Abstract
We determined the genomic organization of 14 clinical strains of Bordetella pertussis isolated over an 18-month period in Alberta, Canada. The maps of these 14 strains, while demonstrating general similarity of gene order, display a number of examples of genomic rearrangements in the form of large chromosomal inversions.
The current revolution taking place in the arena of bacterial genomes, through the acquisition of comprehensive genomic sequence information from a wide variety of bacteria, has given us unprecedented views into the content, structure, and diversity of these genetic structures. In addition, an increasing number of studies which have undertaken the comparative analysis of multiple representatives of the genomes of particular species are leading us away from a view of bacterial genomes as static, monolithic structures and towards the view that they are relatively fluid, plastic structures. This fluidity is manifested not only by the acquisition and loss of genetic information (5, 7, 11) but also by large-scale rearrangements leading to changes in genomic organization (3, 6, 8, 11).
We recently reported a comparative analysis of the genomes of a number of Bordetella pertussis strains in which we observed that the genomic organization of B. pertussis laboratory strains was variable (14). In the current study we have extended our analyses to include a number of B. pertussis isolates from a recent outbreak of whooping cough which occurred in Alberta, Canada, in 1989 to 1991 (2). The maps we derived for these strains document many examples of large-scale chromosomal rearrangements within this group of B. pertussis strains which are representative of natural populations.
The B. pertussis strains characterized in this study were isolated from a whooping cough outbreak which occurred throughout Alberta, Canada, from December 1989 to May 1991. A total of 70 strains from different locations throughout this province were previously analyzed by pulsed-field gel electrophoresis (PFGE) of chromosomal XbaI digests (2). By this procedure, 14 different electrophoretic patterns were distinguishable. Although 15 patterns were initially reported, digestion patterns c and f were later found to be the same (10). Strains representative of these 14 patterns were analyzed to determine their genomic organization. These strains are presented in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Relevant features | Source or reference |
|---|---|---|
| B. pertussis | ||
| 14797 | XbaI PFGE pattern a | 2 |
| 9755 | XbaI PFGE pattern b | 2 |
| 964 | XbaI PFGE pattern c | 2 |
| 9835 | XbaI PFGE pattern d | 2 |
| 6706 | XbaI PFGE pattern e | 2 |
| 12527 | XbaI PFGE pattern g | 2 |
| 14784 | XbaI PFGE pattern h | 2 |
| 2427 | XbaI PFGE pattern i | 2 |
| 8892 | XbaI PFGE pattern j | 2 |
| 3860 | XbaI PFGE pattern k | 2 |
| 11291 | XbaI PFGE pattern l | 2 |
| 12162 | XbaI PFGE pattern m | 2 |
| 12145 | XbaI PFGE pattern n | 2 |
| 8004 | XbaI PFGE pattern o | 2 |
| Plasmid vectors | ||
| pSS1898 | Mapping vector: R6K oriV gen oriT | 14 |
| pSS1914 | Mapping vector: ColE1 oriV kan amp oriT | 14 |
| pSS2070 | pSS1914 with EcoRI site destroyed by addition of NotI site | This study |
| pSS1898 derivativesb | ||
| pSS1919 | 10.0-kb EcoRI fhaB fragment cloned into pSS1898 | 14 |
| pSS1921 | 17.5-kb EcoRI prn fragment cloned into pSS1898 | 14 |
| pSS1923 | 14.0-kb EcoRI cya′ and upstream fragment cloned into pSS1898 | 14 |
| pSS1926 | 4.7-kb EcoRI ptx fragment cloned into pSS1898 | 14 |
| pSS1927 | 17.0-kb EcoRI recA fragment cloned into pSS1898 | 14 |
| pSS1930 | 4-kb EcoRI bvg downstream fragment cloned into pSS1898 | 14 |
| pSS1914 derivativesb | ||
| pSS1931 | 10.0-kb EcoRI fhaB fragment cloned into pSS1914 | 14 |
| pSS1932 | 17.5-kb EcoRI prn fragment cloned into pSS1914 | 14 |
| pSS1933 | 14.0-kb EcoRI cya′ and upstream fragment cloned into pSS1914 | 14 |
| pSS1934 | 4.5-kb EcoRI fimX fragment cloned into pSS1914 | 14 |
| pSS1935 | 4.7-kb EcoRI ptx fragment cloned into pSS1914 | 14 |
| pSS1936 | 17.0-kb EcoRI recA fragment cloned into pSS1914 | 14 |
| pSS1937 | 0.83-kb EcoRI vrg6 fragment cloned into pSS1914 | 14 |
| pSS1938 | 17.5-kb EcoRI fim2 fragment cloned into pSS1914 | 14 |
| pSS1939 | 4.0-kb EcoRI bvg downstream fragment cloned into pSS1914 | 14 |
| pSS1940 | 0.85-kb EcoRI fim3 PCR fragment cloned into pSS1914 | 14 |
| pSS2572 | 7.7-kb EcoRI por fragment cloned into pSS1914 | This study |
| pSS2573 | 2.6-kb EcoRI bpl fragment cloned into pSS1914 | This study |
| pSS2574 | 1.8-kb NotI dnt fragment cloned into pSS2070 | This study |
| pSS2575 | 2.8-kb NotI brkAB fragment cloned into pSS2070 | This study |
| pSS2576 | 1.8-kb EcoRI bfeA fragment cloned into pSS1914 | This study |
| pSS2577 | 6.7-kb EcoRI aroA fragment cloned into pSS1914 | This study |
| pSS2610 | 1.6-kb SalI tcfA fragment cloned into pSS1914 | This study |
| pSS2611 | 1.9-kb SalI ompR/envZ fragment cloned into pSS1914 | This study |
| pSS2612 | 0.84-kb SalI fur fragment cloned into pSS1914 | This study |
Plasmids pSS2572, pSS2574, and pSS2577 contain inserts derived from the previously described pSS1925, pSS2083, and pSS1924, respectively (14). The bpl fragment in pSS2573 was derived from pUCE2a, provided by D. Maskell. The brkAB fragment in pSS2575 was derived from pRF1003, kindly provided by A. Weiss. The bfeA fragment in pSS2576 was derived from pCIX7, kindly provided by B. Beals. The tcfA fragment in pSS2610 was derived from pTF357, kindly provided by T. Finn. The ompR/envZ fragment in pSS2611 was derived from pBP4, kindly provided by D. Maskell. The fur fragment in pSS2612 was derived from pRK/0.8S, kindly provided by S. Armstrong. SalI fragments were first cloned into the SalI site of a plasmid containing a palindromic polylinker and then excised as an EcoRI fragment for cloning into pSS1914.
Derivatives used for mapping.
The “chromosomal surveying” approach to genomic mapping has been described previously (14). This technique relies on homologous recombination to mediate insertion of two independent plasmid suicide vectors, pSS1898 and pSS1914, at chromosomal positions dictated by cloned chromosomal sequences. Both vectors contain the recognition sequence for the intron-encoded restriction enzyme I-SceI, which is not found in the B. pertussis genome. Cleavage of chromosomal DNA prepared from a strain harboring the two suicide plasmids inserted at different genetic loci results in the liberation of a DNA fragment whose size represents the distance between the two loci. Further pairwise distance determinations yields the information required to construct a chromosomal map for the positions of the genes examined.
Each of the 14 isolates was first mapped by using a set of six chromosomal markers (primary markers) cloned into pSS1898 and pSS1914. The chromosomal loci used in this initial phase were bvg (virulence regulation), cya (adenylate cyclase toxin), fha (filamentous hemagglutinin), recA (homologue of E. coli recA), prn (pertactin), and ptx (pertussis toxin). This preliminary analysis allowed the construction of a framework for the placement of additional gene loci. The genetic loci aroA (5-enolpyruvylshikimate-3-phosphate synthase), bfeA (ferric enterobactin receptor), brkAB (complement resistance), bpl (lipooligosaccharide biosynthesis), dnt (dermonecrotic toxin), fim2 (fimbrial subunit, serotype 2), fim3 (fimbrial subunit, serotype 3), fimX (silent fimbrial subunit gene), fur (ferric uptake regulator), ompR/envZ (osmolarity-sensitive gene regulation), por (porin), tcfA (tracheal colonization factor), and vrg6 (vir-repressed gene 6) were localized within this framework following the insertion of each respective pSS1914 derivative into a subset of the strains containing pSS1898 inserted at primary marker locations. To minimize the number of nonfruitful distance determinations (those where the distances were too great to determine by PFGE), an initial assumption of similarity of gene order with B. pertussis Tohama I was made. When this assumption was found to be inaccurate, additional determinations of the distance to other primary markers were made. Gene loci were always mapped with reference to primary markers flanking each side. In all cases, distance determinations between any two markers were made on the same pulsed-field gel for all 14 strains so that differences in intergenic distances were apparent.
Escherichia coli SM10 was used for transfer of all plasmids to the B. pertussis strains under study (12). Selection for transfer and integration of pSS1898 derivatives was done by gentamicin resistance, and counterselection against E. coli donors was by colicin B1 resistance (1). Selection for the subsequent integration of pSS1914 derivatives was by kanamycin resistance, with counterselection by gentamicin resistance. PFGE and I-SceI digestion were performed as previously described (14).
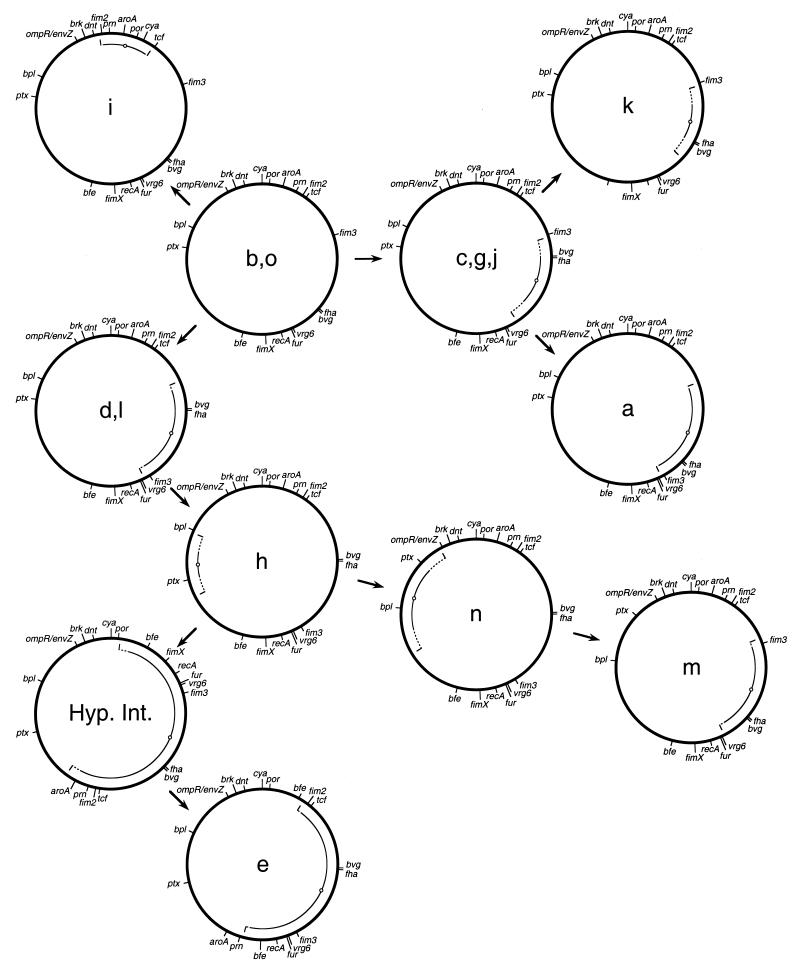
The maps derived using a total of 19 chromosomal markers are shown in Fig. 1. It can be seen that 10 modes of gene organization were observed among the 14 strains analyzed. All the maps were similar in size, with no evidence of large deletions or insertions. The maps show hypothetical interrelationships, with arrows indicating that the maps in question could be derived one from another by inversions of large chromosomal segments. The maps of strain h and strain e could not be related by a single inversion. In this case a hypothetical intermediate is shown, such that h and e could be related by two successive inversions. It should be emphasized that the scheme presented is not the only one which is consistent with these data, and although so depicted here, it is not known that the map for strains b and o is in fact the progenitor of the others.
FIG. 1.
Chromosomal maps of 14 clinical isolates of B. pertussis. Letters designate the XbaI digestion pattern obtained by PFGE (2). Maps show relative positions of 19 genetic loci. Maps were derived by the chromosomal surveying approach previously described and the overall strategy outlined in the text (14). Arrows are used to show hypothetical relationships between maps derived one from the other by inversions. Arcs represent the extent of inverted sequences relative to the hypothetical predecessor. Symbols: solid arc, minimum inversion which is consistent with the differences in the two maps; dotted arc, maximum inversion consistent with the observed differences; small circle on arc, center of inverted sequences. Hyp. Int., hypothetical intermediate.
The maps described for these isolates differ consistently in three ways from that determined for B. pertussis Tohama I (13). While dnt, brkAB, and ompR/envZ have the same configuration with respect to each other, their gene order is reversed relative to that seen for Tohama I. Also, these genes are located very close to the cya locus, unlike Tohama I. This configuration is similar, however, to that seen with B. pertussis 165 and Wellcome 28, in which dnt and cya are proximal (14). An additional difference is the placement of the fur gene. In Tohama I this gene is located between the brkAB and ompR/envZ loci, whereas in the strains mapped here, the fur gene is found between recA and vrg6.
Interestingly, a number of the inversions detected in these isolates involve a region of the chromosome which includes the fha and bvg operons. An inversion of this region was also seen to relate the map of B. pertussis 165 and Wellcome 28 to that of Tohama I (14). Although the extents of these inversions vary, their centers are similar in these cases (a, c, d, e, g, j, k, l, m, n, and the hypothetical intermediate in Fig. 1). The inversions seen on the opposite side of the chromosome (h and n in Fig. 1) can also be viewed as inversions which have approximately the same point as their center. This accounts for all but one of the inversions seen (i in Fig. 1). Previous studies concerning inversions of the chromosome of Salmonella typhimurium have suggested that inversions generally include the origin or terminus of replication (9). By analogy, we would predict that the origin or terminus of replication in these B. pertussis strains would be found at approximately four o’clock on these maps.
The results reported here are relevant to the interpretation of PFGE analysis of bacterial isolates. Changes in PFGE patterns of chromosomal restriction digests have sometimes been assumed to be due to loss or gain of restriction sites due to mutational mechanisms (15). However, our results suggest that chromosomal rearrangements may also be a contributing factor to genome variability detected by this method.
This study provides additional evidence that genomic plasticity is an attribute not just of laboratory strains of B. pertussis but also of natural populations living in association with the human host. Although the molecular mechanisms underlying the genomic rearrangements observed here are unknown, we can form a hypothesis based on earlier studies which examined the nature of chromosomal inversions observed in other bacterial species. Inversions observed in E. coli and S. typhimurium have endpoints within copies of IS5 and rrn loci, respectively (8, 16). In these cases the inversions have apparently been catalyzed by homologous recombination between these islands of homology. B. pertussis is a likely candidate for inversions catalyzed in a similar manner, as approximately 100 copies of a 1-kb insertion sequence, IS481, are estimated to be present in the B. pertussis chromosome (4). Detailed molecular characterization of defined chromosomal rearrangements captured in the laboratory will shed further light on these questions.
REFERENCES
- 1.Brickman T J, Armstrong S K. Colicins B and Ia as novel counterselective agents in interspecies conjugal DNA transfers from colicin-sensitive Escherichia coli donors to other gram-negative recipient species. Gene. 1996;178:39–42. doi: 10.1016/0378-1119(96)00331-9. [DOI] [PubMed] [Google Scholar]
- 2.de Moissac Y R, Ronald S L, Peppler M S. Use of pulsed-field gel electrophoresis for epidemiological study of Bordetella pertussis in a whooping cough outbreak. J Clin Microbiol. 1994;32:398–402. doi: 10.1128/jcm.32.2.398-402.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibbs C P, Meyer T F. Genome plasticity in Neisseria gonorrhoeae. FEMS Microbiol Lett. 1996;145:173–179. doi: 10.1111/j.1574-6968.1996.tb08574.x. [DOI] [PubMed] [Google Scholar]
- 4.Glare E M, Paton J C, Premier R R, Lawrence A J, Nisbet I T. Analysis of a repetitive DNA sequence from Bordetella pertussis and its application to the diagnosis of pertussis using the polymerase chain reaction. J Clin Microbiol. 1990;28:1982–1987. doi: 10.1128/jcm.28.9.1982-1987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groisman E A, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Q, Hirakatsu K, Taylor D E. Variability of gene order in different Helicobacter pylori strains contributes to genome diversity. Mol Microbiol. 1996;20:833–842. doi: 10.1111/j.1365-2958.1996.tb02521.x. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence J G, Roth J R. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S L, Sanderson K E. Highly plastic chromosomal organization in Salmonella typhi. Proc Natl Acad Sci USA. 1996;93:10303–10308. doi: 10.1073/pnas.93.19.10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahan M J, Roth J R. Ability of a bacterial chromosome segment to invert is dictated by included material rather than flanking sequence. Genetics. 1991;129:1021–1032. doi: 10.1093/genetics/129.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peppler, M. S. Personal communication.
- 11.Römling U, Schmidt K D, Tümmler B. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J Mol Biol. 1997;271:386–404. doi: 10.1006/jmbi.1997.1186. [DOI] [PubMed] [Google Scholar]
- 12.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–789. [Google Scholar]
- 13.Stibitz S. Bordetella pertussis strain Tohama I. In: de Bruin F J, Lupski J R, Weinstock G M, editors. Bacterial genomes: physical structure and analysis. New York, N.Y: Chapman & Hall; 1998. [Google Scholar]
- 14.Stibitz S, Yang M-S. Genomic fluidity of Bordetella pertussis assessed by a new method for chromosomal mapping. J Bacteriol. 1997;179:5820–5826. doi: 10.1128/jb.179.18.5820-5826.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Umeda M, Ohtsubo E. Mapping of insertion element IS5 in the Escherichia coli K-12 chromosome: chromosomal rearrangements mediated by IS5. J Mol Biol. 1990;213:229–237. doi: 10.1016/S0022-2836(05)80186-X. [DOI] [PubMed] [Google Scholar]