Abstract
We identified Mg2+-responsive promoters of the phoPQ, mgtA, and mgrB genes of Escherichia coli K-12 by S1 nuclease analysis. Expression of these genes was induced by magnesium limitation and depended on PhoP and PhoQ. The transcription start sites were also determined, which allowed us to find a (T/G)GTTTA direct repeat in their corresponding promoter regions.
PhoP-PhoQ is a two-component regulatory system that controls several virulence properties in the gram-negative bacterium Salmonella typhimurium (6, 8, 11). Recently, extracellular Mg2+ has been identified as a stimulus that affects the PhoP-PhoQ system. The PhoQ protein is a Mg2+ sensor that changes conformation in the presence of periplasmic Mg2+ (6). PhoP is a regulatory protein that is necessary to transcribe some 25 different loci, many of which are essential for growth at low Mg2+ concentration (21).
The PhoP-PhoQ system is also present in Escherichia coli (7, 12), Shigella flexneri, Yersinia enterocolitica, and Yersinia pestis (8), where a basic physiological role in response to Mg2+ starvation has been proposed (6). The PhoP and PhoQ proteins of E. coli and S. typhimurium are 93 and 86% identical, respectively, indicating a high degree of structural and functional similarity (7, 12). Divalent cations seems to bind to an acidic cluster (148EDDDDAE154) of the E. coli PhoQ sensor domain and stabilize a conformation inactive in signaling (23). PhoP-PhoQ in E. coli is a promising system for studying ligand-induced signal transduction because it is one of the few two-component systems whose ligands have been identified. However, the physiological role of the PhoP-PhoQ system in E. coli is not understood. Here, we identified extracellular Mg2+-responsive genes and promoters in E. coli and investigated the regulation of their expression by the PhoP-PhoQ system.
Isolation and identification of Mg2+-responsive genes.
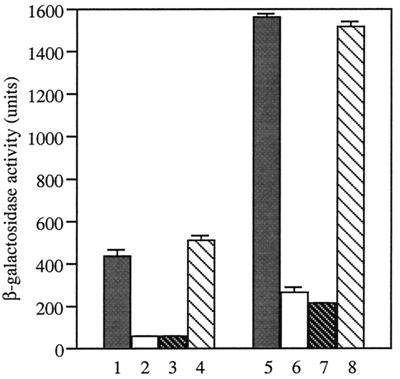
To isolate Mg2+-responsive genes, E. coli MC4100 (Table 1) was infected with λplacMu55 and λpMu507 as described by Bremar et al. (3), and the resulting blue colonies were selected as lac gene transcriptional fusion strains on a Luria-Bertani (LB) medium plate (18) containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg/ml) and kanamycin (30 μg/ml). Bacteria were always grown at 37°C. Among 2,000 independent lacZ gene fusions, Mg2+-responsive clones, which were white on MacConkey plates containing 30 mM MgSO4 and were red in the absence of Mg2+, were selected. A P1 phage lysate prepared from the selected clones was used to infect MC4100 to obtain kanamycin-resistant colonies (MG1301 and MG1601). β-Galactosidase activity of lacZ fusion strains (MG1301 and MG1601) was decreased significantly by 30 mM MgSO4 or 30 mM MgCl2 but not by 30 mM Na2SO4 (Fig. 1), indicating that expression of the fused genes was repressed by higher concentrations of Mg2+. Fusion junctions were sequenced as follows. λplacMu55 specialized transducing phage, which carries various amounts of the adjacent host DNA, was prepared by UV irradiation of the fusion strain (19). Aliquots (1.0 μg of DNA) were sequenced directly with a PRISM Dye Terminator Cycle Sequencing Ready Reaction kit with a Mu C-end primer (MU2; 5′-AATAATCCAATGTCCTCCCGG-3′). The PCR (25 cycles of 96°C for 30 s, 57°C for 30 s, and 60°C for 4 min) identified two Mg2+-responsive genes in E. coli K-12; one (MG1601) was the mgtA gene encoding an ATP-dependent Mg2+ transporter at 96.2 min on the chromosome (positions 8267 to 10963 in GenBank entry AE000495), and the other gene (MG1301) was a newly designated gene, mgrB, located at 41.2 min (complementary to positions 68 to 211 in GenBank entry AE000277).
TABLE 1.
Bacterial strains, phages, and plasmids used in this studya
| Strain or phage | Description | Reference or source |
|---|---|---|
| E. coli strains | ||
| MC4100 | F− Δ(argF-lac)U169 araD139 rpsL150 ptsF25 fibB5301 rbsR deoC relA1 | 4 |
| BD25 | F− Δ(argF-lac)U169 supF supE hsdR galK trpR metB lacY tonA | E. Bremar |
| MQ4007 | MC4100 phoQ608::Tn10dCam | MC4100 × P1 (MG1307)→Camr |
| MP4022 | MC4100 phoP2146::Tn10dCam | MC4100 × P1 (MG1322)→Camr |
| MG1301 | MC4100 mgrB::λplacMu55 | This work |
| MG1307 | MG1301 phoQ608::Tn10dCamb | This work |
| MG1322 | MG1301 phoP2146::Tn10dCamb | This work |
| MG1601 | MC4100 mgtA::λplacMu55 | This work |
| MG1607 | MG1601 phoQ608::Tn10dCam | MG1601 × P1 (MG1307)→Camr |
| MG1622 | MG1601 phoP2146::Tn10dCam | MG1601 × P1 (MG1322)→Camr |
| Bacteriophages | ||
| λplacMu55 | Mu cts62 ner+ A′ am1093 ′uvrD′ MuS′ ′trp′ lacZ+ lacY+ lacA′ Kmr λ imm | 3 |
| λpMu507 | λcIts857 Sam7 Mu A+ B+ | 3 |
| λNK1324 | Tn10dCam cIts857 Pam80 nin5 b522 att− | 13 |
| Plasmids | ||
| pMW119 | Apr, lacZ′, replication origin derived from pSC101 | 2 |
| pHO119 | pMW119, 3.6-kb SalI-EcoRI fragment containing phoP+Q+ from λ phage clone (Kohara clone 239)c | This work |
| pDA51 | PhoP D51A, pHO119 | This work |
| pDN51 | PhoP D51N, pHO119 | This work |
| pHR277 | PhoQ H277R, pHO119 | This work |
| pUC19 | Apr, lacZ′ | 23 |
| pHO19P | pUC19, 560-bp EcoRI-SalI fragment containing phoPQ promoter from pHO119 | This work |
| pMGA19P | pUC19, 820-bp EcoRI-BamHI fragment containing mgtA promoter from λplacMu55 phage harboring mgtA prepared by UV induction of MG1601 | This work |
| pMGB19P | pUC19, 980-bp EcoRI-BamHI fragment containing mgrB promoter from λ phage clone (Kohara clone 335) | This work |
FIG. 1.
Gene expression dependent on Mg2+. E. coli MG1601 (lanes 1 to 4) and MG1301 (lanes 5 to 8) were grown in LB medium and LB medium containing 30 mM MgSO4 (lanes 2 and 6), 30 mM MgCl2 (lanes 3 and 7), or 30 mM Na2SO4 (lanes 4 and 8). An early-log-phase (OD600, 0.4) culture was used to determine β-galactosidase activity (18). Data are the means of triplicate values with standard deviations.
Isolation of phoPQ-defective strains by using Tn10dCam.
To determine that expression of Mg2+-responsive genes was controlled by both PhoP and PhoQ, E. coli MG1301 was infected with λNK1324 grown on E. coli BD25 as described by Kleckner et al. (13). Chloramphenicol-resistant (Camr) colonies (about 30,000) were selected on MacConkey plates containing chloramphenicol (20 μg/ml), from which white colonies were isolated. A P1 phage lysate prepared from one of them was used to infect MG1301, and Camr colonies were isolated. We determined the Tn10dCam insertion site by sequencing the genomic region adjacent to Tn10dCam amplified by thermal asymmetric interlaced PCR (15, 16). Consequently, three phoP mutants (MG1320, MG1322, and MG1323) and four phoQ mutants (MG1303, MG1306, MG1307, and MG1321) were identified. The mutant strains defective in phoP (MG1622) and phoQ (MG1607) were constructed by P1 transduction from MG1322 and MG1307 to MG1601, respectively (Table 1). When β-galactosidase activity was assayed in phoP- and phoQ-defective strains, neither mgtA nor mgrB was expressed, irrespective of Mg2+ concentration (data not shown).
Identification of the promoter region of Mg2+-responsive genes.
We determined the transcriptional start sites of mgtA and mgrB as well as phoPQ by S1 nuclease assay as follows. E. coli cells were grown in LB medium in the absence of MgCl2 overnight, then diluted 100-fold into 20 ml of LB medium in the presence or absence of 30 mM MgCl2, and grown to mid-log phase (optical density at 600 nm [OD600], 0.5 to 0.8) to prepare total RNA (1). The S1 nuclease assay was conducted as described previously (1, 6), with the following modification. The RNA (20 to 100 μg) was mixed with the probe DNA in 50 μl of a hybridization buffer (80% formamide, 20 mM HEPES [pH 6.5], 0.4 M NaCl), incubated at 75°C for 10 min, and then incubated at 37°C overnight. Then 220 μl of H2O and 30 μl of 10× S1 nuclease buffer (0.3 M sodium acetate [pH 4.5], 0.5 M NaCl, 10 mM ZnSO4, 50% glycerol) were added, and the mixture was treated with 50 U of S1 nuclease (Takara) at 37°C for 10 min. The reaction was stopped by adding 300 μl of phenol-chloroform, and the DNAs contained in the aqueous phase were precipitated with ethanol. The precipitate was dissolved in a sequencing loading buffer (80% formamide, 10 mM NaOH, 1 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol) and electrophoresed in a 6 or 4% acrylamide sequencing gel.
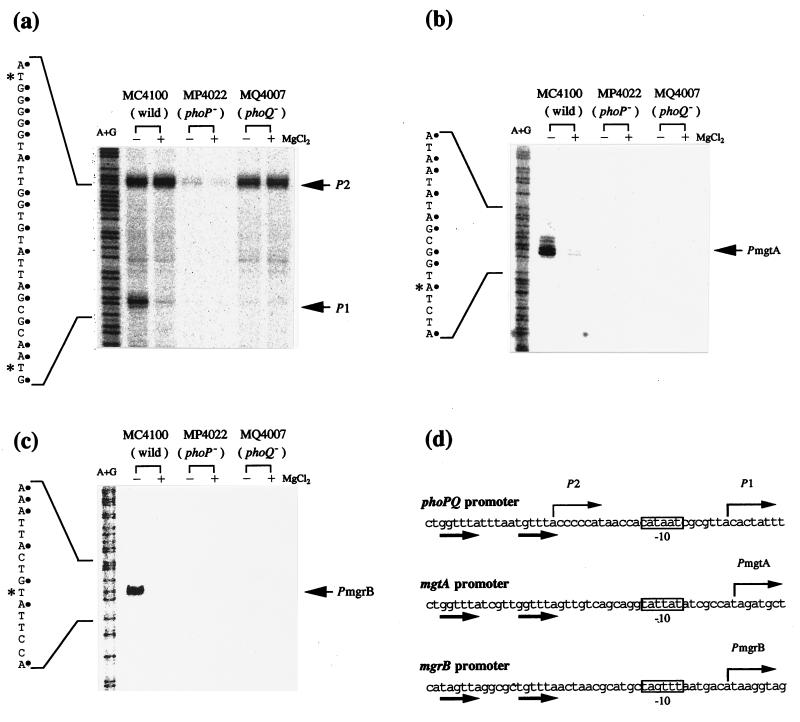
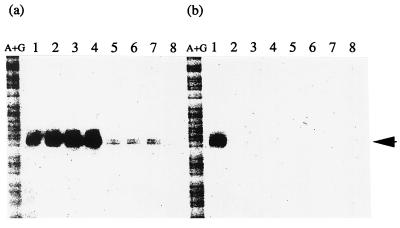
In the E. coli strain (MC4100) grown in LB medium, two transcripts (P1 and P2) of phoPQ were found (Fig. 2a). The transcription of P1 was dependent on extracellular Mg2+ concentration and was decreased in phoP (MP4022) or phoQ (MQ4007) mutants. In MC4100, MP4022, and MQ4007, the P2 transcript was constitutively expressed in the presence or absence of Mg2+. For mgtA and mgrB, only one transcript (PmgtA and PmgrB) was found for each; these transcripts were not detected in MP4022 and MQ4007 (Fig. 2b and c). Gene expression in each of these defective mutants was restored at the lower Mg2+ concentration upon transformation with pHO119 (data not shown). These results indicated that expression of mgtA and mgrB as well as phoPQ is positively controlled by both PhoP and PhoQ in a Mg2+-dependent manner. In addition, mgtA was found to be specifically transcribed during log phase but repressed within 15 min after the addition of 30 mM MgCl2 (Fig. 3).
FIG. 2.
Transcriptional regulation of phoPQ, mgtA, and mgrB. When grown to mid-log phase (OD600, 0.5 to 0.8) in LB medium in the presence (+) or absence (−) of 30 mM MgCl2, cells of the indicated E. coli strains were collected, and then RNA preparation and S1 nuclease assay were done as described in the text. Probes A (SalI-HincII [380-bp] DNA fragment of pHO19P), B (AluI-DdeI [400-bp] fragment of pMGA19P), and C (EcoRV-BstUI [600-bp] fragment of pMGB19P) were used to determine start sites of phoPQ (a), mgtA (b), and mgrB (c), respectively. Electrophoresis was done with a 6% (a) or 4% (b and c) acrylamide sequencing gel. Lanes A+G represent Maxam-Gilbert sequence reactions. P1, P2, PmgtA, and PmgrB point to the corresponding protected transcripts. Transcription start sites are marked with asterisks. (d) DNA sequence (coding strand) around the promoter of phoPQ (complementary to positions 4786 to 4837 of GenBank entry AE000213), mgtA (positions 7961 to 8012 of AE000495), and mgrB (complementary to positions 229 to 280 of AE000277). Thin and bold arrows indicate starts and directions of transcription and direct repeats, respectively. The putative −10 region of each promoter is boxed.
FIG. 3.
Growth-phase-dependent transcription of mgtA. After growth to exponential phase (OD600, 0.3) in LB medium, cells of E. coli MC4100 were further cultivated in the presence (b) or absence (a) of 30 mM MgCl2. Samples were prepared at 0 min (lane 1), 15 min (lane 2), 30 min (lane 3), 1 h (lane 4), 2 h (lane 5), 4 h (lane 6), 6 h (lane 7), and 8 h (lane 8). RNA preparation and S1 nuclease assay were done as described for Fig. 2. Lanes A+G represent Maxam-Gilbert sequence reactions.
The direct repeat (T/G)GTTTA was conserved 25 bp upstream of the transcriptional start site of phoPQ (P1), mgtA, and mgrB (Fig. 2d). This direct repeat is also found in the phoPQ promoter of S. typhimurium (20) and is very similar to one recognized by the regulator PhoB (17). Thus, (T/G)GTTTA is assumed to be a target for PhoP. A search of the entire E. coli genome sequence (4.64 Mb [4a]) for a (T/G)GTTTA-5 bp-(T/G)GTTTA or TAGTTA-5 bp-(T/G)GTTTA motif detected four genes, a bor homolog, ydcD, a fimD homolog, and yrbL (12.5, 33.1, 34.4, and 72 min, respectively, on the chromosome), besides phoP, mgtA, and mgrB. In fact, the promoter of yrbL was Mg2+ responsive (data not shown). In S. typhimurium, this motif was also found in the promoter region of phoP, mgtA, mgtBC, and pagA, which are PhoP-activated genes, but many other PhoP-activated genes lack the consensus sequence (11, 21). Recently, the expression of several genes was reported to be under the direct control of the PmrA-PmrB two-component system, in which PhoP participates by activating pmrAB (9, 22). These results suggested that some genes are regulated indirectly by PhoP through other regulatory factors.
In S. typhimurium, the transcriptional start sites of the phoPQ operon were previously identified (6, 20). Two RNA species were detected in an exponentially grown S. typhimurium culture. These transcripts define two promoters: P1, which requires both PhoP and PhoQ for activity and is regulated by environmental Mg2+; and P2, which remains active in the absence of PhoP and PhoQ (6, 20). We have obtained similar results for the E. coli phoPQ operon.
Identification of amino acid residues involved in the PhoP-PhoQ signaling cascade.
To further examine the regulation by phoPQ, we introduced a point mutation in the coding region with respect to the putative PhoP phosphorylation site Asp51 and the PhoQ autophosphorylation site His277 on pHO119 as follows. Site-directed mutagenesis was performed by using a QuikChange site-directed mutagenesis kit (Stratagene). pDA51 and pDN51 were derived from pHO119, carrying phoP with changes of Asp51 to Ala and Asn, respectively. pHR277 is a derivative of pHO119, carrying phoQ with a change of His277 to Arg. Primers for the site-directed mutagenesis were as follows: pDA51, forward (5′GATATTGCGATTGTCGCTCTCGGATTGCC3′) and reverse (5′GGCAATCCGAGAGCGACAATCGCAATATC3′); pDN51, forward (5′GATATTGCGATTGTCAATCTCGGATTGCC3′) and reverse (5′GGCAATCCGAGATTGACAATCGCAATATC3′); and pHR277, forward (5′CCGACCTGACCCGTAGTCTGAAAACGC3′) and reverse (GCGTTTTCAGACTACGGGTCAGGTCGG3′).
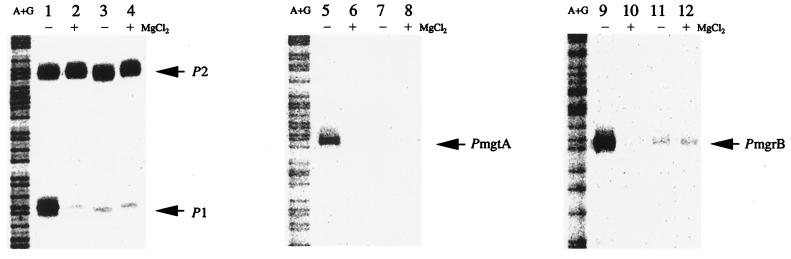
As shown in Fig. 4, transcription of phoPQ (P1), mgtA, and mgrB was not observed when PhoQ H277R was expressed in the phoQ mutant (MQ4007) (Fig. 4, lanes 3, 4, 7, 8, 11, and 12). When the wild-type PhoQ was expressed, expression of phoPQ, mgtA, and mgrB was observed in the absence of Mg2+ but not in the presence of 30 mM Mg2+ (lanes 1, 2, 5, 6, 9, and 10). The same result was obtained for the phoP mutant strain (MP4022) and pDA51 or pDN51 (data not shown). These results demonstrate that Asp51 of PhoP and His277 of PhoQ are indispensable for activation of PhoP-PhoQ signaling and subsequent expression of mgtA and mgrB.
FIG. 4.
Effect of PhoQ H277R on transcription of phoPQ, mgtA, and mgrB. E. coli MQ4007 in the presence of pHO119 (lanes 1, 2, 5, 6, 9, and 10) or pHR277 (lanes 3, 4, 7, 8, 11, and 12) was grown to mid-log phase in LB medium in the presence (+) or absence (−) of 30 mM MgCl2 to prepare RNA. RNA preparation and S1 nuclease assays were performed as described for Fig. 2.
Concluding remarks.
In this study, we tried to isolate directly a Mg2+-responsive lacZ fusion strain of E. coli. Among 2,000 independent lac gene transcriptional fusions, we identified two PhoP-activated genes, mgtA and mgrB. mgtA, encoding an ATP-dependent Mg2+ transporter, is homologous to genes in S. typhimurium, but mgrB is newly designated in this report. mgtA was also found to be specifically transcribed during log phase but repressed through the PhoP-PhoQ system within 15 min after the addition of 30 mM MgCl2. These results suggest that the PhoP-PhoQ-independent P2 promoter of E. coli provides a low intracellular concentration of PhoP and PhoQ when magnesium is in excess, and upon shifting to limited magnesium, autophosphorylated PhoQ serves as the phosphate donor for PhoP. Phospho-PhoP autogenously activates the P1 promoter in the E. coli phoPQ operon, resulting in activation of mgtA gene expression to increase the intracellular Mg2+ concentration. Such a signaling cascade seems to be essential for active growth of E. coli in a low-Mg2+ medium.
Acknowledgments
We thank E. Bremar for providing bacterial strains and phages and K. Yoshida for critical reading of the manuscript. We also thank H. Mori for computer search of PhoP target genes.
This work was financially supported by the Mishima Kaiun Memorial Foundation and a Sasakawa Scientific Research Grant from the Japan Science Society.
REFERENCES
- 1.Aiba H. Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor of its own gene. Cell. 1983;32:141–149. doi: 10.1016/0092-8674(83)90504-4. [DOI] [PubMed] [Google Scholar]
- 2.Bernardi A, Bernardi F. Complete sequence of pSC101. Nucleic Acids Res. 1984;12:9415–9426. doi: 10.1093/nar/12.24.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bremar E, Silhavy T J, Weinstock G M. Transposition of λplacMu is mediated by the A protein altered at its carboxy-terminal end. Gene. 1988;71:177–186. doi: 10.1016/0378-1119(88)90089-3. [DOI] [PubMed] [Google Scholar]
- 4.Casadaban M J. Transposition and fusion of lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 4a.Escherichia coli WWW Home Page. 28 August 1998, revision date. [Online.] http://mol.genes.nig.ac.jp/ecoli/. [16 July 1999, last date accessed.]
- 5.Garcia-Vescovi E, Soncini F C, Groisman E A. The role of the PhoP/PhoQ regulon in Salmonella virulence. Res Microbiol. 1994;145:473–480. doi: 10.1016/0923-2508(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Vescovi E, Soncini F C, Groisman E A. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 7.Groisman E A, Heffron F, Solomon A. Molecular genetic analysis of the Escherichia coli phoP locus. J Bacteriol. 1992;174:486–491. doi: 10.1128/jb.174.2.486-491.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groisman E A, Heffron F. Regulation of Salmonella virulence by two-component regulatory systems. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 319–332. [Google Scholar]
- 9.Gunn J S, Miller S I. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol. 1996;178:6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunn J S, Hohmann E L, Miller S I. Transcriptional regulation of virulence: a PhoQ periplasmic domain mutation results in increased net phosphotransfer to PhoP. J Bacteriol. 1996;178:6369–6373. doi: 10.1128/jb.178.21.6369-6373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hohmann E L, Miller S I. The Salmonella PhoP virulence regulon. In: Torriani-Gorini A, Yagil E, Silver S, editors. Phosphate in microorganisms. Washington, D.C: ASM Press; 1994. pp. 120–125. [Google Scholar]
- 12.Kasahara M, Nakata A, Shinagawa H. Molecular analysis of the Escherichia coli phoP-phoQ operon. J Bacteriol. 1992;174:492–498. doi: 10.1128/jb.174.2.492-498.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 14.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genome library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y-G, Whittier R F. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clone for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y-G, Mitsukawa N, Oosumi T, Whittier R F. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 17.Makino K, Amemura M, Kim S-K, Nakata A, Shinagawa H. Mechanism of transcriptional activation of the phosphate regulon in Escherichia coli. In: Torriani-Gorini A, Yagil E, Silver S, editors. Phosphate in microorganisms. Washington, D.C: ASM Press; 1994. pp. 5–12. [Google Scholar]
- 18.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 19.Roy R N, Mukhopadhyay S, Wei L I C, Schellhorn H E. Isolation and sequencing of gene fusions carried by λplacMu specialized transducing phage. Nucleic Acids Res. 1995;23:3076–3078. doi: 10.1093/nar/23.15.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soncini F C, Vescovi E G, Solomon F, Groisman E A. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J Bacteriol. 1995;177:4364–4371. doi: 10.1128/jb.177.15.4364-4371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soncini F C, Vescovi E G, Solomon F, Groisman E A. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soncini F C, Groisman E A. Two-component regulatory systems can interact to process multiple environmental signals. J Bacteriol. 1996;178:6796–6801. doi: 10.1128/jb.178.23.6796-6801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waldburger C D, Sauer R T. Signal detection by the PhoQ sensor-transmitter. J Biol Chem. 1996;271:26630–26636. doi: 10.1074/jbc.271.43.26630. [DOI] [PubMed] [Google Scholar]
- 24.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]