of using an in vivo method, IVT is a popular and preferred method for producing mRNA on a large-scale [127]. In this study, promoter and termination sequences specific to the T7 polymerase were used for the efficient synthesis of RNA in IVT. Maintaining the homogenous length of the poly(A) tail is a major problem of this method. Most of the time, the poly(A) tails vary in length if the circular plasmid is used. To solve this problem, the pJAZZ-OK® linear plasmid was utilized as a cloning vector [84]. The total length of the recombinant plasmid was 14,977 bp.
All of the results above, as well as previous research on humans as hosts, suggest that the proposed mRNA vaccine candidate, MT. P495, will probably elicit a strong immune response, specific against Mtb. In order to develop a viable Mtb vaccine in the future, this modeled mRNA is an excellent vaccine model that can be readily employed for laboratory testing, including in vitro as well as in vivo studies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge support from Biomedical Research Foundation (BMRF, DU). Authors also acknowledge High Performance Computing Facility support from Centre for Bioinformatics Learning Advancement and Systematics Training (cBLAST), University of Dhaka.
Author contributions
The project was designed by ABMMKI. The data was collected by SS. The analysis was performed by SS and ABMMKI. The draft was written by SS and ABMMKI. All authors reviewed and approved the manuscript.
Funding
The project was not funded by any internal or external organization.
Data availability
The data supporting this article are available in both the article and its Online Supplementary Material.
Declarations
Conflict of interest
All authors declared that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Castro KG. Tuberculosis as an opportunistic disease in persons infected with human immunodeficiency virus. Clin Infect Dis. 1995;21:66–71. doi: 10.1093/clinids/21.Supplement_1.S66. [DOI] [PubMed] [Google Scholar]
- 2.Zhai W, Wu F, Zhang Y, et al. The immune escape mechanisms of Mycobacterium tuberculosis. Int J Mol Sci. 2019 doi: 10.3390/ijms20020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heemskerk D, Caws M, Marais B, Farrar J. Tuberculosis in adults and children. Cham: Springer; 2015. [PubMed] [Google Scholar]
- 4.Shah P, Mistry J, Reche PA, et al. In silico design of Mycobacterium tuberculosis epitope ensemble vaccines. Mol Immunol. 2018;97:56–62. doi: 10.1016/j.molimm.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev. 2003;16:463–496. doi: 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO . Global tuberculosis report 2021: executive summary. Geneva: World Health Organization; 2021. [Google Scholar]
- 7.Houben RMGJ, Dodd PJ. The global burden of latent tuberculosis infection: a re-estimation using mathematical modelling. PLoS Med. 2016;13:1–13. doi: 10.1371/journal.pmed.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitlow E, Mustafa AS, Hanif SNM. An overview of the development of new vaccines for tuberculosis. Vaccines. 2020;8:1–13. doi: 10.3390/vaccines8040586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf AJ, Desvignes L, Linas B, et al. Initiation of the adaptive immune response to Mycobacterium tuberculosis depends on antigen production in the local lymph node, not the lungs. J Exp Med. 2008;205:105–115. doi: 10.1084/jem.20071367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doherty TM, Andersen P. Vaccines for tuberculosis: novel concepts and recent progress. Clin Microbiol Rev. 2005;18:687–702. doi: 10.1128/CMR.18.4.687-702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delogu G, Sali M, Fadda G. The biology of Mycobacterium tuberculosis infection. Mediterr J Hematol Infect Dis. 2013 doi: 10.4084/mjhid.2013.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schrager LK, Vekemens J, Drager N, et al. The status of tuberculosis vaccine development. Lancet Infect Dis. 2020;20:e28–e37. doi: 10.1016/S1473-3099(19)30625-5. [DOI] [PubMed] [Google Scholar]
- 13.Orme IM. Tuberculosis vaccine types and timings. Clin Vaccine Immunol. 2015;22:249–257. doi: 10.1128/CVI.00718-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Day CL, Tameris M, Mansoor N, et al. Induction and regulation of T-cell immunity by the novel tuberculosis vaccine M72/AS01 in South African adults. Am J Respir Crit Care Med. 2013;188:492–502. doi: 10.1164/rccm.201208-1385OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines—a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pascolo S. Messenger RNA-based vaccines. Expert Opin Biol Ther. 2004;4(8):1285–1294. doi: 10.1517/14712598.4.8.1285. [DOI] [PubMed] [Google Scholar]
- 17.Verbeke R, Lentacker I, De Smedt SC, Dewitte H. Three decades of messenger RNA vaccine development. Nano Today. 2019;28:100766. doi: 10.1016/j.nantod.2019.100766. [DOI] [Google Scholar]
- 18.Bian H. The use of bioinformatics for identifying class II-restricted T-cell epitopes. Methods. 2003;29:299–309. doi: 10.1016/S1046-2023(02)00352-3. [DOI] [PubMed] [Google Scholar]
- 19.Sirskyj D, Diaz-Mitoma F, Golshani A, et al. Innovative bioinformatic approaches for developing peptide-based vaccines against hypervariable viruses. Immunol Cell Biol. 2011;89:81–89. doi: 10.1038/icb.2010.65. [DOI] [PubMed] [Google Scholar]
- 20.Jabbar B, Rafique S, Salo-Ahen OMH, et al. Antigenic peptide prediction from E6 and E7 oncoproteins of HPV types 16 and 18 for therapeutic vaccine design using immunoinformatics and MD simulation analysis. Front Immunol. 2018 doi: 10.3389/fimmu.2018.03000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z, Bogdan P, Nazarian S. An in silico deep learning approach to multi-epitope vaccine design: a SARS-CoV-2 case study. Sci Rep. 2021;11:3238. doi: 10.1038/s41598-021-81749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain R, Singh S, Verma SK, Jain A. Genome-wide prediction of potential vaccine candidates for Campylobacter jejuni using reverse vaccinology. Interdiscip Sci Comput Life Sci. 2019;11:337–347. doi: 10.1007/s12539-017-0260-5. [DOI] [PubMed] [Google Scholar]
- 23.Wheeler DL, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:5–12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011 doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waterhouse AM, Procter JB, Martin DMA, et al. JalView Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bateman A. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burley SK, Bhikadiya C, Bi C, et al. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021;49:D437–D451. doi: 10.1093/nar/gkaa1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen MV, Lundegaard C, Lamberth K, et al. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 2007;8:1–12. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters B, Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinform. 2005;6:1–9. doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim Y, Sidney J, Pinilla C, et al. Derivation of an amino acid similarity matrix for peptide:MHC binding and its application as a Bayesian prior. BMC Bioinform. 2009;10:394. doi: 10.1186/1471-2105-10-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vita R, Mahajan S, Overton JA, et al. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019;47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen M, Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinform. 2009;10:296. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007;8:1–7. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dimitrov I, Bangov I, Flower DR, Doytchinova I. AllerTOP vol 2—a server for in silico prediction of allergens. J Mol Model. 2014 doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- 35.Gupta S, Kapoor P, Chaudhary K, et al. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE. 2013 doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bui HH, Sidney J, Li W, et al. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinform. 2007;8:1–6. doi: 10.1186/1471-2105-8-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhanda SK, Gupta S, Vir P, Raghava GP. Prediction of IL4 inducing peptides. Clin Dev Immunol. 2013;2013:263952. doi: 10.1155/2013/263952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagpal G, Usmani SS, Dhanda SK, et al. Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Sci Rep. 2017;7:1–10. doi: 10.1038/srep42851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dhanda SK, Vir P, Raghava GPS. Designing of interferon-gamma inducing MHC class-II binders. Biol Direct. 2013;8:1–15. doi: 10.1186/1745-6150-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bui HH, Sidney J, Dinh K, et al. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinform. 2006;7:1–5. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maupetit J, Derreumaux P, Tufféry P. A fast method for large-scale De Novo peptide and miniprotein structure prediction. J Comput Chem. 2009 doi: 10.1002/jcc.21365. [DOI] [PubMed] [Google Scholar]
- 42.Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson J, Barker DJ, Georgiou X, et al. IPD-IMGT/HLA database. Nucleic Acids Res. 2020;48:D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. doi: 10.1107/s0021889892009944. [DOI] [Google Scholar]
- 45.Williams CJ, Headd JJ, Moriarty NW, et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:407–410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antunes DA, Moll M, Devaurs D, et al. DINC 2.0: a new protein–peptide docking webserver using an incremental approach. Cancer Res. 2017;77:e55–e57. doi: 10.1158/0008-5472.CAN-17-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shahrear S, Islam ABMMK. Immunoinformatics guided modeling of CCHF_GN728, an mRNA-based universal vaccine against Crimean-Congo hemorrhagic fever virus. Comput Biol Med. 2022;140:105098. doi: 10.1016/j.compbiomed.2021.105098. [DOI] [PubMed] [Google Scholar]
- 50.Dassault Systmes (2017) BIOVIA Discovery Studio Dassault Systmes BIOVIA, discovery studio modeling environment, Release 2017. Dassault Systmes
- 51.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 52.Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43–56. doi: 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- 53.Berendsen HJC, Grigera JR, Straatsma TP. The missing term in effective pair potentials. J Phys Chem. 1987;91:6269–6271. doi: 10.1021/j100308a038. [DOI] [Google Scholar]
- 54.Berendsen HJC, Postma JPM, van Gunsteren WF, et al. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 55.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 56.Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 57.Bondi A. van der Waals volumes and radii. J Phys Chem. 1964;68:441–451. doi: 10.1021/j100785a001. [DOI] [Google Scholar]
- 58.Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 59.Harris CR, Millman KJ, van der Walt SJ, et al. Array programming with NumPy. Nature. 2020;585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.R Core Team (2021) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. Accessed 02 May 2022
- 61.Osorio D, Rondón-Villarreal P, Torres R. Peptides: a package for data mining of antimicrobial peptides. R J. 2015;7:4. doi: 10.32614/RJ-2015-001. [DOI] [Google Scholar]
- 62.Ponomarenko J, Bui HH, Li W, et al. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008;9:1–8. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emini EA, Hughes JV, Perlow DS, Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985;55:836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-Q. [DOI] [PubMed] [Google Scholar]
- 66.Karplus PA, Schulz GE. Prediction of chain flexibility in proteins—a tool for the selection of peptide antigens. Naturwissenschaften. 1985;72:212–213. doi: 10.1007/BF01195768. [DOI] [Google Scholar]
- 67.Rapin N, Lund O, Bernaschi M, Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE. 2010 doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gottschalk N, Lang S, Kimmig R, et al. Monocytes and the 38 kDa-antigen of Mycobacterium tuberculosis modulate natural killer cell activity and their cytolysis directed against ovarian cancer cell lines. BMC Cancer. 2012;12:1. doi: 10.1186/1471-2407-12-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Young DB, Garbe TR. Lipoprotein antigens of Mycobacterium tuberculosis. Res Microbiol. 1991;142:55–65. doi: 10.1016/0923-2508(91)90097-T. [DOI] [PubMed] [Google Scholar]
- 70.Liu H, Jiang Y, Dou X, et al. PstS1 polymorphisms of Mycobacterium tuberculosis strains may reflect ongoing immune evasion. Tuberculosis. 2013;93:475–481. doi: 10.1016/j.tube.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 71.Agarwala R, Barrett T, Beck J, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grote A, Hiller K, Scheer M, et al. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:526–531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gruber AR, Lorenz R, Bernhart SH, et al. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:70–74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Magnan CN, Zeller M, Kayala MA, et al. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics. 2010;26:2936–2943. doi: 10.1093/bioinformatics/btq551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gasteiger E, Hoogland C, Gattiker A et al (2005) Protein identification and analysis tools on the ExPASy server. In: The proteomics protocols handbook. Humana Press, Totowa, pp 571–607
- 76.He Y, Xiang Z, Mobley HLT. Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development. J Biomed Biotechnol. 2010 doi: 10.1155/2010/297505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buchan DWA, Jones DT. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 2019;47:W402–W407. doi: 10.1093/nar/gkz297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 79.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:526–531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kozakov D, Hall DR, Xia B, et al. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xue LC, Rodrigues JP, Kastritis PL, et al. PRODIGY: a web server for predicting the binding affinity of protein–protein complexes. Bioinformatics. 2016;32:3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 82.López-Blanco JR, Aliaga JI, Quintana-Ortí ES, Chacón P. iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42:271–276. doi: 10.1093/nar/gku339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jackson NAC, Kester KE, Casimiro D, et al. The promise of mRNA vaccines: a biotech and industrial perspective. NPJ Vaccines. 2020;5:11. doi: 10.1038/s41541-020-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grier AE, Burleigh S, Sahni J, et al. pEVL: a linear plasmid for generating mRNA IVT templates with extended encoded poly(A) sequences. Mol Ther Nucleic Acids. 2016;5:e306. doi: 10.1038/mtna.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Courel M, Clément Y, Bossevain C, et al. GC content shapes mRNA storage and decay in human cells. eLife. 2019 doi: 10.7554/eLife.49708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crampin AC, Glynn JR, Fine PEM. What has Karonga taught us? Tuberculosis studied over three decades. Int J Tuberc Lung Dis. 2009;13:153–164. [PMC free article] [PubMed] [Google Scholar]
- 87.Pollard C, De Koker S, Saelens X, et al. Challenges and advances towards the rational design of mRNA vaccines. Trends Mol Med. 2013;19:705–713. doi: 10.1016/j.molmed.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 88.Iavarone C, O’Hagan DT, Yu D, et al. Mechanism of action of mRNA-based vaccines. Expert Rev Vaccines. 2017;16:871–881. doi: 10.1080/14760584.2017.1355245. [DOI] [PubMed] [Google Scholar]
- 89.Tait DR, Hatherill M, Van Der Meeren O, et al. Final analysis of a trial of M72/AS01 E vaccine to prevent tuberculosis. N Engl J Med. 2019;381:2429–2439. doi: 10.1056/nejmoa1909953. [DOI] [PubMed] [Google Scholar]
- 90.Okoye AA, Picker LJ. CD4+ T-cell depletion in HIV infection: mechanisms of immunological failure. Immunol Rev. 2013;254:54–64. doi: 10.1111/imr.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403–416. doi: 10.1056/nejmoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/nejmoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Voysey M, Clemens SAC, Madhi SA, et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Walsh EE, Frenck RW, Falsey AR, et al. Safety and immunogenicity of two RNA-based COVID-19 vaccine candidates. N Engl J Med. 2020;383:2439–2450. doi: 10.1056/nejmoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sahin U, Muik A, Derhovanessian E, et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 96.Mulligan MJ, Lyke KE, Kitchin N, et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 97.Xiao T, Jiang Y, Li G, et al. Polymorphism of MPT64 and PstS1 in Mycobacterium tuberculosis is not likely to affect relative immune reaction in human. Medicine (US) 2019;98:1–6. doi: 10.1097/MD.0000000000018073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weber CA, Mehta PJ, Ardito M, et al. T cell epitope: Friend or Foe? Immunogenicity of biologics in context. Adv Drug Deliv Rev. 2009;61:965–976. doi: 10.1016/j.addr.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peters B, Bulik S, Tampe R, et al. Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J Immunol. 2003;171:1741–1749. doi: 10.4049/jimmunol.171.4.1741. [DOI] [PubMed] [Google Scholar]
- 100.Arango Duque G, Descoteaux A. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol. 2014 doi: 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vordermeier H-M, Harris DP, Friscia G, et al. T cell repertoire in tuberculosis: selective anergy to an immunodominant epitope of the 38-kDa antigen in patients with active disease. Eur J Immunol. 1992;22:2631–2637. doi: 10.1002/eji.1830221024. [DOI] [PubMed] [Google Scholar]
- 102.Wilkinson KA, Vordermeier MH, Kajtár J, et al. Modulation of peptide specific T cell responses by non-native flanking regions. Mol Immunol. 1997;34:1237–1246. doi: 10.1016/S0161-5890(98)00009-1. [DOI] [PubMed] [Google Scholar]
- 103.Wilkinson RJ, Vordermeier HM, Wilkinson KA, et al. Peptide-specific T cell response to Mycobacterium tuberculosis: clinical spectrum, compartmentalization, and effect of chemotherapy. J Infect Dis. 1998;178:760–768. doi: 10.1086/515336. [DOI] [PubMed] [Google Scholar]
- 104.Wilkinson KA, Vordermeier H, Wilkinson RJ, et al. Synthesis and in vitro T-cell immunogenicity of conjugates with dual specificity: attachment of epitope peptides of 16 and 38 kDa proteins from Mycobacterium tuberculosis to branched polypeptide. Bioconjug Chem. 1998;9:539–547. doi: 10.1021/bc970159+. [DOI] [PubMed] [Google Scholar]
- 105.Venkataprasad N. Induction of cellular immunity to a mycobacterial antigen adsorbed on lamellar particles of lactide polymers. Vaccine. 1999;17:1814–1819. doi: 10.1016/S0264-410X(98)00372-7. [DOI] [PubMed] [Google Scholar]
- 106.Liu J, Chen X, Wang J, et al. Prediction and identification of CD4+ T cell epitope for the protective antigens of Mycobacterium tuberculosis. Medicine (Baltim) 2021;100:e24619. doi: 10.1097/MD.0000000000024619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.da Silva BCM, Grassi MFR, Coutinho R, et al. Mycobacterium tuberculosis epitope-specific interferon-g production in healthy Brazilians reactive and non-reactive to tuberculin skin test. Mem Inst Oswaldo Cruz. 2014;109:999–1004. doi: 10.1590/0074-0276140193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hammond AS, Klein MR, Corrah T, et al. Mycobacterium tuberculosis genome-wide screen exposes multiple CD8+ T cell epitopes. Clin Exp Immunol. 2005;140:109–116. doi: 10.1111/j.1365-2249.2005.02751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shams H, Barnes PF, Weis SE, et al. Human CD8+ T cells recognize epitopes of the 28-kDa hemolysin and the 38-kDa antigen of Mycobacterium tuberculosis. J Leukoc Biol. 2003;74:1008–1014. doi: 10.1189/jlb.0403138. [DOI] [PubMed] [Google Scholar]
- 110.Cho S, Mehra V, Thoma-Uszynski S, et al. Antimicrobial activity of MHC class I-restricted CD8+ T cells in human tuberculosis. Proc Natl Acad Sci USA. 2000;97:12210–12215. doi: 10.1073/pnas.210391497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Watson A, Li H, Ma B, et al. Human antibodies targeting a Mycobacterium transporter protein mediate protection against tuberculosis. Nat Commun. 2021 doi: 10.1038/s41467-021-20930-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gaseitsiwe S, Valentini D, Mahdavifar S, et al. Pattern recognition in pulmonary tuberculosis defined by high content peptide microarray chip analysis representing 61 proteins from M. tuberculosis. PLoS ONE. 2008;3:e3840. doi: 10.1371/journal.pone.0003840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.López-Vidal Y, de León-Rosales SP, Castañón-Arreola M, et al. Response of IFN-γ and IgG to ESAT-6 and 38 kDa recombinant proteins and their peptides from Mycobacterium tuberculosis in tuberculosis patients and asymptomatic household contacts may indicate possible early-stage infection in the latter. Arch Med Res. 2004;35:308–317. doi: 10.1016/j.arcmed.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 114.Sarkar I, Garg R, van Drunen Littel-van den Hurk S. Selection of adjuvants for vaccines targeting specific pathogens. Expert Rev Vaccines. 2019;18:505–521. doi: 10.1080/14760584.2019.1604231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Coccia M, Collignon C, Hervé C, et al. Cellular and molecular synergy in AS01-adjuvanted vaccines results in an early IFNγ response promoting vaccine immunogenicity. NPJ Vaccines. 2017 doi: 10.1038/s41541-017-0027-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee SJ, Shin SJ, Lee MH, et al. A potential protein adjuvant derived from Mycobacterium tuberculosis Rv0652 enhances dendritic cells-based tumor immunotherapy. PLoS ONE. 2014;9:1–11. doi: 10.1371/journal.pone.0104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Holtkamp S, Kreiter S, Selmi A, et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- 118.Zhuang X, Qi Y, Wang M, et al. mRNA vaccines encoding the HA protein of influenza a H1N1 virus delivered by cationic lipid nanoparticles induce protective immune responses in mice. Vaccines. 2020;8:1–17. doi: 10.3390/vaccines8010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zinckgraf JW, Silbart LK. Modulating gene expression using DNA vaccines with different 3′-UTRs influences antibody titer, seroconversion and cytokine profiles. Vaccine. 2003;21:1640–1649. doi: 10.1016/S0264-410X(02)00740-5. [DOI] [PubMed] [Google Scholar]
- 120.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 121.Kou Y, Xu Y, Zhao Z, et al. Tissue plasminogen activator (tPA) signal sequence enhances immunogenicity of MVA-based vaccine against tuberculosis. Immunol Lett. 2017;190:51–57. doi: 10.1016/j.imlet.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 122.Chen X, Zaro JL, Shen WC. Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev. 2013;65:1357–1369. doi: 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Arai R, Wriggers W, Nishikawa Y, et al. Conformations of variably linked chimeric proteins evaluated by synchrotron X-ray small-angle scattering. Proteins Struct Funct Genet. 2004;57:829–838. doi: 10.1002/prot.20244. [DOI] [PubMed] [Google Scholar]
- 124.Chen H, Chen Z, Wu B, et al. Influences of various peptide linkers on the Thermotoga maritima MSB8 nitrilase displayed on the spore surface of Bacillus subtilis. J Mol Microbiol Biotechnol. 2017;27:64–71. doi: 10.1159/000454813. [DOI] [PubMed] [Google Scholar]
- 125.Agallou M, Margaroni M, Kotsakis SD, Karagouni E. A canine-directed chimeric multi-epitope vaccine induced protective immune responses in BALB/C mice infected with Leishmania infantum. Vaccines. 2020;8:1–35. doi: 10.3390/vaccines8030350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Koblan LW, Doman JL, Wilson C, et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol. 2018;36:843–846. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kanwal F, Chen T, Zhang Y, et al. Large-scale in vitro transcription, RNA purification and chemical probing analysis. Cell Physiol Biochem. 2018;48:1915–1927. doi: 10.1159/000492512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this article are available in both the article and its Online Supplementary Material.